Abstract
Mycothiol is a novel thiol produced only by actinomycetes and is the major low-molecular-weight thiol in mycobacteria. Mycothiol was previously shown to be synthesized from 1-d-myo-inosityl-2-amino-2-deoxy-α-d-glucopyranoside by ligation with cysteine followed by acetylation. A novel mycothiol-dependent detoxification enzyme, mycothiol conjugate amidase, was recently identified in Mycobacterium smegmatis and shown to have a homolog, Rv1082, in Mycobacterium tuberculosis. In the present study we found that a protein encoded by the M. tuberculosis open reading frame Rv1170, a homolog of Rv1082, possesses weak mycothiol conjugate amidase activity but shows substantial deacetylation activity with 1-d-myo-inosityl-2-acetamido-2-deoxy-α-d-glucopyranoside (GlcNAc-Ins), a hypothetical mycothiol biosynthetic precursor. The availability of this protein enabled us to develop an assay for GlcNAc-Ins, which was used to demonstrate that GlcNAc-Ins is present in M. smegmatis at a level about twice that of mycothiol. It was shown that GlcNAc-Ins is absent in mycothiol-deficient mutant strain 49 of M. smegmatis and that this strain can concentrate GlcNAc-Ins from the medium and convert it to mycothiol. This demonstrates that GlcNAc-Ins is a key intermediate in the pathway of mycothiol biosynthesis. Assignment of Rv1170 as the gene coding the deacetylase in the M. tuberculosis genome represents the first identification of a gene of the mycothiol biosynthesis pathway. The presence of a large cellular pool of substrate for this enzyme suggests that it may be important in regulating mycothiol biosynthesis.
Most gram-positive bacteria, including many strict aerobes, do not produce glutathione (7, 13), a key component of cellular mechanisms for protection against oxygen toxicity (5). In the search for other thiols in these organisms that might function like glutathione, a major thiol in streptomycetes was discovered (14) and later identified (12, 19, 21) as a novel conjugate of N-acetylcysteine (AcCys) and 1-d-myo-inosityl-2-amino-2-deoxy-α-d-glucopyranoside (GlcN-Ins) producing mycothiol (MSH or AcCys-GlcN-Ins). Bornemann et al. (3) showed that chemically synthesized GlcN-Ins could be converted to Cys-GlcN-Ins and MSH by Mycobacterium smegmatis cell extracts in the presence of ATP, Mg2+, Cys, and acetate or acetyl coenzyme A (acetyl-CoA). They proposed that the final steps of MSH biosynthesis involve ATP-dependent ligation of Cys with GlcN-Ins, followed by acetylation via an acetyltransferase reaction involving acetyl-CoA. Measurements of GlcN-Ins and Cys-GlcN-Ins levels in M. smegmatis (1) and isolation of mutants defective in GlcN-Ins or Cys-GlcN-Ins production support the existence of this pathway (15).
The earlier studies left undefined the biochemical reactions involved in GlcN-Ins formation. By analogy with the established biochemistry for production of the glycosylphosphatidylinositol (GPI) anchor, which contains as a component an isomer of GlcN-Ins with an α(1–6) linkage, one might expect that GlcN-Ins is produced by deacetylation of 1-d-myo-inosityl-2-acetamido-2-deoxy-α-d-glucopyranoside (GlcNAc-Ins) and that the latter is produced by transfer of GlcNAc from UDP-GlcNAc to Ins (6). We present here evidence that GlcNAc-Ins is an intermediate in MSH biosynthesis and is converted to GlcN-Ins by GlcNAc-Ins deacetylase. This deacetylase cleaves an amide bond similar to that hydrolyzed by an MSH-dependent detoxification enzyme, mycothiol S-conjugate amidase, recently identified as being coded by Rv1082 in the Mycobacterium tuberculosis genome (11). We show here that Rv1170, a homolog of Rv1082, codes for a GlcNAc-Ins deacetylase activity involved in the MSH biosynthesis pathway.
MATERIALS AND METHODS
Reagents.
MSH was isolated from M. smegmatis and derivatized with monobromobimane to form the MSH bimane derivative (MSmB), which was purified by high-performance liquid chromatography (HPLC), all as previously described (12). Purified M. smegmatis mycothiol S-conjugate amidase was used to quantitatively hydrolyze MSmB to stereochemically pure GlcN-Ins, and the latter was purified from the other hydrolysis product, AcCySmB, using a Sep-Pak C18 (Waters) cartridge as previously described (11). A 10 mM stock solution of GlcNAc-Ins was prepared by addition of a tenfold excess of acetic anhydride (Fisher) to 10 mM GlcN-Ins in 10 mM NaHCO3 over 20 min while adjusting the pH to 8.5 with NaOH. The reaction was monitored for GlcN-Ins loss as described below to insure that the reaction was complete and that no residual GlcN-Ins was present. This stock solution of GlcNAc-Ins was assayed as described below and used without further purification.
Bacterial stains and culture conditions.
M. smegmatis mc2155 was obtained from W. R. Jacobs, Jr. (Albert Einstein College of Medicine, Bronx, N.Y.) and cultured at 37°C in Middlebrook 7H9 medium supplemented with 0.5% Tween 80 (Fisher) and 0.4% glucose. M. smegmatis mutant 49 is an MSH-deficient mutant derived from mc2155 by chemical mutagenesis and was cultured in Middlebrook 7H9 medium at 37°C as previously described (15).
Assay of MSH and MSH precursors.
MSH and its precursors (cysteine, GlcN-Ins, and Cys-GlcN-Ins) were assayed as described elsewhere (1).
Mycothiol S-conjugate amidase assay.
Mycothiol S-conjugate amidase activity was assayed at 30°C using 30 μM MSmB in 50 mM HEPES (pH 7.0) containing 3 mM 2-mercaptoethanol. The formation of AcCySmB from MSmB was analyzed by HPLC (11).
Cloning and expression of the Rv1170 gene from M. tuberculosis.
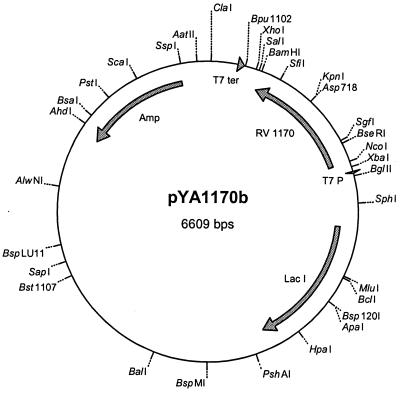
Genomic DNA of M. tuberculosis H37Rv was prepared as described previously (2). The open reading frame Rv1170 was amplified from this DNA with the primers 5′-TAGCCATGGTGTCTGAGACGCCGCG-3′ and 5′-GGATCCCGCGGTGAAGCCCAGAC-3′ containing NcoI and BamHI restriction sites, respectively. PCR was performed with Taq polymerase obtained from Gibco BRL, using 1.5 mM MgCl2 and 5% dimethyl sulfoxide. The 30 cycles of PCR included denaturation at 94°C for 40 s, annealing at 55°C for 1 min, and amplification at 72°C. The PCR products were separated on a 1% agarose gel. The appropriate PCR product was ligated into vector pCR2.1 of the TA cloning kit (Invitrogen) and transformed into Escherichia coli DH5α by standard chemical transformation procedures. Clones containing the vector were selected on plates of Luria-Bertani (LB) agar (20) plus ampicillin (100 μg/ml), and plasmid DNA was digested with the restriction endonucleases NcoI and BamHI (Fermentas). Restriction enzyme-digested plasmids were isolated with a QIAquick gel extraction kit (Qiagen Ltd.). A corresponding digestion was applied to the plasmid pET-22b, and the two products were ligated together with T4 DNA ligase to obtain the plasmid pYA1170. In order to express Rv1170 in E. coli without the pelB leader sequence, the gene from pYA1170 was excised using NcoI and Bpu1102I (Fermentas) and ligated to an aliquot of pET16b cut with NcoI and Bpu1102I to generate the plasmid pYA1170b (Fig. 1). This plasmid was transformed by the heat shock method (20) to competent E. coli BL21(DE3) prepared according to the CaCl2 method (20) and plated on LB agar containing 100 μg of ampicillin/ml. Single colonies were inoculated into 5 ml of LB broth also containing ampicillin (100 μg/ml). After overnight incubation at 37°C with shaking, the individual cultures were diluted 1:100 in the same medium and incubation was continued at 37°C with shaking. Isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 0.4 mM when the A600 reached 0.6, and incubation was continued overnight at 25°C before harvesting by centrifugation at 5,000 × g for 15 min at room temperature. The pellets were lysed by sonication. Proteins were separated by centrifugation (15,000 × g, 4°C, 30 min) into soluble and insoluble fractions. Total proteins were separated by sodium dodecyl sulfate–7.5% polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie blue or transferred to polyvinylidene difluoride membranes (Bio-Rad). The N-terminal amino acid sequence was verified using Edman degradation after separation of samples by SDS-PAGE and electroblotting to a polyvinylidene difluoride membrane.
FIG. 1.
Map of plasmid pYA1170b employed to express Rv1170 in E. coli.
Assay of GlcNAc-Ins.
The assay of GlcNAc-Ins involves deacetylation of the glucosamine moiety prior to labeling of the free amine with AccQ-Fluor (Waters). Recombinant M. tuberculosis Rv1170 in crude extracts of E. coli described above was found to have such activity. E. coli BL21(DE3) carrying the pYA1170b expression plasmid was cultured at 37°C in LB broth with 100 μg of ampicillin/ml to an A600 of 0.6. Isopropyl-β-d-thiogalactopyranoside was added until it reached a concentration of 0.4 mM, and the culture was incubated overnight at 28°C. The bacterial cell pellet was suspended in 50 mM HEPES (pH 7.0) containing 3 mM 2-mercaptoethanol and a 35 μM concentration of each of the protease inhibitors N-α-p-tosyl-l-phenylalanylchloromethyl ketone and N-α-p-tosyl-l-lysinechloromethyl ketone and was then disrupted by sonication. The extract was clarified by centrifugation at 39,000 × g for 30 min at 4°C. Saturated ammonium sulfate was added to the supernatant until it reached 20% by volume, the mixture was allowed to stand 1 h on ice, and the supernatant was clarified by centrifugation. The ammonium sulfate was added until 50% saturation was attained, and the mixture was stored overnight at 4°C. The 20 to 50% ammonium sulfate pellet was collected by centrifugation at 39,000 × g for 30 min at 4°C. This protein pellet (0.69 g) was suspended in 10 ml of the extraction buffer, and the insoluble material was removed by centrifugation at 39,000 × g for 30 min at 4°C. The supernatant was dialyzed overnight at 4°C against 100 volumes of extraction buffer. The supernatant was concentrated in a Millipore UltraFree-15 50-kDa nominal-molecular-weight cutoff centrifugal filter to ∼80 mg of protein per ml. This preparation was frozen in aliquots at −70°C and used as needed for deacetylation of GlcNAc-Ins.
M. smegmatis cell pellets were extracted at 60°C in 50% acetonitrile containing 20 mM HEPES (pH 7.0) with 5 mM N-ethylmaleimide (NEM; Sigma) for 10 min and cooled on ice as described previously for GlcN-Ins analysis (1). The 50% acetonitrile extract was clarified by centrifugation at 14,000 × g for 3 min. The acetonitrile was removed from a 100-μl aliquot of this extract using a Savant Speed Vac, which reduced the volume to <50 μl. The volume was adjusted back to 50 μl with water, and 1 μl of 0.5 M 2-mercaptoethanol (in water) was added to react with any remaining NEM. A 10-μl aliquot of Rv1170 preparation was added to this sample, and the mixture was incubated for 45 min at 30°C. The deacetylase reaction was quenched by adding 60 μl of warm (60°C) 10 mM NEM in acetonitrile, and the sample was incubated for 10 min at 60°C. The extract was iced and clarified by centrifugation at 14,000 × g for 3 min. A 15-μl aliquot of the supernatant was used for amine analysis. It was shown in control experiments that the amount of deacetylase (Rv1170) preparation used exceeded that required for deacetylation of added GlcNAc-Ins by >10-fold. Since the deacetylated sample contains the sum of GlcN-Ins and deacetylated GlcNAc-Ins, the estimate for GlcNAc-Ins is valid only if its content substantially exceeds that of GlcN-Ins.
Recovery experiments for GlcNAc-Ins were conducted with M. smegmatis MSH-deficient mutant 49, in which 50% acetonitrile–NEM extracts were analyzed after the addition of 0.7 and 7.0 μM GlcNAc-Ins. Extracts of mutant 49 contained no detectable GlcN-Ins or MSH (15), so that any GlcN-Ins found in the extracts after treatment with the deacetylase preparation was derived solely from the deacetylation of GlcNAc-Ins.
Uptake and metabolism of GlcNAc-Ins by mutant 49.
M. smegmatis MSH-deficient mutant 49 (15) was cultured in 7H9 Middlebrook medium with 0.4% glucose and 0.5% Tween 80 to log phase (A600 = 0.6), and 10-ml aliquots were transferred to sterile 25-ml Erlenmeyer flasks. Duplicate flasks contained cells only (control), 20 μM myo-inositol, 20 μM glucosamine, 20 μM N-acetylglucosamine, or 17 μM GlcNAc-Ins and were cultured at 37°C and 225 rpm. Samples (∼2.3 × 108 cells) were taken at 2, 6, 19, and 46 h of culture. The cells were pelleted by centrifugation for 2 min at 14,000 × g and were extracted in 50% acetonitrile containing 20 mM HEPES (pH 8.0) and either 2 mM monobromobimane for thiol analysis or 5 mM NEM for amine analysis and thiol control samples. Duplicate cultures were analyzed for GlcN-Ins, GlcNAc-Ins, Cys-GlcN-Ins, cysteine, and MSH. No significant MSH was found in any sample except for the cultures supplemented with GlcNAc-Ins.
The concentration dependence of the uptake of GlcNAc-Ins was examined with cultures of M. smegmatis mutant 49. Mutant 49 was cultured at 37°C for 23 h in 5 ml of 7H9 Middlebrook medium as described above, supplemented with 0, 4, 8, or 21 μM GlcNAc-Ins. The cells were collected by centrifugation and extracted for amine and thiol analysis as described above.
UDP-GlcNAc-Ins:inositol GlcNAc transferase assay.
Late-log-phase M. smegmatis mc2155 cells (1 g [wet weight]) were extracted by sonication in 5 ml of 50 mM HEPES (pH 7.0) containing 3 mM 2-mercaptoethanol and a 35 μM concentration of each of the protease inhibitors N-α-p-tosyl-l-phenylalanylchloromethyl ketone and N-α-p-tosyl-l-lysinechloromethyl ketone. The unfractionated extract was dialyzed against 100 volumes of the extraction buffer for 7 h, all at 4°C. The extract was assayed for formation of GlcN-Ins at 30°C after mixing the dialyzed extract sample with an equal volume of buffer concentrate to give a content of 50 mM HEPES (pH 7.0), 25 mM KCl, 5 mM MnCl2, 5 mM MgCl2, 3 mM 2-mercaptoethanol, and 1 mM ATP. Stock solutions (5 mM) of UDP-GlcNAc (Sigma) and myo-inositol (Sigma) were prepared in water. The extract was assayed for GlcNAc-Ins deacetylase activity with 0.1 mM GlcNAc-Ins as the substrate. The assay of transferase activity was conducted by mixing 2 μl each of 5 mM UDP-GlcNAc and 5 mM myo-inositol stock solutions with 100 μl of extract and treating the mixture as one would for the deacetylase assay. Aliquots (20 μl) were removed and mixed with 20 ml of 5 mM NEM in 50% aqueous acetonitrile at 60°C. The mixture was incubated at 60°C for 10 min and centrifuged, and 15-μl aliquots were analyzed for GlcN-Ins as previously described (1). Duplicate samples were analyzed at 0- and 2-h reaction intervals.
RESULTS
M. smegmatis amidase lacks deacetylase activity.
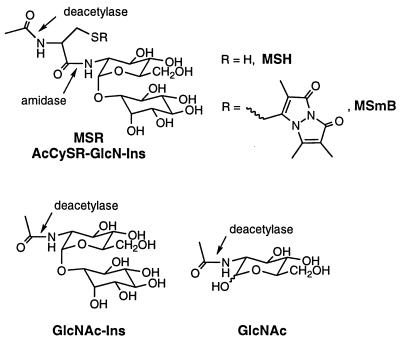
Mycothiol S-conjugate amidase from M. smegmatis, a homolog of the protein encoded by M. tuberculosis Rv1082, cleaves the amide bond linking Cys to GlcN in MSH derivatives having an alkylated sulfur residue (11), and it seemed possible that this enzyme might also cleave the corresponding bond in GlcNAc-Ins, functioning as a deacetylase (Fig. 2). To test this, we prepared GlcNAc-Ins from GlcN-Ins and compared the ability of the amidase purified from M. smegmatis to hydrolyze GlcNAc-Ins with its ability to cleave the monobromobimane conjugate of MSH (MSmB). For a test with 100 μM substrate, the activity measured with MSmB was 4.5 ± 1.0 μmol min−1 mg of protein−1 (11), whereas that measured with GlcNAc-Ins was <1 nmol min−1 mg of protein−1 (n = 4). Thus, the amidase does not exhibit measurable deacetylase activity and cannot serve this function in the MSH biosynthesis pathway of M. smegmatis.
FIG. 2.
Structures of the substrates for assaying amidase and deacetylase activities of MshB (Rv1170) and homologs.
Rv1170 encodes a protein with GlcNAc-Ins deacetylase activity.
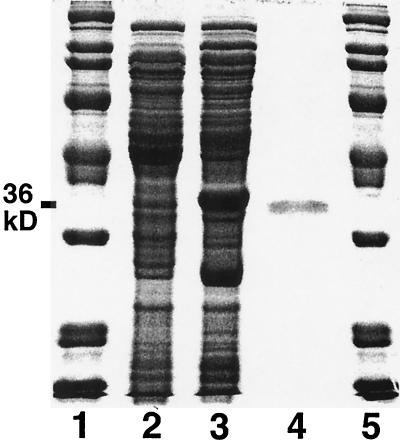
Since the deacetylase and the amidase both cleave an amide bond involving glucosamine, we reasoned that these proteins might be related. We therefore searched the Sanger Centre database for homologs of the 288-amino-acid M. tuberculosis amidase, Rv1082. The closest homolog found was Rv1170, with a length of 304 residues and 36% identity to Rv1082 with homology throughout the sequence. M. tuberculosis open reading frame Rv1170 was PCR cloned and expressed in E. coli by using the expression vector pET16b (Fig. 1). Figure 3 shows that the 20 to 50% saturated ammonium sulfate fraction from E. coli carrying the Rv1170 gene contains elevated levels of a protein of the expected size (36 kDa), as compared to those from E. coli prepared with a blank cloning vector. Another, smaller protein (∼26 kDa) was also present at elevated levels and is speculated to be a degradation product of the deacetylase. The identity of the 36-kDa protein was confirmed by determining the N-terminal amino acid sequence and finding it identical to that predicted for the protein encoded by open reading frame Rv1170.
FIG. 3.
SDS-PAGE of desalted 20 to 50% saturated ammonium sulfate extracts of isopropyl-β-d-thiogalactopyranoside-induced E. coli BL21(DE3) that was transformed with the blank cloning vector (pET16b) (lane 2) or with a vector containing Rv1170 (pYA1170b) (lane 3). Bio-Rad Broad Range molecular mass standards (lanes 1 and 5) and purified M. smegmatis mycothiol S-conjugate amidase (lane 4) are also shown.
Enzyme activity was determined on the 20 to 50% ammonium sulfate fractions after desalting and concentrating (Table 1). Extract from cells expressing Rv1170 exhibited substantial deacetylase activity with GlcNAc-Ins as the substrate, over 300-fold greater than the deacetylase activity determined with GlcNAc and 23-fold greater than the amidase activity measured with MSmB (Fig. 2). Thus, Rv1170 can be putatively identified as the GlcNAc-Ins deacetylase gene in the MSH biosynthesis pathway of M. tuberculosis.
TABLE 1.
Enzyme activity of 20 to 50% saturated ammonium sulfate extracts of E. coli transformed with pYA1170b or with the blank cloning vector pET16b
| Substrate | Activity (nmol min−1 mg of protein−1) (n = 3)
|
|
|---|---|---|
| Rv1170 | Blank vector | |
| MSmBa | 1.2 ± 0.1 | <0.002 |
| MSmBb | <10−4 | NDf |
| GlcNAc-Insc | 27 ± 2 | <0.002 |
| GlcNAcc | 0.087 ± 0.005 | <0.005 |
| MSHd | 0.002 ± 0.001 | ND |
| MSHe | <10−4 | ND |
Analysis for AcCySmB produced by amidase cleavage.
Analysis for production of the deacetylase product CySmB-GlcN-Ins.
Analysis for corresponding amine resulting from deacetylase reaction.
Analysis for AcCys produced by amidase cleavage.
Analysis for production of the deacetylase product Cys-GlcN-Ins.
ND, not determined.
Assay for GlcNAc-Ins.
To confirm that GlcNAc-Ins is present in mycobacteria, an assay for this component was needed. The availability of cloned Rv1170 allowed the development of such an assay. Cell extracts were prepared in warm 50% acetonitrile containing NEM, as employed previously for an assay of GlcN-Ins (1). One portion of the extract was assayed directly for GlcN-Ins content, while a second portion was assayed after treatment with a preparation of expressed Rv1170 under conditions sufficient to convert all GlcNAc-Ins to GlcN-Ins. The difference in these two assay results yields the GlcNAc-Ins content for the sample. In most cases, GlcN-Ins levels are at least an order of magnitude lower than those of GlcNAc-Ins, so this method gives reliable results. Values are reported as micromoles per gram of residual dry weight (RDW) determined on the residue from the 50% acetonitrile extract. The relationship between A600 and RDW was determined for aliquots of mc2155 cells producing ∼30 mg of RDW and found to be 0.42 ± 0.02 mg of RDW per ml of cells with an A600 at 1.00. No variation of >15% in this value was seen between log- and stationary-phase cells.
The method was tested with extracts prepared from log-phase cells of mutant 49 by spiking the extract with GlcNAc-Ins at levels equivalent to 0.13 and 0.013 μmol per g of RDW. The recoveries from triplicate determinations were 97% ± 5% and 132% ± 3%, respectively. At the lowest level of added GlcNAc-Ins, a significant correction had to be applied for background peaks present in unspiked controls which overlapped the GlcN-Ins peak. The greater-than-100% recovery found for the samples spiked at the lowest level indicates that this correction produces a systematic error and thus defines the lower limit of useful sensitivity for this assay.
M. smegmatis mc2155 produces GlcNAc-Ins, but MSH-deficient mutant 49 does not.
M. smegmatis mc2155 cells were examined in exponential and stationary phases for their content of MSH and its precursors (Table 2). M. smegmatis accumulates GlcNAc-Ins to a substantial level, almost twice the MSH content. The GlcN-Ins content was very much lower in log phase and declined further in stationary phase, as had been observed previously (1). When mutant 49 was examined in analogous fashion, the levels of MSH and its precursors were below the limits of detection (see Table 3; [GlcNAc-Ins] = 0). The absence of GlcNAc-Ins in mutant 49 shows that it is defective in an initial step of MSH biosynthesis.
TABLE 2.
Levels of MSH and precursors in M. smegmatis mc2155
| Growth phase | A600 | Content (μmol [g of RDW]−1) (n = 3)
|
||
|---|---|---|---|---|
| MSH | GlcNAc-Ins | GlcN-Ins | ||
| Log | 0.54 | 7.8 ± 0.6 | 14.5 ± 1.2 | 0.2 ± 0.1a |
| Stationary | 3.3 | 6.6 ± 0.6 | 12.6 ± 0.6 | <0.03 |
Best estimate after correction for overlapping peak in HPLC analysis.
TABLE 3.
Levels of MSH and precursors in MSH-deficient strain 49 grown to late log phase (A600, ∼ 2.8) in medium containing various amounts of GlcNAc-Ins
| [GlcNAc-Ins]a (μM) | Content (μmol [g of RDW]−1)
|
||
|---|---|---|---|
| MSH | GlcNAc-Ins | GlcN-Ins | |
| 0 | <0.04 | <0.01 | <0.02 |
| 4 | 0.47 | 0.39 | <0.02 |
| 8 | 0.91 | 0.80 | 0.026 |
| 21 | 2.1 | 1.4 | 0.047 |
In Middlebrook 7H9 medium.
Extracts of M. smegmatis and mutant 49 have deacetylase activity.
If the deacetylase is involved in MSH biosynthesis and if mutant 49 is blocked at an earlier step in the pathway, then strains mc2155 and 49 should both produce deacetylase activity. Centrifuged extracts of exponentially growing cells were assayed in duplicate for GlcN-Ins production with and without the addition of 100 μM GlcNAc-Ins. The background rate for production of GlcN-Ins measured without added GlcNAc-Ins was high for strain mc2155 (6.1 ± 0.1 pmol min−1 mg of protein−1), presumably reflecting the substantial endogenous level of GlcNAc-Ins present in the undialyzed extract. Addition of 100 μM GlcNAc-Ins increased the rate to 19.7 ± 0.4 pmol min−1 mg of protein−1, giving a net rate increase of 13.6 ± 0.5 pmol min−1 mg of protein−1. For mutant 49, which does not contain GlcNAc-Ins, the background rate was <0.06 pmol min−1 mg of protein−1, and the rate with 100 μM GlcNAc-Ins was 16.4 ± 1.4 pmol min−1 mg of protein−1. Thus, the deacetylase activity of strain 49 is essentially the same as that of the parent strain.
MSH-deficient mutant 49 can import GlcNAc-Ins and utilize it to synthesize MSH.
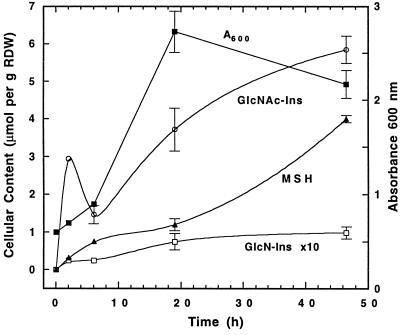
The foregoing results imply that mutant 49 should be able to synthesize MSH if supplied with GlcNAc-Ins. When mutant 49 was grown in media supplemented with GlcNAc-Ins and the cells were analyzed for their content of MSH and MSH precursors, the results established that GlcNAc-Ins is imported and utilized for MSH biosynthesis (Table 3). Levels of MSH and its precursors were found to increase with increasing concentrations of GlcNAc-Ins added to the growth medium. In another experiment, we measured cellular GlcN-Ins, GlcNAc-Ins, cysteine, and MSH levels for strain 49 as a function of growth in medium containing 17 μM GlcNAc-Ins (Fig. 4). Cellular cysteine levels remained unchanged at 0.36 ± 0.08 μmol per g of RDW over the experiment. Cells contained 2.9 μmol per g of RDW of GlcNAc-Ins at the first measurement (2 h), which corresponds to a level approaching millimolar. This occurred prior to the appearance of significant GlcN-Ins in the cell, showing that strain 49 is able to import and concentrate GlcNAc-Ins intact prior to its deacetylation. The level of GlcNAc-Ins appeared to fall at 6 h before rising to an even higher level in stationary phase.
FIG. 4.
Growth of mutant 49 in 7H9 Middlebrook medium containing 17 μM GlcNAc-Ins: MSH and MSH precursor contents are expressed in micromoles per gram of RDW.
The MSH content was measurable as early as 2 h but was about 10-fold less than the GlcNAc-Ins content at that time. The MSH content increased thereafter, and the level at 48 h was about 60% of that measured for the parent strain (Table 2). The level of GlcN-Ins ranged from 0.025 (2 h) to 0.1 (48 h) μmol per g of RDW, values in the range of those measured for mc2155 (Table 2) (1). These results show that exogenous GlcNAc-Ins is efficiently imported by strain 49 and substantially restores its defective biosynthesis of MSH.
Failure to detect GlcNAc-Ins production from UDP-GlcNAc and Ins.
To test whether UDP-GlcNAc could transfer GlcNAc to myo-inositol to produce GlcNAc-Ins, we examined a dialyzed, uncentrifuged, cell extract of M. smegmatis mc2155 for evidence of GlcNAc-Ins formation from these precursors. The extract was assayed for the production of GlcN-Ins from 0.1 mM GlcNAc-Ins, and 22 ± 1 pmol min−1 mg of protein−1 of GlcNAc-Ins deacetylase activity was found. Thus, if GlcNAc-Ins were formed in our assay mixtures, it would be deacetylated by endogenous GlcNAc-Ins deacetylase to GlcN-Ins and assayed as the free amine. When extracts were analyzed for the production of GlcN-Ins from 0.1 mM UDP-GlcNAc and 0.1 mM myo-inositol, GlcN-Ins formation was less than 0.4 pmol min−1 mg of protein−1.
DISCUSSION
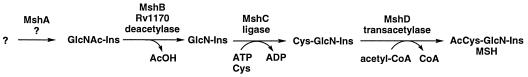
The results presented here further elaborate the pathway involved in MSH biosynthesis. The final two steps in the pathway were postulated by Bornemann et al. (3) to involve the ATP-dependent ligation of Cys with GlcN-Ins to produce Cys-GlcN-Ins, followed by transacetylation of the latter by acetyl-CoA, as shown in Fig. 5. The present studies demonstrate that GlcNAc-Ins is the major intracellular MSH component in M. smegmatis and is converted to GlcN-Ins by GlcNAc-Ins deacetylase. This defines what we propose as the second step in MSH biosynthesis. Assignment of Rv1170 as the gene coding for the deacetylase in the M. tuberculosis genome represents the first gene of the MSH biosynthesis pathway to be identified. Assuming that GlcNAc-Ins is produced by a single enzyme, we propose that the genes for the MSH biosynthesis pathway be designated mshA, mshB, mshC, and mshD, with the corresponding enzymes labeled as shown in Fig. 4. Identification of mshA, mshC, and mshD would be greatly simplified if they were clustered with mshB in a single operon, but inspection of the gene assignments and open reading frames surrounding mshB (Rv1170) in the M. tuberculosis genome (4) indicates that this is not the case.
FIG. 5.
Proposed MSH biosynthesis pathway.
All mycobacteria thus far examined have high MSH content (10) and are expected to have an ortholog of the MSH biosynthesis enzyme MshB (Rv1170). The available unfinished mycobacterial genome databases yield homologs close to Rv1170 for Mycobacterium leprae (Sanger Centre), Mycobacterium bovis (Sanger Centre), M. tuberculosis CDC1551 (The Institute for Genomic Research [TIGR]), M. smegmatis (TIGR), and Mycobacterium avium (TIGR). This indicates that MSH biosynthesis in these organisms utilizes a GlcNAc-Ins deacetylase (MshB) in the same manner as that described here for M. smegmatis. Other actinomycetes that produce MSH are also expected to have a GlcNAc-Ins deacetylase (MshB) gene homologous to Rv1170.
Sequence BLAST searches using the Rv1170 sequence also produced M. tuberculosis homolog Rv1082 (mycothiol conjugate amidase) and the Rv1082 orthologs from M. leprae (Sanger Centre), M. bovis (Sanger Centre), M. tuberculosis CDC1551 (TIGR), M. smegmatis (TIGR), and M. avium (TIGR) as the closest homologs. Sequences with partial homology to Rv1170 were also found in the yeast, rat, and human genome databases. These genes code for PIG-L, the second enzyme in GPI anchor biosynthesis (9), and catalyze the deacetylation of N-acetylglucosaminyl-phosphatidylinositol (GlcNAc-PI). One sequence near the amino-terminal portion of Rv1170, VXAHPDDE, is conserved throughout and is a candidate for the catalytic site based upon the functional similarity of these enzymes. Although the substrate structures differ, all of the these enzymes hydrolyze a C2-amide bond of glucosamine in various disaccharide substrates containing the glucosaminyl-inositol moiety.
Rv1170 (mshB) is the fourth gene of MSH metabolism to be identified. The first was for MSH-dependent formaldehyde dehydrogenase of Amycolatopsis methanolica (8, 16), whose protein sequence is 80% identical to Rv2259 of M. tuberculosis (Sanger Centre). A disulfide reductase (Rv2855) initially designated as a glutathione reductase (4) was cloned and expressed in M. smegmatis and was shown to have specificity for mycothiol disulfide, alternatively designated mycothione (17, 18). The mycothiol S-conjugate amidase gene (Rv1082, mca) was the third gene identified (11), making its homolog GlcNAc-Ins deacetylase (Rv1170, mshB) the fourth.
The results indicate that the deacetylase is rather specific for GlcNAc-Ins and is not a broad-spectrum deacetylase. Removing the Ins residue from the substrate resulted in a more than 300-fold decrease in reactivity (GlcNAc-Ins versus GlcNAc) (Table 1 and Fig. 2). Also, no deacetylation activity was detected with MSH or MSmB as substrates (Table 1), so the AcCys moiety of MSH (Fig. 2) is inert to the deacetylase. This shows that the deacetylase has no significant activity for the degradation of MSH. A more complete evaluation of the substrate specificity of the deacetylase must await its purification to homogeneity.
The deacetylase appears to be the control point for MSH biosynthesis. Thus, there is a very large endogenous pool of its substrate, GlcNAc-Ins (12 to 15 μmol per g of RDW) (Table 2), but quite low levels of its product, GlcN-Ins (≤0.2 μmol per g of RDW) (Table 2), and of the final intermediate, Cys-GlcN-Ins, (<0.006 μmol per g of RDW) leading to MSH. This suggests that substantial quantities of MSH can be produced upon demand from the endogenous pool of GlcNAc-Ins under control of the deacetylase (MshB). Exactly how the activity of the deacetylase is controlled is not clear and will be explored in future studies with the purified enzyme.
Tests for GlcNAc transferase activity producing GlcNAc-Ins with UDP-GlcNAc and myo-inositol as substrates did not yield measurable activity. This suggests that the route to GlcNAc-Ins in mycobacteria may not be analogous to that followed in eucaryotic GPI anchor biosynthesis. Alternatively, the reaction may require some specialized conditions not yet identified. Thus, the detailed nature of the first step of MSH biosynthesis remains to be established.
ACKNOWLEDGMENTS
We thank Mary Ko for technical assistance.
This work was supported by grants to R.C.F. from the National Institute of Alcoholism and Alcohol Abuse (AA11393), the National Institute of Allergy and Infectious Diseases (AI49174), the National Science Foundation (MCB-998150), and the Fogarty International Center (TW00976). Research by Y.A.-G. at the University of British Columbia was supported by the British Columbia Lung Association and the TB Veterans Association.
REFERENCES
- 1.Anderberg S, Newton G L, Fahey R C. Mycothiol biosynthesis and metabolism: cellular levels of potential intermediates in the biosynthesis and degradation of mycothiol. J Biol Chem. 1998;273:30391–30397. doi: 10.1074/jbc.273.46.30391. [DOI] [PubMed] [Google Scholar]
- 2.Av-Gay Y, Jamil S, Drews S J. Expression and characterization of the Mycobacterium tuberculosis serine/threonine protein kinase PknB. Infect Immun. 1999;67:5676–5682. doi: 10.1128/iai.67.11.5676-5682.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bornemann C, Jardine M A, Spies H S C, Steenkamp D J. Biosynthesis of mycothiol: elucidation of the sequence of steps in Mycobacterium smegmatis. Biochem J. 1997;325:623–629. doi: 10.1042/bj3250623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail M A, Rajandream M-A, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 5.Dolphin D, Poulson R, Avramovic O. Glutathione: chemical, biochemical, and medical aspects, parts A and B. New York, N.Y: John Wiley & Sons, Inc.; 1989. [Google Scholar]
- 6.Englund P T. The structure and biosynthesis of glycosyl phosphatidylinositol protein anchors. Annu Rev Biochem. 1993;62:121–138. doi: 10.1146/annurev.bi.62.070193.001005. [DOI] [PubMed] [Google Scholar]
- 7.Fahey R C, Brown W C, Adams W B, Worsham M B. Occurrence of glutathione in bacteria. J Bacteriol. 1978;133:1126–1129. doi: 10.1128/jb.133.3.1126-1129.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Misset-Smits M, van Ophem P W, Sakuda S, Duine J A. Mycothiol, 1-O-(2′-[N-acetyl-L-cysteinyl]amido-2′-deoxy-alpha-d-glucopyranosyl)-d-myo-inositol, is the factor of NAD/factor-dependent formaldehyde dehydrogenase. FEBS Lett. 1997;409:221–222. doi: 10.1016/s0014-5793(97)00510-3. [DOI] [PubMed] [Google Scholar]
- 9.Nakamura N, Inoue N, Watanabe R, Takahashi M, Takeda J, Stevens V L, Kinoshita T. Expression cloning of PIG-L, a candidate N-acetylglucosaminyl-phosphatidylinositol deacetylase. J Biol Chem. 1997;272:15834–15840. doi: 10.1074/jbc.272.25.15834. [DOI] [PubMed] [Google Scholar]
- 10.Newton G L, Arnold K, Price M S, Sherrill C, delCardayré S B, Aharonowitz Y, Cohen G, Davies J, Fahey R C, Davis C. Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J Bacteriol. 1996;178:1990–1995. doi: 10.1128/jb.178.7.1990-1995.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newton G L, Av-Gay Y, Fahey R C. A novel mycothiol-dependent detoxification pathway in mycobacteria involving mycothiol S-conjugate amidase. Biochemistry. 2000;39:10739–10746. doi: 10.1021/bi000356n. [DOI] [PubMed] [Google Scholar]
- 12.Newton G L, Bewley C A, Dwyer T J, Horn R, Aharonowitz Y, Cohen G, Davies J, Faulkner D J, Fahey R C. The structure of U17 isolated from Streptomyces clavuligerus and its properties as an antioxidant thiol. Eur J Biochem. 1995;230:821–825. doi: 10.1111/j.1432-1033.1995.0821h.x. [DOI] [PubMed] [Google Scholar]
- 13.Newton G L, Fahey R C. Glutathione in procaryotes. In: Viña J, editor. Glutathione: metabolism and physiological functions. Boca Raton, Fla: CRC Press; 1989. pp. 69–77. [Google Scholar]
- 14.Newton G L, Fahey R C, Cohen G, Aharonowitz Y. Low-molecular-weight thiols in streptomycetes and their potential role as antioxidants. J Bacteriol. 1993;175:2734–2742. doi: 10.1128/jb.175.9.2734-2742.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newton G L, Unson M, Anderberg S, Aguilera J A, Oh N N, delCardayré S, Av-Gay Y, Fahey R C. Characterization of Mycobacterium smegmatis mutants defective in 1-d-myo-inosityl-2-amino-2-deoxy-α-d-glucopyranoside and mycothiol biosynthesis. Biochem Biophys Res Commun. 1999;255:239–244. doi: 10.1006/bbrc.1999.0156. [DOI] [PubMed] [Google Scholar]
- 16.Norin A, Van Ophem P W, Piersma S R, Persson B, Duine J A, Jornvall H. Mycothiol-dependent formaldehyde dehydrogenase, a prokaryotic medium-chain dehydrogenase/reductase, phylogenetically links different eukaroytic alcohol dehydrogenases—primary structure, conformational modelling and functional correlations. Eur J Biochem. 1997;248:282–289. doi: 10.1111/j.1432-1033.1997.00282.x. [DOI] [PubMed] [Google Scholar]
- 17.Patel M P, Blanchard J S. Expression, purification, and characterization of Mycobacterium tuberculosis mycothione reductase. Biochemistry. 1999;38:11827–11833. doi: 10.1021/bi991025h. [DOI] [PubMed] [Google Scholar]
- 18.Patel M P, Blanchard J S. Synthesis of des-myo-inositol mycothiol and demonstration of a mycobacterial specific reductase activity. J Am Chem Soc. 1998;120:11538–11539. [Google Scholar]
- 19.Sakuda S, Zhou Z-Y, Yamada Y. Structure of a novel disulfide of 2-(N-acetylcysteinyl)amido-2-deoxy-α-d-glucopyranosyl-myo-inositol produced by Streptomyces sp. Biosci Biotech Biochem. 1994;58:1347–1348. doi: 10.1271/bbb.58.1347. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 21.Spies H S, Steenkamp D J. Thiols of intracellular pathogens. Identification of ovothiol A in Leishmania donovani and structural analysis of a novel thiol from Mycobacterium bovis. Eur J Biochem. 1994;224:203–213. doi: 10.1111/j.1432-1033.1994.tb20013.x. [DOI] [PubMed] [Google Scholar]