Abstract
The success of the two mRNA vaccines developed by Moderna and BioNTech during the COVID-19 pandemic increased research interest into the application of mRNA technologies. Compared with the canonical linear mRNA used in these vaccines, circular mRNA has been found to mediate more potent and durable protein expression and demands a simpler manufacturing procedure. However, the application of circular mRNA is still at the initiation stage, and proof of concept for its use as a future medicine or vaccine is required. In the current study, we established a novel type of circular mRNA, termed cmRNA, based on the echovirus 29-derived internal ribosome entry site element and newly designed homology arms and RNA spacers. Our results demonstrated that this type of circular mRNA could mediate strong and durable expression of various types of proteins, compared with typical linear mRNA. Moreover, for the first time, our study demonstrated that direct intratumoral administration of cmRNA encoding a mixture of cytokines achieved successful modulation of intratumoral and systematic anti-tumor immune responses and enhanced anti-programmed cell death protein 1 (PD-1) antibody-induced tumor repression in a syngeneic mouse model. This novel circular mRNA platform is thereby suitable for direct intratumoral administration for cancer therapy.
Keywords: circular mRNA, IRES, cytokines, intratumoral injection, cancer therapy, tumor microenvironment
Graphical abstract
A novel type of circular mRNA, named cmRNA, was designed and its ability to mediate strong and durable protein expression was verified. For the first time, the feasibility of applying cmRNA to intratumoral administration as an effective anti-tumor therapy platform was confirmed.
Introduction
The COVID-19 mRNA vaccines developed by Moderna and BioNTech were granted emergency use authorization by the US Food and Drug Administration (FDA) in 2020, proving that, as a novel class of vaccine, mRNA vaccines meet the requirements for clinical application.1 The COVID-19 mRNA vaccine developed by BioNTech conferred an excellent rate of protection, preventing infection from COVID-19.2,3 Additionally, because of the high titer of neutralizing antibody and the activation of T cell immunity, this mRNA vaccine has been proved to provide protection against COVID-19 variants.4 According to the success of mRNA vaccines during the COVID-19 pandemic, it is believed that mRNA technology may change the paradigm of vaccine and therapeutic development.
To the best of our knowledge, most of the mRNA pipelines that are being evaluated in clinical trials throughout the world are based on typical linear mRNA, which can be generated by RNA polymerase-mediated in vitro transcription (IVT). Linear mRNA consists of a cap structure, 5ʹ and 3ʹ untranslated regions (UTRs), protein encoding region, and poly(A) tail.5 The 5ʹ cap0 or 5ʹ cap1 structure is generated via an enzymatic reaction after the IVT procedure or co-transcriptional incorporation of a cap analog during the IVT procedure.6,7 This capping reaction, which replaces the pre-existing 5ʹ-ppp structure, reduces its immunogenicity and strongly promotes mRNA translational activity by means of the tight interaction between the 5ʹ cap structure and elongation initiation factors.8 The poly(A) tail, which is ∼200–250 bp in length, is generated by poly(A) polymerase after the IVT step and facilitates mRNA stability and finally boosts protein production.9 The 5ʹ and 3ʹ UTRs can be engineered to promote mRNA stability and protein expression.10 To minimize the immunogenicity of mRNA, modified nucleosides can be incorporated into the mRNA strand in the IVT step.11 The complicated structure of linear mRNA determines the complexity of its manufacture, and a relatively low yield may be obtained after a few steps of synthesis and purification.
In addition to linear mRNA, there is emerging research on circular mRNA, which has been proved to mediate potent and durable protein expression in vitro.12 Despite the fact that most circular RNAs found in eukaryotic cells to date are considered non-coding RNAs,13 some circular RNAs are demonstrated to encode and express proteins.14 Most of the circular RNAs that have been identified in eukaryotes are derived from an in vivo back splicing mechanism15 or a rolling circle replication mechanism; for example, hepatitis D virus circular RNA.16 After decades of investigation, different methods for in vitro circularization of RNAs have been developed,17 including enzymatic ligation via T4 ligases, chemical strategies for the ligation of 5ʹ and 3ʹ ends, and a ribozyme strategy by which an intra-molecular covalent bond is formed and circular RNA is generated.
In 1992, Puttaraju and Been developed a group I permuted intron-exon (PIE) system, based on Anabaena group I intron, which they applied to conduct ribozyme-catalyzed in vitro self-splicing to generate exon-exon covalent-ligated circular RNA with high efficiency. In this case, the insertion of foreign nucleic acid sequences into the sites between exons and introns can generate a circular RNA that contains the foreign RNA fragment.18 Wesselhoeft optimized this PIE system by adding homologous arms and spacers to facilitate the generation of large-size circular RNAs and by adding an internal ribosome entry site (IRES) element to mediate efficient protein translation.12 They screened a wide range of viral-origin IRES elements and identified the IRES from coxsackievirus B3 (CVB3) as being highly efficient for cap-independent translation. Importantly, they found that this type of CVB3 IRES-driven circular mRNA conducts almost equal level but more durable protein expression in vitro compared with typical linear mRNA, with or without nucleoside modifications.12 Their further studies indicated that circular mRNA after high-pressure liquid chromatography (HPLC) purification exhibited low mRNA immunogenicity, suggesting that purified circular mRNA may be suitable for in vivo applications such as vaccines or therapeutic medicines.19 However, more evidence is required to approve the clinical usage of circular mRNA, especially from in vivo experiments and clinical trials.
In the current study, we established a novel type of circular mRNA, termed cmRNA, based on echovirus 29 (E29)-derived IRES and newly designed homology arms and RNA spacers. Compared with typical linear mRNA, cmRNA can mediate strong and durable protein expression. Moreover, for the first time, our study demonstrated that circular mRNA encoding a mixture of cytokines can be directly used for intratumoral administration to modulate intratumoral and systemic anti-tumor immune responses, finally leading to an enhancement of anti-programmed cell death protein 1 (PD-1) antibody-induced tumor repression in syngeneic mouse models. Additionally, after optimization of the circular mRNA formulation, a significant improvement in cmRNA-mediated protein expression in vivo was observed. With this optimized formulation, cmRNA resulted in improved anti-tumor effects via intratumoral administration, and elicited significant infiltration as well as activation of CD4+ and CD8+ T cells. In summary, we established cmRNA, a novel circular mRNA platform, and demonstrated that it can be applied for direct intratumoral administration for cancer therapy.
Results
Construction and confirmation of cmRNA, a novel circular mRNA format
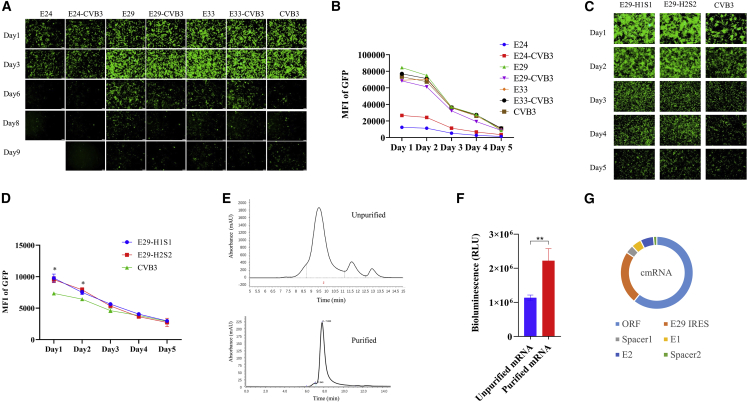
Despite the fact that CVB3 IRES-derived circular mRNA can be converted to protein at high yield, the development of circular mRNA as a therapeutic agent for clinical use (e.g., as a protein replacement therapeutic) requires the expression of a large number of therapeutic proteins, thus it is necessary to identify new IRES elements to enable stronger protein expression. We sought to investigate alternative virus-origin IRES elements for higher and more durable protein expression in mammalian cells. In our study, circular mRNA was generated by a vector that consists of a T7 promoter, the PIE system, various IRES elements, and RNA spacers, which comprise the poly(AC) sequences described by Wesselhoeft.12 The cmRNA was generated, followed by a transcription reaction and a further circularization procedure. As shown in Figures 1A, 1B, and S1A, our results indicated that a series of echovirus-originated IRES elements exhibited significant activity for directing circular RNA translation in the HEK293T cell line, by transfecting equal amounts of different kinds of circular RNAs into HEK293T, and the quantities of RNAs were confirmed by RT-qPCR (Figure S1B). IRES elements originating from echovirus 24 (E24), E29, and echovirus 33 (E33) were found to direct moderate or strong EGFP expression. Among them, E29 was the preferred choice as it directs almost equal protein expression to CVB3 (Figures 1A and 1B), and the expression can last to 11 days (Figure S1A). Furthermore, we designed chimeric IRES components in which the CVB3 core structure was inserted into an echovirus IRES basic framework. The CVB3 core structure is defined as the RNA domain that interacts with the RNA translation complex. However, our results showed that the chimeric IRES did not significantly improve protein expression (Figures 1A and 1B). This may be because the spacers in RNA can facilitate the folding of introns and IRES elements by providing separation between the two RNA elements and this may affect the protein expression of circular mRNA.12 Thus, we also sought to optimize the upstream spacer1 sequence to improve circular mRNA-mediated protein expression. We re-designed the upstream spacers with point mutations for a better separation between intron and IRES, and thereby probably more efficient folding of the IRES was predicted by RNAfoldweb server (http://rna.tbi.univie.ac.at//cgi-bin/RNAWebSuite/RNAfold.cgi).20 The prediction indicated that the new spacer H1S1 facilitated E29 IRES folding with distinct conformational structure compared with the native spacer1 (Figure S1C). Thus, the expression ability of circular RNAs with novel spacers were tested. The downstream spacer2 was unchanged. The EGFP expression mediated by the E29 IRES and two sequences of novel spacer1 (H1S1/H2S2)-based circular mRNA were significantly stronger than the circular mRNA containing the CVB3 IRES and the original spacer1 sequences at 24 and 48 h post transfection (Figures 1C and 1D). Thus, IRES E29 and spacer1 H1S1 were chosen for use in our circular mRNA platform. Collectively, our novel circular mRNA format, termed cmRNA, consists of the E29 IRES, a protein coding region, the upstream spacer1 H1S1, the downstream spacer2, and the Exon2-Exon1 junction (E1-E2) derived from the PIE system (Figure 1G). The circularization reaction can be detected by denatured agarose electrophoresis (Figure S2A) and capillary electrophoresis (CE) (Figure S2B). In the agarose electrophoresis, the circularized RNAs migrate faster than the linear RNA precursor. In CE analysis, the circularized RNAs can be distinguished by their molecular weight, and the circular RNA peak in CE can be quantified and reflects the amount and percentage of circularized RNA. We evaluated the expression of cmRNA with or without HPLC-grade purification in the A549 cell line, which exhibits stronger inflammatory signaling on encountering intracellular foreign RNA than the HEK293T cell line. We found that the purified cmRNA mediated remarkably higher protein expression in A549 cells compared with the unpurified cmRNA (Figures 1E and 1F). The promotion of protein expression may be caused by the removal of immunogenic 5ʹ-ppp containing free-style intron fragments that are generated after the circularization reaction. These results suggested that purification by HPLC is a critical step for cmRNA, whether it is for the elimination of immunogenicity or the promotion of protein expression.
Figure 1.
The development of cmRNA, a novel circular mRNA format
(A) A series of IRES derived from echovirus were tested in circular mRNA to measure their activity in EGFP translation. The image shows the expression of EGFP from day 1 to day 9. (B) Quantification of EGFP expression mediated by cmRNA containing different IRES elements from day 1 to day 5 by fluorescence-activated cell sorting (FACS). (C) EGFP protein expression of circular mRNAs with combinations of IRES E29 and different sequences of new spacer1 were examined from day 1 to day 5 (magnification: 200×; scale bar, 100 μm). (D) Quantification of (C) by FACS. (E) Diagram of HPLC shows the elution of circular mRNA before (up) and after (down) HPLC-SEC purification. (F) Luciferase activity measured 24 h post transfection with cmRNA, with or without HPLC-SEC purification. (G) Scheme for cmRNA. ∗p < 0.05.
cmRNA-directed protein expression in vitro and in vivo
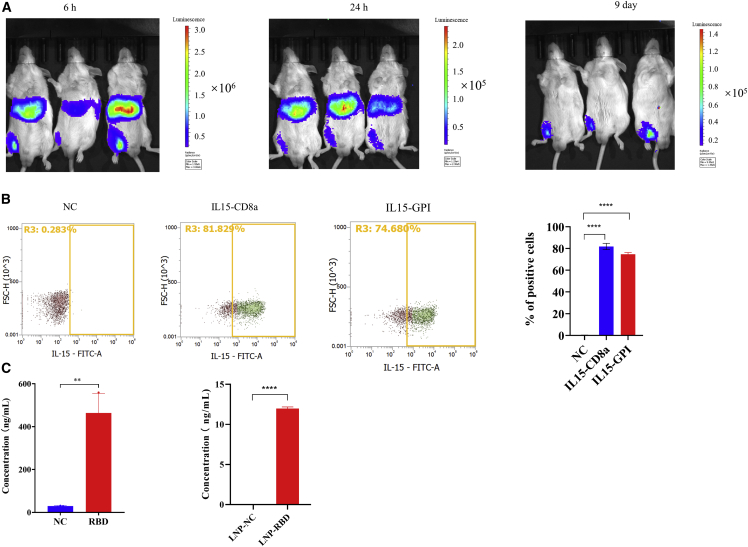
Next, we sought to investigate whether the cmRNA is able to direct the expression of different types of proteins in vitro and in vivo. To delineate the localization of in vivo protein expression, 10 μg of cmRNA encoding luciferase encapsulated by lipid nanoparticles (LNPs) was delivered to mice, and the signal of luciferase bioluminescence was detected by optical in vivo imaging. As shown in Figure 2A, 6 and 24 h after intramuscular injection, potent luciferase activity was detected around the liver and in the muscle at the injection site, suggesting that LNPs mainly delivered cmRNA to the liver and that cmRNA mediated a high level of protein expression. The sharp decrease in luciferase expression in the liver 24 h post injection reflected the high metabolic rate of the liver, as well as the metabolic characteristics of the specific LNPs.21 Importantly, the expression of luciferase was detectable for up to 9 days at the injection site, suggesting that cmRNA mediated durable protein expression in vivo. Due to the different physiological environment, the elimination of LNP and cmRNA in liver is much faster than in muscle. In addition to intracellular proteins (e.g., EGFP and luciferase), secreted protein and transmembrane protein were evaluated in vitro and in vivo. First, to confirm cmRNA-mediated transmembrane protein expression, we evaluated the in vitro expression of interleukin (IL)-15 CD8α and IL-15 GPI, two engineered transmembrane cytokines. As shown in Figure 2B, at 24 h post RNA transfection, IL-15 CD8α and IL-15 GPI were notably expressed on the cell membrane, as detected by flow cytometry, suggesting that cmRNA mediated high and robust expression of these two transmembrane proteins. Next, we investigated whether cmRNA can direct the expression of a secreted protein, namely a homotrimer of the receptor binding domain (RBD) of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), through in vitro transfection. As shown in Figure 2C and 24 h post transfection of the RBD cmRNA into the HEK293T cell line, a high level of secreted recombinant RBD trimer protein was detected in the cell culture supernatant by ELISA. To confirm whether cmRNA can mediate secreted RBD protein expression in vivo, the HPLC-purified RBD cmRNA was encapsulated by LNP and delivered to mice. At 24 h post intramuscular injection of the cmRNA LNP complex, mouse serum was collected and a significant level of RBD trimer protein was detected, suggesting that cmRNA mediated successful RBD protein expression in vivo (Figure 2C). In summary, cmRNA can direct the expression of intracellular, secreted, and transmembrane proteins in vitro. Additionally, the in vivo expression of intracellular protein and secreted protein was verified.
Figure 2.
cmRNA mediated expression of different kinds of proteins in vitro and in vivo
(A) Images show the bioluminescence of firefly luciferase at 6 h, 24 h, and 9 days after intramuscular injection of 10 μg of the LNP-cmRNA complex. (B) Detection of membrane protein IL-15 CD8α and IL-15 GPI expression by FACS. (C) ELISA detection of the secreted form of RBD antigen in the culture supernatant of 293T cells at 24 h post transfection with 0.5 μg of cmRNA, and in mouse serum at 24 h post intramuscular injection with 10 μg of LNP-cmRNA complex. ∗∗p < 0.01, ∗∗∗∗p < 0.0001.
Naked cmRNA can be directly delivered and specifically expressed in tumor tissue by intratumoral administration
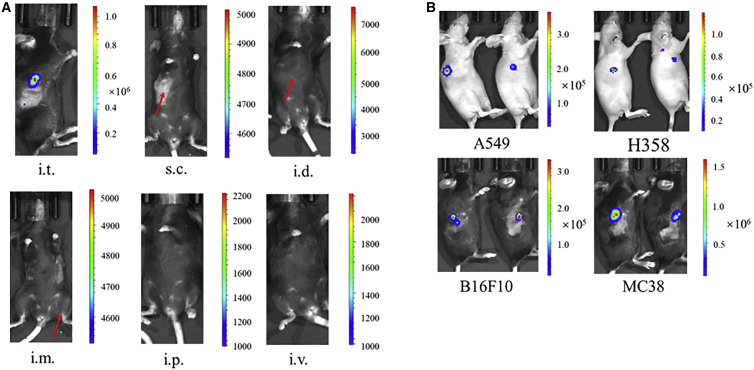
In 2017, Kong et al. reported that cancer cells can spontaneously take up DNA fragments with sizes ranging from 57 to 1,620 bp, which likely reflects the over-activated endocytosis of cancer cells.22 Based on this research, we proposed that circular mRNA may be taken up by cancer cells in a similar way, despite the fact that circular mRNA has a distinct shape and is larger than linear DNA. To test this hypothesis, cmRNA was used for direct intratumoral injection. First, cmRNA encoding luciferase, which was dissolved in PBS, was injected into B16F10 syngeneic tumors in C57BL/6 mice; it was also injected subcutaneously, intradermally, intramuscularly, intraperitoneally, and intravenously for comparison. Significant bioluminescence was specifically detected in the B16F10 tumors after 6 h of cmRNA intratumoral injection (Figure 3A), whereas no bioluminescence was detected with the other injection methods, suggesting that naked cmRNA can be directly delivered and specifically expressed in tumor tissue. According to the report by Kong et al., the direct delivery of naked DNA into cancer cells depends on the characteristics of the cancer cells. Thus, we broadened the types of xenograft tumors tested to include A549 and NCI-H358 non-small-cell lung carcinoma mouse models, as well as a MC38 colon adenocarcinoma mouse model. At 6 h after intratumoral injection of the cmRNA, significant bioluminescence was detected in all of the transplanted tumors (Figure 3B), suggesting that cmRNA was delivered and expressed in the tumor tissue. Thus, the potential for targeting cmRNA to a variety of tumors is promising.
Figure 3.
Intratumoral administration of naked cmRNA directed protein expression in tumors
(A) C57BL/6 mice bearing B16F10 transplanted tumors were injected with 10 μg of naked cmRNA that encodes luciferase via the indicated routes of administration. After 6 h, bioluminescence images were recorded. Scales represent the light intensity in photons per second. i.t., intratumoral; s.c., subcutaneous; i.d., intradermal; i.m., intramuscular; i.p., intraperitoneal; i.v., intravenous. (B) CD-1 nude mice bearing A549 or H358 transplanted tumors and C57BL/6 mice bearing B16F10 or MC38 transplanted tumors were intratumorally injected with 10 μg of naked cmRNA-luciferase. After 6 h, bioluminescence images were recorded. Scales represent the light intensity in photons per second.
Intratumoral cmRNA shows higher and more durable protein expression than linear mRNA
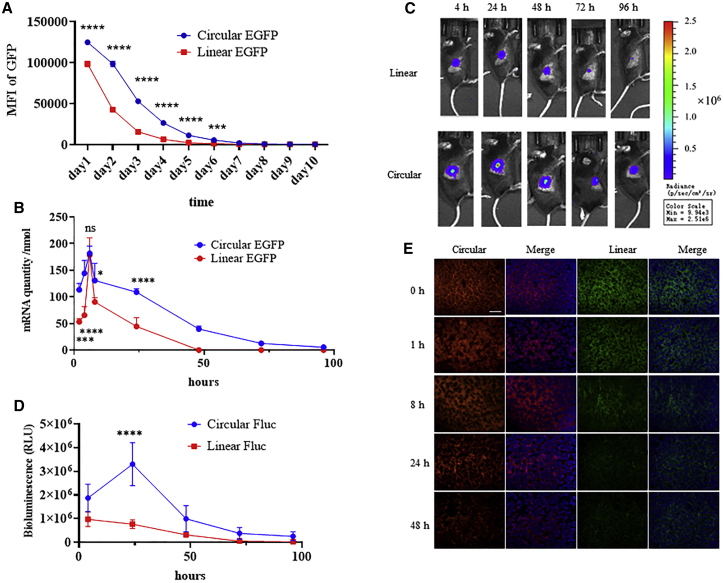
First, protein expression of EGFP by cmRNA and typical linear mRNA were compared in HEK293T cells. As depicted by the in vitro study in Figure 4A, cmRNA mediated higher and more durable protein expression than the typical linear mRNA in the HEK293T cell line. The RNA level inside the cells was also monitored over time by qPCR. As shown in Figure 4B, the cmRNA and linear mRNA both peaked after 6 h post transfection with similar peak value. The elimination rate of cmRNA was much slower. However, it remained to be elucidated whether this phenomenon could be repeated in vivo, especially for the expression of cmRNA in tumor tissues. Thus, we next compared the intratumoral luciferase expression directed by cmRNA and linear mRNA. The linear mRNA was constructed using a typical beta-globin 5ʹ UTR and a tandem beta-globin 3ʹ UTR, the cap1 structure and the poly(A) structure derived from poly(A) polymerase. Intratumoral injection of naked cmRNA or linear mRNA (at equal amounts and each encoding the same luciferase gene) into MC38 tumors in C57BL/6 mice was carried out, and the bioluminescence was measured at the indicated time points. The results indicated that the expression of luciferase can be detected as early as 4 h, and, notably, the bioluminescence level of the cmRNA group was higher than that of the linear mRNA group (Figures 4C and 4D). At the 4-, 24-, 48-, 72-, and 96-h time points, the total bioluminescence signal, calculated according to the area under the curve, was approximately 2-fold higher for cmRNA than for the linear mRNA. Additionally, the bioluminescence level declined more slowly in the cmRNA group, suggesting that cmRNA mediated higher and more durable protein expression in tumor tissue. These results were similar to the in vitro expression data for cmRNA in the HEK293T cell line. Besides protein expression, the mRNA levels were also measured at several time points by RNA fluorescence in situ hybridization (FISH). As shown in Figure 4E, intratumoral injection of the same amount of cmRNA or linear mRNA into B16F10 tumors led to similar initial levels of circular and linear mRNA. However, 24 h after injection, the signal for linear mRNA sharply decreased, whereas the cmRNA signal was maintained, which corresponded to the protein expression level. These results suggested that the long duration of protein expression by cmRNA reflects its high stability rather than unequal RNA uptake of linear versus circular mRNA by tumor cells.
Figure 4.
Comparison of typical linear mRNA and cmRNA for intratumoral protein expression
(A) HEK293T cells were transfected with linear or circular mRNA encoding EGFP. Median fluorescence intensity of EGFP was quantified by FACS at indicated time points. (B) The quantity of linear or circular mRNA was determined by qPCR at indicated time points of (A). (C) C57BL/6 mice bearing MC38 transplanted tumors were intratumorally injected with 10 μg of nucleotide-modified linear luciferase mRNA (Linear Fluc) or cmRNA-luciferase mRNA (Circular Fluc). Bioluminescence images were recorded at the indicated times. (D) Quantification of bioluminescence over time. (E) RNA FISH results of circular mRNA labeled with red probe, linear mRNA with green probe, and DAPI staining (blue) as well as merge images at indicated time points (magnification 400×; scale bar, 25 μm).
Optimization of the cmRNA formulation promoted intratumoral delivery and protein expression
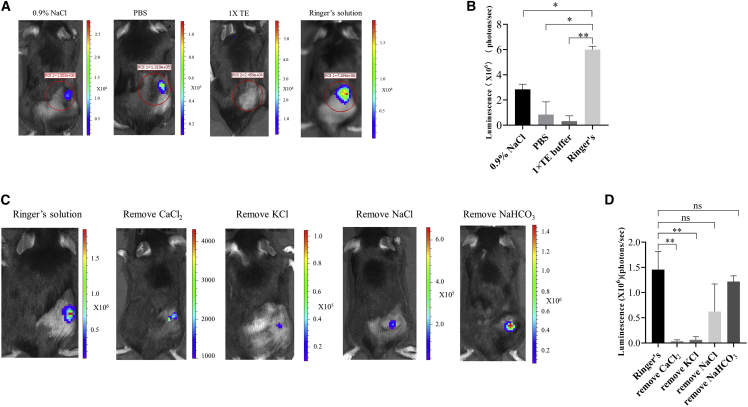
In 2007, Probst et al. reported that Ca2+ affected the delivery of naked linear mRNA into cells via local skin injection.23 The optimization of the mRNA solvent composition led to a ∼10-fold promotion in linear mRNA uptake and protein expression.23 It remained unknown whether the intratumoral delivery of naked circular mRNA could be improved by adjusting the solvent composition. In this study, different compositions were used as cmRNA solvent, including 0.9% NaCl, PBS, 1× TE (Tris-EDTA) buffer, and Ringer’s solution. Bioluminescence was measured 6 h after cmRNA intratumoral administration into B16F10 syngeneic tumors. The results indicated that cmRNA dissolved in Ringer’s solution induced the highest luciferase expression at tumor sites (Figures 5A and 5B). According to the study by Probst et al.,23 the composition of Ringer’s solution, such as the CaCl2, NaCl, KCl, and NaHCO3 content, may be vital for mRNA delivery. Thus, removal of each of these four components individually from the cmRNA solution was performed, and again intratumoral injection into B16F10 tumors was conducted. We found that the removal of CaCl2 or KCl greatly reduced the bioluminescence level following cmRNA injection, and the removal of NaCl and NaHCO3 led to a moderate or mild reduction, respectively (Figures 5C and 5D). These results suggested that CaCl2 and KCl are the two essential components for cmRNA intratumoral delivery.
Figure 5.
Optimization of cmRNA buffer for intratumoral delivery
(A) Representative image of luciferase expression encoded by cmRNA dissolved in different solutions. (B) Quantification of bioluminescence signal of (A). (C) cmRNA dissolved in Ringer’s solution as well as in Ringer’s solutions with the removal of specific components. (D) Quantification of the bioluminescence signal of (C).
cmRNA encoding cytokines facilitated anti-PD-1 antibody-mediated tumor suppression
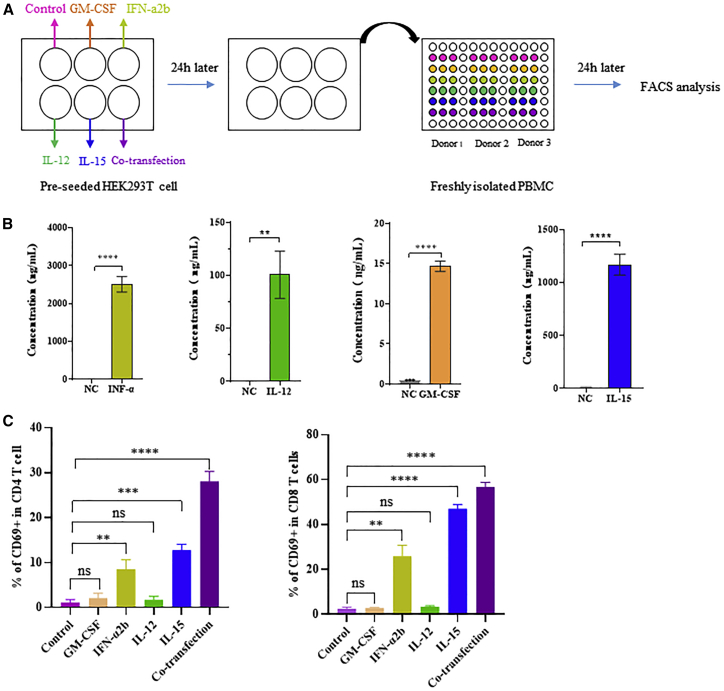
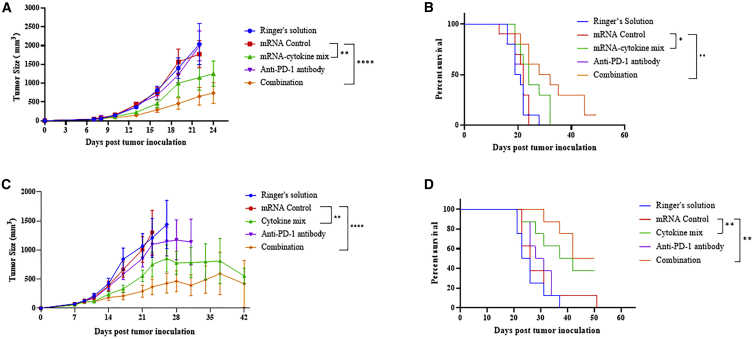
Previous reports indicated that cytokines play critical roles in the regulation of the tumor microenvironment. Here, we employed cmRNAs encoding four different cytokines: active IL-15, a fusion protein of IL-15 and the sushi domain of IL-15Rα; IL-12 single chain (IL-12sc), a fusion protein of p35 and p40; granulocyte macrophage colony-stimulating factor (GM-CSF); and interferon-alpha 2b (IFN-α 2b). After transfecting cmRNAs encoding these four cytokines into HEK293T cells, the expression of the secreted cytokines was quantified by ELISA (Figure 6A). High expression of these cytokines was found 24 h after transfection, compared with the control (the growth medium containing Dulbecco’s modified Eagle’s medium (DMEM) with 10% FBS and 1% penicillin as well as streptomycin) (Figure 6B). Moreover, the co-transfection of the four cmRNAs encoding the respective cytokines was performed and resulted in high expression of the four cytokines respectively (Figure S3). Furthermore, the cell supernatant from each transfection was collected and transferred to peripheral blood mononuclear cells (PBMCs) to assess their activities. We found that incubation with each secreted cytokine resulted in remarkable upregulation of CD69-positive CD4+ and CD8+ T cells in PBMCs (Figure 6C), confirming that individual treatment with each cytokine led to activation of CD4+ and CD8+ T cells in vitro. Significant activation was observed in the group treated with a mixture of secreted cytokines (Figure 6C). Next, cmRNA was dissolved in Ringer’s solution and directly delivered to tumors via intratumoral administration to modulate the tumor microenvironment. A mixture containing 10 μg of each cmRNA was injected intratumorally every 3 days, with or without intraperitoneal administering of anti-PD-1 antibody. Up to 24 days after tumor inoculation, mice with combination treatment presented significant tumor growth inhibition (Figure 7A). Moreover, in the 50-day survival tests, a notable improvement in the final survival rate was found after combination treatment, compared with the negative control or antibody monotherapy, as shown in Figure 7B. These results suggested that mice bearing B16F10 tumors responded well to combination treatment, leading to a significant tumor-suppressive effect and finally resulting in an improved survival rate. Additionally, we evaluated the anti-tumor effect of the cmRNA mixture in the treatment of mice bearing MC-38 tumors. Intratumoral administration of the cmRNA mixture into MC-38 tumors, with Ringer’s solution as solvent, resulted in notable retardation of tumor growth, with either cmRNA monotherapy or combination therapy with anti-PD-1 antibody. Combination treatment of cmRNA and anti-PD-1 antibody presented better control of tumor growth, suggesting that coordination between the cytokine mixture and anti-PD-1 antibody exists (Figure 7C). Importantly, in the 50-day survival tests, a notable improvement in the final survival rate was observed after cmRNA mixture monotherapy or combination treatment (CR 3/8) compared with the negative control or antibody monotherapy (Figure 7D). In summary, these results revealed the strong anti-tumor effect of the cytokine-encoding cmRNA mixture, especially when combined with immune checkpoint blockade agents for tumor therapy.
Figure 6.
In vitro evaluation of cmRNA mediated expression and activity of the four cytokines
(A) Schematic representation of the experimental design. Pre-seeded HEK293T cells were transfected with the indicated cmRNA. After 24 h, the supernatant was collected from each well and centrifuged to remove 293T cells. PBMCs isolated from three healthy donors were seeded into 96-well plates and then resuspended with 100 μL of the indicated supernatant. After 24 h of incubation, PBMCs were stained with fluorophore-conjugated antibodies and analyzed by flow cytometry. (B) The protein concentration in the supernatant of HEK293T cells transfected with indicated cmRNA. (C) The percentage of CD69-positive CD4 T cells among the total CD4 T cells, where each spot represents one blood donor, and the percentage of CD69-positive CD8 T cells among the total CD8 T cells, where each spot represents one blood donor.
Figure 7.
Intratumoral delivered cmRNA encoding cytokines for cancer immunotherapy in B16F10 and MC38 syngeneic mouse model
(A) C57BL/6 mice bearing B16F10 tumors were intratumorally injected with the indicated solution or naked cmRNA and/or intraperitoneally injected with anti-PD-1 antibody. The in vivo anti-tumor activity of mRNA encoding a mixture of four cmRNA cytokines, with/without combination with anti-mouse PD-1 antibody, was evaluated by the tumor growth over time, as well as a survival curve. (B) C57BL/6 mice bearing MC38 tumors were intratumorally injected with the indicated solution or naked cmRNA and/or intraperitoneally injected with anti-PD-1 antibody. The in vivo anti-tumor activity of mRNA encoding a mixture of four cmRNA cytokines, with/without combination with anti-mouse PD-1 antibody, was evaluated by the tumor growth over time, as well as a survival curve. Data plotted are the group mean values for the tumor volume ±SEM. For (A) and (B), n = 10; for (C) and (D), n = 8; ∗∗p < 0.01, ∗∗∗∗p < 0.0001.
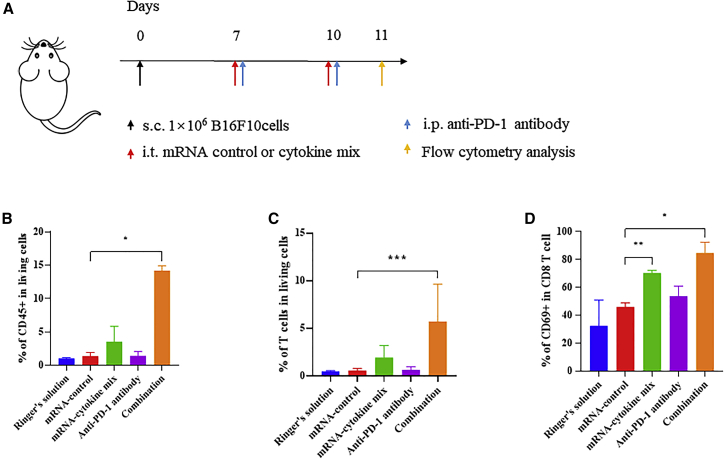
Cytokine-encoding cmRNA suppressed tumor growth by promoting infiltration and activation of T cells
We proposed that the cytokine cmRNA mixture modulates the tumor microenvironment, especially the activation of T cells for tumor suppression. To delineate the cellular mechanism by which the cytokine cmRNA mixture mediates tumor growth suppression, tumor-infiltrated T cells were analyzed by flow cytometry after two doses of treatment (Figure 8A). First, the percentage of infiltrated CD45+ cells was analyzed, and the results indicated that treatment with the cmRNA mixture elicited the promotion of CD45+ cells; importantly, combination treatment with the cmRNA mixture and anti-PD-1 antibody led to a potent elevation in the percentage of CD45+ cells (Figure 8B), suggesting that cmRNA coordinated with anti-PD-1 antibody to facilitate the infiltration of total immune cells into the tumor. We also assessed the percentage of tumor-infiltrated T cells and found that combination treatment resulted in 6% of T cells infiltrating the tumor compared with 0.6% in the anti-PD-1 antibody monotherapy group (Figure 8C). In other words, combination treatment induced T cell infiltration and inflammation in the tumor. The activation of recruited T cells was confirmed by the expression level of the activation marker CD69. As shown in Figure 8D, the percentage of CD69-positive T cells among the total CD4 T cells was increased to 80% following cmRNA monotherapy, as well as cmRNA and anti-PD-1 antibody combination therapy. However, the value was only around 60% for the control and anti-PD-1 antibody monotherapy group. A similar pattern was detected for tumor-infiltrated CD8 T cells. Both cmRNA monotherapy and combination treatment elevated the percentage of CD69-positive cells to values of up to 70% and 80%, respectively. Collectively, not only were more immune cells, especially T cells, recruited to the tumor but also their degree of activation was higher, thus exerting a stronger anti-tumor effect.
Figure 8.
Analysis of the immune modulation by intratumorally delivered cmRNA in mouse tumor model
(A) Experimental timeline for the treatment and analysis of B16F10 tumor-bearing mice. s.c., subcutaneous; i.t., intratumoral; i.p., intraperitoneal. (B) Percentage of CD45-positive cells among the living cells in the tumor. (C) Percentage of CD69-positive cells among the CD4 T cells in the tumor. (D) Percentage of CD69-positive cells among the CD8 T cells in the tumor.
Discussion
The method of generating circular RNA in vitro using the PIE system was developed as early as the 1990s18; however, its application in protein expression to generate circular RNA in vitro has only been realized recently. In fact, the notion of circular RNA-mediated protein expression has only been conceived in recent years. The pioneering work from Wesselhoeft et al. with a series of engineered circular RNAs paved the way for the potential application of circular mRNA as vaccines and therapeutics.12 However, more in vitro and in vivo evidence is required to prove the concept that circular mRNA can be applied to express a variety of proteins, and clinical studies will be required to prove its potential in the development of vaccines and therapeutics. In the current study, we developed a novel format of circular mRNA, termed cmRNA, generated by the PIE system that uses the E29 IRES and novel spacer1 to direct protein translation. Our experimental results demonstrated that cmRNA mediated higher levels of protein translation compared with the circular RNA format by Wesselhoeft et al., as revealed by the high expression of EGFP. Importantly, we found that HPLC-grade RNA purification resulted in notably higher protein expression in an immunogenic sensitive A549 cell line. Considering that immunogenic RNA triggers innate immunity, which may suppress protein expression and cause inflammation in the body,24 the purification of cmRNA by HPLC is crucial for the future development of RNA vaccines and therapeutics. Next, we evaluated whether cmRNA can direct the expression of various types of proteins, such as secreted proteins and transmembrane proteins, both in vitro and in vivo. We proved that cmRNA can direct the in vivo expression of the luciferase intracellular protein in mice, via mRNA delivery by LNP encapsulation. The LNP encapsulation of circular mRNA is similar to that of linear mRNA, and the location of luciferase expression was predominantly in the liver, which was consistent with LNP-mediated linear mRNA delivery.25 Importantly, the expression of luciferase in the muscle at the local injection site lasted 9 days, which is a long duration for mRNA in vivo expression, demonstrating the high stability of cmRNA. Furthermore, a secreted protein, SARS-CoV-2 RBD antigen,26 was encoded by the cmRNA, and significant expression was validated in the HEK-293T cell line as well as in mice, demonstrating that cmRNA directs high expression of secreted protein both in vitro and in vivo. For the evaluation of membrane protein expression, cmRNA directed high levels of expression of IL-15-CD8α or IL-15-GPI membrane proteins in the HEK-293T cell line, suggesting that membrane proteins are suitable expression targets of cmRNA. Collectively, we conclude that the cmRNA platform is sufficient for the expression of various protein types, including intracellular, secreted, and transmembrane proteins, both in vitro and in vivo. Therefore, it is rational to predict that cmRNA has potential for the development of vaccines and therapeutics because of its ability to express a range of protein types, such as antigens, cytokines, antibodies, transmembrane ligands, receptors, and transcription factors.
In 2016, Breckpot and colleagues reported that direct intratumoral injection of a naked linear mRNA mixture encoding CD40L, constitutively active TLR4 and CD70 resulted in the activation of tumor-infiltrated dendritic cells and exerted anti-tumor effects.27 This study suggested that linear mRNA can be directly injected into tumors for protein expression. As previously described, naked DNA can be taken up by cancer cells.22 However, it remained unknown whether naked circular mRNA, a ring-shaped molecule, could be taken up by cancer cells and expressed in tumor tissue. In the current study, we provide evidence to prove that cmRNA, a new circular mRNA format, can be utilized for intratumoral injection. First, our results demonstrated that naked cmRNA mediated high protein expression by direct injection into tumors. Moreover, cmRNA-directed intratumoral expression was found to be more durable than that induced by linear mRNA, which is a significant finding in terms of the application of mRNA as a vaccine or therapeutic agent. This longer duration of mRNA-directed protein expression may (1) facilitate the development of prophylactic vaccines or therapeutic cancer vaccines by increasing the in vivo production of recombinant viral antigens and eliciting higher-titer neutralizing antibodies or stronger antigen-specific T cell activation; (2) benefit the development of mRNA monoclonal or bispecific antibodies by improving the in vivo pharmaceutical kinetics; and (3) facilitate the development of mRNA-based engineered Car-T/natural killer cells by increasing the duration of chimeric antigen receptor expression in T cells or natural killer cells. Using Ringer’s solution in the formulation, cmRNA can be delivered to tumors efficiently, where it can induce strong protein expression. Taken together, these results demonstrated for the first time that a new type of mRNA, naked cmRNA, can be delivered to tumor tissue via intratumoral administration for targeted protein expression. This newly developed naked cmRNA therefore has potential as an RNA platform to develop a variety of intratumoral therapeutics.
By direct injection of immunological regulatory agents into solid tumors, intratumoral immunotherapy is a promising method by which to modulate the tumor microenvironment.28 Several types of therapeutics have shown their potential for clinical application; for example, the oncolytic virus product T-VEC was approved by the FDA in 2015 for the therapy of advanced melanoma.29 T-VEC conducts the lysis of tumor cells, resulting in the release of tumor-derived antigen (TDA), while simultaneously expressing and releasing GM-CSF to activate dendritic cells and T cells to trigger immune responses to tumor antigens, finally inducing distant immune responses and systemic anti-tumor effects. This mechanism of action is designated as in situ vaccination,30 and therapeutic effects are achieved by a combination of immune checkpoint blockade agents.31 Intratumoral injection of mRNA is an emerging area of research that has provided outstanding therapeutic results in recent years. The pipeline of LNP-delivered linear mRNA intratumoral injection developed by Moderna includes mRNA-2905 (IL-12)32and mRNA-2752 (a mixture of IL-23, IL-36γ, and OX40L).33 In preclinical studies, these therapeutic candidates exhibited remarkable anti-tumor effects with a combination of immune checkpoint blockade agents. The basic principle behind the activity of these candidates is the modulation of the tumor microenvironment by boosting immune cell infiltration and activation, ultimately eliciting systemic anti-tumor responses. In the current study, the therapeutic effect of a mixture of circular mRNA encoding cytokines, including IL-15, IL-12, GM-CSF, and IFN-α 2b, was assessed as the proof of concept. Our results revealed that cmRNA directed high-level expression of those cytokines in vitro, as detected by ELISA. Furthermore, the in vitro-expressed cytokines activated immune cells after transferring to PBMCs, demonstrating that cmRNA-derived cytokines are highly active. The intratumoral administration of the cmRNA mixture resulted in remarkable regression of tumor growth in the B16F10 syngeneic mouse tumor model, demonstrating that cmRNA directed the local expression of cytokines in the tumor, thereby suppressing its growth. Further analysis revealed that the proportion as well as the degree of activation of tumor-infiltrated cytotoxic CD8 T cells were upregulated, suggesting that the tumor microenvironment was actually remodeled by locally expressed cytokines, and facilitated anti-PD-1-mediated immune therapy. The “cold” B16F10 tumor was converted to a “hot” tumor after intratumoral injection of the cmRNA-encoded cytokines. These results are consistent with the outcomes of previous studies showing that intratumoral mRNA can exert microenvironmental modulation,33 and suggests that cmRNA is an excellent vector to express immune-modulatory factors intratumorally for cancer therapy. To verify that intratumorally administered cmRNA-encoded cytokines do exhibit a local effect, we performed intratumoral injection of one of the four cmRNAs that encodes IL-12sc and evaluated its expression across different tissues. As shown in Figure S4, IL-12sc expression was mainly restricted to the tumor tissue. The expression of IL-12 in the heart, liver, spleen, lung, and kidney of cmRNA-injected mice was the same as that in control mice. The IL-12 level briefly increased in the serum, but the concentration was 100 times lower than that in the tumor. These results demonstrated that intratumoral administration of cmRNA could direct the targeted expression of cytokines in tumor tissues, which is promising in terms of the clinical safety of cmRNA therapeutics.
In summary, we established a novel form of circular mRNA, termed cmRNA, in which high-level protein expression is driven by E29, an IRES element derived from E29. This type of circular mRNA is more stable and mediates higher and more durable protein expression than linear mRNA. The HPLC-grade purification of cmRNA resulted in the promotion of protein expression in an immunogenic cell line, probably as a result of the removal of immunogenic 5ʹ-ppp intron fragments, suggesting that cmRNA is safe for in vivo use as a potential vaccine or therapeutic agent. Furthermore, from the successful expression of various types of proteins, including intracellular proteins, secreted proteins, and transmembrane proteins, we conclude that cmRNA is a universal vector for protein expression. The intratumoral expression of cmRNA is found to direct higher and more durable protein expression than typical linear mRNA. The intratumoral administration of a cmRNA mixture encoding four cytokines elicited a notable tumor-suppressive effect by boosting the infiltration of T cells to facilitate immune therapy. Collectively, cmRNA has proved to be a potential platform for the development of mRNA vaccines or therapeutics, and may be a preferable RNA vector because it is simple to manufacture and offers high levels of protein expression.
Materials and methods
Gene cloning and vector construction
DNA fragments containing PIE elements, IRES, coding regions, and other elements were chemically synthesized and cloned into a linearized pUC57 plasmid digested with a restriction enzyme. The vector for linear mRNA containing a beta-globin 5ʹ UTR and tandem beta-globin 3ʹ UTR, and the coding regions, were chemically synthesized and cloned into a linearized pUC57 plasmid digested with a restriction enzyme. DNA synthesis and gene cloning were customized and performed by Suzhou Genewitz (Suzhou, China).
cmRNA preparations
cmRNA precursors were synthesized by in vitro transcription from a linearized plasmid DNA template using a Purescribe T7 High Yield RNA Synthesis Kit (CureMed, Suzhou, China). After in vitro transcription, reaction products were treated with DNase I (CureMed) for 15 min. After DNaseI treatment, unmodified linear mRNA was column purified using a GeneJET RNA Purification Kit (Thermo Fisher Scientific, USA). For the generation of cmRNA, RNA was purified, after which guanosine triphosphate (GTP) was added to a final concentration of 2 mM along with a buffer including magnesium (50 mM Tris-HCl, [pH 8.0], 10 mM MgCl2, 1 mM DTT; Thermo Fisher Scientific). RNA was then heated for 15 min to 55°C and purified through a column. RNA was analyzed using 1% agarose gel electrophoresis and an ssRNA ladder (Thermo Fisher Scientific) was used as a standard.
cmRNA purification
For HPLC, RNA was loaded onto a 30 × 300-mm size exclusion column with a particle size of 5 μm and a pore size of 1,000 Å (Sepax Technologies, Suzhou, China) on an SCG (Sepure Instruments) protein purification system (Sepure Instruments, Suzhou, China). Then the column was eluted with RNase-free phosphate buffer (pH 6) and chromatography was performed at a flow rate of 15 mL/min. Chromatograms were recorded at a wavelength of 260 nm. RNA with high purity was collected by peak capture and concentrated, and the buffer was replaced with RNase-free water following ultracentrifugation.
Linear mRNA preparations
The plasmid vector was digested with XbaI for linearization, and transcription was carried out using the HyperScribe All in One mRNA Synthesis Kit (APExBio), with Cap1 analog incorporation and N1-methyl-pseudouridine as the modified nucleoside in the transcription. RNA was purified using a GeneJET RNA Purification Kit (Thermo Fisher Scientific).
RNA fragment analysis
RNA fragments were analyzed using a fragment analyzer (CE) (Agilent, Fragment analyzer system 5200). The parameters were set up as follows: preform prerun (12.0 kV, 30 s), sample injection (8.0 kv, 6 s), separation (4.0 kv, 200 min), and analysis mode (RNA, eukaryotic).
Cell culturing
HEK293T, the murine colon adenocarcinoma cell line MC38, the murine melanoma cell line B16F10, the human non-small-cell lung cancer cell line A549, and NCI-H358 were purchased from Cobier Biosciences (Nanjing, China). NCI-H358 cells were cultured in RPMI 1640 (BI) supplemented with 10% fetal calf serum (BI) and penicillin/streptomycin antibiotics (100 U/mL penicillin, 100 μg/mL streptomycin; Gibco). The other cells were cultured in DMEM (BI) supplemented with 10% fetal calf serum (BI) and penicillin/streptomycin antibiotics (100 U/mL penicillin, 100 μg/mL streptomycin; Gibco). All cells were maintained at 37°C, 5% CO2, and 90% relative humidity.
In vitro transfection of cmRNA
For the cmRNA transfection of HEK293T cells, 1 × 105 cells per well were seeded into 24- or six-well plates. Then, 500 ng of cmRNAs (for the 24-well plates) or 2 μg of cmRNAs (for the six-well plates) were transfected into each well using Lipofectamine MessengerMax (LMRNA003, Invitrogen) according to the manufacturer’s instructions.
RT-qPCR
Total RNA extraction was with a RNeasy Mini Kit (Qiagen, Germany) according to the Quick-Start Protocol. No more than 2 μg of total RNA was subjected to reverse transcription with PrimeScript RT reagent Kit (Takara Bio, Beijing, China). Real-time PCR was performed using Takara Premix Ex Taq (Probe qPCR). PCR was performed in triplicate for each primer set and the cycling conditions were as follows: preheating at 48°C for 15 min, initial denaturation at 95°C for 10 min, followed by 45 cycles at 95°C for 15 s and 60°C for 60 s. Primers and probe were as follows: left primer, 5′-AAACCCATGGGACGCTCTTA-3′; right primer, 5′-TGGGATTAGCCGCATTCAGG-3′; probe, 5′-FAM-AGCTAGTTGGTAGTCCTCCGGCC-TRAMA-3′.
Protein expression analysis
In general, GFP expression was evaluated by fluorescence imaging using an inverted fluorescence microscope (Olympus IX70) and flow cytometry analysis (Attune NXT, Life Technologies). RBD expression was measured using a commercial ELISA kit (SARS-CoV-2 coronavirus spike ELISA kit, Sino Biological). Transmembrane IL-15 expression was measured using fluorescein isothiocyanate (FITC) anti-IL-15 antibody (MA5-23664, Invitrogen) and flow cytometry. Luciferase expression was evaluated by in vivo bioluminescence imaging (IVIS spectrum imaging system, PerkinElmer). The expression of human IFN-a2b, IL-15, GM-CSF, and IL-12sc were determined using commercial ELISA kits (Human IFN-α ELISA kit, Beyotime; Human IL-15 ELISA kit, Abcam; Human GM-CSF ELISA kit, Proteintech; ELISA MAX Deluxe Set Human IL-12 [p70], BioLegend).
Encapsulation of circular mRNA by LNPs
Circular mRNA-LNP complexes were generated through microfluidic devices (Micro&Nano Technologies, Shanghai, China). Purified circular mRNA was dissolved in citric acid buffer, and lipids were dissolved in ethanol. The liquid flow rate was set as 12 mL/min, and mRNA/lipids (v/v) were used at a 3:1 ratio. Circular mRNA-LNP complexes were purified by filtration.
Mice
C57BL/6 female mice and CD-1 nude female mice were purchased from Charles River (Beijing, China) at the age of 6–8 weeks and housed in specific-pathogen-free facilities. All experiments were conducted in accordance with procedures approved by the Institutional Animal Care and Use Committee (IACUC).
In vivo tumor experiments
For tumor implantation, mice were injected with A549, NCI-H358, B16F10, or MC38 (1 × 106 cells per animal) subcutaneously into the right flank. Tumor growth was monitored by measuring two perpendicular diameters with a digital caliper every 2 to 4 days, and tumor volume was calculated according to the modified ellipsoidal formula: V = ½ (length × width2). Intratumoral injections were performed with a 1-mL 29G × 1/2 insulin syringe (BD).
Bioluminescence imaging
Mice were anesthetized in a chamber with 2.5% isoflurane, along with intraperitoneal injections of D-luciferin (150 mg/kg, Promega). Bioluminescence was measured using an IVIS spectrum imaging system (PerkinElmer) while maintaining 2.5% isoflurane in the imaging chamber via a nose cone. Images were captured 10 min after luciferin administration at the indicated time points. The photon flux values (photons per second), corresponding to the region of interest marked around the bioluminescence signal, were analyzed using Living IMAGE 4.5.2 software (Perkin Elmer, USA).
Tumor sample preparation and flow cytometry
Tumors were isolated and dissected into ∼1-mm3 pieces. These pieces were incubated in DMEM containing 1 mg/mL collagenase-IV (Thermo Fisher Scientific) and 100 U/mL DNase I (Novoprotein, China) for 30 min at 37°C with gentle shaking. Dissociated cells were filtered through a 70-μm nylon mesh filter to obtain single cell suspensions. For antibody staining, cells were first stained with eBioscience Fixable Viability Dye eFluor 780 (1:1,000, Thermo Fisher Scientific) and then washed twice with PBS containing 2% FBS and 2 mM EDTA. Next, cells were stained with the following antibodies (at a 1:100 dilution in PBS containing 2% FBS and 2 mM EDTA): APC anti-mouse CD3ε antibody (BioLegend, catalog no. 152306), Brilliant Violet 605 anti-mouse CD4 antibody (BioLegend, catalog no. 100451), Brilliant Violet 421 anti-mouse CD8a (BioLegend, catalog no. 100737), FITC anti-mouse CD69 antibody (BioLegend, catalog no. 104506), and PE anti-mouse CD45 (BioLegend, catalog no. 103106). After incubation for 20 min at 4°C, cells were washed twice with PBS containing 2% FBS and 2 mM EDTA, and analyzed by flow cytometry (Attune NXT, Life Technologies).
PBMC isolation and in vitro studies
Peripheral blood samples (buffy coats) from healthy volunteers were isolated by Ficoll Hypaque gradient separation (Ficoll-Paque-Plus) and washed three times with PBS supplemented with 1 mM EDTA. Freshly isolated PBMCs were seeded into 96-well plates with the density of 500,000 cells/well and treated with a cytokine mixture (collected from different mRNA-transfected 293T supernatants) for 24 h. Then, IFN-γproduction was determined using a human IFN-γELISA kit (Neobioscience, China).
RNA FISH
Tumor tissues injected with mRNA were collected at the indicated time points, and tissue sections were prepared conventionally via the following steps: tissue fixation with 10% neutral buffered formalin, dehydration, embedding in paraffin, and sectioning to a 5-μm thickness. RNA probes were designed based on the mRNA sequences and labeled with digoxin or biotin. The FISH experiment was performed using a commercial RNA FISH kit (Future Biotech, Beijing, China).
In vivo mIL-12p70 quantification
At various protocol-specified endpoint times after mIL-12 mRNA administration, serum was collected from live and terminal bleeds, and organs including the heart, liver, spleen, lung, kidney, lymph node, and tumor were exercised after cardiac perfusion, mechanically disaggregated, and clarified by centrifugation to obtain the organ supernatant. The total protein levels in the serum and organ supernatant were quantified using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. mIL-12 levels in serum and the organ supernatant were quantified using the ELISA MAX Deluxe Set Mouse IL-12 (p70) (BioLegend) according to the manufacturer’s instructions. mIL-12 protein was normalized to the total protein (picograms of mIL-12 per gram of total protein).
Statistical analyses
A two-tailed Student’s t test was used to determine statistical significance for data comparisons at a single time point. Two-way ANOVA was used to determine statistical significance for data comparisons with multiple time points. The Mantel-Cox log rank test was performed to determine statistical significance for the comparison of survival curves. Prism version 8.0 (GraphPad) was used to generate all graphs and to perform statistical analyses. The statistical significance shown for the survival curves represents a comparison of the two survival curves. Statistical significance is denoted as ns (not significant), ∗p < 0.05, ∗∗p < 0.01, or ∗∗∗p < 0.001.
Data availability statement
All data that support the findings of this study are available from the corresponding author upon reasonable request.
Acknowledgments
We thank Mr. Lau Ngai Cheung Patrick for the funding of this project. This study was supported by grants from the China National Center for Biotechnology Development (2021YFC250230). We thank Yingwen Li MSc for support with cmRNA encapsulation in LNPs, and Yuxin Mang BSc and Dan Zheng BSc for their support with animal experiments.
Author contributions
J.Y. and J.Z. designed and performed experiments, analyzed data, and interpreted results. J.S., Y.C., Y.D., Y.T., L.W., M.Z., L.W., N.L., and K.H. performed experiments. Q.H. performed the codon optimization. Z.T., H.T., and Z.S. provided expertise and feedback. A.B. revised manuscript. C.Z. conceived the project, designed experiments, and wrote the manuscript. Z.S. and C.Z co-supervised all the research.
Declaration of interests
One patent has been filed to protect cmRNA as well as its application (inventors Z.S., C.Z., and J.Z.), one patent has been filed to protect the use of circular mRNA administrated intratumorally as anti-tumor therapy (inventors C.Z., J.Y., and Y.T.), and one patent has been filed to protect the use of Ringer’s solution for circular mRNA intratumoral administration (inventors J.Y., Y.T., and J.Z.).
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.09.010.
Contributor Information
Zhenhua Sun, Email: sunzh@purecell.group.
Chijian Zuo, Email: zuocj@purecell.group.
Supplemental information
References
- 1.Oliver S.E., Gargano J.W., Marin M., Wallace M., Curran K.G., Chamberland M., Mcclung N., Campos-Outcalt D., Morgan R.L., Mbaeyi S., et al. The advisory committee on immunization practices' interim recommendation for use of pfizer-BioNTech COVID-19 vaccine - United States, december 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:1922–1924. doi: 10.15585/mmwr.mm6950e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dagan N., Barda N., Kepten E., Miron O., Perchik S., Katz M.A., Hernán M.A., Lipsitch M., Reis B., Balicer R.D. BNT162b2 mRNA covid-19 vaccine in a nationwide mass vaccination setting. N. Engl. J. Med. 2021;384:1412–1423. doi: 10.1056/NEJMoa2101765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jalkanen P., Kolehmainen P., Häkkinen H.K., Huttunen M., Tähtinen P.A., Lundberg R., Maljanen S., Reinholm A., Tauriainen S., Pakkanen S.H., et al. COVID-19 mRNA vaccine induced antibody responses against three SARS-CoV-2 variants. Nat. Commun. 2021;12:3991. doi: 10.1038/s41467-021-24285-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuchs A.L., Neu A., Sprangers R. A general method for rapid and cost-efficient large-scale production of 5' capped RNA. RNA. 2016;22:1454–1466. doi: 10.1261/rna.056614.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henderson J.M., Ujita A., Hill E., Yousif-Rosales S., Smith C., Ko N., Mcreynolds T., Cabral C.R., Escamilla-Powers J.R., Houston M.E. Cap 1 messenger RNA synthesis with Co-transcriptional CleanCap((R)) analog by in vitro transcription. Curr. Protoc. 2021;1:e39. doi: 10.1002/cpz1.39. [DOI] [PubMed] [Google Scholar]
- 8.Koromilas A.E., Lazaris-Karatzas A., Sonenberg N. mRNAs containing extensive secondary structure in their 5' non-coding region translate efficiently in cells overexpressing initiation factor eIF-4E. EMBO J. 1992;11:4153–4158. doi: 10.1002/j.1460-2075.1992.tb05508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallie D.R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 10.Asrani K.H., Farelli J.D., Stahley M.R., Miller R.L., Cheng C.J., Subramanian R.R., Brown J.M. Optimization of mRNA untranslated regions for improved expression of therapeutic mRNA. RNA Biol. 2018;15:756–762. doi: 10.1080/15476286.2018.1450054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Wesselhoeft R.A., Kowalski P.S., Anderson D.G. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun. 2018;9:2629. doi: 10.1038/s41467-018-05096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L., et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 14.Pamudurti N.R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., Ruhe L., Hanan M., Wyler E., Perez-Hernandez D., Ramberger E., et al. Translation of CircRNAs. Mol. Cell. 2017;66:9–21.e7. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L.L., Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macnaughton T.B., Shi S.T., Modahl L.E., Lai M.M.C. Rolling circle replication of hepatitis delta virus RNA is carried out by two different cellular RNA polymerases. J. Virol. 2002;76:3920–3927. doi: 10.1128/JVI.76.8.3920-3927.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Müller S., Appel B. In vitro circularization of RNA. RNA Biol. 2017;14:1018–1027. doi: 10.1080/15476286.2016.1239009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Puttaraju M., Been M.D. Group I permuted intron-exon (PIE) sequences self-splice to produce circular exons. Nucleic Acids Res. 1992;20:5357–5364. doi: 10.1093/nar/20.20.5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wesselhoeft R.A., Kowalski P.S., Parker-Hale F.C., Huang Y., Bisaria N., Anderson D.G. RNA circularization diminishes immunogenicity and can extend translation duration in vivo. Mol. Cell. 2019;74:508–520.e4. doi: 10.1016/j.molcel.2019.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gruber A.R., Lorenz R., Bernhart S.H., Neuböck R., Hofacker I.L. The Vienna RNA websuite. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ke Huang N.L.Y.L., Yuping Liu Q.H.S.G., Ke Wei C.D.C.Z., Sun Z. Delivery of circular mRNA via degradable lipid nanoparticles against SARS-CoV-2 delta variant. bioRxiv. 2022 Preprint at. [Google Scholar]
- 22.Kong Y., Zhang X., Zhao Y., Xue Y., Zhang Y. Uptake of DNA by cancer cells without a transfection reagent. Biol. Res. 2017;50:2. doi: 10.1186/s40659-017-0107-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Probst J., Weide B., Scheel B., Pichler B.J., Hoerr I., Rammensee H.G., Pascolo S. Spontaneous cellular uptake of exogenous messenger RNA in vivo is nucleic acid-specific, saturable and ion dependent. Gene Ther. 2007;14:1175–1180. doi: 10.1038/sj.gt.3302964. [DOI] [PubMed] [Google Scholar]
- 24.Chen N., Xia P., Li S., Zhang T., Wang T.T., Zhu J. RNA sensors of the innate immune system and their detection of pathogens. IUBMB Life. 2017;69:297–304. doi: 10.1002/iub.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hou X., Zaks T., Langer R., Dong Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021;6:1078–1094. doi: 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 27.Van Lint S., Renmans D., Broos K., Goethals L., Maenhout S., Benteyn D., Goyvaerts C., Du Four S., Van der Jeught K., Bialkowski L., et al. Intratumoral delivery of TriMix mRNA results in T-cell activation by cross-presenting dendritic cells. Cancer Immunol. Res. 2016;4:146–156. doi: 10.1158/2326-6066.CIR-15-0163. [DOI] [PubMed] [Google Scholar]
- 28.Hamid O., Ismail R., Puzanov I. Intratumoral immunotherapy-update 2019. Oncol. 2020;25:e423–e438. doi: 10.1634/theoncologist.2019-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harrington K.J., Puzanov I., Hecht J.R., Hodi F.S., Szabo Z., Murugappan S., Kaufman H.L. Clinical development of talimogene laherparepvec (T-VEC): a modified herpes simplex virus type-1-derived oncolytic immunotherapy. Expert Rev. Anticancer Ther. 2015;15:1389–1403. doi: 10.1586/14737140.2015.1115725. [DOI] [PubMed] [Google Scholar]
- 30.Johnson D.B., Puzanov I., Kelley M.C. Talimogene laherparepvec (T-VEC) for the treatment of advanced melanoma. Immunotherapy. 2015;7:611–619. doi: 10.2217/imt.15.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larocca C.A., Leboeuf N.R., Silk A.W., Kaufman H.L. An update on the role of talimogene laherparepvec (T-VEC) in the treatment of melanoma: best practices and future directions. Am. J. Clin. Dermatol. 2020;21:821–832. doi: 10.1007/s40257-020-00554-8. [DOI] [PubMed] [Google Scholar]
- 32.Hewitt S.L., Bailey D., Zielinski J., Apte A., Musenge F., Karp R., Burke S., Garcon F., Mishra A., Gurumurthy S., et al. Intratumoral IL12 mRNA therapy promotes TH1 transformation of the tumor microenvironment. Clin. Cancer Res. 2020;26:6284–6298. doi: 10.1158/1078-0432.CCR-20-0472. [DOI] [PubMed] [Google Scholar]
- 33.Hewitt S.L., Bai A., Bailey D., Ichikawa K., Zielinski J., Karp R., Apte A., Arnold K., Zacharek S.J., Iliou M.S., et al. Durable anticancer immunity from intratumoral administration of IL-23, IL-36gamma, and OX40L mRNAs. Sci. Transl. Med. 2019;11:eaat9143. doi: 10.1126/scitranslmed.aat9143. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data that support the findings of this study are available from the corresponding author upon reasonable request.