Abstract
We report the cloning, sequencing, and characterization of the rpoE homolog in Vibrio angustum S14. The rpoE gene encodes a protein with a predicted molecular mass of 19.4 kDa and has been demonstrated to be present as a single-copy gene by Southern blot analysis. The deduced amino acid sequence of RpoE is most similar to that of the RpoE homolog of Sphingomonas aromaticivorans, ς24, displaying sequence similarity and identity of 63 and 43%, respectively. Northern blot analysis demonstrated the induction of rpoE 6, 12, and 40 min after a temperature shift to 40°C. An rpoE mutant was constructed by gene disruption. There was no difference in viability during logarithmic growth, stationary phase, or carbon starvation between the wild type and the rpoE mutant strain. In contrast, survival of the mutant was impaired following heat shock during exponential growth, as well as after oxidative stress at 24 h of carbon starvation. The mutant exhibited microcolony formation during optimal growth temperatures (22 to 30°C), and cell area measurements revealed an increase in cell volume of the mutant during growth at 30°C, compared to the wild-type strain. Moreover, outer membrane and periplasmic space protein analysis demonstrated many alterations in the protein profiles for the mutant during growth and carbon starvation, as well as following oxidative stress, in comparison with the wild-type strain. It is thereby concluded that RpoE has an extracytoplasmic function and mediates a range of specific responses in stressed as well as unstressed cells of V. angustum S14.
Rapid and efficient adaptation to changes in environmental conditions is required for bacterial replication and survival in natural habitats. The marine bacterium Vibrio angustum S14 produces a highly orchestrated response to starvation and stress conditions, and studies of this organism have provided novel information on adaptive responses (45, 54), including the role of master regulators (11, 42, 44), extracellular signals (55), and regulation of transcript stability essential for the outgrowth response of starved cells (56). Alternative sigma factors play an important role in regulating the transcription of many genes that are induced during stationary phase, starvation, and stress adaptation (16, 58). To examine the role of alternative sigma factors in adaptive responses of V. angustum S14, the identification and characterization of the stress responses mediated by RpoS, the stationary-phase sigma factor, in this organism were sought. By use of an rpoS probe derived from Escherichia coli, several clones from a V. angustum S14 genomic library were isolated. One of these clones encoded another alternative sigma factor, RpoE.
Homologs of rpoE encode proteins that are members of the ςE family, a distinctive subclass of the ς70 type of sigma factors (termed extracytoplasmic-function [ECF] ς factors) (28). In response to the extracellular environment, ECF ς factors have been found to regulate gene expression in diverse bacterial species. RpoE homologs have been implicated as critical in a variety of stress responses. One of the best-studied examples is the role of AlgU in the pathogenicity of Pseudomonas aeruginosa in cystic fibrosis (15). Other, less-characterized examples include the recently reported critical role of the alternative sigma factor, ςE, in the virulence of Salmonella enterica serovar Typhimurium (22), the control of alginate production and tolerance of environmental stress shown by AlgT in the phytopathogen Pseudomonas syringae (24), and the decreased survival of a Mycobacterium smegmatis sigE mutant under conditions of oxidative stress (59), indicating a possible role for ςE in the survival following uptake by macrophages of pathogenic mycobacteria. Reports suggest a role for ECF ς factors in the expression of genes enhancing bacterial adaptation to environmental conditions adverse to growth like heat shock (10, 17, 20, 33, 50, 59), oxidative stress (10, 59, 60), osmotic shock (5), adaptation to cold temperatures and high pressures (7), protection against photolysis (14), acid stress (59), desiccation resistance (39), antibiotic production during stationary phase or at the onset of sporulation (23), and iron limitation (3, 9). More recently, ECF ς factors have also been suggested to be necessary for normal cell wall structure in Streptomyces coelicolor (47), motility behavior under both vegetative and developmental conditions in Myxococcus xanthus (57), and growth at normal temperatures in E. coli (8), indicating a role for these sigma factors in both stressed as well as unstressed environments.
In E. coli, the rpoE gene is induced under conditions leading to the misfolding of proteins in the periplasm and the outer membrane (34, 37). Previously, it has been demonstrated that an extensive overlap exists between the expression profiles of outer membrane and periplasmic proteins during carbon starvation, heat, and ethanol stress in V. angustum S14 (40). These findings suggest that RpoE may play a role in the ability of V. angustum S14 to adapt to environmental stresses. Here, we investigate RpoE-mediated processes in V. angustum S14 by evaluating the role of rpoE in V. angustum S14 during growth, carbon starvation, heat shock, and oxidative stress. This work demonstrates the existence of an rpoE homolog with extracytoplasmic function in V. angustum S14. The rpoE homolog is present as a single-copy gene, which is induced during extreme heat shock, and is involved in survival following heat shock and oxidative stress. This study also provides evidence of a role for rpoE in the protein composition of the outer membrane and periplasm in both stressed and unstressed cells of V. angustum S14.
MATERIALS AND METHODS
Bacterial strains, plasmids, and primers.
The bacterial strains and plasmids used in this study are shown in Table 1, and the primers are listed in Table 2.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Bacterial strains | ||
| V. angustum | ||
| S14 | rpoE+ wild type (CCUG 15956) | 1, 31 |
| S141 | Spontaneous mutant of wild-type S14, Smr | 43 |
| EH1 | S141, rpoE::Kmr | This study |
| E. coli | ||
| XL1-Blue MRF′ | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| XLOLR | As XL1-Blue MRF′ except Su− λr | Stratagene |
| BW20767 | RP4-2tet:Mu-1kan::Tn7 integrant leu-63::IS10 hsdR17 endA1 zbf-5 uidA (ΔMluI):pir+thi | 35 |
| Plasmids | ||
| pGEM-3Z | Cloning vector, Apr | Promega |
| pGP704 | λpir-based suicide vector: Aprori R6K mob RP4 | 36 |
| pBK-CMV | Cloning vector, Kmr | Stratagene |
| pRH324 | rpoS | Gift (R. Hengge-Aronis) |
| pEH8 | pBK-CMV with a 5.4-kb insert containing rpoE isolated from λ-ZAP Express genomic library of S14 | This study |
| pEH13 | pGEM-3Z with a 2.1-kb insert containing rpoE::Kmr from S14 | This study |
| pEH14 | pGP704 with a 2.1-kb insert containing rpoE::Kmr obtained from pEH13 | This study |
TABLE 2.
Primers used in this studya
| Primer | Sequence | Target sequence |
|---|---|---|
| 1F | 5′-GTCAAGGGATCACGGGTAGG-3′ | rpoS on pRH324 |
| 2F | 5′-AGTGCTGGCACGTCGATTCG-3′ | rpoS on pRH324 |
| 1R | 5′-AACACACGCTGTGTGGCTCC-3′ | rpoS on pRH324 |
| 2R | 5′-GCCTCGCTTGAGACTGGCCT-3′ | rpoS on pRH324 |
| C | 5′-TCTAAGTATTCCTGAATCATTAAG-3′ | rpoE on pEH8 |
| F2.mut | 5′-CCACAATATTAATGAGGTTTTAGG-3′ | Upstream of rpoE on pEH8 |
| F3.mut | 5′-ACCTTCTAGCTTCACGCTGCCGC-3′ | nptII on pBSL180 |
| F6.mut | 5′-GATGAATTGCCCGAAGATTAC-3′ | rpoE on pEH8 |
| F10 | 5′-ATGTATCGACTAAAAGCCGAG-3′ | Downstream of rpoE in V. angustum S14 |
| G | 5′-GCATAGAAATGAATTAATTGAAGCG-3′ | rpoE on pEH8 |
| R2 | 5′-ATTATCTGTCGCTCTTTAATAGG-3′ | Upstream of rpoE in V. angustum S14 |
| R2.mut | 5′-AAGGTGCTTTGCGCACTCTCTTT-3′ | rpoE on pEH8 |
| R4.mut | 5′-ACCATAAACCCCAGAGTCCCGCTC-3′ | nptII on pBSL180 |
| R5.mut | 5′-CTTTCAGGGCTGTGTATC-3′ | Downstream of rpoE on pEH8 |
The source for all primers is this study.
Culture media, growth, and starvation conditions.
E. coli strains were grown at 37°C on Luria-Bertani (LB) agar (51). Vibrio strains were grown at 25°C on VNSS agar (32) or LB plates containing NaCl (20 g/liter). LB supplemented with NaCl (15 g/liter) was used for intergenic matings. For liquid cultures of Vibrio strains, culture flasks were inoculated with fresh overnight colonies and grown at 25°C on a rotary shaker in marine minimal medium (MMM) supplemented with glucose (2 g/liter) (43). Growth was monitored by optical density measurements at 610 nm (OD610), and viability was assessed by counting the CFU on appropriate agar plates by the drop plate method (18). Where appropriate, antibiotics were added to the media at the following concentrations: tetracycline, 12.5 μg/ml; kanamycin, 50 μg/ml for E. coli and 75 μg/ml for Vibrio strains; ampicillin, 50 μg/ml; and streptomycin, 100 μg/ml. For RNA isolation and carbon starvation experiments, the culture was grown exponentially for several generations by recurrent dilutions to maximize population homogeneity. Carbon starvation conditions were obtained through the depletion of glucose, which was assessed by monitoring the OD610 of the cultures. Liquid cultures of Vibrio strains grown for other purposes were prepared by subculturing overnight cultures to an OD610 between 0.01 and 0.03.
Isolation of a gene encoding a sigma factor from V. angustum S14.
A previously constructed V. angustum S14 Sau3AI λ-ZAP Express library was screened with an rpoS probe derived from pRH324 by PCR amplification with primers 2F and 2R. The cloned DNA from isolated positive plaques was excised from the λ-ZAP Express vector in pBK-CMV double-stranded phagemids and transformed into E. coli XLOLR selecting for kanamycin resistance. Plasmid preparations obtained from colonies containing pBK phagemid vectors were screened for recombinants by restriction enzyme digestion with the enzymes NotI and PstI and by Southern hybridization with three different rpoS probes amplified by PCR from pRH324, using primers 1F/1R, 2F/2R, and 1F/2R. Positive candidates were further analyzed by DNA sequencing.
Construction of an rpoE mutant.
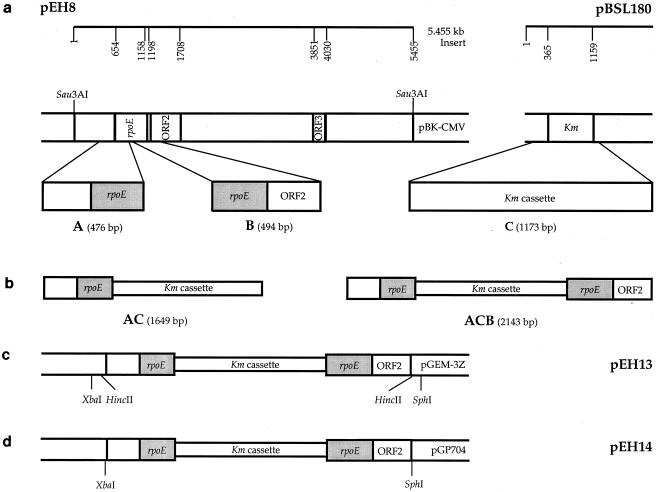
Three PCR products designated A, B, and C were generated. The rpoE PCR products A and B were obtained from pEH8 with primers F2.mut/R2.mut and F6.mut/R5.mut, respectively (Fig. 1a). PCR product C, a kanamycin resistance cassette, was amplified from pBSL180 with primers F3.mut/R4.mut (Fig. 1a). Sequential ligations of PCR products A, C, and B were carried out (Fig. 1b). The resulting 2.143-kbp rpoE::Kmr cassette construct was ligated into the HincII site of pGEM-3Z, yielding pEH13 (Fig. 1c). Plasmid pEH13 was digested with XbaI and SphI, and a 2.143-kbp fragment containing the rpoE::Kmr gene was cloned into the XbaI-SphI sites of pGP704 to generate pEH14 (Fig. 1d). After DNA sequencing, this plasmid was propagated in E. coli BW20767 and transferred to V. angustum S141 by conjugation. Exconjugants were selected on MMM supplemented with kanamycin, counterselecting for the auxotrophic E. coli BW20767 donor strain and the V. angustum S141 recipient without integration of the plasmid into the chromosome. The proportion of V. angustum S141 plasmid-free segregants among the exconjugants was enriched through three consecutive replica platings from selective (kanamycin) to nonselective (without kanamycin) conditions. Double crossover events and/or gene replacements were verified by PCR amplification of genomic DNA using primers R2 and F10, and the occurrence of a single copy of this gene was confirmed by Southern blotting of chromosomal DNA digested with EcoRI and BglI.
FIG. 1.
Schematic representation of the 2.143-kb rpoE::Kmr cassette construction. (a) Generation of PCR products A, B, and C from pEH8 and pBSL180. (b) Illustration of the sequential ligation of PCR products A and C and of AC and B yielding ACB, the 2.143-kb rpoE::Kmr cassette. (c) Cloning of the 2.143-kb rpoE::Kmr cassette (PCR product ACB) into the HincII site of pGEM-3Z generating pEH13. (d) Generation of pEH14 through excision of the 2.143-kb rpoE::Kmr cassette (PCR product ACB) from pEH13 with XbaI and SphI and insertion into the XbaI-SphI sites of pGP704. This plasmid was introduced into V. angustum strain S141 by conjugation for allelic exchange to produce rpoE mutant strain EH1.
Stress assays.
The sensitivity of V. angustum wild-type (S141) and mutant (EH1) strains to carbon starvation, heat, and oxidative stress was investigated in this study.
(i) Carbon starvation.
Exponentially growing cells were carbon starved through the depletion of glucose as described previously. At the onset of starvation, cultures were incubated statically at 25°C and starved for 4 weeks. Starvation survival was assessed at 0, 1, 24, and 48 h and at 1, 2, and 4 weeks.
(ii) Heat shock.
For heat shock survival experiments, exponentially growing cells at 25°C were rapidly shifted to 40°C and exposed to 20-, 40-, and 60-min heat shocks.
(iii) Oxidative stress.
Oxidative stress conditions were established with the reactive oxygen intermediate-generating agent H2O2, which was added to final concentrations of 65 μM and 1 mM to exponentially growing and carbon-starved cells, respectively. Cells were exposed to oxidative stress for 10 min during growth and for 60 min during carbon starvation. Stress survival was assessed by viable count in terms of CFU and expressed as the percentage of surviving cells relative to the initial cell viability.
Heat shock for total RNA isolation.
Exponentially growing cells at 25°C were rapidly shifted to 40°C and exposed to 6-, 12-, and 40-min heat shocks before harvesting and cell lysis for RNA preparation.
RNA isolation and Northern blot analysis.
Total RNA was prepared from V. angustum S141 cultures according to the standard protocol of the RNAgents total RNA isolation system from Promega. For Northern blot analysis, 20 μg of total RNA per sample was fractionated on a 1.8% agarose–formaldehyde gel. RNA was blotted from the gel to HYBOND-N membranes (Amersham), and the RNA was UV cross-linked according to the manufacturer's protocol. A 0.495-kb PCR product, which carries most of the rpoE coding sequence, was obtained from pEH8 using primers G and C and then gel purified. Using this purified PCR product as a template, a 0.495-kb digoxigenin (DIG)-labeled single-stranded DNA probe was generated by PCR amplification with primer C using the PCR DIG probe synthesis kit from Boehringer (Mannheim, Germany). This probe was used at a concentration of 10 ng/ml (DIG Easy Hyb) in hybridization experiments at 42°C with the DIG system for Northern blotting according to the manufacturer's protocol. DIG-labeled RNA was detected on X-ray films by chemiluminescence with CDP-Star as the substrate (Boehringer).
Periplasmic space and outer membrane protein preparation.
To obtain outer membrane proteins, cells were harvested at appropriate time points by centrifugation at 8,000 × g for 6 min at 20°C. Cell pellets were washed with 1 volume of 0.01 M Tris-HCl (pH 7.5), centrifuged, and resuspended in 1/20 volume of cold, 100 mM Tris-HCl (pH 8.0) and 10 mM EDTA. Cells were kept on ice, and cell walls were digested with lysozyme (150 μg/ml) for 10 min. DNA was removed by the addition of MgCl2 (10 mM) and DNase I (50 μg/ml), and spheroplasts were lysed by sonication until clearing of the lysates was observed. After low-speed centrifugation to remove unbroken cells and debris, supernatants were transferred to 1.0-ml thick-walled polycarbonate ultracentrifuge tubes and pelleted by centrifugation for 14 min at 300,000 × g at 4°C (TLA-100.2 fixed-angle rotor; Beckman TL-100 Tabletop Ultracentrifuge). Supernatants containing the total soluble fraction (periplasmic and cytoplasmic proteins) were removed, and pellets were further fractionated into outer and inner membrane proteins by solubilization with 1 ml of 1.67% sodium lauroyl sarcosinate in 11 mM Tris-HCl (pH 7.6) at room temperature for 30 min. The insoluble outer membrane proteins were pelleted by ultracentrifugation as previously described, washed with 1 ml of sarcosyl, recentrifuged, and stored at −80°C for subsequent analysis. To release periplasmic space contents from cells, cells were harvested and washed as described for the outer membrane preparation. Resulting cell pellets were resuspended in 1 ml of sterile distilled water, incubated for 20 min at room temperature, and centrifuged at 15,000 × g for 10 min at 4°C. Supernatants containing the periplasmic space proteins were filter sterilized (pore size, 0.22 μm). Filtrates were desalted and concentrated with Microcon YM-3 spin columns (Amicon) according to the manufacturer's recommendations and stored at −80°C prior to use.
SDS-PAGE.
Pellets containing the outer membrane proteins were resuspended in sterile distilled water, and the protein concentration of outer membrane proteins and periplasmic space proteins was determined using the bicinchoninic acid protein assay kit (Sigma) and a microtiter plate reader at 562 nm (Bio-Rad). The proteins were solubilized in 3 volumes of sodium dodecyl sulfate (SDS) sample buffer and boiled for 4 min. Proteins were separated by discontinuous SDS-polyacrylamide gel electrophoresis (SDS-PAGE) through the use of 12 and 4% polyacrylamide for resolving and stacking gels, respectively. Urea (4 M) was added for the fractionation of outer membrane proteins. The gels were stained with Coomassie brilliant blue R250 (Bio-Rad) to visualize the protein bands.
Protein profile analysis.
Stained gels were analyzed by densitometry using Bio-Rad Multi-Analyst Version 1.0.1. The analysis included protein profiles (optical density versus distance migrated), the percent total content of each protein, and the determination of its molecular mass in kilodaltons. Proteins showing a change in their percent total content of at least 30% in the mutant relative to the wild type were considered to be altered in the analysis of profiles at mid-log phase and at 0 and 24 h of carbon starvation. The role of rpoE in the oxidative stress response of V. angustum S14 was evaluated as a two-step process. Firstly, the oxidative stress-specific proteins for both wild-type and mutant strains were determined independently by establishing the difference in protein expression between stressed and unstressed cells. Secondly, the changes observed specifically during oxidative stress for each strain were then compared for the wild-type and mutant strains. These differences were used to infer the role of rpoE in the oxidative stress response of V. angustum S14.
Cell area measurements.
Cells were grown at 25 and 30°C. Aliquots were withdrawn from culture flasks at OD610 readings of 0.2, 0.4, 0.6, 0.8, and 1.0. Cell samples were fixed immediately by the addition of glutaraldehyde (final concentration, 0.3%) and stored at 4°C. Cell images from fixed samples were captured using a video monitor, and cell area measurements were obtained through computer image analysis using NIH Image 1.61 software.
Statistical analysis.
Cell area measurements from three independent experiments were analyzed using the fixed three-factor analysis of variance followed by the Student Newman-Keuls test. Homogeneity of variances was determined with Cochran's test, and deviations from the Gaussian distribution were determined with the Kolmogorov-Smirnov test.
DNA sequencing and analysis.
Automated DNA sequencing of both strands by dideoxynucleotide chain determination (PE Applied Biosystems) was carried out at the University of New South Wales, Sydney, Australia. Homology searches and alignments were performed with the Basic Local Alignment Search Tool (BLAST) Network Service at the National Center for Biotechnology Information, National Institutes of Health, Bethesda, Md. Protein topology predictions were obtained with the Simple Modular Architecture Research Tool (SMART) (version 3.0).
Nucleotide sequence accession number.
The 5.445-kbp sequence of the V. angustum S14 rpoE locus has been assigned GenBank accession number AF283003.
RESULTS
Isolation of a gene encoding a sigma factor from V. angustum S14.
To examine the role of alternative sigma factors in the carbon starvation response of V. angustum S14, a V. angustum S14 genomic library was screened with a probe targeting the 3′ end of the E. coli rpoS gene. This screen yielded four positive clones. Following in vivo excision, restriction enzyme digestion of the four candidate plasmids with NotI and PstI showed that each plasmid contained cloned DNA of varied sizes. Southern blot analysis revealed that two inserts hybridized to three probes targeting different regions of the E. coli rpoS gene: the 5′ end, the 3′ end, and nearly the entire open reading frame (ORF). An insert of approximately 5 kbp displayed the strongest hybridization signal for all 3 probes. The plasmid containing this insert was designated pEH8 and chosen for further DNA sequencing analysis.
DNA sequence analysis.
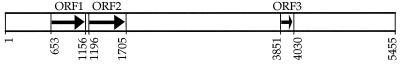
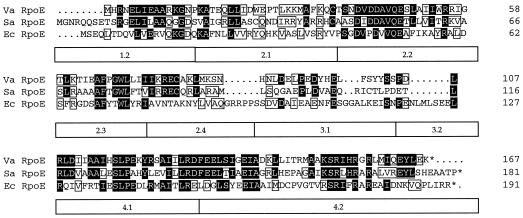
Sequence analysis of the 5.445-kbp insert present on pEH8 revealed three ORFs in the same orientation (Fig. 2). The 167-codon ORF1 encodes a protein with a calculated molecular mass of 19.4 kDa. A global similarity search of protein databases, using the BLAST Network Service, revealed significant similarity to members of the ECF subfamily of transcriptional regulators. The derived amino acid of ORF1 displayed the highest identity and similarity, respectively, to the rpoE homologs of Sphingomonas aromaticivorans (43 and 63%), mycobacteria (31 and 51%) Haemophilus influenzae (27 and 44%), S. enterica serovar Typhimurium (27 and 43%), and E. coli (26 and 43%). An alignment of ORF1 with its close homologs from S. aromaticivorans and E. coli is shown in Fig. 3. Based on these results, the ORF1 gene product was designated the V. angustum S14 RpoE. ORF2, containing 169 codons, is separated from ORF1 by a 39-nucleotide intergenic region without a terminator sequence (Fig. 2). Database searches of the predicted amino acid sequence did not reveal any significant similarities to any previously identified ECF anti-ς factors but showed similarities of 43% to four bacterial transport proteins. Analysis of ORF2 protein topology substantiated the observed sequence similarities, revealing the presence of four transmembrane domains. ORF3 (59 codons) is located 2,143 nucleotides downstream of ORF2 (Fig. 2) and displayed amino acid sequence similarities of 60% to proline-rich cell wall proteins.
FIG. 2.
Schematic representation of the rpoE locus in V. angustum S14. ORF1 shows similarity to rpoE of S. aromaticivorans, ORF2 is a putative inner membrane protein, and ORF3 shows similarity to proline-rich cell wall proteins. The arrows indicate the transcriptional direction of coding regions, and numbers represent the location of ORFs on the 5.455-kbp DNA segment, the sequence of which has been submitted to GenBank (see end of Materials and Methods).
FIG. 3.
Alignment of V. angustum RpoE (Va RpoE) with its close homologs from S. aromaticivorans (Sa RpoE) and E. coli (Ec RpoE). Black shading indicates identical amino acids, boxed residues indicate similar amino acids, and dots represent gaps. Regions of conservation are shown below the alignments (28).
Northern blot analysis.
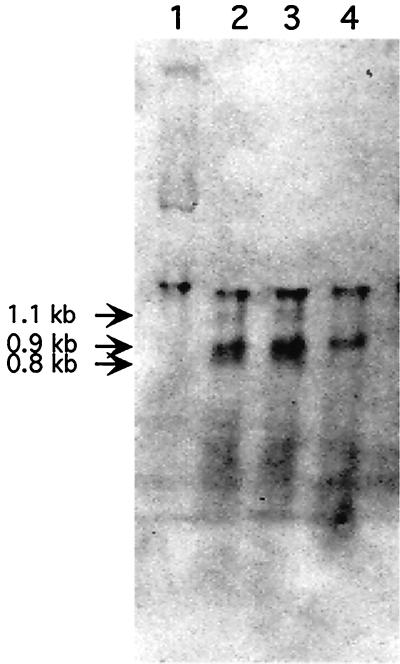
To examine the regulation of rpoE expression, preliminary experiments were carried out to confirm the isogenicity of V. angustum S141 to the wild type and to establish 40°C as the extreme heat shock temperature appropriate for this organism (data not shown). We then proceeded to measure the levels of V. angustum S14 rpoE mRNA before and after heat shock by probing Northern blots with an rpoE-specific probe (Fig. 4). No rpoE transcripts were detectable during growth at 25°C. Three transcripts of approximately 1,100, 900, and 800 nucleotides were observed within 6 and 12 min of the shift to 40°C. The largest and smallest transcripts displayed similar levels of induction after 6 and 12 min at 40°C, whereas the transcript of intermediate size was induced at a slightly higher level after 12 min at 40°C. After a prolonged heat shock of 40 min, only the transcript of intermediate size was expressed. These results show that the extreme heat shock response in V. angustum S14 is under transcriptional regulation and indicate cotranscription of ORF1 and ORF2 for the largest transcript.
FIG. 4.
V. angustum (S14) rpoE mRNA induction by heat shock. RNA was isolated from cells growing in MMM supplemented with glucose at 25°C (lane 1) and at 6, 12, and 40 min after a temperature shift to 40°C (lanes 2, 3, and 4, respectively) and was hybridized with an rpoE-specific probe. (The unmarked top band in all four lanes represents 16S ribosomal RNA, as determined by methylene blue staining of the Northern blot.)
Construction of an rpoE mutant.
To facilitate the characterization of rpoE in V. angustum S14, we replaced the chromosomal gene with the in vitro-generated insertion allele rpoE::kan (Fig. 1) by allelic exchange as described previously, generating rpoE mutant strain EH1 (S141 rpoE::Kmr). The disruption of the rpoE gene in EH1 was confirmed by PCR (data not shown). Southern blot analysis, using the coding region of the V. angustum rpoE gene as a probe, showed that a single copy of this gene was present in V. angustum S141 (data not shown).
The role of rpoE in environmental stress responses.
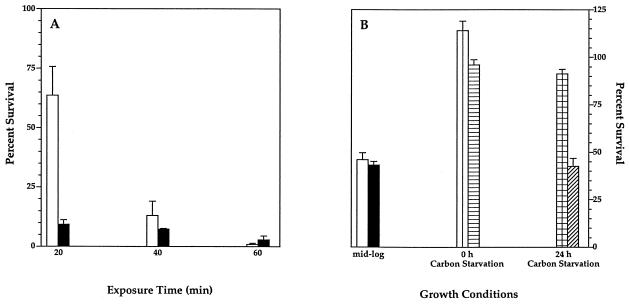
To study the role of rpoE in the environmental stress adaptation of V. angustum S14, survival of the rpoE-disrupted V. angustum EH1 and its parental strain S141 following carbon starvation, heat shock, and oxidative stress was compared (Fig. 5A and B). We found that RpoE plays a role in the survival of heat shock (Fig. 5A) and oxidative stress (Fig. 5B) but not in the survival of carbon starvation (data not shown). RpoE is essential at the onset of heat stress, resulting in a sevenfold decrease in survival of the mutant cells from that of the wild-type cells upon exposure to a 40°C environment for 20 min (Fig. 5A). Prolonged heat stress revealed similar survival rates for mutant and wild-type strains after 40 and 60 min of extreme heat shock (Fig. 5A). Logarithmically growing cells showed similar sensitivity to oxidative stress in mutant and wild-type strains, resulting in a twofold decrease in the survival of both strains (Fig. 5B). At the onset of carbon starvation (t0), the survival of both wild-type and mutant cells was not adversely affected by oxidative stress, as reflected in a negligible reduction or elevation of survival rates in both strains (Fig. 5B). After prolonged carbon starvation, however (t24), the survival rate of the wild type was relatively unchanged, compared to a twofold decrease in the survival of the mutant strain, revealing the specific role of RpoE in the oxidative stress response of cells subjected to carbon starvation for 24 h (Fig. 5B).
FIG. 5.
Increased sensitivity of rpoE mutant V. angustum (EH1) to heat shock (A) and oxidative stress (B). (A) V. angustum wild-type (S141) (white bar) and rpoE mutant (EH1) (black bar) strains were grown to mid-log phase (OD610) in MMM supplemented with glucose at 25°C and were shifted to 40°C. (B) V. angustum wild-type (S141) and rpoE mutant (EH1) cells were exposed to 65 μM H2O2 for 10 min at mid-log phase (S141, white bar; EH1, black bar) and exposed to 1 mM H2O2 for 60 min at both 0 h (S141, vertical stripes; EH1, horizontal stripes) and 24 h (S141, checks; EH1, diagonal stripes) of carbon starvation. Survival was assessed by viable counts of CFU and is expressed as the percentage of surviving cells relative to the initial cell viability. The results presented are mean values of three independent experiments, each carried out in triplicate. The error bars denote the standard deviation.
Effect of the rpoE mutation on outer membrane and periplasmic space protein profiles.
We studied the effect of the rpoE mutation on the protein composition of the cell envelope in logarithmically growing, carbon-starved, and oxidatively stressed cells of the V. angustum wild type (S141) and rpoE mutant strain (EH1) and found that RpoE has an extracytoplasmic function which plays a role during growth as well as environmental stress adaptation.
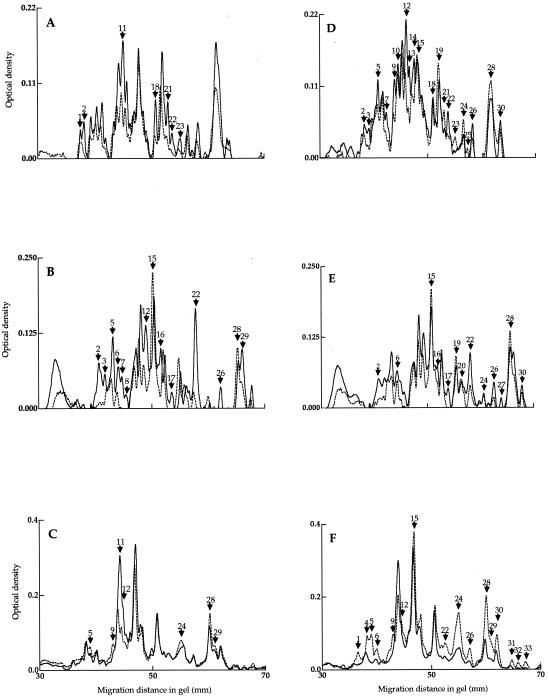
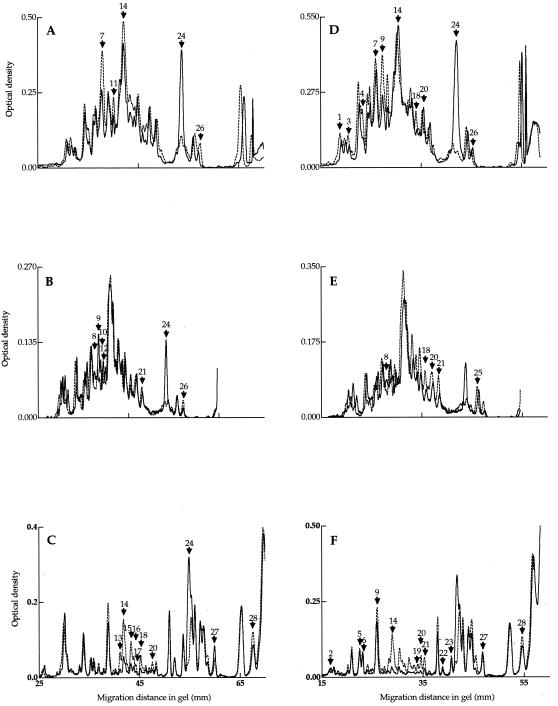
The disruption of rpoE led to increased levels of seven outer membrane proteins in the mutant during exponential growth (Fig. 6A; Table 3). However, the total amount of outer membrane proteins reflected a 40% reduction in the mutant from that in the wild type (data not shown). The levels of four periplasmic space proteins decreased in the rpoE mutant strain during exponential growth (Fig. 7A; Table 3). Interestingly, one periplasmic space protein (ID24) was upregulated by as much as 326% in the mutant compared to the wild type (Fig. 7A; Table 3).
FIG. 6.
SDS-PAGE of outer membrane protein profiles from V. angustum wild-type (S141) (broken line) and rpoE mutant (EH1) (solid line) strains during exponential growth (A), after 0 h (B) and 24 h (C) of carbon starvation, after exposure to oxidative stress (65 μM H2O2, 10 min) at mid-log phase (D), and after carbon starvation (1 mM H2O2, 60 min) at 0 h (E) and 24 h (F). Gels were stained and scanned as described in Materials and Methods. Arrows in panels A, B, and C indicate major differences in the protein profiles between the wild-type and mutant strains. Arrows in panels D, E, and F indicate oxidative stress-specific proteins in the wild-type and mutant strains. Numbers reciprocal to the molecular weight of the protein are assigned. The experiments were carried out in duplicate with insignificant deviations between replicates.
TABLE 3.
Analysis of outer membrane and periplasmic space proteins from logarithmically growing and carbon-starved V. angustum wild-type (S141) and rpoE mutant (EH1) strains
| Time point | Cellular location | No. of proteins: protein identification
|
|
|---|---|---|---|
| Class 1a | Class 2b | ||
| Mid-log phase | OMc | 7: 1, 2, 11, 18, 21, 22, 23 | 0 |
| Pd | 1: 24 | 4: 7, 11, 14, 26 | |
| t0 h | OM | 12: 2, 3, 5, 6, 7, 8, 12, 16, 17, 22, 26, 29 | 2: 15, 28 |
| P | 5: 8, 9, 10, 12, 24 | 2: 21, 26 | |
| t24 h | OM | 2: 11, 12 | 4: 5, 9, 24, 28, 29 |
| P | 2: 24, 27 | 8: 13, 14, 15, 16, 17, 18, 20, 28 | |
Proteins expressed at higher levels in the rpoE mutant strain.
Proteins expressed at lower levels in the rpoE mutant strain.
Outer membrane protein profiles for logarithmically growing and carbon-starved cells, as represented in Fig. 6A to C.
Periplasmic space protein profiles for logarithmically growing and carbon-starved cells, as represented in Fig. 7A to C.
FIG. 7.
SDS-PAGE of periplasmic space protein profiles from V. angustum wild-type (S141) and rpoE mutant (EH1) strains during exponential growth (A), after 0 h (B) and 24 h (C) of carbon starvation, after oxidative stress at mid-log phase (D), and at 0 h (E) and 24 h (F) of carbon starvation. See Fig. 6 legend for experimental methods and symbols.
At the onset of carbon starvation, 14 outer membrane (Fig. 6B; Table 3) and 7 periplasmic space (Fig. 7B; Table 3) proteins were affected in the rpoE mutant. Of these, 12 outer membrane and 5 periplasmic space proteins were upregulated, and 2 outer membrane and 2 periplasmic space proteins were downregulated (Table 3). The most pronounced changes at the onset of starvation were observed in the levels of the outer membrane protein ID22 (Fig. 6B; Table 3) and the periplasmic space protein ID24 (Fig. 7B; Table 3), which increased by 364 and 396%, respectively. In comparison to alteration at t0, at 24 h of carbon starvation, a greater number of proteins of the periplasmic space was altered than in the outer membrane (Table 3). Eight periplasmic space proteins were downregulated, and two were upregulated (Fig. 7C; Table 3). Most notably, periplasmic space protein ID24 (Fig. 7C) was present in the mutant at levels higher than at the onset of starvation. In comparison, this protein was absent in the wild type at 24 h of carbon starvation. In the outer membrane, the prominent changes in the mutant strain at 24 h of carbon starvation were increases of 112 and 60% for proteins ID11 and -12, respectively (Fig. 6C; Table 3).
Oxidative stress-specific proteins were divided into three classes, those restricted to the wild type or the mutant or present in both (Table 4). Specifically, only 2 periplasmic space proteins differed in expression in the mutant, compared to changes in the levels of 10 periplasmic space proteins in the wild type during exponential growth (Fig. 7D; Table 4). A small number of oxidative stress-specific outer membrane proteins was found in the mutant at 24 h of carbon starvation (Fig. 6F; Table 4). In contrast, the wild type showed a pronounced increase in the number of outer membrane oxidative stress-specific proteins after 24 h of carbon starvation (Fig. 6F; Table 4).
TABLE 4.
Analysis of oxidative stress-specific proteins in wild-type (S141) and rpoE mutant (EH1) strains
| Strain | Cellular location | No. of proteins: protein identificationa
|
||
|---|---|---|---|---|
| Mid-log phase | t0 h | t24 h | ||
| Wild-type (S141) | OMb | 13: 2, 3, 5, 9, 13, 15, 18, 22, 23, 25, 26, 28, 30 | 5: 6, 15, 20, 22, 30 | 15: 1, 4, 5, 6, 9, 12, 22, 24, 26, 28, 29, 30, 31, 32, 33 |
| rpoE mutant (EH1) | OM | 17: 3, 5, 7, 9, 10, 12, 13, 14, 19, 21, 22, 23, 24, 25, 26, 28, 30 | 10: 2, 16, 17, 19, 20, 22, 24, 26, 28, 30 | 7: 5, 6, 12, 15, 22, 24, 26 |
| Wild-type (S141) | Pc | 10: 1, 3, 4, 7, 9, 14, 18, 20, 24, 26 | 4: 18, 20, 21, 25 | 10: 2, 5, 6, 9, 19, 20, 21, 22, 23, 27 |
| rpoE mutant (EH1) | P | 2: 14, 26 | 2: 8, 25 | 8: 2, 5, 6, 9, 14, 22, 23, 28 |
Protein designations with bold characters indicate those responsive in both wild-type (S141) and rpoE mutant (EH1) strains.
Outer membrane protein profiles from cells after oxidative stress at mid-log phase and at 0 and 24 h of carbon starvation as represented in Fig. 6D to F.
Periplasmic space protein profiles from cells after oxidative stress at mid-log phase and at 0 and 24 h of carbon starvation as represented in Fig. 7D to F.
These results demonstrate that there is a role for rpoE in the protein composition of the cell envelope during growth, carbon starvation, and carbon starvation-induced cross-protection against oxidative stress and that extensive overlaps exist between the two stress conditions.
Growth characteristics and cell morphology.
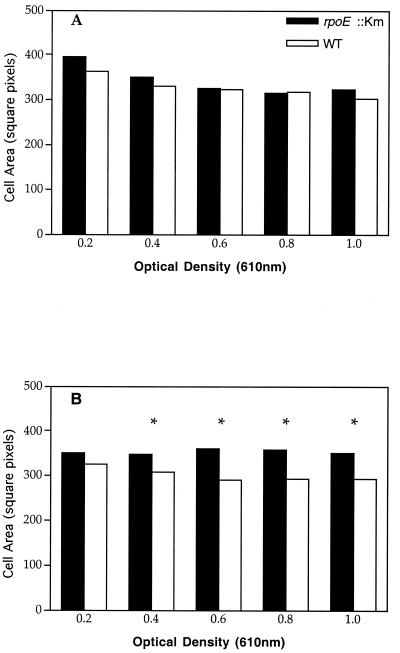
When unstressed rpoE mutant cells were plated on VNSS, they formed microcolonies at optimal growth temperatures (22 to 30°C), which is characteristic of a stressed phenotype for wild-type V. angustum S14 (54). Given the fact that ECF sigma factors have also been demonstrated to have a role in normal and/or unstressed environments, we explored this further. No differences in OD610 values and generation time (60 min) were observed for the growth curves of the mutant and wild type at 25 or 30°C (data not shown). However, marked differences in CFU counts were found for the wild type and mutant at 30°C but not at 25°C (data not shown). Cell area measurements of mutant and wild-type cells were compared. At 25°C, no statistically significant differences between the mutant and wild type were found (Fig. 8A). In contrast, when cells were grown at 30°C, the mutant cells were significantly larger (P < 0.05) than the wild-type cells for OD values between 0.4 and 1.0 (Fig. 8B). These results indicate a role for rpoE in the cellular morphology of V. angustum S14 during growth at optimal temperatures.
FIG. 8.
Effect of growth temperature on cell volume in V. angustum wild-type (S141) and rpoE mutant (EH1) strains. Pictured is a comparison of cell area measurements for the V. angustum wild type (white bar) and rpoE mutant strain (black bar) grown at 25°C (A) and 30°C (B). The results presented are the mean values of three independent experiments, with each data point representing 400 measurements. The asterisk indicates cell area measurements with statistically significant differences (P < 0.05) between the V. angustum wild type (S141) and rpoE mutant strain (EH1), as determined by a three-factor analysis of variance.
DISCUSSION
The ECF family of sigma factors constitutes a diverse but distinct subfamily of the ς70 type of sigma factors (28). This initially small family of alternative sigma factors, characterized through sequence similarity and conservation of extracytoplasmic function (28), has seen a considerable expansion through recent progress in bacterial genome sequencing. This, combined with the recognition that sigma factor regulons cooperate in the management of stress, suggests that ECF sigma factors play an important and diverse role in bacterial stress adaptation.
In the present study, we have demonstrated the existence of the first sigma factor, an RpoE homolog, in V. angustum S14, whose designation is based on its amino acid sequence similarity with the RpoE of other bacteria and its extracytoplasmic function. We have shown that the rpoE gene is induced and required during survival at extreme temperatures. We have also demonstrated the involvement of rpoE in carbon starvation-induced cross-protection against oxidative stress. Moreover, evidence for an extracytoplasmic function of rpoE in the protein composition of the cell envelope of stressed and unstressed cells of V. angustum S14 is provided.
The rpoE gene in V. angustum S14 encodes a protein with a calculated molecular mass of 19.4 kDa. The predicted amino acid sequence of RpoE reveals a high degree of similarity with members of the ECF subfamily of sigma factors (Fig. 3). Sequence conservation among ECF sigma factors is suggested to reflect functional but not phylogenetic relationships with sigma factors falling into functional subgroups, including members from diverse organisms (27). This view is supported by the demonstrated functional equivalence of E. coli ςE and P. aeruginosa ςAlgU (61) and the replacement of E. coli ςFecI by ςSigX from Bacillus subtilis (6). Interestingly, V. angustum S14 RpoE displays the highest similarity in all four regions to the RpoE homolog of another marine organism (Fig. 3) recently classified as S. aromaticivorans (4). The shared environment may substantiate a possible functional equivalence. S. aromaticivorans possesses two plasmids (12) designated pNL1 and pNL2. Surprisingly, the rpoE homolog of S. aromaticivorans is located on the aromatic catabolic plasmid pNL1 (49). This plasmid contains genes encoding proteins associated with functions in plasmid replication, maintenance, transfer, integration, and recombination (49). Conjugative transfer of pNL1 to another Sphingomonas sp. was demonstrated (49), indicating the possibility of horizontal gene transfer of rpoE in the marine environment. In this context, it is interesting to note that conjugative plasmid transfer in a simulated marine environment has been demonstrated for starved V. angustum S14 (13) and that enhanced transfer was found during predation by a microflagellate (46). In contrast to the observed amino acid sequence similarity in all regions between the RpoE homologs of V. angustum S14 and S. aromaticivorans, comparison of the RpoE of V. angustum S14 to that of other RpoE homologs of significant amino acid similarity reveals some divergence in regions 2.4, 3.1, and 3.2 (Fig. 3). The latter two regions are weakly conserved or absent among ECF sigma factors (28). Region 2.4 is implicated in the −10 promoter recognition region (28) and is less conserved among members of different ECF subgroups (28), possibly contributing to altered promoter specificities, which are quite diverse.
Immediately downstream of rpoE is a second ORF in V. angustum S14. Database searches of the predicted amino acid sequence did not reveal any significant similarity to the sequences of any previously identified ECF anti-ς factors. A similar situation has been observed in several mycobacterial species (59) and B. subtilis (20). This could be a reflection of the generally poor sequence similarities observed between ECF anti-ς factors (53). Despite the occurrence of structurally unrelated ECF anti-ς factors, a common theme among the ECF anti-ς factors characterized so far is that they are inner membrane proteins with at least one transmembrane domain which are cotranscribed with their cognate ς factors (21). Analysis of ORF2 topology in V. angustum S14 predicts this gene to encode an inner membrane protein with transmembrane regions, indicating the possibility of an antisigma function of this gene. This is further supported by the cotranscription of ORF1 and ORF2 in V. angustum S14, which is suggested by the size of the largest mRNA transcript detected in Northern hybridization experiments (Fig. 4) and the short intergenic region without a terminator sequence between ORF1 and ORF2 (Fig. 2). Based on these results, it may be suggested that ORF1 and ORF2 in V. angustum S14 are members of an operon, functioning as a sigma factor and antisigma factor, respectively.
The rpoE gene appears to play a role in unstressed cells of V. angustum S14, as suggested by altered phenotypes, such as colony morphology and increased cell volumes at optimal growth temperature (Fig. 8) in the mutant compared to those in the wild-type strain. Importantly, a comparison of outer membrane and periplasmic space proteins obtained from logarithmically growing cells revealed profiles in the mutant altered from those in the wild-type strain, providing evidence of a role for rpoE in unstressed cells of V. angustum S14. Specifically, the percent total content of one periplasmic space protein (ID24) increased 326% in the mutant from that in the wild type (Fig. 7A). The accumulation of this protein may be the result of an increasing number of misfolded outer membrane proteins in the periplasmic space, due to the lack of rpoE regulon-regulated periplasmic chaperones and proteases. A similar observation was made in E. coli, where the lack of Skp, a periplasmic chaperone (which appears to be ςE regulated) in the absence of active DegP (a ςE-regulated periplasmic protease), led to the accumulation of protein aggregates in the periplasm (52). A reduction of 40% in the total amount of outer membrane proteins in the mutant from that in the V. angustum wild type during balanced growth provides further support for this hypothesis.
The induction of rpoE after a temperature shift to 40°C provides experimental evidence for transcriptional regulation of the extreme heat shock response in V. angustum S14. The induction pattern of rpoE, with mRNA levels rapidly increasing 6 min after the temperature shift, peaking at 12 min, and declining at 40 min, is similar to that observed in the induction of htrA (26), a gene of the ςE-controlled heat shock regulon encoding a periplasmic protease (48). Whether the transcripts detected in V. angustum S14 represent three distinct mRNA species or include processed transcripts is unclear at this point.
In agreement with the induction of rpoE during extreme heat shock, disruption of rpoE decreased the survival rate of the isogenic rpoE V. angustum mutant during extreme temperature exposure, as has been shown for E. coli (17), P. aeruginosa (33), M. smegmatis (59), and B. subtilis (20). The heat shock response in V. angustum S14 is poorly understood; characterization is limited to a previously demonstrated sixfold induction of DnaK after a shift to 36°C (19). This study shows that rpoE increases the temperature tolerance of V. angustum S14 to 40°C and suggests the presence of two compartment-specific heat shock responses in V. angustum S14.
While the survival of the mutant was not adversely affected after 24 h of carbon starvation (data not shown), a loss in viability was observed following oxidative stress (Fig. 5B) in cells subjected to 24 h of carbon starvation. This result demonstrates a specific role for rpoE in carbon starvation-induced resistance to oxidative stress in V. angustum S14. This role is further substantiated by the demonstrated extensive overlap of outer membrane and periplasmic space proteins with altered expression during carbon starvation and oxidative stress. Although rpoE does not appear to be essential for carbon starvation survival, an involvement in this response is clearly indicated by altered outer membrane and periplasmic space protein profiles in the mutant at 0 and 24 h of carbon starvation (Table 3). Starvation-induced changes in the outer membrane and periplasmic space, which were suggested to contribute to survival, were reported for V. angustum S14 (2, 29, 30, 31, 41).
The marine environment is frequently limited in nutrients (25). Also, exposure to UV radiation is common for many organisms in this habitat (38). Hence, successful adaptation to starvation and to oxidative stress is crucial for the survival of marine microorganisms. It has previously been demonstrated that starvation-induced cross-protection against oxidative stress is a key feature in the starvation adaptation of V. angustum S14 (L. Gong, unpublished; 42, 55). The data presented in this study show that RpoE is involved in the adaptation of V. angustum S14 to these stresses. RpoE has an extracytoplasmic function, plays a role in growing cells, and most notably has a specific role in carbon starvation-induced cross-protection against oxidative stress.
ACKNOWLEDGMENTS
We thank M. Givskov and S. Rice for valuable discussions, I. Dahllöf for advice on statistical analysis, M. Manefield for assistance in computer image analysis, and R. Hengge-Aronis for supplying plasmid pRH324.
This work was supported by grants from the Australian Research Council.
REFERENCES
- 1.Albertson N H, Nyström T, Kjelleberg S. Exoprotease activity of two marine bacteria during starvation. Appl Environ Microbiol. 1990;56:218–223. doi: 10.1128/aem.56.1.218-223.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albertson N H, Nyström T, Kjelleberg S. Starvation-induced modulations in binding protein-dependent glucose transport by the marine Vibrio sp. S14. FEMS Microbiol Lett. 1990;70:205–210. doi: 10.1111/j.1574-6968.1990.tb13979.x. [DOI] [PubMed] [Google Scholar]
- 3.Angerer A, Enz S, Ochs M, Braun V. Transcriptional regulation of ferric citrate transport in Escherichia coli K-12. FecI belongs to a new subfamily of ς70-type factors that respond to extracytoplasmic stimuli. Mol Microbiol. 1995;18:163–174. doi: 10.1111/j.1365-2958.1995.mmi_18010163.x. [DOI] [PubMed] [Google Scholar]
- 4.Balkwill D L, Drake G R, Reeves R H, Fredrickson J K, White D C, Ringelberg D B, Chandler D P, Romine M F, Kennedy D W, Spadoni C M. Taxonomic study of aromatic-degrading bacteria from deep-terrestrial-subsurface sediments and description of Sphingomonas aromaticivorans sp. nov., Sphingomonas subterranea sp. nov., and Sphingomonas stygia sp. nov. Int J Syst Bacteriol. 1997;47:191–201. doi: 10.1099/00207713-47-1-191. [DOI] [PubMed] [Google Scholar]
- 5.Bianchi A A, Baneyx F. Hyperosmotic shock induces the ς32 and ςE stress regulons of Escherichia coli. Mol Microbiol. 1999;34:1029–1038. doi: 10.1046/j.1365-2958.1999.01664.x. [DOI] [PubMed] [Google Scholar]
- 6.Brutsche S, Braun V. SigX of Bacillus subtilis replaces the ECF sigma factor FecI of Escherichia coli and is inhibited by RsiX. Mol Gen Genet. 1997;256:416–425. doi: 10.1007/s004380050585. [DOI] [PubMed] [Google Scholar]
- 7.Chi E, Bartlett D H. An rpoE-like locus controls outer membrane protein synthesis and growth at cold temperatures and high pressures in the deep-sea bacterium Photobacterium sp. strain SS9. Mol Microbiol. 1995;17:713–726. doi: 10.1111/j.1365-2958.1995.mmi_17040713.x. [DOI] [PubMed] [Google Scholar]
- 8.De Las Peñas A, Connolly L, Gross C A. ςE is an essential sigma factor in Escherichia coli. J Bacteriol. 1997;179:6862–6864. doi: 10.1128/jb.179.21.6862-6864.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enz S, Braun V, Crosa J H. Transcription of the region encoding the ferric dicitrate-transport system in Escherichia coli: similarity between promoters for fecA and for extracytoplasmic function sigma factors. Gene. 1995;163:13–18. doi: 10.1016/0378-1119(95)00380-o. [DOI] [PubMed] [Google Scholar]
- 10.Fernandes N D, Wu Q-L, Kong D, Puyang X, Garg S, Husson R N. A mycobacterial extracytoplasmic sigma factor involved in survival following heat shock and oxidative stress. J Bacteriol. 1999;181:4266–4274. doi: 10.1128/jb.181.14.4266-4274.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flärdh K, Axberg T, Albertson N H, Kjelleberg S. Stringent control during carbon starvation of marine Vibrio sp. strain S14: molecular cloning, nucleotide sequence, and deletion of the relA gene. J Bacteriol. 1994;176:5949–5957. doi: 10.1128/jb.176.19.5949-5957.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fredrickson J K, Brockman F J, Workman D J, Li S W, Stevens T O. Isolation and characterization of a subsurface bacterium capable of growth on toluene, naphthalene, and other aromatic compounds. Appl Environ Microbiol. 1991;57:796–803. doi: 10.1128/aem.57.3.796-803.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodman A E, Hild E, Marshall K C, Hermansson M. Conjugative plasmid transfer between bacteria under simulated marine oligotrophic conditions. Appl Environ Microbiol. 1993;59:1035–1040. doi: 10.1128/aem.59.4.1035-1040.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorham H C, McGowan S J, Robson P R H, Hodgson D A. Light-induced carotenogenesis in Myxococcus xanthus: light-dependent membrane sequestration of ECF sigma factor CarQ by anti-sigma factor CarR. Mol Microbiol. 1996;19:171–186. doi: 10.1046/j.1365-2958.1996.360888.x. [DOI] [PubMed] [Google Scholar]
- 15.Govan J R W, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hengge-Aronis R. Interplay of global regulators and cell physiology in the general stress response of Escherichia coli. Curr Opin Microbiol. 1999;2:148–152. doi: 10.1016/S1369-5274(99)80026-5. [DOI] [PubMed] [Google Scholar]
- 17.Hiratsu K, Amemura M, Nashimoto H, Shinagawa H, Makino K. The rpoE gene of Escherichia coli, which encodes ςE, is essential for bacterial growth at high temperature. J Bacteriol. 1995;177:2918–2922. doi: 10.1128/jb.177.10.2918-2922.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoben H J, Somasegaran P. Comparison of the pour, spread, and drop plate methods for enumeration of Rhizobium spp. in inoculants made from presterilized peat. Appl Environ Microbiol. 1982;44:1246–1247. doi: 10.1128/aem.44.5.1246-1247.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holmquist L, Jouper-Jaan Å, Weichart D, Nelson D R, Kjelleberg S. The induction of stress proteins in three marine Vibrio during carbon starvation. FEMS Microbiol Ecol. 1993;12:185–194. [Google Scholar]
- 20.Huang X, Decatur A, Sorokin A, Helmann J D. The Bacillus subtilis ςX protein is an extracytoplasmic function ς factor contributing to survival at high temperature. J Bacteriol. 1997;179:2915–2921. doi: 10.1128/jb.179.9.2915-2921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes K T, Mathee K. The anti-sigma factors. Annu Rev Microbiol. 1998;52:231–286. doi: 10.1146/annurev.micro.52.1.231. [DOI] [PubMed] [Google Scholar]
- 22.Humphreys S, Stevenson A, Bacon A, Weinhardt A B, Roberts M. The alternative sigma factor, ςE, is critically important for the virulence of Salmonella typhimurium. Infect Immun. 1999;67:1560–1568. doi: 10.1128/iai.67.4.1560-1568.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones G H, Paget M S B, Chamberlin L, Buttner M J. Sigma-E is required for the production of the antibiotic actinomycin in Streptomyces antibioticus. Mol Microbiol. 1997;23:169–178. doi: 10.1046/j.1365-2958.1997.2001566.x. [DOI] [PubMed] [Google Scholar]
- 24.Keith L M W, Bender C L. AlgT (ς22) controls alginate production and tolerance to environmental stress in Pseudomonas syringae. J Bacteriol. 1999;181:7176–7184. doi: 10.1128/jb.181.23.7176-7184.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kjelleberg S, Flärdh K, Nyström T, Moriarty D J W. Growth limitation and starvation of bacteria. In: Ford T, editor. Aquatic microbiology: an ecological approach. Oxford, United Kingdom: Blackwell; 1993. pp. 289–320. [Google Scholar]
- 26.Lipinska B, Sharma S, Georgopoulos C. Sequence analysis and regulation of the htrA gene of Escherichia coli: a ς32-independent mechanism of heat-inducible transcription. Nucleic Acids Res. 1988;16:10053–10067. doi: 10.1093/nar/16.21.10053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lonetto M, Gribskov M, Gross C A. The ς70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lonetto M A, Brown K L, Rudd K E, Buttner M J. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase ς factors involved in the regulation of extracytoplasmic functions. Proc Natl Acad Sci USA. 1994;91:7573–7577. doi: 10.1073/pnas.91.16.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malmcrona-Friberg K, Goodman A, Kjelleberg S. Chemotactic responses of a marine Vibrio sp. strain S14 (CCUG 15956) to low-molecular-weight substances under starvation-survival conditions. Appl Environ Microbiol. 1990;56:3699–3704. doi: 10.1128/aem.56.12.3699-3704.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malmcrona-Friberg K, Tunlid A, Mårdén P, Kjelleberg S, Odham G. Chemical changes in cell envelope and poly-β-hydroxybutyrate during short term starvation of a marine bacterial isolate. Arch Microbiol. 1986;144:340–345. [Google Scholar]
- 31.Mårdén P, Nyström T, Kjelleberg S. Uptake of leucine by a marine Gram-negative heterotrophic bacterium during exposure to starvation conditions. FEMS Microbiol Ecol. 1987;45:233–241. [Google Scholar]
- 32.Mårdén P, Tunlid A, Malmcrona-Friberg K, Odham G, Kjelleberg S. Physiological and morphological changes during short term starvation of marine bacterial isolates. Arch Microbiol. 1985;142:326–332. [Google Scholar]
- 33.Martin D W, Schurr M J, Yu H, Deretic V. Analysis of promoters controlled by the putative sigma factor AlgU regulating conversion to mucoidy in Pseudomonas aeruginosa: relationship to ςE and stress response. J Bacteriol. 1994;176:6688–6696. doi: 10.1128/jb.176.21.6688-6696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mecsas J, Rouvière P E, Erickson J W, Donohue T J, Gross C A. The activity of ςE, an Escherichia coli heat-inducible ς-factor, is modulated by expression of outer membrane proteins. Genes Dev. 1993;7:2618–2628. doi: 10.1101/gad.7.12b.2618. [DOI] [PubMed] [Google Scholar]
- 35.Metcalf W W, Jjang W, Daniels L L, Kim S-K, Haldimann A, Wanner B L. Conditionally replicative and conjugative plasmids carrying lacZα for cloning, mutagenesis, and allele replacement in bacteria. Plasmid. 1996;35:1–13. doi: 10.1006/plas.1996.0001. [DOI] [PubMed] [Google Scholar]
- 36.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Missiakas D, Betton J-M, Raina S. New components of protein folding in extracytoplasmic compartments of Escherichia coli SurA, FkpA and Skp/OmpH. Mol Microbiol. 1996;21:871–884. doi: 10.1046/j.1365-2958.1996.561412.x. [DOI] [PubMed] [Google Scholar]
- 38.Moran M A, Zepp R G. UV radiation effects on microbes and microbial processes. In: Kirchman D L, editor. Microbial ecology of the oceans. New York, N.Y: Wiley-Liss, Inc.; 2000. pp. 201–228. [Google Scholar]
- 39.Moreno S, Nájera R, Guzmán J, Soberón-Chávez G, Espín G. Role of alternative ς factor AlgU in encystment of Azotobacter vinelandii. J Bacteriol. 1998;180:2766–2769. doi: 10.1128/jb.180.10.2766-2769.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nyström T, Albertson N, Kjelleberg S. Synthesis of membrane and periplasmic proteins during starvation of a marine Vibrio sp. J Gen Microbiol. 1988;134:1645–1651. doi: 10.1099/00221287-134-6-1645. [DOI] [PubMed] [Google Scholar]
- 41.Nyström T, Kjelleberg S. Role of protein synthesis in the cell division and starvation induced resistance to autolysis of a marine Vibrio during the initial phase of starvation. J Gen Microbiol. 1989;135:1599–1606. [Google Scholar]
- 42.Östling J, Flärdh K, Kjelleberg S. Isolation of a carbon starvation regulatory mutant in a marine Vibrio strain. J Bacteriol. 1995;177:6978–6982. doi: 10.1128/jb.177.23.6978-6982.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Östling J, Goodman A, Kjelleberg S. Behaviour of IncP-1 plasmids and a miniMu transposon in a marine Vibrio sp.: isolation of starvation inducible lac operon fusions. FEMS Microbiol Ecol. 1991;86:83–94. [Google Scholar]
- 44.Östling J, Holmquist L, Kjelleberg S. Global analysis of the carbon starvation response of a marine Vibrio species with disruptions in genes homologous to relA and spoT. J Bacteriol. 1996;178:4901–4908. doi: 10.1128/jb.178.16.4901-4908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Östling J, McDougald D, Marouga R, Kjelleberg S. Global analysis of physiological responses in marine bacteria. Electrophoresis. 1997;18:1441–1450. doi: 10.1002/elps.1150180819. [DOI] [PubMed] [Google Scholar]
- 46.Otto K, Weichart D, Kjelleberg S. Plasmid transfer between marine Vibrio strains during predation by the heterotrophic microflagellate Cafeteria roenbergensis. Appl Environ Microbiol. 1997;63:749–752. doi: 10.1128/aem.63.2.749-752.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paget M S B, Chamberlin L, Atrih A, Foster S J, Buttner M J. Evidence that the extracytoplasmic function sigma factor ςE is required for normal cell wall structure in Streptomyces coelicolor A3(2) J Bacteriol. 1999;181:204–211. doi: 10.1128/jb.181.1.204-211.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raina S, Missiakas D, Georgopoulos C. The rpoE gene encoding the ςE (ς24) heat shock sigma factor of Escherichia coli. EMBO J. 1995;14:1043–1055. doi: 10.1002/j.1460-2075.1995.tb07085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Romine M F, Stillwell L C, Wong K-K, Thurston S J, Sisk E C, Sensen C, Gaasterland T, Fredrickson S J, Saffer J D. Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J Bacteriol. 1999;181:1585–1602. doi: 10.1128/jb.181.5.1585-1602.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rouvière P E, De Las Peñas A, Mecsas J, Lu C Z, Rudd K E, Gross C A. rpoE, the gene encoding the second heat-shock sigma factor, ςE, in Escherichia coli. EMBO J. 1995;14:1032–1042. doi: 10.1002/j.1460-2075.1995.tb07084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 52.Schäfer U, Beck K, Müller M. Skp, a molecular chaperone of gram-negative bacteria, is required for the formation of soluble periplasmic intermediates of outer membrane proteins. J Biol Chem. 1999;274:24567–24574. doi: 10.1074/jbc.274.35.24567. [DOI] [PubMed] [Google Scholar]
- 53.Schurr M J, Yu H, Martinez-Salazar J M, Boucher J C, Deretic V. Control of AlgU, a member of the ςE-like family of stress sigma factors, by the negative regulators MucA and MucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J Bacteriol. 1996;178:4997–5004. doi: 10.1128/jb.178.16.4997-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Srinivasan S, Kjelleberg S. Cycles of famine and feast: the starvation and outgrowth strategies of a marine Vibrio. J Biosci. 1998;23:501–511. [Google Scholar]
- 55.Srinivasan S, Östling J, Charlton T, De Nys R, Takayama K, Kjelleberg S. Extracellular signal molecule(s) involved in the carbon starvation response of marine Vibrio sp. strain S14. J Bacteriol. 1998;180:201–209. doi: 10.1128/jb.180.2.201-209.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takayama K, Kjelleberg S. The role of RNA stability during bacterial stress responses and starvation. Environ Microbiol. 2000;2:355–365. doi: 10.1046/j.1462-2920.2000.00119.x. [DOI] [PubMed] [Google Scholar]
- 57.Ward M J, Lew H, Treuner-Lange A, Zusman D R. Regulation of motility behavior in Myxococcus xanthus may require an extracytoplasmic-function sigma factor. J Bacteriol. 1998;180:5668–5675. doi: 10.1128/jb.180.21.5668-5675.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wösten M M S M. Eubacterial sigma-factors. FEMS Microbiol Rev. 1998;22:127–150. doi: 10.1111/j.1574-6976.1998.tb00364.x. [DOI] [PubMed] [Google Scholar]
- 59.Wu Q-L, Kong D, Lam K, Husson R N. A mycobacterial extracytoplasmic function sigma factor involved in survival following stress. J Bacteriol. 1997;179:2922–2929. doi: 10.1128/jb.179.9.2922-2929.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu H, Boucher J C, Hibler N S, Deretic V. Virulence properties of Pseudomonas aeruginosa lacking the extreme-stress sigma factor AlgU (ςE) Infect Immun. 1996;64:2774–2781. doi: 10.1128/iai.64.7.2774-2781.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu H, Schurr M J, Deretic V. Functional equivalence of Escherichia coli ςE and Pseudomonas aeruginosa AlgU: E. coli rpoE restores mucoidy and reduces sensitivity to reactive oxygen intermediates in algU mutants of P. aeruginosa. J Bacteriol. 1995;177:3259–3268. doi: 10.1128/jb.177.11.3259-3268.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]