Abstract
Employed for over half a century to study protein synthesis, cycloheximide (CHX, 1) is a small molecule natural product that reversibly inhibits translation elongation. More recently, CHX is commonly used in ribosome profiling – a method for mapping ribosome positions on mRNA genome-wide. Despite CHX’s extensive use, CHX treatment often results in incomplete translation inhibition due to its rapid reversibility, prompting the need for improved reagents. Here we report the concise synthesis of C13-amide functionalized CHX derivatives with increased potencies toward protein synthesis inhibition. Cryogenic electron microscopy (cryo-EM) revealed that C13-aminobenzoyl CHX (8) occupies the same site as CHX to compete with the 3’ end of E-site tRNA. We demonstrate that 8 is superior to CHX for ribosome profiling experiments, enabling more effective capture of ribosome conformations through sustained stabilization of polysomes. Our studies identify powerful chemical reagents to study protein synthesis and reveal the molecular basis of their enhanced potency.
Graphical Abstract
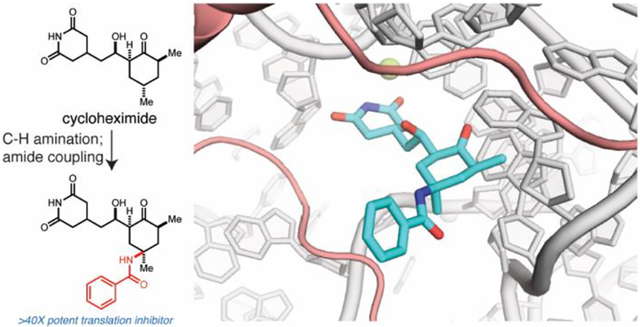
CHX (1) is a natural product inhibitor of the eukaryotic ribosome (Figure 1A), which has been exploited to study protein synthesis.1,2 Pioneering biochemical and structural studies demonstrated that CHX binds the ribosomal exit site (E site).3,4 During translation elongation, the E site accommodates a deacylated tRNA as it moves from the peptidyl site (P site), and CHX occupies a pocket within the E site where the 3’ end of the tRNA resides. These data strongly support the notion that CHX binding would compete with the deacylated tRNA, thus inhibiting the translocation of the ribosome.4,5 Despite this mechanistic understanding of CHX-mediated inhibition, the structural features of CHX responsible for the molecule’s characteristic, rapidly reversible effects on polysome stability remain ill-defined.
Figure 1.
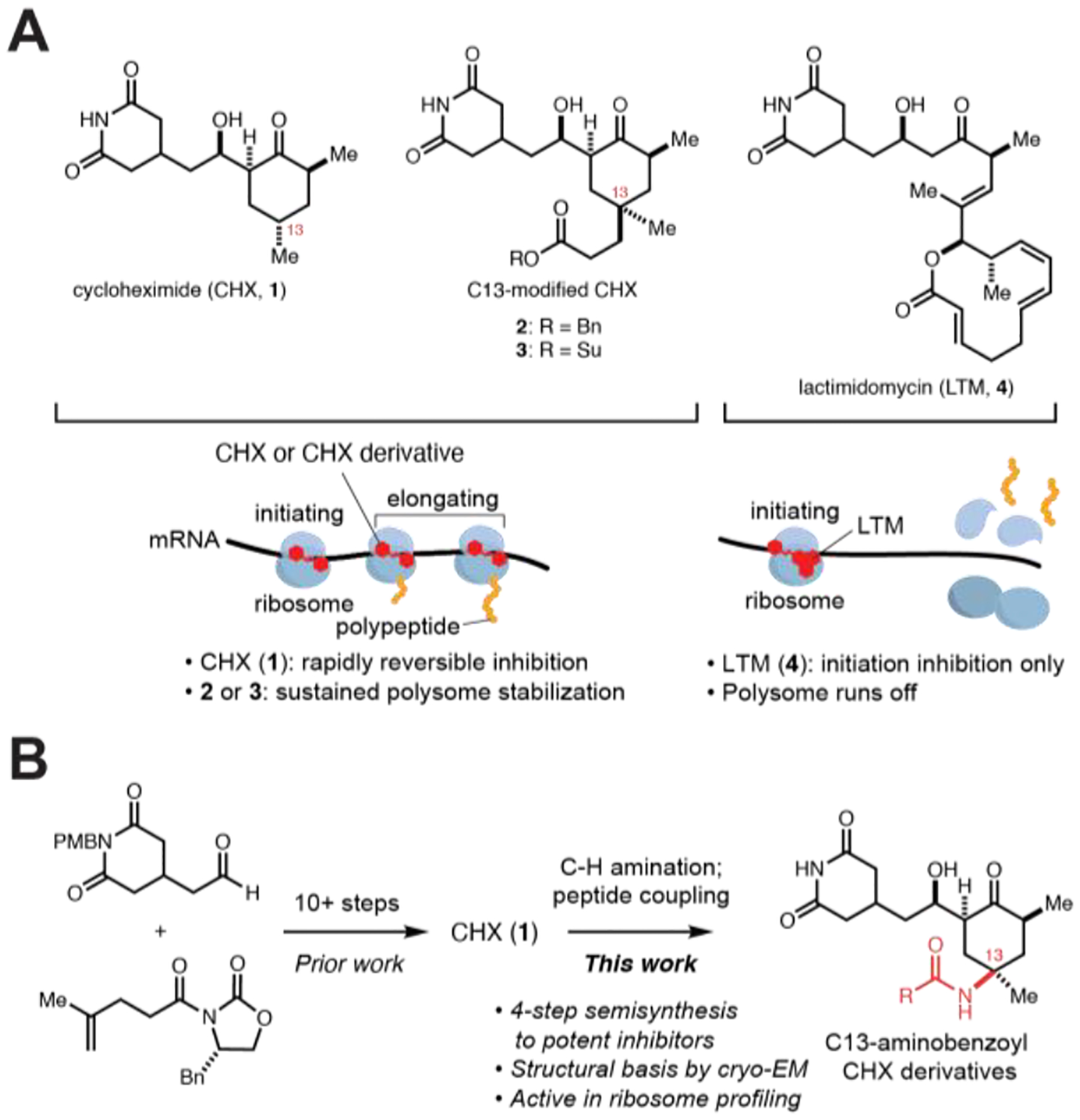
Structures of CHX (1) and C13-substituted CHX derivatives. (A) CHX (1), C13-substituted derivatives (2, 3), and lactimidomycin (LTM, 4) bind the ribosome E site, but have different effects on translation. (B) A semi-synthetic route from 1 allows access to C13-modified derivatives.
CHX is frequently employed for ribosome profiling, where rapid stabilization of polysomes is essential to maintain ribosome positions along transcripts. However, residual translation may still proceed even at high concentrations of CHX, which introduces biases that distort ribosome profiles.6 To overcome these limitations, cocktails of inhibitors including CHX have been recently employed to more faithfully freeze translating ribosomes and capture their conformational state.7,8 Alternatively, more potent derivatives of CHX leading to sustained polysome stabilization may directly overcome some of these limitations as a single agent or be a more effective component of a cocktail.
Motivated by these questions, we completed a total synthesis of CHX that enabled identification of analogs with increased potencies, including benzylester 2 and N-hydroxysuccinimide ester 3 (Figure 1A).9 The relative stereochemistry of the C13-Me group was essential for the enhanced inhibitory activity of the C13-modified derivatives. We speculated that the C13-modified derivatives form additional stabilizing interactions within the binding pocket similarly to lactimidomycin (LTM, 4), another E-site inhibitor that is more potent than CHX but only blocks translation at the initiating codon (Figure 1A).3 However, in contrast to LTM, the C13-modified derivatives can block translation at any position. Existing structural data does not conclusively address how additional C13-substituents increase inhibitor potency while sustaining polysome stabilization. Moreover, while these CHX C13-analogs are promising reagents to study translation, their further exploration is restricted by limitations of the synthetic route (Figure 1B).
Prompted by these limitations, we set out to develop an efficient synthetic route to C13-modified CHX analogs. Inspired by work from Du Bois and coworkers,10 we reasoned that stereospecific C–H functionalization at C13 could directly afford CHX derivatives from the natural product (Figure 1B). Consequently, CHX was protected as TMS-ether 6 and subjected to an intermolecular C–H amination reaction, affording amine 7 as a single diastereomer following global deprotection (Figure 2A). Conveniently, this protocol can be conducted on gram-scale. The amino group was then acylated through peptide coupling with various carboxylic acids to afford CHX C13-amide derivatives. Many of these amide derivatives were effective translation inhibitors, as assessed by a cellular assay for translation that measures incorporation of 2’-O-propargyl puromycin (OPP) (Figure 2B).9,11 In particular, this includes benzamide 8 (IC50=63 nM), which is approximately 40 times more active than CHX (Figure 2C). Derivatives containing simple modifications to the benzamide were not as effective (Figure 2B).
Figure 2.
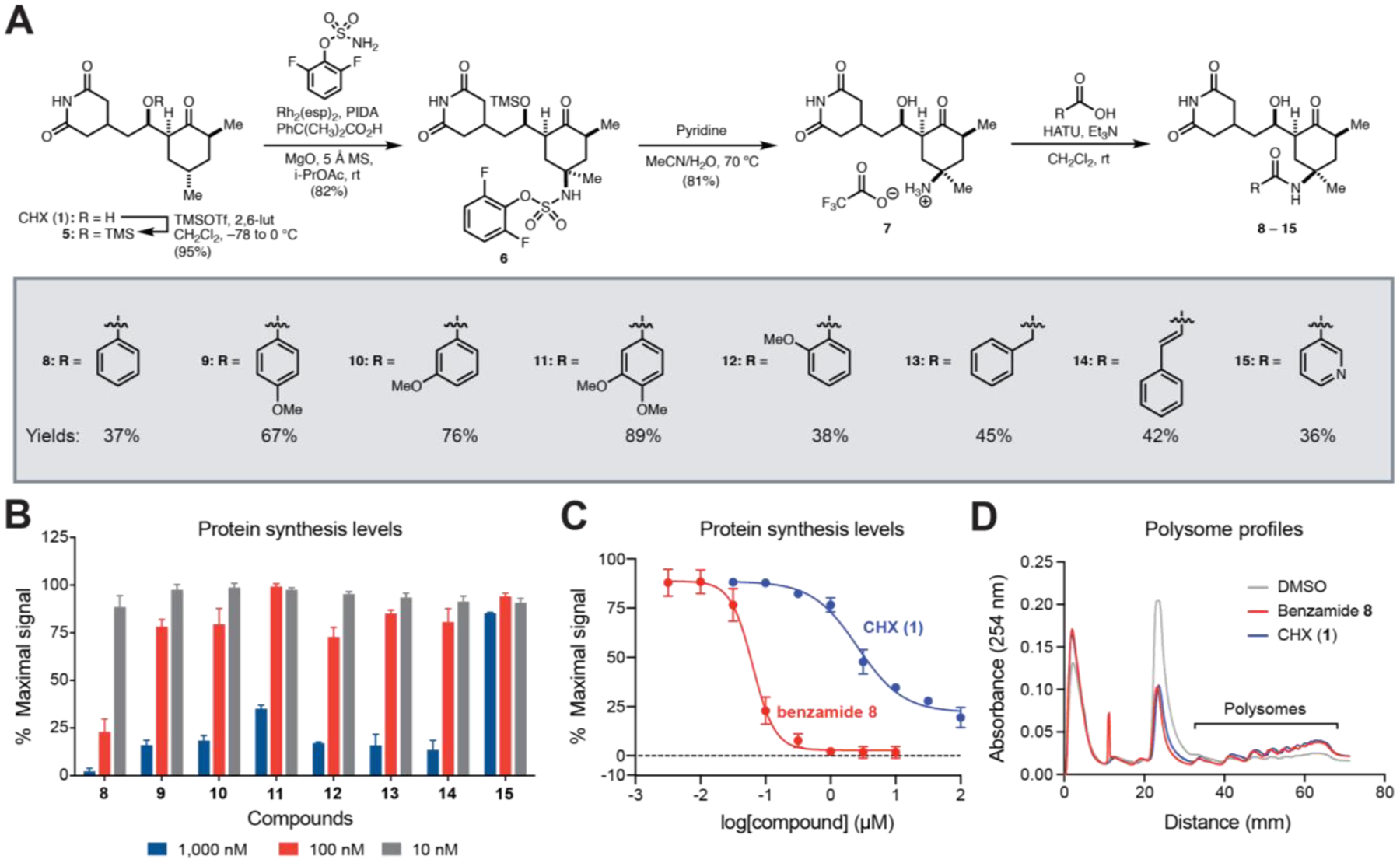
Late-stage C13-C–H amination enables facile synthesis of C13-substituted CHX derivatives that potently inhibit protein synthesis. (A) Semi-synthesis of CHX derivatives 8-15. (B) Relative protein synthesis levels in K562 cells measured by OPP incorporation after treatment with C13-modified CHX derivatives. (C) Dose-response curves show relative protein synthesis levels after treatment with 1 or 8 versus vehicle (0.1 % DMSO v/v). (D) Polysome profiles of 293T cells treated with vehicle, CHX (100 μM), or 8 (100 μM) for 30 min. Error bars represent SE for n=3 for (B) and (C).
In polysome profiling assays, benzamide 8 stabilized polysomes to a similar extent as CHX (Figure 2D). Notably, polysome integrity was achieved with a single-dose treatment of 8 in cells prior to lysis; by contrast, high concentrations of CHX are typically required in all buffers after cell lysis due to its rapidly reversible binding (Figure 1A).6 This robust polysome stabilization suggests that inhibition of translation elongation by 8 is not rapidly reversible (Figure S1, Figure S2).
To investigate the molecular basis of inhibition by benzamide 8, we conducted dimethyl sulfate (DMS) footprinting to determine accessibility of rRNA nucleotides in the presence or absence of compounds. 293T cells pretreated with CHX or 8 before addition of DMS revealed strong protection of C4341, a key residue in the 28S rRNA that H-bonds CHX (Figures 3A). C4341 protection supports that 8 occupies the canonical CHX binding pocket. Comparison between 8- or CHX- pretreatment did not reveal significant differential protection (Figure S3).
Figure 3.
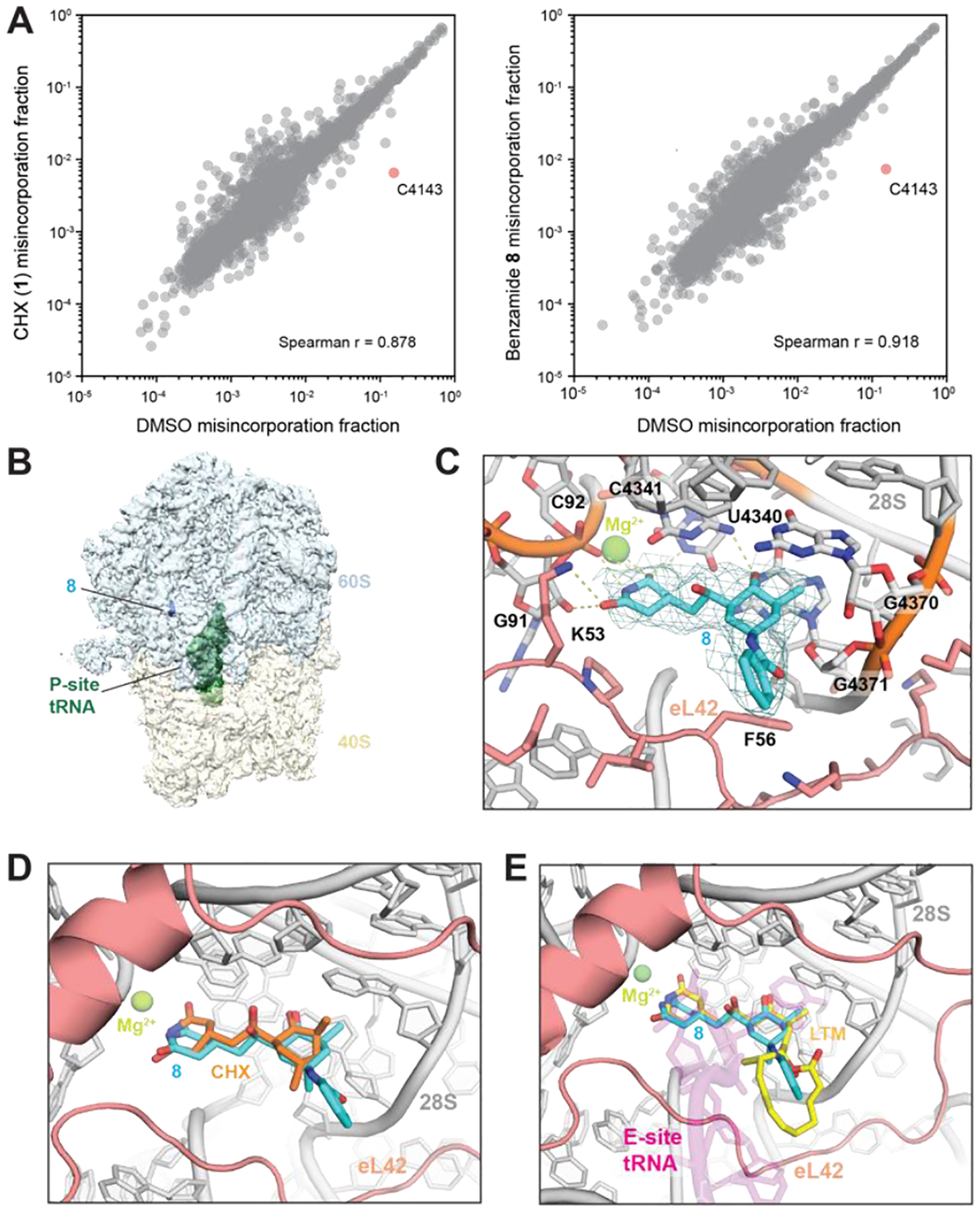
(A) Scatter plots showing DMS-induced mutation rates of 28S and 18S rRNAs comparing pretreatment with CHX (left) or 8 (right) relative to DMSO. (B) Density map overview of non-rotated 80S•8•P-tRNA. (C) Closeup view of the binding site in the 80S•8•P-tRNA model with the map (sigma=2.0). H-bonds are shown as dotted lines. rRNA:gray; eL42:salmon; 8:cyan. Important residues are shown in stick representation. (D) Overlay of the 80S•8•P-tRNA model with the H. sapiens 80S•CHX structure (PDB 5lks). (E) Overlay of the 80S•8•P-tRNA model with the O. cuniculus 80S•P-tRNA•E-tRNA structure (5lzu) and with the S. cerevisiae 80S•LTM structure (4u4r). P-tRNA:magenta; LTM:yellow.
We next used cryo-EM to analyze 8 bound to translating rabbit ribosomes (Table S1, Figure S4).12 Consistent with prior reports of CHX-bound 80S ribosome structures,5 two classes of ribosome particles were observed. One class (approximately two-thirds of particles) contained rotated 80S ribosomes with hybrid P/E and A/P tRNAs. In line with a prior study of CHX,5 the inhibitor is either absent or not visible in this class. The second class (approximately one-third of particles) were classified as non-rotated 80S ribosomes; in this class, a single peptidyl-tRNA occupies the P-site while 8 occupies the canonical CHX-binding site (Figure 3B).
We determined the structure of the non-rotated 80S ribosome bound to 8 at 3.2 Å overall resolution (Figure S4). Density of the CHX-binding pocket is well-resolved, allowing docking of 8 (Figure 3C, Figure S5A). Superposition with the CHX-bound yeast and human ribosomes shows that the binding pocket remains largely in the same conformation (RMSD=0.577 and 0.727 respectively), and that 8 makes similar interactions as CHX with eL42 and the 28S rRNA (Figure 3D, Figure S5B).5 Despite lower local resolution than the rest of the molecule, additional density corresponding to the benzamide of 8 was clearly observed and was not present in a 4.1 Å cryo-EM map of the CHX-bound ribosome (Figure S5A). The benzamide of 8 points toward the space that the tRNA would normally occupy, resembling the positioning of LTM (Figure 3E).4 The macrolactone of LTM is proposed to obstruct LTM binding if the E site is already occupied by tRNA.4 We speculate that the benzamide of 8 may partially recapitulate the interactions made by LTM, but without the full steric demand that only allows LTM to access the E site during the first cycle of translation. Collectively, our structural analyses suggest that the C13-aminobenzoyl group competes more effectively with E-site tRNA than CHX.
To investigate whether 8 may lead to genome-wide differences in ribosome occupancy compared to CHX, we conducted ribosome profiling13 in 293T cells treated with 8 or CHX (representative profile in Figure 4A). Globally, the levels of ribosomes per gene were highly correlated between treatment with 8 and CHX (Figure 4B), consistent with the notion that each inhibitor blocks ribosome elongation. Pairwise comparisons of CHX and benzamide 8 treated-conditions only revealed 23 genes with differential levels of normalized ribosome density out of 20,737 genes total (Figure 4B, Table S2).
Figure 4.
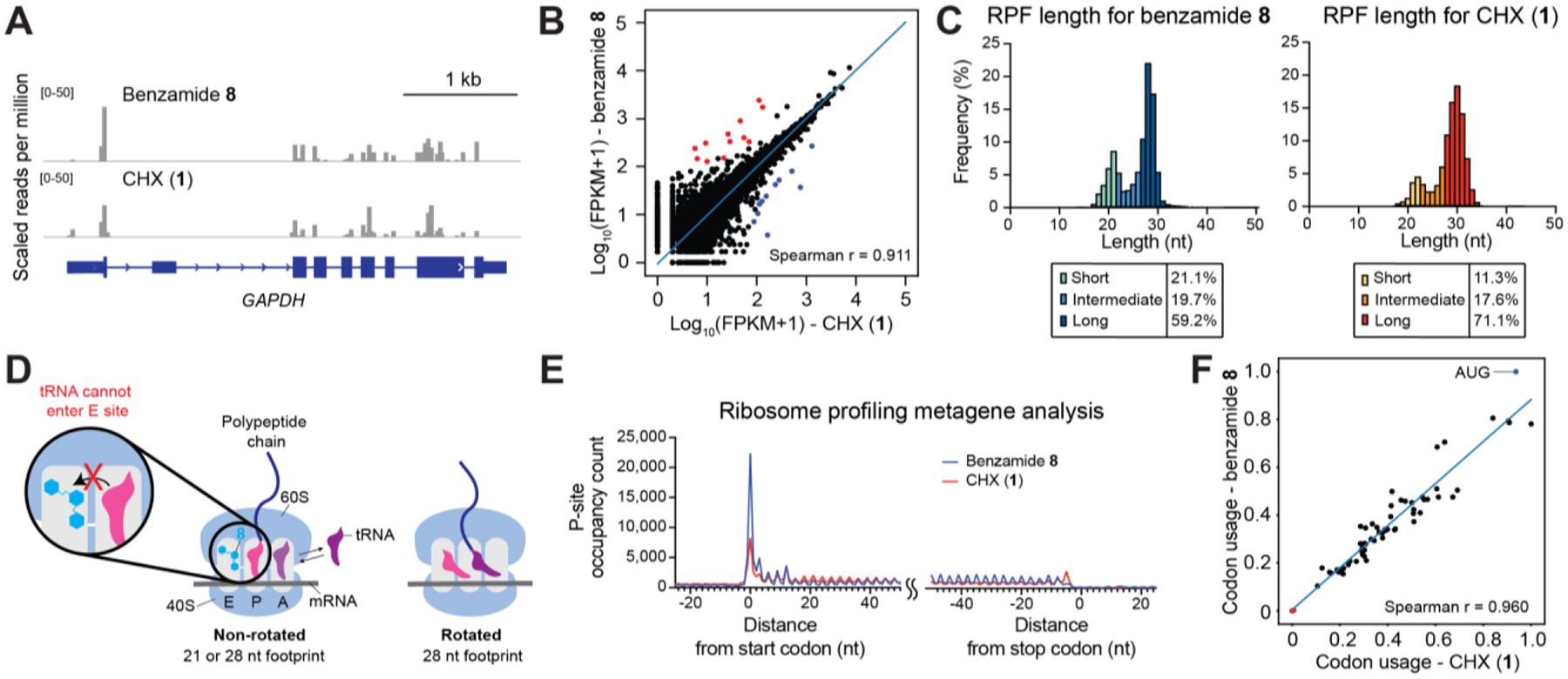
Benzamide 8 is effective in ribosome profiling. (A) Genome browser tracks showing mapped footprints at the GAPDH locus. (B) Scatter plot comparing ribosome density of 8-treated cells and that of CHX-treated cells (FPKM = fragments per kilobase million; Spearman correlation=0.911). Each dot represents a gene with >1 mapped footprint. Out of 20,737 genes, 12 whose mapped footprints were enriched (red) and 11 depleted (blue) in the presence of 8 versus CHX (cutoff: |log2(fold change)|>1, adjusted p-value<0.05). The blue line represents linear regression of log-transformed data. (C) Histogram of footprint length isolated from 8- (left) or CHX-treatment (right). (D) Proposed model where 8 sustainably stabilizes the non-rotated conformation and enriches the 21 nt footprint. By contrast, CHX inhibition is rapidly reversible, allowing transition to the rotated conformation and greater preponderance of longer footprints. (E) Metagene profile showing average P-site occupancy counts of footprints across all detected transcripts. (F) Scatter plot comparing codon usage of 8- or CHX- treated cells (Spearman correlation=0.960). Start codon:blue dot; stop codons: red dots. The blue line represents linear regression.
Ribosome profiling experiments enrich a bimodal distribution of footprint sizes, centered around a smaller 21 nucleotide (nt) and larger 28 nt average fragment size. While ribosome footprints identified in each inhibitor treatment were of comparable size, smaller footprints were preferentially enriched by 8 in comparison to CHX (Figure 4C). A recent study demonstrated that 21 and 28 nt footprints correspond to ribosomes whose 40S A site is unoccupied or occupied by tRNA, respectively.6 While the 28 nt footprint can correspond to ribosomes in either a rotated or non-rotated conformation, the 21 nt footprint corresponds to a non-rotated conformation where the A-site tRNA is absent (Figure 4D). CHX binds the E site of non-rotated ribosomes and is thought to block the deacylated tRNA from entering the E site.3,14,15 In agreement, in our cryo-EM structures the inhibitor-bound ribosomes reside in the non-rotated conformation without an A-site tRNA, which may have dissociated during sample preparation as previously suggested.7 Hence, the 21 nt footprints might arise from CHX- or 8-bound non-rotated ribosomes where the A-site tRNA has dissociated during lysis, while the 28 nt footprints might arise from either inhibitor-bound or unbound ribosomes. Indeed, continued ribosomal movement in lysate even in the presence of CHX is thought to enrich 28 nt footprints.7 Taken together, the enrichment of smaller footprints may reflect the sustained inhibition by 8 in comparison to CHX in cells, which blocks transition to rotated ribosomes (Figure 4D).
Metagenomic analysis of ribosome footprints revealed differences around the start and stop codons (Figure 4E). Ribosome profiles derived from 8-treatment showed increased buildup of reads at the start codon. Thus, consistent with its structure, 8 appears to have an effect intermediate between those of LTM and CHX that allows it to inhibit ribosomes at the start codon more efficiently than CHX without impairing its ability to inhibit elongating ribosomes – as is the case with LTM (Figure 1A and 3E). Metagenomic codon usage analysis also revealed enrichment of the start codons in 8-treated cells compared to CHX-treated cells (Figure 4F). By contrast, CHX-treatment resulted in preferential accumulation of footprints proximal to the stop codon in comparison to 8-treatment (Figure 4E). This accumulation may be attributed to ribosome run-off due to incomplete inhibition, as omitting inhibitor treatment often results in stacking of ribosomes around the stop codon.16,17 These observations are consistent with the notion that 8 may stabilize elongating ribosomes more effectively than CHX. However, the molecular basis of the ribosome profiling differences observed with 8 versus CHX remain to be fully determined.7,16 Altogether, ribosome profiling experiments suggest that treatment with 8 does not introduce systematic differences to protein synthesis relative to CHX treatment and that 8, given its sustained stabilization of polysomes, may be a more effective reagent for this protocol.
In summary, we discovered C13-amide CHX derivatives that inhibit translation elongation with enhanced potencies. Our concise and versatile semi-synthetic route allows facile access to highly functionalized translation inhibitors, which can enable the advancement of both current and novel methodologies to study ribosome dynamics and protein synthesis. In particular, 8 stabilizes polysomes in a sustained manner and acts as a superior reagent in ribosome profiling, more effectively capturing non-rotated ribosome conformations. Through structural resolution of 8-bound ribosomes by cryo-EM, we uncover the binding mode and molecular determinants underlying the increased potency and mechanism of action of the C13-modified CHX derivatives. We anticipate that these derivatives will serve as powerful probes for investigating protein synthesis.
Supplementary Material
ACKNOWLEDGMENT
Y.K. acknowledges funding from a Funai Fellowship. CryoEM data collections were performed at the Molecular Electron Microscopy Suite (MEMS) at Harvard Medical School and at the MRC-LMB cryoEM facility; data processing was supported by SBGrid. We thank Melissa Chambers and Zongli Li for EM support.
Funding Sources
Harvard University Startup Funds, National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) DP2GM137415, Vallee Foundation.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Figures S1–5, tables S1–2, materials and methods, copies of 1H,13C and 19F NMR spectra, and data processing. Structural data have been deposited to the Electron Microscopy Data Bank (EMDB; EMD-23785, EMD-23787) and the RCSB PDB (7MDZ).
The authors declare no competing financial interest.
REFERENCES
- (1).Ingolia NT; Ghaemmaghami S; Newman JR; Weissman JS Genome-Wide Analysis in Vivo of Translation with Nucleotide Resolution Using Ribosome Profiling. Science 2009, 324 (5924), 218–223. 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Ingolia NT; Hussmann JA; Weissman JS Ribosome Profiling: Global Views of Translation. Csh Perspect Biol 2018, 11 (5), a032698. 10.1101/cshperspect.a032698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Schneider-Poetsch T; Ju J; Eyler DE; Dang Y; Bhat S; Merrick WC; Green R; Shen B; Liu JO Inhibition of Eukaryotic Translation Elongation by Cycloheximide and Lactimidomycin. Nat Chem Biol 2010, 6 (3), nchembio.304. 10.1038/nchembio.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).de Loubresse N; Prokhorova I; Holtkamp W; Rodnina MV; Yusupova G; Yusupov M Structural Basis for the Inhibition of the Eukaryotic Ribosome. Nature 2014, 513 (7519), nature13737. 10.1038/nature13737. [DOI] [PubMed] [Google Scholar]
- (5).Myasnikov AG; Natchiar SK; Nebout M; Hazemann I; Imbert V; Khatter H; Peyron J-F; Klaholz BP Structure–Function Insights Reveal the Human Ribosome as a Cancer Target for Antibiotics. Nat Commun 2016, 7 (1), 12856. 10.1038/ncomms12856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Hussmann JA; Patchett S; Johnson A; Sawyer S; Press WH Understanding Biases in Ribosome Profiling Experiments Reveals Signatures of Translation Dynamics in Yeast. Plos Genet 2015, 11 (12), e1005732. 10.1371/journal.pgen.1005732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Wu CC-C; Zinshteyn B; Wehner KA; Green R High-Resolution Ribosome Profiling Defines Discrete Ribosome Elongation States and Translational Regulation during Cellular Stress. Mol Cell 2019, 73 (5), 959–970.e5. 10.1016/j.molcel.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Tesina P; Lessen LN; Buschauer R; Cheng J; Wu CC; Berninghausen O; Buskirk AR; Becker T; Beckmann R; Green R Molecular Mechanism of Translational Stalling by Inhibitory Codon Combinations and Poly(A) Tracts. Embo J 2020, 39 (3), e103365. 10.15252/embj.2019103365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Park Y; Koga Y; Su C; Waterbury AL; Johnny CL; Liau BB Versatile Synthetic Route to Cycloheximide and Analogues That Potently Inhibit Translation Elongation. Angew Chem-ger Edit 2019, 131 (16), 5441–5445. 10.1002/ange.201901386. [DOI] [PubMed] [Google Scholar]
- (10).Roizen JL; Zalatan DN; Du Bois J Selective Intermolecular Amination of C–H Bonds at Tertiary Carbon Centers. Angewandte Chemie Int Ed 2013, 52 (43), 11343–11346. 10.1002/anie.201304238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Liu J; Xu Y; Stoleru D; Salic A Imaging Protein Synthesis in Cells and Tissues with an Alkyne Analog of Puromycin. Proc National Acad Sci 2012, 109 (2), 413–418. 10.1073/pnas.1111561108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Shao S; Murray J; Brown A; Taunton J; Ramakrishnan V; Hegde RS Decoding Mammalian Ribosome-MRNA States by Translational GTPase Complexes. Cell 2016, 167 (5), 1229–1240.e15. 10.1016/j.cell.2016.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Ingolia NT; Brar GA; Rouskin S; McGeachy AM; Weissman JS The Ribosome Profiling Strategy for Monitoring Translation in Vivo by Deep Sequencing of Ribosome-Protected MRNA Fragments. Nat Protoc 2012, 7 (8), nprot.2012.086. 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Budkevich T; Giesebrecht J; Altman RB; Munro JB; Mielke T; Nierhaus KH; Blanchard SC; Spahn CMT Structure and Dynamics of the Mammalian Ribosomal Pretranslocation Complex. Mol Cell 2011, 44 (2), 214–224. 10.1016/j.molcel.2011.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Pellegrino S; Meyer M; Könst ZA; Holm M; Voora VK; Kashinskaya D; Zanette C; Mobley DL; Yusupova G; Vanderwal CD; Blanchard SC; Yusupov M Understanding the Role of Intermolecular Interactions between Lissoclimides and the Eukaryotic Ribosome. Nucleic Acids Res 2019, 47 (6), gkz053-. 10.1093/nar/gkz053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Ingolia NT; Lareau LF; Weissman JS Ribosome Profiling of Mouse Embryonic Stem Cells Reveals the Complexity and Dynamics of Mammalian Proteomes. Cell 2011, 147 (4), 789–802. 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Liu B; Qian S-B Characterizing Inactive Ribosomes in Translational Profiling. Transl Austin Tex 2016, 4 (1), e1138018. 10.1080/21690731.2015.1138018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


