Abstract
CTXφ is a filamentous, lysogenic bacteriophage whose genome encodes cholera toxin, the primary virulence factor produced by Vibrio cholerae. CTX prophages in O1 El Tor and O139 strains of V. cholerae are found within arrays of genetically related elements integrated at a single locus within the V. cholerae large chromosome. The prophages of O1 El Tor and O139 strains generally yield infectious CTXφ. In contrast, O1 classical strains of V. cholerae do not produce CTXφ, although they produce cholera toxin and they contain CTX prophages integrated at two sites. We have identified the second site of CTX prophage integration in O1 classical strains and characterized the classical prophage arrays genetically and functionally. The genes of classical prophages encode functional forms of all of the proteins needed for production of CTXφ. Classical CTX prophages are present either as solitary prophages or as arrays of two truncated, fused prophages. RS1, a genetic element that is closely related to CTXφ and is often interspersed with CTX prophages in El Tor strains, was not detected in classical V. cholerae. Our model for CTXφ production predicts that the CTX prophage arrangements in classical strains will not yield extrachromosomal CTX DNA and thus will not yield virions, and our experimental results confirm this prediction. Thus, failure of O1 classical strains of V. cholerae to produce CTXφ is due to overall deficiencies in the structures of the arrays of classical prophages, rather than to mutations affecting individual CTX prophage genes.
The severe diarrheal disease cholera results from colonization of the human small intestine by pathogenic strains of a gram-negative bacterium, Vibrio cholerae. Cholera has afflicted human populations in many parts of the world for more than a millenium (2). Widespread outbreaks have been common; in the last 200 years alone, seven cholera pandemics have occurred. Most epidemic strains of V. cholerae have been of the O1 serogroup, although in the last 8 years O139 serogroup strains of V. cholerae have also been linked to disease outbreaks (23). The O1 serogroup of V. cholerae has been divided into strains of the classical biotype, thought to have been responsible for the first six cholera pandemics, and the El Tor biotype, which has caused the ongoing seventh pandemic (2). These El Tor and classical strains have traditionally been differentiated in the laboratory with assays of hemolysis, hemagglutination, phage sensitivity, and polymyxin sensitivity and with the Voges-Proskauer reaction (23). Genetic typing of strains has also become possible in recent years. However, despite their phenotypic and genotypic differences, the symptoms of infection with strains of the two O1 biotypes are clinically indistinguishable. The clinical manifestations of cholera are almost entirely due to production of cholera toxin, a potent A-B-type exotoxin that is secreted by pathogenic V. cholerae (10).
The genes that encode cholera toxin, ctxAB, reside within the genome of a filamentous, lysogenic bacteriophage known as CTXφ (22). CTXφ DNA is generally found integrated at either one (El Tor) or two (classical) loci within the V. cholerae genome (13, 16, 22). In El Tor strains, the prophage DNA is usually found in tandem arrays that also include a related genetic element known as RS1 (13, 24). RS1 contains the genes that enable phage DNA replication and integration (rstR, rstA, and rstB) plus an additional gene, rstC, whose function is unknown. RS1 does not contain ctxAB or the other genes of the phage core region, which are thought to produce proteins needed for virion assembly and secretion. Most analyses of the structures of CTX prophage arrays and of CTXφ have used El Tor strains and virions derived from them. Consequently, very little is known about the structure of CTX prophage arrays or the prevalence of RS1 in strains of the classical biotype. Furthermore, although it is known that the two classical prophage insertion sites are on different chromosomes (20), the sequences flanking classical prophages and their precise locations within chromosomal DNA have not been determined.
The CTX prophage-RS1 arrays differ widely among pathogenic O1 and O139 V. cholerae strains. First, there are biotype-specific differences between the phage genomes, in particular between the rstR and flanking sequences from El Tor strains (rstRET) and the corresponding regions of the classical prophage (rstRclass) (11). Second, strains differ by the number of phage genomes and RS1s that they contain and in the relative arrangements of these elements. Numerous patterns have been detected in O1 El Tor strains as well as in O139 strains (4, 13, 21). In contrast, the majority of classical strains have been thought, based on restriction mapping, to share a single arrangement (13). However, a few variant arrangements in classical strains have recently been identified (3) (see below). Thus, there is more heterogeneity among the prophage arrays of O1 classical strains than was initially recognized.
The CTX prophage first found to yield virions resided within an O1 El Tor strain of V. cholerae. Since CTXφ is not a plaque-forming phage, virion formation was detected by the ability of supernatants from a strain containing a kanamycin resistance (Knr)-marked prophage to transduce a recipient strain to Knr. We have subsequently found that O139 strains from Calcutta, India, which contain two distinct prophages (CTXET and CTXcalc) also yield virions and that these virions are produced from both prophages (6). In contrast, we have never detected virions derived from the prophages in a prototypical classical strain of V. cholerae, O395, or a related strain with Knr-marked prophages, O395NT (14). However, classical strains (e.g., O395 and 569B) do produce a high titer of virions following transduction by the Knr-marked El Tor CTXφ (CTXET-Knφ) (22). These data suggest that there is a defect within the classical prophages rather than in chromosomal loci needed for phage replication and/or secretion.
For this study, we have characterized the virion production capacities and genetic structures of classical prophage arrays. We have found that classical strains encode phage proteins capable of mediating both phage DNA replication and virion assembly and that at least some of these genes are constitutively expressed by classical lysogens. We have also found that the genetic element RS1 is not present in classical strains of V. cholerae. The CTX insertion sites in classical strains instead contain either a solitary prophage or a prophage array composed of two truncated, fused prophages. Neither of these arrangements is predicted to yield extrachromosomal CTX phage DNA (7). The inability of classical strains to produce this extrachromosomal replicative form of the classical prophage DNA, which is thought to be an early and critical step in production of virions from CTX prophages (7), probably accounts for the absence of detectable classical CTXφ (CTXclassφ).
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains were cultured in Luria-Bertani medium (LB) at 37°C unless otherwise noted. The antibiotics kanamycin (50 μg/ml) and ampicillin (50 μg/ml) were used as required. The classical strains of V. cholerae analyzed in this study are listed in Table 1. Control strains used were the O139 strain AS207, which has one CTX prophage array (RS1-CTX-CTX-CTX) (6), and the O1 El Tor strain E7946, which also has just one prophage array (RS1-CTX-RS1-CTX-RS1) (13).
TABLE 1.
Classical strains of V. cholerae used in this study
| Strain | Place isolated | Yr isolated | Reference or source |
|---|---|---|---|
| O395 | India | 1964b | 13 |
| C1 | Unknown | 1955 | F. Mooi |
| C14 | Unknown | 1973b | F. Mooi |
| C21 | Pakistan | 1963 | F. Mooi |
| C33 | Egyptb | Unknown | F. Mooi |
| C34 | Egypt | 1949 | F. Mooi |
| CA401 | India | 1953 | 8 |
| GP12 | India | 1971 | G. B. Nair |
| 569Ba | India | 1948b | 13 |
Not analyzed directly; plasmids (pGP2 and pGP3 [13]) derived from this strain were analyzed.
Not known with certainty but believed to be as listed.
Plasmids.
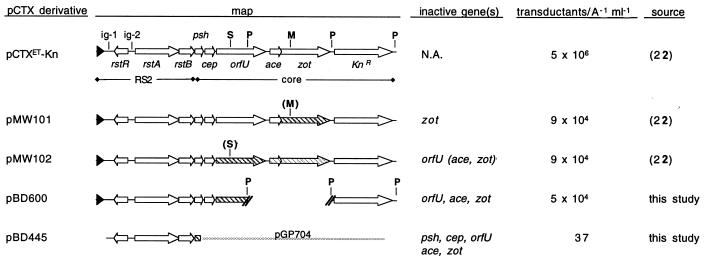
pBD600 was generated by digesting pCTXET-Kn with PstI and religating two of the three resulting restriction fragments (Fig. 1). The insert for pBD445 was amplified from the El Tor strain E7946 by using the primers RstRF (5′CGGCATTATGTTGAGGGGCAGTCGT3′) and psh2 (5′AAATGAGACTAGCAACCGC3′). The resulting PCR product was cloned into the vector pCRII-TOPO (Invitrogen; Carlsbad, Calif.), and the insert was then subcloned on an EcoRI fragment into the suicide vector pGP704 (15) to generate pBD445. The same process was used to generate pBD394 from O395 DNA.
FIG. 1.
Derivatives of pCTXET that do not encode all of the phage proteins required for virion production are packaged into infectious virions by a classical strain of V. cholerae, O395. O395 transformed with wild-type pCTXET-Kn or a plasmid derivative containing an incomplete or mutant phage genome was used as a source of donor supernatant in transduction assays. At least one gene required for production of virions was inactive in or absent from each of the plasmids (except pCTXET-Kn), yet all of the plasmids yielded virions when maintained in O395. Phage titer per milliliter was normalized to the A600 of the donor culture. Titers are average results based on at least three experiments. Sequences that no longer yield functional proteins are marked by diagonal grey lines. (S) and (M) denote SphI and MluI restriction sites that were filled in and religated to create frameshift mutations. It is not known whether the orfU mutation in pMW102 has a polar effect on downstream phage genes (marked with light stripes). Deletion of the PstI (P) fragment results in loss or inactivation of three phage genes. The Knr cassette has been inserted in place of most of ctxAB, which do not contribute to CTXφ production. pGP704 is an Apr suicide vector that cannot replicate independently in V. cholerae. N.A., not applicable.
Transduction assays.
Filter-sterilized supernatants (100 μl) from late-log-phase donor cultures was mixed with the recipient strain O395 (50 μl), which had been cultured at 30°C to induce expression of the CTXφ receptor, TCP. After a 20-min incubation at room temperature, mixtures were plated on LB agar plates containing selective antibiotics. Transduced recipients were identified after growth overnight at 37°C.
PCR.
PCRs for assessing the insertion sites and junction sequences of classical prophages were performed using standard conditions. Primers used were RstBF2 (5′GTGGTGTGTTCCTTGTTTGAC3′), RstRRev (5′CGACCAAGCAAGATAATCGAC3′), TLCF1 (5′TGTCGGAGCTGCTTGGATTAAG3′), Ig1R1 (5′ACGACTGCCCCTCAACATAATG3′), and PyrFF1 (5′GGTGCCTATATCAACTTTCTTGCG3′).
Molecular biology techniques.
DNA probes for Southern blots were labeled with horseradish peroxidase, using the ECL direct nucleic acid labeling and detection system (Amersham Pharmacia, Buckinghamshire, England). Blots were hybridized and washed according to the manufacturer's instructions. Dye-terminator cycle sequencing of DNA was performed by the Tufts Core Facility. Other techniques were performed according to standard protocols (1).
Nucleotide sequence accession number.
The sequence of the junction between adjacent truncated prophages in O395 has been assigned GenBank accession no. AF262318.
RESULTS AND DISCUSSION
Classical prophage core genes encode functional proteins.
To determine which component(s) of the classical CTX prophages is defective, we first tested whether these prophages could complement a variety of mutations in the core region of the CTXET-Knφ genome (Fig. 1). These mutations do not interfere with replication of CTXET-Knφ DNA, but they prevent production of new virions in the absence of supplemental wild-type phage genes (reference 22 and data not shown). The replicative form (also known as pCTX) of each mutant genome was introduced into the classical strain O395, which had previously been shown to maintain newly introduced phage DNA in plasmid form (22). In the O395 strain background, even a pCTXET-Kn derivative lacking three of the five core genes needed for virion assembly (pBD600) readily yielded virions capable of transducing Knr (Fig. 1). Similarly, an ampicillin-resistant (Apr) derivative of pCTX, pBD445, which includes the phage genes rstR, rstA, and rstB but has no intact phage core genes, could also be packaged into virions by O395, although at a much lower frequency. Transduced DNA corresponded to the mutant genomes, not to wild-type genomes generated by recombination between the pCTX derivatives and chromosomal prophage DNA (data not shown). These data suggest that the core genes of the classical prophages encode functional proteins; thus, the defect of these prophages does not appear to lie within their core regions. In addition, these data suggest that all the CTX core genes are at least minimally expressed from the classical prophages, even though CTXclassφ is not produced.
We have not investigated the biological significance of this apparent maintainance of phage protein synthesis in the absence of CTXclassφ production. However, others have proposed that two phage-encoded proteins, Ace and Zot, function as exotoxins in addition to contributing to virion formation (5, 19). It is not yet known how the intracellular fates and functions (i.e., phage related versus toxin related) of individual molecules of Ace and Zot are determined. Consequently, it is not clear whether the failure of classical strains to produce virions, despite production of putative toxic viral proteins, influences the virulence of classical V. cholerae.
Classical prophage RS2 genes encode functional proteins.
In addition to the core-encoded proteins required for CTXφ production, the phage-encoded proteins RstA and RstR are also needed. RstA and RstR mediate replication of phage DNA and regulation of rstA expression, respectively (11, 14). rstRclass has previously been shown to encode a functional protein (11); however, rstAclass, which differs slightly from rstAET, has not been studied. To test for rstAclass function, we cloned a portion of the classical prophage containing rstRclass, rstAclass, rstBclass, and some adjacent sequences into the suicide vector pGP704. The new plasmid, pBD394, is the classical equivalent of the core-negative, pCTXET derivative pBD445 (Fig. 1). Like pBD445, pBD394 was maintained as a plasmid within O395 and within the ΔattRS El Tor vaccine strain Bah-2 (17) (data not shown). Since pGP704 alone cannot replicate in V. cholerae, these data prove that the classical prophage genes encode a functional DNA replication system and, in particular, a functional form of RstA.
The role, if any, of RstB in CTXφ production has not yet been identified; RstB has been shown only to contribute to phage DNA integration (24). However, it appears that a defect in rstBclass does not underlie the lack of virion production from CTX prophages in classical strains of V. cholerae. O395 containing either pBD394 or pCTX with a Mariner transposon inserted in rstB yields infectious virions (data not shown), and such strains can synthesize RstB only from templates within or derived from the classical prophages. Production of virions by O395(pBD394) also demonstrates that pBD394 (and thus classical prophages) contains a phage packaging site; however, specific sequences required for packaging of CTXφ have not yet been identified.
Despite the presence in O395 of an rstA that can mediate replication of phage DNA, we have never detected a replicative form of the classical CTX prophage in any classical strain of V. cholerae (data not shown). In contrast, most El Tor strains produce low but measurable levels of pCTXET, and it appears that production of pCTX is an early step in CTXφ synthesis (7). Initially, we believed that pCTX was generated from a prophage by site-specific recombination between the 17- or 18-bp direct repeats that flank each prophage and RS1, since these are known to mediate integration of CTXφ DNA (16). Consequently, we assessed whether these end repeats (ERs) might have altered sequences in classical strains that prevented site-specific recombination. We subsequently discovered that ERs do not mediate production of CTXφ (7) (see below); however, this sequence analysis of classical ERs did enable us to identify the sites of CTX prophage integration in classical strains and perhaps to account for the previously puzzling finding that classical strains maintain exogenous CTX phage DNA in plasmid form.
Analyses of classical CTX prophage integration sites.
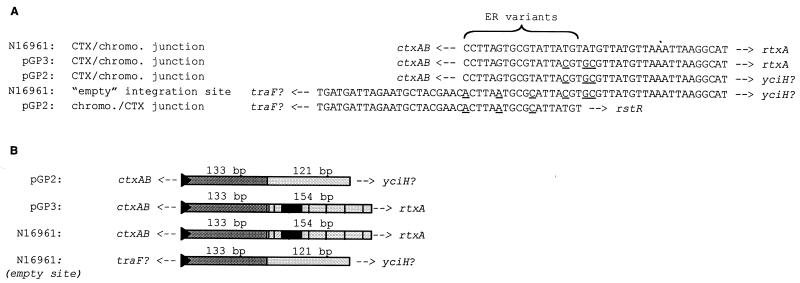
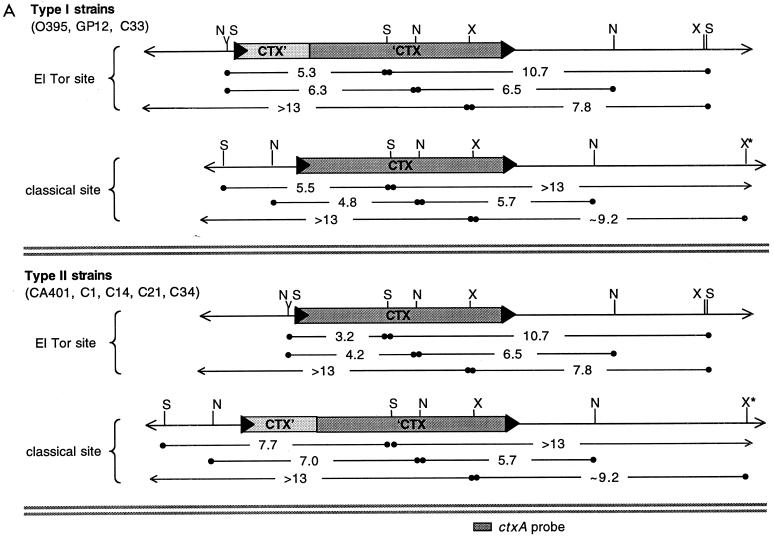
We identified the CTX prophage integration sites in classical strains of V. cholerae by sequencing the junctions between prophage and chromosomal DNA in plasmids (pGP2 and pGP3) derived from the two arrays of the classical strain 569B (13). As expected, we found that one of the two classical prophage arrays (represented by pGP3) is inserted into the chromosome at the same site as the CTX prophage(s) in El Tor strains: the intergenic region between the TLC and RTX gene clusters, which lies on replicon I, the larger of the two V. cholerae chromosomes (9, 12, 18, 20). The other classical prophage array (represented by pGP2) is also inserted into an intergenic region, located on replicon II. Alignment of pGP2-derived sequence with the V. cholerae genome sequence (generated from the El Tor strain N16961 [BLAST search engine for unfinished microbial genomes; http://www.tigr.org/cgi-bin/BlastSearch/blast.cgi?]) revealed that the additional prophage array lies 0.3 kb upstream of a gene encoding a probable ortholog of the translation initiation factor yciH and approximately 1.2 kb upstream of a gene encoding d-Ala–d-Ala ligase. The array is predicted to lie approximately 0.5 kb downstream of an apparent traF ortholog. In the N16961 genome, the sequences corresponding to those found 5′ and 3′ to the pGP2 prophage array form a continuous unit that contains a sequence similar (14 of 18 bp) to an ER (Fig. 2A). This suggests that the extra prophage array in O1 classical strains was inserted into the chromosome by a relatively site specific process, probably one that utilized the standard CTXφ integration machinery. However, it is clear that the alternate insertion site is not a preferred target; we have never detected prophages integrated at this ER variant in El Tor strains.
FIG. 2.
Comparison of the junctional sequences between CTX prophages and adjacent chromosomal DNA in classical and El Tor strains. pGP2 and pGP3 are plasmids derived from the prophages of the classical strain 569B (13). N16961 is the O1 El Tor strain sequenced by the V. cholerae genome project. The empty integration site in N16961 is the locus where the CTX prophage(s) are found on the small chromosome of classical V. cholerae. (A) Analysis of 3′ and 5′ ER sequences. Underlined bases differ from the typical El Tor junction sequences. Known genes and putative homologs that flank junction sequences are indicated. (B) Schematic alignment of sequences downstream of CTX prophage integration sites. Black triangles represent ER variants. Dark grey rectangles represent areas with essentially identical sequences. Light grey rectangles represent areas with similar sequences; the overlaid black lines and boxes represent differences and insertions. Adjacent genes are indicated. The diagram is not drawn to scale.
Our analyses of prophage insertion sites also revealed the sequences of the 3′ ERs following the prophage arrays in pGP2 and pGP3. The final 3′ ER of each phage array in 569B differs by one base pair from the 3′ ER of El Tor prophage arrays. In addition, two bases immediately downstream of these ERs also differ from the El Tor sequence (Fig. 2A). Since the 3′ ER, rather than the 5′ ER, is the integration site for new CTX phage DNA introduced into El Tor strains (6), these differences may account for the previously noted failure of CTXET-Knφ DNA to integrate into the classical chromosome (22).
Surprisingly, other than the 2-bp difference, the intergenic sequences immediately downstream of both classical prophage arrays in 569B were identical to the sequence downstream of the CTX array in O1 El Tor strains for 133 bp. An additional 121 bp of sequence downstream of the two insertion sites were similar (91% identity) (Fig. 2B). This ∼254-bp sequence is also present on the small chromosome of El Tor strains, despite the absence in El Tor strains of a CTX prophage array at this site. Consequently, it seems likely that this ∼254-bp duplication predates the development of the second prophage array in classical strains; it is unlikely to have been generated in conjunction with development of the new prophage array. Nonetheless, the prophage insertion site does mark the boundary of the duplicated region; the DNA sequences upstream of the two insertion sites show no similarity.
Genetic structure of classical CTX prophage arrays.
As mentioned above, we have recently determined that production of extrachromosomal phage DNA (and ultimately of CTXφ) does not depend upon site-specific recombination between the ERs that flank a prophage. Instead, the DNA found in CTXφ is a hybrid sequence that is derived from the DNA of two adjacent chromosomal elements (7). The upstream element contributes all of the CTXφ genome's coding sequences, while the downstream element contributes most of the intergenic sequence between the ER and rstR. Consequently, the presence of tandem elements—either two prophages or a prophage followed by an RS1—is essential for production of CTXφ by a lysogen. We therefore analyzed the structures of the prophage arrays in classical strains to determine whether they contained one of the requisite arrangements. Previously published restriction maps of pGP2 and pGP3 suggested that each insertion site would contain an RS1 followed by a CTX prophage (13). However, our analyses indicate that classical prophages are present either as solitary CTX prophages or as an array of truncated, fused prophages.
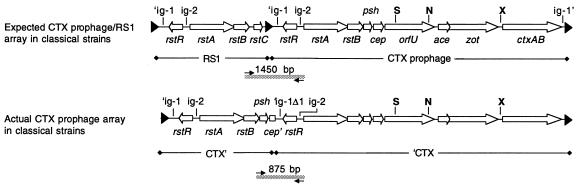
We first attempted to confirm, using PCR, that classical prophage arrays contained an RS1 followed by a CTX prophage, as initially suggested by the restriction maps of pGP2 and pGP3. We therefore generated PCR products expected to span the junction between RS1 and the downstream CTX prophage, using primers complementary to the 3′ end of rstB and to the 5′ end of rstA (Fig. 3). Our templates were the plasmids derived from strain 569B and chromosomal DNA from strain O395. Unexpectedly, all three reactions yielded a band approximately 875 bp long, rather than the predicted product of approximately 1,450 bp (data not shown). Sequence analysis of the PCR product amplified from O395 chromosomal DNA revealed that it contained neither a middle ER sequence nor an RS1-specific sequence. Instead, it consisted of sequence from a 3′ truncated CTX prophage (missing the last 4,400 bp) fused to a 5′-truncated prophage (missing the first 524 bp) (Fig. 3). Thus, the 3′-truncated prophage appeared to contain only rstR, rstA, rstB, psh, and a truncated cep. In contrast, the 5′-truncated prophage lacked most of an intergenic region (′ig-1) normally found upstream of rstR. These data suggested that neither 569B nor O395 contain the expected RS1 element. Furthermore, they indicated that all the prophages within 569B are truncated at either the 5′ or 3′ end.
FIG. 3.
Expected and actual structures of prophage arrays in classical strains of V. cholerae. Instead of an RS1 followed by a CTX prophage, CTX insertion sites that contain tandem elements in classical V. cholerae contain two truncated, fused prophages. The upstream prophage lacks the prophage genes downstream of cep (resulting in cep′), while the downstream prophage lacks ig-1 sequences starting with the ER (resulting in ′ig-1Δ1). The PCR primers used to amplify the sequences between rstB and rstA are shown as small black arrows. The DNA sequence of the unexpected 875-bp PCR product (grey line) was determined. The presence of genes not assayed by DNA sequencing or PCR has been inferred from restriction maps generated with SphI (S), NruI (N), and XbaI (X).
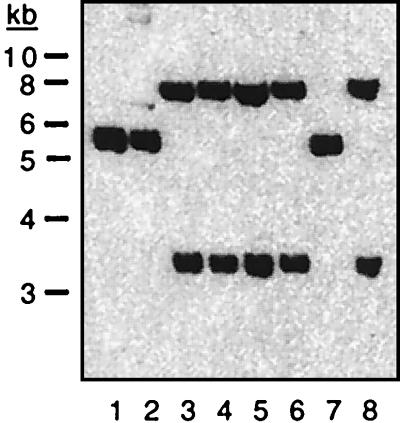
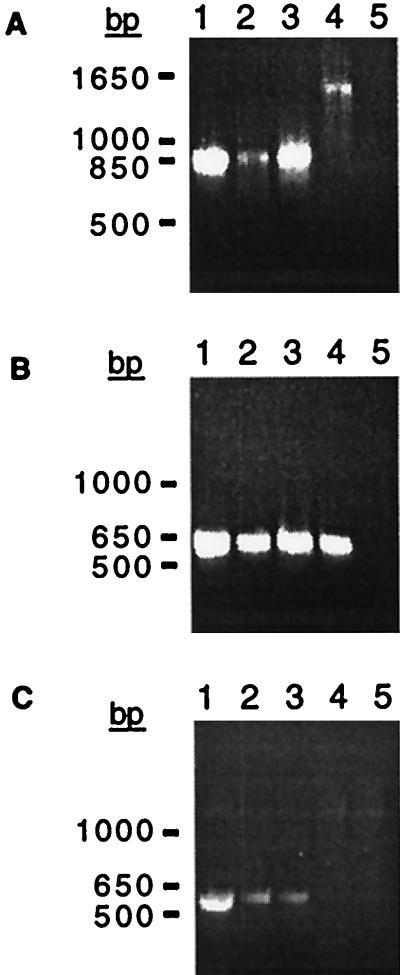
Southern and PCR analyses were performed to determine whether all of our O1 classical strains (clinical isolates obtained over a 20-year period from a variety of regions [Table 1]) lacked RS1 and whether all contained fused arrays of truncated prophages. We found that an rstC probe did not hybridize to DNA from any classical V. cholerae strains under conditions in which rstC was readily detected in El Tor chromosomal DNA (data not shown). Consequently, we suspect that no O1 classical strains contain RS1 and that RS1 is present only in El Tor strains and the closely related O139 strains of V. cholerae. Further hybridization analyses yielded the unexpected finding that our classical strains could be subdivided into two groups based on the restriction maps of their prophage DNA (Fig. 4). Strains O395, GP12, and C33 comprise the first set (type I), while strains C34, C21, C14, C1, and CA401 comprise the second (type II). However, PCR analyses of representative strains demonstrated that both type I and type II strains (i) contain adjacent truncated fused prophages, since the 875-bp PCR product corresponding to fused prophages could be amplified from DNA of both types (Fig. 5A); (ii) contain a CTX prophage or prophage derivative in the El Tor prophage insertion site downstream of TLC, since a product could be amplified with one TLC-specific primer and one prophage-specific primer (Fig. 5B); and (iii) contain a CTX prophage or prophage derivative in the classical strain-specific prophage insertion site downstream of a traF homolog, since a product could be amplified with a chromosome-specific primer and a prophage-specific primer (Fig. 5C). In addition, both types do not appear to contain intact tandem prophages, since no PCR product corresponding to a junction between two intact prophages could be generated from DNA of classical strains (data not shown). Thus, although the two types of classical V. cholerae can be distinguished by their restriction maps, they nonetheless have many features in common.
FIG. 4.
Southern blot of chromosomal DNA from classical V. cholerae strains demonstrating the heterogeneity of CTX prophage arrays. Chromosomal DNA was digested with SphI and probed with rstRclass, which detects only classical prophage DNA. No additional CTX prophage-related sequences were detected with a less specific probe (data not shown). Strains O395, GP12, CA401, C1, C14, C21, C33, and C34 (lanes 1 to 8, respectively) were compared. The single band in lanes 1, 2, and 7 was shown in additional analyses to be a doublet (data not shown).
FIG. 5.
PCR amplification of prophage-prophage and chromosome-prophage junctions in classical V. cholerae. (A) Amplication of the junction between a CTX prophage and an upstream element, using primers RstBF2 and RstRproR (Fig. 3). An ∼875-bp product was generated from DNA of classical strains C34 (lane 1), CA401 (lane 2), and O395 (lane 3), indicating that all contain truncated, fused prophages. An ∼1,450-bp product was amplified from the RS1-CTX prophage junction in the O139 strain AS207 (lane 4), and no product was synthesized in the absence of template DNA (lane 5). (B) Detection of a junction between the chromosomal sequence of TLC and a CTX prophage, using primers TLCF1 and Ig1R1. A ∼600-bp product was amplified from the DNA of classical strains C34 (lane 1), CA401 (lane 2), and O395 (lane 3) as well as the O139 strain AS207 (lane 4); it was not synthesized if template DNA was omitted (lane 5). (C) Detection of a junction between CTX prophages in the classical strain-specific insertion site and upstream chromosomal sequence, using the primers PyrFF1 and Ig1R1. A ∼550-bp product was amplified from DNA of the classical strains O395 (lane 1), C34 (lane 2), and CA401 (lane 3); no product was amplified from AS207 DNA (lane 4), which lacks a CTX prophage at this insertion site, or in the absence of template DNA (lane 5).
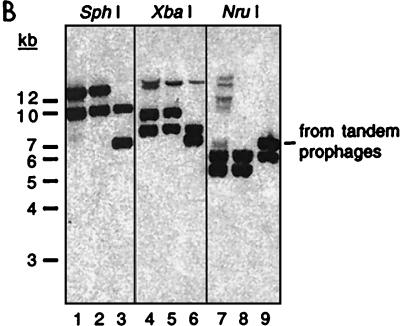
Data from additional Southern blots were combined with DNA sequence data from the V. cholerae genome project to determine the likely structures of the classical CTX prophage arrays. Restriction maps generated with SphI, NruI, BglII, and XbaI were incorporated into the final model. Since the genome data were generated from an El Tor strain of V. cholerae, the conservation in classical V. cholerae of several key diagnostic restriction sites was confirmed by restriction mapping of PCR products amplified from classical templates. These analyses indicated that all of our classical strains contain one array of truncated fused prophages (described above and in Fig. 3) and one site in which a solitary prophage is integrated. Type I strains contain the array of truncated prophages at the El Tor prophage insertion site and the solitary prophage at the classical strain-specific insertion site. In contrast, type II strains have the opposite arrangement: a solitary prophage at the El Tor insertion site and a truncated array at the classical locus (data summarized in Fig. 6A). For numerous restriction enzymes, the ctxAB-containing restriction fragments generated from these two arrangements are identical (Fig. 6); thus, it is not surprising that all classical strains of O1 V. cholerae were initially believed to contain identical prophage arrays. Interestingly, neither type I nor type II strains contain two truncated prophage arrays, although analysis of pGP2 and pGP3 indicated that classical strain 569B contains such arrays at both prophage integration sites. We hypothesize that homologous recombination between sequences within the prophage arrays can result in conversion of one array, via deletion, into a solitary prophage. Preliminary analyses suggest that such a conversion has occurred in an auxotrophic derivative of 569B, strain RV508, since restriction maps of RV508 prophages resemble those of type II classical strains (data not shown). It is possible that all of our classical strains initially had fused truncated prophages at both insertion sites, and that type I and type II strains differ now due to their modification by different recombination events. These type I and type II strains are the first natural isolates of V. cholerae that we have found to contain solitary prophages; prophages in El Tor and O139 strains are always flanked by at least one intact related element.
FIG. 6.
Restriction mapping of the CTX prophage insertions sites in classical biotype strains. (A) Summary of the restriction fragments detected on Southern blots of DNA from type I and type II classical strains digested with SphI (S), NruI (N), and XbaI (X) and probed with various prophage-derived probes. The precise sizes of restriction fragments (in kilobases) were derived from the V. cholerae genome database as well as laboratory sequence data. Band sizes on blots could not be determined as precisely, but all except one were consistent with sequence-derived sizes. X★ denotes an XbaI site that is farther downstream than expected; classical biotypes apparently lack an XbaI site that is present in this region in El Tor strains. The location of the ctxA probe used for panel B is shown at the bottom. (B) ctxAB-spanning restriction fragments from diverse classical strains of V. cholerae are identical. A Southern blot of DNAs digested with SphI, XbaI, and NruI, as indicated, was probed with a ctxA probe. Each enzyme produced equal-size fragments from the classical strains C21 (lanes 1, 4, and 7) and O395 (lanes 2, 5, and 8). AS207 DNA (lanes 3, 6, and 9) had one fragment in common with the classical DNAs for each enzyme. Digests of classical DNA do not have the band with a constant size (∼7 kb), indicative of tandem prophages, that is produced from AS207 DNA. The AS207 prophage array was previously shown to contain an RS1 followed by three tandem CTX prophages. Faint bands in a few lanes resulted from incomplete digestion of DNA.
The restriction analyses confirmed the conclusion, initially based on PCR analyses, that classical strains do not contain intact tandem prophages. Southern blots of classical V. cholerae DNA digested with multiple restriction enzymes always lacked the prophage-length band that is the hallmark of tandem prophages (Fig. 6B). In addition, DNA sequence analysis revealed that the downstream prophages in the arrays of truncated classical prophages lack most, if not all, of the sequence typically contributed to CTXφ by the downstream element in an array that produces virions. Consequently, classical strains do not contain any arrays of elements that we expect to yield extrachromosomal CTX DNA and CTXclassφ. The finding that neither pCTX, pCTX variants, nor CTXclassφ is produced by classical strains of V. cholerae is consistent with, and provides additional support for, our new model describing production of CTXφ.
In conclusion, our analyses have revealed that all of the proteins required for CTXφ production can be generated from the prophages within classical strains of V. cholerae. Many of these proteins are synthesized by classical strains under typical laboratory growth conditions, although some (e.g., Cep) are probably produced at low levels that can be limiting for virion assembly. Thus, it seems likely that phage genes are expressed to some degree even when pCTX and CTXφ are not produced. CTX prophages are located at two sites within the genome of classical O1 V. cholerae: at the insertion site occupied in El Tor strains (on chromosome I between TLC and the RTX gene cluster) and within an intergenic region of chromosome II, near the gene encoding d-Ala–d-Ala ligase. Classical prophages are present either as solitary elements or as truncated elements within arrays, and the prophage arrays never contain RS1. Thus, the prophages in classical strains lack the necessary chromosomal arrangements (i.e., tandem prophages or a prophage followed by an RS1) for generation of pCTX and CTXφ, and this fact apparently underlies the failure of classical V. cholerae to produce classical CTXφ.
ACKNOWLEDGMENTS
We thank J. Mekalanos and G. Pearson for pGP2 and pGP3. We are grateful to A. Camilli and to Waldor lab colleagues for helpful suggestions and critical reading of the manuscript. We thank A. Kane and the New England Medical Center GRASP Center for preparation of plates and media.
This work was supported by NIH grant AI-42347 to M.K.W., NIH grant P30DK-34928 to the NEMC GRASP Digestive Center, and NIH grant GM20483-01 to B.M.D. M.K.W. is a Pew Scholar in the Biomedical Sciences and an Assistant Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Ausubel F M R, Brent, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Wiley-Interscience; 1990. [Google Scholar]
- 2.Barua D. History of cholera. In: Barua D, Greenough W B, editors. Cholera. New York, N.Y: Plenum Medical Book Co.; 1992. pp. 1–36. [Google Scholar]
- 3.Basu A, Mukhopadhyay A K, Garg P, Chakraborty S, Ramamurthy T, Yamasaki S, Takeda Y, Nair G B. Diversity in the arrangement of the CTX prophages in classical strains of Vibrio cholerae O1. FEMS Microbiol Lett. 2000;182:35–40. doi: 10.1111/j.1574-6968.2000.tb08869.x. [DOI] [PubMed] [Google Scholar]
- 4.Basu A, Mukhopadhyay A K, Sharma C, Jyot J, Gupta N, Ghosh A, Bhattacharya S K, Takeda Y, Faruque A S, Albert M J, Balakrish Nair G. Heterogeneity in the organization of the CTX genetic element in strains of Vibrio cholerae O139 Bengal isolated from Calcutta, India and Dhaka, Bangladesh and its possible link to the dissimilar incidence of O139 cholera in the two locales. Microb Pathog. 1998;24:175–183. doi: 10.1006/mpat.1997.0186. [DOI] [PubMed] [Google Scholar]
- 5.Baudry B, Fasano A, Ketley J, Kaper J B. Cloning of a gene (zot) encoding a new toxin produced by Vibrio cholerae. Infect Immun. 1992;60:428–434. doi: 10.1128/iai.60.2.428-434.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis B M, Kimsey H H, Chang W, Waldor M K. The Vibrio cholerae O139 Calcutta bacteriophage CTXφ is infectious and encodes a novel repressor. J Bacteriol. 1999;181:6779–6787. doi: 10.1128/jb.181.21.6779-6787.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis B M, Waldor M K. CTXφ contains a hybrid genome derived from tandemly integrated elements. Proc Natl Acad Sci USA. 2000;97:8572–8577. doi: 10.1073/pnas.140109997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanne L F, Finkelstein R A. Characterization and distribution of the hemagglutinins produced by Vibrio cholerae. Infect Immun. 1982;36:209–214. doi: 10.1128/iai.36.1.209-214.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heidelberg J F, Eisen J A, Nelson W C, Clayton R A, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Umayam L, Gill S R, Nelson K E, Read T D, Tettelin H, Richardson D, Ermolaeva M D, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann R D, Nierman W C, White O, Salzberg S L, Smith H O, Colwell R R, Mekalanos J J, Venter J C, Fraser C M. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–484. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaper J B, Morris J G, Jr, Levine M M. Cholera. Clin Microbiol Rev. 1995;8:48–86. doi: 10.1128/cmr.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimsey H H, Waldor M K. CTXφ immunity: application in the development of cholera vaccines. Proc Natl Acad Sci USA. 1998;95:7035–7039. doi: 10.1073/pnas.95.12.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin W, Fullner K J, Clayton R, Sexton J A, Rogers M B, Calia K E, Calderwood S B, Fraser C, Mekalanos J J. Identification of a Vibrio cholerae RTX toxin gene cluster that is tightly linked to the cholera toxin prophage. Proc Natl Acad Sci USA. 1999;96:1071–1076. doi: 10.1073/pnas.96.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mekalanos J J. Duplication and amplification of toxin genes in Vibrio cholerae. Cell. 1983;35:253–263. doi: 10.1016/0092-8674(83)90228-3. [DOI] [PubMed] [Google Scholar]
- 14.Mekalanos J J, Swartz D J, Pearson G D, Harford N, Groyne F, de Wilde M. Cholera toxin genes: nucleotide sequence, deletion analysis and vaccine development. Nature. 1983;306:551–557. doi: 10.1038/306551a0. [DOI] [PubMed] [Google Scholar]
- 15.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pearson G D, Woods A, Chiang S L, Mekalanos J J. CTX genetic element encodes a site-specific recombination system and an intestinal colonization factor. Proc Natl Acad Sci USA. 1993;90:3750–3754. doi: 10.1073/pnas.90.8.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearson G D N. Doctoral thesis. Cambridge, Mass: Harvard University Medical School; 1989. [Google Scholar]
- 18.Rubin E J, Lin W, Mekalanos J J, Waldor M K. Replication and integration of a Vibrio cholerae cryptic plasmid linked to the CTX prophage. Mol Microbiol. 1998;28:1247–1254. doi: 10.1046/j.1365-2958.1998.00889.x. [DOI] [PubMed] [Google Scholar]
- 19.Trucksis M, Galen J E, Michalski J, Fasano A, Kaper J B. Accessory cholera enterotoxin (Ace), the third toxin of a Vibrio cholerae virulence cassette. Proc Natl Acad Sci USA. 1993;90:5267–5271. doi: 10.1073/pnas.90.11.5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trucksis M, Michalski J, Deng Y K, Kaper J B. The Vibrio cholerae genome contains two unique circular chromosomes. Proc Natl Acad Sci USA. 1998;95:14464–14469. doi: 10.1073/pnas.95.24.14464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waldor M K, Mekalanos J J. Emergence of a new cholera pandemic: molecular analysis of virulence determinants in Vibrio cholerae O139 and development of a live vaccine prototype. J Infect Dis. 1994;170:278–283. doi: 10.1093/infdis/170.2.278. [DOI] [PubMed] [Google Scholar]
- 22.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 23.Waldor M K, Mekalanos J J. Vibrio cholerae: molecular pathogenesis, immune response, and vaccine development. In: Paradise L J, editor. Enteric infections and immunity. New York, N.Y: Plenum Press; 1996. pp. 37–56. [Google Scholar]
- 24.Waldor M K, Rubin E J, Pearson G D, Kimsey H, Mekalanos J J. Regulation, replication, and integration functions of the Vibrio cholerae CTXφ are encoded by region RS2. Mol Microbiol. 1997;24:917–926. doi: 10.1046/j.1365-2958.1997.3911758.x. [DOI] [PubMed] [Google Scholar]