Summary
Background
Genetically distinct viral variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have been recorded since January 2020. The introduction of global vaccine programs has contributed to lower COVID-19 hospitalisation and mortality rates, particularly in developed countries. In late 2021, Omicron BA.1 emerged, with substantially altered genetic differences and clinical effects from other variants of concern. Shortly after dominating global spread in early 2022, BA.1 was supplanted by the genetically distinct Omicron lineage BA.2. A sub-lineage of BA.2, designated BA.5, presently has an outgrowth advantage over BA.2 and other BA.2 sub-lineages. Here we study the neutralisation of Omicron BA.1, BA.2 and BA.5 and pre-Omicron variants using a range of vaccine and convalescent sera and therapeutic monoclonal antibodies using a live virus neutralisation assay. Using primary nasopharyngeal swabs, we also tested the relative fitness of BA.5 compared to pre-Omicron and Omicron viral lineages in their ability to use the ACE2-TMPRSS2 pathway.
Methods
Using low passage clinical isolates of Clade A.2.2, Beta, Delta, BA.1, BA.2 and BA.5, we determined humoral neutralisation in vitro in vaccinated and convalescent cohorts, using concentrated human IgG pooled from thousands of plasma donors, and licensed monoclonal antibody therapies. We then determined infectivity to particle ratios in primary nasopharyngeal samples and expanded low passage isolates in a genetically engineered ACE2/TMPRSS2 cell line in the presence and absence of the TMPRSS2 inhibitor Nafamostat.
Findings
Peak responses to 3 doses of BNT162b2 vaccine were associated with a 9-fold reduction in neutralisation for Omicron lineages BA.1, BA.2 and BA.5. Concentrated pooled human IgG from convalescent and vaccinated donors and BNT162b2 vaccination with BA.1 breakthrough infections were associated with greater breadth of neutralisation, although the potency was still reduced 7-fold across all Omicron lineages. Testing of clinical grade antibodies revealed a 14.3-fold reduction using Evusheld and 16.8-fold reduction using Sotrovimab for the BA.5. Whilst the infectivity of BA.1 and BA.2 was attenuated in ACE2/TMPRSS2 entry, BA.5 was observed to be equivalent to that of an early 2020 circulating clade and had greater sensitivity to the TMPRSS2 inhibitor Nafamostat.
Interpretation
Observations support all Omicron variants to significantly escape neutralising antibodies across a range of vaccination and/or convalescent responses. Potency of therapeutic monoclonal antibodies is also reduced and differs across Omicron lineages. The key difference of BA.5 from other Omicron sub-variants is the reversion in tropism back to using the well-known ACE2-TMPRSS2 pathway, utilised efficiently by pre-Omicron lineages. Monitoring if these changes influence transmission and/or disease severity will be key for ongoing tracking and management of Omicron waves globally.
Funding
This work was primarily supported by Australian Medical Foundation research grants MRF2005760 (ST, GM & WDR), MRF2001684 (ADK and ST) and Medical Research Future Fund Antiviral Development Call grant (WDR), Medical Research Future Fund COVID-19 grant (MRFF2001684, ADK & SGT) and the New South Wales Health COVID-19 Research Grants Round 2 (SGT).
Keywords: SARS-CoV-2, Omicron BA.1, BA.2, BA.5, ACE2, TMPRSS2, Neutralising antibodies
Research in context.
Evidence before this study
As the Omicron lineage BA.2 supplanted the initial BA.1 Omicron wave, a sub-lineage of BA.2, designated BA.5, appeared globally with a distinct out-growth advantage. Initial observations of antibody neutralisation using viral pseudotyping observed a continuum of results, with several studies showing either similar or increased fold evasion against a range of convalescent and/or vaccine responses and therapeutic monoclonal antibodies.
Added value of this study
Using both primary nasopharyngeal swabs and low passage clinical isolates we determined neutralisation potency from: i. convalescent and BNT162b2 vaccination; ii. 3 dose BNT162b2 vaccination; iii. 2 and 3 dose BNT162b2 vaccination and recovery from BA.1 infection; iv. pooled panels of antibodies from 135,677 US plasma donors acquired following the peak of the Delta wave from August to November 2021 and v. clinical grade therapeutics Evusheld and Sotrovimab. In addition, we measured the viral fitness in both primary swabs and expanded clinical isolates using an ACE2-TMPRSS2 cell line. To the best of our knowledge, this represents the first study that utilises clinical isolates to determine efficiency of entry through the ACE2-TMPRSS2 pathway.
Implications of all the available evidence
Using clinically derived low passage Omicron isolates, we observe similar results to pseudotyping observations where 3 dose BNT162b2 vaccination provides similar reduced potency across BA.1, BA.2 and BA.5 lineages. BA.1 breakthrough infections in vaccinated donors increases neutralisation titres but does not significantly increase neutralisation breadth across Omicron BA.1, BA.2 and BA.5. Neutralisation at the population level shows that antibodies derived from 135,677 donations at the time of the US Delta wave also provides sufficient breadth to target all Omicron lineages at equivalent potency. Increased efficiency of BA.5 using the ACE2-TMPRSS2 will need to be monitored and further investigations are required to understand if it is associated with the observed outgrowth advantage and/or increased disease severity of Omicron BA.5.
Alt-text: Unlabelled box
Introduction
At the beginning of November 2021, the VOC Omicron BA.1 surged globally with close to 4 million infections per day reported by mid-January 2022. This variant was then supplanted by the genetically divergent BA.2 Omicron lineage, which represented over 80% of cases reported worldwide by mid-April 2022. In June 2022, three lineages derived from BA.2 had started to dominate, which included BA.2.12.1, BA.4 and BA.5. BA.4 and BA.5 share amino acid substitutions (compared to BA.2) L452R, F486V, and R493Q in the Spike receptor binding domain (RBD) whereas BA.2.12.1 is the only variant with the L452Q change. Across several areas globally, BA.5 has a growth advantage over BA.2.12.1 and BA.4. The growth advantage of any variant must consider many variables, including the prevalence of infection and/or vaccine coverage and the time from that latter antigenic exposure. In addition to the population level of immunity, the path of viral entry and changes thereof may significantly influence viral tropism and subsequent disease severity even within previously vaccinated populations. For instance, Delta had significant tropism for the ACE2-TMPRSS2 pathway, which is associated with infection of the lung and increased disease severity in animal models.1, 2, 3, 4, 5 In contrast, Omicron BA.1 diverged from this pathway with a tropism trajectory towards the upper respiratory tract.3 The change in tropism is presently hypothesised to be the switch from TMPRSS2 to another serine or cysteine protease, present at the plasma membrane or enriched within the endolysosomal compartment.3,6, 7, 8 Whilst BA.2 has similar tropism to BA.1, a recent pre-print on BA.5 and related lineages bearing L452 polymorphisms highlight a shift in tropism back to pre-Omicron lineages, with increased disease severity in animal models (pre-print).9
Recently we developed a rapid and sensitive platform for the isolation and characterisation of SARS-CoV-2 variants with respect to their relative transmission threat in previously infected and vaccinated populations.6 This platform rapidly feeds back three key observations with respect to early characterisation of viral variants in primary nasopharyngeal samples. Firstly, it enables neutralisation studies on primary clinical viral isolates. Secondly, it determines which immunotherapeutics retain potency. Finally, it can resolve subtle changes in tropism towards or away from the ACE2-TMPRSS2 pathway by the increase or decrease of viral infectivity to particle ratios. In the latter setting, this system showed increased usage of TMPRSS2 by Delta in primary nasopharyngeal swabs and demonstrated the decreased use of TMPRSS2 by Omicron BA.1. As this can be done with diagnostic primary samples, it can reveal tropism changes when a variant starts expanding within a community.
Through using individual serum samples from 74 patients recruited to ADAPT, a community-based cohort of approximately 200 patients followed from the time of diagnosis throughout all waves of infection in Australia, we tested a continuum of responses including 3 dose vaccination, infection and vaccination and three dose vaccination followed by Omicron infection. To assess breadth across variants, we tested neutralisation potency to A.2.2, Beta and Delta alongside the Omicron lineages BA.1, BA.2 and BA.5. We then tested 13 polyclonal human IgG batches that constitute pools of thousands of primarily US plasma donors collected in late 2021 prior to the global Omicron BA.1 wave (Figure 1e). This latter analysis establishes the extent of immune evasion at the population level at that time, as the IgG is comprised of all plasma donors irrespective if they are convalescent and/or vaccinated. Alongside patient sera, we also tested clinical grade Sotrovimab and Evusheld for changes in potency across the aforementioned variants. Finally, with overlapping waves of BA.2 and BA.5 infection within Australia, we then determined the infectivity to particle ratios of virus within primary nasopharyngeal swabs and furthermore established the mode of entry of BA.5 versus other Omicron and pre-Omicron lineages.
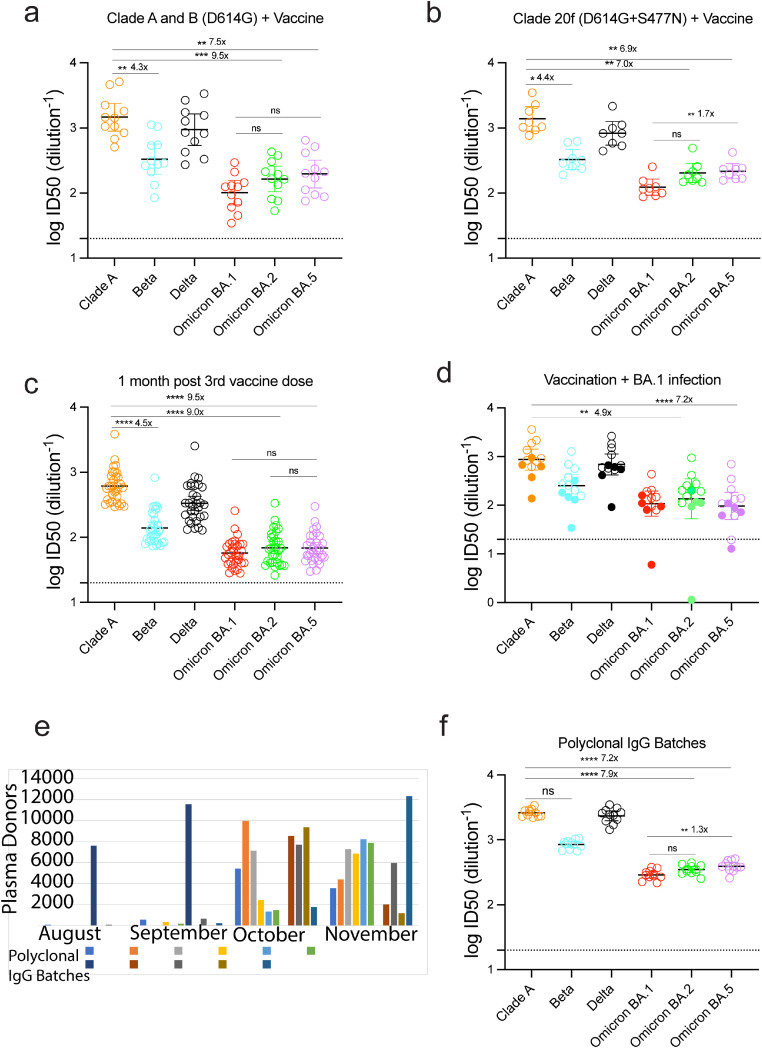
Figure 1.
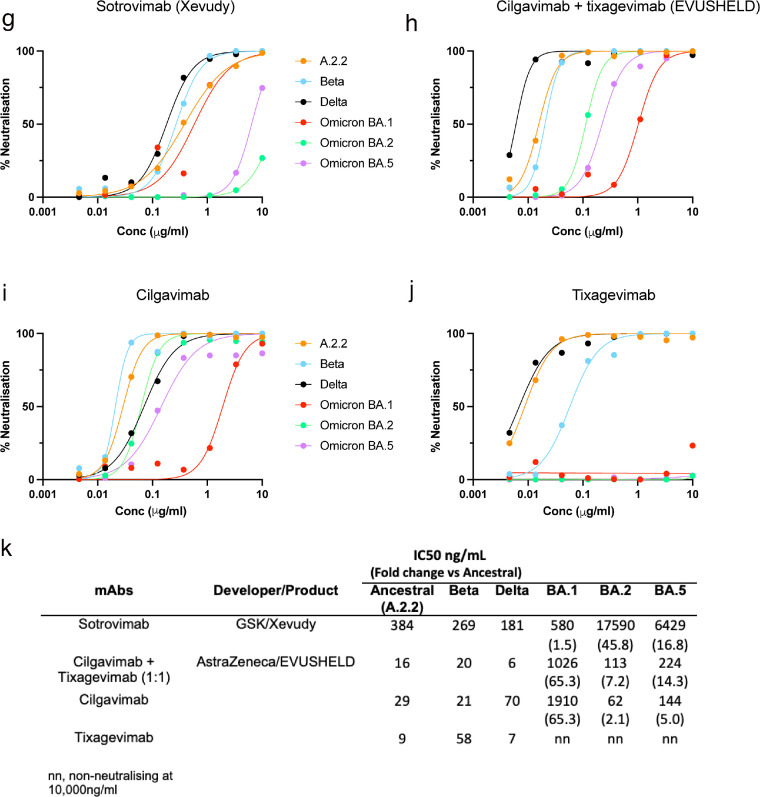
Humoral neutralisation of clinical SARS-CoV-2 variants in convalescent and vaccinated donors, and pooled concentrated human IgG plasma samples. Neutralisation assays were performed in a high-throughput format in HAT-24 cells using live virus isolates from the variants of concern Delta (B.1.617.2), Beta (B.1.351), Omicron BA.1, Omicron BA.2 and Omicron BA.5 and the ancestral Wuhan-like virus with the original D614 background (A.2.2) as a control. ID50 neutralisation titres presented for 6 live variants for vaccinated donors from (a) Clade A and B (D614G) (First wave; n=10), (b) Clade 20F (D614G + S477N) (Second wave; n=7), (c) Healthy donors one month after third dose of vaccination (n=31), (d) Vaccinated (Open circles; n=9) or unvaccinated donors (Solid circles; n=5) infected with Omicron BA.1, (e) Number of United States based plasma donors per month sourced for polyclonal immunoglobulins (Poly-Ig; on average, greater than 10,000 donors were pooled for each batch tested) and (f) Concentrated polyclonal IgG from either convalescent and vaccinated donors. Data in (a-d and f) indicates the mean ID50 of technical replicates for individual samples (Circles) with the geometric mean and 95% confidence interval shown for each variant. Dotted lines show the limit of detection (LOD). Fold change reductions in ID50 neutralisation titres compare variants of concern to the ancestral variant and Omicron BA.1 where indicated. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 for Kruskal Wallis test with Dunn's multiple comparison test. Data in (g-k) show the neutralisation activity of monoclonal antibodies (g) Sotrovimab, (h) Cilgavimab and Tixagevimab cocktail, (i) Cilgavimab alone and (j) Tixagevimab alone, against ancestral (k) A.2.2, Beta, Delta, Omicron BA.1, Omicron BA.2 and Omicron BA.5. IC50 values (ng/µL) and fold change are relative to ancestral A.2.2 for all monoclonal antibodies against Omicron BA.1, BA.2 and BA.5. Antibodies used herein were clinical grade batches.
Methods
Human sera
The ADAPT cohort is composed of RT-PCR–confirmed convalescent individuals (including some subsequently vaccinated) recruited in Australia since 2020.10 Serum samples from healthy volunteers vaccinated with ChAdOx1 and BNT162b2 were collected 4 weeks post-third dose vaccination.
Ethics
All human serum samples were obtained with written informed consent from the participants (2020/ETH00964; 2020/ETH02068; 2019/ETH03336; 2021/ETH00180). All primary isolates used herein were obtained from de-identified remnant diagnostic swabs that had completed all diagnostic testing under approval by the New South Wales Chief Health Officer following independent scientific review and as outlined in the ADAPT ethics protocol 2020/ETH00964.
Other immunoglobulin products
Clinical grade Sotrovimab (62.5 mg/mL; NDC 0173-0901-86) was kindly provided by GSK Healthcare while clinical grade Cilgavimab and Tixagevimab (100 mg/mL each; AstraZeneca) were kindly provided by Dr Sarah Sasson (Kirby Institute, UNSW). Cilgavimab and Tixagevimab were mixed in equal volumes to generate the monoclonal antibody cocktail Evusheld. All monoclonal antibodies were tested at a starting concentration of 10 µg/mL and diluted two-fold in an eight step dilution series.
Polyclonal immunoglobulin preparations
The immunoglobulins used herein were purified using the licensed and fully validated immunoglobulin manufacturing process used for Privigen,11 notionally similar to others.12 Thirteen Poly-Ig batches were manufactured using the Privigen process11 and included US plasma collected by plasmapheresis from a mixture of vaccinated (SARS-CoV-2 mRNA vaccines), convalescent and non-convalescent donors (source plasma, n between 9495-23,667 per batch) collected in the September 2021. The WHO international reference standard for SARS-CoV-2 neutralisation (NIBSC 20/136) was obtained from NIBSC.13
Cell culture
HEK293T cells stably expressing human ACE2 and TMPRSS2 were generated by lentiviral transductions as previously described.6,10 A highly permissive clone (HAT-24) was identified through clonal selection and used for this study. The HAT-24 line has been extensively cross-validated with the VeroE6 cell line.6 HAT-24 and VeroE6-TMPRSS2 cells (CellBank Australia, JCRB1819) were cultured in Dulbecco's Modified Eagle Medium (Gibco, 11995073) containing 10% foetal bovine serum (Gibco, 10099141; DMEM-10%FBS) and VeroE6 cells (ATCC® CRL-1586™) in Minimal Essential Medium (Sigma Aldrich, M4655) containing 10% FBS and 1% penicillin-streptomycin (Gibco, 15140122; MEM-10%FBS). All cells were incubated at 37°C, 5% CO2 and >90% relative humidity. VeroE6-TMPRSS2 cell line authentication was performed as previously described.14 The STR profiling for authentication of HAT-24 was done as previously described.6 Both cell lines tested negative for mycoplasma.
Viral isolation, propagation, and titration
All laboratory work involving infectious SARS-CoV-2 occurred under biosafety level 3 (BSL-3) conditions. Diagnostic respiratory specimens testing positive for SARS-CoV-2 (RT-qPCR, Seegene Allplex SARS-CoV-2) were sterile-filtered through 0.22 µm column-filters at 10,000 x g and serially diluted (1:3) on HAT-24 cells (104 cells/well in 96-well plates). Upon confirmation of cytopathic effect by light microscopy, 300 μL pooled culture supernatant from infected wells (passage 1) were added to VeroE6-TMPRSS2 cells in a 6-well plate (0.5 × 106 cells/well in 2 mL MEM2%) and incubated for 48 h. The supernatant was cleared by centrifugation (2000 x g for 5 minutes), frozen at -80°C (passage 2), then thawed and titrated to determine median 50% Tissue Culture Infectious Dose (TCID50/mL) on VeroE6-TMPRSS2 cells according to the Spearman-Karber method.15 Viral stocks used in this study correspond to passage 3 virus, which were generated by infecting VeroE6-TMPRSS2 cells at MOI=0.025 and incubating for 24 h before collecting, clearing, and freezing the supernatant as above. Sequence identity and integrity were confirmed for both passage 1 and passage 3 virus via whole-genome viral sequencing using an amplicon-based Illumina sequencing approach, as previously described.16 The latter was also used in parallel for sequencing of primary nasopharyngeal swabs. For a list of the viral variants used in this study see Supplementary Table S4. Passage 3 stocks were titrated by serial dilution (1:5) in DMEM-5%FBS, mixing with HAT-24 cells live-stained with 5% v/v nuclear dye (Invitrogen, R37605) at 1.6 × 104 cells/well in 384-well plates, incubating for 20 h, and determining whole-well nuclei counts with an IN Cell Analyzer high-content microscope and IN Carta analysis software (Cytiva, USA). Data was normalised to generate sigmoidal dose-response curves (average counts for mock-infected controls = 100%, and average counts for highest viral concentration = 0%) and median Virus Effective (VE50) values were obtained with GraphPad Prism software.
Rapid high-content SARS-CoV-2 microneutralisation assay with HAT-24 cells (R-20)
Human sera or monoclonal antibodies were serially diluted (1:2 series starting at 1:10 for sera and 20 µg/mL for antibodies) in DMEM-5%FBS and mixed in duplicate with an equal volume of SARS-CoV-2 virus solution standardised at 2xVE50. After 1 h of virus–serum coincubation at 37°C, 40 μL were added to an equal volume of nuclear-stained HAT-24 cells pre-plated in 384-well plates as above. Plates were incubated for 20 h before enumerating nuclear counts with a high-content fluorescence microscopy system as indicated above. The % neutralisation was calculated with the formula: %N = (D-(1-Q)) × 100/D as previously described.10 Briefly, “Q” is a well's nuclei count divided by the average count for uninfected controls (defined as having 100% neutralisation) and D = 1-Q for the average count of positive infection controls (defined as having 0% neutralisation). Sigmoidal dose-response curves and IC50 values (reciprocal dilution at which 50% neutralisation is achieved) were obtained with GraphPad Prism software. Neutralisation assays with VeroE6 cells were performed exactly as described above excepting that; input virus solution was standardised at 1.25 × 104 TCID50/mL, cells were seeded at 5 × 103 cells/well in MEM-2%FBS (final MOI = 0.05), plates were incubated for 72 h, and cells were stained with nuclear dye only 2 h before imaging.
Statistics
Statistical analyses were performed using GraphPad Prism 9 for all experiments and details have been presented in the figure legends. No statistical methods were used to predetermine sample size. The experiments were not randomised, and the investigators were not blinded to allocation during experiments and assessment of outcomes.
Role of funders
Funding bodies did not contribute to study design, data collection, data analysis or writing of the manuscript. Study design, data collection, data analysis and data interpretation were performed by investigators at Kirby Institute; writing and review of the manuscript was completed by the authors.
Results
Humoral evasion for Omicron lineages BA.1, BA.2 and BA.5 relative to early clade A, Beta and Delta variants
The ability of Omicron lineages and other variants (Beta and Delta) to evade neutralising antibody responses was assessed using our rapid 20-hour live virus neutralisation platform (R-20).6 Initially, we examined neutralisation responses in sera from convalescent donors from early clade infections who had subsequently been vaccinated with either the BNT162b or ChAdOx1 vaccine (Figure 1a and b and Supplementary Table 1; presented together as a single “vaccinated” group). While this vaccinated group was observed to have potent neutralisation titres to the ancestral strain, we observed a 7 to 15-fold reduction in neutralisation against all Omicron sub-lineages (15.6-fold for BA.1, p<0.0001; 9.5-fold for BA.2, p<0.001; 7.5-fold for BA.5, p<0.01; Kruskal Wallis with Dunn's correction for multiple comparisons) compared to a 1.5 and 5-fold decrease observed for Delta and Beta, respectively (p>0.05). Similar reductions were also observed for triple vaccinated SARS-CoV-2 naïve donors (Figure 1c; Supplementary Table 2). We have previously tested these sera against Omicron BA.1 and observed greater fold reductions.6 This minor discrepancy can be accounted for by changes in viral expansion, which currently uses the VeroE6-TMPRSS2 cell line and a 24-hour incubation period (compared to previously used VeroE6 cells across a 48-hour incubation). The use of VeroE6-TMPRSS2 cells enables virus to be harvested within a 24-hour window, thus limiting the accumulation of non-viable viral particles (the half-life of SARS-CoV-2 we have calculated at room temperature to be 1.4 days).
Similar results were obtained when we tested neutralisation responses against thirteen polyclonal human IgG batches comprised of more than ten thousand pooled plasma donors per batch collected during late 2021, following the peak of the Delta wave, although prior to the BA.1 Omicron wave (Figure 1f; Supplementary Table 1). There was a 7 to 9-fold reduction in neutralisation against Omicron lineages (9.4-fold for BA.1, p<0.0001; 7.9-fold for BA.2, p<0.0001 and 7.2-fold for BA.5, p<0.001; Kruskal Wallis with Dunn's correction for multiple comparisons) compared with 3.2-fold (p>0.05) and 1.1-fold decrease (p>0.05) for Beta and Delta, respectively. Across most patient samples, we observed no significant differences across the Omicron lineages BA.1, BA.2 and BA.5.
Whilst the above samples give a population snapshot of antibody potency and breadth from donors in 2021, we then turned to serum samples collected from the ADAPT cohort, composite of triple vaccinated donors then infected during the BA.1 wave. Given those infected globally with BA.1 were largely the vaccinated population, our aim was to determine the potential increase in breadth towards Omicron lineages following BA.1 infection. Across all donors, we observed greater neutralisation potency to pre-Omicron viral variants with neutralisation titres to Omicron lineages similar to that observed with SARS-CoV-2 naïve triple vaccinated donors (Figure 1d; Supplementary Table 1). Two donors did reach significant titres to all Omicron lineages, but they were lower than their observed responses to pre-Omicron variants.
Evusheld and Sotrovimab activity against Omicron clinical BA.2 versus BA.5 isolates
Current, clinically utilised monoclonal antibodies (mAbs) in Evusheld and Sotrovimab were assessed for neutralisation primarily against the Omicron variants BA.2 and BA.5, as they currently represent the dominant variants within the community. These therapies may need to be used in individuals that have not mounted a vaccine response from therapy-induced or pre-existing immunodeficiencies. The potency of Evusheld against BA.2 and BA.5 was reduced by 7.2 and 14.3-fold, respectively, when compared to the ancestral clade A.2.2 variant. In comparison, the potency of Sotrovimab against BA.2 and BA.5 was reduced 46-fold and 16.8-fold, respectively (Figure 1g-k). This shift from BA.2 to BA.5 increases the potency of Sotrovimab by 3-fold, whilst decreasing the potency of Evusheld by 2-fold.
Increased infectivity of the BA.5 Omicron variant through more efficient use of TMPRSS2
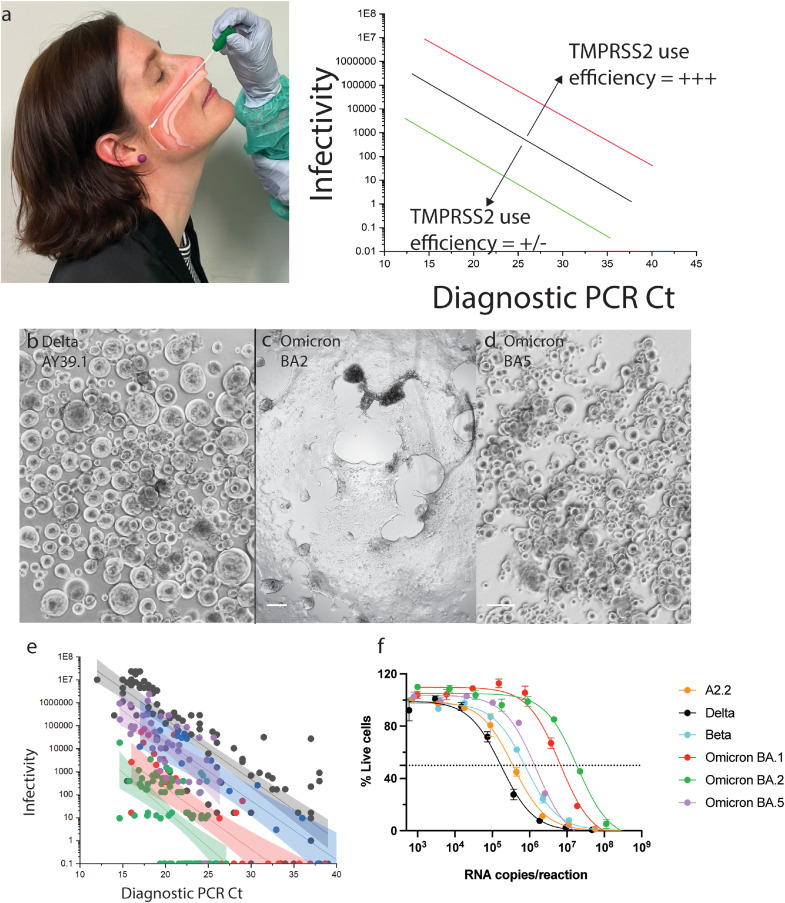
In the pre-Omicron era, we developed a hyper-permissive ACE2-TMPRSS2 HEK293T cell line (HAT-24) with culture sensitivity approaching that of diagnostic PCR.6,10 In brief, this cell line was the product of iterative genetic addition of ACE2 and then TMPRSS2 in the HEK 293T cell line. A cell clone (clone 24) was identified that sustained significant viral cytopathic effects within 6 to 8 hours post-infection. The phenotype of this cell was then used in three primary methods to rapidly characterise emerging variants. Firstly, during the Delta wave, virus could be cultured in 75% of clinical swabs with diagnostic PCR Ct values above 306 (i.e. the cell line enabled variant isolation at low viral loads). Secondly, we curated clinical samples that were PCR positive and cryopreserved them at –80°C within 24 hours. Using this material, we inoculated this cell line at limiting dilutions and after 96 hours in culture determined 50% Tissue Culture Infectious Dose (TCID50). Using the latter value as a measure of viral infectivity (TCID50/mL), we then generated linear regressions of infectivity (TCID50/mL) versus particle number (viral load-diagnostic PCR Ct value) across clinical specimens with increasing levels of virus. The final method established with this clone involved using this in combination with high-content microscopy and analysis platforms. In this latter setting, we observed the rapid formation of viral syncytia which led to viral dose-dependent reductions in cell nuclei numbers overnight. This was the result of rapid viral replication resulting in cell-cell fusion and the concentration of cellular nuclei into viral syncytia. Pre-labelling of cells with live nuclei stain could then be used as a surrogate to enumerate viral replication through syncytia formation.
Using the abovementioned approaches, we could readily resolve the increased infectivity to particle ratio of an early circulating variant against that of a variant with increased or decreased efficiency in using the ACE2-TMPRSS2 pathway (see Delta Figure 2a). This can be observed through an upward or downward shift in the linear regression established by determining the infectivity (TCID50/mL) of primary nasopharyngeal samples with decreasing viral loads (increasing diagnostic PCR Ct value). Omicron BA.1 and BA.2 use the ACE2-TMPRSS2 pathway inefficiently and this can rapidly be resolved using primary nasopharyngeal samples through a downward shift compared to Delta (Figure 2a). Following the BA.2 wave in Australia, Omicron BA.5 then supplanted and became the dominant variant over winter in 2022. Using the above cell line, we collected samples when both BA.2 and BA.5 were highly prevalent within the community. This concurrent collection strategy controlled for any immunity in the community that could influence the results. Testing of these BA.2 and BA.5 virus swab samples revealed BA.2 could be distinguished from BA.5 based on the cytopathic effects observed alone (Figure 2c-d). Similar to BA.1, BA.2 was poorly infectious in the HAT-24 ACE2-TMPRSS2 cell line, resulting in the development of limited cytopathic effects after three days of culture (Figure 2c), even whilst using samples with high viral loads (diagnostic PCR Ct values lower than 20). In contrast, BA.5 mediated extensive cytopathic effects (Figure 2d) and was also significantly more infectious per diagnostic Ct value compared to the co-circulating BA.2 parental variant. These differences in Omicron sub-variants were revealed in their linear regression, in which BA.5 is comparable to an early circulating variant from 2020 yet its infectivity relative to viral load is lower when compared to Delta (Figure 2e). However, for the latter comparisons, we need to consider these early isolates were tested when the community in Australia was primarily unvaccinated. To further investigate the increase in infectivity per Ct value, we expanded pre-Omicron and Omicron variants at the same time using the same VeroE6-TMPRSS2 cell line. This cell line overexpresses TMPRSS2, allowing variants that poorly utilise TMPRSS2 to be expanded under similar conditions over 24 hours.14 With respect to RNA copies per mL, this cell line generated viral stocks with similar particle numbers (Supplementary Table S3). Delta was the only exception as it exhibited greater levels of cytopathic effect (Figure 2d) whilst producing lower particle numbers (Supplementary Table S3). When testing these samples within the HAT-24 cell line and accounting for particle input, we observed similar infectivity to particle ratios as those established using primary clinical swabs (Figure 2f). To understand the mechanism whereby the HAT-24 cell line can readily rank primary isolates based on increasingly efficient TMPRSS2 use, we determined the expression of TMPRSS2 in this cell line versus that of the SARS-CoV-2 permissive cell line Calu3. In this setting, we observed the HAT-24 cell line abundantly expresses ACE2 but expresses low levels of TMPRSS2. In contrast, Calu3 cells and the VeroE6-TMPRSS2 cell line that we used to expand isolates were ACE2 low but TMPRSS2 high (Supplementary Figure S1). Therefore, we concluded that the rate limiting entry step for the HAT-24 line is TMPRSS2 and when variants poorly utilise this serine protease, it can be readily resolved in culture even with primary swab samples.
Figure 2.
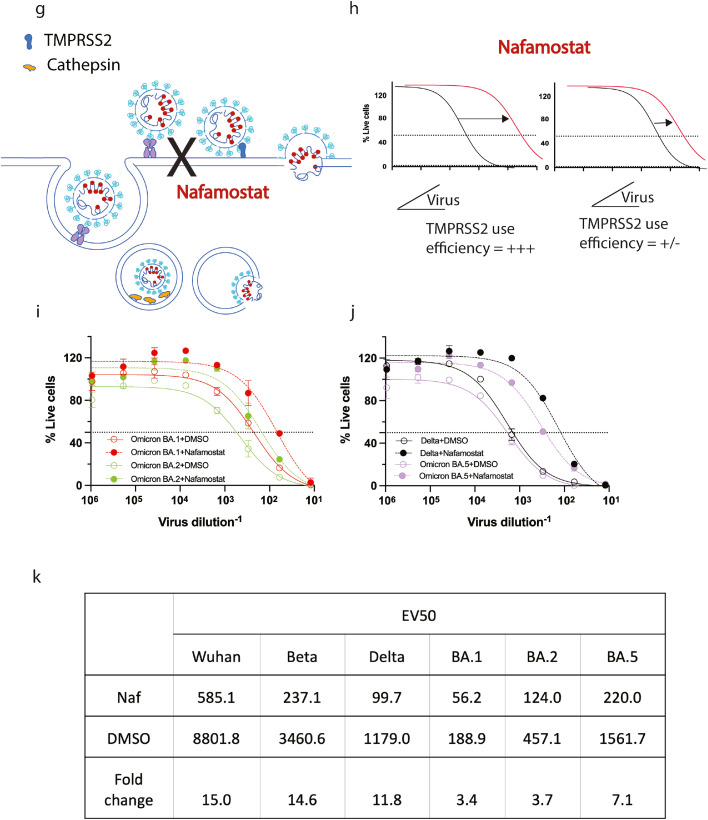
Changing tropism of the BA.5 variant relative to BA.1, BA.2 and pre-Omicron variants. (a) Shows the shift in linear regression between virus infectivity and diagnostic PCR Ct values gives a measure of TMPRSS2 use by individual variants. Photo in (a) is not a participant in this study and consents to the inclusion of this photo. (b) Schematic showing the effect of TMPRSS2 inhibitor Nafamostat on virus entry at the cell membrane. (c) Efficiency of TMPRSS2 usage by the virus can be determined by observing a shift in VE50 when virus is titrated in the presence of saturating levels of Nafamostat. (d-f) Primary nasopharyngeal swabs were used to inoculate the HAT-24 cell line. All swabs are representative of high viral loads from diagnostic PCR Ct values of 17 to 19. Cultures of (d) AY39.1 (one of the last detected Delta lineages), (e) BA.2 and (f) BA.5 imaged 72 hours post-infection. Of note, only BA.2 and BA.5 were from samples collected at the same time period. Scale bars in d-f, 100 µm. Images in (d-f) are representative of >20 primary samples with high viral loads. Data in (f) is representative of three independent low passage expansions of the 6 isolates used. (g) Infectivity (TCID50/mL) is presented against original diagnostic PCR Ct values for BA.2 and BA.5. Shading represents the 95% confidence intervals for each linear regression. As a comparison, earlier tested samples from Delta and an early 2020 circulating clade are also presented. (h) Three early clade variants (A.2.2, Beta and Delta) and three Omicron sub-lineages were grown within a 24-hour time frame under identical culture conditions (i.e. expanded at the same time, with the same MOI and harvest time). Virion particle counts were then determined using quantitative RT-PCR. Titres were then determined overnight using the R-20 assay to establish infectivity per variant per viral particle. In brief, the HAT-24 cell line rapidly develops cytopathic effects overnight, in a dose-dependent manner that can be enumerated by nuclei counts using high-content microscopy. Infectivity can then be determined by calculating 50% death of cells within the culture (VE50). Each point and error bar represent the mean and standard deviation respectively of four technical replicates. Virus titrations of (i) BA.1, BA.2 and (j) Delta, BA.5 were carried out in the presence of 20 µM Nafamostat (Dashed line) or DMSO (Solid line) and (k) change in VE50 values was used as a measure of TMPRSS2 sensitivity. Data is representative of two independent experiments (N = 2).
To confirm that the increase in infectivity in the HAT-24 cell line was based on be TMPRSS2 usage by BA.5, we utilised the well characterised TMPRSS2 inhibitor Nafamostat. Using the HAT-24 cell line, we could not resolve dose-dependent Nafamostat inhibition for Omicron lineages and this was consistent with our previous observations of Omicron BA.1.6 As all SARS-CoV-2 isolates can also enter through endocytosis, which is independent of TMPRSS2, the lack of resolution of variants that poorly use TMPRSS2 is expected and consistent with our prior observations. In an attempt to resolve TMPRSS2 use by Omicron lineages we established a different approach. This was achieved by performing titrations of virus stocks in the presence of saturating amounts of Nafamostat (20 µM) (Figure 2g-h). Delta and Omicron (BA.1 and BA.2) were used as controls in this setting as they represent variants with efficient and inefficient TMPRSS2 use, respectively.3,17 In this setting, the greater the drop of the viral titre in the presence of the TMPRSS2 inhibitor Nafamostat, the greater the infectivity per particle attributed to TMPRSS2 use. Using this approach, we observed an 11-fold drop in infectivity for Delta versus a 3-fold drop seen for both BA.1 and BA.2 (Figure 2k) with Nafamostat. This is consistent with Delta's efficient use of TMPRSS2 for viral entry compared to the poor TMPRSS2 use by Omicron lineages BA.1 and BA.2. For BA.5, we observed a 7.0-fold drop in infectivity in presence of Nafamostat. This confirms BA.5 has an increased infectivity to particle ratio over other Omicron lineages in the HAT-24 line which is primarily related to more efficient TMPRSS2 use. Importantly, extensive observations by independent teams for pre-Omicron and Omicron lineages BA.1 and BA.2 readily supports the changing tropism of the virus, ranging from primary cultures to a diverse continuum of animal models.3,7,18,19 Our current observations on the tropism change in primary swabs and low passage BA.5 isolates is also consistent with a recent pre-print showing increased replication in lung tissue and greater disease severity in animal models using BA.5 (pre-print).9 Whilst other recent animal studies have not observed BA.5 to replicate in the lung efficiently, a pre-print by Uraki and colleagues did observe another BA.2 sub-lineage BA.2.75 to have an increased tropism for lung tissue (pre-print).20 Thus both in vivo animal studies have highlighted two BA.2 sub-lineages have regained tropism towards that of pre-Omicron clades. Whilst a change in tropism can be mediated by TMPRSS2 use, it is important to note that tropism and viral fitness can be influenced by changes in the genome outside of the Spike glycoprotein. More efficient evasion of innate cellular factors can also facilitate viral replication in viral genes such as Orf6 and nucleocapsid.21,22 Although, it must be noted the cell line used in the study lacks many innate viral restriction pathways.23
Discussion
Omicron BA.5, like other Omicron lineages, represents a continuing challenge for present vaccine strategies. Omicron lineages all share the ability to significantly evade both convalescent and vaccine antibody responses. The structural changes in Spike reveal the hallmark of Omicron lineage evasion is through interprotomer RBD-RBD packing that culminates in stabilisation of the RBD closed state.24 Using clinical isolates, we confirm findings from early seminal contributions.25, 26, 27, 28, 29, 30 Observations using pseudovirus have seen greater antibody evasion of BA.5 compared to its parent BA.2 following different vaccine schedules. Importantly herein, we observe the peak responses following three doses of BNT162b2 to generate antibody neutralising breadth to equally cover BA.2 and BA.5 and this is consistent with that recently observed by Bowen and colleagues.30 Peak antibody responses from breakthrough Omicron BA.1 infections following two or three doses of the vaccine BNT162b2 also enables sufficient breadth to equally neutralise BA.2 and BA.5, which is consistent with observations observed by Hachmann and colleagues.31
All Omicron variants contain significant substitutions within their RBD site relative to the ancestral variant. Therefore, fluctuations in potency in antibodies against the RBD (e.g. Evusheld) are to be expected. Furthermore, the Evusheld binding sites cover the unique RBD polymorphisms (L452R, F486V, and R493Q) that distinguish the BA.5 spike glycoprotein from BA.2. In contrast, the class 3 antibody Sotrovimab targets a highly conserved region of the sarbecovirus RBD32 and based on its neutralising epitope should retain activity against both BA.2 and BA.5. Many recent studies on Omicron lineages BA.1, BA.2 and BA.5 have studied the potency of clinical monoclonal antibodies in detail26, 27, 28,33,34 and whilst each study has demonstrated potency reductions, they vary with respect to fold reductions per variant tested. In the context of the latter work, the fold reductions for Sotrovimab and Evusheld herein are within twice of that of the median of the above cited studies. Herein we restricted our tests to that on clinical grade monoclonal therapies which included Evusheld (Cilgavimab and Tixagevimab) and Sotrovimab against a low passage panel of clinical isolates. With no detectable activity towards BA.2 even at the highest dose tested and now detectable activity towards BA.5, the above RBD changes outside of the Sotrovimab epitope support a conformation change in the Spike glycoprotein that now renders BA.5 susceptible to neutralisation. The recurrent emergence of new variants and the persistence of a high rate of infection around the world highlight the extreme relevance of treatments, especially for immunocompromised subjects. Whilst the retention of neutralising activity by Sotrovimab and Evusheld against Omicron BA.5 is promising, it is of lower potency compared to the early viral variants it was originally designed and tested against. The development of new and improved monoclonal antibody modalities and alternative therapies is still urgently warranted. As with treatment of other RNA viruses (e.g. HIV-1 and HCV), combination therapies will likely limit the appearance of therapeutic resistant variants that can arise within the clinic.
Whilst many clinical studies have observed lower disease severity following infection in vaccinated populations, the contribution of the Omicron variant versus the vaccine response was not readily clear.19 Whilst BA.1 and BA.2 are different in their transmissibility, their tropism towards cells of the upper respiratory tract is very similar. Mechanistically this change in tropism that moved efficient viral replication from the lung to the bronchus is presently hypothesised to be a consequence of alternate protease usage away from the serine protease TMPRSS2.3,6,7,18 Curiously, the efficient use of TMPRSS2 is associated with prior furin cleavage of the S1 and S2 domains of Spike.3,17 A recent pre-print demonstrated that the furin cleavage site is retained in Omicron lineages and acts as a functional substrate for furin when expressed as a separate peptide from the Spike (pre-print).35 Therefore, conformation changes within the Spike of BA.1 and BA.2 would be consistent with the furin cleavage site being held in a conformation with limited furin accessibility. In contrast, the small changes in the RBD of BA.5 appear to encourage furin cleavage in recent studies using pseudotyped viral platforms.36 This is consistent with observations for more efficient TMPRSS2 use, both herein and in animal models observing increased infection of the lung and increased disease severity.37 Curiously, a pre-print by Uraki and colleagues demonstrates that the other BA.2 sub-lineage BA.2.75 also appears to be taking a similar phenotypic trajectory to that of BA.5 (pre-print).20 The change in BA.5 provides the virus greater antibody evasion potential, changed its tropism and increased transmission potential in the community. Whilst observations of disease severity initially in South Africa were comparable with other Omicron lineages,38 the increasing prevalence of BA.5 globally will now enable observations of disease severity across a range of immune backgrounds. In a preliminary pre-print report from Denmark, the vaccine efficacy for BA.5 versus BA.2 is similar and consistent with the breadth being sustained for BA.5 after three doses of mRNA vaccines like BNT162b2 (pre-print).39 Unfortunately, as we have observed herein, prior Omicron infections in those who are triple vaccinated have primarily increased potency responses to early pre-Omicron variants and have done little to increase breadth to Omicron lineages. Moving forward, vaccine strategies that can rapidly come online to increase breadth to current variants or induce responses that can future proof communities to emerging variants would be more pragmatic. In addition, whilst BA.5 appears to have evolved tropism towards pre-Omicron variant entry pathways, we will now need to look closely towards future variant trajectories and importantly if their tropism aligns with greater disease severity. BA.5 and potentially other emerging BA.2 sub-lineages may indeed have an outgrowth advantage through better TMPRSS2 use. As observed herein, BA.5 has not reached the tropism of TMPRSS2 use exhibited by that of Delta. Although, if increased TMPRSS2 use is key to ranking variants for potential disease severity in the community, assays both herein and elsewhere will be able to resolve this changing TMPRSS2 tropism as soon as variants appear within the community through analysis of viral infectivity in primary nasopharyngeal swabs across TMPRSS2 cell lines.
Contributors
Assay was developed and performed by A. Aggarwal., A.O.S. and S.G.T. Viruses were propagated by A. Aggarwal., A. Akerman., V. Milogiannakis., G. Walker, A.O.S and S.G.T. Experiments were performed by A. Aggarwal., A. Akerman., V. Milogiannakis., M.R.S., G. Walker., A. Kindinger., and S.G.T. Additional research support including sequencing, mAbs and serum samples were provided by T. Angelovich., S.A.C., N. Roth., S. Manni., T. Hauser., T. Barnes., M. Yeang., T. Jean., C. S. P. Foster., D. Christ., A. Condylios, M. Wong., A.C.H., M.L.M., D. Darley., M. Churchill., D. Stark., G. Matthews., and W. D. Rawlinson. Study analysis was performed by A. Aggarwal., A. Akerman., V. Milogiannakis., G. Walker., and S.G.T. Data was verified by A. Aggarwal., A. Akerman., V. Milogiannakis., G. Walker and S.G.T. The manuscript was drafted by A. Aggarwal., A. Akerman., V. Milogiannakis., and S.G.T., All authors provided editorial support for the published version. The study was supervised by G. Matthews., W. D. Rawlinson., A.D.K. and S.G.T. All authors read and approved the final version of the manuscript.
Data sharing statement
Source data for generating the main Figure 1 are available in the online version of the paper. Any other data are available on request.
Declaration of interests
A.O.S contributed to this manuscript in his capacity as adjunct associate lecturer (from January 2022) at the University of New South Wales. Since January 2022, A.O.S is an employee of GlaxoSmithKline Australia (medical science liaison for COVID-19).
Acknowledgements
We sincerely thank the efforts of Sara Stinca, Kelly LoPresti, Peter Gomme and Michael Jorgensen from CSL for logistics and administration with the use of pooled polyclonal antibodies.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104270.
Appendix. Supplementary materials
References
- 1.Rajah MM, Hubert M, Bishop E, et al. SARS-CoV-2 Alpha, Beta, and Delta variants display enhanced Spike-mediated syncytia formation. EMBO J. 2021;40(24) doi: 10.15252/embj.2021108944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao H, Lu L, Peng Z, et al. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with Delta variant in TMPRSS2-expressed cells. Emerg Microbes Infect. 2022;11(1):277–283. doi: 10.1080/22221751.2021.2023329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meng B, Abdullahi A, Ferreira I, et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature. 2022;603(7902):706–714. doi: 10.1038/s41586-022-04474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halfmann PJ, Iida S, Iwatsuki-Horimoto K, et al. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature. 2022;603(7902):687–692. doi: 10.1038/s41586-022-04441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki R, Yamasoba D, Kimura I, et al. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature. 2022;603(7902):700–705. doi: 10.1038/s41586-022-04462-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aggarwal A, Stella AO, Walker G, et al. Platform for isolation and characterization of SARS-CoV-2 variants enables rapid characterization of Omicron in Australia. Nat Microbiol. 2022;7(6):896–908. doi: 10.1038/s41564-022-01135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hui KPY, Ho JCW, Cheung MC, et al. SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature. 2022;603(7902):715–720. doi: 10.1038/s41586-022-04479-6. [DOI] [PubMed] [Google Scholar]
- 8.Dejnirattisai W, Huo J, Zhou D, et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185(3):4674–4684.e15. doi: 10.1016/j.cell.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kimura I, Yamasoba D, Tamura T, et al. Virological characteristics of the novel SARS-CoV-2 Omicron variants including BA.2.12.1, BA.4 and BA.5. bioRxiv. 2022 doi: 10.1101/2022.05.26.493539. [DOI] [Google Scholar]
- 10.Tea F, Ospina Stella A, Aggarwal A, et al. SARS-CoV-2 neutralizing antibodies: longevity, breadth, and evasion by emerging viral variants. PLoS Med. 2021;18(7) doi: 10.1371/journal.pmed.1003656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stucki M, Boschetti N, Schafer W, et al. Investigations of prion and virus safety of a new liquid IVIG product. Biologicals. 2008;36(4):239–247. doi: 10.1016/j.biologicals.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 12.Karbiener M, Farcet MR, Schwaiger J, et al. Plasma from post-COVID-19 and COVID-19-vaccinated donors results in highly potent SARS-CoV-2 neutralization by intravenous immunoglobulins. J Infect Dis. 2021;(Online ahead of print) doi: 10.1093/infdis/jiab482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kristiansen PA, Page M, Bernasconi V, et al. WHO International Standard for anti-SARS-CoV-2 immunoglobulin. Lancet. 2021;397(10282):1347–1348. doi: 10.1016/S0140-6736(21)00527-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amarilla AA, Sng JDJ, Parry R, et al. A versatile reverse genetics platform for SARS-CoV-2 and other positive-strand RNA viruses. Nat Commun. 2021;12(1):3431. doi: 10.1038/s41467-021-23779-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramakrishnan MA. Determination of 50% endpoint titer using a simple formula. World J Virol. 2016;5(2):85–86. doi: 10.5501/wjv.v5.i2.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bull RA, Adikari TN, Ferguson JM, et al. Analytical validity of nanopore sequencing for rapid SARS-CoV-2 genome analysis. Nat Commun. 2020;11(1):6272. doi: 10.1038/s41467-020-20075-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mlcochova P, Kemp SA, Dhar MS, et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021;599(7883):114–119. doi: 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shuai H, Chan JF, Hu B, et al. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature. 2022;603(7902):693–699. doi: 10.1038/s41586-022-04442-5. [DOI] [PubMed] [Google Scholar]
- 19.Kozlov M. Omicron's feeble attack on the lungs could make it less dangerous. Nature. 2022;601(7892):177. doi: 10.1038/d41586-022-00007-8. [DOI] [PubMed] [Google Scholar]
- 20.Uraki R, Iida S, Halfmann PJ, et al. Characterization of SARS-CoV-2 Omicron BA.2.75 clinical isolates. bioRxiv. 2022 doi: 10.1101/2022.08.26.505450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Syed AM, Taha TY, Tabata T, et al. Rapid assessment of SARS-CoV-2-evolved variants using virus-like particles. Science. 2021;374(6575):1626–1632. doi: 10.1126/science.abl6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thorne LG, Bouhaddou M, Reuschl AK, et al. Evolution of enhanced innate immune evasion by SARS-CoV-2. Nature. 2022;602(7897):487–495. doi: 10.1038/s41586-021-04352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferreira CB, Sumner RP, Rodriguez-Plata MT, et al. Lentiviral vector production titer is not limited in HEK293T by induced intracellular innate immunity. Mol Ther Methods Clin Dev. 2020;17:209–219. doi: 10.1016/j.omtm.2019.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stalls V, Lindenberger J, Gobeil SM, et al. Cryo-EM structures of SARS-CoV-2 Omicron BA.2 spike. Cell Rep. 2022;39(13) doi: 10.1016/j.celrep.2022.111009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gruell H, Vanshylla K, Korenkov M, et al. SARS-CoV-2 Omicron sublineages exhibit distinct antibody escape patterns. Cell Host Microbe. 2022;(Online ahead of print) doi: 10.1016/j.chom.2022.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Touret F, Baronti C, Pastorino B, et al. In vitro activity of therapeutic antibodies against SARS-CoV-2 Omicron BA.1, BA.2 and BA.5. Sci Rep. 2022;12(1):12609. doi: 10.1038/s41598-022-16964-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takashita E, Yamayoshi S, Simon V, et al. Efficacy of antibodies and antiviral drugs against Omicron BA.2.12.1, BA.4, and BA.5 subvariants. N Engl J Med. 2022;387(5):468–470. doi: 10.1056/NEJMc2207519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Q, Guo Y, Iketani S, et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4, & BA.5. Nature. 2022;608(7923):603–608. doi: 10.1038/s41586-022-05053-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tuekprakhon A, Nutalai R, Dijokaite-Guraliuc A, et al. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell. 2022;185(14):2422–2433.e13. doi: 10.1016/j.cell.2022.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bowen JE, Addetia A, Dang HV, et al. Omicron spike function and neutralizing activity elicited by a comprehensive panel of vaccines. Science. 2022;377:890–894. doi: 10.1126/science.abq0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hachmann NP, Miller J, Collier AY, et al. Neutralization escape by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4, and BA.5. N Engl J Med. 2022;387(1):86–88. doi: 10.1056/NEJMc2206576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pinto D, Park YJ, Beltramello M, et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583(7815):290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 33.Cao Y, Yisimayi A, Jian F, et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022;608(7923):593–602. doi: 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arora P, Kempf A, Nehlmeier I, et al. Augmented neutralisation resistance of emerging omicron subvariants BA.2.12.1, BA.4, and BA.5. Lancet Infect Dis. 2022;22(8):1117–1118. doi: 10.1016/S1473-3099(22)00422-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lubinski B, Jaimes JA, Whittaker GR. Intrinsic furin-mediated cleavability of the spike S1/S2 site from SARS-CoV-2 variant B.1.1.529 (Omicron) bioRxiv. 2022;26 doi: 10.1101/2022.04.20.488969. [DOI] [Google Scholar]
- 36.Zhang Y, Zhang T, Fang Y, Liu J, Ye Q, Ding L. SARS-CoV-2 spike L452R mutation increases Omicron variant fusogenicity and infectivity as well as host glycolysis. Signal Transduct Target Ther. 2022;7(1):76. doi: 10.1038/s41392-022-00941-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balint G, Voros-Horvath B, Szechenyi A. Omicron: increased transmissibility and decreased pathogenicity. Signal Transduct Target Ther. 2022;7(1):151. doi: 10.1038/s41392-022-01009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tegally H, Moir M, Everatt J, et al. Emergence of SARS-CoV-2 Omicron lineages BA.4 and BA.5 in South Africa. Nat Med. 2022 doi: 10.1038/s41591-022-01911-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hansen CH, Friis NU, Bager P, et al. Risk of reinfection, vaccine protection, and severity of infection with the BA.5 Omicron subvariant: a Danish Nation-Wide Population-Based Study. 2022. Available at SSRN: https://ssrn.com/abstract=4165630 or 10.2139/ssrn.4165630. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.