Abstract
Pseudomonas aeruginosa strains that cause chronic pulmonary infections in cystic fibrosis patients typically undergo mucoid conversion. The mucoid phenotype indicates alginate overproduction and is often due to defects in MucA, an antisigma factor that controls the activity of sigma-22 (AlgT [also called AlgU]), which is required for the activation of genes for alginate biosynthesis. In this study we hypothesized that mucoid conversion may be part of a larger response that activates genes other than those for alginate synthesis. To address this, a two-dimensional (2-D) gel analysis was employed to compare total proteins in strain PAO1 to those of its mucA22 derivative, PDO300, in order to identify protein levels enhanced by mucoid conversion. Six proteins that were clearly more abundant in the mucoid strain were observed. The amino termini of such proteins were determined and used to identify the gene products in the genomic database. Proteins involved in alginate biosynthesis were expected among these, and two (AlgA and AlgD) were identified. This result verified that the 2-D gel approach could identify gene products under sigma-22 control and upregulated by mucA mutation. Two other protein spots were also clearly upregulated in the mucA22 background, and these were identified as porin F (an outer membrane protein) and a homologue of DsbA (a disulfide bond isomerase). Single-copy gene fusions were constructed to test whether these proteins were enhanced in the mucoid strain due to increased transcription. The oprF-lacZ fusion showed little difference in levels of expression in the two strains. However, the dsbA-lacZ fusion showed two- to threefold higher expression in PDO300 than in PAO1, suggesting that its promoter was upregulated by the deregulation of sigma-22 activity. A dsbA-null mutant was constructed in PAO1 and shown to have defects predicted for a cell with reduced disulfide bond isomerase activity, namely, reduction in periplasmic alkaline phosphatase activity, increased sensitivity to dithiothreitol, reduced type IV pilin-mediated twitching motility, and reduced accumulation of extracellular proteases, including elastase. Although efficient secretion of elastase in the dsbA mutant was still demonstrable, the elastase produced appeared to be unstable, possibly as a result of mispaired disulfide bonds. Disruption of dsbA in the mucoid PDO300 background did not affect alginate production. Thus, even though dsbA is coregulated with mucoid conversion, it was not required for alginate production. This suggests that mucA mutation, which deregulates sigma-22, results in a global response that includes other factors in addition to increasing the production of alginate.
Pseudomonas aeruginosa is a common opportunistic pathogen that can cause fatal illnesses in a variety of patients, including those suffering from cystic fibrosis (CF), burn wounds, tissue injury, and immunosuppressive therapy. In CF patients, thick mucus accumulates in the airways, making them highly susceptible to chronic pulmonary infection with P. aeruginosa. The P. aeruginosa strains isolated from CF patients also have an unusual mucoid colony morphology due to the overproduction of alginate, an exopolysaccharide of O-acetylated d-mannuronate and l-guluronate (13). Alginate is a virulence factor that confers antiphagocytic and adherence properties (13).
The chromosome of P. aeruginosa contains an operon of 12 genes encoding enzymes for alginate biosynthesis (8). The promoter of this operon, PalgD, is under direct or indirect control by several transcriptional regulators, including AlgB, AlgR, AlgZ, and RpoS (5, 36, 39). PalgD is also specifically recognized by the alternative sigma factor ς22, encoded by algT (also called algU) (10). ς22 is a member of the extracytoplasmic function (ECF) sigma factor family characterized by their responsiveness to extracytoplasmic stimuli (10, 26). A similar ECF sigma factor in Escherichia coli called ςE is involved in the stress response to extreme heat shock, and it regulates several promoters (31). ECF sigma factors are often controlled by an antisigma factor. In P. aeruginosa, MucA is an antisigma factor for ς22 that affects transcriptional activity (32). MucA is an inner membrane protein that probably interacts with periplasmic MucB to control the activity of ς22 in the cytoplasm (23). Mutations in MucA are typically seen in CF isolates and are usually responsible for conversion of P. aeruginosa strains to the mucoid phenotype (21). ς22 shows increased activity and protein level in cells where MucA is defective (23). Mucoid conversion due to mutations in mucA has also been observed as an in vitro response to activated human polymorphonucleocytes and to hydrogen peroxide, suggesting that alginate production may be a stress response to toxic oxygen by-products (22).
The role of ς22 in P. aeruginosa under free-living conditions in the environment is unclear, but understanding its role may provide important clues for deciphering its regulation of alginate production in the CF patient lung. The ς22 control system appears to involve a membrane-bound signal transduction complex including MucA and MucB (23). Its similarity to other ECF sigma factors suggests that ς22 may be part of a general stress response. Mucoid conversion is apparently due to ς22 deregulation, which occurs through adaptive mutations that cause defects in the antisigma factor MucA. This conversion appears to be a short-circuit approach to activating ς22 activity and bypasses the need for a signal. If ς22 is normally part of a general stress response, then ς22 deregulation during mucoid conversion (ie., mucA mutation) may promote the expression of proteins in addition to those for alginate biosynthesis.
To examine the hypothesis that ς22 controls a stress response network, we undertook a proteome analysis of P. aeruginosa. The proteome refers to the set of proteins expressed by the genome under a defined condition and/or genetic background. The entire sequence of the P. aeruginosa genome recently became available (35), which provided a critical tool for an effective proteome analysis. In general, the use of proteomics has also been fostered by technological developments, like two-dimensional (2-D) gel electrophoresis, to simplify the separation of complex mixtures of proteins and by methods for protein identification, such as amino-terminal sequence analysis and mass spectrometry. These advances in technology, used in combination with the generation of total genomic and protein databases, have spurred proteomic research and led to comprehensive proteome analyses of several bacterial species, including Bacillus subtilis (2), Haemophilus influenzae (20), and E. coli (29).
In the study described here, we used 2-D gel analyses to compare the proteomes of wild-type P. aeruginosa strain PAO1 to its mucoid derivative PDO300 (22). This mucoid strain was constructed to have the mucA22 allele, which is the mucoid conversion mutation seen in most isolates from CF patients (6), including the CF strain FRD1 used in many studies from this laboratory (10). Total proteins from the wild-type strain and isogenic mucA22 strain were each subjected to a 2-D gel analysis, and the constellations of total proteins from the strains were compared. Several proteins appeared to be more intense as a result of increased ς22 activity. Amino acid sequence analyses revealed not only proteins for alginate production but also previously unidentified ones, including disulfide bond isomerase (DsbA). Interestingly, the dsbA gene in P. aeruginosa was recently shown to play a role in virulence in the Caenorhabditis elegans infection model (37). We used a mutant analysis to examine the roles of DsbA in the expression of several virulence factors, including alginate.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains used in this study are listed in Table 1. The P. aeruginosa strains used in this study were PAO1, a prototypic wild-type strain, and its isogenic mucA22 derivative PDO300, which produces alginate (22). E. coli strains HB101 (proA2 leuB6 thi-l lacYI hsdR hsdM recA13 supE44 rpsL20) and DH10B [F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 endA1 recA1 deoR Δ(ara, leu)7697 araD139 galU galK nupG rpsL λ−) were used as cloning hosts. The P. aeruginosa and E. coli strains were grown routinely in L broth (Sigma). Plasmids were transferred to P. aeruginosa by triparental mating mediated by pRK2013 with antibiotic selection on Pseudomonas Isolation Agar (Difco) mixed 1:1 with L agar. The antibiotics and concentrations used for selecting P. aeruginosa were carbenicillin at 300 μg/ml, tetracycline at 100 μg/ml, gentamicin at 300 μg/ml, and kanamycin at 1 mg/ml. For plasmid selections in E. coli, ampicillin was used at 100 μg/ml, tetracycline was used at 25 μg/ml, kanamycin was used at 40 μg/ml, and gentamicin was used at 30 μg/ml.
TABLE 1.
P. aeruginosa strains and plasmids used in this study
| P. aeruginosa strain or plasmid | Genotype or phenotypea | Reference or source |
|---|---|---|
| Strains | ||
| PAO1 | Wild-type prototroph, Alg− | 15 |
| PDO300 | PAO1 with mucA22, Alg+ | 22 |
| PDO310 | PAO1 with dsbA::Gmr | This study |
| PDO311 | PDO300 with dsbA::Gmr, Alg+ | This study |
| Plasmids | ||
| pRK2013 | ColE1-Tra(RK2)+ Kmr | Lab collection |
| pMC1871 | ColE1 LacZ (deletion of 1st to 8th amino acid), Tetr | Pharmacia |
| pMS7 | pMC1871 derivative with 1.2-kb mob-cos fragment from pSF4 | This study |
| pUJ10 | ColE1 lacZα Apr | 9 |
| pMS122 | pMS7 with a 1.2-kb trp-lacZ fragment replacement from pUJ10 | This study |
| pMS180 | pMS122 with 0.9-kb dsbA-lacZ (transcriptional) | This study |
| pMS181 | pMS122 with 2.1-kb clpP-lacZ (transcriptional) | This study |
| pMS182 | pMS122 with 1.1-kb oprF-lacZ (transcriptional) | This study |
| pZerOTM-2.1 | ColE1 lacZα f1 ori Kmr | Invitrogen |
| pLS159 | pZerO-2.1 with 1.0-kb dsbA | This study |
| pLS159-oriT | pLS159 carrying oriT in the BamHI site | This study |
| pLS168 | pLS159 with dsbA-Gmr (nonpolar) | This study |
Abbreviations: Apr, ampicillin resistance; Tetr, tetracycline resistance; Kmr, kanamycin resistance; Gmr, gentamicin resistance; Alg+, alginate overproduction.
Nucleic acid manipulations.
The plasmids used and constructed in this study are listed in Table 1. The DNA manipulations used for generating the plasmid constructs were carried out as described elsewhere (3). PCR was performed using Pfu polymerase (Stratagene) according to the instructions of the manufacturer, with DNA templates extracted from overnight cultures of PAO1 as previously described (36). Reagents required for DNA manipulations such as restriction enzymes were obtained from Boehringer Mannhiem, New England Biolabs, or Promega. Plasmid DNA purification from E. coli was performed using columns and reagents from Qiagen, Inc.
2-D gel electrophoresis and analysis.
To prepare whole-cell lysates, strains PAO1 and PDO300 were grown under identical conditions in L broth to an A600 of 0.8 to 1.0. Cells were harvested by centrifugation (13,000 × g, 30 mm) and broken in an osmotic lysis buffer (Kendrick Laboratories, Inc., Madison, Wis.). Protein concentration was determined by the Bradford method. 2-D gel electrophoresis of cell proteins was performed using the method of O'Farrell (28) at Kendrick Laboratories, Inc., and the gel was stained with Coomassie blue. To characterize a spot of interest, the 2-D gel was trans-blotted onto a polyvinylidene difluoride membrane, and an amino-terminal sequence analysis was performed (Biotechnology Center, St. Jude Children's Research Hospital). The sequence obtained was used as a query in a BLAST analysis of the P. aeruginosa genome through the National Center for Biotechnology Information.
Construction of lacZ transcriptional fusions.
The translational lacZ fusion vector pMC1871 (Pharmacia) was made mobilizable by inserting a 2-kb EcoRI fragment containing oriT and cos from pSF4 (34) into the ScaI site, resulting in pMS7. To convert it to a transcriptional fusion vector, pMS7 was digested with SmaI and SacI to remove a portion of the 5′ end of lacZ, which was replaced with a 1.2-kb fragment carrying a similar 5′ end of lacZ from the transcriptional fusion vector pUJ10 (9). The new lacZ transcriptional fusion vector, pMS122, carried a ribosome binding site and a multiple cloning site at the 5′ end of lacZ. To generate the various transcriptional gene fusions described in this study, approximately 1 kb of genomic DNA of each selected gene was amplified by PCR from a overnight culture of PAO1 and cloned into pMS122 using ends compatible with SmaI and BglII (pMS180 through pMS182) (Table 1). The fusions were then mobilized into the P. aeruginosa strains PAO1 and PDO300 by triparental mating with selection for tetracycline resistance to integrate such fusions into the chromosome by homologous recombination. This process resulted in a single copy of lacZ placed under the control of the native promoter of the targeted gene. Integration of the transcriptional fusions was verified by PCR. An assay of β-galactosidase activity in lysates of P. aeruginosa strains was performed as previously described (23).
Construction of a dsbA mutant of strain PAO1.
The sequence of dsbA was obtained from the P. aeruginosa genomic DNA sequence database (Pathogenesis Corp.). This sequence was PCR amplified from PAO1 templates using primers dsbA1 (TGCACTGATCGCTGCGTAGCAC) and dsbA1036 (CGTCCGCCATCGCTACAATGCT). The 1-kb fragment was cloned into pZERO 2.1 (Invitrogen) at the EcoRV site to obtain pLS159. A gentamicin resistance cassette from pUC-GM (33) as an SmaI fragment was inserted into the unique MluI site within the dsbA coding sequence in pLS159. A resulting plasmid containing the gentamicin cassette in the same orientation as the dsbA coding sequence (pLS168) was used in this study. An origin of transfer (oriT) fragment from pSF4 (34) was cloned into pLS168 at the unique BamHI site to generate pLS204. To construct a dsbA::Gm mutant, pLS204 was introduced into PAO1 by triparental mating with selection for gentamicin resistance. Colonies were then screened for kanamycin sensitivity, indicating double-crossover events leading to loss of the vector. Presumptive dsbA::Gm mutants were verified by PCR to contain a gentamicin cartridge insertion in the dsbA gene, and the dsbA::Gm mutant PDO310 was chosen at random for further characterization. Mutant complementation was performed by introducing in trans pLS159-oriT containing dsbA+ from P. aeruginosa.
Assays.
Production of casein-degrading proteases by P. aeruginosa strains was determined by patching strains onto skim milk agar plates that contained a 1:1 mixture of 4% skim milk and 2× L agar (Sigma). After incubation for 18 h at 30°C, the zone of clearance from the edge of growth was measured. Assay for the protease elastase in 18-h culture supernatants was performed as described previously (25) using an Elastin-Congo red conjugate (Sigma). Elastase activity was represented as the change in the optical density at 495 nm (OD495) per mg of protein. Elastase protein was also examined by an immunoblot analysis as described previously (25). Alkaline phosphatase activity in periplasmic extracts was determined as previously described (7), with activity represented as a change in OD420 per mg of protein. To assess type IV pilin function, the ability of P. aeruginosa to move on a solid substratum by twitching motility was determined as described previously (1). Briefly, thin 1% L agar plates were stab inoculated to the bottom of the plate with a wire loop and incubated at 37°C for 24 h. The plate was then flooded with 0.05% Coomassie blue, and the diameter of the translucent zone of growth on the bottom of the agar was measured. To compare alginate production levels, the strains to be tested were grown in L broth under identical conditions for 20 h and alginate contents in the supernatants were determined as described previously (19).
RESULTS
Effects of mucA22 mutation on the proteome.
The major global effects of ς22 deregulation in P. aeruginosa due to a defective antisigma factor, MucA, were determined by a proteome analysis. Strain PAO1 and its mucA22 derivative PDO300 were grown under identical conditions in L broth to the late logarithmic phase of growth. Total proteins were extracted, resolved by 2-D gel electrophoresis, and stained with Coomassie blue (Fig. 1). A comparison of the protein profiles revealed six protein spots that were obviously more intense in the mucA22 mutant (PDO300) than in the parent strain, suggesting that the genes encoding these proteins may be positively controlled directly or indirectly by ς22. The proteins upregulated as a result of mucA mutation were designated by using their molecular weights (in thousands) and isoelectric points (pIs), and they are listed in Table 2. Interestingly, three protein spots were more intense in PAO1 than in PDO300, suggesting that they were under negative control by ς22 (Table 2). The six most prominent proteins, upregulated as a result of the mucA22 mutation, were subjected to amino-terminal sequencing, and sequence data were obtained from all but two spots. The four proteins that produced amino acid sequence data could be positively identified in the genomic database of P. aeruginosa (Table 2). Spots 50/4.5 and 45/5.4 were revealed to be phosphomannose isomerase (AlgA) and GDP-mannose dehydrogenase (AlgD), respectively. These are enzymes involved in alginate biosynthesis, and all are encoded by the algD operon, which contains 12 genes for alginate biosynthesis. The identification of such proteins was expected because they are encoded by the algD operon, which is directly under ς22 control and upregulated by mucA mutation in mucoid strains. This result verified that the 2-D gel approach could identify proteins under ς22 control. Spot 30/4.4 was porin F (11), a major outer-membrane protein in P. aeruginosa. Spot 23/6.2 was found to be a homologue of E. coli DsbA, a periplasmic disulfide bond isomerase (27). One spot next to DsbA labeled 22.5/6.2 appeared to have the same intensity in both strains and, when analyzed as a negative control, was shown to be a homologue of E. coli ClpP (30), a periplasmic serine protease.
FIG. 1.
2-D gel electrophoresis patterns of proteins isolated from cells of P. aeruginosa strain PAO1 (top) and its isogenic mucA22 derivative PDO300 (bottom). Cells were grown in L broth with aeration and collected in late logarithmic phase of growth, and total proteins were extracted for a 2-D gel analysis. The gels were stained with Coomassie blue. The arrowhead points to the internal standard, tropomyosin (32.7 kDa, pI 5.2). The molecular mass standard lines are from myosin (220 kDa), phosphorylase A (94 kDa), catalase (60 kDa), actin (43 kDa), carbonic anhydrase (29 kDa), and lysozyme (14 kDa). The spots that appeared more intense in one strain than in the other were marked with arrows and given the designations described in Table 2. Gels were trans-blotted onto polyvinylidene difluoride membranes, and spots were subjected to an amino-terminal sequence analysis for identification (Table 2).
TABLE 2.
Effect of the mucA22 mutation (PDO300) on proteins expressed in P. aeruginosa PAO1 as observed by 2-D gel analysis of total proteins
| Protein designation (molecular weight/pI)a | Expression in mucA22 | N-terminal sequenceb | Identical protein or homologue |
|---|---|---|---|
| 50/4.5 | Upregulated | MIPVILSX(G)GSGSRLWPX(L)SRK | AlgA |
| 45/5.4 | Upregulated | MRISIFGLGYVGAVX(C)AGX(C)LS | AlgD |
| 23/6.2 | Upregulated | DDYTAGKEYVELSRPVRXYD | DsbA |
| 30/4.4 | Upregulated | QSQNSVEIEAFGKRYFTDSV | Porin F |
| 28/4.5 | Upregulated | Blocked | Unknown |
| 14/4.6 | Upregulated | Blocked | Unknown |
| 90/4.4 | Downregulated | ND | Unknown |
| 85/5.0 | Downregulated | ND | Unknown |
| 50/6.2 | Downregulated | ND | Unknown |
| 22.5/6.2 | Unchanged | AGYLVPMVIEQTSRGERXAQ | ClpP |
Proteins were designated by their apparent molecular weight (in thousands) over their isoelectric point (pI) from the 2-D gel.
Residues shown as X could not be determined in the amino-terminal analysis, but the appropriate residue from the genome sequence is shown in parentheses. Some proteins did not produce sequences and were presumed to be blocked. ND, not done.
Effects of mucA22 mutation on promoter activities for genes encoding porin F, ClpP, and DsbA.
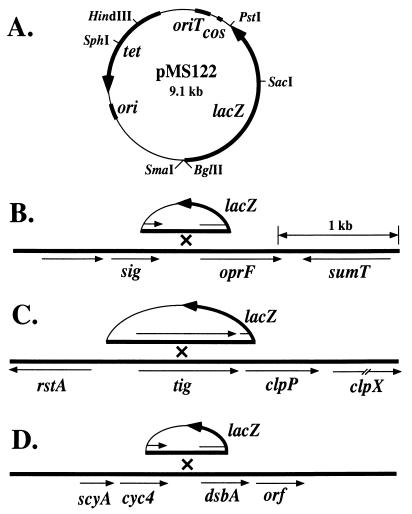
The higher protein levels of porin F and DsbA seen in the mucA22 strain may be due to increased transcription. To test this, single-copy lacZ fusions of their respective genes were constructed with ClpP included as a negative control. Genomic database information was also examined to identify the flanking open reading frames for each gene and its neighboring genes. The oprF monocistronic gene for porin F is clustered between genes encoding homologues of sumT, which encodes a uroporphyrin biosynthetic enzyme in P. fluorescens, and a gene (sig) for an ECF family sigma factor (Fig. 2). The clpP gene was flanked upstream by tig, which encodes a homologue of the E. coli trigger factor, and downstream by clpX, which encodes a homologue of a protease in E. coli. Interestingly, the local genomic organization of clpP in P. aeruginosa was similar to that found in E. coli. The dsbA gene was flanked upstream by the gene cyc4, which encodes cytochrome c4, and downstream by an open reading frame encoding a protein of unknown function. The 5′ end of each gene and about 1 kb of upstream DNA were generated by PCR amplification. Each fragment was then cloned in the transcribing orientation into a lacZ transcriptional fusion vector (pMS122) (Fig. 2) that was constructed in this study. The plasmids generated (pMS180, pMS181, and pMS182) (Table 1) were conjugated into PAO1 and PDO300 with selection for the plasmid-borne tetracycline resistance marker. This process resulted in the integration of the transcriptional fusions into the chromosome by homologous recombination, which placed a single copy of lacZ under the control of the promoters of the respective genes (Fig. 2). These PAO1 and PDO300 derivatives were grown in L broth under the same conditions as those used for the cultures prepared for 2-D gels (i.e., an OD600 of 1.0), and cells were then harvested for β-galactosidase assays. All fusions were active, and the results of the analysis are shown in Table 3. As expected, the transcriptional activity of clpP-lacZ was approximately the same in both strain backgrounds. Interestingly, the expression of the oprF-lacZ fusion was not observed to increase significantly as a result of the mucA22 background even though the porin F protein concentration was dramatically increased in PDO300 (Fig. 1). However, dsbA transcription was two- to threefold higher in the mucA22 background than in the wild type, which correlated with DsbA protein levels. This finding suggested that dsbA is transcriptionally controlled, directly or indirectly, by deregulated ς22.
FIG. 2.
Strategy for the analysis of the transcriptional regulation of oprF and dsbA, which encode proteins enhanced by mucA22, and of clpP, which was constitutive. (A) Map of the lacZ transcriptional fusion vector pMS122 used to construct integrative lacZ gene fusions in P. aeruginosa PAO1 and PDO300. Genes indicated are lacZ (β-galactosidase), cos (cohesive ends of lambda), oriT (origin of transfer from RK2), tet (tetracycline resistance), and ori (the ColE1 origin of replication). Below are shown the genes flanking oprF, clpP, and dsbA, which were obtained from the genomic sequence analysis (35) of strain PAO1. (B) Depiction of a fragment in pMS122 containing the 5′ end of oprF integrated into the chromosome by homologous recombination to generate an oprF-lacZ fusion. sig encodes an ECF sigma factor, and sumT encodes a homologue of a uroporphyrin biosynthetic enzyme in P. fluorescens. (C) Depiction of a fragment in pMS122 containing the 5′ end of clpP integrated into the chromosome by homologous recombination to generate a clpP-lacZ fusion. rstA encodes a transcriptional regulatory protein in E. coli, and clpP and clpX encode homologues of proteases in E. coli. (D) Depiction of a fragment in pMS122 containing the 5′ of dsbA integrated into the chromosome by homologous recombination to generate a dsbA-lacZ fusion. scyA encodes a homologue of the mono-heme c-type cytochrome in Shewnaella putrefaciens, cyc4 encodes cytochrome c4, and orf is an open reading frame that encodes a protein of unknown function.
TABLE 3.
Effect of the mucA22 mutation on the expression of single-copy lacZ transcriptional fusions to the genes for ClpP, porin F, and DsbA in P. aeruginosaa
| Strain | Genotype | Transcriptional activity of:
|
||
|---|---|---|---|---|
| clpP-lacZ | oprF-lacZ | dsbA-lacZ | ||
| PAO1 (Alg−) | Wild type | 25 ± 6 | 120 ± 3 | 30 ± 5 |
| PDO300 (Alg+) | mucA22 | 23 ± 7 | 110 ± 5 | 90 ± 4 |
PAO1 and its mucA22 derivative PDO300 (Alg+, alginate overproducing) had derivatives of pMS122 integrated into the chromosome by homologous recombination to form the transcriptional fusions clpP-lacZ, oprF-lacZ, and dsbA-lacZ (Fig. 2). Strains were grown in L broth to an OD600 of 1.0, and cell extracts were then subjected to β-galactosidase assays, with transcriptional activity defined as follows: (β-galactosidase activity OD420 × 1,000)/(reaction time [min] × vol of cells [ml] × OD600) (23). Data are the averages of results from four experiments ± standard errors.
Mutation in dsbA results in reduced alkaline phosphatase and dithiothreitol sensitivity.
An amino acid sequence alignment of the DsbA proteins from E. coli and P. aeruginosa showed 25% identity and 29% similarity (data not shown). This relatively low level of homology between DsbA proteins from different species is not uncommon (27). Both proteins showed alignment of a thioredoxin fold motif (CPHC) that is considered a characteristic of DsbA (27). To test whether the putative DsbA in P. aeruginosa has disulfide bond isomerase activity, as it does in E. coli, we constructed a dsbA::Gm mutant of strain PAO1 named PDO310 by gene replacement. A PCR analysis verified that it contained a chromosomal gentamicin cartridge insertion in the dsbA gene (data not shown). Such a dsbA mutant was expected to exhibit pleiotropic defects as a result of protein misfolding in the periplasm due to the defect in disulfide bond isomerase activity. In E. coli, dsbA mutants are reduced in periplasmic alkaline phosphatase activity due to a misfolding of this protein and mispairing of disulfide bonds (4). In P. aeruginosa, alkaline phosphatase in the periplasm is repressed under normal growth conditions but can be derepressed by reducing the phosphate in the growth medium (12). Under phosphate-limiting growth conditions, the periplasmic extract of PAO1 contained about 300 U of alkaline phosphatase per mg of protein whereas the dsbA mutant (PDO310) contained about 10-fold less, which was consistent with the predicted phenotype (Table 4). Another characteristic of a dsbA mutation in E. coli is increased sensitivity to dithiothreitol. On L agar containing 8 mM dithiothreitol, wild-type PAO1 grew normally but PDO310 grew poorly and colonies were small after overnight incubation (Table 4). PDO310 did not exhibit any obvious morphological change or growth rate defect in L broth as a result of the DsbA− defect.
TABLE 4.
Phenotypic effects of the dsbA::Gm mutation in P. aeruginosa PAO1
| Test | Test result with:
|
|
|---|---|---|
| PAO1 (wild type) | PDO310 (dsbA::Gm) | |
| Alkaline phosphatase | 300 U/ml | 30 U/ml |
| Dithiothreitol sensitivity | Resistant | Sensitive |
| Zone of casein degradation (width) | 3 mm | <1 mm |
| Elastase activity | 150 U | 15 U |
| Type IV pilin-mediated twitching motility (distance) | 21 mm | 12 mm |
| Alginate (in a mucA22 background)a | 150 μg/ml | 150 μg/ml |
Alginate production was tested in PDO300, the mucA22 derivative of PAO1, and in PDO311, the dsbA::Gm derivative of PDO300.
Mutation in dsbA caused defects in protease production.
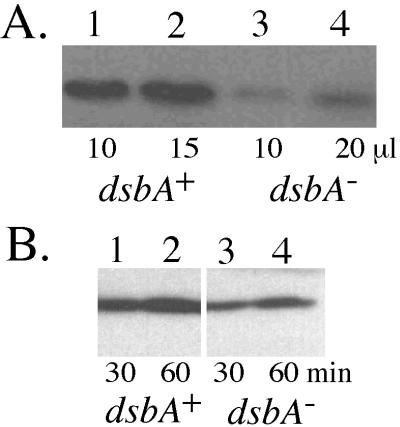
In general, DsbA is involved in the maturation of proteins containing disulfide bonds that are transported across the inner membrane (27). Much of the virulence of P. aeruginosa as an opportunistic pathogen is attributed to its ability to secrete toxic and degradative proteins that cross the inner and outer membranes and must be folded properly before entering the extracellular environment. We examined whether the secretion of proteases, which serve as major virulence factors of P. aeruginosa, were affected by the dsbA mutation. Several proteases have casein-degrading activity, and so the ability to form zones of casein clearing on agar plates containing skimmed milk was tested. Wild-type PAO1 formed a cleared zone with a width of 3 mm from the edge of bacterial growth, but the dsbA mutant PDO310 produced a zone with a width of less than 1 mm (Table 4). PDO310 (pLS159-oriT) carried dsbA in trans on an integrative plasmid to complement the chromosomal dsbA::Gm mutation, and the ability to produce proteases that degraded casein was completely restored. Thus, production of proteases was generally compromised by the loss of DsbA in the cell. The most active protease produced by P. aeruginosa is a zinc-metallo protease called elastase (also known as LasB protease or pseudolysin). Elastase requires a propeptide chaperone for proper folding (24), and it also contains two disulfide bonds (38), suggesting that DsbA may also be required for proper folding. Compared to wild-type PAO1, the dsbA::Gm mutant PDO310 exhibited a greater than 10-fold reduction in elastolytic activity (Table 4). Elastolytic activity in PDO310 was restored with dsbA in trans. Since elastolytic activity is due to LasA protease as well as elastase (17), we also examined elastase protein levels in the culture supernatants by immunoblot analysis. This analysis (Fig. 3A) demonstrated that much less elastase protein accumulated in an 18-h culture supernatant of the dsbA mutant than in the wild type. As a test for the ability to secrete elastase protein, which may be unstable over time, washed cells were tested for the ability to release the protein after just 30 and 60 min into fresh medium as previously described (18). Under these conditions (Fig. 3B), the difference between the wild type and the dsbA mutant was less striking, suggesting that DsbA has more effect on elastase folding and stability than on the type II secretion apparatus which is required for elastase secretion.
FIG. 3.
Examination of elastase protein levels produced in PAO1 and its dsbA mutant derivative PDO310 by Western blot analysis with rabbit antielastase antibody. (A) To evaluate the accumulation of elastase in the stationary-phase cultures, strains were grown 18 h in L broth and cells were removed. Lanes 1 and 2 show the reaction to PAO1 using 10 and 15 μl of supernatant, respectively. Lanes 3 and 4 show the reaction to PDO310 using 10 and 20 μl of supernatant, respectively. (B) To evaluate the secretion of elastase, which may be unstable in protease-rich stationary-phase culture supernatants, cells collected as described above were incubated in fresh L broth for 30 or 60 min and harvested and equal volumes of supernatant were tested from PAO1 (lanes 1 and 2) and from PDO310 (lanes 3 and 4).
Effect of dsbA mutation on type IV pilin and alginate production.
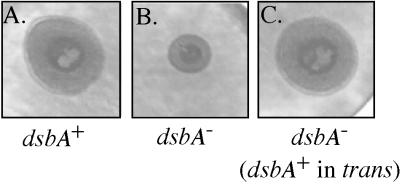
Type IV fimbriae function as an important adhesion for P. aeruginosa in pathogenesis and especially adhere to epithelial cell surfaces (14). These cylindrical pilus filaments (∼2.5 by 0.005 μm in size) are multifunctional retractile structures. In addition to being required for adherence, they are required for a form of bacterial motility over a solid surface known as twitching motility (14). The pilus is a homopolymer of pilin protein (15 kDa) that has an intrachain disulfide loop that contains an epithelial cell binding domain (14). When we tested the role of DsbA in type IV pilin function by examining twitching motility, the dsbA mutant produced a zone of motility (Fig. 4) that was about 60% of that of wild-type PAO1 (Table 4).
FIG. 4.
Demonstration of the effect of dsbA mutation on type IV pilus-mediated twitching motility. Twitching motility was assayed by measuring bacterial migration under agar and visualized by staining with Coomassie blue. PAO1 typically produced a zone of growth with a diameter of 21 ± 1.5 mm. The dsbA::Gm mutant (PDO310) produced cleared zones with a width of 12 ± 1.0 mm but was complemented with dsbA in trans, which produced a zone of 20.5 ± 1.0 mm.
Elevated DsbA activity was associated with alginate overproduction, the typical phenotype of a mucA22 mutant, suggesting that elevated DsbA may be involved in alginate overproduction. It has been proposed that alginate synthesis and/or secretion may occur through a polymer assembly complex of proteins located in the periplasm (16), and DsbA may be required to fold the members of this complex that contain disulfide bonds. To test this, we constructed a dsbA mutant in the mucA22 background of strain PDO300, thus forming PDO311, which also showed a mucoid phenotype on L agar plates. When culture supernatants of these two strains were tested, both contained about 150 μg of alginate per ml. Thus, alginate production and accumulation under these conditions were not affected by dsbA mutation. This finding suggests that the alginate polymer assembly complex does not require DsbA activity.
DISCUSSION
The goal of this study was to further our understanding of the changes that occur in the proteome of P. aeruginosa when it undergoes mucoid conversion, which is common during pulmonary infection of CF patients. In this study we hypothesized that mucoid conversion (i.e., overproduction of alginate) may be part of a larger stress response mechanism. It was recently shown that production of this polymer can occur in response to the toxic oxygen by-products of phagocytic cells (22). A common mutation that leads to the mucoid phenotype is the mucA22 allele, which is often seen in CF isolates (6, 21) and was seen in an in vitro study where P. aeruginosa biofilms were treated with activated polymorphonucleocytes or H2O2 (23). MucA is an inner membrane protein and antisigma factor that probably interacts with periplasmic MucB to control the activity of ς22 in the cytoplasm (23). Mutation in MucA appears to increase ς22 activity and possibly its stability, which leads to increased expression of ς22-dependent promoters (23). The mucA22 allele has a single base pair deletion that leads to premature truncation of a periplasmic portion of the protein but retains its single transmembrane domain. Recent studies of ECF sigma factors, to which group ς22 belongs, suggest that they can belong to global stress networks. To address this possibility with respect to ς22, we used the combination of 2-D gel analysis of proteins and the ability to identify the amino terminus of a protein on the basis of a predicted gene product in the genome database as a powerful method of analyzing global expression patterns.
Our 2-D gel analysis of proteins in total cell extracts was able to resolve about 200 proteins in PAO1, which is probably much less than the steady state of protein expression expected from a genome of this size. Thus, our analysis was limited to only well-expressed proteins. We compared the proteome of wild-type strain PAO1 to that of a mucA22 isogenic mutant by 2-D gel electrophoresis. Our resolution was high enough to detect six protein spots that were clearly more abundant in the mucA22 strain where ς22 activity was high. The recently completed sequence of the 6.3-Mbp chromosome of P. aeruginosa (35) then provided the fundamental information for a functional genomic study. It was unfortunate that two of these proteins did not produce sequence information, and this was probably due to amino-terminal blockage. Other technologies to overcome this problem are still being pursued. Interestingly, there were also three proteins whose levels were reduced in the mucA22 mutant, suggesting that ς22 may control a negative regulator or repressor protein. A future study will address the nature of these repressed proteins. To our knowledge, this is the first report of a 2-D gel proteome analysis of P. aeruginosa where a wild-type strain was compared to that of a regulatory mutant.
Two proteins that were overexpressed in the mucA22 mutant were shown to be encoded by the alginate biosynthetic operon: AlgA and AlgD. This was an important positive control to show that this method for proteome analysis was effective. Although 2 alginate biosynthesis proteins encoded by the algD operon were identified by this method to be under deregulated ς22 control, there were 10 other proteins encoded by the same operon that were not detected. Some of these enzymes have basic pIs that cannot readily be resolved in the pI gradient of 4 to 8 used in our 2-D gels, and these include AlgL (pI 8.6), AlgI (pI 8.9), AlgG (pI 8.9), AlgF (pI 9.5), and Alg44 (pI 9.0). The other alginate biosynthetic proteins not detected were probably expressed at levels too low for detection or were obscured by other proteins. This result demonstrates some of the limitations of this method for analyzing global expression patterns.
Two other proteins overexpressed in the mucA22 strain were porin F and DsbA. Single-copy transcriptional fusions to the promoters of their respective genes were analyzed in wild-type and mucA22 backgrounds. The dsbA-lacZ fusion showed two- to threefold increased activity in the mucA22 background. As a control, a clpP-lacZ fusion activity which reflected protein levels in the cell was constant. It was interesting that the gene fusion to oprF, which encodes porin F, did not show evidence of enhanced transcription, which may explain the increased protein level following mucoid conversion. Porin F has two disulfide bonds, and so its presentation may be enhanced by increased levels of DsbA in the mucA22 background. Alternatively, since ς22 is an ECF sigma factor, it may be responsible for other mechanisms that stabilize proteins associated with the surface of the cell.
Our subsequent experiments focused on DsbA. The dsbA gene in P. aeruginosa appears to encode a precursor DsbA of approximately 23 kDa with a pI of 6.2. The genomic sequence information on the open reading frame for dsbA revealed that the amino-terminal sequence corresponded to the mature protein after cleavage of a signal sequence of 22 amino acid residues. This signal sequence contained a typical core of hydrophobic amino acids followed by Ala-X-Ala, which served as a typical cleavage site for signal peptidase. The deduced protein sequence of P. aeruginosa DsbA was aligned by the Lipmann-Pearson protocol to its homologue from E. coli, which showed that the two sequences were similar by 29%. Both proteins showed alignment of a thioredoxin fold motif (CPHC) that is considered a characteristic of DsbA (27).
Mutants defective in dsbA were generated in the PAO1 and PDO300 strain backgrounds to test the role of P. aeruginosa dsbA in protein folding and production of virulence factors. We anticipated from studies done with other organisms that a dsbA mutation in P. aeruginosa was likely to produce pleiotropic effects. In an E. coli dsbA mutant, alkaline phosphatase activity is reduced (4). In addition, dsbA mutation also attenuates the virulence of certain pathogenic bacteria, such as Shigella flexneri (40). We looked for similar phenotypic defects in the P. aeruginosa dsbA::Gm mutants constructed here. Compared to the activity in the wild-type strain PAO1, dsbA mutation resulted in about 10-fold less alkaline phosphatase activity in the periplasmic extracts and also reduced accumulation of secreted proteases. We focused on the secretion of elastase, primarily because it contains two disulfide bonds in the mature protein (Cys-30—Cys-58 and Cys-270—Cys-297) (38). Elastase activity in the culture supernatant was about 10-fold lower in the dsbA mutant than in wild-type PAO1. This finding suggested that elastase was either not well secreted or in the supernatant in an inactive form due to misfolding. The examination of elastase in stationary-phase culture supernatants using a Western blot analysis showed reduced levels of elastase, suggesting that the dsbA mutation compromised elastase accumulation. However, examination of 30-min washed-cell cultures, which can detect unstable secreted proteins (18), showed that elastase was secreted. Thus, the lack of elastase accumulation in the supernatant over time probably reflects its instability in the absence of proper folding by DsbA. A mutation in dsbA gene was recently shown to play a role in P. aeruginosa virulence in the Caenorhabditis elegans infection model (37), which may be due to the activity of DsbA in the accumulation of extracellular virulence factors like elastase.
We also examined twitching motility, which is mediated by type IV fimbriae. These fimbriae are composed of pilin subunits that contain an intrastrand disulfide loop (DSL). It has been shown that insertions in the DSL do not compromise assembly of the pilin subunits in the outer membrane but that they do affect binding to epithelial cells (14). We hypothesized that a dsbA mutation was likely to affect the formation of DSL and hence the ability of pilin to bind a solid surface, which would affect the twitching motility phenotype. Indeed, our results showed that a dsbA mutant exhibited a twitching motility range that was about 60% of that of the wild type.
Since elevated DsbA protein levels and dsbA gene transcription were associated with the mucoid phenotype of a mucA22 strain, we were curious as to whether elevated DsbA was important for alginate overproduction. A periplasmic complex of proteins appears to be required for alginate secretion (16), and some of these proteins (e.g., AlgK and AlgL) contain disulfide bonds. Thus, we expected that the presence of DsbA would be important for alginate production. Instead, we found that the complete loss of DsbA had little effect on alginate production under normal laboratory conditions. This finding suggests that the components of the complex necessary for polymer secretion can be maintained by other oxidoreductases in a dsbA mutant of P. aeruginosa.
Overall, this study demonstrates the power of a proteome analysis to discover the role of a regulator like ς22 in global protein expression. Our discovery that dsbA, a gene apparently unrelated to alginate production, is coregulated with mucoid conversion is further evidence that ς22 is part of a global stress response that includes more than alginate production. Further studies will examine whether dsbA is under direct or indirect control of ς22 and the role of other regulators associated with alginate production.
ACKNOWLEDGMENTS
We thank Sang-Jin Suh and for helpful discussions, and Joanne Johnston for assistance with several assays during the course of these studies. We also acknowledge the Pathogenesis Corporation for supplying genomic sequence data at www.pseudomonas.com from the P. aeruginosa genome sequencing project.
This work was supported by Public Health Service grants AI-19146 and AI-26187 from the National Institute of Allergy and Infectious Diseases (D.E.O.) and in part by Veterans Administration Medical Research funds (D.E.O.).
REFERENCES
- 1.Alm R, Mattick J. Identification of a gene, pilV, required for type 4 fimbrial biogenesis in Pseudomonas aeruginosa, whose product possesses a pre-pilin-like leader sequence. Mol Microbiol. 1995;16:485–496. doi: 10.1111/j.1365-2958.1995.tb02413.x. [DOI] [PubMed] [Google Scholar]
- 2.Antelmann H, Bernhardt J, Schmid R, Mach H, Volker U, Hecker M. First steps from a two-dimensional protein index towards a response-regulation map for Bacillus subtilis. Electrophoresis. 1997;18:1451–1463. doi: 10.1002/elps.1150180820. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: Greene Publishing Associates, Inc., and John Wiley & Sons, Inc.; 1993. [Google Scholar]
- 4.Bardwell J, McGovern K, Beckwith J. Identification of a protein required for disulfide bond formation in vivo. Cell. 1991;67:581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- 5.Baynham P, Wozniak D. Identification and characterization of AlgZ, an AlgT-dependent DNA-binding protein required for Pseudomonas aeruginosa algD transcription. Mol Microbiol. 1996;22:97–108. doi: 10.1111/j.1365-2958.1996.tb02659.x. [DOI] [PubMed] [Google Scholar]
- 6.Boucher J C, Yu H, Mudd M H, Deretic V. Mucoid Pseudomonas aeruginosa in cystic fibrosis: characterization of muc mutations in clinical isolates and analysis of clearance in a mouse model of respiratory infection. Infect Immun. 1997;65:3838–3846. doi: 10.1128/iai.65.9.3838-3846.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng K J, Ingram J M, Costerton J W. Interactions of alkaline phosphatase and the cell wall of Pseudomonas aeruginosa. J Bacteriol. 1971;107:325–336. doi: 10.1128/jb.107.1.325-336.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chitnis C E, Ohman D E. Genetic analysis of the alginate biosynthetic gene cluster of Pseudomonas aeruginosa shows evidence of an operonic structure. Mol Microbiol. 1993;8:583–590. doi: 10.1111/j.1365-2958.1993.tb01602.x. [DOI] [PubMed] [Google Scholar]
- 9.de Lorenzo V, Herrero M, Jakubzik U, Timmis K. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeVries C A, Ohman D E. Mucoid to nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in algT, encoding a putative alternative sigma factor, and shows evidence for autoregulation. J Bacteriol. 1994;176:6677–6687. doi: 10.1128/jb.176.21.6677-6687.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duchêne M, Schweizer A, Lottspeieh F, Krauss G, Marget M, Vogel K, von Specht B-U, Domdey H. Sequence and transcriptional start site of the Pseudomonas aeruginosa outer membrane porin protein F gene. J Bacteriol. 1988;170:155–162. doi: 10.1128/jb.170.1.155-162.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filloux A, Bally M, Soscia C, Murgier M, Lazdunski A. Phosphate regulation in Pseudomonas aeruginosa: cloning of the alkaline phosphatase gene and identification of phoB- and phoR-like genes. Mol Gen Genet. 1988;212:510–513. doi: 10.1007/BF00330857. [DOI] [PubMed] [Google Scholar]
- 13.Govan J R W, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahn H P. The type-4 pilus in the major virulence-associated adhesion of Pseudomonas aeruginosa—a review. Gene. 1997;192:99–108. doi: 10.1016/s0378-1119(97)00116-9. [DOI] [PubMed] [Google Scholar]
- 15.Holloway B W, Krishnapillai V, Morgan A F. Chromosomal genetics of Pseudomonas. Microbiol Rev. 1979;43:73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jain S, Ohman D E. Deletion of algK in mucoid Pseudomonas aeruginosa blocks alginate polymer formation and results in uronic acid secretion. J Bacteriol. 1998;180:634–641. doi: 10.1128/jb.180.3.634-641.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kessler E, Safrin M, Abrams W R, Rosenbloom J, Ohman D E. Inhibitors and specificity of Pseudomonas aeruginosa LasA. J Biol Chem. 1997;272:9884–9889. doi: 10.1074/jbc.272.15.9884. [DOI] [PubMed] [Google Scholar]
- 18.Kessler E, Safrin M, Gustin J K, Ohman D E. Elastase and LasA protease of Pseudomonas aeruginosa are secreted with their propeptides. J Biol Chem. 1999;273:30225–30231. doi: 10.1074/jbc.273.46.30225. [DOI] [PubMed] [Google Scholar]
- 19.Knutson C A, Jeanes A. A new modification of the carbazole analysis: application to heteropolysaccharides. Anal Biochem. 1968;24:470–481. doi: 10.1016/0003-2697(68)90154-1. [DOI] [PubMed] [Google Scholar]
- 20.Link A, Hays L, Carmack E, Yates J., III Identifying the major proteome components of Haemophilus influenzae type-strain NCTC 8143. Electrophoresis. 1997;18:1314–1334. doi: 10.1002/elps.1150180808. [DOI] [PubMed] [Google Scholar]
- 21.Martin D W, Schurr M J, Mudd M H, Govan J R W, Holloway B W, Deretic V. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc Natl Acad Sci USA. 1993;90:8377–8381. doi: 10.1073/pnas.90.18.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathee K, Ciofu O, Sternberg C K, Lindum P, Campbell J, Jensen P, Johnsen A, Givskov M, Ohman D, Molin S, Høiby N, Kharazmi A. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology. 1999;145:1349–1357. doi: 10.1099/13500872-145-6-1349. [DOI] [PubMed] [Google Scholar]
- 23.Mathee K, McPherson C J, Ohman D E. Posttranslational control of the algT (algU)-encoded ς22 for expression of the alginate regulon in Pseudomonas aeruginosa and localization of its antagonist proteins MucA and MucB (AlgN) J Bacteriol. 1997;179:3711–3720. doi: 10.1128/jb.179.11.3711-3720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McIver K S, Kessler E, Olson J C, Ohman D E. The elastase propeptide functions as an intramolecular chaperone required for elastase activity and secretion in Pseudomonas aeruginosa. Mol Microbiol. 1995;18:877–889. doi: 10.1111/j.1365-2958.1995.18050877.x. [DOI] [PubMed] [Google Scholar]
- 25.McIver K S, Olson J C, Ohman D E. Pseudomonas aeruginosa lasB1 mutants produce an elastase, substituted at active-site His-223, that is defective in activity, processing, and secretion. J Bacteriol. 1993;175:4008–4015. doi: 10.1128/jb.175.13.4008-4015.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Missiakas D, Raina S. The extracytoplasmic function sigma factors: role and regulation. Mol Microbiol. 1998;28:1059–1066. doi: 10.1046/j.1365-2958.1998.00865.x. [DOI] [PubMed] [Google Scholar]
- 27.Missiakas D, Raina S. Protein folding in the bacterial periplasm. J Bacteriol. 1997;179:2465–2471. doi: 10.1128/jb.179.8.2465-2471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Farrel P. High-resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 29.Pasquali C, Frutiger S, Wilkins M R, Hughes G J, Appel R D, Bairoch A, Schaller D, Sanchez J-C, Hochstrasser D F. Two-dimensional gel electrophoresis of Escherichia coli homogenates: the Escherichia coli SWISS-2DPAGE database. Electrophoresis. 1996;17:547–555. doi: 10.1002/elps.1150170325. [DOI] [PubMed] [Google Scholar]
- 30.Porankiewicz J, Wang J, Clarke A K. New insights into the ATP-dependent Clp protease: Escherichia coli and beyond. Mol Microbiol. 1999;32:449–458. doi: 10.1046/j.1365-2958.1999.01357.x. [DOI] [PubMed] [Google Scholar]
- 31.Rouviere P E, De Las Penas A, Mecsas J, Lu C Z, Rudd K E, Gross C A. rpoE, the gene encoding the second heat-shock sigma factor, ςE, in Escherichia coli. EMBO J. 1995;14:1032–1042. doi: 10.1002/j.1460-2075.1995.tb07084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schurr M J, Yu H, Martinez-Salazar J M, Boucher J C, Deretic V. Control of AlgU, a member of the sigma E-like family of stress sigma factors, by the negative regulators MucA and MucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J Bacteriol. 1996;178:4997–5004. doi: 10.1128/jb.178.16.4997-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schweizer H P. Small broad-host-range gentamicin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques. 1993;15:831–833. [PubMed] [Google Scholar]
- 34.Selvaraj G, Fong Y C, Iyer V N. A portable DNA sequence carrying the cohesive site (cos) of bacteriophage λ and the mob (mobilization) region of the broad-host-range plasmid RK2: a module for the construction of new cosmids. Gene. 1984;32:235–241. doi: 10.1016/0378-1119(84)90051-9. [DOI] [PubMed] [Google Scholar]
- 35.Stover C, Pham X, Erwin A, Mizoguchi S, Warrener P, Hickey M, Brinkman F, Hufnagle W, Kowalik D, Lagrou M, Garber R, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody L, Coulter S, Folger K, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong G K-S, Wu Z, Paulsenk I, Reizer J, Saier M, Hancock R, Lory S, Olson M. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 36.Suh S-J, Silo-Suh L, Woods D, Hassett D, West S, Ohman D. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J Bacteriol. 1999;181:3890–3897. doi: 10.1128/jb.181.13.3890-3897.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan M-W, Rahme L, Sternberg J, Tompkins R, Ausubel F. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc Natl Acad Sci USA. 1999;96:2408–2413. doi: 10.1073/pnas.96.5.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thayer M M, Flaherty K M, McKay D B. Three-dimensional structure of the elastase of Pseudomonas aeruginosa at 1.5-Å resolution. J Biol Chem. 1991;266:2864–2871. doi: 10.2210/pdb1ezm/pdb. [DOI] [PubMed] [Google Scholar]
- 39.Wozniak D J, Ohman D E. Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB, and algD reveals a hierarchy of alginate gene expression which is modulated by algT. J Bacteriol. 1994;176:6007–6014. doi: 10.1128/jb.176.19.6007-6014.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu J. Inactivation of DsbA, but not DsbC and DsbD, affects intracellular survival and virulence of Shigella flexneri. Infect Immun. 1998;66:3909–3917. doi: 10.1128/iai.66.8.3909-3917.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]