Abstract
The Saccharomyces cerevisiae ICL1 gene encodes isocitrate lyase, an essential enzyme for growth on ethanol and acetate. Previous studies have demonstrated that the highly homologous ICL2 gene (YPR006c) is transcribed during the growth of wild-type cells on ethanol. However, even when multiple copies are introduced, ICL2 cannot complement the growth defect of icl1 null mutants. It has therefore been suggested that ICL2 encodes a nonsense mRNA or nonfunctional protein. In the methylcitrate cycle of propionyl-coenzyme A metabolism, 2-methylisocitrate is converted to succinate and pyruvate, a reaction similar to that catalyzed by isocitrate lyase. To investigate whether ICL2 encodes a specific 2-methylisocitrate lyase, isocitrate lyase and 2-methylisocitrate lyase activities were assayed in cell extracts of wild-type S. cerevisiae and of isogenic icl1, icl2, and icl1 icl2 null mutants. Isocitrate lyase activity was absent in icl1 and icl1 icl2 null mutants, whereas in contrast, 2-methylisocitrate lyase activity was detected in the wild type and single icl mutants but not in the icl1 icl2 mutant. This demonstrated that ICL2 encodes a specific 2-methylisocitrate lyase and that the ICL1-encoded isocitrate lyase exhibits a low but significant activity with 2-methylisocitrate. Subcellular fractionation studies and experiments with an ICL2-green fluorescent protein fusion demonstrated that the ICL2-encoded 2-methylisocitrate lyase is located in the mitochondrial matrix. Similar to that of ICL1, transcription of ICL2 is subject to glucose catabolite repression. In glucose-limited cultures, growth with threonine as a nitrogen source resulted in a ca. threefold induction of ICL2 mRNA levels and of 2-methylisocitrate lyase activity in cell extracts relative to cultures grown with ammonia as the nitrogen source. This is consistent with an involvement of the 2-methylcitrate cycle in threonine catabolism.
The complete sequencing of the Saccharomyces cerevisiae genome has yielded a large number of open reading frames with unknown function (11). Some of the newly discovered open reading frames exhibited a strong homology with known yeast genes and were demonstrated to encode hitherto-unknown isoenzymes. An example is the PYK2 gene, which encodes a pyruvate kinase isoenzyme but can restore growth of pyk1 null mutants on glucose only when overexpressed (3). In other cases, the biochemical function of the proteins (if any) encoded by the homologous open reading frames remains unknown.
An intriguing case is presented by the ICL2 gene. This gene (YPR006c) exhibits a substantial sequence similarity with ICL1 (13), the unique S. cerevisiae structural gene encoding isocitrate lyase (38% identity at the amino acid level). Isocitrate lyase is a key enzyme of the glyoxylate cycle. As this pathway is essential for growth on acetate and ethanol, icl1 null mutants are unable to grow on ethanol or acetate (9, 28). ICL2 is transcribed in ethanol-grown cultures of wild-type S. cerevisiae, and experiments with ICL2-lacZ fusions indicated that its transcriptional regulation is similar to that of ICL1 (13). However, even the introduction of multiple copies of ICL2 cannot complement the growth deficiency of icl1 null mutants (13). This indicates that ICL2 does not encode a functional isocitrate lyase. Since, furthermore, icl2 null mutants have not been found to exhibit a discernible phenotype (13), the physiological function of ICL2 has so far remained an enigma.
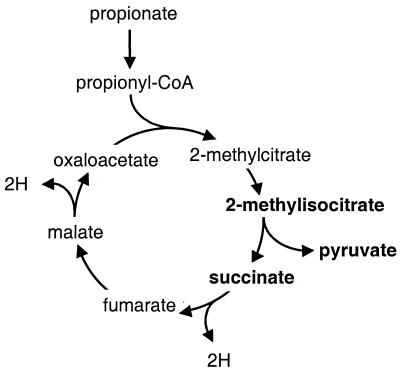
A reaction analogous to the conversion of isocitrate to glyoxylate and succinate occurs in the metabolism of propionyl-coenzyme A (CoA) via the 2-methylcitrate cycle (Fig. 1). This pathway was first discovered in alkane- and lipid-metabolizing yeasts (31, 32). An erroneous assumption about the pathway of propionyl-CoA metabolism in S. cerevisiae has caused some confusion. Based on the assumption that propionyl-CoA metabolism in this yeast involves methyl-malonyl-CoA as an intermediate, results from experiments with 13C-labeled propionate were interpreted as proof of the occurrence of tight channeling of tricarboxylic acid (TCA) cycle intermediates (30). It was later shown that, instead, the methylcitrate cycle is the key pathway of propionate metabolism (23). The methylcitrate cycle is initiated by the synthesis of 2-methylcitrate from propionyl-CoA and oxaloacetate. 2-Methylcitrate is then converted into 2-methylisocitrate, which is subsequently split into pyruvate and succinate. The latter reaction is very similar to the conversion of isocitrate to succinate and glyoxylate, the reaction catalyzed by the ICL1-encoded isocitrate lyase.
FIG. 1.
2-Methylcitrate cycle of propionyl-CoA metabolism (31, 32). The reaction catalyzed by 2-methylisocitrate lyase is shown in bold.
The physiological function of the methylcitrate cycle in S. cerevisiae is not entirely clear. S. cerevisiae cannot grow on propionate as a sole carbon source, but in aerobic sugar-limited chemostat cultures, propionate can be cometabolized (23). It is also conceivable that this pathway may be involved in the degradation of the carbon skeletons of certain amino acids. For example, oxidative decarboxylation of 2-ketobutyrate, an intermediate in threonine catabolism, yields propionyl-CoA (19).
In the present study, we tested the hypothesis that the S. cerevisiae ICL2 gene encodes a specific 2-methylisocitrate lyase. Furthermore, the regulation of ICL2 expression and the subcellular localization and physiological function of Icl2p were investigated.
MATERIALS AND METHODS
Yeast strains and maintenance.
All S. cerevisiae strains used in this study are prototrophic members of the CEN.PK series and are described in Table 1. The strains were grown to stationary phase at 30°C in shake flask cultures on YPD medium (Difco yeast extract, 10 g per liter; Difco peptone, 20 g per liter; glucose, 20 g per liter). Subsequently, glycerol (20%, vol/vol) was added and 2-ml aliquots were stored at −70°C in sterile vials. Precultures were inoculated directly from these frozen stocks.
TABLE 1.
S. cerevisiae strains used in this studya
| Strain | Genotype |
|---|---|
| CEN.PK113-7D | MATa URA3 HIS3 LEU2 TRP1 MAL2-8c SUC2 |
| CEN.PK122 | MATa/MATα URA3/URA3 HIS3/HIS3 LEU2/LEU2 TRP1/TRP1 MAL2-8c/MAL2-8c SUC2/SUC2 |
| CEN.PK229-4D | URA3 HIS3 LEU2 TRP1 MAL2-8c SUC2 icl1Δ |
| CEN.PK288-1C | URA3 HIS3 LEU2 TRP1 MAL2-8c SUC2 icl2Δ |
| CEN.PK321-2D | URA3 HIS3 LEU2 TRP1 MAL2-8c SUC2 icl1Δ icl2Δ |
| CEN.PK525-7D | MATa URA3 HIS3 LEU2 TRP1 MAL2-8c SUC2 ICL1 ICL2::yEGFP3-loxP-KanMX-loxP |
All strains listed can be obtained from EUROSCARF (European Saccharomyces cerevisiae Archive for Functional Analysis) at http://www.rz.uni-frankfurt.de/FB /fb16/mikro/euroscarf/index.html.
Construction of null mutants.
Haploid S. cerevisiae null mutants were constructed by replacing the gene(s) of interest with a kanamycin resistance gene (the kanMX module) according to the PCR-based method of Wach et al. (39) as described previously (17). Mating type and replacement of genes by the kanMX module were verified by PCR as described previously (17). The sequences of the primers that were used for deletion (S1 and S2) and verification (A1, A4, K1, and K2) are listed in Table 2.
TABLE 2.
Oligonucleotides used for construction of disruption cassettes (S1 and S2) and as primers for analytical PCR of deletion mutants (A1-K1 and A4-K2)a
| Gene | Oligonucleotide | Sequence |
|---|---|---|
| ICL1 | S1 | TGCCTATCCCCGTTGGAAATACGAAGAACGATTTTGCAGCAGCTGAAGCTTCGTACGC |
| ICL1 | S2 | TTCTTTACGCCATTTTCTTTGAATTGATCTTCTGTGACACGCATAGGCCACTAGTGGATCTG |
| ICL1 | A1 | GGAGAACTTCTAGCACGTTG |
| ICL1 | A2 | CGGCAGCATCTGCATCTAG |
| ICL1 | A3 | TCTGGTGCGGAGTACATCG |
| ICL1 | A4 | GTGTGTGTACATGTATGCGG |
| ICL2 | S1 | GGAACACTGAAGAAGCTGGTTTTGTCATCAGACAAGTCACCAGCTGAAGCTTCGTACGC |
| ICL2 | S2 | AAATTGCGTTTCGGTAAAGCTCTCACCAGATGTACTTAAAGCATAGGCCACTAGTGGATCTG |
| ICL2 | A1 | CAAGTAGTACCAGGAGTCACG |
| ICL2 | A2 | CTTGATTCCGAGAACACGAGG |
| ICL2 | A3 | CAGCTATGGTCTGGTGCAG |
| ICL2 | A4 | GAGAGTGTAGTCATGCAATCC |
| kanMX | K1 | GGATGTATGGGCTAAATGTACG |
| kanMX | K2 | GTTTCATTTGATGCTCGATGAG |
Mineral medium.
The mineral medium used for batch and chemostat experiments contained the following per liter of demineralized water: (NH4)2SO4, 5 g; KH2PO4, 3 g; MgSO4 · 7H2O, 0.5 g; EDTA, 15 mg; ZnSO4 · 7H2O, 4.5 mg; CoCl2 · 6H2O, 0.3 mg; MnCl2 · 2H2O, 0.84 mg; CuSO4 · 5H2O, 0.3 mg; CaCl2 · 2H2O, 4.5 mg; FeSO4 · 7H2O, 3.0 mg; Na2MoO4 · 2H2O, 0.4 mg; H3BO3, 1.0 mg; KI, 0.1 mg; and silicone antifoam (BDH), 0.15 ml. After autoclaving (120°C, 20 min), the medium was cooled to room temperature. Subsequently, filter-sterilized vitamins were added to the following final concentrations (per liter): biotin, 0.05 mg; calcium pantothenate, 1.0 mg; nicotinic acid, 1.0 mg; myo-inositol, 25.0 mg; thiamine-HCl, 1.0 mg; pyridoxol-HCl, 1.0 mg; and para-aminobenzoic acid, 0.2 mg. Glucose was sterilized separately for 20 min at 110°C, and ethanol was added without separate sterilization. Carbon substrates were added to a concentration of 250 mM carbon unless indicated otherwise. When l-threonine was used as a nitrogen source, the ammonium sulfate was replaced by an equimolar amount (based on nitrogen content) of this amino acid. To compensate for the reduced sulfate content of these media, 6.6 g of K2SO4 per liter was added as well. The mineral medium used for ammonium-limited chemostat cultivation contained a fivefold-reduced concentration of (NH4)2SO4 (1.0 g per liter) and a fivefold-increased glucose concentration (37.5 g per liter, corresponding to 1.25 M carbon) and was supplemented with 5.3 g of K2SO4 per liter. Threonine-limited cultures were grown on the same medium with 1.8 g of l-threonine per liter instead of ammonium sulfate.
Shake flask cultivation.
Shake flask cultures were grown at 30°C in 500-ml Erlenmeyer flasks on an orbital shaker (200 rpm). Precultures were prepared by inoculating 100 ml of YPD medium with a frozen-stock culture. After 48 h of incubation, a 1-ml sample was inoculated in a 500-ml Erlenmeyer flask containing 100 ml of mineral medium (pH 5.5). The composition of the mineral medium was as specified above, with the following modifications: ammonium sulfate was omitted and 30 mmol of l-aspartate and 62 mmol of ethanol per liter were added as carbon/nitrogen and carbon sources, respectively. After 24 h of incubation, a 2-ml sample was used to inoculate a second 100-ml culture on the same medium. This was again incubated for 24 h and subsequently used to prepare cell extracts for enzyme assays.
Chemostat cultivation.
Chemostat cultivation was performed at 30°C in laboratory fermentors (Applikon, Schiedam, The Netherlands). The working volume of the cultures was kept at 1.0 liters by a peristaltic effluent pump coupled to an electrical level sensor; the stirrer speed was 800 rpm. The pH was kept constant at 5.0 by an ADI 1020 biocontroller via the automatic addition of 2 mol of KOH per liter. The gas flow through the cultures was maintained at 0.5 liters per min using a Brooks 5876 mass flow controller. The dissolved oxygen concentration was monitored with an autoclavable oxygen electrode (no. 34 100 3002; Ingold). Cultures were assumed to be in steady state when, after a change of growth conditions, at least five volume changes had passed and two subsequent samples taken at an interval of at least one volume change gave identical results for dry weight, CO2 production rate, and O2 consumption rate. Cultures were checked for purity using phase-contrast microscopy (×1,000 magnification) and by plating on YPD agar plates.
Gas analysis.
The exhaust gas was cooled in a condenser (4°C) and dried in a Perma Pure dryer (PD-625-12P). O2 consumption was determined with a Servomex 1100A oxygen analyzer (Taylor Servomex Co., Crowborough, United Kingdom). CO2 production by the cultures was determined with a Beckman model 864 infrared detector. The CO2 production and O2 consumption rates were calculated according to the method of van Urk et al. (37).
Determination of culture dry weight.
The dry weight of 10.0-ml culture samples was determined using 0.45-μm-pore-size nitrocellulose filters (Gelman Sciences) and a microwave oven (22). Duplicate samples varied by less than 1%.
Metabolite analysis.
The concentration of glucose in reservoir media and supernatants was determined enzymatically using the UV method for d-glucose (no. 716 251; Boehringer Mannheim). The concentration of ethanol was determined enzymatically (38) using Hansenula polymorpha alcohol oxidase (kindly provided by Bird Engineering, Schiedam, The Netherlands). The concentration of other metabolites (organic acids and glycerol) were detected by high-performance liquid chromatography (Waters Alliance coupled to a dual-wavelength-absorbance and refractive-index detector) analysis on an Aminex HPX-87H column (Bio-Rad). The column was eluted at 60°C with 0.5 g of H2SO4 per liter at a flow rate of 0.6 ml · min−1.
Preparation of cell extracts.
Cells from shake flask cultures or chemostat cultures (ca. 80 mg [dry weight]) were harvested by centrifugation (4,000 × g, 10 min), washed once with 100 mM potassium phosphate buffer, pH 7.5, containing 2 mM MgCl2 and 1 mM dithiothreitol (4°C), and resuspended in the same buffer. Cells were disrupted by sonication with 0.7-mm glass beads (0°C, 3 min; 30-s bursts with 30-s cooling intervals) using an MSE sonicator (150-W output, 7- to 8-μm peak-to-peak amplitude). Whole cells and debris were removed by centrifugation at 20,000 × g for 20 min at 4°C. The clear supernatant was used as the cell extract. Protein concentrations in cell extracts were determined by the Lowry method. Bovine serum albumin (fatty acid free; Sigma, St. Louis, Mo.) was used as a standard.
Enzyme assays.
Enzyme activities were assayed at 30°C in a Hitachi model 100-60 spectrophotometer with freshly prepared cell extracts. The following enzymes were assayed according to previously published procedures: glucose-6-phosphate-dehydrogenase (EC 1.1.1.49 [22]), cytochrome-c oxidase (EC 1.9.3.1 [8]), citrate synthase and isocitrate lyase (EC 4.1.3.7 and EC 4.1.3.1 [6]), and catalase (EC 1.11.1.6 [35]). 2-Methylisocitrate lyase activity was assayed in a reaction mixture (1 ml) containing potassium phosphate buffer (pH 7.0) (100 μmol), phenylhydrazine (4 μmol), cysteine (2.5 μmol), MgCl2 (2.5 μmol), and cell extract. The reaction was started by addition of 2-methylisocitrate (2 μmol). The molar extinction coefficient of pyruvate phenylhydrazone was experimentally determined to be 12 mM−1 · cm−1 (data not shown). Control experiments in which the rate of pyruvate production from 2-methylisocitrate was coupled to a NADH-linked lactate dehydrogenase gave identical specific activities (data not shown). In all enzyme assays, reaction rates were linearly proportional to the amount of cell extract added.
Northern experiments.
Total RNA was extracted as previously described (26). Amplified PCR fragments of ACT1, ICL1, and ICL2 were used as probes for Northern analysis. A ready-to-go DNA labeling kit (Pharmacia Biotech Europe) was used for DNA [α-32P]dCTP radiolabeling. Total RNA (30 μg/lane) was separated on 1.2% agarose-formaldehyde gels, blotted to an Amersham Hybond-N membrane, and hybridized overnight at 42°C (27). After being washed, membranes were exposed to Kodak X-Omat MR film and incubated with an intensifying screen at −70°C.
Isolation of organelles.
An organellar fraction was isolated from aerobic, glucose-limited chemostat cultures grown with threonine as the nitrogen source. The procedure, which involved differential centrifugation of cell homogenates obtained by controlled lysis of spheroplasts, has been described previously (17). To investigate the latency of enzymes in organellar fractions, enzyme activities were first measured in the presence of 0.65 mol of sorbitol per liter to osmotically stabilize the organelles. Subsequently, 0.1% Triton X-100 was added to disrupt the organelles and to measure intraorganellar enzyme activity. In independent control experiments, organelles were disrupted by sonication (150-W output, 30 s).
Sucrose density gradient centrifugation of organellar fractions.
Cells were harvested by centrifugation at room temperature (1,600 × g, 5 min). For the generation of spheroplasts, cells were resuspended (0.1 g [wet weight] · ml−1) in 0.1 M Tris buffer (pH 9.3) containing 10 mM dithiothreitol and incubated at 30°C for 10 min. After centrifugation (1,600 × g, 5 min), cells were washed once in a 50 mM potassium phosphate buffer (pH 7.2) containing 1.2 M sorbitol, resuspended in the same buffer containing 0.5 mg of Zymolyase 20T (ICN Biomedicals BV, Zoetermeer, The Netherlands) per ml, and incubated at 30°C for approximately 30 min. All subsequent steps were performed at 4°C. Spheroplasts were collected by centrifugation (2,800 × g, 7 min), washed in 5 mM morpholineethanesulfonic acid (MES) buffer (pH 5.5) containing 1.2 M sorbitol, and osmotically lysed by resuspension in 5 mM MES buffer (pH 5.5) containing 0.8 M sorbitol, 1 mM phenylmethylsulfonyl fluoride, and 2.5 mg of leupeptin per ml. After homogenization using a Potter-Elvehjem homogenizer, the suspension was adjusted to 1.2 M sorbitol by addition of 5 mM MES buffer (pH 5.5) containing 3.0 M sorbitol. The homogenate was subjected to two consecutive centrifugation runs (2,000 × g, 10 min, and 7,800 × g, 15 min). The resulting postnuclear supernatant was centrifuged (30,000 × g, 30 min) in order to obtain an organellar fraction, which was subsequently resuspended in 5 mM MES buffer (pH 5.5) containing 35% (wt/vol) sucrose. The organellar fraction was loaded onto a discontinuous sucrose density gradient consisting of 5 mM MES buffer (pH 5.5) containing 65% (wt/vol) sucrose (5 ml), 50% (wt/vol) sucrose (6 ml), 45% (wt/vol) sucrose (6 ml), 40% (wt/vol) sucrose (6 ml), sample, and 25% (wt/vol) sucrose (2-ml overlay). After centrifugation of the gradient in a vertical rotor (30,000 × g, 3 h), fractions of approximately 1.25 ml were taken from the gradient, starting at the highest sucrose concentration.
Experiments with an Icl-GFP fusion.
To fuse the C terminus of Icl2p with the green fluorescent protein (GFP) gene, the yEGFP2-loxP-KanMX-loxP cassette from pUG30 (kindly provided by H. Hegemann, University of Düsseldorf, Düsseldorf, Germany) was amplified by PCR. The resulting construct, with ends homologous to the 3′ end of the ICL2 gene, was then integrated at the ICL2 locus of the wild-type strain, CEN.PK113-7D. Analysis of cells by fluorescence microscopy (using a Zeiss fluorescence microscope) and fixation and preparation for electron microscopy were performed as described previously (1, 40). Immunolabeling was performed on ultrathin sections of unicryl-embedded cells using specific antibodies against GFP and gold-conjugated goat anti-rabbit antibodies (40).
Sequence analysis.
A search for a potential mitochondrial targeting sequence in Icl2p was performed with the PcGene software package (version 6.80; Intelligenetics, Mountain View, Calif.). This program is based on the method of Gavel and von Heijne (10).
RESULTS
ICL2 encodes a specific 2-methylisocitrate lyase.
To investigate whether the ICL2 gene encodes a specific 2-methylisocitrate lyase, activities of isocitrate lyase and 2-methylisocitrate lyase were determined in cell extracts of wild-type S. cerevisiae and isogenic icl1Δ, icl2Δ, and icl1Δ icl2Δ mutants. All strains were grown on ethanol in shake flask cultures to induce ICL2 transcription (13). Aspartate was used as the sole nitrogen source and as an additional carbon source to circumvent the growth deficiency of icl1 null mutants on ethanol-ammonia media (9, 28).
Cell extracts of the wild-type strain exhibited substantial activities of isocitrate lyase as well as 2-methylisocitrate lyase (Table 3). Inactivation of the ICL1 gene completely abolished isocitrate lyase activity, confirming the earlier report that ICL2 does not encode a functional isocitrate lyase (13). When, in addition to ICL1, ICL2 also was deleted, the resulting strain completely lacked 2-methylisocitrate lyase activity as well as isocitrate lyase activity (Table 3). An icl2 null mutant, which still exhibited high isocitrate lyase activity, also retained a residual activity of 2-methylisocitrate lyase (Table 3). These results demonstrate that the S. cerevisiae ICL2 gene encodes a specific 2-methylisocitrate lyase and that the ICL1-encoded enzyme can utilize both isocitrate and 2-methylisocitrate as substrates.
TABLE 3.
Activity of isocitrate lyase and 2-methylisocitrate lyase in cell extracts of wild-type S. cerevisiae and of isogenic null mutants lacking ICL1, ICL2, or botha
| Strain | Genotype | Activity (μmol · min−1 · mg of protein−1) of:
|
me-ICL/ICL | |
|---|---|---|---|---|
| ICL | me-ICL | |||
| CEN.PK113-7D | ICL1 ICL2 | 0.10 | 0.046 | 0.5 |
| CEN.PK 229-4D | icl1Δ ICL2 | <0.002 | 0.075 | >37 |
| CEN.PK 321-2D | icl1Δ icl2Δ | <0.002 | <0.002 | NAb |
| CEN.PK 288-1C | ICL1 icl2Δ | 0.083 | 0.016 | 0.2 |
All strains were grown in shake flask cultures on ethanol-aspartate medium. Data are from one set of shake flask cultures; activities in independent replicate experiments differed by less than 20%. ICL, isocitrate lyase; me-ICL, 2-methylisocitrate.
NA, not applicable.
ICL2 expression is repressed by glucose and induced by threonine.
Regulation of ICL2 expression was studied in steady-state chemostat cultures of the wild-type strain, S. cerevisiae CEN.PK113-7D, grown at a fixed specific growth rate of 0.10 h−1, by standard Northern analysis and by assaying the levels of isocitrate lyase and 2-methylisocitrate lyase in cell extracts.
Consistent with the results of shake flask cultures (Table 3), aerobic ethanol-limited chemostat cultures exhibited a substantial activity of isocitrate lyase as well as of 2-methylisocitrate lyase (Table 4). Under this cultivation condition, both ICL genes yielded a clearly detectable signal on Northern blots (Fig. 2). Aerobic cultivation on glucose as the growth-limiting substrate yielded a circa 15-fold-lower isocitrate lyase activity than growth on ethanol (Table 4) and on Northern blots yielded only a faint signal of the ICL1 transcript (Fig. 2). The activity of 2-methylisocitrate lyase in glucose-limited chemostat cultures was circa threefold lower than that in the ethanol-limited cultures, consistent with a similar difference in ICL2 transcript levels (Table 4 and Fig. 2). Both enzyme activities, as well as the ICL1 and ICL2 transcripts, were undetectable in cultures grown on glucose with ammonium sulfate as the growth-limiting nutrient (Table 4 and Fig. 2). Apparently, the high residual glucose concentrations in such cultures (28.7 g per liter) (data not shown) led to glucose catabolite repression of both ICL genes. Neither enzyme activity nor transcripts were detected in anaerobic glucose-limited cultures.
TABLE 4.
Activities of isocitrate lyase and 2-methylisocitrate lyase in cell extracts of S. cerevisiae CEN.PK113-7D in steady-state chemostat culturesa
| Carbon source | Nitrogen source | Activity (μmol · min−1 · mg of protein−1) of:
|
me-ICL/ICL | |
|---|---|---|---|---|
| ICL | me-ICL | |||
| Ethanol | Ammonia | 0.15 ± 0.02 | 0.09 ± 0.00 | 0.63 |
| Glucose | Ammonia | 0.01 ± 0.00 | 0.03 ± 0.00 | 3 |
| Glucose | Ammonia | <0.002 | <0.002 | NAb |
| Glucose (anaerobic) | Ammonia | <0.002 | <0.002 | NA |
| Glucose | Threonine | <0.002 | 0.10 ± 0.01 | >50 |
| Glucose | Threonine | <0.002 | <0.002 | NA |
The growth-limiting substrates are printed in bold. Unless otherwise indicated, cultures were aerobic (dissolved oxygen concentration above 60% of air saturation). Data are presented as the ± the standard deviations of two independent chemostat cultivations. D = 0.10 h−1.
NA, not applicable.
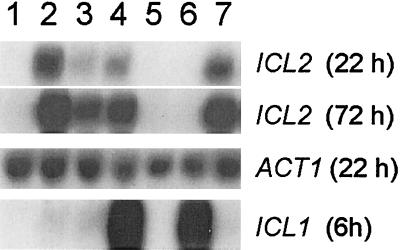
FIG. 2.
Transcriptional regulation of ICL1 and ICL2 in chemostat cultures. Northern blots of mRNA isolated from chemostat cultures (D = 0.10 h−1) grown under various nutrient limitation regimens were hybridized with ICL1 and ICL2 probes as well as with an ACT1 reference probe. Numbers indicate growth conditions as follows: 1, aerobic, threonine-limited cultivation of the wild type with glucose as the carbon source; 2, aerobic, glucose-limited cultivation of the wild type with threonine as the nitrogen source; 3, aerobic, glucose-limited cultivation of the wild type with ammonia as the nitrogen source; 4, aerobic, ethanol-limited cultivation of the wild type with ammonia as the nitrogen source; 5, anaerobic, glucose-limited cultivation of the wild type with ammonia as the nitrogen source; 6, aerobic, ethanol-limited cultivation of the icl2Δ mutant with ammonia as the nitrogen source; 7, aerobic, glucose-limited cultivation of the wild type with threonine as the nitrogen source (independent duplicate of experiment under condition 2). Times in parentheses are autoradiography exposure times.
Propionyl-CoA, the substrate of the 2-methylcitrate cycle in which 2-methylisocitrate lyase participates, may be formed as an intermediate of threonine catabolism. Indeed, glucose-limited cultivation with threonine instead of ammonia as the nitrogen source led to a circa threefold increase of activity of 2-methylisocitrate lyase in cell extracts and of the ICL2 transcript level (Table 4 and Fig. 2). No inducing effect of threonine was observed for expression of ICL1 (Table 4 and Fig. 2). No ICL2 transcript or 2-methylisocitrate lyase activity was detectable during threonine-limited growth on glucose (Table 4 and Fig. 2). As the residual glucose concentration in these cultures, was 13.7 g per liter, this indicates that induction of ICL2 expression by threonine was overruled by glucose catabolite repression.
2-Methylisocitrate lyase is a mitochondrial matrix protein.
Initial subcellular fractionation experiments were performed by differential centrifugation of cell homogenates prepared from glucose-limited chemostat cultures grown with threonine as the nitrogen source. As discussed above, the 2-methylisocitrate lyase in such cultures was exclusively encoded by the ICL2 gene (Table 4). In four independent experiments, 60 to 80% of the 2-methylisocitrate lyase activity in the cell homogenates was recovered in the particulate fraction. A similar incomplete recovery in the particulate fraction was found for citrate synthase, which in S. cerevisiae is known to be confined to mitochondria and/or microbody matrices. For both enzymes, 20 to 40% of the total activity was recovered in the soluble fraction of the homogenates. As the mitochondrial inner membrane protein cytochrome-c oxidase in the same experiments was completely (>90%) recovered in the particulate fraction, the most probable explanation for the incomplete recovery of citrate synthase is that damage to some organelles during the fractionation experiments led to some leakage of matrix enzymes. The cytosolic marker enzyme glucose-6-phosphate dehydrogenase was exclusively recovered in the cytosol.
2-Methylisocitrate lyase activity in organellar fractions exhibited latency. When organelles were osmotically stabilized during the enzyme assays, only a very low activity was measured. This activity increased 18- to 20-fold when organelles were disrupted prior to the enzyme assays by 30 s of sonication or by addition of Triton X-100. This indicated that 2-methylisocitrate lyase is located in an organellar matrix.
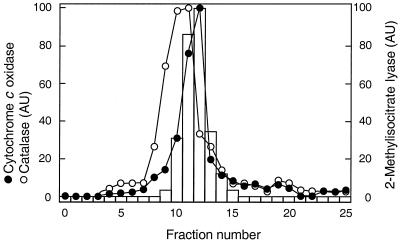
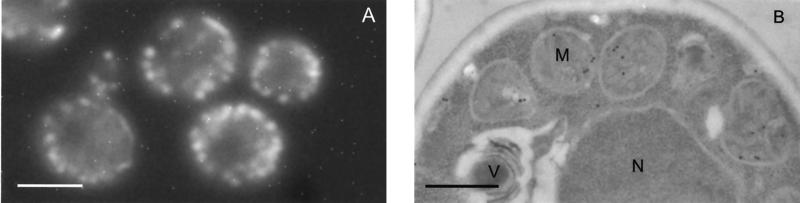
To investigate in which organelle 2-methylisocitrate lyase is located, the particulate fraction of a cell homogenate was subjected to sucrose density gradient centrifugation. In the density gradient centrifugation, catalase and cytochrome-c oxidase were used as marker proteins for the microbody and mitochondrial fractions, respectively (7, 35). These marker proteins showed clearly different sedimentation patterns, with catalase sedimenting at higher average sucrose concentrations than cytochrome-c oxidase (Fig. 3). The sedimentation pattern of 2-methylisocitrate lyase perfectly matched that of the mitochondrial marker enzyme cytochrome-c oxidase, indicating that the ICL2 gene product is a mitochondrial matrix enzyme. To verify this conclusion, the C-terminal end of Icl2p was fused to GFP. Fluorescence microscopy of intact cells as well as immunogold labeling of thin sections confirmed that the fusion protein was targeted to the mitochondrial matrix (Fig. 4). Indeed, analysis of the predicted protein sequence of Icl2p (10) revealed a potential N-terminal mitochondrial transit peptide from positions 1 to 32.
FIG. 3.
Sedimentation patterns of 2-methylisocitrate lyase in sucrose density gradients. An organellar fraction isolated from a glucose-limited, aerobic chemostat culture (D = 0.10 h−1) grown with threonine as the nitrogen source was subjected to sucrose density gradient centrifugation (see Materials and Methods). Fraction 1 corresponds to the bottom fraction of the gradient. Cytochrome-c oxidase and catalase were used as mitochondrial and microbody marker enzymes, respectively. Enzymes are plotted as a percentage of the activity in the peak fraction. A replicate experiment yielded the same results (data not shown). AU, arbitrary units.
FIG. 4.
Localization of an Icl1-GFP fusion. S. cerevisiae CEN.PK525-7D (ICL2::yEGFP3) was pregrown on ethanol-aspartate medium. (A) Fluorescence microscopy. Bar, 2 μm. (B) Electron micrograph of a thin section, labeled with anti-GFP antiserum and goat anti-rabbit antibodies linked to gold particles. Abbreviations: M, mitochondria; N, nucleus; V, vacuole. Specific labeling is exclusively located on the mitochondrial matrix. Bar, 0.5 μm.
DISCUSSION
Since its discovery in alkane- and lipid-metabolizing yeasts (31, 32), the methylcitrate cycle has also been shown to be the key pathway of propionyl-CoA metabolism in the Enterobacteriaceae Escherichia coli and Salmonella enterica serovar Typhimurium (14, 16, 33). Biochemical evidence indicates the existence in lipid- and alkane-metabolizing yeasts of dedicated methylcitrate cycle enzymes different from their counterparts in the TCA and glyoxylate cycles (34). In Enterobacteriaceae, propionate metabolism operons harbor genes encoding key enzymes of the pathway (propionyl-CoA synthetase, 2-methylcitrate synthase, an aconitase, and a 2-methylisocitrate lyase) (14, 33).
The identification of a specific 2-methylisocitrate lyase in S. cerevisiae was surprising, as previous studies seemed to indicate that, in this yeast, the reactions of propionate metabolism can be catalyzed by enzymes involved in the analogous reactions of acetyl-CoA metabolism. In S. cerevisiae, activation of propionate to propionyl-CoA can be catalyzed by the ACS1-encoded isoenzyme of acetyl-CoA synthetase (36), and propionyl-CoA is a substrate for the acetylcarnitine transferase shuttle (21). Likely candidate genes for the 2-methylcitrate synthase-encoding gene are CIT1, CIT2, and CIT3, which all encode active citrate synthase isoenzymes (15). In glucose-limited chemostat cultures fed with increasing concentrations of propionate as a cosubstrate, induction of 2-methylcitrate synthase activity was paralleled by an increase in citrate synthase activity (23). This is consistent with the involvement of one or more of the citrate synthase isoenzymes in the 2-methylcitrate synthase reaction. Candidate structural genes for the aconitase-like enzyme of the 2-methylcitrate cycle include ACO1 (the structural gene for aconitase), YJL200c (the predicted polypeptide product of which exhibits 55% amino acid identity with ACO1 [25]), and LYS4 (the structural gene for homocitrate dehydratase).
McFadden et al. (18) reported that the activities of purified isocitrate lyase from S. cerevisiae with 2-methylisocitrate and isocitrate exhibited a 1-to-5 ratio. This is in good agreement with the ratio of 2-methylisocitrate lyase and isocitrate lyase activities found in cell extracts of an icl2 null mutant (Table 3). Even though the ICL1-encoded isocitrate lyase can catalyze the conversion of 2-methylisocitrate to succinate and pyruvate, our data indicate that S. cerevisiae contains a specific 2-methylisocitrate lyase encoded by ICL2. A factor that may have contributed to the evolution of a highly specific mitochondrial 2-methylisocitrate lyase in S. cerevisiae is that any isocitrate lyase activity of Icl2p could lead to intramitochondrial accumulation of glyoxylate. Glyoxylate, the product of the isocitrate lyase reaction, is an inhibitor of yeast citrate synthase (12). The presence of a mitochondrial 2-methylisocitrate lyase also prevents inhibition of TCA cycle enzymes by the C7 intermediates 2-methylcitrate and 2-methylisocitrate, an effect described for human and bovine TCA cycle enzymes (2, 5).
In contrast to the other microorganisms in which the methylcitrate cycle has been studied, S. cerevisiae cannot grow on propionate as the sole carbon source (23). This is peculiar, since the 2-methylcitrate cycle oxidizes propionyl-CoA to pyruvate (Fig. 1), which does support growth of S. cerevisiae. Growth on propionate requires that part of the pyruvate formed in the 2-methylcitrate cycle be carboxylated to oxaloacetate, a key precursor in biosynthesis. Since, in contrast to the mitochondrial localization of Icl2p, both isoenzymes of pyruvate carboxylase are exclusively cytosolic (24), this requires export of pyruvate from the mitochondria. Biochemical evidence demonstrates that mitochondrial pyruvate import in S. cerevisiae is a carrier-mediated process (20), but it is unknown whether pyruvate transport is reversible in vivo. If pyruvate transport across the mitochondrial inner membrane is unidirectional (i.e., catalyzing only the import of pyruvate from the cytosol), the mitochondrial localization of Icl2p might preclude growth on propionate as the sole carbon source.
The induction of ICL2 expression in glucose-limited cultures (Fig. 2 and Table 4) grown with l-threonine as the nitrogen source suggests that the main physiological role of the methylcitrate cycle in S. cerevisiae is the metabolism of endogenous propionyl-CoA. Conversion of threonine into propionyl-CoA is initiated by its deamination to 2-oxobutyrate, catalyzed by threonine dehydratase (encoded by the CHA1 gene [4]). The subsequent oxidative decarboxylation of 2-oxobutyrate to propionyl-CoA can be catalyzed by the yeast mitochondrial branched-chain 2-oxo-acid dehydrogenase complex (29). From a physiological perspective, it is not illogical that ICL2 is subject to glucose catabolite repression: efficient recovery of the carbon skeletons of amino acids is unlikely to be a major advantage under conditions of glucose excess. Whether additional potential sources of propionyl-CoA other than threonine catabolism, such as the catabolism of isoleucine or β-oxidation of odd-chain fatty acids (19), also feed the methylcitrate cycle of S. cerevisiae remains to be investigated.
ACKNOWLEDGMENTS
We thank C. Kennedy for providing us with 2-methylisocitrate, C. Gancedo for his suggestion to grow the mutant strains on ethanol-aspartate mixtures, J. Kiel for help with the sequence interpretation, and I. Keizer for microscopical analysis.
REFERENCES
- 1.Baerends R J, Faber K N, Kram A M, Kiel J A, van der Klei I J, Veenhuis M. A stretch of positively charged amino acids at the N terminus of Hansenula polymorpha Pex3p is involved in incorporation of the protein into the peroxisomal membrane. J Biol Chem. 2000;275:9986–9995. doi: 10.1074/jbc.275.14.9986. [DOI] [PubMed] [Google Scholar]
- 2.Beach R L, Aogaichi T, Plaut G W. Identification of d-threo-alpha-methylisocitrate as sterochemically specific substrate for bovine heart aconitase and inhibitor of TPN-linked isocitrate dehydrogenase. J Biol Chem. 1977;252:2702–2709. [PubMed] [Google Scholar]
- 3.Boles E, Schulte F, Miosga T, Freidel K, Schlüter E, Zimmermann F K, Hollenberg C P, Heinisch J J. Characterization of a glucose-repressed pyruvate kinase (Pyk2p) in Saccharomyces cerevisiae that is catalytically insensitive to fructose-1,6-bisphosphate. J Bacteriol. 1997;179:2987–2993. doi: 10.1128/jb.179.9.2987-2993.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bornaes C, Petersen J G, Holmberg S. Serine and threonine catabolism in Saccharomyces cerevisiae: the CHA1 polypeptide is homologous with other serine and threonine dehydratases. Genetics. 1992;131:531–539. doi: 10.1093/genetics/131.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheema-Dhadli S, Leznoff C C, Halperin M L. Effect of 2-methylcitrate on citrate metabolism: implications for the management of patients with propionic acidemia and methylmalonic aciduria. Pediatr Res. 1975;9:905–908. doi: 10.1203/00006450-197512000-00008. [DOI] [PubMed] [Google Scholar]
- 6.de Jong-Gubbels P, Vanrolleghem P, Heijnen S, van Dijken J P, Pronk J T. Regulation of carbon metabolism in chemostat cultures of Saccharomyces cerevisiae grown on mixtures of glucose and ethanol. Yeast. 1995;11:407–418. doi: 10.1002/yea.320110503. [DOI] [PubMed] [Google Scholar]
- 7.de Vries S, Marres C A M. The mitochondrial respiratory chain of yeast. Structure and biosynthesis and the role in cellular metabolism. Biochim Biophys Acta. 1987;895:205–239. doi: 10.1016/s0304-4173(87)80003-4. [DOI] [PubMed] [Google Scholar]
- 8.Douma A C, Veenhuis M, de Koning W, Evers M, Harder W. Dihydroxyacetone phosphate synthase is localized in the peroxisomal matrix of methanol-grown Hansenula polymorpha. Arch Microbiol. 1985;143:237–243. [Google Scholar]
- 9.Fernandez E, Moreno F, Rodicio R. The ICL1 gene from Saccaaromyces cerevisiae. Eur J Biochem. 1992;204:983–990. doi: 10.1111/j.1432-1033.1992.tb16720.x. [DOI] [PubMed] [Google Scholar]
- 10.Gavel Y, von Heijne G. Cleavage-site motifs in mitochondrial targeting peptides. Protein Eng. 1990;4:33–37. doi: 10.1093/protein/4.1.33. [DOI] [PubMed] [Google Scholar]
- 11.Goffeau A, Barrell B G, Bussey H, Davis R W, Dujon B, Feldmann H, Galibert F, Hoheisel J D, Jacq C, Johnston M, Louis E J, Mewes H W, Murakami Y, Philippsen P, Tettelin H, Oliver S G. Life with 6000 genes. Science. 1996;274:546–573. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez A, Rodriguez R, Folch J, Soberon M, Olivera H. Coordinated regulation of ammonium assimilation and carbon catabolism by glyoxylate in Saccharomyces cerevisiae. J Gen Microbiol. 1987;133:2497–2501. doi: 10.1099/00221287-133-9-2497. [DOI] [PubMed] [Google Scholar]
- 13.Heinisch J J, Valdes E, Alvarez J, Rodicio R. Molecular genetics of ICL2, encoding a non-functional isocitrate lyase in Saccharomyces cerevisiae. Yeast. 1996;12:1285–1295. doi: 10.1002/(SICI)1097-0061(199610)12:13%3C1285::AID-YEA5%3E3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 14.Horswill A R, Escalante-Semerena J C. Salmonella typhimurium LT2 catabolizes propionate via the 2-methylcitric acid cycle. J Bacteriol. 1999;181:5615–5623. doi: 10.1128/jb.181.18.5615-5623.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia Y K, Becam A M, Herbert C J. The CIT3 gene of Saccharomyces cerevisiae encodes a second mitochondrial isoform of citrate synthase. Mol Microbiol. 1997;24:53–59. doi: 10.1046/j.1365-2958.1997.3011669.x. [DOI] [PubMed] [Google Scholar]
- 16.London R E, Allen D L, Gabel S A, DeRose E F. Carbon-13 nuclear magnetic resonance study of metabolism of propionate by Escherichia coli. J Bacteriol. 1999;181:3562–3570. doi: 10.1128/jb.181.11.3562-3570.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luttik M A H, Overkamp K M, Kötter P, de Vries S, van Dijken J P, Pronk J T. The Saccharomyces cerevisiae NDE1 and NDE2 genes encode separate mitochondrial NADH dehydrogenases catalyzing the oxidation of cytosolic NADH. J Biol Chem. 1998;273:24529–24534. doi: 10.1074/jbc.273.38.24529. [DOI] [PubMed] [Google Scholar]
- 18.McFadden B, Rose I A, Williams J O. Production of pyruvate and succinate by action of isocitrate lyase on α-methylisocitrate. Arch Biochem Biophys. 1972;148:84–88. doi: 10.1016/0003-9861(72)90118-x. [DOI] [PubMed] [Google Scholar]
- 19.Metzler D E. Biochemistry: the chemical reactions of living cells. New York, N.Y: Academic Press, Inc.; 1977. [Google Scholar]
- 20.Naleçz M J, Naleçz K A, Azzi A. Purification and functional characteristics of the pyruvate (monocarboxylate) carrier from baker's yeast mitochondria (Saccharomyces cerevisiae) Biochim Biophys Acta. 1991;1079:87–95. doi: 10.1016/0167-4838(91)90028-x. [DOI] [PubMed] [Google Scholar]
- 21.Palmieri L, Lasorsa F L, Iacobazzi V, Runswick M J, Palmieri F, Walker J E. Identification of the mitochondrial carnitine carrier in Saccharomyces cerevisiae. FEBS Lett. 1999;462:472–476. doi: 10.1016/s0014-5793(99)01555-0. [DOI] [PubMed] [Google Scholar]
- 22.Postma E, Verduyn C, Scheffers W A, van Dijken J P. Enzymatic analysis of the Crabtree effect in glucose-limited chemostat cultures of Saccharomyces cerevisiae. Appl Environ Microbiol. 1989;55:468–477. doi: 10.1128/aem.55.2.468-477.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pronk J T, van der Linden-Beuman A, Verduyn C, Scheffers W A, van Dijken J P. Propionate metabolism in Saccharomyces cerevisiae: implications for the metabolon hypothesis. Microbiology. 1994;140:717–722. doi: 10.1099/00221287-140-4-717. [DOI] [PubMed] [Google Scholar]
- 24.Pronk J T, Steensma H Y, van Dijken J P. Pyruvate metabolism in Saccharomyces cerevisiae. Yeast. 1996;12:1607–1633. doi: 10.1002/(sici)1097-0061(199612)12:16<1607::aid-yea70>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 25.Purnelle B, Coster F, Goffeau A. The sequence of a 36 kb segment on the left arm of yeast chromosome X identifies 24 open reading frames including NUC1, PRP21 (SPP91), CDC6, CRY2, the gene for S24, a homologue to the aconitase gene ACO1 and two homologues to chromosome III genes. Yeast. 1994;10:1235–1249. doi: 10.1002/yea.320100912. [DOI] [PubMed] [Google Scholar]
- 26.Rose M D, Winston F, Hieter P. Methods in yeast genetics. A laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Schöler A, Schüller H J. Structure and regulation of the isocitrate lyase gene from the yeast Saccharomyces cerevisiae. Curr Genet. 1993;23:375–381. doi: 10.1007/BF00312621. [DOI] [PubMed] [Google Scholar]
- 29.Sinclair D A, Dawes I W, Dickinson J R. Purification and characterization of the branched-chain alpha-ketoacid dehydrogenase complex from Saccharomyces cerevisiae. Biochem Mol Biol Int. 1993;31:911–922. [PubMed] [Google Scholar]
- 30.Sumegi B, Sherry A D, Malloy C R. Channeling of TCA cycle intermediates in cultured Saccharomyces cerevisiae. Biochemistry. 1990;29:9106–9110. doi: 10.1021/bi00491a002. [DOI] [PubMed] [Google Scholar]
- 31.Tabuchi T, Serisawa N. A hypothetical cyclic pathway for the metabolism of odd-carbon n-alkanes or propionyl-CoA via seven-carbon tricarboxylic acids in yeasts. Agric Biol Chem. 1975;39:1055–1061. [Google Scholar]
- 32.Tabuchi T, Uchiyama H. Methylcitrate condensing and methylisocitrate cleaving enzymes: evidence for the pathway of oxidation of propionyl-CoA to pyruvate via C7-tricarboxylic acids. Agric Biol Chem. 1975;39:2035–2042. [Google Scholar]
- 33.Textor S, Wendisch V F, de Graaf A A, Müller U, Linder M I, Linder D, Buckel W. Propionate oxidation in Escherichia coli: evidence for the operation of a methylcitrate cycle in bacteria. Arch Microbiol. 1997;168:428–436. doi: 10.1007/s002030050518. [DOI] [PubMed] [Google Scholar]
- 34.Uchiyama H, Ando M, Toyonaka Y, Tabuchi T. Subcellular localization of the methycitric-acid-cycle enzymes in propionate-metabolism of Yarrowia lipolytica. Eur J Biochem. 1982;125:523–527. doi: 10.1111/j.1432-1033.1982.tb06713.x. [DOI] [PubMed] [Google Scholar]
- 35.Ueda M, Tanaka A, Fukui S. Enhancement of carnitine acetyltransferase synthesis in alkane-grown cells and propionate-grown cells and propionate-grown cells of Candida tropicalis. Arch Microbiol. 1985;141:29–31. doi: 10.1007/BF00446735. [DOI] [PubMed] [Google Scholar]
- 36.van den Berg M A, de Jong-Gubbels P, Kortland C J, van Dijken J P, Pronk J T, Steensma H Y. The two acetyl-coenzyme A synthetases of Saccharomyces cerevisiae differ with respect to kinetic properties and transcriptional regulation. J Biol Chem. 1996;271:28953–28959. doi: 10.1074/jbc.271.46.28953. [DOI] [PubMed] [Google Scholar]
- 37.van Urk H, Mak P R, Scheffers W A, van Dijken J P. Metabolic responses of Saccharomyces cerevisiae CBS8066 and Candida utilis CBS621 upon transition from glucose limitation to glucose excess. Yeast. 1988;4:283–291. doi: 10.1002/yea.320040406. [DOI] [PubMed] [Google Scholar]
- 38.Verduyn C, van Dijken J P, Scheffers W A. Colorimetric alcohol assays with alcohol oxidase. J Microbiol Methods. 1984;2:15–25. [Google Scholar]
- 39.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 40.Waterham H R, Titorenko V I, Haima P, Cregg J M, Harder W, Veenhuis M. The Hansenula polymorpha PER1 gene is essential for peroxisome biogenesis and encodes a peroxisomal matrix protein with both carboxy- and amino-terminal targeting signals. J Cell Biol. 1994;127:737–749. doi: 10.1083/jcb.127.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]