Abstract
The coronavirus SARS-CoV-2 has caused a health care crisis all over the world since the end of 2019. Although vaccines and neutralizing antibodies have been developed, rapidly emerging variants usually display stronger immune escape ability and can better surpass vaccine protection. Therefore, it is still vital to find proper treatment strategies. To date, antiviral drugs against SARS-CoV-2 have mainly focused on proteases or polymerases. Notably, noncanonical nucleic acid structures called G-quadruplexes (G4s) have been identified in many viruses in recent years, and numerous G4 ligands have been developed. During this pandemic, literature on SARS-CoV-2 G4s is rapidly accumulating. Here, we first summarize the recent progress in the identification of SARS-CoV-2 G4s and their intervention by ligands. We then introduce the potential interacting proteins of SARS-CoV-2 G4s from both the virus and the host that may regulate G4 functions. The innovative strategy to use G4s as a diagnostic tool in SARS-CoV-2 detection is also reviewed. Finally, we discuss some key questions to be addressed in the future.
Keywords: G-quadruplex, SARS-CoV-2, Targeting
1. Introduction
In late 2019, a novel coronavirus infection was discovered, which soon caused large-scale infection and outbreaks around the world. The WHO has released the official name of the disease as COVID-19, and the virus has been designated severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). SARS-CoV-2 caused a global health care crisis. There have been 599,071,265 confirmed cases of COVID-19, including 6,467,023 deaths reported to WHO as of 4:12 pm CEST, 30 August 2022 (https://covid19.who.int/). Patients present with fever, dry cough, fatigue, dyspnea, mild upper respiratory tract illness, severe viral pneumonia, and even death [1], [2], [3], which are similar features in other SARS-CoV infections [4], [5]. In addition to SARS-CoV in 2003 [6] and MERS-CoV in 2012 [7], this is the third pandemic in which the coronavirus has caused human social unrest and economic loss during the past 20 years. It is noteworthy that the SARS-CoV-2 genome undergoes persistent and rapid mutations. Since the beginning of 2022, Omicron has started to spread all over the world, displaying a stronger immune escape ability than Delta and better-surpassing vaccine protection. Even now, new variants of Omicron are still emerging. Therefore, it is still vital to find proper therapeutic strategies against SARS-CoV-2.
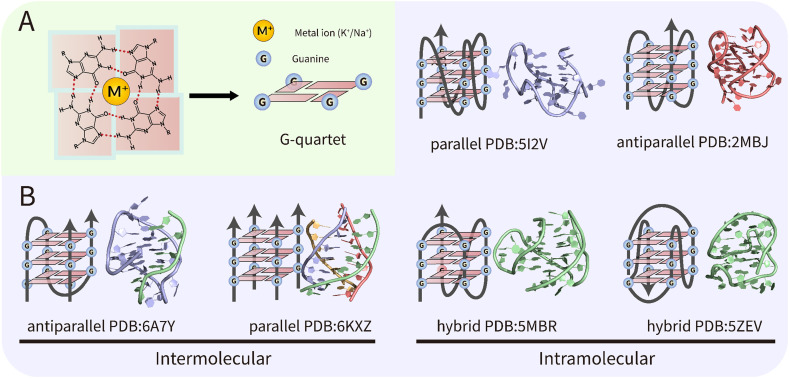
Although the genomes of the viruses undergo rapid mutations, the secondary structural elements are relatively conserved and therefore can be used as potential antiviral targets. In 1953, Watson and Crick established the B-form double-helical DNA model, opening the era of modern molecular biology [8]. However, non-B-form secondary structures such as Z-DNA [9], cruciform DNA [10], triplexes [11], and G-quadruplexes (G4s) [12] have also been found in the genome of many species with essential biological functions, among which G4s have attracted great attention in recent years. As a special type of nucleic acid structure formed by G-rich DNA or RNA sequences, G4s are stacked through the planar G-quartets formed by Hoogsteen bonds and further stabilized by ions (Fig. 1A). These structures can be further classified as intramolecular G4s or intermolecular G4s with different topologies depending on the chain orientation (Fig. 1B). Sequencing of different genomes showed that the location of these putative G-quadruplex-forming sequences (PQSs) is not random. For example, in mammalian cells, telomere G4 consisting of tandemly arranged TTAGGG repeats is the most abundant [13]. In addition, more than 90 % of human DNA replication origins contain G4s, which may be a major determinant involved in the regulation of human cell replication [14]. PQSs are also enriched in proto-oncogenes in humans [15], [16]. Overall, the presence of G4s is involved in many cellular functions, such as DNA replication, gene expression, and telomere preservation. Interested readers can refer to some excellent reviews [15], [16], [17], [18].
Fig. 1.
Structures and topologies of G-quadruplexes (G4s). (A) structure of G-quartet. (B) Schematic representation and examples of intramolecular or intermolecular G4s with different topologies.
Due to their essential biological functions and globular shape similar to proteins, G4s have been developed into anticancer drug targets. While early studies mainly blocked telomere elongation by targeting telomere G4s in cancer cells [19], [20], subsequent research has focused on inhibiting the expression of individual oncogenes by ligands, e.g., 3,8,10-trisubstituted isoalloxazines show selective binding to the c-kit oncogene promoter G4, thereby inhibiting c-kit oncogene expression [21]. Due to the ubiquitous presence of G4s in proto-oncogenes, targeting multiple G4s to simultaneously inhibit the expression of multiple oncogenes is also plausible. The above pieces of evidence suggest that a similar strategy may be applied in antiviral therapy because PQSs are also widely distributed in viral genomes with essential regulatory functions. In particular, a comprehensive analysis of PQSs in the genome of known viruses that can infect humans showed that the occurrence and location of PQSs are features characteristic of each virus class and family [22]. In this context, several recent reviews have systematically summarized the function of viral G4s and the potential of targeting viral G4s in antiviral therapy [23], [24], [25].
We noticed that over the past two years since the emergence of COVID-19, many predictions of SARS-CoV-2 PQSs have been made, and some experimentally validated; however, the results seem to vary due to the differences in the prediction software or algorithm used. Therefore, it is necessary to summarize and compare these data. In this review, we first introduce recent studies on SARS-CoV-2 G4s and their targeting by ligands. We searched for the literature related to this topic by the keywords SARS-CoV-2 and G-quadruplex. Most of the studies up to now are included. Then, the potential interacting protein partners of SARS-CoV-2 G4s that may regulate G4 structures are comprehensively reviewed. These results were mainly searched and selected by the keywords SARS-CoV-2, RNA, and interactome. We also discuss the innovative strategy to use G4s as a diagnostic tool in SARS-CoV-2 detection. The related studies were obtained by the keywords SARS-CoV-2, G-quadruplex, detection, or diagnosis. Finally, we present some key questions to be addressed in the future.
2. Potential G4s in the SARS-CoV-2 genome and their features
In cells infected with SARS-CoV-2, viral RNA accounts for 65.4 % of the total RNA, which suppresses host gene expression [26]. As a β-coronavirus, SARS-CoV-2 possesses a 29.9-kb-long positive-strand ssRNA genome, which is one of the largest genomes in all RNA viruses. Complex secondary structural features are present in the SARS-CoV-2 genome in vivo; interestingly, some are conserved across β-coronaviruses. For instance, Huston et al. reported the complete secondary structures of the SARS-CoV-2 genome using SHAPE-MaP data obtained in living cells, showing many long stretches of short, locally folded stem loops [27]. The protein-coding sections are among the most well-structured regions, suggesting that complex molecular architectures not only appear to be conserved across the β-coronavirus family but also play a functional role in the virus life cycle [27], [28]. Manfredonia et al. proved that approximately 10 % of secondary structural features in the SARS-CoV-2 genome show significant covariation and may be relevant to conserved functions. The 3D modeling of these segments further allowed for the identification of putative druggable pockets [29]. Increasing evidence suggests that large-scale structures in SARS-CoV-2 represent important elements in their cellular interactions, which may contribute to their persistence and transmissibility [30]. Meanwhile, disruption of some conserved structures results in inhibition of viral growth [27]. These discoveries together indicate that the secondary structural features of the SARS-CoV-2 genome may play essential roles in the virus life cycle.
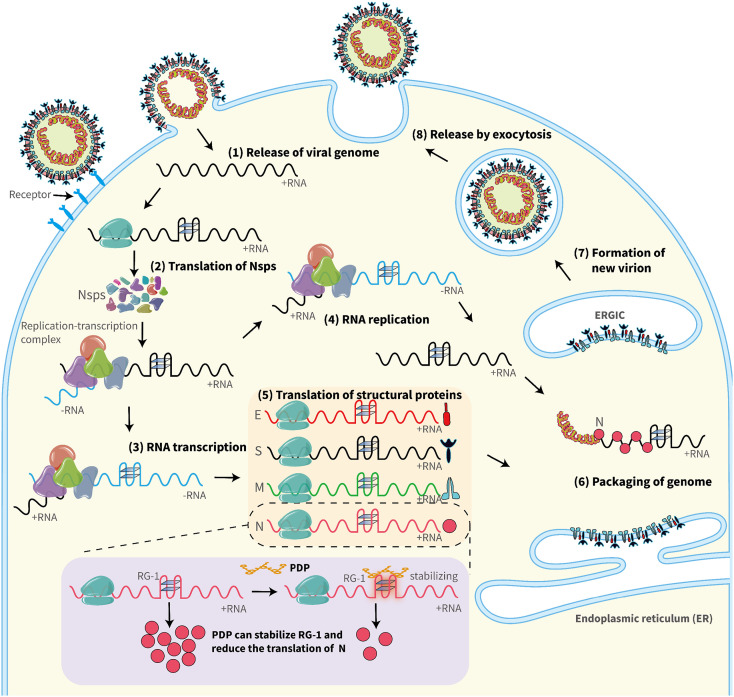
As one of the most important noncanonical nucleic acid secondary structures, G4s may be ideal targets in developing innovative drugs against coronaviruses. Therefore, since the very beginning of COVID-19, many studies have focused on PQSs in the SARS-CoV-2 genome. Cui et al. identified 14 PQSs in the positive RNA strand with runs of G2NxG2NyG2NzG2 and loop size from 0 to 12 by the quadruplex forming G-rich sequences mapper (QGRS). Their results further suggested that compared to SARS-CoV, there are fewer PQSs in SARS-CoV-2, which may account for the faster replication rate of SARS-CoV-2 [31]. Panera et al. identified 25 PQSs using QGRS with loop sizes of PQSs from 0 to 36 [32]. Ji et al. not only identified the same 25 results but also found that the PQSs at genome positions 353, 644, 2714, 3467, 8687, 10,261, 13,385, 14,947, 15,448, 24,215, 24,268, 26,746, 28,781, and 28,903 are well conserved in the coronavirus family [33]. Later, several studies used other bioinformatics prediction tools (G4CatchAll, pqsfinder, G4Hunter Web, G4screener) and identified some new PQSs, which appeared in the negative strand [34], [35]. Researchers also used G4-iM Grinder to analyze the SARS-CoV-2 genome and found a total of 323 PQSs. Although most of them scored poorly, 7 PQSs were scored over 30 with the highest probability of G4 formation, and 4 PQSs were conserved in SARS-coronaviruses and Bat-CoV [36]. Recently, Josué Carvalho et al. analyzed more than 200,000 SARS-CoV-2 genome sequences from five continents and found PQSs at positions 1574, 13,385, 24,215, 24,268, and 25,197 with a high conservation level above 98 %. Besides, PQS at position 28,903 in the N protein-coding sequence revealed 90.82 % conservation [37]. The potential roles played by the RNA G4s in the SARS-CoV-2 life cycle are illustrated in Fig. 2 , including translation of Nsps and structural proteins, RNA transcription, RNA replication, and packaging of the genome.
Fig. 2.
Possible roles of RNA G4s in the SARS-CoV-2 life cycle. Different steps of viral infection are illustrated. (1) Release of the viral genome; (2) Translation of Nsps; (3) RNA transcription; (4) RNA replication; (5) Translation of structural proteins; (6) Packaging of the genome; (7) Formation of new virion; (8) Release by exocytosis. Virus G4s may be involved in regulating the efficiency of steps 2–6 and may become a target for antiviral therapy. For instance, the G4-specific ligand PDP can stabilize RG-1 (at genome position 28,903) and reduce the translation of the nucleocapsid (N) protein.
In addition to bioinformatic prediction, some PQSs have been confirmed by circular dichroism (CD) spectroscopy analysis, nuclear magnetic resonance (NMR), fluorescence turn-on assays, multiple other biophysical techniques, and molecular biology assays [31], [32], [33], [36], [38], [39]. In particular, the first calculated G4 structure of RG-1 at position 28,903 was recently reported by a multiscale approach combining quantum and classical molecular modeling [40]. We summarize the PQSs that have been predicted or verified in Table 1 . Importantly, all of them contain two G-quartets once folded. The conserved PQSs among coronaviruses are shown in Table 2 . Due to differences in prediction software or algorithms, there are some inconsistencies among these results. Therefore, these data will require rigorous experimental verification.
Table 1.
PQSs in the SARS-CoV-2 genome.
| G4 position | Gene | Sequence | QGRS mapper Score | G4Hunter score | G4-iM grinder Score | Reference of prediction | Confirmed by reference | |
|---|---|---|---|---|---|---|---|---|
| 1 | +236 | 5′-UTR | GGUUUCGUCCGGGUGUGACCGAAAGGUAAGAUGG | [31] | ||||
| 2 | +353 | Nsp1 | GGCUUUGGAGACUCCGUGGAGGAGG | 16 | 0.64 | [31], [32], [33], [34], [35] | [31] | |
| 3 | +359 | Nsp1 | GGAGACUCCGUGGAGGAGG | 30 | [34], [36] | |||
| 4 | +370 | Nsp1 | GGAGGAGGUCUUAUCAGAGG | 30 | [36] | |||
| 5 | +545 | Nsp1 | GGCAUUCAGUACGGUCGUAGUGGUGAGACACUUGG | [31] | ||||
| 6 | +644 | Nsp1 | GGUAAUAAAGGAGCUGGUGG | 15 | 0.8 | 30 | [31], [32], [33], [34], [35], [36] | [31], [39] |
| 7 | +1463 | Nsp2 | GGUGGUCGCACUAUUGCCUUUGGAGG | 6 | 0.423 | [32], [33], [35] | ||
| 8 | +1574 | Nsp2 | GGUGUUGUUGGAGAAGGUUCCGAAGG | 18 | 0.615 | [31], [32], [33], [35], [37] | ||
| 9 | +2714 | Nsp2 | GGCGGUGCACCAACAAAGGUUACUUUUGG | 10 | 0.31 | [31], [32], [33], [35] | ||
| 10 | +3467 | Nsp3 | GGAGGAGGUGUUGCAGG | 15 | 1 | 34 | [31], [32], [33], [34], [35], [36] | [36], [39] |
| 11 | +4162 | Nsp3 | GGUUAUACCUACUAAAAAGGCUGGUGG | 6 | 0.37 | [32], [33], [35] | ||
| 12 | +4255 | Nsp3 | GGGUCAGGGUUUAAAUGGUUACACUGUAGAGGAGG | 31 | [31], [36] | |||
| 13 | +4261 | Nsp3 | GGGUUUAAAUGGUUACACUGUAGAGGAGG | 10 | 0.933 | [32], [33], [34], [35] | ||
| 14 | +4262 | Nsp3 | GGUUUAAAUGGUUACACUGUAGAGGAGG | 10 | [31], [34] | |||
| 15 | +5036 | Nsp3 | GGACAACAGUUUGGUCCAACUUAUUUGGAUGG | [31] | ||||
| 16 | +8687 | Nsp4 | GGAUACAAGGCUAUUGAUGGUGG | 14 | 0.652 | [31], [32], [33], [34], [35] | ||
| 17 | +10,058 | Nsp5 | GGUUUUAGAAAAAUGGCAUUCCCAUCUGGUAAAGUUGAGG | [31] | ||||
| 18 | +10,255 | Nsp5 | GGUACAGGCUGGUAAUGUUCAACUCAGG | [34] | ||||
| 19 | +10,261 | Nsp5 | GGCUGGUAAUGUUCAACUCAGGGUUAUUGG | 9 | 0.6 | [32], [33], [34], [35] | ||
| 20 | +13,385 | Nsp10 | GGUAUGUGGAAAGGUUAUGG | 19 | 1.048 | 31 | [31], [32], [33], [34], [35], [36], [37] | [31], [33], [38] |
| 21 | +14,947 | Nsp12 | GGUUUUCCAUUUAAUAAAUGGGGUAAGG | 4 | 0.714 | [32], [33], [35] | ||
| 22 | +15,208 | Nsp12 | GGAACAAGCAAAUUCUAUGGUGGUUGG | 6 | 0.678 | [32], [33], [35] | ||
| 23 | +15,448 | Nsp12 | GGCGGUUCACUAUAUGUUAAACCAGGUGG | 3 | 0.345 | [32], [33], [35] | ||
| 24 | +18,296 | Nsp14 | GGAUUGGCUUCGAUGUCGAGGGG | 9 | 1.043 | [31], [32], [33], [34], [35] | ||
| 25 | +20,869 | Nsp16 | GGUGCUGGUUCUGAUAAAGGAGUUGCACCAGG | [31] | ||||
| 26 | +22,316 | S | GGUGAUUCUUCUUCAGGUUGGACAGCUGG | 10 | 0.448 | [32], [33], [35] | ||
| 27 | +24,215 | S | GGUUGGACCUUUGGUGCAGG | 17 | 0.6 | [31], [32], [33], [35], [37] | [31] | |
| 28 | +24,268 | S | GGCUUAUAGGUUUAAUGGUAUUGG | 19 | 0.625 | [31], [32], [33], [34], [35], [37] | [31], [33] | |
| 29 | +25,197 | S | GGCCAUGGUACAUUUGGCUAGG | 17 | 0.455 | [31], [32], [33], [34], [35], [37] | [31] | |
| 30 | +25,951 | ORF3a | GGUGGUUAUACUGAAAAAUGGGAAUCUGG | 8 | 0.69 | [32], [33], [34], [35] | ||
| 31 | +26,746 | M | GGAUCACCGGUGGAAUUGCUAUCGCAAUGG | 7 | 0.333 | [32], [33], [34], [35] | ||
| 32 | +28,613 | N | GGAACUGGGCCAGAAGCUGGACUUCCCUAUGG | [31] | ||||
| 33 | +28,781 | N | GGCUUCUACGCAGAAGGGAGCAGAGGCGG | 9 | 0.655 | [32], [33], [35] | ||
| 34 | +28,903 | N | GGCUGGCAAUGGCGG | 18 | 0.867 | 34 | [31], [32], [33], [35], [36], [37] | [31], [36], [38], [39] |
| 35 | +29,123 | N | GGAAAUUUUGGGGACCAGG | 14 | 1.053 | [31], [32], [33], [35] | ||
| 36 | +29,234 | N | GGCAUGGAAGUCACACCUUCGGGAACGUGG | 11 | 0.467 | [32], [33], [35] | ||
| 37 | +29,254 | N | GGGAACGUGGUUGACCUACACAGGUGCCAUCAAAUUGG | [31] | ||||
| 38 | −165 | GGCCUCGGUGAAAAUGUGGUGG | 13 | 0.591 | [31], [35] | |||
| 39 | −1591 | GGGGUGCAUUUCGCUGAUUUUGGGG | 1.280 | [35] | ||||
| 40 | −2987 | GGUCUGGUCAGAAUAGUGCCAUGGAGUGG | 9 | 0.483 | [34], [35] | |||
| 41 | −6822 | GGUUGGUAACCAACACCAUUAGUGGGUUGG | 6 | 0.433 | [35] | |||
| 42 | −11,440 | GGCGGUGGUUUAGCACUAACUCUGG | 7 | 0.48 | [35] | |||
| 43 | −13,136 | GGUUAAGUGGUGGUCUAGG | 16 | 0.842 | [31], [35] | |||
| 44 | −13,963 | GGAUCUGGGUAAGGAAGG | 19 | 1.111 | [31], [34], [35] | [39] | ||
| 45 | −16,623 | GGAUUUGGAUGAUCUAUGUGGCAACGG | 14 | 0.556 | [31], [35] | |||
| 46 | −19,865 | GGUGAUAGAGGUUUGUGGUGG | [34] | |||||
| 47 | −19,865 | GGUGAUAGAGGUUUGUGGUGGUUGG | 19 | 0.92 | [31], [34], [35] | |||
| 48 | −19,874 | GGUUUGUGGUGGUUGG | [34] | |||||
| 49 | −23,877 | GGAUAUGGUUGGUUUGG | 19 | 0.941 | [31], [34], [35] | [39] | ||
| 50 | −25,003 | GGUGGAAUGUGGUAGG | 17 | 1.063 | [31], [34], [35] | |||
| 51 | −27,432 | GGGGCUUUUAGAGGCAUGAGUAGG | 13 | 1.042 | [31], [34], [35] | |||
| 52 | −29,867 | GGUUGGUUUGUUACCUGGGAAGG | 13 | 0.783 | [31], [34], [35] |
Table 2.
Conserved PQSs in the SARS-CoV-2 genome.
| G4 position | Gene | Sequence | Found in other members of the Coronaviridae family | Reference | |
|---|---|---|---|---|---|
| 1 | +353 | Nsp1 | GGCUUUGGAGACUCCGUGGAGGAGG | SARS-CoV | [31], [33] |
| Bat-CoV | |||||
| 2 | +644 | Nsp1 | GGUAAUAAAGGAGCUGGUGG | SARS-CoV | [31], [33] |
| Bat-CoV | |||||
| BtRl-BetaCoV | |||||
| BtRs-BetaCoV | |||||
| BtRf-BetaCoV | |||||
| Rhinolophus-affinis-coronavirus | |||||
| 3 | +2714 | Nsp2 | GGCGGUGCACCAACAAAGGUUACUUUUGG | Bat-CoV | [33] |
| 4 | +3467 | Nsp3 | GGAGGAGGUGUUGCAGG | BtRt-BetaCoV | [33] |
| 5 | +8687 | Nsp4 | GGAUACAAGGCUAUUGAUGGUGG | Bat-CoV | [33] |
| 6 | +10,261 | Nsp5 | GGCUGGUAAUGUUCAACUCAGGGUUAUUGG | Bat-CoV | [33] |
| 7 | +13,385 | Nsp10 | GGUAUGUGGAAAGGUUAUGG | SARS-CoV | [31], [33], [36] |
| Bat-CoV | |||||
| Rhinolophus-affinis-coronavirus | |||||
| 8 | +14,947 | Nsp12 | GGUUUUCCAUUUAAUAAAUGGGGUAAGG | SARS-CoV | [33] |
| Bat-CoV | |||||
| BtRs-BetaCoV | |||||
| Rhinolophus-affinis-coronavirus | |||||
| 9 | +15,448 | Nsp12 | GGCGGUUCACUAUAUGUUAAACCAGGUGG | SARS-CoV | [33] |
| Bat-CoV | |||||
| BtRl-BetaCoV | |||||
| BtRs-BetaCoV | |||||
| Rhinolophus-affinis-coronavirus | |||||
| 10 | +24,215 | S | GGUUGGACCUUUGGUGCAGG | SARS-CoV | [31], [33] |
| Bat CoV | |||||
| 11 | +24,268 | S | GGCUUAUAGGUUUAAUGGUAUUGG | SARS-CoV | [31], [33] |
| Bat-CoV | |||||
| BtRf-BetaCoV | |||||
| Rhinolophus-affinis-coronavirus | |||||
| 12 | +25,197 | S | GGCCAUGGUACAUUUGGCUAGG | SARS-CoV | [31] |
| Bat-CoV | |||||
| 13 | +26,746 | M | GGAUCACCGGUGGAAUUGCUAUCGCAAUGG | Bat-CoV | [33] |
| 14 | +28,781 | N | GGCUUCUACGCAGAAGGGAGCAGAGGCGG | SARS-CoV | [33] |
| Bat-CoV | |||||
| BtRs-BetaCoV | |||||
| Rhinolophus-affinis-coronavirus | |||||
| 15 | +28,903 | N | GGCUGGCAAUGGCGG | SARS-CoV | [31], [33] |
| Bat-CoV |
3. Targeting RNA G4s in SARS-CoV-2 by small-molecule compounds and natural products
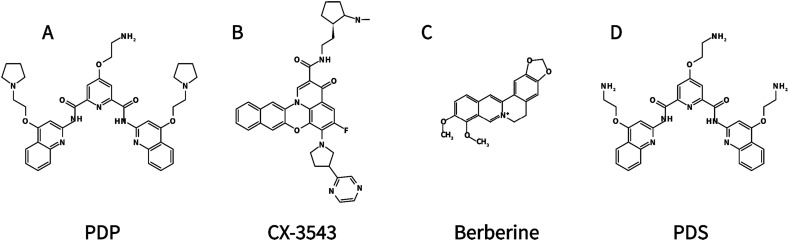
Recently, Zhao et al. used multiple approaches, such as fluorescence turn-on assays, CD, NMR, and fluorescence resonance energy transfer (FRET), to show that RG-1 at the coding region of the SARS-CoV-2 N protein, indeed forms a G4 structure in vitro [38]. In the following experiment, for the first time, PQS in the SARS-CoV-2 genome was verified to fold into stable unimolecular G4 structures in live cells. It is worth noting that G4-specific compounds, such as PDP (pyridostatin)(Fig. 3A), a bisquinolinium derivative that inhibited HCV replication in cells with a half-maximum inhibitory concentration (IC50) of 1.2 μM [41], can stabilize RG-1 and significantly reduce the expression levels of the N protein not only in vitro but also in vivo (Fig. 2) [38]. For instance, the HeLa cells pre-transfected with Cy5-labeled RG-1-WT and RG-1-Mut were treated with 4 μM PDP. As a result, the colocalization of RG-1-WT and G4 antibody was significantly increased, but not for RG-1-Mut, suggesting the promotion of G4 structure formation by PDP. The authors further found that 4 μM PDP treatment can inhibit the protein expression of Flag-N-RG-1-WT in both HeLa and HEK293T cells to ~1/5 of the control without PDP (~1.0 to ~0.2). However, the protein expression of Flag-N-RG-1-Mut was unchanged upon PDP treatment. These results indicate that PDP can inhibit N protein expression by targeting RG-1. The related equilibrium dissociation constant KD and IC50 values remain to be determined in the future.
Fig. 3.
Chemical structures of (A) PDP, (B) CX-3543, (C) Berberine, and (D) PDS.
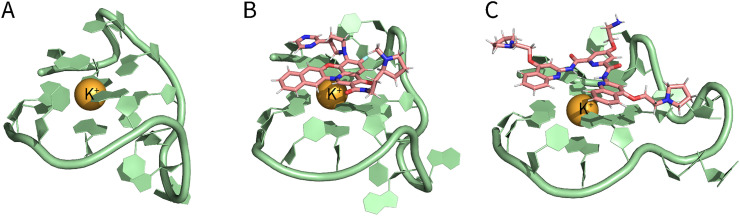
Classical molecular dynamics trajectories (MD) are consistent with the experimental data in confirming the folding of RG-1 into a parallel G4 conformation composed of two rigid tetrads and a flexible peripheral loop (Fig. 4A). Moreover, Miclot et al. further obtained docking complexes of PDP and CX-3543 (also referred to as quarfloxin, a G4 ligand derived from fluoroquinolones) (Fig. 3B) with RG-1 (Fig. 4B-C). The binding takes place by π-π stacking of the ligands on top of the G-quartet, and it is mostly driven by dispersion and hydrophobic interactions. There is no significant energetic barrier or steric hindrance, which greatly facilitates the recruitment of the targeting ligands. Therefore, both G4 ligands can potentially be seen as valuable compounds to stabilize G4 arrangements, particularly the parallel conformation of RG-1 [40].
Fig. 4.
Structure of RG-1 and its molecular docking with ligands. (A) Calculated structure of RG-1. (B) Molecular docking of RG-1 and CX-3543. (C) Molecular docking of RG-1 and PDP. (Structural data from the study by Miclot et al. [40]).
Researchers have been striving to find effective G4-specific ligands to target G4s in SARS-CoV-2. Berberine is a planar molecule with an extended π-delocalized system (Fig. 3C), which can interact with G-quartets of G4 via π-π stacking interactions [42]. In addition, Berberine is not fluorescent in the aqueous solution, whereas a strong fluorescence increase can be observed upon binding to G4. As a common folk medicine used for hundreds of years in China, berberine has shown antiviral, anti-allergic, and anti-inflammatory properties [43]. Quite recently, the binding properties of berberine to RG-1 were characterized [44]. Oliva et al. designed a titration experiment that kept the concentration of RG-1 constant, while the concentration of berberine was varied. The reported data indicate that berberine can interact with RG-1 with binding constants of 2.8 ± 0.3 × 104 M−1 in NaCl and 1.48 ± 0.3 × 104 M−1 in KCl. Further results showed that two berberine molecules are bound to one RG-1. In addition, RG-1 adopts a parallel conformation in both the ligand-free and ligand-bound states. All the above evidence suggests that, as therapeutic targets, SARS-CoV-2 G4s may provide a promising direction for humans to fight against SARS-CoV-2 (Table 3 ).
Table 3.
The small-molecule compound used in G4 targeting for SARS-CoV-2 treatment.
| Ligand | targeting G4 | G4 origin | G4 sequence | Effect | Reference |
|---|---|---|---|---|---|
| PDP | RG-1 | SARS-CoV-2 N protein coding sequence | GGCUGGCAAUGGCGG | It stabilizes the RG-1 and significantly reduces the expression levels of the SARS-CoV-2 N protein. | [38] |
| Berberine | RG-1 | SARS-CoV-2 N protein coding sequence | GGCUGGCAAUGGCGG | It interacts with RG-1 with binding constants of 2.8 ± 0.3 × 104 M−1 in NaCl, and 1.48 ± 0.3 × 104 M−1 in KCl | [44] |
| PDS | PQS-675 | Host TMPRSS2 coding sequence | GGGCGGGCGGCCUGCAGGGACAUGGG | It inhibits SARS-CoV-2 entry in cells and hinders SARS-CoV-2 infection in vivo. | [46] |
In addition to the G4s in the SARS-CoV-2 genome, the host G4s that regulate the expression of crucial proteins for virus entry can also be targeted for antiviral therapy. For instance, the transmembrane serine protease TMPRSS2 can cleave the hemagglutinin of many subtypes of influenza virus and the spike proteins of coronavirus [45]. In particular, TMPRSS2 has been identified as one of the essential host determinants for SARS-CoV-2 infection [46]. In the promoter of the human Tmprss2 gene, there is a G-rich tract capable of forming a G4 structure in the presence of K+, significantly affecting the transcription level of Tmprss2 [45]. Moreover, in the open reading frame, an RNA G4 motif appears, inhibiting Tmprss2 translation and SARS-CoV-2 entry in cells. The G4-stabilizer PDS (pyridostatin) (Fig. 3D), which promoted the folding of telomeric G4 with a KD of 490 ± 80 nM [47], also binds RNA G4 named PQS-675 in Tmprss2 with the KD of 604.9 ± 2.84 nM. Furthermore, PDS can attenuate the infection of SARS-CoV-2 pseudoviruses in human lung cells, as shown by the fact that the entry efficiency with 50 μM PDS treatment is only about 10 % of the control. The half-maximal effective concentration (EC50) is between 2 and 5 μM. In addition to PDS, G4-stabilizer cPDS and TMPyP4 also significantly reduce pseudovirus entry efficiency with the EC50 around 50 μM and 20 μM, respectively. In mouse models, researchers found that PDS-treated mice have a significantly decreased entry of pseudoviruses (~20 %) compared with saline-treated mice. Immunohistochemical, ELISA, and western blot experiments further showed that PDS administration led to a decrease in TMPRSS2 protein levels in the lungs [46]. These results together indicated that Tmprss2 RNA G4 is a potential target for SARS-CoV-2 inhibition (Table 3).
4. Regulation of SARS-CoV-2 G4s by proteins
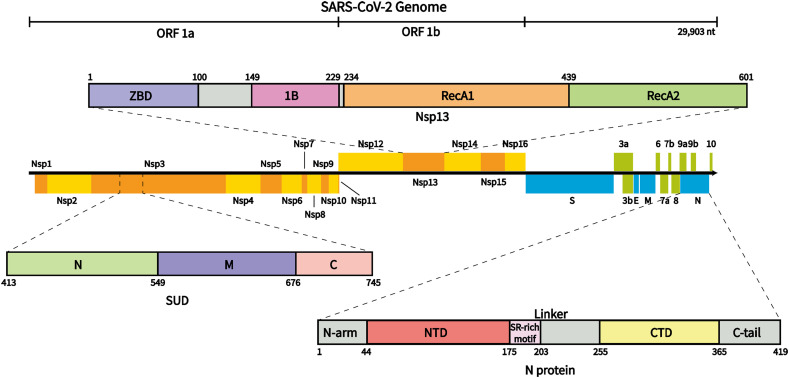
As shown in Fig. 5 , the SARS-CoV-2 genome contains 14 open reading frames (ORFs). ORF1a/ORF1b encodes nonstructural proteins (NSPs) 1–16, most of which are enzymes related to virus replication and transcription. At the 3′ end, the genome encodes 4 structural proteins, spike (S), envelope (E), membrane (M), and nucleocapsid (N), which play vital roles in viral structure integrity. The genome also encodes 9 accessory factors [48]. It is worth noting that some of these proteins have been reported to interact with or have the potential to interact with RNA G4s.
Fig. 5.
SARS-CoV-2 genome organization and the schematic domain structure of SUD, Nsp13, and N protein. ORF1a/ORF1b encodes Nsps 1–16; at the 3′ end, the genome encodes 4 structural proteins, spike (S), envelope (E), membrane (M), nucleocapsid (N), and 9 accessory factors. SUD contains SUD-N, SUD-M, and SUD-C domains; Nsp13 contains a zinc-binding domain (ZBD), 1B domain, two RecA-like domains, and a stalk domain; N protein contains two structural domains, N-NTD, N-CTD, and three intrinsically disordered regions.
4.1. SUD in Nsp3
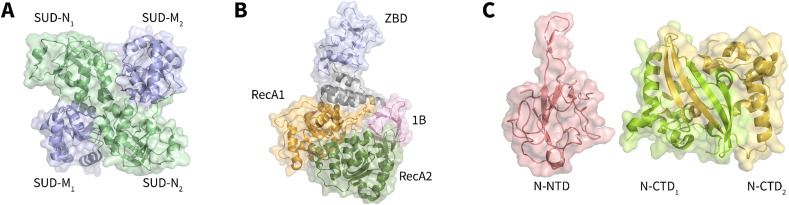
Nsp3 is the largest Nsp in SARS coronaviruses, consisting of 1922 amino acid residues [49], [50]. As part of the SARS coronavirus replicase/transcriptase complex, Nsp3 plays an important role in the formation and activity of the viral replication/transcription complex (RTC) [51]. Nsp3 consists of at least seven domains, one of which is the SARS-Unique Domain (SUD) (Fig. 5). X-ray crystallography and NMR spectroscopy indicate that the SUD comprises two macrodomains and one frataxin-like domain, named SUD-N, SUD-M, and SUD-C, respectively. SUD-C is not stable in full-length SUD; thus, SUD-N and SUD-M constitute the core domain. The full-length SUD has 338 amino acid residues, while SUD-NM (SUD-core) consists of 264 amino acid residues. The 3D structure shows that SARS-CoV SUD-NM is in a head-to-tail dimer form (Fig. 6A) [52], [53], [54]. It is worth noting that SUD is not conserved among coronaviruses that can infect humans. The SUD was only found in SARS-CoV and SARS-CoV-2, with an amino acid identity of 75 % and similarity of 95 % [55]; however, it was incomplete in MERS-CoV and absent in other less pathogenic coronaviruses [34], [55], [56]. Therefore, the SUD may play an important role in the strong pathogenicity of SARS coronaviruses.
Fig. 6.
Crystal structures of (A) SARS-CoV SUD dimer (PDB: 2W2G), (B) SARS-CoV-2 Nsp13 (PDB: 6ZSL), (C) SARS-CoV-2 N protein (PDB: 7CDZ/7CE0).
Tan et al. showed that the SARS-CoV SUD or its subdomains SUD-N and SUD-M can bind oligo(G) stretches known to form G4s, and the affinity was enhanced by the addition of K+, revealing the SUD as a G4-binding protein for the first time [52], [53]. More recently, Lavigne et al. confirmed that the SUD is also present in the Nsp3 of SARS-CoV-2. The authors further showed that the SARS-CoV-2 SUD can interact with both DNA and RNA G4s. Because this interaction can be disrupted by G4 ligands, the authors then proposed that inhibitors of their interactions may be used as potential antiviral compounds [55]. Molecular simulations further showed that RNA G4 can favor and stabilize the dimerization of the SUD, which may be related to its biological role in assisting the SARS virus to bypass the protective response of the host [57]. However, previous studies have not clarified the effect of SUD binding on G4s. Whether the SUD promotes G4 formation, stabilizes, or disrupts G4 structures is still unknown.
At the functional level, SUD/G4 interaction first has a significant influence on the activity of SARS-CoV RTC, in which the SUD-M domain is crucial while SUD-N is dispensable [58]. This observation may be consistent with the findings of Tan et al., who showed that mutations of selected lysine residues on the surface of SUD-N led to a reduction in G4 binding, whereas mutations in SUD-M abolished it [53]. Importantly, these amino acids are conserved in SARS-CoV-2 [55], suggesting that the replication of the SARS-CoV-2 genome may also require the SUD, especially SUD-M, to interact with G4s. Second, as Nsp3 has not been reported to enter the nucleus, the host targets of the SUD are most likely RNA G4s. Many RNA G4s reside in both the non-translated and translated regions of mRNAs coding for host cell proteins involved in apoptosis or signal transduction [51], [58], [59], [60], [61], [62], [63]. These proteins can induce controlled cellular death of infected cells, thereby slowing or preventing infection, or promoting cell survival by producing antiviral cytokines [62]. However, the binding of the viral SUD to mRNA G4s necessary for the production of the above signaling factors may impair the apoptosis/survival response pathways and then allow massive cell infection [57]. Altogether, the SUD may regulate both viral replication and the host immune response by binding to RNA G4s.
In addition to G4s, the SUD can also directly interact with host cell proteins. For instance, the interaction between the SUD and PLpro increases the stability of RCHY1 and augments RCHY1-mediated p53 ubiquitination. Consequently, p53 degradation is upregulated, thereby promoting viral replication [64]. A recent study showed that both SARS-CoV and SARS-CoV-2 SUDs interact with human poly(A)-binding protein (PAPB)-interacting protein 1 (Paip1) [56]. This interaction enhances SARS-CoV RNA translation but not the translation of host proteins, indicating that the virus takes advantage of the host translation machinery for its benefit [56]. Lavigne et al. believed that the SUD/Paip1 interaction may be involved in the selective translation regulation of G4-containing mRNAs in infected cells [55]. SUDs combined with G4s may recruit Paip1 and interfere positively or negatively with the recruitment of other translation factors, given that the SUD binding site to Paip1 is different from that to G4 [55]. Therefore, the SUD/Paip1 interaction may offer a new antiviral target for SARS-CoV-2. In addition, the SARS-CoV SUD modulated NLRP3 inflammasome-dependent CXCL10-mediated pulmonary inflammation [65]. However, the molecular mechanism has not yet been characterized. Perhaps there are other undiscovered SUD-interacting proteins in the host cell, and more work needs to be done to clarify the biological functions of the SUD, in particular, SUD-G4 interactions. Altogether, we think that targeting the SUD seems reasonable given its specificity in coronaviruses and its high G4 binding affinity.
4.2. Nsp13
Coronavirus Nsp13 is a superfamily 1 helicase (SF1) with NTPase activity [66], [67] that can unwind both RNA and DNA substrates in the 5′-to-3′ direction [68]. Recently, SARS-CoV-2 Nsp13 was also shown to have the same activities [66]. Nsp13 is very conserved in evolution. The SARS-CoV-2 Nsp13 contains 601 aa, with only one amino acid substitution to SARS-CoV (V570 in SARS-CoV-2 to I570 in SARS-CoV) [69]. As shown in Fig. 6B, the pyramid-shaped SARS-CoV-2 Nsp13 contains two typical SF1 helicase RecA domains, an N-terminal zinc-binding domain (ZBD), a stalk, and a 1B domain specific to coronavirus helicases [70], [71]. Six key residues, K288, S289, D374, E375, Q404, and R567, which cluster in the base cleft between the RecA1 and RecA2 domains, are involved in NTP hydrolysis [70].
In the host cell, Nsp13 localizes to the endoplasmic reticulum or an endoplasmic reticulum-derived membrane compartment, suggesting that it is likely the core of the coronavirus replication machinery [72]. This is consistent with the structure of a stable Nsp13-replication-transcription complex determined by cryo-electron microscopy (cryo-EM) to 3.5-Å nominal resolution [69]. Further study demonstrated that Nsp13 could act in the 5′-3′ direction on tRNA to disrupt stable RNA secondary structures or downstream RNA binding proteins, both of which could be significant impediments to RNA elongation [69]. In addition to its roles in replication, Nsp13 binds and inhibits TBK1 phosphorylation, resulting in decreased IRF3 activation and IFN-β production. The above evidence suggests that Nsp13 is also involved in viral evasion of immune responses [73].
Shum et al. used exponential enrichment (SELEX) as an in vitro selection strategy to isolate DNA sequences that have a high affinity for Nsp13. They found that most of the sequences were guanosine rich and likely to fold into G4 structures [74]. This may indicate a binding preference of Nsp13 for G4. Recently, Ji et al. used microscale thermophoresis (MST) to measure the interaction between Nsp13 and G4s at positions 13,385 and 24,268 within the SARS-CoV-2 genome. Their results showed that Nsp13 interacts with G4 at 13385 and G4 at 24268 with KD values of 0.79 ± 0.14 and 0.37 ± 0.08 μM, respectively. Subsequently, molecular docking showed that the guanine bases in G4s deviated significantly from their expected placement in both cases. These guanine bases are predicted to be flipped away from the G-quartets, thus reflecting the unfolding of SARS-CoV-2 G4 structures by Nsp13 [33].
It is worth noting that since the outbreak of SARS-CoV in 2003, research on the development of inhibitors of Nsp13 has emerged. Bananins [75], dihydroxychromone derivatives [76], SSYA10–001, and a 1,2,4 triazole [77] have all been reported to be potent inhibitors of SARS-CoV Nsp13. Recently, lumacaftor and cepharanthine were shown to inhibit SARS-CoV-2 Nsp13 ATPase activity [78]. Natural flavonoids were also identified as selective inhibitors of SARS-CoV-2 Nsp13 [79]. Together, these pieces of evidence reflect that targeting Nsp13 could be one of the major areas of anti-SARS-CoV-2 research.
4.3. N protein
Nucleocapsid (N) protein can bind to the SARS-CoV-2 genome and package the RNA into the ribonucleoprotein (RNP) complex, which plays a vital role during viral self-assembly [80]. As shown in Fig. 5, β-coronavirus N proteins share a common overall domain organization, including the RNA-binding domain (N-NTD) and dimerization domain (N-CTD) that are separated by short regions with a high predicted disorder [81], [82]. The SARS-CoV-2 N protein contains 419 aa [48]. Huang et al. solved the 3D structure of the N-terminal domain of the SARS-CoV N protein, which consists of a five-stranded β-sheet with a folding topology distinct from other RNA-binding proteins. The ssRNA binds to the protein surface at the junction between a flexible positively charged beta-hairpin and the core structure [83]. Recently, Peng et al. solved the crystal structures of SARS-CoV-2 N-NTD and N-CTD at 1.8 Å and 1.5 Å resolution, respectively (Fig. 6C). Both structures show conserved features from other coronavirus N proteins. The N-NTD presents a right-handed fist shape and consists of a four-stranded antiparallel-sheet core subdomain. The protruding loops are positively charged, providing a putative site for RNA binding [82]. The N-CTD displays a compact, intertwined dimer similar to that of related coronaviruses, including SARS-CoV [81], [84].
The N protein can recognize packaging signals to mediate coronavirus genomic RNA packaging into virions [85], [86]. In addition, many other RNA processing, RNA metabolism, and transcriptional regulatory proteins have been identified as SARS-CoV-2 N protein-interacting partners, which suggests that the N protein may be involved in multiple viral RNA processes [87]. Interestingly, several recent studies revealed that SARS-CoV-2 N protein and RNA underwent liquid-liquid phase separation depending on the length and concentration of ssRNA [88], [89]. Moreover, the SARS-CoV-2 N protein can be recruited to phase-separated assemblies formed by human proteins, providing a potential mechanism for the role of the N protein in SARS-CoV-2 viral genome packaging [89]. In addition to genome packaging, SARS-CoV N protein can inhibit S phase progression in mammalian cells [90] and antagonize immune regulation by targeting the initial step of the IFN-β induction pathway [91]. SARS-CoV-2 N protein can directly interact with NLRP3 to promote NLRP3 inflammasome activation, leading to aggravation of lung injury [92]. In addition, as viral suppressors of RNAi, both SARS-CoV-2 and SARS-CoV N proteins antagonize the RNAi pathway to protect viral RNA in cells [93], [94]. These findings all indicate the importance of the N protein and its perspectives for drug design as well as vaccine development.
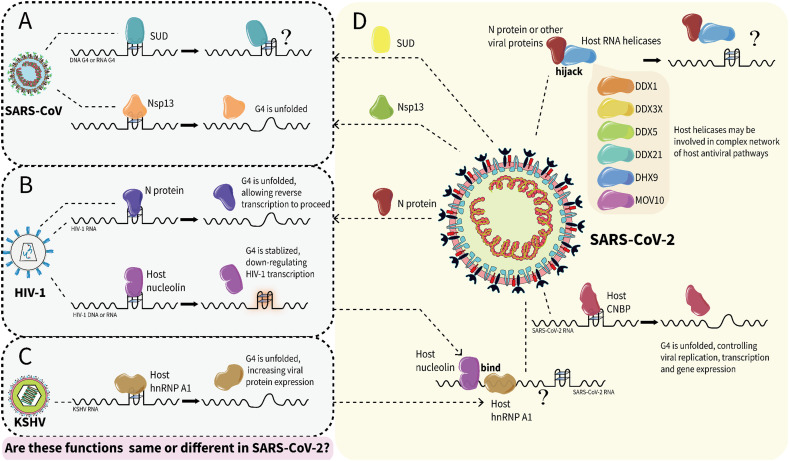
In infected cells, SARS-CoV-2 and SARS-CoV N proteins localize exclusively in the cytoplasm [95], [96]. During viral self-assembly, RNA needs to be packaged by N protein, while during viral infection, RNA needs to be released (Fig. 2). Therefore, the N protein may play an important role in remodeling and maintaining the RNA structures in the virus. In this regard, HIV-1 nucleocapsid protein (NCp7) was reported to readily destabilize and unfold DNA G4 [97]. Butovskaya et al. further demonstrated that NCp7 binds and unfolds HIV-1 RNA G4s and promotes DNA/RNA duplex formation, allowing reverse transcription to proceed [98] (Fig. 7 ). These studies also suggest a possible role of the SARS-CoV-2 N in regulating RNA G4; however, this speculation still needs to be verified by experiments.
Fig. 7.
The speculative interactions between SARS-CoV-2 G4s and proteins based on previous studies of other viruses. (A) In SARS-CoV, SUD binds G4; Nsp13 binds and unfolds RNA G4. (B) In HIV-1, N protein binds and unfolds RNA G4; host nucleolin binds and stabilizes G4. (C) In KSHV, host hnRNP A1 binds and unfolds RNA G4. (D) In SARS-CoV-2, viral N protein hijack host helicases; host CNBP binds and unfolds RNA G4; both nucleolin and hnRNP A1 bind to SARS-CoV-2 RNA. It is unknown whether these proteins in SARS-CoV-2 may have similar or different functions compared to other viruses.
During viral infection, some host helicases, especially members of the DEAD-box family act as proviral factors [99]. Recently, Squeglia et al. showed that hijacking of the DDX helicase by SARS-CoV-2 is expected to act as a proviral process by enhancing key steps in the viral life cycle [100]. Ciccosanti et al. further identified DDX3X and DHX9 as the interactome of the SARS-CoV-2 N protein and suggested that the N protein recruits DDX3X to the replication complex for virus production [101]. Researchers have also directly observed the association of DDX21 with the SARS-CoV-2 N protein, but how this interaction affects viral replication and infection is still unknown [102]. As DDX3X [103], DDX21 [104], and DHX9 [105] have all been shown to bind RNA G4s, these studies may implicate the interplay of N proteins with G4s, either through their own binding activity or through the hijacking of helicases (Fig. 7).
4.4. Cellular G4-binding proteins
Coronaviruses need host factors to facilitate their replication [106], [107], and SARS-CoV-2 has developed multiple strategies to interact with a complex network of host antiviral pathways [108], [109]. Recently, several studies have focused on the interactome between host proteins and the SARS-CoV-2 genome. For instance, Lee et al. identified 109 host factors that directly bind to SARS-CoV-2 RNA, in which they delineated 17 antiviral and 8 proviral RNA binding proteins (RBPs) hijacked by the virus [110]. Kamel et al. showed that SARS-CoV-2 infection profoundly remodels the cellular RNA-bound proteome and identified the proteins directly interacting with viral RNA, uncovering dozens of cellular RNA-binding proteins and 6 viral proteins [111]. Schmidt et al. identified up to 104 human proteins that directly and specifically bind to SARS-CoV-2 RNA in infected cells, 38 of which are unique SARS-CoV-2 RNA binders [112]. Flynn et al. identified 309 host proteins that bind SARS-CoV-2 RNA during infection [113]. Most of them serve as antiviral factors with a functional impact on host cell survival, suggesting that the initial response of the host to viral infection is to identify viral RNA and control the viral life cycle [113]. Taken together, a large number of cellular proteins bind to SARS-CoV-2 RNA in infected cells, highlighting viral RNA as a hub for complex host-virus interactions.
It is worth noting that some of the RBPs, such as nucleolin, hnRNPs, and helicases, that appeared in the SARS-CoV-2 genome interactome have already been reported to interact with G4s in other viruses (Fig. 7). For instance, nucleolin interacts with RNA G4 formed in EBNA1 mRNA to restrict EBNA1 expression in EBV-infected human cells [114] and can also stabilize G4s in the LTR promoter to silence HIV-1 viral transcription in HIV-1-infected cells [115]. Meanwhile, many hnRNPs appear in the host interactome of SARS-CoV-2 RNA, including A0/A1/A2B1/A3/AB/D/H3/L/M [110], [112], [113]. In HIV-1-infected cells, hnRNP A2B1 unfolds LTR promoter G4s, thereby enhancing transcription of the HIV-1 genome [116]. In KSHV-infected cells, G4 in LANA1 mRNA inhibits its translation, while host hnRNP A1 increases LANA1 expression by selectively binding and unfolding G4s [117]. These studies suggest that nucleolin and hnRNPs may also have a similar effect on SARS-CoV-2. In addition, the interactome of SARS-CoV-2 RNA contains many host helicases, including DDX5/6/21/23/38B/42/46, DHX36/39B, MOV10, UPF1, and the previously discussed DDX3X, DDX21, DHX9, which can be hijacked by the N protein [110], [112], [113]. These RNA helicases may play an important role in regulating viral RNA G4s [118], [119]. Finally, it is worth noting that in the study of Schmidt et al., CNBP was shown to be the most enriched SARS-CoV-2 RNA-binding protein, even surpassing N, S, M, and all Nsp proteins [112]. CNBP has also been reported to bind and unfold G4 formed in the SARS-CoV-2 genome in vitro [39]. Thus, CNBP appears to be crucial for SARS-CoV-2, and the available evidence seems to point to its unfolding activity on SARS-CoV-2 G4s, just as the helicase does. Altogether, we found these potential G4 regulators in the RNA interactome of SARS-CoV-2, and further studies are needed to uncover their detailed functions.
Here, we summarized the potential G4-binding proteins from both the virus and the host in Table 4 . Now, this is a rapidly developing research field. When we were almost finished with this review, Mukherjee et al. reported modulation of the conformational space of RG-1 G4 in SARS-CoV-2 by cellular components and two amyloidogenic peptides, α-Syn and hIAPP, which are involved in Parkinson's disease and type-II diabetes [148]. These peptides stabilized RG-1 folding in a sequence-specific way, which was different from their interaction with human telomeric G4. The specific interaction may alter the expression profiles of disease-modifying genes and contribute to the development of specific ligands for intervention. Therefore, we look forward to the discovery of more specific interactions between SARS-CoV-2 G4 and proteins, which could be studied in-depth and applied to the treatment of SARS-CoV-2 in the future.
Table 4.
Potential G4-interacting proteins in the interactome of SARS-CoV-2 genome.
| Origin | Protein | Bind G4s | Unfold G4s | Stabilize G4s | Bind SARS-CoV-2 genome | Inhibitor | Reference | |
|---|---|---|---|---|---|---|---|---|
| Virus | SARS-CoV | SUD | ✓ | [52], [53] | ||||
| SARS-CoV-2 | SUD | ✓ | [55] | |||||
| Nsp13 | ✓ | ✓ | ✓ | [33], [75], [76], [77] | ||||
| N | ✓ | ✓ | [112], [120] | |||||
| Host | hnRNPs | hnRNP A1 | ✓ | ✓ | ✓ | [112], [113], [117] | ||
| hnRNP A2 | ✓ | ✓ | ✓ | [113], [121] | ||||
| hnRNP AB | ✓ | ✓ | ✓ | [112], [113], [122] | ||||
| hnRNP A2B1 | ✓ | ✓ | ✓ | [111], [112], [113], [116] | ||||
| hnRNP D | ✓ | ✓ | ✓ | [113], [123], [124] | ||||
| hnRNP F | ✓ | ✓ | ✓ | [113], [125], [126] | ||||
| hnRNP H | ✓ | ✓ | ✓ | [113], [126] | ||||
| hnRNP P2 | ✓ | ✓ | [113], [127] | |||||
| RNA Helicases | DDX1 | ✓ | ✓ | ✓ | [112], [128] | |||
| DDX3X | ✓ | ✓ | ✓ | [103], [111], [112], [113], [129], [130] | ||||
| DDX5 | ✓ | ✓ | ✓ | [113], [131] | ||||
| DDX21 | ✓ | ✓ | [104] | |||||
| DDX42 | ✓ | [132] | ||||||
| DHX9 | ✓ | ✓ | ✓ | [105], [113] | ||||
| DHX36 | ✓ | ✓ | [105], [133] | |||||
| MOV10 | ✓ | ✓ | ✓ | [112], [134] | ||||
| Other proteins | CNBP | ✓ | ✓ | ✓ | ✓ | [112], [113], [135], [136], [137], [138] | ||
| Nucleolin | ✓ | ✓ | ✓ | ✓ | [113], [139], [140], [141], [142], [143], [144], [145], [146], [147] | |||
5. G4s as biosensors for SARS-CoV-2 detection
In addition to targeting SARS-CoV-2 G4s for antiviral therapy, we further discuss the possibility of using G4s for viral detection. Due to their high sensitivity, high stability, convenient operation, and low cost, G4 structures have been applied as novel biosensors to detect the surrounding conditions, including metal ions, pH, molecular crowding, etc. based on their conformational changes. The signal can be visualized by G4-specific dye molecules; alternatively, G4s combine with hemin to form the active DNAzyme, which then catalyzes the reaction between H2O2 and ABTS, leading to a color change for visualization [149]. It is worth noting that G4 structures have also been employed in the detection of pathogen genes as signal elements (Fig. 8 ). For example, Guo et al. reported a label-free biosensor for HIV-1 detection based on ligand-responsive G4 formation [150]. The target DNA formed a hybrid with the beacon, and then the released strand folded into G4 and could be visualized by NMM (Fig. 8A). Target DNA detection in HBV using PCR amplification was also established based on the sensitive G4 probe [151]. In addition, the ultrasensitive detection of HIV and HCV with G4-specific fluorescent or colorimetric probes as signal carriers have been realized using a DNA cascaded multiple amplification strategy [152].
Fig. 8.
The G4-based biosensor is a potential tool to detect the target gene and target protein. (A) The target gene formed a hybrid with the beacon, and then the released strand folded into G4 and could be visualized either by the fluorescent ligand or by the colorimetric method. (B—C) The binding of the target protein triggers the rapid switching in the G4-aptamer structure and induces the folding or unfolding of G4.
In addition to their application in gene detection, G4-based aptamers have been developed against protein targets as recognition elements for diagnostic purposes (Fig. 8B-C). As a proof of concept, the thrombin binding aptamer (TBA) is the most well-studied G4-aptamer in protein detection due to its high affinity and specificity to the target. The thrombin binding triggers the rapid switching in the aptamer structure and induces the folding of an antiparallel G4 structure that can be detected by a variety of means (Fig. 8B) [153]. Using the first phase II clinical trial aptamer, AS1411, which can specifically bind nucleolin on the surface of cancer cells, an electrochemical sensor that can achieve label-free cancer cell detection was established [154]. The G4-aptamer was also applied to detect Staphylococcus aureus by targeting the presenting protein A on the cell surface, expanding the application of G4-aptamers as an analytical tool for the specific recognition and rapid detection of pathogens [155].
Considering the existence of PQSs in the SARS-CoV-2 genome, a G4-based biosensor has been proposed for SARS-CoV-2 detection in a recent review [156]. With the aid of molecular beacons comprising G4 sequences that can be folded after combining with the target gene, the SARS-CoV-2 gene may be detected as described in the above examples. In addition, identifying and utilizing the conserved PQSs in the SARS-CoV-2 genome may allow simple and direct virus detection (Table 5 ). For example, Josué Carvalho et al. designed two molecular beacons based on conserved SARS-CoV-2 G4s at positions 1574 and 24,268 to develop a faster, more sensitive, and inexpensive SARS-CoV-2 infection detection assay [37]. Recently, a highly conserved antiparallel DNA G4 sequence was identified from the SARS-CoV-2 genome, with unique loop compositions forming a distinct recognition motif. This sequence changes from the duplex form to the G4 structure in a pH-dependent manner. Thereafter, the authors developed a reliable fluorometric detection of SARS-CoV-2 by targeting this DNA G4 through pH-triggered conformational changes [157]. Furthermore, G4-based aptamers against proteins have been used in disease diagnosis and therapy [153]. As there are specific interactions between G4s and SARS-CoV-2-encoded proteins, such as Nsp13 and the SUD, G4-based aptamers may also be applied in SARS-CoV-2 detection. Recently, a 44-nt G4-forming aptamer was identified against the spike protein in SARS-CoV-2 [158], further reflecting the possibility of utilizing a G4-aptamer for SARS-CoV-2 detection (Table 5).
Table 5.
| G4 origin | G4 sequence | Principle | Effect | The possible limitation to users |
|---|---|---|---|---|
| Two molecular beacons were designed based on the complementary sequences from SARS-CoV-2 RNA to detect G4s within the SARS-CoV-2 genome from human samples. |
1.GGTGTTGTTGGAGAAGGTTCCGAAGG 2.GGCTTATAGGTTTAATGGTATTGG |
Detect the G4 regions in ORF1ab and S genes in SARS-CoV-2 RNA by the molecular beacons based on their conformational rearrangements. | A fast (total duration of 2 h 20 min including amplification and fluorescence reading stages) and simple way of detecting SARS-CoV-2 in clinical samples. | Although the sensitivity (96.3 %) was similar to the current methods, the specificity (46.9 %) was still lower than the reference RT-PCR. |
| A conserved G4 DNA was obtained after reverse transcription and amplification from genomic SARS-CoV-2 RNA. | GGTATGTGGAAAGGTTATGG | Detect target G4 DNA by a fluorogenic probe named BTMA within an exclusive pH window (3.5–4.0). | A reliable strategy for a fluorogenic organic molecule-based platform to diagnose COVID-19 clinical samples. | A specific and noncommercial fluorescent probe is required, which was developed by the authors. |
| A 44-mer G4 aptamer against spike trimer antigen of SARS-CoV-2. | TGGGAGCCTGGGACATAGTGGGGAAAGAGGGGAAGAGTGGGTCT | The G4 aptamer binds tightly and selectively to the spike antigen of SARS-CoV-2. | 91 % sensitivity and 98 % specificity between SARS-CoV-2 infected individuals from the non-infected individuals. | Despite the low cost (~$2–3 USD/test), the aptamer-based assay delivers sample-to-answer within 5 h. |
6. Conclusion and perspective
The G4 structure, as a promising drug target, has been widely applied in studies of disease treatment, such as cancer [159], parasite infections [160], neurodegenerative disorders [161], and virus infections [25]. In this review, we focus on G4s in SARS-CoV-2. Over the last two years, studies on the genomic PQSs of SARS-CoV-2 have emerged rapidly, reflecting that the G4 structure has received great attention in virus-related fields. We summarize all the reported SARS-CoV-2 PQSs to date in Table 1, Table 2. These PQSs still require in-depth structural and functional characterization.
In addition to the G4 structure itself, we also comprehensively introduce the G4-interacting proteins of SARS-CoV-2 from both the virus and the host cell, as summarized in Table 4. The SARS-CoV-2 SUD (Nsp3), Nsp13, and N proteins are the most significant virus-encoded G4 regulators. In addition, the host proteins that have been confirmed to be members of the SARS-CoV-2 RNA interactome and previously shown to act as G4 regulators in other viruses, such as helicases, hnRNPs, nucleolin, and CNBP, are also presented because these proteins may serve as potential drug targets to interfere with the normal functions of viral G4s. As the structures of some G4-binding proteins are already known, it is feasible to design inhibitors against these proteins to inhibit their interactions with G4s. Finally, we introduce the application of G4s as a possible approach to detect SARS-CoV-2.
It is worth noting that research in this area has just begun, and many questions are waiting to be resolved, including the following:
-
1)
There are many predictions about the PQSs in SARS-CoV-2, but only some of them have been experimentally verified in vitro. The 3D structures of these G4s as well as their folding dynamics remain to be solved. Furthermore, whether all these PQSs can fold in vivo is also unclear.
-
2)
RNA G4s may act as cis-acting elements in the process of SARS-CoV-2 transcription, replication, translation, and genome packaging, as shown in Fig. 2. However, the exact functions of RNA G4s in SARS-CoV-2 need to be further explored experimentally.
-
3)
Trans-acting factors are required to resolve the RNA G4s in the viral life cycle, which are likely to be helicases. However, it is unknown whether host helicases or viral Nsp13 perform this function or whether there are more complex regulatory interactions between them. Whether host helicases are directly hijacked by viral RNA G4s or recruited through the N protein to viral RNA G4s is also not clear.
-
4)
The identification and functional analysis of SARS-CoV-2 RNA G4-interacting proteins in vivo is needed. In particular, for host proteins, it is important to determine whether they are proviral or antiviral.
-
5)
Using PDP to stabilize SARS-CoV-2 RG-1 and reduce N protein levels provided some clues regarding mechanisms of interference with viral G4s [38]. Whether the ligands can be effective in virus treatment still needs further verification. Moreover, as both viruses and hosts contain a large number of PQSs, designing ligands specific for virus G4s without affecting the normal function of cells is particularly important. This requires an in-depth study of the structures and folding mechanism of SARS-CoV-2 G4s.
-
6)
In addition to SARS-CoV-2 RNA G4s, targeting G4-related proteins by inhibitors or G4-aptamers is also a plausible strategy for antiviral therapy.
Funding
This work was supported by the National Natural Science Foundation of China (32071225), and the Natural Science Basic Research Program of Shaanxi (2020JQ-251).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to thank Dr. Fang-Yuan Teng at the Affiliated Hospital of Southwest Medical University, Dr. Wen-Qiang Wu at Henan University, and Ms. Hai-Hong Li at Northwest A&F University for insightful discussions.
Data availability
Data will be made available on request.
References
- 1.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in WuhanChina. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hui D.S., Madani T.A., Ntoumi F., Kock R., Dar O., Ippolito G., McHugh T.D., Memish Z.A., Drosten C., Zumla A., Petersen E., E I.A. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health - The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 5.Rabaan A.A., Al-Ahmed S.H., Haque S., Sah R., Tiwari R., Malik Y.S., Dhama K., Yatoo M.I., Bonilla-Aldana D.K., Rodriguez-Morales A.J. SARS-CoV-2, SARS-CoV, and MERS-COV: a comparative overview. Infez. Med. 2020;28(2):174–184. [PubMed] [Google Scholar]
- 6.Zhong N.S., Zheng B.J., Li Y.M., Poon Z.H.Xie, Chan K.H., Li P.H., Tan S.Y., Chang Q., Xie J.P., Liu X.Q., Xu J., Li D.X., Yuen K.Y., Peiris Y.Guan. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet. 2003;362(9393):1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MERS-CoV: a global challenge. Lancet. 2013;381(9882):1960. doi: 10.1016/S0140-6736(13)61184-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watson J.D., Crick F.H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171(4356):737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 9.Wang A.H., Quigley G.J., Kolpak F.J., Crawford J.L., van Boom J.H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- 10.Panayotatos N., Wells R.D. Cruciform structures in supercoiled DNA. Nature. 1981;289(5797):466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- 11.Soyfer V.N., Potaman V.N. The Discovery of Triple-Stranded Nucleic Acids. Springer; New York: 1996. pp. 1–46. [Google Scholar]
- 12.Sen D., Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334(6180):364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 13.Maizels N., Gray L.T. The G4 genome. PLoS Genet. 2013;9(4) doi: 10.1371/journal.pgen.1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Besnard E., Babled A., Lapasset L., Milhavet O., Parrinello H., Dantec C., Marin J.M., Lemaitre J.M. Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat. Struct. Mol. Biol. 2012;19(8):837–844. doi: 10.1038/nsmb.2339. [DOI] [PubMed] [Google Scholar]
- 15.Eddy J., Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34(14):3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rhodes D., Lipps H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015;43(18):8627–8637. doi: 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansel-Hertsch R., Di Antonio M., Balasubramanian S. DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 2017;18(5):279–284. doi: 10.1038/nrm.2017.3. [DOI] [PubMed] [Google Scholar]
- 18.Varshney D., Spiegel J., Zyner K., Tannahill D., Balasubramanian S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020;21(8):459–474. doi: 10.1038/s41580-020-0236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun D., Thompson B., Cathers B.E., Salazar M., Kerwin S.M., Trent J.O., Jenkins T.C., Neidle S., Hurley L.H. Inhibition of human telomerase by a G-quadruplex-interactive compound. J. Med. Chem. 1997;40(14):2113–2116. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- 20.Neidle S., Parkinson G. Telomere maintenance as a target for anticancer drug discovery. Nat. Rev. Drug Discov. 2002;1(5):383–393. doi: 10.1038/nrd793. [DOI] [PubMed] [Google Scholar]
- 21.Bejugam M., Sewitz S., Shirude P.S., Rodriguez R., Shahid R., Balasubramanian S. Trisubstituted isoalloxazines as a new class of G-quadruplex binding ligands: small molecule regulation of c-kit oncogene expression. J. Am. Chem. Soc. 2007;129(43):12926–12927. doi: 10.1021/ja075881p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavezzo E., Berselli M., Frasson I., Perrone R., Palu G., Brazzale A.R., Richter S.N., Toppo S. G-quadruplex forming sequences in the genome of all known human viruses: a comprehensive guide. PLoS Comput. Biol. 2018;14(12) doi: 10.1371/journal.pcbi.1006675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruggiero E., Zanin I., Terreri M., Richter S.N. G-quadruplex targeting in the fight against viruses: an update. Int. J. Mol. Sci. 2021;22(20) doi: 10.3390/ijms222010984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abiri A., Lavigne M., Rezaei M., Nikzad S., Zare P., Mergny J.L., Rahimi H.R. Unlocking G-quadruplexes as antiviral targets. Pharmacol. Rev. 2021;73(3):897–923. doi: 10.1124/pharmrev.120.000230. [DOI] [PubMed] [Google Scholar]
- 25.Ruggiero E., Richter S.N. G-quadruplexes and G-quadruplex ligands: targets and tools in antiviral therapy. Nucleic Acids Res. 2018;46(7):3270–3283. doi: 10.1093/nar/gky187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181(4):914–921. doi: 10.1016/j.cell.2020.04.011. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huston N.C., Wan H., Strine M.S., de Cesaris Araujo Tavares R., Wilen C.B., Pyle A.M. Comprehensive in vivo secondary structure of the SARS-CoV-2 genome reveals novel regulatory motifs and mechanisms. Mol. Cell. 2021;81(3):584–598. doi: 10.1016/j.molcel.2020.12.041. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manfredonia I., Incarnato D. Structure and regulation of coronavirus genomes: state-of-the-art and novel insights from SARS-CoV-2 studies. Biochem. Soc. Trans. 2021;49(1):341–352. doi: 10.1042/BST20200670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manfredonia I., Nithin C., Ponce-Salvatierra A., Ghosh P., Wirecki T.K., Marinus T., Ogando N.S., Snijder E.J., van Hemert M.J., Bujnicki J.M., Incarnato D. Genome-wide mapping of SARS-CoV-2 RNA structures identifies therapeutically-relevant elements. Nucleic Acids Res. 2020;48(22):12436–12452. doi: 10.1093/nar/gkaa1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simmonds P. Pervasive RNA secondary structure in the genomes of SARS-CoV-2 and other coronaviruses. MBio. 2020;11(6) doi: 10.1128/mBio.01661-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cui H., Zhang L. G-quadruplexes are present in human coronaviruses including SARS-CoV-2. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.567317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Panera N., Tozzi A.E., Alisi A. The G-Quadruplex/Helicase world as a potential antiviral approach against COVID-19. Drugs. 2020;80(10):941–946. doi: 10.1007/s40265-020-01321-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ji D., Juhas M., Tsang C.M., Kwok C.K., Li Y., Zhang Y. Discovery of G-quadruplex-forming sequences in SARS-CoV-2. Brief. Bioinform. 2021;22(2):1150–1160. doi: 10.1093/bib/bbaa114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang R., Xiao K., Gu Y., Liu H., Sun X. Whole genome identification of potential G-quadruplexes and analysis of the G-quadruplex binding domain for SARS-CoV-2. Front. Genet. 2020;11 doi: 10.3389/fgene.2020.587829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bartas M., Brazda V., Bohalova N., Cantara A., Volna A., Stachurova T., Malachova K., Jagelska E.B., Porubiakova O., Cerven J., Pecinka P. In-depth bioinformatic analyses of nidovirales including human SARS-CoV-2, SARS-CoV, MERS-CoV viruses suggest important roles of non-canonical nucleic acid structures in their lifecycles. Front. Microbiol. 2020;11:1583. doi: 10.3389/fmicb.2020.01583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belmonte-Reche E., Serrano-Chacon I., Gonzalez C., Gallo J., Banobre-Lopez M. Potential G-quadruplexes and i-motifs in the SARS-CoV-2. PLoS One. 2021;16(6) doi: 10.1371/journal.pone.0250654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carvalho J., Lopes-Nunes J., Figueiredo J., Santos T., Miranda A., Riscado M., Sousa F., Duarte A.P., Socorro S., Tomaz C.T., Felgueiras M., Teixeira R., Faria C., Cruz C. Molecular Beacon assay development for severe acute respiratory syndrome coronavirus 2 detection. Sensors (Basel) 2021;21(21) doi: 10.3390/s21217015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao C., Qin G., Niu J., Wang Z., Wang C., Ren J., Qu X. Targeting RNA G-quadruplex in SARS-CoV-2: a promising therapeutic target for COVID-19? Angew. Chem. Int. Ed. Engl. 2021;60(1):432–438. doi: 10.1002/anie.202011419. [DOI] [PubMed] [Google Scholar]
- 39.Bezzi G., Piga E.J., Binolfi A., Armas P. CNBP binds and unfolds in vitro G-quadruplexes formed in the SARS-CoV-2 positive and negative genome strands. Int. J. Mol. Sci. 2021;22(5) doi: 10.3390/ijms22052614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miclot T., Hognon C., Bignon E., Terenzi A., Marazzi M., Barone G., Monari A. Structure and dynamics of RNA guanine quadruplexes in SARS-CoV-2 Genome. Original strategies against emerging viruses. J. Phys. Chem. Lett. 2021;12(42):10277–10283. doi: 10.1021/acs.jpclett.1c03071. [DOI] [PubMed] [Google Scholar]
- 41.Wang S.R., Min Y.Q., Wang J.Q., Liu C.X., Fu B.S., Wu F., Wu L.Y., Qiao Z.X., Song Y.Y., Xu G.H., Wu Z.G., Huang G., Peng N.F., Huang R., Mao W.X., Peng S., Chen Y.Q., Zhu Y., Tian T., Zhang X.L., Zhou X. A highly conserved G-rich consensus sequence in hepatitis C virus core gene represents a new anti-hepatitis C target. Sci. Adv. 2016;2(4) doi: 10.1126/sciadv.1501535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arora A., Balasubramanian C., Kumar N., Agrawal S., Ojha R.P., Maiti S. Binding of berberine to human telomeric quadruplex - spectroscopic, calorimetric and molecular modeling studies. FEBS J. 2008;275(15):3971–3983. doi: 10.1111/j.1742-4658.2008.06541.x. [DOI] [PubMed] [Google Scholar]
- 43.Warowicka A., Nawrot R., Gozdzicka-Jozefiak A. Antiviral activity of berberine. Arch. Virol. 2020;165(9):1935–1945. doi: 10.1007/s00705-020-04706-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oliva R., Mukherjee S., Manisegaran M., Campanile M., Del Vecchio P., Petraccone L., Winter R. Binding properties of RNA quadruplex of SARS-CoV-2 to berberine compared to telomeric DNA quadruplex. Int. J. Mol. Sci. 2022;23(10) doi: 10.3390/ijms23105690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen L.W., Qian M.Q., Yu K., Narva S., Yu F., Wu Y.L., Zhang W. Inhibition of influenza a virus propagation by benzoselenoxanthenes stabilizing TMPRSS2 gene G-quadruplex and hence down-regulating TMPRSS2 expression. Sci. Rep. 2020;10(1):7635. doi: 10.1038/s41598-020-64368-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu G., Du W., Sang X., Tong Q., Wang Y., Chen G., Yuan Y., Jiang L., Cheng W., Liu D., Tian Y., Fu X. RNA G-quadruplex in TMPRSS2 reduces SARS-CoV-2 infection. Nat. Commun. 2022;13(1):1444. doi: 10.1038/s41467-022-29135-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koirala D., Dhakal S., Ashbridge B., Sannohe Y., Rodriguez R., Sugiyama H., Balasubramanian S., Mao H. A single-molecule platform for investigation of interactions between G-quadruplexes and small-molecule ligands. Nat. Chem. 2011;3(10):782–787. doi: 10.1038/nchem.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331(5):991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S., Weissbrich B., Snijder E.J., Rabenau H., Doerr H.W., Gorbalenya A.E., Ziebuhr J. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84(Pt 9):2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 51.Lei J., Kusov Y., Hilgenfeld R. Nsp3 of coronaviruses: structures and functions of a large multi-domain protein. Antivir. Res. 2018;149:58–74. doi: 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tan J., Kusov Y., Mutschall D., Tech S., Nagarajan K., Hilgenfeld R., Schmidt C.L. The "SARS-unique domain" (SUD) of SARS coronavirus is an oligo(G)-binding protein. Biochem. Biophys. Res. Commun. 2007;364(4):877–882. doi: 10.1016/j.bbrc.2007.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tan J., Vonrhein C., Smart O.S., Bricogne G., Bollati M., Kusov Y., Hansen G., Mesters J.R., Schmidt C.L., Hilgenfeld R. The SARS-unique domain (SUD) of SARS coronavirus contains two macrodomains that bind G-quadruplexes. PLoS Pathog. 2009;5(5) doi: 10.1371/journal.ppat.1000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson M.A., Chatterjee A., Neuman B.W., Wuthrich K. SARS coronavirus unique domain: three-domain molecular architecture in solution and RNA binding. J. Mol. Biol. 2010;400(4):724–742. doi: 10.1016/j.jmb.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lavigne M., Helynck O., Rigolet P., Boudria-Souilah R., Nowakowski M., Baron B., Brule S., Hoos S., Raynal B., Guittat L., Beauvineau C., Petres S., Granzhan A., Guillon J., Pratviel G., Teulade-Fichou M.P., England P., Mergny J.L., Munier-Lehmann H. SARS-CoV-2 Nsp3 unique domain SUD interacts with guanine quadruplexes and G4-ligands inhibit this interaction. Nucleic Acids Res. 2021;49(13):7695–7712. doi: 10.1093/nar/gkab571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lei J., Ma-Lauer Y., Han Y., Thoms M., Buschauer R., Jores J., Thiel V., Beckmann R., Deng W., Leonhardt H., Hilgenfeld R., von Brunn A. The SARS-unique domain (SUD) of SARS-CoV and SARS-CoV-2 interacts with human Paip1 to enhance viral RNA translation. EMBO J. 2021;40(11) doi: 10.15252/embj.2019102277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hognon C., Miclot T., Garci A.I.C., Frances-Monerris A., Grandemange S., Terenzi A., Marazzi M., Barone G., Monari A. Role of RNA guanine quadruplexes in favoring the dimerization of SARS unique domain in coronaviruses. J. Phys. Chem. Lett. 2020;11(14):5661–5667. doi: 10.1021/acs.jpclett.0c01097. [DOI] [PubMed] [Google Scholar]
- 58.Kusov Y., Tan J., Alvarez E., Enjuanes L., Hilgenfeld R. A G-quadruplex-binding macrodomain within the "SARS-unique domain" is essential for the activity of the SARS-coronavirus replication-transcription complex. Virology. 2015;484:313–322. doi: 10.1016/j.virol.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brazda V., Fojta M. The rich world of p53 DNA binding targets: the role of DNA structure. Int. J. Mol. Sci. 2019;20(22) doi: 10.3390/ijms20225605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wei J.S., Whiteford C.C., Cenacchi N., Son C.G., Khan J. BBC3 mediates fenretinide-induced cell death in neuroblastoma. Oncogene. 2005;24(54):7976–7983. doi: 10.1038/sj.onc.1208947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garcia-Saez I., Tcherniuk S., Kozielski F. The structure of human neuronal Rab6B in the active and inactive form. Acta Crystallogr. D Biol. Crystallogr. 2006;62(Pt 7):725–733. doi: 10.1107/S0907444906015319. [DOI] [PubMed] [Google Scholar]
- 62.Mizutani T. Signal transduction in SARS-CoV-infected cells. Ann. N. Y. Acad. Sci. 2007;1102:86–95. doi: 10.1196/annals.1408.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jin G., Klika A., Callahan M., Faga B., Danzig J., Jiang Z., Li X., Stark G.R., Harrington J., Sherf B. Identification of a human NF-kappaB-activating protein, TAB3. Proc. Natl. Acad. Sci. U. S. A. 2004;101(7):2028–2033. doi: 10.1073/pnas.0307314101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ma-Lauer Y., Carbajo-Lozoya J., Hein M.Y., Muller M.A., Deng W., Lei J., Meyer B., Kusov Y., von Brunn B., Bairad D.R., Hunten S., Drosten C., Hermeking H., Leonhardt H., Mann M., Hilgenfeld R., von Brunn A. p53 down-regulates SARS coronavirus replication and is targeted by the SARS-unique domain and PLpro via E3 ubiquitin ligase RCHY1. Proc. Natl. Acad. Sci. U. S. A. 2016;113(35):E5192–E5201. doi: 10.1073/pnas.1603435113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chang Y.S., Ko B.H., Ju J.C., Chang H.H., Huang S.H., Lin C.W. SARS unique domain (SUD) of severe acute respiratory syndrome coronavirus induces NLRP3 inflammasome-dependent CXCL10-mediated pulmonary inflammation. Int. J. Mol. Sci. 2020;21(9) doi: 10.3390/ijms21093179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shu T., Huang M., Wu D., Ren Y., Zhang X., Han Y., Mu J., Wang R., Qiu Y., Zhang D.Y., Zhou X. SARS-Coronavirus-2 Nsp13 possesses NTPase and RNA helicase activities that can be inhibited by bismuth salts. Virol. Sin. 2020;35(3):321–329. doi: 10.1007/s12250-020-00242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ivanov K.A., Ziebuhr J. Human coronavirus 229E nonstructural protein 13: characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5′-triphosphatase activities. J. Virol. 2004;78(14):7833–7838. doi: 10.1128/JVI.78.14.7833-7838.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Seybert A., Hegyi A., Siddell S.G., Ziebuhr J. The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5′-to-3′ polarity. RNA. 2000;6(7):1056–1068. doi: 10.1017/s1355838200000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen J., Malone B., Llewellyn E., Grasso M., Shelton P.M.M., Olinares P.D.B., Maruthi K., Eng E.T., Vatandaslar H., Chait B.T., Kapoor T.M., Darst S.A., Campbell E.A. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell. 2020;182(6):1560–1573. doi: 10.1016/j.cell.2020.07.033. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jia Z., Yan L., Ren Z., Wu L., Wang J., Guo J., Zheng L., Ming Z., Zhang L., Lou Z., Rao Z. Delicate structural coordination of the severe acute respiratory syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019;47(12):6538–6550. doi: 10.1093/nar/gkz409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Newman J.A., Douangamath A., Yadzani S., Yosaatmadja Y., Aimon A., Brandao-Neto J., Dunnett L., Gorrie-Stone T., Skyner R., Fearon D., Schapira M., von Delft F., Gileadi O. Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase. Nat. Commun. 2021;12(1):4848. doi: 10.1038/s41467-021-25166-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78(11):5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xia H., Cao Z., Xie X., Zhang X., Chen J.Y., Wang H., Menachery V.D., Rajsbaum R., Shi P.Y. Evasion of type I interferon by SARS-CoV-2. Cell Rep. 2020;33(1) doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shum K.T., Tanner J.A. Differential inhibitory activities and stabilisation of DNA aptamers against the SARS coronavirus helicase. Chembiochem. 2008;9(18):3037–3045. doi: 10.1002/cbic.200800491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tanner J.A., Zheng B.J., Zhou J., Watt R.M., Jiang J.Q., Wong K.L., Lin Y.P., Lu L.Y., He M.L., Kung H.F., Kesel A.J., Huang J.D. The adamantane-derived bananins are potent inhibitors of the helicase activities and replication of SARS coronavirus. Chem Biol. 2005;12(3):303–311. doi: 10.1016/j.chembiol.2005.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee C., Lee J.M., Lee N.R., Kim D.E., Jeong Y.J., Chong Y. Investigation of the pharmacophore space of severe acute respiratory syndrome coronavirus (SARS-CoV) NTPase/helicase by dihydroxychromone derivatives. Bioorg. Med. Chem. Lett. 2009;19(16):4538–4541. doi: 10.1016/j.bmcl.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Adedeji A.O., Singh K., Calcaterra N.E., DeDiego M.L., Enjuanes L., Weiss S., Sarafianos S.G. Severe acute respiratory syndrome coronavirus replication inhibitor that interferes with the nucleic acid unwinding of the viral helicase. Antimicrob. Agents Chemother. 2012;56(9):4718–4728. doi: 10.1128/AAC.00957-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.White M.A., Lin W., Cheng X. Discovery of COVID-19 inhibitors targeting the SARS-CoV-2 Nsp13 helicase. J. Phys. Chem. Lett. 2020;11(21):9144–9151. doi: 10.1021/acs.jpclett.0c02421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Corona A., Wycisk K., Talarico C., Manelfi C., Milia J., Cannalire R., Esposito F., Gribbon P., Zaliani A., Iaconis D., Beccari A.R., Summa V., Nowotny M., Tramontano E. Natural compounds inhibit SARS-CoV-2 nsp13 unwinding and ATPase enzyme activities. ACS Pharmacol. Transl. Sci. 2022;5(4):226–239. doi: 10.1021/acsptsci.1c00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chang C.K., Hou M.H., Chang C.F., Hsiao C.D., Huang T.H. The SARS coronavirus nucleocapsid protein–forms and functions. Antivir. Res. 2014;103:39–50. doi: 10.1016/j.antiviral.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ye Q., West A.M.V., Silletti S., Corbett K.D. bioRxiv; 2020. Architecture and Self-assembly of the SARS-CoV-2 Nucleocapsid Protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Peng Y., Du N., Lei Y., Dorje S., Qi J., Luo T., Gao G.F., Song H. Structures of the SARS-CoV-2 nucleocapsid and their perspectives for drug design. EMBO J. 2020;39(20) doi: 10.15252/embj.2020105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang Q., Yu L., Petros A.M., Gunasekera A., Liu Z., Xu N., Hajduk P., Mack J., Fesik S.W., Olejniczak E.T. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry. 2004;43(20):6059–6063. doi: 10.1021/bi036155b. [DOI] [PubMed] [Google Scholar]
- 84.Chang C.K., Sue S.C., Yu T.H., Hsieh C.M., Tsai C.K., Chiang Y.C., Lee S.J., Hsiao H.H., Wu W.J., Chang W.L., Lin C.H., Huang T.H. Modular organization of SARS coronavirus nucleocapsid protein. J. Biomed. Sci. 2006;13(1):59–72. doi: 10.1007/s11373-005-9035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Masters P.S. Coronavirus genomic RNA packaging. Virology. 2019;537:198–207. doi: 10.1016/j.virol.2019.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hsieh P.K., Chang S.C., Huang C.C., Lee T.T., Hsiao C.W., Kou Y.H., Chen I.Y., Chang C.K., Huang T.H., Chang M.F. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J. Virol. 2005;79(22):13848–13855. doi: 10.1128/JVI.79.22.13848-13855.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen Z., Wang C., Feng X., Nie L., Tang M., Zhang H., Xiong Y., Swisher S.K., Srivastava M., Chen J. Interactomes of SARS-CoV-2 and human coronaviruses reveal host factors potentially affecting pathogenesis. EMBO J. 2021;40(17) doi: 10.15252/embj.2021107776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen H., Cui Y., Han X., Hu W., Sun M., Zhang Y., Wang P.H., Song G., Chen W., Lou J. Liquid-liquid phase separation by SARS-CoV-2 nucleocapsid protein and RNA. Cell Res. 2020;30(12):1143–1145. doi: 10.1038/s41422-020-00408-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Perdikari T.M., Murthy A.C., Ryan V.H., Watters S., Naik M.T., Fawzi N.L. SARS-CoV-2 nucleocapsid protein phase-separates with RNA and with human hnRNPs. EMBO J. 2020;39(24) doi: 10.15252/embj.2020106478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Surjit M., Liu B., Chow V.T., Lal S.K. The nucleocapsid protein of severe acute respiratory syndrome-coronavirus inhibits the activity of cyclin-cyclin-dependent kinase complex and blocks S phase progression in mammalian cells. J. Biol. Chem. 2006;281(16):10669–10681. doi: 10.1074/jbc.M509233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lu X., Pan J., Tao J., Guo D. SARS-CoV nucleocapsid protein antagonizes IFN-beta response by targeting initial step of IFN-beta induction pathway, and its C-terminal region is critical for the antagonism. Virus Genes. 2011;42(1):37–45. doi: 10.1007/s11262-010-0544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pan P., Shen M., Yu Z., Ge W., Chen K., Tian M., Xiao F., Wang Z., Wang J., Jia Y., Wang W., Wan P., Zhang J., Chen W., Lei Z., Chen X., Luo Z., Zhang Q., Xu M., Li G., Li Y., Wu J. SARS-CoV-2 N protein promotes NLRP3 inflammasome activation to induce hyperinflammation. Nat. Commun. 2021;12(1):4664. doi: 10.1038/s41467-021-25015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mu J., Xu J., Zhang L., Shu T., Wu D., Huang M., Ren Y., Li X., Geng Q., Xu Y., Qiu Y., Zhou X. SARS-CoV-2-encoded nucleocapsid protein acts as a viral suppressor of RNA interference in cells. Sci. China Life Sci. 2020;63(9):1413–1416. doi: 10.1007/s11427-020-1692-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cui L., Wang H., Ji Y., Yang J., Xu S., Huang X., Wang Z., Qin L., Tien P., Zhou X., Guo D., Chen Y. The nucleocapsid protein of coronaviruses acts as a viral suppressor of RNA silencing in mammalian cells. J. Virol. 2015;89(17):9029–9043. doi: 10.1128/JVI.01331-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gordon D.E., Hiatt J., Bouhaddou M., Rezelj V.V., Ulferts S., Braberg H., Jureka A.S., Obernier K., Guo J.Z., Batra J., Kaake R.M., Weckstein A.R., Owens T.W., Gupta M., Pourmal S., Titus E.W., Cakir M., Soucheray M., McGregor M., Cakir Z., Jang G., O'Meara M.J., Tummino T.A., Zhang Z., Foussard H., Rojc A., Zhou Y., Kuchenov D., Huttenhain R., Xu J., Eckhardt M., Swaney D.L., Fabius J.M., Ummadi M., Tutuncuoglu B., Rathore U., Modak M., Haas P., Haas K.M., Naing Z.Z.C., Pulido E.H., Shi Y., Barrio-Hernandez I., Memon D., Petsalaki E., Dunham A., Marrero M.C., Burke D., Koh C., Vallet T., Silvas J.A., Azumaya C.M., Billesbolle C., Brilot A.F., Campbell M.G., Diallo A., Dickinson M.S., Diwanji D., Herrera N., Hoppe N., Kratochvil H.T., Liu Y., Merz G.E., Moritz M., Nguyen H.C., Nowotny C., Puchades C., Rizo A.N., Schulze-Gahmen U., Smith A.M., Sun M., Young I.D., Zhao J., Asarnow D., Biel J., Bowen A., Braxton J.R., Chen J., Chio C.M., Chio U.S., Deshpande I., Doan L., Faust B., Flores S., Jin M., Kim K., Lam V.L., Li F., Li J., Li Y.L., Li Y., Liu X., Lo M., Lopez K.E., Melo A.A., Moss F.R., 3rd, Nguyen P., Paulino J., Pawar K.I., Peters J.K., Pospiech T.H., Jr., Safari M., Jr., Sangwan S., Jr., Schaefer K., Jr., Thomas P.V., Jr., Thwin A.C., Jr., Trenker R., Jr., Tse E., Jr., Tsui T.K.M., Jr., Wang F., Jr., Whitis N., Jr., Yu Z., Jr., Zhang K., Jr., Zhang Y., Jr., Zhou F., Jr., Saltzberg D., Jr., Consortium Q.S.B., Jr., Hodder A.J., Jr., Shun-Shion A.S., Jr., Williams D.M., Jr., White K.M., Jr., Rosales R., Jr., Kehrer T., Jr., Miorin L., Jr., Moreno E., Jr., Patel A.H., Jr., Rihn S., Jr., Khalid M.M., Jr., Vallejo-Gracia A., Jr., Fozouni P., Jr., Simoneau C.R., Jr., Roth T.L., Jr., Wu D., Jr., Karim M.A., Jr., Ghoussaini M., Jr., Dunham I., Jr., Berardi F., Jr., Weigang S., Jr., Chazal M., Jr., Park J., Jr., Logue J., Jr., McGrath M., Jr., Weston S., Jr., Haupt R., Jr., Hastie C.J., Jr., Elliott M., Jr., Brown F., Jr., Burness K.A., Jr., Reid E., Jr., Dorward M., Jr., Johnson C., Jr., Wilkinson S.G., Jr., Geyer A., Jr., Giesel D.M., Jr., Baillie C., Jr., Raggett S., Jr., Leech H., Jr., Toth R., Jr., Goodman N., Jr., Keough K.C., Jr., Lind A.L., Jr., Zoonomia C., Jr., Klesh R.J., Jr., Hemphill K.R., Jr., Carlson-Stevermer J., Jr., Oki J., Jr., Holden K., Jr., Maures T., Jr., Pollard K.S., Jr., Sali A., Jr., Agard D.A., Jr., Cheng Y., Jr., Fraser J.S., Jr., Frost A., Jr., Jura N., Jr., Kortemme T., Jr., Manglik A., Jr., Southworth D.R., Jr., Stroud R.M., Jr., Alessi D.R., Jr., Davies P., Jr., Frieman M.B., Jr., Ideker T., Jr., Abate C., Jr., Jouvenet N., Jr., Kochs G., Jr., Shoichet B., Jr., Ott M., Palmarini M., Shokat K.M., Garcia-Sastre A., Rassen J.A., Grosse R., Rosenberg O.S., Verba K.A., Basler C.F., Vignuzzi M., Peden A.A., Beltrao P., Krogan N.J. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science. 2020;370(6521) doi: 10.1126/science.abe9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.You J., Dove B.K., Enjuanes L., DeDiego M.L., Alvarez E., Howell G., Heinen P., Zambon M., Hiscox J.A. Subcellular localization of the severe acute respiratory syndrome coronavirus nucleocapsid protein. J. Gen. Virol. 2005;86(Pt 12):3303–3310. doi: 10.1099/vir.0.81076-0. [DOI] [PubMed] [Google Scholar]
- 97.Kankia B.I., Barany G., Musier-Forsyth K. Unfolding of DNA quadruplexes induced by HIV-1 nucleocapsid protein. Nucleic Acids Res. 2005;33(14):4395–4403. doi: 10.1093/nar/gki741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Butovskaya E., Solda P., Scalabrin M., Nadai M., Richter S.N. HIV-1 nucleocapsid protein unfolds stable RNA G-quadruplexes in the viral genome and is inhibited by G-quadruplex ligands. ACS Infect. Dis. 2019;5(12):2127–2135. doi: 10.1021/acsinfecdis.9b00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Taschuk F., Cherry S. DEAD-box helicases: sensors, regulators, and effectors for antiviral defense. Viruses. 2020;12(2) doi: 10.3390/v12020181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Squeglia F., Romano M., Ruggiero A., Maga G., Berisio R. Host DDX helicases as possible SARS-CoV-2 proviral factors: a structural overview of their hijacking through multiple viral proteins. Front. Chem. 2020;8 doi: 10.3389/fchem.2020.602162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ciccosanti F., Di Rienzo M., Romagnoli A., Colavita F., Refolo G., Castilletti C., Agrati C., Brai A., Manetti F., Botta L., Capobianchi M.R., Ippolito G., Piacentini M., Fimia G.M. Proteomic analysis identifies the RNA helicase DDX3X as a host target against SARS-CoV-2 infection. Antivir. Res. 2021;190 doi: 10.1016/j.antiviral.2021.105064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Huttenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O'Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu-Ozturk D., Wang H.Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d'Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., Garcia-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Herdy B., Mayer C., Varshney D., Marsico G., Murat P., Taylor C., D'Santos C., Tannahill D., Balasubramanian S. Analysis of NRAS RNA G-quadruplex binding proteins reveals DDX3X as a novel interactor of cellular G-quadruplex containing transcripts. Nucleic Acids Res. 2018;46(21):11592–11604. doi: 10.1093/nar/gky861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McRae E.K.S., Booy E.P., Moya-Torres A., Ezzati P., Stetefeld J., McKenna S.A. Human DDX21 binds and unwinds RNA guanine quadruplexes. Nucleic Acids Res. 2017;45(11):6656–6668. doi: 10.1093/nar/gkx380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Murat P., Marsico G., Herdy B., Ghanbarian A.T., Portella G., Balasubramanian S. RNA G-quadruplexes at upstream open reading frames cause DHX36- and DHX9-dependent translation of human mRNAs. Genome Biol. 2018;19(1):229. doi: 10.1186/s13059-018-1602-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.de Wilde A.H., Snijder E.J., Kikkert M., van Hemert M.J. Host factors in coronavirus replication. Curr. Top. Microbiol. Immunol. 2018;419:1–42. doi: 10.1007/82_2017_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fung T.S., Liu D.X. Human coronavirus: host-pathogen interaction. Annu. Rev. Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- 108.Sa Ribero M., Jouvenet N., Dreux M., Nisole S. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 2020;16(7) doi: 10.1371/journal.ppat.1008737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z., Wang C., Wang Y., Li L., Ren L., Guo F., Zhao Z., Zhou Z., Xiang Z., Wang J. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11(1):3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lee S., Lee Y.S., Choi Y., Son A., Park Y., Lee K.M., Kim J., Kim J.S., Kim V.N. The SARS-CoV-2 RNA interactome. Mol. Cell. 2021;81(13):2838–2850. doi: 10.1016/j.molcel.2021.04.022. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kamel W., Noerenberg M., Cerikan B., Chen H., Jarvelin A.I., Kammoun M., Lee J.Y., Shuai N., Garcia-Moreno M., Andrejeva A., Deery M.J., Johnson N., Neufeldt C.J., Cortese M., Knight M.L., Lilley K.S., Martinez J., Davis I., Bartenschlager R., Mohammed S., Castello A. Global analysis of protein-RNA interactions in SARS-CoV-2-infected cells reveals key regulators of infection. Mol. Cell. 2021;81(13):2851–2867. doi: 10.1016/j.molcel.2021.05.023. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schmidt N., Lareau C.A., Keshishian H., Ganskih S., Schneider C., Hennig T., Melanson R., Werner S., Wei Y., Zimmer M., Ade J., Kirschner L., Zielinski S., Dolken L., Lander E.S., Caliskan N., Fischer U., Vogel J., Carr S.A., Bodem J., Munschauer M. The SARS-CoV-2 RNA-protein interactome in infected human cells. Nat. Microbiol. 2021;6(3):339–353. doi: 10.1038/s41564-020-00846-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Flynn R.A., Belk J.A., Qi Y., Yasumoto Y., Wei J., Alfajaro M.M., Shi Q., Mumbach M.R., Limaye A., DeWeirdt P.C., Schmitz C.O., Parker K.R., Woo E., Chang H.Y., Horvath T.L., Carette J.E., Bertozzi C.R., Wilen C.B., Satpathy A.T. Discovery and functional interrogation of SARS-CoV-2 RNA-host protein interactions. Cell. 2021;184(9):2394–2411. doi: 10.1016/j.cell.2021.03.012. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lista M.J., Martins R.P., Billant O., Contesse M.A., Findakly S., Pochard P., Daskalogianni C., Beauvineau C., Guetta C., Jamin C., Teulade-Fichou M.P., Fahraeus R., Voisset C., Blondel M. Nucleolin directly mediates epstein-barr virus immune evasion through binding to G-quadruplexes of EBNA1 mRNA. Nat. Commun. 2017;8:16043. doi: 10.1038/ncomms16043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tosoni E., Frasson I., Scalabrin M., Perrone R., Butovskaya E., Nadai M., Palu G., Fabris D., Richter S.N. Nucleolin stabilizes G-quadruplex structures folded by the LTR promoter and silences HIV-1 viral transcription. Nucleic Acids Res. 2015;43(18):8884–8897. doi: 10.1093/nar/gkv897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Scalabrin M., Frasson I., Ruggiero E., Perrone R., Tosoni E., Lago S., Tassinari M., Palu G., Richter S.N. The cellular protein hnRNP A2/B1 enhances HIV-1 transcription by unfolding LTR promoter G-quadruplexes. Sci. Rep. 2017;7:45244. doi: 10.1038/srep45244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dabral P., Babu J., Zareie A., Verma S.C. LANA and hnRNP A1 regulate the translation of LANA mRNA through G-quadruplexes. J. Virol. 2020;94(3) doi: 10.1128/JVI.01508-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mendoza O., Bourdoncle A., Boule J.B., Brosh R.M., Jr., Mergny J.L. G-quadruplexes and helicases. Nucleic Acids Res. 2016;44(5):1989–2006. doi: 10.1093/nar/gkw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Teng F.Y., Jiang Z.Z., Guo M., Tan X.Z., Chen F., Xi X.G., Xu Y. G-quadruplex DNA: a novel target for drug design. Cell. Mol. Life Sci. 2021;78(19–20):6557–6583. doi: 10.1007/s00018-021-03921-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang Y.T., Long X.Y., Ding X., Fan S.R., Cai J.Y., Yang B.J., Zhang X.F., Luo R.H., Yang L., Ruan T., Ren J., Jing C.X., Zheng Y.T., Hao X.J., Chen D.Z. Novel nucleocapsid protein-targeting phenanthridine inhibitors of SARS-CoV-2. Eur. J. Med. Chem. 2022;227 doi: 10.1016/j.ejmech.2021.113966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Khateb S., Weisman-Shomer P., Hershco I., Loeb L.A., Fry M. Destabilization of tetraplex structures of the fragile X repeat sequence (CGG)n is mediated by homolog-conserved domains in three members of the hnRNP family. Nucleic Acids Res. 2004;32(14):4145–4154. doi: 10.1093/nar/gkh745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Weisman-Shomer P., Cohen E., Fry M. Distinct domains in the CArG-box binding factor a destabilize tetraplex forms of the fragile X expanded sequence d(CGG)n. Nucleic Acids Res. 2002;30(17):3672–3681. doi: 10.1093/nar/gkf506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dempsey L.A., Sun H., Hanakahi L.A., Maizels N. G4 DNA binding by LR1 and its subunits, nucleolin and hnRNP D, a role for G-G pairing in immunoglobulin switch recombination. J. Biol. Chem. 1999;274(2):1066–1071. doi: 10.1074/jbc.274.2.1066. [DOI] [PubMed] [Google Scholar]
- 124.Enokizono Y., Konishi Y., Nagata K., Ouhashi K., Uesugi S., Ishikawa F., Katahira M. Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D. J. Biol. Chem. 2005;280(19):18862–18870. doi: 10.1074/jbc.M411822200. [DOI] [PubMed] [Google Scholar]
- 125.Huang H., Zhang J., Harvey S.E., Hu X., Cheng C. RNA G-quadruplex secondary structure promotes alternative splicing via the RNA-binding protein hnRNPF. Genes Dev. 2017;31(22):2296–2309. doi: 10.1101/gad.305862.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Decorsiere A., Cayrel A., Vagner S., Millevoi S. Essential role for the interaction between hnRNP H/F and a G quadruplex in maintaining p53 pre-mRNA 3′-end processing and function during DNA damage. Genes Dev. 2011;25(3):220–225. doi: 10.1101/gad.607011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Imperatore J.A., McAninch D.S., Valdez-Sinon A.N., Bassell G.J., Mihailescu M.R. FUS recognizes G quadruplex structures within neuronal mRNAs. Front. Mol. Biosci. 2020;7:6. doi: 10.3389/fmolb.2020.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.de Almeida C.Ribeiro, Dhir S., Dhir A., Moghaddam A.E., Sattentau Q., Meinhart A., Proudfoot N.J. RNA helicase DDX1 converts RNA G-quadruplex structures into R-loops to promote IgH class switch recombination. Mol. Cell. 2018;70(4):650–662. doi: 10.1016/j.molcel.2018.04.001. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yang S.N.Y., Atkinson S.C., Audsley M.D., Heaton S.M., Jans D.A., Borg N.A. RK-33 is a broad-Spectrum antiviral agent that targets DEAD-box RNA helicase DDX3X. Cells. 2020;9(1) doi: 10.3390/cells9010170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Brai A., Martelli F., Riva V., Garbelli A., Fazi R., Zamperini C., Pollutri A., Falsitta L., Ronzini S., Maccari L., Maga G., Giannecchini S., Botta M. DDX3X helicase inhibitors as a new strategy to fight the West Nile virus infection. J. Med. Chem. 2019;62(5):2333–2347. doi: 10.1021/acs.jmedchem.8b01403. [DOI] [PubMed] [Google Scholar]
- 131.Wu G., Xing Z., Tran E.J., Yang D. DDX5 helicase resolves G-quadruplex and is involved in MYC gene transcriptional activation. Proc. Natl. Acad. Sci. U. S. A. 2019;116(41):20453–20461. doi: 10.1073/pnas.1909047116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zyner K.G., Mulhearn D.S., Adhikari S., Martinez Cuesta S., Di Antonio M., Erard N., Hannon G.J., Tannahill D., Balasubramanian S. Genetic interactions of G-quadruplexes in humans. Elife. 2019;8 doi: 10.7554/eLife.46793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Vaughn J.P., Creacy S.D., Routh E.D., Joyner-Butt C., Jenkins G.S., Pauli S., Nagamine Y., Akman S.A. The DEXH protein product of the DHX36 gene is the major source of tetramolecular quadruplex G4-DNA resolving activity in HeLa cell lysates. J. Biol. Chem. 2005;280(46):38117–38120. doi: 10.1074/jbc.C500348200. [DOI] [PubMed] [Google Scholar]
- 134.Kenny P.J., Kim M., Skariah G., Nielsen J., Lannom M.C., Ceman S. The FMRP-MOV10 complex: a translational regulatory switch modulated by G-quadruplexes. Nucleic Acids Res. 2020;48(2):862–878. doi: 10.1093/nar/gkz1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Borgognone M., Armas P., Calcaterra N.B. Cellular nucleic-acid-binding protein, a transcriptional enhancer of c-myc, promotes the formation of parallel G-quadruplexes. Biochem. J. 2010;428(3):491–498. doi: 10.1042/BJ20100038. [DOI] [PubMed] [Google Scholar]
- 136.Benhalevy D., Gupta S.K., Danan C.H., Ghosal S., Sun H.W., Kazemier H.G., Paeschke K., Hafner M., Juranek S.A. The human CCHC-type zinc finger nucleic acid-binding protein binds G-rich elements in target mRNA coding sequences and promotes translation. Cell Rep. 2017;18(12):2979–2990. doi: 10.1016/j.celrep.2017.02.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.David A.P., Pipier A., Pascutti F., Binolfi A., Weiner A.M.J., Challier E., Heckel S., Calsou P., Gomez D., Calcaterra N.B., Armas P. CNBP controls transcription by unfolding DNA G-quadruplex structures. Nucleic Acids Res. 2019;47(15):7901–7913. doi: 10.1093/nar/gkz527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Leipheimer J., Bloom A.L.M., Baumstark T., Panepinto J.C. CNBP homologues Gis2 and Znf9 interact with a putative G-quadruplex-forming 3′ untranslated region, altering polysome association and stress tolerance in Cryptococcus neoformans. MSphere. 2018;3(4) doi: 10.1128/mSphere.00201-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hanakahi L.A., Sun H., Maizels N. High affinity interactions of nucleolin with G-G-paired rDNA. J. Biol. Chem. 1999;274(22):15908–15912. doi: 10.1074/jbc.274.22.15908. [DOI] [PubMed] [Google Scholar]
- 140.Gonzalez V., Guo K., Hurley L., Sun D. Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein. J. Biol. Chem. 2009;284(35):23622–23635. doi: 10.1074/jbc.M109.018028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gonzalez V., Hurley L.H. The C-terminus of nucleolin promotes the formation of the c-MYC G-quadruplex and inhibits c-MYC promoter activity. Biochemistry. 2010;49(45):9706–9714. doi: 10.1021/bi100509s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Saha A., Duchambon P., Masson V., Loew D., Bombard S., Teulade-Fichou M.P. Nucleolin discriminates drastically between long-loop and short-loop quadruplexes. Biochemistry. 2020;59(12):1261–1272. doi: 10.1021/acs.biochem.9b01094. [DOI] [PubMed] [Google Scholar]
- 143.Miranti C.K., Moore S., Kim Y., Chappeta V.R., Wu K., De B., Gokhale V., Hurley L.H., Reyes-Reyes E.M. Nucleolin represses transcription of the androgen receptor gene through a G-quadruplex. Oncotarget. 2020;11(19):1758–1776. doi: 10.18632/oncotarget.27589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Figueiredo J., Miranda A., Lopes-Nunes J., Carvalho J., Alexandre D., Valente S., Mergny J.L., Cruz C. Targeting nucleolin by RNA G-quadruplex-forming motif. Biochem. Pharmacol. 2021;189 doi: 10.1016/j.bcp.2021.114418. [DOI] [PubMed] [Google Scholar]
- 145.Santos T., Miranda A., Campello M.P.C., Paulo A., Salgado G., Cabrita E.J., Cruz C. Recognition of nucleolin through interaction with RNA G-quadruplex. Biochem. Pharmacol. 2021;189 doi: 10.1016/j.bcp.2020.114208. [DOI] [PubMed] [Google Scholar]
- 146.Xu C., Wang Y., Tu Q., Zhang Z., Chen M., Mwangi J., Li Y., Jin Y., Zhao X., Lai R. Targeting surface nucleolin induces autophagy-dependent cell death in pancreatic cancer via AMPK activation. Oncogene. 2019;38(11):1832–1844. doi: 10.1038/s41388-018-0556-x. [DOI] [PubMed] [Google Scholar]
- 147.Romano S., Fonseca N., Simoes S., Goncalves J., Moreira J.N. Nucleolin-based targeting strategies for cancer therapy: from targeted drug delivery to cytotoxic ligands. Drug Discov. Today. 2019;24(10):1985–2001. doi: 10.1016/j.drudis.2019.06.018. [DOI] [PubMed] [Google Scholar]
- 148.Mukherjee S.K., Knop J.M., Winter R. Modulation of the conformational space of SARS-CoV-2 RNA quadruplex RG-1 by cellular components and the amyloidogenic peptides alpha-synuclein and hIAPP. Chemistry. 2022;28(9) doi: 10.1002/chem.202104182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Nishio M., Tsukakoshi K., Ikebukuro K. G-quadruplex: flexible conformational changes by cations, pH, crowding and its applications to biosensing. Biosens. Bioelectron. 2021;178 doi: 10.1016/j.bios.2021.113030. [DOI] [PubMed] [Google Scholar]
- 150.Guo Y., Xu P., Hu H., Zhou X., Hu J. A label-free biosensor for DNA detection based on ligand-responsive G-quadruplex formation. Talanta. 2013;114:138–142. doi: 10.1016/j.talanta.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 151.Li Z., Zou S., Wu S., Miao X., Ma D.L. Polymerase chain reaction-based ultrasensitive detection of HBV DNA via G-quadruplex selective iridium(III) complex luminescent probe. Talanta. 2021;221 doi: 10.1016/j.talanta.2020.121661. [DOI] [PubMed] [Google Scholar]
- 152.Wang K., Fan D., Liu Y., Dong S. Cascaded multiple amplification strategy for ultrasensitive detection of HIV/HCV virus DNA. Biosens Bioelectron. 2017;87:116–121. doi: 10.1016/j.bios.2016.08.017. [DOI] [PubMed] [Google Scholar]
- 153.Platella C., Riccardi C., Montesarchio D., Roviello G.N., Musumeci D. G-quadruplex-based aptamers against protein targets in therapy and diagnostics. Biochim Biophys Acta Gen Subj. 2017;1861(5 Pt B):1429–1447. doi: 10.1016/j.bbagen.2016.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Feng L., Chen Y., Ren J., Qu X. A graphene functionalized electrochemical aptasensor for selective label-free detection of cancer cells. Biomaterials. 2011;32(11):2930–2937. doi: 10.1016/j.biomaterials.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 155.Stoltenburg R., Krafcikova P., Viglasky V., Strehlitz B. G-quadruplex aptamer targeting protein a and its capability to detect Staphylococcus aureus demonstrated by ELONA. Sci. Rep. 2016;6:33812. doi: 10.1038/srep33812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Xi H., Juhas M., Zhang Y. G-quadruplex based biosensor: a potential tool for SARS-CoV-2 detection. Biosens. Bioelectron. 2020;167 doi: 10.1016/j.bios.2020.112494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Pratihar S., Agrawal R., Pal V.K., Singh A., Govindaraju T. Reliable fluorometric detection of SARS-CoV-2 by targeting the G-quadruplex through pH-triggered conformational polymorphism. ACS Sens. 2022;7(2):453–459. doi: 10.1021/acssensors.1c02113. [DOI] [PubMed] [Google Scholar]
- 158.Gupta A., Anand A., Jain N., Goswami S., Anantharaj A., Patil S., Singh R., Kumar A., Shrivastava T., Bhatnagar S., Medigeshi G.R., Sharma T.K., D.B.T.I.C.f.C.-Research A novel G-quadruplex aptamer-based spike trimeric antigen test for the detection of SARS-CoV-2. Mol Ther Nucleic Acids. 2021;26:321–332. doi: 10.1016/j.omtn.2021.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Carvalho J., Mergny J.L., Salgado G.F., Queiroz J.A., Cruz C. G-quadruplex, friend or foe: the role of the G-quartet in anticancer strategies. Trends Mol. Med. 2020;26(9):848–861. doi: 10.1016/j.molmed.2020.05.002. [DOI] [PubMed] [Google Scholar]
- 160.Dumetz F., Merrick C.J. Parasitic protozoa: unusual roles for G-quadruplexes in early-diverging eukaryotes. Molecules. 2019;24(7) doi: 10.3390/molecules24071339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wang E., Thombre R., Shah Y., Latanich R., Wang J. G-quadruplexes as pathogenic drivers in neurodegenerative disorders. Nucleic Acids Res. 2021;49(9):4816–4830. doi: 10.1093/nar/gkab164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.