Summary
Omicron has become the globally dominant severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant, creating additional challenges due to its ability to evade neutralization. Here, we report that neutralizing antibodies against Omicron variants are undetected following COVID-19 infection with ancestral or past SARS-CoV-2 variant viruses or after two-dose mRNA vaccination. Compared with two-dose vaccination, a three-dose vaccination course induces broad neutralizing antibody responses with improved durability against different SARS-CoV-2 variants, although neutralizing antibody titers against Omicron remain low. Intriguingly, among individuals with three-dose vaccination, Omicron breakthrough infection substantially augments serum neutralizing activity against a broad spectrum of SARS-CoV-2 variants, including Omicron variants BA.1, BA.2, and BA.5. Additionally, after Omicron breakthrough infection, memory T cells respond to the spike proteins of both ancestral and Omicron SARS-CoV-2 by producing cytokines with polyfunctionality. These results suggest that Omicron breakthrough infection following three-dose mRNA vaccination induces pan-SARS-CoV-2 immunity that may protect against emerging SARS-CoV-2 variants of concern.
Keywords: SARS-CoV-2, recovered patient, breakthrough infection, ancestral, D614G, variants of concern, Omicron BA.1, Omicron BA.2, cross-neutralization, mRNA vaccine, T cell immune response
Graphical abstract

Highlights
-
•
Early SARS-CoV-2 variant infections do not elicit NAbs against Omicron variants
-
•
Three-dose vaccination induces broad, but variant-specific, NAbs against SARS-CoV-2
-
•
Omicron breakthrough infection elicits pan-SARS-CoV-2 humoral immunity
-
•
T cell immunity dose not differ upon stimulation with ancestral or Omicron spike
Jeong et al. investigate the efficacies of the NAb elicited by SARS-CoV-2 infection or mRNA vaccination. Through the longitudinal analysis, the authors demonstrate that although cross-reactivity of the NAb induced by SARS-CoV-2 infection or two-dose vaccination is variant specific, Omicron breakthrough infection following booster vaccination can elicit NAb against pan-SARS-CoV-2.
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has led to over 507 million confirmed cases and 6.1 million deaths, and the disease continues to surge in many parts of the world. The continuing evolution of SARS-CoV-2 has given rise to several novel variants.1 Notably, the Alpha (B.1.1.7), Beta (B.1.351), Gamma (P1), Delta (B.1.617.2), and Omicron (B.1.1.529) variants are characterized by sets of mutations, raising concerns about possible immune evasion and increased transmissibility. Following the Delta variant SARS-CoV-2 surge in 2021, Omicron achieved worldwide predominance in a remarkably short time period due to its high transmissibility.2
One main target of antibody responses to coronaviruses is the spike (S) protein—the surface glycoprotein that mediates virus attachment to the host receptor. Neutralizing antibodies (NAb) targeting the SARS-CoV-2 S protein can prevent viral entry into host cells by blocking receptor binding or membrane fusion. Although the N-terminal domain of the S protein plays a role in NAb binding,3 the receptor-binding domain (RBD) of the S1 subunit of the S protein is the immunodominant target for highly potent and specific NAbs.4 Mutations in these sites can decrease the avidity of NAbs for the S protein.5 The Omicron (B.1.1.529) variant exhibits over 30 mutations in the S protein, including at least 15 mutations in the RBD.6 These substantial changes in the S protein gene of the Omicron variant raised concerns regarding the effectiveness of the current COVID-19 vaccines and antibody therapeutics,7,8 which were developed based on the S protein of the ancestral Wuhan-1 strain. In fact, many vaccinated persons have experienced breakthrough infections by Omicron variants, including BA.1, BA.2, or BA.5.9,10 The rapid surge of vaccine breakthrough infections by Omicron variants has raised questions about the effectiveness of the antiviral immunity elicited by the current COVID-19 vaccines or by prior infection with SARS-CoV-2 variants.
Investigation of the cross-reactivity among SARS-CoV-2 variants will help identify possible changes in the efficacies of SARS-CoV-2 vaccines and antibody therapeutics against newly emerging SARS-CoV-2 variants. In this study, we obtained sera from unvaccinated individuals infected with ancestral, D614G, or Omicron SARS-CoV-2 and from individuals previously vaccinated with two or three doses of COVID-19 mRNA vaccine. We evaluated the cross-neutralizing profiles of these sera against recent SARS-CoV-2 strains, including the ancestral L clade and D614G lineage, and the Alpha, Beta, Delta, Omicron BA.1, Omicron BA.2, and Omicron BA.5 SARS-CoV-2 variants. Cross-neutralizing tests were also conducted using sera from healthcare workers with breakthrough Omicron SARS-CoV-2 infection. In parallel, we also examined the memory T cell responses to Omicron breakthrough infections.
Results
Cross-neutralizing activity in unvaccinated COVID-19 patients
During the early stage of the COVID-19 pandemic in South Korea in 2020, SARS-CoV-2 was isolated from 21 COVID-19 patients. The viral clade was identified by whole-genome sequencing: 9 cases were the L and S clades of SARS-CoV-2 (ancestral group), and 12 were D614G SARS-CoV-2 variants (D614G group). For serologic analysis, patient serum samples were obtained immediately after RT-PCR confirmation (the day of hospital admission [day 1]), as well as 2 weeks later (14 days from initial hospitalization [day 14]). Additionally, paired serum samples were obtained from nine unvaccinated Omicron (BA.1)-recovered patients at the day of diagnosis (day 1) and 14 days (day 14) from initial RT-PCR positivity (Table S1). To assess patients’ previous SARS-CoV-2 infection history, we conducted a nucleocapsid protein (NP)-based enzyme-linked immunosorbent assay (ELISA). The ELISA results for all participants (21 ancestral-/D614G- and 9 Omicron-recovered patients) confirmed no antibody responses against NP protein on the day of hospital admission, while all sera were positive at 14 days from diagnosis (Figures 1B, 1E, and 1H; NP ELISA). These findings confirmed that the patients had no prior infection with other SARS-CoV-2 variants.
Figure 1.
Serum-neutralizing activity against recent SARS-CoV-2 strains and variants in unvaccinated COVID-19 patients
(A, D, and G) Sera samples were obtained from unvaccinated patients recovered from COVID-19 infection with (A) the ancestral L (n = 9), (D) D614G (n = 12), or (G) Omicron variant (n = 9).
(B, E, and H) Direct infection of the patients was validated via SARS-CoV-2 NP-specific ELISA using sera collected on the day of confirmation (day 1) and at 14 days (day 14) from the initial positive RT-PCR result.
(C, F, and I) Serum-neutralizing activity against live SARS-CoV-2 ancestral L, D614G, and indicated variants, in samples from unvaccinated patients infected with SARS-CoV-2 (C) ancestral L, (F) D614G, and (I) Omicron BA.1.
Violin plots represent the geometric mean titer of the neutralizing antibody. The geometric mean titers and fold changes of neutralizing activity are indicated above the violin plots. Dashed lines indicate the mean. Each dotted line represents a single individual. The nonparametric Wilcoxon matched-pairs signed rank test was used to analyze the statistical significance (ns, not significant). The neutralizing activity tests were repeated three times independently.
To compare longitudinal NAb activity among recovered COVID-19 patients, we selected day 1 and day 14 serum specimens from each group and performed a microneutralization antibody assay (NAb) using live SARS-CoV-2 of each reference strain or variant (ancestral L clade, D614G-like, Alpha, Beta, Delta, and Omicron BA.1, BA.2, and BA.5). The sera from ancestral-recovered patients (Figure 1A) showed high NAb titers (GMT-NAb 34.29) against L clade virus and were fairly cross-reactive with the D614G (GMT-NAb 6.30), Alpha (GMT-NAb 7.35), and Delta (GMT-NAb 7.35) variants. However, no cross-reactivity was observed against the Beta or Omicron BA.1, BA.2, and BA.5 variants (Figure 1C). The sera from D614G-recovered patients (Figure 1D) exhibited the highest NAb titers against the D614G virus and moderate cross-reactivity against the L clade, Alpha, Beta, and Delta variants (Figure 1F). Again, the sera of D614G-recovered patients showed no cross-reactivity against the Omicron BA.1, BA.2 or BA.5 variants (Figure 1F). Sera from Omicron-recovered patients showed compelling titers of NAb against the Omicron BA.1 variant. Although some sera showed titers of NAb against BA.2 variants, these NAb titers were significantly lower than the titers of NAb against BA.1, and we observed no cross-reactivity against Omicron BA.5. Moreover, Omicron-recovered patients (Figure 1G) showed marginal cross-reactivity against D614G (GMT-NAb 6.80) and the Beta variant (GMT-NAb 5.83) but no cross-reactivity against the earlier ancestral (L clade) or the Alpha and Delta variants (Figure 1I). These results demonstrate that distinct epitopes are required for neutralization of the early strains of SARS-CoV-2 compared with the Omicron variants. Furthermore, even among Omicron variants, we observed antigenic differences between the BA.1 and BA.5 variants.
Longitudinal evaluation of the cross-neutralizing efficacy of vaccines
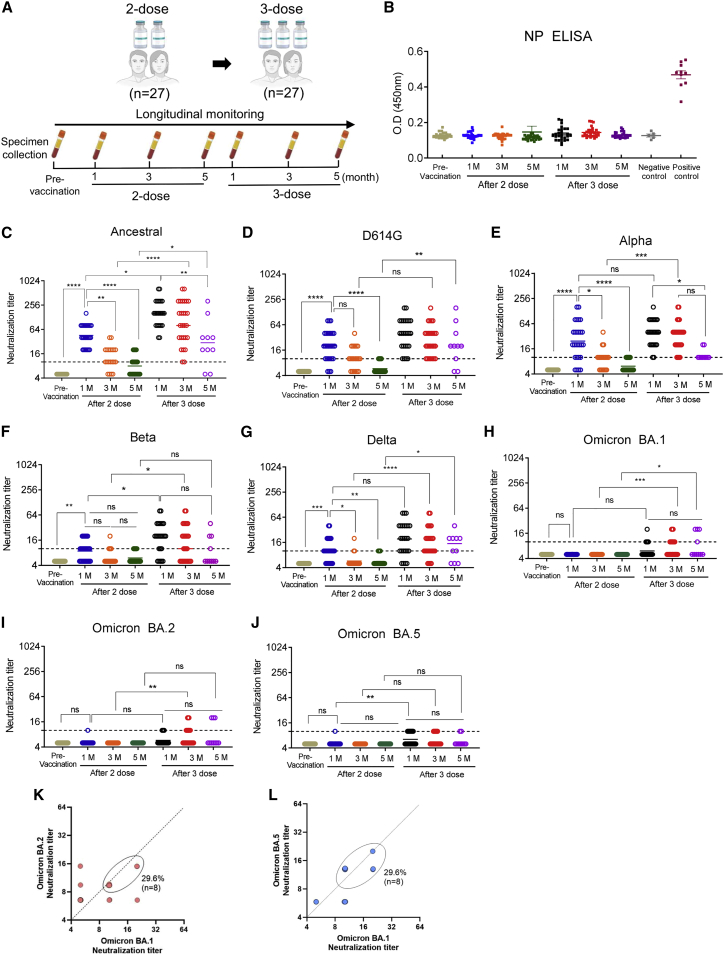
Despite high COVID-19 vaccination rates, several countries have reported massive numbers of breakthrough infections with Omicron variants,10,11 raising concerns about the relatively short period of vaccine-induced humoral immunity against recent variants. Therefore, we sought to evaluate the cross-neutralizing titers and the duration of antibody presence elicited by current COVID-19 mRNA vaccines. We obtained serial serum samples from healthcare workers following two-dose vaccination and booster vaccination with mRNA COVID-19 vaccines (vaccination cohort, n = 27; Figure 2A). Sera were collected at 1, 3, and 5 months after the second vaccination dose and at 1, 3, and 5 months after booster vaccination (Figure 2). NP ELISA confirmed that these vaccinees had no previous infection history (Figure 2B).
Figure 2.
Longitudinal evaluation of serum neutralization against SARS-CoV-2 strains and variants after vaccination with two or three doses
(A) Schematic image of the sera sampling from the vaccinees (n = 27). Healthcare workers were serially sampled at 1, 3, and 5 months after their second dose and at 1, 3, and 5 months after their third dose of COVID-19 mRNA vaccine.
(B) ELISA analysis using anti-SARS-CoV-2 NP immunoglobulin G (IgG) antibodies was performed to verify that the vaccinees were not infected with SARS-CoV-2 prior to vaccination.
(C–J) Serum samples serially collected from two- or three-dose vaccinated individuals were assessed for cross-neutralizing activity against (C) SARS-CoV-2 ancestral (WIV-04; L), (D) D614G, (E) Alpha, (F) Beta, (G) Delta, (H) Omicron BA.1, (I) Omicron BA.2, and (J) Omicron BA.5.
(K and L) Cross-neutralizing activity against both Omicron BA.1 and BA.2 (K) or BA.1 and BA.5 (L) was evaluated in three-dose vaccinated individuals (after 3 months).
Dashed line indicates the first serum dilution (1:10) of the NAb assay. Statistical analysis was performed using the Kruskal-Wallis test. ns, not significant; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. The neutralizing activity tests were repeated three times independently.
At one month after the second dose of a COVID-19 vaccine, GMT-NAb against ancestral, D614G, Alpha, Beta, and Delta strains were relatively high compared with pre-vaccination (Figures 2C–2G). However, no cross-neutralizing titer was detected against the Omicron BA.1, BA.2, and BA.5 variants (Figure 2). In two-dose vaccinated individuals, the antibody titers against all tested virus clades were decreased at 3 months after the last vaccination, and, surprisingly, the NAb level had returned to almost baseline at 5 months. These results indicate considerable and rapid waning of serum neutralizing activity following two-dose mRNA vaccination (Figures 2 and S1). Compared with two-dose vaccination, booster vaccination yielded markedly increased GMT-NAb titers against all tested viruses, including Omicron variants (Figures 2 and S1, two-dose versus three-dose). Furthermore, unlike after two-dose vaccination, after the third vaccination, high GMT-NAb levels persisted for 3 months, and marginal but detectable cross-neutralizing antibodies were maintained until 5 months, demonstrating the beneficial effect of three-dose vaccination (Figure 2, after three doses). Interestingly, with regards to cross-reactivity against Omicron variants, following the third vaccine dose, about 40% of individuals (11 out of 27) exhibited detectable NAb activity against Omicron variants, although at levels lower than against other strains (Figures 2H–2J). Among individuals with three-dose vaccination, 29.6% exhibited cross-neutralizing activity against Omicron BA.1, BA.2, and BA.5 after 3 months (Figures 2K–L). Altogether, these results indicate that three-dose booster vaccination enhanced the durability and longevity of NAb responses against a broad spectrum of SARS-CoV-2 variants.
Cross-neutralizing antibodies induced by Omicron breakthrough infection
During this study, in an additional breakthrough infection cohort group, we observed 50 cases of Omicron breakthrough infection among individuals with two (n = 25) (Figures 3A–3C) and three (n = 25) doses of mRNA vaccines (Figures 3D–3F). To investigate NAb responses and the status of cross-neutralizing immunity, we measured GMT-NAb against reference variants of SARS-CoV-2 using paired serum samples collected at two time points: 1–3 months prior to Omicron breakthrough infection, and after recovery (14 days after diagnosis of virus infection). Compared with the baseline NAb titers (pre-Omicron infection), the two-dose or three-dose vaccinated group showed much higher titers of GMT-NAb against the ancestral L, D614G, Alpha, and Delta variants compared with those against the Beta and Omicron variants (Table 1). In the two-dose vaccinated breakthrough infection cohort, Omicron variant infection led to significantly (>4-fold) increased GMT-NAb titers against the Omicron BA.1 and BA.2 variants (Figure 3C), which were comparable to the GMT-NAb titers against Omicron BA.1 in the unvaccinated Omicron infection group (Figure 1I; Table 1). However, only 48% (12/25) of two-dose vaccinated individuals showed moderately increased GMT-NAb titers (10.57) against Omicron BA.5. Intriguingly, all individuals in the two-dose vaccinated breakthrough infection cohort showed significantly increased GMT-NAbs against the ancestral (5.3-fold), D614G (5.1-fold), and Alpha (5.6-fold) variants and moderately increased NAbs against the Beta (2.9-fold) and Delta (2.2-fold) variants (Figure 3C; Table 1). Compared with the unvaccinated infection group (G1), the two-dose vaccinated breakthrough infection group (G2) exhibited a significantly increased geometric mean fold-rise (GMFR) against Omicron BA.2 but not against BA.1 or BA.5 (Table 1).
Figure 3.
Enhanced cross-neutralizing activity against a broad range of SARS-CoV-2 variants following Omicron breakthrough infections in COVID-19 vaccine-boosted individuals
(A and D) Schematic image of Omicron breakthrough infections among COVID-19 vaccinees (n = 25), with (A) two doses (n = 25) or (D) three doses (n = 25).
(B and E) Omicron BA.1 infections were verified by SARS-CoV-2 NP-specific ELISA,with (B) two doses and (E) three doses.
(C and F) Measurements of serum neutralizing activity against ancestral, D614G, and five VOCs of SARS-CoV-2 in the serum of Omicron breakthrough infected individuals with (C) two-dose or (F) three-dose vaccination.
Violin plots represent the geometric mean titer of the neutralizing antibody. Geometric mean titers and fold changes of neutralizing activity are indicated above the violin plots. In the violin plots, middle dashed lines indicate the mean, and outer dashed horizontal lines indicate the interquartile range. Each dotted line represents a single individual. Statistical significance was analyzed with the nonparametric Wilcoxon matched-pairs signed rank test. Neutralization titers lower than 1:10 were considered negative. The neutralizing activity tests were repeated three times independently.
Table 1.
Paired geometric mean titers of neutralizing antibody after Omicron variant infection among unvaccinated and two-dose or three-dose vaccinated individuals
| SARS-CoV-2 strain | G1 (n = 9) |
G2 (n = 25) |
G3 (n = 25) |
pa | pb | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| GMT |
GMFR (95% CI) | GMT |
GMFR (95% CI) | GMT |
GMFR (95% CI) | ||||||
| Pre | Post | Pre | Post | Pre | Post | ||||||
| Ancestral | 5.0 | 5.0 | 1.0 (–) | 12.5 | 65.9 | 5.3 (3.5–8.0) | 40.0 | 160.0 | 4.0 (2.5–6.4) | Gr1≠Gr2≠Gr3∗ | Gr1≠Gr2∗∗, Gr1≠Gr3∗∗ |
| D614G | 5.0 | 6.8 | 1.4 (0.9–2.2) | 10.6 | 54.3 | 5.1 (3.4–7.8) | 28.7 | 128.2 | 4.5 (2.6–7.6) | Gr1≠Gr3∗∗, Gr2≠Gr3∗∗ | Gr1≠Gr2∗∗, Gr1≠Gr3∗ |
| Alpha | 5.0 | 5.0 | 1.0 (–) | 8.0 | 44.7 | 5.6 (3.6–8.7) | 26.4 | 108.5 | 4.1 (2.7–6.2) | Gr1≠Gr3∗∗, Gr2≠Gr3∗∗ | Gr1≠Gr2∗∗, Gr1≠Gr3∗∗ |
| Beta | 5.0 | 5.8 | 1.2 (0.9–1.5) | 5.9 | 17.4 | 2.9 (1.8–4.9) | 10.9 | 32.0 | 2.9 (1.9–4.7) | Gr1≠Gr3∗∗, Gr2≠Gr3∗∗ | – |
| Delta | 5.0 | 5.0 | 1.0 (–) | 6.8 | 14.7 | 2.2 (1.4–3.5) | 18.4 | 82.2 | 4.5 (2.7–7.3) | Gr1≠Gr3∗∗, Gr2≠Gr3∗∗ | Gr1≠Gr3∗∗ |
| Omicron BA.1 | 5.0 | 20.0 | 4.0 (1.9–8.5) | 5.1 | 31.2 | 6.1 (4.2–8.8) | 6.4 | 49.9 | 7.8 (5.6–10.8) | Gr2≠Gr3∗ | – |
| Omicron BA.2 | 5.0 | 5.8 | 1.2 (0.9–1.5) | 5.0 | 22.3 | 4.5 (3.0–6.6) | 5.3 | 34.8 | 6.7 (5.0–8.6) | – | Gr1≠Gr2∗∗, Gr1≠Gr3∗∗ |
| Omicron BA.5 | 5.0 | 5.0 | 1.0 (–) | 5.0 | 10.6 | 2.1 (1.4–3.2) | 5.1 | 18.9 | 3.7 (2.3–5.8) | – | Gr1≠Gr3∗∗ |
∗p < 0.05; ∗∗p < 0.01. CI, confidence interval; G1, individuals not vaccinated against COVID-19; G2: two-dose COVID-19 vaccinated individuals; G3: three-dose COVID-19 vaccinated individuals; GMT, geometric mean titer; GMFR, geometric mean fold-rise.
Comparison of pre-Omicron infection GMT by group using Kruskal-Wallis test with Bonferroni correction as a post-hoc analysis. Only groups that showed statistical significance are indicated.
Comparison of ratios by group using the Kruskal-Wallis test with Bonferroni correction as a post-hoc analysis. Only groups that showed statistical significance are indicated.
The three-dose vaccinated Omicron breakthrough infection group (G3) showed significantly increased titers of GMT-NAb against all tested SARS-CoV-2 strains, including three Omicron variants (Figure 3F; Table 1). All patients in G3 showed NAb titers in the range of 10–160 against Omicron variants (Figure 3F), which were much higher compared with in the unvaccinated infection group (G1) (Table 1). Furthermore, G3 showed much higher titers of GMT-NAb against the ancestral L, D614G, Alpha, and Delta variants compared with against the Omicron variants, as well as a relatively lower (2.9-fold) increase of GMT-NAb against the Beta variant (Figure 3F; Table 1). Compared with G1, G3 showed a significantly higher GMFR against BA.2 and BA.5 (Table 1). Altogether, these data demonstrate that breakthrough infection by Omicron variants can enhance the antibody responses against multiple SARS-CoV-2 variants, including Omicron BA.1, BA.2, and BA.5.
Memory T cell response following Omicron breakthrough infection
Finally, we investigated CD4+ and CD8+ T cell responses to the ancestral SARS-CoV-2 and Omicron variant S proteins after Omicron breakthrough infection. To this end, we performed intracellular cytokine staining (ICS) assays for interferon gamma (IFN-γ), tumor necrosis factor (TNF), and interleukin-2 (IL-2) in stimulated peripheral blood mononuclear cells (PBMCs) obtained from three-dose vaccinated individuals who had suffered Omicron breakthrough infection. Stimulation was performed using an overlapping peptide (OLP) pool generated from the S protein of the ancestral Wuhan-1 strain or the Omicron variant (Figure 4). The frequency of cytokine-producing CD4+ T cells did not differ following stimulation with ancestral spike versus the Omicron spike (Figures 4A and 4B). The CD8+ T cell responses were relatively weak compared with CD4+ T cell responses (Figure 4C). Similarly, the frequency of cytokine-producing CD8+ T cells did not differ following stimulation with the ancestral spike versus Omicron spike (Figure 4D). In investigations of CD4+ T cells, we also analyzed polyfunctionality, which plays a critical role in host protection during viral infection. Representative flow cytometry plots show that a proportion of CD4+ T cells simultaneously produced IFN-γ, TNF, and IL-2 in response to both ancestral and Omicron spike (Figure 4E). The frequencies of polyfunctional (triple-positive or double-positive) CD4+ T cells did not significantly differ between T cells stimulated with the ancestral versus Omicron spike (Figure 4F). Polyfunctionality analysis using pie graphs (Figure 4G) and every possible combination of functions (Figure 4H) revealed no significant difference between CD4+ T cells responding to the ancestral versus Omicron spike. Taken together, these data indicate that following the Omicron breakthrough infections, memory T cells produced cytokines with polyfunctionality equally against both ancestral and Omicron spike.
Figure 4.
Memory T cell responses against the ancestral and Omicron spikes after Omicron breakthrough infection
Memory T cell responses were assessed in individuals who suffered Omicron breakthrough infection after receiving a third booster shot with COVID-19 mRNA vaccines (n = 18). PBMCs were obtained during a convalescence period, at 10–30 days after symptom onset. PBMCs were subjected to intracellular cytokine staining to examine the frequency of CD4+ or CD8+ T cells responding to the ancestral spike or the Omicron spike.
(A and C) Representative flow cytometry plots show the cytokine-producing cells among CD4+ (A) or CD8+ (C) T cells.
(B and D) The frequencies of cytokine-producing CD4+ (B) or CD8+ (D) T cells against the ancestral spike and the Omicron spike are compared. Data are presented as mean ± SD. p values were calculated using a two-tailed unpaired Mann-Whitney U test.
(E) Representative flow cytometry plots show polyfunctional cells among CD4+ T cells.
(F) The percentage of polyfunctional T cells among cytokine-producing CD4+ T cells is presented.
(G) Pie graphs represent the fraction of cells positive for a given number of functions among CD4+ T cells with any type of function. Permutation test was performed with 10,000 permutations.
(H) Detailed analyses of polyfunctionality are presented with every possible combination of functions. Data are presented as mean ± SD. p values were calculated using the two-tailed unpaired Mann-Whitney U test.
We also examined CD4+ and CD8+ T cell responses using PBMCs obtained from three-dose vaccinated individuals without breakthrough infection. Although there was no difference in the frequency of TNF+ or IL-2+ cells among CD4+ T cells, the frequency of IFN-γ+ cells among CD4+ T cells was significantly lower following stimulation with the Omicron spike versus the ancestral spike (Figure S2A). The frequency of cytokine-producing CD8+ T cells did not significantly differ following stimulation with ancestral spike versus Omicron spike (Figure S2B). Polyfunctionality analysis revealed no significant difference between CD4+ T cells responding to the ancestral versus Omicron spike (Figures S2C–S2E). To compare the contribution of natural infection versus vaccination in eliciting polyfunctional T cell responses, we also examined CD4+ and CD8+ T cell responses using PBMCs obtained from Omicron-infected patients without previous vaccination. The results revealed that the frequency of cytokine-producing CD4+ (Figure S2F) or CD8+ (Figure S2G) T cells did not significantly differ following stimulation with ancestral spike versus Omicron spike. Furthermore, the polyfunctionality analysis revealed no significant difference between CD4+ T cells responding to the ancestral spike versus Omicron spike (Figures S2H and S2I).
Discussion
The large number of mutations in the Omicron variant of SARS-CoV-2, especially within the S protein,11,12 has enabled its explosive global spread, prompting concerns about waning efficacy of the current COVID-19 vaccines and antibody therapeutics. Omicron comprises several sublineages, including BA.1, BA.2, BA.3, BA.4, and BA.5.13 Currently, the most prevalent are BA.2 and BA.5, and BA.2.12 has also recently shown rapid global spread. These sublineages have exhibited substantial escape from vaccination-induced NAbs.14 Although they share numerous mutations, these two sublineages are differentiated by 20 mutations in the S protein.15
In the present study, we found that the sera of patients who have recovered from ancestral or D614G infections exhibited moderate cross-reactivity against the Alpha, Beta, and Delta variants but no cross-reactivity against the BA.1, BA.2, and BA.5 Omicron variants. In contrast, sera from patients who recovered from Omicron infection exhibited limited cross-reactivity against the D614G, Alpha, and Delta variants.
While current vaccines have shown efficacy in controlling the global spread of COVID-19, emerging SARS-CoV-2 variants possess highly mutated antigenic epitopes that can evade the antiviral immunity elicited by vaccination or prior exposure to the early SARS-CoV-2 variants. Indeed, here we demonstrated that infection with ancestral/D614G viruses, or two-dose vaccination with the current COVID-19 mRNA vaccine, induced NAbs against major SARS-CoV-2 variants of concern (VOCs). However, we observed low titers of NAbs against Omicron variants and a rapid waning of serum neutralizing activity following two doses of mRNA vaccine. In contrast, three-dose vaccination induced a broad range of cross-NAb responses against recent VOCs, as well as ancestral strain, and these NAbs were maintained until 5 months after the last vaccination. Importantly, the titers of NAb against Omicron BA.1, BA.2, and BA.5 were considerably increased and exhibited longer potency following booster vaccination, although this markedly enhanced anti-Omicron NAb activity was only observed in about 40% of individuals. Several recent papers have reported boosted Omicron NAb titers in nearly 100% of individuals.16,17 However, those studies were conducted using a pseudovirus assay system rather than real live variants. Although our present assay method may have lacked sensitivity compared with a pseudovirus system that contains only the S protein of each SARS-CoV-2 variant, we believe that our data may indicate the actual NAb titers against live recent SARS-CoV-2 variants, which contained all other surface proteins in addition to the S protein.
Our results showed that in cases of natural Omicron BA.1 infection without vaccination, the titers of cross-neutralizing antibodies against BA.2 and BA.5 were significantly lower than those against BA.1, suggesting some antigenic differences among these three variants. On the other hand, Omicron breakthrough infections following two-dose or three-dose vaccination did not show markedly different titers of NAb against BA.1 versus BA.2. Furthermore, Omicron infection following three-dose vaccination induced robust NAbs that are broadly reactive and more potently protective. Thus, the current findings suggest that a high level and broad spectrum of NAbs are elicited by Omicron breakthrough infection following three-dose vaccination and that these antibodies may neutralize both former and novel SARS-CoV-2 variants.
Although still under debate, studies suggest that prior infection with common seasonal human coronaviruses (hCoVs) impacts the severity of SARS-CoV-2 infection.18,19 Furthermore, antibodies reactive against the β-hCoV hCoV-OC43 can be generated upon SARS-CoV-2 infection20,21 and SARS-CoV-2 mRNA vaccinations.22 A recent study of individuals with fatal COVID-19 suggests that the back boosting of hCoV-OC43 antibodies is associated with a compromised de novo SARS-CoV-2 response.18,23,24 The boosting of hCoV antibodies upon infection with the antigenically distinct SARS-CoV-2 is consistent with the doctrine of “original antigenic sin,” which was first proposed by Thomas Francis in 1960 to describe influenza virus antibody responses.25 In the current study, we evaluated whether infection with the Omicron variant could boost antibodies against earlier SARS-CoV-2 strains. Surprisingly, Omicron breakthrough cases in individuals previously vaccinated against COVID-19 resulted in a stronger antibody response against not only Omicron but also against a broad spectrum of SARS-CoV-2 variants. Notably, this phenomenon was stronger among patients with three-dose vaccination compared with two-dose vaccination. Our results suggest that vaccine-induced antibody responses may induce broad-spectrum immunity by increasing the titer of NAbs against ancestral strains following Omicron infection. In addition, our observations were not consistent with the original antigenic sin; antibody response against Omicron was not impaired in the Omicron-breakthrough infection cases.
Interestingly, our data showed that although the mRNA vaccine encodes the ancestral SARS-CoV-2 spike gene, three-dose booster vaccination could elicit cross-neutralizing antibodies against broad-range SARS-CoV-2 strains and variants, including Omicron variants. There may be several possible mechanisms underlying this phenomenon. Since the mRNA COVID-19 vaccine encodes a full-length ancestral spike gene, some generated antibodies may recognize conserved epitopes that are commonly possessed by a broad range of SARS-CoV-2 VOCs, and these antibodies could be amplified by booster vaccination. Moreover, since mRNA vaccination could generate variant cross-binding memory B cells,26, 27, 28, 29, 30 it is reasonable to speculate that somatic hypermutation occurring in memory B cells generated by the 1st and 2nd vaccination could be recalled and amplified by the 3rd dose booster vaccination, eventually potentiating cross-neutralizing ability against a broad range of SARS-CoV-2 strains and variants. It would be intriguing if a booster vaccination could generate NAbs against SARS-CoV-2 variants that may appear in the future or even against an old SARS-CoV-1 variant. Nevertheless, these results suggest that booster vaccination with the current mRNA vaccine could induce a considerable boosting effect, with a longer period of NAb responses against a broad spectrum of SARS-CoV-2 variants, including Omicron variants.
In the current study, we also examined memory T cell responses against the Omicron variant among individuals who had recovered from Omicron breakthrough infection after three-dose COVID-19 vaccination. Memory T cell responses against the Omicron variant have also been previously examined among individuals immunized with a COVID-19 vaccine but without Omicron breakthrough infection. Those studies revealed that vaccine-induced T cells substantially responded to both the ancestral and Omicron spikes, although the T cell responses against the Omicron spike were slightly decreased compared with T cell responses against the ancestral spike.31, 32, 33 In contrast, our current data showed comparable T cell responses against the Omicron and ancestral spikes, without any significant difference, among individuals recovered from Omicron breakthrough infection. This indicates that Omicron breakthrough infection may elevate T cell responses against Omicron-specific epitopes. It was recently demonstrated that memory T cells contribute to host protection against SARS-CoV-2 infection, particularly in cases with insufficient humoral immune response.34, 35, 36, 37 Given that memory T cells reduce viral titers and lung pathology in SARS-CoV-2 infection,37 individuals who have recovered from Omicron breakthrough infections may experience only mild disease even if they are re-infected with other variants that escape NAbs.
Our results show that Omicron infection after two- or three-dose vaccination substantially boosts the potency and breadth of the NAb response. Notably, recent studies have reported that many breakthrough infection cases among vaccinated individuals are either asymptomatic or mildly symptomatic,38,39 but patients who recover from Omicron breakthrough infection still mount relatively high NAb titers against the Omicron variant.40 As massive vaccinations lead to increased asymptomatic or mild cases, further studies are needed to clarify the underlying mechanism of how enhanced antibody responses against the broad spectrum of SARS-CoV-2 strains and variants, including Omicron, are elicited following the Omicron breakthrough infection. Although two-dose vaccination alone elicits a NAb response to prevent further infection, this induced immunity is short lived. In contrast, the third dose of an mRNA vaccine induces a strong boosting effect, resulting in the maintenance of a longer NAb response against a broad spectrum of SARS-CoV-2 variants. Concomitantly, our findings support the hypothesis that the Omicron variant is capable of potent immune escape and shows little cross-reactivity with earlier variants. Notably, it is possible that both two-dose and three-dose vaccination provide protection against severe Omicron infection, as Omicron breakthrough infection generally leads to a mild course of illness. Moreover, in boosted individuals, Omicron induces significantly increased NAb titers against ancestral strains, which may confer broad-spectrum antibody responses that might be effective against novel SARS-CoV-2 variants. Overall, these findings suggest that multiple exposures to the SARS-CoV-2 S protein improve the elicitation of highly reactive NAbs against a broad spectrum of SARS-CoV-2 viruses, justifying booster vaccination for enhancing or maintaining vaccine effectiveness.
Limitations of the study
This study has several limitations, including a relatively small sample size, the issue of sensitivity of the NAb assay, and the retrospective study design (Tables S1 and S2). Specifically, the relatively small sample size may limit the generalizability of our findings, thus studies with a larger and more diverse population are warranted. Further, the present study may show lower NAb titers compared with other several studies, which used pseudovirus-based assays. Although there is an issue with the sensitivity of the NAb assay in our study, we used live viruses for each SARS-CoV-2 strain as opposed to other studies and tried to evaluate the actual cross-neutralizing titers against live SARS-CoV-2 variants, which contained all other surface proteins other than the S protein. In this regard, more precise comparison studies to evaluate the sensitivity of NAb assay using different platforms are needed for further studies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD3 BV786(Clone UCHT1) | BD Biosciences | Cat #: 565491 |
| CD4 BV650 (clone SK3) | BD Biosciences | Cat#: 563875 |
| CD8 APC-H7 (clone SK1) | BD Biosciences | Cat# 560179 |
| CD14 BV510 (clone MφP9) | BD Biosciences | Cat# 563079 |
| CD19 BV510 (clone SJ25C1) | BD Biosciences | Cat# 562947 |
| IFN-γ APC (clone B27) | BD Biosciences | Cat# 554702 |
| IL-2 PE (clone MQ1-17H12) | ThermoFisher | Cat# 12-7029-42 |
| TNF FITC (clone MAb11) | ThermoFisher | Cat# 11-7349-82 |
| anti-human IgG(H + L) polyclonal antibody, HRP conjugate | ThermoFisher | Cat # 31420 |
| Purified anti-human CD28/49d | BD Biosciences | Cat # 347690 |
| Bacterial and virus strains | ||
| hCoV-19/South Korea/NMC-nCoV-07/2021 | GISAID | EPI_ISL_10189535 |
| hCoV-19/South Korea/NMC-nCoV-08/2021 | GISAID | EPI_ISL_10189542 |
| hCoV-19/South Korea/NMC-nCoV-09/2021 | GISAID | EPI_ISL_10189545 |
| hCoV-19/South Korea/NMC-nCoV-11/2021 | GISAID | EPI_ISL_10189547 |
| hCoV-19/South Korea/CBNU-nCoV-10/2020 | GISAID | EPI_ISL_10189661 |
| hCoV-19/South Korea/CBNU-nCoV-50/2021 | GISAID | EPI_ISL_10187719 |
| hCoV-19/South Korea/CBNU-nCoV-55/2021 | GISAID | EPI_ISL_12179149 |
| hCoV-19/South Korea/KDCA61371/2022 | GISAID | EPI_ISL_13086516 |
| hCoV-19/South Africa/CERI-KRISP-K032284/2021 | GISAID | EPI_ISL_6699770 |
| Biological samples | ||
| Human sera sample obtained from COVID-19 patients | National Medical Center | www.nmc.or.kr |
| Human sera sample obtained from COVID-19 patients | Chungbuk National University Hospital | www.chnuh.or.kr |
| Human sera sample obtained from COVID-19 patients | Korea University Guro Hospital | www.guro.kumc.or.kr |
| Human sera sample obtained from vaccinated healthcare workers | Chungbuk National University Hospital | www.chnuh.or.kr |
| Human sera sample obtained from vaccinated healthcare workers | Korea University Guro Hospital | www.guro.kumc.or.kr |
| Human PBMC samples obtained from COVID-19 patients | Seoul National University Bundang Hospital | www.snubh.org |
| Human PBMC samples obtained from COVID-19 patients | Korea University Guro Hospital | www.guro.kumc.or.kr |
| Chemicals, peptides, and recombinant proteins | ||
| SARS-CoV-2 NP Protein (B.1.1.529) | Acrobiosystem | Cat #: NUN-C52Ht |
| SARS-CoV-2 (Spike Glycoprotein) | peptides&elephants | LB01792 |
| Omicron full length B.1.1.529 SCoV2 (Spike Glycoprotein) | peptides&elephants | LB02000 |
| Critical commercial assays | ||
| SARS-CoV-2 (2019-nCoV) Nucleocapsid Detection ELISA Kit | SinoBiological | Cat #: KIT40588 |
| 3,3′,5,5′-Tetramethylbenzidine (TMB) substrate solution | ThermoFisher | Cat #: N301 |
| Experimental models: Cell lines | ||
| Vero E6 | ATCC | Vero C1008 |
| Software and algorithms | ||
| MiniSeq system | Illumina | www.illumina.com |
| QIAGEN Bioinformatics CLC Workbench program | CLC bio | www.digitalinsights.qiagen.com |
| Graphpad Prism 9 | Graphpad | www.graphpad.com |
| FACSDiva 9 | BD Bioscience | www.bdbiosciences.com |
| FlowJo 10 | FlowJo | www.flowjo.com |
| SPICE v.6.1. | SPICE | https://niaid.github.io/spice/ |
| BioRender | BioRender | www.biorender.com |
| Other | ||
| iMark Microplate Absorbance Reader | Bio-Rad | Cat # 1681130 |
Resource availability
Lead contact
Additional information and requests for resources and reagents should be directed to the lead contact, Young Ki Choi (choiki55@chungbuk.ac.kr).
Materials availability
All SARS-CoV-2 viruses used, and unique/stable reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Experimental model and subject details
Patients and specimens
During the early stage of the COVID-19 pandemic in South Korea in 2020, ancestral and D614G SARS-CoV-2 were isolated from 21 COVID-19 patients [11 men and 10 women; mean age, 52.09 years; standard deviation, 18 years] from National Medical Center (NMC) and Chungbuk National University Hospital (CBNUH) (Table S1). The viral clades were confirmed by whole-genome sequencing. For serologic analysis, patient serum samples were obtained shortly after RT-PCR confirmation (the day of hospital admission, D1), as well as 14 days after the diagnosis (D14). Paired serum samples were also obtained from 9 unvaccinated Omicron (BA.1)-recovered patients on the day of infection confirmation (D1) and 14 days (D14) from initial RT-PCR positivity. Disease severity ranged from asymptomatic to mild COVID-19. Furthermore, to evaluate the neutralizing antibody duration and booster dose effects following mRNA vaccination, we recruited healthcare workers (HCW) [15 men and 12 women; mean age, 40 years; standard deviation, 8.06 years] from CBNUH and Korea University Guro Hospital, who received SARS-CoV-2 mRNA vaccines (vaccination cohort, n = 27). We serially collected their sera beginning at 1, 3, and 5 months after two-dose vaccination, and at 1, 3, and 5 months after their booster shot (third dose).
To evaluate the immune response, the Omicron breakthrough infection cohort (n = 50) was sampled before and after Omicron infection. Table S2 describes the characteristics of the breakthrough infection cohort. In the two-dose vaccine breakthrough group, 19 patients were immunized with two-dose of BNT162b2, 6 patients were immunized with two doses of mRNA-1273 mRNA vaccine. Among the three dose breakthrough infection individuals, two patients were immunized with ChAdOx1 – ChAdOx1 – BNT162b2, 12 patients with three-dose of mRNA-1273, and 11 patients with BNT162b2. From the two-dose and three-dose vaccinated individuals, pre-infection serum samples were collected a median of 30 days before Omicron infection. For T-cell analysis, we selected 18 individuals from the breakthrough infection cohort (n = 18; 6 males, 12 females; mean age, 30.5 years; age range, 25–47 years). We also recruited three-dose vaccinated individuals without breakthrough infection (n = 38; 12 males, 26 females; mean age, 36.5 years; age range, 24–57 years; days after the third shot, 21–31 days; 20 with three doses of BNT162b2, 18 with ChAdOx1–ChAdOx1–BNT162b2) from CBNUH and Seoul National University Bundang Hospital, and Omicron-infected patients without previous vaccination (n = 9; 5 males, 4 females; mean age, 72.5 years; age range, 52–90 years; days after symptom onset, 6–15 days). Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation using Lymphocyte Separation Medium (Corning). The isolated PBMCs were cryopreserved in fetal bovine serum (Corning) with 10% dimethyl sulfoxide (Sigma-Aldrich) until use. All participants provided informed consent. This study was conducted according to the principles of the Declaration of Helsinki.
Ethics statement
This study was conducted in accordance with the study protocol approved by the Institutional Review Board of Chungbuk National University Hospital (IRB No. 2020-03-036, 2021-02-010), National Medical Center (IRB No. H- 2002-111-002), the institutional review board of Korea University Guro Hospital (2021GR0099, 2021GR0274) and Seoul National University Bundang Hospital (B-2102-669-302). Informed consent was obtained from all patients or their guardians. All SARS-CoV-2 handling experiments were carried out in a BSL3 facility at Chungbuk National University (approval number CBNUA-1352-20-02).
Method details
Viral isolation and genome sequencing
SARS-CoV-2 viruses were isolated from nasal swab specimens of confirmed COVID-19 patients hospitalized between February 2020 and March 2022. To identify the virus clades, complete SARS-CoV-2 genome cDNA libraries were prepared using the next-generation sequencing (NGS) method with the Nextera XT DNA Library Preparation Kit (Illumina, CA, USA) with an average insert size of 300 bp. Sequencing runs were performed on a MiniSeq system (Illumina, CA, USA) using a MiniSeq mid-output kit, yielding 2 × 150 nucleotide paired-end reads.
To identify each corresponding SARS-CoV-2 clade, the complete genome sequences were compared to the reference genome sequence of Wuhan-Hu-1 (GenBank accession number MN908947.3, also known as GISAID Clade ‘L’). Based on full-length sequences, we selected a reference virus for each variant (ancestral, D614G, Alpha, Beta, Delta, and BA.1 and BA.2 Omicron). Whole-genome sequences for each virus are available in GISAID (accession numbers: EPI_ISL_1069194, EPI_ISL_10189661, EPI_ISL_10189535, EPI_ISL_10189542, EPI_ISL_10189547, EPI_ISL_10187719, EPI_ISL_12179149). Alignment of each clade and recent variant genome sequences was performed using the QIAGEN Bioinformatics CLC Workbench program (version 10.1.1; CLC bio, Denmark). According to the sequences, we divided the patients into three groups: the ancestral virus-recovered group, D614G virus-recovered group, and Omicron virus-recovered group.
Microneutralization assay
Neutralizing antibodies against the ancestral SARS-CoV-2 strain, D614G strain, Alpha, Beta, Delta, and BA.1 and BA.2 Omicron variants were assayed in the collected serum samples (Table 1). The microneutralization assay (NAb) of serum specimens was conducted using Vero E6 cells (ATCC #: Vero C1008). Briefly, serum specimens were inactivated at 56°C for 30 min. Then 50 μL of a two-fold serially diluted serum sample (starting from 1:10), was mixed with 50 μL of the 100 median tissue culture infectious dose (TCID50) of each SARS-CoV-2 strain. These mixtures were incubated at 37°C for 1 h to neutralize the infectious virus and then transferred to VeroE6 cell monolayers. The cells were incubated at 37°C under 5% CO2 for 4 days and monitored. Gentian violet staining (1%) was performed to stain and fix the cell culture layer. The neutralizing dilution of each serum sample was checked by identifying the well with the highest serum dilution causing at least a 50% reduction in cytopathic effect (CPE), which was taken as the NAb titer. A dilution equal to 1:10 or above was considered neutralizing. When a neutralizing antibody titer was not detected, it was recorded as 5.
Enzyme-linked immunosorbent assay (ELISA)
Briefly, the ELISA for total Ab detection was developed based on direct immunoassay using recombinant SARS-CoV-2-NP protein produced in E. coli. Flat-bottom Immuno plates (SPL Life Sciences) were coated with SARS-CoV-2-NP (1 μg/mL) in a bicarbonate buffer, overnight at 4°C. The plates were subsequently washed three times with PBS containing 0.05% Tween 20 (PBST), and then blocking buffer (3% BSA in PBS) was added at 150 μL per well. The plates were incubated for 3 h at room temperature, and then washed three times with PBST. Human sera were prepared at a dilution of 1:200 in PBS, and added at 100 μL volume per well. The plates were incubated for 3 h at 37°C, and then washed five times with PBST. Secondary antibody anti-human all IgH + L polyclonal antibody-HRP conjugate was added at a dilution of 1:10,000 in PBST, followed by a 1-h incubation at 37°C. The plates were washed seven times with PBST. Finally, 50 μL of 3,3′,5,5′-Tetramethylbenzidine (TMB) (Seracare, Milford, MA, USA) was added for 10 min for color development, and then the reaction was stopped with addition of 50 μL of 1 N sulfuric acid (H2SO4). The iMark Microplate Absorbance Reader (Bio-Rad, USA) was used to measure the optical density at 450 nm (OD450).
Intracellular cytokine staining
Cryopreserved PBMCs were thawed, rested overnight at 37°C, and cultured in the presence of OLP pools (1 μg/mL for each peptide; Peptides & Elephants GmbH) of the spike protein of the ancestral strain (GenBank: NM908947) or Omicron variant (hCoV-19/South Africa/CERI-KRISP-K032284/2021; EPI_ISL_6699770 in GISAID) and anti-human CD28 and CD49d monoclonal antibodies (1 μg/mL each; BD Biosciences) for 6 h at 37°C. Negative controls were cultured in the presence of DMSO and anti-CD28/anti-CD49d antibodies. At 1 h after the initial stimulation, brefeldin A (GolgiPlug, BD Biosciences) and monensin (GolgiStop, BD Biosciences) were added. The cells were stained with fluorochrome-conjugated antibodies for specific surface markers, for 10 min at 4°C. Dead cells were excluded using LIVE/DEAD dye (Invitrogen). For intracellular staining, cells were fixed and permeabilized using the FoxP3 staining buffer kit (Invitrogen), and stained for cytokines for 30 min at 4°C. The following monoclonal antibodies were used for multi-color flow cytometry: anti-CD3 BV786 (clone UCHT1, cat# 565491, 1:100), anti-CD4 BV650 (clone SK3, cat# 563875, 1:100), anti-CD8 APC-H7 (clone SK1, cat# 560179, 1:100), anti-CD14 BV510 (clone MφP9, cat# 563079, 1:100), anti-CD19 BV510 (clone SJ25C1, cat# 562947, 1:100), and anti-IFN-γ APC (clone B27, cat# 554702, 1:100) from BD Biosciences; and anti-IL-2 PE (clone MQ1-17H12, cat# 12-7029-42, 1:100) and anti-TNF FITC (clone MAb11, cat# 11-7349-82, 1:100) from ThermoFisher. Multi-color flow cytometry was performed using an LSRII instrument with FACSDiva (BD Biosciences). The data were analyzed using FlowJo software (FlowJo).
Quantification and statistical analysis
Statistics analysis
The statistical experimental analyses of unvaccinated COVID-19 patient samples and omicron breakthrough infections in COVID-19 vaccinated people sera were assessed by nonparametric Wilcoxon matched-pairs signed rank test (Figures 1 and 3). Longitudinal evaluation of serum sample, we use the Kruskal-Wallis test (Figure 2). Statistical analyses were performed using GraphPad Prism 9.3.1(GraphPad Software, La Jolla, CA). Differences between groups were analyzed by nonparametric (Wilcoxon matched-paired signed rank or Mann–Whitney U) tests. To determine the differences in the pie charts, permutation tests in the SPICE software were used (10,000 permutations) (Figure 4). Statistical details of our experiments and analyses can be found in each figure legends.
Acknowledgments
We thank the study participants and coworkers at the Laboratory Medicine, Division of Pulmonary and Critical Care Medicine, Department of Internal Medicine at NMC. We thank B. Judd and P. Kretchmer for proofreading the article. This work was supported by the Institute for Basic Science (IBS), Korea (IBS-R801-D1 and IBS-R801-D2); National Medical Center (grant no. NMC2020-MS-02); Korea National Institute of Health; and Korea Disease Control and Prevention Agency (2021ER260300). This work was also financially supported by the Research Year of Chungbuk National University in 2021 for H.W.J.
Author contributions
Conceptualization, H.W.J., J.Y.S., E.-C.S., and Y.K.C.; methodology, H.W.J., S.-M.K., M.K.J., J.-S.Y., E.-H.K., S.-G.J., and J.G.; investigation, H.W.J., S.-M.K., M.K.J., J.Y.N., J.-S.Y., E.-H.K., Y.-I.K., S.-G.J., J.G., M.A.C., R.R., J.H.C., H.-S.K., J.H.K., J.U., C.K., J.-S.P., Y.K., B.S.C., S.J., J.Y.C., K.-H.S., Y.-D.K., J.Y.S., E.-C.S., and Y.K.C.; visualization, H.W.J., S.-M.K., M.K.J., and J.-S.Y.; funding acquisition, E.-C.S., Y.K.C., J.Y.S., and J.-S.P.; writing – original draft, H.W.J., S.-M.K., M.K.J., J.-S.Y., E.-C.S., and Y.K.C.; writing – review & editing, H.W.J., S.-M.K., M.K.J., J.Y.N., J.-S.Y., J.Y.S., E.-C.S., and Y.K.C.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We worked to ensure gender balance and to ensure ethnic or other types of diversity in the recruitment of human subjects. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
Published: October 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2022.100764.
Contributor Information
Joon Young Song, Email: infection@korea.ac.kr.
Eui-Cheol Shin, Email: ecshin@kaist.ac.kr.
Young Ki Choi, Email: choiki55@ibs.re.kr.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This study did not generate any new codes.
-
•
Any additional information required to reanalyze the data reported in this work is available from the lead contact upon request.
References
- 1.WHO . 2022. WHO Coronavirus (COVID-19) Dashboard.http://covid19.who.int [Google Scholar]
- 2.Pascarella S., Ciccozzi M., Bianchi M., Benvenuto D., Cauda R., Cassone A. The electrostatic potential of the omicron variant spike is higher than in Delta and delta-plus variants: a hint to higher transmissibility? J. Med. Virol. 2022;94:1277–1280. doi: 10.1002/jmv.27528. [DOI] [PubMed] [Google Scholar]
- 3.Dai L., Gao G.F. Viral targets for vaccines against COVID-19. Nat. Rev. Immunol. 2021;21:73–82. doi: 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim C., Ryu D.K., Lee J., Kim Y.I., Seo J.M., Kim Y.G., Jeong J.H., Kim M., Kim J.I., Kim P., et al. A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein. Nat. Commun. 2021;12:288. doi: 10.1038/s41467-020-20602-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rees-Spear C., Muir L., Griffith S.A., Heaney J., Aldon Y., Snitselaar J.L., Thomas P., Graham C., Seow J., Lee N., et al. The effect of spike mutations on SARS-CoV-2 neutralization. Cell Rep. 2021;34:108890. doi: 10.1016/j.celrep.2021.108890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.VanBlargan L.A., Errico J.M., Halfmann P.J., Zost S.J., Crowe J.E., Purcell L.A., Kawaoka Y., Corti D., Fremont D.H., Diamond M.S. An infectious SARS-CoV-2 B. 1.1. 529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat. Med. 2022;28:490–495. doi: 10.1038/s41591-021-01678-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffmann M., Krüger N., Schulz S., Cossmann A., Rocha C., Kempf A., Nehlmeier I., Graichen L., Moldenhauer A.S., Winkler M.S., et al. The Omicron variant is highly resistant against antibody-mediated neutralization: implications for control of the COVID-19 pandemic. Cell. 2022;185:447–456.e11. doi: 10.1016/j.cell.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu L., Iketani S., Guo Y., Chan J.F.W., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2022;602:676–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- 9.Servellita V., Syed A.M., Morris M.K., Brazer N., Saldhi P., Garcia-Knight M., Sreekumar B., Khalid M.M., Ciling A., Chen P.Y., et al. Neutralizing immunity in vaccine breakthrough infections from the SARS-CoV-2 Omicron and Delta variants. Cell. 2022;185:1539–1548.e5. doi: 10.1016/j.cell.2022.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan K., Karim F., Ganga Y., Bernstein M., Jule Z., Reedoy K., Cele S., Lustig G., Amoako D., Wolter N., et al. Omicron BA. 4/BA. 5 escape neutralizing immunity elicited by BA. 1 infection. Nat. Commun. 2022;13:4686. doi: 10.1038/s41467-022-32396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pulliam J.R.C., van Schalkwyk C., Govender N., von Gottberg A., Cohen C., Groome M.J., Dushoff J., Mlisana K., Moultrie H. Increased risk of SARS-CoV-2 reinfection associated with emergence of the Omicron variant in South Africa. Science. 2022;376:eabn4947. doi: 10.1126/science.abn4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.WHO Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern. http://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern
- 13.Viana R., Moyo S., Amoako D.G., Tegally H., Scheepers C., Althaus C.L., Anyaneji U.J., Bester P.A., Boni M.F., Chand M., et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022;603:679–686. doi: 10.1038/s41586-022-04411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao Y., Yisimayi A., Jian F., Song W., Xiao T., Wang L., Du S., Wang J., Li Q., Chen X., et al. BA. 2.12. 1, BA. 4 and BA. 5 escape antibodies elicited by Omicron infection. Nature. 2022;608:593–602. doi: 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cele S., Jackson L., Khoury D.S., Khan K., Moyo-Gwete T., Tegally H., San J.E., Cromer D., Scheepers C., Amoako D.G., et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022;602:654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans J.P., Zeng C., Carlin C., Lozanski G., Saif L.J., Oltz E.M., Gumina R.J., Liu S.L. Neutralizing antibody responses elicited by SARS-CoV-2 mRNA vaccination wane over time and are boosted by breakthrough infection. Sci. Transl. Med. 2022;14:eabn8057. doi: 10.1126/scitranslmed.abn8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chu L., Vrbicky K., Montefiori D., Huang W., Nestorova B., Chang Y., Carfi A., Edwards D.K., Oestreicher J., Legault H., et al. Immune response to SARS-CoV-2 after a booster of mRNA-1273: an open-label phase 2 trial. Nat. Med. 2022;28:1042–1049. doi: 10.1038/s41591-022-01739-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng K.W., Faulkner N., Cornish G.H., Rosa A., Harvey R., Hussain S., Ulferts R., Earl C., Wrobel A.G., Benton D.J., et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370:1339–1343. doi: 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wratil P.R., Schmacke N.A., Karakoc B., Dulovic A., Junker D., Becker M., Rothbauer U., Osterman A., Spaeth P.M., Ruhle A., et al. Evidence for increased SARS-CoV-2 susceptibility and COVID-19 severity related to pre-existing immunity to seasonal coronaviruses. Cell Rep. 2021;37:110169. doi: 10.1016/j.celrep.2021.110169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson E.M., Goodwin E.C., Verma A., Arevalo C.P., Bolton M.J., Weirick M.E., Gouma S., McAllister C.M., Christensen S.R., Weaver J., et al. Seasonal human coronavirus antibodies are boosted upon SARS-CoV-2 infection but not associated with protection. Cell. 2021;184:1858–1864.e10. doi: 10.1016/j.cell.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo L., Wang Y., Kang L., Hu Y., Wang L., Zhong J., Chen H., Ren L., Gu X., Wang G., et al. Cross-reactive antibody against human coronavirus OC43 spike protein correlates with disease severity in COVID-19 patients: a retrospective study. Emerg. Microbes Infect. 2021;10:664–676. doi: 10.1080/22221751.2021.1905488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narowski T.M., Raphel K., Adams L.E., Huang J., Vielot N.A., Jadi R., de Silva A.M., Baric R.S., Lafleur J.E., Premkumar L. SARS-CoV-2 mRNA vaccine induces robust specific and cross-reactive IgG and unequal neutralizing antibodies in naïve and previously infected recipients. Cell Rep. 2022;38:110336. doi: 10.1016/j.celrep.2022.110336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaplonek P., Wang C., Bartsch Y., Fischinger S., Gorman M.J., Bowman K., Kang J., Dayal D., Martin P., Nowak R.P., et al. Early cross-coronavirus reactive signatures of humoral immunity against COVID-19. Sci. Immunol. 2021;6:eabj2901. doi: 10.1126/sciimmunol.abj2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamaguchi T., Shinagawa T., Kobata H., Nakagawa H. Immunity against seasonal human coronavirus OC43 mitigates fatal deterioration of COVID-19. Int. J. Infect. Dis. 2021;109:261–268. doi: 10.1016/j.ijid.2021.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monto A.S., Malosh R.E., Petrie J.G., Martin E.T. The doctrine of original antigenic sin: separating good from evil. J. Infect. Dis. 2017;215:1782–1788. doi: 10.1093/infdis/jix173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goel R.R., Painter M.M., Apostolidis S.A., Mathew D., Meng W., Rosenfeld A.M., Lundgreen K.A., Reynaldi A., Khoury D.S., Pattekar A., et al. mRNA vaccines induce durable immune memory to SARS-CoV-2 and variants of concern. Science. 2021;374:abm0829. doi: 10.1126/science.abm0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turner J.S., O'Halloran J.A., Kalaidina E., Kim W., Schmitz A.J., Zhou J.Q., Lei T., Thapa M., Chen R.E., Case J.B., et al. SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses. Nature. 2021;596:109–113. doi: 10.1038/s41586-021-03738-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muecksch F., Weisblum Y., Barnes C.O., Schmidt F., Schaefer-Babajew D., Wang Z., C Lorenzi J.C., Flyak A.I., DeLaitsch A.T., Huey-Tubman K.E., et al. Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations. Immunity. 2021;54:1853–1868.e7. doi: 10.1016/j.immuni.2021.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moriyama S., Adachi Y., Sato T., Tonouchi K., Sun L., Fukushi S., Yamada S., Kinoshita H., Nojima K., Kanno T., et al. Temporal maturation of neutralizing antibodies in COVID-19 convalescent individuals improves potency and breadth to circulating SARS-CoV-2 variants. Immunity. 2021;54:1841–1852.e4. doi: 10.1016/j.immuni.2021.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., Cho A., Jankovic M., Schaefer-Babajew D., Oliveira T.Y., et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi S.J., Kim D.U., Noh J.Y., Kim S., Park S.H., Jeong H.W., Shin E.C. T cell epitopes in SARS-CoV-2 proteins are substantially conserved in the Omicron variant. Cell. Mol. Immunol. 2022;19:447–448. doi: 10.1038/s41423-022-00838-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keeton R., Tincho M.B., Ngomti A., Baguma R., Benede N., Suzuki A., Khan K., Cele S., Bernstein M., Karim F., et al. T cell responses to SARS-CoV-2 spike cross-recognize Omicron. Nature. 2022;603:488–492. doi: 10.1038/s41586-022-04460-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naranbhai V., Nathan A., Kaseke C., Berrios C., Khatri A., Choi S., Getz M.A., Tano-Menka R., Ofoman O., Gayton A., et al. T cell reactivity to the SARS-CoV-2 Omicron variant is preserved in most but not all prior infected and vaccinated individuals. medRxiv. 2022 doi: 10.1101/2022.01.04.21268586. Preprint at. [DOI] [Google Scholar]
- 34.Bange E.M., Han N.A., Wileyto P., Kim J.Y., Gouma S., Robinson J., Greenplate A.R., Hwee M.A., Porterfield F., Owoyemi O., et al. CD8+ T cells contribute to survival in patients with COVID-19 and hematologic cancer. Nat. Med. 2021;27:1280–1289. doi: 10.1038/s41591-021-01386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McMahan K., Yu J., Mercado N.B., Loos C., Tostanoski L.H., Chandrashekar A., Liu J., Peter L., Atyeo C., Zhu A., et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noh J.Y., Jeong H.W., Kim J.H., Shin E.-C. T cell-oriented strategies for controlling the COVID-19 pandemic. Nat. Rev. Immunol. 2021;21:687–688. doi: 10.1038/s41577-021-00625-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhuang Z., Lai X., Sun J., Chen Z., Zhang Z., Dai J., Liu D., Li Y., Li F., Wang Y., et al. Mapping and role of T cell response in SARS-CoV-2–infected mice. J. Exp. Med. 2021;218:e20202187. doi: 10.1084/jem.20202187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lipsitch M., Krammer F., Regev-Yochay G., Lustig Y., Balicer R.D. SARS-CoV-2 breakthrough infections in vaccinated individuals: measurement, causes and impact. Nat. Rev. Immunol. 2022;22:57–65. doi: 10.1038/s41577-021-00662-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bergwerk M., Gonen T., Lustig Y., Amit S., Lipsitch M., Cohen C., Mandelboim M., Levin E.G., Rubin C., Indenbaum V., et al. Covid-19 breakthrough infections in vaccinated health care workers. N. Engl. J. Med. 2021;385:1474–1484. doi: 10.1056/NEJMoa2109072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng S.M.S., Mok C.K.P., Leung Y.W.Y., Ng S.S., Chan K.C.K., Ko F.W., Chen C., Yiu K., Lam B.H.S., Lau E.H.Y., et al. Neutralizing antibodies against the SARS-CoV-2 Omicron variant BA. 1 following homologous and heterologous CoronaVac or BNT162b2 vaccination. Nat. Med. 2022;28:486–489. doi: 10.1038/s41591-022-01704-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This study did not generate any new codes.
-
•
Any additional information required to reanalyze the data reported in this work is available from the lead contact upon request.