Abstract
A superfamily of cyclic amidohydrolases, including dihydropyrimidinase, allantoinase, hydantoinase, and dihydroorotase, all of which are involved in the metabolism of purine and pyrimidine rings, was recently proposed based on the rigidly conserved structural domains in identical positions of the related enzymes. With these conserved domains, two putative cyclic amidohydrolase genes from Escherichia coli, flanked by related genes, were identified and characterized. From the genome sequence of E. coli, the allB gene and a putative open reading frame, tentatively designated as a hyuA (for hydantoin-utilizing enzyme) gene, were predicted to express hydrolases. In contrast to allB, high-level expression of hyuA in E. coli of a single protein was unsuccessful even under various induction conditions. We expressed HyuA as a maltose binding protein fusion protein and AllB in its native form and then purified each of them by conventional procedures. allB was found to encode a tetrameric allantoinase (453 amino acids) which specifically hydrolyzes the purine metabolite allantoin to allantoic acid. Another open reading frame, hyuA, located near 64.4 min on the physical map and known as a UUG start, coded for d-stereospecific phenylhydantoinase (465 amino acids) which is a homotetramer. As a novel enzyme belonging to a cyclic amidohydrolase superfamily, E. coli phenylhydantoinase exhibited a distinct activity toward the hydantoin derivative with an aromatic side chain at the 5′ position but did not readily hydrolyze the simple cyclic ureides. The deduced amino acid sequence of the novel phenylhydantoinase shared a significant homology (>45%) with those of allantoinase and dihydropyrimidinase, but its functional role still remains to be elucidated. Despite the unclear physiological function of HyuA, its presence, along with the allantoin-utilizing AllB, strongly suggested that the cyclic ureides might be utilized as nutrient sources in E. coli.
A superfamily of cyclic amidohydrolases (EC 3.5.2), including hydantoinase, dihydropyrimidinase, allantoinase, and dihydroorotase, has been recently proposed based on the functional and structural similarity of the related enzymes, providing evidence for an evolutionary common ancestor in related amidohydrolases (1, 21, 23). This family of enzymes is involved in the metabolism of pyrimidines and purines, sharing the property of hydrolyzing the cyclic amide bond of each substrate to the corresponding N-carbamyl amino acids (16, 35). Dihydropyrimidinase and allantoinase catalyze the degradation of pyrimidines and purines, respectively, while hydantoinase, a microbial counterpart of dihydropyrimidinase, is also supposed to be involved in pyrimidine degradation. On the other hand, dihydroorotase participates in the de novo synthesis of pyrimidines (16). In a recent report, guanine deaminase from human kidney showed a common structural motif conserved in the family of cyclic amidohydrolases (46), expanding the family to more divergent enzymes acting on the purine and pyrimidine rings.
Comparative analyses with the enzymes belonging to this superfamily revealed that the related enzymes possess very similar biochemical and structural properties in terms of the quaternary structure, oligomerization, metal dependency, and apparent reaction conditions, along with a striking similarity in the primary and secondary structures of the enzymes (19, 21). In addition, the enzymes were found to have a number of conserved motifs, such as PGXIDXHXH, which is responsible for binding metal (19, 21, 46, 47). However, the approach undertaken to classify and predict the properties for the cyclic amidohydrolase family of enzymes has been mainly based on the substrate specificity and the sequence-based alignment using the protein database. Such studies have not provided enough information about the exact function of these enzymes in vivo. For instance, the narrow substrate specificity of the dihydroorotase confirmed its unique functional role in the de novo synthesis of pyrimidines (18). Other enzymes belonging to the superfamily exhibited the highest affinity for their own substrates and also showed a rather low level of activity with other substrates. Allantoinase, eukaryotic dihydropyrimidinase, microbial hydantoinase, and imidase were found to hydrolyze cyclic ureides such as allantoin, dihydropyrimidine, and hydantoin derivatives, while also showing rather low but distinct levels of activity towards other substrates (14, 21, 22, 28, 33). Therefore, to elucidate the functional role and substitution of the related enzymes in vivo, it is necessary to undertake genetic study in vivo. Functional complementation among the enzymes and defects due to the deleted gene segments are expected to address the above points. These findings might also provide some information regarding the utilization of purine or pyrimidine bases in various organisms (25, 41).
Recently, research on the cyclic amidohydrolase family of enzymes has been proliferating (13, 24, 32, 45, 46). Most of these results are derived from sequence-based motifs and biochemical characterizations, mainly due to the absence of established genetic data or lack of attention to the physiological role of the enzyme. In order to determine their exact cellular functions from a subtle functional heterogeneity, it is crucial to identify and characterize the related enzymes in a strain where the corresponding genes can be easily manipulated. In this paper, we describe the identification of the two genes from Escherichia coli encoding the enzymes belonging to a cyclic amidohydrolase superfamily. From the biochemical characteristics of the expressed enzymes, one was confirmed to be allantoinase, and the other, in accord with its unique substrate specificity, was considered a novel enzyme and named phenylhydantoinase. Our results confirmed the recent report (11) that a gene cluster spanning the region of the allB gene is involved in the utilization of purines in E. coli. We present here the physicochemical and catalytic properties of two cyclic amidohydrolase enzymes of E. coli.
MATERIALS AND METHODS
Strains and media.
Derivatives of E. coli K-12 were used as a source for the two cyclic amidohydrolase genes. E. coli cells were grown at 37°C in Luria-Bertani (LB) broth supplemented with ampicillin (50 μg/ml) when needed.
Protein database search and sequence alignment.
The alignment of sequences deduced from the two putative amidohydrolase genes of E. coli was performed by hierarchical clustering of the individual sequences based on the pair-wise similarity scores. The identity of discrete amino acid sequences common in related enzymes was analyzed by the CLUSTAL W program (36). Further analyses were performed by visual inspection for the detailed comparison of the sequence alignment. To predict the distribution of the secondary structural elements in the enzyme sequences, Chou and Fasman's algorithms (8) were applied to each enzyme sequence by using a program from the National Center for Biotechnology Information.
Cloning and expression of cyclic amidohydrolases from E. coli.
Chromosomal DNA was isolated from E. coli by using a genomic DNA purification kit (Promega) and cloned by using standard recombinant DNA techniques. Two sets of primers were designed to span the genes encoding for the putative hydantoin-utilizing enzyme (hyuA, 1,398 bp) and the allantoinase (allB, 1,362 bp) of E. coli. The two primers for the hydantoin-utilizing enzyme are designated HYUN (5′-GGAGAATTCTTGGAGTTTGCTATGCGCGTA-3′) and HYUC (5′-TGGCTGCAGTTAGAGCACGGGAGGGACAAA-3′). The two primers for allantoinase are designated ALLN (5′-AGGAATTCGTTATGTCTTTTGATTTAATCATT-3′) and ALLC (5′-GGGGATCCTTACTGCTGATGTTTAAGGATAA-3′). Restriction sites, EcoRI/PstI for HyuA and EcoRI/BamHI for AllB, were introduced into the N- and C-terminal primers, respectively. The amplified DNA fragments encoding the putative hydrolases AllB and HyuA were cloned into the EcoRI/BamHI and EcoRI/PstI sites of pTrc99A, respectively, yielding the corresponding plasmids pTAB and pTHY.
Preparation of MBP-HyuA fusion protein.
The pMAL vector (New England Biolabs) was used to express the product of the putative amidohydrolase gene hyuA as a fusion protein with the product of the E. coli maltose binding protein (MBP) gene according to the procedures of the manufacturer. The hyuA gene was amplified by PCR from the E. coli strain by using the primers. HYUN and HYUC, and then cloned into the EcoRI and PstI sites of pMAL-c2X (New England Biolabs). The resulting construct (pMHY) was confirmed by DNA sequencing.
Overexpression and purification of the two cyclic amidohydrolases.
All E. coli strains harboring the plasmids pTAB, pTHY, and pMHY were prepared by transforming E. coli JM109 with the individual plasmid. Each overnight culture (10 ml) was inoculated into 500 ml of LB medium containing ampicillin (50 μg/ml) and then grown at 37°C. When the optical density of the cells at 600 nm reached about 0.5 to 0.6, the cloned gene was induced by the addition of 0.5 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG). After an additional 2-h incubation period, the cells were harvested by centrifugation at 8,000 × g for 10 min, and the resulting pellets were resuspended in a total volume of 20 ml of Tris-HCl buffer (20 mM; pH 8.0) containing 1 mM dithiotreitol (DTT) and 1 mM phenylmethylsulfonyl fluoride. The suspended cells were sonicated, and the supernatant was obtained after centrifugation at 18,000 × g for 30 min. The resulting solution was assayed directly for enzyme activity and protein expression by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). For biochemical characterizations, the protein was further purified to apparent homogeneity by the standard chromatographic procedures described below.
The putative allantoinase (AllB) was purified as previously reported with slight modification (21). All purification steps were conducted at room temperature. The supernatant (20 ml) was loaded onto a column of Resource Q (5 ml) equilibrated with 20 mM Tris-HCl (pH 8.0) buffer containing 5% glycerol on a fast protein liquid chromatography system system (Pharmacia). The column was washed with 10 volumes of the same buffer and eluted with a linear gradient of 0 to 0.5 M NaCl. The active fractions were pooled and concentrated using a Centriprep 10 (Amicon) filter. The concentrated protein solution was loaded onto a Superose 12 gel filtration column equilibrated with 20 mM Tris-HCl (pH 8.0) buffer containing 150 mM NaCl. The eluted enzyme was concentrated by dialysis against 20 mM Tris-HCl (pH 8.0) buffer containing 5% glycerol.
The MBP fusion protein in the soluble fraction was absorbed onto amylose resin columns (New England Biolabs) and then eluted with a buffer containing 10 mM maltose and 200 mM NaCl. The affinity column-purified fusion protein was further concentrated by using a Centriprep 10 filter. The fused enzyme was separated from the MBP by treatment with factor Xa for 40 h at 8 to 10°C. Then the cleaved enzyme was isolated from the MBP by reapplying it onto the amylose resin and concentrated by dialysis against 20 mM Tris-HCl (pH 8.0) buffer containing 5% glycerol.
Immunoprecipitation and N-terminal sequence analysis.
For the immunoprecipitation of the putative cyclic amidohydrolases from E. coli, an antibody raised against the structurally related d-hydantoinase from Bacillus stearothermophilus SD1 was used (19). Cells (5 ml) were harvested, washed with phosphate-buffered saline solution, and then lysed in 0.5 ml of buffer containing 10 mM Tris-HCl (pH 7.5), 1% Nonidet P-40, 1 mM EDTA, 0.15 M NaCl, 1 mM phenylmethylsulfonyl fluoride, and 1 mM iodoacetamide. E. coli JM109 cell lysates were pretreated with the preimmune serum followed by the addition of protein A-Sepharose. The putative cyclic amidohydrolases were precipitated by adding rabbit anti-d-hydantoinase-coupled protein A-Sepharose for 1 h at 4°C. The precipitates were washed three times with lysis buffer. For the SDS-PAGE analysis and N-terminal sequence analysis, the immunoprecipitated enzyme was eluted from the resin using an elution buffer containing 100 mM carbonate (pH 10.5) and 2 M NaCl and then concentrated by using a Centricon 30 filter. Half of the purified enzyme was analyzed on a 9% denatured polyacrylamide gel.
After SDS-PAGE of the purified enzymes (5 μg), proteins were transferred to a polyvinylidine difluoride membrane loaded in a Blot Cartridge (Applied Biosystems). The membrane was fully washed with deionized distilled water and then stained with Coomassie brilliant blue R-250. Polyvinylidene difluoride-blotted protein bands corresponding to the putative amidohydrolases were sequenced with a protein sequencer (model 610A; Applied Biosystems).
Oligomeric structure analysis and enzyme assay.
The oligomeric structures of the enzymes were determined on a fast protein liquid chromatography system (Pharmacia) with a gel filtration column (Superose-12 HR10/30). The flow rate of the mobile phase containing 20 mM Tris-HCl and 150 mM NaCl was 0.3 ml/min. The column was calibrated using native protein markers (Pharmacia), and a molecular mass standard curve was established by plotting the elution profile of the protein markers (Pharmacia) versus the known molecular masses on semilog paper.
The activities of the putative allantoinase and hydantoin-utilizing enzyme were determined at 40°C for 30 min with constant shaking. The enzyme reaction was carried out with the purified enzyme (10 μg) in 1 ml of reaction mixture containing 100 mM Tris-HCl (pH 8.0), 0.5 mM DTT, and 10 mM cyclic ureide. A decrease in the concentration of the cyclic ureide used as a substrate and an increase in the production of the corresponding N-carbamyl compound were analyzed, respectively, by high-performance liquid chromatography and thin-layer chromatography (TLC) (33, 40). The amount of product formed was also determined by using either high-performance liquid chromatography or the color reagent p-dimethylaminobenzaldehyde (20). All assays were carried out in duplicate. One unit of enzyme activity was defined as the amount of enzyme required to hydrolyze 1 μmol of cyclic ureide per min under the specified conditions. Kinetic parameters and metal dependency were determined according to previously reported methods (20, 31). Chiral TLC (Merck) was used to analyze the stereospecificity of the enzyme as reported in a previous work (20).
RESULTS
Identification of the two cyclic amidohydrolases from E. coli.
Although the utilization of a cyclic ureide (allantoin) as a nitrogen source by E. coli was first suggested by Vogels and van der Drift (38), the detailed genetic and biochemical studies have become a recent subject. Accordingly, little information on the utilization of other cyclic ureides such as hydantoin derivatives and pyrimidines has been revealed. Therefore, we tested various E. coli strains as potential cloning hosts for the expression of the pyrimidine or purine-catabolizing enzymes, dihydropyrimidinase and allantoinase. Most of the E. coli strains grown in minimal medium supplemented with hydantoin as the sole nitrogen source, even after 40 h of cultivation on agar plates, did not grow sufficiently, but distinctive colonies were observed. Minimal medium supplemented with allantoin also supported the basal growth of the E. coli strains (data not shown). In light of this finding, further experimental observations that the crude extract of E. coli cells exhibited hydantoin- and allantoin-hydrolyzing activities strongly suggested the presence of cyclic ureide-utilizing enzymes in E. coli.
We previously analyzed the primary and secondary structures of the functionally related enzymes at the molecular level and found that the similarity of the enzymes is sufficient to define a novel cyclic amidohydrolase superfamily (21). In a further finding, several regions in the primary and the secondary structures were found to be rigidly conserved in identical positions over the entire sequences. The striking structural similarity of these regions strongly supported the idea that the conserved regions might play a critical role in the structure or function of this family of enzymes. Consistent with this view, further evidence was obtained from the scanning of the conserved regions on the SwissProt protein database. Query sequences were designed from the regions of the alignment with high stringency (percentage of fixed residues in the sequences), always avoiding gaps to reduce fortuitous matching results. Interestingly, alignment revealed that the conserved regions have a large percentage of homology with the known cyclic amidohydrolase family enzymes, suggesting that such arranged domains over a significant length of the protein backbone are crucial to the structure and function of the related enzymes (16, 21).
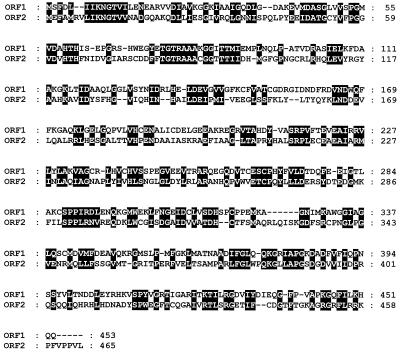
In order to find a possible candidate gene encoding the putative cyclic amidohydrolase, we aligned the conserved regions of this family of enzymes with the complete genome sequence of E. coli (2). The scanning result comprised three sequences of open reading frames (ORFs). One was already identified and characterized as pyrC, an E. coli dihydroorotase gene (43), another was designated allB. The allB gene was recently identified as encoding an allantoinase (11), but no biochemical information was reported. The other gene was predicted to encode a putative amidohydrolase. As shown in Fig. 1, a reliable assignment of the homologous regions, comprising an entire ORF, was revealed. ORF1, known as allB, was found in the gcl-ylbC intergenic region, and ORF2 (Gene Bank accession no. U28375.1), not previously classified and tentatively designated hyuA (encoding a putative hydantoin-utilizing enzyme), was located at the pbl-lysS intergenic region of the E. coli chromosome.
FIG. 1.
Amino acid sequence alignment of the two putative cyclic amidohydrolases from E. coli. Identical and conserved amino acid residues are indicated by black boxes.
Characterization of the two hydrolase genes allB and hyuA.
The ORFs of allB and hyuA have a very similar GC content, with values close to the overall GC content determined for the E. coli chromosome. The consensus sequence PGXI(V)DXHXH, commonly conserved in the superfamily enzymes, was also found. Analyses of the spanning regions of the E. coli allB and hyuA genes revealed several ORFs with deduced amino acid sequences, but most of their gene products have not been characterized yet.
The amino acid sequence deduced from the allB nucleotide sequence comprised a protein consisting of 453 amino acid residues with a calculated molecular mass of 50.4 kDa, while the nucleotide sequence deduced from hyuA encoded a polypeptide of 465 amino acid residues with a calculated molecular mass of 51.5 kDa. The putative metal binding site, previously identified from other enzymes in this family, was found in both the region (57 to 61 residues) of AllB and the region (61 to 65 residues) of HyuA. There was a high degree of amino acid identity between these two enzymes, and the similarity increased up to 46.1% when the conservative substitution was considered. BLAST (basic local alignment search tool) analysis with the allB and hyuA sequences revealed homologous genes encoding other cyclic amidohydrolase family enzymes. Especially, the predicted ORFs displayed a high homology (22 to 41%) to allantoinases, dihydropyrimidinases, and hydantoinases which were isolated from various eukaryotic and prokaryotic sources (5, 6, 13, 19, 26). At the genetic level, thus, two ORFs were considered to encode enzymes belonging to a cyclic amidohydrolase superfamily.
Cloning and expression of the allB and hyuA of E. coli.
To identify the proteins encoded by the predicted genes allB and hyuA, the corresponding genes were amplified from E. coli chromosomal DNA by PCR. The transformed E. coli JM109 cells harboring pTAB and pTHY were screened on the activity-staining plates (19). Each plate was supplemented with a typical substrate (hydantoin, allantoin, dihydrouracil, or dihydroorotate) of this family of enzymes. As expected, E. coli cells transformed with pTAB clearly exhibited enzyme activity on the purine metabolite allantoin, producing the allantoic acid. The specific enzyme activity of cells induced in LB medium was much higher (>500 fold) than that of the wild-type cells. However, E. coli cells harboring the pTHY did not show any apparent activity on the activity-staining plate containing the typical substrate. We thus continued assaying with other possible substrates which are structurally analogous to cyclic ureides and detected distinct activity in a reaction mixture containing hydroxyphenylhydantoin as a substrate. Based on this observation, we tentatively designated HyuA as phenylhydantoinase by taking into consideration its apparent activity toward phenylhydantoin. Cell extracts of the recombinant E. coli showed a minor protein band (<0.5%), corresponding to HyuA, even under various induction conditions.
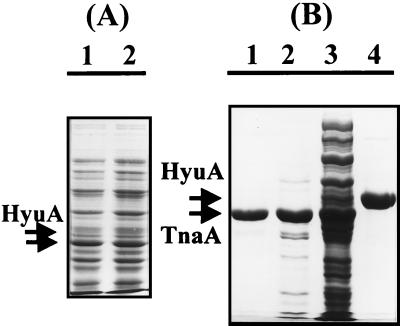
In an attempt to increase the in vivo expression level of HyuA by E. coli transformed with pTHY, we observed an unexpected protein band which is expressed at high levels concomitantly with HyuA (Fig. 2A). This protein apparently showed a faster migration rate than HyuA, and its expression level reached about 20 to 25% of the total cell protein as shown in lane 3 of Fig. 2B. But the expression level of the HyuA under identical conditions accounted for only 0.1 to 0.5% of the total protein. When other E. coli strains, MC4100, XL1-BLUE, and HB101, were tested as hosts, a similar expression pattern was observed. For additional differentiation, we attempted immunoaffinity precipitation with an antibody raised against the d-hydantoinase from B. stearothermophilus SD1. As shown in lane 4 of Fig. 2B, the HyuA produced by E. coli was reactive and precipitated with the antibody. However, no interaction was observed between the overexpressed protein and the antibody, which indicates that the coexpressed protein did not result from the fragmentation of the E. coli HyuA. For further identification, the coexpressed protein was purified from E. coli AllB from a small-scale culture by identical procedures, and then the N-terminal amino acid residues were sequenced (Fig. 2B, lanes 1 to 3). The resulting sequence, MENFKHLPEPFRIRV, was an exact match with the sequence of E. coli tryptophanase, TnaA (12). The tentatively designated phenylhydantoinase revealed the expected sequence MEFAMRVLIKNGTVVNADGQ.
FIG. 2.
SDS-PAGE analysis of the protein coexpressed at high levels when HyuA was expressed by induction. (A) E. coli cells harboring pTHY were induced with 0.5 mM IPTG at 37°C, and an aliquot of the crude extract was analyzed via SDS-PAGE after induction for 30 min (lane 1) and 1 h (lane 2). The proteins concomitantly overexpressed at high levels are indicated by arrows. (B) Small-scale purification for N-terminal sequence analysis of E. coli TnaA and HyuA. Lane 1, purified TnaA eluted from the gel filtration column; lane 2, eluted fraction from the ion-exchange column; lane 3, whole-cell extract; lane 4, immunoaffinity-purified HyuA. For the affinity purification of HyuA, the immunoprecipitated enzyme was eluted from the resin column and then concentrated with a Centricon 30 filter. After SDS-PAGE, the gel was stained with Coomassie brilliant blue. Arrows indicate HyuA and TnaA, respectively.
Interestingly, we observed that the expression level of the HyuA was significantly increased when the enzyme was expressed with the fused MBP (Fig. 3A). To determine whether the fusion protein expressed by the malE-hyuA genes is intrinsically functional in E. coli, we measured the enzyme activity of the induced cells in LB medium. As a result, extracts of the cells expressing the MBP-HyuA protein exhibited a 350- to 600-fold-higher increase in activity toward hydroxyphenylhydantoin than that of the wild-type cells. In control experiments, the considerable activity and protein band were not detected in wild-type E. coli cells.
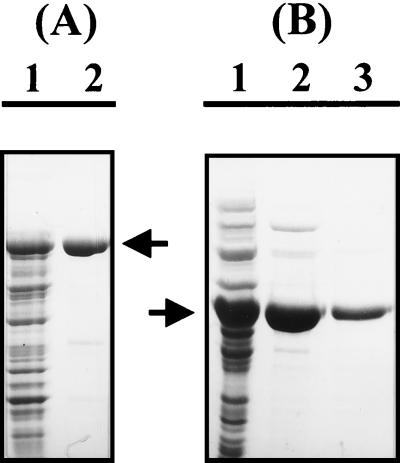
FIG. 3.
SDS-PAGE analysis of MBP-HyuA and AllB. Aliquots (5 μl each) of the protein samples were analyzed on a 10% polyacrylamide gel under denaturing conditions. (A) Analysis of the MBP-HyuA fusion protein. Lane 1, whole-cell extracts after induction for 2 h at 37°C; lane 2, purified MBP-HyuA fusion protein (95 kDa). The arrow indicates the MBP-HyuA fusion protein. (B) Analysis of purified AllB. Lane 1, whole-cell extract; lane 2, fractions eluted from the ion-exchange chromatography column; lane 3, purified AllB from the active fractions after gel filtration column chromatography. The arrow indicates AllB.
Purification of AllB and HyuA.
To confirm whether the enzymes expressed by the hyuA and allB genes indeed belong to a cyclic amidohydrolase family, we purified the two enzymes for further biochemical characterization.
In the case of AllB, further purification was carried out with cell extracts from a 500-ml culture broth. Allantoinase activity was detected mainly in the supernatant fraction, with negligible activity (<5%) found in the cell pellet. Crude extract was appropriately concentrated and subjected to SDS-PAGE analysis. As shown in Fig. 3B, a distinct band corresponding to a molecular mass of about 52 kDa, which was not detected in the wild-type E. coli strain, appeared. After native-gel electrophoresis, activity staining was performed by overlaying the protein gel with an agarose gel containing allantoin as a substrate (19). Consequently, a bright yellow color developed at the overexpressed-band position. After clarification by filtration, the crude cellular extract was loaded onto an ion-exchange chromatography column. The active fractions were collected, concentrated, and further resolved with a Superose-12 gel filtration column. A single distinct band was detected after SDS-PAGE, and the N-terminal amino acid sequence of this purified enzyme was found to be identical to that of AllB. The purification procedure and yields are summarized in Table 1.
TABLE 1.
Purification procedures and yields of allantoinase from E. coli
| Step | Total protein (mg) | Sp. act. (U/mg) | Purification (fold) |
|---|---|---|---|
| Crude extract | 148.2 | 0.13 | 1.0 |
| Protamine sulfate | 129.5 | 0.15 | 1.2 |
| Resource Q | 2.7 | 4.32 | 33.2 |
| Gel filtration | 2.1 | 5.72 | 44 |
In the case of HyuA, the enzyme was first purified as a fusion protein with MBP. Upon IPTG induction, E. coli cells significantly overproduced the corresponding fusion protein MBP-HyuA. SDS-PAGE analysis of the crude extracts indicated that the fusion protein accounted for 20 to 25% of the total cell protein and was mainly detected in the soluble fraction (80 to 85%). The fusion protein was purified with an amylose resin affinity column and then concentrated (Fig. 3A). About 4 mg of the MBP-HyuA fusion protein was recovered from a 200-ml culture of the induced cells. For further purification, the fusion protein was cleaved with factor Xa and then reapplied onto the amylose resin. Gel electrophoresis under denaturing conditions showed a homogeneous enzyme with the expected molecular mass of 51 to 53 kDa. The resulting functional HyuA was concentrated and stored in 20 mM Tris-HCl (pH 7.5) buffer containing 1 mM DTT and 20% glycerol at −20°C for further analysis.
Characterizations of AllB and HyuA. (i) Relative molecular mass and oligomeric structure.
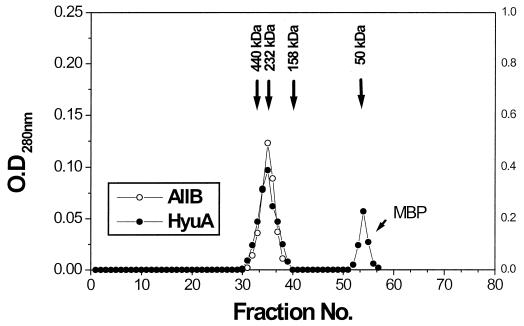
From the Superose-12 gel filtration chromatography column fractions, two enzymes were observed to coelute with an apparent molecular mass between 200 to 230 kDa (Fig. 4). The respective subunit masses were determined to be 50.5 kDa for AllB and 51.5 kDa for HyuA. Thus, the quaternary structure for each of these enzymes was predicted to be a homotetramer. The cross-linking experiments with sulfosuccinimidyl 4-(N-maleimidomethyl)-cyclohexane-1-carboxylate and 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride also supported the prediction that the quaternary structure of both enzymes was tetrameric. The molecular mass and oligomeric structure of AllB and HyuA were found to be similar to those of cyclic amidohydrolases from a variety of other sources (3, 6, 17, 21, 22, 46).
FIG. 4.
Gel filtration analysis of the purified HyuA and AllB. The purified enzyme ranging from 35 to 125 μg was analyzed on a Superose-12 gel filtration column. Open and closed circles, respectively, indicate the elution profiles of AllB and HyuA. The native size of each protein was estimated from the elution profiles of standard protein markers: blue dextran, 2,000 kDa; ferritin, 440 kDa; catalase, 232 kDa; aldolase, 158 kDa; Fab fragment, 50 kDa; and MBP, 42 kDa. All experiments were repeated three times at different protein concentrations. The shift in elution time was negligible (<0.2 min). OD, optical density.
(ii) Optimum pH and temperature.
The pH and temperature optimum for the hydrolysis of the cyclic ureides (allantoin and hydroxyphenylhydantoin) were determined. The temperature dependency of AllB and HyuA showed a similar profile, ranging from 20 to 55°C, and the resulting optimum temperature was 40 to 45°C and 45 to 50°C for AllB and HyuA, respectively.
The pH dependency was analyzed in 0.1 mM boric acid-NaOH, 0.1 mM Tris-HCl, and 0.1 mM sodium phosphate buffer systems at pHs ranging from 8.5 to 10.5, 7.5 to 8.5, and 5.5 to 7.5, respectively. The catabolic activity of AllB and HyuA showed an optimum pH of about 7.5 to 8.0 and 8.0 to 8.5, respectively.
(iii) Effects of divalent metal ions.
It has been known that the activity of the cyclic amidohydrolase family of enzymes is partly or completely affected by the presence of their cofactors (21, 30, 35). We tested various cofactors, such as ATP, NAD(P)H, metal ions, and reducing agents. As expected, except for the divalent metal ions, the other tested cofactors had a negligible effect on the enzyme activity. To determine the specificity with respect to the metal requirement, the enzyme solution (0.1 mg) was first dialyzed with the chelating agent EDTA against metal-free buffer (20 mM Tris [pH 7.5]). Enzyme activity was then determined in the presence of different metal ions under standard assay conditions. As shown in Table 2, a slight decrease in the activity of the EDTA-treated E. coli allantoinase AllB was observed. The addition of divalent metal ions such as Co2+, Ni2+, and Mn2+ enhanced allantoinase activity, while the metal ion Cu2+ severely inhibited the activity of this enzyme. An inhibitory or stimulatory effect was not observed even at a high concentration of metal ions, up to 1 mM (data not shown).
TABLE 2.
Metal dependency of phenylhydantoinase and allantoinase from E. coli
| Metal ion or assaya | Concn (μM) | Relative enzyme activity (%)
|
|
|---|---|---|---|
| Phenylhydantoinase | Allantoinase | ||
| Control | 3.4 | 71.3 | |
| EDTA | 5,000 | NDb | 54.9 |
| Cu2+ | 5 | 2.6 | |
| 50 | ND | 11.4 | |
| 500 | ND | 2.0 | |
| Mn2+ | 5 | 28.2 | |
| 50 | 59.2 | 58.9 | |
| 500 | 100 | 89.4 | |
| Mg2+ | 5 | 3.9 | |
| 50 | 6.8 | 52.7 | |
| 500 | 20.6 | 56.0 | |
| Fe2+ | 5 | 3.7 | |
| 50 | 4.6 | 41.8 | |
| 500 | 15.9 | 43.6 | |
| Zn2+ | 5 | 3.1 | |
| 50 | ND | 35.1 | |
| 500 | ND | 27.2 | |
| Co2+ | 5 | 23.2 | |
| 50 | 56.1 | 89.9 | |
| 500 | 45.8 | 100 | |
| Ca2+ | 5 | 3.9 | |
| 50 | 11.5 | 58.4 | |
| 500 | 20.7 | 68.1 | |
| Ni2+ | 5 | 8.2 | |
| 50 | 29.8 | 89.3 | |
| 500 | 25.4 | 91.7 | |
Preincubation with metal ion and enzyme assay was carried out as described in Materials and Methods. Each value represents the average of two independent assays.
ND, not detected under specified experimental conditions.
The E. coli HyuA, in contrast to the AllB, was quite unstable and showed a strong dependency on the metal ions. Treatment with a chelating agent and preincubation in the assay solution without metal ions resulted in the complete loss of enzyme activity, as shown in Table 2. The addition of either 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (3 mM) or diethylpyrocarbonate (2 mM) also led to complete loss of activity, which suggests that histidyl or aspartyl residues play a crucial role in the catalysis of the phenylhydantoinase. When the metal ion Mn2+ was added exogenously, the activity was fully restored. Other divalent ions, such as Co2+, Ni2+, Ca2+, Mg2+, and Fe2+, could be substituted for Mn2+, partly recovering the catalytic activity. The enzyme activity increased with increasing concentrations of Mn2+ and reached a maximum at concentrations higher than 0.5 mM. At lower concentrations (5 and 50 μM), the metal dependency was not changed significantly. The effect of monovalent cations on the enzyme activity was negligible.
(iv) Substrate specificity.
Table 3 shows the specific activities of the purified enzymes AllB and HyuA toward various cyclic ureides including four typical substrates of the superfamily enzymes. Both enzymes were found to possess a distinct substrate specificity, although a minor level of activity for the analogous compounds was observed. Interestingly, these two enzymes displayed a relatively narrow substrate spectrum compared to that of other enzymes belonging to the same family. Based on the activity exerted by the AllB of E. coli, this enzyme was presumed to be a typical allantoinase. Under standard reaction conditions (pH 8.0, 40°C), AllB showed the highest level of activity toward allantoin among various compounds tested (6.59 U/mg of protein), and only minimally detectable activity was observed for hydantoin, isopropylhydantoin, and 5-bromouracil. The Km and Vmax values were calculated to be 4.2 mM and 6.7 μmol min−1 mg of protein−1 for allantoin, respectively, from the double reciprocal plot of Lineweaver-Burk, resulting in a catalytic efficiency (Vmax/Km) of 1.6 μmol min−1mg of protein−1 mM−1.
TABLE 3.
Specific activities of phenylhydantoinase and allantoinase from E. coli for different substrates
| Substrate | Enzyme activity (U)a
|
|
|---|---|---|
| Phenylhydantoinase | Allantoinase | |
| Hydroxyphenylhydantoin | 3.29 | ND |
| Phenylhydantoin | 2.93 | ND |
| Isopropylhydantoin | 0.85 | 0.02 |
| Hydantoin | 0.37 | 0.12 |
| Dihydrothymine | NDb | ND |
| Uracil | ND | ND |
| 5-Bromouracil | 0.08 | 0.01 |
| Dihydrouracil | ND | ND |
| Dihydroorotate | ND | ND |
| Allantoin | ND | 6.59 |
Enzyme activity was determined by adding the purified enzyme (10 μg) to the reaction mixture containing 1 ml of 100 mM Tris-HCl (pH 8.0) buffer and 10 mM substrate. The reaction products were analyzed by using either HPLC or the colorimetric method as described in Materials and Methods. Each value represents the average of three independent assays.
ND, not detected under specified experimental conditions.
In the case of HyuA, this enzyme hydrolyzed various hydantoin derivatives, exhibiting the highest levels of activity toward hydroxyphenylhydantoin and phenylhydantoin. The Km and Vmax values were determined to be 32.8 mM and 12.6 μmol min−1 mg of protein−1 for hydroxyphenylhydantoin and 7.8 mM and 3.3 μmol min−1 mg of protein−1 for phenylhydantoin. The HyuA of E. coli also hydrolyzed hydantoin with a Km value of 138 mM and a Vmax value of 0.15 μmol min−1 mg of protein−1, but the catalytic efficiency (Vmax/Km) for hydantoin was decreased by a factor of 420, compared to that toward phenylhydantoin. Other hydantoin derivatives with an aliphatic or no side chain at their 5′ position resulted in a much lower level of activity than those with aromatic side chains at the 5′ position. Chiral TLC and the d-carbamylase-coupled enzyme assay confirmed the d-stereospecific conversion of hydantoin derivatives by HyuA (data not shown). No detectable activity was found for the other cyclic ureides tested in this study. Even under reaction conditions of high-enzyme loading for a prolonged reaction time, HyuA did not show the activity expected for the typical substrates utilized by allantoinase, dihydroorotase, and dihydropyrimidinase. Based on the above observations, HyuA was designated phenylhydantoinase.
DISCUSSION
We report here the functional expression and characterization of two enzymes, allantoinase and phenylhydantoinase in E. coli, belonging to a cyclic amidohydrolase superfamily. Recently, a cyclic amidohydrolase superfamily was identified and found to share quite similar structural and catalytic properties regarding the cyclic amide bond in the pyrimidine and purine rings (16, 21, 30). As a member of this family, dihydroorotase is known to play a pivotal role in the de novo synthesis of pyrimidines, and this enzyme has been well characterized at the biochemical and molecular levels (1, 4, 7, 18, 29, 37, 42, 43, 47). However, relatively little attention, especially at the genetic level, has been devoted to the other enzymes belonging to this family, such as allantoinase, dihydropyrimidinase, and hydantoinase (25, 41). With the complete genome sequences available from a variety of sources, it is very interesting that the putative cyclic amidohydrolases are ubiquitously distributed in eukaryotic and prokaryotic cells. Elucidation of the exact role and investigation of the functional defect in genetically manipulated cells could address some crucial information regarding the nucleotide metabolism in organisms.
The metabolism regarding the use and reductive degradation of allantoin has been reported in amphibians (15), Saccharomyces cerevisiae (5, 9, 10), Pseudomonas aeruginosa (17), and in fish and the livers of invertebrates (27). Although the end products of purine metabolism are different from species to species, a common pathway for the degradation of purines to urate is known to exist in all species. Among the enzymes involved in the purine catabolic pathway, allantoinase is the first enzyme required for the hydrolysis of allantoin to allantoic acid. In P. aeruginosa (17) and S. cerevisiae (9), allantoin was found to be an energy source as well as a source of carbon and nitrogen. Therefore, production of allantoinase was dependent on the nature of the supplemented nitrogen and carbon sources and subject to catabolite repression. This suggests that allantoin is utilized as a sole nitrogen or carbon source in these organisms.
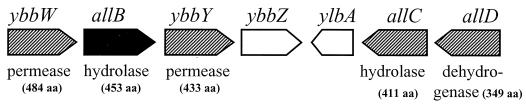
Although the catabolism of purine (allantoin) as a nitrogen source in E. coli was first proposed by Vogels and van der Drift (38), the involved genes and corresponding enzymes have not been characterized in detail. Recently, a gene cluster in E. coli involved in the catabolic degradation of allantoin was recognized, and the gene o453 expressing allantoinase was designated allB (11). However, little information on the gene and its product has been revealed. In this work, we confirmed that the allB gene of E. coli indeed encodes allantoinase and that its gene product has properties quite similar to those of other known allantoinases. These results indicate that allantoin is utilized as a sole nitrogen source when either added exogenously or produced from urate in vivo. In addition, the presence of allantoinase in E. coli strongly suggests that other relevant enzymes exist, including uricase (which catabolizes urate to allantoin) and allantoinase permease (which facilitates the transport of allantoin). These expectations were strongly supported by the presence of a gene cluster found in the flanking region of allB (Fig. 5), as recently identified and proposed by Cusa et al. (11).
FIG. 5.
Genetic organization of the ORFs flanking E. coli allB. Open arrows indicate the proposed direction and extension of the putative ORFs. Labeled numbers indicate the amino acid residues translated and/or deduced from the corresponding nucleotide sequences.
Analyses of the DNA sequence flanking the E. coli allB gene revealed several ORFs with deduced amino acid sequences similar to those of previously characterized enzymes, especially those involved in the catabolic degradation of allantoin such as allantoinase, allantoate amidohydrolase, and ureidoglycolate dehydrogenase. From the above observations, although the biochemical characteristics remain to be elucidated, we are convinced that the flanking region of allB might play a crucial role in the purine metabolic pathway of E. coli. This is supported by our observation that allantoin is utilized as a sole nitrogen source in E. coli. Further genetic and biochemical studies are expected to elucidate its physiological role in vivo.
The HyuA of E. coli, described herein as a novel family of enzymes, shared a great deal of biochemical and structural properties with other enzymes in the superfamily. The most striking resemblance was found in the closely related hydantoinase family of enzymes. The experimental result that the HyuA shows cross-activity to the antibody raised against the d-hydantoinase from B. stearothermophilus SD1 (19) supported the close relationship between the d-hydantoinase and E. coli HyuA. Further evidence for this close relationship is exhibited by the identical preference in stereospecificity for d-enantiomers. From the current literature and experimental results, d-hydantoinase has been suggested to be a microbial counterpart of eukaryotic dihydropyrimidinase (3), a key enzyme in the catabolism of uracil and thymine. This concept was derived from the observation that the dihydropyrimidinase isolated from calf liver catalyzes not only dihydropyrimidines, but also the structurally related hydantoin derivatives (39). Based on this property, microbial d-hydantoinase has gained considerable attention for its ability to synthesize the less commonly occurring d-amino acids (34, 44). E. coli HyuA, however, showed some novel properties in that its activity is absolutely dependent on metal ions and it has no activity on dihydropyrimidines. Current information on E. coli HyuA strongly suggests that the enzyme has a different functional role from the previously known d-hydantoinase, as observed by Runser and Meyer (31). Despite the indisputable observation that it has distinct activity on the aromatic hydantoin derivatives (that is, on an unnatural substrate), the physiological function of the enzyme is still unknown. However, a possible link was provided from our observation that the E. coli HyuA strongly stimulates the concomitant expression of TnaA. Additional physiological functions and biochemical relationships with other cyclic amidohydrolase family enzymes remain to be elucidated.
REFERENCES
- 1.Aleksenko A, Liu W, Gojkovic Z, Nielsen J, Piskur J. Structural and transcriptional analysis of the pyrABCN, pyrD and pyrF genes in Aspergillus nidulans and the evolutionary origin of fungal dihydroorotases. Mol Microbiol. 1999;33:599–611. doi: 10.1046/j.1365-2958.1999.01507.x. [DOI] [PubMed] [Google Scholar]
- 2.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 3.Brooks K P, Jones E A, Kim B D, Sander E G. Bovine liver dihydropyrimidine amidohydrolase. Purification, properties, and characterization as a zinc metalloenzyme. Arch Biochem Biophys. 1983;226:469–483. doi: 10.1016/0003-9861(83)90316-8. [DOI] [PubMed] [Google Scholar]
- 4.Brown D C, Collins K D. Dihydroorotase from Escherichia coli. Substitution of Co(II) for the active site Zn(II) J Biol Chem. 1991;266:1597–1604. [PubMed] [Google Scholar]
- 5.Buckholz R G, Cooper T G. The allantoinase (DAL1) gene of Saccharomyces cerevisiae. Yeast. 1991;7:913–923. doi: 10.1002/yea.320070903. [DOI] [PubMed] [Google Scholar]
- 6.Chien H R, Jih Y L, Yang W Y, Hsu W H. Identification of the open reading frame for the Pseudomonas putidad-hydantoinase gene and expression of the gene in Escherichia coli. Biochim Biophys Acta. 1998;1395:68–77. doi: 10.1016/s0167-4781(97)00097-3. [DOI] [PubMed] [Google Scholar]
- 7.Choi K Y, Zalkin H. Regulation of Escherichia coli pyrC by the purine regulon repressor protein. J Bacteriol. 1990;172:3201–3207. doi: 10.1128/jb.172.6.3201-3207.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chou P Y, Fasman G D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- 9.Cooper T G. Allantoin degradation by Saccharomyces cerevisiae—a model system for gene regulation and metabolic integration. Adv Enzymol Relat Areas Mol Biol. 1984;56:91–139. doi: 10.1002/9780470123027.ch2. [DOI] [PubMed] [Google Scholar]
- 10.Cooper T G, Gorski M, Turoscy V. A cluster of three genes responsible for allantoin degradation in Saccharomyces cerevisiae. Genetics. 1979;92:383–396. doi: 10.1093/genetics/92.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cusa E, Obradors N, Baldoma L, Badia J, Aguilar J. Genetic analysis of a chromosomal region containing genes required for assimilation of allantoin nitrogen and linked glyoxylate metabolism in Escherichia coli. J Bacteriol. 1999;181:7479–7484. doi: 10.1128/jb.181.24.7479-7484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deeley M C, Yanofsky C J. Nucleotide sequence of the structural gene for tryptophanase of Escherichia coli K-12. J Bacteriol. 1981;147:787–796. doi: 10.1128/jb.147.3.787-796.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamajima N, Matsuda K, Sakata S, Tamaki N, Sasaki M, Nonaka M. A novel gene family defined by human dihydropyrimidinase and three related proteins with differential tissue distribution. Gene. 1996;180:157–163. doi: 10.1016/s0378-1119(96)00445-3. [DOI] [PubMed] [Google Scholar]
- 14.Hasinoff B B. Stereoselective hydrolysis of ICRF-187 (dexrazoxane) and ICRF-186 by dihydropyrimidine amidohydrolase. Chirality. 1994;6:213–215. doi: 10.1002/chir.530060309. [DOI] [PubMed] [Google Scholar]
- 15.Hayashi S, Jain S, Chu R, Alvares K, Xu B, Erfurth F, Usuda N, Rao M S, Reddy S K, Noguchi T, Reddy J K, Yeldandi A V. Amphibian allantoinase. Molecular cloning, tissue distribution, and functional expression. J Biol Chem. 1994;269:12269–12276. [PubMed] [Google Scholar]
- 16.Holm L, Sander C. An evolutionary treasure: unification of a broad set of amidohydrolases related to urease. Proteins. 1997;28:72–82. [PubMed] [Google Scholar]
- 17.Janssen D B, Smits R A, van der Drift C. Allantoinase from Pseudomonas aeruginosa. Purification, properties and immunochemical characterization of its in vivo inactivation. Biochim Biophys Acta. 1982;718:212–219. doi: 10.1016/0304-4165(82)90221-5. [DOI] [PubMed] [Google Scholar]
- 18.Jones M E. Pyrimidine nucleotide biosynthesis in animals: genes, enzymes, and regulation of UMP biosynthesis. Annu Rev Biochem. 1980;49:253–279. doi: 10.1146/annurev.bi.49.070180.001345. [DOI] [PubMed] [Google Scholar]
- 19.Kim G J, Park J H, Lee D C, Ro H S, Kim H S. Primary structure, sequence analysis, and expression of the thermostable d-hydantoinase from Bacillus stearothermophilus SD1. Mol Gen Genet. 1997;255:152–156. doi: 10.1007/pl00008610. [DOI] [PubMed] [Google Scholar]
- 20.Kim G J, Kim H S. Optimization of the enzymatic synthesis of d-p-hydroxyphenylglycine from dl-5-substituted hydantoin using d-hydantoinase and N-carbamylase. Enzyme Microb Technol. 1995;17:63–67. [Google Scholar]
- 21.Kim G J, Kim H S. Identification of the structural similarity in the functionally related amidohydrolases acting on the cyclic amide ring. Biochem J. 1998;330:295–302. doi: 10.1042/bj3300295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luksa V, Starkuviene V, Starkuviene B, Dagys R. Purification and characterization of the d-hydantoinase from Bacillus circulans. Appl Biochem Biotechnol. 1997;62:219–231. doi: 10.1007/BF02787998. [DOI] [PubMed] [Google Scholar]
- 23.May O, Habenicht A, Mattes R, Syldatk C, Siemann M. Molecular evolution of hydantoinases. Biol Chem. 1998;379:743–747. [PubMed] [Google Scholar]
- 24.May O, Siemann M, Pietzsch M, Kiess M, Mattes R, Syldatk C. Substrate-dependent enantioselectivity of a novel hydantoinase from Arthrobacter aurescens DSM 3745: purification and characterization as a new member of cyclic amidases. J Biotechnol. 1998;61:1–13. doi: 10.1016/s0168-1656(98)00005-4. [DOI] [PubMed] [Google Scholar]
- 25.Moriwaki Y, Yamamoto T, Higashino K. Enzymes involved in purine metabolism—a review of histochemical localization and functional implications. Histol Histopathol. 1999;14:1321–1340. doi: 10.14670/HH-14.1321. [DOI] [PubMed] [Google Scholar]
- 26.Mukohara Y, Ishikawa T, Watabe K, Nakamura H. A thermostable hydantoinase of Bacillus stearothermophilus NS1122A: cloning, sequencing, and high expression of the enzyme gene, and some properties of the expressed enzyme. Biosci Biotechnol Biochem. 1994;58:1621–1626. doi: 10.1271/bbb.58.1621. [DOI] [PubMed] [Google Scholar]
- 27.Noguchi T, Fujiwara S, Hayashi S. Evolution of allantoinase and allantoicase involved in urate degradation in liver peroxisomes. A rapid purification of amphibian allantoinase and allantoicase complex, its subunit locations of the two enzymes, and its comparison with fish allantoinase and allantoicase. J Biol Chem. 1986;261:4221–4223. [PubMed] [Google Scholar]
- 28.Ogawa J, Soong C L, Honda M, Shimizu S. Imidase, a dihydropyrimidinase-like enzyme involved in the metabolism of cyclic imides. Eur J Biochem. 1997;243:322–327. doi: 10.1111/j.1432-1033.1997.0322a.x. [DOI] [PubMed] [Google Scholar]
- 29.Ogawa J, Shimizu S. Purification and characterization of dihydroorotase from Pseudomonas putida. Arch Microbiol. 1995;164:353–357. doi: 10.1007/BF02529982. [DOI] [PubMed] [Google Scholar]
- 30.Ogawa J, Shimizu S. Diversity and versatility of microbial hydantoin-transforming enzymes. J Mol Catal B (Enzym) 1997;2:163–176. [Google Scholar]
- 31.Runser S M, Meyer P C. Purification and biochemical characterization of the hydantoin hydrolyzing enzyme from Agrobacterium sp. I-671: a hydantoinase with no 5,6-dihydropyrimidine amidohydrolase activity. Eur J Biochem. 1993;213:1315–1324. doi: 10.1111/j.1432-1033.1993.tb17883.x. [DOI] [PubMed] [Google Scholar]
- 32.Sadowsky M J, Tong Z, de Souza M, Wackett L P. AtzC is a new member of the amidohydrolase protein superfamily and is homologous to other atrazine-metabolizing enzymes. J Bacteriol. 1998;180:152–158. doi: 10.1128/jb.180.1.152-158.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soong C L, Ogawa J, Honda M, Shimizu S. Cyclic-imide-hydrolyzing activity of d-hydantoinase from Blastobacter sp. strain A17p-4. Appl Environ Microbiol. 1999;65:1459–1462. doi: 10.1128/aem.65.4.1459-1462.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Syldatk C, Laüfer A, Müller R, Höke H. Production of optically pure d- and l-amino acids by bioconversion of dl-5-monosubstituted hydantoin derivatives. Adv Biochem Eng Biotechnol. 1990;41:29–75. [Google Scholar]
- 35.Syldatk C, May O, Altenbuchner J, Mattes R, Siemann M. Microbial hydantoinases—industrial enzymes from the origin of life? Appl Microbiol Biotechnol. 1999;51:293–309. doi: 10.1007/s002530051395. [DOI] [PubMed] [Google Scholar]
- 36.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van de Casteele, Chen M P, Roovers M, Legrain C, Glansdorff N. Structure and expression of a pyrimidine gene cluster from the extreme thermophile Thermus strain ZO5. J Bacteriol. 1997;179:3470–3481. doi: 10.1128/jb.179.11.3470-3481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vogels G D, van der Drift C. Degradation of purines and pyrimidines by microorganisms. Bacteriol Rev. 1970;40:403–468. doi: 10.1128/br.40.2.403-468.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wallach D P, Grisolia S. The purification and properties of hydropyrimidine hydrase. J Biol Chem. 1957;226:277–288. [PubMed] [Google Scholar]
- 40.Watabe K, Ishikawa T, Mukohara Y, Nakamura H. Cloning and sequencing of the genes involved in the conversion of 5-substituted hydantoins to the corresponding l-amino acids from the native plasmid of Pseudomonas sp. strain NS671. J Bacteriol. 1992;174:962–969. doi: 10.1128/jb.174.3.962-969.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.West T P. Pyrimidine base and ribonucleoside utilization by the Pseudomonas alcaligenes group. Antonie Leeuwenhoek. 1991;59:263–268. doi: 10.1007/BF00583679. [DOI] [PubMed] [Google Scholar]
- 42.Wilson H R, Archer C D, Liu J K, Turnbough C L., Jr Translational control of pyrC expression mediated by nucleotide-sensitive selection of transcriptional start sites in Escherichia coli. J Bacteriol. 1992;174:514–524. doi: 10.1128/jb.174.2.514-524.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson H R, Chan P T, Turnbough C L., Jr Nucleotide sequence and expression of the pyrC gene of Escherichia coli K-12. J Bacteriol. 1987;169:3051–3058. doi: 10.1128/jb.169.7.3051-3058.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamada H, Shimizu S. Microbial and enzymatic processes for the production of biologically and chemically useful compounds. Angew Chem Int Ed Engl. 1988;27:622–642. [Google Scholar]
- 45.Yang Y S, Ramaswamy S, Jakoby W B. Rat liver imidase. J Biol Chem. 1993;268:10870–10875. [PubMed] [Google Scholar]
- 46.Yuan G, Bin J C, McKay D J, Snyder F F. Cloning and characterization of human guanine deaminase. Purification and partial amino acid sequence of the mouse protein. J Biol Chem. 1999;274:8175–8180. doi: 10.1074/jbc.274.12.8175. [DOI] [PubMed] [Google Scholar]
- 47.Zimmermann B H, Kemling N M, Evans D R. Function of conserved histidine residues in mammalian dihydroorotase. Biochemistry. 1995;34:7038–7046. doi: 10.1021/bi00021a015. [DOI] [PubMed] [Google Scholar]