Abstract
Vanilla planifolia, the species cultivated to produce one of the world’s most popular flavors, is highly prone to partial genome endoreplication, which leads to highly unbalanced DNA content in cells. We report here the first molecular evidence of partial endoreplication at the chromosome scale by the assembly and annotation of an accurate haplotype-phased genome of V. planifolia. Cytogenetic data demonstrated that the diploid genome size is 4.09 Gb, with 16 chromosome pairs, although aneuploid cells are frequently observed. Using PacBio HiFi and optical mapping, we assembled and phased a diploid genome of 3.4 Gb with a scaffold N50 of 1.2 Mb and 59 128 predicted protein-coding genes. The atypical k-mer frequencies and the uneven sequencing depth observed agreed with our expectation of unbalanced genome representation. Sixty-seven percent of the genes were scattered over only 30% of the genome, putatively linking gene-rich regions and the endoreplication phenomenon. By contrast, low-coverage regions (non-endoreplicated) were rich in repeated elements but also contained 33% of the annotated genes. Furthermore, this assembly showed distinct haplotype-specific sequencing depth variation patterns, suggesting complex molecular regulation of endoreplication along the chromosomes. This high-quality, anchored assembly represents 83% of the estimated V. planifolia genome. It provides a significant step toward the elucidation of this complex genome. To support post-genomics efforts, we developed the Vanilla Genome Hub, a user-friendly integrated web portal that enables centralized access to high-throughput genomic and other omics data and interoperable use of bioinformatics tools.
Keywords: vanilla, whole-genome sequencing, optical mapping, partial endoreplication, genome hub
The genome of the orchid Vanilla planifolia (4.09 Gb; 16 pairs of chromosomes) is very prone to partial endoreplication (PE), which leads to a very unbalanced DNA content in cells. This study reports the first molecular evidence for PE at the chromosome scale through the assembly and annotation of an accurate haplotype-phased genome of V. planifolia. Distinct haplotype-specific sequencing depth variation patterns suggest complex molecular regulation of endoreplication along chromosomes. To facilitate post-genomics efforts, an integrated public and user-friendly web portal (the Vanilla Genome Hub) has been developed.
Introduction
Endoreplication, characterized by a series of DNA replications in the nucleus without mitotic cell division, is found in a large number of both animal and plant species (Lee et al., 2009). During regular endoreplication, each step of this mechanism leads to a two-fold increase in nuclear DNA content in somatic cells (2C, 4C, 8C, 16C, etc.), where 1C corresponds to the DNA content of the non-replicated holoploid chromosome set. Endoreplication is very common in plants and is related to various biological processes, such as plant development and growth, and occurs in response to biotic and abiotic stresses (Bourdon et al., 2012; Lang and Schnittger, 2020). This phenomenon depends on the type of tissue and its stage of development, suggesting involvement in cell differentiation and maintenance of the final stage of differentiation (Bhosale et al., 2018). The molecular mechanisms involved in regular endoreplication have been particularly well studied in Arabidopsis thaliana over the past few years. A downregulation of mitotic activity caused by mitotic cyclin-dependent kinase (CDK)–cyclin complexes has been shown to be directly involved in the control of endoreplication (Lang and Schnittger, 2020).
In many orchid species, measurements of genomic content by flow cytometry (FCM) have not agreed with the commonly accepted model of complete endoreplication. In this case, nuclear DNA content in endoreplicated cells was present at less than twice the 2C cell content. Because this ratio was constant for a given Vanilla species, whatever the cell ploidy level, it was suggested that the nuclear DNA could be categorized into two parts: the P fraction, subject to endoreplication, and the F fraction, not endoreplicated (Brown et al., 2017). These fractions are constant in all cells undergoing partial endoreplication (PE), which suggests the fine regulation of genome rearrangements. The fact that the gametes are haploid also suggests the presence of molecular mechanisms that enable the isolation of the holoploid genome. This type of endoreplication, which appears to be specific to the Orchidaceae lineage in plants, has been successively termed “progressively PE” (Bory et al., 2008; Trávníček et al., 2015; Hřibová et al., 2016), strict PE (Brown et al., 2017), and more recently, PE (Chumová et al., 2021; Trávníček et al., 2019). To be in line with the latest works and to harmonize the terminology for this phenomenon, the term PE will be used in this work. To date, PE has been observed in all species studied within the genus Vanilla (Bory et al., 2008; Brown et al., 2017; Lepers-Andrzejewski et al., 2011; Trávníček et al., 2015).
Vanilla planifolia G. Jackson is an emblematic orchid cultivated for its fruit (pod) fragrance. Pods contain many aromatic compounds, particularly vanillin in high proportion (Perez-Silva et al., 2006). In this species, diploid nuclei (2C) are found mainly in nodal tissues (with PE up to 32E), whereas the nuclei of mature leaf cells contain a low 2C fraction and show PE up to 64E (Brown et al., 2017). The F fraction was estimated to be 71.6% of the genome, whereas the P fraction (28.4%) could be duplicated up to 64E. In addition, the proportion of the non-endoreplicated (F) genome varies greatly from species to species. It is very high in Vanilla pompona (F = 81%) but rather low in Vanilla mexicana (F = 17%) (Brown et al., 2017). Several studies on orchids have also shown that species prone to PE have a larger genome than those prone to conventional endoreplication (Trávníček et al., 2015, 2019; Chumová et al., 2021). Nevertheless, the molecular mechanisms involved in PE are not yet elucidated.
A chromosome-scaled, phased V. planifolia genome (Daphna cultivar) was recently reported, highlighting haplotype differences and one ancestral whole-genome duplication shared by all sequenced orchids (Hasing et al., 2020). However, the 1.5 Gb size of the assembled genome was far from the V. planifolia genome size, estimated to be about 4 Gb using FCM measurement (Bory et al., 2008; Lepers-Andrzejewski et al., 2011), suggesting that the Daphna genome assembly may be highly incomplete. As mentioned by Hasing et al. (2020), the reason for the genome size discrepancy between FCM and assembly results remains to be elucidated. With about 65% of the V. planifolia genome missing in the Daphna assembly, we hypothesize that the missing part of the genome corresponds mainly to the F (non-endoreplicated) fraction (71.6%) (Brown et al., 2017), whose lower representation results in a lower sequencing depth.
Here, we address this issue by developing an approach that combines FCM, cytogenetics, and whole-genome sequencing using the most recently developed technologies (Supplemental Figure 1) with a tissue that is enriched in the 2C fraction (nodes), resulting in a reduced P/F ratio and therefore a greater proportion of the F fraction. We demonstrate that the genome size discrepancy was due to the occurrence of PE, for which further knowledge at the chromosome scale was gained from this study. We present the most complete version to date of a high-quality, chromosome-level phased genome of V. planifolia using a traditional vanilla cultivar from the Indian Ocean region (CR0040). Our results are shared through a web portal that facilitates data access, use, and analyses by a wide community.
Results
Genome size, ploidy level, and chromosome content
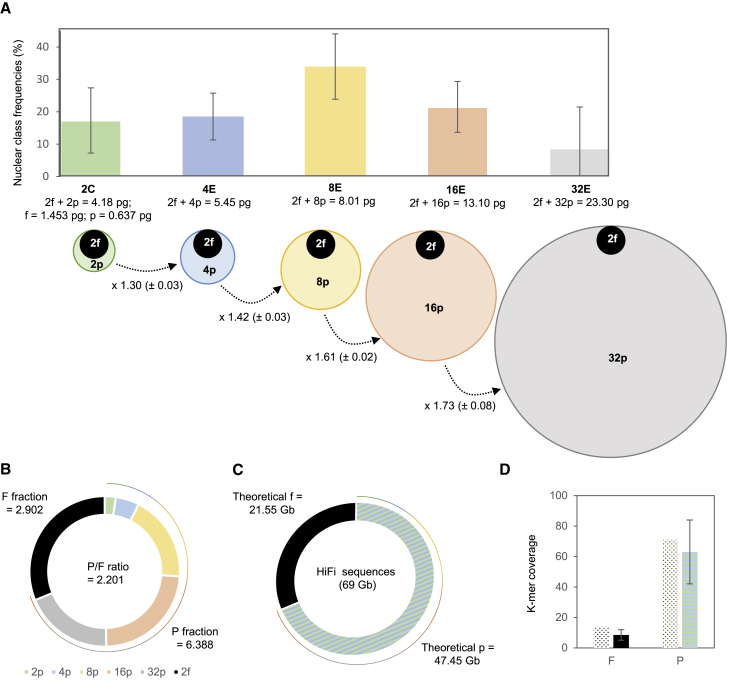
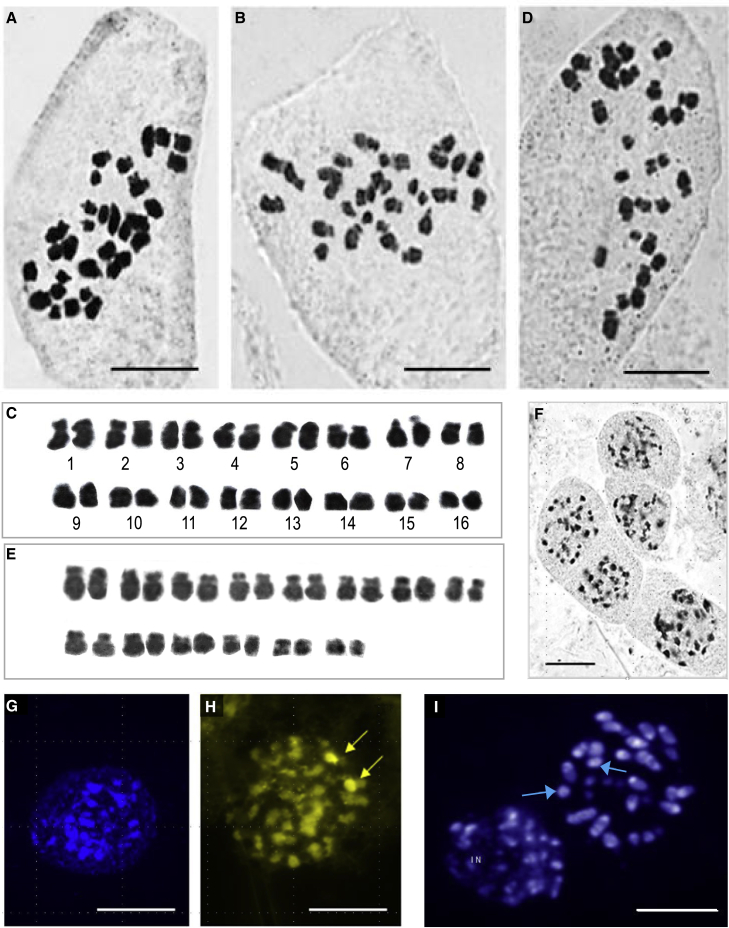
The 2C genome size of V. planifolia CR0040, a traditional vanilla cultivar from La Reunion island (Supplemental Note 1), was estimated in nodal tissues to be 4.18 ± 0.08 pg by FCM (Supplemental Note 1), corresponding to 4.09 Gb (Doležel et al., 2003). To estimate PE levels, the fluorescence ratio of DNA content between consecutive peaks of endoreplication levels was estimated (Supplemental Note 1; Supplemental Figure 2; Supplemental Table 1). Results showed no significant differences (calculated t-values of 1.116, 1.900, 0.935, and 0.365 compared with Student table t-value [α = 0.05] of 2.131) between the PE pattern of CR0040 and those of other V. planifolia cultivars, such as CR1110 (2C = 4.16 ± 0.04 pg), studied by Brown et al. (2017). The replicated fraction P was also calculated (P = 30.5% ± 3.2%). The equivalent amount 2p was then P × 2C = 1.275 pg, which meant that the absolute quantity p was 0.637 pg, and the absolute quantity f of fixed amount was 1.453 pg (Figure 1A and 1B; Supplemental Note 1). The karyotype of V. planifolia obtained by cytogenetics approaches (Supplemental Note 1) appeared to be of bimodal type, composed of 16 both large and small chromosome pairs (Figures 2A–2C), although aneuploid cells were frequently observed, such as those with only 28 chromosomes (Figure 2D and 2E). V. planifolia chromosomes possess important portions of telomeric and pericentromeric heterochromatin, which made the determination of their morphology difficult. In the interphase nuclei, this heterochromatin was present in the form of numerous chromocenters that were clearly visible after staining with both orcein (Figure 2F) and DAPI (Figure 2G). This type of heterochromatin is unspecific, whereas heterochromatin linked to rRNA genes is rich in G-C bases. Only one locus (two spots) of rDNA (18S-5.8S-26S) was present in the genome of V. planifolia (Figure 2H, arrows), evidenced after chromomycin (CMA3) staining. After Hoechst 33258 staining, our results also revealed that AT-rich DNA regions were more common than GC-rich regions in the V. planifolia chromosomes and that some chromosomes were entirely or almost entirely heterochromatinized (Figure 2I).
Figure 1.
Endoreplicated and non-endoreplicated fractions in the CR0040 Vanilla planifolia genome.
(A) The histogram represents the distribution of nuclei in V. planifolia nodal tissues according to the partial endoreplication state of cells, from 2C (green) to 4E (blue), 8E (yellow), 16E (orange), and 32E (gray). The disks below represent the endoreplicated (colored) and non-endoreplicated (black) DNA content for each class of nuclei, proportionally to their mass (pg). The lowercase f and p denote the respective DNA quantities of the F fraction (fixed proportion of the haploid genome that cannot endoreplicate) and the P fraction (part that participates in endoreplication). The mean and the standard deviation (SD) of the interpeak ratio have been indicated below the dotted arrows.
(B) F and P fractions and P/F ratio values obtained by flow cytometry and detailed for the P fraction for each nuclear class (2C, green; 4E, blue; 8E, yellow; 16E, orange; and 32E, gray).
(C) Theoretical F and P fractions expected from HiFi sequencing and from flow-cytometry data.
(D) Theoretical (dotted) and experimental k-mer coverages for F (black) and P (hatched) fractions.
Figure 2.
Cytogenetic analysis of Vanilla planifolia CR0040.
(A–D) Orcein staining: (A and B) mitotic metaphases with 2n = 32 chromosomes; (C) karyotype corresponding to (B); (D) hypoaneuploid mitotic metaphase with 2n = 28 chromosomes; (E) karyotype corresponding to (D); (F) interphase nuclei showing heterochromatic chromocenters; (G) DAPI-stained interphase nucleus showing unspecific heterochromatin; (H) chromomycin fluorochrome staining with two CMA+ regions (arrows) corresponding to rDNA sites; (I) Hoechst-stained AT-rich DNA in metaphase and interphase nucleus (IN), with two fully heterochromatinized chromosomes (arrows). Scale bars represent 10 μm.
Whole-genome assembly and k-mer analysis
CR0040 genome sequencing produced 69 Gb of Pacific Biosciences (PacBio) HiFi long reads, 147 Gb of Oxford Nanopore Technology (ONT) long reads, and 200 Gb of Illumina 10X Genomics short reads (Supplemental Note 1; Supplemental Table 2). These DNA sequencing (DNA-seq) reads were assembled using different bioinformatics pipelines. The best result was obtained using only high-quality HiFi long reads (Supplemental Note 2; Supplemental Tables 3 and 4). Contigs from the HiFi read assembly were scaffolded with optical maps to obtain a final phased assembly of 3.4 Gb (1.5 Gb for haplotype A and 1.9 Gb for haplotype B), representing around 83% of the expected genome size. One third of the assembly could be anchored onto 14 chromosomes using published Daphna chromosomes as references (Hasing et al., 2020). Unfortunately, no data could help to organize the remaining contigs into the two missing chromosomes. Therefore, the remaining two-thirds correspond to unanchored additional sequences that were compiled into two unknown random pseudomolecules, A0 and B0. The final assembly comprised 24 534 contigs with a contig N50 length of 924 kb. The lengths of the 14 chromosomes ranged from 73.5 Mb (Chr01) to 20 Mb (Chr14). Main genome assembly statistics are synthesized in Table 1.
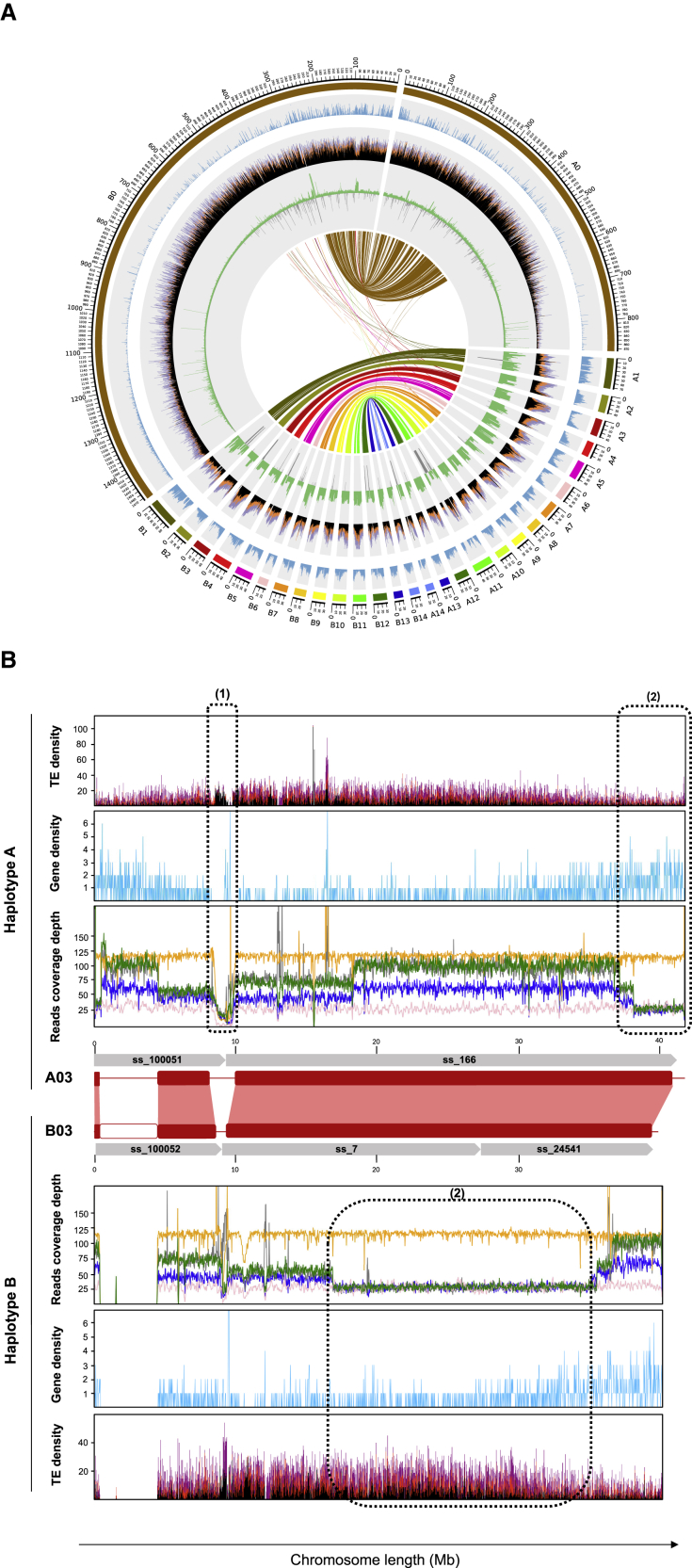
Figure 4.
Overview of the assembled vanilla genome.
(A) Circos plot of the genomic content along V. planifolia haplotypes A and B and the relationship between them. All tracks are divided into 500 kb genomic windows. From the outside to the inside of the circular representation, ideograms of 28 chromosomes and two random mosaic chromosomes that contain the unanchored scaffolds are shown. Gene density (blue) and interspersed repeat RepeatMasker hit density (black: retroelements; orange: long terminal repeat/Copia; purple: long terminal repeat/Gypsy) are shown. Sequencing depth was obtained by mapping CR0040 PacBio HiFi reads on the assembly (green) and N density (gray). Syntenic blocks across haplotypes are connected by lines in the innermost part of the figure.
(B) Sequencing depth along the CR0040 A03 and B03 chromosomes (red rectangles) obtained by mapping Daphna Illumina (yellow) and ONT (pink) reads and CR0040 PacBio HiFi (blue), Nanopore (green), and Illumina (gray) reads onto the CR0040 assembly. Synteny between homologous chromosomes is represented by red boxes. Gaps (N stretches) that explain sudden drops in sequencing depth are shown with white blocks. (1) Low level of sequencing depth for all data is shown. (2) Inverted level of sequencing depth for CR0040 between haplotypes A and B and constant level of sequencing depth for both Daphna haplotypes are shown. Gene and retrotransposon distributions along the chromosomes are represented by a blue line chart and a stacked histogram (Copia: red; Gypsy: purple; other retrotransposons: black), respectively.
Table 1.
HiFi assembly and annotation statistics of the diploid CR0040 genome
| Total assembly size (Gb) | 3.4 |
| Total contig number | 24 534 |
| Contig N50 length (Mb) | 0.924 |
| Maximum contig length (Mb) | 31 |
| GC content (%) | 31.6 |
| Number of protein-coding genes | 59 128 |
| Benchmarking Universal Single-Copy Orthologs completeness (%) | 93.2 |
| Total of interspersed repeats (%) | 47.0 |
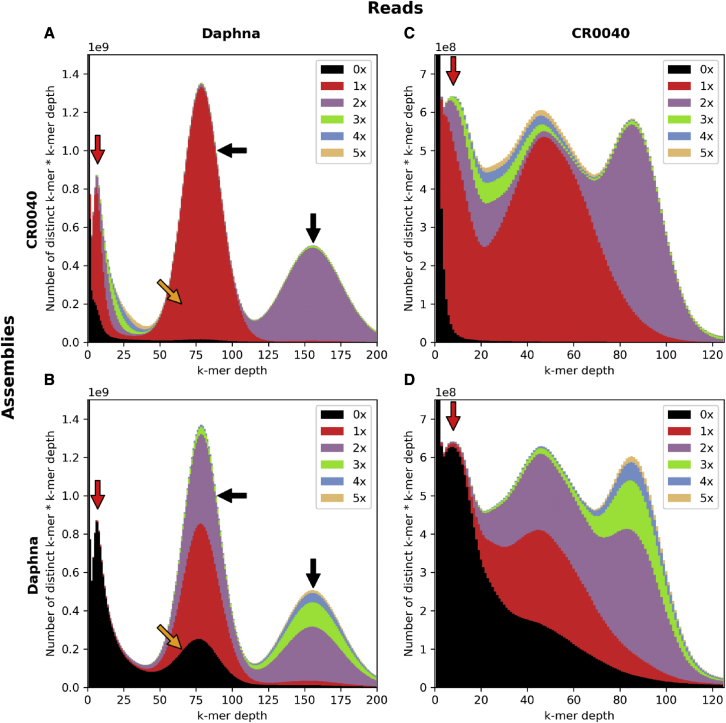
In order to understand how PE affects the assembly, a k-mer analysis was produced. The results should reflect the sequencing coverage of the different genome fractions present in our raw data and assembly. In brief, the reads were split into overlapping k-mers (47-mers in our case). K-mers were then sorted and occurrences counted. These counts were then used to produce a histogram. A spectra-cn plot was used to compare the k-mers found in the reads versus the k-mers found in the assembly (Supplemental Figure 3). The x axis gives the number of times a given k-mer was found in all the reads, reflecting the coverage of the k-mer. The y axis gives a value representing the number of k-mers that were found a specified number of times (x axis value). Interestingly, two k-mer distributions were centered at 42× and 84×, representing a classical diploid distribution with heterozygous and homozygous k-mer content. We assumed that these peaks represented the k-mers of the endoreplicated fraction with a high sequencing depth due to higher representation. Remarkably, the graph also showed an additional k-mer distribution centered around 10× (Supplemental Figure 3, red arrow). This distribution could easily be mistaken for an erroneous k-mer distribution, but we assumed that it represented non-endoreplicated k-mers of the V. planifolia genome, with low-sequencing depth due to lower representation.
To validate the assembly and compare it with the already published reference, we produced four k-mer spectra-cn plots showing k-mer distributions of Daphna Illumina reads and CR0040 HiFi reads colored both with the Daphna and CR0040 assemblies (Figure 3). A spectra-cn plot enables comparison of the k-mers found in the reads versus the k-mers found in the assembly. The k-mer histogram from the reads is colored based on the number of times each k-mer is found in the assembly. For a heterozygous diploid assembly, we expected to find two distributions: on the left, the heterozygous distribution, which should be colored in red because each k-mer is only found once in the assembly, and on the right, the homozygous distribution, which is purple because the corresponding k-mers are found twice in the assembly. The black area at the far left of the diagram corresponds to k-mers that include sequencing errors; these are found a limited number of times in the reads and never in the assembly. Daphna Illumina sequencing, being deeper, resulted in better separation between the homozygous (80× sequencing depth) and heterozygous (160× depth) k-mer fractions in the spectra-cn graph compared with CR0040 (Figure 3A and 3B against 3C and 3D). The same pattern occurred for CR0040 HiFi data around 45× and 90× (Figure 3C). The differences between Figure 3A and 3C come from the sequencing depth and the type of tissue used: mature leaves with a higher proportion of the P fraction for Daphna and nodal tissues with a lower P/F ratio for CR0040. The k-mer distribution of the non-endoreplicated fraction (low coverage) was not found in the Daphna assembly (black area left of Figure 3B and 3D) but is mostly present in the CR0040 assembly. Regarding the completeness of the Daphna reference assembly, the spectra-cn plots (Figure 3B and 3D) showed that part of the heterozygous fraction was missing (orange arrows), and some k-mers were in overrepresented copies (>2×) in both heterozygous and homozygous fractions (Figure 3B, black arrows). The spectra-cn diagram also showed heterozygous content present two or more times instead of once in this assembly (Figure 3, black arrows), which could indicate spurious duplications. As a whole, our CR0040 genome assembly is close in size to the FCM estimate and has the expected k-mer diploid profile, with a well-represented non-endoreplicated fraction (Figure 1C and 1D).
Figure 3.
Assembly k-mer content comparison between CR0040 PacBio HiFi long reads and Daphna Illumina short reads using spectra-cn graph.
(A–D) The x axis represents k-mer multiplicity (counts), and the y axis indicates the number of distinct k-mers multiplied by their counts. Because of different sequencing depths between read sets, the y axis upper values are 109 for (A) and (B) and 108 for (C) and (D). The area colors indicate the number of k-mer copies found in the assembly (black: 0× or missing k-mers, red: 1×, purple: 2×, green: 3×, blue: 4×, and orange: 5×). Four spectra-cn plots are presented: (A) Daphna reads versus CR0040 assembly, (B) Daphna reads versus Daphna assembly, (C) CR0040 reads versus CR0040 assembly, and (D) CR0040 reads versus Daphna assembly. The red arrows point toward a low-coverage k-mer distribution not expected in a diploid genome assembly spectra-cn graph. The black arrows point toward the heterozygous (on the left) and homozygous (on the right) k-mer distributions expected in a diploid genome assembly. The orange arrows point toward missing k-mers in the heterozygous k-mer distribution. The lower the black distribution at this location, the fewer k-mers are missing in the assembly.
Gene and transposable element annotation
The assembled genome supplemented with transcriptomic data from nine distinct tissues made it possible to identify 59 128 protein-coding genes (26 392 for haplotype A and 32 736 for haplotype B), 90.31% of which could be associated with a function (Supplemental Note 3; Supplemental Tables 5–10). Sixty-seven percent of the predicted genes were anchored onto the 14 chromosome pairs and the remaining 33% onto the two random mosaic chromosomes that were constructed from the unanchored scaffolds and contigs (Figure 4A, blue distributions). We estimated the annotation completeness at 93.2% with the Benchmarking Universal Single-Copy Orthologs approach using the Viridiplantae database. In total, 72% of the assembly consisted of repeats, including single-sequence repeats (15.4%), and 9.7% of other low-complexity regions (Supplemental Note 3; Supplemental Table 11). A high content of retrotransposons was found (41.5%), whereas the content of DNA transposons was low (1.4%). The long terminal repeat retrotransposon content was richer in Gypsy (9.7%; Figure 4A, purple distributions) than in Copia (6.1%; Figure 4A, orange distribution), although a number of annotated retrotransposons (12.5%) were not more precisely classified. The two random mosaic chromosomes were enriched in repeats and showed low gene density and low sequencing depth (Figure 4A, green distributions, and Supplemental Table 12). Indeed, compared with the 14 chromosome sequences, the unanchored regions showed higher proportions of long interspersed nuclear element sequences (8% and 14.05%), and this was true for both haplotypes. By contrast, DNA transposons (3.14% and 0.93%), short interspersed nuclear elements (0.12% and 0.05%), and long terminal repeats (21.67% and 15.57%) represented a larger part of the 14 chromosome sequences than of the unanchored regions. The biggest difference in unanchored regions was observed for unclassified retrotransposons, which represented 16.76% of the unanchored sequences versus 3.28% of the 14 chromosome sequences. Main genome annotation statistics are synthesized in Table 1.
V. planifolia pangenomics and whole-genome duplication
The comparison of the four mosaic haplotypes from the two V. planifolia cultivars, CR0040 and Daphna (Supplemental Tables 12 and 13), showed that the 14 pseudomolecules of CR0040 were shorter and contained fewer genes than those of Daphna and that a large number of regions in the CR0040 pseudomolecules (haplotype A or B) were not located in the Daphna pseudomolecules (Supplemental Figure 4). Pan-genomic analysis of the orthogroups from proteomes derived from the 14 chromosomes only (Supplemental Figure 5; Supplemental Table 14; Supplemental Note 4) indicated that the core genome was composed of 14 210 families and 77 692 genes (35 972 CR0040 and 41 720 Daphna). The dispensable genome of CR0040 contains 1266 families and 3613 genes specific to CR0040. The dispensable genome of Daphna contains 3997 Daphna-specific families and 13 645 genes. Finally, we looked at the expansion or reduction of gene families in relation to six proteomes (CR0040, Daphna, Phalaenopsis equestris, Phalaenopsis aphrodite, A. thaliana, and Oryza sativa; Supplemental Figure 6). From an orchid perspective, the expansion number for the orchid node is rather low (+36), whereas the Daphna-specific number is rather high (A +1841 and B +1943) compared with CR0040 (A +418 and B +826).
To identify whole-genome duplications (WGDs), pairwise genome synteny analyses between CR0040, Daphna, and P. aphrodite and within themselves were carried out (Supplemental Figures 7 and 8; Supplemental Notes 4 and 6). The CR0040 haplotype A dot plot validated at least one pan-orchid WGD (α°, the origin of the paleo-allotetraploid) previously found by Hasing et al. (2020). An additional dot plot diagonal and dS peak suggested a second WGD, possibly the tau of Monocots (τm).
Figure 6.
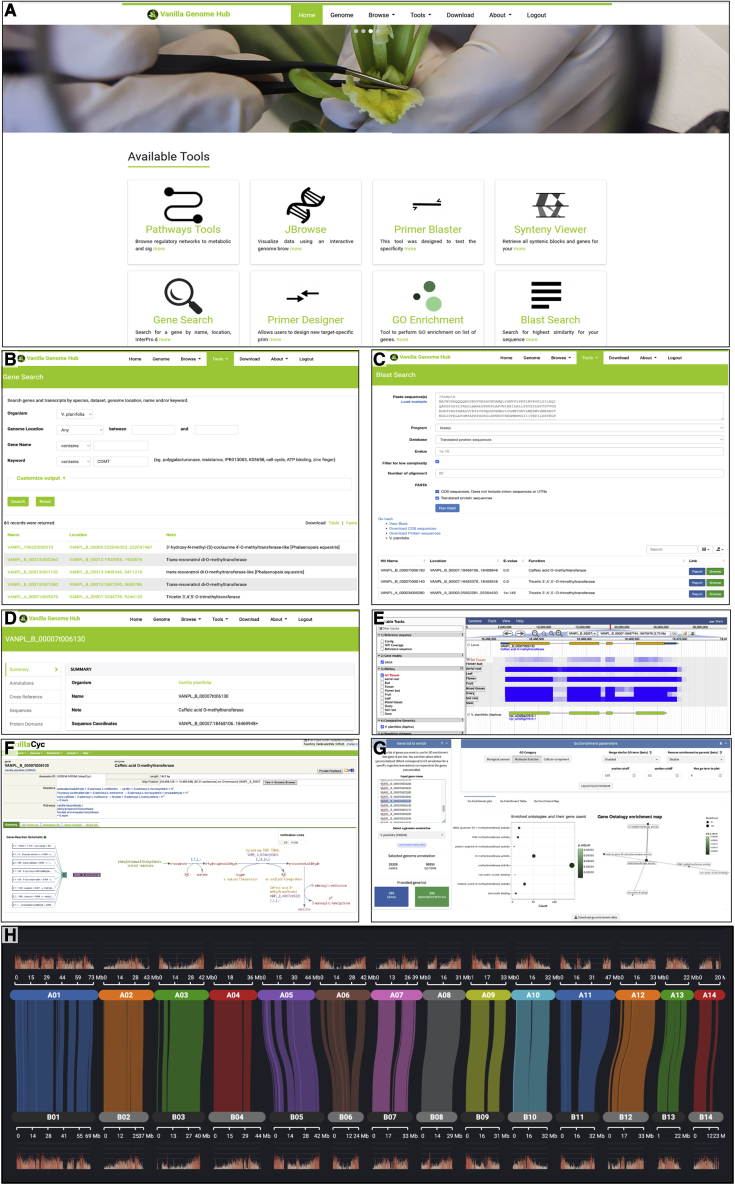
Overview (screen shots) of some interoperable vanilla genome analysis tools integrated into the Vanilla Genome Hub.
(A) Main menu.
(B) Gene search (Tripal MegaSearch).
(C) Sequence homology search (Blast).
(D) Gene report (Tripal).
(E) Genome Browser (JBrowse).
(F) Metabolic pathway visualization (Pathway Tools).
(G) Gene Ontology enrichment (DIANE).
(H) Comparison of genomic sequences (SynVisio).
Detection of non-endoreplicated regions
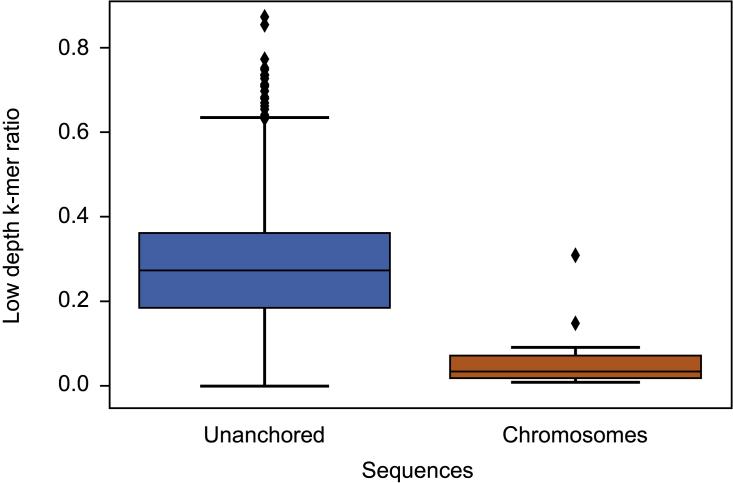
PE induces highly unbalanced DNA representation with a P/F DNA ratio ranging from 3 to 10, depending on the tissue. This was reflected in our assembly by highly variable sequencing depth (Figure 4A, green lane). The two random mosaic chromosomes that showed a low sequencing depth at most loci may therefore contain a large part of the non-endoreplicated F fraction of the genome. It is likely that a large number of unanchored sequences originate from the two fully heterochromatinized chromosomes observed in the interphase nuclei (Figure 2I), possibly chromosome pairs 15 or 16. The remaining unanchored sequences should correspond to missing fractions in the anchored chromosomes. Interestingly, the sequencing reads that mapped to the CR0040 and Daphna assemblies also showed intra-chromosomal sequencing depth variations (Figure 4B). These patterns were consistent, regardless of the technology used. To observe this phenomenon globally on all chromosomes and genomes with all technologies (HiFi, ONT, and Illumina), sequencing depth analysis tools were used and manual validation performed (Supplemental Note 5; Supplemental Tables 15 and 16). Two patterns of sequencing depth variation were identified along all chromosomes. The first one (indicated with a dotted box labeled “1” in Figure 4B and Supplemental Figure 9) corresponded to a sharp decrease in sequencing depth for both cultivars, with all sequencing technologies, which dropped down from 45×–120× to less than 20×. Surprisingly, this pattern occurred independently on the two haplotypes. A total of 37 very low-coverage regions with this pattern (from 0.4 to 6 Mb in length) were identified along the chromosomes (24 in haplotype A and 13 in haplotype B) for a cumulative size of 60.1 Mb. In a large portion of these regions, we found low gene density and high repeat density. This pattern could correspond to non-endoreplicated regions present in both the Daphna and CR0040 genomes. The fact that these patterns are systematically located at junctions between super-scaffolds is consistent with the decrease in sequencing depth caused by non-endoreplication, which impaired the assembly of the endoreplicated regions located on either side. The second pattern (indicated with a dotted box labeled “2” in Figure 4B and Supplemental Figure 9) corresponded to 36 regions (from 1.2 Mb to 20 Mb in length; cumulative size of 207.2 Mb) with segmental sequencing depth variation present in CR0040 (with HiFi, ONT, and Illumina) but not in Daphna (with ONT and Illumina). Furthermore, these variations were syntenic along the two haplotypes, but the direction of variation was inverted between the two phases. Their respective levels of sequencing depth differ by a factor of about three in CR0040. The cause of these apparently coordinated sequencing depth inversions between CR0040 haplotypes remains unclear. After analyzing the locations of these k-mers in the CR0040 assembly, it appeared that these low depth k-mers (between 5× and 15×) were mostly present in the unanchored part of the genome compared to the chromosome sequences (Figure 5), with median ratio values equal to 0.27 and 0.036, respectively, showing a significant difference (Wilcoxon-Mann-Whitney test; p = 4e−13). However, chromosomes 7A and 6B were outliers, showing also a high proportion of low-depth k-mers. In addition, the distribution of these k-mers along the genome was globally consistent with the areas identified, except for some discrepancies (Supplemental Figure 10). Chromosomes 6B and 7A showed strong signals in terms of low-depth k-mer proportions, as already pointed out in Figure 5. Indeed, on chromosome 6B, k-mers of this type were positioned on nearly all the assembled sequence, whereas they were localized on approximately half of the assembled chromosome 7A sequence.
Figure 5.
Ratio of k-mers within unanchored and anchored CR0040 genome.
This boxplot shows the ratio of k-mers with a depth less than 15 in our HiFi reads within unanchored sequences (blue) and within chromosomes (orange).
Orthologs of cell cycle regulator genes involved in A. thaliana endoreplication
A search for orthologs of the CDK and cyclin (Cyc) families of A. thaliana, involved in the regular endoreplication mechanism, showed that representatives of these two families were indeed found in the proteomes of CR0040 and P. aphrodite (Supplemental Table 17). However, the number of genes encoding CDKs and Cycs found via the orthogroups approach was lower for these two species. For example, the gene encoding Cyc-D3-1 in A. thaliana (At4g34160) was part of a species-specific orthogroup that contained some other D-type Cyc genes. Regarding genes encoding regulatory proteins of the CDK–Cyc complexes, all of them had orthologs in both orchids. However, it appeared that these multigenic families were slightly under-represented in the CR0040 gene annotation compared with those of A. thaliana and P. aphrodite. Finally, an imbalance between the A and B haplotypes was observed for Fizzy-related proteins and CDK inhibitor (Krp) orthologs.
Vanilla Genome Hub
The Vanilla Genome Hub (VGH) (https://vanilla-genome-hub.cirad.fr) has been developed to support post-genomics efforts. It centralizes vanilla genomic information with a set of user-friendly interconnected modules and interfaces for the analysis and visualization of genomic data. From the main menu of the VGH (Supplemental Note 6; Figure 6A), the search for genes of interest to biologists is simplified using the interoperable system by the identification of paralogous genes using keywords and sequence homology (Figure 6B and 6C) and the production of an information report with gene name, gene localization, and polypeptide function (Figure 6D). The genome browser was built to offer tracks of supplemental information, such as GC content, gene structure, gene expression, DNA-seq depth, and repeat composition to support the identification of new genes of interest (Figure 6E and Supplemental Figure 11). A metabolic pathway reconstruction and visualization tool enables the identification of annotated genes involved in pathways (Figure 6F). A Gene Ontology enrichment tool enables testing and visualization of enrichment according to a Gene Ontology category of a gene group (Figure 6G). Finally, comparative analysis at the genome scale is supported by an interactive multiscale synteny visualization (Figure 6H).
Discussion
Flow cytometry and cytogenetic data validate genome size and chromosome content
Genome size, ploidy level, and chromosome content of the V. planifolia CR0040 cultivar were validated by FCM and cytogenetic analyses. The estimated size of 4.09 Gb indicated a ploidy level similar to those of other traditional diploid V. planifolia cultivars (Bory et al., 2008; Lepers-Andrzejewski et al., 2011). Estimation of endoreplication levels confirmed PE, as previously described in V. planifolia (Brown et al., 2017). This species was shown to exhibit diploidized meiotic chromosome pairing with 16 bivalents (Bory, 2007). This result demonstrates the complete diploidization of this supposed segmental paleo-allotetraploid (Ravindran, 1979; Nair and Ravindran, 1994). The same meiotic observation was also performed for Vanilla × tahitensis by Lepers-Andrzejewski et al. (2011). Aneuploid chromosome numbers were frequently observed in mitotic metaphases of V. planifolia (Nair and Ravindran, 1994; Bory et al., 2008), possibly owing to the observed mitotic associations that could lead to unequal anaphase separation. This may lead to errors in the evaluation of basic chromosome number, as was the case in a recent paper in which the authors considered that the basic number was x = 14 (Hasing et al., 2020). The phenomenon of aneuploidy apparently occurs only in somatic cells, whereas meiosis appears to be regular, with a stable number of chromosomes (Bory, 2007). Although the CR0040 assembly is more complete than that of Daphna, only 14 pseudomolecules were obtained because CR0040 scaffolds were anchored on the 14 Daphna pseudomolecules. Chromosomes 15 and 16 are probably non-endoreplicated and present in the unanchored part (CR0040_A0 and CR0040_B0).
PE hinders whole-genome assembly
Given the CR0040 diploid genome size estimate of 4.09 Gb by FCM, our genome assembly represented around 83% of the expected genome size and was twice the size of the Daphna genome published previously (Hasing et al., 2020). This difference could be explained by the fact that the CR0040 genome was assembled from HiFi reads that enabled the assembly of repetitive regions in different contigs despite their low sequencing depth. We were thus able to assemble a greater number of repeated sequences that may correspond to a large fraction of the non-endoreplicated genome and were missing from the Daphna genome assembly. The biological reality of this hypothesis is reinforced by the consistency of the k-mer depth profiles and sequencing depth patterns that resulted from the mapping of reads from different sequencing technologies (HiFi, 10X, and ONT) tested in this study for CR0040 (Supplemental Figure 9). A k-mer spectra-cn diagram is an efficient tool for visually comparing the k-mer compositions of reads and assemblies. Such diagrams are used to validate diploid or haploid assembly quality (Yen et al., 2020). The k-mer spectra-cn diagram clearly shows a general diploid pattern, with a heterozygous distribution containing only k-mers present once in the assembly and a homozygous distribution containing, as expected, only k-mers present twice in the assembly. Unexpectedly for a diploid genome, this figure includes a third distribution that is located in the low-coverage area of the diagram. The color pattern shows clearly that these k-mers present in low frequencies (5–15 times) are also present in our assembly. These k-mers represent the non-repeated fraction of low-coverage sections of the assembly, which are mainly located in the unanchored sequences but are also present in low-coverage sections of other chromosomes. Even if the unanchored sequences are mainly built of repeats, they also harbor genes and other non-repeated blocks, and these portions are large enough in terms of k-mers to generate this unexpected k-mer distribution in the spectra-cn plot. These k-mers are not present in the public V. planifolia Daphna assembly, and therefore, the corresponding distribution is black in Figure 3B.
Molecular signatures of partial endoreplication
The abundance of interspersed repeats detected in CR0040 was consistent with already mentioned data in other orchids, such as P. equestris (Cai et al., 2015) and P. aphrodite (Chao et al., 2018), and in other lineages, like the Oryza genus (Stein et al., 2018). The high content of retrotransposons and low content of DNA transposons were in the range of what has been found for different orchids (Cai et al., 2015; Chao et al., 2018). High repeat content was found in candidate non-endoreplicated regions, which is in agreement with previous descriptions in other orchids (Chumová et al., 2021). Furthermore, some types of repeats may be preferentially found in non-endoreplicated regions, as shown by differences in repeat proportions, particularly retrotransposon proportions, between assembled chromosomes and unanchored sequences. Thus, long interspersed nuclear elements, for example, occupy a larger portion of the unanchored regions than of the 14 chromosomes, even though these regions are overrepresented in the present assembly. However, the lack of a more detailed annotation of the retrotransposon class hampers the search for a potential preferential distribution of repeat families between endoreplicated and non-endoreplicated regions. It is therefore crucial to better annotate these repeats in order to determine exactly which kinds are found preferentially in the two fractions of the genome. On the other hand, the distribution of genes in the genome shows the opposite trend, with approximately two-thirds of the protein-coding sequences localized in the anchored region.
The distinct sequencing depth profiles observed between CR0040 and Daphna probably reflected a tissue-specific endoreplication pattern. Indeed, the nodes used to sequence the CR0040 genome are growing and differentiating tissues, whereas the leaves used to sequence the Daphna genome are composed of fully differentiated cells. The irregular haplotype-specific endoreplication pattern (segmental or not) observed in CR0040 could thus result from a peculiar physiological activity. Whatever the reason, this intriguing pattern suggests a complex and fine regulation of PE at the chromosome level, which deserves further study. Although no previous study has demonstrated the mechanisms underlying PE in orchids, many works have focused on the regulation of regular endoreplication found in a large number of plant species and well analyzed in tomato and Arabidopsis (Lang and Schnittger, 2020). The common mechanism that triggers endoreplication is a downregulation of mitotic CDK activity to suppress mitosis and a fine regulation of this activity throughout the induced endocycle, with an alternation between high and low activity levels at specific checkpoints in order to maintain the replication process (De Veylder et al., 2011; Shimotohno et al., 2021). CDK controls cell-cycle progression and mitosis entry via its phosphorylation activity, which is activated by association with CYC proteins. Recently, Inada et al. (2021) demonstrated the involvement of actin and actin-binding protein in the regulation of A. thaliana endoreplication. The present whole-genome analysis made it possible to identify orthologous CDK, Cycs, CDK-activators and repressors, and actin depolymerizing factors in V. planifolia CR0040. A first step in understanding orchid PE would therefore be to further analyze these molecular regulators. Indeed, the recognition of orthologs and paralogs in large gene families, such as the CDK–Cyc complex, is challenging and requires deeper investigation by high-quality manual annotation of the genes of interest (Vaattovaara et al., 2019).
Although PE seems specific to orchids in plants (Trávníček et al., 2015), this phenomenon of under-represented genomic regions is well known in metazoans. Ciliates, such as Paramecium tetraurelia and Tetrahymena thermophile, show programmed DNA elimination following endoreplication in their macronucleus (MAC), involving chromosome fragmentation and elimination of specific sequences called internal eliminated sequences (IESs) (Bracht et al., 2013; Sellis et al., 2021). However, FCM approaches in Ludisia discolor, an orchid subject to PE, have ruled out the possibility of such DNA elimination and favor the hypothesis of under-replication (Hřibová et al., 2016). Under-replication has also been studied in several organisms, such as Drosophila, in which it has been proposed that a reduction in the expression of genes involved in DNA replication may lead to a slower mitosis S phase and an incomplete replication of genomic regions during late S phase (Lilly and Spradling, 1996). Molecular mechanisms described in Drosophila highlighted an inhibition of replication fork progression involving Rif1 protein, which interacts with the SUUR protein (Munden et al., 2018; Armstrong et al., 2019).
Finally, cytogenetic studies using in situ hybridization techniques (fluorescence in situ hybridization [FISH] and genomic in situ hybridization) could also be used to increase our knowledge of the molecular signatures of PE (Younis et al., 2015). A recent advance in FISH is the development of probes based on synthetic oligonucleotides specific to repetitive sequences or to particular chromosome regions (Jiang, 2019). This new generation of FISH probes in plants has been applied to species with sequenced genomes, such as Zea and Cucumis species (Han et al., 2015; Martins et al., 2019; Braz et al., 2020; Zhang et al., 2021a). Endoreplicated versus non-endoreplicated genomic regions could be used to synthesize oligo-based FISH probes specific to each fraction in order to precisely locate these PE signatures on chromosomes. The genomic in situ hybridization technique uses the total genomic DNA of a species, in contrast to FISH. We hypothesize that hybridizing the total DNA of highly endoreplicated nuclei (16E and 32E) to CR0040 chromosomes would induce a more intense hybridization signal in endoreplicated regions, thus enabling us to identify non-endoreplicated areas that showed little hybridization.
Impact of technologies on whole-genome evolution analysis
The strategy of combining optical mapping with HiFi long-read sequencing for the CR0040 genome assembly resulted in a haplotype A with 14 pseudomolecules of better quality and with fewer scaffolding errors than the Daphna haplotype A, which was built with Hi-C and ONT technologies (Hasing et al., 2020). Indeed, comparisons between the Daphna and CR0040 A haplotypes revealed a dual-haplotype conservation problem in the Daphna phased assembly, which is reflected in the Daphna Hi-C scaffolding. The use of HiFi long reads and optical maps enabled more accurate haplotype separation, as shown in previous works (Matthews et al., 2018; Du et al., 2020). In the case of CR0040, not only did HiFi enable better assembly of non-endoreplicated regions but also Hifiasm allowed better separation of haplotypes. These improvements were therefore necessary to better solve the sequencing of the complex vanilla genome, with a high rate of heterozygosity (Ho V. planifolia cultivars = 0.362; Favre et al., 2022) and subjected to PE (Brown et al., 2017). However, this dual-haplotype conservation problem observed in Daphna, and not in CR0040, impacted comparative pan-genomics analyses and distorted the results obtained. Thus, the differences observed between the two V. planifolia genomes (number of paralogs, numbers of gene families with expansions and contractions, and complete and duplicated Benchmarking Universal Single-Copy Orthologs scores) could be explained by these mosaic assembly problems and therefore by an incorrect separation between haplotypes A and B.
Monocot genome evolution analyses were carried out using the high-quality haplotype A sequence of CR0040. Dot plot results were in agreement with the fact that V. planifolia is a diploidized paleo-polyploid species with a primary basic chromosome number x = 8 and a secondary basic number x = 16, as described for the whole Vanilla genus (Felix and Guerra, 2005). Moreover, only one locus (two spots) of rDNA (18S-5.8S-26S) was identified in the genome of V. planifolia by cytogenetic approaches, which provides additional evidence of an ancient diploidization of this supposed segmental paleo-allotetraploid. Finally, two WGDs, possibly corresponding to α° and τm, were highlighted, as also described for the Dendrobium chrysotoxum chromosome-scale genome assembly (Zhang et al., 2021b).
Efficiency of an integrative approach combining cytogenetics with high-quality whole-genome sequencing
In this study, we confirmed the size and structure of the V. planifolia genome using both cytogenetics and nuclear DNA-seq methods. The particular phenomenon of PE at play in many orchids has been explored at the chromosome level for the first time in plants, to our knowledge. Our data showed that the non-endoreplicated sequences are very predominantly made up of repeated sequences. This confirmed, at the genomic level, previous findings in orchids by Chumová et al. (2021) based on a phylogenetics generalized least squares model and by Brown et al. (2017), who used nuclei imaging to demonstrate that in Vanilla, on the other hand, the endoreplicated part was transcribed. We nevertheless revealed that 33% of the 59 128 annotated protein-coding genes were present in the two random mosaic chromosomes, corresponding mainly to the non-endoreplicated part, as shown by low sequencing depth. In addition, a thorough examination of sequencing depths along anchored chromosomes with three different technologies revealed 73 regions whose different endoreplication levels vary with haploid phase, half of which may be linked to tissue type (leaves versus nodes). This last conclusion remains to be confirmed with DNA-seq from different tissues of the same cultivar. This work constitutes considerable progress in our understanding of V. planifolia genomics and sheds light on the most relevant methodologies for further deciphering this complex genome and the PE phenomenon. The VGH was built to help the community to address major unresolved questions about vanilla, such as PE, biosynthesis of aromatic compounds, and resistance to pathogens. We are working on a new version of the vanilla nuclear genome sequence that will be improved in terms of haplotype separation, chromosome reconstruction, and gene and repeat element annotation in order to further investigate the molecular mechanisms of PE with appropriate plant material, bio-technologies, and bioinformatics tools.
Methods
Cytometry, cytogenetics, and DNA sequencing
A traditional vanilla cultivar (CR0040) from Reunion Island was used in this study (Supplemental Note 1). FCM and cytogenetics studies were performed using protocols described in Supplemental Note 1. High-molecular-weight DNA and ultra-high-molecular-weight DNA were extracted from node tissues and sequenced using PacBio HiFi, ONT, and Illumina technologies (Supplemental Note 1). Optical genome maps were produced using the Bionano Genomics protocol and the Saphyr G1 System (Supplemental Note 1).
Genome assembly and analysis
HiFi reads were assembled into contigs using Hifiasm 0.13 with default parameters (Cheng et al., 2021). The hybrid scaffolding between DNA contigs and optical genome maps was performed using the hybrid Scaffold pipeline of Bionano Genomics with default parameters. These scaffolds were phased into two haplotypes using in-house scripts, and the unscaffolded contigs were phased using purge dups (https://github.com/dfguan/purge_dups). Then, pseudomolecules were reconstructed using alignments of the phased assembly on Daphna chromosomes (Hasing et al., 2020; Supplemental Note 2). The assembly quality was estimated with QUAST 5.1.0 (Gurevich et al., 2013) and using the approach of Benchmarking Universal Single-Copy Orthologs (version 5.0.0) (Simao et al., 2015; Supplemental Note 2). The k-mer analysis was performed with kat 2.4.2 using the comp tool (Mapleson et al., 2017). The plot script was slightly modified to project on the y axis the number of distinct k-mers multiplied by the k-mer multiplicity instead of just the number of distinct k-mers. In parallel, k-mers of size 47 with a depth between 5 and 15 were extracted within PacBio sequences using Jellyfish 2.3.0 (Marcais and Kingsford, 2011). These k-mers were repositioned on our reference using the tool “query_per_sequence” (https://github.com/gmarcais/Jellyfish/tree/master/examples/query_per_sequence), and the ratio of these k-mers was computed among each sequence of our genome. These sequences were split between chromosomes and unanchored sequences, and the repartition of the k-mer ratio was drawn using the python seaborn library (https://seaborn.pydata.org/).
Structural and functional genome annotation
Automatic gene prediction was performed on CR0040 contigs with the EuGene Eukaryotic Pipeline (EGNEP version 1.5) (Sallet et al., 2019; Supplemental Note 3). Transcriptomic data from CR0040 were produced using RNA sequencing of nine organs with Illumina technology (Supplemental Note 3). In addition, gene expression profiles and putative novel isoforms were identified with StringTie v.2.0.3 (Kim et al., 2019; Supplemental Note 3). Transcriptomic data from V. planifolia cultivars (CR0040, Daphna [NCBI BioProjects: PRJNA668740 and PRJNA633886], and an unspecified cultivar [NCBI GEO: GSE134155]); proteomic data from V. planifolia Daphna (Hasing et al., 2020), P. equestris (NCBI BioProject: PRJNA382149), and the Liliopsida class (Swissprot: 2020_06); and a custom orchid-specific statistical model for splice-site detection were used for this analysis (Supplemental Note 3). Functions were assigned through InterProScan domain searches as well as similarity searches against the UniProt/Swissprot and UniProt/TrEMBL databases (BlastP). Gene Ontology terms were assigned through InterProScan (Jones et al., 2014) results, and enzyme classification numbers were predicted by combining the tools PRIAM (Claudel-Renard et al., 2003) and BlastKOALA (Kanehisa et al., 2016).
Repeats were first identified using RepeatModeler v.2.0.1 (Flynn et al., 2020), RepeatScout v.1.0.5, and transposable element genes predicted from EGNEP annotation and then classified with REPET v.3.0 (Flutre et al., 2011) and PASTEC v.2.0 (Hoede et al., 2014) according to Wicker’s transposable element classification (Wicker et al., 2007). After cleaning steps (see details in Supplemental Note 3), repeats were clustered using CD-HIT v.4.8.1 (Fu et al., 2012) to produce two banks of repeats. The CR0040 genome was then annotated for repeats using previous banks, RepeatMasker v.4.1.1 (Tarailo-Graovac and Chen, 2009), and bedtools intersect v.2.29.2 (Quinlan and Hall, 2010).
Genomic comparisons and reconstruction of gene families
To compare the 14 haplotype A chromosomes of both vanilla cultivars, check the completeness of the Vanilla genome, and study the pan-orchid α° WGD, a series of analyses were performed with the CoGe SynMap pipeline as described in Supplemental Note 4. Gene family reconstruction was performed using OrthoFinder2 (v.2.4.0) (Emms and Kelly, 2019). Genes known to be involved in cell cycle control in A. thaliana, such as Cycs, CDKs, and known regulators of these genes, were searched in the CR0040 and P. aphrodite proteomes with a combination of BlastP searches and orthogroups. This analysis was applied to CDK-A and B types as well as Cyc-A, B, and D types. Regulators of these genes included CDK inhibitor (KRP), transcriptional repressor ILP1, WEE1, actin depolymerizing factor, and Fizzy-related proteins.
Detection of non-endoreplicated genomic regions
Reads from each sequencing technology used in this study (HiFi, ONT, and Illumina reads from CR0040), as well as ONT and Illumina reads from Daphna, were mapped onto the CR0040 assembly. Illumina short reads and long reads (HiFi and ONT) were mapped onto the CR0040 assembly using BWA-MEM2 (Vasimuddin et al., 2019) and Minimap2 (Li, 2018), respectively. Sequencing depths were averaged for genomic windows of 20 kb. To detect sequencing depth bias and limit the risk of detecting false positives, the mean sequencing depth for every 20 successive 20-kb windows was computed using Illumina reads for Daphna and using long reads (HiFi and ONT) for CR0040. Identified regions were manually validated and refined by visualization of sequencing depth drops for each CR0040 chromosome and for all available sequencing datasets (see details in Supplemental Note 5).
VGH
The VGH was constructed using the Tripal system, a specific toolkit for the construction of online community genomic databases, by integrating the GMOD Chado database schema and the Drupal open-source platform (https://www.drupal.org/). The VGH implements a set of interconnected modules and user-friendly interfaces (details in Supplemental Note 6).
Data availability
The chromosome assembly and accompanying data received the following identifiers in NCBI: BioProject (with SRA database) ID: PRJNA753216 (haplotype A) and PRJNA754028 (haplotype B) BioSample (node) SAMN20691751.
RNA sequencing data are readily accessible on the NCBI portal: BioSamples SAMN20691786 (fruit), SAMN20691787 (leaf), SAMN20691788 (flower), SAMN20691789 (stem), SAMN20691790 (soil root), SAMN20691791 (aerial root), SAMN20691792 (bud), SAMN20691793 (flower bud), SAMN20691794 (ovary), SAMN20691795 (mixed tissues) and SRA: SRR15411867 (mixed tissues), SRR15411868 (ovary), SRR15411869 (flower bud), SRR15411870 (bud), SRR15411871 (aerial root), SRR15411872 (soil root), SRR15411873 (stem), SRR15411874 (flower), SRR15411875 (leaf), and SRR15411876 (fruit)
In addition, these data and various exploration tools are accessible at VGH (https://vanilla-genome-hub.cirad.fr/).
Funding
This work was supported by grants from Eurovanille and V. Mane Fils companies. The research was co-funded by the Centre de Coopération Internationale en Recherche Agronomique pour le Développement (CIRAD), the Université de La Réunion (UR), the Institut National de Recherche pour l’Agriculture l’Alimentation et l’Environnement (INRAE), the Centre National de la Recherche Scientifique (CNRS), and the Etablissement Vanille de Tahiti (EVT). This work was also supported by grants from the European Regional Development Fund (ERDF), the Conseil Régional de la Réunion, and the Conseil Départemental de la Réunion. This work was supported by France Génomique National infrastructure, funded as part of “Investissement d’avenir” program managed by Agence Nationale pour la Recherche (contrat ANR-10-INBS-09) and has also benefited from Imagerie-Gif core facility supported by l’Agence Nationale de la Recherche (ANR-11-EQPX-0029/Morphoscope, ANR-10-INBS-04/FranceBioImaging; ANR-11-IDEX-0003-02/Saclay Plant Sciences).
Author contributions
C.J., M.D., M.G., and P.B. contributed to conceptualization of the study. C.C., C.J., G.S., M.B., M.D., O.B., S.B., and W.M. designed the experiments. M.B. and S.S.-Y. performed flow cytometry and cytogenetic experiments and analyses. L.B. and J.Z. contributed to the funding of the research, monitored the progress of the work, and supported the researchers throughout the project. C.C., C.L.-R., O.B., and W.M. performed nucleic acid preparation and sequencing. C.C., C.J., C.K., G.D., G.S., Q.P., S.B., and W.M. performed sequence analyses and assemblies. C.C., G.D., Q.P., S.B., and S.L.-A. performed genome annotation and built the genome hub. C.C., C.J., C.K., M.G., Q.P., and W.M. outlined the manuscript and wrote first drafts. C.C., C.J., C.K., C.L.-R., G.D., G.S., M.B., M.D., M.G., P.B., Q.P., S.B., S.S.-Y., and W.M. provided input and revisions to the manuscript.
Acknowledgments
We are grateful to Jean Bernard Dijoux and Katia Jade for preparing the plant material and to the Plant Protection Platform (3P, IBISA) for lab facilities and access to plant resources (BRC Vatel). We acknowledge the SouthGreen Bioinformatics Platform (http://www.southgreen.fr/) for access to computational resources and the GeT-PlaGe platform (INRAE, Toulouse, France) for the use of sequencing facilities. Finally, the authors would like to thank the reviewers for their suggestions, which helped to improve the manuscript. No conflict of interest is declared.
Published: May 5, 2022
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information can be found online at Plant Communications Online.
Contributor Information
Gaetan Droc, Email: gaetan.droc@cirad.fr.
Michel Grisoni, Email: michel.grisoni@cirad.fr.
Cyril Jourda, Email: cyril.jourda@cirad.fr.
Supplemental information
References
- Armstrong R.L., Penke T., Chao S.K., Gentile G.M., Strahl B.D., Matera A.G., McKay D.J., Duronio R.J. H3K9 promotes under-replication of pericentromeric heterochromatin in Drosophila salivary gland polytene chromosomes. Genes. 2019;10:93. doi: 10.3390/genes10020093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhosale R., Boudolf V., Cuevas F., Lu R., Eekhout T., Hu Z.B., Van Isterdael G., Lambert G.M., Xu F., Nowack M.K., et al. A spatiotemporal DNA endoploidy map of the Arabidopsis root reveals roles for the endocycle in root development and stress adaptation. Plant Cell. 2018;30:2330–2351. doi: 10.1105/tpc.17.00983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bory S. Université de La Réunion; France: 2007. Diversity of Vanilla planifolia in the Indian Ocean and its Related Species : Genetics, Cytogenetics and Epigenetics Aspect. [Google Scholar]
- Bory S., Catrice O., Brown S., Leitch I.J., Gigant R., Chiroleu F., Grisoni M., Duval M.F., Besse P. Natural polyploidy in Vanilla planifolia (Orchidaceae) Genome. 2008;51:816–826. doi: 10.1139/G08-068. [DOI] [PubMed] [Google Scholar]
- Bourdon M., Pirrello J., Cheniclet C., Coriton O., Bourge M., Brown S., Moise A., Peypelut M., Rouyere V., Renaudin J.P., et al. Evidence for karyoplasmic homeostasis during endoreduplication and a ploidy-dependent increase in gene transcription during tomato fruit growth. Development. 2012;139:3817–3826. doi: 10.1242/dev.084053. [DOI] [PubMed] [Google Scholar]
- Bracht J.R., Fang W., Goldman A.D., Dolzhenko E., Stein E.M., Landweber L.F. Genomes on the edge: programmed genome instability in ciliates. Cell. 2013;152:406–416. doi: 10.1016/j.cell.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braz G.T., do Vale Martins L., Zhang T., Albert P.S., Birchler J.A., Jiang J.M. A universal chromosome identification system for maize and wild Zea species. Chromosome Res. 2020;28:183–194. doi: 10.1007/s10577-020-09630-5. [DOI] [PubMed] [Google Scholar]
- Brown S.C., Bourge M., Maunoury N., Wong M., Wolfe Bianchi M., Lepers-Andrzejewski S., Besse P., Siljak-Yakovlev S., Dron M., Satiat-Jeunemaître B. DNA remodeling by strict partial endoreplication in orchids, an original process in the plant kingdom. Genome Biol. Evol. 2017;9:1051–1071. doi: 10.1093/gbe/evx063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J., Liu X., Vanneste K., Proost S., Tsai W.C., Liu K.W., Chen L.J., He Y., Xu Q., Bian C., et al. The genome sequence of the orchid Phalaenopsis equestris. Nat. Genet. 2015;47:65–72. doi: 10.1038/ng.3149. [DOI] [PubMed] [Google Scholar]
- Chao Y.T., Chen W.C., Chen C.Y., Ho H.Y., Yeh C.H., Kuo Y.T., Su C.L., Yen S.H., Hsueh H.Y., Yeh J.H., et al. Chromosome-level assembly, genetic and physical mapping of Phalaenopsis aphrodite genome provides new insights into species adaptation and resources for orchid breeding. Plant Biotechnol. J. 2018;16:2027–2041. doi: 10.1111/pbi.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Concepcion G.T., Feng X., Zhang H., Li H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods. 2021;18:170–175. doi: 10.1038/s41592-020-01056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumová Z., Záveská E., Hloušková P., Ponert J., Schmidt P.A., Čertner M., Mandáková T., Trávníček P. Repeat proliferation and partial endoreplication jointly shape the patterns of genome size evolution in orchids. Plant J. Cel. Mol. Biol. 2021;107:511–524. doi: 10.1111/tpj.15306. [DOI] [PubMed] [Google Scholar]
- Claudel-Renard C., Chevalet C., Faraut T., Kahn D. Enzyme-specific profiles for genome annotation: PRIAM. Nucleic Acids Res. 2003;31:6633–6639. doi: 10.1093/nar/gkg847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L., Larkin J.C., Schnittger A. Molecular control and function of endoreplication in development and physiology. Trends Plant Sci. 2011;16:624–634. doi: 10.1016/j.tplants.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Doležel J., Bartoš J., Voglmayr H., Greilhuber J. Letter to the editor. Cytom. Part A. 2003;51A:127–128. doi: 10.1002/cyto.a.10013. [DOI] [PubMed] [Google Scholar]
- Du K., Stock M., Kneitz S., Klopp C., Woltering J.M., Adolfi M.C., Feron R., Prokopov D., Makunin A., Kichigin I., et al. The sterlet sturgeon genome sequence and the mechanisms of segmental rediploidization. Nat. Ecol. Evol. 2020;4:841–852. doi: 10.1038/s41559-020-1166-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms D.M., Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20 doi: 10.1186/S13059-019-1832-Y. 20Artn. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre F., Jourda C., Grisoni M., Piet Q., Rivallan R., Dijoux J.B., Hascoat J., Lepers-Andrzejewski S., Besse P., Charron C. A genome-wide assessment of the genetic diversity, evolution and relationships with allied species of the clonally propagated crop Vanilla planifolia Jacks. Ex Andrews. Genet. Resour. Crop Ev. 2022 doi: 10.1007/s10722-022-01362-1. [DOI] [Google Scholar]
- Felix L.P., Guerra M. Basic chromosome numbers of terrestrial orchids. Plant Syst. Evol. 2005;254:131–148. doi: 10.1007/s00606-004-0200-9. [DOI] [Google Scholar]
- Flutre T., Duprat E., Feuillet C., Quesneville H. Considering transposable element diversification in de novo annotation approaches. Plos One. 2011;6:e16526. doi: 10.1371/journal.pone.0016526. 6ARTN. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn J.M., Hubley R., Goubert C., Rosen J., Clark A.G., Feschotte C., Smit A.F. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci. U S A. 2020;117:9451–9457. doi: 10.1073/pnas.1921046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L.M., Niu B.F., Zhu Z.W., Wu S.T., Li W.Z. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y.H., Zhang T., Thammapichai P., Weng Y.Q., Jiang J.M. Chromosome-specific painting in Cucumis species using bulked oligonucleotides. Genetics. 2015;200:771–779. doi: 10.1534/genetics.115.177642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasing T., Tang H.B., Brym M., Khazi F., Huang T.F., Chambers A.H. A phased Vanilla planifolia genome enables genetic improvement of flavour and production. Nat. Food. 2020;1:811–819. doi: 10.1038/s43016-020-00197-2. [DOI] [PubMed] [Google Scholar]
- Hoede C., Arnoux S., Moisset M., Chaumier T., Inizan O., Jamilloux V., Quesneville H. PASTEC: an automatic transposable element classification tool. PLoS One. 2014;9:e91929. doi: 10.1371/journal.pone.0091929. 9ARTN e91929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hřibová E., Holušová K., Trávníček P., Petrovská B., Ponert J., Šimková H., Kubátová B., Jersáková J., Čurn V., Suda J., et al. The enigma of progressively partial endoreplication: new insights provided by flow cytometry and next-generation sequencing. Genome Biol. Evol. 2016;8:1996–2005. doi: 10.1093/gbe/evw141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada N., Takahashi N., Umeda M. Arabidopsis thaliana subclass I ACTIN DEPOLYMERIZING FACTORs and vegetative ACTIN2/8 are novel regulators of endoreplication. J. Plant Res. 2021;134:1291–1300. doi: 10.1007/s10265-021-01333-0. [DOI] [PubMed] [Google Scholar]
- Jiang J.M. Fluorescence in situ hybridization in plants: recent developments and future applications. Chromosome Res. 2019;27:153–165. doi: 10.1007/s10577-019-09607-z. [DOI] [PubMed] [Google Scholar]
- Jones P., Binns D., Chang H.Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G., et al. InterProScan 5: genome-scale protein function classification. Bioinformatics (Oxford, England) 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Sato Y., Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016;428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang L., Schnittger A. Endoreplication - a means to an end in cell growth and stress response. Curr. Opin. Plant Biol. 2020;54:85–92. doi: 10.1016/j.pbi.2020.02.006. [DOI] [PubMed] [Google Scholar]
- Lee H.O., Davidson J.M., Duronio R.J. Endoreplication: polyploidy with purpose. Genes Dev. 2009;23:2461–2477. doi: 10.1101/gad.1829209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepers-Andrzejewski S., Siljak-Yakovlev S., Brown S.C., Wong M., Dron M. Diversity and dynamics of plant genome size: an example of polysomaty from a cytogenetic study of Tahitian vanilla (Vanilla ×tahitensis, Orchidaceae) Am. J. Bot. 2011;98:986–997. doi: 10.3732/ajb.1000415. [DOI] [PubMed] [Google Scholar]
- Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly M.A., Spradling A.C. The Drosophila endocycle is controlled by Cyclin E and lacks a checkpoint ensuring S-phase completion. Genes Dev. 1996;10:2514–2526. doi: 10.1101/gad.10.19.2514. [DOI] [PubMed] [Google Scholar]
- Mapleson D., Garcia Accinelli G., Kettleborough G., Wright J., Clavijo B.J. KAT: a K-mer analysis toolkit to quality control NGS datasets and genome assemblies. Bioinformatics. 2017;33 doi: 10.1093/bioinformatics/btw663. btw663–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcais G., Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- do Vale Martins L., Yu F., Zhao H.N., Dennison T., Lauter N., Wang H.Y., Deng Z.H., Thompson A., Semrau K., Rouillard J.M., et al. Meiotic crossovers characterized by haplotype-specific chromosome painting in maize. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-12646-z. 10Artn. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews B.J., Dudchenko O., Kingan S.B., Koren S., Antoshechkin I., Crawford J.E., Glassford W.J., Herre M., Redmond S.N., Rose N.H., et al. Improved reference genome of Aedes aegypti informs arbovirus vector control. Nature. 2018;563:501–507. doi: 10.1038/s41586-018-0692-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munden A., Rong Z., Sun A., Gangula R., Mallal S., Nordman J.T. Rif1 inhibits replication fork progression and controls DNA copy number in Drosophila. eLife. 2018;7:e39140. doi: 10.7554/eLife.39140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair R.R., Ravindran P.N. Somatic association of chromosomes and other mitotic abnormalities in Vanilla planifolia (andrews) Caryologia. 1994;47:65–73. doi: 10.1080/00087114.1994.10797284. [DOI] [Google Scholar]
- Perez-Silva A., Odoux E., Brat P., Ribeyre F., Rodriguez-Jimenes G., Robles-Olvera V., Garcia-Alvarado M.A., Gunata Z. GC-MS and GC-olfactometry analysis of aroma compounds in a representative organic aroma extract from cured vanilla (Vanilla planifolia G. Jackson) beans. Food Chem. 2006;99:728–735. doi: 10.1016/j.foodchem.2005.08.050. [DOI] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindran P.N. Nuclear behavior in the sterile pollen of Vanilla planifolia (andrews) Cytologia. 1979;44:391–396. doi: 10.1508/cytologia.44.391. [DOI] [Google Scholar]
- Sallet E., Gouzy J., Schiex T. EuGene: an automated integrative gene finder for eukaryotes and prokaryotes. Gene Prediction: Methods Protoc. 2019;1962:97–120. doi: 10.1007/978-1-4939-9173-0_6. [DOI] [PubMed] [Google Scholar]
- Sellis D., Guérin F., Arnaiz O., Pett W., Lerat E., Boggetto N., Krenek S., Berendonk T., Couloux A., Aury J.M., et al. Massive colonization of protein-coding exons by selfish genetic elements in Paramecium germline genomes. PLoS Biol. 2021;19:e3001309. doi: 10.1371/journal.pbio.3001309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotohno A., Aki S.S., Takahashi N., Umeda M. Regulation of the plant cell cycle in response to hormones and the environment. Annu. Rev. Plant Biol. 2021;72:273–296. doi: 10.1146/annurev-arplant-080720-103739. [DOI] [PubMed] [Google Scholar]
- Simao F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Stein J.C., Yu Y., Copetti D., Zwickl D.J., Zhang L., Zhang C., Chougule K., Gao D., Iwata A., Goicoechea J.L., et al. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat. Genet. 2018;50:285–296. doi: 10.1038/s41588-018-0040-0. [DOI] [PubMed] [Google Scholar]
- Tarailo-Graovac M., Chen N. In: Current Protocols in Bioinformatics. Baxevanis A.D., editor. 2009. Using RepeatMasker to identify repetitive elements in genomic sequences. [DOI] [PubMed] [Google Scholar]
- Trávníček P., Čertner M., Ponert J., Chumová Z., Jersáková J., Suda J. Diversity in genome size and GC content shows adaptive potential in orchids and is closely linked to partial endoreplication, plant life-history traits and climatic conditions. New Phytol. 2019;224:1642–1656. doi: 10.1111/nph.15996. [DOI] [PubMed] [Google Scholar]
- Trávníček P., Ponert J., Urfus T., Jersáková J., Vrána J., Hřibová E., Doležel J., Suda J. Challenges of flow-cytometric estimation of nuclear genome size in orchids, a plant group with both whole-genome and progressively partial endoreplication. Cytometry Part A. Journal Int. Soc. Anal. Cytol. 2015;87:958–966. doi: 10.1002/cyto.a.22681. [DOI] [PubMed] [Google Scholar]
- Vaattovaara A., Leppälä J., Salojärvi J., Wrzaczek M. High-throughput sequencing data and the impact of plant gene annotation quality. J. Exp. Bot. 2019;70:1069–1076. doi: 10.1093/jxb/ery434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasimuddin M., Misra S., Li H., Aluru S. Efficient architecture-aware acceleration of BWA-MEM for multicore systems. Int. Parall Distrib P. 2019:314–324. doi: 10.1109/Ipdps.2019.00041. [DOI] [Google Scholar]
- Wicker T., Sabot F., Hua-Van A., Bennetzen J.L., Capy P., Chalhoub B., Flavell A., Leroy P., Morgante M., Panaud O., et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007;8:973–982. doi: 10.1038/nrg2165. nrg2165 [pii] [DOI] [PubMed] [Google Scholar]
- Yen E.C., McCarthy S.A., Galarza J.A., Generalovic T.N., Pelan S., Nguyen P., Meier J.I., Warren I.A., Mappes J., Durbin R., et al. A haplotype-resolved, de novo genome assembly for the wood tiger moth (Arctia plantaginis) through trio binning. GigaScience. 2020;9:giaa088. doi: 10.1093/gigascience/giaa088. ARTN. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younis A., Ramzan F., Hwang Y.J., Lim K.B. FISH and GISH: molecular cytogenetic tools and their applications in ornamental plants. Plant Cell Rep. 2015;34:1477–1488. doi: 10.1007/s00299-015-1828-3. [DOI] [PubMed] [Google Scholar]
- Zhang T., Liu G.Q., Zhao H.N., Braz G.T., Jiang J.M. Chorus2: design of genome-scale oligonucleotide-based probes for fluorescence in situ hybridization. Plant Biotechnol. J. 2021;19:1967–1978. doi: 10.1111/pbi.13610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.X., Zhang G.Q., Zhang D.Y., Liu X.D., Xu X.Y., Sun W.H., Yu X., Zhu X.E., Wang Z.W., Zhao X., et al. Chromosome-scale assembly of the Dendrobium chrysotoxum genome enhances the understanding of orchid evolution. Hortic. Res-england. 2021;8 doi: 10.1038/s41438-021-00621-z. 8 artn. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The chromosome assembly and accompanying data received the following identifiers in NCBI: BioProject (with SRA database) ID: PRJNA753216 (haplotype A) and PRJNA754028 (haplotype B) BioSample (node) SAMN20691751.
RNA sequencing data are readily accessible on the NCBI portal: BioSamples SAMN20691786 (fruit), SAMN20691787 (leaf), SAMN20691788 (flower), SAMN20691789 (stem), SAMN20691790 (soil root), SAMN20691791 (aerial root), SAMN20691792 (bud), SAMN20691793 (flower bud), SAMN20691794 (ovary), SAMN20691795 (mixed tissues) and SRA: SRR15411867 (mixed tissues), SRR15411868 (ovary), SRR15411869 (flower bud), SRR15411870 (bud), SRR15411871 (aerial root), SRR15411872 (soil root), SRR15411873 (stem), SRR15411874 (flower), SRR15411875 (leaf), and SRR15411876 (fruit)
In addition, these data and various exploration tools are accessible at VGH (https://vanilla-genome-hub.cirad.fr/).