Abstract
When attacked by pathogens, plants need to reallocate energy from growth to defense to fend off the invaders, frequently incurring growth penalties. This phenomenon is known as the growth–defense tradeoff and is orchestrated by a hardwired transcriptional network. Altering key factors involved in this network has the potential to increase disease resistance without growth or yield loss, but the mechanisms underlying such changes require further investigation. By conducting a genome-wide association study (GWAS) of leaves infected by the hemi-biotrophic bacterial pathogen Pseudomonas syringae pv. tomato (Pst) DC3000, we discovered that the Arabidopsis transcription factor REPLUMLESS (RPL) is necessary for bacterial resistance. More importantly, RPL functions in promoting both disease resistance and growth. Transcriptome analysis revealed a cluster of genes in the GRETCHEN HAGEN 3 (GH3) family that were significantly upregulated in rpl mutants, leading to the accumulation of indole-3-acetic acid-aspartic acid (IAA-Asp). Consistent with this observation, transcripts of virulence effector genes were activated by IAA-Asp accumulated in the rpl mutants. We found that RPL protein could directly bind to GH3 promoters and repress their expression. RPL also repressed flavonol synthesis by directly repressing CHI expression and thus activated the auxin transport pathway, which promotes plant growth. Therefore, RPL plays an important role in plant immunity and functions in the auxin pathway to optimize Arabidopsis growth and defense.
Key words: natural variation, GWAS, REPLUMLESS, bacterial resistance, auxin
This study reports that the Arabidopsis transcription factor RPL identified by GWAS is involved in both plant growth and defense. RPL promotes plant growth by repressing expression of CHI and promotes plant defense by repressing expression of the GH3 gene family. A missense SNP that causes a K233T substitution in RPL may underlie bacterial resistance variation among ecotypes.
Introduction
In nature, a wide array of organisms, including pathogens, insects, and animals, can invade or destroy plants. Therefore, the ability to activate defense responses quickly and properly is crucial for plant survival. However, plant defense responses vary among species (Todesco et al., 2010). One explanation for this phenomenon is energy competition, in which the resources obtained from the environment are limited, and excessive activation of defense responses incurs yield and growth penalties (Coley et al., 1985; Brown, 2002; Liu et al., 2019). The utilization of resistance (R) genes is a major strategy in crop breeding to improve disease resistance. Although the pyramiding of R genes generates multiple-race resistance, the growth and yield of crops are significantly reduced (Nelson et al., 2018; Liu et al., 2019). In Arabidopsis, previous work has documented that introduction of the single R gene RESISTANCE TO PSEUDOMONAS (RPM1) to ecotype Bla-2 leads to a dramatic reduction in biomass (Grant et al., 1995; Tian et al., 2003). This effect on plant biomass is best explained by costs incurred from constitutive defenses in a pathogen-free environment (Mauricio, 1998; Heil and Baldwin, 2002; Tian et al., 2003). In addition, activation of growth or defense responses in plants may lead to competition for a limited supply of co-factors (Huot et al., 2014). Bacterial flagellin and elongation factor Tu (EF-Tu) are detected by FLAGELLIN-SENSING 2 (FLS2) and the EF-Tu receptor (EFR), respectively, to initiate immune responses in Arabidopsis. This recognition mechanism requires the co-factor BRI1-ASSOCIATED RECEPTOR KINASE 1 (BAK1) protein, which is also involved in brassinosteroid (BR) signaling (Gomez-Gomez and Boller, 2000; Jones and Dangl, 2006; Zipfel et al., 2006). The competition for co-receptor BAK1 results in antagonistic regulation of defense and growth (Chandran et al., 2014; Huot et al., 2014). Complex interactions have also been observed in the plant hormone network (Spoel and Dong, 2008; Robert-Seilaniantz et al., 2011; Pieterse et al., 2012). Activation of the salicylic acid (SA) signaling pathway in Arabidopsis reduces growth by stabilizing IAA/AUX, repressor proteins in the auxin pathway (Wang et al., 2007). In addition, jasmonates (JAs) can repress gibberellin (GA)-dependent growth (Yan et al., 2007; Hou et al., 2010) and can also influence growth by affecting the distribution of auxin (Yang et al., 2012). Conversely, certain pathogens secrete free IAA into host cells to repress SA-mediated resistance, thus promoting pathogen proliferation (Kazan and Manners, 2009). Atypical DP-E2F-like 1 (DEL1), an Arabidopsis cell cycle regulator, can repress the expression of Enhanced Disease Susceptibility 5 (EDS5), a transporter of SA, to retard SA-dependent resistance (Chandran et al., 2014; Rekhter et al., 2019; Torrens-Spence et al., 2019).
If resources are limited, it seems that the prioritization of resources for defense responses must reduce the resources available for growth; however, this relationship may be uncoupled under particular circumstances. Recent studies have demonstrated that rewiring of growth and defense signaling pathways can uncouple tradeoffs in growth and defense (Coley et al., 1985; Tian et al., 2003; Campos et al., 2016). For example, an Arabidopsis mutant (jazQ phyB) lacking a quintet of JAZ transcriptional repressors and the photoreceptor phytochrome B (phyB) displays increased biomass and resistance to insects (Campos et al., 2016). However, depletion of 10 JAZ repressors (JAZ1–7, 9, 10, and 13) in plants results in robust defense responses and retarded vegetative growth due to altered carbon partitioning (Guo et al., 2018). In addition, allyl glucosinolate (GSL), an Arabidopsis thaliana defense metabolite, may affect both plant growth and defense through a series of genes including HB2 and HB4 (Francisco et al., 2016). In rice, the transcription factor gene Broad-Spectrum Resistance-Digu 1 (Bsr-d1) and a nucleotide-binding oligomerization domain-like receptor (NLR) pair, Pyricularia-Gumei Resistant (PigmR) and Pyricularia-Gumei Susceptible (PigmS), mediate blast resistance without reducing yield (Deng et al., 2017; Li et al., 2017). The insertion of pathogen-responsive upstream open reading frames (ORFs) of TBF1 to control AtNPR1 translation in transgenic rice conferred broad-spectrum resistance without a yield penalty (Xu et al., 2017). Furthermore, a single transcription factor gene, Ideal Plant Architecture 1 (IPA1), can promote rice resistance and yield by activating a different cluster of genes (Wang et al., 2018).
These studies demonstrated that growth-defense tradeoffs are regulated by three main mechanisms: intelligent resource allocation decisions, ecological evolutionary developmental configurations, and a series of transcriptional networks that control plant development in complex environments (Campos et al., 2016; Francisco et al., 2016; Guo et al., 2018; Ballaré and Austin, 2019; Sestari and Campos, 2021).
To identify new components involved in growth-defense tradeoffs, we performed a genome-wide association study (GWAS) and revealed that the gene REPLUMLESS (RPL), also named PENNYWISE (PNY) or BELLRINGER (BLR) (Heil and Baldwin, 2002; Byrne et al., 2003; Roeder et al., 2003), is involved in regulating both plant growth and immunity in A. thaliana. Genetic depletion of RPL confers susceptibility to Pseudomonas syringae pv. tomato (Pst) DC3000 infection and retards growth, implying that RPL protein is a common factor involved in plant immunity and growth. Additional transcriptome analysis revealed that the auxin metabolism pathway is altered in rpl mutants, leading to the accumulation of IAA-Asp and bacterial effectors and thereby promoting bacterial proliferation. In the absence of pathogens, RPL represses flavonol biosynthesis, and the reduced accumulation of flavonols facilitates auxin transport to promote plant growth.
Results
RPL identified by genome-wide association mapping is necessary for plant growth and defense
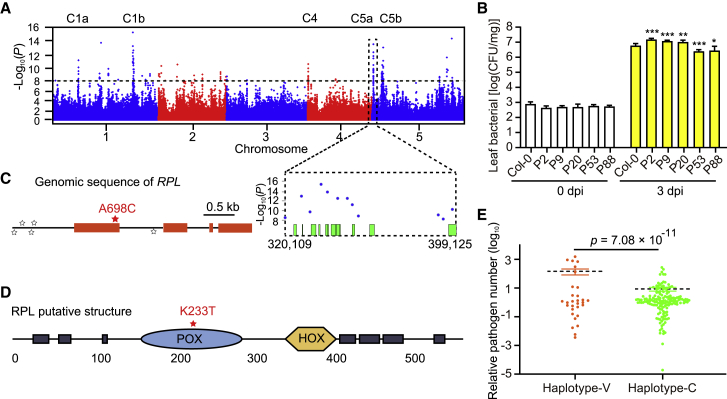
To identify the key factors involved in plant growth and defense, we investigated growth and immunity-related phenotypes in a natural population of Arabidopsis. Growth metrics included rosette diameter, rosette fresh weight, and rosette dry weight, whereas immunity was assessed by inoculating Arabidopsis ecotypes with Pst DC3000, followed by detection of the expression levels of PR1, a marker gene downstream of SA, at 24 h post infiltration (hpi) and pathogen proliferation number at 5 days post infiltration (dpi). Phenotypic data were collected from more than 300 worldwide Arabidopsis accessions (see details in Supplemental Table 1), for which single nucleotide polymorphism (SNP) datasets were downloaded from the 1001 Genomes Project or obtained by whole-genome resequencing in this study. Principal component analysis (PCA) found no significant difference between these two SNP datasets (Supplemental Figure 1). Accordingly, we merged the two SNP datasets and performed genome-wide association (GWA) mapping for each phenotype with SNPs based on a mixed linear model approach in GEMMA software (Zhou and Stephens, 2014). No significant SNP met the threshold for suggestive evidence of association (Q < 0.05, Bonferroni test), which corresponded to a p value of 4.54 × 10−8 in GWA mapping of all growth phenotypes (Supplemental Figure 2A–2C). GWA mapping of PR1 expression levels at 24 hpi identified only a few significant SNPs (Supplemental Figure 2D). GWA mapping of bacterial number at 5 dpi identified a large number of significant SNPs distributed in three chromosomes and some significant peaks, which were designated C1a, C1b, C4, C5a, and C5b, dispersed on chromosomes 1, 4, and 5 (Figure 1A; Supplemental Table 2). The most significantly associated SNP (marker SNP) of each peak was located at position 7340680 for C1a, 23075387 for C1b, 171923 for C4, 348803 for C5a, and 2940316 for C5b on their corresponding chromosomes.
Figure 1.
Genome-wide association (GWA) mapping of plant pathogen number.
(A) Manhattan plots of GWA results for bacterial accumulation at 5 dpi. The chromosomes are shown in different colors. The horizontal black dashed line corresponds to a nominal 0.05 significance threshold after a Bonferroni test (Q < 0.05). Candidate genes selected in these genomic regions are shown on the bottom.
(B) Immunity phenotypes of mutants of five candidate genes. Data are shown as mean ± SEM (n = 8–12). The results are representative of three independent experiments. Statistical analysis was performed via two-tailed Student’s t-test compared with Col-0. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(C) GWA signals for plant bacterial resistance of RPL in C5a at 320.1–399.1 kb on chromosome 5. Star size indicates the significance of SNPs, and the red star indicates the most significant SNP (position 396451, A698C).
(D) The putative domain of RPL predicted by SMART (https://smart.embl-heidelberg.de/smart/show_motifs.pl). The SNP caused a K233T mutation in the putative POX domain.
(E) Haplotype analysis of bacterial resistance for Arabidopsis accessions divided by non-synonymous SNPs. Haplotype-C (green) represents the reference (Col-0) allele. Haplotype-V (orange, Ha-P-13) represents the variant vulnerable to bacterial infection.
To identify the causal gene underlying the association, 61 genes within approximately 5-kb regions surrounding several of the most significant SNPs and 27 genes with missense SNPs at each GWA peak were chosen as candidate genes. Six of the candidate genes were present in both candidate lists. Thus, 82 candidate genes, including eight long non-coding RNAs, were identified from these genomic regions and were designated in the pathogen number (P) series in numerical order. Functional annotation analyses of these genes revealed enrichment in transcriptional regulation, chromatin remodeling, cellular metabolism, stress response, and so on. We obtained mutants (T-DNA insertion) for these candidate genes and examined bacterial growth at 3 dpi. Three mutant lines (P2, P9, and P20) supported more bacterial growth, whereas another two mutant lines (P53 and P88) supported less bacterial growth (Figure 1B).
Among the tested mutants, only one candidate gene (P9) mutant exhibited a growth defect (Supplemental Figure 3). The P9 mutation was located around the GWA peak C5a (320.1–399.1 kb). The P9 candidate gene, RPL, encodes a BEL1-like TALE homeodomain (BLH) transcription factor that controls multiple processes involved in plant development, including meristem morphogenesis and floral development (Byrne et al., 2003; Roeder et al., 2003; Smith and Hake, 2003; Bencivenga et al., 2016). Six significant SNPs from the GWA mapping were dispersed in the genomic region of RPL (Figure 1C). The SNP causing missense variation is A698C in the exon region of RPL, which causes an amino acid substitution (K233T) in the putative POX domain of the RPL protein (Figure 1D). Using haplotype analysis of this non-synonymous SNP (position 396451), we classified Arabidopsis ecotypes in this study into two haplogroups, haplotype-V and haplotype-C, which implies that this missense SNP may play an important role in bacterial resistance variation among ecotypes (Figure 1E).
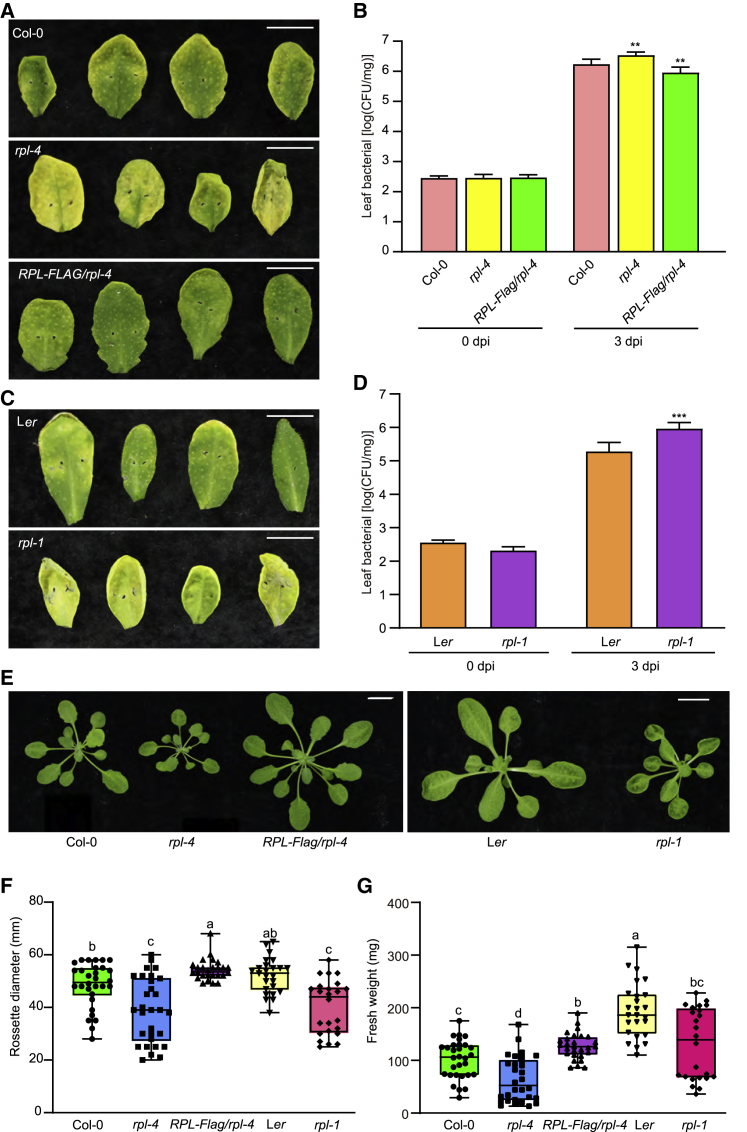
Next, to verify the roles of RPL in immunity and growth, rpl-1 (Ler background) and rpl-4 (Col-0 background) mutants (Supplemental Figure 4) and RPL-FLAG/rpl-4 transgenic plants were inoculated with Pst DC3000. Compared with the wild type, bacterial number was significantly increased in rpl-4 at 3 dpi but severely reduced in the RPL-FLAG/rpl-4 overexpression line (Figures 2A and 2B). Similar results were also observed in rpl-1 mutants (Figures 2C and 2D). In addition, the rosette diameter of rpl-4 was smaller than that of Col-0, and the rosette fresh weight of rpl-4 was also significantly reduced, whereas the RPL-FLAG/rpl-4 overexpression line had increased biomass (Figures 2E–2G). The rpl-1 mutant displayed a similar growth defect phenotype (Figures 2E–2G). Based on these data, we concluded that RPL is necessary for both immunity and growth in different Arabidopsis accessions and thus acts as a positive regulator of plant growth and defense.
Figure 2.
RPL contributes to both disease resistance and plant growth.
(A) Defense phenotypes of Col-0, rpl-4, and RPL-FLAG/rpl-4 at 3 dpi. The scale bar represents 1 cm.
(B) Pathogen numbers of Col-0, rpl-4, and RPL-FLAG/rpl-4 after inoculation days as indicated. Data are shown as mean ± SEM (n = 8–12). Statistical analysis was performed via two-tailed Student’s t-test. ∗∗p < 0.01.
(C) Defense phenotypes of Ler and rpl-1 at 3 dpi. The scale bar represents 1 cm.
(D) Pathogen numbers of Ler and rpl-1 after inoculation days as indicated. Data are shown as mean ± SEM (n = 8–12). Statistical analysis was performed via two-tailed Student’s t-test. ∗∗∗p < 0.001.
(E) Growth phenotypes of Col-0, rpl-4, RPL-FLAG/rpl-4, Ler, and rpl-1 after 3 weeks of growth. The scale bar represents 1 cm.
(F and G) (F) Rosette diameter and (G) fresh weight of Col-0, rpl-4, RPL-FLAG/rpl-4, Ler, and rpl-1 after 3 weeks of growth. Data are shown as mean ± SEM (n > 25). Statistical analysis was performed by one-way ANOVA with Brown–Forsythe and Welch’s test (significance was set to p < 0.05). Different letters indicate significant differences in rosette diameter or fresh weight. The results in (B), (D), (F), and (G) are representative of three independent experiments.
RPL represses the expression of GH3 genes to contribute to plant immunity
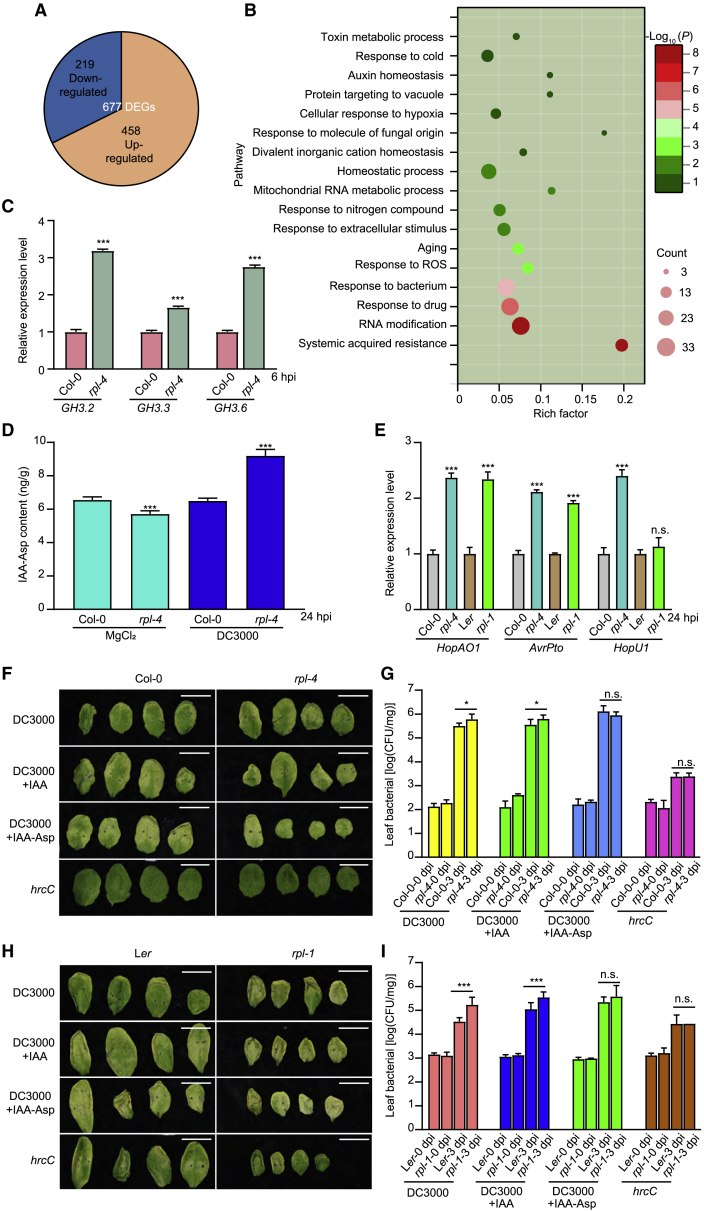
To identify the mechanism through which the transcription factor RPL modulates plant growth and defense, we performed RNA sequencing (RNA-seq) of Col-0 and rpl-4 after inoculation with Pst DC3000. To identify the best time point after bacterial infection for RNA-seq, we first detected the expression pattern of RPL after inoculation and found that RPL transcript abundance peaked at 6 hpi (Supplemental Figure 5A). Thus, we chose 6 hpi for RNA-seq and identified 677 differentially expressed genes (DEGs) between mutant and wild-type plants. Among these DEGs, 219 genes were downregulated in rpl-4 relative to Col-0, whereas 458 genes were upregulated (Figure 3A; Supplemental Table 3). This implies that RPL mainly functions as a repressor in transcriptional regulation, which is consistent with the results of a previous study (Bencivenga et al., 2016). Gene Ontology (GO) analysis of the upregulated DEGs showed enrichment of 17 biological pathways (Figure 3B). Among them, three pathways were associated with defense responses, including “response to bacterium,” “systemic acquired resistance” (related to the SA pathway), and “auxin homeostasis.” GO analysis of the downregulated DEGs showed that most enriched terms were related to the JA pathway, including “response to oomycetes,” “response to wounding,” and “response to herbivore” (Supplemental Figure 5B). Accordingly, we conducted analysis to determine whether SA and JA play a role in RPL-regulated bacterial resistance. We detected the levels of SA and JA in rpl-4 mutant and Col-0 plants at 24 hpi and found that the SA level was slightly lower in rpl-4 than in Col-0 (p = 0.0441; Supplemental Figure 5C), whereas the JA level did not differ between Col-0 and rpl-4 (Supplemental Figure 5D). Thus, the JA and SA pathways may not play a major role in RPL-mediated defense responses.
Figure 3.
RPL regulates plant immunity through GH3 genes.
(A) Venn diagram of DEGs between rpl-4 and Col-0 after Pst DC3000 infection. Among the 677 DEGs in rpl-4 mutant compared with wild-type plants, 219 genes were downregulated and 458 genes were upregulated.
(B) GO analysis of upregulated DEGs in rpl-4 compared with Col-0. Count represents the number of enriched DEGs in each cluster, and color represents the fold change.
(C) Relative expression of GH3 family genes in Col-0 and rpl-4 at 6 hpi. Data are shown as mean ± SEM (n = 3). Statistical analysis was performed via two-tailed Student’s t-test compared with Col-0. ∗∗∗p < 0.001.
(D) Indole-3-acetic acid-aspartic acid (IAA-Asp) levels in Col-0 and the rpl-4 mutant. Data are shown as mean ± SEM (n = 4). Statistical analysis was performed via two-tailed Student’s t-test compared with Col-0. ∗∗∗p < 0.001.
(E) Relative expression of genes encoding effectors secreted by Pst DC3000 in Col-0 and rpl-4 at 24 hpi. Data are shown as mean ± SEM (n = 3). Statistical analysis was performed via two-tailed Student’s t-test compared with Col-0. ∗∗∗p < 0.001; n.s., no significance.
(F) Defense phenotype of Col-0 and rpl-4. The combination of Pst DC3000, indole-3-acetic acid (IAA), IAA-Asp, and T3SS-defective Pst DC3000 hrcC (hrcC) is annotated. The scale bar is 1 cm. (G) Pathogen numbers of Col-0 and rpl-4 after inoculation days as indicated. Data are shown as mean ± SEM (n = 8–12). Statistical analysis was performed via two-tailed Student’s t test compared with Col-0. ∗p < 0.05; n.s., no significance. (H) Defense phenotype of Ler and rpl-1. The combination of Pst DC3000, IAA, IAA-Asp, and Pst DC3000 hrcC is annotated. The scale bar is 1 cm. (I) Pathogen numbers of Ler and rpl-1 after inoculation days as indicated. Data are shown as mean ± SEM (n = 8–12). Statistical analysis was performed via two-tailed Student’s t test compared with Col-0. ∗∗∗p < 0.001; n.s., no significance.
Next, we focused on the GO term “auxin homeostasis”, for which three of four enriched genes belonged to the GRETCHEN HAGEN 3 (GH3) gene family. The GH3 gene family encodes IAA-amido synthases that conjugate amino acids to the active form of IAA (Staswick et al., 2005). Among these products, IAA-Asp can facilitate pathogen proliferation by promoting the expression of pathogen effector genes such as HopA O1, HopU1, and AvrPto (Bogdanove and Martin, 2000; Gonzalez-Lamothe et al., 2012; Nicaise et al., 2013; Castaneda-Ojeda et al., 2017). Consistent with the RNA-seq data, the expression levels of three GH3 genes, GH3.2, GH3.3, and GH3.6, were significantly increased in rpl-4 (Figure 3C). Based on the hypothesis that RPL regulates immunity by modulating expression of GH3 gene family members, we measured the content of endogenous IAA-Asp and detected effector translocation and the expression levels of bacterial virulence genes. Twenty-four hours after inoculation with Pst D3000, the content of endogenous IAA-Asp was significantly higher in rpl-4 than in Col-0 owing to elevated expression levels of GH3 genes (Figure 3D). As expected, the expression levels of HopA O1, HopU1, and AvrPto were significantly increased in rpl-1 and rpl-4 after inoculation with Pst D3000 (Figure 3E and Supplemental Figure 6A). In addition, significant differences in cyclic AMP (cAMP) production between the wild type and rpl mutants were observed after inoculation with Pst DC3000 carrying plasmids expressing AvrPto-Cya fusions (Supplemental Figure 6E), which implied that RPL regulated the translocation and expression of pathogen effectors. Furthermore, exogenous treatment with IAA-Asp, but not IAA, together with Pst DC3000, eliminated differences in the expression levels of effector genes and the difference in susceptibility to bacterial infection between rpl mutants and wild-type plants (Figures 3F–3I and Supplemental Figures 6B–6D). Consistent with this result, elevated translocation of the AvrPto effector was eliminated by exogenous treatment with IAA-Asp (Supplemental Figure 6E).
Bacterial accumulation and the expression levels of effector proteins were significantly decreased in the gh3.2 mutants but not in the gh3.6 mutants (Supplemental Figures 7A and 7B), which was consistent with a previous study (Gonzalez-Lamothe et al., 2012). In agreement with the results above, translocation of AvrPto was significantly reduced in the gh3.2 mutant (Supplemental Figure 7C). In addition, the gh3.2 mutant, but not the gh3.6 mutant, had a larger rosette diameter and greater fresh weight compared with the wild-type line (Supplemental Figures 7D and 7E).
Based on the results described above, we concluded that RPL promotes plant immunity, possibly by repressing expression of the GH3 gene family, especially GH3.2, to reduce IAA-Asp accumulation, thus inhibiting the expression and translocation of pathogenic effector proteins and preventing proliferation of pathogens.
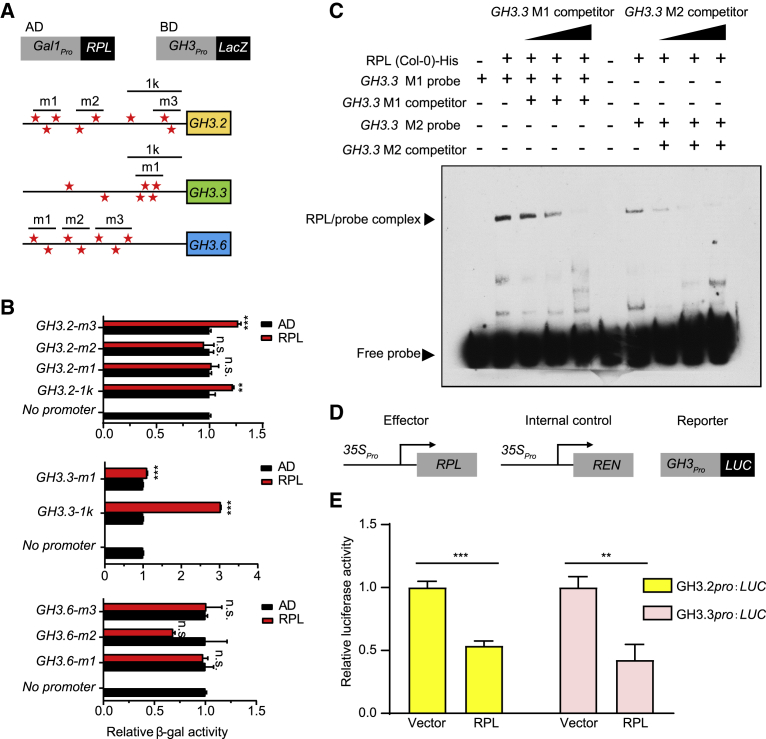
RPL directly binds to the promoters of the GH3 gene family
Next, we determined whether RPL repressed the expression of GH3 family members via direct binding to their promoters. RPL is a BLH transcription factor that recognizes conserved motifs containing TGAC/T (Bencivenga et al., 2016). First, we tested whether RPL interacts with the GH3.2 promoter using yeast one-hybrid (Y1H) assays. We investigated putative RPL binding motifs on the 3-kb promoter region of GH3.2, and we found that several regions contained such motifs, which we designated GH3.2-1K, GH3.2-m1, GH3.2-m2, and GH3.2-m3 (Figure 4A). Measurement of relative β-galactosidase (β-gal) activity revealed that RPL could bind directly to the GH3.2 promoter in Y1H assays (Figure 4B). Assessment of the binding activity of RPL to GH3.3 and GH3.6 showed that RPL could bind to the promoter of GH3.3, but not that of GH3.6, in Y1H assays (Figures 4A and 4B). To confirm the binding activity of RPL, we expressed His-tagged recombinant RPL protein (Supplemental Figure 8A) and used it to perform electrophoretic mobility shift assays (EMSAs). As the concentration of the competitor increased, the binding activity of RPL with two GH3.3 probes containing the potential RPL binding motif decreased, indicating that RPL directly and specifically interacted with probes derived from the GH3.3 promoter region (Figure 4C). Furthermore, we expressed and purified a natural variant of RPL-K233T (His-tag) derived from Ha-P-13 (a representative ecotype of haplotype-V) for binding activity experiments (Supplemental Figure 8B). Interestingly, this natural variant of RPL-K233T (Ha-P-13) displayed a weaker binding activity with the probe in comparison with RPL (Col-0) in vitro (Supplemental Figure 8C), consistent with the lower bacterial resistance of the Arabidopsis ecotype carrying the variant RPL allele. In addition to in vitro binding assays, we performed dual luciferase assays in Arabidopsis protoplasts by transiently co-expressing RPL and pGH3.2-LUC (Figure 4D). The LUC signal was decreased by almost 50% when both RPL and pGH3.2-LUC were co-transformed, which suggested that RPL interacts with the GH3.2 promoter and represses GH3.2 expression in vivo (Figure 4E). The same result was observed for the GH3.3 promoter (Figures 4D and 4E).
Figure 4.
RPL directly binds to the promoters of the GH3 gene family.
(A) Schematic representation of various constructs used in Y1H and promoters of GH3 containing several potential RPL binding motifs. The stars represent the positions of potential RPL binding motifs.
(B) RPL binding activity in Y1H analysis. Data are shown as mean ± SEM (n = 3). Statistical analysis was performed via two-tailed Student’s t-test compared with Col-0. ∗∗p < 0.01; ∗∗∗p < 0.001; n.s., no significance.
(C) EMSA data showed that recombinant RPL protein directly bound to probes derived from the GH3.3 promoter region in vitro. M1 and M2 are two different probes listed in Supplemental Table 5.
(D) Schematic representation of various constructs used in the transient transfection assay in Arabidopsis protoplasts.
LUC, firefly luciferase; REN, renilla luciferase.
(E) Relative LUC expression levels driven by GH3pro in Arabidopsis protoplasts. Gene expression levels were quantified as ratios of LUC/REN enzyme activities. Data are shown as mean ± SEM (n = 3). Statistical analysis was performed via two-tailed Student’s t-test compared with Col-0. ∗∗p < 0.01; ∗∗∗p < 0.001; n.s.; no significance. The results in (B) and (E) are representative of three independent experiments.
RPL represses CHI expression to promote plant growth
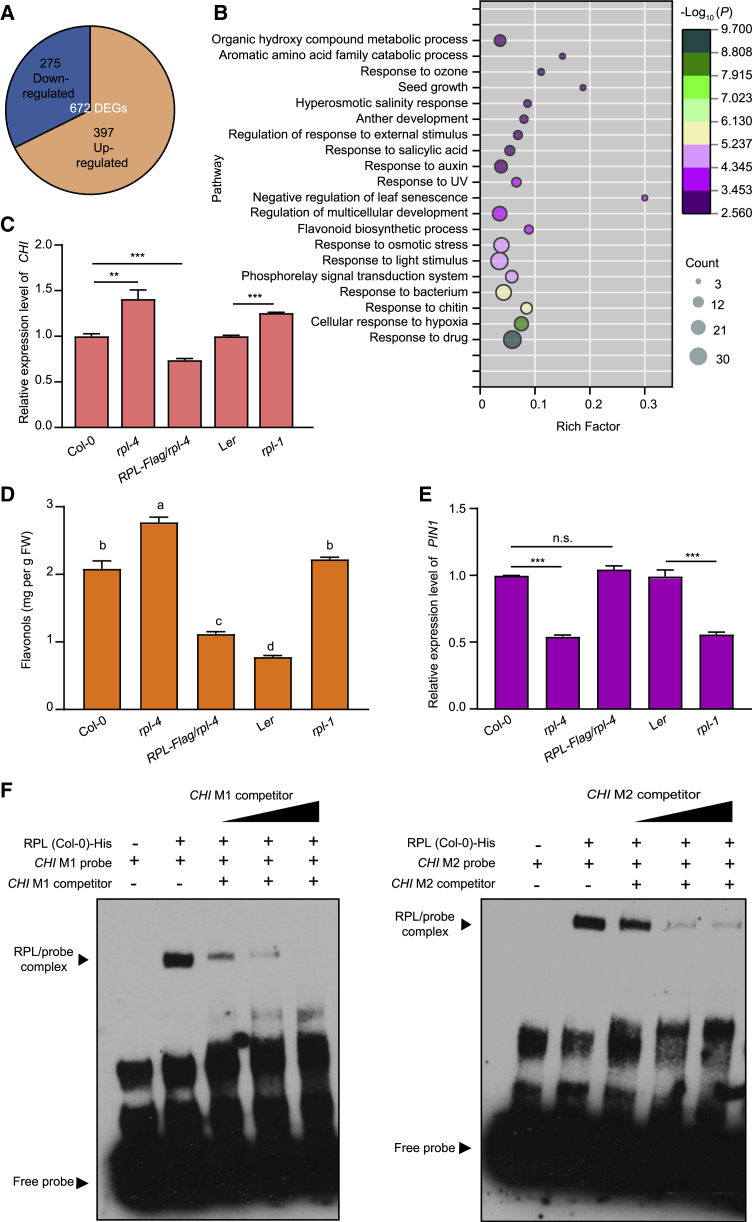
In parallel, we also performed RNA-seq for Col-0 and rpl-4 without bacterial infection and identified 672 DEGs, including 397 upregulated genes and 275 downregulated genes in the rpl-4 mutant (Figure 5A; Supplemental Table 4). GO analysis of the upregulated DEGs revealed 20 enriched terms, three of which, “response to salicylic acid,” “response to auxin,” and “flavonoid biosynthetic process,” are related to plant hormones (Figure 5B). However, the list of GO terms enriched in the set of DEGs identified here did not include GH3-related genes (Figures 5A and 5B). Previous studies have reported that activation of defense-related hormone pathways may retard growth (Tian et al., 2003; Todesco et al., 2010; Huot et al., 2014). As rpl-4 displayed reduced biomass, we first detected the expression levels of marker genes in the SA and JA pathways, and we found no significant changes in the expression levels of these genes in rpl-4 compared with wild-type Col-0 (Supplemental Figures 9A and 9B). This result implied that the reduced growth of rpl-4 did not result from inappropriate activation of defense pathways in the absence of pathogen infection. Reduced plant growth could also have been a result of flavonoid accumulation leading to inhibition of auxin transport (Sharma et al., 2020). Because the flavonoid biosynthetic process pathway was activated in the rpl-4 mutants, we detected the expression levels of some key genes involved in flavonoid biosynthesis (Mehrtens et al., 2005; Pandey et al., 2014). Significantly increased expression levels of CHALCONE ISOMERASE (CHI) and two flavonoid synthesis-related genes, At4G14090 and At2G41040, but not CHALCONE SYNTHASE (CHS), FLAVONOID-3-HYDROXYLASE (F3H), or FLAVONOL SYNTHASE 1 (FLS1), were observed in the rpl-4 mutants (Figure 5C and Supplemental Figure 9C), suggesting that RPL contributes to plant growth mainly by repressing the gene expression of CHI.
Figure 5.
RPL promotes plant growth through the flavonoid synthesis pathway.
(A) Venn diagram of DEGs between rpl-4 and Col-0 without bacterial infection. Among the 672 DEGs in rpl-4 mutants compared with wild-type plants, 275 genes were downregulated and 397 genes were upregulated.
(B) GO analysis of upregulated DEGs in rpl-4 compared with Col-0. Count represents the number of enriched DEGs in each cluster, and color represents the fold change.
(C) Relative expression levels of CHI in rpl mutants and the RPL-Flag/rpl-4 transgenic line. Data are shown as mean ± SEM (n = 3). Statistical analysis was performed via two-tailed Student’s t-test compared with the wild type. ∗∗p < 0.01, ∗∗∗p < 0.001.
(D) The abundance of flavonols in rpl mutants and the overexpression line RPL-Flag/rpl-4. Data are shown as mean ± SEM (n = 3). Statistical analysis was performed by one-way ANOVA with Brown–Forsythe and Welch’s test (significance was set to p < 0.05).
(E) Relative expression levels of PIN1 in rpl mutants and the RPL-Flag/rpl-4 transgenic line. Data are shown as mean ± SEM (n = 3). Statistical analysis was performed via two-tailed Student’s t-test compared with the wild type. ∗∗∗p < 0.001; n.s., no significance.
(F) EMSA data showed that recombinant RPL protein directly bound to probes derived from the CHI promoter region in vitro. The results in (C)–(E) are representative of three independent experiments.
Considering that CHI is necessary for flavonoid synthesis, we measured total flavonols in mutants and overexpression lines to assess the role of RPL in the accumulation of this metabolite. Compared with wild-type plants, rpl-4 and rpl-1 plants had significantly higher flavonol levels, whereas those of RPL-Flag/rpl-4 plants were dramatically reduced (Figure 5D). As flavonoids affect auxin transport (Sharma et al., 2020), we detected the expression levels of auxin transport-related genes such as PIN-FORMED1 (PIN1), AUX1, and ATP-BINDING CASSETTE SUBFAMILY B (ABCB19). Compared with wild-type plants, the expression levels of these genes were reduced in both rpl-1 and rpl-4 mutants (Figure 5E, Supplemental Figures 10A and 10B). To test whether RPL directly binds to the promoter region of CHI, we used His-tagged recombinant RPL protein to perform EMSA, which showed that RPL possessed specific binding activity with two probes derived from the promoter region of CHI (Figure 5F). To further confirm the relationship between flavonoids and plant growth, we measured the growth phenotypes of two flavonoid-related mutants, chi (GK176H30) and f3h (GK292E08). The rosette diameters of chi and f3h plants were larger than that of Col-0, and the rosette fresh weights of chi and f3h plants were also increased significantly (Supplemental Figure 11).
Taken together, our results revealed that RPL physically interacts with and represses the activity of promoters of GH3, which inhibits the synthesis of IAA-Asp and finally leads to repression of bacterial effector proteins to promote bacterial resistance. On the other hand, RPL physically interacts with and represses the activity of the promoter of CHI, a key gene in the flavonoid synthesis pathway, which results in reduced flavonol levels and finally leads to increased expression of auxin transport genes to promote plant growth.
Discussion
Upon pathogen invasion, some plants activate defense responses quickly and properly, but this process often incurs growth penalties. Recent studies have shown that this growth–defense tradeoff can be reconciled by key regulators in a hardwired transcriptional network. In this study, we built a large dataset including the natural variation of five phenotypes related to both plant growth and immunity in Arabidopsis. Using GWAS analysis, we identified RPL, a novel gene regulating plant growth and defense. RPL encodes a BLH transcription factor that not only regulates plant growth and development but also participates in plant defense, according to the results of our study. Plants lacking RPL show impaired resistance and reduced growth, whereas overexpression of RPL promotes plant biomass and defense. The expression of RPL is regulated by pathogen infection, yet the upstream regulator of RPL is still unknown. Corresponding to its expression pattern, overexpression of RPL can reduce bacterial proliferation. Further analysis suggests that RPL contributes to plant immunity primarily through pathways other than the SA and JA pathways, which is beneficial because activation of those pathways always results in a growth penalty (Coley et al., 1985; Brown, 2002; Liu et al., 2019). Furthermore, our study provides a possible link between RPL and the auxin metabolism pathway via the GH3 gene family. GH3 genes encode IAA-amido synthases that conjugate Asp with IAA to form IAA-Asp, which can facilitate bacterial proliferation by promoting the expression of virulence genes. Correspondingly, following exogenous treatment with IAA-Asp, but not IAA, together with Pst DC3000, the expression levels of effectors and the pathogen numbers in rpl mutants were similar to those in the wild type. Using Y1H, EMSA, and dual luciferase assays, we demonstrated that RPL recognizes conserved motifs in the promoters of GH3 genes, thereby repressing their expression to regulate plant immunity. We also suggest that the flavonoid biosynthesis pathway is involved in RPL-regulated plant growth, in which CHI plays an important role. Compared with the wild type, rpl mutants had significantly higher flavonol levels, which may have led to reduced expression of downstream auxin transport genes such as AUX1, PIN1, and ABCB19. Consistent with these results, flavonol biosynthetic mutants such as chi and f3h show enhanced growth (Supplemental Figure 11). Future studies should assess whether these mutants recover the growth defect of the rpl mutants. Furthermore, transcriptomic and proteomic analyses of a chi mutant in tomato showed that flavonoid deficiency probably impairs terpenoid biosynthesis, which is important for preventing biotic threats such as arthropod herbivores (Sugimoto et al., 2022). Therefore, RPL-directed CHI expression may be involved in both plant defense and growth in tomato.
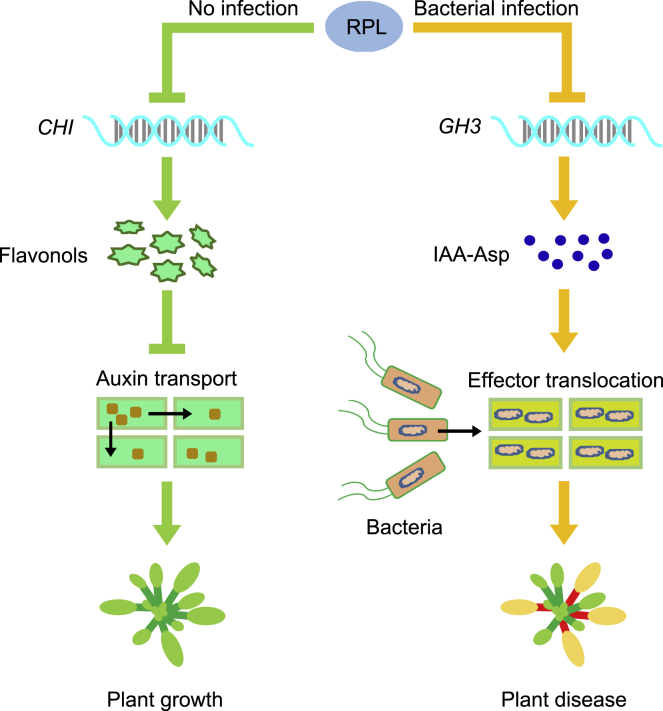
In summary, we propose a model in which RPL functions in plant growth and disease resistance (Figure 6). When there is no pathogen, RPL, as a transcription factor, directly represses gene expression of CHI, reducing flavonol accumulation and thus promoting downstream auxin transport to promote growth. After infection by bacteria such as Pst DC3000, RPL directly binds to the promoters of GH3-related genes and represses their expression, which results in reduction in the abundance of IAA-Asp and inhibition of the translocation of pathogen virulence effectors, thus conferring bacterial resistance. Therefore, RPL plays a critical role in promoting both plant growth and defense.
Figure 6.
Proposed working model of RPL contributing to both plant growth and disease resistance.
In healthy plants, RPL directly binds to the promoter of CHI and represses its gene expression, reducing flavonol accumulation, which promotes auxin transport to facilitate plant growth. When plants are invaded by bacteria such as Pst DC3000, RPL directly represses gene expression of GH3, resulting in reduced abundance of IAA-Asp, which inhibits expression and translocation of pathogen virulence effectors and reduces plant disease.
Our RNA-seq data from pathogen-infected leaves show that RPL is involved in leaf resistance. Previous studies showed that RPL transcripts are most abundant in flowers and inflorescences, and RPL can bind to the promoters of genes that function in meristem development, organ patterning, and growth (Byrne et al., 2003; Roeder et al., 2003; Smith and Hake, 2003; Bencivenga et al., 2016). This implies that RPL may be involved in flower and inflorescence resistance via a molecular mechanism different from those involved in bacterial resistance. Thus, RPL may have an organ-specific function in the regulation of plant growth and immunity, which merits further study. As reported in a previous study, IPA1 can promote rice resistance and yield by activating different clusters of genes through different phosphorylation sites (Wang et al., 2018). Here, we show that RPL can also promote both defense and growth according to environmental cues. The activation of immunity pathways during pathogen infection may be mediated by post-translational modification or by accumulation of RPL protein, as the RPL expression level increased upon infection. The exact mechanism that regulates the switch from growth to defense by RPL protein remains a promising subject for future studies.
The EMSA assay results show that a natural RPL variant (Ha-P-13, carrying a missense variation of A698C) shows decreased binding activity to the promoter containing the conserved motifs (Supplemental Figure 7C). The significant SNP (A698C) in the coding region of the POX domain causes a missense variation in RPL that reduces its promotor-binding activity, suggesting that the POX domain is crucial for RPL protein function. Furthermore, this non-synonymous SNP can be used to divide Arabidopsis ecotypes into two distinct groups with differing bacterial resistance, which implies that it plays a key role in natural variation. The variant RPL protein is harmful to plant survival in the environment when pathogens are common, and this variant SNP should remain rare if selection pressure is sufficiently strong. Consistent with this speculation, the variant SNP was found in only 13.4% of ecotypes (30 of 224). However, we cannot exclude the possibility that it has beneficial effects in specific environments. It is tempting to speculate how this missense mutation affects RPL’s function. Finally, homologs of RPL in other crops may be good targets for breeding programs aimed at producing varieties with increased disease resistance and yield.
Methods
Plant materials and growth conditions
A total of 308 Arabidopsis ecotypes were used for GWAS, all of which were purchased from the Arabidopsis Biological Resource Center (ABRC), as shown in Supplemental Table 1. The mutants, including rpl-4 (SALK_098505C), gh3.2 (SALK_037520C), gh3.6 (SALK_013458C), chi (GK176H30), and f3h (GK292E08), were in the Col-0 background, and rpl-1 was in the Ler background (Byrne et al., 2003; Roeder et al., 2003; Smith and Hake, 2003; Bencivenga et al., 2016). RPL-FLAG/rpl-4 was a 3× FLAG-tagged overexpression line in the rpl-4 background. Plants were grown on Murashige and Skoog medium containing 1% sucrose at 22°C under white light (100 μmol m−2 s−1, 16 h light/8 h dark). Plants for pathogen inoculation were grown in soil under 8-h light and 16-h dark conditions in light chambers where light, temperature, and humidity were controlled, and all plants were grown at a controlled temperature (22°C ± 0.2°C) with 65% relative humidity. Plants for protoplast transient expression assays were grown in soil under 12-h light and 12-h dark conditions in light chambers, and all plants were grown at a controlled temperature (22°C ± 0.2°C day and 18°C ± 0.2°C night) with 65% relative humidity.
Bacterial inoculation assay
Pst DC3000 or T3SS-defective Pst DC3000 hrcC mutant bacteria were grown in King’s B medium (10 g l−1 protease peptone, 1.5 g l−1 K2HPO4, 10 ml l−1 glycine, pH 7.0) supplemented with 25 mg l−1 rifampicin at 28°C for 2–3 days. Fully expanded A. thaliana leaves (3–4 weeks old) were chosen for the inoculation assays. A suspension of bacterial cells (optical density [OD] = 0.0004) in 10 mM MgCl2 was infiltrated into the apoplastic spaces through the abaxial leaf surface using a 1-ml needleless syringe. At 3 or 5 dpi, each sample (including four leaves from independent plants) was weighed and collected into a 2-ml microcentrifuge tube, then ground in 1 ml of 10 mM MgCl2. For each genotype, 8–12 samples were collected. The ground material was diluted 105 times (for Pst DC3000 inoculation) or 103 times (for Pst DC3000 hrcC mutants), and 45 μl of each diluted sample was spread onto King’s B medium supplemented with 25 mg l−1 rifampicin. The plates were grown for 2 days at 28°C, after which the leaf bacterial number was measured and calculated as the pathogen number.
For treatment with IAA and IAA-Asp, Arabidopsis plant leaves (3–4 weeks old) were infiltrated with a solution containing 10 mM MgCl2, 100 μM IAA, or 100 μM IAA-Asp together with Pst DC3000 (OD = 0.0004).
GWA mapping
GWA mapping was performed as previously reported (Ren et al., 2019). In general, the indicated phenotypes of A. thaliana accessions were grown for 4 weeks with a short-day photoperiod and subsequently selected for GWAS. First, SNPs of 224 ecotypes were obtained from the Arabidopsis 1001 Genome Project website (205 ecotypes, http://1001genomes.org/) and from the resequencing data (19 ecotypes, provided in this study, SRP344532). As for resequencing data, SNP detection was performed using the Genome Analysis Toolkit (GATK, version 3.80) to minimize the negative influence of sequencing errors in gene identification (Cao et al., 2016). The reads around indels were realigned after alignment with the Burrows-Wheeler Alignment Tool (BWA, version 0.1.17). Realignment was performed using GATK in two steps. First, the RealignerTargetCreator package was used to identify regions that required realignment. Second, IndelRealigner was used to realign the identified regions, which generated a realigned BAM file for each accession. In addition, SNPs were called at the population level with the GATK tool. The SNP confidence score was set to greater than 30, and the parameter -stand_call_conf was set to 30; a total of 1 218 492 SNPs were reported. These SNPs were merged with SNPs downloaded from the 1001 Genomes Project and then imputed using BEAGLE (version 4.0). A total of 1 100 004 SNPs were retained by filtering positions with a minor allele frequency <0.05 and multiple allele loci. PCA of the entire SNP dataset found no significant difference between these two datasets. GWAS was performed using a univariate mixed linear model method in GEMMA software with default parameters (Zhou and Stephens, 2014). The p values from the GEMMA output were Bonferroni corrected, and adjusted p values (Q values) <0.05 were considered significant.
Plasmid construction and plant transformation
To generate GH3-related constructs for Y1H assays, promoter fragments (as indicated in Figure 4) were amplified and inserted into the pLacZi vector (Lin et al., 2007). To generate pB42AD-RPL constructs for Y1H assays, fragments containing the full-length RPL CDS were amplified and inserted into the pB42AD vector (Clontech) using a recombinant method. For purification of RPL-related recombinant proteins, full-length RPL CDS fragments amplified from Col-0 cDNA and Ha-P-13 cDNA were inserted separately into the pET28b vector (EMD Biosciences).
To generate the GH3pro-LUC constructs for dual luciferase assays, DNA fragments 876 bp and 980 bp upstream of the 5′ untranslated region of GH3.2 and GH3.3, respectively, were separately amplified and cloned into the pGreenII0800-LUC vector (Hellens et al., 2005). For the RPL construct for dual luciferase assays, a fragment containing the full-length RPL CDS was amplified and cloned into the pRI101 vector.
To generate an RPL overexpression line in the rpl-4 background, the full-length RPL CDS was amplified and inserted into the pCAMBIA1307 vector under the control of the CaMV35S promoter. The plasmids were then transformed into plants using Agrobacterium GV3101 via the floral dipping method. Transformants were selected on MS medium containing hygromycin. All primers used for cloning and genotyping are listed in Supplemental Table 5.
Whole-transcriptome RNA sequencing
Three- or 4-week-old leaves from Col-0 and rpl-4 were collected after inoculation with Pst DC3000 or MgCl2 and extracted with a Qiagen RNA extraction kit according to the manufacturer’s instructions. The Illumina NovaSeq 6000 sequencing platform was used for high-throughput sequencing with a sequencing depth of 40×. HISAT2 was used to align RNA-seq sequences to the Col-0 reference genome (TAIR 10). Fully matched sequences were used for subsequent studies. Genes with false discovery rate (FDR) <0.05 were defined as DEGs using edgeR (a Bioconductor package for differential expression analysis of digital gene expression data). DEGs were identified using the criterion fold change >2. There were three biological replicates for each genotype and condition, and each biological replicate was pooled from 10 different individual plants. GO enrichment was analyzed using the online tool Metascape (http://metascape.org/gp/).
RT-qPCR
Unless otherwise stated, total RNA was extracted from leaves as indicated using the RNeasy plant mini kit (Qiagen; kit ID, 74904). Total RNA (1–3 μg) was reverse transcribed using SuperScript II (Invitrogen; kit ID, 18064014). The qPCR was performed on a 7500 Fast Real-Time PCR System (ABI) using TB Green mix (Takara; kit ID, RR430A). ACTIN2 was used as an internal control. The primers used for RT-qPCR are listed in Supplemental Table 5.
Determination of endogenous SA, JA, and IAA-Asp
Mature and fully expanded leaves of plants (3–4 weeks old) were inoculated with Pst DC3000. At 24 hpi, each sample containing 10 infected leaves from independent plants was collected. For detection of SA and JA, samples were ground in liquid nitrogen and spiked with 200 mg of grinding powder mixed with 1 ml of ethyl acetate. For detection of IAA-Asp, samples were ground in liquid nitrogen and spiked with 1 g of grinding powder mixed with 2 ml of extraction buffer (methanol:acetonitrile:double-distilled water [ddH2O] = 2:2:1). Later, phytohormone extraction and quantification were performed via high-pressure liquid chromatography (HPLC)-tandem mass spectrometry (MS/MS) (LCMS-8040 system, Shimadzu, Tokyo, Japan). Three or four replicates were collected for statistical analyses.
Translocation assay
Translocation assays were performed as described previously (Crabill et al., 2010). Pst DC3000 and Pst DC3000 hrcC transformants carrying plasmids expressing AvrPto-Cya fusions were inoculated into fully expanded leaves using a 1-ml syringe. The bacterial density was adjusted to OD600 = 0.5 in a solution of 10 mM MgCl2 and 100 μM isopropyl β-d-1-thiogalactopyranoside (IPTG). Four leaf disks around 0.8 cm2 in total area were collected at 6 hpi or 24 hpi and extracted with 250 μl of 0.1 M HCl at −20°C overnight. The samples were diluted to 5 ng/μl of total protein using the Bradford assay (Bio-Rad). Finally, cAMP levels were determined using the cAMP-Glo Assay kit according to the manufacturer’s directions (PROMEGA; kit ID, V1502).
Quantification of total flavonols
Total flavonol quantification assays were performed as described previously (Sharma et al., 2020). In brief, 3-week-old rosette leaves (<200 mg per sample) were ground and extracted in 1 ml of 80% methanol at 4°C for 2 h with gentle shaking. Later, the liquid mixture was centrifuged at 12 000 g for 12 min at 4°C. About 0.3 ml of each supernatant was transferred to a new 5-ml tube, after which 0.9 ml of methanol was added, and the mixture was then mixed with 0.06 ml of aluminum chloride (10% water solution), 0.06 ml of potassium acetate (1 M), and 1.68 ml of ddH2O. The mixture was mixed gently upside-down and incubated for 30 min at room temperature. After incubation, the absorbance of each sample was measured at 415 nm. A standard curve was obtained using rutin as the standard. The total flavonol content in mature leaves was calculated based on the rutin standard curve.
Y1H assay
Y1H assays were performed as described previously (Li et al., 2020). In brief, pB42AD-RPL (effector) and pLacZi-GH3pro (reporter) were co-transformed into yeast strain EGY48, and the transformants were plated on minimal synthetic defined base with -Ura/-Trp dropout (DO) mix and X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) for blue color development. Quantification of β-galactosidase activity was performed as described in the Yeast Protocols Handbook (BD Clontech).
EMSA assay
DNA-protein interactions were characterized via EMSA using bacterially purified RPL proteins and 5′-biotin-labeled probes as shown in Supplemental Table 5. RPL (Col-0)-His and RPL (Ha-P-13)-His were purified with Nickel Nitrilotriacetate (Ni-NTA) beads. EMSA was performed with kits using the manufacturer’s instructions (Thermo Scientific; kit ID, 20148).
Protoplast transient expression assays
Protoplasts from Arabidopsis mesophyll cells were prepared and transformed as described previously (Li et al., 2020). The effector plasmid expressing RPL-GFP and reporter plasmids carrying internal control 35S:REN and GH3pro:LUC were co-transformed into protoplasts (4-week-old A. thaliana leaves) and incubated under constant weak light for 12–16 h. The protoplasts were harvested, and the luminescent signals of LUC and REN were detected with the Dual-Glo Luciferase Assay System (Promega). The LUC expression levels were quantified as the ratio of LUC/REN enzyme activities.
Quantification and statistical analysis
For each related experiment, the number of replicates (n) and the p values are indicated in the figure legends or the results. Statistical significance was calculated using a two-tailed Student’s t-test or one-way ANOVA with Brown–Forsythe and Welch’s test (significance was set at p <0.05) with GraphPad Prism9, as indicated in the legends.
Funding
This study was supported by grants from the National Natural Science Foundation of China (31871221 and 31621001), the State Key Laboratory of Protein and Plant Gene Research, and the Peking-Tsinghua Center for Life Sciences (to X.W.D.).
Author contributions
G.H. and X.W.D. designed the project. M.X., J.L., A.J., and C.X. performed the phenotyping. M.X. performed other experiments. X.W. performed population genetic analyses, GWA mapping, and RNA-seq analysis. M.X., G.H., and X.W.D. prepared this manuscript.
Acknowledgments
The authors would like to thank Dr. Jian-Min Zhou for providing Pst DC3000 and Pst DC3000 carrying constructs related to the translocation assay, Dr. Xiu-Fang Xin for providing the T3SS-defective Pst DC3000 hrcC mutant, Dr. Robert Sablowski for providing rpl-1 and Ler plant seeds, Dr. Shangwei Zhong for providing chi and f3h plant seeds, and Dr. Jie Zhang and Dr. Jun-Jie Ling for helpful comments. No conflict of interest is declared.
Published: June 26, 2022
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Contributor Information
Xing Wang Deng, Email: deng@pku.edu.cn.
Guangming He, Email: heguangming@pku.edu.cn.
Accession numbers
TAIR and NCBI accession numbers for all genes examined in this study are listed in Supplemental Table 6.
Supplemental information
References
- Ballaré C.L., Austin A.T. Recalculating growth and defense strategies under competition: key roles of photoreceptors and jasmonates. J. Exp. Bot. 2019;70:3425–3434. doi: 10.1093/jxb/erz237. [DOI] [PubMed] [Google Scholar]
- Bencivenga S., Serrano-Mislata A., Bush M., Fox S., Sablowski R. Control of oriented tissue growth through repression of organ boundary genes promotes stem morphogenesis. Dev. Cell. 2016;39:198–208. doi: 10.1016/j.devcel.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanove A.J., Martin G.B. AvrPto-dependent Pto-interacting proteins and AvrPto-interacting proteins in tomato. Proc. Natl. Acad. Sci. USA. 2000;97:8836–8840. doi: 10.1073/pnas.97.16.8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.K.M. Yield penalties of disease resistance in crops. Curr. Opin. Plant Biol. 2002;5:339–344. doi: 10.1016/s1369-5266(02)00270-4. [DOI] [PubMed] [Google Scholar]
- Byrne M.E., Groover A.T., Fontana J.R., Martienssen R.A. Phyllotactic pattern and stem cell fate are determined by the Arabidopsis homeobox gene BEHRINGER. Development. 2003;130:3941–3950. doi: 10.1242/dev.00620. [DOI] [PubMed] [Google Scholar]
- Campos M.L., Yoshida Y., Major I.T., de Oliveira Ferreira D., Weraduwage S.M., Froehlich J.E., Johnson B.F., Kramer D.M., Jander G., Sharkey T.D., Howe G.A. Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat. Commun. 2016;7:12570. doi: 10.1038/ncomms12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao K., Zhou Z.K., Wang Q., Guo J., Zhao P., Zhu G.R., Fang W.C., Chen C.W., Wang X.W., Wang X.L., et al. Genome-wide association study of 12 agronomic traits in peach. Nat. Commun. 2016;7:13246. doi: 10.1038/ncomms13246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castañeda-Ojeda M.P., Moreno-Pérez A., Ramos C., López-Solanilla E. Suppression of plant immune responses by the Pseudomonas savastanoi pv. savastanoi NCPPB 3335 type III effector tyrosine phosphatases HopAO1 and HopAO2. Front. Plant Sci. 2017;8:680. doi: 10.3389/fpls.2017.00680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran D., Rickert J., Huang Y.X., Steinwand M.A., Marr S.K., Wildermuth M.C. Atypical E2F transcriptional repressor DEL1 acts at the intersection of plant growth and immunity by controlling the hormone salicylic acid. Cell Host Microbe. 2014;15:506–513. doi: 10.1016/j.chom.2014.03.007. [DOI] [PubMed] [Google Scholar]
- Coley P.D., Bryant J.P., Chapin F.S., 3rd Resource availability and plant antiherbivore defense. Science. 1985;230:895–899. doi: 10.1126/science.230.4728.895. [DOI] [PubMed] [Google Scholar]
- Crabill E., Joe A., Block A., van Rooyen J.M., Alfano J.R. Plant immunity directly or indirectly restricts the injection of type III effectors by the Pseudomonas syringae type III secretion system. Plant Physiol. 2010;154:233–244. doi: 10.1104/pp.110.159723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y.W., Zhai K.R., Xie Z., Yang D.Y., Zhu X.D., Liu J.Z., Wang X., Qin P., Yang Y.Z., Zhang G.M., et al. Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science. 2017;355:962–965. doi: 10.1126/science.aai8898. [DOI] [PubMed] [Google Scholar]
- Francisco M., Joseph B., Caligagan H., Li B., Corwin J.A., Lin C., Kerwin R.E., Burow M., Kliebenstein D.J. Genome wide association mapping in Arabidopsis thaliana identifies novel genes involved in linking allyl glucosinolate to altered biomass and defense. Front. Plant Sci. 2016;7:1010. doi: 10.3389/fpls.2016.01010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gomez L., Boller T. FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell. 2000;5:1003–1011. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- González-Lamothe R., El Oirdi M., Brisson N., Bouarab K. The conjugated auxin indole-3-acetic acid-aspartic acid promotes plant disease development. Plant Cell. 2012;24:762–777. doi: 10.1105/tpc.111.095190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant M.R., Godiard L., Straube E., Ashfield T., Lewald J., Sattler A., Innes R.W., Dangl J.L. Structure of the Arabidopsis Rpm1 gene enabling dual-specificity disease resistance. Science. 1995;269:843–846. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- Guo Q., Yoshida Y., Major I.T., Wang K., Sugimoto K., Kapali G., Havko N.E., Benning C., Howe G.A. JAZ repressors of metabolic defense promote growth and reproductive fitness in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2018;115:E10768–E10777. doi: 10.1073/pnas.1811828115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil M., Baldwin I.T. Fitness costs of induced resistance: emerging experimental support for a slippery concept. Trends Plant Sci. 2002;7:61–67. doi: 10.1016/s1360-1385(01)02186-0. [DOI] [PubMed] [Google Scholar]
- Hellens R.P., Allan A.C., Friel E.N., Bolitho K., Grafton K., Templeton M.D., Karunairetnam S., Gleave A.P., Laing W.A. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods. 2005;1:13. doi: 10.1186/1746-4811-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X.L., Lee L.Y.C., Xia K.F., Yan Y., Yen Y.Y., Yu H. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell. 2010;19:884–894. doi: 10.1016/j.devcel.2010.10.024. [DOI] [PubMed] [Google Scholar]
- Huot B., Yao J., Montgomery B.L., He S.Y. Growth-defense tradeoffs in plants: a balancing act to optimize fitness. Mol. Plant. 2014;7:1267–1287. doi: 10.1093/mp/ssu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D.G., Dangl J.L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Kazan K., Manners J.M. Linking development to defense: auxin in plant-pathogen interactions. Trends Plant Sci. 2009;14:373–382. doi: 10.1016/j.tplants.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Li J., Terzaghi W., Gong Y.Y., Li C.R., Ling J.J., Fan Y.Y., Qin N.X., Gong X.Q., Zhu D.M., Deng X.W. Modulation of BIN2 kinase activity by HY5 controls hypocotyl elongation in the light. Nat. Commun. 2020;11:1592. doi: 10.1038/s41467-020-15394-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.T., Zhu Z.W., Chern M.S., Yin J.J., Yang C., Ran L., Cheng M.P., He M., Wang K., Wang J., et al. A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell. 2017;170:114–126.e15. doi: 10.1016/j.cell.2017.06.008. [DOI] [PubMed] [Google Scholar]
- Lin R.C., Ding L., Casola C., Ripoll D.R., Feschotte C., Wang H.Y. Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science. 2007;318:1302–1305. doi: 10.1126/science.1146281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.M., Shi Z.Y., Zhang X.H., Wang M.X., Zhang L., Zheng K.Z., Liu J.Y., Hu X.M., Di C.R., Qian Q., et al. Inducible overexpression of Ideal Plant Architecture1 improves both yield and disease resistance in rice. Native Plants. 2019;5:389–400. doi: 10.1038/s41477-019-0383-2. [DOI] [PubMed] [Google Scholar]
- Mauricio R. Costs of resistance to natural enemies in field populations of the annual plant Arabidopsis thaliana. Am. Nat. 1998;151:20–28. doi: 10.1086/286099. [DOI] [PubMed] [Google Scholar]
- Mehrtens F., Kranz H., Bednarek P., Weisshaar B. The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 2005;138:1083–1096. doi: 10.1104/pp.104.058032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson R., Wiesner-Hanks T., Wisser R., Balint-Kurti P. Navigating complexity to breed disease-resistant crops. Nat. Rev. Genet. 2018;19:21–33. doi: 10.1038/nrg.2017.82. [DOI] [PubMed] [Google Scholar]
- Nicaise V., Joe A., Jeong B.R., Korneli C., Boutrot F., Westedt I., Staiger D., Alfano J.R., Zipfel C. Pseudomonas HopU1 modulates plant immune receptor levels by blocking the interaction of their mRNAs with GRP7. EMBO J. 2013;32:701–712. doi: 10.1038/emboj.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A., Misra P., Bhambhani S., Bhatia C., Trivedi P.K. Expression of Arabidopsis MYB transcription factor, AtMYB111, in tobacco requires light to modulate flavonol content. Sci. Rep. 2014;4:5018. doi: 10.1038/srep05018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse C.M.J., Van der Does D., Zamioudis C., Leon-Reyes A., Van Wees S.C.M. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012;28:489–521. doi: 10.1146/annurev-cellbio-092910-154055. [DOI] [PubMed] [Google Scholar]
- Rekhter D., Lüdke D., Ding Y.L., Feussner K., Zienkiewicz K., Lipka V., Wiermer M., Zhang Y.L., Feussner I. Isochorismate-derived biosynthesis of the plant stress hormone salicylic acid. Science. 2019;365:498–502. doi: 10.1126/science.aaw1720. [DOI] [PubMed] [Google Scholar]
- Ren D., Wang X., Yang M., Yang L., He G., Deng X.W. A new regulator of seed size control in Arabidopsis identified by a genome-wide association study. New Phytol. 2019;222:895–906. doi: 10.1111/nph.15642. [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A., Grant M., Jones J.D.G. Hormone crosstalk in plant disease and defense: more than just JASMONATE-SALICYLATE antagonism. Annu. Rev. Phytopathol. 2011;49:317–343. doi: 10.1146/annurev-phyto-073009-114447. [DOI] [PubMed] [Google Scholar]
- Roeder A.H.K., Ferrándiz C., Ferrandiz C., Yanofsky M.F. The role of the REPLEVIESS homeodomain protein in patterning the Arabidopsis fruit. Curr. Biol. 2003;13:1630–1635. doi: 10.1016/j.cub.2003.08.027. [DOI] [PubMed] [Google Scholar]
- Sestari I., Campos M.L. Into a dilemma of plants: the antagonism between chemical defenses and growth. Plant Mol. Biol. 2021;109:469–482. doi: 10.1007/s11103-021-01213-0. [DOI] [PubMed] [Google Scholar]
- Sharma A., Badola P.K., Bhatia C., Sharma D., Trivedi P.K. Primary transcript of miR858 encodes regulatory peptide and controls flavonoid biosynthesis and development in Arabidopsis. Native Plants. 2020;6:1262–1274. doi: 10.1038/s41477-020-00769-x. [DOI] [PubMed] [Google Scholar]
- Smith H.M.S., Hake S. The interaction of two homeobox genes, BREVIPEDICELLATUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. Plant Cell. 2003;15:1717–1727. doi: 10.1105/tpc.012856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel S.H., Dong X.N. Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe. 2008;3:348–351. doi: 10.1016/j.chom.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Staswick P.E., Serban B., Rowe M., Tiryaki I., Maldonado M.T., Maldonado M.C., Suza W. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell. 2005;17:616–627. doi: 10.1105/tpc.104.026690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K., Zager J.J., Aubin B.S., Lange B.M., Howe G.A. Flavonoid deficiency disrupts redox homeostasis and terpenoid biosynthesis in glandular trichomes of tomato. Plant Physiol. 2022;188:1450–1468. doi: 10.1093/plphys/kiab488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D., Traw M.B., Chen J.Q., Kreitman M., Bergelson J. Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature. 2003;423:74–77. doi: 10.1038/nature01588. [DOI] [PubMed] [Google Scholar]
- Todesco M., Balasubramanian S., Hu T.T., Traw M.B., Horton M., Epple P., Kuhns C., Sureshkumar S., Schwartz C., Lanz C., et al. Natural allelic variation underlying a major fitness trade-off in Arabidopsis thaliana. Nature. 2010;465:632–636. doi: 10.1038/nature09083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrens-Spence M.P., Bobokalonova A., Carballo V., Glinkerman C.M., Pluskal T., Shen A., Weng J.K. PBS3 and EPS1 complete salicylic acid biosynthesis from iIsochorismate in Arabidopsis. Mol. Plant. 2019;12:1577–1586. doi: 10.1016/j.molp.2019.11.005. [DOI] [PubMed] [Google Scholar]
- Wang D., Pajerowska-Mukhtar K., Culler A.H., Dong X.N. Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 2007;17:1784–1790. doi: 10.1016/j.cub.2007.09.025. [DOI] [PubMed] [Google Scholar]
- Wang J., Zhou L., Shi H., Chern M., Yu H., Yi H., He M., Yin J.J., Zhu X.B., Li Y., et al. A single transcription factor promotes both yield and immunity in rice. Science. 2018;361:1026–1028. doi: 10.1126/science.aat7675. [DOI] [PubMed] [Google Scholar]
- Xu G.Y., Uan M.Y., Ai C.R., Liu L.J., Zhuang E., Karapetyan S., Wang S., Dong X.N. uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature. 2017;545:491–494. doi: 10.1038/nature22372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Stolz S., Chetelat A., Reymond P., Pagni M., Dubugnon L., Farmer E.E. A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell. 2007;19:2470–2483. doi: 10.1105/tpc.107.050708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D.L., Yao J., Mei C.S., Tong X.H., Zeng L.J., Li Q., Xiao L.T., Sun T.P., Li J.G., Deng X.W., et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA. 2012;109:1192–1200. doi: 10.1073/pnas.1201616109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Stephens M. Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nat. Methods. 2014;11:407–409. doi: 10.1038/nmeth.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J.D.G., Boller T., Felix G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.