Abstract
This study reports the identification of the rice open reading frame Semi-Dwarf in chr8 (SD8) that encodes a putative ortholog of Arabidopsis thaliana ABCB1. Genome editing of SD8 leads to optimized rice architecture by reducing plant height and flag-leaf angle without yield penalty. Rice SD8 knockouts may also have the potential for increased yield under high density planting.
Keywords: ABCB1, SD8, genome editing, dwarf, leaf angle, rice
Dear editor,
In the 1960s, the use of semi-dwarf rice and wheat varieties ushered in the “Green Revolution,” leading to reduced lodging and increased harvest index. In rice, essentially all modern semi-dwarf varieties carry a specific null mutation or weak alleles of Semi-Dwarf1 (SD1), which encodes a GA20-2 oxidase in the gibberellin biosynthetic pathway (Monna et al., 2002; Sasaki et al., 2002; Spielmeyer et al., 2002). In addition to gibberellins, other plant hormones such as brassinosteroids, strigolactones, and auxin also function in reducing rice height (Ferrero-Serrano et al., 2019). However, many dwarf or semi-dwarf mutants have not been widely used in rice-breeding programs because they adversely impact grain yield (Ferrero-Serrano et al., 2019). Moreover, the flag leaf has a higher photosynthetic capacity than lower canopy leaves, which allows for greater interception of light. Rice yield is closely related to the flag leaf because it contributes about 50% of the assimilates used to fill the grain with starch (Dong et al., 2018). Crops with erect flag leaves can grow at higher plant densities without compensatory reductions in photosynthesis, leading to increased grain yield. Therefore, dwarfing and leaf erectness have been breeding targets for several decades, as components of ideal plant architecture. Identification of genes that moderately reduce rice height (semi-dwarfing) and optimize rice architecture without yield penalty is still highly desirable.
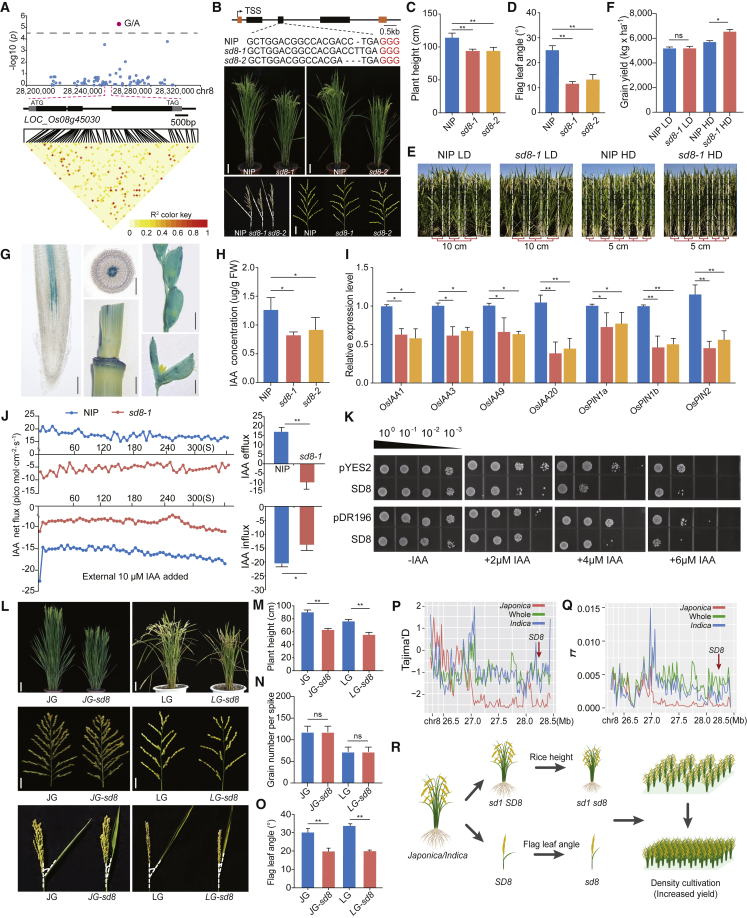
Using data from a previous genome-wide association study, we analyzed single-nucleotide polymorphisms (SNPs) associated with rice height in the 3000 rice genomes dataset (Alexandrov et al., 2015; Wang et al., 2018) and successfully identified one predicted open reading frame, Semi-Dwarf in chr8 (SD8, LOC_Os08g45030), in a 50-kb interval (28,270,000–28,280,000) of chromosome 8 (Figure 1A). Through phylogenetic analysis, we found that SD8 encodes a putative ortholog of Arabidopsis thaliana ATP Binding Cassette B1 (ABCB1)/P-glycoprotein1 (Noh et al., 2003; Geisler et al., 2005). To investigate the biological functions of SD8 in rice, we used CRISPR-Cas9-mediated gene editing to obtain two knockout (KO) lines in the Nipponbare (NIP) background (Figure 1B). Phenotypically, sd8-1 (one-bp insertion mutant) and sd8-2 (two-bp deletion mutant) plants had moderately reduced height due to shorter internode lengths, as well as a smaller flag-leaf angle, and thus displayed optimized plant architecture (Figures 1B–1D; supplemental Figure 1). sd8 mutant phenotypes could be rescued in transgenic complementation lines (supplemental Figure 2). Notably, there were no significant phenotypic differences between NIP and the two sd8 mutants in seven yield-related traits (supplemental Figures 2E–2I and 3). Because of the desirable possibility that the combination of semi-dwarf height and leaf angle in sd8 could increase production yields under dense planting, we investigated yields of NIP and sd8-1 in paddy-field plots at two planting densities. In the high-density plots (560,000 plants/ha), sd8-1 mutants and NIP plants showed yield increases of ∼20.6% and ∼10%, respectively, compared with those grown in low-density plots. There was no significant difference in yield between genotypes grown in the low-density plots (280,000 plants/ha) (Figures 1E and 1F; supplemental Figure 4). Collectively, these data revealed that loss of SD8 function could optimize rice architecture by reducing plant height and flag-leaf angle without yield penalty and that SD8 KOs may even have the potential for increased yield under high-density planting.
Figure 1.
SD8 knockouts showed reduced plant height and flag-leaf angle without yield penalty.
(A) Identification of a putative open reading frame on chromosome 8 (LOC_08g45030) associated with plant height based on re-analysis of SNPs in the 3000 rice genomes dataset (Alexandrov et al., 2015).
(B) Gross phenotypes of SD8 KO lines in the Nipponbare (NIP) background obtained using CRISPR-Cas9 gene editing. Top panel: mutation sites in the two knockout lines (sd8-1 and sd8-2). Bottom panels: height, flag-leaf angle, and panicle morphology in NIP, sd8-1, and sd8-2 plants.
(C and D) Comparison of plant height (C) and flag-leaf angle (D) between NIP, sd8-1, and sd8-2 plants.
(E) Representative NIP and sd8-1 plants grown under different planting densities. Seeds from NIP and sd8-1 were grown at high (5 × 20 cm) and low (10 × 20 cm) planting densities.
(F) Grain yield of NIP and sd8-1 plants grown at high and low planting densities. ns, not significant. ∗p < 0.01 (Student’s t-test).
(G) Glucuronidase staining in roots of 7-day-old seedlings, internodes at the early heading stage, glumes at the early heading stage, and glumes at the late heading stage. Scale bars: 1 mm.
(H) Gas chromatography–mass spectroscopy analysis of endogenous free IAA concentrations in NIP, sd8-1, and sd8-2 seedlings.
(I) Relative expression levels of OsIAA1/3/9/20 and OsPIN1a/1b/2 in aerial tissues of 3-week-old NIP, sd8-1, and sd8-2 seedlings.
(J) Time course analysis of IAA efflux and net influx in the primary root meristem of 7-day-old NIP and sd8-1 seedlings as measured continuously for 5 min by the scanning ion-selective electrode technique. IAA influx was measured in the presence of 10 μM exogenous IAA. Columns represent the mean net influx rates averaged over the entire 5-min observation window (±SE, n = 6–10 plants). ∗p < 0.05 (one-way analysis of variance).
(K) SD8 functionality assays for auxin acquisition in the IAA-sensitive yeast strain yap1-1. The growth status is shown for yap1-1 cells expressing empty vectors (pYES2 and pDR196) and SD8 on SD-U medium without uracil supplemented with 2, 3, 4, or 6 μM IAA. Serial dilutions (1:10) of yeast cells were spotted onto SD-U solid medium containing 2% galactose or glucose, then incubated at 30°C for 4 to 6 days.
(L) Gross phenotypes of SD8 KO lines in the Jingeng818 and Longgeng31 backgrounds.
(M–O) Quantitative analysis of plant height and flag-leaf angle in wild-type and SD8 KO lines in the Jingeng818 and Longgeng31 backgrounds.
(P and Q) Tajima’s D and nucleotide diversity (π) values for a ∼2-Mb genomic region flanking SD8 in the 3000 rice genomes dataset.
(R) A model for loss of SD8 function with and without SD1 in which plant height is reduced but yield is increased under high-density planting.
Consistent with sd8 mutant phenotypes, β-glucuronidase reporter assays and quantitative real-time PCR indicated that SD8 was primarily expressed in the internode (Figure 1G; supplemental Figure 5A). SD8 also showed differences in expression among seven japonica and indica cultivars (supplemental Figure 5B). We observed that SD8 was localized in the plasma membrane (supplemental Figure 5C). In plants, ABCB1 homologs are known to mediate cellular efflux of indole-3-acetic acid (IAA) and to regulate polar auxin transport (Multani et al., 2003; Noh et al., 2003; Geisler et al., 2005). We therefore measured the endogenous IAA content in NIP, sd8-1, and sd8-2 seedlings. IAA levels were significantly lower in sd8 than in NIP seedlings (Figure 1H). Moreover, we found that the shortened plant height and reduced leaf-angle phenotypes of sd8 mutants could be rescued by applying exogenous IAA (supplemental Figure 6). Consistent with the observed reduction in auxin concentration, sd8 mutants had reduced expression of genes in the auxin signaling pathway, including OsPIN1a/1b/2 and OsIAA3/9/20 (Figure 1I).
To further investigate whether SD8 modulated auxin transport in rice, we measured IAA flux speed in NIP and sd8-1 seedlings. IAA efflux and influx currents were significantly lower in sd8-1 than in NIP, both with and without IAA treatment (Figure 1J), suggesting that loss of SD8 function affected IAA flux currents. In addition, we used a previously reported assay to measure auxin acquisition in the IAA-sensitive yeast strain yap1-1 (Yang et al., 2020) and found that SD8 indeed promoted IAA accumulation in yeast, resulting in a stronger suppression of IAA-induced growth (Figure 1K).
To determine whether SD8 had similar biological functions and loss-of-function mutant phenotypes in diverse rice varieties, we created CRISPR-Cas9-edited SD8 KO mutants in two key elite cultivars in the japonica background, Jingeng818 (JG) and Longgeng31 (LG). Similar to the SD8 KO plants in the NIP background, we observed a remarkable decrease in height and flag-leaf angle in JG-sd8 and LG-sd8 but no differences in the examined yield-related traits (Figures 1L–1O; supplemental Figure 7). We detected a considerable decrease in IAA content in these KO lines, and auxin-responsive gene expression was reduced in JG-sd8 and LG-sd8 (supplemental Figure 8). We also knocked out SD8 in the indica rice cultivars 93-11, YexiangB (YX), Nongxiang32, and Yuzhenxiang. Similar to the SD8 KO plants in the japonica background (NIP, JG, and LG), the KO lines in indica backgrounds also exhibited semi-dwarf phenotypes (Figure S9) and significant decreases in IAA content (Figure S10). Together, these results showed that loss of SD8 function in different backgrounds could indeed reduce rice height and flag-leaf angle, suggesting an essential role for SD8 in the optimization of rice architecture.
Analyses of SNPs and haplotypes (Haps) have become a major strategy for understanding evolutionary relationships and phenotypic variations, and these methods have breeding applications in rice (Wang et al., 2018). In the 3000 rice genomes dataset (Alexandrov et al., 2015), we identified 14 Haps using 16 SNPs in SD8 (supplemental Figure 11A). The Hap frequencies differed significantly between the indica and japonica subspecies (supplemental Figures 11B and 11C). Next, we revealed significant differences in rice height among the top five most frequent Haps; Hap4 showed a significantly lower mean height than the other four Haps (supplemental Figure 11D). Based on Hap frequencies in SD8, we found that the japonica population had significantly higher Tajima’s D and π (nucleotide diversity) values than the indica population in the ∼2-Mb region flanking SD8 (Figures 1P and 1Q). These data indicated that SD8 has undergone strong balancing selection in the japonica subpopulation, suggesting that there is considerable potential for using SD8 to balance increased productivity and reduced height.
The discovery of the semi-dwarfism gene SD1 enabled the introduction of dwarfism to breeding programs in the 1960s, a major scientific advance for the rice Green Revolution. SD1 has undergone significant artificial selection in japonica and indica rice cultivars (Asano et al., 2011), and different mutant alleles of SD1 have been used separately for rice breeding in the japonica or indica background to produce semi-dwarf cultivars. The indica cultivars contain loss-of-function SD1 mutations, and the japonica cultivars contain weak alleles. Based on previous reports, japonica cultivars NIP and LG contain weak sd1 mutant alleles (sd1-EQ type), and indica cultivars YX and 9311 are considered to contain loss-of-function mutant alleles (sd1-d allele type for YX, and dwarf sd1-9311 type for 9311) (Asano et al., 2011; Wu et al., 2018). Despite the many advantages of sd1 as a source of dwarfism and lodging resistance, its widespread use has been revealed to have other associated negative effects. It has been reported that mutation of SD1 has negative effects on spikelet number per panicle, panicle length, and branch number, eventually resulting in reduced yield (Murai et al., 2002; Su et al., 2021). Like mutation of SD1 in modern indica and japonica cultivars, we propose that genome editing of SD8 may have similar potential for reducing the height of indica and japonica rice cultivars (Figure 1Q). In addition, sd8 mutation could also reduce flag-leaf angle without yield penalty. Thus, we believe that SD8 could be an alternative dwarfing gene for rice-breeding programs and that it has more potential to further reduce rice height or even increase yield under high-density planting. The application of sophisticated genome-editing technology to SD8 enabled us to develop sustainable rice varieties with optimized architecture and without yield penalty. This approach has the potential to revolutionize direct-seeding strategies for green-agriculture cultivation of rice.
The Arabidopsis abcb1 mutant does not show a dwarf phenotype, in contrast to the semi-dwarf and reduced flag-leaf-angle phenotypes of sd8 in rice (Noh et al., 2003; Geisler et al., 2005). Although mutation of ABCB1 homologs in the monocots maize (br2) and sorghum (dw3) causes a severe dwarf phenotype, grain yield is also severely reduced (Multani et al., 2003), which may hinder the application of ABCB1 homologs to the breeding of semi-dwarf plants. It will be crucial to determine whether ABCB1 homologs have conserved functional effects on auxin transport but different KO phenotypes in various plant lineages.
Funding
This work was supported by grants from the National Natural Science Foundation of China (32130080) to X.G., the Central Public-interest Scientific Institution Basal Research Fund (Y2022PT22) to X.G., the Agricultural Science and Technology Innovation Program (CAAS-ZDRW202109), the Hainan Yazhou Bay Seed Laboratory (B21HJ0223) to X.G., and the Key Technologies R & D Program of Tianjin (18YFZCNC01210) to S.Y.
Author contributions
X.G., S.Y., and L.Y. supervised the project. R.Q., P.Z., and Q.L. performed most of the experiments. Y.W. and W.G. analyzed the data. Z.D. and X.L. assisted with the experiments. P.Z. and X.G. wrote the manuscript. P.Z., R.Q., and X.G. revised the manuscript. All authors read and approved the final manuscript.
Acknowledgments
No conflict of interest is declared.
Published: June 10, 2022
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Contributor Information
Liwen Yang, Email: yangliwen@caas.cn.
Shuangyong Yan, Email: bioysy@139.com.
Xiaofeng Gu, Email: guxiaofeng@caas.cn.
Supplemental information
References
- Alexandrov N., Tai S., Wang W., Mansueto L., Palis K., Fuentes R.R., Ulat V.J., Chebotarov D., Zhang G., Li Z., et al. SNP-Seek database of SNPs derived from 3000 rice genomes. Nucleic Acids Res. 2015;43:D1023–D1027. doi: 10.1093/nar/gku1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K., Yamasaki M., Takuno S., Miura K., Katagiri S., Ito T., Doi K., Wu J., Ebana K., Matsumoto T., et al. Artificial selection for a green revolution gene during japonica rice domestication. Proc. Natl. Acad. Sci. U S A. 2011;108:11034–11039. doi: 10.1073/pnas.1019490108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H.J., Zhao H., Li S.L., Han Z.M., Hu G., Liu C., Yang G.Y., Wang G.W., Xie W.B., Xing Y.Z. Genome-wide association studies reveal that members of bHLH subfamily 16 share a conserved function in regulating flag leaf angle in rice (Oryza sativa) PLoS Genet. 2018;14:e1007323. doi: 10.1371/journal.pgen.1007323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrero-Serrano Á., Cantos C., Assmann S.M. The role of dwarfing traits in historical and modern agriculture with a focus on rice. Cold Spring Harb. Perspect. Biol. 2019;11:a034645. doi: 10.1101/cshperspect.a034645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M., Blakeslee J.J., Bouchard R., Lee O.R., Vincenzetti V., Bandyopadhyay A., Titapiwatanakun B., Peer W.A., Bailly A., Richards E.L., et al. Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J. 2005;44:179–194. doi: 10.1111/j.1365-313x.2005.02519.x. [DOI] [PubMed] [Google Scholar]
- Multani D.S., Briggs S.P., Chamberlin M.A., Blakeslee J.J., Murphy A.S., Johal G.S. Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science. 2003;302:81–84. doi: 10.1126/science.1086072. [DOI] [PubMed] [Google Scholar]
- Monna L., Kitazawa N., Yoshino R., Suzuki J., Masuda H., Maehara Y., Tanji M., Sato M., Nasu S., Minobe Y. Positional cloning of rice semidwarfing gene, sd-1: rice "green revolution gene" encodes a mutant enzyme involved in gibberellin synthesis. DNA Res. 2002;9:11–17. doi: 10.1093/dnares/9.1.11. [DOI] [PubMed] [Google Scholar]
- Murai M., Takamure I., Sato S., Tokutome T., Sato Y. Effects of the dwarfing gene originating from 'Dee-geo-woo-gen' on yield and its related traits in rice. Breed Sci. 2002;52:95–100. doi: 10.1270/jsbbs.52.95. [DOI] [Google Scholar]
- Noh B., Bandyopadhyay A., Peer W.A., Spalding E.P., Murphy A.S. Enhanced gravi- and phototropism in plant mdr mutants mislocalizing the auxin efflux protein PIN1. Nature. 2003;423:999–1002. doi: 10.1038/nature01716. [DOI] [PubMed] [Google Scholar]
- Spielmeyer W., Ellis M.H., Chandler P.M. Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl. Acad. Sci. U S A. 2002;99:9043–9048. doi: 10.1073/pnas.132266399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A., Ashikari M., Ueguchi-Tanaka M., Itoh H., Nishimura A., Swapan D., Ishiyama K., Saito T., Kobayashi M., Khush G.S., et al. Green revolution: a mutant gibberellin-synthesis gene in rice. Nature. 2002;416:701–702. doi: 10.1038/416701a. [DOI] [PubMed] [Google Scholar]
- Su S., Hong J., Chen X., Zhang C., Chen M., Luo Z., Chang S., Bai S., Liang W., Liu Q., et al. Gibberellins orchestrate panicle architecture mediated by DELLA-KNOX signaling in rice. Plant Biotechnol. J. 2021;19:2304–2318. doi: 10.1111/pbi.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Mauleon R., Hu Z., Chebotarov D., Tai S., Wu Z., Li M., Zheng T., Fuentes R.R., Zhang F., et al. Genomic variation in 3, 010 diverse accessions of Asian cultivated rice. Nature. 2018;557:43–49. doi: 10.1038/s41586-018-0063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Tang D., Liu K., Miao C., Zhuo X., Li Y., Tan X., Sun M., Luo Q., Cheng Z. Characterization of a new semi-dominant dwarf allele of SLR1 and its potential application in hybrid rice breeding. J. Exp. Bot. 2018;69:4703–4713. doi: 10.1093/jxb/ery243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Feng H., Zhang S., Xiao H., Hu Q., Chen G., Xuan W., Moran N., Murphy A., Yu L., et al. The potassium transporter OsHAK5 alters rice architecture via ATP-dependent transmembrane auxin fluxes. Plant Commun. 2020;1:100052. doi: 10.1016/j.xplc.2020.100052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.