Abstract
Coenzyme Q (CoQ) is a conserved redox-active lipid that has a wide distribution across the domains of life. CoQ plays a key role in the oxidative electron transfer chain and serves as a crucial antioxidant in cellular membranes. Our understanding of CoQ biosynthesis in eukaryotes has come mostly from studies of yeast. Recently, significant advances have been made in understanding CoQ biosynthesis in plants. Unique mitochondrial flavin-dependent monooxygenase and benzenoid ring precursor biosynthetic pathways have been discovered, providing new insights into the diversity of CoQ biosynthetic pathways and the evolution of phototrophic eukaryotes. We summarize research progress on CoQ biosynthesis and regulation in plants and recent efforts to increase the CoQ content in plant foods.
Keywords: coenzyme Q, 4-hydroxybenzoic acid, mitochondria, biofortification, plant metabolism
Coenzyme Q (CoQ) is an essential electron-carrying lipid that functions in oxidative phosphorylation to generate adenosine triphosphate (ATP) and also serves as a lipophilic antioxidant in cellular membranes. Recent investigations have provided new insights into the biosynthetic routes of its aromatic precursor and have identified a unique hydroxylase that catalyzes the penultimate step of plant CoQ biosynthesis.
Introduction
Coenzyme Q (CoQ), also known as ubiquinone, is an essential electron transporter in the oxidative respiratory chain that generates adenosine triphosphate (ATP). CoQ is synthesized by nearly all eukaryotes and some proteobacteria. Structurally, CoQ is composed of a benzoquinone head group attached to a polyisoprenoid tail whose number of isoprene units varies among species: 10 (CoQ10) in humans and some crops (such as tomato and soybean), CoQ9 in Arabidopsis thaliana and rice, CoQ8 in Escherichia coli, and CoQ6 in yeast (Saccharomyces cerevisiae). The quinone head group of CoQ can exist in three oxidation states: the fully oxidized form (CoQ, ubiquinone), the semi-oxidized form with one electron (CoQH·, ubisemiquinone), and the fully reduced form (CoQH2, ubiquinol).
In eukaryotes, CoQ is a central component in mitochondrial oxidative phosphorylation, mediating the electron transfer from complex I (NADH: ubiquinone oxidoreductase) and II (succinate dehydrogenase) to complex III (cytochrome bc1 oxidoreductase). CoQ also serves as the electron acceptor for several other mitochondrial inner-membrane dehydrogenases (Banerjee et al., 2021) involved in pyrimidine biosynthesis (Evans and Guy, 2004), sulfide detoxification (Zhang et al., 2008; Ziosi et al., 2017), fatty acid β-oxidation, branched-chain amino acid oxidation (Watmough and Frerman, 2010), and so on. CoQ is also a lipid-soluble antioxidant in all cellular compartments (Baschiera et al., 2021). Recently, CoQ was found to be a cofactor of ferroptosis suppressor protein 1, which reduces CoQ to CoQH2 to suppress ferroptosis (Bersuker et al., 2019; Doll et al., 2019). Because it is essential to human health and important in disease prevention and recovery (Cirilli et al., 2021), CoQ is among the most widely consumed dietary supplements (Arenas-Jal et al., 2020).
Biochemical characterizations of CoQ biosynthetic enzymes in eukaryotes have been focused on the yeast model S. cerevisiae. In most cases, human functional orthologs were identified through restoration of the respective yeast mutants. Progress in the characterization of CoQ biosynthesis in yeast, humans, and prokaryotes has been reviewed recently (Stefely and Pagliarini, 2017; Awad et al., 2018; Wang and Hekimi, 2019; Abby et al., 2020). The S. cerevisiae and human genes required for CoQ biosynthesis are written in capital letters. The corresponding proteins in humans are in uppercase letters, whereas in S. cerevisiae, they are written with only the first letter capitalized. We capitalize only the first letter for both genes and proteins of plants in this review: for example, human gene COQ3, human protein COQ3, yeast gene COQ3, yeast protein Coq3, Arabidopsis gene Coq3, and Arabidopsis protein Coq3. The prokaryotic proteins of CoQ biosynthesis are typically named with a prefix of “Ubi.”
The CoQ biosynthetic pathway can be divided into three parts: formation of the aromatic ring precursor, biosynthesis of the polyisoprenoid tail, and modifications of the aromatic ring (Figure 1). Although the eukaryotic CoQ biosynthetic pathway has not been fully defined to date, some components have been found to be conserved across fungi, metazoans, and plants (Toda et al., 2014). Most plants are photo-autotrophic; although photophosphorylation in the chloroplast is the major source of ATP supply, the mitochondrial oxidative respiratory chain is indispensable for plant survival. In recent years, plants have been reported to have special routes for generating the head group and a unique enzyme in the terminal stage. Here, we outline the CoQ biosynthetic pathway in plants and summarize progress in plant CoQ enhancement.
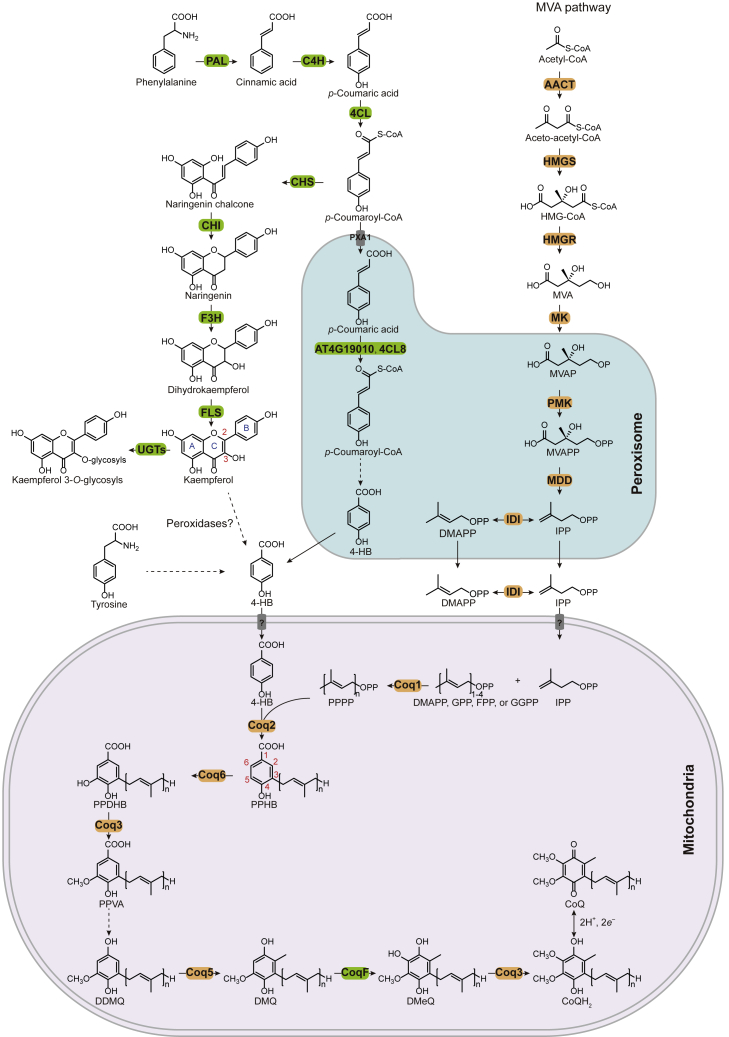
Figure 1.
The plant CoQ biosynthetic pathway.
The plant-specific enzymes are shown in a green background. The B-ring of kaempferol contributes to the 4-HB pool. For 4-HB biosynthesis, 4-HB, 4-hydroxybenzoic acid; PAL, phenylalanine ammonia-lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; FLS, flavonol synthase; PXA1, peroxisomal ABC transporter one; and UGTs, uridine diphosphate glycosyltransferases. For the MVA pathway, HMG-CoA, 3-hydroxy-3-methylglutaryl-CoA; MVA, mevalonate; MVAP, mevalonate 5-phosphate; MVAPP, mevalonate diphosphate; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; AACT, acetoacetyl-CoA thiolase; HMGS, 3-hydroxy-3-methylglutaryl-CoA synthase; HMGR, 3-hydroxy-3-methylglutaryl-CoA reductase; MK, mevalonate kinase; PMK, phosphomevalonate kinase; MDD, mevalonate diphosphate decarboxylase; and IDI, isopentenyl diphosphate isomerase. For the CoQ pathway in mitochondria, GPP, geranyl diphosphate; FPP, farnesyl diphosphate; GGPP, geranylgeranyl diphosphate; PPPP, polyprenyl-pyrophosphate; PPHB, polyprenyl-hydroxybenzoate; PPDHB, polyprenyl-dihydroxybenzoate; PPVA, polyprenyl-vanillic acid; DDMQ, demethoxy-demethyl-coenzyme Q; DMQ, demethoxy-coenzyme Q; and DMeQ, demethyl-coenzyme Q.
Aromatic ring precursor biosynthesis
4-hydroxybenzoic acid (4-HB) is the classic benzene quinone ring precursor of CoQ in eukaryotes and prokaryotes. The origins of 4-HB in different species have been reviewed recently (Fernandez-Del-Rio and Clarke, 2021). Here, we discuss 4-HB biosynthetic pathways in plants.
Plants synthesize all proteinogenic amino acids themselves. In plants, 4-HB is derived from either L-phenylalanine or L-tyrosine via independent pathways. Investigations using 13C-labeled compounds demonstrated that phenylalanine is a 2- to 5-fold better precursor than tyrosine in Arabidopsis (Block et al., 2014; Soubeyrand et al., 2019). Phenylalanine is deaminated by phenylalanine ammonia-lyase (PAL) and hydroxylated by cinnamate 4-hydroxylase (C4H), producing p-coumaric acid in the cytosol (Table 1). The knockout mutant of C4H appears to be unable to convert phenylalanine into CoQ (Soubeyrand et al., 2018). Two routes have been shown to convert p-coumaric acid into 4-HB. The predominant route is the β-oxidative metabolism of p-coumaric acid, unique to plants, in which p-coumaric acid is imported into peroxisomes by peroxisomal ATP-binding cassette transporter 1 (PXA1), which also transports other substrates for β-oxidation, including fatty acids and a wide range of bioactive molecules (Bussell et al., 2014; Theodoulou et al., 2005; Zolman et al., 2001). PXA1 has fatty acyl-coenzyme A (CoA) thioesterase activity (De Marcos Lousa et al., 2013), suggesting that the substrates of PXA1 are transported as CoA esters. In peroxisomes, 4-coumarate CoA ligases (AT4G19010 and 4CL8 in Arabidopsis) catalyze the formation of p-coumaroyl-CoA, initiating β-oxidative side chain shortening. The subsequent steps have been proposed to be similar to the shortening of cinnamoyl-CoA to benzoic acid, involving cinnamoyl-CoA hydratase and dehydrogenase and 3-ketoacyl-CoA thiolase (Widhalm and Dudareva, 2015). However, experimental evidence that these enzymes are responsible for the production of 4-HB is lacking. Loss of function of PXA1 or AT4G19010 resulted in a 55%–65% decrease in CoQ content in Arabidopsis (Block et al., 2014), suggesting that β-oxidation of p-coumaric acid is the dominant route (Figure 1).
Table 1.
Genes involved in CoQ biosynthesis in Arabidopsis.
| Gene | AGI | Protein | Function | References |
|---|---|---|---|---|
| C4H | AT2G30490 | cinnamate 4-hydroxylase | 4-HB biosynthesis from phenylalanine | (Block et al., 2014) |
| PXA1 | AT4G39850 | peroxisomal ABC transporter 1 | (Block et al., 2014) | |
| AT4G19010 | AT4G19010 | peroxisomal 4-coumarate CoA ligase | 4-HB biosynthesis from phenylalanine | (Block et al., 2014) |
| 4CL8 | AT5G38120 | peroxisomal 4-coumarate CoA ligase | 4-HB biosynthesis from phenylalanine | (Soubeyrand et al., 2019) |
| 4CL3 | AT1G65060 | cytosolic 4-coumarate CoA ligase | 4-HB biosynthesis from phenylalanine | (Soubeyrand et al., 2018) |
| CHS | AT5G13930 | chalcone synthase | 4-HB biosynthesis from phenylalanine | (Soubeyrand et al., 2018) |
| F3H | AT3G51240 | flavanone 3-hydroxylase | 4-HB biosynthesis from phenylalanine | (Soubeyrand et al., 2018) |
| IDI1 | AT5G16440 | isopentenyl diphosphate isomerase | isoprenoid biosynthesis | (Okada et al., 2008; Phillips et al., 2008) |
| IDI2 | AT3G02780 | isopentenyl diphosphate isomerase | isoprenoid biosynthesis | (Okada et al., 2008; Phillips et al., 2008) |
| FPS1 | AT5G47770 | farnesyl diphosphate synthase | isoprenoid biosynthesis | (Closa et al., 2010; Manzano et al., 2016) |
| FPS2 | AT4G17190 | farnesyl diphosphate synthase | isoprenoid biosynthesis | (Closa et al., 2010; Manzano et al., 2016) |
| Coq1 | AT2G34630 | polyprenyl diphosphate synthase | isoprene polymerization | (Ducluzeau et al., 2012) |
| Coq2 | AT4G23660 | 4-hydroxybenzoate polyprenyl diphosphate transferase | C3 prenylation | (Okada et al., 2004) |
| Coq3 | AT2G30920 | SAM-dependent methyltransferase | O methylations | (Avelange-Macherel and Joyard, 1998) |
| Coq5 | AT5G57300 | SAM-dependent methyltransferase | C2 methylation | (Toda et al., 2014) |
| Coq6 | AT3G24200 | flavin-dependent monooxygenase | C5 hydroxylation | (Latimer et al., 2021) |
| CoqF | AT1G24340 | flavin-dependent monooxygenase | C6 hydroxylation | (Latimer et al., 2021) |
| Coq4 | AT2G03690 | scaffold protein? | (Toda et al., 2014) | |
| Coq8 | AT4G01660 | ATPase | (Toda et al., 2014) | |
| Coq9a | AT1G19140 | isoprene lipid-binding protein | (Toda et al., 2014) | |
| Coq11Aa | AT5G10730 | atypical short chain dehydrogenase and reductase | (Xu et al., 2021) | |
| Coq11Ba | AT5G15910 | atypical short chain dehydrogenase and reductase | (Xu et al., 2021) |
Putative, lacking experimental supporting data.
In another pathway, p-coumaric acid enters flavonoid biosynthesis in the cytosol, as covered and specifically discussed in a recent review (Berger et al., 2022). Genetic analyses and isotopic labeling experiments showed that 4-HB can be derived from kaempferol. First, 4CL3 catalyzes the formation of p-coumaroyl-CoA. Chalcone synthase (CHS), the first committed enzyme in flavonoid biosynthesis, carries out the condensation of three malonyl-CoA with p-coumaroyl-CoA, leading to the formation of naringenin chalcone, which is converted to kaempferol via the action of chalcone isomerase, flavanone 3-hydroxylase (F3H), and flavonol synthase. Knockout mutants of 4CL3, CHS, and F3H displayed a 15%–25% reduction in CoQ content. Then, the B-ring of kaempferol is proposed to be cleaved by heme-dependent peroxidases to generate 4-HB. This reaction requires a free hydroxyl group on C3 as well as a C2–C3 double bond in the C-ring, as naringenin, dihydrokaempferol, and kaempferol 3-β-D-glucopyranoside failed to release 4-HB in vitro. The candidate peroxidases have not been identified but are likely to be heme peroxidases because the activity was abolished by azide (Soubeyrand et al., 2018).
In plants, tyrosine also gives rise to 4-HB, but the steps and responsible enzymes have not been identified. Although tyrosine serves as the source of 4-HB in yeast and mammals, the tyrosine-to-4-HB pathway has not been fully defined in any eukaryotes. The first and last reactions in yeast have been characterized (Payet et al., 2016; Stefely et al., 2016). First, the deamination of tyrosine to 4-hydroxyphenylpyruvate (4-HPP) is catalyzed by five redundant aminotransferases (Aro8, Aro9, Bat2, Bna3, and Aat2) (Robinson et al., 2021), and the final step is achieved by aldehyde dehydrogenase Hfd1, which oxidizes 4-hydroxybenzaldehyde to 4-HB. Plants have functional tyrosine aminotransferase (Wang et al., 2016, 2019), but the in planta role of tyrosine aminotransferase in 4-HB biosynthesis has not been demonstrated. Hfd1 belongs to the aldehyde dehydrogenase family, and the human enzyme ALDH3A1 restored CoQ biosynthesis of the yeast Δhfd1 strain (Payet et al., 2016). It is still unknown whether the last step is also conserved in plants. A recent report showed that hydroxyphenylpyruvate dioxygenase-like catalyzes the second reaction in mitochondria of human cells, converting 4-hydroxyphenylpyruvate to 4-hydroxymandelate (Banh et al., 2021). However, Arabidopsis has no homolog of hydroxyphenylpyruvate dioxygenase-like. Moreover, it remains unclear whether plant 4-HB production is completed in the mitochondria and how the aromatic ring precursor is transported across the mitochondrial inner membrane.
Polyisoprenoid tail biosynthesis
In plants, the isoprene subunits for the CoQ side chain are generated through the mevalonate (MVA) pathway, which also produces precursors for sesquiterpene, triterpene, sterol, and brassinosteroid biosynthesis (Zhou and Pichersky, 2020; Pu et al., 2021). The mechanism underlying the import of isoprene units into the mitochondria remains uncharacterized.
The polyisoprenoid tail is synthesized in mitochondria by Coq1, a trans-polyprenyl diphosphate synthase. The Arabidopsis homozygous transfer DNA knockout mutants of AtCoq1 (also known as AtSPS3) are embryo lethal (Ducluzeau et al., 2012). Polyprenyl diphosphate synthases catalyze the condensations of isopentenyl diphosphate with allylic substrates. In vitro enzyme assays showed that Coq1 can use dimethylallyl diphosphate, geranyl diphosphate, farnesyl diphosphate (FPP), or geranylgeranyl diphosphate as its allylic diphosphate primers (Ohara et al., 2010; Hsieh et al., 2011). Although the substrates used in planta remain uncertain, Arabidopsis loss-of-function mutants of FPS1 or FPS2 (FPP synthase) had a moderate reduction in CoQ9 contents (Closa et al., 2010), suggesting that FPP is a potential substrate. Arabidopsis contains five genes encoding eranylgeranyl diphosphate synthase (GGPPS), and GGPPS1 (AT1G49530) was shown to be targeted to the mitochondria (Zhu et al., 1997; Okada et al., 2000). It is still unclear whether these GGPPSs are responsible for the biosynthesis of CoQ (Ruiz-Sola et al., 2016), and the means by which the isoprenyl diphosphate substrates are transported into the mitochondria awaits elucidation.
Coq1 is believed to determine the side-chain length of CoQ, which varies among species. Based on its final product, Coq1 can be named hexaprenyl diphosphate synthase, octaprenyl diphosphate synthase, solanesyl diphosphate synthase, and decaprenyl diphosphate synthase, which generate chains of 6 (C30), 8 (C40), 9 (C45), and 10 (C50) isoprene units, respectively. In plants (Table 2), several species of Poaceae, Cucurbitaceae, and Asteraceae have been shown to synthesize mainly CoQ9, whereas CoQ10 is predominant in Fabaceae and Solanaceae (Threlfall and Whistance, 1970; Ikeda and Kagei, 1979). Species in the same family may produce different CoQs. For example, in Brassicaceae, A. thaliana contains primarily CoQ9, but cauliflower (Brassica oleracea var. botrytis) accumulates CoQ10 (Mattila and Kumpulainen, 2001). The mechanism by which Coq1 determines the product length remains enigmatic, although studies in bacteria have proposed a “single-floor” or “double-floor” model for octaprenyl diphosphate synthases (Guo et al., 2004; Han et al., 2015).
Table 2.
Predominant forms of CoQ in different plant species.
| Family | Species | Common name | Predominant form of CoQ | References |
|---|---|---|---|---|
| Poaceae | Oryza sativa | Rice | CoQ9 | (Ikeda and Kagei, 1979) |
| Poaceae | Triticum aestivum | Wheat | CoQ9 | (Ikeda and Kagei, 1979) |
| Poaceae | Zea mays | Maize | CoQ9 | (Threlfall and Whistance, 1970) |
| Cucurbitaceae | Cucumis melo | Muskmelon | CoQ9 | (Threlfall and Whistance, 1970) |
| Cucurbitaceae | Cucumis sativus | Cucumber | CoQ9 | (Threlfall and Whistance, 1970) |
| Asteraceae | Lactuca sativa | Cultivated lettuce | CoQ9 | (Threlfall and Whistance, 1970) |
| Asteraceae | Cichorium intybus | Chicory | CoQ9 | (Threlfall and Whistance, 1970) |
| Ericaceae | Vaccinium vitis-idaea | Cowberry | CoQ9 | (Mattila and Kumpulainen, 2001) |
| Brassicaceae | Arabidopsis thaliana | Thale cress | CoQ9 | (Xu et al., 2021) |
| Brassicaceae | Brassica oleracea var. botrytis | Cauliflower | CoQ10 | (Mattila and Kumpulainen, 2001) |
| Brassicaceae | Brassica rapa subsp. pekinensis | Chinese cabbage | CoQ10 | (Kettawan et al., 2007) |
| Fabaceae | Glycine max | Soybean | CoQ10 | (Ikeda and Kagei, 1979) |
| Fabaceae | Pisum sativum | Pea | CoQ10 | (Mattila and Kumpulainen, 2001) |
| Fabaceae | Arachis hypogaea | Peanut | CoQ10 | (Ikeda and Kagei, 1979) |
| Solanaceae | Solanum lycopersicum | Tomato | CoQ10 | (Mattila and Kumpulainen, 2001) |
| Solanaceae | Solanum tuberosum | Potato | CoQ10 | (Mattila and Kumpulainen, 2001) |
| Solanaceae | Solanum melongena | Eggplant | CoQ10 | (Kettawan et al., 2007) |
| Apiaceae | Daucus carota | Carrot | CoQ10 | (Mattila and Kumpulainen, 2001) |
| Apiaceae | Petroselinum crispum | Parsley | CoQ10 | (Kettawan et al., 2007) |
| Rosaceae | Malus domestica | Apple | CoQ10 | (Mattila and Kumpulainen, 2001) |
| Rutaceae | Citrus clementina | Clementine | CoQ10 | (Mattila and Kumpulainen, 2001) |
Aromatic ring modifications
The aromatic head is decorated into a fully substituted benzoquinone ring in mitochondria by one prenylation, one decarboxylation, three hydroxylations, and three methylations. First, the isoprene tail is attached to 4-HB, and then the ring is hydroxylated at C5, followed by O-methylation. After sequential decarboxylation and hydroxylation at C1, the ring is further modified via C2 methylation, C6 hydroxylation, and O methylation (Figure 1). The order of these reactions in eukaryotes is still debatable (Acosta Lopez et al., 2019; Fernandez-Del-Rio and Clarke, 2021). Among the enzymes characterized, four (Coq2, Coq3, Coq5, and Coq6) are conserved across plants, fungi, and mammals. The recently identified CoqF is a unique flavin-dependent monooxygenase prevalent in plants and green algae but distinct from its counterpart in fungi and Metazoa. Besides these enzymes, a number of proteins without a clear catalytic role are also involved in CoQ biosynthesis.
Prenylation
Coq2, 4-hydroxybenzoate polyprenyl diphosphate transferase, transfers the polyisoprenoid chain to the 4-HB ring (Figure 1), generating the first lipophilic CoQ intermediate. Arabidopsis Coq2 (also known as AtPPT1) was able to complement the yeast coq2 deletion strain, and the knockout mutant of Arabidopsis was embryo lethal at an early stage (Okada et al., 2004).
Hydroxylations
CoQ biosynthesis requires hydroxylation on three positions of the aromatic ring: C1, C5, and C6. In E. coli, C1 hydroxylation is catalyzed by UbiH, a group A flavin-dependent monooxygenase (Pelosi et al., 2016). The C1-hydroxylase is unidentified in all eukaryotes.
The C5 hydroxylation is mediated by Coq6, a flavin-dependent monooxygenase that is conserved in eukaryotes, including plants (Toda et al., 2014). In the yeast and E. coli mutant strains in which the C5-hydroxylase-encoding gene was disrupted, CoQ production was recovered when Arabidopsis Coq6 was introduced (Latimer et al., 2021); however, the function of AtCoq6 in planta has yet to be characterized.
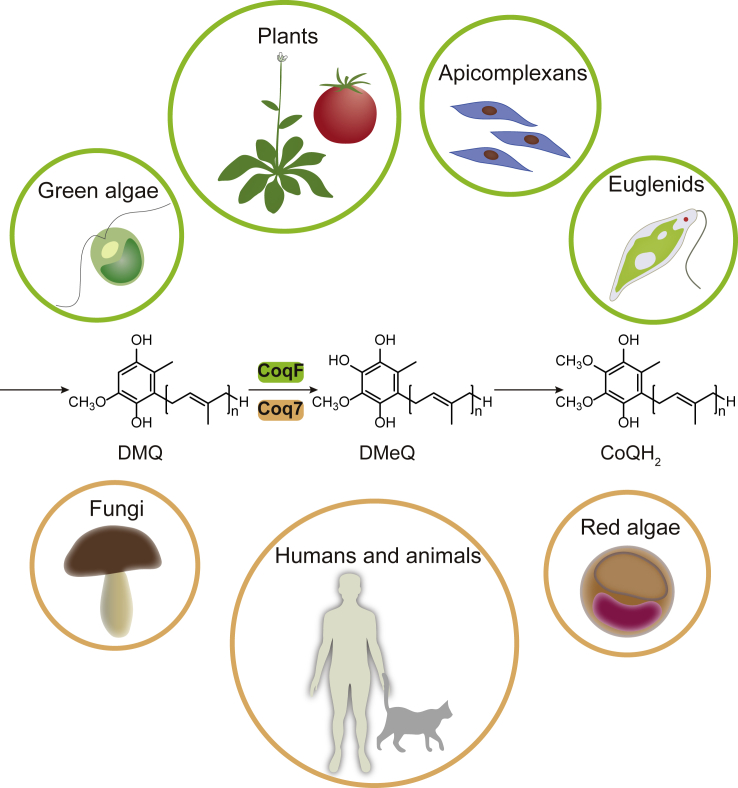
In yeast and humans, the C6 hydroxylation is catalyzed by Coq7, a di-iron protein. However, plants do not have a Coq7 homolog. Recently, two teams independently reported that plants use a unique enzyme, which we named CoqF, to complete this last hydroxylation step (Latimer et al., 2021; Xu et al., 2021). CoqF restored CoQ biosynthesis when expressed in the yeast coq7 mutant and the E. coli ubiF (encoding C6 hydroxylase in some Gammaproteobacteria; Pelosi et al., 2016) mutant. Insertional mutations of AtCoqF resulted in embryo lethality that could be rescued by either human COQ7 or E. coli UbiF. In addition, suppression of CoqF expression in Nicotiana benthamiana via virus-induced gene silencing (VIGS) resulted in CoQ deficiency. CoqF is a unique flavin-dependent monooxygenase, in that it has a C-terminal extension (∼200 amino acids), distinct from other flavin-dependent enzymes previously identified to participate in CoQ biosynthesis. Besides being ubiquitous in land plants, CoqF is also widely distributed in several other major groups of eukaryotes, such as green algae, Cryptista, Haptista, Stramenopiles, Alveolata, and Rhizaria, and it occurs sporadically in other eukaryotic domains owing to lateral gene transfers, providing an excellent marker for distinguishing eukaryotes (Xu et al., 2021). Notably, apicomplexan parasites such as Plasmodium falciparum and Toxoplasma gondii all have CoqF instead of Coq7 (Figure 2). Thus, CoqF represents an interesting antiparasitic drug target for further exploration.
Figure 2.
The diversity of CoQ biosynthesis in eukaryotes: Two different enzymes catalyze the penultimate step.
Coq7, a di-iron monooxygenase, occurs in major lineages of eukaryotes, including Metazoa (animals), fungi, red algae, etc. The recently discovered flavin-dependent monooxygenase CoqF catalyzes this reaction in land plants, green algae, apicomplexans, euglenids, and some others. For details, see Xu et al. (2021).
Methylations
There are two methoxyl groups at C5 and C6 of the benzoquinone ring, both of which are methylated by Coq3 in eukaryotes (Poon et al., 1999). It is likely that these reactions require the substrate in reduced form (quinol). Arabidopsis Coq3 restored CoQ production when expressed in a yeast coq3 deletion mutant (Avelange-Macherel and Joyard, 1998). A transfer DNA insertion line of AtCoq3 was characterized as an embryo-defective mutant with an early stage of arrest by the SeedGenes project (Meinke, 2020).
Coq5 catalyzes the C2 methylation (C methylation) in eukaryotes. Arabidopsis Coq5 was shown to be functional in the fission yeast Schizosaccharomyces pombe (Toda et al., 2014), but its function in planta has not been confirmed.
Decarboxylation
In E. coli, the decarboxylation is catalyzed by the decarboxylase UbiD (Cox et al., 1969), supported by flavin prenyltransferase UbiX that produces a highly modified flavin cofactor for UbiD activity (White et al., 2015). The C1 decarboxylation in eukaryotes is still a mystery.
Other proteins
Besides the enzymes discussed above, several yeast proteins have been found to be required for efficient biosynthesis of CoQ, including Coq4, Coq8, Coq9, and Coq11; their homologs all exist in Arabidopsis.
Coq4 is thought to act as a scaffold protein that organizes the CoQ biosynthetic complex (Marbois et al., 2009), but the precise mechanisms are unclear. Coq8 is a member of an atypical kinase family, and it exhibits a conserved ATPase activity that is activated by CoQ-like phenolic compounds and cardiolipin-containing liposomes (Reidenbach et al., 2018). Overexpression of Coq8 in yeast coq-null mutants stabilized the remaining CoQ biosynthesis polypeptides (Xie et al., 2011, 2012). Expression of Arabidopsis Coq4 and Coq8 recovered CoQ biosynthesis in the S. pombe coq4- and coq8-null mutants, respectively (Toda et al., 2014).
Coq9 is a lipid-binding protein. In human models, it has been proposed that COQ9 may act as a “lipid presenter” to deliver intermediates directly to COQ7 (Lohman et al., 2019). For instance, COQ9 physically interacts with COQ7 and is required for C6 hydroxylation catalyzed by COQ7 (Lohman et al., 2014, 2019). However, plants use the flavin-dependent CoqF instead of the di-iron Coq7 as the C6 hydroxylase, and the Arabidopsis homolog of Coq9 failed to complement the S. pombe coq9 deletion (Toda et al., 2014). Whether and how Coq9 participates in plant CoQ biosynthesis is an open question.
Coq11, a member of the atypical short-chain dehydrogenase and reductase superfamily, was identified as a constituent of the CoQ biosynthetic complex via affinity purification of tagged Coq proteins in yeast (Allan et al., 2015). The genome of Arabidopsis encodes several homologs of Coq11, two of which (Coq11A and Coq11B) show co-expression with CoQ biosynthesis (Xu et al., 2021). Again, their function has not been demonstrated.
Biosynthetic complex
Enzymes that act downstream of polyprenyl-hydroxybenzoate and additional associated proteins form a biosynthetic complex known as the CoQ-synthome or Complex Q. In yeast, Coq3–Coq9 and Coq11 are members of the complex. The CoQ-synthome also exists in humans (Floyd et al., 2016). In E. coli, seven Ubi proteins form a stable metabolon that synthesizes ubiquinone in the cytosol (Hajj Chehade et al., 2019). In plants, such a complex awaits exploration.
The evolution of plant CoQ biosynthesis
Plants have evolved the unique ability to synthesize 4-HB from phenylalanine via two parallel routes. The phenylpropanoid pathway also serves as a starting point for the production of a variety of metabolites such as lignin, flavonoids, coumarins, and lignans. A recent analysis of the evolutionary history of key enzymes in the phenylpropanoid pathway showed that the enzymes involved in making p-coumaroyl-CoA from phenylalanine (PAL, C4H, and 4CL), are generally present across Embryophyta (de Vries et al., 2021). Homologs of the genes encoding PAL and 4CL can even be found in streptophyte algae, the algal sisters of land plants. The main route that opens up after the synthesis of p-coumaroyl-CoA is β-oxidative metabolism in the peroxisomes. In Arabidopsis, two peroxisomal 4CLs (AT4G19010 and 4CL8) have been identified to participate in this pathway. In the acyl-activating enzyme superfamily, AT4G19010 and 4CL8 belong to clade V (Shockey and Browse, 2011), which also contains several members that exhibit high activities toward fatty acids (Kienow et al., 2008). A phylogenetic analysis showed that the clade V members of Arabidopsis fell into a clade that included sequences from major lineages of land plants. It would be interesting to see whether peroxisomal 4CLs are widely distributed in plants. The second route is the conversion of p-coumaric acid into kaempferol. Among flavonoids, kaempferol is a flavonol that is widely present in land plant species ranging from bryophytes and ferns to seed plants, often in the form of glycosides (Iwashina and Murai, 2013). Notably, mammalian cells also have the ability to use exogenous kaempferol as a precursor for CoQ biosynthesis (Fernandez-Del-Rio et al., 2017). The mechanism of release of a CoQ ring precursor from kaempferol is likely to be conserved between plants and mammals (Fernandez-Del-Rio et al., 2020).
Unlike animals and fungi, in which the C6 hydroxylation is catalyzed by Coq7, plants use CoqF (Figure 2). Although the two enzymes are functional counterparts, they are evolutionarily unrelated and have different origins: Coq7 is a di-iron hydroxylase, whereas CoqF belongs to an isolated subfamily of flavoenzymes. It has been proposed that CoqF emerged early during eukaryotic diversification and then became dominant among photosynthetic and related organisms. Both CoqF and Coq7 were found in Prasinodermophyta and Chlorophyta, but usually in different sublineages. In streptophyte algae and land plants, CoqF is the sole C6 hydroxylase, implying an adaptive advantage of this unique flavoenzyme in aerobic respiration during plant terrestrialization (Xu et al., 2021).
The bioactive compound shikonin and its derivatives are a group of red-pigmented naphthoquinones produced in many members of the family Boraginaceae. The shikonin and CoQ pathways share precursors and contain similar biochemical architectures, and evolutionary links between CoQ and shikonin biosynthesis have been identified in red gromwell, Lithospermum erythrorhizon (Auber et al., 2020; Suttiyut et al., 2022). The p-hydroxybenzoate:geranyltransferase genes and several gene candidates for shikonin biosynthesis were found to have evolved via duplication of the CoQ pathway genes. Further investigation of CoQ biosynthesis is likely to help gene discovery for the biosynthesis of shikonins and other quinone-bearing specialized metabolites.
Regulation of CoQ biosynthesis
Regulation of CoQ biosynthesis in yeast and mammals has been reviewed recently, and Coq7 seems to be a key regulatory hub that integrates endogenous and environmental signals (Villalba and Navas, 2021). Far less is known about the regulation of CoQ biosynthesis in plants. Since plants use a distinct flavoenzyme in place of Coq7, it is likely that plants employ a different regulatory mechanism.
According to the Arabidopsis Electronic Fluorescent Pictograph Browser (Winter et al., 2007), Coq genes are highly expressed in seeds. Congruently, CoQ is distributed throughout the plant and more abundant in seeds (Xu et al., 2021). Analysis of Arabidopsis demonstrated the co-expression of genes involved in CoQ biosynthesis in mitochondria (Ducluzeau et al., 2012). In addition, genes involved in 4-HB biosynthesis (AT4G19010, C4H, 4CL3, CHS, CHI, F3H, and FLS1) and the MVA pathway (HMGR1, HMGR2, and HMGS) in the cytosol were also co-expressed with the Coq genes (Ducluzeau et al., 2012; Soubeyrand et al., 2018). It will be interesting to analyze the regulatory mechanisms at the transcriptional level.
It remains elusive how plants synthesize CoQ in response to environmental stimuli. A recent report showed that continuous high-light treatments promoted the de novo biosynthesis of CoQ in Arabidopsis (Soubeyrand et al., 2019); the underlying signaling pathway is worthy of investigation.
CoQ10 biofortification
CoQ is endogenously synthesized in the human body, and CoQ levels decrease as people age (Ernster and Forsmark-Andree, 1993). In addition, certain cholesterol-lowering drugs, such as statins, inhibit 3-hydroxy-3-methylglutaryl-CoA reductase activity, resulting in a decrease in CoQ (Bhagavan and Chopra, 2006). Dietary intake from food is another source of CoQ; however, this supply is not always sufficient because plant-based foods, particularly vegetables, fruits, and cereals, are generally low in CoQ contents (Pravst et al., 2010; Parmar et al., 2015). In addition, many plants synthesize CoQ9 instead of CoQ10 as the principal CoQ molecule. Thus, there is a great need for engineering CoQ10 production in plants. Major strategies used for CoQ10 biofortification include increasing precursor availability and overexpressing rate-limiting enzymes in mitochondria.
Increasing the precursor supply
4-HB serves as the skeleton for the CoQ head group. When 4-HB was fed to Arabidopsis seedlings, CoQ accumulated to as much as 150% of the control level (Soubeyrand et al., 2021). Overexpression of AT4G19010 or 4CL8, which encode 4-coumarate CoA ligases responsible for 4-HB production in peroxisomes, increased CoQ accumulation to ∼150% of the wild-type level (Block et al., 2014; Soubeyrand et al., 2019). In knockout mutants of genes encoding kaempferol 3-O-glycosyltransferase that restricts the supply of 4-HB, CoQ content was elevated to 160% of the wild-type level (Soubeyrand et al., 2021). These results indicated that 4-HB supply limits plant CoQ biosynthesis.
Precursors of the isoprene tail are produced from the cytosolic MVA pathway. In Nicotiana tabacum, expression of a bacterial phosphomevalonate decarboxylase that increases available isopentenyl phosphate resulted in increased production of MVA-derived terpenoids; however, CoQ accumulation was not affected (Henry et al., 2018). One reason might be that the isoprenoid subunits produced in the cytosol were not efficiently transported to mitochondria.
Overcoming downstream rate-limiting steps
In mitochondria, transfer of the polyprenyl chain to 4-HB by Coq2 is considered to be a rate-limiting step in CoQ biosynthesis. Overexpression of Coq2 in Salvia miltiorrhiza resulted in up to a 3-fold increase in CoQ content (Liu et al., 2019). Heterologous expression of a yeast Coq2 in tobacco led to elevation of CoQ content, which was two times higher than that of the wild-type control (Ohara et al., 2004).
Recently, encouraging progress was achieved in tomato, which showed a 7-fold increase in CoQ10 production following overexpression of four genes encoding Arabidopsis 3-hydroxy-3-methylglutaryl-CoA reductase, E. coli chorismate pyruvate-lyase UbiC (which catalyzes the removal of pyruvate from chorismate to produce 4-HB), and tobacco Coq1 and Coq2, all driven by the tomato-fruit-specific E8 promoter (Fan et al., 2021). The results provide evidence that Coq1 and Coq2, which catalyze the decaprenyl chain formation and attachment in mitochondria, play a key role in determining the final CoQ10 yield.
Modifying the side-chain length
Another problem encountered in engineering CoQ in plants is that most cereal crops, as well as some vegetables and fruits, predominantly produce CoQ9, whereas human mitochondria synthesize CoQ10. Side-chain length has been shown to be a critical factor for CoQ biological activity, and the efficacies of CoQ analogs for medical purposes have been reviewed recently (Suarez-Rivero et al., 2021). Wang and Hekimi generated conditional Coq7 (also known as Mclk1) knockout mouse embryonic fibroblasts in which CoQ9 was undetectable and exogenously applied CoQs of varying isoprenoid chain length. CoQ9, which is the original species of CoQ in mice, appeared to improve respiratory chain activity more effectively than other CoQs tested (Wang and Hekimi, 2013). Another investigation, based on the kinetics of bovine respiratory complex I catalysis with a series of CoQs of different isoprenoid side-chain lengths (from 1 to 10 units), suggested that CoQ10 has both the highest binding affinity and the fastest binding rate (Fedor et al., 2017).
The length of the side chain is determined by Coq1. In rice and Panicum meyerianum, expression of decaprenyl diphosphate synthase from Gluconobacter suboxydans modified the length of the CoQ side chain from 9 to 10 isoprene units (Takahashi et al., 2006, 2009, 2010; Seo et al., 2011). The mechanism of Coq1-mediated chain-length determination is, however, poorly understood. Characterization of the catalytic steps will facilitate the development of CoQ10-enriched crops by selection of appropriate natural variations and genome editing.
Future perspectives
CoQ biosynthesis has been studied mainly in yeast and human cells, and thus our understanding of the diversity of the CoQ biosynthetic pathway in eukaryotes is highly limited. Recent investigations in plants have identified several unique enzymes in the plant CoQ biosynthetic pathway. It will be of great interest to search for the enzymes and cofactors that act in the steps that have not been identified in plants, and this should help us to understand the evolution of oxidative respiration from bacteria to eukaryotes.
In plants, CoQ is important for growth and development. Arabidopsis mutants lacking Coq1, Coq2, Coq3, or CoqF are embryo lethal (Avelange-Macherel and Joyard, 1998; Okada et al., 2004; Ducluzeau et al., 2012; Xu et al., 2021). Because CoQ is an essential component in mitochondrial oxidative phosphorylation, plant-specific enzymes of CoQ biosynthesis are potential targets for herbicide development.
At the moment, microbial fermentation is the major industrial source of CoQ10. Enhancement in plant foods can be a cost-effective and environmentally friendly strategy for improving CoQ10 supply. However, to date, little attention has been paid to CoQ10 biofortification, and current engineering strategies have had a relatively modest impact on CoQ10 production in plants. Recently, a genome-wide genetic screen of yeast identified 30 previously unknown regulators of CoQ accumulation; phospholipid metabolism was confirmed to be a key player, as deficiency in phosphatidylethanolamine methylation resulted in a 5-fold increase in CoQ (Ayer et al., 2021). It would be particularly interesting to test whether some of these regulators are functionally conserved in plants. A better understanding of the ubiquinone biosynthetic pathway and its regulatory mechanisms in plants will facilitate the breeding of CoQ10-enriched crop cultivars, hopefully in the near future.
Funding
This work was supported by the National Key R&D Program of China (grant no. 2020YFA0907900), the National Natural Science Foundation of China (grant nos. 32070338 and 31788103), and the Special Fund for Scientific Research of Shanghai Landscaping & City Appearance Administrative Bureau (grant no. G222414).
Author contributions
J.-J.X. and X.-Y.C. wrote the manuscript. M.H. and L.Y. contributed revisions.
Acknowledgments
No conflict of interest is declared.
Published: May 25, 2022
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
References
- Abby S.S., Kazemzadeh K., Vragniau C., Pelosi L., Pierrel F. Advances in bacterial pathways for the biosynthesis of ubiquinone. Biochim. Biophys. Acta Bioenerg. 2020;1861:148259. doi: 10.1016/j.bbabio.2020.148259. [DOI] [PubMed] [Google Scholar]
- Acosta Lopez M.J., Trevisson E., Canton M., Vazquez-Fonseca L., Morbidoni V., Baschiera E., Frasson C., Pelosi L., Rascalou B., Desbats M.A., et al. Vanillic acid restores coenzyme Q biosynthesis and ATP production in human cells lacking COQ6. Oxid. Med. Cell. Longev. 2019;2019:1–11. doi: 10.1155/2019/3904905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan C.M., Awad A.M., Johnson J.S., Shirasaki D.I., Wang C., Blaby-Haas C.E., Merchant S.S., Loo J.A., Clarke C.F. Identification of Coq11, a new coenzyme Q biosynthetic protein in the CoQ-synthome in Saccharomyces cerevisiae. J. Biol. Chem. 2015;290:7517–7534. doi: 10.1074/jbc.M114.633131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas-Jal M., Sune-Negre J.M., Garcia-Montoya E. Coenzyme Q10 supplementation: efficacy, safety, and formulation challenges. Compr. Rev. Food Sci. Food Saf. 2020;19:574–594. doi: 10.1111/1541-4337.12539. [DOI] [PubMed] [Google Scholar]
- Auber R.P., Suttiyut T., McCoy R.M., Ghaste M., Crook J.W., Pendleton A.L., Widhalm J.R., Wisecaver J.H. Hybrid de novo genome assembly of red gromwell (Lithospermum erythrorhizon) reveals evolutionary insight into shikonin biosynthesis. Hortic. Res. 2020;7:82. doi: 10.1038/s41438-020-0301-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avelange-Macherel M.H., Joyard J. Cloning and functional expression of AtCOQ3, the Arabidopsis homologue of the yeast COQ3 gene, encoding a methyltransferase from plant mitochondria involved in ubiquinone biosynthesis. Plant J. 1998;14:203–213. doi: 10.1046/j.1365-313x.1998.00109.x. [DOI] [PubMed] [Google Scholar]
- Awad A.M., Bradley M.C., Fernandez-Del-Rio L., Nag A., Tsui H.S., Clarke C.F. Coenzyme Q10 deficiencies: pathways in yeast and humans. Essays Biochem. 2018;62:361–376. doi: 10.1042/EBC20170106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayer A., Fazakerley D.J., Suarna C., Maghzal G.J., Sheipouri D., Lee K.J., Bradley M.C., Fernandez-Del-Rio L., Tumanov S., Kong S.M., et al. Genetic screening reveals phospholipid metabolism as a key regulator of the biosynthesis of the redox-active lipid coenzyme Q. Redox Biol. 2021;46:102127. doi: 10.1016/j.redox.2021.102127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee R., Purhonen J., Kallijarvi J. The mitochondrial coenzyme Q junction and complex III: biochemistry and pathophysiology. FEBS J. 2021 doi: 10.1111/febs.16164. [DOI] [PubMed] [Google Scholar]
- Banh R.S., Kim E.S., Spillier Q., Biancur D.E., Yamamoto K., Sohn A.S.W., Shi G., Jones D.R., Kimmelman A.C., Pacold M.E. The polar oxy-metabolome reveals the 4-hydroxymandelate CoQ10 synthesis pathway. Nature. 2021;597:420–425. doi: 10.1038/s41586-021-03865-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baschiera E., Sorrentino U., Calderan C., Desbats M.A., Salviati L. The multiple roles of coenzyme Q in cellular homeostasis and their relevance for the pathogenesis of coenzyme Q deficiency. Free Radic. Biol. Med. 2021;166:277–286. doi: 10.1016/j.freeradbiomed.2021.02.039. [DOI] [PubMed] [Google Scholar]
- Berger A., Latimer S., Stutts L.R., Soubeyrand E., Block A.K., Basset G.J. Kaempferol as a precursor for ubiquinone (coenzyme Q) biosynthesis: an atypical node between specialized metabolism and primary metabolism. Curr. Opin. Plant Biol. 2022;66:102165. doi: 10.1016/j.pbi.2021.102165. [DOI] [PubMed] [Google Scholar]
- Bersuker K., Hendricks J.M., Li Z., Magtanong L., Ford B., Tang P.H., Roberts M.A., Tong B., Maimone T.J., Zoncu R., et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019;575:688–692. doi: 10.1038/s41586-019-1705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagavan H.N., Chopra R.K. Coenzyme Q10: absorption, tissue uptake, metabolism and pharmacokinetics. Free Radic. Res. 2006;40:445–453. doi: 10.1080/10715760600617843. [DOI] [PubMed] [Google Scholar]
- Block A., Widhalm J.R., Fatihi A., Cahoon R.E., Wamboldt Y., Elowsky C., Mackenzie S.A., Cahoon E.B., Chapple C., Dudareva N., et al. The origin and biosynthesis of the benzenoid moiety of ubiquinone (coenzyme Q) in Arabidopsis. Plant Cell. 2014;26:1938–1948. doi: 10.1105/tpc.114.125807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussell J.D., Reichelt M., Wiszniewski A.A., Gershenzon J., Smith S.M. Peroxisomal ATP-binding cassette transporter COMATOSE and the multifunctional protein abnormal INFLORESCENCE MERISTEM are required for the production of benzoylated metabolites in Arabidopsis seeds. Plant Physiol. 2014;164:48–54. doi: 10.1104/pp.113.229807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirilli I., Damiani E., Dludla P.V., Hargreaves I., Marcheggiani F., Millichap L.E., Orlando P., Silvestri S., Tiano L. Role of coenzyme Q10 in health and disease: an update on the last 10 Years (2010-2020) Antioxidants. 2021;10:1325. doi: 10.3390/antiox10081325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Closa M., Vranova E., Bortolotti C., Bigler L., Arro M., Ferrer A., Gruissem W. The Arabidopsis thaliana FPP synthase isozymes have overlapping and specific functions in isoprenoid biosynthesis, and complete loss of FPP synthase activity causes early developmental arrest. Plant J. 2010;63:512–525. doi: 10.1111/j.1365-313X.2010.04253.x. [DOI] [PubMed] [Google Scholar]
- Cox G.B., Young I.G., McCann L.M., Gibson F. Biosynthesis of ubiquinone in Escherichia coli K-12: location of genes affecting the metabolism of 3-octaprenyl-4-hydroxybenzoic acid and 2-octaprenylphenol. J. Bacteriol. 1969;99:450–458. doi: 10.1128/jb.99.2.450-458.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marcos Lousa C., van Roermund C.W.T., Postis V.L.G., Dietrich D., Kerr I.D., Wanders R.J.A., Baldwin S.A., Baker A., Theodoulou F.L. Intrinsic acyl-CoA thioesterase activity of a peroxisomal ATP binding cassette transporter is required for transport and metabolism of fatty acids. Proc. Natl. Acad. Sci. USA. 2013;110:1279–1284. doi: 10.1073/pnas.1218034110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries S., Furst-Jansen J.M.R., Irisarri I., Dhabalia Ashok A., Ischebeck T., Feussner K., Abreu I.N., Petersen M., Feussner I., de Vries J. The evolution of the phenylpropanoid pathway entailed pronounced radiations and divergences of enzyme families. Plant J. 2021;107:975–1002. doi: 10.1111/tpj.15387. [DOI] [PubMed] [Google Scholar]
- Doll S., Freitas F.P., Shah R., Aldrovandi M., da Silva M.C., Ingold I., Goya Grocin A., Xavier da Silva T.N., Panzilius E., Scheel C.H., et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature. 2019;575:693–698. doi: 10.1038/s41586-019-1707-0. [DOI] [PubMed] [Google Scholar]
- Ducluzeau A.L., Wamboldt Y., Elowsky C.G., Mackenzie S.A., Schuurink R.C., Basset G.J. Gene network reconstruction identifies the authentic trans-prenyl diphosphate synthase that makes the solanesyl moiety of ubiquinone-9 in Arabidopsis. Plant J. 2012;69:366–375. doi: 10.1111/j.1365-313X.2011.04796.x. [DOI] [PubMed] [Google Scholar]
- Ernster L., Forsmark-Andree P. Ubiquinol: an endogenous antioxidant in aerobic organisms. Clin. Invest. 1993;71:S60–S65. doi: 10.1007/BF00226842. [DOI] [PubMed] [Google Scholar]
- Evans D.R., Guy H.I. Mammalian pyrimidine biosynthesis: fresh insights into an ancient pathway. J. Biol. Chem. 2004;279:33035–33038. doi: 10.1074/jbc.R400007200. [DOI] [PubMed] [Google Scholar]
- Fan H., Liu Y., Li C.Y., Jiang Y., Song J.J., Yang L., Zhao Q., Hu Y.H., Chen X.Y., Xu J.J. Engineering high coenzyme Q10 tomato. Metab. Eng. 2021;68:86–93. doi: 10.1016/j.ymben.2021.09.007. [DOI] [PubMed] [Google Scholar]
- Fedor J.G., Jones A.J.Y., Di Luca A., Kaila V.R.I., Hirst J. Correlating kinetic and structural data on ubiquinone binding and reduction by respiratory complex I. Proc. Natl. Acad. Sci. USA. 2017;114:12737–12742. doi: 10.1073/pnas.1714074114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Del-Rio L., Clarke C.F. Coenzyme Q biosynthesis: an update on the origins of the benzenoid ring and discovery of new ring precursors. Metabolites. 2021;11:385. doi: 10.3390/metabo11060385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Del-Rio L., Soubeyrand E., Basset G.J., Clarke C.F. Metabolism of the flavonol kaempferol in kidney cells liberates the B-ring to enter coenzyme Q biosynthesis. Molecules. 2020;25:2955. doi: 10.3390/molecules25132955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Del-Rio L., Nag A., Gutierrez Casado E., Ariza J., Awad A.M., Joseph A.I., Kwon O., Verdin E., de Cabo R., Schneider C., et al. Kaempferol increases levels of coenzyme Q in kidney cells and serves as a biosynthetic ring precursor. Free Radic. Biol. Med. 2017;110:176–187. doi: 10.1016/j.freeradbiomed.2017.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd B.J., Wilkerson E.M., Veling M.T., Minogue C.E., Xia C., Beebe E.T., Wrobel R.L., Cho H., Kremer L.S., Alston C.L., et al. Mitochondrial protein interaction mapping identifies regulators of respiratory chain function. Mol. Cell. 2016;63:621–632. doi: 10.1016/j.molcel.2016.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R.T., Kuo C.J., Ko T.P., Chou C.C., Liang P.H., Wang A.H.J. A molecular ruler for chain elongation catalyzed by octaprenyl pyrophosphate synthase and its structure-based engineering to produce unprecedented long chain trans-prenyl products. Biochemistry. 2004;43:7678–7686. doi: 10.1021/bi036336d. [DOI] [PubMed] [Google Scholar]
- Hajj Chehade M., Pelosi L., Fyfe C.D., Loiseau L., Rascalou B., Brugiere S., Kazemzadeh K., Vo C.D.T., Ciccone L., Aussel L., et al. A soluble metabolon synthesizes the isoprenoid lipid ubiquinone. Cell Chem. Biol. 2019;26:482–492.e7. doi: 10.1016/j.chembiol.2018.12.001. [DOI] [PubMed] [Google Scholar]
- Han X., Chen C.C., Kuo C.J., Huang C.H., Zheng Y., Ko T.P., Zhu Z., Feng X., Wang K., Oldfield E., et al. Crystal structures of ligand-bound octaprenyl pyrophosphate synthase from Escherichia coli reveal the catalytic and chain-length determining mechanisms. Proteins. 2015;83:37–45. doi: 10.1002/prot.24618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry L.K., Thomas S.T., Widhalm J.R., Lynch J.H., Davis T.C., Kessler S.A., Bohlmann J., Noel J.P., Dudareva N. Contribution of isopentenyl phosphate to plant terpenoid metabolism. Nat. Plants. 2018;4:721–729. doi: 10.1038/s41477-018-0220-z. [DOI] [PubMed] [Google Scholar]
- Hsieh F.L., Chang T.H., Ko T.P., Wang A.H.J. Structure and mechanism of an Arabidopsis medium/long-chain-length prenyl pyrophosphate synthase. Plant Physiol. 2011;155:1079–1090. doi: 10.1104/pp.110.168799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M., Kagei K. Ubiquinone content of eight plant species in cell culture. Phytochemistry. 1979;18:1577–1578. doi: 10.1016/S0031-9422(00)98506-6. [DOI] [Google Scholar]
- Iwashina T., Murai Y. Distribution of kaempferol glycosides and their function in plants. Chem. Phys. Res. J. 2013;6:271–303. [Google Scholar]
- Kettawan A., Kunthida C., Takahashi T., Kishi T., Chikazawa J., Sakata Y., Yano E., Watabe K., Yamamoto Y., Okamoto T. The quality control assessment of commercially available coenzyme q(10)-containing dietary and health supplements in Japan. J. Clin. Biochem. Nutr. 2007;41:124–131. doi: 10.3164/jcbn.2007017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienow L., Schneider K., Bartsch M., Stuible H.P., Weng H., Miersch O., Wasternack C., Kombrink E. Jasmonates meet fatty acids: functional analysis of a new acyl-coenzyme A synthetase family from Arabidopsis thaliana. J. Exp. Bot. 2008;59:403–419. doi: 10.1093/jxb/erm325. [DOI] [PubMed] [Google Scholar]
- Latimer S., Keene S.A., Stutts L.R., Berger A., Bernert A.C., Soubeyrand E., Wright J., Clarke C.F., Block A.K., Colquhoun T.A., et al. A dedicated flavin-dependent monooxygenase catalyzes the hydroxylation of demethoxyubiquinone into ubiquinone (coenzyme Q) in Arabidopsis. J. Biol. Chem. 2021;297:101283. doi: 10.1016/j.jbc.2021.101283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Chen X., Wang M., Lu S. SmPPT, a 4-hydroxybenzoate polyprenyl diphosphate transferase gene involved in ubiquinone biosynthesis, confers salt tolerance in Salvia miltiorrhiza. Plant Cell Rep. 2019;38:1527–1540. doi: 10.1007/s00299-019-02463-5. [DOI] [PubMed] [Google Scholar]
- Lohman D.C., Aydin D., Von Bank H.C., Smith R.W., Linke V., Weisenhorn E., McDevitt M.T., Hutchins P., Wilkerson E.M., Wancewicz B., et al. An isoprene lipid-binding protein promotes eukaryotic coenzyme Q biosynthesis. Mol. Cell. 2019;73:763–774.e10. doi: 10.1016/j.molcel.2018.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman D.C., Forouhar F., Beebe E.T., Stefely M.S., Minogue C.E., Ulbrich A., Stefely J.A., Sukumar S., Luna-Sanchez M., Jochem A., et al. Mitochondrial COQ9 is a lipid-binding protein that associates with COQ7 to enable coenzyme Q biosynthesis. Proc. Natl. Acad. Sci. USA. 2014;111:E4697–E4705. doi: 10.1073/pnas.1413128111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano D., Andrade P., Caudepón D., Altabella T., Arró M., Ferrer A. Suppressing farnesyl diphosphate synthase alters chloroplast development and triggers sterol-dependent induction of jasmonate- and Fe-related responses. Plant Physiol. 2016;172:93–117. doi: 10.1104/pp.16.00431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbois B., Gin P., Gulmezian M., Clarke C.F. The yeast Coq4 polypeptide organizes a mitochondrial protein complex essential for coenzyme Q biosynthesis. Biochim. Biophys. Acta. 2009;1791:69–75. doi: 10.1016/j.bbalip.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila P., Kumpulainen J. Coenzymes Q9and Q10: contents in foods and dietary intake. J. Food Compos. Anal. 2001;14:409–417. doi: 10.1006/jfca.2000.0983. [DOI] [Google Scholar]
- Meinke D.W. Genome-wide identification of EMBRYO-DEFECTIVE (EMB) genes required for growth and development in Arabidopsis. New Phytol. 2020;226:306–325. doi: 10.1111/nph.16071. [DOI] [PubMed] [Google Scholar]
- Ohara K., Sasaki K., Yazaki K. Two solanesyl diphosphate synthases with different subcellular localizations and their respective physiological roles in Oryza sativa. J. Exp. Bot. 2010;61:2683–2692. doi: 10.1093/jxb/erq103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara K., Kokado Y., Yamamoto H., Sato F., Yazaki K. Engineering of ubiquinone biosynthesis using the yeast coq2 gene confers oxidative stress tolerance in transgenic tobacco. Plant J. 2004;40:734–743. doi: 10.1111/j.1365-313X.2004.02246.x. [DOI] [PubMed] [Google Scholar]
- Okada K., Saito T., Nakagawa T., Kawamukai M., Kamiya Y. Five geranylgeranyl diphosphate synthases expressed in different organs are localized into three subcellular compartments in Arabidopsis. Plant Physiol. 2000;122:1045–1056. doi: 10.1104/pp.122.4.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K., Kasahara H., Yamaguchi S., Kawaide H., Kamiya Y., Nojiri H., Yamane H. Genetic evidence for the role of isopentenyl diphosphate isomerases in the mevalonate pathway and plant development in Arabidopsis. Plant Cell Physiol. 2008;49:604–616. doi: 10.1093/pcp/pcn032. [DOI] [PubMed] [Google Scholar]
- Okada K., Ohara K., Yazaki K., Nozaki K., Uchida N., Kawamukai M., Nojiri H., Yamane H. The AtPPT1 gene encoding 4-hydroxybenzoate polyprenyl diphosphate transferase in ubiquinone biosynthesis is required for embryo development in Arabidopsis thaliana. Plant Mol. Biol. 2004;55:567–577. doi: 10.1007/s11103-004-1298-4. [DOI] [PubMed] [Google Scholar]
- Parmar S.S., Jaiwal A., Dhankher O.P., Jaiwal P.K. Coenzyme Q10 production in plants: current status and future prospects. Crit. Rev. Biotechnol. 2015;35:152–164. doi: 10.3109/07388551.2013.823594. [DOI] [PubMed] [Google Scholar]
- Payet L.A., Leroux M., Willison J.C., Kihara A., Pelosi L., Pierrel F. Mechanistic details of early steps in coenzyme Q biosynthesis pathway in yeast. Cell Chem. Biol. 2016;23:1241–1250. doi: 10.1016/j.chembiol.2016.08.008. [DOI] [PubMed] [Google Scholar]
- Pelosi L., Ducluzeau A.-L., Loiseau L., Barras F., Schneider D., Junier I., Pierrel F., Dorrestein P.C. Evolution of ubiquinone biosynthesis: multiple proteobacterial enzymes with various regioselectivities to catalyze three contiguous aromatic hydroxylation reactions. mSystems. 2016;1:1–16. doi: 10.1128/mSystems.00091-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M.A., D'Auria J.C., Gershenzon J., Pichersky E. The Arabidopsis thaliana type I isopentenyl diphosphate isomerases are targeted to multiple subcellular compartments and have overlapping functions in isoprenoid biosynthesis. Plant Cell. 2008;20:677–696. doi: 10.1105/tpc.107.053926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon W.W., Barkovich R.J., Hsu A.Y., Frankel A., Lee P.T., Shepherd J.N., Myles D.C., Clarke C.F. Yeast and rat Coq3 and Escherichia coli UbiG polypeptides catalyze both O-methyltransferase steps in coenzyme Q biosynthesis. J. Biol. Chem. 1999;274:21665–21672. doi: 10.1074/jbc.274.31.21665. [DOI] [PubMed] [Google Scholar]
- Pravst I., Zmitek K., Zmitek J. Coenzyme Q10 contents in foods and fortification strategies. Crit. Rev. Food Sci. Nutr. 2010;50:269–280. doi: 10.1080/10408390902773037. [DOI] [PubMed] [Google Scholar]
- Pu X., Dong X., Li Q., Chen Z., Liu L. An update on the function and regulation of methylerythritol phosphate and mevalonate pathways and their evolutionary dynamics. J. Integr. Plant Biol. 2021;63:1211–1226. doi: 10.1111/jipb.13076. [DOI] [PubMed] [Google Scholar]
- Reidenbach A.G., Kemmerer Z.A., Aydin D., Jochem A., McDevitt M.T., Hutchins P.D., Stark J.L., Stefely J.A., Reddy T., Hebert A.S., et al. Conserved lipid and small-molecule modulation of COQ8 reveals regulation of the ancient kinase-like UbiB family. Cell Chem. Biol. 2018;25:154–165.e11. doi: 10.1016/j.chembiol.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson K.P., Jochem A., Johnson S.E., Reddy T.R., Russell J.D., Coon J.J., Pagliarini D.J. Defining intermediates and redundancies in coenzyme Q precursor biosynthesis. J. Biol. Chem. 2021;296:100643. doi: 10.1016/j.jbc.2021.100643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Sola M.A., Barja M.V., Manzano D., Llorente B., Schipper B., Beekwilder J., Rodriguez-Concepcion M. A single Arabidopsis gene encodes two differentially targeted geranylgeranyl diphosphate synthase isoforms. Plant Physiol. 2016;172:1393–1402. doi: 10.1104/pp.16.01392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M.S., Takahashi S., Kadowaki K., Kawamukai M., Takahara M., Takamizo T. Expression of CoQ10-producing ddsA transgene by efficient Agrobacterium-mediated transformation in Panicum meyerianum. Plant Cell Tissue Organ Cult. 2011;107:325–332. doi: 10.1007/s11240-011-9984-9. [DOI] [Google Scholar]
- Shockey J., Browse J. Genome-level and biochemical diversity of the acyl-activating enzyme superfamily in plants. Plant J. 2011;66:143–160. doi: 10.1111/j.1365-313X.2011.04512.x. [DOI] [PubMed] [Google Scholar]
- Soubeyrand E., Johnson T.S., Latimer S., Block A., Kim J., Colquhoun T.A., Butelli E., Martin C., Wilson M.A., Basset G.J. The peroxidative cleavage of kaempferol contributes to the biosynthesis of the benzenoid moiety of ubiquinone in plants. Plant Cell. 2018;30:2910–2921. doi: 10.1105/tpc.18.00688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubeyrand E., Kelly M., Keene S.A., Bernert A.C., Latimer S., Johnson T.S., Elowsky C., Colquhoun T.A., Block A.K., Basset G.J. Arabidopsis 4-COUMAROYL-COA LIGASE 8 contributes to the biosynthesis of the benzenoid ring of coenzyme Q in peroxisomes. Biochem. J. 2019;476:3521–3532. doi: 10.1042/BCJ20190688. [DOI] [PubMed] [Google Scholar]
- Soubeyrand E., Latimer S., Bernert A.C., Keene S.A., Johnson T.S., Shin D., Block A.K., Colquhoun T.A., Schaffner A.R., Kim J., et al. 3-O-glycosylation of kaempferol restricts the supply of the benzenoid precursor of ubiquinone (Coenzyme Q) in Arabidopsis thaliana. Phytochemistry. 2021;186:112738. doi: 10.1016/j.phytochem.2021.112738. [DOI] [PubMed] [Google Scholar]
- Stefely J.A., Pagliarini D.J. Biochemistry of mitochondrial coenzyme Q biosynthesis. Trends Biochem. Sci. 2017;42:824–843. doi: 10.1016/j.tibs.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefely J.A., Kwiecien N.W., Freiberger E.C., Richards A.L., Jochem A., Rush M.J.P., Ulbrich A., Robinson K.P., Hutchins P.D., Veling M.T., et al. Mitochondrial protein functions elucidated by multi-omic mass spectrometry profiling. Nat. Biotechnol. 2016;34:1191–1197. doi: 10.1038/nbt.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Rivero J.M., Pastor-Maldonado C.J., Povea-Cabello S., Alvarez-Cordoba M., Villalon-Garcia I., Munuera-Cabeza M., Suarez-Carrillo A., Talaveron-Rey M., Sanchez-Alcazar J.A. Coenzyme Q10 analogues: benefits and challenges for therapeutics. Antioxidants. 2021;10:236. doi: 10.3390/antiox10020236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttiyut T., Auber R.P., Ghaste M., Kane C.N., McAdam S.A.M., Wisecaver J.H., Widhalm J.R. Integrative analysis of the shikonin metabolic network identifies new gene connections and reveals evolutionary insight into shikonin biosynthesis. Hortic. Res. 2022;10:uhab087. doi: 10.1093/hr/uhab087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S., Ogiyama Y., Kusano H., Shimada H., Kawamukai M., Kadowaki K. Metabolic engineering of coenzyme Q by modification of isoprenoid side chain in plant. FEBS Lett. 2006;580:955–959. doi: 10.1016/j.febslet.2006.01.023. [DOI] [PubMed] [Google Scholar]
- Takahashi S., Ohtani T., Satoh H., Nakamura Y., Kawamukai M., Kadowaki K. Development of coenzyme Q10-enriched rice using sugary and shrunken mutants. Biosci. Biotechnol. Biochem. 2010;74:182–184. doi: 10.1271/bbb.90562. [DOI] [PubMed] [Google Scholar]
- Takahashi S., Ohtani T., Iida S., Sunohara Y., Matsushita K., Maeda H., Tanetani Y., Kawai K., Kawamukai M., Kadowaki K.-i. Development of CoQ10-enriched rice from giant embryo lines. Breed. Sci. 2009;59:321–326. doi: 10.1270/jsbbs.59.321. [DOI] [Google Scholar]
- Theodoulou F.L., Job K., Slocombe S.P., Footitt S., Holdsworth M., Baker A., Larson T.R., Graham I.A. Jasmonic acid levels are reduced in COMATOSE ATP-binding cassette transporter mutants. Implications for transport of jasmonate precursors into peroxisomes. Plant Physiol. 2005;137:835–840. doi: 10.1104/pp.105.059352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threlfall D.R., Whistance G.R. Biosynthesis of ubiquinone—a search for polyprenyl phenol and quinone precursors. Phytochemistry. 1970;9:355–359. doi: 10.1016/s0031-9422(00)85147-x. [DOI] [Google Scholar]
- Toda T., Hayashi K., Ogiyama Y., Yokomi K., Nakagawa T., Kaino T., Kawamukai M. Functional conservation of coenzyme Q biosynthetic genes among yeasts, plants, and humans. PLoS One. 2014;9:e99038. doi: 10.1371/journal.pone.0099038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalba J.M., Navas P. Regulation of coenzyme Q biosynthesis pathway in eukaryotes. Free Radic. Biol. Med. 2021;165:312–323. doi: 10.1016/j.freeradbiomed.2021.01.055. [DOI] [PubMed] [Google Scholar]
- Wang M., Toda K., Maeda H.A. Biochemical properties and subcellular localization of tyrosine aminotransferases in Arabidopsis thaliana. Phytochemistry. 2016;132:16–25. doi: 10.1016/j.phytochem.2016.09.007. [DOI] [PubMed] [Google Scholar]
- Wang M., Toda K., Block A., Maeda H.A. TAT1 and TAT2 tyrosine aminotransferases have both distinct and shared functions in tyrosine metabolism and degradation in Arabidopsis thaliana. J. Biol. Chem. 2019;294:3563–3576. doi: 10.1074/jbc.RA118.006539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Hekimi S. Mitochondrial respiration without ubiquinone biosynthesis. Hum. Mol. Genet. 2013;22:4768–4783. doi: 10.1093/hmg/ddt330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Hekimi S. The complexity of making ubiquinone. Trends Endocrinol. Metabol. 2019;30:929–943. doi: 10.1016/j.tem.2019.08.009. [DOI] [PubMed] [Google Scholar]
- Watmough N.J., Frerman F.E. The electron transfer flavoprotein: ubiquinone oxidoreductases. Biochim. Biophys. Acta Bioenerg. 2010;1797:1910–1916. doi: 10.1016/j.bbabio.2010.10.007. [DOI] [PubMed] [Google Scholar]
- White M.D., Payne K.A., Fisher K., Marshall S.A., Parker D., Rattray N.J., Trivedi D.K., Goodacre R., Rigby S.E., Scrutton N.S., et al. UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis. Nature. 2015;522:502–506. doi: 10.1038/nature14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widhalm J.R., Dudareva N. A familiar ring to it: biosynthesis of plant benzoic acids. Mol. Plant. 2015;8:83–97. doi: 10.1016/j.molp.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. An "Electronic Fluorescent Pictograph" browser for exploring and analyzing large-scale biological data sets. PLoS One. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L.X., Hsieh E.J., Watanabe S., Allan C.M., Chen J.Y., Tran U.C., Clarke C.F. Expression of the human atypical kinase ADCK3 rescues coenzyme Q biosynthesis and phosphorylation of Coq polypeptides in yeast coq8 mutants. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2011;1811:348–360. doi: 10.1016/j.bbalip.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L.X., Ozeir M., Tang J.Y., Chen J.Y., Jaquinod S.K., Fontecave M., Clarke C.F., Pierrel F. Overexpression of the Coq8 kinase in Saccharomyces cerevisiae coq null mutants allows for accumulation of diagnostic intermediates of the coenzyme Q6 biosynthetic pathway. J. Biol. Chem. 2012;287:23571–23581. doi: 10.1074/jbc.M112.360354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J.J., Zhang X.F., Jiang Y., Fan H., Li J.X., Li C.Y., Zhao Q., Yang L., Hu Y.H., Martin C., et al. A unique flavoenzyme operates in ubiquinone biosynthesis in photosynthesis-related eukaryotes. Sci. Adv. 2021;7:eabl3594. doi: 10.1126/sciadv.abl3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Wakitani S., Hayashi K., Miki R., Kawamukai M. High production of sulfide in coenzyme Q deficient fission yeast. Biofactors. 2008;32:91–98. doi: 10.1002/biof.5520320111. [DOI] [PubMed] [Google Scholar]
- Zhou F., Pichersky E. More is better: the diversity of terpene metabolism in plants. Curr. Opin. Plant Biol. 2020;55:1–10. doi: 10.1016/j.pbi.2020.01.005. [DOI] [PubMed] [Google Scholar]
- Zhu X.F., Suzuki K., Saito T., Okada K., Tanaka K., Nakagawa T., Matsuda H., Kawamukai M. Geranylgeranyl pyrophosphate synthase encoded by the newly isolated gene GGPS6 from Arabidopsis thaliana is localized in mitochondria. Plant Mol. Biol. 1997;35:331–341. doi: 10.1023/a:1005898805326. [DOI] [PubMed] [Google Scholar]
- Ziosi M., Di Meo I., Kleiner G., Gao X.H., Barca E., Sanchez-Quintero M.J., Tadesse S., Jiang H., Qiao C., Rodenburg R.J., et al. Coenzyme Q deficiency causes impairment of the sulfide oxidation pathway. EMBO Mol. Med. 2017;9:96–111. doi: 10.15252/emmm.201606356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman B.K., Silva I.D., Bartel B. The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid beta-oxidation. Plant Physiol. 2001;127:1266–1278. doi: 10.1104/pp.010550. [DOI] [PMC free article] [PubMed] [Google Scholar]