Abstract
Crop wild relatives are an important reservoir of natural biodiversity. However, incorporating wild genetic diversity into breeding programs is often hampered by reproductive barriers and a lack of accurate genomic information. We assembled a high-quality, accurately centromere-anchored genome of Gossypium anomalum, a stress-tolerant wild cotton species. We provided a strategy to discover and transfer agronomically valuable genes from wild diploid species to tetraploid cotton cultivars. With a (Gossypium hirsutum × G. anomalum)2 hexaploid as a bridge parent, we developed a set of 74 diploid chromosome segment substitution lines (CSSLs) of the wild cotton species G. anomalum in the G. hirsutum background. This set of CSSLs included 70 homozygous substitutions and four heterozygous substitutions, and it collectively contained about 72.22% of the G. anomalum genome. Twenty-four quantitative trait loci associated with plant height, yield, and fiber qualities were detected on 15 substitution segments. Integrating the reference genome with agronomic trait evaluation of the CSSLs enabled location and cloning of two G. anomalum genes that encode peroxiredoxin and putative callose synthase 8, respectively, conferring drought tolerance and improving fiber strength. We have demonstrated the power of a high-quality wild-species reference genome for identifying agronomically valuable alleles to facilitate interspecific introgression breeding in crops.
Keywords: wild diploid species, Gossypium anomalum, genome, chromosome segment substitution lines, drought tolerance, fiber strength
This study reports the assembly of a high-quality, accurately centromere-anchored genome of Gossypium anomalum, a diploid stress-tolerant wild Gossypium species indigenous to Africa, and the development of a set of 74 G. anomalum chromosome segment substitution lines (CSSLs) in the G. hirsutum background. By integrating the reference genome with agronomic trait evaluation of the CSSLs, two G. anomalum genes conferring drought tolerance and improving fiber strength are readily located and cloned.
Introduction
The world is expected to reach a projected population of 9.8 billion in 2050 and 11.2 billion in 2100 (www.un.org), but increases in crop yields are not keeping pace with the concomitant growing demand. The narrow genetic base of modern crops has resulted in a yield plateau from crop breeding (Tanksley and McCouch, 1997). Crop wild relatives represent both raw material for breeding and a valuable source of diversity that can be used to improve the adaptation and agricultural performance of modern crop cultivars. However, the key challenge in using wild diversity is overcoming inherent difficulties in distant hybridization such as cross-incompatibility and the sterility of F1 hybrids (Zhang and Batley, 2020). Another obstacle is the dual lack of efficient introgression strategies and genomic information, which greatly hinders the wide use of wild species in breeding programs. Genome assemblies are expected to provide increased opportunities for revealing wild-species-derived genetic variation (Bredeson et al., 2016) and introducing disease resistance (Szymanski et al., 2020; Wang et al., 2020), plant architecture for high yield (Stein et al., 2018; Tian et al., 2019; Mamidi et al., 2020), and quality (Szymanski et al., 2020) into cultivated varieties. It is now possible to rapidly discover and clone agronomic genes from crop wild relatives and engineer them into domesticated varieties with the help of their reference genome. Therefore, wild relatives of modern crops can be a rich resource to mine for useful variants lost during domestication (Stein et al., 2018; Hake and Richardson, 2019).
Gossypium anomalum (B1B1, 2n = 2x = 26), a stress-tolerant diploid wild Gossypium species, grows widely in arid to extremely arid parts of the southern Sahara, from the Sudan to the upper reaches of the Baraka River valley in Eritrea (Silow, 1941). It offers a rich source of breeding potential for desirable traits such as good fiber quality, immunity to certain bacterial diseases, resistance to insect pests, tolerance of water deficit, and cytoplasmic male sterility (Mehetre, 2010; Newaskar et al., 2013). Here, we assembled a high-quality, accurately centromere-anchored genome of G. anomalum and developed a set of 74 chromosome segment substitution lines (CSSLs) of G. anomalum in the G. hirsutum background. This genome resource enabled the discovery of genes for drought tolerance and high fiber quality (Supplemental Figure 1). This wild-species genome assembly and CSSL development will aid our understanding of the extent of hybridization between wild and domesticated populations and of wild relative diversity, and it will allow the identification of genomic regions for additional introgression breeding to improve domesticated cotton.
Results
Assembly, annotation, and evolution of the G. anomalum genome
We generated 82.68 Gb (∼64×) of high-quality long reads using the PacBio SMART platform (Supplemental Table 1). After correction using 132.61 Gb (∼103×) of Illumina paired-end data, the PacBio long reads were assembled into 611 contigs that captured 1.20 Gb of the G. anomalum genome, 363 of which were too short (total length 9.35 Mb) to be then validated and scaffolded using BioNano optical maps (Supplemental Tables 1–3). A total of 249 816 916 valid high-throughput chromosome conformation capture (Hi-C) reads were used to categorize, order, and orient these scaffolds (Supplemental Figure 2; Supplemental Tables 2 and 4). The final assembly comprised 364 scaffolds (N50 = 99.19 Mb), spanning 1.21 Gb and accounting for ∼93.66% of the estimated genome (total size 1.29 Gb based on K-mer distribution analysis and 1.35 Gb by flow-cytometry analysis), 351 of which were short scaffolds (total length 9.58 Mb) and 13 were super-scaffolds representing the complete set of G. anomalum pseudo-chromosomes and comprising 99.21% of the total assembled sequence (Table 1, Supplemental Figure 3, and Supplemental Tables 2, 5, and 6).
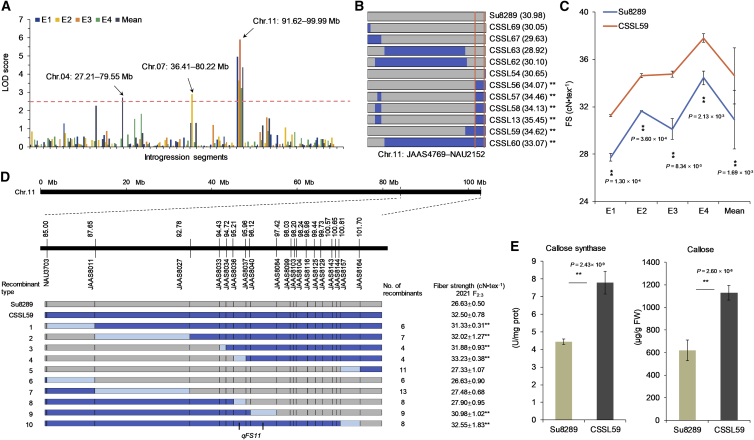
Figure 4.
Identification of valuable substitution segment associated with fiber strength and characterization of the candidate gene on Chr.11 of G. anomalum.
(A) QTL distribution for the fiber strength trait in multiple environments. The x axis represents all introgression segments and the y axis the logarithm of odds (LOD) score; the dashed line indicates the threshold value of 2.5, and the physical locations of QTLs are denoted by arrows.
(B) Graphical genotypes of SSR interval for fiber strength on Chr.11 and the phenotype of corresponding CSSLs. The orange line indicates the G. anomalum locus qFS11 mapped to the interval of JAAS4769–NAU2152 on Chr.11.
(C) Fiber strength phenotypic values of Su8289 and CSSL59 in multiple environments. ∗∗P < 0.01, Student’s t-test.
(D) Graphical genotypes and fiber strength values of Su8289, CSSL59, and 76 recombinants from the F2:3 population. Gray portions represent the genotype of Su8289, blue portions represent the genotype of CSSL59, and light blue represents regions where crossover has occurred. The table on the right indicates mean fiber strength values for the recombinant classes collected from Hainan Island. ∗∗P < 0.01, Student’s t-test.
(E) Callose synthase activity and callose content in fibers from CSSL59 and Su8289 at 20 DPA. ∗∗P < 0.01, Student’s t-test.
Table 1.
Summary of the final genome assembly and annotation for G. anomalum
| Category | G. anomalum | G. anomalum (Grover et al., 2021) |
|---|---|---|
| Sequenced genome size (bp) | 1 208 248 306 | 1 193 340 424 |
| Anchored chromosome size (bp) | 1 198 670 087 | 1 191 544 213 |
| Percentage of anchoring (%) | 99.21 | 99.85 |
| Contig N50 (Mb) | 7.78 | 10.8 |
| Scaffold N50 (bp) | 99 188 525 | 97 682 888 |
| GC content (%) | 34.25 | 34.27 |
| Complete BUSCOs (%) | 99.01 | 97.1 |
| Number of annotated genes | 42 752 | 37 830 |
| Percentage of TEs (%) | 62.59 | 47.90 |
Significant correlation was observed between the linkage and physical maps (Zhai et al., 2015) (Supplemental Figure 4). The assembly’s completeness in genic regions was supported by the identification of 2303 (99.01%) of the 2326 BUSCO groups (Simão et al., 2015) and 242 (97.58%) of the 248 core eukaryotic genes in the CEGMA v2.5 database (Parra et al., 2007) (Supplemental Tables 7 and 8). Evaluation of assembly continuity on the basis of repeat sequences yielded a long terminal repeat (LTR) Assembly Index (LAI) value (Ou et al., 2018) of 15.71. The Illumina short-read data and transcripts derived from RNA sequencing (RNA-seq) of various tissues were aligned to the genome with mapping ratios of 97.25% and 88.65% (>500 bp), respectively (Supplemental Tables 9 and 10). Assembly of centromeric regions was evaluated by chromatin immunoprecipitation and sequencing (ChIP-seq) using cotton CenH3 antibodies (Bi et al., 2020) (Supplemental Figure 5A–5C). Unique prominent ChIP-seq peaks were observed from each chromosome assembly, ranging from 0.96 to 1.89 Mb in length (Figure 1A and Supplemental Figure 5D).
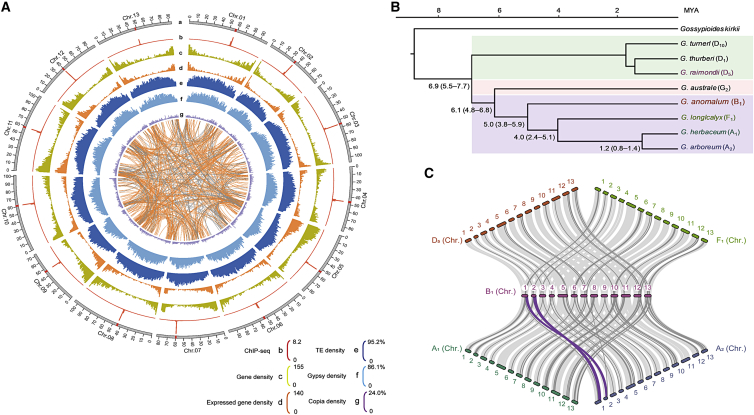
Figure 1.
Overview and evolution of the G. anomalum genome.
(A) Chromosomal characterization of the G. anomalum genome. a, centromere distribution in each chromosome; b, centromere on each chromosome by CenH3 ChIP-seq mapping; c, gene density in each chromosome; d, genes expressed in at least one tissue (root, stem, leaf, and flower); e, f, g, transposable element (TE), Gypsy, and Copia retrotransposon density, respectively, on each chromosome. The inner lines show syntenic blocks among the 13 chromosomes.
(B) Phylogenetic analysis of eight diploid cotton species and Gossypioides kirkii.
(C) Analysis of synteny among diploid cotton genomes. Light gray indicates syntenic regions, dark gray indicates inversions, and purple indicates translocations.
A total of 42 752 high-confidence protein-coding gene models were predicted in G. anomalum (Figure 1A, Table 1, and Supplemental Table 11), similar to previous predictions of published diploid Gossypium species (Paterson et al., 2012; Wang et al., 2012b; Li et al., 2014; Du et al., 2018; Udall et al., 2019a; Cai et al., 2020; Grover et al., 2020; Huang et al., 2020). Approximately 97.29% of the genes identified in G. anomalum were annotated in the Swiss-Prot, NR, KEGG, InterPro, Gene Ontology (GO), or Pfam database (Supplemental Table 12). In addition, 262 microRNAs (miRNAs), 1085 tRNAs, 774 rRNAs, and 6064 small nuclear RNAs (snRNAs) were predicted in G. anomalum (Supplemental Table 13). Transposable elements (TEs) comprising a total of 756.28 Mb accounted for 62.59% of the total genome (Figure 1A, Table 1, and Supplemental Table 14). Compared with the published G. anomalum genome (Grover et al., 2021), our assembled genome had a little larger genome size and scaffold N50, more annotated genes, a larger percentage of TEs, slightly higher complete BUSCO ratio, and 13 anchored centromeres but a similar anchoring percentage, GC content, and CEGMA ratio, a lower contig N50, and more contigs (Table 1, Supplemental Figure 5, and Supplemental Tables 6–8).
From the molecular phylogenetic tree, G. anomalum and its close relatives G. longicalyx (F1) (Grover et al., 2020), G. herbaceum (A1) (Huang et al., 2020), and G. arboreum (A2) (Du et al., 2018) were estimated to have diverged ∼5.0 (3.8–5.9) million years ago (MYA); likewise, the divergence time for G. anomalum and G. australe (G2) (Cai et al., 2020) was determined to be ∼6.1 (4.8–6.8) MYA. In addition, their common ancestor diverged from G. turneri (D10) (Udall et al., 2019a), G. thurberi (D1) (Grover et al., 2019), and G. raimondii (D5) (Udall et al., 2019a) at around ∼6.9 (5.5–7.7) MYA (Figure 1B). The LTR activity of G. anomalum increased continuously from 8.0 MYA until about 0.5 MYA, and it apparently had higher LTR retrotransposition activity than G. raimondii (D5) (Udall et al., 2019a) but lower activity than G. herbaceum (A1) (Huang et al., 2020), G. arboreum (A2) (Du et al., 2018), and G. australe (G2) (Cai et al., 2020) (Supplemental Figure 6). This finding is consistent with the species’ different genome sizes (Hawkins et al., 2006).
In one-to-one matching of syntenic blocks, approximately 86.88% of the G. anomalum genome matched with 89.99% of the G. raimondii (D5) genome (Udall et al., 2019a), 89.51% with 87.21% of G. longicalyx (F1) (Grover et al., 2020), 80.41% with 72.11% of G. herbaceum (A1) (Huang et al., 2020), 77.42% with 69.49% of G. arboreum (A2) (Du et al., 2018), and 65.21% with 64.63% of G. australe (G2) (Cai et al., 2020) (Figure 1C and Supplemental Figure 7). These results suggest that the overall collinearities of G. anomalum with G. raimondii (D5) and G. longicalyx (F1) are more conserved than those with other species. There were at least 13 inversion events spanning a total of 85.65 Mb that occurred across nine chromosomes (Chr.02–05, 07, and 10–13) between G. anomalum and G. raimondii (D5) (Udall et al., 2019a), (Figure 1C and Supplemental Table 15). A much greater degree of chromosomal rearrangement, 129.78 Mb, 184.77 Mb, 153.10 Mb, and 146.49 Mb, was detected between G. anomalum and G. longicalyx (F1) (Grover et al., 2020), G. herbaceum (A1) (Huang et al., 2020), G. arboreum (A2) (Du et al., 2018), and G. australe (G2) (Cai et al., 2020), respectively (Figure 1C, Supplemental Figure 7, and Supplemental Table 15).
Development of the CSSL population
To transfer valuable genes that control important agronomic traits from G. anomalum into G. hirsutum, a fertile hexaploid hybrid (AADDBB)1 was successfully developed by first crossing G. hirsutum cv. 86-1 × G. anomalum and then inducing chromosome doubling (Zhang et al., 2014). This hexaploid was further backcrossed with G. hirsutum cv. Su8289 to develop the CSSL population (Supplemental Figure 8). The major problem in developing a cotton wild relative CSSL population is the selection of recombinants, which occur at low frequency. In the BC2F1 segregation generation, 50 recombination types from 357 recombination events were identified among 384 BC2F1 individuals, and the recombination events occurred only between G. anomalum and the At genome of G. hirsutum (Zhai et al., 2015). In the present study, only 36 recombination types produced viable BC3F1 seeds. To recover any of the missing donor segments and obtain as many recombination types as possible, alien addition lines of 13 G. anomalum chromosomes were also backcrossed to Su8289 in the BC2F1 and BC3F1 generations. Marker-assisted selection (MAS) was conducted in each proceeding generation (Supplemental Table 16). A summary of population sizes during five successive generations during the establishment of the CSSL population is provided in Supplemental Table 17.
In the BC3F1 generation, 40 recombination types were obtained from 4331 BC3F1 individuals. Compared with BC2F1, nine new recombination types were identified on five chromosomes, and five recombination types were lost during the backcross process on the other five chromosomes (Supplemental Table 18). In the BC4F1 generation, 56 recombination types were obtained from 8540 BC4F1 individuals. Compared with BC3F1, 18 new recombination types were detected on eight chromosomes, and two recombination types were lost on Chr.04 (Supplemental Table 18). In the BC4F2 generation, 51 recombination types, including 45 homozygous genotypes and six heterozygous genotypes, were obtained from 4543 BC4F2 individuals. Compared with BC4F1, five new recombination types were detected on Chr.06, Chr.09, and Chr.11 (Supplemental Table 18), and 10 recombination types on 6 chromosomes were lost in BC4F2. We assumed that some homozygous genotypes had low gamete or zygote viability. In the BC4F3 generation, homozygous candidate substitution lines were investigated again to confirm the homozygous exotic genotype of each line; in the BC4F3 and BC4F4 generations, heterozygous substitution lines were further analyzed to identify homozygous substitution segments. A total of 53 recombination types, including 47 homozygous substitutions and 6 heterozygous substitutions, were obtained from 1533 BC4F3 individuals. Compared with BC4F2, two new recombination types were detected on Chr.05 and Chr.11 (Supplemental Table 18).
In the BC4F4 generation, all recombination types were subjected to a whole-genome survey with 230 markers evenly distributed across the G. anomalum genome. A total of 74 CSSLs were obtained, including 70 homozygous substitutions and four heterozygous substitutions. These four heterozygous CSSLs remain in the final CSSL set, as they are expected to be of further use in fine mapping significant quantitative trait locus (QTL) regions. Genome-wide scanning enabled us to detect an additional 21 substitution lines, most of which carry more than one introgression segment in addition to the target introgression (Supplemental Table 18).
Among the 74 CSSLs numbered from CSSL1 to CSSL74, 41, 26, and 7 CSSLs had one, two, and three substitution segments of the donor parents, respectively (Supplemental Table 19). CSSL24 on Chr.03 and Chr.08, CSSL37 on Chr.05, CSSL43 on Chr.06, and CSSL51 on Chr.09 still carried heterozygous substitution segments even when they were selfed two or three times. The cover length of substitution segments among different CSSLs ranged from 4.75 cM (CSSL17) to 267.45 cM (CSSL43), with an average of 72.09 cM. The total cover length was 1668.69 cM, and the coverage rate was about 72.22% of the G. anomalum genome (Figure 2A and Supplemental Table 20). Uneven distribution among the 13 chromosomes was observed: Chr.01 and Chr.11 of G. anomalum were completely represented by 20 and 28 different CSSLs, respectively, whereas Chr.03 and Chr.07 were only represented by one CSSL, and Chr.10 had the lowest coverage of only 17.89%. The distribution of all the substitution segments among the CSSL population is shown in Figure 2A and 2B.
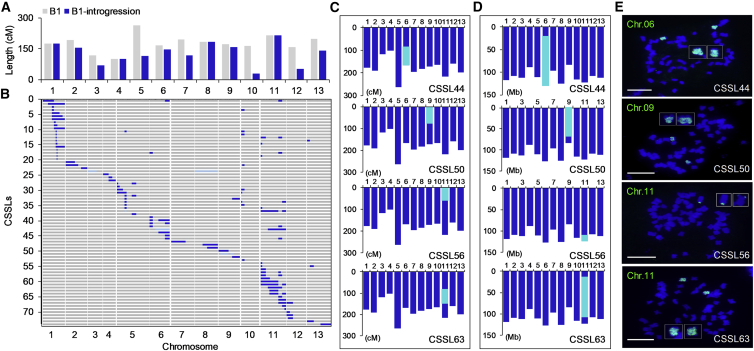
Figure 2.
Characterization of the CSSL population derived from G. anomalum and introgression segments identified in CSSL44, CSSL50, CSSL56, and CSSL63 based on SSR markers, resequencing, and FISH experiments.
(A) Genome coverage of substitution segments in the CSSL population.
(B) Chromosomal distribution of introgression segments identified from the 74 CSSLs and genome coverage of substitution segments in the CSSL population. Gray represents the genotype of the recurrent parent Su8289, blue represents the genotype of the donor parent G. anomalum, and light blue represents heterozygous regions. On the vertical axis are the 74 CSSLs, arranged from top to bottom, and on the horizontal axis are the 13 chromosomes of the G. hirsutum At1 genome, arranged from left to right.
(C–E) Introgression segments identified by SSR markers (C), resequencing (D), and FISH experiments (E). Introgression segments are indicated by light-blue boxes. Scale bars, 10 μm.
To verify the accuracy of introgression segment identifications using simple sequence repeat (SSR) markers, six CSSLs were resequenced and verified for the presence of corresponding G. anomalum segments (Figure 2C and 2D, Supplemental Figure 9, and Supplemental Table 21). Four of these six CSSLs (CSSL44, CSSL50, CSSL56, and CSSL63) were further confirmed by fluorescence in situ hybridization (FISH) using G. anomalum-specific oligo-FISH probes. FISH signals from corresponding G. anomalum chromosomes were detected, and signal coverage was consistent with the results of SSR and resequencing (Figure 2E and Supplemental Figure 10).
Agronomically valuable QTLs identified in G. anomalum
A total of nine quantitative traits, including plant height (PH), three yield traits (boll seed index [SI], boll weight [BW], and lint percentage [LP]), and five fiber quality traits (fiber length [FL], fiber strength [FS], micronaire [MIC], fiber uniformity [FU], and fiber elongation [FE]), were investigated in four environments (Supplemental Table 22). The genetic coefficient of variation (GCV) of the nine traits ranged from 0.60% (FU in E3) to 17.21% (BW in E4), indicating that various degrees of genetic variation existed in all of the traits. The relatively low GCVs were related to the relatively high genetic background recovery rate of Su8289. The heritability of traits ranged from 35.91% (FE in Joint) to 98.49% (MIC in E4). The mean values of most traits were similar to those of the recurrent parent, indicating that transgressive segregation occurred in both positive and negative directions in the CSSL population. Therefore, CSSLs with significant differences from Su8289 could be found at both sides for most traits in Supplemental Figure 11, and these CSSLs might carry elite genes from G. anomalum.
Twenty-four QTLs were detected on 15 substitution segments of six chromosomes (Supplemental Table 23). The total cover length of the 15 segments was 402.25 cM, accounting for 24.11% of the total cover length of all the substitution segments. Of these 24 QTLs, four were associated with PH, seven with yield traits (four with BW and three with LP), and 13 with fiber quality traits (three with FL, three with FS, two with MIC, two with FU, and three with FE) (Supplemental Table 23). Some QTLs were located on the same substituted segments; for example, four different QTLs (qBW11-1, qLP11-2, qFL11, and qFU11) all mapped to the interval NAU3234–NAU2877 on Chr.11, and two QTLs (qBW01 and qFL01-2) were both located on the same interval as the linked marker JAAS0392 on Chr.01 (Supplemental Table 23). Of the 24 QTLs, 13 for 7 traits (PH, LP, FL, FS, MIC, FU, and FE) were identified as valuable loci involving 12 different substitution segments on Chr.01, Chr.04, Chr.05, and Chr.11. Two valuable QTLs, qFL01-1 and qFE01, were located on a single G. anomalum segment on Chr.01, indicating that this segment could increase FL and elongation simultaneously (Supplemental Table 23).
Causal gene conferring drought tolerance in G. anomalum
Through the transfer of genomic fragments associated with distinct agronomic traits into cultivars in CSSLs, it is possible to rapidly discover and clone agronomic genes from crop wild relatives that have been sequenced. G. anomalum is distinctly characterized by its extreme drought tolerance due to adaptation to an extremely arid environment (Figure 3A); accordingly, we attempted to identify G. ANOMALUM DROUGHT TOLERANCE (GADT) gene(s) from the CSSLs by combining substitution mapping, expression profiling analyses, and functional validation by virus-induced gene silencing (VIGS). Drought tolerance of all CSSLs was assessed using 20% polyethylene glycol (PEG) treatment, and three introgression lines (CSSL29, CSSL30, and CSSL31) showed tolerance to PEG stress at the seedling stage (Figure 3B; Supplemental Figure 12A and 12B). Interestingly, these three CSSLs carried overlapping substitution segments on Chr.05 anchored by SSR markers JAAS6365 and JAAS5604. Moreover, another introgression line, CSSL28, harbored a G. anomalum segment anchored by JAAS6365–JAAS0803 that also overlapped with the interval anchored by JAAS6365 and JAAS5604. PEG treatment demonstrated that CSSL28 was drought sensitive (Supplemental Figure 12C). These results indicate that one or more genes involved in drought tolerance were located in the JAAS0803–JAAS5604 interval (Chr.05: 1 415 831 bp to 2 878 211 bp) (Supplemental Figure 12D). This interval of the G. anomalum genome contained 192 genes.
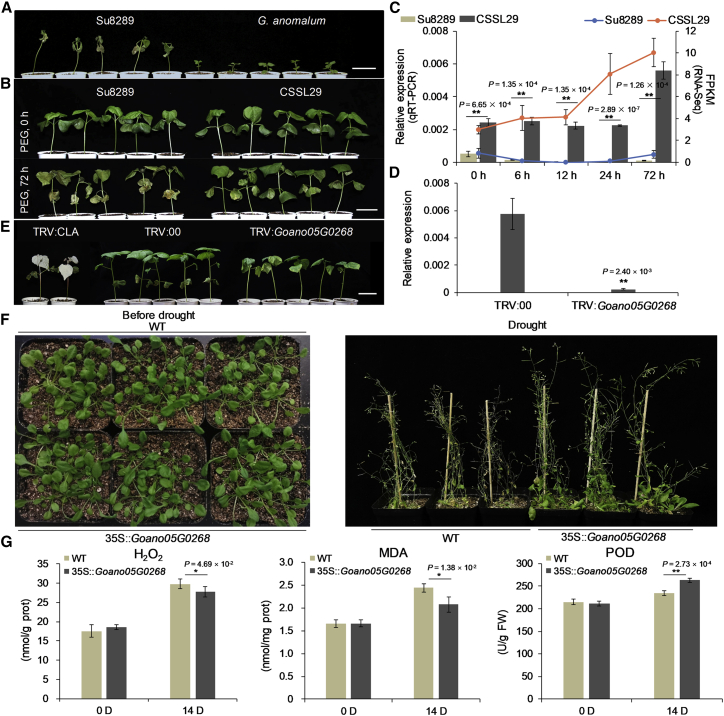
Figure 3.
Drought tolerance of CSSL29, functional verification of Goano05G0268 by VIGS in CSSL29, and ectopic expression in Arabidopsis.
(A) Phenotypic comparison of Su8289 and G. anomalum in response to water deficit. Scale bar, 5 cm.
(B) Phenotypic comparison of Su8289 and CSSL29 in response to drought stress using 20% PEG treatment. Photographs were taken at 0 and 72 h after PEG treatment. Scale bar, 5 cm.
(C) Expression level of Goano05G0268 in Su8289 and CSSL29 under PEG stress at 0, 6, 12, 24, and 72 h. The left y axis shows the relative expression according to qRT–PCR, and the right y axis shows the FPKM (fragments per kilobase per million mapped reads) value obtained from RNA-seq data. ∗∗P < 0.01, Student’s t-test.
(D) Transcript levels of Goano05G0268 in leaves from CSSL29 plants infected with pTRV2 and pTRV2-Goano05G0268 under PEG stress at 72 h, evaluated by qRT–PCR. ∗∗P < 0.01, Student’s t-test.
(E) Phenotypes of CSSL29 plants infected with pTRV2, pTRV2-Goano05G0268, and pTRV2-CLA. Scale bar, 5 cm.
(F) Phenotypic comparison of Arabidopsis EE and WT seedlings after 25 days of drought stress.
(G) Endogenous H2O2 content, MDA content, and POD activity of Arabidopsis EE and WT lines before and after 14 days of drought stress. ∗∗P < 0.01, Student’s t-test.
To exploit genes in the JAAS0803–JAAS5604 interval involved in drought tolerance, a global analysis of transcriptome dynamics was performed to compare CSSL29 and its recurrent parent Su8289, both grown under 20% PEG stress conditions (Supplemental Table 24). Twenty genes in the interval were upregulated in CSSL29 relative to Su8289 under PEG stress at any time point (6, 12, 24, and 72 h) (Supplemental Table 25). Upon integrating these upregulated genes with functional annotations of their Arabidopsis orthologs, nine genes were validated by qRT–PCR and their functional relevance further confirmed by VIGS (Figure 3C, 3D, and 3E; Supplemental Figures 13 and 14; Supplemental Tables 26 and 27). The expression level of nine genes in VIGS-silenced plants at 72 h of PEG stress was much lower than that in control plants (Figure 3D; Supplemental Figure 14). Plants in which the peroxiredoxin gene Goano05G0268 was silenced were more sensitive to PEG stress at 72 h than TRV:00 plants, whereas plants in which any of the other eight genes were silenced showed no significant sensitivity to PEG stress (Figure 3E; Supplemental Figure 14). Hydrogen peroxide (H2O2) and malondialdehyde (MDA) content and peroxidase (POD) activity were measured in TRV:00 and TRV:Goano05G0268 cotton plants under PEG stress at 0 and 72 h. Goano05G0268-silenced plants had higher (P < 0.01) H2O2 and MDA content but lower (P < 0.01) POD activity than TRV:00 plants under PEG stress at 72 h (Supplemental Figure 15).
Ectopic expression of Goano05G0268 modulates drought response in Arabidopsis (Figure 3F and 3G; Supplemental Figure 16). Three ectopic expression (EE) transgenic lines of Goano05G0268 grown on Murashige and Skoog (MS) medium with 200 and 250 mM mannitol had longer roots (P < 0.05) than wild-type (WT) lines (Supplemental Figure 16B and 16C). These lines had relatively lower (P < 0.01) H2O2 and MDA content but higher (P < 0.01) POD activity than WT lines to modulate drought-tolerant phenotypes (Figure 3F and 3G). Goano05G0268 encodes a peroxiredoxin protein, which participates in protection against oxidative damage and plays a role in plant responses to drought stress (Rey et al., 2005; Dietz, 2007; Hernández et al., 2012; Marquez-Garcia et al., 2015; AbdElgawad et al., 2020). Therefore, Goano05G0268 may be involved in drought response and might play an important role in drought-adapted evolution.
The Goano05G0268 gene in CSSL29 and its orthologous gene GH_A05G0249 (Hu et al., 2019) (GhGADT) in Su8289 were isolated by PCR amplification (Supplemental Table 28). Sequence analysis revealed that the Su8289-derived GhGADT contained one deletion (408 bp), three insertions (2549 bp, 159 bp, and 2 bp) in the second intron, and 11 SNPs within exons 1, 2, and 3 (Supplemental Figure 17), leading to the conclusion that this gene likely became pseudogenized in Su8289 over the course of evolution. The Goano05G0268 gene in G. anomalum was also aligned with its orthologs in other allotetraploid cotton species, such as G. hirsutum L. acc. TM-1 (Wang et al., 2019), G. barbadense L. cv. Hai7124 (Hu et al., 2019), G. barbadense L. accession 3-79 (Wang et al., 2019), G. tomentosum (Chen et al., 2020), G. mustelinum (Chen et al., 2020), and G. darwinii (Chen et al., 2020). The results were similar to those of the alignment of G. anomalum and Su8289, that is, Goano05G0268 orthologs in these allotetraploid cotton species contained a ∼2500-bp insertion and other small deletions and insertions in the second intron and different numbers of SNPs within exons 1, 2, and 3 (Supplemental Figure 18 and Supplemental Table 29).
Candidate gene for improved fiber strength in G. anomalum
Among the two elite QTLs identified as associated with FS, the G. anomalum locus qFS11 mapped to the JAAS4769–NAU2152 interval on Chr.11 was consistently detected across all four environments (Figure 4A and Supplemental Table 30). Six CSSLs carrying the overlapping segment (CSSL13, CSSL56, CSSL57, CSSL58, CSSL59, and CSSL60) all showed significantly (P < 0.01) greater FS values than the recurrent parent Su8289; CSSLs lacking that segment showed no significant difference from Su8289, indicating a tight association between the introgression segment and greater FS (Figure 4B and 4C; Supplemental Figure 19). To mine the G. ANOMALUM FIBER STRENGTH (GAFS) gene, we performed fine mapping of qFS11 in an F2:3 population constructed from CSSL59 × Su8289 using data collected at Hainan Island in 2021. Substitution mapping narrowed down qFS11 to the 1.19-Mb interval of JAAS8037–JAAS8040, which contained 49 candidate genes (Figure 4D and Supplemental Table 31). We then conducted a transcriptomic analysis of fibers from CSSL59, CSSL13, and Su8289 at 20 days post anthesis (DPA) (Supplemental Table 32), when the fiber elongates and the secondary cell wall (SCW) thickens. Twelve genes located in the qFS11 interval JAAS8037–JAAS8040 showed upregulated expression in fibers from CSSL59 and CSSL13 relative to those from Su8289 (Supplemental Table 33).
Of those 12 genes, nine contained probable function-altering variations relative to their homologs in G. hirsutum, whether from transcript splicing, frameshift, gain of stop codon, or loss of stop codon (Supplemental Table 34). These nine genes were verified by qRT–PCR (Supplemental Figure 19 and Supplemental Table 35). Among them, one gene, Goano11G3883 encoding PUTATIVE CALLOSE SYNTHASE 8 PROTEIN (PCSY8P), was significantly upregulated at 20 DPA in CSSL13, CSSL56, CSSL57, CSSL58, CSSL59, and CSSL60, all of which carried the same substituted segment JAAS4769–NAU2152 (Supplemental Figure 20). The orthologous gene GH_A11G3315 in G. hirsutum (Hu et al., 2019) contains two deletions (2 bp and 13 bp) within exon 30 that result in a frameshift (Supplemental Figure 21A). The protein encoded by GH_A11G3315 has five amino acids deleted from the Glucan_synthase domain (Supplemental Figure 21B). We amplified the partial sequence of this gene containing structural variation from CSSL59 and Su8289 and confirmed their sequence differences (Supplemental Figure 22 and Supplemental Table 36).
The SCW thickening stage, characterized by the synthesis and accumulation of cellulose, is a key period that determines FS (Meinert and Delmer, 1977; Hsieh et al., 2000; Kim and Triplett, 2001). PCSY8P is involved in the biosynthesis of callose ((1→3)-β-D-glucan), an intermediate in cellulose biosynthesis (Meier et al., 1981; Brown et al., 1996; Salnikov et al., 2003; Ruan et al., 2004). Callose synthase activity and relative callose content were significantly higher (P < 0.01) in CSSL59 than in Su8289 (Figure 4E). Therefore, Goano11G3883 appears to be a candidate for the GAFS gene that improves FS. The GAFS gene may have been pseudogenized in the A subgenome chromosome of G. hirsutum during evolution.
Discussion
High-quality assembly of a wild-species genome will greatly facilitate interspecific introgression breeding
It is well known that wild relatives of modern crops are rich resources to mine for useful variants lost during domestication (Hake and Richardson, 2019). Obtaining a high-quality reference genome is an essential step in understanding the evolution, origin, and domestication of wild and cultivated species and enables the best utilization of genetic resources and the improvement of agronomic traits in modern plant breeding (Stein et al., 2018; Mamidi et al., 2020; Szymanski et al., 2020). In this study, we assembled a high-quality genome of G. anomalum based on PacBio Illumina, BioNano, and Hi-C technology. The quality of our assembly and the recently published genome of G. anomalum (Grover et al., 2021) are comparable, as we used similar approaches (PacBio + Illumina + BioNano + Hi-C for our assembly, PacBio + Hi-C for Grover et al., 2021) for genome sequencing and assembly. The different results may arise from differences in analytical methods and criteria. The two genomes can complement each other in reference and utility value for the cotton community.
During the domestication and improvement of cultivated species, particularly during the formation of allopolyploid species, some agronomic genes lose their function or become defunctionalized because of genome shock; these can be recovered by transferring the original functional gene from a crop wild relative into domesticated stock. However, interspecific hybridization between cultivated species and wild species from the tertiary gene pool is often challenging, hampered by reproductive barriers and a lack of genomic information (Wendel et al., 2010). Here, we have developed a strategy to transfer elite genes from a wild diploid cotton species to tetraploid cultivars by developing CSSLs, performing transcriptome and sequence variation analysis, and identifying causal genes integrated with the reference genome. We report that introgressive G. anomalum genes encoding peroxiredoxin and putative callose synthase 8 can confer drought tolerance and improve FS in upland cotton. Such transfers of original functional genes from wild or progenitor species into G. hirsutum along with the corresponding agronomic trait, such as FS, will be very important and useful in interspecific introgression breeding to improve yield and quality.
Development of a first set of CSSLs derived from a diploid wild species and their application in cotton breeding
In previous reports, the tetraploid wild species G. tomentosum (AADD)3, G. mustelinum (AADD)4, and G. darwinii (AADD)5, were used as donor parents to create CSSLs (Wang et al., 2012a; Chandnani et al., 2017; Keerio et al., 2018). There are no previous reports on creating a CSSL library with a diploid wild cotton species as a donor parent owing to crossability inhibition and limited recombination between chromosomes of wild and cultivated plants. In this study, a fertile hexaploid hybrid (AADDBB)1 developed in our previous studies was further backcrossed three times with G. hirsutum cv. Su8289 and selfed three times. MAS was applied in every generation. Finally, a set of 74 CSSLs in the G. hirsutum background were developed. This G. anomalum CSSL population is comparable with those developed for other crop species with regard to the “quality” of introgression lines, e.g., relatively high exotic genome coverage (72.22%) and a high grade of pureness (41 out of 74 lines possess a single exotic introgression segment). These lines will be powerful materials for QTL identification, fine mapping, map-based cloning, and ultimately breeding utilization.
Cotton breeders have long recognized that low-performing wild cotton species can contribute agronomically favorable alleles. Successful examples of utilizing cotton wild species in cotton-breeding history include the use of G. harknessii (D2-2) as the source of cytoplasmic male sterility (Meyer, 1973), the use of G. thurberi (D1) to improve fiber quality (Culp and Harrell, 1973; Culp et al., 1979), and the use of G. aridum (D4) to provide resistance to reniform nematode (Sacks and Robinson, 2009). In this study, agronomically positive QTL alleles from G. anomalum were identified for drought tolerance, fiber quality (e.g., FL, FS), and yield traits (LP), despite G. anomalum having short seed hairiness. Linkage drag is one of the main factors that affect the utilization of wild species in breeding programs. We observed that exotic chromosome fragments were associated with deleterious effects on some traits such as BW and LP. For example, 28 of 74 CSSLs showed significantly lower BW than the recurrent parent Su8289 (Supplemental Figure 11). Linkage drag between fiber quality and yield traits was observed for G. anomalum loci on Chr.01 and Chr.11 (Supplemental Table 23). However, CSSL17 contained a positive FL QTL at a small introgression fragment of Chr.01 and showed no detectable negative effects on BW and LP, indicating that further genetic dissection of the target region could break linkage drag. Disruption of deleterious linkages and reduction of pleiotropic effects could be achieved by fine mapping of target QTL regions or editing negative genes using CRISPR technology.
Methods
Plant materials
Highly homozygous G. anomalum plants were cultivated at Lishui Plant Experiment Station, Jiangsu Academy of Agricultural Sciences (JAAS), China. Young leaves from a single plant were harvested and frozen immediately in liquid nitrogen for extraction of genomic DNA.
PacBio sequencing
An improved cetyltrimethylammonium bromide (CTAB) method was used to extract the genomic DNA of G. anomalum. Size selection was carried out on more than 5 μg of sheared and concentrated DNA by the BluePippin system. Approximately 40-kb SMRTbell libraries were prepared according to the protocol released by PacBio. A total of 14 single-molecule, real-time (SMRT) cells were run on the PacBio Sequel system, and 82.75 Gb of polymerase reads were produced.
Illumina sequencing
A total of 1.5 μg of G. anomalum genomic DNA was used as input material for sample preparation. Sequencing libraries were generated using the TruSeq Nano DNA HT Sample Preparation Kit (Illumina, USA) following the manufacturer’s recommendations; index codes were added in order to attribute sequences to each sample. DNA samples were fragmented by sonication to sizes of 230 bp, 350 bp, and 500 bp, and the fragments were end-polished, A-tailed, and ligated with the full-length adapter for Illumina sequencing with further PCR amplification. PCR products were purified (AMPure XP system), and libraries were analyzed for size distribution and quantification with an Agilent 2100 Bioanalyzer and real-time PCR, respectively. The libraries were sequenced on an Illumina HiSeq platform, producing 46.80 Gb, 41.38 Gb, and 44.92 Gb of raw data, respectively, for each fragment size.
De novo assembly
The G. anomalum PacBio data were first corrected by the Fast Alignment and CONsensus (Falcon) sense method (option correctedErrorRate = 0.025). High-quality pre-assembled reads were then used for genome assembly via Falcon using the Overlap-Layout-Consensus algorithm. Quiver software was used to compute the genomic consensus and to call variants relative to the reference. The resulting contigs were then used to map the Illumina short reads with BWA (version 0.7.15-r1140) (Li and Durbin, 2009) and were polished using Pilon (version 1.22) with default parameters (Walker et al., 2014).
Genome assembly improvement with BioNano optical maps
A highly homozygous G. anomalum plant was cultivated in a greenhouse at the JAAS. Young leaves were collected after 4 days of dark treatment. High-molecular-weight DNA was isolated and labeled according to the BioNano protocol with the single-stranded nicking endonuclease Nt.BssSI (Lam et al., 2012). After labeling and staining, the complete genome-specific marker library was constructed and loaded onto the Loading Saphyr array. The stretched DNA molecules were imaged on a BioNano Irys system, and the raw image data were converted into bnx files. After filtering on the Label SNR filter (threshold: 3.5–20), molecule length (more than 150 kb), and label density, a total of 291.88 Gb of single-molecule data were produced. High-quality labeled single molecules were pairwise aligned, clustered, and assembled into contigs according to the BioNano assembly pipeline (Lam et al., 2012; Cao et al., 2014), yielding a physical map with a total length of 1.29 Gb. Hybrid scaffolding was performed with the assembled PacBio contigs and BioNano optical maps using BioNano Solve. To detect the best matches and potential reciprocal scaffolding of each dataset, BioNano genome nick-based maps were compared with in silico nick maps of the genome sequence. If any sites conflicted between the genome sequence and optical maps, both were broken at those sites and reassembled using Hybrid Scaffold software.
Hi-C library construction and chromosome assembly
Hi-C library construction for G. anomalum was performed according to the Hi-C procedure (Servant et al., 2015). The Hi-C library was controlled for quality and sequenced on an Illumina HiSeq X Ten sequencer. After filtering, we obtained 249 816 916 valid read pairs for the high-quality assembly of G. anomalum. The 396 assembled scaffolds were separately broken into fragments with an average length of 50 kb, and valid Hi-C read pairs were mapped to these fragments using BWA (Li and Durbin, 2009). Uniquely mapped reads were retained and used for assembly with LACHESIS (Burton et al., 2013). Any two fragments that showed inconsistent connections with information from the raw scaffolds were checked manually, and these corrected scaffolds were then assembled by LACHESIS. Finally, 364 scaffolds with a total length of 1.21 Gb were anchored, and 13 super-scaffolds (99.19%) were oriented to the respective 13 high-quality groups of G. anomalum.
The completeness of the genome assembly was evaluated using the Benchmarking Universal Single-Copy Orthologs (BUSCO) dataset (Simão et al., 2015) and the Core Eukaryotic Genes Mapping Approach (CEGMA) version 2.5 with default settings (Parra et al., 2007). The LAI was used to evaluate the continuity of repeat element sequences throughout the assembly (Ou et al., 2018). In addition, assembly accuracy and completeness were also supported by alignment via BLAT of Illumina short-read data and transcripts derived from RNA-seq of different tissues (Kent, 2002).
Identification of centromeric regions by CenH3 ChIP
ChIP experiments were performed according to a published protocol (Nagaki et al., 2004). A total of 47.8 and 22.9 million CenH3 ChIP-seq and genomic control reads (150 bp) were generated, respectively, 17.8 and 2.8 million of which were mapped to unique sites in the G. anomalum genome assembly using Bowtie 2. Read density was calculated for each genomic window by dividing the total number of uniquely mapped reads by the total number of mapped nucleotides. The density of ChIP-seq reads in each window was normalized using the density of input reads. CenH3 domains were detected with SICER version 1.1 (Zang et al., 2009) using the following parameters: 200-bp windows, required fold change (ChIP/control) ≥5 and false discovery rate (FDR) <0.01, and allowed gaps of 400 bp.
Repeat sequence and non-coding RNA prediction
A de novo TE library was first constructed using LTR_FINDER (Xu and Wang, 2007), RepeatScout (Price et al., 2005), and PILER-DF (Edgar and Myers, 2005). TEs were then identified with RepeatMasker (version 4.0.6) (Chen, 2004) run against the de novo TE library and the Repbase database (version 20.01) (Jurka et al., 2005). Tandem Repeats Finder was used to search for tandem repeats in the genome (Benson, 1999). The tRNAscan-SE program was used to predict tRNA fragments, and INFERNAL was used against the Rfam database (release 9.1) to identify rRNA, miRNA, and snRNA fragments (Griffiths-Jones et al., 2005). All intact LTR-retrotransposons (LTRs) in cotton genome species were used to calculate the insertion time using the formula time = Ks/2r, where Ks is the synonymous substitutions per synonymous site and r is the rate of nucleotide substitution (which was set to 7 × 10−9) (Li et al., 2014).
Protein-coding gene prediction
De novo, homology-based, and RNA-seq-based predictions were used to annotate protein-coding genes in the G. anomalum genome. De novo prediction used the programs AUGUSTUS (version 3.0.2) (Stanke and Waack, 2003), GENSCAN (version 1.0) (Burge and Karlin, 1997), geneid (version 1.3) (Blanco et al., 2007), GlimmerHMM (version 3.0.2) (Majoros et al., 2004), and SNAP (Korf, 2004). For identification by homology, protein sequences from six plant species—Arabidopsis thaliana (Arabidopsis Genome Initiative, 2000), G. arboreum (CRI) (Du et al., 2018), G. hirsutum (NAU) (Zhang et al., 2015), G. raimondii (JGI) (Paterson et al., 2012), Populus trichocarpa (Tuskan et al., 2006), and Theobroma cacao (Argout et al., 2011)—were aligned against the repeat-masked G. anomalum genome using tblastn (E-value ≤1e−5). BLAT (Kent, 2002) and GeneWise (version 2.4.1) (Birney et al., 2004) were used to predict gene models based on the aligned sequences. Finally, RNA-seq-based predictions were made on the basis of two methods for assembling the data into unique transcript sequences: mapping the RNA-seq data to the G. anomalum genome using TopHat (version 2.0.8) and cufflinks (version 2.1.1), and using Trinity to assemble the RNA-seq data followed by PASA to model the gene structures (Haas et al., 2003). Consensus gene models were generated by merging the de novo predictions, protein alignments, and transcript data using EVidenceModeler (EVM) (Haas et al., 2008). Finally, the gene models generated by EVM were adjusted using PASA based on the transcript assembly, yielding 42 752 predicted protein-coding genes.
Functional annotation of protein-coding genes was performed according to the best BLAST hit by BLASTP (E-value ≤1e−5) against Swiss-Prot (Boeckmann et al., 2003), Pfam, and NCBI non-redundant (NR) protein databases. Motifs and domains were annotated by searching against InterPro (version 29.0) (Zdobnov and Apweiler, 2001). GO terms (Ashburner et al., 2000) for each gene were obtained from the corresponding InterPro description. Pathways in which the protein-coding genes might be involved were assigned by performing BLAST searches (McGinnis and Madden, 2004) against the KEGG database (release 53) Kanehisa and Goto, 2000) (E-value ≤1e−5). In all, 41 592 protein-coding genes were annotated, accounting for 96.5% of the predicted genes.
Phylogenetic tree construction and gene synteny analysis
We identified single-copy gene families using the OrthoFinder package based on protein sequences of eight diploid cotton species and Gossypioides kirkii (Udall et al., 2019b). The corresponding CDS sequences of proteins from single-copy gene families were extracted and aligned using MAFF with default parameters (Rozewicki et al., 2019). The alignments were then concatenated into a super matrix, which was used for phylogenetic tree reconstruction via maximum-likelihood methods implemented in RAxML with a GTRGAMMA substitution model (Stamatakis, 2014). Divergence times among these species were estimated using the MCMCtree program (Yang, 1997). The collinearity between cotton species was estimated using the MCScanX package (Wang et al., 2012c).
Development and molecular characterization of the CSSL population
From the BC2F1 generation produced in 2013, recombinants carrying as few substitution segments as possible were selected to make successive backcrosses with Su8289 to produce the BC4F1 generation in 2015. In parallel, to obtain as many recombination types as possible, alien addition lines of 13 G. anomalum chromosomes were also backcrossed with the BC2F1 and BC3F1 generations. The BC4F1 plants were subsequently selfed three times to achieve stable and homozygous recombined lines.
To efficiently monitor G. anomalum substitution segments and reduce labor cost as much as possible, 130 G. anomalum-specific markers (7–14 markers per chromosome) were used to track the target substitution segments (marker names listed in Supplemental Table 37) in each generation. These markers were selected as being near the recombination breakpoints in the BC2F1 generation and represented genomic regions delimited by specific recombination events. MAS for foreground selection was used to confirm the presence of target substitution segments in each generation. In the BC4F3 generation, homozygous candidate substitution lines were investigated again to confirm the homozygous exotic genotype of each line and were then propagated until BC4F4 to obtain a sufficient number of seeds for phenotype studies.
In the BC4F4 generation, all recombination types were subjected to foreground and background genotyping with 230 markers evenly distributed across the G. anomalum genome. The size of the substitution segment in each finished CSSL was calculated, with the midpoint of two adjacent markers with different genotypes being considered the endpoint of a substitution segment (Young and Tanksley, 1989). Graphical genotype analysis was performed using GGT 2.0 software (van Berloo, 2008).
All generations except BC4F3 were planted at the Experiment Station for Plant Science (JAAS), Nanjing, China (N31°36′ E119°10′); BC4F3 plants were grown at Nanbin Farm, Sanya, China (N18°21′ E109°10′).
SSR marker analysis
Genomic DNA was extracted from fresh leaves using a modified CTAB method for cotton. PCR reactions were performed using a Applied Biosystems 2720 thermal cycler (Thermo Fisher Scientific, MA, USA) in a 10-μL volume containing 7.5 μL of PCR Master Mix (1×, TsingKe Biotech, Beijing, China), 0.5 μL of each primer (100 μM), and 1.5 μL of DNA template (20 ng). For the collection of genotypic data, banding patterns were marked as follows: the same band type as G. hirsutum was marked as “1;” the same band type as G. anomalum was marked as “2;” the same band type as the F1 was marked as “3;” and missing and unclear band types were marked as “0.”
Resequencing of CSSLs and Su8289 and identification of SNPs
Leaves from six CSSLs (CSSL29, CSSL44, CSSL50, CSSL56, CSSL59, and CSSL63) and Su8289 were collected from the Lishui Plant Experiment Station, JAAS, China. DNA was extracted from leaves using an improved CTAB method, and at least 1.5 μg of DNA was used for library construction via the TruSeq Library Construction Kit with an insert size of about 350 bp. All libraries were sequenced on the Illumina HiSeq platform with genome coverage of at least 50× for Su8289, 30× for CSSL63, and 10× for other CSSLs (Supplemental Table 21). After trimming of low-quality bases using Trimmomatic (version 0.32), the clean data were mapped to the A subgenome of G. hirsutum TM-1 using BWA (Li and Durbin, 2009). All uniquely mapped reads were extracted and SNP identification performed using GATK (version 3.1.1) (McKenna et al., 2010) and SAMtools (version 0.1.19) (Li et al., 2009).
Development of G. anomalum chromosome type-specific oligo-FISH probes and FISH
Oligo-FISH probes were designed using Chorus2 based on a previously published pipeline (Albert et al., 2019). In brief, single-copy oligos (45 nt) were generated from Chr.06, Chr.09, and Chr.11 of G. anomalum with the parameters of chorus “-l 45 -homology 75 -step 5.” Repetitive oligos were then filtered using G. anomalum genomic sequence data. In total, 117 685, 117 287, and 156 457 oligos were generated from G. anomalum Chr.06, Chr.09, and Chr.11, respectively. To distinguish G. anomalum Chr.06, Chr.09, and Chr.11 in CSSL lines, we developed oligo probes specific to G. anomalum following a previously published protocol (do Vale Martins et al., 2019). Oligos from G. anomalum Chr.06, Chr.09, and Chr.11 were first mapped to the G. hirsutum reference genome and then classified into three sets on the basis of that mapping: oligos that were identical to G. hirsutum, oligos that were completely different (PAV oligos), and oligos that contained mismatches and/or indels (SNP oligos). Finally, all PAV oligos and SNP oligos were selected as haplotype-specific oligo-FISH probes for G. anomalum. This process yielded 266 PAV and 8 861 SNP oligos, 314 PAV and 7 185 SNP oligos, and 259 PAV and 10 597 SNP oligos for G. anomalum Chr.06, Chr.09, and Chr.11, respectively. All oligos were synthesized by MYcroarray (Ann Arbor, MI, USA).
FISH was performed as previously described with several modifications (Meng et al., 2020). Mitotic chromosome spreads were prepared using root tips, and slides with good metaphase chromosomes were selected for the FISH experiment. After hybridization, chromosomes were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, USA), and the signal was detected directly under an Olympus BX63 fluorescence microscope. Chromosome and FISH signals were captured using cellSens Dimension 1.9 software with an Olympus DP80 CCD camera. Final image adjustments were performed using Adobe Photoshop CC software.
Phenotypic identification of the CSSL population
The CSSL population and Su8289 were planted in a completely randomized block design with three replications at the Experiment Station for Plant Science (JAAS), Nanjing, China in 2017 (designated E1), 2018 (E2), and 2019 (E4). In each environment, the seeds were first sown in plug trays to ensure good seedling emergence and growth rate. Seedlings were transplanted to the field after 30 days, with one row per replication and 10 plants per row. Plants were separated by 50 cm and rows by 100 cm. Su8289 was planted among every tenth row and used as a boundary. The lines were also planted in a completely randomized block design with four replications at the National Crop Seeding Farm, Kuerle, China (N41°50′ E85°48′) in 2018 (E3). In this environment, the seeds were sown directly on mulch film with one row per replication and 30 plants per row. The rows were 4 m in length with 40 cm between rows. Field management was carried out according to the local cotton production management model in each environment.
PH was measured as the length from the cotyledon node to the apical bud at maturity in environments E1 and E2, and the average of 10 plants per row was taken as the corresponding phenotypic value. Thirty mature cotton bolls, fully open and from the interior middle of the plants, were selected to investigate BW, LP, and SI in E1, E2, E3, and E4 (SI was determined only in E2 and E3). Approximately 15 g of lint from each sample was used for quality testing of FL, FS, MIC, FU, and FE at the Supervision, Inspection and Test Center of Cotton Quality, Ministry of Agriculture and Rural Affairs, Anyang, China.
Descriptive analysis of phenotypes was performed using PROC MEANS in SAS/STAT version 9.1.2 (SAS Institute, NC, USA).
Identification of substitution segments associated with agronomic traits
A likelihood ratio test based on stepwise regression (RSTEP-LRT) in QTL IciMapping 4.1 (Li et al., 2007) was used to detect associations between substitution segments and traits in each environment with the statistical threshold of LOD (likelihood of odds) >2.5. RSTEP-LRT is suitable for QTL mapping in a non-idealized CSSL population in which each line carries two or more segments from the donor parent. The stepwise regression was used to select the most important segments for the trait of interest, and the likelihood ratio test was used to calculate the LOD score of each chromosome segment (Wang et al., 2006). The QTL designations begin with “q” followed by abbreviations of trait name, chromosome name, and serial number.
Fine mapping of the genes for fiber strength
The hybridization of CSSL59 × Su8289 was performed in 2019. The F1 seeds were planted and self-pollinated in Hainan Island. The F2 population of 2168 individuals was grown at Lishui Plant Experiment Station, JAAS, China in 2020. Nineteen polymorphic SSR markers were used to genotype all the F2 plants, and 81 recombinants were identified (Supplemental Table 31). The seeds of recombinants in the F2 population were harvested separately, and these seeds of each F2 individual generated F2:3 families, which were planted in Hainan Island. The fiber quality of homozygous recombinants in the F2:3 families was tested at the Supervision, Inspection and Test Center of Cotton Quality, Ministry of Agriculture and Rural Affairs, Anyang, China.
Transcriptome analysis
For the drought tolerance test, CSSL29 and Su8289 were germinated in soil and then irrigated with 20% PEG6000 solution at the seedling stage (14 days after germination). Leaves were collected at 0, 6, 12, 24, and 72 h after PEG treatment. Fiber samples at 20 DPA were collected from CSSL13, CSSL59, and Su8289. Three biological replicates of total RNA for each sample were extracted using a plant RNA purification kit (Omega, Beijing, China) following the manufacturer’s instructions. Libraries were constructed using the Illumina TruSeq Stranded RNA Library Preparation Kit and then sequenced on an Illumina HiSeq platform (150 bp paired-end). We used the DESeq2 package (http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html) to identify genes that were differentially expressed across samples or groups. We defined genes as significantly differentially expressed when comparison yielded a fold change ≥ 2 and an FDR < 0.05.
Quantitative RT–PCR analysis
To estimate the validity of RNA-seq technology for expression profile analysis and screen the candidate genes involved in PEG stress and fiber development, total RNA was extracted from leaves at 0, 6, 12, 24, and 72 h after PEG treatment and from fiber samples at 20 DPA and then reverse transcribed into cDNA. Endogenous cotton histone-3 (AF024716) was used as an internal standard to normalize the total amount of cDNA in each reaction. Gene-specific primers corresponding to the candidate genes were designed with Beacon Designer software (Supplemental Tables 26 and 35). The qRT–PCR experiment was conducted using the TB Green Premix Ex Taq (Tli RNaseH Plus) kit (TaKaRa), and three biological replicates of all reactions were run on a QuantStudio 5 Real-Time PCR System. Relative transcript levels were computed using the 2–ΔCt method, where ΔCt is the difference in Ct values between the control histone-3 gene (AF024716) products and the target gene products.
Virus-induced gene-silencing assay
For knockdown of Goano05G0164, Goano05G0170, Goano05G0207, Goano05G0222, Goano05G0235, Goano05G0236, Goano05G0268, Goano05G0301, and Goano05G0319, approximately 300-bp fragments of these genes were PCR-amplified from CSSL29 cDNA. The primers used are given in Supplemental Table 27. The resulting PCR products were recombined into pTRV2 to produce VIGS vectors, after which pTRV1 and recombinant pTRV2 vectors were separately introduced into Agrobacterium strain GV3101 by means of electroporation (Bio-Rad, Hercules, CA, USA); bacteria harboring each vector were then mixed by equal volume and incubated for 3 h at 28°C. Infiltration of CSSL29 seedlings with mature cotyledons but without a visible true leaf (7 days after germination) was then performed by inserting the combined Agrobacterium suspension into the cotyledons via a syringe. The plants were grown at 23°C with a 16 h/8 h (light/dark) cycle and a relative humidity of 60%. VIGS effectiveness was assessed by generating a TRV:GbCLA construct using the G. anomalum CLA1 gene as described previously. The VIGS experiments were repeated at least three times with more than five plants in each repeat.
Drought stress tolerance assays in Arabidopsis
We used the floral dip method (Clough and Bent, 1998) to generate EE lines of Goano05G0268 in Arabidopsis and selected the positive lines on MS basal salt mixture medium with 50 mg/l kanamycin. The positive lines were further confirmed by PCR amplification (Supplemental Figure 16A). Three EE (T1 generation) and WT lines were grown on MS medium containing 0, 200, and 250 mM mannitol. The root length was measured after 8 days. To assess drought stress during seedling development, the WT and EE lines (T0 generation) were well watered for 21 days and then treated without supplemental water in the soil to place the plants under drought stress. After 25 days, all of the WT plants wilted and lost vitality owing to severe drought stress, whereas most EE plants remained alive (Figure 3F).
Determination of physiological and biochemical indexes
Callose content (Keppler and Novacky, 1987) and callose synthase activity in fibers of CSSL59 and Su8289 at 20 DPA were measured using the aniline blue staining method. POD activity and H2O2 and MDA content in TRV:00 and TRV:Goano05G0268 cotton plants under PEG stress and in EE and WT lines of Arabidopsis under drought stress were measured with assay kits (JianCheng, Nanjing, China).
Funding
National Natural Science Foundation of China (31471545, 32171986, 32100494, and 32070544), Jiangsu Agricultural Science and Technology Innovation Fund (CX (20) 3139), and Leading Innovative and Entrepreneur Team Introduction Program of Zhejiang (2019R01002).
Author contributions
X.S. conceptualized the project. X.S., T.Z., K.W., Z.X., J.C., and S.M. conceived and designed the project. Z.X., S.M., P.X., C.Z., and F.H. constructed the CSSL population. Z.X. and S.M. collected the plant materials and prepared DNA and RNA for Illumina and PacBio SMRT sequencing. Z.X., S.M., and J.C. performed genome assembly and annotation. S.M. and Z.X. analyzed the BioNano and Hi-C data. J.C. analyzed the genome evolution of G. anomalum and identified SNPs in CSSL and Su8289. Z.M. and Y.Z. identified centromeric regions by CenH3 ChIP-seq mapping. X.S., Z.X., S.M., Y.Q., T.W., X.Z., J.L., J.G., W.N., X.C., and S.W. performed the field experiments and identified the phenotypes of the CSSL population. Z.X., S.M., and Y.S. performed fine mapping of genes for fiber strength. Z.X., S.M., Q.G., L.Z., Y.S., J.Z., J.X., W.J., N.W., and X.L. analyzed genes related to fiber development and drought stress. J.C., Z.X., and S.M. carried out data submission. Z.X., X.S., T.Z., S.M., and K.W. wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgments
No conflict of interest declared.
Published: June 22, 2022
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Contributor Information
Kai Wang, Email: kwang5@126.com.
Tianzhen Zhang, Email: cotton@zju.edu.cn.
Xinlian Shen, Email: xlshen68@126.com.
Accession numbers
The G. anomalum genome assembly and annotation data are available at https://www.cottongen.org/. All raw sequencing data generated in the current study are deposited in the BioProject database of NCBI and the National Genomics Data Center under accession numbers PRJNA697836 and PRJCA004607, respectively. PacBio, Illumina paired-end, Hi-C, and ChIP-seq data of G. anomalum have been deposited in the Sequence Read Archive (SRA) under accession numbers SRR19241842–SRR19241849, SRR19241851–SRR19241856, SRR19241861, SRR19241872, SRR19241876–SRR19241877, SRR19241883, SRR19241894, SRR19241902, SRR19241905, SRR19241916–SRR19241917, and SRR19426739. The BioNano file ID is SUPPF_0000004289. All of the RNA-seq data are available at the SRA under accession numbers SRR19241840–SRR19241841, SRR19241850, SRR19241857–SRR19241860, SRR19241862–SRR19241871, SRR19241873–SRR19241875, SRR19241879–SRR19241882, SRR19241884–SRR19241893, SRR19241895, SRR19241903–SRR19241904, and SRR19241906–SRR19241915. The resequencing data for CSSLs and Su8289 are available under at the SRA under accession numbers SRR19261769, SRR19241896–SRR19241897, SRR19241899–SRR19241901, and SRR19241878.
Supplemental information
References
- AbdElgawad H., Avramova V., Baggerman G., Van Raemdonck G., Valkenborg D., Van Ostade X., Guisez Y., Prinsen E., Asard H., Van den Ende W., et al. Starch biosynthesis contributes to the maintenance of photosynthesis and leaf growth under drought stress in maize. Plant Cell Environ. 2020;43:2254–2271. doi: 10.1111/pce.13813. [DOI] [PubMed] [Google Scholar]
- Albert P.S., Zhang T., Semrau K., Rouillard J.M., Kao Y.H., Wang C.J.R., Danilova T.V., Jiang J.M., Birchler J.A. Whole-chromosome paints in maize reveal rearrangements, nuclear domains, and chromosomal relationships. Proc. Natl. Acad. Sci. USA. 2019;116:1679–1685. doi: 10.1073/pnas.1813957116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Argout X., Salse J., Aury J.M., Guiltinan M.J., Droc G., Gouzy J., Allegre M., Chaparro C., Legavre T., Maximova S.N., et al. The genome of Theobroma cacao. Nat. Genet. 2011;43:101–108. doi: 10.1038/ng.736. [DOI] [PubMed] [Google Scholar]
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/Nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi Y., Zhao Q., Yan W., Li M., Liu Y., Cheng C., Zhang L., Yu X., Li J., Qian C., et al. Flexible chromosome painting based on multiplex PCR of oligonucleotides and its application for comparative chromosome analyses in Cucumis. Plant J. 2020;102:178–186. doi: 10.1111/tpj.14600. [DOI] [PubMed] [Google Scholar]
- Birney E., Clamp M., Durbin R. GeneWise and genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco E., Parra G., Guigó R. Using geneid to identify genes. Curr. Protoc. Bioinformatics. 2007;Chapter 4:Unit 4.3. doi: 10.1002/0471250953.bi0403s18. [DOI] [PubMed] [Google Scholar]
- Boeckmann B., Bairoch A., Apweiler R., Blatter M.C., Estreicher A., Gasteiger E., Martin M.J., Michoud K., O'Donovan C., Phan I., et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredeson J.V., Lyons J.B., Prochnik S.E., Wu G.A., Ha C.M., Edsinger-Gonzales E., Grimwood J., Schmutz J., Rabbi I.Y., Egesi C., et al. Sequencing wild and cultivated cassava and related species reveals extensive interspecific hybridization and genetic diversity. Nat. Biotechnol. 2016;34:562–570. doi: 10.1038/nbt.3535. [DOI] [PubMed] [Google Scholar]
- Brown R.M., Saxena I.M., Kudlicka K. Cellulose biosynthesis in higher plants. Trends Plant Sci. 1996;1:149–156. doi: 10.1016/S1360-1385(96)80050-1. [DOI] [Google Scholar]
- Burge C., Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Burton J.N., Adey A., Patwardhan R.P., Qiu R., Kitzman J.O., Shendure J. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat. Biotechnol. 2013;31:1119–1125. doi: 10.1038/nbt.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Cai X., Wang Q., Wang P., Zhang Y., Cai C., Xu Y., Wang K., Zhou Z., Wang C., et al. Genome sequencing of the Australian wild diploid species Gossypium australe highlights disease resistance and delayed gland morphogenesis. Plant Biotechnol. J. 2020;18:814–828. doi: 10.1111/pbi.13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Hastie A.R., Cao D., Lam E.T., Sun Y., Huang H., Liu X., Lin L., Andrews W., Chan S., et al. Rapid detection of structural variation in a human genome using nanochannel-based genome mapping technology. GigaScience. 2014;3:34. doi: 10.1186/2047-217X-3-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandnani R., Wang B., Draye X., Rainville L.K., Auckland S., Zhuang Z., Lubbers E.L., May O.L., Chee P.W., Paterson A.H. Segregation distortion and genome-wide digenic interactions affect transmission of introgressed chromatin from wild cotton species. Theor. Appl. Genet. 2017;130:2219–2230. doi: 10.1007/s00122-017-2952-y. [DOI] [PubMed] [Google Scholar]
- Chen Z.J., Sreedasyam A., Ando A., Song Q.X., De Santiago L.M., Hulse-Kemp A.M., Ding M.Q., Ye W.X., Kirkbride R.C., Jenkins J., et al. Genomic diversifications of five Gossypium allopolyploid species and their impact on cotton improvement. Nat. Genet. 2020;52:525–533. doi: 10.1038/s41588-020-0614-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinformatics. 2004;Chapter 4 doi: 10.1002/0471250953.bi0410s05. Unit 4.10. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Culp T.W., Harrell D.C. Breeding methods for improving yield and fiber quality of upland cotton (Gossypium hirsutum L.) Crop Sci. 1973;13:686–689. doi: 10.2135/cropsci1973.0011183X001300060030x. [DOI] [Google Scholar]
- Culp T.W., Harrell D.C., Kerr T. Some genetic implications in the transfer of high fiber strength genes to upland cotton. Crop Sci. 1979;19:481–484. doi: 10.2135/cropsci1979.0011183X001900040013x. [DOI] [Google Scholar]
- Dietz K.J. In: Peroxiredoxin Systems. Flohé L., Harris J.R., editors. Springer; 2007. The dual function of plant peroxiredoxins in antioxidant defence and redox signaling; pp. 267–294. [DOI] [PubMed] [Google Scholar]
- do Vale Martins L., Yu F., Zhao H., Dennison T., Lauter N., Wang H., Deng Z., Thompson A., Semrau K., Rouillard J.M., et al. Meiotic crossovers characterized by haplotype-specific chromosome painting in maize. Nat. Commun. 2019;10:4604. doi: 10.1038/s41467-019-12646-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X., Huang G., He S., Yang Z., Sun G., Ma X., Li N., Zhang X., Sun J., Liu M., et al. Resequencing of 243 diploid cotton accessions based on an updated A genome identifies the genetic basis of key agronomic traits. Nat. Genet. 2018;50:796–802. doi: 10.1038/s41588-018-0116-x. [DOI] [PubMed] [Google Scholar]
- Edgar R.C., Myers E.W. PILER: identification and classification of genomic repeats. Bioinformatics. 2005;21:i152–i158. doi: 10.1093/bioinformatics/bti1003. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S., Moxon S., Marshall M., Khanna A., Eddy S.R., Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover C.E., Arick M.A., II, Thrash A., Conover J.L., Sanders W.S., Peterson D.G., Frelichowski J.E., Scheffler J.A., Scheffler B.E., Wendel J.F. Insights into the evolution of the New World diploid cottons (Gossypium, subgenus Houzingenia) based on genome sequencing. Genome Biol. Evol. 2019;11:53–71. doi: 10.1093/gbe/evy256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover C.E., Pan M., Yuan D., Arick M.A., Hu G., Brase L., Stelly D.M., Lu Z., Schmitz R.J., Peterson D.G., et al. The Gossypium longicalyx genome as a resource for cotton breeding and evolution. G3 (Bethesda) 2020;10:1457–1467. doi: 10.1534/g3.120.401050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover C.E., Yuan D., Arick M.A., Miller E.R., Hu G., Peterson D.G., Wendel J.F., Udall J.A. The Gossypium anomalum genome as a resource for cotton improvement and evolutionary analysis of hybrid incompatibility. G3 (Bethesda) 2021;11:jkab319. doi: 10.1093/g3journal/jkab319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B.J., Delcher A.L., Mount S.M., Wortman J.R., Smith R.K., Hannick L.I., Maiti R., Ronning C.M., Rusch D.B., Town C.D., et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003;31:5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B.J., Salzberg S.L., Zhu W., Pertea M., Allen J.E., Orvis J., White O., Buell C.R., Wortman J.R. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008;9:R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake S., Richardson A. Using wild relatives to improve maize. Science. 2019;365:640–641. doi: 10.1126/science.aay5299. [DOI] [PubMed] [Google Scholar]
- Hawkins J.S., Kim H., Nason J.D., Wing R.A., Wendel J.F. Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Res. 2006;16:1252–1261. doi: 10.1101/gr.5282906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández I., Cela J., Alegre L., Munné-Bosch S. In: Plant Responses to Drought Stress. Aroca R., editor. Springer; 2012. Antioxidant defenses against drought stress; pp. 231–258. [Google Scholar]
- Hsieh Y.L., Hu X.P., Wang A.J. Single fiber strength variations of developing cotton fibers - strength and structure of G. hirsutum and G. barbedense. Textil. Res. J. 2000;70:682–690. doi: 10.1177/004051750007000805. [DOI] [Google Scholar]
- Hu Y., Chen J., Fang L., Zhang Z., Ma W., Niu Y., Ju L., Deng J., Zhao T., Lian J., et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 2019;51:739–748. doi: 10.1038/s41588-019-0371-5. [DOI] [PubMed] [Google Scholar]
- Huang G., Wu Z., Percy R.G., Bai M., Li Y., Frelichowski J.E., Hu J., Wang K., Yu J.Z., Zhu Y. Genome sequence of Gossypium herbaceum and genome updates of Gossypium arboreum and Gossypium hirsutum provide insights into cotton A-genome evolution. Nat. Genet. 2020;52:516–524. doi: 10.1038/s41588-020-0607-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J. Repbase update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/Nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keerio A.A., Shen C., Nie Y., Ahmed M.M., Zhang X., Lin Z. QTL mapping for fiber quality and yield traits based on introgression lines derived from Gossypium hirsutum x G. tomentosum. Int. J. Mol. Sci. 2018;19:243. doi: 10.3390/Ijms19010243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J. BLAT--The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppler L.D., Novacky A. The initiation of membrane lipid-peroxidation during bacteria-induced hypersensitive reaction. Physiol. Mol. Plant Pathol. 1987;30:233–245. doi: 10.1016/0885-5765(87)90037-3. [DOI] [Google Scholar]
- Kim H.J., Triplett B.A. Cotton fiber growth in planta and in vitro. models for plant cell elongation and cell wall biogenesis. Plant Physiol. 2001;127:1361–1366. doi: 10.1104/pp.010724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. Gene finding in novel genomes. BMC Bioinf. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E.T., Hastie A., Lin C., Ehrlich D., Das S.K., Austin M.D., Deshpande P., Cao H., Nagarajan N., Xiao M., et al. Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat. Biotechnol. 2012;30:771–776. doi: 10.1038/nbt.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Fan G., Wang K., Sun F., Yuan Y., Song G., Li Q., Ma Z., Lu C., Zou C., et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 2014;46:567–572. doi: 10.1038/ng.2987. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Genome Project Data Processing S. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Ye G., Wang J. A modified algorithm for the improvement of composite interval mapping. Genetics. 2007;175:361–374. doi: 10.1534/genetics.106.066811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majoros W.H., Pertea M., Salzberg S.L. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics. 2004;20:2878–2879. doi: 10.1093/bioinformatics/bth315. [DOI] [PubMed] [Google Scholar]
- Mamidi S., Healey A., Huang P., Grimwood J., Jenkins J., Barry K., Sreedasyam A., Shu S., Lovell J.T., Feldman M., et al. A genome resource for green millet Setaria viridis enables discovery of agronomically valuable loci. Nat. Biotechnol. 2020;38:1203–1210. doi: 10.1038/s41587-020-0681-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez-Garcia B., Shaw D., Cooper J.W., Karpinska B., Quain M.D., Makgopa E.M., Kunert K., Foyer C.H. Redox markers for drought-induced nodule senescence, a process occurring after drought-induced senescence of the lowest leaves in soybean (Glycine max) Ann. Bot. 2015;116:497–510. doi: 10.1093/aob/mcv030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis S., Madden T.L. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehetre S.S. Wild Gossypium anomalum: a unique source of fibre fineness and strength. Curr. Sci. 2010;5:e11405. doi: 10.1371/journal.pone.0011405. [DOI] [Google Scholar]
- Meier H., Buchs L., Buchala A.J., Homewood T. (1→3)-β-D-Glucan (callose) is a probable intermediate in biosynthesis of cellulose of cotton fibres. Nature. 1981;289:821–822. doi: 10.1038/289821a0. [DOI] [Google Scholar]
- Meinert M.C., Delmer D.P. Changes in biochemical composition of the cell wall of the cotton fiber during development. Plant Physiol. 1977;59:1088–1097. doi: 10.1104/pp.59.6.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Z., Han J., Lin Y., Zhao Y., Lin Q., Ma X., Wang J., Zhang M., Zhang L., Yang Q., et al. Characterization of a Saccharum spontaneum with a basic chromosome number of x = 10 provides new insights on genome evolution in genus Saccharum. Theor. Appl. Genet. 2020;133:187–199. doi: 10.1007/s00122-019-03450-w. [DOI] [PubMed] [Google Scholar]
- Meyer V.G. Proceedings of Beltwide Cotton Production Research Conference. 1973. Fertility-restorer genes for cytoplasmic male sterility from Gossypium harknessii. [Google Scholar]
- Nagaki K., Cheng Z., Ouyang S., Talbert P.B., Kim M., Jones K.M., Henikoff S., Buell C.R., Jiang J. Sequencing of a rice centromere uncovers active genes. Nat. Genet. 2004;36:138–145. doi: 10.1038/ng1289. [DOI] [PubMed] [Google Scholar]
- Newaskar G.S., Chimote V.P., Mehetre S.S., Jadhav A.S. Interspecific hybridization in Gossypium L.: characterization of progenies with different ploidy-confirmed multigenomic backgrounds. Plant Breed. 2013;132:211–216. doi: 10.1111/pbr.12031. [DOI] [Google Scholar]
- Ou S., Chen J., Jiang N. Assessing genome assembly quality using the LTR Assembly Index (LAI) Nucleic Acids Res. 2018;46:e126. doi: 10.1093/nar/gky730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G., Bradnam K., Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- Paterson A.H., Wendel J.F., Gundlach H., Guo H., Jenkins J., Jin D., Llewellyn D., Showmaker K.C., Shu S.Q., Udall J., et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature. 2012;492:423–427. doi: 10.1038/nature11798. [DOI] [PubMed] [Google Scholar]
- Price A.L., Jones N.C., Pevzner P.A. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21:i351–i358. doi: 10.1093/bioinformatics/bti1018. [DOI] [PubMed] [Google Scholar]
- Rey P., Cuiné S., Eymery F., Garin J., Court M., Jacquot J.P., Rouhier N., Broin M. Analysis of the proteins targeted by CDSP32, a plastidic thioredoxin participating in oxidative stress responses. Plant J. 2005;41:31–42. doi: 10.1111/j.1365-313X.2004.02271.x. [DOI] [PubMed] [Google Scholar]
- Rozewicki J., Li S., Amada K.M., Standley D.M., Katoh K. MAFFT-DASH: integrated protein sequence and structural alignment. Nucleic Acids Res. 2019;47:W5–W10. doi: 10.1093/nar/gkz342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y.L., Xu S.M., White R., Furbank R.T. Genotypic and developmental evidence for the role of plasmodesmatal regulation in cotton fiber elongation mediated by callose turnover. Plant Physiol. 2004;136:4104–4113. doi: 10.1104/pp.104.051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacks E.J., Robinson A.F. Introgression of resistance to reniform nematode (Rotylenchulus reniformis) into upland cotton (Gossypium hirsutum) from Gossypium arboreum and a G. hirsutum/Gossypium aridum bridging line. Field Crop. Res. 2009;112:1–6. doi: 10.1016/j.fcr.2009.01.006. [DOI] [Google Scholar]
- Salnikov V.V., Grimson M.J., Seagull R.W., Haigler C.H. Localization of sucrose synthase and callose in freeze-substituted secondary-wall-stage cotton fibers. Protoplasma. 2003;221:175–184. doi: 10.1007/s00709-002-0079-7. [DOI] [PubMed] [Google Scholar]
- Servant N., Varoquaux N., Lajoie B.R., Viara E., Chen C.J., Vert J.P., Heard E., Dekker J., Barillot E. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16:259. doi: 10.1186/S13059-015-0831-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silow R.A. The comparative genetics of Gossypium anomalum and the cultivated Asiatic cottons. J. Genet. 1941;42:259–358. doi: 10.1007/bf02982878. [DOI] [Google Scholar]
- Simão F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M., Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19(Suppl 2):ii215–ii225. doi: 10.1093/bioinformatics/btg1080. [DOI] [PubMed] [Google Scholar]
- Stein J.C., Yu Y., Copetti D., Zwickl D.J., Zhang L., Zhang C., Chougule K., Gao D., Iwata A., Goicoechea J.L., et al. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat. Genet. 2018;50:285–296. doi: 10.1038/s41588-018-0040-0. [DOI] [PubMed] [Google Scholar]
- Szymański J., Bocobza S., Panda S., Sonawane P., Cárdenas P.D., Lashbrooke J., Kamble A., Shahaf N., Meir S., Bovy A., et al. Analysis of wild tomato introgression lines elucidates the genetic basis of transcriptome and metabolome variation underlying fruit traits and pathogen response. Nat. Genet. 2020;52:1111–1121. doi: 10.1038/s41588-020-0690-6. [DOI] [PubMed] [Google Scholar]
- Tanksley S.D., McCouch S.R. Seed banks and molecular maps: unlocking genetic potential from the wild. Science. 1997;277:1063–1066. doi: 10.1126/science.277.5329.1063. [DOI] [PubMed] [Google Scholar]
- Tian J., Wang C., Xia J., Wu L., Xu G., Wu W., Li D., Qin W., Han X., Chen Q., et al. Teosinte ligule allele narrows plant architecture and enhances high-density maize yields. Science. 2019;365:658–664. doi: 10.1126/science.aax5482. [DOI] [PubMed] [Google Scholar]
- Tuskan G.A., Difazio S., Jansson S., Bohlmann J., Grigoriev I., Hellsten U., Putnam N., Ralph S., Rombauts S., Salamov A., et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Udall J.A., Long E., Hanson C., Yuan D., Ramaraj T., Conover J.L., Gong L., Arick M.A., Grover C.E., Peterson D.G., et al. De novo genome sequence assemblies of Gossypium raimondii and Gossypium turneri. G3 (Bethesda) 2019;9:3079–3085. doi: 10.1534/g3.119.400392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udall J.A., Long E., Ramaraj T., Conover J.L., Yuan D., Grover C.E., Gong L., Arick M.A., 2nd, Masonbrink R.E., Peterson D.G., et al. The genome sequence of Gossypioides kirkii illustrates a descending dysploidy in plants. Front. Plant Sci. 2019;10:1541. doi: 10.3389/fpls.2019.01541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berloo R. GGT 2.0: versatile software for visualization and analysis of genetic data. J. Hered. 2008;99:232–236. doi: 10.1093/jhered/esm109. [DOI] [PubMed] [Google Scholar]
- Walker B.J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., Cuomo C.A., Zeng Q.D., Wortman J., Young S.K., et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Nie Y., Lin Z., Zhang X., Liu J., Bai J. Molecular diversity, genomic constitution, and QTL mapping of fiber quality by mapped SSRs in introgression lines derived from Gossypium hirsutum x G. darwinii Watt. Theor. Appl. Genet. 2012;125:1263–1274. doi: 10.1007/s00122-012-1911-x. [DOI] [PubMed] [Google Scholar]
- Wang H., Sun S., Ge W., Zhao L., Hou B., Wang K., Lyu Z., Chen L., Xu S., Guo J., et al. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science. 2020;368:eaba5435. doi: 10.1126/science.aba5435. [DOI] [PubMed] [Google Scholar]
- Wang J., Wan X., Crossa J., Crouch J., Weng J., Zhai H., Wan J. QTL mapping of grain length in rice (Oryza sativa L.) using chromosome segment substitution lines. Genet. Res. 2006;88:93–104. doi: 10.1017/s0016672306008408. [DOI] [PubMed] [Google Scholar]
- Wang K., Wang Z., Li F., Ye W., Wang J., Song G., Yue Z., Cong L., Shang H., Zhu S., et al. The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 2012;44:1098–1103. doi: 10.1038/ng.2371. [DOI] [PubMed] [Google Scholar]
- Wang M., Tu L., Yuan D., Zhu D., Shen C., Li J., Liu F., Pei L., Wang P., Zhao G., et al. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat. Genet. 2019;51:224–229. doi: 10.1038/s41588-018-0282-x. [DOI] [PubMed] [Google Scholar]
- Wang Y., Tang H., Debarry J.D., Tan X., Li J., Wang X., Lee T.H., Jin H., Marler B., Guo H., et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel J.F., Brubaker C.L., Seelanan T. In: Physiology of Cotton. Stewart J.M., Oosterhuis D.M., Heitholt J.J., Mauney J.R., editors. Springer; 2010. The origin and evolution of Gossypium; pp. 1–18. [Google Scholar]
- Xu Z., Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007;35:W265–W268. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Young N.D., Tanksley S.D. Restriction fragment length polymorphism maps and the concept of graphical genotypes. Theor. Appl. Genet. 1989;77:95–101. doi: 10.1007/Bf00292322. [DOI] [PubMed] [Google Scholar]
- Zang C., Schones D.E., Zeng C., Cui K., Zhao K., Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov E.M., Apweiler R. InterProScan--an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- Zhai C., Xu P., Zhang X., Guo Q., Zhang X., Xu Z., Shen X. Development of Gossypium anomalum-derived microsatellite markers and their use for genome-wide identification of recombination between the G. anomalum and G. hirsutum genomes. Theor. Appl. Genet. 2015;128:1531–1540. doi: 10.1007/s00122-015-2528-7. [DOI] [PubMed] [Google Scholar]
- Zhang F., Batley J. Exploring the application of wild species for crop improvement in a changing climate. Curr. Opin. Plant Biol. 2020;56:218–222. doi: 10.1016/j.pbi.2019.12.013. [DOI] [PubMed] [Google Scholar]
- Zhang T., Hu Y., Jiang W., Fang L., Guan X., Chen J., Zhang J., Saski C.A., Scheffler B.E., Stelly D.M., et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015;33:531–537. doi: 10.1038/nbt.3207. [DOI] [PubMed] [Google Scholar]
- Zhang X., Zhai C., He L., Guo Q., Zhang X., Xu P., Su H., Gong Y., Ni W., Shen X. Morphological, cytological and molecular analyses of a synthetic hexaploid derived from an interspecific hybrid between Gossypium hirsutum and Gossypium anomalum. Crop J. 2014;2:272–277. doi: 10.1016/j.cj.2014.06.009. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.