Abstract
Covid-19 is now one of the most incredibly intense and severe illnesses of the twentieth century. Covid-19 has already endangered the lives of millions of people worldwide due to its acute pulmonary effects. Image-based diagnostic techniques like X-ray, CT, and ultrasound are commonly employed to get a quick and reliable clinical condition. Covid-19 identification out of such clinical scans is exceedingly time-consuming, labor-intensive, and susceptible to silly intervention. As a result, radiography imaging approaches using Deep Learning (DL) are consistently employed to achieve great results. Various artificial intelligence-based systems have been developed for the early prediction of coronavirus using radiography pictures. Specific DL methods such as CNN and RNN noticeably extract extremely critical characteristics, primarily in diagnostic imaging. Recent coronavirus studies have used these techniques to utilize radiography image scans significantly. The disease, as well as the present pandemic, was studied using public and private data. A total of 64 pre-trained and custom DL models concerning imaging modality as taxonomies are selected from the studied articles. The constraints relevant to DL-based techniques are the sample selection, network architecture, training with minimal annotated database, and security issues. This includes evaluating causal agents, pathophysiology, immunological reactions, and epidemiological illness. DL-based Covid-19 detection systems are the key focus of this review article. Covid-19 work is intended to be accelerated as a result of this study.
Keywords: Coronavirus (Covid-19), Deep Machine Learning, Diagnosis, Convolutional neural network (CNN), Disease detection and classification, Radiography Images (X-ray, CT, Ultrasound)
Introduction
Covid-19, which began in Wuhan, China [1], has afflicted all nationals worldwide since December 2019 [2, 3]. As of July 15, 2022, the WHO received 557,917,904 affirmed cases of Covid-19, totaling 6,358,899 mortalities. As of July 11, 2022, 12,130,881,147 vaccines had been administered [4, 5] globally. According to the Ministry of Health and Family Welfare [6], 43,730,071 affirmed cases[5], 140,760 active cases, 525,660 deaths, and 1,99,71,61,438 vaccines had been administered in India by July 16, 2021.
SARSCoV2 is supporter of Coronaviridae and Nidovirales groups [7]. Coronavirinae is divided into four sub-groups: Humanoid coronavirus belongs to alpha; SARS belongs to beta [8] containing humanoid coronavirus and MERS-CoV; viruses of whales and poultry belong to gamma; viruses from pigs and birds belong to the delta. SARSCoV2 [9] goes to beta composed of two extremely bacterium viruses such as SARS-CoV [10] and MERS-CoV [11]. A human contaminating beta Covid-19 measures SARSCoV2. The ethnological examination of SARSCoV2-DNA[12] shows that the disease is severely interconnected to two bat animals. Resulting in SARS similar coronavirus composed in eastern China(2018) and hereditarily different from SARS and MERS-CoV [13]. Using the DNA orders of SARSCoV2 and SARS-CoV, an additional reading [14] found that the virus is further correlated to bat category virus, as previously noticed within Rhinolophus in the Yunnan area, by 96.2% DNA order uniqueness.
In the RT-PCR study [15], the disease was spotted beginning in the upper respiratory tract (URT) and lower respiratory tract (LRT) samples on two to three days of indication. The virus-related burden was improved from day three to five in the LRT sample. The Rapid Diagnostic Test (RDT) is an antigen-finding assessment that can yield outcomes in thirty minutes [16]. The sensitivity of RT-PCR [17] experiments was poor (60–70%); however, it improved significantly over time [18]. Also, it has various defies like false positives, little compassion, costly, and needs specialists to test. As the Covid-19 patient number increases, there is an utmost necessity to create a fast testing technique that is precise and cheaper [19].
The coronavirus spreads through droplets [20] of air by coughing, sneezing, cold, and talk [21] from their nose or mouth, dispersing and affecting those nearby people's lungs. The transferred droplets into the lungs through the respiratory system started killing lung cells. This Covid-19 virus disturbs the lung system of a patient & produces patchy white shades in the respiratory part of the lung [22]. In some instances & recent studies show that individuals with no symptoms but are infected with the virus [4] also play a part in the virus spread and need to detect the coronavirus-affected portion of the lungs [23].
The finding of Covid-19 using AI methods and notably DL will support the identification of disease in the upcoming days, which will imitate in growing chances of patients' speedy recovery globally [24]. As a result, the healthcare system's work strain will be relieved worldwide [25]. A few papers have validated that CT could be a compassionate diagnostic examination to identify Covid-19 pneumonia. Also, early PCR testing is more sensitive for Covid-19 pneumonia detection than late PCR testing to assess its severity. According to a WHO report, contaminated zones used chest CT to screen Covid-19 positive individuals [26]. However, other studies hypothesize that chest X-ray holds a high potential to ease the organization of patients with Covid-19 [27]. This article elaborates on the various examples of DL-based Covid-19 detection systems with the following key contributions:
-
(i)
Recent research literature collection and classification based on the development of X-ray, CT, and multimodal radiography images.
-
(ii)
Create a taxonomy of the examined research based on pre-trained and custom models concerning image modality.
-
(iii)
Discussion on the challenging facets of current developments in DL-based Covid-19 diagnostic systems
-
(iv)
Providing direction for future research to further develop an effective and reliable Covid-19 detection system.
-
(v)
Detailed experimental and performance analysis of 64 Covid-19 detection systems based on DL.
The remaining sections of this review study are separated into subsequent sections. Section 2 classifies the taxonomy of the analyzed systems based on neural networks, classification-based tasks, and datasets that can be understood and analyzed quantitatively. Covid-19 diagnosis employing X-ray, CT, and multimodal imaging modalities using pre-trained, hybrid, and custom-trained experimental architecture with DTL are discussed in Sects. 3, 4 and 5. Section 6 discusses challenges and potential future trends. Finally, closing remarks have been concluded in Sect. 7. Performance metrics are provided in Appendix A, and Appendix B lists abbreviations utilized throughout the paper.
Classification of Deep Learning-Based Covid-19 Diagnosis Systems
This literature review emphasizes the Covid-19 prediction and classification with medical imaging modalities using DL techniques. ML and DL methods can resolve multifaceted complications by gaining insight knowledge from simple representations. The critical structure that has resulted in widespread neural network approaches is the ability to learn precise depictions. As a result, numerous layers are used in a stacked sequence of CNN, and the benefits of gaining in-depth knowledge are maximized for optimal model performance [28]. DL systems are broadly utilized in medical diagnosis systems [29] like biomedicine, competent healthcare, and medical image analysis [30]. The taxonomy used from the selected studied systems is grouped into CNN architecture, class-based task formulation, and three radiography modalities (X-ray, CT, and multimodal); each is linked to a pre-trained and custom DL model.
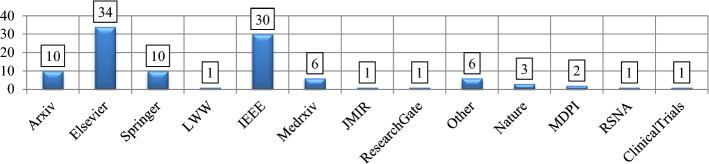
The primary task was to conduct a literature review and search for relevant published work for this paper. The literature review is a collection of principles or methods for providing a solution to the investigator's queries. The PRISMA [31] framework was utilized for this review. The article screening metrics included a heading, keywords, aims, competence requirements, datasets sources, search, measurement, synthesizing results, summarization of proofs, challenges, and conclusions. This methodology (Fig. 1) aimed to provide clear steps in DL, Covid-19, and classification. A set of inclusion criteria was determined to find the articles, and query patterns were analyzed on Elsevier, IEEEXplore, ScienceDirect, LWW, tandfonline, nature, RSNA, MDPI, SpringerLink, ArXiv, MedRxiv, ResearchGate, and other publications till July 2021. Most popular keywords were used like “Deep Learning,” “Covid-19”, “Diagnosis,” and “Radiological Imaging” for obtaining the articles. Figure 2 shows the total articles published/indexed on DL techniques on Covid-19 by several databases.
Fig. 1.
PRISMA methodology used for research review
Fig. 2.
Selected published papers on Covid-19 diagnosis using DL
CNN Architecture
DL is broadly used in various domains, such as finding abnormalities, object detection, and cataloging [32] in the biomedical arena using pre-trained or personalized models [33]. Creating a fresh CNN model is a challenging and slow process. However, pre-trained models are quicker with additional functionalities than creating a programmer-defined or custom model. Pre-trained networks are generally employed for feature mining, transfer learning, and classification [10].
Several pre-trained models used in transfer learning(TL) are built for the scale of CNN[34], 2DCNN [35], GAN [36], CNNRNN, CNNLSTM [37] and hybrid networks like FCONet [38], COVID-CheXNet [39], CovidCTNet [40], COVIDetection-Net [41], COVIDX-Net [42], DRE-Net [43]. Other custom networks like EDL-COVID [44], CVDNet [45], COVIDSDNet [46], LDC-Net [47] etc., have been developed for the Covid-19 automatic detection system.
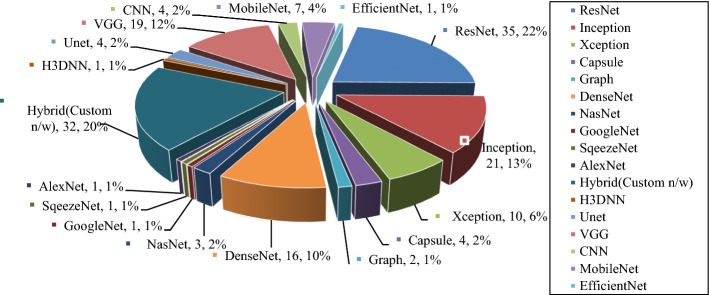
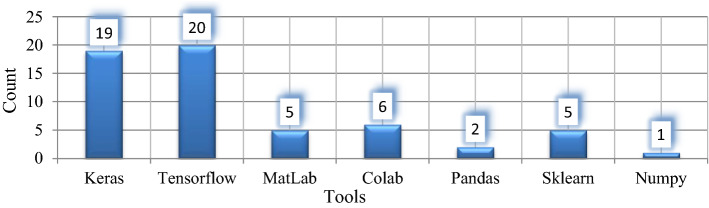
Along with large and heavy pre-trained networks, the lightweight networks also perform a critical role in detecting Covid-19 like CoVNet-19 [48], MKs-ELM-DNN [49], UNet + + [1], LDC-Net [47] that can be easily deployed on resource constraint platforms. ResNet [50], GoogleNet [51], Inception [52], DenseNet[53], NasNet[54], Xception [55], AlexNet [28], SqueezeNet [56], VGG [57] etc. are the pre-trained models applied for the classification/detection of Covid-19 from radiological pictures in the studied articles. Fully automated systems [27] were utilized to minimize processing time from blurry samples and reject those samples with misclassified lung regions for Covid-19 detection. Figure 3 reveals the quantitative analysis of DL models used for Covid-19 detection systems. Among all pre-trained DL models, ResNet was utilized by 35 reviewed systems, 21 reviewed systems used Inception, and VGG was used by 19 reviewed systems, whereas 32 Covid-19 detection systems used hybrid and custom networks. Figure 4 depicts that Keras and TensorFlow are the commonly used tools for Covid-19 detection in the experimental setup (analysis from Table 4).
Fig. 3.
Deep learning models used in the studied articles
Fig. 4.
DL tools used for Covid-19 detection
Table 4.
Summary report of work done on experimental setup in the studied papers
| Author | Model | Layer | Kernel Size | Pool size | Stride, Batch Size | Image Size |
|---|---|---|---|---|---|---|
| Panwar et al. [57] | VGG-19 | Conv:16, Maxpool:5, FCNN:3, | 3 × 3 | 2 × 2 | Batch size:16, Stride:2 | 512 × 512 |
| Nath et al. [34] | CNN | Conv:6, BN:6, Pool:4 | 3 × 3 | 2 × 2 | Stride:2 | 256 × 256 |
| Kassani et al. [97] | MobileNet, DenseNet, Xception, ResNet, InceptionV3, InceptionRes-NetV2, VGGNet, NASNet | Standard layer | 331 × 331NASNetLarge, 224 × 224NASNetMobile, 600 × 450 | |||
| Hussain et al. [93] | CoroDet | Conv:9, pool:9, dense:2, ft:1,LR:1 | Batch size:10 | 256 × 256 | ||
| Gilanie et al. [135] | CNN | Conv:8, pool:2, fc:4 | 3 × 3 | 2 × 2 | Batch size:128 | 512 × 512 |
| Silva et al. [106] | EfficientNet | Conv:3, Mbconv:7 | 3 × 3 | Batch size:32 | 104 × 153, 484 × 416 | |
| Turkoglu[49] | MKs-ELM-DNN | DenseNet201:3 STD layer block + ELM | Batch size:100 | 224 × 224 | ||
| Horry et al. [101] | VGG16/19, Resnet50, Inception V3, Xception, InceptionResNet, DenseNet, NASNetLarge | Standard layer | Batch size:2 and 16 | 224 × 224( VGG), 299 × 299 (Inception) | ||
| Dutta et al. [112] | CNN, Inception V3 | Batch size:32 | ||||
| Mertyüz et al. [51] | VGG-16, ResNet, GoogleNet, | Standard layer | 3 × 3 | 1 × 1 | Batch size:8 | |
| Ko et al. [38] | FCONet, VGG16, ResNet-50, Inception-v3, Xception | Standard layer, fc:2 | Batch size:32 | 256 × 256 | ||
| Alazab et al. [67] | VGG16 | Standard layer | 3 × 3 | 1 × 1 | Batch size:25 | 224 × 224 |
| Sharma et at [68] | VGG, MobileNet, Xception, DenseNet, InceptionResNet | Standard layer | Batch size:8 | 224 × 224 | ||
| Apostolopoulos et al. [66] | VGG19, MobileNetv2, Inception, Xception, Inception-ResNetv2 | Standard layer | Batch size:64 | 200 × 266 | ||
| Wu et al. [115] | Covid-AL | 3 × 3 | 1 × 1 | Batch size:30 | 352 × 320 | |
| Al-Waisy et al. [39] | COVID-CheXNet (HRNet, ResNet34) | 7 × 7 | 3 × 3 | Batch size:100 | ||
| Haghanifar et al. [65] | COVID-CXNet | 224 × 224 | ||||
| Sarker et al. [69] | COVID-DenseNet | DenseB:4, TraLayer:3 | Batch size:5 | 224 × 224 | ||
| Wang et al. [70] | COVID-Net | 7 × 7 to 1 × 1 | batch size:64 | |||
| Mangal et al. [63] | CovidAID(ChexNet, Covid-Net) | Batch size:16 | 224 × 224 | |||
| Javaheri et al. [40] | CovidCTNet(BCDU-Net, U Net) | 3DConv:10 3DMaxPool:5, Dense:2 | 128 × 128 | |||
| Elkorany et al. [41] | COVIDetection-Net (ShuffleNet, SqueezeNet) | 300 × 300 | ||||
| Tabik et al. [46] | COVIDSDNet | Batch size:16 | ||||
| Ucar et al. [103] | COVIDiagnosis-Net (Bayes-SqueezeNet) | 3 × 3 | 1 × 1 | Batch size:32 | 227 × 227 | |
| Hemdan et al. [42] | COVIDX-Net (VGG19, DenseNet201, InceptionV3,ResNetV2, InceptionResNetV2, Xception, MobileNetV2) | Standard layer | Batch size:7 | 224 × 224 | ||
| Kedia et al. [48] | CoVNet-19 (DenseNet121, VGG16) | Dense layer: 32 | Batch size:32 | 224 × 224 | ||
| Ouchicha et al. [45] | CVDNet | Conv:9, max pool:9, concat:1, ftn:1,fc:3 | 5 × 5, | 2 × 2 | Batch size:8 | 256 × 256 |
| Javor et al. [91] | ResNet50 | Standard Layer | Batch size:32 | 448 × 448 | ||
| Ismael et al. [98] | ResNet18, ResNet50, ResNet101, VGG16, VGG19 | Conv:5, ReLU:5, BN:5 | 3 × 3 | 1 × 1 | 224 × 224 | |
| Rohila et al. [124] | ReCOV-101 (ResNet50, ResNet101,DenseNet169, DenseNet201) | Conv2D:23, pool:1 | 3 × 3 | 2 × 2 | 224 × 224 | |
| Padma et al. [35] | 2DCNN | 2 × 2 | ||||
| Jain et al. [64] | Inception V3, Xception, ResNeXt | Standard Layer | 128 × 128 | |||
| Anwar et al. [107] | EfficientNet B4 | Standard Layer | Batch size:16 | 348 × 348 | ||
| Sethi et al. [104] | Inception V3, ResNet50, MobileNet, Xception | Standard Layer | Batch size:32 | |||
| Ying et al. [43] | DRE-Net (ResNet50) | Batch size:15 | 512 × 512 | |||
| Jiang et al. [36] | VGG, ResNet, Inception-v3, DenseNet, InceptionResNetv2, | Standard Layer | Batch size:4 | 512 × 512 | ||
| Yang et al. [92] | DenseNet | DenseBlock:4, pool:1, linear:1 | Batch size:32 | |||
| Serener et al. [111] | ResNet50, ResNet18, MobileNetV2, VGG, SqueezeNet, AlexNet, DenseNet121 | 224 × 224 | ||||
| Basu et al. [28] | DETL(AlexNet, VGGNet, ResNet50) | Alex:8, Vgg:16, Res:50 | 11 × 11 | 3 × 3 | ||
| Wang et al. [95] | RestNet50, ResNet101, ResNet152 | Batch size:64 | ||||
| Arellano, Ramos [53] | DenseNet121 | Standard Layer | ||||
| Voulodimos et al. [96] | FCN-8, U-Net | 3 × 3 | 1 × 1 | 630 × 630 | ||
| Chen et al. [1] | ResNet50, Unet + + | 512 × 512 | ||||
| Wu et al. [50] | ResNet50 | Standard Layer | Batch size:4 | 256 × 256 | ||
| Minaee et al. [56] | Deep-COVID (ResNet 18, ResNet50, Squeeze Net, DenseNet-121) | 3 × 3 | 1 × 1 | Batch size:20 | 224 × 224 | |
| Demir[99] | DeepCoroNet | 11 layers | Batch size:6 | 100 × 100 | ||
| Perumal et al. [23] | Resnet50, VGG16, InceptionV3 | Standard layer | 3 × 3 | 2 × 2 | Stride:1, Batch size:250 | 226 × 226 |
| Sheykhivand et al. [52] | Inception V4 | Conv:4,pool:4, lstm:2: fc:2 | 4 × 4 | 1 × 1 | Batch size:10 | 224 × 224 |
| Mishra et al. [55] | CovAI-Net (Inception, DenseNet, Xception) | Conv:8, pool:4, d:2, | 7 × 7 | 3 × 3 | Stride:2, Batch size:32 | 224 × 224 |
| Shah et al. [108] | CTNet-10 (DenseNet-169, VGG-16, ResNet-50, InceptionV3, VGG-19) | Conv:5, pool:3, fc:3 | Batch size:32 | 128 × 128 CTNet10, 224 × 224 VGG19 | ||
| Sakib et al. [100] | DL-CRC | Batch size:8 | ||||
| Tang et al. [44] | EDL-COVID | 6 layers of CovidNet | 3 × 3 | 1 × 1 | Batch size:64 | |
| Saha et al. [94] | EMCNet(AlexNet, VGG 16, Inception, ResNet-50) | Conv:6, pool:5, ft:1, db:6, fc:2, | 7 × 7 | 3 × 3 | Batch size:32 | 224 × 224 |
| Khan et al. [102] | H3DNN(3D ResNets, C3D, 3DDenseNets, I3D, LRCN) | Conv:9, pool: 6, incep:9,fc:2 | 7 × 7 | 3 × 3 | Batch size:2 | 224 × 224 |
| Gupta et al. [54] | InstaCovNet-19 ( NasNet, (InceptionV3, Xception, ResNet101, MobileNetV2) | 3 × 3 | 1 × 1 | Batch size:16 | 224 × 224 |
| Author | Learning Rate | Epoch | Performance | Activation function | Optimizer | Software |
|---|---|---|---|---|---|---|
| Panwar et al. [57] | 47 | Accuracy:95.61%, Sensitivity:94.04%, Specificity:95.86% | Softmax | Adam | ||
| Nath et al. [34] | 0.001 | 15 | Accuracy X-ray:99.68%, CT:71.81% | ReLU | SGDM, Adam, RmsProp | Matlab 2019b |
| Kassani et al. [97] | Accuracy:99% | Keras package with Tensor flow | ||||
| Hussain et al. [93] | 0.0001 | 50 | Accuracy: 99.1%, Precision:99.27%, Recall:98.17%, F1-score:98.51% | Sigmoid, ReLU, Leaky ReLU | Adam | Google Colab Keras, Tensorflow 2.0 |
| Gilanie et al. [135] | 0.01 | 25 | Accuracy:96.68%, Specificity:95.65%, Sensitivity:96.24% | ReLU, Softmax | MATLAB 2018b | |
| Silva et al. [106] | 0.001 | 20 | Accuracy:87.6%, F1-score:86.19%, AUC:90.5% | ReLu, Sigmoid | Adam | TensorFlow Keras |
| Turkoglu[49] | 100 | ReLUELM, PReLU-ELM, TanhRL | ||||
| Horry et al. [101] | 0.001, 0.00001 | 10 | Precision: 86% for X-ray, 100% for LUS, 84% for CT scans | Keras APIs with a TensorFlow 2 | ||
| Dutta et al. [112] | 0.00003 | 31 | Accuracy:84% | Sigmoid | RMSprop | Google Colab TensorFlow |
| Mertyüz et al. [51] | 0.00001 | 20 | Accuracy:96.90%, Sensitivity:95.45%, Specificity:100% | ReLu, Sigmoid | Anaconda Spyder, Keras | |
| Ko et al. [38] | 0.0001 | optimal epoch | Sensitivity:99.58%, Specificity:100.00%, Accuracy:99.87% | SoftMax | Adam | TensorFlow package, Keras |
| Alazab et al. [67] | 0.1 | 25 | F-Measure:99% | Pandas, Scikit, TensorFlow | ||
| Sharma et at [68] | 0.0001 | 100 | Accuracy:98%, Sensitivity:100%, Specificity:100%, Precision:100%, | Relu | Adam | sklearn, tensorflow, keras |
| Apostolopoulos et al. [66] | 10 | Accuracy:96.78%, Sensitivity:98.66%, Specificity:96.46% | Relu | Adam | ||
| Wu et al. [115] | 0.001 | 30 | Accuracy:86.60%, Precision:96.20%, AUC:96.80% | Softmax | ||
| Al-Waisy et al. [39] | 0.01 | 10, 20 | Accuracy:99.99%, Sensitivity:99.98%, Specificity:100%, Precision:100%, F1-score:99.99%, MSE:0.011%, RMSE:0.012% | Adam, SGD | ||
| Haghanifar et al. [65] | 0.0001 | 100 | Accuracy:98.68%, F-score:94% | Sigmoid | Adam | |
| Sarker et al. [69] | 0.00001 | Accuracy:94%, Precision:94%, Recall:94%, F-score:94% | Softmax | Adam | ||
| Wang et al. [70] | 0.0002 | 22 | Accuracy:93.3%, Sensitivity:100%, Precision:80% | Adam | Keras, Tensor- Flow | |
| Mangal et al. [63] | 0.0001 | 30 | Accuracy:99.94%, Sensitivity100%, Precision:93.80% | Sigmoid | Adam | |
| Javaheri et al. [40] | 0.001 | Accuracy:91.66%, Sensitivity:87.5%, Specificity:94%, AUC:95% | Adam | |||
| Elkorany et al. [41] | Recall:94.45%, Specificity:98.15%, Precision:94.42%, F1-score:94.4% | Softmax | ||||
| Tabik et al. [46] | 0.0002 | 100 | Accuracy:76.18%, Sensitivity:72.59%, Specificity:78.67%, F1-score:75.71%, | Softmax | SGD, Adam | |
| Ucar et al. [103] | 0.0004 | 35 | Accuracy:98.26%, Specificity:99.13%, F1-score:98.25% | ReLU | MATLAB | |
| Hemdan et al. [42] | 0.001 | 50 | Accuracy:90%, F1-score:91% | SGD | Keras package with TensorFlow2 | |
| Kedia et al. [48] | 0.001 | 5 to 10 | Accuracy:99.71%, | ReLU, Softmax | Adam | Keras,TensorFlow2 Scikit-Learn |
| Ouchicha et al. [45] | Adaptive LR | 20 | Accuracy:96.69%, Precision:96.72% Recall:96.84%, F1-score:96.68% | Softmax | Adam | |
| Javor et al. [91] | 17 | Accuracy:95.6%, Sensitivity:99.3%, Specificity:75.8% | ||||
| Ismael et al. [98] | 0.01 | 10 | Accuracy:94.7%, Sensitivity:91.00%, Specificity:98.89%, F1-score94.79%, AUC:99.90% | ReLU | SGDM | MATLAB |
| Rohila et al. [124] | 0.0001 | 100 | Accuracy:94.9% | ReLU | Adam, SGD | |
| Padma et al. [35] | Accuracy: 99.2%, Sensitivity:99.1%, Specificity:98.8%, Precision:100% | ReLU | ||||
| Jain et al. [64] | 0.001 | 14 | Accuracy:97.97% | LeakyReLU | Adam | |
| Anwar et al. [107] | 0.0001 | 25 | Accuracy:89.7, F1-score:89.6%, AUC:89.5% | Adam | Google colab | |
| Sethi et al. [104] | 200 | Accuracy:98.6%, Specificity:99.3%, Precision:87.8%, Sensitivity:87.8%, F1-score:87.8% | Adadelta | Google Colab | ||
| Ying et al. [43] | Accuracy:98.6%, Specificity:99.3%, Precision:87.8%, Sensitivity:87.8% | |||||
| Jiang et al. [36] | 0.0002 | Accuracy:98.92%, Recall:97.80%, Precision:100.00%, F1-score:98.89% | Adam | TensorFlow | ||
| Yang et al. [92] | 0.9 | 20 | Accuracy:95%, Sensitivity:100%, Specificity:90%, F1-score:95%, AUC:99% | SGD | Sklearn 0.22.1 | |
| Serener et al. [111] | Accuracy:89%, Sensitivity:98%, Specificity:86%, AUC:95% | SGD | ||||
| Basu et al. [28] | 0.0001 | 100 | Accurary:99% | ReLU | Adam, SGD | TensorFlow, Keras |
| Wang et al. [95] | 0.0001 | 50 | Accuracy:96.1% | Softmax | ||
| Arellano, Ramos [53] | Sensitivity:89.47%, Specificity:100% | ReLU | Gradient Descent | Pandas, seaborn, matplotlib, Keras, | ||
| Voulodimos et al. [96] | Accuracy:99%, Recall:89%, Precision:91%, F1-Score:89% | Keras, TensorFlow Google Colab | ||||
| Chen et al. [1] | Accuracy:96%, Sensitivity:98%, Specificity:94%, PPV:94.23%, NPV:97.92% | Keras | ||||
| Wu et al. [50] | 0.00001 | Accuracy:76%, AUC:81.9%, Sensitivity:81.1%, Specificity:61.5% | rmsprop | Keras 2.1.6 | ||
| Minaee et al. [56] | 0.0001 | 100 | Sensitivity:98%, Specificity:92.9% | SoftMax | Adam | |
| Demir[99] | 0.001 | 150 | Accuracy:100%, Sensitivity:100%, Specificity:100% | SoftMax | SGDM | MATLAB 2020a |
| Perumal et al. [23] | Accuracy:93.8% | ReLu | Adam | |||
| Sheykhivand et al. [52] | 0.001 | 200 | Accuracy:99.5%, Sensitivity100%, Specificity:99.02% | Leaky-Relu | RMSProp | Keras, Tensorflow |
| Mishra et al. [55] | 0.001 | 60 | Accuracry:98.31%, Precision:100%, Sensitivity:96.74%, Specificity:100%, F1-score:98.34%, PPV:100% | ReLU, Softmax | Adam | Tensorflow, Keras |
| Shah et al. [108] | 0.0001 | 30 | CTNet Accuracy:82.1%, VGG19 Accuracy: 94.52% | Sigmoid | RMSProp, Adam | Google Colab |
| Sakib et al. [100] | 0.001 | 100 | Accuracy:93.94%, AUC:95.25% | Leaky-Relu | Adagrad | Keras, TensorFlow |
| Tang et al. [44] | 0:002 | 50 | Accuracy:95%, Sensitivity:96.0%, PPV:94.1% | TensorFlow 2.0.0 | ||
| Saha et al. [94] | 0.0001 | 50 | Accuracy:98.91%, Precision:100%, Recall:97.82%, F1-score:98.89% | ReLu | ||
| Khan et al. [102] | 0.00001 | 1000 | Accuracy:85% | Softmax | Adam | Keras TensorFlow |
| Gupta et al. [54] | Reduced L.R | Accuracy:99.53%, Precision:100%, Recall:99%, F1-score:99% | SoftMax, Relu | Adam | NumPy, Scikit-Learn, TensorFlow 2 |
Deep transfer learning [58] firstly trains the CNN for a particular job utilizing a bulky dataset like Image Net. The dataset must have at least 5500 or more samples per class, i.e., data availability is the utmost key aspect for early training to effectively take out the essential features and obtain advantageous network parameters. Afterward, the initial practical training of CNN is ready to handle and manage the new data and mining features as it is established on facts and information grown from initial training [20]. Figure 5 shows the DL-based general flow used in Covid-19 detection systems. In DCNN, TL can be achieved in two methods.
Fig. 5.
DL-based Covid-19 diagnosis flow
(1) It contains feature mining with TL. The innovative CNN model [59] is served as a feature miner, and a novel classifier is trained on topmost. The already trained network also remembers its ideal structure and its learned features. Learned features from this network are provided to new classifiers learned for the particular job at hand.
(2) It contains structural alteration to already trained models to improve outcomes [60]. Generally, several network units in such prototypes are exchanged with newly adjusted layers, in-tune features, and only allow for a particular job. Primarily fully connected (FC) layers [47, 61] in the newest pre-trained network structure are exchanged with a unique FC wherein weights are prepared arbitrarily. The convolutional regions [61] are locked to realm of the selective filtering learned by convolutional regions. This means that backpropagation is only possible up to totally connected layers, as these layer weights are arbitrary [29]. This technique permits FC layers to study the structures and shapes from extremely judgmental and affluent-featured convolutional layers [62]. Further, FC layers gain the structures and shapes of a newly given image set; the complete prototype is allowable to train with an actual lesser learning rate to attain enough precision on the new image.
Class-Based Task Formulation
We have categorized the studied Covid-19 detection articles using DL from binary to multi-class classifications. Because each label signifies one or more class types. DL networks are used to create network models that helps in diagnosis of multi-class(Normal, BacterialPneumonia, ViralPneumonia, Covid19) [41, 63], 3-class(Healthy, Covid19, Pneumonia) [48, 64], (CovidPneumonia, nonCovidPneumonia, Normal) [65], (BacterialViralPneumonia, Covid19, Normal) [66] and 2-class(Normal, Covid-19) [67, 68], (Covid19, nonCovid) [69] classification. Figure 6 shows the radio image type, number of systems, and proportion of radiography images, including X-ray, CT, and multimodal types in studied papers as a dataset. As indicated in Fig. 6, 36 examined techniques (56%) used X-ray modality, while 20 examined methods (31%) used CT scans and the remaining eight systems (13%) used multimodal as a data source.
Fig. 6.
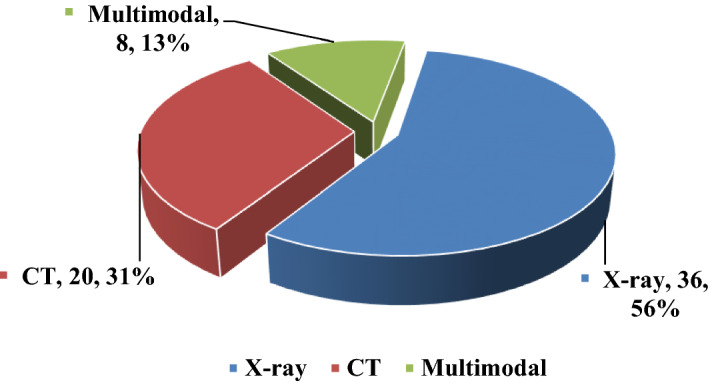
Distribution of the three-radiography imaging modality
Datasets
From studied papers, a total of 35 different datasets are recovered. An outline of these datasets is produced in Table 1. Some authors successfully generated a new clean dataset of Covid-19 [70] from various institutes, hospitals, and research centers and shared it publically on a common platform. The publicly shared online platforms are Kaggle [71–74], Github [75, 76], SIRM [77], Mendeley [78, 79], Radiopedia [80], IEEE-Dataport [81], NIH [82], Eurorad [83], wiki-cancerimagingarchive [84, 85], Harvard [86], social media (Twitter [87] and Instagram [88]), physionet[89], stanfordmlgroup [90] etc. to open community. While others did not disclose the dataset on a public platform to control the privacy policies of hospitals, research institutes, and patients, they have only used the dataset for experiments [1, 36, 39, 43, 91, 92]. Each tuple from Table 1 states the data source name, image resolution, imaging mode, type, images with class, URL (address to access the dataset), reference number, and authors using the same dataset. According to our findings, Covid-19 X-ray by Cohen [72] is the most frequently used dataset in 23 studies.
Table 1.
Covid-19 datasets and repositories used in the studied articles
| Data Source Name | Mode | Type | Images | URL | Data set | Corresponding Author’s |
|---|---|---|---|---|---|---|
| Github: Agchung | X-Ray, CT | JPG | 55(Covid19) CT, X-Ray | 1)”https://github.com/agchung/Figure1-COVID-chestxray-dataset/tree/master/images”, 2)”https://github.com/agchung/Actualmed- COVID- chestxray-dataset“ | [75] | [46, 48, 69, 70, 93, 94] |
| RICORD data set (open survey by the RSNA) | X-Ray, CT | DICOM | 1000 CXRI and 240 thoracic CT scans | 1)”https://radiopaedia.org/articles/imaging-data-sets-artificial-intelligence”, 2)”https://radiopaedia.org/articles/Covid-19-4?lang=gb#article-images“ | [80] | [55, 65, 94–96] |
| SIRM-Covid19 Database | X-Ray, CT | JPG | 115(Covid19) | 1)”https://sirm.org/category/Covid-19/”, 2)”www.sirm.org/category/senza-categoria/Covid-19/“ | [77] | [28, 38, 45, 55, 65, 94, 97] |
| Kaggle: Chest Pneumonia X-Ray Images | X-Ray | JPG | 5856 (Pneumonia) | “https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia “ | [71] | [28, 41, 45, 52, 63, 68, 94, 98–100] |
| NIH Chest X-rays (ChestX-ray14 pulmonary disease, ChestX-Ray8 | X-Ray | PNG | 112,000 (Other than Covid Diseases), Summers, Ronald (NIH /CC / DRD), Enterprise Owner NIHC Center | 1)”https://www.kaggle.com/nih-chest-xrays/data?select=Data_Entry_2017.csv”, 2)”https://nihcc.app.box.com/v/ChestXray-NIHCC“ | [82] | [23, 46, 47, 53, 54, 65, 94, 99–102] |
| Github: covid-chestxray-dataset (Cohen) | X-Ray, CT | JPG, PNG | 930(Covid19), MERS, SARS | “https://github.com/ieee8023/covid-chestxray-dataset “ | [72] | [34, 35, 41, 42, 45, 48, 52, 56, 57, 63, 66, 68–70, 93–95, 97, 98, 100, 103, 104] |
| SARSCOV2 CT_Scan Dataset (Hospital in Sao Paulo, Brazil) | CT | PNG | 1252(Covid19), 1230 CT-samples of Non-Covid Patients | “https://www.kaggle.com/plameneduardo/sarscov2-ctscan-dataset “ | [105] | [57, 93, 106] |
| Daniel Kermany et al.: Labeled OCT and X-Ray Images | CT, X-Ray | 5856 Samples of Pneumonia, Normal patients | “https://data.mendeley.com/datasets/rscbjbr9sj/2 “ | [78] | [23, 57, 63, 65, 103] | |
| COVID-CT: CT dataset about Covid19 (Xingyi Yang) | CT | JPG | 349 CT images, Clinical findings of Covid19 216 patients | “https://github.com/UCSD-AI4H/COVID-CT “ | [76] | [34, 38, 49, 93, 101, 106–108] |
| Figure 1 Covid-19 clinical cases | X-Ray | PNG | 35 Covid19 | “https://www.figure1.com/Covid-19-clinical-cases” “ | [109] | [65] |
| Kaggle: Covid19 Radio Database: X-ray (Md. E. H. Chowdhxury) | X-Ray | PNG | 3616 Covid19 | “https://www.kaggle.com/tawsifurrahman/covid19-radiography-database “ | [73] | [34, 48, 51–54, 70, 93] |
| Radiological Society of North America | X-Ray | DICOM | 9555 Pneumonia RSNA Pneumonia | “https://www.kaggle.com/c/rsna-pneumonia-detection-challenge/ “ | [110] | [46, 69, 70] |
| IEEEDataport: CCAP-CT data sets from multicenter hospitals | CT | JPG | 42 chest CT scans of Covid19 for test 4 Covid19 | “https://ieee-dataport.org/documents/ccap “ | [81] | [111] |
| Kaggle Covid-19 X rays, CT snapshots | X-Ray, CT | JPEG | 79 X-Ray, 16 CT Covid19 images | “https://www.kaggle.com/andrewmvd/convid19-x-rays “ | [74] | [48, 52, 66, 112] |
| kaggle Covid19 chest Xray: Covid19 data collection Bachrr | X-Ray | JPEG | 357 Covid19 Chest X-Ray images | “https://www.kaggle.com/bachrr/covid-chest-xray “ | [113] | [98, 99] |
| Github arthursdays HKBU-HPML-Covid-19 CT dataset (Clean-CC-CCII) | CT | JPEG | 340,190 Slices of CT images with Covid19 | “https://github.com/arthursdays/HKBU_HPML_Covid-19 “ | [114] | [115] |
| Khoong WH. Covid-19 x-ray dataset | X-Ray | Covid19, Pneumonia | “https://www.kaggle.com/khoongweihao/covid19-xray-dataset-train-test-sets“ | [116] | [93] | |
| Sajid N. Covid19 Patients lungs xray images | X-Ray | 10,000 | 10,000 Normal Patients | “https://www.kaggle.com/nabeelsajid917/Covid-19-x-ray-10000-images “ | [117] | [67, 93] |
| Daniel Kermany et al.: Large Dataset Labeled OCT, X-Ray | X-Ray, CT | CNV, DME, DRUSEN, and NORMAL (ZhangLabData) | “https://data.mendeley.com/datasets/rscbjbr9sj/3 “ | [79] | [48, 66] | |
| POCOVID-Net data set (POCUS) | Ultrasound | > 200 LUS videos (Convex, Linear) | Images:22(Covid19), 22(bacterial pneumonia, 15(healthy), viral pneumonia; Videos:115(Covid19), 51(bacterial pneumonia), 75(healthy), 6(viral pneumonia) | “https://github.com/jannisborn/covid19_ultrasound/tree/master/data “ | [118] | [101] |
| The Cancer Imaging Archive: [dataset] | X-Ray | DICOM | Thoracic capacity, pleural effusion segmentations | “https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=68551327#685513274dc5f53338634b35a3500cbed18472e0 “ | [84] | [115] |
| Eurorad imaging database | X-Ray, CT | JPEG | Covid-19 X-Ray and CT Image modalities | “https://www.eurorad.org/advanced-search?search=COVID” | [83] | [65] |
| Twitter: Chest Imaging database | X-Ray, CT | JPEG, PNG | Normal, Covid, etc | “https://twitter.com/ChestImaging/ “ | [87] | [28, 65] |
| Instagram: Chest Imaging database | X-Ray, CT | JPEG, PNG | Chest X-Ray of different disease | 1)”www.instagram.com/theradiologistpage” 2)”www.instagram.com/radiology_case_reports/“ | [88] | [65] |
| Github: Covid-19 image repository (Winther) | X-Ray | PNG | 243 Covid19 repository (IDIR Hannover, Germany) | “https://github.com/ml-workgroup/Covid-19-image-repository” | [119] | [65] |
| COVIDx Dataset[70]: Collection of [72, 73, 75, 110] | X-Ray | PNG | 358 CXR images Covid19, 8066 normal, 5,538 nonCovid pneumonia | “https://github.com/lindawangg/COVID-Net “ | [120] | [44, 70] |
| X-Ray, CT Dataset | X-Ray, CT | PNG | Normal, Bacteria, Viral, Covid19 | “https://github.com/zeeshannisar/Covid-19 “ | [121] | [63] |
| LUNGx SPIEAAPM -NCI Lung Nodule Class | CT | DICOM | CT Lung Nodule Images | “https://wiki.cancerimagingarchive.net/display/Public/LUNGx+SPIE-AAPM-NCI + Lung + Nodule + Classification + Challenge “ | [85] | [40] |
| MIMIC-CXR DatabaseV2.0 | X-Ray | JPEG | 377,110 images other than Covid19 | “https://physionet.org/content/mimic-cxr/2.0.0/ | [89] | [46] |
| Pad Chest | X-Ray | PNG | 160,000 X-Ray other than Covid19 | “https://bimcv.cipf.es/bimcv-projects/padchest/ | [122] | [46] |
| MosMedData: Results of CT, Signs of Covid-19 | CT | JPG | 1110 patients lung parenchyma 0, 25%, 50%, 75% radiological signs of viral Pneumonia (Covid19), without signs(normal) | “https://mosmed.ai/datasets/covid19_1110/ | [123] | [124] |
| Kaggle: Chest X-ray | X-Ray | JPG | Covid19, Pneumonia, normal | “https://www.kaggle.com/prashant268/chest-xray-covid19-pneumonia | [125] | [64] |
| Github: Covid-19 image repository | X-Ray | PNG | 243 Covid Images | “https://github.com/ml-workgroup/covid-19-image-repository | [126] | [28] |
| CheXpert Dataset: Stanford University Medical Center | X-Ray | JPG | 224,316 chest XRay of 65,240 patients | “https://stanfordmlgroup.github.io/competitions/chexpert/ | [90] | [54, 56] |
| Covid-19-CT-Dataset: Harvard Dataverse, SM Mostafavi Dataset | CT | 1000 images | 1000 + Patients Confirmed Covid19 | “https://dataverse.harvard.edu/dataset.xhtml?persistentId=doi:10.7910/DVN/6ACUZJ | [86] | [102] |
Diagnosis Based on X-Ray Images
Pre-trained Deep Learning Models
Hemdan et al. [42] in Mar. 2020, created a system to detect Covid-19 using the pre-trained models of CNN from chest X-ray images(CXRI). The proposed system was named as COVIDX-Net, uses 7 pre-trained variants of DCNN (InceptionV3, ResNetV2, Xception, and MobileNetV2, VGG19, InceptionResNetV2, DenseNet201). They collected a dataset from a public repository [72]. The dataset consists of 25 samples for each class (healthy, Covid-19). All images were resized from 1112 × 624 and 2170 × 1953 to 224 × 224 pixels. The data set was partitioned into 80:20% for training and testing. The investigational performance exposed VGG19, DenseNet achieved the highest results over other simulations with an F1-score of 90% and 91%, respectively, whereas InceptionV3 obtained the lowest accuracy of 0.00%. Mangal et al. [63] in Apr. 2020, proposed CovidAID system using pre-trained CheXNet prototypical to categorize a frontal-view of CXR image to normal, bacterialPneumonia, viralPneumonia, and Covid19 classes. CovidAID builds upon CheXNet, which detects pneumonia from CXRI. CheXNet is 121-layer DenseNet constructed structure and skilled on the ChestXRay14 largest dataset. The assembled dataset from 4 public online repositories [71, 72, 78, 121] and separated into training(70%), testing(20%), and validation(10%) sets. The dataset contains 6011 samples, including 155 for Covid-19, 1493 for viral pneumonia, 2780 for bacterial pneumonia, and 1583 for normal classes. The experimental results obtained an overall accuracy of 87.2% for 4-class classification and 90.5% for ternary class. For Covid-19 class the achieved accuracy, sensitivity, PPV for binary classification is 99.97%, 100%, 96.80% and for 4-class is 99.94%,100%, 93.80%. To visualize the region of interest, they have generated saliency maps for COVID-AID model predictions using RISE (Randomized input sampling for explanation).
Alazab et al. [67] in May 2020, the proposed identification of the Covid-19 model using VGG16 from CXRI. The original dataset was collected from a single source [117] which contains 128 images, including 28 images for the healthy class and 70 images for the Covid19 class. After applying data expansion, the total images reached 1000(500 for healthy and 500 for Covid-19 class). 80%:20% ratio partitioned for testing and training dataset. The proposed system improved F-measure from 0.95 to 0.99 for the original and augmented data sets, respectively. Apostolopoulos et al. [66] in Jun. 2020, modeled an architecture for the spontaneous identification of Covid-19 individuals by employing the perception of DTL with five different pre-trained CNNs (Inception, Xception, InceptionResNetv2, VGG19, MobileNetV2). They assembled the dataset from 3 public online platforms [72, 74, 79] in two forms Dataset_1 and Dataset_2. The system considered (Dataset_1) 1427 CXRI comprising Covid-19(224), common-bacterial pneumonia(700), and healthy(504) cases. In Dataset_2, 224 photos of Covid-19, 714 images of common-bacterial, viral-pneumonia, and 504 images of healthy. The dataset was distributed using ten-fold cross-validation. The projected model using MobileNet-V2 attained the highest performance over a Dataset_2 binary classification with a 96.78% accuracy, 98.66% sensitivity, 96.46% specificity, and 3-class classification accuracy of 94.72%. Haghanifar et al. [65] in Jul. 2020, anticipated a framework to identify Covid-19 using TL pattern, CheXNet model is used to develop COVID-CXNet. The collected dataset was assembled from 9 different public online platforms [77, 78, 80, 82, 83, 87, 88, 109, 119] consist of 780 X-ray samples of Covid19 pneumonia, 3500 for nonCovid pneumonia, 3500 samples of normal class. Images were downsized to 320 × 320 resolutions. CheXNet was created on DenseNet architecture, and it has trained on frontal CXR images. The proposed COVID-CXNet was adjusted on the Covid-19 CXR data set 431 layers and 7 million parameters. This system consists of a lung separation unit to increase the model localization of lung irregularities. Covid-19 pneumonia class achieved the highest performance from a base model with 98.68% accuracy, 94% F1-score, and COVID-CXNet_v1 with 99.04% accuracy, 96% F1-score. For hierarchical multiclass, COVID-CXNet attained an 87.21% accuracy and a 92% F-score. COVID-CXNet used Grad-CAM for visualizing the results.
Sethi et al. [104] created a Covid-19 recommender system using DL from CXRI in Jul. 2020. The proposed structure uses four deep CNN architectures: Inception V3, ResNet50, MobileNet, and Xception. The collected dataset from a single repository [72] contains 320 ‘Covid + ve’ and 5928 non-Covid19 samples. The dataset splitting with a 75:25 ratio for training and testing sets. For the validation set, 30% of the training set was utilized. The MobileNet achieved the highest 98.6% accuracy, 99.3% specificity, 87.8% precision, 87.8% sensitivity, and 87.8% F1-score from all four models. Ucar et al. [103] in Jul. 2020, projected a network structure for an immediate investigation of Covid-19 built on deepBayesSqueezeNet(COVIDiagnosisNet) built on CNN pretrained SqueezeNet model. The dataset was collected from 2 public repositories [72, 78]. Data augmentation was employed due to the low CXRI of Covid-19(70 samples), and the sample size reached 1536 images. After augmentation, the total image samples moved to 4602. The data splitting into training(60%) set, testing set(20%), and validation set(20%) and labeled into 1536 images for each Covid, normal, and pneumonia class. The experimental results revealed that deepBayesSqueezeNet achieved complete accuracy of 98.26%, specificity of 99.13%, and F1-measure of 98.25%. Mertyuz et al. [51] in Oct. 2020, the proposed Covid-19 prognosis from CXRI using 3 DCNN variants(VGG-16, ResNet, GoogleNet). The dataset collected from a public platform [73] contains Covid-19 positive(219), normal(1341), and viral pneumonia(1345) images. The proposed system achieves accuracy, sensitivity, specificity for VGG-16 network 95.87%, 97.73%, and 99.63%; using ResNet network 96.90%, 95.45%, and 100%; using the GoogleNet network 95.18%, 86.36%, and 100% respectively. Sharma et al. [68] in Oct. 2020, proposed Covid-19 screening with residual attention network using X-ray samples. The system uses 10 variations of CNN(VGG16, InceptionResNet, Xception, VGG19, MobileNet, MobileNetV2, DenseNet121, DenseNet201, NASNet, Vanilla Residual Attention Network (RAN)). They assembled a dataset from 2 online public platforms [71, 72], containing 120 images for Covid and 119 for normal class. The dataset divided into 70% for training (167 images), 20% for testing (50 images), and 30% for validation (22 images) sets. For non-linear dimensionality reduction images, the UMAP (Uniform Manifold Approximation and Projection) technique was applied. The residualAttNet-based proposed system achieves 98% accuracy, 100% sensitivity and specificity, precision, and recall metrics on a validation set.
The innovative Domain Extension Transfer (DETL) algorithm, developed by Basu et al. [28] in Dec. 2020, based on TL to screen Covid-19 from the CXR. DETL used three pre-trained versions of TL(AlexNet, VGGNet, and ResNet50). The datasets were collected from 4 online repositories [71, 77, 87, 126]. Data-A was formed based on [82] to classify normal and diseased classes, and Data-B classified pneumonia, normal, other diseases, and Covid-19 classes. Other disease classes contain “Atelectasis, Cardiomegaly, Infiltration, Effusion, Nodule, and Mass” type image. The proposed DETL model structure consists of AlexNet with eight layers, VGGNet with 16 layers, and ResNet has 50 layers. The accuracy attained with fivefold cross-validation on Data-B for AlexNet, VGGNet, and ResNet of 82.98%, 90.13%, and 85.98%, respectively. VGGNet correctly classified on Data-A with the best validation accuracy of 99% Covid and 100% normal. Sarker et al. [69], in Jan. 2021, proposed a COVID-DenseNet, a DTL-based framework for classifying Covid-19 from healthy and pneumonia individuals. The collected dataset from 3 public platforms [72, 75, 110] are divided into 80%:10%:10% ratio for train, test and validation set for binary classification (Covid19, NonCovid19) and 3 class classification (Covid19, pneumonia, and normal). The originally assembled dataset contains normal(8851), pneumonia(6045), and Covid-19(238 images) class. Data augmentation was employed to upsurge the samples of Covid class(11,416 images). The system achieved 96% accuracy for the binary and 94% for the 3-class classification. Patient-wise, tenfold cross-validation attained an average accuracy of 92.91% for 3-class classification to validate the consistency of the proposed model. Using Grad-CAM highlighted the image regions in making a prediction. Kedia et al. [48] in Feb. 2021, proposed a Stacked-Ensemble(SE) model to sense Covid-19 + Ve patients using CXRI. The proposed model(CoVNet-19) is a two-level stacked ensemble machine learning structure that understands similar facts in altered methods. The Phase-1 structure of CoVNet-19 joined two pre-trained DCNNs(VGG19 and DenseNet121). Mutually networks remained independently trained on composing datasets to achieve the classification work. CXR image of size 224 × 224 as the input given to CovNet-19 model. In phase-2 of CoVNet-19, the SVM classifier's SE structure was trained to accomplish binary and 3-class classification. The system was trained on the extracted features from the phase-1 structure. The assembled dataset from five different public repositories [72–75, 79] initially contains Covid19(798 frames), normal(2241 frames), and pneumonia(2345 frames) class. Later data augmentation was applied on Covid images, and the sample size increased to 1628. The proposed model hyperparameters were adjusted distinctly for 3-class (Covid19, normal, and pneumonia) and binary classification (Covid19, nonCovid19). Experimental results revealed 99.71% accuracy for binary and 98.28% for 3-class classification.
Ismael et al. [98] in Feb. 2021, proposed a DCNN approach to categorize coronavirus from healthy CXR images. They used five variants of pre-trained CNNs(VGG16, ResNet101, VGG19, ResNet18, and ResNet50) to extract features. SVM uses kernel functions like quadratic, cubic, linear, and gaussian for deep feature classification. Dataset was collected from 3 online public repositories [71, 72, 113], which contain 380 frames comprising 180 for Covid19 and 200 for normal/healthy classes. Dataset was partitioned into train and test sets for 75%:25% ratios. All CXRs were rescaled to 224 × 224 pixels. The deep features extracted with a linear-kernel function of the ResNet50 model and SVM classification generated 94.7% accuracy, 91% sensitivity, 98.89% specificity, 94.79% F1score, and 99.90% AUC, which was the highest among all obtained results. VGG16 achieved the lowest accuracy(i.e., 85.26%). Jain et al. [64] in Mar. 2021, proposed DL-based discovery & investigation of Covid-19 on CXR images. The system used 3 pretrained variants:InceptionV3, Xception, and ResNeXt. The dataset collected from the Kaggle repository [125] contains 6432 PA-views of CXRI, including training(5467) and validation(965), healthy(1583), Covid(576), and pneumonia(4263 images) classes. Experimental result analysis shows that Xception offered an uppermost accuracy of 97.97% for detecting Covid-19 than the other two networks. Elkorany and Elsharkawy [41] in Apr. 2021, developed a tailored Covid-19 detection called COVIDetectionNet from CXRI. The COVIDetectionNet was built on the architecture of ShuffleNet, SqueezeNet for extracting the features, and multi-class, SVM for recognition and classification. The combined features are fed to MSVM. The dataset contains 1200 CXRs composed of two public online repositories [71, 72] and classified into 300 CXRI for Covid, normal, bacterialPneumonia, and viralPneumonia classes. The dataset was partitioned into training(80%) and testing(20%). The uppermost 100% finding accuracy was attained for the binary class. For 3-class classification (Covid19, normal, pneumonia) achieved 99.72% accuracy and 94.44% for 4-class classification (3-class + bacterialPneumonia, viralPneumonia). For 4-class classification, the system achieved performance metrics in a recall, specificity, precision, and F1-score of 94.45%, 98.15%, 94.42%, and 94.4%, respectively.
Hybrid and Custom Deep Learning Models
Padma et al. [35] in Sept. 2020, the proposed DL-based diagnostic tool for detecting Covid-19 using 2DCNN. For mining features and categorization, the 2D-CNN was implemented. The planned technique fine-tuned for optical features like texture and sharpness was measured to find Covid-19 using CXRI. The data set was collected from GitHub [72], comprising 60 images(30 for normal and 30 for COVID positive class). The dataset was divided into training(80%) and testing(20%). The experimental results for the training set achieved a 99.2% accuracy, 98.3% validation accuracy, 0.3% loss, 99.1% sensitivity, 98.8% specificity, and 100% precision was attained by spotting Covid-19 imageries. Ouchicha et al. [45] in Nov. 2020, suggested an innovative DCNN-based CVDNet model for the finding of Covid-19. The CVDNet architecture consists of 2 parallel networks with 9-convolutional, 9-max-pools, 1-concatenation, 1-flattened, 3-FC with Softmax and ReLU layers. The dataset was assembled from 3 online public repositories [71, 72, 77] consisting of 2905 CXRI, containing 219 for Covid-19, 1345 for viral-pneumonia, and 1341 for normal class. The dataset was partitioned into fivefold cross-validation(training:70%, validation:10%, and testing:20%). In general, CVDNet model accomplished a typical accuracy of 97.20%, 96.73%, and 96.58% for 3-class classification, i.e., Covid-19, normal, and viral-pneumonia, respectively. For fivefold cross-validation results, a precision of 96.72%, accuracy of 96.69%, recall of 96.84%, and F1 score of 96.68%. Al-Waisy et al. [39] in Nov. 2020, presented the Covid-19 CXR virus detection framework using hybrid DL (COVID-CheXNet). Initially, the CXRI was enhanced, and the noise level was reduced by local contrasting histogram adaptation and Butterworth band-pass filters. After combining the results acquired after two altered DL models based on a ResNet34 & high-resolution network structure are trained with a high-range data set. The experimented system(COVID-CheXNet) for Covid-19 detection achieved 99.99% accuracy, 99.98% sensitivity, 100% specificity, 100% precision, and 99.99% F1-score. Also, the weighted sum rule was used to calculate MSE of 0.011% and RMSE of 0.012% at the score level. Tabik et al. [46] in Dec. 2020, developed the CNN-based COVIDSDNet method. COVIDSDNet chains data expansion, segmentation, and conversions altogether with a suitable CNN for implication. They collected the dataset from 5 different online public repositories [75, 82, 89, 110, 122] named COVIDGR1.0 comprised 426 Covid-19(+ ve) and 426 Covid-19(− ve) PA-views. Because of excessive variations between multiple executions, implemented fivefold cross-validations in all trials. Experimental configuration utilizes 80% for training, and 20% for testing(COVIDGR1.0 dataset). In the initial phase, the COVIDNetA, COVIDNetB, and COVIDNetC models were trained on the COVIDx dataset; in the second phase, COVIDSDNet was retrained on our COVIDGR. The experiment obtains 72.59% sensitivity, 78.67% specificity, 75.71% F1-score, and 76.18% accuracy.
To identify Covid-19, Wang et al. [70] in Dec. 2020, suggested a DCNN structure. The projection-expansion-projection-extension (PEPX) design was used in COVID-Net construction. They used a human–machine collaborative system design in the initial step. In COVID-Net, the implemented approach blends a human-driven fundamental system architecture prototype with a machine-driven screening tool in the second step. They assembled a dataset from 4 public online repositories [72, 73, 75, 110]. They introduced the same dataset named COVIDx [120] with open access, containing 13,975 CXR images through 13,870 patients. The assembled CXR dataset was categorized into Covid-19(358 images), normal(8066 images), nonCovid19-pneumonia(5538 images). COVID-Net model achieved 93.3% test accuracy. COVID-Net achieved sensitivity of 73.9%, 93.1%, 81.9%, 100.0%; precision of 95.1%, 87.1%, 67%, 80.0% for four-class classification(normal, bacterial, nonCovid19 viral, and Covid19 viral classes) respectively.
A combination of graph neural network(GNN) and CNN based(i.e., CNN + 2L-GCN) SARS-Net was proposed by Kumar et al. [127] in 2021 for detecting Covid-19 abnormalities. The COVIDx[120] dataset was used for training(90%), val(10%), test(10%) the model. The SARS-Net achieved an accuracy of 97.60% and a sensitivity of 92.90%. An agent-based simulation was proposed by Chopra et al. [128] in 2021 for vaccine administration. A Graph Neural Network(GNN) based DeepABM-COVID framework utilizes agent, interaction, infection, and progression modules. They presented a sample for results on delaying the second dose of the mRNA vaccine and presented recommendations on when this strategy could be usefully adopted. The author suggests DeepABM is a scalable and efficient. A lightweight IoT feature vector-based custom DCNN, Raspberry Pi deployed Covid-19 detection CAD-GUI-tool application proposed by Bhosale et al. [47] in May 2022. LDC-net was trained on 5-different datasets[82]. The 76%:12%:12% CXRs ratio samples allocated for train:test:val sets. The Proposed LDC-Net not only classifies Covid-19 but also other eight lung diseases for the X-ray radiography images. The IoT deployed application trained large X-ray samples(10,800). The system attained the highest 99.28% accuracy and a minimum error-rate of 4.83%. The LDC-Net took 0.136 s to test individual CXR. Zanwar et al. [61], in Feb 2022, proposed a custom DCNN-based Covid-19 classification using CXRs. The proposed system utilizes the Cohen datasets containing normal(1540), Covid(1520), and Pneumonia(1560) labels. Implemented DCNN-based system attained the 96.59% recognition rate for pneumonia disease. A multi-channel capsule(MLCN) based architecture was proposed for the detection of Covid-19 by Sridhar and Sanagavarapu [129] in April 2021. MLCN utilizes 1678(healthy) and 902(Covid-19) CXR samples for feature extraction and testing. MLCN attained an accuracy of 96.8%. A ensemble VGGCapsNet(Capsule Network + VGG16) proposed by Tiwari et al. [130] for 3-class classification. The proposed CapsNet utilizes 219(Covid-19), 1345(Pnuemonia), and 1341(Normal) for feature extraction. It utilizes primaryCap + XRayCaps architecture for disease classification. CapsNet attained overall 92% accuracy for Covid-19 label.
Diagnosis Based on Computed Tomography (CT) Images
Pre-Trained Deep Learning Models
Yang et al. [92] in Apr. 2020, created a DL-based coronavirus detection approach from HRCT images using DenseNet. The pre-trained network uses a 3-layer block with dense, global average pooling and an FC-layer. They collected a dataset of confirmed Covid-19 patients from Shanghai hospitals containing 295 HRCT images, including healthy (149) and Covid (146) classes. The dataset was partitioned to train (45%), test (45%), and validation (10%) sets. The experiment shows the performance with the threshold of 0.8 for accuracy, sensitivity, specificity, F1-score, and AUC of 95%, 100%, 90%, 95%, and 99%, respectively. Silva et al. [106] in Sept. 2020, vote casting and different datasets strategies offered a DL approach for selecting Covid-19. However, data inclines to current frames of variable superiority, which might originate in other CT mechanisms. Replicating circumstances of the nations and towns with data origin. In this system, the data from assumed individuals have categorized a cluster into a polling mechanism. The suggested method is verified on two datasets and a cross-dataset study of Covid-19 CT scrutiny and divided into [76, 105] forms. The input CT pictures from a given patient are categorized as a cluster in polling. The system uses exploited and extended version of EfficientNet(a version of synthetic DNN). The assembled dataset partitioned randomly into training(80%) and test(20%) set contains 3294 CT images, including 1601 for Covid19 and 1693 for nonCovid19 class. The individual execution was attained for COVID-CT [105] with an 87.60% accuracy; SARS-CoV-2 CT-scan [76] with 98.99% accuracy. The suggested method achieved an 87.6% accuracy, 86.19% F1-measure, and 90.5% AUC. However, the cross-dataset performance has shown that accuracy decreases to 56.16% in the most delicate assessment setting. Anwar et al. [107] in Nov. 2020, proposed DL-based diagnoses of Covid-19 through CT-scan using a variant of DNN(EfficientNet B4). The proposed system uses three different learning rates: reducing the learning-rate(reducing on a plateau), recurring, and constant learning-rate. The dataset collected from a single online public source [76] contains 1494 CT images, including 702 scans for Covid and 792 for nonCovid class. The data expansion technique was applied to upsurge the quantity of the samples. No separate validation dataset was used in this study because of the limited dataset. The fivefold cross-validation is practiced such that test data is expected in each fold. EfficientNet DL architecture offered an 89.7% accuracy, 89.6% F1-score, and 89.5% AUC. The reducing learning rate approach achieved the highest 0.9 F1-score. Whereas recurring learning-rate, constant learning-rate achieved F1-score of 0.86, 0.82 respectively.
A DL-derived ML-classifier developed by Javor et al. [9] in Dec. 2020 for Covid-19 grouping. They collected a private dataset of 6868 CT samples, including 3102 for Covid19(+ Ve) and 3766 Covid19(-Ve). The dataset was separated into 80:20 training & validation sets. For testing, 90 images of 90 patients, including 45 Covid-19(+ Ve) patients, were selected randomly, and 45 negative patients were chosen manually. Further, photos were resized to 448 × 448 pixels. The proposed model reached a complete accuracy of 98.6%, a sensitivity of 99.3%, and a specificity of 75.8%. Ko et al. [38] in 2020, established a 2D-DL framework using CT images to diagnose Covid-19 pneumonia and discriminate it from nonCovid pneumonia and non–pneumonia. TL was used to create FCONet (FastTrack Covid-19 classification network), constructed by utilizing pre-trained DL models(VGG16, ResNet-50, Inception-v3, and Xception). They have assembled the dataset from 2 hospitals and two public platforms [76, 77], containing 3993 CT images. After data augmentation, the entire image set contains 31,940 CT-images, including Covid-19 pneumonia(9550), other pneumonia(10,860), normal lung(7890), and lung-cancer(3550). At a ratio of 80%:20%, the dataset was separated into training and testing sets. Among four pretrained models of FCONet, ResNet50 revealed outstanding execution with 99.58% sensitivity, 100% specificity, and 99.87% accuracy. Compared to other exterior testing data sets with poor CT scans, ResNet-50 attained the uppermost 96.97% accuracy, followed by Xception, Inception-v3, and VGG16 with 90.71%, 89.38%, and 87.12%, respectively. Dutta et al. [112] in Jan. 2021, suggested a Covid-19 recognition system through TL with multilayer DCNN(Inception V3) from CT images. The dataset was picked from Kaggle [74] with binary classification for ‘Covid + ve’ and ‘Covid-ve’ classes. For the training set, 279 images of ‘Covid + ve’ and 279 of ‘Covid-ve.’ 70 images for each ‘Covid + ve’ and ‘Covid-ve’ class for the validation set. The dataset was partitioned into two parts: training and validation, with an 80:20% ratio. The last few layers of the model were swapped with a customized DNN, with four customized layers, including the flattening layer, dense layer, dropout, and sigmoid activation function. The projected structure with 84% accuracy outperformed in this classification task.
Javaheri et al. [40] in Feb. 2021, created CovidCTNet, a DL open-source technique for recognizing Covid-19 from CT scans. In CovidCTNet framework, the compound preprocessing steps on CT samples were applied using the BCDU-Net structural design. The BCDUNet was designed based on U-Net. They collected a dataset from a public repository [85] and five medical centers in Iran. The BCDUNet differentiates Covid-19 from CAP in addition to supplementary lung illnesses. The assembled dataset contains 89,145 CT images, including 32,230 samples of affirmed Covid19, 25,699 of CAP, and 31,216 with healthy lungs or other illnesses. All CT slices were resized from 512 × 512 to 128 × 128. The dataset is divided into 90% for training and 10% for testing. CovidCTNet system attained a 91.66% accuracy, 87.5% sensitivity, 94% specificity, and 95% AUC based on experimental data. Jiang et al. [36] in Feb. 2021, suggested recognizing Covid-19 from thoracic CT images using DL. VGG16, ResNet50, Inception-v3, InceptionResNetv2, and DenseNet169 are the five extensively utilized DL pre-trained models employed in this system. The originally collected dataset comprises Covid-19(349) and non-Covid-19(397) CT pictures collected from preprints and 888 lung cancer CT scans from LUNA16. CycleGAN was used for synthetic image generation. Then datasets were classified into Covid, nonCovid, and lungCancer, containing 1300 CT images in each class. All samples were resized to 512 × 512 pixels and 66.33:33.33% dataset splitting ratio for training(1000 images) and testing(300 images) set. The experiment shows the DenseNet169 attain the finest performance with all the measures, including accuracy, recall, precision, F1-Score for the synthetic dataset of 98.92%, 97.80%, 100.00%, 98.89%, and real dataset of 98.09%, 97.80%, 97.37% 97.92% respectively.
The proposed approach (ReCOV-101) by Rohila et al. [124] in Jun. 2021 utilizes complete lung CT scans to detect the possibilities of Covid-19 contamination. They used pre-trained DCNN models are ResNet-50, ResNet-101, DenseNet-169, DenseNet-201. A DCNN with ResNet-101 as a pillar for ReCOV-101 resolves the vanishing gradient's difficulties using frisk connections. It avoids training from tiny layers and attaches straight to the output layer. The performance affecting layer is skipped by regularization. The dataset was collected from MosMedData [123] with partitioning based on the degree of seriousness of the Covid-19 contamination (GGO participation of the lung parenchyma from 25%, 50%, 75% degree). Dataset CT-0 contains normal lung tissue; CT-1 contains parenchyma less than 25%; CT-2 contains parenchyma between 25 and 50%; CT-3 contains parenchyma between 50 and 75%; CT-4 contains parenchyma exceeding 75%. The collected dataset contains 1105 images, including 250(normal) images for CT-0 class and 856(Covid) slices from CT-1 to CT-4 class. The dataset is distributed into 60:20:20 ratios for training, validation, and testing. The proposed structure achieved uppermost performance by ResNet101 using Adam-1 with 94.9% accuracy.
Hybrid and Custom Deep Learning Models
Ying et al. [43] in Feb. 2020, created a DL-based CT diagnoses system called as DeepPneumonia. The private datasets were collected from 3 hospitals and medical institutes. The data set was separated into randomized divisions of 60%:10%:30% for train, validate, and test sets. The DRENet structure was built using ResNet-50, in which the FPN is responsible for extracting top K data features from every picture. The proposed DRENet achieved an 86% accuracy, 95% AUC, 96% recall, 79% precision, and 87% F1-score. Turkoglu[49], in Jan. 2021, proposed a novel multiple kernels ELM (Extreme Learning Machine) built on a DCNN named MKs-ELM-DNN for recognition of Covid-19. The features were retrieved from CT using a DenseNet201 structure. An online public database [76] comprising Covid19 and nonCovid labels are employed to calculate ELM-classifier's architecture performance based on different activation approaches like ReLU-ELM, PReLU-ELM, and TanhReLU-ELM. At last, class annotation is defined as the voting technique aimed at estimating outcomes. After applying the data augmentation technique, the 746 CT images (349 and 397 for no-findings and Covid-19, respectively) expanded to 3730 images. At the same time, the extended dataset enclosed 1745 samples of Covid-19 and 1985 nonCovid19 classes. The highest accuracy achieved by the ReLU-ELM activation based on Multiple Kernels-ELM for data augmented(No) and augmented(Yes) of 87.02% and 96.75%, respectively. The MKs-ELM-DNN classification attained 98.36% accuracy, 98.28% sensitivity, 98.44% specificity, and 98.22% precision, 98.25% F1-score, and 98.36% AUC for Covid-19 recognition.
Wu et al. [115] in Feb. 2021, suggested a hybrid structure called COVID-AL for diagnoses of Covid-19 using weakly supervised DL from CT images. They assembled a dataset from 3 public online platforms [84, 114] and Ronneberger for lung region segmentation. The system considered 962 CT images considering 304 for Coronavirus-pneumonia, 316 for pneumonia, and 342 for normal class. The data splitting into the form of training (70%), testing(10%), and validation(20%). The proposed network design of a 2D U-Net for the lung area segmentation and a 3D residual network for the finding of Covid-19. In four steps of downsampling, the encoder of network segmentation retrieves image features by twice convolutional and pooling layers. The decoder of the segmentation network avoids connection to add features in the same phase. The system evaluation of COVID-AL offers 86.60% accuracy, 96.20% precision, and 96.80% AUC.
In another study by Tiwari et al. [131] developed lightweight applications in combination with modified TL models(VGG, DenseNet, ResNet, MobileNet) and Capsule networks using CT images. All the modifications were made based on CNN + PrimaryCaps + CTscanCaps layers sequences using DL-varients. The highest 99% classification accuracy was attained by MobileCapsNet. Modi et al. [132], in July 2021, suggested a capsule network for the classification of CT scan images performing the detection of Covid-19. The suggested DOCN was initially trained on 360 Covid-19 scans, and 397 CT scans for other diseases and healthy subjects. DOCN utilises 3-Conv + 3-Capsule layers. The system attained binary classification accuracy of 98%, sensitivity of 81%, and specificity of 98.4%.
Apart from the above description, we have tabulated the remaining systems in Table 2; which highlights the key aspects, such as data sources, training models, class-wise number of images, data partitioning, and the performance metrics of the examined DL-based Covid-19 diagnostic systems from X-ray, CT frames utilizing pre, hybrid, and custom-made networks with DTL.
Table 2.
Remaining summary of DL-based Covid-19 X-Ray, CT diagnosis systems
| Author | Type | Training Model | Resol-ution | Total Images |
|---|---|---|---|---|
| Wang et al. [95] | Pre-trained (XRay) | RestNet50, ResNet101, ResNet152 | 18,567 | |
| Arellan,Ramos [53] | Pre-trained (XRay) | DenseNet121 | 38 | |
| Minaee et al. [56] | Pre-trained (XRay) | Deep-COVID (ResNet18,ResNet50, SqueezeNet, DenseNet-121) | 5420 | |
| Demir [99] | Custom (XRay) | DeepCoroNet | 100 × 100 | 1061 |
| Sheykhivand et al. [52] | Pre-trained (XRay) | Inception V4 | 224 × 224 | 11,383 |
| Mishra et al. [55] | Pre-trained (XRay) | CovAI-Net (Inception, DenseNet, Xception) | 224 × 224 | 1878 |
| Sakib et al. [100] | Custom (XRay) | DL-CRC | 2905 | |
| Tang et al. [44] | Custom (XRay) | EDL-COVID | 15,477 | |
| Saha et al. [94] | Pre-trained (XRay) | EMCNet(AlexNet, VGG 16, Inception, and ResNet-50) | 224 × 224 | 4600 |
| Gupta et al. [54] | Hybrid(XRay) | InstaCovNet-19 ( InceptionV3,NasNet, Xception,Mobile NetV2, ResNet101) | 224 × 224 | 3047 |
| Vaid et al. [134] | Pre-trained (XRay) | VGG19 | 224 × 224 | 545 |
| Bhosale et al. [47] | Custom (XRay) | LDC-Net (IoT based) | 1024 × 1024 | 10,800 |
| Serener et al. [111] | Pre-trained (CT) | ResNet-50, ResNet-18, MobileNetV2, VGG,AlexNet,SqueezeNet, DenseNet121 | 224 × 224 | 1005 |
| Voulodimos et al. [96] | Pre-trained (CT) | FCN-8, U-Net | 630 × 630 | 939 |
| Chen et al. [1] | Pre-trained (CT) | ResNet50, Unet + + | 512 × 512 | 80,030 |
| Wu et al. [50] | Pre-trained (CT) | ResNet50 | 256 × 256 | 495 |
| Shah et al. [108] | Custom, Pre-trained (CT) | CTNet-10,DenseNet 169, VGG16/19, ResNet50,InceptionV3, | 128 × 128 to 224 × 224 | 812 |
| Khan et al. [102] | Hybrid (CT) | H3DNN(3DResNet, C3D, 3D DenseNet, I3D, LRCN) | 224 × 224 | 880 |
| Author | Classes | Partition | Performance | Data Source | Time |
|---|---|---|---|---|---|
| Wang et al. [95] | 8851(Normal), 9576(Pneumonia), 140(Covid19) | NA | Accuracy:96.1% | [72, 80] | NA |
| Arellan,Ramos [53] | 19(Covid + Ve), 19(Covid-Ve) | NA | Accuracy:99%, Recall:89%,Precision:91%,Fscore: 89% | [73, 82] | NA |
| Minaee et al. [56] | 5000(Normal), 420(Covid19) | Random | Sensitivity:98%, Specificity:92.9% | [72, 90] | NA |
| Demir [99] | 361(Covid19), 200(Normal), 500(Pneumonia) | Training:80%, Testing:20% | Accuracy:100%, Sensitivity:100%, Specificity:100% | [71, 82, 113], | Train: 53 M 22 s |
| Sheykhivand et al. [52] | 2923(Healthy), 2842(Covid19), 2778(Bacterial), 2840(Viral) | Training:70%, Testing:10%, Validation:20% | Accuracy:99.5%, Sensitivity100%, Specificity:99.02% | [71–74] | Test: 3 s |
| Mishra et al. [55] | 570(Pneumonia), 630(Non-pneumonia), 369(Covid19 +), 309(Covid19 -) | Random | Accuracry:98.31%, Precision:100%, Sensitivity:96.74%, Specificity:100%, F1-Score:98.34% | [77, 80], ESR | NA |
| Sakib et al. [100] | 219 (Covid19 +), 1341(Normal), 1345(pneumonia) | fivefold cross validation | Accuracy:93.94%, AUC:95.25% | [71, 72, 82] | NA |
| Tang et al. [44] | 6053(Pneumonia), 8851(Normal), 573(Covid19) | Accuracy:95%, Sensitivity:96.0%, PPV:94.1% | [120] | 1 s Exec | |
| Saha et al. [94] | 2300(Covid19), 2300(Normal) | Training:70%, Testing:10%, Validation:20% | Accuracy:98.91%, Precision:100%, Recall:97.82%, F1-score:98.89% | [71, 72, 75, 77, 80, 82, 133], | NA |
| Gupta et al. [54] | 1345(Pneumonia), 1341(Normal), 361(Covid19) | Training:80%, Testing:20% | Accuracy:99.53%, Precision:100%, Recall:99%, F1-Score:99% | [73, 82, 90] | NA |
| Vaid et al. [134] | 181(Covid19), 364(NoFinding) | Training:80%, Testing:20%, Validation:20% | Accuracy:96.3% | [72, 82] | NA |
| Bhosale et al. [47] | Covid-19, other 8 lung diseases | Train:76%, Test:12%, Val:12% | Acc:96.3%,Recall: 96.78%,Fscore:96.77%, AUC:98.18% | [82] and other 4 datasets | 0.136 s |
| Serener et al. [111] | 397(Mycoplasma Pneumonia), 145 (ViralPneumonia), 463(Covid19) | Random | Accuracy:89%, Sensitivity:98%, Specificity:86%, AUC:95% | [81] | NA |
| Voulodimos et al. [96] | 447(Covid-negative), 492(Covid-positive) | Training:85%, Validation:15% | Accuracy:99%, Recall:89%, Precision:91%, F1-Score: 89% | [80] | NA |
| Chen et al. [1] | 49,089(Covid19), 30,941(Normal) | Random | Accuracy:96%, Sensitivity:98%, Specificity:94%, PPV:94.23%, NPV:97.92% | Renmin Wuhan Univ., Qianjiang Hospital, China | NA |
| Wu et al. [50] | 368(Covid19), 127(other pneumonia) | Training:80%, Testring:10%, Validation:10% | Accuracy:76%, AUC:81.9%, Sensitivity:81.1%, Specificity:61.5% | China Medical Univ., BYH in China | < 5 s |
| Shah et al. [108] | 349(Covid19 confirmed), 463(nonCovid19) | Training:80%, Testring:10%, Validation:10% | CTNet Accuracy:82.1%, VGG19 Accuracy: 94.52% | [76] | Train: 130 s, Test:0.9, Exe: 0.01233 s |
| Khan et al. [102] | 417(Covid19), 463(Non-Covid19) | NA | Accuracy:85% | [82, 86] | NA |
NA indicates the corresponding author did not disclose the parameter value
Diagnosis Based On Multimodal Radiography Images
Kassani et al. [97] in Apr. 2020 suggested a DL-based automatic recognition of coronavirus infection from binary(X-ray, CT) modalities. The system used 8 DCNN variants(MobileNet, DenseNet, Xception, ResNet, InceptionV3, InceptionRes-NetV2, VGGNet, NASNet). Obtained features were fed to an ML classifier to separate the subjects, either Covid-19 or other. This method avoids work definite in data pre-processing. They assembled a multisource dataset [72, 77] containing Covid-19 positive frames(117:X-ray, 20:CT) and healthy frames(117:X-ray, 20:CT). The DenseNet121 feature mining using a bagging tree attained the most satisfactory performance at 99% categorization accuracy. The 2nd most extraordinary learner was a hybrid of a ResNet50 feature extractor trained by LightGBM with 98% precision. Horry et al. [101] in Aug. 2020, developed a Covid-19 method of identification employing seven versions of CNNs from three(X-ray, ultrasound, CT) radiography modalities images (a multimodal image classification). The used different CNN techniques are: VGG16/VGG19, Resnet50V2, InceptionV3, Xception, InceptionResNetV2, DenseNet121, and NASNetLarge. The experimentation used four public datasets[76, 82, 118]; a total of 62,476 images are chosen from the source dataset, including 140 for Covid-19, 320 for pneumonia, 60,361 for normal images of X-ray; 349 for Covid-19, 397 for nonCovid images of CT; 399 of Covid-19, 275 of pneumonia, 235 of normal images for the ultrasound. The X-ray dataset has been selected to eliminate a certain solitary image that was wrongly marked. After being curated, the X-ray set contains 139(Covid-19), 190(pneumonia), and 400(normal) samples. After applying data augmentation, the complete dataset reached to 34,560 samples, including Covid-19(2920), pneumonia(2920), normal(5840) for X-ray; Covid-19(6000), nonCovid(6000) for CT; and Covid-19(2720), pneumonia(2720), normal(5440) for the ultrasound. Further, N-CLAHE improves luminosity and sharpness. A picture is redrawn to the default classifier with 224 × 224 for VGG and 299 × 299 for InceptionV3. The data partitions were randomly selected for train and test sets. Overall results show that ultrasound images deliver higher recognition accuracy than X-ray & CT modalities. The VGG19 achieves significant Covid-19 recognition alongside pneumonia or normal, although three modalities approached with the precision of 86% for X-ray, 100% for ultrasound, and 84% for CT. Nath et al. [34] in Oct. 2020, introduced DNN structure to sense Covid-19 for 2-class (Covid, nonCovid) and 3-class (Covid, nonCovid, pneumonia) classification from X-ray and CT. The data set is collected from 3 public platforms [72, 73, 76]. CXR dataset includes Covid19 (219), normal(1341), viralPneumonia(1345); 349 for Covid, and 397 for nonCovid classes of CT modality. Further data set separated to 80:20 proportion for training and testing. CNN-model has been constructed with the organization of 24-layers. SGD with momentum optimizer was utilized for both datasets with LR = 0.001. It achieves 99.68% and 71.81% accuracy for X-ray and CT, respectively.
Panwar et al. [57] in Nov. 2020, suggested a new DTL algorithm for binary classification and tested it on three diverse radiology online datasets [72, 78, 105] of X-ray and CT modalities. The system was an altered version of DCNN called VGG16. The structure comprises 19-weighted layers of DCNN in VGG19. Experiments used the PA-view of CXRI to examine the lungs better. The experiment attained an 95.61% accuracy, 94.04% sensitivity, 95.86% specificity, 95% F1-score, and 96% recall. The weights acquired after network training through CT treatment would also answer X-ray significantly. The achieved outcomes disclose that the individual identified with pneumonia had additional probabilities of becoming confirmed as a false-positive by the projected network. Alakus et al. [37] in Nov. 2020, proposed DL-based Covid-19 recognition system. The suggested system was trained on six neural network variants(ANN, CNN, LSTM, RNN) and 2 hybrid models(CNNLSTM, CNNRNN). Using laboratory decisions, X-ray, and CT images, the prediction was learned for Covid-19 infection with DL models. The laboratory findings are: hematocrit, hemoglobin, platelets, red blood cells, lymphocytes, leukocytes, basophils, eosinophils, monocytes, serum glucose, neutrophils, urea, C-reactive protein, creatinine, potassium, sodium, alanine transaminase, aspartate transaminase. The single dataset contains 520 no-findings and 80 Covid19 patients. The experimented DTL properties include a batch size of 512, 256, 32, 16 with a learning rate of 0.001 and epoch 250 with activation function ReLu and SGD optimizer. The proposed experiment results were categorized into two methods: data split and model performance. LSTM gives the highest assessment outcomes with tenfold cross-validation 86.66% correctness, 91.89% F1-measure, 86.75% exactness, 99.42% recall, 62.50% AUC. CNNLSTM gives the highest assessment results among all DL models with train-test(80:20) divided method are 92.30% accuracy, 93% F1-measure, 92.35% exactness, 93.68% recall, and 90% AUC.
For Covid-19 automatic detection, Hussain et al. [93] in Jan. 2021, constructed a new CNN model called CoroDet using primary X-ray and CT scanning photographs. The system considered 7 public online repositories [72, 73, 75, 76, 105, 116, 117] by combining, modifying, and preparing COVID-R dataset. The Covid-R dataset prepared based on binary class(Covid-19 and normal), 3-class(Covid-19, pneumonia, and normal), and 4-class(Covid-19, pneumonia-bacterial, pneumonia-viral, and normal) classification. Binary classification is stated on Covid-19(500) and normal (800) images. For the 3-class classification, pneumonia-bacteria images are 800 and binary class samples. For 4-class classification, pneumonia-viral and pneumonia-bacteria samples are 400 each and binary class samples. System attained an 99.1% accuracy, 99.27% precision, 98.17% recall, 98.51% F1-score for 2-class classification; 94.2% accuracy, 95.37% precision, 97.47% recall, 98.62% F1-score for 3-class classification; and 91.2% accuracy, 94.27% precision, 96.17% recall, 97.51% F1-score for 4-class classification. The authors have consulted with clinicians to understand the difference between each class of CXRI. Perumal et al. [23] in Jan. 2021, introduced a new DL approach for classifying distinct pulmonary illnesses containing Covid-19 from CXR and CT scans. Perumal proved that Covid-19 is considerably like viral pneumonia lung infection using TL. The knowledge adapted from the training model for spotting viral pneumonia could be applied to Covid-19 detection. Haralick features were used for feature extraction to emphasize only the ROI to detect Covid-19. The noise impedance from infected areas and tissues makes it challenging to sense the images' atypical features. The proposed model uses three pre-trained networks(VGG16, ResNet50, and InceptionV3). The dataset was downloaded from 2 public repositories (NIH [82], Mendeley [78]), contains pulmonary disease(81,176), bacterial-pneumonia(2538), viral-pneumonia(1345), normal (1349), Covid-19(205) of CXRI; and 202 for Covid-19 CT class. The images were resized to 256 × 256. The HE (to increase the contrast) and Weiner filters(to eliminate the noise) were used to enhance the image quality. The analysis shows that 385 Covid-19 images were properly categorized under Covid-19 class, and 22 images were wrongly categorized under non-viral pneumonia class out of 407 samples. These results conclude that Covid-19 is related to viral-pneumonia. The misclassification rate was 0.012 for viral pneumonia. Testing pneumonia model with Covid-19 dataset, the VGG16 offers the highest accuracy of 93.8%.
A CNN technique for categorizing healthy people from Covid-19 and pneumonia has been created by Gilanie et al. [135] in Apr. 2021. The model was trained on 3-classes (normal, pneumonia, and Covid). The system used three openly accessible and locally created datasets (Radiology Dept. BVHB, Pakistan). The datasets under experiment consist of 15,108 images, including 7021 X-ray, CT scans of each normal and pneumonia class, and 1066(539 X-ray, 527 CT) frames of Covid-19 class. During system design, each image shrank to 256 × 256. Dataset was separated into 60%, 20%, and 20% for train, cross-validation, and test sets. The projected method attained an average of 96.68% accuracy, 95.65% specificity, and 96.24% sensitivity. The quantitative information for multimodal imaging is listed in Table 3.
Table 3.
Summary of DL-based Covid-19 multimodal diagnosis systems
| Author | Type | Training Model | Resolu-tion | X-Ray | CT | Ultrasound |
|---|---|---|---|---|---|---|
| Panwar et al. [57] | Pre-trained | VGG-19 | 512 × 512 pixels | 800(Covid19), 800(nonCovid19) | Covid Positive (1252) | NA |
| Nath et al. [34] | Pre-trained | CNN | 256 × 256 | 219(Covid19), 1341(Normal), 1345(Viral Pneumonia) | 349(Covid-19), 397( NonCovid19) | NA |
| Kassani et al. [97] | Pre-trained | MobileNet, DenseNet, Xception, ResNet, InceptionV3, InceptionRes-NetV2, VGGNet, NASNet | 600 × 450 | 117(Covid-19), 117(Healthy) | 20(Covid-19), 20(Healthy) | NA |
| Hussain et al. [93] | Custom | CoroDet | 256 × 256 | 2843(COVID-19), 3108(Normal), 1439(Pneumonia Viral + Bacteria) | 2843(Covid-19), 3108 (Normal), 1439 (Pneumonia Viral + Bacterial) | NA |
| Gilanie et al. [135] | Pre-trained | CNN | 512 × 512 | 4021(Normal), 4021(Pneumonia), 539(Covid-19) | 3000 (Normal), 3000 (Pneumonia), 527 (Covid19) | NA |
| Horry et al. [101] | Pre-trained | VGG16/VGG19, Resnet50, Inception V3, Xception, InceptionResNet, DenseNet, and NASnetlarge | 224 × 224 for VGG16/19 and 299 × 299 for InceptionV3 | 140 (Covid-19), 320 (Pneumonia), 60,361 (Normal) | 349(Covid-19), 397(NonCOVID) | 399(Covid-19), 275 (Pneumonia), 235 (Normal) |
| Perumal et al. [23] | Pre-trained | Resnet50, VGG16, InceptionV3 | 226 × 226 | 81,176(Pulmonary), 2,538(BacterialPneumonia), 1,345(ViralPneumonia), 1349(Normal), 205(Covid-19 CXR), 202(Covid-19 CT) | NA | Pulmonary, Bacterial Pneumonia, Viral pneumonia, Normal, Covid-19 |
| Author | Classes | Partition | Performance | Data Source | Time |
|---|---|---|---|---|---|
| Panwar et al. [57] | Pneumonia, nonCovid19, Normal, Covid19 | Train:64% Validate:20%, Test:16% | Accuracy:95.61%, Sensitivity:94.04%, Specificity:95.86%, Precision:95%, F1-score:95%, | [180–182] | NA |
| Nath et al. [34] | Covid19, Normal, Pneumonia | Train:80%, Test:20% | Accuracy X-ray:99.68%, CT:71.81% | [72, 73, 76] | NA |
| Kassani et al. [97] | Covid19, Normal, | tenfold cross-validation | Accuracy:99.00% | [72, 77] | 0.028 s |
| Hussain et al. [93] | Covid19, PneumoniaBacterial, Pneumoniaviral, Normal | fivefold cross-validation | Accuracy:99.1%, Precision:99.27%, Recall:98.17%, F1-score:98.51% | [72, 73, 75, 76, 105, 116, 117] | NA |
| Gilanie et al. [135] | Normal, Pneumonia, Covid-19 | Train:60% Validate:20%, Test:20% | Accuracy:96.68%, Specificity:95.65%, Sensitivity:96.24% | [72, 80, 110], Radiology Dept. BVHB Pakistan | NA |
| Horry et al. [101] | Normal, Pneumonia, Covid-19 and other nonCovid-19 | Train:80%, Test:20% | Precision:86%(X-ray), 100%(LUS), 84%(CT) | [76, 82, 118] | NA |
| Perumal et al. [23] | Random | Accuracy:93.8% | [78, 82] | NA |
Imaging Segmentation for Covid-19 Diagnosis
Voulodimos et al. [96] in May 2020, proposed DL-based classification & semantic-segmentation labeling of diseased lung regions for Covid-19 from CT scans. Suggested system used FCN-8 and U-Net for Covid region segmentation [136]. Given an input as a CT image, the FCN8 tends to create new coarse borders. Alternatively, U-Net provides smaller slicker regions than the original annotated region. The dataset was collected from radiopaedia [80] for the experiment consisting of 939 cross-sectional images, including 447 CT slices annotated as ‘–Ve’ and 492 as ‘ + Ve.' From the collected dataset, 85% were utilized for training and validation, 15% for testing. Among the total dataset, 90% were used for training and 10% for the validation set. The system reached 99% accuracy, 89% recall, 91% precision, and 89% F1-score on the validation set and offered the usual execution time per image between 0.01 s to 0.018 s.
Abdel-Basset et al. [137] in Jan. 2021, a dual-path DL network was demonstrated for partial-monitored FewShot Segmentation for Covid-19 (FSS-2019-nCov) contamination. FSS-nCov delivers a precise division of Covid-19 contamination from the insufficient number of labeled images. Each path covers encoder-decoder(ED) architecture to extract features while preserving the channel of Covid-19 CT slices. The ED structure comprises an encoder, context enrichment, and decoder. The FSS-nCov uses RestNet34 for feature extraction at encoder. They introduced a Smoothed-Atrous-Convolution block, Multiscale-Pyramid-Pooling block, and an adaptive Recombination-Recalibration(RR) unit, allowing rigorous information sharing between tracks. The experimental results achieve the dice similarity coefficient (DSC) of 79.8%, the sensitivity of 80.3%, specificity of 98.6%, and Mean Absolute Error (MEA) of 6.5%.
Open Discussion, Challenges, and Future Trends
Open Discussion
The primary objective of this analysis is to find the most vital DL model for detecting, segmenting, and classifying Covid-19 using CNN variants. Table 4 summarizes individual systems' outcomes and compares different DL experimental setup work reported in the studied articles. In the comparative analysis of experimental results from Table 4, each row represents the number of layers used in the model, the kernel size, pool size, stride, batch size, image size, learning rate, number of epochs, performance evaluation metrics, activation function, optimizer, and libraries used for Covid-19 detection. Throughout the study, 64 systems were examined, with 36 systems based on X-ray images, 20 on CT, and 8 on multimodal imaging systems. Most systems used multisource data, and only a few used a single data source among the studied articles. The utilization and demand of enormous datasets with good maximum-likelihood diagnosis, such as RT-PCR, confirm Covid-19, and outcomes such as death and discharge duration are common to DL techniques for imaging Covid-19 [138]. Integrating medical data in time series containing recurrent samples and blood tests may significantly boost training datasets. Some researchers and radiological studies have taken the initiative to release Covid-19 datasets on public platforms [71–82, 136].
Most Covid-19 detection systems assessed accuracy, sensitivity, specificity, and F1-score as performance evaluation metrics (Appendix A). From individual image modality the highest 100% accuracy achieved by [99], 100% specificity achieved by [38, 39, 51, 53, 55, 68], 100% sensitivity (recall) achieved by [52, 63, 68, 70, 92], 100% precision achieved by [35, 36, 39, 54, 55, 68], 99.99% AUC by [98], and the best F1-score of 99.99% attained by [39]. Whereas 2-mode imaging (X-ray, CT) has the highest of 99.68% accuracy achieved by [97], 98.17% recall, and 99.27% precision achieved by [93]. Along with 3-mode imaging (X-ray, CT, ultrasound), the highest 100% precision is achieved [101]. In the case of 4-class classification 99.94% accuracy achieved by CovidAid [63], for 3-class classification 99.72% accuracy achieved by CovidDetection-Net [41], and for binary classification 99.97 accuracy achieved by [63]. In a custom network, the highest accuracy of 99.99% was attained by Covid-ChexNet[39]. The X-ray [72] dataset is the most frequently used dataset by 23 times (analysis from Table 1). The highest time taken by the hybrid EDL-COVID [60] model was the fastest detection speed of 342.92 s, whereas the least execution time was 0.013 and 0.014 s by [47] and [139]. However, a custom model has consequences like computation efficiency, layer size, epochs, training parameters, number of layers, etc.
Compared to the custom models, most evaluated techniques outscored the pre-trained network. Because different data sizes were used practically in each investigation, the efficiency of the developed models varied (regarding the data source) and hence is not comparable. In terms of imaging modality comparison, the X-ray modality outperformed, followed by CT and ultrasound.
However, one instance was the pneumonia analysis from CT [138], which intended to autonomously detect and measure aberrant structures throughout the chest, allowing the detailed examination of non-contrast chest CT scans for scientific purposes. This process detects pneumonia-related lungs, divisions, and anomalies. It also evaluates more significant anomalies, which have been linked to severe conditions. These findings could assess the degree and progress of anomalies in Covid-19 patients [138]. These evaluation methods should be deployed at the remote console, i.e., the clinicians' viewing desk. Attempts are being taken to ensure that the right user experience and interoperability for such concepts occur using limited processing power, hardware, and the internet [138]. The overwhelming bulk of cutting-edge DNN is trained on 2D drawings. CT and MRI, 3D scams certainly contribute to the root issue. Because standard DL networks are not fine-tuned, history is invaluable when implementing DL models on such imageries [140]. However, some solutions used a large sample dataset for other diseases, whereas Covid-19 instances are limited. Across the study, binary to multi-classifications is considered. The Covid-19 detection systems [41, 42, 46, 72, 91, 112] are managed to get an equal quantity of samples in each class to achieve the highest accuracy.
Numerous publications ignored highlighting the diagnosis efficacy of their technique, even though distinguishing severe Covid-19 instances versus regular chest X-rays may not have been challenging [62, 141]. Based on the degree of severity, Covid-19 affected lungs were classified as severe vs. non-severe [142], and severity [143], normal vs. severe [144] only. In [16], the dataset was considered for model training to find the severity of the affected lungs, but the results were not satisfactory. Furthermore, there is no reason why investigators preferred a particular network structure over another and did not equate their outcome if some other CNN design was chosen [141]. The study [145] proves how dropout weights can affect the uncertainty of the DL model of Covid-19 prediction. Along with disease classification, Alazab [146] et al. predicted the Covid-19 outbreak using LSTM and time-series analysis for coastal areas. Suggested research proves that the Covid-19 outbreak in coastal regions is higher than in non-coastal.
The medical observations [45] for Covid-19 CT scans are radial, symmetrical, sub-pleural, multichannel, posterior, frontal, and middle expansion of airways, broncho-vascular thinning (thickness of the in lesion), traction bronchiectasis, a bizarre pavement look (GGOs & inter-/intra-lobular septum thickness). Likewise, the following aspects are seen in pneumonia sufferers' CT images: Reticular transparency, centralized dispersion of GGO, unilateral, and More widespread dispersion across the Bronchovascular link (Vascular, Bronchial wall thickness). CAM, GradCAM [65] shows an important role in object detection for visualization of Covid-19 affected area on lung from radiograph images and bounding box [136]. A lightweight RaspberryPi-based GUI application (LDC-Net) [47] may be deployed with radiography machines(X-ray, CT, Ultrasound), which further may help physicians for diagnosis assistance.
However, in the case of Ultrasound imaging for Covid-19 respiratory periodic inspection and advancement of Covid-19 illness is visible as B-lines oxygenation from the initial phases to severe phases [101]. Also, radiological indicators in LUS could be exploited in Covid-19 client treatment [27]. A 4-level grading scheme with a scale from zero to three for coarse disease assessment was developed. Grade 0 denotes the existence of a consistent pleural lining followed by horizontal artifacts known as A-lines [147] that represent an excellent lung area. Grade 1 means the earliest evidence of irregularity, i.e., the advent of pleural-line changes associated with vertical artifacts. Grades 2 & 3 indicate a more progressive pathologic condition, with modest or substantial consolidations occurring accordingly. Lastly, a grade of 3 confirms the value of a larger hyperechogenic region [136] underneath the pleural area, known as the “white lung.” In [148], LUS imaging identification signs were collected from radiologist practitioners, such as healthy lung(with horizontal A-lines), pneumonia infected(alveolar consolidations), and SARS-CoV-2 infected(sub-pleural consolidation and a focal B-line) cases. B-lines are the most common pathological sign of LUS, and they are caused by pulmonary edoema or non-cardiac sources of interstitial diseases [149]. The Covid-19 LUS image has patchy B-lines, Pleural lines fragmented/irregular, and sub-pleural consolidation as common sign findings [150]. Unlike an X-ray or CT scan, LUS does not use ionizing radiation[151].
The study shows that the dataset used for the experiment was divided into ratios (hold-back, e.g., 80:10:10 for training, testing, and validation) and fivefold or tenfold cross-validation [60] techniques. The investigative presentation was assessed and equated to professional radiologists on an independent testing dataset [91]. To increase the sample size, default transformations and custom augmentations are offered by the fastai2 [91]. Issues like ample storage, noisy images, classification of images, and image retrieval tend to be tedious for developing a DL-based system, and to overcome such problems, CBIR(Content-Based Image Retrieval) [152] was created. Further, similar issues can be solved using ML-based classification algorithms such as Decision Tree, SVM, Naive Bayes, and KNN [153].
Challenges and Future Trends
There are numerous obstacles using DL methods to discover new coronavirus. Although, DL with Covid-19 detection from lung ultrasound, CT, and X-ray images demonstrates promising outcomes. At the same time, the DL technique requires a large amount of dataset and computation to develop a robust diagnosis system and train the networks.
The available dataset is noisy [151], blurry, unnecessary content like catalog scripts, symbols, and manufacturer-specific user interface[150], and not accurately labeled data. The developed system uses internet downloaded data, so there is a high possibility of data duplication, missing data, weakly labeled dataset [154], or limited labeled dataset [155]. Articles referred [46, 47, 63, 65, 69, 70, 93, 94, 101] for the study using different multisource datasets for their experimentation. Because of this intention, it is pretty problematic to determine which system harvests the most satisfactory result. With a small amount of data, the DL architecture may over-fit, which may degrade the performance of the developed system. To expand the sample-size of Covid-19 class from limited datasets, various authors applied data augmentation [38, 48, 49, 69, 101, 103].
Cross-Dataset Performance The significant model performance assessment for different dataset-based efficiency of learned features by CNN at training from a particular source and testing it on another data source. This results in the sensitivity and specificity generating many varying quantitative results with a difference of 10 to 40%, which has been achieved by[141] for AlexNet, ResNet, VGG, Squeeze Net, and DenseNet. In [106], the cross-dataset analysis results in accuracy drops. In a clinical decision, more excellent and more different data sets are taken to assess the approaches in a real-world situation. Multiple authors used the same dataset for testing and training, so the generated system accuracy is very high, but this is a severe issue about inspecting the efficiency of feature maps generated by CNN on the particular dataset and testing it on features from a diverse source [141]. Reducing features in the system [39] is not an intelligent approach while training a model because it leads to lesser accuracy in the validation and test phase.
Severity Level Recognition of Covid-19 Covid-19 severity levels such as mild-Covid, moderate-Covid, and severe-Covid are used to build triage systems with high clinical value [156]. Apart from severity, the detection of Covid-19 in pregnancy and childbirth [24] did not aggravate the course of symptoms or CT characteristics of Covid-19. The suggested platform's efficiency was not evaluated to that of radiologists.
Custom Models In a multiclassification problem (with three or even more categories), the set of features in the CoVNet-19 feature matrix using a single DCNN could be raised to 64 or 128. CoVNet19 [48] can be optimized and changed into a lighter version since the author employed a stacked ensemble system with two DCNNs and an SVC network [48]. The authors state that they have used a global filter to sense local-features using a filter dimension of 3 × 3, but there is a possibility to spot features lost by the used filters. CheXNet [39] contains flaws, like the number of sample unpredictability regarding data sequencing variations and sensitivity to adversarial attacks. Using CheXNet to improve pulmonary abnormalities identification in CXRs necessitates the creation of ensemble methods that are presently prone to overfitting due to many pictures from Covid-19 infected cells [65].
Ninety percent of referred articles focused on developing standalone Covid-19 detection systems, including computer-aided detection (CAD) [157, 158], which runs on a single machine. So there could be a separate provision for developing public web or enterprise applications for global data collection, processing, and detecting Covid-19. Nevertheless, few developed web applications [10] on the cloud [159] where the doctor can able to upload the X-ray, CT image to the application and provide the doctors with a simple Covid-19(Yes) or Covid-19(No) as a result [97]. Finding the severity of patients using radiography images is also a challenging task. Also, it is hard to find (as per the dataset) the most common age ratio of Covid19 ‘ + Ve’ patients is? Even for the gender/geography ratio [160]. No author has categorized the Covid-19 phases (1st wave, second wave, third wave, delta coronavirus) based on the finding of Covid-19.
In addition, medical investigations would have been needed to prove the remarkable precisions reported in the literature studied article [161]. The clinical evaluation of the developed system should be verified in collaboration with hospitals and medical research centers. After generating the CoroDet [93] system, the DL researcher approached the radiologist to understand the effects of coronavirus and pneumonia on the lung segment [93] and collected the necessary information. It’s hard to suggest an excellent yet concise DL network for the screening of Covid-19. For the large storage capacity of a vast dataset, the big data on the cloud[159] plays a significant role in DL [162]. Because LUS pictures are blurry and data filtration, Fourier processing, and deconvolution are used to upgrade the effectiveness of radiography images. CLAHE, HE, and AHE were used, and the picture with contrast limited AHE [65] was employed before the workflow to improve the clarity of ultrasound images [101]. A U-Net-based semantics separation [163] can also distinguish pulmonary pixels from the body tissues and background [65]. Data fusion enables us to mix several kinds of data to enhance the grouping accuracy of the model. Developing a web application that accepts radiological lung pictures as a source and produces a probability of Covid-19 or pneumonia occurrence and a heatmap emphasizing the likely contaminated areas [69]. We believe that DL-based quantification can help address patients with worsening respiratory status and moderate or severe infection of Covid-19.
Technical Limitations
See Table 5.
Table 5.
Technical limitations for COVID-19 diagnosis
| Sr.No | Technical Limitation | Description |
|---|---|---|
| (a) | Covid-19 detection where limited data are available | The system developed by [42, 63, 63, 67, 68, 98, 103] utilized less than 200 Covid-19 samples. The DL architecture may over-fit with a tiny dataset, lowering the performance of the developed system. Data augmentation offers to expand the sample size of the Covid-19 class from limited datasets to overcome the tiny dataset issue |
| (b) | Covid-19 detection under big-data situation | Decentralized tensors are used by DL to process data. Using Named Entity Recognition and Relation Extraction tasks, DL accelerates the building of Knowledge Graphs[162]. It would be impractical to conduct these activities personally on massive data such as collecting biomedical samples. They can make use of massive data by scaling up GPU-computing. An extensive data collection necessitates the most significant computer resources to train and test the DL model |
| (c) | Automatic detection of a single case involving multiple diseases | Disease classification using a DL-based system in various diseases like merging the samples of viral, bacterial, mycoplasma[164], and Covid-19-pneumonia in a single class (as pneumonia) can lead to false prediction[93]. We can separately train them in individual classes to improve several disease classification accuracy |
| (d) | Manage uncertainties in automatic detection | It is critical to assess their efficiency before using DL systems. The accuracy obtained by these systems is uncertain because they are susceptible to noise and incorrect model interpretation, and the inductive implications inherent in cases of uncertainty[165]. Therefore, it is extremely desired to describe uncertainty in any AI-based platform in a trusted and robust environment. Bayesian DL is used to create a resilient model trained on small and large datasets to manage uncertainties. Other methods include Monte Carlo dropout, Markov chain Monte Carlo, variational inference, Bayesian active learning, Bayes via backpropagation, variational autoencoders, and the gaussian procedure |
| (e) | Percentage of training and testing samples | The volume of data required for learning is determined by the model's complexity. Consequently, the number of training samples needed for a well-performing system is ten times the number of model parameters[166]. We can partition the training, testing, and validation dataset based on constant ratios and random and specific cross-fold validation techniques. Data partitioning is based on the following factors: different label classification, augmented training data, pre-trained weights to initialize the lower layers of the model, and batch normalization |
| (g) | Diagnosis performance using DL | Ghoshal and Tucker [145] explored why dropweights-based Bayesian-CNN can measure volatility in DL strategies to enhance the diagnoses efficiency of the human–machine pairing utilizing the openly accessible Covid-19 chest X-ray and found that classification volatility is highly associated with recognition reliability in decision making. The dropweights range varies from 0.1 to 0.5 for experimentation. They attained predictive entropy of 0.9952 and bayesian active learning disagreement of 0.8980 for 0.1 dropweights |
| (h) | Covid-19 variants detection using Radiography images | Apart from above limitations the Covid-19 variant detection [47] or classification (Alpha, Beta, Delta, Omicron, Deltacron, BA1/2 etc.) from radiography images is another challenge. However Covid-19 variants detection could be possible due to genome sequence data |
Conclusion
This paper provides the recent DL techniques utilized to classify Covid-19 from different lung and chest imaging modalities. However, subsequent X-ray, CT, and ultrasound studies for Covid-19 classification are limited. Widely accepted changes in pulmonary lesion patterns could be witnessed, including GGO and high mortality convergence in the comparatively initial phases. Datasets used in various experiments are provided and discussed in Sect. 2. Its significant barriers to the current methods are underlined in Sect. 6. The identical set of data accumulated by Cohen [72] is used in most studies. The implemented DL-based methods for detecting Covid-19 in the literature indicate impressive outcomes. Even after the remarkable results, there is indeed much space for enhancement.
In most cases, a limited set of images for diseased samples with Covid-19 is used. It is necessary to create public and diversified data sources. Radiology professionals must authenticate the data sets and categorize them with the respective lung illness abnormalities. The majority of the existing methodologies used binary classification, although various other factors can cause pneumonia. GAN’s encouraging outcomes are worthy of additional research. Also, there is a possibility of developing a standard Covid-19 detection system, where multimodal radiography images could be accepted; instead of creating an individual system for each modality. The blended utilization of AI and radiography imaging modalities can compensate for limited hospital facilities while assisting in the definitive screening and diagnostic forecasting of Covid-19. Healthcare professionals and software developers, on the other hand, should regularly interact and use their equally skilled to verify the utility of DL methods. We are enthusiastic about these frameworks' great versatility and assume their fundamental constraints could be resolved. We keep hoping that this beginning will assist the audience in narrowing their focus and pursuing such implementations for the classification of Covid-19 variants(Alpha, Beta, Delta, Omicron, Deltacron, etc.) using medical imaging [47] with deep learning techniques.
Appendix A: Performance Metrics
When creating a classification model, we must assess how well it predicts using a confusion matrix. To evaluate it, a true positive (TP) is an outcome in which the model adequately predicts the positive class. Likewise, a true negative (TN) is an outcome in which the model predicts the negative class adequately. A false positive (FP) results in which the model forecasts the positive class inaccurately. A false negative (FN) results in which the model forecasts the negative class inaccurately.
Appendix B
Abbreviations used.
| Abbreviations | Referred To |
|---|---|
| ANN | Artificial Neural Networks |
| AUC | Area Under the Curve |
| CAD | Computer Assisted Diagnosis |
| CAM | Class Activation Map |
| CAP | Community-Acquired Pneumonia |
| CNN | Convolutional Neural Network |
| Covid-19 | Corona Virus Disease |
| CT | Computed Tomography |
| CXR | Chest X-Ray |
| DCNN | Deep Convolutional Neural Network |
| DL | Deep Learning |
| GAN | Generative Adversarial Network |
| GGO | Ground Glass Opacity |
| LSTM | Long Short Term Memory |
| LUS | Lung Ultrasound |
| MERS-CoV | Middle East Respiratory Syndrome Coronavirus |
| ML | Machine Learning |
| MSE | Mean Squared Error |
| NPV | Negative Predictive Value |
| PA | Posteroanterior |
| PCR | Polymerase Chain Reaction |
| PPV | Positive Predictive Value |
| ResNet | Residual Network |
| RMSE | Root Mean Square Error |
| RNN | Recurrent Neural Networks |
| RT-PCR | Reverse Transcription-Polymerase Chain Reaction |
| SARS | Severe Acute Respiratory Syndrome |
| SARS-COV-2 | Severe Acute Respiratory Syndrome Coronavirus 2 |
| SVM | Support Vector Machines |
| TL | Transfer Learning |
Author Contribution
YHB: review and editing, writing manuscript, conceptualization, visualization, and formal analysis. Dr. KSP: supervision.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yogesh H. Bhosale, Email: yogeshbhosale988@gmail.com
K. Sridhar Patnaik, Email: kspatnaik@bitmesra.ac.in.
References
- 1.Chen J. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography. Scientific Rep. 2020 doi: 10.1101/2020.02.25.20021568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed M, Ahmad JJPC, Rodrigues GJ, Din S. A deep learning-based social distance monitoring framework for COVID-19. Sustain Cities Soc. 2021;65:102571. doi: 10.1016/j.scs.2020.102571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abdelminaam DS, Ismail FH, Taha M, Taha A, Houssein EH, Nabil A. CoAID-DEEP: an optimized intelligent framework for automated detecting COVID-19 misleading information on Twitter. IEEE Access. 2021;9:27840–27867. doi: 10.1109/ACCESS.2021.3058066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO Dashboard (2022) (Online). Accessed on 16 July 2022 [Online] https://covid19.who.int
- 5.Real time database and live updates of Covid-19 cases (2022). Accessed on 16 July 2022. [Online] https://www.worldometers.info/coronavirus/
- 6.Ministry HFW COVID Report (2021). Accessed on Jul. 16 2021. [Online]. Available: https://www.mohfw.gov.in/
- 7.Chauhan N, Soni S, Gupta A, Aslam M, Jain U. Interpretative immune targets and contemporary position for vaccine development against SARS-CoV-2: a systematic review. J Med Virol. 2020 doi: 10.1002/jmv.26709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajaraman S, Antani S (2020) Training deep learning algorithms with weakly labeled pneumonia chest X-ray data for COVID-19 detection Radiol Imag preprint, 10.1101/2020.05.04.20090803.
- 9.Rajaraman S, Siegelman J, Alderson PO, Folio LS, Folio LR, Antani SK. Iteratively pruned deep learning ensembles for COVID-19 detection in chest X-rays. IEEE Access. 2020;8:115041–115050. doi: 10.1109/ACCESS.2020.3003810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guellil MS et al. (2020) WEB predictor COVIDz: deep learning for COVID-19 disease detection from chest X-rays In: 2020 International Conference on Decision Aid Sciences and Application (DASA), Sakheer, Bahrain, Nov. 2020, pp 601–606 10.1109/DASA51403.2020.9317291
- 11.Sethy PK, Behera SK, Ratha PK, Biswas P. Detection of coronavirus disease (COVID-19) based on deep features and support vector machine. Int J Math Eng Manag Sci. 2020;5(4):643–651. doi: 10.33889/IJMEMS.2020.5.4.052. [DOI] [Google Scholar]
- 12.Chauhan N, Soni S, Jain U. Optimizing testing regimes for the detection of COVID-19 in children and older adults. Expert Rev Mol Diagn. 2021;21(10):999–1016. doi: 10.1080/14737159.2021.1962708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salehi W, Baglat P, Gupta G. Review on machine and deep learning models for the detection and prediction of Coronavirus. Mater Today Proc. 2020;33:3896–3901. doi: 10.1016/j.matpr.2020.06.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harapan H, et al. Coronavirus disease 2019 (COVID-19): a literature review. J Infect Public Health. 2020;13(5):667–673. doi: 10.1016/j.jiph.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tahamtan A, Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev Mol Diagn. 2020;20(5):453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alghamdi HS, Amoudi G, Elhag S, Saeedi K, Nasser J. Deep learning approaches for detecting COVID-19 from chest X-ray images: a survey. IEEE Access. 2021;9:20235–20254. doi: 10.1109/ACCESS.2021.3054484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chauhan N, Soni S, Gupta A, Jain U. New and developing diagnostic platforms for COVID-19: a systematic review. Expert Rev Mol Diagn. 2020;20(9):971–983. doi: 10.1080/14737159.2020.1816466. [DOI] [PubMed] [Google Scholar]
- 18.Wu D, et al. Severity and consolidation quantification of COVID-19 from CT images using deep learning based on hybrid weak labels. IEEE J Biomed Health Inform. 2020;24(12):3529–3538. doi: 10.1109/JBHI.2020.3030224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ibrahim U, Ozsoz M, Serte S, Al-Turjman F, Yakoi PS. Pneumonia classification using deep learning from chest X-ray images during COVID-19. Cogn Comput. 2021 doi: 10.1007/s12559-020-09787-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El Asnaoui K, Chawki Y. Using X-ray images and deep learning for automated detection of coronavirus disease. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1767212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmed I, Ahmad M, Jeon G. Social distance monitoring framework using deep learning architecture to control infection transmission of COVID-19 pandemic. Sustain Cities Soc. 2021;69:102777. doi: 10.1016/j.scs.2021.102777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medhi K, Jamil M, Hussain I (2020) Automatic detection of COVID-19 infection from chest X-ray using deep learning Health Inform preprint 10.1101/2020.05.10.20097063
- 23.Perumal V, Narayanan V, Rajasekar SJS. Detection of COVID-19 using CXR and CT images using transfer learning and Haralick features. Appl Intell. 2021;51(1):341–358. doi: 10.1007/s10489-020-01831-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong D, et al. The role of imaging in the detection and management of COVID-19: a review. IEEE Rev Biomed Eng. 2021;14:16–29. doi: 10.1109/RBME.2020.2990959. [DOI] [PubMed] [Google Scholar]
- 25.Loey M, Manogaran G, Khalifa NEM. A deep transfer learning model with classical data augmentation and CGAN to detect COVID-19 from chest CT radiography digital images. Neural Comput Appl. 2020 doi: 10.1007/s00521-020-05437-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.COVID-19-Use of chest imaging in COVID-19 (2020). A Rapid Advice Guide (11 June 2020). Accessed: Apr. 10, 2022. [Online]. https://apps.who.int/iris/bitstream/handle/10665/332336/WHO-2019-nCoV-Clinical-Radiology_imaging-2020.1-eng.pdf
- 27.Fontanellaz M, et al. A deep-learning diagnostic support system for the detection of COVID-19 using chest radiographs: a multireader validation study. Invest Radiol. 2021;56(6):348–356. doi: 10.1097/RLI.0000000000000748. [DOI] [PubMed] [Google Scholar]
- 28.Basu S, Mitra S, Saha N (2020) Deep learning for screening COVID-19 using chest X-Ray images, In: 2020 IEEE Symposium Series on Computational Intelligence (SSCI), Canberra, ACT, Australia, 2020, pp 2521–2527 10.1109/SSCI47803.2020.9308571
- 29.Li L, et al. Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: evaluation of the diagnostic accuracy. Radiology. 2020;296(2):E65–E71. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Islam MdM, Karray F, Alhajj R, Zeng J. A review on deep learning techniques for the diagnosis of novel coronavirus (COVID-19) IEEE Access. 2021;9:30551–30572. doi: 10.1109/ACCESS.2021.3058537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.PRISMA (2022). Transparent Reporting of Systematic Reviews and Meta-Analyses. Accessed: Apr. 10, 2022. [Online]. Available: http://www.prisma-statement.org/
- 32.Fangoh M and Selim S (2020) Using CNN-XGBoost deep networks for COVID-19 detection in chest X-ray images, In 2020 15th International Conference on Computer Engineering and Systems (ICCES), Cairo, Egypt, 2020, pp 1–7 10.1109/ICCES51560.2020.9334600
- 33.Karhan Z and Akal F (2020) Covid-19 classification using deep learning in chest X-Ray images, In: 2020 medical technologies Congress (TIPTEKNO), Antalya, Turkey, 2020, pp 1–4 10.1109/TIPTEKNO50054.2020.9299315
- 34.Nath MK, Kanhe A and Mishra M (2020) A novel deep learning approach for classification of COVID-19 Images, In 2020 IEEE 5th International Conference on Computing Communication and Automation (ICCCA), Greater Noida, India, 2020, pp 752–757 10.1109/ICCCA49541.2020.9250907
- 35.Padma T, Kumari CU (2020) Deep learning based chest X-ray image as a diagnostic tool for COVID-19 In 2020 International Conference on Smart Electronics and Communication (ICOSEC), Trichy, India, 2020, pp 589–592 10.1109/ICOSEC49089.2020.9215257
- 36.Jiang H, Tang S, Liu W, Zhang Y. Deep learning for COVID-19 chest CT (computed tomography) image analysis: a lesson from lung cancer. Comput Struct Biotechnol J. 2021;19:1391–1399. doi: 10.1016/j.csbj.2021.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alakus TB, Turkoglu I. Comparison of deep learning approaches to predict COVID-19 infection. Chaos Solitons Fract. 2020;140:110120. doi: 10.1016/j.chaos.2020.110120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ko H, et al. COVID-19 Pneumonia diagnosis using a simple 2D deep learning framework with a single chest CT image: model development and validation. J Med Internet Res. 2020;22(6):e19569. doi: 10.2196/19569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Al-Waisy AS, et al. COVID-CheXNet: hybrid deep learning framework for identifying COVID-19 virus in chest X-rays images. Soft Comput. 2020 doi: 10.1007/s00500-020-05424-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Javaheri T, et al. CovidCTNet: an open-source deep learning approach to diagnose covid-19 using small cohort of CT images. npj Digit Med. 2021;4(1):29. doi: 10.1038/s41746-021-00399-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elkorany S, Elsharkawy ZF. COVIDetection-Net: a tailored COVID-19 detection from chest radiography images using deep learning. Optik. 2021;231:166405. doi: 10.1016/j.ijleo.2021.166405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.EED Hemdan, MA Shouman, ME Karar COVIDX-Net: a framework of deep learning classifiers to diagnose COVID-19 in X-ray images p 14 arxiv.org/abs/2003.11055
- 43.Song Y et al (2021) Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT 1000 images. IEEE/ACM transactions on computational biology and bioinformatics, 18(6):2775 – 2780. 10.1109/TCBB.2021.3065361 01 Nov-Dec. [DOI] [PMC free article] [PubMed]
- 44.Tang S, et al. EDL-COVID: ensemble deep learning for COVID-19 cases detection from chest X-Ray images. IEEE Trans. Ind. Inf. 2021 doi: 10.1109/TII.2021.3057683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ouchicha C, Ammor O, Meknassi M. CVDNet: A novel deep learning architecture for detection of coronavirus (Covid-19) from chest x-ray images. Chaos Solitons Fract. 2020;140:110245. doi: 10.1016/j.chaos.2020.110245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tabik S, et al. COVIDGR dataset and COVID-SDNet methodology for predicting COVID-19 based on chest X-ray images. IEEE J Biomed Health Inform. 2020;24(12):3595–3605. doi: 10.1109/JBHI.2020.3037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhosale YH and Sridhar Patnaik K (2022) IoT deployable lightweight deep learning application for COVID-19 detection with lung diseases using RaspberryPi, 2022 International Conference on IoT and Blockchain Technology (ICIBT), 10.1109/ICIBT52874.2022.9807725
- 48.Kedia P, Katarya R. CoVNet-19: a deep learning model for the detection and analysis of COVID-19 patients. Appl Soft Comput. 2021;104:107184. doi: 10.1016/j.asoc.2021.107184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turkoglu M. COVID-19 detection system using chest CT images and multiple kernels-extreme learning machine based on deep neural network. IRBM. 2021 doi: 10.1016/j.irbm.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu X, et al. Deep learning-based multi-view fusion model for screening 2019 novel coronavirus pneumonia: a multicentre study. Eur J Radiol. 2020;128:109041. doi: 10.1016/j.ejrad.2020.109041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mertyuz I, Mertyuz T, Tasar B, Yakut O (2020) Covid-19 disease diagnosis from radiology data with deep learning algorithms, In 2020 4th International Symposium on Multidisciplinary Studies and Innovative Technologies (ISMSIT), Istanbul, Turkey, 1–4. 10.1109/ISMSIT50672.2020.9255380
- 52.Sheykhivand S, et al. Developing an efficient deep neural network for automatic detection of COVID-19 using chest X-ray images. Alex Eng J. 2021;60(3):2885–2903. doi: 10.1016/j.aej.2021.01.011. [DOI] [Google Scholar]
- 53.MC Arellano, OE Ramos (2020) Deep Learning Model to Identify COVID-19 Cases from Chest Radiographs, In 2020 IEEE XXVII International Conference on Electronics, Electrical Engineering and Computing (INTERCON), Lima, Peru, 1–4. 10.1109/INTERCON50315.2020.9220237
- 54.Gupta A, Gupta S, Katarya R. InstaCovNet-19: A deep learning classification model for the detection of COVID-19 patients using Chest X-ray. Appl Soft Comput. 2021;99:106859. doi: 10.1016/j.asoc.2020.106859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.M. Mishra, V. Parashar, and R. Shimpi, “Development and evaluation of an AI System for early detection of Covid-19 pneumonia using X-ray (Student Consortium),” in 2020 IEEE Sixth International Conference on Multimedia Big Data (BigMM), New Delhi, India, Sep. 2020, pp. 292–296. doi: 10.1109/BigMM50055.2020.00051.
- 56.Minaee S, Kafieh R, Sonka M, Yazdani S, Jamalipour Soufi G (2020) Deep-COVID: Predicting COVID-19 from chest X-ray images using deep transfer learning, Med Image Anal, 65: 101794, 10.1016/j.media.2020.101794 [DOI] [PMC free article] [PubMed]
- 57.Panwar H, Gupta PK, Siddiqui MK, Morales-Menendez R, Bhardwaj P, Singh V. A deep learning and grad-CAM based color visualization approach for fast detection of COVID-19 cases using chest X-ray and CT-Scan images. Chaos Solitons Fractals. 2020;140:110190. doi: 10.1016/j.chaos.2020.110190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Karthik R, Menaka R, Hariharan M. Learning distinctive filters for COVID-19 detection from chest X-ray using shuffled residual CNN. Appl Soft Comput. 2021;99:106744. doi: 10.1016/j.asoc.2020.106744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mukherjee H, Ghosh S, Dhar A, Obaidullah SM, Santosh KC, Roy K (2021) Shallow convolutional neural network for COVID-19 outbreak screening using chest X-rays, Cogn Comput, 10.1007/s12559-020-09775-9 [DOI] [PMC free article] [PubMed]
- 60.Zhou T, Lu H, Yang Z, Qiu S, Huo B, Dong Y. The ensemble deep learning model for novel COVID-19 on CT images. Appl Soft Comput. 2021;98:106885. doi: 10.1016/j.asoc.2020.106885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bhosale YH, Zanwar S, Ahmed Z, Nakrani M, Bhuyar D, Shinde U (2022) Deep convolutional neural network based Covid-19 classification from radiology X-Ray images for IoT enabled devices,” 2022 8th International Conference on Advanced Computing and Communication Systems (ICACCS), pp. 1398–1402, 10.1109/ICACCS54159.2022.9785113
- 62.J. Zhang et al. (2021) Viral Pneumonia Screening on Chest X-ray Images Using Confidence-Aware Anomaly Detection, arXiv:2003.12338 [cs, eess], Dec. 2020, Accessed: Jun. 10 2021. [Online]. Available: http://arxiv.org/abs/2003.12338
- 63.Mangal et al. (2021) “CovidAID: COVID-19 Detection Using Chest X-Ray,” arXiv:2004.09803 [cs, eess], Apr. 2020, Accessed: Jun. 10, 2021. [Online]. Available: http://arxiv.org/abs/2004.09803
- 64.Jain R, Gupta M, Taneja S, Hemanth DJ. Deep learning based detection and analysis of COVID-19 on chest X-ray images. Appl Intell. 2021;51(3):1690–1700. doi: 10.1007/s10489-020-01902-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haghanifar A, Majdabadi MM, Choi Y, Deivalakshmi S, Ko S (2021) COVID-CXNet: Detecting COVID-19 in Frontal Chest X-ray Images using Deep Learning, arXiv:2006.13807 [cs, eess], Jul. 2020, Accessed: Jun. 10, 2021. [Online]. Available: http://arxiv.org/abs/2006.13807 [DOI] [PMC free article] [PubMed]
- 66.Apostolopoulos D, Mpesiana TA. Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020;43(2):635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alazab M, Awajan A, Mesleh A, Abraham A, Jatana V, Alhyari S COVID-19 Prediction and Detection Using Deep Learning, p. 15. [Online]. Available: https://www.researchgate.net/publication/341980921
- 68.Sharma V, Dyreson C (2021) COVID-19 Screening Using Residual Attention Network an Artificial Intelligence Approach, arXiv:2006.16106 [cs, eess], Oct. 2020, Accessed: Jun. 10, 2021. [Online]. Available: http://arxiv.org/abs/2006.16106
- 69.Sarker L, Islam MM, Hannan T, Ahmed Z, COVID-DenseNet: A deep learning architecture to detect COVID-19 from chest radiology images, Math Comput Sci, 10.20944/preprints202005.0151.v1
- 70.Wang L, Lin ZQ, Wong A. COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci Rep. 2020;10(1):19549. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kaggle chest x-ray repository. Accessed on Jun. 14 2021 https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia.
- 72.GITHub: covid-chestxray-dataset. Accessed on Jun. 14 2021https://github.com/ieee8023/covid-chestxray-dataset
- 73.COVID-19 Radiography Database: Chest X-ray. Accessed on 17 Jun 2021. https://www.kaggle.com/tawsifurrahman/covid19-radiography-database
- 74.COVID-19 X rays and CT snapshots of CONVID-19 patients Kaggle dataset. Accessed on 22 June 2020. https://www.kaggle.com/andrewmvd/convid19-x-rays
- 75.COVID-19 chest X-ray Database. Accessed on Jun. 14 2021 https://github.com/agchung
- 76.COVID-CT-Dataset: a CT scan dataset about COVID-19. Accessed on 16 Jun 2021 https://github.com/UCSD-AI4H/COVID-CT
- 77.COVID-19 database SIRM. Accessed on Jun. 14 2021 https://www.sirm.org/en/category/articles/covid-19-database/
- 78.Daniel Kermany et al. (2021) Labeled Optical Coherence Tomography (OCT) and Chest X-Ray Images for Classification. Accessed on Jun. 15 2021 https://data.mendeley.com/datasets/rscbjbr9sj/2
- 79.Daniel Kermany et al.: Large dataset of labeled optical coherence tomography (OCT) and Chest X-Ray Images. Accessed on Jun. 24 2021 https://data.mendeley.com/datasets/rscbjbr9sj/3
- 80.Covid-19 Database. Accessed on Jun. 14 2021 https://radiopaedia.org/
- 81.IEEEDataport: CCAP-CT data sets from multi-centre hospitals included five categories. Accessed on 21 June 2020. https://ieee-dataport.org/documents/ccap
- 82.NIH chest X-rays | Kaggle. Accessed on Jun. 14 2021 https://www.kaggle.com/nih-chest-xrays/data?select=Data_Entry_2017.csv
- 83.Eurorad imaging databse. Accessed on Jun. 28 2021 https://www.eurorad.org/advanced-search?search=COVID
- 84.Kiser, et al. Data from the thoracic volume and pleural effusion segmentations in diseased lungs for benchmarking chest CT processing pipelines. Cancer Imag Archive. 2020 doi: 10.7937/tcia.2020.6c7y-gq39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Armato SG III et al. (2021) SPIE-AAPM-NCI lung nodule classification challenge dataset. Cancer Imaging Archive. Accessed on Jun. 29 2021. https://wiki.cancerimagingarchive.net/display/Public/LUNGx+SPIE-AAPM-NCI+Lung+Nodule+Classification+Challenge
- 86.COVID19-CT-Dataset (2021) An open-access chest CT image repository of 1000+ Patients with Confirmed COVID-19 Diagnosis. Accessed on Jul. 06 2021 https://dataverse.harvard.edu/dataset.xhtml?persistentId=doi:10.7910/DVN/6ACUZJ [DOI] [PMC free article] [PubMed]
- 87.Twitter: Chest Imaging database. Accessed on Jun. 28 2021 https://twitter.com/ChestImaging/
- 88.Instagram: Chest Imaging databse. Accessed on Jun. 28 2021 https://www.instagram.com/theradiologistpage/, https://www.instagram.com/radiology_case_reports/
- 89.MIMIC-CXR Database v2.0.0. Accessed on Jun. 29 2021 https://physionet.org/content/mimic-cxr/2.0.0/
- 90.CheXpert Dataset. Accessed on Jun. 30 2021 https://stanfordmlgroup.github.io/competitions/chexpert/
- 91.Javor D, Kaplan H, Kaplan A, Puchner SB, Krestan C, Baltzer P. Deep learning analysis provides accurate COVID-19 diagnosis on chest computed tomography. Eur J Radiol. 2020;133:109402. doi: 10.1016/j.ejrad.2020.109402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang S, et al. Deep learning for detecting corona virus disease 2019 (COVID-19) on high-resolution computed tomography: a pilot study. Ann Transl Med. 2020;8(7):450–450. doi: 10.21037/atm.2020.03.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hussain E, Hasan M, Rahman MA, Lee I, Tamanna T, Parvez MZ. CoroDet: A deep learning based classification for COVID-19 detection using chest X-ray images. Chaos Solitons Fractals. 2021;142:110495. doi: 10.1016/j.chaos.2020.110495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Saha P, Sadi MS, Islam MdM. EMCNet: Automated COVID-19 diagnosis from X-ray images using convolutional neural network and ensemble of machine learning classifiers. Inf Med Unlocked. 2021;22:100505. doi: 10.1016/j.imu.2020.100505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang N, Liu H, Xu C (2020) Deep learning for the detection of COVID-19 using transfer learning and model integration, In 2020 IEEE 10th International Conference on Electronics Information and Emergency Communication (ICEIEC), Beijing, China, 281–284. 10.1109/ICEIEC49280.2020.9152329.
- 96.A Voulodimos, Protopapadakis E, Katsamenis I, Doulamis A, Doulamis N (2020) Deep learning models for COVID-19 infected area segmentation in CT images, Health Inf, 10.1101/2020.05.08.20094664 [DOI] [PMC free article] [PubMed]
- 97.Kassani SH, Kassasni PH, Wesolowski MJ, Schneider KA, Deters R (2020) Automatic detection of coronavirus disease (COVID-19) in X-ray and CT images: a machine learning-based approach, arXiv:2004.10641 [cs, eess], Apr. 2020, Accessed: Jun 10, 2021 [Online]. Available: http://arxiv.org/abs/2004.10641 [DOI] [PMC free article] [PubMed]
- 98.Ismael M, Şengür A. Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Syst Appl. 2021;164:114054. doi: 10.1016/j.eswa.2020.114054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Demir F. DeepCoroNet: a deep LSTM approach for automated detection of COVID-19 cases from chest X-ray images. Appl Soft Comput. 2021;103:107160. doi: 10.1016/j.asoc.2021.107160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Serte S, Serener A (2020) Discerning COVID-19 from mycoplasma and viral pneumonia on CT images via deep learning, In: 2020 4th International Symposium on Multidisciplinary Studies and Innovative Technologies (ISMSIT), Istanbul, Turkey, Oct 2020, pp 1–5 10.1109/ISMSIT50672.2020.9254970
- 101.Horry MJ, et al. COVID-19 detection through transfer learning using multimodal imaging data. IEEE Access. 2020;8:149808–149824. doi: 10.1109/ACCESS.2020.3016780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Khan A, Shafiq S, Kumar R, Kumar J, Haq AU (2020) H3DNN: 3D deep learning based detection of COVID-19 virus using lungs computed tomography,” In: 2020 17th International Computer Conference on Wavelet Active Media Technology and Information Processing (ICCWAMTIP), Chengdu, China, 2020, pp 183–186 10.1109/ICCWAMTIP51612.2020.9317357
- 103.Ucar F, Korkmaz D. COVIDiagnosis-net: deep bayes-squeezenet based diagnosis of the coronavirus disease 2019 (COVID-19) from X-ray images. Med Hypotheses. 2020;140:109761. doi: 10.1016/j.mehy.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sethi R, Mehrotra M, Sethi D (2020) Deep learning based diagnosis recommendation for COVID-19 using chest X-rays images, In: 2020 Second International Conference on Inventive Research in Computing Applications (ICIRCA), Coimbatore, India, Jul 2020, pp 1–4 10.1109/ICIRCA48905.2020.9183278.
- 105.GITHub: SARS-COV-2 Ct-scan dataset (2021). Accessed on Jun. 15 2021 https://www.kaggle.com/plameneduardo/sarscov2-ctscan-dataset
- 106.Silva P, et al. COVID-19 detection in CT images with deep learning: a voting-based scheme and cross-datasets analysis. Inform Med Unlocked. 2020;20:100427. doi: 10.1016/j.imu.2020.100427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Anwar T and Zakir S (2020) Deep learning based diagnosis of COVID-19 using chest CT-scan images, In: 2020 IEEE 23rd International Multitopic Conference (INMIC), Bahawalpur, Pakistan, Nov 2020, pp 1–5 10.1109/INMIC50486.2020.9318212
- 108.Shah V, Keniya R, Shridharani A, Punjabi M, Shah J, Mehendale N. Diagnosis of COVID-19 using CT scan images and deep learning techniques. Emerg Radiol. 2021;28(3):497–505. doi: 10.1007/s10140-020-01886-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Figure1 covid-19 clinical cases. Accessed on 17 Jun 2021 https://www.figure1.com/covid-19-clinical-cases
- 110.Radiological Society of North America (2021). RSNA Pneumonia Detection Challenge. Accessed on 17 Jun 2021 https://www.kaggle.com/c/rsna-pneumonia-detection-challenge
- 111.Serener A and Serte S (2020) Deep learning for mycoplasma pneumonia discrimination from pneumonias like COVID-19, In: 2020 4th International Symposium on Multidisciplinary Studies and Innovative Technologies (ISMSIT), Istanbul, Turkey, Oct. 2020, pp 1–5 10.1109/ISMSIT50672.2020.9254561
- 112.Dutta P, Roy T, and Anjum N (2021) COVID-19 detection using transfer learning with convolutional neural network, In: 2021 2nd International Conference on Robotics, Electrical and Signal Processing Techniques (ICREST), DHAKA, Bangladesh, Jan. 2021, pp 429–432: 10.1109/ICREST51555.2021.9331029
- 113.kaggle COVID-19 chest XRay: COVID-19 image data collection (Bachrr). Accessed on 22 June 2020.https://www.kaggle.com/bachrr/covid-chest-xray
- 114.Github arthursdays HKBU-HPML-COVID-19 CT dataset. Accessed on 22 June 2020. https://github.com/arthursdays/HKBU_HPML_COVID-19
- 115.Wu X, Chen C, Zhong M, Wang J, Shi J. COVID-AL: the diagnosis of COVID-19 with deep active learning. Med Image Anal. 2021;68:101913. doi: 10.1016/j.media.2020.101913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Khoong WH (2020) COVID-19 x-ray dataset (train & test sets) with COVID-19CNN pneumonia detector. Apr 2020 https://www.kaggle.com/khoongweihao/covid19-xray-dataset-train-test-sets
- 117.Sajid N. COVID-19 Patients lungs x ray images 10000. https://www.kaggle.com/nabeelsajid917/covid-19-x-ray-10000-images
- 118.Born et al (2021). POCOVID-Net data set. Accessed on Jun. 26 2021. https://github.com/jannisborn/covid19_ultrasound/tree/master/data
- 119.Covid-19 image repository (2021). Accessed on Jun. 28 2021 https://github.com/ml-workgroup/covid-19-image-repository
- 120.COVIDx Dataset (2021). Accessed on Jun. 29 2021 https://github.com/lindawangg/COVID-Net
- 121.Nisar Z.: X-Ray, CT Dataset. Accessed on Jun. 29 2021 https://github.com/zeeshannisar/COVID-19
- 122.PadChest: a large chest x-ray image dataset with multi-label annotated reports. Accessed on Jun. 29 2021 https://bimcv.cipf.es/bimcv-projects/padchest/ [DOI] [PubMed]
- 123.MosMedData: results of computed tomography of the chest with signs of COVID-19. Accessed on Jun. 30 2021 https://mosmed.ai/datasets/covid19_1110/
- 124.Rohila VS, Gupta N, Kaul A, Sharma DK. Deep learning assisted COVID-19 detection using full CT-scans. IoT. 2021;14:100377. doi: 10.1016/j.iot.2021.100377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Patel P (2021) Chest X-ray(Covid-19 & Pneumonia). Accessed on Jun. 30 2021 https://www.kaggle.com/prashant268/chest-xray-covid19-pneumonia
- 126.SARS-CoV-2 X-Ray and CT image dataset (2021). Accessed on Jun. 30 2021 https://radiopaedia.org/search?utf8=%E2%9C%93&q=covid&scope=all&lang=us
- 127.Kumar A, Tripathi AR, Satapathy SC, Zhang YD (2022) SARS-Net: COVID-19 detection from chest X-rays by combining graph convolutional network and convolutional neural network, Pattern Recognition, 122: 108255, ISSN 0031-3203, 10.1016/j.patcog.2021.108255 [DOI] [PMC free article] [PubMed]
- 128.Chopra A, Gel E, Subramanian J, Krishnamurthy B, Romero-Brufau S, Pasupathy KS, Kingsley TC, Raskar R (2021) DeepABM: scalable, efficient and differentiable agent-based simulations via graph neural networks. Proceedings of the winter simulation conference. 10.48550/arXiv.2110.04421 9 Oct 2021
- 129.Sridhar S, Sanagavarapu S (2021) Multi-lane capsule network architecture for detection of COVID-19, 2021 2nd International Conference on Intelligent Engineering and Management (ICIEM), 2021, pp 385–390 10.1109/ICIEM51511.2021.9445363
- 130.Tiwari S, Jain, A (2021) Convolutional capsule network for COVID-19 detection using radiography images. Int J Imaging Syst Technol 31: 525– 539. 10.1002/ima.22566 [DOI] [PMC free article] [PubMed]
- 131.Tiwari S, Jain A. A lightweight capsule network architecture for detection of COVID-19 from lung CT scans. Int J Imaging Syst Technol. 2022 doi: 10.1002/ima.22706,32,2,(419-434). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Modi S, Guhathakurta R, Praveen S, Tyagi S, Bansod SN (2021) Detail-oriented capsule network for classification of CT scan images performing the detection of COVID-19, Materials Today: Proceedings, ISSN 2214–7853, 10.1016/j.matpr.2021.07.367 [DOI] [PMC free article] [PubMed]
- 133.The cancer imaging archive (TCIA) (2021). Accessed on Jun. 14 2021 https://www.cancerimagingarchive.net/
- 134.Vaid S, Kalantar R, Bhandari M. Deep learning COVID-19 detection bias: accuracy through artificial intelligence. Int Orthop (SICOT) 2020;44:1539–1542. doi: 10.1007/s00264-020-04609-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gilanie G, et al. Coronavirus (COVID-19) detection from chest radiology images using convolutional neural networks. Biomed Signal Process Control. 2021;66:102490. doi: 10.1016/j.bspc.2021.102490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bhosale YH (2020) “Digitization of households with population using cluster and list sampling frame in aerial images”, ISSN (Online) 2456–3293 www.oaijse.com, 5(2): 22–26
- 137.Abdel-Basset M, Chang V, Hawash H, Chakrabortty RK, Ryan M. FSS-2019-nCov: a deep learning architecture for semi-supervised few-shot segmentation of COVID-19 infection. Knowl Based Syst. 2021;212:106647. doi: 10.1016/j.knosys.2020.106647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Desai SB, Pareek A, Lungren MP. Deep learning and its role in COVID-19 medical imaging. Intel Based Med. 2020;3–4:100013. doi: 10.1016/j.ibmed.2020.100013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Panahi H, Rafiei A, Rezaee A. FCOD: fast COVID-19 detector based on deep learning techniques. Inform Med Unlocked. 2021;22:100506. doi: 10.1016/j.imu.2020.100506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bhattacharya S, et al. Deep learning and medical image processing for coronavirus (COVID-19) pandemic: a survey. Sustain Cities Soc. 2021;65:102589. doi: 10.1016/j.scs.2020.102589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Majeed T, Rashid R, Ali D and Asaad A (2020) Covid-19 detection using CNN transfer learning from X-ray Images, 10, 2020, 10.1101/2020.05.12.20098954
- 142.Gozes O, Frid-Adar M, Sagie N, Zhang H, Ji W and Greenspan H (2021) Coronavirus detection and analysis on chest CT with deep learning, arXiv:2004.02640 [cs, eess], Apr. 2020, Accessed: Jun. 10, 2021 [Online] Available: http://arxiv.org/abs/2004.02640
- 143.Abdani SR, Zulkifley MA, Hani Zulkifley N (2020) A lightweight deep learning model for COVID-19 detection, In: 2020 IEEE Symposium on Industrial Electronics & Applications (ISIEA), TBD, Malaysia, Jul. 2020, pp 1–5 10.1109/ISIEA49364.2020.9188133
- 144.Abbas A, Abdelsamea MM, Gaber MM (2021) Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network Appl Intell 51(2): 854–864 10.1007/s10489-020-01829-7 [DOI] [PMC free article] [PubMed]
- 145.Ghoshal B and Tucker A (2020) Estimating uncertainty and interpretability in deep learning for coronavirus (COVID-19) detection arXiv:2003.10769v2 [eess.IV]
- 146.Alazab M, Awajan A, Mesleh A, Abraham A, Jatana V, Alhyari S (2020) COVID-19 Prediction and detection using deep learning ISSN 2150–7988 12: 168–181
- 147.Soldati et al. (2020) Simple, safe, same: lung ultrasound for COVID-19 (LUSCOVID19), ClinicalTrials.gov Identifier: NCT04322487
- 148.Born J, et al. Accelerating detection of lung pathologies with explainable ultrasound image analysis. Appl Sci. 2021;11(2):672. doi: 10.3390/app11020672. [DOI] [Google Scholar]
- 149.Buda N, Segura-Grau E, Cylwik J, Wełnicki M. Lung ultrasound in the diagnosis of COVID-19 infection - a case series and review of the literature. Adv Med Sci. 2020;65(2):378–385. doi: 10.1016/j.advms.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Arntfield R et al. (2020) Development of a deep learning classifier to accurately distinguish COVID-19 from look-a-like pathology on lung ultrasound, Respiratory Medicine, preprint, 10.1101/2020.10.13.20212258
- 151.Roy S, et al. Deep learning for classification and localization of COVID-19 markers in point-of-care lung ultrasound. IEEE Trans Med Imag. 2020;39(8):2676–2687. doi: 10.1109/TMI.2020.2994459. [DOI] [PubMed] [Google Scholar]
- 152.Zhong, et al. Deep metric learning-based image retrieval system for chest radiograph and its clinical applications in COVID-19. Med Image Anal. 2021;70:101993. doi: 10.1016/j.media.2021.101993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Khan MA, et al. Prediction of COVID-19 - Pneumonia based on selected deep features and one class kernel extreme learning machine. Comput Electr Eng. 2021;90:106960. doi: 10.1016/j.compeleceng.2020.106960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hu S, et al. Weakly supervised deep learning for COVID-19 infection detection and classification from CT images. IEEE Access. 2020;8:118869–118883. doi: 10.1109/ACCESS.2020.3005510. [DOI] [Google Scholar]
- 155.Ghoshal B and Tucker A (2020) Estimating uncertainty and interpretability in deep learning for coronavirus (COVID-19) detection arXiv:2003.10769 [cs, eess, stat], Accessed: Jun. 10, 2021 [Online] Available: http://arxiv.org/abs/2003.10769
- 156.Waheed M, Goyal D, Gupta A, Khanna A, Al-Turjman F, Pinheiro PR. CovidGAN: data augmentation using auxiliary classifier GAN for improved covid-19 detection. IEEE Access. 2020;8:91916–91923. doi: 10.1109/ACCESS.2020.2994762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hwang J, Kim H, Yoon SH, Goo JM, Park CM. Implementation of a deep learning-based computer-aided detection system for the interpretation of chest radiographs in patients suspected for COVID-19. Korean J Radiol. 2020;21(10):1150. doi: 10.3348/kjr.2020.0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Horry MJ et al. (2020) X-ray image based COVID-19 detection using pre-trained deep learning models engrXiv, preprint 10.31224/osf.io/wx89s.
- 159.El-Rashidy N, El-Sappagh S, Islam SMR, El-Bakry HM, Abdelrazek S. End-To-End deep learning framework for coronavirus (COVID-19) detection and monitoring. Electronics. 2020;9(9):1439. doi: 10.3390/electronics9091439. [DOI] [Google Scholar]
- 160.Channa A, Popescu N and ur R Malik N (2020) Robust technique to detect COVID-19 using chest X-ray images,” In: 2020 International Conference on e-Health and Bioengineering (EHB), Iasi, Romania, Oct 2020, pp 1–6 10.1109/EHB50910.2020.9280216
- 161.Bassi PRAS, Attux R. A deep convolutional neural network for COVID-19 detection using chest X-rays. Res Biomed Eng. 2021 doi: 10.1007/s42600-021-00132-9. [DOI] [Google Scholar]
- 162.Shorten C, Khoshgoftaar TM, Furht B. Deep learning applications for COVID-19. J Big Data. 2021;8:18. doi: 10.1186/s40537-020-00392-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Rajendra Acharya U. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Computers Biol Med. 2020;121:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Soni S, Pudake, RN, Jain, U, Chauhan, N. (2021) A systematic review on SARS-CoV-2- associated fungal coinfections. J Med Virol 94: 99–109. 10.1002/jmv.27358 [DOI] [PMC free article] [PubMed]
- 165.Abdar M et al. (2021) Review of uncertainty quantification in deep learning: techniques, applications and challenges. arXiv:2011.06225v4
- 166.Juba B and HS Le (2018) Precision-recall versus accuracy and the role of large data sets, Association for the Advancement of Artificial Intelligence
- 167.Shoeibi et al. (2021) Automated detection and forecasting of COVID-19 using deep learning techniques: a review,” arXiv:2007.10785 [cs, eess], Jul. 2020, Accessed: Jun. 10, 2021. [Online] Available: http://arxiv.org/abs/2007.10785