Abstract
As a family of short noncoding RNAs, MicroRNAs have been identified as possible biomarkers for cancer discovery and assist in therapy control due to their epigenetic involvement in gene expression and other cellular biological processes. In the present review, the evidence for reaching the clinical effect and the molecular mechanism of miR-942 in various kinds of cancer is amassed. Dysregulation of miR-942 amounts in different kinds of malignancies, as bladder cancer, esophageal squamous cell carcinoma, breast cancer, cervical cancer, gastric cancer, colorectal cancer, Kaposi's sarcoma, melanoma, Hepatocellular carcinoma, nonsmall-cell lung cancer, oral squamous cell carcinoma, osteosarcoma, ovarian cancer, pancreatic ductal adenocarcinoma, renal cell carcinoma, and prostate cancer has stated a considerable increase or decrease in its level indicating its function as oncogene or tumor suppressor. MiR-942 is included in cell proliferation, migration, and invasion through cell cycle pathways, including pathways of transforming growth factor-beta signaling pathways, Wnt pathway, JAK/STAT pathway, PI3K/AKT pathway, apoptosis pathway, hippo signaling pathway, lectin pathway, interferon-gamma signaling, signaling by G-protein coupled receptor, developmental genes, nuclear factor-kappa B pathway, Mesodermal commitment pathway, and T-cell receptor signaling in cancer. An important biomarker, MiR-942 is a potential candidate for prediction in several cancers. The present investigation introduced miR-942 as a prognostic marker for early discovery of tumor progression, metastasis, and development.
Keywords: Biomarker, cancer, microRNA, microRNA-942, miR-942
Introduction
About 2% of humans' genome is comprised twenty-thousand protein-coding genes. Not <70% of the sequences are recommended for transcription into the RNAs that are often noncoding RNAs (ncRNAs).[1,2,3,4,5] According to the modern advancements in sequencing approaches and large-scale genome sequencing, the long (>200 nucleotides) and short ncRNAs (<20 nucleotides) are proposed as the essential regulators in the human's genome.[6,7,8] MicroRNAs are noncoding, endogenous, single-stranded, and small molecules having a regulatory effect on the mammalian genome and the human genome that are capable of encoding approximately a thousand types of them. MicroRNAs are about 22–24 nucleotides long and are located in eukaryotes.[9] The main duty of MicroRNAs is regulating posttranscription that is conducted by the interaction with mRNA and silencing the target gene. MicroRNAs are mainly the result of the intron sections of other genes, and such genes are mostly transcribed via RNA polymerase II.[10] It should be noted that further studies are to be conducted to emphasize the discovery of new biological events associated with carcinogenesis and innovative therapeutic targets including microRNAs. The expression of microRNAs differs in different tissues in different types of cancer. It could remain constant, be increased, or decreased.[11]
MiR-942 of the human body consists of two homologous miRNAs on human chromosome 1, hsa-miR-942-5p and hsa-miR-942-3p. MiR-942-5p is the product of the 5' arm of miR-942 hairpin and is more suitable to be a potential diagnosis and prognosis biomarker in Cancer but not therapeutic targets. However, miR-942-3p is the product of the 3' arm of miR-942 and it might act as an oncogenic factor.[12,13] The secondary structure predictions of miR-942 with a minimum free-energy-56.70 kcal/mol and dot-bracket notation have been plotted through the Rfold webserver (provided from Vienna package) available at http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold. cgi, as can be seen in Figure 1. A conservative evolution was observed in MiR-942 of humans, mice, flies, and other varieties of which comparable sequence and structure exist. The regulation of MiR-942 is significant in the biological process for the physiological equilibrium between various systems in the human body. As reported in some studies, MiR-942 appears to improve cell proliferation and invasion while it can induce apoptosis in multiple cancers.[14,15,16,17,18] In the present review, the evidence for reaching the molecular mechanism and clinical significance of the miR-942 is amassed in various kinds of cancers.
Figure 1.

Prediction of Optimal Secondary structure of the has-miR-942 (EPS format) with -56.70 kcal/mol with its dot-bracket notation using the Rfold web server. The sequence of this microRNA: ATTAGGAGAG TATCTTCTCTGTTTTGGCCA TGTGTGT ACTCACAGCC CCTCACACATGGCCGAAACAGAGAAGTTACTTTCCTAAT
miR-942 variations in several tumors
According to several investigations, miR-942 has been dysregulated in diverse human tumors such as bladder cancer,[12] breast cancer,[15,16] cervical cancer,[13,17] colorectal cancer (CRC),[18,19,20,21] esophageal squamous cell carcinoma (ESCC),[22] gastric cancer,[23] hepatocellular carcinoma (HCC),[24,25,26,27] Kaposi's sarcoma,[28] melanoma,[29] nonsmall-cell lung cancer (NSCLC),[30,31,32] oral squamous cell carcinoma (OSCC),[33] osteosarcoma,[34] ovarian cancer (OC),[35,36] pancreatic ductal adenocarcinoma (PDAC),[37] renal cell carcinoma (RCC),[38,39,40] as well as prostate cancer[41,42] [Table 1].
Table 1.
Functional characterization of miR-942 in cancers
| Cancer type | Expression | Related gene | Clinical features | Role | Ref. |
|---|---|---|---|---|---|
| Bladder cancer | ↑ | GAS1, LATS2 | Cell proliferation; migration; invasion; metastasis; inhibition of apoptosis | OG | [12] |
| Breast cancer | ↑ | FOXA2, SOCS3 | Cell viability; proliferation; migration; invasion; inhibition of apoptosis; shorter survival rate | OG | [15,16] |
| Cervical cancer | ↓ | AKT1, GFI1 | Cell proliferation; invasion; EMT | TSG | [13,17] |
| Colorectal cancer | ↑ | APC, DLG2, BATF2, CCBE1 | Cell proliferation; metastasis | OG | [18,19,20,21104] |
| Esophageal squamous cell carcinoma | ↑ | sFRP4, GSK3β, TLE1 | Cell proliferation; differentiation; migration | OG | [22] |
| Gastric cancer | ↑ | NFKBIA | Cell proliferation; invasion | OG | [23] |
| Hepatocellular caarcinoma | ↑ | RRM2B, MBL2, ALX4, ISG12a, PPARγ, BAMBI, GFI1 | Proliferation; inhibition of apoptosis; colony formation; migration, invasion; metastasis | OG | [24,25,26,27] |
| Kaposi’s sarcoma | ↑ | IκBα | Tumor development | OG | [28] |
| Melanoma | ↑ | DKK3 | Cell proliferation; migration; invasion; inhibition of apoptosis; cell viability | OG | [29] |
| Nonsmall-cell lung cancer | ↑ | BARX2, ZNF471 | Cell migration; invasion; angiogenesis; metastasis | OG | [30,31,32] |
| Oral squamous cell carcinoma | ↓ | LTBP2 | Cell proliferation; migration; invasion; high T stage; advanced TNM stage | TSG | [33] |
| Osteosarcoma | ↑ | STAT | Cell proliferation; induce apoptosis | OG | [34] |
| Ovary cancer | ↓ | CUL4B, EPSTI1 | Cell proliferation; apoptosis; migration; invasion | TSG | [35,36] |
| Pancreatic cancer | ↓ | ANK1, GDNF, PAX6 | Cell growth; clonogenic ability; migration, invasion; liver metastasis; apoptosis resistance | TSG | [37] |
| Renal cell carcinoma | ↑ | FOXO3, SALL1, METAP1, DCAF11 | Metastasis | OG | [38,39,40] |
| Prostate cancer | ↓ | - | High grade tumor | TSG | [41,42] |
↓: Downregulate, ↑: Upregulate, TSG: Tumor suppressor gene, OG: Oncogene, EMT: Epithelial-mesenchymal transition
Bladder Cancer
As a significantly popular urological malignancy, bladder cancer has been known as a prevalent universal cancer so that its occurrence is increasing. Although various treatments have been provided, a 5-year survival rate of bladder cancer cases is yet reached.[43] Almost 33% to 75% of the BC patients are not capable of responding to therapy owing to metastasis or disease relapse. Furthermore, biomarkers are considered as the surrogate markers that are capable of enhancing or declining the clinicians' suspect of the additional clinically significant issues, such as cancer episode, recurrence, expansion, or the cases losses could or could not occur and or particular therapies can be capable of declining risks of the events.[44] Wang et al., indicated that by hindering the expression of large tumor suppressor 2, i.e., LASTS2 a TAZ inhibitor, TAZ-induced miR-942-3p expression up-regulation leads to the amplification of upstream signaling. The effects on cell proliferation, angiogenesis, EMT, glycolysis and, ROS levels induced by TAZ knockdown were attenuated by MiR-942-3p. An innovative positive feedback loop were identified between TAZ and miR-942-3p regulating biological functions in bladder cancer cells by GAS1 expression showing TAZ, miR-942-3p, and GAS1 as possible therapeutic options for bladder cancer therapy.[12]
Breast Cancer
According to several studies, breast cancer is accounted for the most prevalent malignancy and a significant reason for universal death of women involving 23% of all new cases and 14% of all cancer deaths in 2008.[45] Although notable progress has been achieved in the early prognosis and targeted therapies, the precise reason for such disease has been under investigation so far. Therefore, the discovery of molecular mechanism (s) of genetic tumors and the development of such diseases are of great importance. In other words, promoting reliable biomarkers for the prediction of drug responses and tumor progression are the top priorities.[46,47] Based on an investigation conducted by Zhang et al., although miR-942 had great expression in breast cancer, the cell viability, proliferation, migration, and invasion of breast cancer were considerably suppressed by its low expression. However, such low expression led to an increase in cell apoptosis. The induction of down-regulation of N-cadherin and Snail and up-regulation of E-cadherin was associated with the low-expression of miR-942. FOXA2, which was verified as the direct target gene for miR-942 and was low-expressed in breast cancer, partially reversed the impact of overexpressed miR-942 on promoting cell viability, migration, proliferation, invasion, proliferation, and suppressed cell apoptosis. Lower expression of FOXA2 and higher expression of miR-942 resulted in a less survival rate in breast cancer cases. The progression of breast cancer was promoted by MiR-942 via downregulating the expression of FOXA2.[15] Li et al. demonstrated that sponging miR-942, hsa_circ_0001785 can regulate the SOCS3 in breast cancer cells.[16]
Cervical Cancer
In 2018, with an expected 570,000 patients and 311,000 mortalities around the world, cervical cancer has been accounted for the fourth-most prevalent cancer and the fourth major cause of death by cancer in females.[48] However, nearly 85% of cervical cancer mortalities happen in developing or underdeveloped countries, and with an 18-fold higher death rate in low-income and middle-income countries in comparison to wealthier countries.[49] Furthermore, the infection caused by the human papillomavirus is known as one of the major reasons for such a disease. Nevertheless, most of such infections have been known to be self-evident.[50,51] Ou et al., demonstrated that by sponging miR-942-5p, circ-AKT1 up-regulates AKT1. CircRNA-AKT1's sequestration of miR-942-5p to up-regulate AKT1 and promote cervical cancer progression can be a new molecular target to improve cervical cancer treatment.[13] Zhang et al., confirmed that lncRNA HCG11 can be capable of binding to miR-942-5p directly. Furthermore, the invasion and growth of cervical cancer cells were suppressed through inhibition of miR-942-5p with the growth factor-independent transcription repressor 1 (GFI1) gene as the target gene of miR-942-5p.[17]
Colorectal Cancer
CRC is accounted for the third most prevalent variety of cancer in humans around the world with more than 1.2 million new subjects per annum[52] Although researchers have achieved significant progress in acquiring awareness of the precise molecular mechanism and advances in CRC therapy in radiation therapy, chemotherapy, surgical resection, diagnosis of CRC, there has yet been poor overall survival and no significant improvement has not yet been obtained.[45,53] Shan et al., found that there is a negative relation between miR-942 and Linc00675 and that miR-942 is highly expressed in clinical CRC tissues. Furthermore, they stated the prevention of Wnt/β-catenin signaling in the Linc00675/miR-942-regulated pathway in CRC cells.[18] The function of the Wnt signaling pathway in carcinogenesis and embryonic development has been identified. It has been considered to have a major impact on the initiation and progression of CRC. Another study demonstrated that miR-942 up-regulated the Wnt signaling by directly targeting APC, a tumor suppressor in the same pathway.[19] Li et al., revealed that miR-942-5p was notably upregulated in human CRC tissues and cell lines. Furthermore, miR-942-5p could directly target DLG2 mRNA leading to the enhancement of CRC cells malignancy phenotypes. They also found that DLG2 overexpression enhanced YAP phosphorylation, a critical downstream effector of DLG2. The tumor-suppressive capacity of YAP in CRC cell lines was demonstrated.[20] Yu et al. stated that miR-942-5p expression was clearly up-regulated in CRC tissues or cells in comparison to the control groups. Furthermore, they demonstrated that circ_0005927 was a sponge of miR-942-5p, and miR-942-5p bound to BATF2.[21] Zhou et al., confirmed that miR-942-5p exerts oncogenic actions in CRC by targeting CCBE1 and identified miR-942-5p as a potential clinical biomarker for CRC diagnosis and therapy.[54]
Esophageal Squamous Cell Carcinoma
Very often, esophageal cancer does not respond to the current therapeutic approaches and therefore, has poor outcomes. The annual rate of new esophageal cancer is around 400,000 cases worldwide, being the eighth most prevalent cancer and the sixth most prevalent cause of cancer-associated deaths. For most of the 20th century, squamous cell cancer (SCC) comprised the vast majority of esophageal cancers globally.[55] Ge et al., reported that in ESCC, miR-942 is significantly up-regulated and that higher miR-942 levels are correlated with weak diagnosis in ESCC cases. MiR-942 directly targeting sFRP4, GSK3 β and, TLE1, multiple level negative regulators of Wnt/β-catenin signaling cascade, miR-942 upregulate the Wnt/β-catenin signaling activity. Furthermore, their findings revealed that by directly binding to the miR-942 promoter, c-myc promotes its expression. Taken together, the oncogenic effect of miR-942 in ESCC established in this study suggests miR-942 as an efficient therapeutic option for ESCC.[22]
Gastric Cancer
Based on several studies, gastric cancer accounts for the second principal cause of cancer-associated deaths. Despite the efficacy of the initial treatment of Gastric cancer, the prolonged survival rate of such cancer is yet limited since there have not been appropriate biomarkers for initial gastric cancer (GC) determination. Although CA125 and carcinoembryonic antigen (CEA) have been recognized as markers in clinical studies, they lack enough sensitivity and specificity even in the case of integrated use. Therefore, there is an urgent need for novel molecular diagnoses for the initial discovery of GC.[56] Lu et al., stated circ-CEP85 L can be capable of binding to miR-942-5p directly. Furthermore, rescue trials demonstrated that the invasion and proliferation of gastric cancer cells may be inhibited by circ-CEP85 L through sponging miR-942-5p. Conclusively, they verified the effective reversal of NFKBIA-induced circ-CEP85 L overexpression inhibition by down-regulation of miR-942-5p.[23]
Hepatocellular Carcinoma
HCC, known as the 3rd major reason for world cancer-associated mortality leading to nearly 600,000 deaths per annum. Furthermore, HCC has been associated with the infections caused by hepatitis B and C virus. Moreover, weaker diagnosis and recurrence rates have been largely associated with intracerebral and bone metastasis in the initial HCC.[57] Despite successful progress in the HCC therapeutic approaches, including chemotherapy, ablation, and hepatectomy, due to the unlimited proliferation and distant metastasis of HCC, patients' outcomes are still unfavorable.[58] The association between the initiation and progression of HCC with multiple genetic mutations, activation of oncogenes, or inactivation of antioncogenes is well understood.[59] Therefore, identifying promising new therapeutic options can facilitate the early diagnosis and therapy of HCC. Zhang et al., showed the significant upregulation in HCC. The high expression of miR-942 was significantly correlated with lymphatic metastasis, serum alanine transaminase level, tumor size, and T stage. Furthermore, the high expression of miR-942 was related to shorter disease-free survival time and overall survival in HCC patients. RRM2B was proven to be a target gene of miR-942. The malignant phenotypes of Huh7 and MHCC97H cell lines were markedly promoted by miR-942 mimickers, while the opposite effect was observed with its inhibitor. Egr-1 and PTEN, markers of epithelial-mesenchymal transition and matrix metalloproteinases are among downstream genes of RRM2B that can be regulated by miR-942. Finally, these researchers proposed miR-942 as a potential biomarker for HCC, making its inhibitor a possible therapeutic option for treating such a fatal disease.[24] Xu et al., found that compared to normal tissues, the level is elevated in HCC cell lines and tissues. They also showed that miR-942-3p expression is related to the pathological step and tumor node metastasis step, making it an independent predictor of weak survival in HCC cases. Furthermore, mannose-binding lectin 2 (MBL2) has been known as a direct target of miR-942-3p, having an inverse correlation with miR-942-3p expression and improper survival in HCC cases. Naturally, MBL2 restoration inhibited t HCC progression and attenuated the miR-942-3p-induced tumor-promoting effects. In conclusion, by targeting MBL2 and acting as an oncogenic factor in HCC, miR-942-3p can be a possible marker for HCC cases.[25] Xu et al. demonstrated that an increased miR-942-5p was evident in HCC cells and tissues. Moreover, the cell growth of HCC was hindered by the up-regulation of miR-942-5p via targeting ALX4.[26] Lu et al., reported the promoted progression of HCC with miR-942-5p overexpression, which was reversed by the upregulation of GFI1.[27] In liver cancer cells and tissues, there is a negative relation between miR-942 and the expression of interferon-stimulated gene 12a (ISG12a). Forced expression of miR-942 in TRAIL-sensitive cells turns the TRAIL-sensitive phenotype into a resistant one by significantly reducing endogenous ISG12a levels. Downregulation of ISG12a by miR-942 is required for maintaining the TRAIL-resistant phenotype of cancer cells in favor of cancer cell survival. Thus, miR-942 could be an innovative potential marker of response to medications with serious implications in designing novel therapeutic agents targeting TRAIL-resistant tumors.[60] Further investigations revealed the effect of miR-942 on the diagnosis and prognosis of liver diseases and infections that may develop to HCC in the future. MiR-942 expression was decreased by HCV infection in HLCZ01 cells and miR-942 was negatively associated with ISG12a expression in both liver biopsies and HCV-infected cells.[61] Tao et al., stated that miR-942 expression was up-regulated inactivated, hepatic stellate cells (HSCs) and correlated inversely with activin membrane-bound inhibitor (BAMBI) expression in liver fibrosis progression. MiR-942 expression is induced in HSCs by two significant drivers of liver fibrosis and inflammation, namely transforming growth factor-beta (TGF-β) and lipopolysaccharide (LPS), via Smad2/3 respective NF-κB/p50 binding to the miR-942 promoter. In conclusion, in human liver fibrosis, HSC activation is mediated by the TGF-β and LPS-induced miR-942 via down-regulation of BAMBI. Their study provided new insights into the molecular mechanisms of HSC activation and their role in liver fibrosis.[62] By targeting the peroxisome proliferator-activated receptor-gamma (PPARγ) 3'UTR, miR-942 negatively regulates PPARγ expression. TGFβ1-induced HSC activation is promoted by PPARγ inhibition, and this effect is blocked after inhibiting the miR-942. Furthermore, chiefly represented in fibrous septa, there is a negative correlation between miR-942 and PPARγ in liver fibrosis. PPARγ targeted by miR-942 decreases HSC activation in human hepatic fibrosis.[63]
Kaposi's Sarcoma
Kaposi sarcoma has been identified as a soft tissue tumor that is prevalent among humans of different races and ages with several types and presentations. In its most popular type, Kaposi sarcoma is experienced in subjects with immunosuppression, including acquired immunodeficiency syndrome (AIDs) or immunosuppression owing to organ transplantation.[64] Kaposi's sarcoma (KS) is caused by its related herpesvirus, known as Kaposi's sarcoma-associated herpesvirus (KSHV), or human herpesvirus 8. AIDS-associated KS (AIDS-KS) is still clinically challenging in patients from the United States and sub-Saharan Africa, such as a subset of AIDS cases on highly active antiretroviral therapy (HAART).[65,66,67,68] There is also a link between KSHV infection and two AIDS-associated B-cell lymphoproliferative diseases, including a subset of multicentric Castleman's disease (MCD) and primary effusion lymphoma (PEL).[69,70] Yan et al., found that miR-942-5p suppression relieves IκBα expression and reduces Vpr inhibition of Kaposi's sarcoma-associated herpesvirus (KSHV) lytic replication, while miR-942-5p overexpression enhances Vpr inhibition of KSHV lytic replication. Collectively, their findings demonstrate that, via activating NF-κB signaling through up-regulation of a cellular miRNA to target IκBα, internalized HIV-1 Vpr inhibits KSHV lytic replication. Based on these findings, the significant impact of Vpr on KSHV's life cycle is evident.[28]
Melanoma
Melanoma is a kind of aggressive skin cancer, which accounted for nearly 73% of skin cancer-related death and is usually resulted from direct exposure to the Sun's ultraviolet radiation.[71,72] The incidence of melanoma is gradually increasing worldwide in the past years.[73] Current therapies of melanoma are surgery, chemotherapy, and target therapy.[74] However, the prognosis of melanoma is still poor with only 10%–15% of the 10-year survival rate in metastatic melanoma.[75] Therefore, a better understanding of melanoma pathogenesis needs to be investigated to improve the prognosis. Zhang et al., studies the upregulation of miR-942-5p in melanoma cells and tissues which was considerably related to a weak diagnosis. The Wnt/β-catenin pathway can be activated via MiR-942-5p by targeting 3'-UTR of DKK3, leading to melanoma cell proliferation, invasion, and migration, which promotes the progression of melanoma. Based on their findings, miR-942-5p can be a biomarker for the prognosis and diagnosis of melanoma.[29]
Non-small-Cell Lung Cancer
NSCLC is regarded as the most prevalent lung cancer and the most fatal malignancy worldwide. Based on several investigations, lung cancer is accounted for the most prevalent reason for global fatality and considered approximately 1.5 million losses in 2012.[76] The disease accounts for yearly death of 353,000 in Europe, showing about 20% of total cancer deaths.[77] The principal kind of lung cancer is known as NSCLC involving almost 80% of lung cancer patients. NSCLC is accounted for a weak prognosis following chemotherapy. Therefore, prognostic biomarkers and molecular targets are of great importance for lung cancer.[78] Yang et al. showed the relatively high expression of miR-942 in human NSCLC tissues and cells. In vitro assays expressed the promotion of cell migration, invasion, and angiogenesis by miR-942 overexpression. By inhibiting BARX2, miR-942 increases EMT-associated proteins, N-cadherin, and vimentin, while reduces the expression of E-cadherin. According to their investigation, directly targeting BARX2, miR-942 induces EMT-related metastasis providing a possible therapeutic approach for NSCLC.[30] Wang et al. showed that LIFR-AS1 is a pivotal miR-942-5p-interacting lncRNA. MiR-942-5p overexpression reduces LIFR-AS1 in NSCLC cells. LIFR-AS1 can sponge miR-942-5p, leading to the repression of ZNF471.[31] Zhou et al., revealed that serum miR-942 expression amounts were notably up-regulated in NSCLC. In addition, serum miR-942 was superior to CEA, CYFRA21-1, and SCCA for the initial prognosis of NSCLC. The integration of serum miR-601 and miR-942 improved the effectiveness of identifying early-stage NSCLC. Taken together, serum miR-942 can be capable of acting as a hopeful molecular marker for the early prognosis of NSCLC and its diagnosis prediction.[32]
Oral Squamous Cell Carcinoma
OSCC represents the most malignant neoplasm in oral cancer with a mortality rate of more than 50%. The OSCC is multistep neoplasia initially developed from mild oral epithelial hyperplasia to dysplasia followed by carcinoma in situ.[79] OSCC is rated among the 6th most frequent oral malignancies, with a yearly prevalence of more than 500,000 cases.[80] OSCC alone is considered responsible for more than 90% of oral cancers cases and has the highest rate of mortality globally.[81,82,83] Wang et al., indicated that by accelerating EMT and phosphorylation of PI3K/Akt/mTOR signaling pathway components, the circEPSTI1/miR-942-5p/LTBP2 axis influences oral squamous, cell carcinoma, cell invasion, and proliferation.[33]
Osteosarcoma
Osteosarcoma is known as the most prevalent bone cancer in infants, adolescents, and young adults[84] and mostly around the remodeling and bone growth regions. Based on new studies, the possible cause of osteosarcoma is the genetic and epigenetic alterations disrupting mesenchymal stem cells for modifying into osteoblasts.[85] Therapy outcomes have recently been updated considerably for osteosarcoma,[86] leading to a notable advancement in osteosarcoma cases with a 5-year survival rate (60%–70%). Nevertheless, outcomes have yet been unknown, and recurrence of the pulmonary metastasis after chemotherapy and surgery is the reason for the mortality in most subjects.[87,88] Sun et al., stated that circ_0001649 acts as a sponge absorbing miR-942 to suppress osteosarcoma cell proliferation, induce apoptosis, and hinder the STAT pathway.[34]
Ovarian Cancer
OC is a malignant tumor of the female reproductive system, with epithelial OC ranking first with respect to mortality among gynecological cancers.[89] The main treatment strategies for OC are platinum-based chemotherapy and surgery.[90] The occurrence of OC increases by gradual aging.[91] The diagnosis of OC is mostly made at advanced stages owing to the shortage of specific early clinical symptoms.[92] Hence, investigating the molecular mechanisms of OC progression can contribute to the discovery of efficient tumor markers for OC diagnosis and therapy monitoring. Du et al. revealed that miR-942-5p level was reduced in OC cells and tissues. Moreover, up-regulation of miR-942-5p impeded OC cell growth via targeting CUL4B.[35] Xie et al. found that by sponging miR-942, circEPSTI1 regulates EPSTI1 expression and OC development.[36]
Pancreatic Ductal Adenocarcinoma
PDAC, the most prevalent neoplastic disorder of the pancreas, accounts for more than 90% of the whole pancreatic malignancies.[93] Currently, the 5-year overall survival of PDAC is lower than 8% making it the fourth most prevalent reason for cancer-related mortalities around the world.[94] Although numerous investigations have recently been conducted on PDAC, not enough effectiveness has been achieved by chemotherapy, radiation, and therapeutic regimens.[95] Therefore, the only therapy remains surgery which only 15–20 percent of PDAC cases could be eligible for effective surgery at the presentation time.[96] Wong et al. identified 169 circRNAs in PDAC cells differentially expressed in comparison to nontumor human pancreatic ductal epithelial cells. Compared with the others, circFOXK2 was identified as significantly upregulated in PDAC cells and 63% of primary tumors (53 of 84). Multiple miRNA binding sites were found in circFOXK2, functioning as a sponge for miR-942 and promoting ANK1, GDNF, and PAX6 expression.[37]
Renal Cell Carcinoma
RCC is accounted for the most prevalent malignant solid tumor in adults. A total of 63,920 new kidney and renal pelvis cancer cases and 13,860 associated deaths were expected to happen in the US in 2014.[97] With surgical resection remaining the best curative therapeutic strategy for RCC, nearly 20%–30% of cases undergo local and/or distant disease recurrence.[98] Furthermore, at the time of the initial prognosis, metastases are found in up to 30%.[99] MiR-942 has been one of the most reliable predictors of effectiveness. In this innovative paracrine mechanism, MMP-9 and VEGF secretion are up-regulated by high levels of miR-942 in MRCC cells for enhancing endothelial migration and sunitinib resistance.[38] Chen et al. stated that lncRNA LINC00461 is capable of acting as a miR-942 ceRNA and affecting the survival of cases of RCC by controlling the expression of SALL1, METAP1, and DCAF11.[39] In another study, Luo et al., in acute kidney injury expressed that miR-942-5p expression was decreased in the LPS-treated HK-2 cells, and miR-942-5p overexpression is capable of inhibiting LPS-induced inflammation and apoptosis of HK-2 cells by targeting FOXO3.[40]
Prostate Cancer
As the most prevalent cancer in men, prostate cancer is observed in one out of every nine men at the age of 65.[100] Despite the efficiency of surgical operation and radiotherapy for most cases, prediction in the individuals experiencing higher degrees of the disease is limited.[101] the affected cells have a high susceptibility to metabolic variations.[102,103] McDonald et al., stated that the expression of miR-942 decreases in high-grade compared to low-grade prostate cancer cases at biopsy.[41] Li et al., demonstrated that miR-942-5p were repressed in PC3 cells but upregulated in DU145 cells.[42]
MiR-942 Regulatory Mechanisms
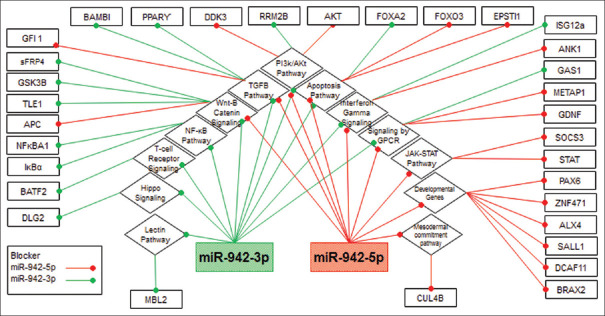
Figure 2 shows the influence of miR-942 on molecular pathways and the related interaction with other genes. MiR-942 is mainly related to carcinogenesis and cancer progression by well-known pathways, such as TGF-β signaling pathways, Wnt pathway, JAK/STAT pathway, PI3K/AKT pathway, Apoptosis pathway, Hippo signaling pathway, Lectin Pathway, Interferon-gamma signaling, Signaling by G-protein coupled receptor, Developmental genes, nuclear factor-kappa B pathway, Mesodermal commitment pathway, and T-cell receptor signaling in cancer, All the mentioned pathways have a relationship with each other. Furthermore, they have a significant influence on the progression, proliferation, or rapid growth, migrating and, invading the cancer cells, as well as cell differentiation in a variety of tumors.
Figure 2.
The overview of miR-942 contribution in different cellular pathways, which mediated mechanisms involved in cancer progression. Totally, miR-942 working as oncogene to regulate genes such as Apoptosis, which by own have interactions with cell cycle control. In the other side, miR-942 have a hand in controlling the pathways of transforming growth factor-beta signaling pathways, Wnt pathway, JAK/STAT pathway, PI3K/AKT pathway, hippo signaling pathway, lectin pathway, interferon gamma signaling, signaling by GPCR, developmental genes, nuclear factor kappa B pathway, Mesodermal commitment pathway, and T-cell receptor signaling, which targeted by miR-942 and also involved in carcinogenesis, and other pathways which all of them function in cell proliferation and oncogenesis. peroxisome proliferator-activated receptor gamma IκBα
As can be seen in Table 2, MiR-942 participates in several pathways. The results of our bioinformatics work using DIANA-miRPath based on Tarbase, microT-CDS, TargetScan and KEGG pathway show that miR-942 is associated with various other pathways including ECM-receptor interaction, thyroid hormone synthesis, thyroid hormone signaling pathway, arrhythmogenic right ventricular cardiomyopathy, TGF-beta signaling pathway, hippo signaling pathway, vascular smooth muscle contraction, and Lysine degradation which targeted by miR-942-3p and-5p and also included in carcinogenesis.
Table 2.
Interactions derived from databases in different pathways for hsa-miR-942-5p and-3p
| KEGG pathway | Related genes | |
|---|---|---|
|
| ||
| miR-942-5p | miR-942-3p | |
| ECM-receptor interaction | COL4A5, COL24A1, ITGB6, COL6A6, DAG1, ITGA10, COL11A2, LAMC1, ITGA7, FN1, CD47 | - |
| Thyroid hormone synthesis | ADCY7, TPO, PAX8, PLCB1, HSP90B1, TG, ITPR2, ADCY4 | ATP1B1, PLCB1, |
| Thyroid hormone signaling pathway | NRAS, RAF1, THRA, PLCZ1, MED1, PLCB1, PLCG1, NCOA2, THRB, PLCE1, PFKFB2, ATP2A2, FOXO1, CREBBP, SIN3A | ACTB, ATP1B1, PLCB1, TSC2, DIO2 |
| ARVC | CACNG8, TCF7L2, ITGB6, PKP2, CTNNA1, SLC8A1, DAG1, ITGA10, ITGA7, CTNNA3, LMNA, ATP2A2 | ACTB, SLC8A1 |
| TGF-β signaling pathway | SMAD2, INHBA, ZFYVE16, SMAD4, ACVR1C, BMPR1A, BAMBI, CREBBP, BMPR2 | CDKN2B, ZFYVE16, SKP1 |
| Hippo signaling pathway | YWHAH, SMAD2, BTRC, PPP2R2C, TCF7L2, CCND2, GLI2, WNT2B, WNT3, AMOT, TP53BP2, SMAD4, CTNNA1, NKD1, RASSF6, BMPR1A, CTNNA3, WNT9B, FBXW11, SERPINE1, BMPR2 | ACTB, YWHAZ, PPP1CB |
| Vascular smooth muscle contraction | ADCY7, GUCY1A3, ROCK2, PLA2G4F, RAF1, KCNMA1, SPECC1L-ADORA2A, PLCB1, PPP1R12A, GUCY1A2, PRKG1, ITPR2, ADCY4, MYLK | GUCY1B3, ACTG2, PLCB1, MYLK3, PPP1CB |
| Lysine degradation | WHSC1L1, SETD7, PLOD2, ASH1L, KMT2E, KMT2C, EHMT1 | - |
ARVC: Arrhythmogenic right ventricular cardiomyopathy, TGF-β: Transforming growth factor-beta
Conclusion
MicroRNAs make significant contributions toward the regulation of genes, and microRNA dysregulation could be regarded as a distinctive feature in cancer. According to several investigations, miR-942 is categorized in the ncRNAs contributing chiefly to the invasion and migration steps, and in the proliferation or rapid development of cancer cells. According to the majority of the studies, unlike miR-942-3p is an oncogene, miR-942-5p acts as one of the tumor suppressors. The mechanism through which miR-942 up-regulation or downregulation involves in the tumor expansion as well as carcinogenesis is complicated. Furthermore, it includes various levels that mostly having a significant effect on cell proliferation and hence tumor growth despite its unknown features. According to the results of prior investigations, variations in expressing miR-942 are firmly associated with the severity of cancer tumor development. Hence, they may be potential candidates for measuring the degree of cancer, prognosis, response to therapy, and even as a possible therapeutic option for solid tumors. Serum miR-942 levels have a direct connection with the progression of the disease and reduced survival rate. Considering the functional mechanisms of miR-942 in cellular pathways, it is promising for the diagnosis of the disease progression, stage, as well as the prognosis and measuring the effectiveness of therapy. Moreover, it shows considerable potential for increasing miR-942 expression as a therapeutic option as effective as anti-tumor medications. According to the obtained results, miR-942 is proposed as a potential candidate for a prognostic marker for early discovery of tumor growth, metastasis, and progression.
Materials and Methods
As previously discussed, two strategies were adopted by the authors. In the first strategy, all the published articles after the year 2000 were surveys in PubMed, Scopus, Embase, Cochrane, and Google Scholar databases using keywords of MIRN942, MiRNA942, Hsa-Mir-942, MicroRNA 942, and Hsa-miR-942. The relation to the biological mechanism (s) and expression of miR-942 in growing tumors in any type of cancer was selected as inclusion criteria.
In addition, the second strategy tried to find the targeted genes by miR-942 and relative pathways involved in carcinogenesis in the base of prediction in relative of databases specific for microRNAs such as Software available in DIANA Tools (http://diana.imis.athenainnovation.gr/) including MicroT–CDS as well as Tarbase and also Target Scan (http://www.targetscan.org/), miRDB (http://www.mirdb.org/), miRwalk (http://mirwalk.umm.uniheidelberg.de/), ENCORI (http://starbase.sysu.edu.cn/) and finally miRPathDP (https://mpd.bioinf.uni-sb.de/). In the first phase of the search, we found 107624 genes, which were predicted to interact with miR-942. The filtering was performed on the basis of calculated scores in each database and inclusion in carcinogenesis. Ultimately, we reached 15399 predicted genes that were repeated at least in two databases and had a correlation with carcinogenicity. Then filtered genes were selected in the base of their repeats in three and four databases or studied in scientific articles [Figure 3]. The pathways chose according to the KEGG database (available at www.genome.jp/kegg/pathway.html).
Figure 3.
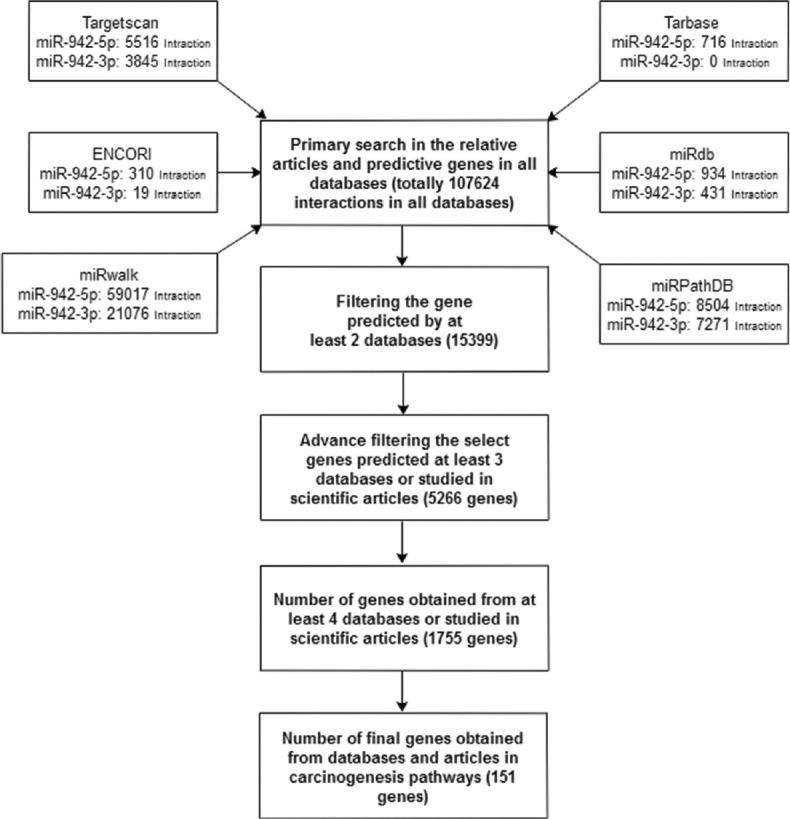
The strategy of search the genes targeted miR-942 in cancers
Ethical approval
This study was completed without any testing on human or animal models by any of the authors.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–9. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 3.Zhu S, Li W, Liu J, Chen CH, Liao Q, Xu P, et al. Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR-Cas9 library. Nat Biotechnol. 2016;34:1279–86. doi: 10.1038/nbt.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kambara H, Niazi F, Kostadinova L, Moonka DK, Siegel CT, Post AB, et al. Negative regulation of the interferon response by an interferon-induced long non-coding RNA. Nucleic Acids Res. 2014;42:10668–80. doi: 10.1093/nar/gku713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Z, Shen J, Chan MT, Wu WK. TUG 1: A pivotal oncogenic long non-coding RNA of human cancers. Cell Prolif. 2016;49:471–5. doi: 10.1111/cpr.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang X, Ma N, Wang D, Li F, He R, Li D, et al. Metformin inhibits tumor growth by regulating multiple miRNAs in human cholangiocarcinoma. Oncotarget. 2015;6:3178–94. doi: 10.18632/oncotarget.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vance KW, Sansom SN, Lee S, Chalei V, Kong L, Cooper SE, et al. The long non-coding RNA Paupar regulates the expression of both local and distal genes. EMBO J. 2014;33:296–311. doi: 10.1002/embj.201386225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang B, Song JH, Cheng Y, Abraham JM, Ibrahim S, Sun Z, et al. Long non-coding antisense RNA KRT7-AS is activated in gastric cancers and supports cancer cell progression by increasing KRT7 expression. Oncogene. 2016;35:4927–36. doi: 10.1038/onc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ambros V. microRNAs: Tiny regulators with great potential. Cell. 2001;107:823–6. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 10.Zhao Y, Srivastava D. A developmental view of microRNA function. Trends Biochem Sci. 2007;32:189–97. doi: 10.1016/j.tibs.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321–33. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang F, Fan M, Zhou X, Yu Y, Cai Y, Wu H, et al. A positive feedback loop between TAZ and miR-942-3p modulates proliferation, angiogenesis, epithelial-mesenchymal transition process, glycometabolism and ROS homeostasis in human bladder cancer. J Exp Clin Cancer Res. 2021;40:44. doi: 10.1186/s13046-021-01846-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ou R, Mo L, Tang H, Leng S, Zhu H, Zhao L, et al. circRNA-AKT1 sequesters miR-942-5p to upregulate AKT1 and promote cervical cancer progression. Mol Ther Nucleic Acids. 2020;20:308–22. doi: 10.1016/j.omtn.2020.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guerra Martins dos Santos Assunção JA. Evolutionary Analysis of Animal microRNAs (Doctoral Dissertation, University of Cambridge) 2013 [Google Scholar]
- 15.Zhang J, Zhang Z, Sun J, Ma Q, Zhao W, Chen X, et al. MiR-942 regulates the function of breast cancer cell by targeting FOXA2. Biosci Rep. 2019;39:BSR20192298. doi: 10.1042/BSR20192298. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Li Z, Zheng J, Lin W, Weng J, Hong W, Zou J, et al. Circular RNA hsa_circ_0001785 inhibits the proliferation, migration and invasion of breast cancer cells in vitro and in vivo by sponging miR-942 to upregulate SOCS3. Cell Cycle. 2020;19:2811–25. doi: 10.1080/15384101.2020.1824717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Zhang J, Mao L, Li X. Long noncoding RNA HCG11 inhibited growth and invasion in cervical cancer by sponging miR-942-5p and targeting GFI1. Cancer Med. 2020;9:7062–71. doi: 10.1002/cam4.3203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Shan Z, An N, Qin J, Yang J, Sun H, Yang W. Long non-coding RNA Linc00675 suppresses cell proliferation and metastasis in colorectal cancer via acting on miR-942 and Wnt/β-catenin signaling. Biomed Pharmacother. 2018;101:769–76. doi: 10.1016/j.biopha.2018.02.123. [DOI] [PubMed] [Google Scholar]
- 19.Fasihi A, Soltani BM, Ranjbaran ZS, Bahonar S, Norouzi R, Nasiri S. Hsa-miR-942 fingerprint in colorectal cancer through Wnt signaling pathway. Gene. 2019;712:143958. doi: 10.1016/j.gene.2019.143958. [DOI] [PubMed] [Google Scholar]
- 20.Li S, Yan G, Liu W, Li C, Wang X. Circ0106714 inhibits tumorigenesis of colorectal cancer by sponging miR-942-5p and releasing DLG2 via Hippo-YAP signaling. Mol Carcinog. 2020;59:1323–42. doi: 10.1002/mc.23259. [DOI] [PubMed] [Google Scholar]
- 21.Yu C, Li D, Yan Q, Wang Y, Yang X, Zhang S, et al. Circ_0005927 inhibits the progression of colorectal cancer by regulating miR-942-5p/BATF2 axis. Cancer Manag Res. 2021;13:2295–306. doi: 10.2147/CMAR.S281377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ge C, Wu S, Wang W, Liu Z, Zhang J, Wang Z, et al. miR-942 promotes cancer stem cell-like traits in esophageal squamous cell carcinoma through activation of Wnt/β-catenin signalling pathway. Oncotarget. 2015;6:10964. doi: 10.18632/oncotarget.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu J, Wang YH, Huang XY, Xie JW, Wang JB, Lin JX, et al. circ-CEP85L suppresses the proliferation and invasion of gastric cancer by regulating NFKBIA expression via miR-942-5p. J Cell Physiol. 2020;235:6287–99. doi: 10.1002/jcp.29556. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Q, Zhu B, Qian J, Wang K, Zhou J. miR-942 promotes proliferation and metastasis of hepatocellular carcinoma cells by inhibiting RRM2B. Onco Targets Ther. 2019;12:8367–78. doi: 10.2147/OTT.S207549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu CY, Dong JF, Chen ZQ, Ding GS, Fu ZR. MiR-942-3p promotes the proliferation and invasion of hepatocellular carcinoma cells by targeting MBL2. Cancer Control. 2019;26:1073274819846593. doi: 10.1177/1073274819846593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Q, Zhou L, Yang G, Meng F, Wan Y, Wang L, et al. Overexpression of circ_0001445 decelerates hepatocellular carcinoma progression by regulating miR-942-5p/ALX4 axis. Biotechnol Lett. 2020;42:2735–47. doi: 10.1007/s10529-020-02985-z. [DOI] [PubMed] [Google Scholar]
- 27.Lu L, Li S, Zhang Y, Luo Z, Chen Y, Ma J, et al. GFI1-mediated upregulation of LINC00675 as a ceRNA restrains hepatocellular carcinoma metastasis by sponging miR-942-5p. Front Oncol. 2020;10:607593. doi: 10.3389/fonc.2020.607593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan Q, Shen C, Qin J, Li W, Hu M, Lu H, et al. HIV-1 Vpr inhibits Kaposi's sarcoma-associated herpesvirus lytic replication by inducing microrna miR-942-5p and activating NF-κB signaling. J Virol. 2016;90:8739–53. doi: 10.1128/JVI.00797-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang W, Mao K, Liu S, Xu Y, Ren J. miR-942-5p promotes the proliferation and invasion of human melanoma cells by targeting DKK3. J Recept Signal Transduct Res. 2021;41:180–7. doi: 10.1080/10799893.2020.1804280. [DOI] [PubMed] [Google Scholar]
- 30.Yang F, Shao C, Wei K, Jing X, Qin Z, Shi Y, et al. miR-942 promotes tumor migration, invasion, and angiogenesis by regulating EMT via BARX2 in non-small-cell lung cancer. J Cell Physiol. 2019;234:23596–607. doi: 10.1002/jcp.28928. [DOI] [PubMed] [Google Scholar]
- 31.Wang Q, Wu J, Huang H, Jiang Y, Huang Y, Fang H, et al. lncRNA LIFR-AS1 suppresses invasion and metastasis of non-small cell lung cancer via the miR-942-5p/ZNF471 axis. Cancer Cell Int. 2020;20:180. doi: 10.1186/s12935-020-01228-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou C, Chen Z, Zhao L, Zhao W, Zhu Y, Liu J, et al. A novel circulating miRNA-based signature for the early diagnosis and prognosis prediction of non-small-cell lung cancer. J Clin Lab Anal. 2020;34:e23505. doi: 10.1002/jcla.23505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Jiang C, Li N, Wang F, Xu Y, Shen Z, et al. The circEPSTI1/mir-942-5p/LTBP2 axis regulates the progression of OSCC in the background of OSF via EMT and the PI3K/Akt/mTOR pathway. Cell Death Dis. 2020;11:682. doi: 10.1038/s41419-020-02851-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun D, Zhu D. Circular RNA hsa_circ_0001649 suppresses the growth of osteosarcoma cells via sponging multiple miRNAs. Cell Cycle. 2020;19:2631–43. doi: 10.1080/15384101.2020.1814026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du Z, Wang L, Xia Y. Circ_0015756 promotes the progression of ovarian cancer by regulating miR-942-5p/CUL4B pathway. Cancer Cell Int. 2020;20:572. doi: 10.1186/s12935-020-01666-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie J, Wang S, Li G, Zhao X, Jiang F, Liu J, et al. circEPSTI1 regulates ovarian cancer progression via decoying miR-942. J Cell Mol Med. 2019;23:3597–602. doi: 10.1111/jcmm.14260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong CH, Lou UK, Li Y, Chan SL, Tong JH, To KF, et al. CircFOXK2 promotes growth and metastasis of pancreatic ductal adenocarcinoma by complexing with RNA-binding proteins and sponging MiR-942. Cancer Res. 2020;80:2138–49. doi: 10.1158/0008-5472.CAN-19-3268. [DOI] [PubMed] [Google Scholar]
- 38.Prior C, Perez-Gracia JL, Garcia-Donas J, Rodriguez-Antona C, Guruceaga E, Esteban E, et al. Identification of tissue microRNAs predictive of sunitinib activity in patients with metastatic renal cell carcinoma. PLoS One. 2014;9:e86263. doi: 10.1371/journal.pone.0086263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Y, He J, Su C, Wang H, Chen Y, Guo W, et al. LINC00461 affects the survival of patients with renal cell carcinoma by acting as a competing endogenous RNA for microRNA942. Oncol Rep. 2019;42:1924–34. doi: 10.3892/or.2019.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo N, Gao HM, Wang YQ, Li HJ, Li Y. MiR-942-5p alleviates septic acute kidney injury by targeting FOXO3. Eur Rev Med Pharmacol Sci. 2020;24:6237–44. doi: 10.26355/eurrev_202006_21521. [DOI] [PubMed] [Google Scholar]
- 41.McDonald AC, Vira M, Walter V, Shen J, Raman JD, Sanda MG, et al. Circulating microRNAs in plasma among men with low-grade and high-grade prostate cancer at prostate biopsy. Prostate. 2019;79:961–8. doi: 10.1002/pros.23803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Z, Zheng J, Xia Q, He X, Bao J, Chen Z, et al. Identification of specific long non-coding ribonucleic acid signatures and regulatory networks in prostate cancer in fine-needle aspiration biopsies. Front Genet. 2020;11:62. doi: 10.3389/fgene.2020.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu J, Wang J. HMGN5 expression in bladder cancer tissue and its role on prognosis. Eur Rev Med Pharmacol Sci. 2018;22:970–5. doi: 10.26355/eurrev_201802_14378. [DOI] [PubMed] [Google Scholar]
- 44.Chamie K, Litwin MS, Bassett JC, Daskivich TJ, Lai J, Hanley JM, et al. Recurrence of high-risk bladder cancer: A population-based analysis. Cancer. 2013;119:3219–27. doi: 10.1002/cncr.28147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 46.Liu W, Schaffer L, Herrs N, Chollet C, Taylor S. Improved sleep after Qigong exercise in breast cancer survivors: A pilot study. Asia Pac J Oncol Nurs. 2015;2:232–9. doi: 10.4103/2347-5625.170537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kam J, Kam J, Mann GB, Phillips C, Wentworth JM, King J, et al. Solitary pituitary metastasis from HER2-positive breast cancer. Asia Pac J Clin Oncol. 2017;13:e181–4. doi: 10.1111/ajco.12353. [DOI] [PubMed] [Google Scholar]
- 48.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 49.Prabhu M, Eckert LO. Development of World Health Organization (WHO) recommendations for appropriate clinical trial endpoints for next-generation Human Papillomavirus (HPV) vaccines. Papillomavirus Res. 2016;2:185–9. doi: 10.1016/j.pvr.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodríguez AC, Schiffman M, Herrero R, Wacholder S, Hildesheim A, Castle PE, et al. Rapid clearance of human papillomavirus and implications for clinical focus on persistent infections. J Natl Cancer Inst. 2008;100:513–7. doi: 10.1093/jnci/djn044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, et al. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci U S A. 2005;102:16426–31. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blum HE. Hepatocellular carcinoma: Therapy and prevention. World J Gastroenterol. 2005;11:7391–400. doi: 10.3748/wjg.v11.i47.7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sostres C, Gargallo CJ, Lanas A. Aspirin, cyclooxygenase inhibition and colorectal cancer. World J Gastrointest Pharmacol Ther. 2014;5:40–9. doi: 10.4292/wjgpt.v5.i1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou L, Chen Q, Wu J, Yang J, Yin H, Tian J, et al. miR-942-5p inhibits proliferation, metastasis, and epithelial-mesenchymal transition in colorectal cancer by targeting CCBE1. Biomed Res Int. 2021;2021:9951405. doi: 10.1155/2021/9951405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parkin DM, Bray F. Evaluation of data quality in the cancer registry: Principles and methods Part II.Completeness. Eur J Cancer. 2009;45:756–64. doi: 10.1016/j.ejca.2008.11.033. [DOI] [PubMed] [Google Scholar]
- 56.Sitarz R, Skierucha M, Mielko J, Offerhaus GJ, Maciejewski R, Polkowski WP. Gastric cancer: Epidemiology, prevention, classification, and treatment. Cancer Manag Res. 2018;10:239–48. doi: 10.2147/CMAR.S149619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Invernizzi F, Viganò M, Grossi G, Lampertico P. The prognosis and management of inactive HBV carriers. Liver Int. 2016;36(1):100–4. doi: 10.1111/liv.13006. [DOI] [PubMed] [Google Scholar]
- 58.Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301–14. doi: 10.1016/S0140-6736(18)30010-2. [DOI] [PubMed] [Google Scholar]
- 59.Lee JS. The mutational landscape of hepatocellular carcinoma. Clin Mol Hepatol. 2015;21:220–9. doi: 10.3350/cmh.2015.21.3.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu N, Zuo C, Wang X, Chen T, Yang D, Wang J, et al. miR-942 decreases TRAIL-induced apoptosis through ISG12a downregulation and is regulated by AKT. Oncotarget. 2014;5:4959–71. doi: 10.18632/oncotarget.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang D, Meng X, Xue B, Liu N, Wang X, Zhu H. MiR-942 mediates hepatitis C virus-induced apoptosis via regulation of ISG12a. PLoS One. 2014;9:e94501. doi: 10.1371/journal.pone.0094501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tao L, Xue D, Shen D, Ma W, Zhang J, Wang X, et al. MicroRNA-942 mediates hepatic stellate cell activation by regulating BAMBI expression in human liver fibrosis. Arch Toxicol. 2018;92:2935–46. doi: 10.1007/s00204-018-2278-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tao L, Wu L, Zhang W, Ma WT, Yang GY, Zhang J, et al. Peroxisome proliferator-activated receptor γ inhibits hepatic stellate cell activation regulated by miR-942 in chronic hepatitis B liver fibrosis. Life Sci. 2020;253:117572. doi: 10.1016/j.lfs.2020.117572. [DOI] [PubMed] [Google Scholar]
- 64.Stănescu L, Foarfă C, Georgescu AC, Georgescu I. Kaposi's sarcoma associated with AIDS. Rom J Morphol Embryol. 2007;48:181–7. [PubMed] [Google Scholar]
- 65.Ganem D. KSHV and the pathogenesis of Kaposi sarcoma: Listening to human biology and medicine. J Clin Invest. 2010;120:939–49. doi: 10.1172/JCI40567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mesri EA, Cesarman E, Boshoff C. Kaposi's sarcoma and its associated herpesvirus. Nat Rev Cancer. 2010;10:707–19. doi: 10.1038/nrc2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gates AE, Kaplan LD. AIDS malignancies in the era of highly active antiretroviral therapy. Oncology (Williston Park) 2002;16:657–65. [PubMed] [Google Scholar]
- 68.Fan C, Moews PC, Walsh CT, Knox JR. Vancomycin resistance: Structure of D-alanine: D-alanine ligase at 2.3 A resolution. Science. 1994;266:439–43. doi: 10.1126/science.7939684. [DOI] [PubMed] [Google Scholar]
- 69.Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–91. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 70.Zuur AF, Ieno EN, Walker NJ, Saveliev AA, Smith GM. In: Statistics for Biology and Health. New York: Springer; 2009. Ebooks Corporation. Mixed effects models and extensions in ecology with R [Internet] [Google Scholar]
- 71.Carr S, Smith C, Wernberg J. Epidemiology and risk factors of melanoma. Surg Clin North Am. 2020;100:1–12. doi: 10.1016/j.suc.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 72.Owens B. Melanoma. Nature. 2014;515:S109. doi: 10.1038/515S109a. [DOI] [PubMed] [Google Scholar]
- 73.Rastrelli M, Tropea S, Rossi CR, Alaibac M. Melanoma: Epidemiology, risk factors, pathogenesis, diagnosis and classification. In Vivo. 2014;28:1005–11. [PubMed] [Google Scholar]
- 74.Varrone F, Caputo E. The miRNAs role in melanoma and in its resistance to therapy. Int J Mol Sci. 2020;21:878. doi: 10.3390/ijms21030878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.O'Neill CH, Scoggins CR. Melanoma. J Surg Oncol. 2019;120:873–81. doi: 10.1002/jso.25604. [DOI] [PubMed] [Google Scholar]
- 76.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 77.van Meerbeeck JP, Fennell DA, De Ruysscher DK. Small-cell lung cancer. Lancet. 2011;378:1741–55. doi: 10.1016/S0140-6736(11)60165-7. [DOI] [PubMed] [Google Scholar]
- 78.Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, et al. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries in 2012. Eur J Cancer. 2013;49:1374–403. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 79.Yu JS, Chen YT, Chiang WF, Hsiao YC, Chu LJ, See LC, et al. Saliva protein biomarkers to detect oral squamous cell carcinoma in a high-risk population in Taiwan. Proc Natl Acad Sci U S A. 2016;113:11549–54. doi: 10.1073/pnas.1612368113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Giovannacci I, Vescovi P, Manfredi M, Meleti M. Non-invasive visual tools for diagnosis of oral cancer and dysplasia: A systematic review. Med Oral Patol Oral Cir Bucal. 2016;21:e305–15. doi: 10.4317/medoral.20996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Maleki D, Ghojazadeh M, Mahmoudi SS, Mahmoudi SM, Pournaghi-Azar F, Torab A, et al. Epidemiology of oral cancer in Iran: A systematic review. Asian Pac J Cancer Prev. 2015;16:5427–32. doi: 10.7314/apjcp.2015.16.13.5427. [DOI] [PubMed] [Google Scholar]
- 82.Liu D, Zhao X, Zeng X, Dan H, Chen Q. Non-invasive techniques for detection and diagnosis of oral potentially malignant disorders. Tohoku J Exp Med. 2016;238:165–77. doi: 10.1620/tjem.238.165. [DOI] [PubMed] [Google Scholar]
- 83.Rivera C. Essentials of oral cancer. Int J Clin Exp Pathol. 2015;8:11884–94. [PMC free article] [PubMed] [Google Scholar]
- 84.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: Data from the Surveillance, Epidemiology, and End Results Program. Cancer. 2009;115:1531–43. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yan K, Gao J, Yang T, Ma Q, Qiu X, Fan Q, et al. MicroRNA-34a inhibits the proliferation and metastasis of osteosarcoma cells both in vitro and in vivo. PLoS One. 2012;7:e33778. doi: 10.1371/journal.pone.0033778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Amankwah EK, Conley AP, Reed DR. Epidemiology and therapies for metastatic sarcoma. Clin Epidemiol. 2013;5:147–62. doi: 10.2147/CLEP.S28390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bacci G, Briccoli A, Rocca M, Ferrari S, Donati D, Longhi A, et al. Neoadjuvant chemotherapy for osteosarcoma of the extremities with metastases at presentation: Recent experience at the Rizzoli Institute in 57 patients treated with cisplatin, doxorubicin, and a high dose of methotrexate and ifosfamide. Ann Oncol. 2003;14:1126–34. doi: 10.1093/annonc/mdg286. [DOI] [PubMed] [Google Scholar]
- 88.Rainusso N, Wang LL, Yustein JT. The adolescent and young adult with cancer: State of the art – Bone tumors. Curr Oncol Rep. 2013;15:296–307. doi: 10.1007/s11912-013-0321-9. [DOI] [PubMed] [Google Scholar]
- 89.Jayson GC, Kohn EC, Kitchener HC, Ledermann JA. Ovarian cancer. Lancet. 2014;384:1376–88. doi: 10.1016/S0140-6736(13)62146-7. [DOI] [PubMed] [Google Scholar]
- 90.Matulonis UA, Sood AK, Fallowfield L, Howitt BE, Sehouli J, Karlan BY. Ovarian cancer. Nat Rev Dis Primers. 2016;2:1–22. doi: 10.1038/nrdp.2016.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tew WP, Muss HB, Kimmick GG, Von Gruenigen VE, Lichtman SM. Breast and ovarian cancer in the older woman. J Clin Oncol. 2014;32:2553–61. doi: 10.1200/JCO.2014.55.3073. [DOI] [PubMed] [Google Scholar]
- 92.Longuespée R, Boyon C, Desmons A, Vinatier D, Leblanc E, Farré I, et al. Ovarian cancer molecular pathology. Cancer Metastasis Rev. 2012;31:713–32. doi: 10.1007/s10555-012-9383-7. [DOI] [PubMed] [Google Scholar]
- 93.Kleeff J, Costello E, Jackson R, Halloran C, Greenhalf W, Ghaneh P, et al. The impact of diabetes mellitus on survival following resection and adjuvant chemotherapy for pancreatic cancer. Br J Cancer. 2016;115:887–94. doi: 10.1038/bjc.2016.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 95.Neoptolemos JP, Stocken DD, Friess H, Bassi C, Dunn JA, Hickey H, et al. A randomized trial of chemoradiotherapy and chemotherapy after resection of pancreatic cancer. N Engl J Med. 2004;350:1200–10. doi: 10.1056/NEJMoa032295. [DOI] [PubMed] [Google Scholar]
- 96.Simianu VV, Zyromski NJ, Nakeeb A, Lillemoe KD. Pancreatic cancer: Progress made. Acta Oncol. 2010;49:407–17. doi: 10.3109/02841860903447051. [DOI] [PubMed] [Google Scholar]
- 97.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 98.Rathmell WK, Godley PA. Recent updates in renal cell carcinoma. Curr Opin Oncol. 2010;22:250–6. doi: 10.1097/CCO.0b013e328337a5d2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Janzen NK, Kim HL, Figlin RA, Belldegrun AS. Surveillance after radical or partial nephrectomy for localized renal cell carcinoma and management of recurrent disease. Urol Clin North Am. 2003;30:843–52. doi: 10.1016/s0094-0143(03)00056-9. [DOI] [PubMed] [Google Scholar]
- 100.Khan AP, Rajendiran TM, Ateeq B, Asangani IA, Athanikar JN, Yocum AK, et al. The role of sarcosine metabolism in prostate cancer progression. Neoplasia. 2013;15:491–501. doi: 10.1593/neo.13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Costello LC, Franklin RB. The clinical relevance of the metabolism of prostate cancer; zinc and tumor suppression: Connecting the dots. Mol Cancer. 2006;5:17. doi: 10.1186/1476-4598-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sadeghi RN, Karami-Tehrani F, Salami S. Targeting prostate cancer cell metabolism: Impact of hexokinase and CPT-1 enzymes. Tumour Biol. 2015;36:2893–905. doi: 10.1007/s13277-014-2919-4. [DOI] [PubMed] [Google Scholar]
- 103.Ben Sahra I, Laurent K, Giuliano S, Larbret F, Ponzio G, Gounon P, et al. Targeting cancer cell metabolism: The combination of metformin and 2-deoxyglucose induces p53-dependent apoptosis in prostate cancer cells. Cancer Res. 2010;70:2465–75. doi: 10.1158/0008-5472.CAN-09-2782. [DOI] [PubMed] [Google Scholar]