Abstract
A gene encoding a putative GTP-binding protein, a TrmE homologue that is highly conserved in both prokaryotes and eukaryotes, was cloned from Thermotoga maritima, a hyperthermophilic bacterium. T. maritima TrmE was overexpressed in Escherichia coli and purified. TrmE has a GTPase activity but no ATPase activity. The GTPase activity can be competed with GTP, GDP, and dGTP but not with GMP, ATP, CTP, or UTP. Km and kcat at 70°C were 833 μM and 9.3 min−1, respectively. Our results indicate that TrmE is a GTP-binding protein with a very high intrinsic GTP hydrolysis rate. We also propose that TrmE homologues constitute a novel subfamily of the GTPase superfamily.
Proteins possessing GTP-binding and GTPase activities play essential roles in cell proliferation in both prokaryotes and eukaryotes. Their functions are very diversified and include involvement in protein translation (e.g., EF-Tu), signal transduction (e.g., small G proteins), cell growth (e.g., RAS), vesicular transport (e.g., RAB), and protein translocation across membranes (e.g., SRP), etc. (3, 4, 14). They all contain well-conserved motifs—G-1, G-3, and G-4—which are important for GTP-binding activity (4). Another motif, G-2, is not involved in binding to GTP but is involved in interaction with an effector molecule (3, 4). G-2 sequences are well conserved within each subfamily in the large GTPase family. Most GTP-binding proteins possess a very high intrinsic GTP hydrolysis rate, while their GTPase activities are highly stimulated by effector molecules through their binding to the G-2 motif. In general, the ratio of the active GTP-bound form to the inactive GDP-bound form of a GTP-binding protein is crucial for its functional regulation, and this process includes GTP hydrolysis that results from conformational changes of the GTP-binding protein (14).
Analysis of the Escherichia coli genome revealed that it contains a GTP-binding protein encoded by the gene originally designated thdF (1, 5). Recently, this gene was shown to be essential in E. coli and involved in tRNA modification and was thus redesignated trmE (6). The E. coli TrmE protein contains plausible GTP-binding motifs G-1 to G-4 consisting of 454 amino acid residues (6). To date, more than 20 proteins homologous to TrmE have been found in prokaryotes and eukaryotes (Fig. 1). Notably, all eubacteria, but not archaea, whose entire genomes have been sequenced contain TrmE; in addition, a TrmE homologue exists in expressed sequence tags from humans (data not shown). A TrmE homologue in Saccharomyces cerevisiae, MSS1, has been proposed to be a GTPase and is thought to be involved in translational regulation in mitochondria (9, 10). In this study we cloned the trmE gene, highly homologous to the E. coli trmE gene, from the hyperthermophilic bacterium Thermotoga maritima (19), and its product was highly purified. Its biochemical characterization revealed that T. maritima TrmE is indeed a GTP-binding protein having a very high intrinsic GTPase activity at 80°C.
FIG. 1.
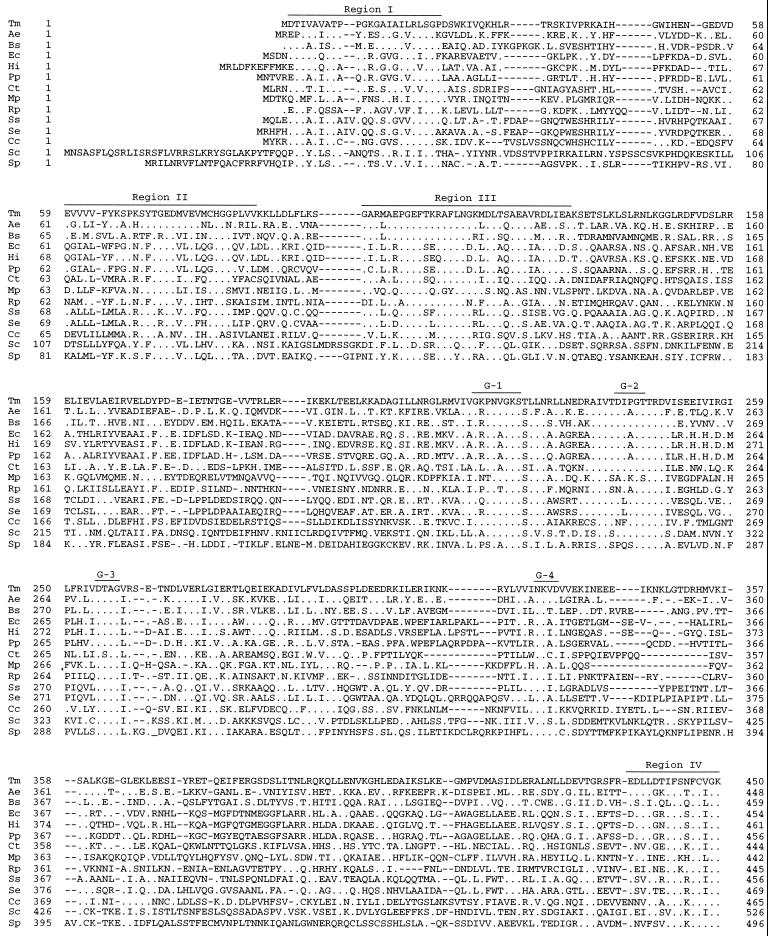
Amino acid sequence alignments of TrmE homologues. A homology search was carried out by the BLAST program (2). Residues identical to those of T. maritima TrmE are shown as dots, and gaps are indicated by dashes. The GTP-binding motifs G-1 to G-4 and well-conserved regions I to IV are indicated by bars above the sequences. Tm, T. maritima (National Center for Biotechnology Information [NCBI] protein database accession no. AAD35356.1); Ae, Aquifex aeolicus (AAC06992.1); Bs, Bacillus subtilis (CAA44403.1); Ec, E. coli (AAC76729); Hi, Haemophilus influenzae (AAC22664.1); Pp, Pseudomonas putida (CAA44418.1); Ct, Chlamydia trachomatis (AAC68293.1); Mp, Mycoplasma pneumoniae (AAB95794); Rp, Rickettsia prowazekii (CAA15187); Ss, Synechocystis sp. strain PCC6803 (BAA17896); Se, Synechococcus elongatus (CAB46651.1); Cc, Cyanidium caldarium (chloroplast genome) (AAF12952); Sc, S. cerevisiae (CAA49238.1); Sp, Schizosaccharomyces pombe (CAB60697).
The trmE gene encodes a GTP-binding protein.
GTP-binding motifs G-1 to G-4 are well conserved in all TrmE homologues (Fig. 1), strongly indicating that T. maritima TrmE is a GTP-binding protein and possesses a GTP hydrolysis activity. Motif G-2 is thought to be involved in binding to an effector molecule but not in GTP binding. The consensus sequence for G-2 is limited to TrmE homologues and cannot be applied to other subfamilies of GTP-binding proteins (data not shown), suggesting that TrmE homologues constitute a novel subfamily of the GTPase superfamily and that TrmE probably interacts with a specific effector molecule, which could regulate the GTPase activity of TrmE. The GTP-binding motifs are located within the third quarter of this protein from the N-terminal end (Fig. 1).
Besides GTP-binding motifs, at least four well-conserved regions (regions I, II, III, and IV) can be assigned (Fig. 1). Although their functional significance is unknown at present, these regions are likely to be related to the TrmE-specific function. Only the GTP-binding domains G-1 to G-4 are involved in GTP binding and GTPase activity in E. coli TrmE (6); therefore, regions I to IV are unlikely to be involved in GTP binding and GTPase activity in T. maritima. It is, however, possible that they are associated with the regulation of GTP binding and GTPase activity and/or that they might be involved in tRNA modification, since trmE mutants exhibit deficiency in biosynthesis of 5-methylaminomethyl-2-thiouridine of tRNA (11). As three E. coli GTPases—Ffh, EF-G, and Era—are known to bind to RNA (8, 12, 13, 15, 17, 18, 21, 22) and their RNA binding is closely associated with their GTPase activities, the TrmE GTPase may also be closely associated with its tRNA modification activity. It is interesting that the C-terminal sequence CVGK in region IV seems to be a consensus sequence, CAAX, where A represents an aliphatic amino acid residue and X represents any amino acid residue, for isoprenylation in the Ras protein (23), although no isoprenylation has been reported and no genes involved in isoprenylation have been identified in the prokaryotes so far. It is also interesting that E. coli TrmE has been shown to be localized in both the cytoplasm and the inner membrane (6).
During the process of a homology search for TrmE, we found another homologous protein, which has been registered as TM1446 in the T. maritima genome (19). This protein contains two tandem repeats of the GTP-binding domain of TrmE, but its homologues, unlike those of TrmE, are found only in the prokaryotes (data not shown).
Purification of T. maritima TrmE.
To clone the trmE gene from T. maritima, the genomic DNA of T. maritima (a generous gift from Francis E. Jenney, Jr., University of Georgia) was used as a template for PCR. Primers 9541 (5′-AGACAACATATGGATACCATTGTCGCTGTAG-3′ [an NdeI site is underlined]) and 9540 (5′-CCAAGCTTTCATTTTCCAACGCAGAAA-3′ [a HindIII site is underlined]) were used. PCR was carried out with 30 cycles of amplification of 1 min at 95°C, 2 min at 50°C, and 2 min at 72°C. The PCR product was digested with NdeI and HindIII and cloned into the NdeI-HindIII site of pET17b, yielding pET-TmTrmE. The DNA sequences of the inserted fragment were confirmed.
Plasmid pET-TmTrmE was introduced into E. coli BL21(DE3) cells containing the ndk::Cmr mutation (16). The ndk disruption strain was used to avoid contamination of GTPase with nucleoside diphosphate kinase in the T. maritima TrmE preparation. Transformed cells were grown at 37°C to mid-exponential phase in 3 liters of M9 medium supplemented with 0.2% Casamino Acids and 50 μg of ampicillin per ml. TrmE was induced for 5 h in the presence of 1 mM isopropyl-β-thiogalactopyranoside. TrmE was well expressed as approximately 40% of the total cellular protein (data not shown). The cells were harvested by centrifugation, washed with 10 mM bis-Tris buffer (pH 7.0), and resuspended in 45 ml of the same buffer. The cells were broken by two passes through a French press at 9,000 lb/in2, followed by centrifugation at 8,000 × g for 10 min to remove cell debris and unbroken cells and by ultracentrifugation at 100,000 × g for 1 h to remove membrane and insoluble fractions. The soluble fraction was treated at 70°C for 10 min to denature E. coli proteins and centrifuged at 12,000 × g for 20 min to remove denatured proteins. Ammonium sulfate was then added to the resulting soluble fraction to 80% saturation. The solution was kept on ice for 1 h and centrifuged at 16,000 × g for 20 min to obtain the precipitates.
The ammonium sulfate precipitate was solubilized in 20 ml of 20 mM Tris-HCl (pH 8.0) containing 25 mM NaCl and 5 mM β-mercaptoethanol (β-ME), and the resulting solution was then dialyzed twice against 2 liters of the same buffer. The dialyzed sample was loaded onto a Q-Sepharose anion-exchange column (Pharmacia) (2.5 by 14 cm) which had been equilibrated with the same buffer. TrmE was eluted with the same buffer using a gradient of 0.025 to 1 M NaCl. Fractions containing TrmE were pooled and precipitated with ammonium sulfate at 80% saturation.
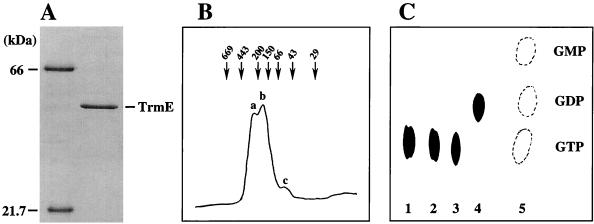
The ammonium sulfate precipitate was solubilized in 10 ml of 10 mM potassium phosphate buffer (pH 7.0) containing 50 mM NaCl and 5 mM β-ME, and the solution was dialyzed twice against 2 liters of the same buffer. The dialyzed sample was then loaded onto a hydroxylapatite column (Bio-Rad) (2.5 by 5 cm) which had been equilibrated with the same buffer. TrmE was eluted with the same buffer using a gradient of 10 to 500 mM potassium phosphate. Fractions containing TrmE were pooled and precipitated with ammonium sulfate at 80% saturation. The precipitate was solubilized in 2 ml of 20 mM Tris-HCl (pH 7.5) containing 50 mM NaCl, 5 mM β-ME, and 10% glycerol, and the solution was dialyzed twice against 1 liter of the same buffer. The dialyzed sample was then loaded onto a Blue column (Pharmacia) (1.0 by 30 cm) which had been equilibrated with the same buffer. TrmE was eluted with the same buffer using a gradient of 0.05 to 1.5 M NaCl. Fractions containing TrmE were pooled and precipitated with ammonium sulfate at 80% saturation. The precipitate was solubilized in 2 ml of 20 mM Tris-HCl (pH 7.5) containing 50 mM NaCl, 5 mM β-ME, and 10% glycerol, and the solution was dialyzed twice against 1 liter of the same buffer. TrmE protein thus obtained was approximately 95% pure as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis (Fig. 2A). Typically, 20 mg of purified TrmE was obtained from a 3-liter culture.
FIG. 2.
Purification, oligomer formation, and GTPase activity of TrmE. (A) Purified TrmE was analyzed by sodium dodecyl sulfate–12.5% polyacrylamide gel electrophoresis, and the gel was stained with Coomassie brilliant blue. Lane 1, bovine serum albumin (66 kDa) and trypsin inhibitor (21.7 kDa) as molecular mass standards; lane 2, purified TrmE (2 μg). (B) Purified TrmE was applied to a Superdex G200 (Pharmacia) column which had been equilibrated with 20 mM KPO4 buffer (pH 8.0) containing 50 mM NaCl. Thyroglobulin (669 kDa), apoferritin (443 kDa), β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumin (66 kDa), ovalbumin (43 kDa), and carbonic anhydrase (29 kDa) were used as molecular mass standards. Peaks at 260, 160, and 52 kDa are designated a, b, and c, respectively, as shown. (C) Hydrolytic activity from GTP to GDP of TrmE. The GTPase assay was carried out in a 50-μl reaction mixture of 50 mM Tris-HCl (pH 9.0) containing 200 mM KCl, 5 mM MgCl2, 1 mM DTT, 10 μM [α-32P]GTP, and 10 μg of purified TrmE protein at 70°C for 10 min. The reaction was terminated by transfer of 5 μl of samples to 10 μl of ice-cold 20 mM EDTA. Portions of the terminated reaction mixture were spotted onto a polyethyleneimine-cellulose thin-layer chromatography plate, which was developed in 0.75 M KH2PO4 (pH 3.65). The plate was autoradiographed to identify hydrolyzed products of GTP. Spots corresponding to GTP, GDP, and GMP (lane 5) were identified by UV shadowing. Lanes 1 and 2, incubations without TrmE for 0 and 60 min, respectively; lanes 3 and 4, incubations with 10 μg of TrmE for 0 and 60 min, respectively.
Interestingly, when the purified T. maritima TrmE (50,650 Da) was applied to a gel filtration column, it was eluted into three peaks (Fig. 2B): peak a at 260 kDa, peak b at 160 kDa, and peak c at 52 kDa. This indicates that TrmE mostly exists as oligomers in solution: primarily as a trimer (peak b) and a hexamer (peak a), with a minor fraction as a monomer (peak c). E. coli TrmE (49,200 Da) is also known to be eluted from a gel filtration column in a range of 50 to 250 kDa (6). Thus, oligomerization might be a common characteristic of the TrmE subfamily, and it is possible that at least one of the four well-conserved regions (regions I to IV) is involved in the oligomerization.
GTPase activity of TrmE.
In order to demonstrate that the purified T. maritima TrmE protein is indeed a GTP-binding protein and possesses a GTPase activity, purified TrmE was incubated for 60 min at 70°C with [α-32P]GTP and the reaction mixture was analyzed by thin-layer chromatography. As shown in Fig. 2C, GTP was converted to GDP by TrmE, indicating that T. maritima TrmE has a GTPase activity.
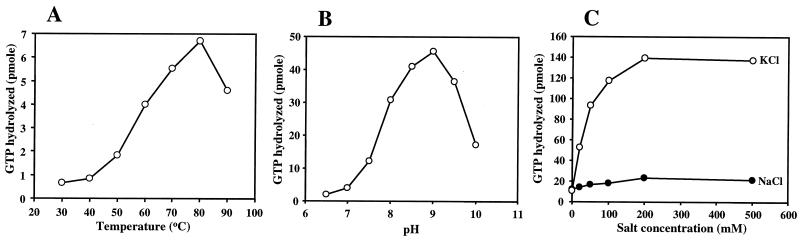
Next, we determined the optimum conditions for GTPase activity. GTP hydrolysis occurred linearly with up to 30 μg of protein per reaction mixture (data not shown). With 10 μg of protein, GTP hydrolysis occurred linearly for up to 60 min of incubation (data not shown). To determine the optimal reaction temperature, GTP hydrolysis assays were carried out at 30, 40, 50, 60, 70, 80, and 90°C. As shown in Fig. 3A, maximum activity was found at 80°C. However, the higher temperature caused a higher background: 0.7 and 1.9% of input GTP was hydrolyzed without protein in 10 min at 80 and 90°C, respectively, while less than 0.3% was hydrolyzed at other temperatures. Therefore, all of the reactions described below were carried out at 70°C.
FIG. 3.
Effects of reaction temperature, pH, and salt concentration on the GTPase activity of TrmE. (A) The GTPase assay was carried out in a 50-μl reaction mixture of 50 mM Tris-HCl (pH 7.5) containing 5 mM MgCl2, 1 mM DTT, 10 μM [γ-32P]GTP, and 10 μg of TrmE for 10 min at different temperatures. The GTPase assay reaction was stopped by adding activated charcoal followed by centrifugation, and the release of 32Pi in the supernatant was assayed using a liquid scintillation counter. (B) The GTPase assay was carried out in a 50-μl reaction mixture. Between pH 6.5 and 9.0, the mixture contained 50 mM Tris-HCl; at pH 9.5 and 10, the mixture contained 50 mM 3-(cyclohexylamino)-2-hydroxy-1-propanesulfonic acid. Both buffers contained 5 mM MgCl2, 1 mM DTT, 10 μM [γ-32P]GTP, and 10 μg of TrmE, and the reaction was carried out for 10 min at 70°C. (C) The GTPase assay was carried out in a 50-μl reaction mixture of 50 mM Tris-HCl (pH 7.5) containing 5 mM MgCl2, 1 mM DTT, 10 μM [γ-32P]GTP, 10 μg of TrmE, and different concentrations (0 to 500 mM) of KCl or NaCl for 10 min at 70°C. The reaction was carried out at least twice, and the average value for each point was used. Background values (without protein) were subtracted.
The effects of pH, salt, and Mg2+ on GTPase activity were subsequently examined. TrmE was found to prefer alkaline conditions, with the optimum activity at pH 9.0, while it had almost no activity at neutral and acidic pHs (Fig. 3B). The TrmE GTPase was found to be specifically activated by KCl and, as shown in Fig. 3C, at a sevenfold-higher rate at 200 mM KCl than at 200 mM NaCl. At present, we do not know whether these in vitro characteristics of pH and K+ concentration reflect in vivo conditions. The Mg2+ ion is also required for GTPase activity, which reached its highest level at greater than 1 mM Mg2+ (data not shown).
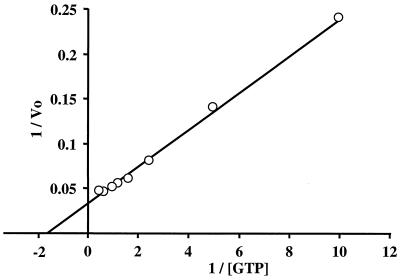
Thus, the optimum conditions for TrmE GTPase activity were determined to be as follows: the reaction is carried out at 70°C for 10 min in 50 mM Tris-HCl (pH 9.0) containing 200 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol (DTT), and 10 μM GTP with 10 μg of TrmE in a 50-μl reaction mixture. Biochemical parameters of the TrmE GTPase activity were determined under optimum conditions. Using GTP concentrations from 0.001 to 3 mM, the Lineweaver-Burk plot was determined (Fig. 4). Km and Vmax for the GTPase activity of TrmE were estimated to be 833 μM and 37 μM/min, respectively. kcat was calculated to be 9.3 min−1. This indicates that T. maritima TrmE has a very high intrinsic GTP hydrolysis rate. Note that the concentration of GTP in exponentially growing bacterial cells is about 1 mM (20). E. coli TrmE also shows a very high intrinsic GTP hydrolysis rate (Km, 378 μM; kcat, 26 min−1) (6), which is comparable to T. maritima TrmE, suggesting that the very high intrinsic GTP hydrolysis rate might be a common characteristic among TrmE proteins from various species. It would be interesting to know if the hydrolysis rate is modulated by an effector molecule in the cells.
FIG. 4.
Lineweaver-Burk plot of TrmE GTPase activity. The GTPase assay was carried out in a 50-μl reaction mixture of 50 mM Tris-HCl (pH 9.0) containing 200 mM KCl, 5 mM MgCl2, 1 mM DTT, 10 μg of TrmE, and different concentrations (0.001 to 3 mM) of GTP for 10 min at 70°C. Each point is the average of at least two experiments. Background values (without protein) were subtracted. Vo, initial rate of reaction.
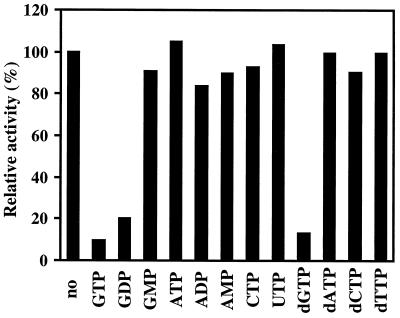
To determine the substrate specificity, competition experiments were carried out with various nucleotides added at a 300-fold excess over the GTP used in the reaction. As shown in Fig. 5, only GTP, GDP, and dGTP were found to compete GTP hydrolysis. The high substrate specificity of TrmE to guanine nucleotides was further confirmed by the fact that TrmE has no detectable ATPase activity under the conditions used for the GTPase assay (data not shown). Note that GTP binding of E. coli TrmE was shown to be competed with GTP, GDP, and dGTP (6).
FIG. 5.
GTPase assay in the presence of competitors. The GTPase assay was carried out in a 50-μl reaction mixture of 50 mM Tris-HCl (pH 9.0) containing 200 mM KCl, 5 mM MgCl2, 1 mM DTT, 10 μM [γ-32P]GTP, and 10 μg of TrmE for 10 min at 70°C in the presence of each competitor at a final concentration of 3 mM. Each point is the average of at least two independent experiments. Competitors used are shown, and activities are relative to the value with no competitor (100%).
Conclusions.
We have demonstrated that T. maritima TrmE possesses a GTPase activity having a very high intrinsic GTP hydrolysis rate. Although its Km value is quite high (833 μM) in comparison with the Km value of E. coli Era (10 μM) (7), the TrmE GTPase activity is likely to be significantly regulated by its effector molecule(s) in the cells. The identification of such an effector molecule is important for our understanding of TrmE function, which is known to be essential for cell growth in E. coli (6).
Acknowledgments
K. Yamanaka and J. Hwang contributed equally to this work.
We are very grateful to F. E. Jenney, Jr., for providing genomic DNA of T. maritima.
REFERENCES
- 1.Alam K Y, Clark D P. Molecular cloning and sequence of the thdF gene, which is involved in thiophene and furan oxidation by Escherichia coli. J Bacteriol. 1991;173:6018–6024. doi: 10.1128/jb.173.19.6018-6024.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourene H R, Sanders D A, McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990;348:125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- 4.Bourene H R, Sanders D A, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 5.Burland V, Plunkett III G, Daniels D L, Blattner F R. DNA sequence and analysis of 136 kilobases of the Escherichia coligenome: organizational symmetry around the origin of replication. Genomics. 1993;16:551–561. doi: 10.1006/geno.1993.1230. [DOI] [PubMed] [Google Scholar]
- 6.Cabedo H, Macian F, Villarroya M, Escudero J, Martinez-Vicente M, Knecht E, Armengod M E. The Escherichia coli trmE (mnmE) gene, involved in tRNA modification, codes for an evolutionarily conserved GTPase with unusual biochemical properties. EMBO J. 1999;18:7063–7076. doi: 10.1093/emboj/18.24.7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen S-M, Takiff H E, Barber A M, Dubois G C, Bardwell J C A, Court D L. Expression and characterization of RNaseIII and Era proteins: products of the rnc operon of Escherichia coli. J Biol Chem. 1990;265:2888–2895. [PubMed] [Google Scholar]
- 8.Chen X, Court D L, Ji X. Crystal structure of ERA: a GTPase-dependent cell cycle regulator containing an RNA binding motif. Proc Natl Acad Sci USA. 1999;96:8396–8401. doi: 10.1073/pnas.96.15.8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colby G, Wu M, Tzagoloff A. MTO1 codes for a mitochondrial protein required for respiration in paromomycin-resistant mutants of Saccharomyces cerevisiae. J Biol Chem. 1998;273:27945–27952. doi: 10.1074/jbc.273.43.27945. [DOI] [PubMed] [Google Scholar]
- 10.Decoster E, Vassal A, Faye G. MSS1, a nuclear-encoded mitochondrial GTPase involved in the expression of COX1 subunit of cytochrome coxidase. J Mol Biol. 1993;232:79–88. doi: 10.1006/jmbi.1993.1371. [DOI] [PubMed] [Google Scholar]
- 11.Elseviers D, Petrullo L A, Gallagher P. Novel E. colimutants deficient in biosynthesis of 5-methylaminomethyl-2-thiouridine. Nucleic Acids Res. 1984;12:3521–3534. doi: 10.1093/nar/12.8.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farmery M, Macao B, Larsson T, Samuelsson T. Binding of GTP and GDP induces a significant conformational change in the GTPase domain of Ffh, a bacterial homologue of the SRP 54 kDa subunit. Biochim Biophys Acta. 1998;1385:61–68. doi: 10.1016/s0167-4838(98)00045-4. [DOI] [PubMed] [Google Scholar]
- 13.Johnstone B H, Handler A A, Chao D K, Nguyen V, Smith M, Ryu S Y, Simons E L, Anderson P E, Simons R W. The widely conserved Era G-protein contains an RNA-binding domain required for Era function in vivo. Mol Microbiol. 1999;36:1118–1131. doi: 10.1046/j.1365-2958.1999.01553.x. [DOI] [PubMed] [Google Scholar]
- 14.Kjeldgaard M, Nyborg J, Clark B F C. The GTP binding motif: variations on a theme. FASEB J. 1996;10:1347–1368. [PubMed] [Google Scholar]
- 15.Kurita K, Honda K, Suzuma S, Takamatsu H, Nakamura K, Yamane K. Identification of a region of Bacillus subtilisFfh, homologue of mammalian SRP54 protein, that is essential for binding to small cytoplasmic RNA. J Biol Chem. 1996;271:13140–13146. doi: 10.1074/jbc.271.22.13140. [DOI] [PubMed] [Google Scholar]
- 16.Lu Q, Inouye M. Adenylate kinase complements nucleoside diphosphate kinase deficiency in nucleotide metabolism. Proc Natl Acad Sci USA. 1996;93:5720–5725. doi: 10.1073/pnas.93.12.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meier T I, Peery R B, Jaskunas S R, Zhao G. 16S rRNA is bound to Era of Streptococcus pneumoniae. J Bacteriol. 1999;181:5242–5249. doi: 10.1128/jb.181.17.5242-5249.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller J D, Bernstein H D, Walter P. Interaction of E. coliFfh/4.5S ribonucleoprotein and FtsY mimics that of mammalian signal recognition particle and its receptor. Nature. 1994;356:657–659. doi: 10.1038/367657a0. [DOI] [PubMed] [Google Scholar]
- 19.Nelson K E, Clayton R A, Gill S R, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Nelson W C, Ketchum K A, McDonald L, Utterback T R, Malek J A, Linher K D, Garrett M M, Stewart A M, Cotton M D, Pratt M S, Phillips C A, Richardson D, Heidelberg J, Sutton G G, Fleischmann R D, White O, Salzberg S L, Smith H O, Venter J C, Fraser C M. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 20.Neuhard J, Nygaard P. Purines and pyrimidines. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1987. pp. 445–473. [Google Scholar]
- 21.Sayed A, Matsuyama S, Inouye M. Era, an essential Escherichia colismall G-protein, binds to the 30S ribosomal subunit. Biochem Biophys Res Commun. 1999;264:51–54. doi: 10.1006/bbrc.1999.1471. [DOI] [PubMed] [Google Scholar]
- 22.Shibata T, Fuji Y, Nakamura Y, Nakamura K, Yamane K. Identification of protein synthesis elongation factor G as a 4.5 S RNA-binding protein in Escherichia coli. J Biol Chem. 1996;271:13162–13168. doi: 10.1074/jbc.271.22.13162. [DOI] [PubMed] [Google Scholar]
- 23.Willumsen B M, Norris K, Papageorge A G, Hubbert N L, Lowy D R. Harvey murine sarcoma virus p21 ras protein: biological and biochemical significance of the cysteine nearest the carboxy terminus. EMBO J. 1984;3:2581–2585. doi: 10.1002/j.1460-2075.1984.tb02177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]