Summary
Although glucagon secretion is perturbed in both T1D and T2D, the pathophysiological changes in individual pancreatic alpha cells are still obscure. Using recently curated single-cell RNASeq data from T1D or T2D donors and their controls, we identified alpha cell transcriptomic alterations consistent with both common and discrete pathways. Although alterations in alpha cell identity gene (ARX, MAFB) expression were conserved, cytokine-regulated genes and genes involved in glucagon biosynthesis and processing were up-regulated in T1D. Conversely, mitochondrial genes associated with ROS (COX7B, NQO2) were dysregulated in T2D. Additionally, T1D alpha cells displayed altered expression of autoimmune-induced ER stress genes (ERLEC1, HSP90), whilst those from T2D subjects showed modified glycolytic and citrate cycle gene (LDHA?, PDHB, PDK4) expression. Thus, despite conserved alterations related to loss of function, alpha cells display disease-specific gene signatures which may be secondary to the main pathogenic events in each disease, namely immune- or metabolism-mediated-stress, in T1D and T2D, respectively.
Subject areas: Biological sciences, bioinformatics, transcriptomics
Graphical abstract
Highlights
-
•
Transcriptomic profiles of alpha cells display differences between T1D and T2D
-
•
Dysregulation of some alpha cell identity genes was found in both conditions
-
•
In T1D alpha cells displayed signatures of increased ER stress
-
•
T2D alpha cells show increased ROS defense and depression of central metabolism
Biological sciences; Bioinformatics; Transcriptomics.
Introduction
Both types 1 (T1D) and 2 diabetes (T2D) are characterized by varying degrees of pancreatic beta cell failure (Eizirik et al., 2020; Marchetti et al., 2020). This is paralleled by the dysfunction of alpha cells, which in T1D may contribute to insulin-induced hypoglycemia and in T2D, at least at the initial phases of the disease, to hyperglycemia (Brissova et al., 2018; Gromada et al., 2018).
Alpha and beta cells are intermingled in human pancreatic islets (Bosco et al., 2010) and there is a crosstalk between these cells that regulates at least in part their function (Campbell and Newgard, 2021). It is thus conceivable that the reduced functional beta cell mass in T1D and T2D impacts alpha cells and contributes to their dysfunction in each disease. Alternatively, it may be that mechanisms inherent to each disease, i.e. a predominance of autoimmunity and consequent islet inflammation in T1D as compared to severe metabolic stress in T2D (Eizirik et al., 2020), directly impair the alpha cells.
Differentiated cells trigger diverse adaptive responses that are determined by the stress to which they are exposed. For instance, beta cells exposed to pro-inflammatory cytokines trigger branches of the unfolded protein response that are different from the ones triggered in response to the metabolic stressor palmitate, and the global gene signatures of islets obtained from patients affected by T1D or T2D are markedly different (Eizirik et al., 2020). The cellular responses to diverse stresses may leave gene expression footprints—particularly in long-lived cells such as human alpha and beta cells—that can be detected by RNA sequencing. Examination of these footprints may enable to the differentiation of the principal cause(s) of the alpha cell stress present in T1D and T2D. If the leading cause of alpha cell stress is the relative or absolute loss of neighboring beta cells, and the deficiency of insulin leads to hyperglycemia, we may expect to find similar gene signatures on alpha cells from patients with T1D or T2D; on the other hand, if the stress is disease-specific then alpha cells should show different signatures in each case, for instance, immune-induced stress in T1D and more metabolic changes T2D.
To test these hypotheses we presently used recently curated human islet single-cell transcriptomic data from control donors or individuals affected by either T1D or T2D that are publicly available (Kaestner et al., 2019). The results indicate similar patterns, but also major divergences between the gene expression signatures present in alpha cells from patients with T1D or T2D, arguing in favor of disease-specific mechanisms leading to alpha cell dysfunction in each case.
Results
Assembling single-cell datasets from the Human Pancreas Analysis Program (HPAP) database
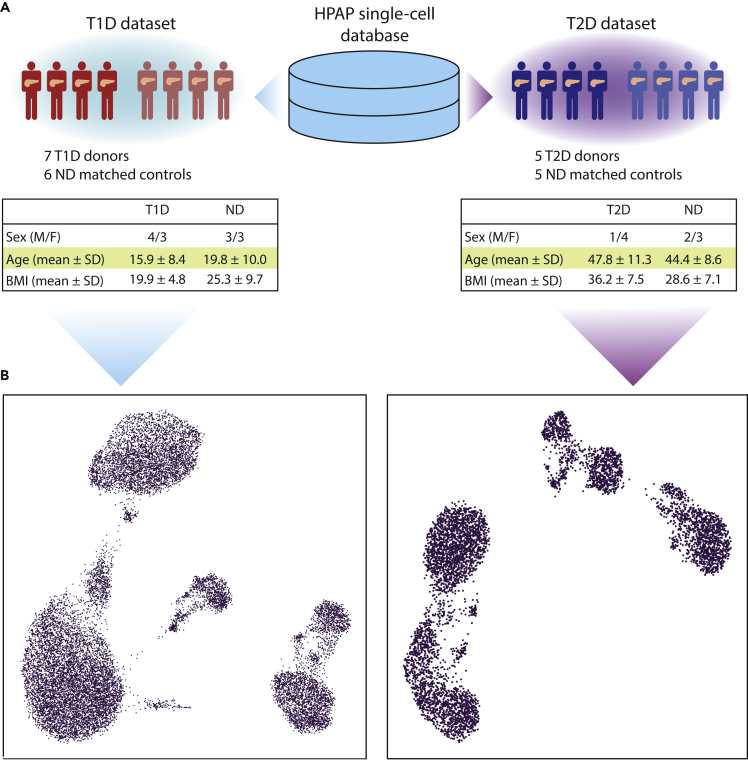
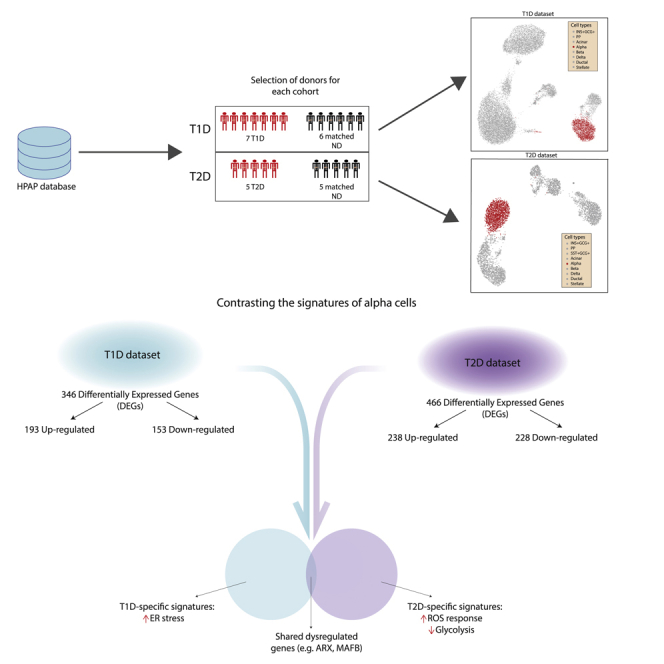
The populations of T1D and T2D donors are characterized by different clinical features, with the most prominent differences being in age and body mass index (BMI), which usually are higher in T2D (American Diabetes Association, 2010). For this reason, separate control groups were assembled to match (as much as possible) the clinical features of the affected donors, controlling: (i) single-cell technology used to assess transcriptomes, ii) age, iii) BMI, and iiii) sex. Normoglycemic donors showing positivity toward pancreatic auto-antibodies were excluded. After the selection of control donors on the basis of such criteria, the T1D dataset included 7 diabetic donors and 6 controls, whereas T2D contained 5 affected donors and 5 controls (Figure 1A). The features of the donors included in this study are reported in Table S1.
Figure 1.
T1D and T2D single-cell transcriptomics datasets
(A) Single-cell transcriptomic samples were obtained from the database of the Human Pancreas Analysis Program (HPAP) and divided among two datasets, T1D and T2D. Samples from non-diabetic donors were assigned to a dataset according to donor and technical features in order to match those of the T1D and T2D samples. The tables report the donor features of T1D and T2D datasets.
(B) The UMAP plots provide a representation of the T1D (left) and T2D (right) datasets. Each dot corresponds to the transcriptomic profile of a single cell projected in a two-dimensional space (UMAP1, UMAP2).
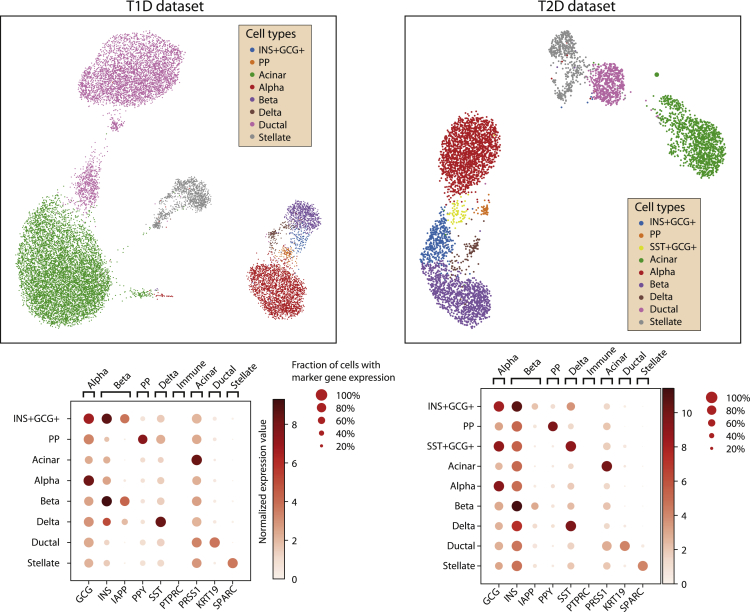
Analysis of raw sequencing data and exclusion of low-quality cells and genes (Figure S1) delivered a T1D dataset comprising 17,503 cells and 31,890 genes, and a T2D dataset with 5,772 cells and 32,221 genes. The quality control (QC) thresholds are reported in Table S2. The datasets have been corrected for technical covariates (single-cell technology and sequencing batch) and visualized in a Uniform Manifold Approximation Projection (UMAP) space (Figures 1B, S2, and S3). Clusters of single cells were identified with the Louvain algorithm and annotated considering the expression of marker genes (Figure 2). The genes highly expressed in the annotated clusters were compared with the marker genes reported in PanglaoDB (Franzén et al., 2019) to confirm the cell type annotation. Some marker genes displayed low expression levels across different cell types, an effect likely owing to ambient RNA contamination that requires the application of specific tools (such as DecontX, SoupX, EmptyDrops) (Lun et al., 2019; Yang et al., 2020; Young and Behjati, 2020). As the contamination levels were of low magnitude, we decided not to apply such correction to the expression matrices.
Figure 2.
Cell type annotation of T1D and T2D datasets
The single cells in the UMAP plots of T1D (left) and T2D (right) datasets have been clustered and annotated to a pancreatic cell type (top) based on the expression of known marker genes (bottom).
The number of cells for each cell type in each dataset is reported in Table 1. Alpha cells were then analyzed separately in each dataset to find transcriptomic alterations potentially linked to diabetes.
Table 1.
Cell type distributions in the T1D and T2D datasets
| INS + GCG+ | SST + GCG+ | PP | Acinar | Alpha | Beta | Delta | Ductal | Stellate | |
|---|---|---|---|---|---|---|---|---|---|
| T1D | 140 | 0 | 97 | 7909 | 2362 | 736 | 145 | 4996 | 1118 |
| T2D | 382 | 61 | 43 | 1246 | 1757 | 1098 | 102 | 564 | 518 |
The table reports the number of cells assigned to each cell type in the T1D and T2D datasets.
The transcriptional signatures of alpha cells in T1D
The T1D dataset comprised 2,362 alpha cells, collectively expressing 6,265 genes (after filtering). A more detailed breakdown of the number of cells at the level of individual, single-cell technology used, and diabetes status is reported in Table S3. After removing cells from individuals with less than 50 alpha cells, the resulting dataset, including 2,225 alpha cells, was analyzed with MAST to identify genes differentially expressed in T1D cells.
A total of 346 differentially expressed genes (DEGs) were identified, of which 193 were up-regulated and 153 were down-regulated versus cells from the control, normoglycemic group (Table S4). A pattern observed among the DEGs was the over-expression of genes involved in Reactive Oxygen Species (ROS) response (PRNP, NDUFA6, NDUFB4, GLRX, and TXN), and protein folding stability via chaperone activity (HSP9, HSP90AA1, and HSP90AB1). We also observed overexpression of IL-8 and HLA-A, genes downstream of the transcription factors (TFs) NF-κB and STAT1/STAT2 that are regulated by the pro-inflammatory cytokines IL-1β and types I and II interferons (IFNs) and participate in the immune system-islet cell dialogue present in T1D (Eizirik et al., 2020). Importantly, these genes were not overexpressed in alpha cells from patients affected by T2D (see later in discussion). The over-expression of DDIT3 (also known as CHOP), a key mediator of ER stress induced-beta cell death (Eizirik et al., 2008), fits with this scenario. Another trend of pathophysiological relevance was the significant downregulation of genes implicated in endocrine function, namely PCSK2, CHGA, SRP14, and PAK3. These genes contribute to proglucagon peptide maturation and eventual exocytosis, and their decreased expression could contribute to reduced glucagon secretion from alpha cells in human T1D (Gerich et al., 1973). Thus, in PCSK2 gene-null mice lacking the prohormone convertase in alpha cells, proglucagon maturation to the mature hormone is blocked (Furuta et al., 2001). Of interest, there was also down-regulation (−2.5-fold change) of PCSK1N, a specific inhibitor of PCSK1.
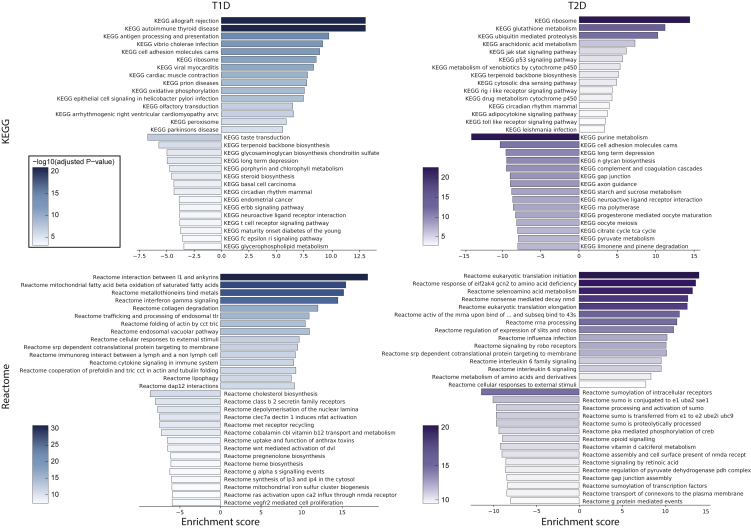
A comprehensive investigation focusing on the functional signatures enriched in T1D was performed with an enrichment test as implemented in MAST, using six different collections (mSIGDB, Reactome, KEGG, GO-MF, GO-BP, GO-CC) encompassing broad (i.e. mSIGDB, KEGG) or very specific (i.e. GO) functional terms (Table S5). Collectively, there were a total of 1,539 significantly enriched terms, with 1,159 positively and 380 negatively enriched. Of these, 1,207 were GO terms (159 MF, 896 BP, 152 CC), whereas mSIGDB, KEGG, and Reactome presented 22, 49, and 261 enriched terms. Some of these terms were consistent with the patterns observed with the results of the differential expression analysis, in that functional categories related to ROS and unfolded protein response (UPR) and exposure to immune mediators were consistently enriched in the different datasets, thus reinforcing the view that T1D alpha cells endure higher levels of these stresses. To better highlight the most important signatures of T1D alpha cells, the enrichment results were ranked according to the obtained significance (Figure3). The top three KEGG categories positively enriched were related to immunity (ALLOGRAFT REJECTION, AUTOIMMUNE THYROID DISEASE, ANTIGEN PROCESSING, AND PRESENTATION), consistently with the inflammation associated with islets in T1D (Eizirik et al., 2020). The most positively enriched KEGG terms also included RIBOSOME and OXIDATIVE PHOSPHORYLATION, both associated with increased ROS production. Mitochondrial activity is a major source of ROS, whereas oxidative stress impairs protein biosynthesis and modifies rRNA (Shcherbik and Pestov, 2019), requiring an increased turnover of these molecules by activating mechanisms to repair or recycle damaged molecules. Pathways associated with immunity and ROS production/response were among the most positively enriched pathways in all databases. In Reactome we found, related to immunity, a positive enrichment of interferon-gamma signaling, endosomal/vacuolar pathway of antigen presentation, and trafficking/processing of endosomal toll-like receptor (TLR). Related to ROS there is an increase in mitochondrial fatty acid oxidation, metallothionein ROS scavenging, and actin folding. In mSIGDB hallmark, the top ten pathways included allograft rejection, oxidative phosphorylation, apoptosis, interferon gamma response, unfolded protein response, and reactive oxygen species. The top three GO biological processes were negative regulation of cell killing, negative regulation of leukocyte mediated cytotoxicity, and positive regulation of T-cell mediated cytotoxicity. The other highly enriched pathways include positive regulation of protein depolymerization, positive regulation of actin filament depolymerization, and negative regulation of IRE1-mediated unfolded protein response.
Figure 3.
Comparison of most significantly enriched pathways in T1D and T2D
Gene-set enrichment analysis was performed for T1D (left) and T2D (right) datasets. The barplots report the normalized enrichment score (bar length) and the significance (bar color) of the most significant 15 positively and negatively enriched pathways from KEGG (top) and Reactome (bottom).
The transcriptional signatures of alpha cells in T2D
After filtering out low-quality cells, in the T2D dataset, there were 1,757 alpha cells expressing a total of 6,872 genes. Following the approach used for the T1D dataset, we detailed the number of alpha cells at the levels of individual, single-cell technology, and diabetes status (Table S3). Two donors had a low number of cells and were excluded from the differential expression analysis; the remaining cells (1,718) were analyzed with MAST.
There were 466 genes differentially expressed in T2D versus control alpha cells, 238 up-regulated and 228 down-regulated (Table S6). Among the over-expressed DEGs, there were activators of stress and apoptosis mediated by p53, i.e. CCNK, JUN, BTG1, GADD45A, GADD45B, XPC, CDKN2AIP, DDIT3, TXNIP, and ATF3. Of these, XPC, GADD45A, GADD45B, and CDKN2AIP respond to DNA oxidative damage (Hasan et al., 2009; Salvador et al., 2013; Wang et al., 2012), while DDIT3 is activated by endoplasmic reticulum (ER) stress (Eizirik et al., 2008; Ohoka et al., 2005; Yamaguchi and Wang, 2004). TXNIP and ATF3 are instead modulated by glucose concentration and play a role in both apoptosis and hormone secretion. TXNIP controls a pathway (miR-204/MafA/insulin) that reduces insulin expression (Xu et al., 2013), whereas ATF3 increases the expression of proglucagon, as well as regulating the expression of genes related to apoptosis (such as GADD45, BNIP3, and NOXA).
The over-expression of such genes, and others related to ROS (SBNO2, EGLN2, MBP) and the unfolded protein response (UPR; EEF2, ATF4, HERPUD1) suggests that alpha cells in T2D are subject to oxidative stress, a hallmark of toxicity induced by elevating glucose levels (Gromada et al., 2018; Kawahito et al., 2009). The downregulation of genes involved in pyruvate metabolism (LDHA, PDHB, PDK4) and oxidative phosphorylation (COX7B, NQO2, SUCLA2, UQCR10, SLC25A4) is also consistent with oxidative stress pathways, in that these may affect ROS production by mitochondria.
Of functional relevance to hormone biosynthesis, the genes encoding prohormone convertases PCSK1 and PCSK2 were both down-regulated, as well as SCGN and SYT13, involved in hormone storage and exocytosis, respectively (Andersson et al., 2012; Yang et al., 2016), and the transcription factor MAFB, crucial for preproglucagon gene expression and the production and release of the hormone (Katoh et al., 2018).
As for the T1D dataset, a gene set enrichment analysis was performed to identify functions and pathways significantly enriched in the T2D alpha cells (Figure 3). Overall, 1,973 significantly enriched terms were identified, of which 494 were positively and 1,479 were negatively enriched (Table S7). Most of these (1,552) were Gene Ontology (GO) terms (207 MF, 1158 BP, 187 CC), while the enriched terms in mSIGDB, KEGG, and Reactome were 12, 99, and 310, respectively.
The 6 mSIGDB pathways positively enriched ("INTERFERON GAMMA RESPONSE," "INTERFERON ALPHA RESPONSE," "P53 PATHWAY," "ALLOGRAFT REJECTION," "TNFA SIGNALING VIA NFKB," "APOPTOSIS”) are associated to inflammation, whereas the negatively enriched terms are mostly involved in the energetic metabolism ("OXIDATIVE PHOSPHORYLATION," "ADIPOGENESIS," "GLYCOLYSIS”). This trend is consistent in the other sets: for instance, in KEGG the terms such as “P53 SIGNALING PATHWAY” and “TOLL LIKE RECEPTOR SIGNALING PATHWAY” are positively enriched, while negatively enriched ones included metabolic terms such as "PYRUVATE METABOLISM," "KEGG STARCH AND SUCROSE METABOLISM" and "CITRATE CYCLE TCA CYCLE." Similarly, among the Reactome positively enriched pathways there are "INTERFERON GAMMA SIGNALING,” "INFLAMMASOMES” and "ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC,” while the negatively enriched ones include "REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX," "THE CITRIC ACID TCA CYCLE AND RESPIRATORY ELECTRON TRANSPORT" and "GLUCOSE METABOLISM." Of interest, we report a number of negatively enriched terms specifically linked to alpha cell function: "GLUCAGON SIGNALING IN METABOLIC REGULATION," "PKA ACTIVATION IN GLUCAGON SIGNALLING," "GLUCAGON LIKE PEPTIDE 1 GLP1 REGULATES INSULIN SECRETION" and "GLUCAGON TYPE LIGAND RECEPTORS," among the Reactome terms; "CELLULAR RESPONSE TO GLUCAGON STIMULUS" and "RESPONSE TO GLUCAGON" among the GO-BP terms.
Contrasting the alpha cell signatures of T1D and T2D
By analyzing the changes in gene expression in alpha cells separately in T1D and T2D the corresponding transcriptomic signatures were identified and described, revealing a number of similarities, such as upregulated DEGs involved in the stress response or the downregulation of genes relevant for the secretory function. To quantify more precisely the extent to which the two series overlap and to underscore their differences, a systematic comparison of the obtained results was undertaken next.
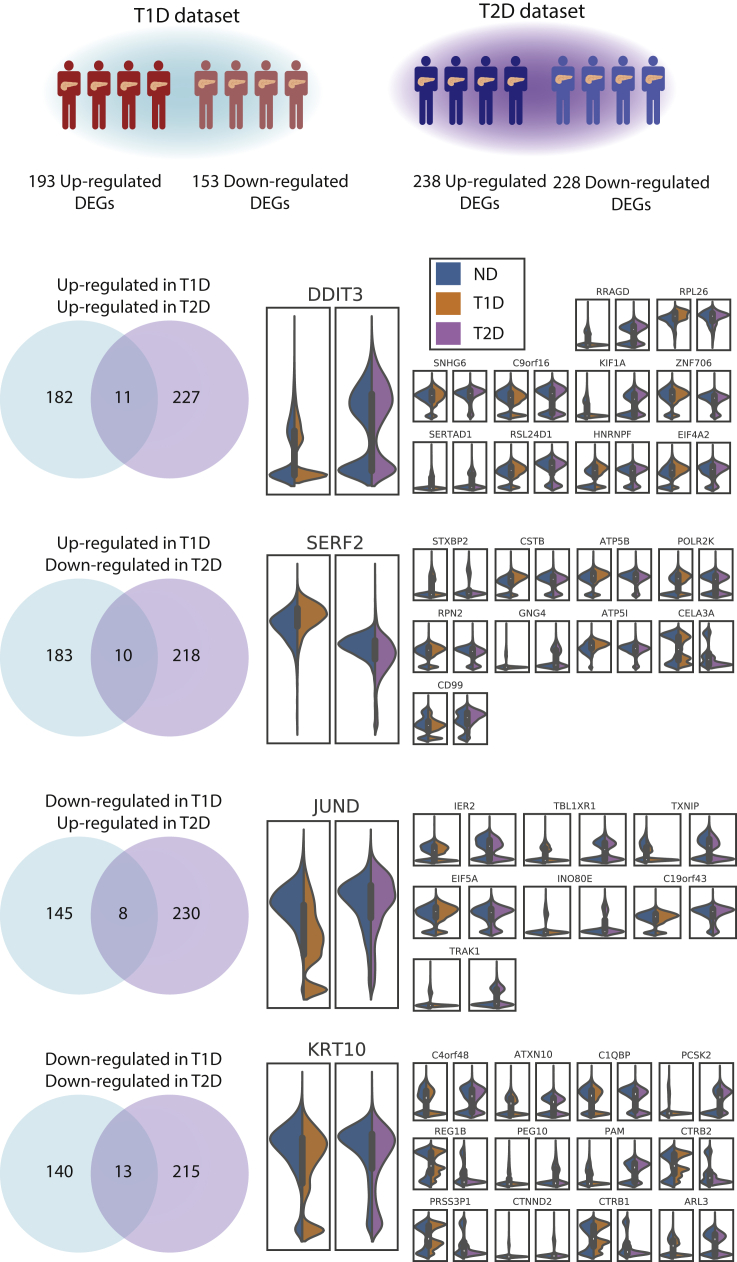
In T1D and T2D there were a total of 770 DEGs (Table S8). Of these, 42 were shared, while DEGs present only in T1D and T2D were 304 and 424, respectively. Considering only up-regulated genes, there were 420 DEGs, of which 11 were shared, 182 were T1D-specific and 227 were T2D-specific (Figure 4). The shared up-regulated genes are EIF4A2, DDIT3, RRAGD, RPL26, SNHG6, C9orf16, KIF1A, ZNF706, SERTAD1, RSL24D1, and HNRNPF. For the down-regulated genes, from a total of 368 DEGs, there were 13 shared, 140 T1D-specific, and 215 T2D-specific. The down-regulated DEGs in common are C4orf48, KRT10, C1QBP, PCSK2, REG1B, PEG10, PAM, CTRB2, PRSS3P1, CTNND2, CTRB1, ARL3, and ATXN10. The overlap of DEGs regulated in different directions in T1D and T2D was assessed as well. There were 10 genes in common between the T1D up-regulated and the T2D down-regulated DEGs (STXBP2, CSTB, ATP5B, SERF2, RPN2, GNG4, ATP5I, CELA3A, CD99, and POLR2K), while the shared genes between the T1D down-regulated and the T2D up-regulated were 8 (IER2, JUND, TXNIP, EIF5A, INO80E, C19orf43, TRAK1, and TBL1XR1). Finally, as DEGs were identified based both on the significance and fold-change thresholds, there are several genes that have a significant change but with fold-change below the threshold. Although these genes have not been reported as DEGs, some could provide relevant information. For instance, genes related to alpha cell identity (notably the TFs ARX and MAFB) were significantly down-regulated in both T1D and T2D, and, although their fold-change fell below a 0.5 threshold, this provides evidence that both diseases are associated with the alteration of alpha cell identity and function. Other TFs linked to alpha cell development and function (Gromada et al., 2007), i.e. RFX6, PAX6, FOXA2, NEUROD1, and ISL1, were found significantly dysregulated in T1D: while the change in RFX6 expression was minimal, all the other TFs were up-regulated. In T2D we found a significant change only for ISL1, whereas all the other TFs were down-regulated, although with low significance.
Figure 4.
Comparison of differentially expressed genes in T1D and T2D alpha cells
Alpha cells from T1D (top left) and T2D (top right) datasets were separately analyzed contrasting the expression signatures of cells from affected and ND individuals. The differentially expressed genes (DEGs) were compared to identify shared and dataset-specific DEGs. The violin plots report the expression values of these such genes. It should be noted that the expression values reported are not corrected for the effect of covariates.
Examining the extent of the overlap between Gene Set Enrichment Analysis (GSEA) results, from a total of 2,974 terms significantly enriched either in T1D or T2D (Table S9), 506 were shared, 1,033 were unique to T1D and 1,435 to T2D. Considering only the positively enriched terms (in T1D, T2D, or both), there were 1,475 terms, 171 of which, in common; the negatively enriched ones were 1,781 in total, with 53 shared. There were 282 terms enriched in different directions: 261 of these were positively enriched in T1D and negatively in T2D, whereas 21 were shared between positively enriched in T2D and negatively in T1D. A breakdown of the overlapping terms in T1D and T2D (in different directions, and their combinations) across the different datasets used for GSEA is reported in Table S9.
Although the signatures retrieved for T1D and T2D indicate a partial overlap of stress response genes, we found some remarkable differences between the two series. In T1D, genes linked with ER stress and UPR are either significantly up-regulated (ERLEC1, HSP90) or with a significant change that is below the fold-change threshold, we used (ATF6, ER mannosidase I, TRAM1), an effect that was not observed in T2D (Figure S4). Concerning the insulin signaling pathway, the overall trend was for repression, which was more marked in T1D as compared to T2D (Figure S5). For instance, the downregulation of PI3K is doubled in T1D with respect to T2D, whereas IRS2 is up-regulated in T1D and down-regulated in T2D.
Another major difference between T1D and T2D involved metabolism i.e. glycolysis, citrate cycle, and oxidative phosphorylation (Figure S6). Notably, in T1D we observed little changes in the expression of genes involved in glycolysis and citrate cycle, but increased levels of those contributing to oxidative phosphorylation, while these pathways were repressed in T2D: most of the genes involved in glycolysis were down-regulated, and four with high significance (PKM, ENO1, ALDH2, TPI1) and three with high significance and fold-change (LDHA, ALDH9A1, and PDHB); strikingly, most of the citrate cycle genes were down-regulated in alpha cells from T2D donors, with six genes displaying a significant change; among the complexes involved in oxidative phosphorylation, genes contributing in complex III were the most negatively affected, with almost all genes being under-expressed in diseased alpha cells.
Discussion
Analysis of single-cell data sets provides a unique opportunity to better understand the changes at the level of the alpha cell which drive dysregulated glucagon secretion in T1D and T2D. In the present work, we used publicly available single-cell transcriptomic data from human islets to test whether the dysfunction of alpha cells in T1D and T2D had common bases or if the transcriptional signatures are more disease-specific. This revealed both shared features (i.e. down-regulation of genes related to alpha cell identity and function, and the up-regulation of stress response mechanisms and inflammation signatures) and, more importantly, disease-specific stress pathways. In T1D, several of the alpha cell alterations were traced back to ER stress, a process associated with chronic inflammation and autoimmune diseases. In T2D, up-regulation of ROS defense mechanisms (SBNO2, EGLN2, MBP) was accompanied by the modification of the central metabolism, with the repression of genes involved in glycolysis (LDHA, PDHB, PDK4), citrate cycle, and mitochondrial respiration (COX7B, NQO2, SUCLA2, UQCR10, SLC25A4). We note that some of these gene expression changes in T2D alpha cells are akin to findings reported in a recent study using a separate data set (Dai et al., 2022) which highlighted the alteration of mitochondrial respiratory complex genes in T2D and the inhibitory role of H2O2 in glucagon secretion. However, no data were provided in the latter report on alpha cells from patients with T1D, nor was any comparison made of changes in the two disease types.
We speculate that in the T2D down-regulation of glucose catabolism is adaptive to ROS stress, not only by leading to a decrease of radicals from oxidative phosphorylation but also by redirecting glucose flux to Pentose Phosphate Pathway, increasing NADPH to improve ROS scavenging (Mullarky and Cantley, 2015). However, the relationship here is complex as lowered LDHA (favoring pyruvate flux into mitochondria) and PDK4 (favoring PDH dephosphorylation and activation) would seem likely to oppose the impact of lowered PDHB activate (decreasing PDH-E1 levels and conversion of pyruvate into acetyl-CoA) (Figure S4). Measurements of the corresponding gene products at the protein level will be important to substantiate these mRNA-based findings (Figure S4). Of note, glucose oxidation is lowered in islets from T2D subjects (Del Guerra et al., 2005) though the relative contribution of changes in beta and alpha cells is not established. We also report that, among the respiratory chain genes, the most negatively down-regulated were those involved in mitochondrial complex III, a major producer of ROS with implications for cellular transduction (Bleier and Dröse, 2013). Additionally, and complementing the above mechanism, a decreased ability of mitochondria to synthesize ATP in response to elevated glucose concentrations (Ravier and Rutter, 2005) may contribute to the failure of the alpha cell in T2D to efficiently shut down glucagon secretion as glucose concentrations rise, consistent with other recent findings (Knudsen et al., 2019).
In T1D we observed signatures of ER stress, with most of the genes involved in ER protein processing being up-regulated (Figure S6). Considering that in T1D we did not observe repression of the central metabolism (Figure S4), the partial overlap of ROS response between T1D and T2D could be owing to the fact that ER stress increases the ROS cellular levels (Zeeshan et al., 2016). Related to ROS production, oxidative phosphorylation is enhanced in T1D, a striking difference with respect to T2D alpha cells in which the pathway is repressed. Finally, alpha cells in T1D display signatures of inflammation, including markers of cytokine exposure, with multiple related pathways among the most significantly enriched.
Besides a stress response, we reported for T1D the down-regulation of a number of genes involved with hormone secretion, with a likely relevance in the pathophysiological context. The apparent repression of PCSK2 transcription is consistent with reduced glucagon production, as well as the decreased expression of PCSK1N, a repressor of PCSK1. As this gene is involved with glucagon-like peptide 1 (GLP-1) production from glucagon (Rouillé et al., 1997), the pattern we observed would be consistent with increased endogenous production of GLP-1 and lower glucagon maturation. Also, the down-regulation of CHGA, encoding chromogranin A, implies impaired hormone secretion with a role in secretory granule biogenesis (Kim et al., 2001).
Another important aspect investigated is the extent to which genes involved in alpha cell identity changed in the islet from diabetic donors. We also assessed the expression of specific transcription factors involved with alpha cell development and function, i.e. RFX6, PAX6, FOXA2, FOXA1, NEUROD1, ISL1, and BRN4 (Gromada et al., 2007). These genes were differently regulated in alpha cells from T1D and T2D: while in T1D we found an up-regulation of PAX6, FOXA2, NEUROD1, and ISL1, in T2D there was a trend for all these genes to be down-regulated. These directionally opposing changes may thus contribute to the differences in overall gene expression in alpha cells in T1D versus T2D.
Finally, we found dysregulated genes that were previously associated with T1D and/or T2D but whose function in alpha cells remains unclear: for instance, the genes CTRB1 and CTRB2, down-regulated in both T1D and T2D, are genetically associated with T1D as well as incretin responsiveness in T2D (Barrett et al., 2009; ’t Hart et al., 2013). Further investigating such genes could be useful to determine their relevance in the context of alpha cell dysfunction and the mechanisms linking them to diabetes.
In conclusion, alpha cells in T1D and T2D display limited shared signatures, with the most notable common changes being the reduced expression of MAFB and ARX. We note that an earlier study (Russell et al., 2020) has explored the impact on the differentiation of pluripotent stem cells (PSCs) into islet endocrine cells of MAFB inactivation. Importantly the defective progenitors displayed lower rates of conversion to beta compared to alpha and delta cells, with GCG expression not significantly affected in MAFB null progenitors. Thus, changes in MAFB are unlikely to be the major driver of altered gene expression in alpha cells in T1D.
On the other hand, it is possible that there are other shared changes that were missed owing to the limitations of our study (see later in discussion). On the other hand, alpha cells present important disease-specific signatures in T1D and T2D, suggesting that these signatures and the consequent alpha cell dysfunction are secondary to the main pathogenic events characteristic to each disease, namely immune-mediated- or metabolic-mediated-stress, respectively, in T1D and T2D.
Limitations of the study
The present results should be considered within the context of the limitations associated with single-cell transcriptomics, particularly related to the limited number of genes than can be detected by this technique (i.e. around 5.000 genes for scRNAseq vs > 100.000 transcripts detected by deep sequencing bulk RNAseq) (Colli et al., 2020; Mawla and Huising, 2019). Other issues include relatively small sample size and variability over donors’ clinical features. As the availability of humanì islet single-cell transcriptomic data grows further, it will be important to validate the present findings with a larger number of samples and include proteomics and Western blot analysis to confirm the translation of the differentially regulated transcripts.
The alterations we described as associated with T1D and T2D could include not only pathogenic but also compensatory mechanisms. For instance, in T2D we reported a significant down-regulation of a number of factors associated with alpha cell development and function (PAX6, FOXA2, NEUROD1, and ISL1). Interestingly, a previous study identified a subpopulation of alpha cells in mice and T2D donors in which the expression of these genes was inversely correlated with alpha cell function (Dai et al., 2022). It is thus conceivable that the differential expression we reported represents a compensatory mechanism.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Single-cell RNA seq data | The Human Pancreas Analysis Program - HPAP | HPAP-012 |
| Single-cell RNA seq data | The Human Pancreas Analysis Program - HPAP | HPAP-034 |
| Single-cell RNA seq data | The Human Pancreas Analysis Program - HPAP | HPAP-036 |
| Single-cell RNA seq data | The Human Pancreas Analysis Program - HPAP | HPAP-039 |
| Single-cell RNA seq data | The Human Pancreas Analysis Program - HPAP | HPAP-052 |
| Single-cell RNA seq data | The Human Pancreas Analysis Program - HPAP | HPAP-056 |
| Single-cell RNA seq data | The Human Pancreas Analysis Program - HPAP | HPAP-015 |
| Single-cell RNA seq data | The Human Pancreas Analysis Program - HPAP | HPAP-020 |
| Single-cell RNA seq data | The Human Pancreas Analysis Program - HPAP | HPAP-021 |
| Single-cell RNA seq data | The Human Pancreas Analysis Program - HPAP | HPAP-023 |
| Single-cell RNA seq data | The Human Pancreas Analysis Program - HPAP | HPAP-028 |
| Single-cell RNA seq data | The Human Pancreas Analysis Program - HPAP | HPAP-032 |
| Single-cell RNA seq data | The Human Pancreas Analysis Program - HPAP | HPAP-055 |
| Single-cell RNA seq data | The Human Pancreas Analysis Program - HPAP | HPAP-053 |
| Single-cell RNA seq data | The Human Pancreas Analysis Program - HPAP | HPAP-054 |
| Single-cell RNA seq data | The Human Pancreas Analysis Program - HPAP | HPAP-059 |
| Single-cell RNA seq data | The Human Pancreas Analysis Program - HPAP | HPAP-006 |
| Single-cell RNA seq data | The Human Pancreas Analysis Program - HPAP | HPAP-014 |
| Single-cell RNA seq data | The Human Pancreas Analysis Program - HPAP | HPAP-051 |
| Single-cell RNA seq data | The Human Pancreas Analysis Program - HPAP | HPAP-057 |
| Single-cell RNA seq data | The Human Pancreas Analysis Program - HPAP | HPAP-058 |
| Single-cell RNA seq data | The Human Pancreas Analysis Program - HPAP | HPAP-001 |
| Single-cell RNA seq data | The Human Pancreas Analysis Program - HPAP | HPAP-007 |
| Software and algorithms | ||
| STAR 2.7.3a | https://github.com/alexdobin/STAR | |
| Custom python and R software used to generate results | https://github.com/EBosi/scPanBetaT2D | scPanBetaT2D |
| Other | ||
| Human reference genome with ENSEMBL annotation | http://ftp.ensembl.org/pub/grch37/ | GRCh37 (release 87) |
| GO Biological Process gene sets | mSigDB 7.4 | c5.go.bp.v7.2.symbols.gmt |
| GO Molecular Function gene sets | mSigDB 7.4 | c5.go.mf.v7.2.symbols.gmt |
| GO Cellular Component gene sets | mSigDB 7.4 | c5.go.cc.v7.2.symbols.gmt |
| Reactome gene sets | mSigDB 7.4 | c2.cp.reactome.v7.2.symbols.gmt |
| KEGG gene sets | mSigDB 7.4 | c2.cp.kegg.v7.2.symbols.gmt |
| mSigDB Hallmark gene sets | mSigDB 7.4 | h.all.v7.2.symbols.gmt |
Resource availability
Lead contact
Further information and requests should be directed to and will be fulfilled by the lead contact, Emanuele Bosi (bosiemanuele@gmail.com).
Materials availability
This study did not generate new unique reagents.
Method details
Analysis of single-cell data and integration with the other datasets
Fastq files and the corresponding metadata were downloaded from the database of the Human Pancreas Analysis Program (HPAP) (https://hpap.pmacs.upenn.edu). Reads were aligned using STAR 2.7.3a (Dobin et al., 2013) against the human reference genome GRCh37 (Ensembl 87 annotation) with different parameters according to the technology used for library production. For the reads from libraries prepared with Fluidigm 800 cell IFC, the mapping parameters were “--quantMode TranscriptomeSAM GeneCounts, --outSAMmultNmax −1”; for the other samples, prepared using 10X with chemistry Single Cell 3′ v2 or v3, the parameters used were “--soloType Droplet --soloUMIfiltering MultiGeneUMI --soloCBmatchWLtype 1MM_multi_pseudocounts --outSAMmultNmax −1 --outSAMtype BAM SortedByCoordinate --quantMode TranscriptomeSAM GeneCounts”. The arguments of two parameters, “--soloCBwhitelist” and “--soloUMIlen“, were different for the 10X chemistry kits: “737K-august-2016.txt” and “10” for v2; “3M-february-2018.txt” and “12” for v3.
After the read mapping step, read count tables were analyzed with ad-hoc python scripts implementing the toolbox Scanpy (Wolf et al., 2018). In particular, cell-wise and gene-wise metrics were computed for QC to define excluding criteria of low-quality cells, similarly as previously done (Bosi et al., 2020) and in line with existing guidelines (Luecken and Theis, 2019). The parameters considered for each cell were: i) the number of genes with at least one read mapped (expressed genes); ii) the number of reads mapped to genes (counts); iii) the ratio of reads mapped on mitochondrial genes (mitochondrial fraction). Additionally, genes with detected expression in three or fewer cells were excluded from the analyses. These values were used to exclude cells with high counts and expressed genes (likely representing multiplets), and with high mitochondrial fraction and low expressed genes (indicative of lysed cells). These variables and their covariation were considered separately for each sample, as recommended when the distributions of QC covariates differ between samples (Luecken and Theis, 2019), and as done in previous studies (e.g. (Plasschaert et al., 2018)). These distributions allowed to define, separately for each sample, threshold values to flag the cells with at least one QC metric below the threshold as “low quality”. A QC scheme reporting the strategy used to define the thresholds is reported in Figure S1.
After QC, the processed samples were concatenated and the counts normalized to a total of 10,000 for each cell, then log-transformed. The dispersion of each gene with respect to its mean value was computed in the integrated dataset to annotate highly variable genes using the homonymous Scanpy function. The resulting set of genes displaying high variability was used to perform dataset integration with Batch Balanced KNN (BBKNN) (Polański et al., 2020), similarly as done by Park and colleagues (Park et al., 2020). Briefly, BBKNN was first used with the HPAP id as batch key, followed by a Louvain clustering (Lu et al., 2015) on the corrected dataset. Then, the obtained clusters were used as biological covariates to perform a ridge regression on the adjusted data, followed by a further BBKNN correction. The transcriptomes of single-cells were visualized using UMAP.
Single-cell clusters were identified with the Louvain modularity algorithm (Lu et al., 2015) as implemented in Scanpy (https://github.com/vtraag/louvain-igraph) with resolution = 0.5, with further sub-clustering for groups of interest. The genes with cluster-specific expression were found with the “rank_genes_groups” function of Scanpy, curating their association with known cell types using literature information and gene expression markers reported in PanglaoDB (Franzén et al., 2019).
Quantification and statistical analysis
Differential expression and gene set enrichment analysis
The DEGs in T1D and T2D as compared to the corresponding controls were identified using MAST (Finak et al., 2015) with the following mixed effect model: Counts ∼ Diabetes + ngenes + Technology + Race + Sex + BMI + Age, including also a random effect estimated for each individual (Individual). Since the estimation of such an effect may be hampered by a low number of observations, individuals with ≤50 alpha cells (after filtering for QC criteria) were excluded from the analysis. Counts is the matrix of raw count data, filtered of genes being expressed in less than 20% of the cells with the filterLowExpressedGenes function, ngenes is a variable encoding the number of expressed genes and Diabetes is a 2-level factor (Disease, ND) indicating the diabetes status of the donor. Genes with corrected p value (Benjamini-Hochberg, FDR) lower than 0.05 and absolute fold-change (FC) greater or equal than 0.5 were considered as DEGs. The robustness of the results obtained with MAST was assessed by obtaining a “pooled” control group by aggregating the transcriptomes from ND donors of both groups (i.e the ones compared against respectively T1D and T2D).
GSEA was performed for the following datasets: i) Gene Ontology (GO), separately for Biological Process (GO BP), Molecular Function (GO MF) and Cellular Component (GO CC) (Ashburner et al., 2000; The Gene Ontology Consortium, 2019); ii) KEGG (Kanehisa, 2019; Kanehisa et al., 2019; Kanehisa and Goto, 2000); iii) Reactome 2016 (Jassal et al., 2020); the Hallmark collection from mSigDB (Liberzon et al., 2015). All gene sets were obtained from mSigDB (Subramanian et al., 2005) (version 7.4) and used to perform GSEA on the hurdle distributions obtained with MAST: the “gseaAfterBoot” function has been used on 50 distributions (bootstraps) derived with the “bootVcov1” function.
Acknowledgments
This work was supported by non-profit organisations and public bodies for funding of scientific research conducted within the European Union: the Innovative Medicines Initiative 2 Joint Undertaking, RHAPSODY [115881 to EB, DLE, GAR, PM], INNODIA [115797 to EB, DLE, PM] and INNODIA HARVEST [945268 to DLE, PM] - this Joint Undertaking receives support from the Union’s Horizon 2020 research and innovation programme, “EFPIA,” “JDRF” and “The Leona M. and Harry B. Helmsley Charitable Trust" (INNODIA, INNODIA HARVEST), the "EFPIA" and the Swiss State Secretariat for Education, Research and Innovation under contract number 16.0097 (RHAPSODY); the European External Action Service Horizon 2020 research and innovation programme, project T2DSystems [667191 to EB, DLE, PM]; the Walloon Region through the FRFS-WELBIO Fund for Strategic Fundamental Research [grant numbers CR-2015A-06s, CR-2019C-04 to DLE]; the Welbio-Fonds National de la Recherche Scientifique, Belgium and Dutch Diabetes Fonds, Holland [2018.10.002 to DLE]; the Brussels Capital Region-Innoviris project Diatype [2017-PFS-24 to DLE]. GAR was supported by a Wellcome Trust Investigator Award (212625/Z/18/Z), MRC Programme grant (MR/R022259/1) and a start-up grant from the CR-CHUM, Université de Montréal.
This article used data acquired from the Human Pancreas Analysis Program (HPAP-RRID:SCR_016202) Database (https://hpap.pmacs.upenn.edu), a Human Islet Research Network (RRID:SCR_014393) consortium (UC4-DK-112217, U01-DK-123594, UC4-DK-112232, and U01-DK-123716).
Author contributions
EB, PM, and DLE conceptualized the work and designed the analyses. EB analyzed the data. EB, PM, GR, and DLE interpreted and discussed the results and prepared the article.
Declaration of interests
The authors declare no competing interests.
Published: October 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105056.
Supplemental information
Data and code availability
-
•
This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table.
-
•
The data and code used for this work has been archived in the GitHub repository at https://github.com/EBosi/scPanAlpha.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- American Diabetes Association Diagnosis and classification of diabetes mellitus. Diabetes Care. 2010;33:S62–S69. doi: 10.2337/dc10-S062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson S.A., Olsson A.H., Esguerra J.L.S., Heimann E., Ladenvall C., Edlund A., Salehi A., Taneera J., Degerman E., Groop L., et al. Reduced insulin secretion correlates with decreased expression of exocytotic genes in pancreatic islets from patients with type 2 diabetes. Mol. Cell. Endocrinol. 2012;364:36–45. doi: 10.1016/j.mce.2012.08.009. [DOI] [PubMed] [Google Scholar]
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J.C., Clayton D.G., Concannon P., Akolkar B., Cooper J.D., Erlich H.A., Julier C., Morahan G., Nerup J., Nierras C., et al. Type 1 Diabetes Genetics Consortium Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleier L., Dröse S. Superoxide generation by complex III: from mechanistic rationales to functional consequences. Biochim. Biophys. Acta. 2013;1827:1320–1331. doi: 10.1016/j.bbabio.2012.12.002. [DOI] [PubMed] [Google Scholar]
- Bosco D., Armanet M., Morel P., Niclauss N., Sgroi A., Muller Y.D., Giovannoni L., Parnaud G., Berney T. Unique arrangement of alpha- and beta-cells in human islets of Langerhans. Diabetes. 2010;59:1202–1210. doi: 10.2337/db09-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosi E., Marselli L., De Luca C., Suleiman M., Tesi M., Ibberson M., Eizirik D.L., Cnop M., Marchetti P. Integration of single-cell datasets reveals novel transcriptomic signatures of β-cells in human type 2 diabetes. NAR Genom. Bioinform. 2020;2:lqaa097. doi: 10.1093/nargab/lqaa097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brissova M., Haliyur R., Saunders D., Shrestha S., Dai C., Blodgett D.M., Bottino R., Campbell-Thompson M., Aramandla R., Poffenberger G., et al. α cell function and gene expression are compromised in type 1 diabetes. Cell Rep. 2018;22:2667–2676. doi: 10.1016/j.celrep.2018.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J.E., Newgard C.B. Mechanisms controlling pancreatic islet cell function in insulin secretion. Nat. Rev. Mol. Cell Biol. 2021;22:142–158. doi: 10.1038/s41580-020-00317-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colli M.L., Ramos-Rodríguez M., Nakayasu E.S., Alvelos M.I., Lopes M., Hill J.L.E., Turatsinze J.-V., Coomans de Brachène A., Russell M.A., Raurell-Vila H., et al. An integrated multi-omics approach identifies the landscape of interferon-α-mediated responses of human pancreatic beta cells. Nat. Commun. 2020;11:2584. doi: 10.1038/s41467-020-16327-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X.-Q., Camunas-Soler J., Briant L.J.B., Dos Santos T., Spigelman A.F., Walker E.M., Arrojo E Drigo R., Bautista A., Jones R.C., Avrahami D., et al. Heterogenous impairment of α cell function in type 2 diabetes is linked to cell maturation state. Cell Metabol. 2022;34:256–268.e5. doi: 10.1016/j.cmet.2021.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Guerra S., Lupi R., Marselli L., Masini M., Bugliani M., Sbrana S., Torri S., Pollera M., Boggi U., Mosca F., et al. Functional and molecular defects of pancreatic islets in human type 2 diabetes. Diabetes. 2005;54:727–735. doi: 10.2337/diabetes.54.3.727. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eizirik D.L., Cardozo A.K., Cnop M. The role for endoplasmic reticulum stress in diabetes mellitus. Endocr. Rev. 2008;29:42–61. doi: 10.1210/er.2007-0015. [DOI] [PubMed] [Google Scholar]
- Eizirik D.L., Pasquali L., Cnop M. Pancreatic β-cells in type 1 and type 2 diabetes mellitus: different pathways to failure. Nat. Rev. Endocrinol. 2020;16:349–362. doi: 10.1038/s41574-020-0355-7. [DOI] [PubMed] [Google Scholar]
- Finak G., McDavid A., Yajima M., Deng J., Gersuk V., Shalek A.K., Slichter C.K., Miller H.W., McElrath M.J., Prlic M., et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzén O., Gan L.-M., Björkegren J.L.M. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database. 2019;2019 doi: 10.1093/database/baz046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta M., Zhou A., Webb G., Carroll R., Ravazzola M., Orci L., Steiner D.F. Severe defect in proglucagon processing in islet A-cells of prohormone convertase 2 null mice. J. Biol. Chem. 2001;276:27197–27202. doi: 10.1074/jbc.M103362200. [DOI] [PubMed] [Google Scholar]
- Gerich J.E., Langlois M., Noacco C., Karam J.H., Forsham P.H. Lack of glucagon response to hypoglycemia in diabetes: evidence for an intrinsic pancreatic alpha cell defect. Science. 1973;182:171–173. doi: 10.1126/science.182.4108.171. [DOI] [PubMed] [Google Scholar]
- Gromada J., Chabosseau P., Rutter G.A. The α-cell in diabetes mellitus. Nat. Rev. Endocrinol. 2018;14:694–704. doi: 10.1038/s41574-018-0097-y. [DOI] [PubMed] [Google Scholar]
- Gromada J., Franklin I., Wollheim C.B. Alpha-cells of the endocrine pancreas: 35 years of research but the enigma remains. Endocr. Rev. 2007;28:84–116. doi: 10.1210/er.2006-0007. [DOI] [PubMed] [Google Scholar]
- Hasan K., Cheung C., Kaul Z., Shah N., Sakaushi S., Sugimoto K., Oka S., Kaul S.C., Wadhwa R. CARF Is a vital dual regulator of cellular senescence and apoptosis. J. Biol. Chem. 2009;284:1664–1672. doi: 10.1074/jbc.M805778200. [DOI] [PubMed] [Google Scholar]
- Jassal B., Matthews L., Viteri G., Gong C., Lorente P., Fabregat A., Sidiropoulos K., Cook J., Gillespie M., Haw R., et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020;48:D498–D503. doi: 10.1093/nar/gkz1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaestner K.H., Powers A.C., Naji A., HPAP Consortium. Atkinson M.A. NIH initiative to improve understanding of the pancreas, islet, and autoimmunity in type 1 diabetes: the human pancreas analysis Program (HPAP) Diabetes. 2019;68:1394–1402. doi: 10.2337/db19-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28:1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Sato Y., Furumichi M., Morishima K., Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019;47:D590–D595. doi: 10.1093/nar/gky962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M.C., Jung Y., Ugboma C.M., Shimbo M., Kuno A., Basha W.A., Kudo T., Oishi H., Takahashi S. MafB is critical for glucagon production and secretion in mouse pancreatic α cells in vivo. Mol. Cell Biol. 2018;38:e00504-17. doi: 10.1128/MCB.00504-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahito S., Kitahata H., Oshita S. Problems associated with glucose toxicity: role of hyperglycemia-induced oxidative stress. World J. 2009 doi: 10.3748/wjg.15.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T., Tao-Cheng J.-H., Eiden L.E., Loh Y.P. Chromogranin A, an “On/Off” switch controlling dense-core secretory granule biogenesis. Cell. 2001;106:499–509. doi: 10.1016/s0092-8674(01)00459-7. [DOI] [PubMed] [Google Scholar]
- Knudsen J.G., Hamilton A., Ramracheya R., Tarasov A.I., Brereton M., Haythorne E., Chibalina M.V., Spégel P., Mulder H., Zhang Q., et al. Dysregulation of glucagon secretion by hyperglycemia-induced sodium-dependent reduction of ATP production. Cell Metabol. 2019;29:430–442.e4. doi: 10.1016/j.cmet.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luecken M.D., Theis F.J. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol. Syst. Biol. 2019;15:e8746. doi: 10.15252/msb.20188746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Halappanavar M., Kalyanaraman A. Parallel heuristics for scalable community detection. Parallel Comput. 2015;47:19–37. [Google Scholar]
- Lun A.T.L., Riesenfeld S., Andrews T., Dao T.P., Gomes T., Participants in the 1st Human Cell Atlas Jamboree EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 2019;20:63. doi: 10.1186/s13059-019-1662-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti P., Suleiman M., De Luca C., Baronti W., Bosi E., Tesi M., Marselli L. Seminars in Cell & Developmental Biology. Elsevier; 2020. A direct look at the dysfunction and pathology of the β cells in human type 2 diabetes; pp. 83–93. [DOI] [PubMed] [Google Scholar]
- Mawla A.M., Huising M.O. Navigating the depths and avoiding the shallows of pancreatic islet cell transcriptomes. Diabetes. 2019;68:1380–1393. doi: 10.2337/dbi18-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullarky E., Cantley L.C. Diverting glycolysis to combat oxidative stress. Innovat. Med. 2015:3–23. [PubMed] [Google Scholar]
- Ohoka N., Yoshii S., Hattori T., Onozaki K., Hayashi H. TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J. 2005;24:1243–1255. doi: 10.1038/sj.emboj.7600596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.-E., Botting R.A., Domínguez Conde C., Popescu D.-M., Lavaert M., Kunz D.J., Goh I., Stephenson E., Ragazzini R., Tuck E., et al. A cell atlas of human thymic development defines T cell repertoire formation. Science. 2020;367:eaay3224. doi: 10.1126/science.aay3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasschaert L.W., Žilionis R., Choo-Wing R., Savova V., Knehr J., Roma G., Klein A.M., Jaffe A.B. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature. 2018;560:377–381. doi: 10.1038/s41586-018-0394-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polański K., Young M.D., Miao Z., Meyer K.B., Teichmann S.A., Park J.-E. BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics. 2020;36:964–965. doi: 10.1093/bioinformatics/btz625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravier M.A., Rutter G.A. Glucose or insulin, but not zinc ions, inhibit glucagon secretion from mouse pancreatic alpha-cells. Diabetes. 2005;54:1789–1797. doi: 10.2337/diabetes.54.6.1789. [DOI] [PubMed] [Google Scholar]
- Rouillé Y., Kantengwa S., Irminger J.-C., Halban P.A. Role of the prohormone convertase PC3 in the processing of proglucagon to glucagon-like peptide 1. J. Biol. Chem. 1997;272:32810–32816. doi: 10.1074/jbc.272.52.32810. [DOI] [PubMed] [Google Scholar]
- Russell R., Carnese P.P., Hennings T.G., Walker E.M., Russ H.A., Liu J.S., Giacometti S., Stein R., Hebrok M. Loss of the transcription factor MAFB limits β-cell derivation from human PSCs. Nat. Commun. 2020;11:2742. doi: 10.1038/s41467-020-16550-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador J.M., Brown-Clay J.D., Fornace A.J., Jr. Gadd45 in stress signaling, cell cycle control, and apoptosis. Adv. Exp. Med. Biol. 2013;793:1–19. doi: 10.1007/978-1-4614-8289-5_1. [DOI] [PubMed] [Google Scholar]
- Shcherbik N., Pestov D.G. The impact of oxidative stress on ribosomes: from injury to regulation. Cells. 2019;8 doi: 10.3390/cells8111379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ’t Hart L.M., Fritsche A., Nijpels G., van Leeuwen N., Donnelly L.A., Dekker J.M., Alssema M., Fadista J., Carlotti F., Gjesing A.P., et al. The CTRB1/2 locus affects diabetes susceptibility and treatment via the incretin pathway. Diabetes. 2013;62:3275–3281. doi: 10.2337/db13-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47:D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.-E., Han C., Zhang B., Sabapathy K., Wani A.A. Nucleotide excision repair factor XPC enhances DNA damage-induced apoptosis by downregulating the antiapoptotic short isoform of caspase-2. Cancer Res. 2012;72:666–675. doi: 10.1158/0008-5472.CAN-11-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf F.A., Angerer P., Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Chen J., Jing G., Shalev A. Thioredoxin-interacting protein regulates insulin transcription through microRNA-204. Nat. Med. 2013;19:1141–1146. doi: 10.1038/nm.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H., Wang H.-G. CHOP is involved in endoplasmic reticulum stress-induced apoptosis by enhancing DR5 expression in human carcinoma cells. J. Biol. Chem. 2004;279:45495–45502. doi: 10.1074/jbc.M406933200. [DOI] [PubMed] [Google Scholar]
- Yang S., Corbett S.E., Koga Y., Wang Z., Johnson W.E., Yajima M., Campbell J.D. Decontamination of ambient RNA in single-cell RNA-seq with DecontX. Genome Biol. 2020;21:57. doi: 10.1186/s13059-020-1950-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.-Y., Lee J.-J., Lee J.-H., Lee K., Oh S.H., Lim Y.-M., Lee M.-S., Lee K.-J. Secretagogin affects insulin secretion in pancreatic β-cells by regulating actin dynamics and focal adhesion. Biochem. J. 2016;473:1791–1803. doi: 10.1042/BCJ20160137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.D., Behjati S. SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. GigaScience. 2020;9:giaa151. doi: 10.1093/gigascience/giaa151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeshan H.M.A., Lee G.H., Kim H.-R., Chae H.-J. Endoplasmic reticulum stress and associated ROS. Int. J. Mol. Sci. 2016;17:327. doi: 10.3390/ijms17030327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table.
-
•
The data and code used for this work has been archived in the GitHub repository at https://github.com/EBosi/scPanAlpha.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.