Abstract
Pharmacogenomic testing can be an effective tool to enhance medication safety and efficacy. Pharmacogenomically actionable medications are widely used, and approximately 90–95% of individuals have an actionable genotype for at least one pharmacogene. For pharmacogenomic testing to have the greatest impact on medication safety and clinical care, genetic information should be made available at the time of prescribing (preemptive testing). However, the use of preemptive pharmacogenomic testing is associated with some logistical concerns, such as consistent reimbursement, processes for reporting preemptive results over an individual’s lifetime, and result portability. Lessons can be learned from institutions that have implemented preemptive pharmacogenomic testing. In this review, we discuss the rationale and best practices for implementing pharmacogenomics preemptively.
Keywords: pharmacogenetics, pharmacogenomics, precision medicine, individualized medicine, personalized medicine, clinical decision support, genomic medicine
INTRODUCTION
Pharmacogenomics is the study of how an individual’s genetic makeup influences their response to medications. Although the role of genetics in drug response has been studied for decades, broad pharmacogenomic testing has only recently been integrated into prescribing decisions. Resources from groups such as the Clinical Pharmacogenetics Implementation Consortium (CPIC) and the Dutch Pharmacogenetics Working Group (DPWG) have allowed for more informed integration of pharmacogenomics into prescribing decisions (9, 25, 91, 111, 112), but different perspectives on the evidence needed for implementation of drug–gene pairs exist (23, 67, 87). Furthermore, pharmacogenomics implementation approaches vary regarding the timing of when the test should be ordered. Some advocate that pharmacogenomic testing should be reactive and obtained only for certain medications prior to prescribing or after a patient has had an adverse reaction to the medication or is failing therapy. Others advocate for using a preemptive pharmacogenomic testing approach as a prevention and medication safety tool. In this review, we compare and contrast reactive and preemptive testing and discuss the rationale for implementing pharmacogenomics preemptively along with best practices for using preemptive pharmacogenomics as a preventive tool.
EVIDENCE AND RESOURCES FOR PHARMACOGENOMIC TESTING
The level of evidence needed to support the use of pharmacogenomics in routine clinical practice remains controversial (23, 67, 87). Although randomized clinical trials (RCTs) are considered the gold standard for evidence-based medicine, many argue that, for pharmacogenomics, RCTs are unnecessary and may be impractical because of their cost and the number of patients needed. In some cases, RCTs may even be unethical (23, 67, 87). Many decisions made for patient care are not based on RCT data, with notable examples being dosing adjustments based on renal dysfunction, liver dysfunction, drug interactions, and other patient-specific characteristics. For example, regulatory label requirements do not mandate that RCTs show equivalent clinical outcomes for dosing adjustments based on renal function (129).
In the absence of RCTs, pragmatic trials and observational studies are often used as evidence supporting the use of pharmacogenomics in patient care. Although observational data are prone to bias, they do have advantages over RCTs: Pragmatic and observational trials are conducted in the context of clinical practice, making them more generalizable, and they may be less costly than RCTs (12). Furthermore, the evidence paradigm for medication use and regulation is increasingly embracing data beyond RCTs (88, 105, 107, 129b). For example, as part of the 21st Century Cures Act, the US Food and Drug Administration (FDA) was tasked to consider how to use real-world evidence (which typically consists of observational data) to support new indications and other post approval data requirements, and the FDA released a framework for use of these data (88, 105, 107, 129b). Finally, pharmacogenomics should be viewed as a patient safety intervention, and RCTs are not always performed for many widely used patient safety strategies (23, 111).
Current resources, such as CPIC and DPWG guidelines, provide recommendations for how to use available pharmacogenomic test results and do not focus on whether ordering a pharmacogenomic test is required. In most cases, institutions implementing pharmacogenomics use the CPIC and/or DPWG guidelines to guide gene–drug selection for implementation and prescribing recommendations (31c, 38). CPIC guidelines are based on published evidence, adhere to National Academy of Medicine standards for clinical guidelines, and are updated, freely available, and peer reviewed. Furthermore, the data needed for many steps in clinical implementation can be downloaded from the CPIC database (25, 31a).
CHARACTERISTICS OF PHARMACOGENOMIC TESTING
Reactive Testing
Reactive testing is prompted by a medication prescribing decision, either one that has already taken place or one that is pending (Table 1). Reactive testing has the advantage that it is simple and consistent with what is often routine practice: Consider a therapeutic intervention and do appropriate testing to decide whether the intervention should be undertaken. Reactive pharmacogenomic tests are often single-gene tests because most actionable pharmacogenomic decisions are based on evidence regarding variants in a single gene. Because the results are not yet available at the time of prescribing, the initial prescription may need to change after the results are known. Depending on the turnaround time, this delay could be problematic, as it may be difficult to revise the prescription in a timely fashion if the test results require a change in pharmacotherapy. Quickly changing a prescription may be especially complex in an ambulatory setting after the prescription has been ordered and dispensed. For some medications, harm may come to the patient because of either drug toxicity or a lack of efficacy while the test result is still pending, when the patient could be receiving inappropriate therapy. Single-gene tests are inefficient in that panel testing for all actionable pharmacogenes can be performed at just slightly higher costs and using the same DNA as single-gene testing (147). Reactive testing is contrasted with preemptive testing, in which test results are available prior to medication prescribing decisions, often by using multigene arrays or DNA sequencing technologies (36, 38).
Table 1.
Common features of reactive versus preemptive pharmacogenomic testing
| Feature | Reactive pharmacogenomic testing | Preemptive pharmacogenomic testing |
|---|---|---|
| Timing of ordering | Ordered as drug therapy is initiated or being contemplated | Ordered independently of medication use |
| Turnaround time | Often ~5–7 business days; can be a point-of-care test (only available for a limited set of genes; use is not widespread) | Test result already available in the electronic health record at the time of prescribing; results can be reused as other medications are prescribed |
| Prescriber knowledge about pharmacogenomics | Requires knowledge about which test to order | Clinical decision support alerts prompt prescriber for action |
| Cost | Routinely found to be cost effective or cost saving, but expensive relative to the potential benefit of one therapeutic decision | Approximately equivalent to the cost of two single-gene tests, and provides cost savings over years with continued use of genetic information for medication use |
| General testing platform used | Often single gene (one pharmacogenomic test result) | Often array based (multiple pharmacogenomic test results) |
Point-of-care testing.
Point-of-care testing is a type of reactive testing. Technically, point-of-care testing is performed at or near the patient (e.g., blood glucose monitoring tests). In some acute care settings, it may be important for a quick turnaround time to physically perform the test using an instrument that is in a clinic or inpatient unit. There are a few examples, such as anticipating the need for clopidogrel antiplatelet therapy after percutaneous coronary intervention, when the availability of quick turnaround times in or near the coronary procedure room could facilitate prescribing decisions needed the same day as the procedure (31, 101, 104, 113). Point-of-care testing has also been employed for warfarin (analyzing variants in CYP2C9 and VKORC1), another example when the decision to prescribe and dose the medication is somewhat urgent (44). Point-of-care testing has also been proposed for HLA-B*57:01 with abacavir and for TPMT with thiopurines (140). Issues of technical performance for point-of-care devices have been reviewed (137).
Single-gene testing.
Most reactive pharmacogenomic tests are done for a single gene, or occasionally two or three genes, and are ordered in the context of prescribing a specific medication or class of medications (45). The techniques used to interrogate the single gene can vary, from interrogating only a handful of variants in that gene to complete gene sequencing. Because germline pharmacogenomic tests for an individual can be performed from DNA extracted from a single sample (usually blood or saliva) without regard to timing, sending multiple samples for interrogation of multiple single genes over a patient’s lifetime is inherently redundant and an inefficient use of the patient’s DNA sample (147). Some clinical laboratories even market medication-specific pharmacogenomic tests, treating tests for the same gene as different tests depending on the medication being contemplated; for example, a CYP2D6 tamoxifen test may be marketed separately from a CYP2D6 antidepressant test or a CYP2D6-containing pain panel, even though the gene test itself is identical. Some clinical laboratories have their sample tracking and validation procedures linked to obtaining a new sample for each pharmacogenomic test, and reimbursement incentives favor obtaining a new sample for each gene test (72, 143, 147). Thus, single-gene tests continue to dominate the clinical landscape of genomic testing.
Single-gene tests imply a practice model in which genes are contemplated one at a time as each new medication is considered for the patient. With adequate interpretation and workflows, single-gene test results may be reused over time for multiple affected medications that may be ordered for the patient over time.
Single-gene tests can be ordered preemptively (pre-prescription). Examples include testing for DPYD before starting fluoropyrimidine-based chemotherapy (57), testing for CYP2C9 and VKORC1 before prescribing warfarin (80), and testing for HLA before prescribing abacavir or carbamazepine (146). Despite their inefficiency compared with multigene panels, single-gene tests have been shown to be cost effective or cost saving (14). The cost effectiveness is frequently sensitive to pharmacogenomic test cost, which continues to decline, and the adverse effects that might be caused without testing are expensive to manage or treat, such as severe neutropenia in patients with cancer and increased cardiovascular adverse events in patients with acute coronary syndrome (33, 148).
Preemptive Testing
Preemptive testing means that the test result is available in the medical record pre-prescription; this could involve a single-gene test ordered preemptively because the patient is likely to need an affected medication in the future (e.g., TPMT testing for all patients with an autoimmune disease likely to need a thiopurine), but more commonly the test result is available preemptively because it was included in a broad panel of multiple genes that has already been performed (38) (Table 1). Preemptive testing has several advantages over reactive, single-gene testing (1, 114, 120). One is efficiency: Ordering a single array of multiple actionable pharmacogenes avoids the need to perform many single-gene tests, and often the cost of a multigene test is not much greater than the cost of a single-gene test. This is partly because some of the cost of genetic testing is ascribed to obtaining the patient sample and isolating DNA. Another advantage is preemptive multigene testing avoids the delay that would be incurred by reactive single-gene testing: Instead of needing to order a gene test and then wait for the individual results, the results are already available in the patient’s medical record at the time of the prescribing decision.
It should be mentioned that pharmacogenomic testing may be initiated by the patient, especially as direct-to-consumer testing becomes more common. A major challenge for the healthcare system will be to accommodate incorporating relevant information from patient-initiated testing into the clinical workflow (43, 50, 54).
Multiple types of pharmacogenomic testing panels are available from clinical laboratories. These can include commercially available arrays, bundles of tests performed with a single technology (e.g., PCR), whole-exome or whole-genome sequencing, or hybrid models using a combination of technologies. The panels can be agnostic with regard to medication category (e.g., panels containing all actionable pharmacogenes) or can be disease specific (e.g., genes relevant for treating colon cancer, breast cancer, cardiovascular disease, or psychiatric disorders). Cancer gene panels that might include the somatic cancer-specific lesions that drive certain targeted therapies as well as likely important germline pharmacogenes have been proposed (71). It should be noted that a challenge for those offering panel testing (via an array or via DNA sequencing) is that several pharmacogenes present technical challenges, including HLA genes and CYP2D6, with the latter being subject to common partial- or whole-gene duplications and rearrangements (34, 70, 100, 117).
Although panel-based testing is accompanied by the complications of providing interpretations for many genes, and the costs incurred and methods needed to update interpretations over time have not been fully addressed, even limited panels have some evidence for cost effectiveness relative to usual care. For example, a preemptive panel with 30 test-return days that included the gene–drug pairs CYP2C19–clopidogrel, CYP2C9/VKORC1–warfarin, DPYD–fluoropyrimidines, and SLCO1B1–statins was found to be cost effective compared with usual care (33, 147).
Panel-based tests, particularly those based on DNA sequencing, present the problem that clinicians must decide how to handle the inclusion of variants whose actionability is uncertain. Certain institutions mask the results of genes or variants whose actionability is uncertain, whereas others return results for all tested variants to the electronic health record (EHR) (56). Some have argued that pharmacogenomic panel-based clinical testing should be restricted to clinically validated variants and that comprehensive testing should be used only for research purposes (79). Moreover, some panels include not only variants of unknown significance but even genes of unknown actionability. Returning such results to the medical record could place clinicians in the difficult position of having unimportant results obscure the important ones or of failing to act on results that might in the future become actionable. There are multiple descriptions of how panel-based preemptive testing has been handled at early-adopter sites (36, 78, 101, 121, 132). Some centers return all results into the patient’s medical record, while others use an approach of returning results of pharmacogenes that have clinical recommendations for actionability in the CPIC or DPWG guidelines.
Rationale for Preemptive Pharmacogenomic Testing
Evidence for widespread use of pharmacogenomically actionable medications is now overwhelming. For example, examination of more than 70 million patient insurance records in the United States found that approximately half of all patients 40–64 years of age received at least one pharmacogenomically actionable medication and more than 25% received two or more actionable medications over only a 4-year period (118). A study of 56 actionable medications in more than 50,000 patients at Vanderbilt University Medical Center estimated that 65% of patients received a high-risk medication within a 5-year time period (119). An analysis of claims data in Austria for more than 6.7 million patients found that 72% of patients over 65 years of age received at least one actionable medication in a 2-year period (77). In Singapore, based primarily on 69 medications that were rated as CPIC level A or B (actionable), the 5-year risk of receiving any new pharmacogenomically actionable medication was approximately 43%, with the risk being higher for older patients and higher for Malays and Indians than Chinese (27). Of more than 1.3 million South Koreans, 47.4% were prescribed a pharmacogenomically high-risk medication in a 1-year period (74). More than 84% of nursing home residents used pharmacogenomically high-risk medications (134). Of more than 7.7 million American veterans, 54.8% received at least one CPIC level A drug (actionable) over a 6-year period, with the most common gene–drug pairs being SLCO1B1–simvastatin, CYP2D6–tramadol, and CYP2C9–warfarin or VKORC1–warfarin (28). In a British primary care setting, 58% of patients were prescribed at least one high-risk medication over a 2-year period, but that increased to 80% over 20 years, with exposure increasing with age (75).
Despite the very high frequency of pharmacogenomically high-risk medication use in the general population, strategies to identify groups of patients with the highest likelihood of receiving a high-risk medication have been proposed to identify subsets in whom array-based preemptive genotyping would have the biggest payoff (123). Grouping patients by clinic type and age in a US population, the percentage receiving a high-risk medication ranged from 12.7% in the outpatient pediatric psychiatry group to 75.7% in adult inpatients over 50 years of age (59).
The rationale for widespread panel-based preemptive testing is simple: Pharmacogenomically actionable medications are widely used in both inpatient and outpatient settings, and every individual (regardless of race or genetic ancestry) is extremely likely to harbor at least one actionable variant (Figure 1). Moreover, with results available preemptively, turnaround time for returning test results is not an issue (120). With preemptive testing, genotype results reside in EHRs and can be available immediately when a high-risk medication is ordered. More than 98% of American whites and blacks are estimated to have at least one high-risk actionable genotype (38). More than 99% of 44,000 Estonians had at least one high-risk genotype (117), with a similar percentage found at the Mayo Clinic in the United States (68). In studies that considered both medication use and the presence of actionable genotypes and analyzed more than 3 million patients receiving more than 3.6 million new prescriptions in the Netherlands, approximately 24% of new prescriptions involving 45 actionable medications represented an actionable gene–drug interaction (2, 10, 11).
Figure 1.
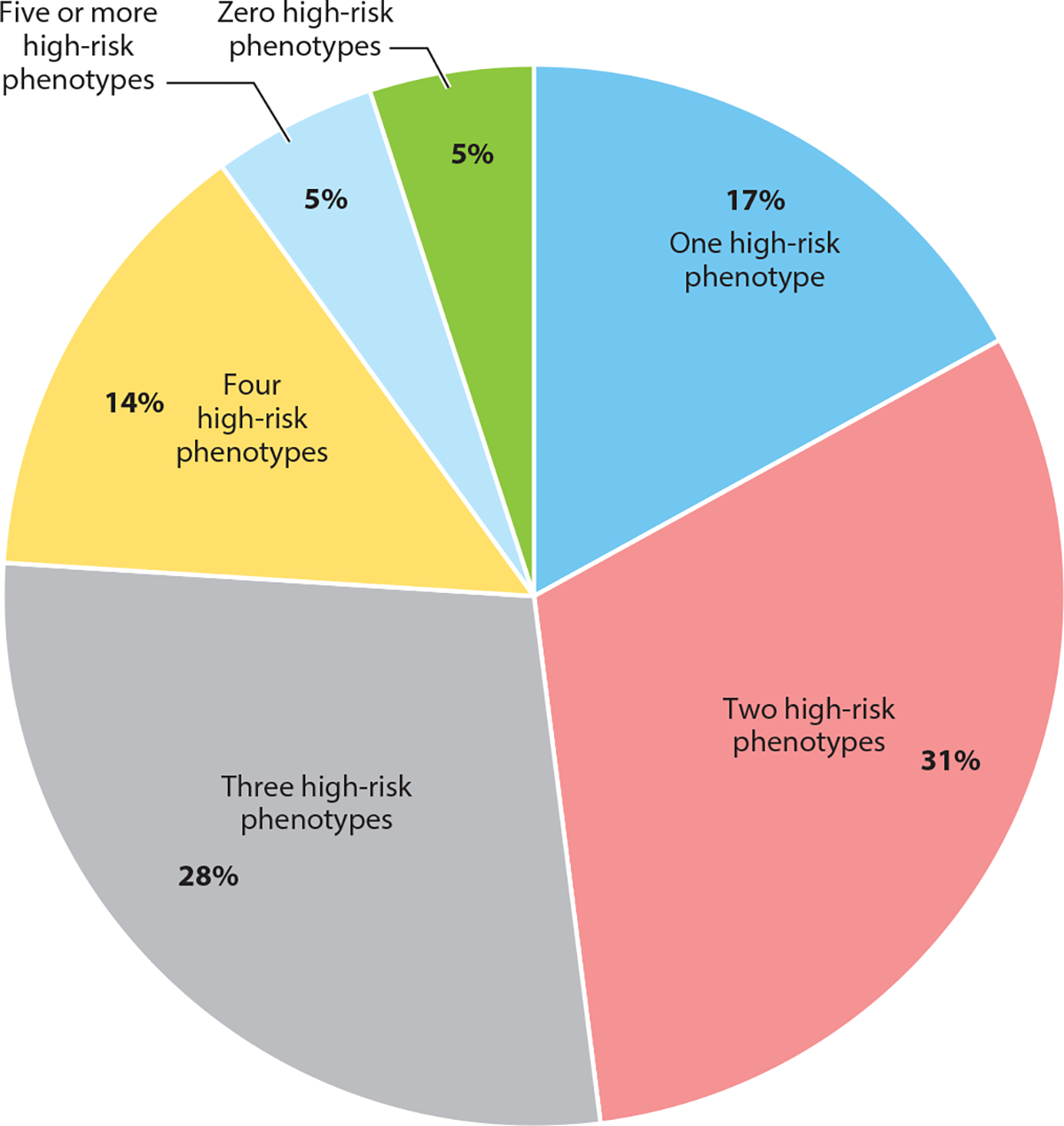
Proportions of patients preemptively genotyped at St. Jude Children’s Research Hospital with high-risk phenotypes. Data are from the PG4KDS clinical trial and are shown as of November 1, 2021. The study genotyped 5,912 patients for the following 13 pharmacogenes: CACNA1S, CYP2B6, CYP2C19, CYP2C9, CYP2D6, CYP3A5, DPYD, G6PD, mt-RNR1, RYR1, SLCO1B1, TPMT, and UGT1A1. Patients diagnosed with acute lymphoblastic leukemia were also genotyped for NUDT15. A high-risk phenotype is a result that has implications for the use of at least one medication.
Thus, theoretically, essentially all patients would benefit from having multigene preemptive pharmacogenomic tests. Whereas payers may be willing to pay for single-gene tests, largely because they see this as likely to be a limited market, many report being skeptical of the costs involved in universal panel-based preemptive testing (72). There is limited evidence for the cost effectiveness of a preemptive approach (142, 147, 148). In addition to the costs associated with genotyping and with the delivery of results and clinical decision support (CDS), some have described potential disadvantages of array-based preemptive testing. For example, there is a school of thought that says that having multiple pharmacogenomic test results makes it too complicated for clinicians to interpret the results (76). A preemptive testing approach also requires an excellent healthcare system with integration of laboratory results and electronic prescribing practices. Sustainability is a clear challenge: The problems of how to update interpretations over time (6, 56) and how to continue to make results follow patients as they age and progress in and out of multiple healthcare systems have not been successfully addressed. Some describe the liability involved in providing a large number of results when a portion of these results may not be used or adequately interpreted (19). Testing more genes and variants also increases the theoretical danger of having incidental findings that may impact disease risk and may not have been anticipated by the patient or provider at the time of testing (54, 81). Examples of incidental pharmacogenomic findings include discovery of Klinefelter syndrome based on males having two G6PD (X chromosome) alleles and discovery of Gilbert syndrome based on UGT1A1 genotype test results.
Cascade genetic testing—the practice of extending testing to at-risk family members of an individual determined to have a high-risk genotype—may be indicated for select pharmacogenes (127). The genes that are most amenable to this approach are those that have autosomal dominant inheritance (placing family members at very high or even certain risk for having a high-risk genotype); the frequency of use of the affected medications may also affect the decision to implement cascade genetic testing of family members. For example, certain RYR1 and CACNA1S variants are associated with a predisposition to develop malignant hyperthermia with the administration of certain inhaled anesthetics or succinylcholine. Because these two genes exhibit autosomal dominant inheritance, the observation of a high-risk RYR1 and CACNA1S genotype in a patient may result in offering cascade genetic testing to family members (53).
Using whole-genome sequencing for preemptive pharmacogenomic testing instead of array-based techniques has been proposed. The challenges of generating large numbers of false negative or false positive results with genome sequencing have been presented (122). On the other hand, some studies have reported promising levels of concordance between genome sequencing results and microarrays (110, 145), so that one possible strategy is for centers to leverage genome sequencing already performed for other reasons to yield actionable pharmacogenomic results.
WORKFLOW PRACTICES OF INCORPORATING PHARMACOGENOMIC TEST RESULTS INTO ELECTRONIC HEALTH RECORDS
For pharmacogenomic testing to be most useful, structured genetic test results (generally organized by gene) should be delivered into the EHR and coupled with CDS; structured results are particularly needed for multigene test results (6). Because the testing may have predated the relevant prescribing decision by months or even years, systems that can find past pharmacogenomic results and link the information with current prescribing decisions are needed. Thus far, workflows have been reported for 19 programs (86) and vary among institutions (6). However, there are several commonalities among these published workflows (6, 18, 21, 26, 35, 37, 58, 63, 66, 85, 86, 89, 94, 95, 106, 116, 124–126, 131).
Each clinical workflow encompasses the steps from ordering the pharmacogenomic test to making informed prescribing decisions to educating patients (Figure 2). Standardized terms for test names and test results facilitate standardizing the workflow and improving the interoperability of using the results across healthcare settings (24, 83). Some implementers propose combining disease-risk genes (e.g., those included on the American College of Medical Genetics and Genomics list of disease-causing variants and pharmacogenomic genes in one workflow (92), while others have separate workflows (7). Most workflows incorporate both active CDS presented at the time a high-risk medication is ordered and passive CDS in the form of a static written interpretive consultation (13, 61). Workflows have been described for point-of-care multigene implementations in ambulatory clinical settings (62, 69), as well as for panel-based preemptive testing in a large health center (85) and testing in cancer clinics (90). Some workflows include clinician and patient education (103). Toolboxes for workflows have been provided by the Implementing Genomics in Practice (IGNITE) network (39). Some sites set up a special pharmacogenomics clinic that incorporates patient encounters for testing and return of results (8, 37). Most sites incorporate pharmacogenomic testing into routine care workflows, and different considerations exist depending on the genes, medications, setting of care (i.e., ambulatory or inpatient), patient population, and other factors (21, 30, 106).
Figure 2.
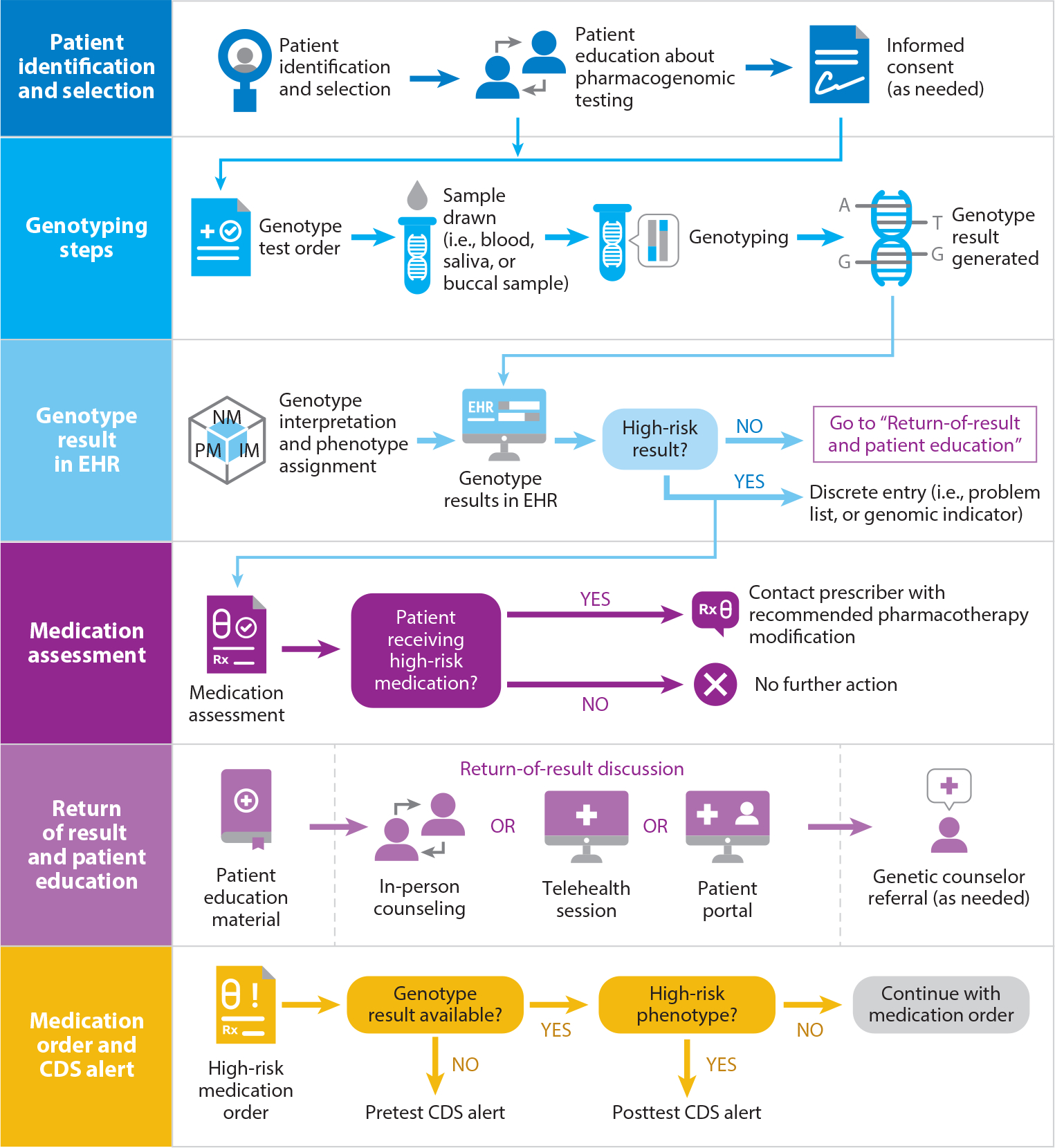
Preemptive pharmacogenomics service workflow process. Abbreviations: CDS, clinical decision support; EHR, electronic health record.
Some implementers establish a pharmacogenomics oversight committee, often as a subcommittee of a pharmacy and therapeutics committee, which can help prioritize the genes and medications for incorporation into the clinical workflow and can have external approval of CDS language (6, 13). Workflows can incorporate processes to distinguish clinically actionable results from other results, to ensure that only a portion of interrogated genetic results are reported or acted upon (8). Pharmacogenomic results can be incidental results from whole-genome or whole-exome sequencing (99), and workflows have been proposed based on using next-generation DNA sequencing methods (51).
Considerations for Identifying a Genetic Testing Laboratory and Interpreting Results
An important step in implementing preemptive pharmacogenomics is identifying an appropriate laboratory to perform the genetic test (135). When implementing preemptive testing using a multigene panel, consideration should be given to what genes are applicable to the clinical setting, what variants for each gene are interrogated, how much the testing costs, what type of sample is needed, how long the turnaround time will be, and how results will be reported. Many programs across the United States that have implemented pharmacogenomics rely on the CPIC guidelines to aid in gene and variant selection (136). Considerations for selecting a laboratory have been reviewed (135).
It is important to translate the genotype data into a clinical phenotype (e.g., CYP2D6*4/*4 to CYP2D6 poor metabolizer), as this phenotype designation often drives therapeutic decisions. For example, CPIC guideline recommendations are generally built around phenotype groups rather than individual diplotypes. Furthermore, an advantage of this approach is that CDS alerts can be coded to alert from a limited number of phenotype assignments as opposed to multiple individual diplotype calls. For example, CYP2D6 has more than 10,000 possible diplotypes but only four primary phenotypes. A system that provides alerts for all 10,000 diplotypes would be time consuming to build and maintain and is not necessary. Grouping diplotypes into clinical phenotypes for the purpose of designing CDS rules has been standard for pharmacogenomics implementation across multiple sites (38) (Figure 3).
Figure 3.
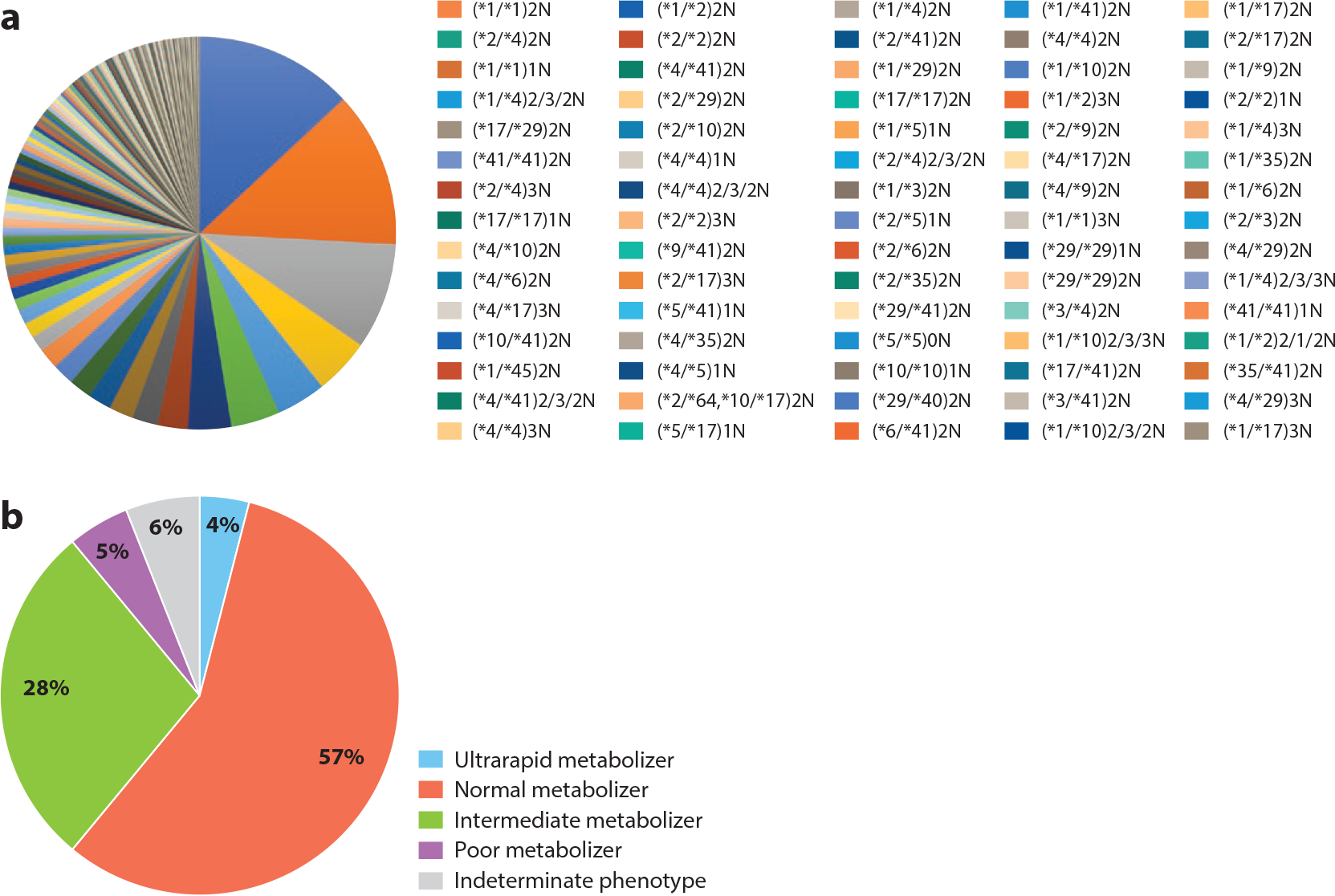
Demonstration of the reduction in complexity of pharmacogenomic result interpretation by utilizing phenotype instead of genotype. In this example, 406 unique CYP2D6 genotype (diplotype) results (panel a) can be classified into four distinct phenotypes (panel b), with a small percentage left as indeterminate. This approach simplifies result interpretation for clinicians and enables clinical decision alerts to be triggered based on phenotype instead of diplotype.
Returning Pharmacogenomic Results and Incorporating Genetic Test Results into Electronic Health Records
For clinical pharmacogenomics to be truly used as a preventive service, test results must be incorporated into EHRs with CDS, so that results may be used to individualize therapy at any future time point (61, 65, 139). Most EHR vendors do not have a pharmacogenomics framework built into their system as a core function, and the major medication knowledge vendors used in EHRs are slowly adding pharmacogenomic content. Therefore, currently, incorporating pharmacogenomics into EHRs with CDS requires careful integration of pharmacogenomic knowledge from external resources (such as CPIC guidelines) combined with configuration of the EHR vendor’s custom clinical rule capabilities. The CPIC Informatics Working Group provides informatics resources with each CPIC guideline (64) that include point-of-care CDS workflows, example text for decision support alerts, and templates for interpretive consultations. Most successful clinical implementation programs have relied on the integration of pharmacogenomic results and clinical phenotypes as discrete elements of EHRs to prompt CDS alerts that notify prescribers of special consideration for pharmacotherapy (13, 20, 22, 60, 63, 85). Some level of customization of the health system’s EHR instance may be required to fully support pharmacogenomic CDS. While health systems may chafe at customization of their EHR system, innovations for both genomics and patient safety often require some new approaches and a higher level of informatics support (42, 108). The growth in the number of healthcare systems implementing pharmacogenomic CDS using different EHR vendors demonstrates that the approach is feasible and can be scaled to more health systems (15, 21, 52, 89, 102, 109, 131, 139).
There are two main categories of CDS, both of which have value for preemptive pharmacogenomics. Instead of expecting clinicians to search for pharmacogenomic results in the EHR system (passive CDS), or sometimes even to search for the results in a separate system, interruptive alerts that guide clinicians to make changes should also be used (active CDS). Many programs have published on the different types of CDS alerts used in pharmacogenomics as well as the differences in how they are used across implementer sites (73, 139), which are briefly reviewed below.
Passive clinical decision support.
Information can be passively made available to the ordering clinician as a genetic test interpretation, generally in the form of a written consultation from a pharmacogenomics expert (61). While useful, passive CDS interpretations have limitations: (a) passive CDS tools rely first on the prescriber remembering the potential for a gene–drug interaction at the point of care and knowing the location of the pharmacogenomic test results in the EHR system, (b) pharmacogenomic interpretation reports consist of static consultations and may not be updated regularly to reflect new prescribing information (e.g., a newly published pharmacogenomic guideline), and (c) the location of pharmacogenomic results differs across EHR systems, and finding a location that works for all has been challenging (32).
Active clinical decision support.
Active or interruptive CDS alerts stop the clinician’s workflow during the medication ordering phase to inform them of the availability of pharmacogenomic information (i.e., a pretest alert) or to present pharmacotherapy recommendations on a high-risk pharmacogenomic test result (i.e., a posttest result).
Pretest alerts inform prescribers that they are attempting to order a medication affected by pharmacogenomic variation but the associated genotype test has not yet been ordered, the result is still pending, or the result has not been recorded using standardized terminology that the CDS system uses to identify the absence or presence of pharmacogenomic results. This type of alert generally informs clinicians of the potential risks associated with prescribing the medicine without a known genotype, provides recommendations for alternative medications to use or select, and offers the option of ordering the genotype test, often from within the alert window itself (13). Alternative agent selection in the alert text should consider the applicable medication formulary system and patient populations who are usually prescribed the medicine. For example, a pretest alert presented to a clinician trying to order codeine for treatment of pain should ideally recommend alternative analgesic medications that are part of the formulary system of the hospital and would have the same route of administration or formulation that the clinician was attempting to prescribe. A benefit to preemptive pharmacogenomic testing is that pretest alerts are infrequent if a process is in place to test patients before a medication is ordered. In many cases, the pharmacogenomic test results should already be in the patient’s EHR.
Posttest alerts interrupt the normal medication ordering or dispensing process, usually by presenting a new window explaining the need to modify pharmacotherapy based on the patient’s high-risk pharmacogenomic test result (high-risk phenotype). Ideally, the posttest alert should have a concise message to the clinician that explains the risk if the patient were to receive the medication at the dose ordered. The alert should provide clear alternatives and functions to guide the prescriber to these alternatives. Like pretest alerts, it is important to tailor the selection of alternative agents to the institution’s medication formulary system and ideally to match the route of administration and formulation of the medication that was originally intended to be prescribed. Drug–drug interaction alerts can also be combined with gene–drug information, as in the case of paroxetine, which is a strong inhibitor of CYP2D6. The coadministration of paroxetine may lead to a clinical CYP2D6 phenotype that is different from the genotype-defined phenotype, a process referred to as phenoconversion (16, 29). Additionally, multifactorial alert rules can be created to incorporate other clinical elements that influence medication dosing and selection, such as age, route of administration, previous medication use, previous therapeutic drug monitoring, or hepatic and renal dysfunction (61). In some cases, sophisticated alert logic and other features within the EHR can decrease the need for a posttest alert, thus decreasing the chance of clinician alert fatigue (61).
Result Portability
Pharmacogenomic test results are relevant across a patient’s life span, with use of high-risk pharmacogenomic medications tending to increase with age (75, 118). Processes need to be developed to make it routine for pharmacogenomic results to be available to clinicians at any future encounter that may start or change medication therapy. The fragmented and reactive nature of healthcare in many countries creates challenges to the optimal use of pharmacogenomic information over time. There is a need for systems that will exchange results and/or a registry of pharmacogenomic results, akin to vaccination registries.
The challenges of creating a workflow that allows for long-term sustainability and handoffs with providers outside the original implementation program have been described (130). Some suggest embracing patients as the keepers of their pharmacogenomic results, creating tools such as QR-code-compliant pharmacogenomic passports that patients can present to various providers (133). This issue of result portability remains one of the biggest and toughest barriers to positioning pharmacogenomics as a routine preventive measure.
Pharmacogenomic Education
As pharmacogenomics moves from single-gene reactive tests to more comprehensive panels preemptively ordered or directly obtained by consumers, prescribers may be faced with incorporating a genetic test result into a therapeutic selection and counselling a patient for a test they did not order.. One of the most cited barriers to implementing pharmacogenomics is a lack of knowledge or education of clinicians and patients to support the interpretation of results (17, 47).
Healthcare provider education strategies.
Preemptive programs have addressed the knowledge gap in pharmacogenomics education by developing comprehensive educational programs aimed at clinicians, including physicians, pharmacists, advanced practice nurses, and others. These programs fall under several categories: just-in-time education and resources for clinical implementers, education in professional schools, postdoctoral training (e.g., residencies, fellowships, and graduate programs), and certification programs.
Structured onsite educational in-services for clinicians are often recognized as crucial when establishing a pharmacogenomics program (38, 66, 86, 103). Educational in-services can be tailored to pharmacists, to a relevant specialty within a health system, or as general education across all clinical staff. Once a preemptive program is launched, the most immediate and real-time form of provider education includes just-in-time education embedded in provider workflows (46, 144). Methods of just-in-time education include active CDS presented to the prescriber within the clinical workflow, which provides clinicians with information required to interpret an individual patient’s pharmacogenomic results. Active CDS is enhanced by passive CDS, as discussed above. A review of pharmacogenomics programs found that individual programs have also included messages to clinicians’ inboxes, dedicated webpages, and on-site in-services for providers (86). Combining institutional education with access to available resources such as CPIC guidelines is key in educating the workforce so that pharmacogenomic results can be used once they have been implemented at an institution.
Accreditation standards for some schools of medicine and pharmacy now include pharmacogenomics as foundational content in predoctoral education. Including pharmacogenomic concepts in medical and pharmacy school curricula will ensure that graduates are poised to integrate pharmacogenomics into patient care. For practicing clinicians, continuing education programs focused on therapeutic areas or practice settings are offered. The Inter-Society Coordinating Committee for Practitioner Education in Genomics, supported by the National Human Genome Research Institute, is a collaborative group aimed at improving healthcare provider genomics education by developing educational resources (96). Competencies are also well established, especially for pharmacists (49, 115). In addition, the National Human Genome Research Institute’s Genetics/Genomics Competency Center website (https://www.genomicseducation.net) contains freely accessible genomics-related competency material for healthcare professionals.
Despite the increased presence of pharmacogenomics education and training for clinicians in medical and pharmacy school curricula, there is a need for some clinicians to complete specialty training via residencies, fellowships, or graduate programs (98, 141). Significant expansion of preemptive pharmacogenomics programs has led to novel pharmacist-led pharmacogenomic services and outpatient clinics and thus has created a need for leadership roles at the level of individual health systems. Accordingly, the American Society of Health-System Pharmacists has developed accreditation standards for specialty postgraduate year-two pharmacy residencies in Clinical Pharmacogenomics (3, 55). In the United States, nine pharmacogenomics residency programs either have been accredited by the American Society of Health-System Pharmacists are have preaccreditation status, and additional programs continue to apply for accreditation (4). The accreditation standards ensure that pharmacists who successfully complete an accredited postgraduate year-two clinical pharmacogenomics residency are prepared for advanced patient care, academic positions, or other specialized positions in the field. Fellowships and graduate programs in pharmacogenomics are other avenues of specialized training in the field. Several established pharmacogenomics programs and professional organizations offer certification in pharmacogenomics for practitioners. Certificate programs are most often offered as continuing education modules that may be self-paced and focus on principles of pharmacogenomics and applying them at a patient care level.
Patient education strategies.
Educating patients about their individual results, which have lifelong implications, is an important aspect of a preemptive pharmacogenomics program. Preemptive programs have published several methods for patient education (86). Methods for educating patients about their individual results vary. General education about genetic tests can occur at the time that the pharmacogenomic testing is being ordered (66, 86, 138). At the time that results are placed in a patient’s EHR, individualized patient education can be included in the workflow (Figure 2). Patients may be provided with information about their results in person, in writing or via a patient portal (38, 66, 84). Programs may provide information about medications and pharmacogenes through a dedicated webpage (e.g., 128) or via educational videos (66, 93).
ECONOMIC AND REIMBURSEMENT CHALLENGES FOR PREEMPTIVE PHARMACOGENOMIC TESTING
Although the cost of a pharmacogenomic test can vary across different testing companies and platforms, many single-gene tests range from approximately $100 to $500, while a multigene panel can cost approximately the same or twice as much as a single-gene test (5). Even though a multigene panel would be less expensive than multiple single-gene tests, many payers are reluctant to reimburse for the multigene panels that would be used for preemptive pharmacogenomics implementation. Clinical utility is a requirement for coverage and reimbursement, and demonstrating clinical utility for preemptive testing can be difficult, as it is unclear exactly how the test results would be used clinically at the time the test is ordered (e.g., a patient may not take a medication that would be affected by pharmacogenomic test results for some time). Proving clinical utility for reactive testing is more straightforward, as the test is being ordered with a medication in mind and prior to administration of a high-risk medication, but even reimbursement for single-gene tests remains a challenge.
However, more consistent reimbursement for pharmacogenomics in the United States may be on the horizon (40). Recently, building on previous decisions from public and private payers, local coverage determinations for the Molecular Diagnostic Services (MolDx) program were released, adding expanded coverage of pharmacogenomic tests for Medicare patients in select states and mention that “pharmacogenomic tests are indicated when medications are being considered for use (or already being administered) that are medically necessary, appropriate, and approved for use in the patient’s condition and are known to have a gene(s)-drug interaction that has been demonstrated to be clinically actionable as defined by the FDA (pharmacogenomic information required for safe drug administration) or Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines” (129a). The new local coverage determinations now allow for coverage of all CPIC level A and B drugs [prescribing action recommended by CPIC (31b)] and gene–drug pairs listed in FDA labeling. More importantly for preemptive pharmacogenomic testing, the local coverage determination does cover multigene tests, albeit only if the patient is receiving or may receive a medication covered by at least one gene on the panel (40).
The cost of the pharmacogenomic test is only one financial aspect of preemptive pharmacogenomics implementation. As discussed above, a multidisciplinary team of clinicians, informaticians, and laboratory specialists is required to adequately implement preemptive pharmacogenomics in an EHR with CDS. Once the infrastructure has been set up in the EHR, monitoring of evolving evidence for pharmacogenomics and guidelines will need to continue on an ongoing basis, and the CDS will need to be updated in the EHR (56, 85).
A DECADE OF PREEMPTIVE PHARMACOGENOMICS AT ST. JUDE CHILDREN’S RESEARCH HOSPITAL
St. Jude Children’s Research Hospital established an institution-wide preemptive pharmacogenomics program through the PG4KDS protocol in May 2011 (66, 128). Patients and their families meet with a member of the hospital’s Clinical Pharmacogenomics Service, who explains to them what pharmacogenomics is and have a consent discussion where they indicate whether they agree to have the child genotyped. As of October 2021, 95% of patients (6,171 out of 6,508) approached for consent have agreed to genotyping. Reasons for refusal of genotyping vary and include concern over extra blood collection, enrollment in too many clinical trials, distrust of DNA research, and privacy concerns. Genomic DNA is obtained from a blood sample, and genotyping is performed by RPRD Diagnostics using the PharmacoScan array from Thermo Fisher Scientific.
The results of 14 pharmacogenes are used to guide therapy using 66 drugs (Figures 4 and 5) for all patients who have been genotyped. Pharmacogenomic test results are displayed in a dedicated pharmacogenomics specialty flow-sheet tab and are presented independently of the date that the tests were performed. When pharmacogenomic test results are returned in the EHR system, a written consultation assigning a phenotype and explaining the implications of the result is generated and verified by a trained pharmacist (60). The consultation notes are a passive decision support tool and concisely convey useful information to clinicians who may not have deep knowledge of pharmacogenomics. The interpretive consultation language is also broad enough to be useful over time as new medications related to that gene are implemented. The genotype result and consultation note are viewable in the patient’s online portal. For all patients with a high-risk diplotype, a member of the Clinical Pharmacogenomics Service also assesses the medications that the patient is receiving to address therapy changes that may be necessary based on the patient’s pharmacogenomic profile.
Figure 4.
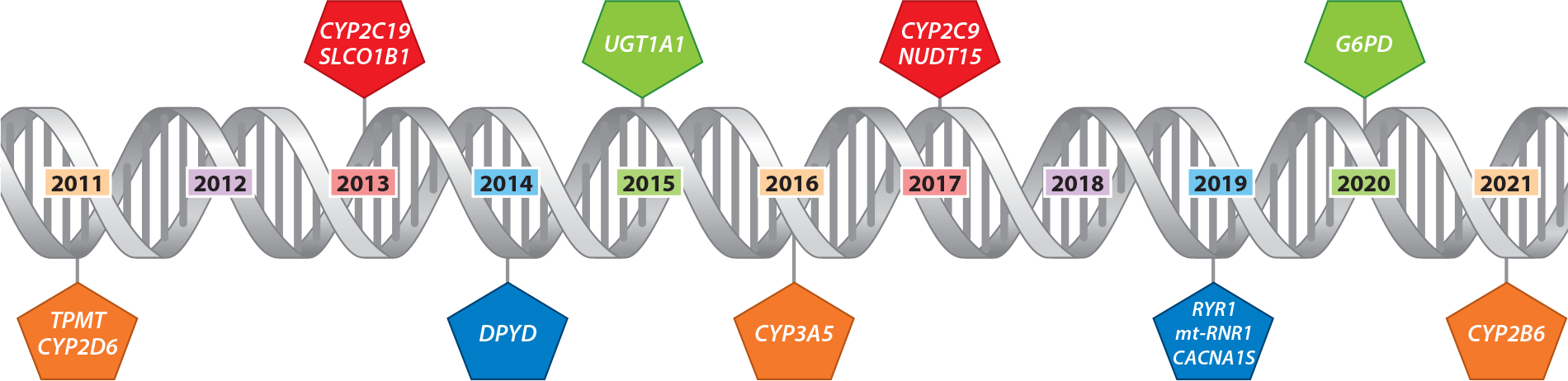
Example of a preemptive implementation time line for pharmacogene testing, showing the rollout of gene test results over time in St. Jude Children Research Hospital’s PG4KDS study. All gene test resultswere interrogated starting in 2011 but were placed in the EHR system as CDS for at least one drug affected by that gene had been implemented in the EHR systemAll gene–drug pair rollouts were approved by the hospital’s Pharmacogenomics Oversight Committee. Abbreviations: CDS, clinical decision support; EHR, electronic health record.
Figure 5.
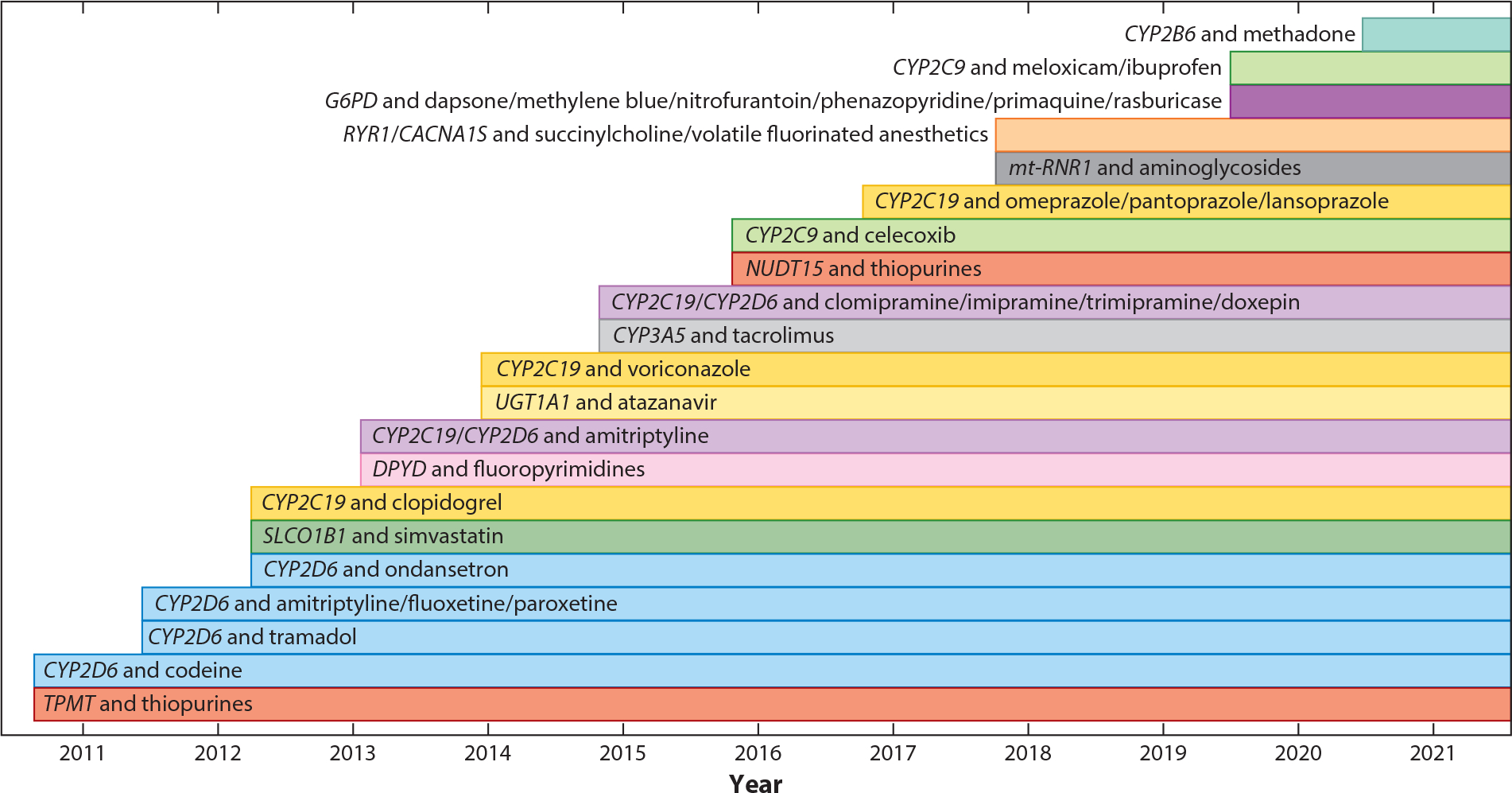
Genes and medications implemented at St. Jude Children’s Research Hospital. Test results for some genes (e.g., CYP2C19; yellow bars) were implemented as early as 2013, but CDS for additional drugs was implemented over time for different drugs (clopidogrel in 2013, voriconazole in 2015, and proton pump inhibitors in 2017). Abbreviation: CDS, clinical decision support.
When a patient is prescribed a high-risk medication and has a corresponding high-risk phenotype, structured and discrete data in the EHR facilitate presenting an interruptive CDS alert to the prescriber (13). The alert language is provided in a standard format that recommends a dosage change or selection of an alternative regimen and reminds the prescriber that more information is available in the EHR or by contacting a pharmacist (61). The clinician acceptance of the pharmacotherapy modification recommendation is greater than 90% for CYP2D6–codeine (48) and TPMT–thiopurines (13). As pharmacogenomic knowledge develops and evolves, some results require updating and reinterpretation (56). For example, prior to 2017, patients with a CYP2C19 genotype of *1/*17 were assigned a CYP2C19 ultrarapid metabolizer phenotype. As new evidence emerged about the functional status of CYP2C19 enzymes in patients with this genotype, the phenotype assignment was changed to a rapid metabolizer phenotype. This update in phenotype assignment has implications for medication therapy recommendations; for example, initial dosing recommendations for omeprazole differ for a CYP2C19 rapid metabolizer compared with a CYP2C19 ultrarapid metabolizer (82).
The hospital’s Pharmacogenomics Oversight Committee provides oversight of the activities of the Clinical Pharmacogenomics Service. This committee, chaired by the principal investigator of the PG4KDS protocol, meets at least quarterly and is structured as a subcommittee of the Pharmacy and Therapeutics Committee, which reports to the Medical Executive Committee. It includes clinician champions representing all major clinical services across the institution, informatics specialists, an external pathologist, and an external advisor who is an internationally recognized leader in the clinical implementation of pharmacogenomics. Beyond connecting pharmacogenomics implementation to medication use, the committee approves the priority of future gene–drug implementations and the wording, logic, and rules of active CDS alerts and rules.
Pharmacogenomics is established as a patient safety strategy at St. Jude. Consistent with the institutional approach to transparently share quality and patient safety data across the institution, pharmacogenomics is included as a quality indicator, and data are available to the entire institution. A primary metric is the proportion of thiopurine-naive patients diagnosed with acute lymphoblastic leukemia for whom both TPMT and NUDT15 (as of April 2017) genotypes are placed in the EHR before thiopurine therapy is initiated (Figure 6). Approximately 22 new thiopurine-naive patients are started on thiopurine therapy each quarter at St. Jude. Our goal is that 100% of these patients will receive pharmacogenomics-guided dosing at the time thiopurine therapy is initiated. For rare cases of patients who did not have TPMT or NUDT15 genotype results available before thiopurine was prescribed, we investigate the cause of the missed genotyping and implement improvement actions based on the findings. Examples of improvements made based on this ongoing data monitoring include adding an order for TPMT/NUDT15 genotyping in the prebuilt order sets of all patients newly diagnosed with leukemia and changing the name of the test from thiopurine genotype to TPMT/NUDT15 genotype, thereby allowing clinicians to more easily find the order. These data metrics are reported quarterly to the Pharmacogenomics Oversight Committee and incorporated into the hospital’s medication safety dashboard.
Figure 6.
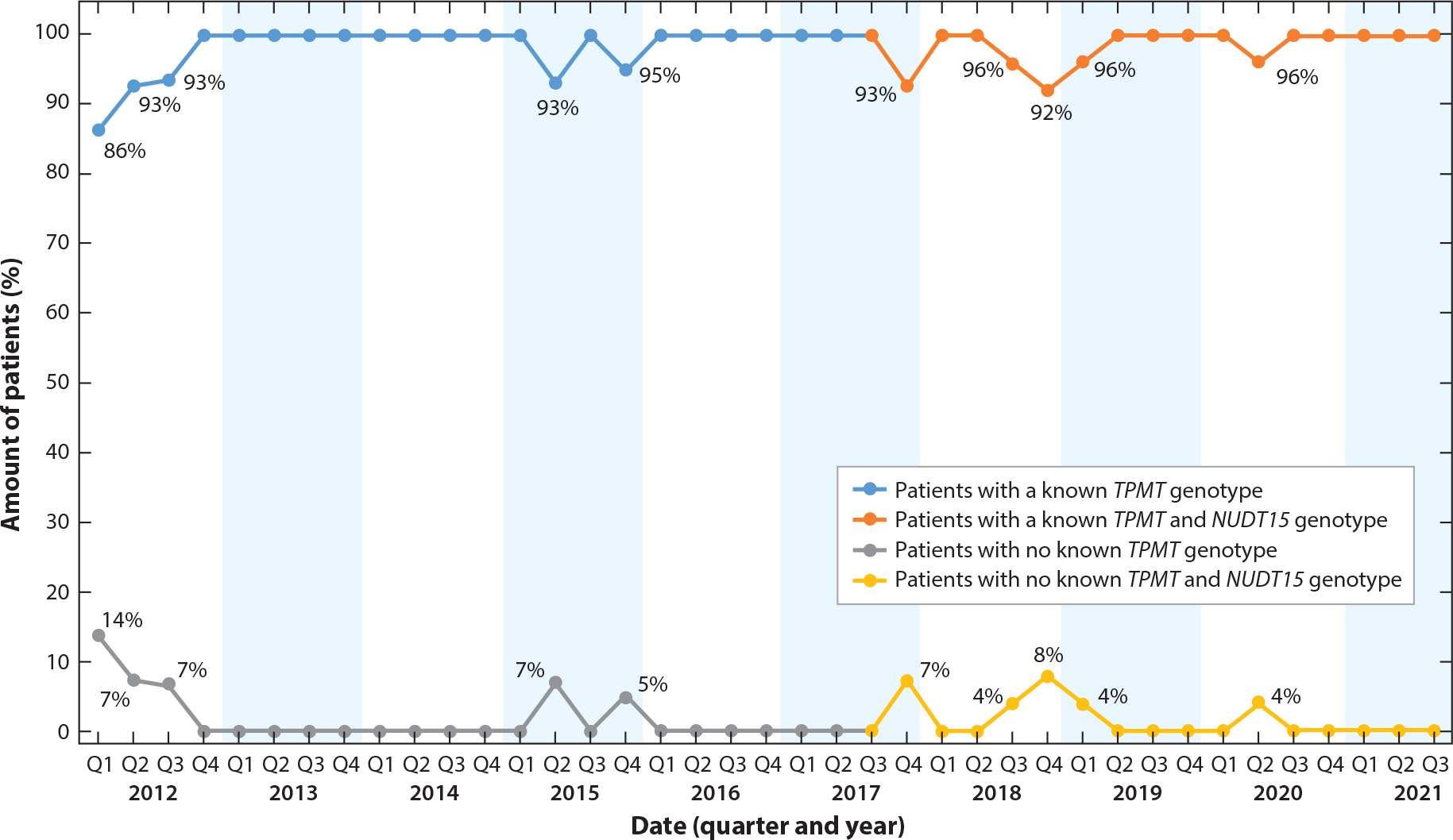
Percentage of thiopurine-naive patients diagnosed with acute lymphocytic leukemia who had a known TPMT and NUDT15 genotype prior to initiating thiopurine therapy at St. Jude Children’s Research Hospital, by quarter. TPMT genotyping was implemented prior to 2012; NUDT15 genotyping was implemented clinically in 2017. The goal quality metric is 100% for the top curve and 0% for the bottom curve. Approximately 22 patients per quarter were included in this metric.
ADVANCING PREEMPTIVE PHARMACOGENOMICS INTO PRACTICE
Examples of successful preemptive pharmacogenomics programs exist across practice settings, and some of these programs have been in place for over a decade (6, 18, 21, 26, 35, 37, 58, 63, 66, 85, 86, 89, 94, 95, 106, 116, 124–126, 131). Use of medications that are impacted by pharmacogenomic tests is common. Likewise, genetic variation that can result in adverse medication outcomes is common. The necessary technology—both testing and informatics—is well established. However, gaps in knowledge, incomplete reimbursement, the need for processes for reporting of preemptive results over time, and poor result portability across healthcare encounters are tangible barriers that need to be addressed in order to fully embrace a preemptive approach to pharmacogenomics. Continued standardization and use of established resources such as CPIC’s can advance implementing preemptive pharmacogenomics (24). One barrier is that some clinicians and payers consider some amount of trial and error to be an inevitable part of medication use, especially for less expensive medications (72). To embrace preemptive pharmacogenomics, this mind set must shift from accepting poor prescribing outcomes toward preventing harm and using all available information to select the right medication and dose for the patient from the first prescription.
ACKNOWLEDGMENTS
We would like to thank the Biomedical Communications Department at St. Jude Children’s Research Hospital and Jenny Nguyen, PharmD, for assistance with figures. This work was funded in part by the American Lebanese Syrian Associated Charities and by National Institutes of Health grant U24HG010135.
Footnotes
DISCLOSURE STATEMENT
K.E.C., M.V.R, and J.M.H. receive funding from the NIH to support CPIC (U24HG010135). All other authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Abul-Husn NS, Owusu Obeng A, Sanderson SC, Gottesman O, Scott SA. 2014. Implementation and utilization of genetic testing in personalized medicine. Pharmacogenom. Pers. Med 7:227–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alshabeeb MA, Deneer VHM, Khan A, Asselbergs FW. 2019. Use of pharmacogenetic drugs by the Dutch population. Front. Genet 10:567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Am. Soc. Health-Syst. Pharm. 2018. Required competency areas, goals and objectives for postgraduate year two (PGY2) clinical pharmacogenomics pharmacy residencies. Resid. Program Doc., Am. Soc. Health-Syst. Pharm, Bethesda, MD. https://www.ashp.org/-/media/assets/professional-development/residencies/docs/pgy2-clinical-pharmacogenomics-cago.pdf
- 4.Am. Soc. Health-Syst. Pharm. 2021. Residency directory. American Society of Health-System Pharmacists. https://accreditation.ashp.org/directory/#/program/residency [Google Scholar]
- 5.Anderson HD, Crooks KR, Kao DP, Aquilante CL. 2020. The landscape of pharmacogenetic testing in a US managed care population. Genet. Med. 22:1247–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aquilante CL, Kao DP, Trinkley KE, Lin CT, Crooks KR, et al. 2020. Clinical implementation of pharmacogenomics via a health system-wide research biobank: the University of Colorado experience. Pharmacogenomics 21:375–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arwood MJ, Chumnumwat S, Cavallari LH, Nutescu EA, Duarte JD. 2016. Implementing pharmacogenomics at your institution: establishment and overcoming implementation challenges. Clin. Transl. Sci. 9:233–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arwood MJ, Dietrich EA, Duong BQ, Smith DM, Cook K, et al. 2020. Design and early implementation successes and challenges of a pharmacogenetics consult clinic. J. Clin. Med. 9:2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bank PCD, Caudle KE, Swen JJ, Gammal RS, Whirl-Carrillo M, et al. 2018. Comparison of the guidelines of the Clinical Pharmacogenetics Implementation Consortium and the Dutch Pharmacogenetics Working Group. Clin. Pharmacol. Ther. 103:599–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bank PCD, Swen JJ, Guchelaar HJ. 2019. Estimated nationwide impact of implementing a preemptive pharmacogenetic panel approach to guide drug prescribing in primary care in The Netherlands. BMC Med. 17:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bank PCD, Swen JJ, Schaap RD, Klootwijk DB, Baak-Pablo R, Guchelaar HJ. 2019. A pilot study of the implementation of pharmacogenomic pharmacist initiated pre-emptive testing in primary care. Eur. J. Hum. Genet. 27:1532–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barnish MS, Turner S. 2017. The value of pragmatic and observational studies in health care and public health. Pragmat. Obs. Res. 8:49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bell GC, Crews KR, Wilkinson MR, Haidar CE, Hicks JK, et al. 2014. Development and use of active clinical decision support for preemptive pharmacogenomics. J. Am. Med. Inform. Assoc. 21:e93–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berm EJ, Looff M, Wilffert B, Boersma C, Annemans L, et al. 2016. Economic evaluations of pharmacogenetic and pharmacogenomic screening tests: a systematic review. Second update of the literature. PLOS ONE 11:e0146262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blagec K, Koopmann R, Crommentuijn-van Rhenen M, Holsappel I, van der Wouden CH, et al. 2018. Implementing pharmacogenomics decision support across seven European countries: the Ubiquitous Pharmacogenomics (U-PGx) project. J. Am. Med. Inform. Assoc 25:893–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blagec K, Kuch W, Samwald M. 2017. The importance of gene-drug-drug-interactions in pharmacogenomics decision support: an analysis based on Austrian claims data. Stud. Health Technol. Inform. 236:121–27 [PubMed] [Google Scholar]
- 17.Borden BA, Galecki P, Wellmann R, Danahey K, Lee SM, et al. 2019. Assessment of provider-perceived barriers to clinical use of pharmacogenomics during participation in an institutional implementation study. Pharmacogenet. Genom. 29:31–38 [DOI] [PubMed] [Google Scholar]
- 18.Borobia AM, Dapia I, Tong HY, Arias P, Munoz M, et al. 2018. Clinical implementation of pharmacogenetic testing in a hospital of the Spanish national health system: strategy and experience over 3 years. Clin. Transl. Sci. 11:189–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brothers KB, Langanke M, Erdmann P. 2013. Implications of the incidentalome for clinical pharmacogenomics. Pharmacogenomics 14:1353–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caraballo PJ, Bielinski SJ, St. Sauver JL, Weinshilboum RM. 2017. Electronic medical record-integrated pharmacogenomics and related clinical decision support concepts. Clin. Pharmacol. Ther. 102:254–64 [DOI] [PubMed] [Google Scholar]
- 21.Caraballo PJ, Hodge LS, Bielinski SJ, Stewart AK, Farrugia G, et al. 2017. Multidisciplinary model to implement pharmacogenomics at the point of care. Genet. Med. 19:421–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caraballo PJ, Sutton JA, Giri J, Wright JA, Nicholson WT, et al. 2020. Integrating pharmacogenomics into the electronic health record by implementing genomic indicators. J. Am. Med. Inform. Assoc. 27:154–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caudle KE, Gammal RS, Whirl-Carrillo M, Hoffman JM, Relling MV, Klein TE. 2016. Evidence and resources to implement pharmacogenetic knowledge for precision medicine. Am. J. Health Syst. Pharm. 73:1977–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caudle KE, Keeling NJ, Klein TE, Whirl-Carrillo M, Pratt VM, Hoffman JM. 2018. Standardization can accelerate the adoption of pharmacogenomics: current status and the path forward. Pharmacogenomics 19:847–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caudle KE, Klein TE, Hoffman JM, Muller DJ, Whirl-Carrillo M, et al. 2014. Incorporation of pharmacogenomics into routine clinical practice: the Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline development process. Curr. Drug Metab. 15:209–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cavallari LH, Weitzel KW, Elsey AR, Liu X, Mosley SA, et al. 2017. Institutional profile: University of Florida Health Personalized Medicine Program. Pharmacogenomics 18:421–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan SL, Liew HZW, Nguyen F, Thumboo J, Chow WC, Sung C. 2021. Prescription patterns of outpatients and the potential of multiplexed pharmacogenomic testing. Br. J. Clin. Pharmacol. 87:886–94 [DOI] [PubMed] [Google Scholar]
- 28.Chanfreau-Coffinier C, Hull LE, Lynch JA, DuVall SL, Damrauer SM, et al. 2019. Projected prevalence of actionable pharmacogenetic variants and level A drugs prescribed among US Veterans Health Administration pharmacy users. JAMA Netw. Open 2:e195345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cicali EJ, Elchynski AL, Cook KJ, Houder JT, Thomas CD, et al. 2021. How to integrate CYP2D6 phenoconversion into clinical pharmacogenetics: a tutorial. Clin. Pharmacol. Ther. 110:677–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cicali EJ, Weitzel KW, Elsey AR, Orlando FA, Vinson M, et al. 2019. Challenges and lessons learned from clinical pharmacogenetic implementation of multiple gene-drug pairs across ambulatory care settings. Genet. Med. 21:2264–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Claassens DMF, Vos GJA, Bergmeijer TO, Hermanides RS, van’t Hof AWJ, et al. 2019. A genotype-guided strategy for oral P2Y12 inhibitors in primary PCI. N. Engl. J. Med. 381:1621–31 [DOI] [PubMed] [Google Scholar]
- 31a.Clin. Pharmacogenet. Implement. Consort. 2020. API and database. Clinical Pharmacogenetics Implementation Consortium. https://cpicpgx.org/api-and-database [Google Scholar]
- 31b.Clin. Pharmacogenet. Implement. Consort. 2021. Genes-drugs. Clinical Pharmacogenetics Implementation Consortium. https://cpicpgx.org/implementation [Google Scholar]
- 31c.Clin. Pharmacogenet. Implement. Consort. 2021. Implementation. Clinical Pharmacogenetics Implementation Consortium. https://cpicpgx.org/implementation [Google Scholar]
- 32.Collins T, Power K, McCallie D Jr., Owings R 2019. Finding a place for pharmacogenetics in the electronic health record. Clin. Pharmacol. Ther. 106:295–97 [DOI] [PubMed] [Google Scholar]
- 33.Cortejoso L, Garcia-Gonzalez X, Garcia MI, Garcia-Alfonso P, Sanjurjo M, Lopez-Fernandez LA. 2016. Cost-effectiveness of screening for DPYD polymorphisms to prevent neutropenia in cancer patients treated with fluoropyrimidines. Pharmacogenomics 17:979–84 [DOI] [PubMed] [Google Scholar]
- 34.Crews KR, Monte AA, Huddart R, Caudle KE, Kharasch ED, et al. 2021. Clinical Pharmacogenetics Implementation Consortium guideline for CYP2D6, OPRM1, and COMT genotypes and select opioid therapy. Clin. Pharmacol. Ther. 110:888–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dressler LG, Bell GC, Ruch KD, Retamal JD, Krug PB, Paulus RA. 2018. Implementing a personalized medicine program in a community health system. Pharmacogenomics 19:1345–56 [DOI] [PubMed] [Google Scholar]
- 36.Duarte JD, Dalton R, Elchynski AL, Smith DM, Cicali EJ, et al. 2021. Multisite investigation of strategies for the clinical implementation of pre-emptive pharmacogenetic testing. Genet. Med. 23:2335–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dunnenberger HM, Biszewski M, Bell GC, Sereika A, May H, et al. 2016. Implementation of a multidisciplinary pharmacogenomics clinic in a community health system. Am. J. Health Syst. Pharm. 73:1956–66 [DOI] [PubMed] [Google Scholar]
- 38.Dunnenberger HM, Crews KR, Hoffman JM, Caudle KE, Broeckel U, et al. 2015. Preemptive clinical pharmacogenetics implementation: current programs in five US medical centers. Annu. Rev. Pharmacol. Toxicol. 55:89–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duong BQ, Arwood MJ, Hicks JK, Beitelshees AL, Franchi F, et al. 2020. Development of customizable implementation guides to support clinical adoption of pharmacogenomics: experiences of the Implementing GeNomics In pracTicE (IGNITE) Network. Pharmacogenom. Pers. Med 13:217–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Empey PE, Pratt VM, Hoffman JM, Caudle KE, Klein TE. 2021. Expanding evidence leads to new pharmacogenomics payer coverage. Genet. Med. 23:830–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feldman SS, Buchalter S, Hayes LW. 2018. Health information technology in healthcare quality and patient safety: literature review. JMIR Med. Inform. 6:e10264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Filipski KK, Murphy JD, Helzlsouer KJ. 2017. Updating the landscape of direct-to-consumer pharmacogenomic testing. Pharmacogenom. Pers. Med. 10:229–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fitzgerald G, Prince C, Downing J, Reynolds J, Zhang JE, et al. 2019. Processes and barriers to implementation of point-of-care genotype-guided dosing of warfarin into UK outpatient anticoagulation clinics. Pharmacogenomics 20:599–608 [DOI] [PubMed] [Google Scholar]
- 45.Flockhart DA, Skaar T, Berlin DS, Klein TE, Nguyen AT. 2009. Clinically available pharmacogenomics tests. Clin. Pharmacol. Ther. 86:109–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Freimuth RR, Formea CM, Hoffman JM, Matey E, Peterson JF, Boyce RD. 2017. Implementing genomic clinical decision support for drug-based precision medicine. CPT Pharmacometr. Syst. Pharmacol. 6:153–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frigon MP, Blackburn ME, Dubois-Bouchard C, Gagnon AL, Tardif S, Tremblay K. 2019. Pharmacogenetic testing in primary care practice: opinions of physicians, pharmacists and patients. Pharmacogenomics 20:589–98 [DOI] [PubMed] [Google Scholar]
- 48.Gammal RS, Crews KR, Haidar CE, Hoffman JM, Baker DK, et al. 2016. Pharmacogenetics for safe codeine use in sickle cell disease. Pediatrics 138:e20153479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gammal RS, Lee YM, Petry NJ, Iwuchukwu O, Hoffman JM, et al. 2021. Pharmacists leading the way to precision medicine: updates to the core pharmacist competencies in genomics. Am. J. Pharm. Educ:8634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gammal RS, Mayes J, Caudle KE. 2019. Ready or not, here it comes: direct-to-consumer pharmacogenomic testing and its implications for community pharmacists. J. Am. Pharm. Assoc. 59:646–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giannopoulou E, Katsila T, Mitropoulou C, Tsermpini EE, Patrinos GP. 2019. Integrating next-generation sequencing in the clinical pharmacogenomics workflow. Front. Pharmacol. 10:384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goldspiel BR, Flegel WA, DiPatrizio G, Sissung T, Adams SD, et al. 2014. Integrating pharmacogenetic information and clinical decision support into the electronic health record. J. Am. Med. Inform. Assoc. 21:522–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gonsalves SG, Dirksen RT, Sangkuhl K, Pulk R, Alvarellos M, et al. 2019. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for the use of potent volatile anesthetic agents and succinylcholine in the context of RYR1 or CACNA1S genotypes. Clin. Pharmacol. Ther. 105:1338–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haga SB, Moaddeb J. 2014. Comparison of delivery strategies for pharmacogenetic testing services. Pharmacogenet. Genom. 24:139–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haidar CE, Petry N, Oxencis C, Douglas JS, Hoffman JM. 2021. ASHP statement on the pharmacist’s role in clinical pharmacogenomics. Am. J. Health Syst. Pharm. In press. 10.1093/ajhp/zxab339 [DOI] [PubMed] [Google Scholar]
- 56.Haidar CE, Relling MV, Hoffman JM. 2019. Preemptively precise: returning and updating pharmacogenetic test results to realize the benefits of preemptive testing. Clin. Pharmacol. Ther. 106:942–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hamzic S, Aebi S, Joerger M, Montemurro M, Ansari M, et al. 2020. Fluoropyrimidine chemotherapy: recommendations for DPYD genotyping and therapeutic drug monitoring of the Swiss Group of Pharmacogenomics and Personalised Therapy. Swiss Med. Wkly. 150:w20375. [DOI] [PubMed] [Google Scholar]
- 58.Harada S, Zhou Y, Duncan S, Armstead AR, Coshatt GM, et al. 2017. Precision medicine at the University of Alabama at Birmingham: laying the foundational processes through implementation of genotype-guided antiplatelet therapy. Clin. Pharmacol. Ther. 102:493–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heise CW, Gallo T, Curry SC, Woosley RL. 2020. Identification of populations likely to benefit from pharmacogenomic testing. Pharmacogenet. Genom. 30:91–95 [DOI] [PubMed] [Google Scholar]
- 60.Hicks JK, Crews KR, Hoffman JM, Kornegay NM, Wilkinson MR, et al. 2012. A clinician-driven automated system for integration of pharmacogenetic interpretations into an electronic medical record. Clin. Pharmacol. Ther. 92:563–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hicks JK, Dunnenberger HM, Gumpper KF, Haidar CE, Hoffman JM. 2016. Integrating pharmacogenomics into electronic health records with clinical decision support. Am. J. Health Syst. Pharm. 73:1967–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hicks JK, Sangkuhl K, Swen JJ, Ellingrod VL, Muller DJ, et al. 2017. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin. Pharmacol. Ther. 102:37–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hicks JK, Stowe D, Willner MA, Wai M, Daly T, et al. 2016. Implementation of clinical pharmacogenomics within a large health system: from electronic health record decision support to consultation services. Pharmacotherapy 36:940–48 [DOI] [PubMed] [Google Scholar]
- 64.Hoffman JM, Dunnenberger HM, Kevin Hicks J, Caudle KE, Whirl Carrillo M, et al. 2016. Developing knowledge resources to support precision medicine: principles from the Clinical Pharmacogenetics Implementation Consortium (CPIC). J. Am. Med. Inform. Assoc 23:796–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hoffman JM, Flynn AJ, Juskewitch JE, Freimuth RR. 2020. biomedical data science and informatics challenges to implementing pharmacogenomics with electronic health records. Annu. Rev. Biomed. Data Sci. 3:289–314 [Google Scholar]
- 66.Hoffman JM, Haidar CE, Wilkinson MR, Crews KR, Baker DK, et al. 2014. PG4KDS: a model for the clinical implementation of pre-emptive pharmacogenetics. Am. J. Med. Genet. C 166C:45–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huddart R, Sangkuhl K, Whirl-Carrillo M, Klein TE. 2019. Are randomized controlled trials necessary to establish the value of implementing pharmacogenomics in the clinic? Clin. Pharmacol. Ther. 106:284–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ji Y, Skierka JM, Blommel JH, Moore BE, VanCuyk DL, et al. 2016. Preemptive Pharmacogenomic testing for precision medicine: a comprehensive analysis of five actionable pharmacogenomic genes using next-generation DNA sequencing and a customized CYP2D6 genotyping cascade. J. Mol. Diagn. 18:438–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jorgensen AL, Prince C, Fitzgerald G, Hanson A, Downing J, et al. 2019. Implementation of genotype-guided dosing of warfarin with point-of-care genetic testing in three UK clinics: a matched cohort study. BMC Med. 17:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Karnes JH, Rettie AE, Somogyi AA, Huddart R, Fohner AE, et al. 2021. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2C9 and HLA-B genotypes and phenytoin dosing: 2020 update. Clin. Pharmacol. Ther. 109:302–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kasi PM, Koep T, Schnettler E, Shahjehan F, Kamatham V, et al. 2019. Feasibility of integrating panel-based pharmacogenomics testing for chemotherapy and supportive care in patients with colorectal cancer. Technol. Cancer Res. Treat. 18. 10.1177/1533033819893667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Keeling NJ, Rosenthal MM, West-Strum D, Patel AS, Haidar CE, Hoffman JM. 2019. Preemptive pharmacogenetic testing: exploring the knowledge and perspectives of US payers. Genet. Med. 21:1224–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Khelifi M, Tarczy-Hornoch P, Devine EB, Pratt W. 2017. Design recommendations for pharmacogenomics clinical decision support systems. AMIA Jt. Summits Transl. Sci. Proc 2017:237–46 [PMC free article] [PubMed] [Google Scholar]
- 74.Kim GJ, Lee SY, Park JH, Ryu BY, Kim JH. 2017. Role of preemptive genotyping in preventing serious adverse drug events in South Korean patients. Drug Saf. 40:65–80 [DOI] [PubMed] [Google Scholar]
- 75.Kimpton JE, Carey IM, Threapleton CJD, Robinson A, Harris T, et al. 2019. Longitudinal exposure of English primary care patients to pharmacogenomic drugs: an analysis to inform design of pre-emptive pharmacogenomic testing. Br. J. Clin. Pharmacol. 85:2734–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koch BC, van Schaik RH, van Gelder T, Mathijssen RH (Rotterdam Clin. Pharmacol.-Pharmacogenet. Group). 2013. Doubt about the feasibility of preemptive genotyping. Clin. Pharmacol. Ther. 93:233. [DOI] [PubMed] [Google Scholar]
- 77.Kuch W, Rinner C, Gall W, Samwald M. 2016. How many patients could benefit from pre-emptive pharmacogenomic testing and decision support? a retrospective study based on nationwide Austrian claims data. Stud. Health Technol. Inform. 223:253–58 [PubMed] [Google Scholar]
- 78.Kullo IJ, Haddad R, Prows CA, Holm I, Sanderson SC, et al. 2014. Return of results in the genomic medicine projects of the eMERGE network. Front. Genet. 5:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lauschke VM, Ingelman-Sundberg M. 2016. Requirements for comprehensive pharmacogenetic genotyping platforms. Pharmacogenomics 17:917–24 [DOI] [PubMed] [Google Scholar]
- 80.Leary E, Brilliant M, Peissig P, Griesbach S. 2019. Preliminary outcomes of preemptive warfarin pharmacogenetic testing at a large rural healthcare center. Am. J. Health Syst. Pharm. 76:387–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee EM, Xu K, Mosbrook E, Links A, Guzman J, et al. 2016. Pharmacogenomic incidental findings in 308 families: the NIH Undiagnosed Diseases Program experience. Genet. Med. 18:1303–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lima JJ, Thomas CD, Barbarino J, Desta Z, Van Driest SL, et al. 2021. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2C19 and proton pump inhibitor dosing. Clin. Pharmacol. Ther. 109:1417–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Linan MK, Sottara D, Freimuth RR. 2015. Creating shareable clinical decision support rules for a pharmacogenomics clinical guideline using structured knowledge representation. AMIA Annu. Symp. Proc. 2015:1985–94 [PMC free article] [PubMed] [Google Scholar]
- 84.Lipschultz E, Danahey K, Truong TM, Schierer E, Volchenboum SL, et al. 2021. Creation of a pharmacogenomics patient portal complementary to an existing institutional provider-facing clinical decision support system. JAMIA Open 4:ooab067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu M, Vnencak-Jones CL, Roland BP, Gatto CL, Mathe JL, et al. 2021. A tutorial for pharmacogenomics implementation through end-to-end clinical decision support based on ten years of experience from PREDICT. Clin. Pharmacol. Ther. 109:101–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Luczak T, Brown SJ, Armbruster D, Hundertmark M, Brown J, Stenehjem D. 2021. Strategies and settings of clinical pharmacogenetic implementation: a scoping review of pharmacogenetics programs. Pharmacogenomics 22:345–64 [DOI] [PubMed] [Google Scholar]
- 87.Luzum JA, Petry N, Taylor AK, Van Driest SL, Dunnenberger HM, Cavallari LH. 2021. Moving pharmacogenetics into practice: It’s all about the evidence! Clin. Pharmacol. Ther. 110:649–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mahendraratnam N, Mercon K, Gill M, Benzing L, McClellan MB. 2022. Understanding use of real-world data and real-world evidence to support regulatory decisions on medical product effectiveness. Clin. Pharmacol. Ther. 111:150–54 [DOI] [PubMed] [Google Scholar]
- 89.Manzi SF, Fusaro VA, Chadwick L, Brownstein C, Clinton C, et al. 2017. Creating a scalable clinical pharmacogenomics service with automated interpretation and medical record result integration – experience from a pediatric tertiary care facility. J. Am. Med. Inform. Assoc. 24:74–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Marrero RJ, Cicali EJ, Arwood MJ, Eddy E, DeRemer D, et al. 2020. How to transition from single-gene pharmacogenetic testing to preemptive panel-based testing: a tutorial. Clin. Pharmacol. Ther. 108:557–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Matic M, Nijenhuis M, Soree B, de Boer-Veger NJ, Buunk AM, et al. 2021. Dutch Pharmacogenetics Working Group (DPWG) guideline for the gene-drug interaction between CYP2D6 and opioids (codeine, tramadol and oxycodone). Eur. J. Hum. Genet. 10.1038/s41431-021-00920-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mehandziska S, Stajkovska A, Stavrevska M, Jakovleva K, Janevska M, et al. 2020. Workflow for the implementation of precision genomics in healthcare. Front. Genet. 11:619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mills R, Ensinger M, Callanan N, Haga SB. 2017. Development and initial assessment of a patient education video about pharmacogenetics. J. Pers. Med. 7:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mitropoulos K, Al Jaibeji H, Forero DA, Laissue P, Wonkam A, et al. 2015. Success stories in genomic medicine from resource-limited countries. Hum. Genom. 9:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Natasha P, Baye J, Aifaoui A, Wilke RA, Lupu RA, et al. 2019. Implementation of wide-scale pharmacogenetic testing in primary care. Pharmacogenomics 20:903–13 [DOI] [PubMed] [Google Scholar]
- 96.Natl. Hum. Genome Res. Inst. 2021. Educational resources. National Human Genome Research Institute. https://www.genome.gov/About-Genomics/Educational-Resources [Google Scholar]
- 98.Nickola TJ, Green JS, Harralson AF, O’Brien TJ. 2012. The current and future state of pharmacogenomics medical education in the USA. Pharmacogenomics 13:1419–25 [DOI] [PubMed] [Google Scholar]
- 99.Nishimura AA, Shirts BH, Dorschner MO, Amendola LM, Smith JW, et al. 2015. Development of clinical decision support alerts for pharmacogenomic incidental findings from exome sequencing. Genet. Med. 17:939–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nofziger C, Turner AJ, Sangkuhl K, Whirl-Carrillo M, Agundez JAG, et al. 2020. PharmVar GeneFocus: CYP2D6. Clin. Pharmacol. Ther. 107:154–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.O’Donnell PH, Danahey K, Jacobs M, Wadhwa NR, Yuen S, et al. 2014. Adoption of a clinical pharmacogenomics implementation program during outpatient care—initial results of the University of Chicago “1,200 Patients Project.” Am. J. Med. Genet. C 166C:68–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.O’Donnell PH, Wadhwa N, Danahey K, Borden BA, Lee SM, et al. 2017. Pharmacogenomics-based point-of-care clinical decision support significantly alters drug prescribing. Clin. Pharmacol. Ther. 102:859–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pasternak AL, Ward KM, Ateya MB, Choe HM, Thompson AN, et al. 2020. Establishment of a pharmacogenetics service focused on optimizing existing pharmacogenetic testing at a large academic health center. J. Pers. Med. 10:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Petrek M, Kocourkova L, Zizkova V, Nosek Z, Taborsky M, Petrkova J. 2016. Characterization of three CYP2C19 gene variants by MassARRAY and point of care techniques: experience from a Czech centre. Arch. Immunol. Ther. Exp. 64:99–107 [DOI] [PubMed] [Google Scholar]
- 105.Rahmatian D, Tadrous M. 2021. Old dog with new tricks: an introduction to real-world evidence for pharmacists. Am. J. Health Syst. Pharm. 78:2277–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ramsey LB, Prows CA, Zhang K, Saldana SN, Sorter MT, et al. 2019. Implementation of pharmacogenetics at Cincinnati Children’s Hospital Medical Center: lessons learned over 14 years of personalizing medicine. Clin. Pharmacol. Ther. 105:49–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Raphael MJ, Gyawali B, Booth CM. 2020. Real-world evidence and regulatory drug approval. Nat. Rev. Clin. Oncol. 17:271–72 [DOI] [PubMed] [Google Scholar]
- 108.Rasmussen LV, Smith ME, Almaraz F, Persell SD, Rasmussen-Torvik LJ, et al. 2019. An ancillary genomics system to support the return of pharmacogenomic results. J. Am. Med. Inform. Assoc. 26:306–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rasmussen-Torvik LJ, Stallings SC, Gordon AS, Almoguera B, Basford MA, et al. 2014. Design and anticipated outcomes of the eMERGE-PGx project: a multicenter pilot for preemptive pharmacogenomics in electronic health record systems. Clin. Pharmacol. Ther. 96:482–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Reisberg S, Krebs K, Lepamets M, Kals M, Magi R, et al. 2019. Translating genotype data of 44,000 biobank participants into clinical pharmacogenetic recommendations: challenges and solutions. Genet. Med. 21:1345–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Relling MV, Klein TE. 2011. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin. Pharmacol. Ther. 89:464–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Relling MV, Klein TE, Gammal RS, Whirl-Carrillo M, Hoffman JM, Caudle KE. 2020. The Clinical Pharmacogenetics Implementation Consortium: 10 years later. Clin. Pharmacol. Ther. 107:171–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Roberts JD, Wells GA, Le May MR, Labinaz M, Glover C, et al. 2012. Point-of-care genetic testing for personalisation of antiplatelet treatment (RAPID GENE): a prospective, randomised, proof-of-concept trial. Lancet 379:1705–11 [DOI] [PubMed] [Google Scholar]
- 114.Roden DM, Van Driest SL, Mosley JD, Wells QS, Robinson JR, et al. 2018. Benefit of preemptive pharmacogenetic information on clinical outcome. Clin. Pharmacol. Ther. 103:787–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Roederer MW, Kuo GM, Kisor DF, Frye RF, Hoffman JM, et al. 2017. Pharmacogenomics competencies in pharmacy practice: a blueprint for change. J. Am. Pharm. Assoc. 2003 57:120–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rosenman MB, Decker B, Levy KD, Holmes AM, Pratt VM, Eadon MT. 2017. Lessons learned when introducing pharmacogenomic panel testing into clinical practice. Value Health 20:54–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Saito Y, Stamp LK, Caudle KE, Hershfield MS, McDonagh EM, et al. 2016. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for human leukocyte antigen B (HLA-B) genotype and allopurinol dosing: 2015 update. Clin. Pharmacol. Ther. 99:36–37. Correction. 2013. Clin. Pharmacol. Ther. 93:153–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Samwald M, Xu H, Blagec K, Empey PE, Malone DC, et al. 2016. Incidence of exposure of Patients in the United States to multiple drugs for which pharmacogenomic guidelines are available. PLOS ONE 11:e0164972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Schildcrout JS, Denny JC, Bowton E, Gregg W, Pulley JM, et al. 2012. Optimizing drug outcomes through pharmacogenetics: a case for preemptive genotyping. Clin. Pharmacol. Ther. 92:235–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Scott SA. 2013. Clinical pharmacogenomics: opportunities and challenges at point of care. Clin. Pharmacol. Ther. 93:33–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Scott SA, Owusu Obeng A, Botton MR, Yang Y, Scott ER, et al. 2017. Institutional profile: translational pharmacogenomics at the Icahn School of Medicine at Mount Sinai. Pharmacogenomics 18:1381–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shahandeh A, Johnstone DM, Atkins JR, Sontag JM, Heidari M, et al. 2016. Advantages of array-based technologies for pre-emptive pharmacogenomics testing. Microarrays 5:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shi Y, Graves JA, Garbett SP, Zhou Z, Marathi R, et al. 2019. A decision-theoretic approach to panel-based, preemptive genotyping. MDM Policy Pract 4. 10.1177/2381468319864337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shotelersuk V, Tongsima S, Pithukpakorn M, Eu-Ahsunthornwattana J, Mahasirimongkol S. 2019. Precision medicine in Thailand. Am. J. Med. Genet. C 181:245–53 [DOI] [PubMed] [Google Scholar]
- 125.Shuldiner AR, Palmer K, Pakyz RE, Alestock TD, Maloney KA, et al. 2014. Implementation of pharmacogenetics: the University of Maryland Personalized Anti-platelet Pharmacogenetics Program. Am. J. Med. Genet. C 166C:76–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sissung TM, McKeeby JW, Patel J, Lertora JJ, Kumar P, et al. 2017. Pharmacogenomics implementation at the National Institutes of Health Clinical Center. J. Clin. Pharmacol. 57 (Suppl. 10):S67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Srinivasan S, Won NY, Dotson WD, Wright ST, Roberts MC. 2020. Barriers and facilitators for cascade testing in genetic conditions: a systematic review. Eur. J. Hum. Genet. 28:1631–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.St. Jude Child. Res. Hosp. 2021. PG4KDS: clinical implementation of pharmacogenetics. St. Jude Children’s Research Hospital. https://www.stjude.org/pg4kds [Google Scholar]
- 129a.US Cent. Medicare Medicaid Serv. 2020. MolDX: pharmacogenomics testing. Local Cover. Determ. L38294, US Cent. Medicare Medicaid Serv., Baltimore, MD. https://www.cms.gov/medicare-coverage-database/view/lcd.aspx?LCDId=38294&ver=16 [Google Scholar]
- 129b.US Food Drug Adm. 2018. Framework for FDA’s Real-World Evidence Program. Rep, US Food Drug Adm., Silver Spring, MD. https://www.fda.gov/media/120060/download [Google Scholar]
- 129.US Food Drug Adm. 2021. Prescription drug labeling resources. US Food and Drug Adminstration. https://www.fda.gov/drugs/laws-acts-and-rules/prescription-drug-labeling-resources [Google Scholar]
- 130.Unertl KM, Jaffa H, Field JR, Price L, Peterson JF. 2015. Clinician perspectives on using pharmacogenomics in clinical practice. Pers. Med. 12:339–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.van der Wouden CH, Bank PCD, Ozokcu K, Swen JJ, Guchelaar HJ. 2019. Pharmacist-initiated pre-emptive pharmacogenetic panel testing with clinical decision support in primary care: record of PGx results and real-world impact. Genes 10:416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.van der Wouden CH, Cambon-Thomsen A, Cecchin E, Cheung KC, Davila-Fajardo CL, et al. 2017. Implementing pharmacogenomics in Europe: design and implementation strategy of the Ubiquitous Pharmacogenomics Consortium. Clin. Pharmacol. Ther. 101:341–58 [DOI] [PubMed] [Google Scholar]
- 133.van der Wouden CH, van Rhenen MH, Jama WOM, Ingelman-Sundberg M, Lauschke VM, et al. 2019. Development of the PGx-Passport: a panel of actionable germline genetic variants for pre-emptive pharmacogenetic testing. Clin. Pharmacol. Ther. 106:866–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Vermehren C, Sogaard Nielsen R, Jorgensen S, Drastrup AM, Westergaard N. 2020. Drug use among nursing home residents in Denmark for drugs having pharmacogenomics based (PGx) dosing guidelines: potential for preemptive PGx testing. J. Pers. Med 10:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Vo TT, Bell GC, Owusu Obeng A, Hicks JK, Dunnenberger HM. 2017. Pharmacogenomics implementation: considerations for selecting a reference laboratory. Pharmacotherapy 37:1014–22 [DOI] [PubMed] [Google Scholar]
- 136.Volpi S, Bult CJ, Chisholm RL, Deverka PA, Ginsburg GS, et al. 2018. Research directions in the clinical implementation of pharmacogenomics: an overview of US programs and projects. Clin. Pharmacol. Ther. 103:778–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Vu CL, Chan J, Todaro M, Skafidas S, Kwan P. 2015. Point-of-care molecular diagnostic devices: an overview. Pharmacogenomics 16:1399–409 [DOI] [PubMed] [Google Scholar]
- 138.Wake DT, Bell GC, Gregornik DB, Ho TT, Dunnenberger HM. 2021. Synthesis of major pharmacogenomics pretest counseling themes: a multisite comparison. Pharmacogenomics 22:165–76 [DOI] [PubMed] [Google Scholar]
- 139.Wake DT, Smith DM, Kazi S, Dunnenberger HM. 2021. Pharmacogenomic clinical decision support: a review, how-to guide, and future vision. Clin. Pharmacol. Ther. 10.1002/cpt.2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wallner J, Beck IA, Panpradist N, Ruth PS, Valenzuela-Ponce H, et al. 2021. Rapid, near point-of-care assay for HLA-B*57:01 genotype associated with severe hypersensitivity reaction to abacavir. AIDS Res. Hum. Retroviruses. In press. 10.1089/aid.2021.0103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Weitzel KW, Aquilante CL, Johnson S, Kisor DF, Empey PE. 2016. Educational strategies to enable expansion of pharmacogenomics-based care. Am. J. Health Syst. Pharm. 73:1986–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Weitzel KW, Cavallari LH, Lesko LJ. 2017. Preemptive panel-based pharmacogenetic testing: The time is now. Pharm. Res. 34:1551–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Weitzel KW, Elsey AR, Langaee TY, Burkley B, Nessl DR, et al. 2014. Clinical pharmacogenetics implementation: approaches, successes, and challenges. Am. J. Med. Genet. C 166C:56–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Williams MS. 2019. Early lessons from the implementation of genomic medicine programs. Annu. Rev. Genom. Hum. Genet. 20:389–411 [DOI] [PubMed] [Google Scholar]
- 145.Yang W, Wu G, Broeckel U, Smith CA, Turner V, et al. 2016. Comparison of genome sequencing and clinical genotyping for pharmacogenes. Clin. Pharmacol. Ther. 100:380–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhou Y, Krebs K, Milani L, Lauschke VM. 2021. Global frequencies of clinically important HLA alleles and their implications for the cost-effectiveness of preemptive pharmacogenetic testing. Clin. Pharmacol. Ther. 109:160–74 [DOI] [PubMed] [Google Scholar]
- 147.Zhu Y, Moriarty JP, Swanson KM, Takahashi PY, Bielinski SJ, et al. 2021. A model-based cost-effectiveness analysis of pharmacogenomic panel testing in cardiovascular disease management: preemptive, reactive, or none? Genet. Med. 23:461–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhu Y, Swanson KM, Rojas RL, Wang Z, St. Sauver JL, et al. 2020. Systematic review of the evidence on the cost-effectiveness of pharmacogenomics-guided treatment for cardiovascular diseases. Genet. Med. 22:475–86 [DOI] [PMC free article] [PubMed] [Google Scholar]


