Abstract
A major 30.5-kb cluster of nif and associated genes of Acetobacter diazotrophicus (syn. Gluconacetobacter diazotrophicus), a nitrogen-fixing endophyte of sugarcane, was sequenced and analyzed. This cluster represents the largest assembly of contiguous nif-fix and associated genes so far characterized in any diazotrophic bacterial species. Northern blots and promoter sequence analysis indicated that the genes are organized into eight transcriptional units. The overall arrangement of genes is most like that of the nif-fix cluster in Azospirillum brasilense, while the individual gene products are more similar to those in species of Rhizobiaceae or in Rhodobacter capsulatus.
Biological nitrogen fixation occurs in species of more than 100 genera distributed among several of the major phylogenetic divisions of prokaryotes (Eubacteria and Archaea) (25). Sequence and mutational analyses of the genes necessary for nitrogen fixation (nif) in many diazotrophs indicate that their products have common structures and functions, while the degree of linkage and arrangement of specific nif and associated genes vary considerably (5, 8, 17). In addition, nif genes and genes involved in plant invasion and nitrogen fixation effectiveness, such as nod and fix in species of Rhizobiaceae, are often linked.
The identification of nitrogen-fixing bacteria with endophytic habitats raises the possibility of a new classification of symbiosis (3). The relationship of a proteobacterial α group member, Acetobacter diazotrophicus (syn. Gluconacetobacter diazotrophicus), with sugarcane represents a promising model system for the study of an association between a monocot and an endophytic nitrogen-fixing bacterium (13, 22). The ability of A. diazotrophicus to enhance sugarcane growth has been documented, and while the benefit to plant growth might be due at least in part to the transfer of bacterially fixed N, another plant growth-stimulating factor(s) is indicated, possibly auxin production by A. diazotrophicus (22). Because of its potential agronomic use and unique status as the only diazotrophic species of Acetobacter so far identified, it was of interest to isolate and characterize genes that are involved in nitrogen fixation and regulation.
Identification of a major cluster of nif and associated genes.
A genomic library of A. diazotrophicus constructed in the wide-host-range cosmid pLAFR3 (Tetr) (4) was transferred by conjugation from Escherichia coli to several different nif mutants of Azotobacter vinelandii (12). Two mutants that yielded Nif+ Tetr transconjugants were DJ71 (nifV) and DJ35 (nifE), and several transconjugants of both strains carried the same cosmid, pAD71, the 22-kb insert fragment of which carries nifD through mcpA (Fig. 1). One end of the insert fragment contained sequences identical to a portion of the nifHDK region cloned previously (M. Sevilla and C. Kennedy, submitted for publication) and also to a portion of pAD101, another cosmid from the pLAFR3 library, isolated by its ability to complement nifA mutants of A. vinelandii (24). A third cosmid, pAD102, which also complemented nifA mutants, provided the region further upstream of nifA.
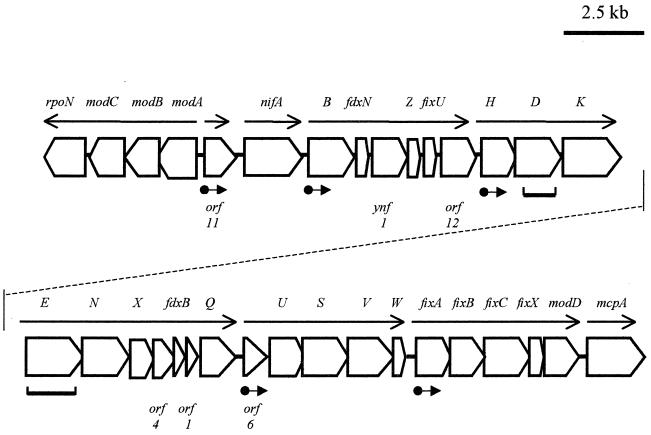
FIG. 1.
Major nif gene map of A. diazotrophieus. Arrows indicate the positions and direction for transcription initiation sites. •→, ς54-, NifA-dependent promoters. The thick lines underneath represent the DNA fragments used as probes in Northern analysis.
Thirty-two open reading frames (ORFs) were identified in the>30 kbp of sequence obtained (Fig. 1). Gene characteristics, potential regulatory sequences, and gene products are summarized in Table 1. Genes not known to be directly involved in nitrogenase structure, function, or regulation of gene expression include mcpA, encoding a methyl-accepting chemotaxis protein that responds to extracellular signals for chemotactic responses (19), and modABCD, which encode a high-affinity molybdate transport system in E. coli (9), Staphylococcus carnosus (20), and A. vinelandii (18). FixABCX may comprise an electron transfer chain, and, while the fixABCX genes were first identified in Rhizobium meliloti (6) and subsequently in other diazotrophs, some or all are also present in the genomes of E. coli (fixABCX) and Mycobacterium tuberculosis (fixAB), neither of which is a diazotroph. The Fix proteins in E. coli are required for carnitine breakdown, an anaerobic function related to respiration (2), while their function in other organisms is unknown (7). Also of unknown function but similar to gene products in other diazotrophs are the ferredoxins FdxN and FdxB and the products of ORFs 1, 4, and 6, named on the basis of ORF numbers originally assigned. The only ORF without significant similarity to known genes is the one between rpoN and modC, orf11. The nif-fix and associated gene cluster characterized here for A. diazotrophicus is unique in representing the largest single grouping of genes required for nitrogenase structure and function, nif and fix gene regulation (nifA and rpoN), and associated functions (molybdenum uptake and electron transfer) found in any diazotroph so far studied.
TABLE 1.
Comparison of nif-associated gene products
| Genea | Function | Regulatory feature | Product size (kDa) | Organisms with most similar gene productsb (% amino acid identity c) |
|---|---|---|---|---|
| rpoN | ς54 | 51.3 | B. japonicum (34), A. caulinodans (31), C. crescentus (30) | |
| modC | Molybdenum transport; ATP-binding protein | 36.8 | R. capsulatus(48), A. vinelandii (47), E. coli (46) | |
| modB | Molybdenum transport; permease protein | 24.7 | R. capsulatus (57), A. vinelandii (40), A. aeolicus (35) | |
| modA | Molybdenum transport; molybdate-binding periplasmic protein | 26.9 | R. capsulatus (42), A. vinelandii (40), A. aeolicus (32) | |
| orf11 | Unknown | NifA-, ςn-binding sites | 20.2 | |
| nifA | Transcriptional activator | NifA-binding sites | 62.2 | A. caulinodans (51), B. japonicum (50), R. etli (47) |
| nifB | Fe-Mo cofactor synthesis | NifA-, ςn-binding sites | 51.9 | R. capsulatus (67), R. meliloti (67), Rhizobium sp. strain NGR234 (66) |
| fdxN | Ferredoxin | 6.8 | R. palustris (77), Rhizobium sp. strain NGR234 (67), R. meliloti (67) | |
| ynfl | Unknown | 26.4 | R. capsulatus (31) | |
| nifZ | Maturation and activation | 6.9 | Rhizobium sp. strain NGR234 (63), R. capsulatus (59), E. agglomerans (47) | |
| fixU | Unknown | 8.6 | R. leguminosarum (56), Rhizobium sp. strain NGR234 (55) | |
| orfl2 | Unknown | 36.4 | A. brasilense (42), A. vinelandii (28) | |
| nifH | Nitrogenase structure; Fe protein | NifA-, ςn-binding site | 31.9 | R. phaseoli (89), Rhizobium sp. strain NGR234 (85), R. meliloti (85) |
| nifD | Nitrogenase structure; Fe-Mo protein alpha subunit | 55.9 | Rhizobium sp. strain NGR234 (80), P. rhizobium (75), H. seropedicae (75) | |
| nifK | Nitrogenase structure; Fe-Mo protein beta subunit | 57.2 | Rhizobium sp. strain NGR234 (71), B. japonicum (66), T. ferrooxidans (62) | |
| nifE | Fe-Mo cofactor synthesis | ςn-binding site | 51.8 | B. japonicum (60), Rhizobium sp. strain NGR234 (58), R. capsulatus (57) |
| nifN | Fe-Mo cofactor synthesis | 49.5 | R. meliloti (50), Rhizobium sp. strain NGR234 (49), B. japonicum (46) | |
| nifX | Fe-Mo cofactor synthesis | 17.4 | Rhizobium sp. strain NGR234 (55), R. capsulatus (46), H. seropedicae (37) | |
| orf4 | Unknown | 16.9 | A. caulinodans (53), Frankia spp. (50), F. alni (45) | |
| orf1 | Unknown | 7.6 | R. capsulatus (50), Rhizobium sp. strain NGR234 (48), Anabgena PCC7120 (39) | |
| fdxB | Ferredoxin | 9.9 | R. capsulatus (41), Rhizobium sp. strain NGR234 (41), P. boryanum (34) | |
| nifQ | Fe-Mo cofactor synthesis | ρ-independent terminator | 22.9 | R. capsulatus (41), A. vinelandii (40), Rhizobium sp. strain NGR234 (33) |
| orf6 | Unknown | NifA-, ςn-binding sites | 10.9 | R. capsulatus (48), R. sphaeroides (46), B. japonicum (45) |
| nifU | Maturation and activation; assembly of iron-sulfur clusters | 32.9 | A. vinelandii (55), A. brasilense (53), A. chroococcum (52) | |
| nifS | Maturation and activation; homodimeric cysteine desulfurase | 43.0 | A. vinelandii (59), Rhizobium sp. strain NGR234 (59), R. sphaeroides (59) | |
| nifV | Fe-Mo cofactor synthesis; homocitrate synthase | 92.2 | Z. mobilis (61), R. sphaeroides (51), F. alni (49) | |
| nifW | Maturation and activation; oxygen protection of the Mo-Fe protein | ρ-independent terminator | 12.0 | A. caulinodans (48), Rhizobium sp. strain NGR234 (42), A. brasilense (41) |
| fixA | Electron transfer; electron transfer flavoprotein beta subunit | NifA-, ςn-binding sites | 30.7 | A. caulinodans (69), Rhizobium sp. strain NGR234 (67), B. japonicum (66) |
| fixB | Electron transfer; electron transfer flavoprotein alpha subunit | 38.9 | A. caulinodans (68), Rhizobium sp. strain NGR234 (67), B. japonicum (67) | |
| fixC | Electron transfer; electron transfer flavoprotein-quinone oxidoreductase | 48.1 | Rhizobium sp. strain NGR234 (63), B. japonicum (62), A. caulinodans (61) | |
| fixX | Electron transfer; ferredoxin-like protein | 10.7 | R. leguminosarum (73), R. meliloti (71), Rhizobium sp. strain NGR234 (70) | |
| modD | Molybdenum transport | 26.7 | Z. mobilis (51), R. capsulatus (41), E. coli (39) | |
| mcpA | Chemotaxis | 69.3 | C. crescentus (63), R. sphaeroides (51), R. leguminosarum (44) |
Organization of gene expression is indicated by arrows in Fig. 1.
Organism in which the gene product most similar to that of A. diazotrophicus was found. Organisms: Aquifex aeolicust, Azorhizobium caulinodans, Azotobacter chroococcum, Azotobacter vinelandii, Bradyrhizobium japonicum, Caulobacter crescentus, Escherichia coli, Frankia alni, Herbaspirillum seropedicae, Parasponia rhizobium, Plectonema boryamum, Rhizobium etli, Rhizobium leguminosarum, Rhizobium meliloti, Rhizobium phaseoli, Rhodobacter capsulatus, Rhodobacter sphaeroides, Rhodopseudomonas palustris, Thiobacillus ferrooxidans, and Zymomonas mobilis.
Identity of the deduced A. diazotrophicus gene product sequence to the gene product of the three organisms to which it is most related.
Comparison of organization of nif genes and gene products.
The individual A. diazotrophicus gene products are generally most similar to those found in other α group proteobacteria, with 17 gene products being most like those in members of the Rhizobiaceae and 9 gene products being most closely related to Rhodobacter capsulatus proteins (Table 1). NifU and NifS were most similar to the gene products of Azotobacter species, members of the γ group of proteobacteria. McpA was most similar to the mcpA gene product of the unrelated Caulobacter crescentus (67% identity).
Although none of the individual gene products were most similar to those of Azospirillum species, the overall organization of genes in the A. diazotrophicus nif-fix cluster is most like that of Azospirillum brasilense. These are the only two diazotrophs so far characterized that have an mcpA-like gene associated with the nif-fix cluster (J. Frazzon and I. S. Schrank, personal communication; also this work). The McpA protein is involved in chemotaxis in several organisms. Because both A. diazotrophicus and A. brasilense are found naturally associated with monocot plants, it will be of interest to determine whether McpA is responsible for chemotactic responses to plant exudates for signaling or nutrition. An mcpA mutant strain of A. diazotrophicus lost chemotaxis toward a wide range of attractant stimuli (unpublished result).
Differences between the clusters are that nifA and nifB of A. brasilense are not linked to the other nif genes and nifQ, nifW, and fdxB are absent from the latter (8, 15). nifY is not found in the A. diazotrophicus cluster, but its requirement for nitrogen fixation is uncertain in other diazotrophs; it may be involved in Fe-Mo cofactor insertion into nitrogenase enzyme or in fixed N sensing (11, 23). One portion of the nif-fix cluster of A. diazotrophicus is more like that of R. capsulatus (nifE to nifW) than to A. brasilense, although no ORF1-like gene is found in the latter. The nifU gene in R. capsulatus is truncated compared to most other nif genes, encoding only the C-terminal end of NifU (16).
Transcriptional and translational organization of the cluster.
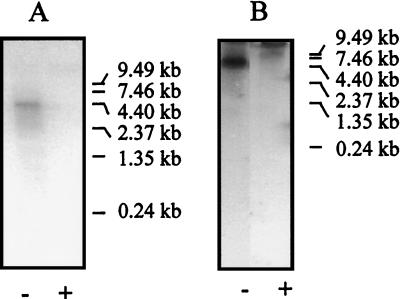
The transcriptional and translational organization of genes in the nif-fix cluster of A. diazotrophicus shows several interesting features. Northern analysis of mRNA was successful in identifying the cotranscription of nifHDK, as occurs in most other diazotrophs, according to size of transcripts hybridizing to a nifD probe, and also of nifENX orf4 orfl fdxB nifQ, indicated by hybridization of RNA to a nifE probe (Fig. 1 and 2). The former was predicted by sequence analysis, which revealed ςn- and NifA-binding sites upstream of nifH (Sevilla and Kennedy, submitted) required, respectively, for nif promoter recognition and for nif gene transcriptional activation in all other proteobacterial diazotrophs. A ςn recognition sequences but no NifA recognition sequence was found upstream of nifE. Northern blots hybridized with [32P]dCTP-labeled orf6, nifU, or fixA probes showed only smears or broad bands after autoradiography, suggesting rapid degradation of mRNA or a very low level of transcription. The putative transcriptional organization of the other genes was suggested by sequence analysis (Fig. 1). Locations of possibly significant ςn, NifA recognition sites upstream of genes and of potential transcription terminators downstream of genes are given in Table 1.
FIG. 2.
Northern analysis with probes for nifD (A) and nifE (B). Probes were radiolabeleled with [α-32P]dCTP by random priming (Stratagene). −, cultures grown with low concentrations of fixed N (0.5 mM NH4); +, cultures grown with high concentrations of fixed N (10 mM NH4).
An unusual degree of overlap between the 3′ and 5′ ends of adjacent genes, indicating translational coupling, was found for nifN-nifX (30 bp). Other cases of overlap were found for fixC-fixX (1 bp), rpoN-modC (3 bp), modC-modB (3 bp), modB-modA (3 bp), nifZ-fixU (3 bp), and fixU-orfl (3 bp). Several other cases of translational coupling are found between nif and/or related genes in other diazotrophs, including one large overlap of 34 bp between nifN and nifX in the archeal diazotroph Methanococcus maripaludis and smaller overlaps in M. maripaludis (nifD-nifK, 7 bp) (14), A. vinelandii (orf8-nifW, 3 bp; nifZ-nifM, 10 bp; nifM-orf9, 7 bp) (12), and Klebsiella pneumoniae (nifB-nifQ, 1 bp; nifL-nifA, 4 bp; nifZ-nifM, 4 bp; and nifN-nifX, 14 bp) (1). Of the A. diazotrophicus genes described here, only nifN and modC had the translational initiation codon GTG. This initiation codon also occurs in several genes in various diazotrophs, for example, nifB and nifX of Frankia alni and ORF1 of Plectonema boryanum PCC 73110 (10, 21).
Characterization of genes related to nitrogen fixation and/or plant colonization is important for elucidating these processes in endophytic diazotrophs. This work examines a major cluster of genes that are certainly or potentially important for the ability of A. diazotrophicus to fix nitrogen inside its plant host. This knowledge may be relevant for efforts to use this organism or others like it, such as the endophyte Herbaspirillum seropedicae, to better benefit growth of sugarcane and possibly other monocot plants.
Nucleotide sequence accession number.
Sequences in previous GenBank files have been compiled and combined with new data under accession no. AF030414.
Acknowledgments
This work was supported by the National Science Foundation (IBN-9728184) and by Deutsche (Germany), ME1254/3-1. S.L. was partially sponsored by Chunbuk National University in Korea (overseas scholarship).
REFERENCES
- 1.Arnold W, Rump A, Klipp W, Priefer U B, Puhler A. Nucleotide sequence of a 24,206 base-pair DNA fragment carrying the entire nitrogen fixation gene cluster of Klebsiella pneumoniae. J Mol Biol. 1988;203:715–738. doi: 10.1016/0022-2836(88)90205-7. [DOI] [PubMed] [Google Scholar]
- 2.Buchet A, Nasser W, Eichler K, Mandrand-Berthelot M A. Positive co-regulation of the Escherichia coli carnitine pathway cai and fix operons by CRP and the CaiF activator. Mol Microbiol. 1999;34:562–575. doi: 10.1046/j.1365-2958.1999.01622.x. [DOI] [PubMed] [Google Scholar]
- 3.Cavalcante V A, Dobereiner J. A new acid-tolerant nitrogen-fixing bacterium associated with sugarcane. Plant Soil. 1988;108:23–31. [Google Scholar]
- 4.Daniels M J, Barber C E, Turner P C, Sawczyc M K, Byrde R J W, Fielding A H. Cloning of genes involved in pathogenicity of Xanthomonas campestris pv. campestris showing altered pathogenicity. EMBO J. 1984;3:3323–3327. doi: 10.1002/j.1460-2075.1984.tb02298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dean D R, Jacobson M R. Biochemical genetics of nitrogenase. In: Stacey G, Evans H J, Burris R, editors. Biological nitrogen fixation. New York, N.Y: Chapman & Hall; 1992. pp. 763–834. [Google Scholar]
- 6.Earl C D, Ronson C W, Ausubel F M. Genetic and structural analysis of the Rhizobium meliloti fixA, fixB, fixC, and fixX genes. J Bacteriol. 1987;169:1127–1136. doi: 10.1128/jb.169.3.1127-1136.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eichler K, Buchet A, Bourgis F, Kleber H P, Mandrand-Berthelot M A. The fix Escherichia coli region contains four genes related to carnitine metabolism. J Basic Microbiol. 1995;35:217–227. doi: 10.1002/jobm.3620350404. [DOI] [PubMed] [Google Scholar]
- 8.Frazzon J, Schrank I S. Sequencing and complementation analysis of the nifUSV genes from Azospirillum brasilense. FEMS Microbiol Lett. 1998;159:151–158. doi: 10.1111/j.1574-6968.1998.tb12854.x. [DOI] [PubMed] [Google Scholar]
- 9.Grunden A M, Ray R M, Rosentel J K, Healy F G, Shanmugam K T. Repression of the Escherichia coli modABCD (molybdate transport) operon by ModE. J Bacteriol. 1996;178:735–744. doi: 10.1128/jb.178.3.735-744.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harriott O T, Hosted T J, Benson D R. Sequences of nifX, nifW, nifZ, nifB and two ORF in the Frankia nitrogen fixation gene cluster. Gene. 1995;161:63–67. doi: 10.1016/0378-1119(95)00300-u. [DOI] [PubMed] [Google Scholar]
- 11.Homer M J, Paustian T D, Shah V K, Roberts G P. The nifY product of Klebsiella pneumoniae is associated with apodinitrogenase and dissociates upon activation with the iron-molybdenum cofactor. J Bacteriol. 1993;175:4907–4910. doi: 10.1128/jb.175.15.4907-4910.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobson M R, Brigle K E, Bennett L, Setterquist R A, Wilson R A, Cash V L, Beynon J, Newton W E, Dean D R. Physical and genetic map of the major nif gene cluster from Azotobacter vinelandii. J Bacteriol. 1989;171:1017–1027. doi: 10.1128/jb.171.2.1017-1027.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.James E K, Reis V M, Olivares F L, Baldani J I, Dobereiner J. Infection of sugar cane by nitrogen-fixing bacterium Acetobacter diazotrophicus. J Exp Bot. 1994;45:757–766. [Google Scholar]
- 14.Kessler P S, Blank C, Leigh J A. The nif gene operon of the methanogenic archaeon Methanococcus maripaludis. J Bacteriol. 1998;180:1504–1511. doi: 10.1128/jb.180.6.1504-1511.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang Y Y, Kaminski P A, Elmerich C. Identification of a nifA-like regulatory gene of Azospirillum brasilense-Sp7 expressed under conditions of nitrogen fixation and in the presence of air and ammonia. Mol Microbiol. 1991;5:2735–2744. doi: 10.1111/j.1365-2958.1991.tb01982.x. [DOI] [PubMed] [Google Scholar]
- 16.Masepohl B, Angermuller S, Hennecke S, Hubner P, Moreno-Vivian C, Klipp W. Nucleotide sequence and genetic analysis of the Rhodobacter capsulatus ORF6-nifU1SVW gene region: possible role of NifW in homocitrate processing. Mol Gen Genet. 1993;238:369–382. doi: 10.1007/BF00291996. [DOI] [PubMed] [Google Scholar]
- 17.Masepohl B, Klipp W. Organization and regulation of genes encoding the molybdenum nitrogenase and the alternative nitrogenase in Rhodobacter capsulatus. Arch Microbiol. 1996;165:80–90. [Google Scholar]
- 18.Mouncey N J, Mitchenall L A, Pau R N. Mutational analysis of genes of the mod locus involved in molybdenum transport, homeostasis, and processing in Azotobacter vinelandii. J Bacteriol. 1995;177:5294–5302. doi: 10.1128/jb.177.18.5294-5302.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mowbray S L. Bacterial chemoreceptors: recent progress in structure and function. Mol Cell. 1999;9:115–118. [PubMed] [Google Scholar]
- 20.Neubauer H, Pantel I, Lindgren P E, Gotz F. Characterization of the molybdate transport system ModABC of Staphylococcus carnosus. Arch Microbiol. 1999;172:109–115. doi: 10.1007/s002030050747. [DOI] [PubMed] [Google Scholar]
- 21.Schrautemeier B, Cassing A, Bohme H. Characterization of the genome region encoding a fdxH-type ferredoxin and a new 2[4Fe-4S] ferredoxin from the nonheterocystous, nitrogen-fixing cyanobacterium Plectonema boryanum PCC 73110. J Bacteriol. 1994;176:1037–1046. doi: 10.1128/jb.176.4.1037-1046.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sevilla M, de Oliveira A L, Baldani I, Kennedy C. Contributions of the bacterial endophyte Acetobacter diazotrophicus to sugarcane nutrition: a preliminary study. Symbiosis. 1998;25:181–192. [Google Scholar]
- 23.Simon H M, Gosink M M, Roberts G P. Importance of cis determinants and nitrogenase activity in regulated stability of the Klebsiella pneumoniae nitrogenase structural gene mRNA. J Bacteriol. 1999;181:3751–3760. doi: 10.1128/jb.181.12.3751-3760.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teixeira K R S, Wulling M, Morgan T, Galler R, Zellerman E M, Baldani J I, Kennedy C, Meletzus D. Molecular analysis of the chromosomal region encoding the nifA and nifB genes of Acetobacter diazotrophicus. FEMS Microbiol Lett. 1999;71:521–530. [PubMed] [Google Scholar]
- 25.Young J P W. Phylogenetic classification of nitrogen-fixing organisms. In: Stacey G, Burris R H, Evans H J, editors. Biological nitrogen fixation. New York, N.Y: Chapman & Hall; 1992. pp. 43–86. [Google Scholar]