Abstract
ClpB is a highly conserved heat shock protein that is essential for thermotolerance in bacteria and eukaryotes. One distinctive feature of all bacterial clpB genes is the dual translation of a truncated 79-kDa form (ClpB-79) in addition to the full-length 93-kDa protein (ClpB-93). To investigate the currently unknown function of ClpB-79, we have examined the ability of the two different-sized ClpB homologues from the cyanobacterium Synechococcus sp. strain PCC 7942 to confer thermotolerance. We show that the ClpB-79 form has the same capacity as ClpB-93 to confer thermotolerance and that the ClpB-79 protein contributes ca. one-third of the total thermotolerance developed in wild-type Synechococcus, the first in vivo demonstration of a functional role for ClpB-79 in bacteria.
Changing environmental conditions elicit the synthesis of new types of cellular proteins in all organisms. This conserved molecular response is best exemplified by high-temperature stress, during which specific groups of polypeptides known as heat shock proteins (HSPs) are rapidly induced. Many of these HSPs are now known to function as molecular chaperones and are part of larger protein families that include constitutive members. Protein denaturation and aggregation are the major types of cellular damage that result from high temperatures, and HSP chaperones respond by preventing aggregation, assisting refolding, and targeting misfolded protein for degradation (13). The activity of such chaperones is essential for cell survival during heat shock and for subsequent recovery.
ClpB is a heat shock-inducible representative of the HSP100/Clp family of oligomeric ATPases. The family can be divided into several types based on specific sequence signatures and other structural characteristics (17). Most member proteins are large (79 to 105 kDa) and contain two distinct nucleotide-binding domains separated by a spacer region of variable length (ClpA-E), whereas some are smaller (50 kDa) and have only one such domain (ClpX and ClpY). The various HSP100/Clp proteins are thought to function as molecular chaperones, with a common mechanism of dismantling multimeric protein complexes, as shown for ClpA and -X in Escherichia coli (7, 22).
ClpB is an HSP found in almost all organisms studied to date. Separate nuclear genes encode two different-sized ClpB proteins in yeast, a 78-kDa protein localized in mitochondria (6) and a 104-kDa protein localized primarily in the cytosol (16). Both ClpB proteins function as molecular chaperones. Along with DnaK (HSP70), mitochondrial ClpB helps prevent protein denaturation and aggregation at high temperatures (18, 19), whereas cytosolic ClpB (HSP104) disassembles large protein aggregates that accumulate at extreme high temperatures (14). Cytosolic ClpB also acts in concert with DnaK or DnaJ to promote the refolding of the once aggregated polypeptides (4). Such functions are thought to underlie the necessity of cytosolic ClpB for the acquisition of thermotolerance in yeast (16), which is the tolerance developed to a normally lethal high temperature by being preconditioned to a more permissive high temperature.
Like the cytosolic homologue in yeast, prokaryotic ClpB also facilitates resolubilization of protein aggregates during heat shock and cooperates with DnaK to enable their renaturation (5, 9, 10). Despite this functional similarity, however, bacterial and eukaryotic ClpB proteins have one striking and highly conserved difference. Although like in eukaryotes, two different-sized forms of ClpB occur in eubacteria (79 and 93 kDa), both proteins originate from a single gene as a result of an internal translational initiation site within the clpB transcript (2, 21). To date, the specific function of the truncated 79-kDa form (ClpB-79) remains unknown, although in E. coli, it is thought to have a regulatory role in the activity of the full-length ClpB-93 protein (12). To investigate the roles of ClpB-79 and -93 more closely, we have examined the involvement of each ClpB form in the development of thermotolerance in cyanobacteria. The model for this study was the ClpB1 protein in the unicellular strain Synechococcus sp. strain PCC 7942 (hereafter referred to as Synechococcus), which, like the HSP104 protein in yeast, confers thermotolerance (2)—the first and so far only such example in bacteria.
Site-directed mutagenesis of Synechococcus clpB1 gene.
To test the function of the two different-sized ClpB1 proteins in Synechococcus, we used site-directed mutagenesis to prepare strains that synthesize only the full-length (ClpB1-93) or truncated (ClpB1-79) forms. Three plasmid constructs were made. In the first, pClpB93, the Val160 start codon for ClpB1-79 synthesis was changed in the clpB1 gene from GTG to GTT to prevent translation initiation, but maintain the amino acid identity. For the second construct, pClpB79, the start Met1 codon (ATG) was changed to a Thr (ACG) to prevent ClpB1-93 synthesis. Similar base alterations were earlier shown to prevent synthesis of the selected ClpB in E. coli without affecting the translation of the other form (12). A control construct was also made (pClpB-c) containing the intact native clpB1 gene that produces both ClpB1-93 and -79 proteins. For all three constructs, a 200-bp region containing the heat-inducible promoter of the native clpB1 gene was included upstream of the clpB1 genes. A chloramphenicol resistance gene was also ligated upstream of the promoter region as a selectable marker. The three clpB1 constructs were transformed separately into cultures of the clpB1 null mutant (ΔclpB1) and integrated by recombination into a phenotypically neutral site locus (NSL) of the chromosome (1). Plasmid insertion into the NSL and its complete segregation were confirmed for each transformed strain by Southern blotting (data not shown). Strains successfully transformed with either the pClpB93, pClpB79, or pClpB-c constructs were termed ANB1-93, ANB1-79, or ANB1-c, respectively.
High-temperature induction of ClpB1 proteins.
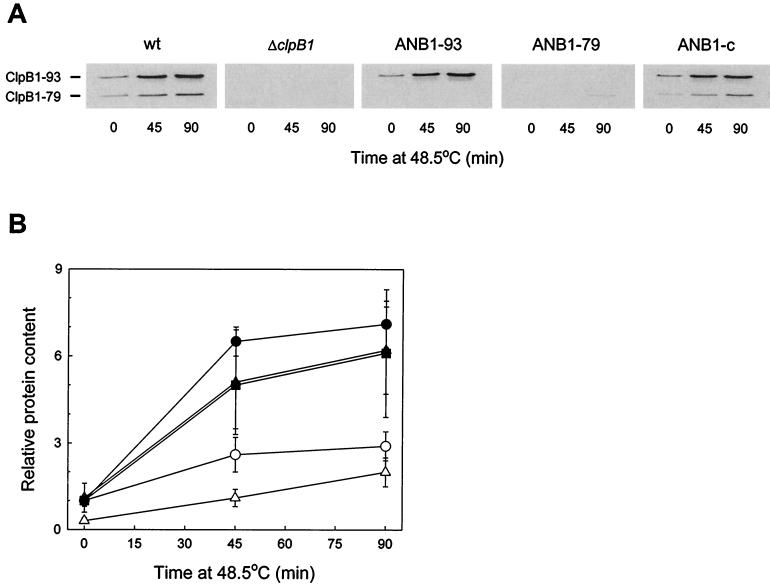
All cyanobacterial strains were grown in BG-11 medium as previously described (3). Each of the three new strains (ANB1-93, ANB1-79, and ANB1-c) exhibited no phenotypic differences (growth rate, pigment composition, or cell morphology) from wild-type Synechococcus or the original ΔclpB1 strain under standard growth conditions. To test the heat shock inducibility of the clpB1 constructs, the three ANB1 strains along with the wild-type and ΔclpB1 strains were shifted from 37 to 48.5°C for 90 min, with all other growth parameters kept constant. Using an antibody specific for the C-terminal region of Synechococcus ClpB1 (15), the levels of ClpB1-93 and/or -79 were analyzed in cell extracts taken at 45-min intervals as previously described (3). As shown in Fig. 1A, wild-type Synechococcus induced both ClpB1-93 and -79 proteins during the heat shift, consistent with the results shown previously (2). On average, there was a six- to sevenfold increase in ClpB1-93 protein during the high-temperature treatment, concomitant to a two- to threefold rise in ClpB1-79 content (Fig. 1B). A similar induction profile for both ClpB1 proteins occurred in the ANB1-c complementation strain. The level of ClpB1 protein in ANB1-c, however, was consistently 80 to 90% of that in the wild type, suggesting that the 200-bp clpB1 promoter region used in the plasmid constructs confers this degree of native clpB1 gene expression.
FIG. 1.
Heat shock induction of ClpB1 forms in wild-type Synechococcus, ΔclpB1, ANB1-93, ANB1-79, and ANB1-c cultures. (A) Immunoblot detection of ClpB1-93 and -79 proteins using a polyclonal antibody against the C terminus of Synechococcus ClpB1. The figure shows a representative result from one of four replicates. (B) Quantification of ClpB1-93 (●) and ClpB1-79 (○) in wild-type Synechococcus, ClpB1-93 (▴) and ClpB1-79 (▵) in ANB1-c, and ClpB1-93 (■) in ANB1-93. Values for each ClpB1 protein were plotted relative to the amount of ClpB1-93 in wild-type Synechococcus at time zero (37°C control), which was set at 1. Levels of ClpB1-79 protein in strain ANB1-79 are not shown, since the protein was only detectable in the 90-min heat-shocked sample. All values represent averages ± standard errors (n = 4).
For the two site-directed mutant strains, the level of ClpB1-93 induction in ANB1-93 mirrored that in the complementation ANB1-c strain, whereas no truncated ClpB1-79 protein was observed (Fig. 1B). This confirmed that the single base change to Val160 blocked ClpB1-79 synthesis in Synechococcus without affecting ClpB1-93 synthesis, as did the corresponding alteration to the clpB gene in E. coli (12). In contrast, the ANB1-79 strain produced considerably less ClpB1-79 protein during the heat shock than both ANB1-c and wild-type Synechococcus, with ClpB1-79 protein only just being detectable after 90 min at 48.5°C (10 to 20% of the wild-type level). This indicated that mutagenesis of the Met1 codon not only prevented ClpB1-93 synthesis but it also significantly reduced the translation of ClpB1-79 protein initiated at the Val160 codon. Such an inhibition of ClpB-79 synthesis was not observed for the corresponding base change to the E. coli clpB gene (12).
ClpB1-93 confers two-thirds of total thermotolerance acquired by Synechococcus.
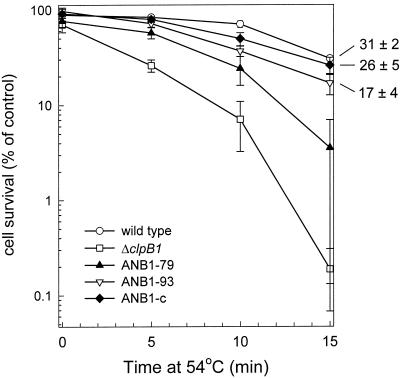
The ability of the ANB1-93 and -79 strains to develop thermotolerance was directly compared to that of ANB1-c, ΔclpB1, and wild-type Synechococcus. As described previously (3), the thermotolerance assay involved shifting each strain from the 37°C growth temperature to either the severe temperature of 54°C for 15 min or first preconditioning them at 48.5°C for 90 min prior to the 54°C shift. Acquisition of thermotolerance was determined by cell survival measurements, with samples taken at selected time points. As shown in Fig. 2, the loss of ClpB1 protein greatly reduced the degree of thermotolerance acquired by the ΔclpB1 strain (0.2% ± 0.1% cell survival after 15 min at 54°C) relative to the wild type (31% ± 2%), consistent with our earlier findings (2). In comparison, the pClpB1-c construct conferred ca. 85% of wild-type thermotolerance to the complementation strain ANB1-c (26% ± 5%), matching the relative level of ClpB1 protein induction in this strain. In ANB1-93, induction of only the ClpB1-93 protein also conferred a high degree of thermotolerance (17% ± 4%) (Fig. 2), but one-third less than the ANB1-c strain despite identical levels of ClpB1-93 protein (Fig. 1B). The ANB1-79 strain also developed thermotolerance, although only slightly more than the original ΔclpB1 mutant, again consistent with the low induction of ClpB1-79 protein in ANB1-79. Apart from thermotolerance, no significant differences in cell viability were observed between the strains during the direct shift from 37 to 54°C or during the 90-min pretreatment at 48.5°C (data not shown).
FIG. 2.
Development of thermotolerance in wild-type Synechococcus (○), ΔclpB1 (□), ANB1-93 (▿), ANB1-79 (▴), and ANB1-c (⧫) cultures grown at 37°C, preheated at 48.5°C for 90 min, and then shifted to 54°C for 15 min. Average numbers of viable cells after each temperature treatment are expressed as percentages of the 37°C control value (100%). Values represent averages ± standard errors (n = 3).
Increased levels of ClpB1-79 during heat shock.
The difference in thermotolerance acquired by the ANB1-c and ANB1-93 strains and the ability of the ANB1-79 strain to develop thermotolerance above that of ΔclpB1 both suggest that the truncated ClpB1-79 protein confers some degree of heat resistance to Synechococcus. Because of the low level of ClpB1-79 protein synthesis and developed thermotolerance in ANB1-79, however, we prepared a second construct for ClpB1-79 synthesis to clarify this point. In the pClpB1-79a plasmid, the 5′ region of clpB1 from Met1 to Val160 was deleted and the Val codon was changed to a MetATG. The resulting construct therefore should express only a truncated clpB1 transcript for ClpB1-79 synthesis under the control of the 200-bp clpB1 promoter fragment. This construct was transformed into the NSL of the ΔclpB1 strain, and the resulting strain was called ANB1-79a. Correct insertion of pClpB1-79a into the NSL of ANB1-79a and its complete segregation were confirmed by Southern blotting (data not shown).
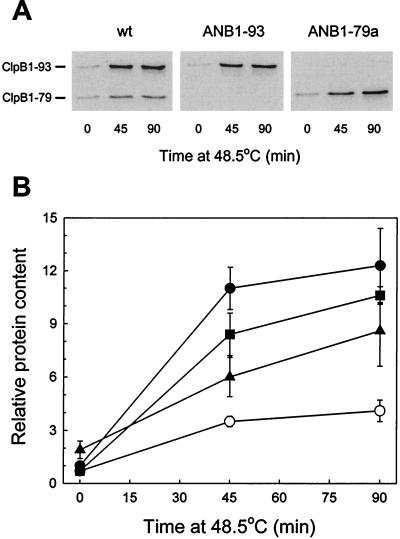
To test whether the new ANB1-79a strain produced increased amounts of ClpB1-79 protein, high-temperature shifts were simultaneously performed with cultures of ANB1-79a along with ANB1-93, the ΔclpB1 strain, and wild-type Synechococcus. In these heat shock experiments, the induction profiles for ClpB1 in the wild type and ANB1-93 were similar to that of the earlier experiments, except that the relative level of induction was consistently higher in both strains (Fig. 3). However, as observed earlier, the heat shock induction of ClpB1-93 in ANB1-93 was 80 to 90% of that in the wild type. For the new ANB1-79a strain, the amount of ClpB1-79 protein induced during the high-temperature treatment was considerably higher than that in the previous ANB1-79 strain. Compared to the induction of ClpB1-93 in ANB1-93, the increase in ClpB1-79 was only slightly lower in ANB1-79a (Fig. 3).
FIG. 3.
Increased heat shock induction of ClpB1-79 protein in ANB1-79a strain. Wild-type Synechococcus (wt), ΔclpB1, ANB1-93, and ANB1-79a cultures were shifted from 37 to 48.5°C for 90 min. (A and B) Immunoblot detection (A) and quantification (B) of ClpB1-93 and -79 proteins. Levels of ClpB1-93 (●) and ClpB1-79 (○) in wild-type Synechococcus, ClpB1-93 (■) in ANB1-93, and ClpB1-79 (▴) in ANB1-79a were plotted relative to the 37°C control value for ClpB1-93 protein in wild-type Synechococcus, which was set at 1. (A) The figure shows a representative result from one of three independent replicates. (B) All values represent averages ± standard errors (n = 3).
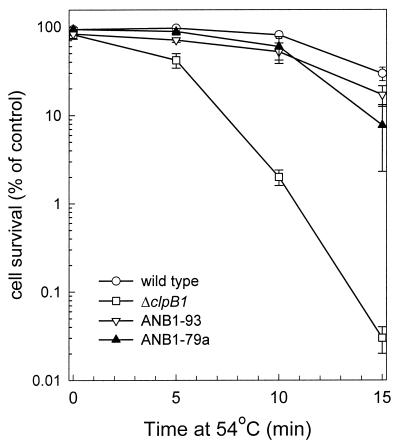
With the ANB1-79a strain producing greater amounts of ClpB1-79 protein than the first ANB1-79 strain, we tested its ability to develop thermotolerance compared to ANB1-93. For each replicate experiment, ANB1-93 and -79a cultures were simultaneously used along with the wild-type and ΔclpB1 strains. As shown in Fig. 4, the new ANB1-79a strain developed a high level of thermotolerance, many times greater than that of the previous ANB1-79 strain. Indeed, the degree of thermotolerance acquired by ANB1-79a was not significantly different from that of ANB1-93 throughout the assay time course, indicating the ClpB1-79 protein has the same capacity of ClpB1-93 to restore thermotolerance to the ΔclpB1 mutant.
FIG. 4.
Increased thermotolerance acquired by strain ANB1-79a. Synechococcus wild-type (○), ΔclpB1 (□), ANB1-93 (▿), and ANB1-79a (▴) cultures grown at 37°C were preheated at 48.5°C for 90 min and then shifted to 54°C for 15 min. Average numbers of viable cells after each temperature treatment are expressed as percentages of the 37°C control value (100%). Values represent averages ± standard errors (n = 3).
ClpB1-79 contributes to the development of thermotolerance in Synechococcus.
We have clearly demonstrated in this study that the truncated form of ClpB1 has the same capacity as the full-length ClpB1 protein to confer thermotolerance and that loss of ClpB1-79 reduced the total thermotolerance developed in Synechococcus. Given the relative proportion of both proteins produced from the native clpB1 gene, these results suggest that ca. one-third of the thermotolerance developed in wild-type Synechococcus derives from the ClpB1-79 protein. To our knowledge, this is the first in vivo evidence of the shorter ClpB protein having an active, functional role in bacteria at high temperatures.
The cotranslation of the ClpB-79 protein along with the full-length ClpB-93 is a characteristic of all known eubacterial clpB genes, but it is a feature absent for the eukaryotic homologues. Both prokaryotic forms are synthesized in coordinate amounts under various stress conditions (21), and yet the reason for the two different-sized ClpB proteins in eubacteria has so far remained unresolved. Studies of the two ClpB forms in E. coli have suggested that the ClpB-79 protein has a regulatory effect on the full-length ClpB-93. Although both ClpB proteins exhibited similar basal ATPase activity, ClpB-79 lacked the enhanced activity stimulated by the addition of casein, suggesting it lacked one or more sites responsible for binding of protein substrates (12). Moreover, this protein-enhanced ATPase activity of ClpB-93 was increasingly inhibited by the incorporation of the truncated ClpB-79 into the ClpB-93 oligomer. It was proposed that the truncated ClpB-79 could function as a regulatory protein, controlling the protein-activated ATPase activity of ClpB-93 and hence its function during high-temperature stresses (12).
Given the evidence of this study that the truncated ClpB protein has an identical capacity to confer thermotolerance as the full-length protein, the suggestion that ClpB-79 acts as a regulatory protein during heat shock is questionable. Although a regulatory role cannot be excluded at this stage, the ClpB-79 protein would appear to enhance the cell's ability to develop thermotolerance rather than restrict it, as was demonstrated by the higher degree of thermotolerance developed in the ANB1-c strain compared to that of ANB1-93. Precedence for an active ClpB of similar size to prokaryotic ClpB-79 exists in yeast in the form of the mitochondrial ClpB homologue (HSP78). As for ClpB-79, HSP78 has only a minimal N-terminal region extending upstream of the first nucleotide-binding domain (6). Studies have shown that HSP78 cooperates with mitochondrial HSP70 to both prevent aggregation of misfolded matrix proteins (8) and dissolve protein aggregates that form at extreme high temperatures (19). During severe heat shock, HSP78 is essential for maintenance of mitochondrial genome integrity and respiratory competence and for the reactivation of mitochondrial protein synthesis (19). Moreover, when expressed and localized in the cytosol, HSP78 can partially substitute for cytosolic ClpB (HSP104) in conferring cellular thermotolerance (19), suggesting that like the two forms of bacterial ClpB proteins, HSP78 and HSP104 in yeast have conserved modes of action.
The role of ClpB as a molecular chaperone during heat shock appears to be highly conserved in bacteria and eukaryotes. In yeast, both mitochondrial and cytosolic forms of ClpB promote the resolubilization of heat-denatured proteins that form large aggregates, initiating their refolding in cooperation with the DnaK chaperone system (4, 19). Similar interactions between ClpB and the DnaK system in recovering aggregated proteins also occur in bacteria (9, 10, 23). Like for yeast HSP104, it is presumably such chaperone activity by ClpB1 that confers thermotolerance to Synechococcus. If so, then the capacity of ClpB1-93 and -79 to confer thermotolerance infers they both function to disassemble heat-induced protein aggregates. Furthermore, a heat-inducible DnaK homologue has been identified in Synechococcus (11), and it is likely that this chaperone functions cooperatively with the ClpB1 proteins during the development of thermotolerance.
Given the active involvement of ClpB1-79 in the acquisition of thermotolerance in Synechococcus, it would appear that full-length ClpB has at least two separate protein binding domains. One would be located in the N-terminal domain and be responsible for the casein-stimulated ATPase activity, as demonstrated for the E. coli ClpB protein, whereas the other would be located elsewhere and be present in both ClpB-93 and -79 proteins. The latter domain would be the one involved in recognition and binding of heat-induced protein aggregates and thus conferring thermotolerance to organisms like Synechococcus during severe heat stress. A possible site for this second protein-binding domain would be the C-terminal domain, downstream of the second ATP-binding domain. In this region, a specific domain for protein binding has been proposed for Clp/HSP100 proteins, including ClpB (20). This domain, termed “sensor- and substrate discrimination,” or SSD, has been shown to bind several protein substrates, although the specificity differs between different Clp/HSP100 proteins. Recognition and binding of protein aggregates by such a C-terminal SSD domain within ClpB would be consistent with the equivalent capacities of ClpB1-93 and -79 to confer thermotolerance as reported in this study.
Acknowledgments
We thank Ewa Miskiewicz for technical assistance.
This research was supported by the Swedish Natural Science Research Council and the Foundation for Strategic Research (Centre for Forestry Biotechnology and Chemistry).
REFERENCES
- 1.Bustos S A, Golden S S. Light-regulated expression of the psbD gene family in Synechococcus sp. PCC 7942: evidence for the role of duplicated psbD genes in cyanobacteria. Mol Gen Genet. 1992;232:221–230. doi: 10.1007/BF00280000. [DOI] [PubMed] [Google Scholar]
- 2.Eriksson M-J, Clarke A K. The heat shock protein ClpB mediates the development of thermotolerance in the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol. 1996;178:4839–4846. doi: 10.1128/jb.178.16.4839-4846.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eriksson M-J, Clarke A K. The E. coli heat shock protein ClpB restores acquired thermotolerance to a cyanobacterial clpB deletion mutant. Cell Stress Chaperones. 2000;5:255–264. doi: 10.1379/1466-1268(2000)005<0255:techsp>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glover J R, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 5.Goloubinoff P, Mogk A, Zvi A P B, Tomoyasu T, Bukau B. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc Natl Acad Sci USA. 1999;96:13732–13737. doi: 10.1073/pnas.96.24.13732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leonhardt S A, Fearon K, Danese P N, Mason T L. HSP78 encodes a yeast mitochondrial heat shock protein in the Clp family of ATP-dependent proteases. Mol Cell Biol. 1993;13:6304–6313. doi: 10.1128/mcb.13.10.6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levchenko I, Luo L, Baker T A. Disassembly of the Mu transposase tetramer by the ClpX chaperone. Genes Dev. 1995;9:2399–2408. doi: 10.1101/gad.9.19.2399. [DOI] [PubMed] [Google Scholar]
- 8.Moczko M, Schönfisch B, Voos W, Pfanner N, Rassow J. The mitochondrial ClpB homolog Hsp78 cooperates with matrix Hsp70 in maintenance of mitochondrial function. J Mol Biol. 1995;254:538–543. doi: 10.1006/jmbi.1995.0636. [DOI] [PubMed] [Google Scholar]
- 9.Mogk A, Tomoyasu T, Goloubinoff P, Rüdiger S, Röder D, Langen H, Bukau B. Identification of thermolabile Escherichia coli proteins: prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 1999;18:6934–6949. doi: 10.1093/emboj/18.24.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Motohashi K, Watanabe Y, Yohda M, Yoshida M. Heat-inactivated proteins are rescued by the DnaK · J-GrpE set and ClpB chaperones. Proc Natl Acad Sci USA. 1999;96:7184–7189. doi: 10.1073/pnas.96.13.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nimura K, Yoshikawa H, Takahashi H. Identification of dnaK multigene family in Synechococcus sp. PCC7942. Biochem Biophys Res Commun. 1994;201:466–471. doi: 10.1006/bbrc.1994.1724. [DOI] [PubMed] [Google Scholar]
- 12.Park S K, Kim K I, Woo K M, Seol J H, Tanaka K, Ichihara A, Ha D B, Chung C H. Site-directed mutagenesis of the dual translational initiation sites of the clpB gene of Escherichia coli and characterization of its gene products. J Biol Chem. 1993;268:20170–20174. [PubMed] [Google Scholar]
- 13.Parsell D A, Lindquist S. The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu Rev Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- 14.Parsell D A, Kowal A S, Singer M A, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 15.Porankiewicz J, Clarke A K. Induction of the heat shock protein ClpB affects cold acclimation in the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol. 1997;179:5111–5117. doi: 10.1128/jb.179.16.5111-5117.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanchez Y, Lindquist S L. HSP104 required for induced thermotolerance. Science. 1990;248:1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- 17.Schirmer E C, Glover J R, Singer M A, Lindquist S. HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem Sci. 1996;21:289–295. [PubMed] [Google Scholar]
- 18.Schmitt M, Neupert W, Langer T. Hsp78, a Clp homologue within mitochondria, can substitute for chaperone functions of mt-hsp70. EMBO J. 1995;14:3434–3444. doi: 10.1002/j.1460-2075.1995.tb07349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmitt M, Neupert W, Langer T. The molecular chaperone Hsp78 confers compartment-specific thermotolerance to mitochondria. J Cell Biol. 1996;134:1375–1386. doi: 10.1083/jcb.134.6.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith C K, Baker T A, Sauer R T. Lon and Clp family proteases and chaperones share homologous substrate-recognition domains. Proc Natl Acad Sci USA. 1999;96:6678–6682. doi: 10.1073/pnas.96.12.6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Squires C L, Pedersen S, Ross B M, Squires C. ClpB is the Escherichia coli heat shock protein F84.1. J Bacteriol. 1991;173:4254–4262. doi: 10.1128/jb.173.14.4254-4262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wickner S, Gottesman S, Skowyra D, Hoskins J, McKenney K, Maurizi M R. A molecular chaperone, ClpA, functions like DnaK and DnaJ. Proc Natl Acad Sci USA. 1994;91:12218–12222. doi: 10.1073/pnas.91.25.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zolkiewski M. ClpB cooperates with DnaK, DnaJ, and GrpE in suppressing protein aggregation. J Biol Chem. 1999;274:28083–28086. doi: 10.1074/jbc.274.40.28083. [DOI] [PubMed] [Google Scholar]