Abstract
In this issue of Cell Stem Cell, Xu et al. and Yu et al. use low-input epigenetic profiling techniques to map H3K9me3 deposition in early human development. They reveal stage-specific H3K9me3 deposition on retrotransposons, which may play crucial cis-regulatory roles in early development.
During early mammalian preimplantation development, a series of fascinating events occurs that ensure proper development of the embryo. These events include the fusion of the gametes, a switch from oocyte control of the transcriptome to the zygote via zygotic genome activation, exit from totipotency, and the emergence of the inner cell mass (ICM) and the extraembryonic lineages. The epigenetic reset that takes place during this process leads to a dynamic reactivation of retrotransposons, which is crucial for normal development (Peaston et al., 2004). However, given the potential for retrotransposons to remobilize and cause genotoxicity, their expression must be tightly constrained (Leung et al., 2012). The histone modification histone 3 lysine 9 trimethylation (H3K9me3) has emerged as a key player in silencing retrotransposons by inducing heterochromatin formation (Nicetto et al., 2019).
The importance of H3K9me3 in early development is underscored by genetic studies that show that disruption of H3K9me3 writers leads to embryonic lethality in mice (Nicetto et al., 2019). Interestingly, promoting H3K9me3 deposition by overexpressing Suv39h1 (another H3K9me3 writer) also causes developmental arrest (Burton et al., 2020). This finding suggests that the precise regulation of H3K9me3 is essential for normal mouse development. To explore the dynamics of H3K9me3 in early development, immunostaining experiments have been carried out in early mouse embryos (Liu et al., 2004). However, these studies are limited by the imaging resolution and do not reveal H3K9me3-deposited loci. A previous study to measure the genome-wide distribution and dynamics of H3K9me3 was carried out in mice by Wang et al. (2018). The authors showed that H3K9me3 was established on Long Terminal Repeat (LTR) retrotransposons to transcriptionally silence them following DNA demethylation (Wang et al., 2018). However, studies to evaluate the H3K9me3 status are limited in humans due to technical and ethical reasons.
In this issue of Cell Stem Cell, two groups investigate the role of H3K9me3 in human preimplantation development. Xu et al. (2022) used “cleavage under targets and release using nuclease” (Cut&Run) and Yu et al. (2022) used ultra-low input native ChIP-seq, both on human embryos. Remarkably, Xu et al. found that H3K9me3 bucks the trend of epigenetic marks during preimplantation development: unlike histone 3 lysine 27 trimethylation (H3K27me3) and histone 3 lysine 4 trimethylation (H3K4me3), which drastically reset reaching their lowest levels at the 8C stage, H3K9me3 levels continually increase, starting after fertilization and continuing past the 8C stage (Figure 1).
Figure 1. Unique kinetics of H3K9me3 in early human development.
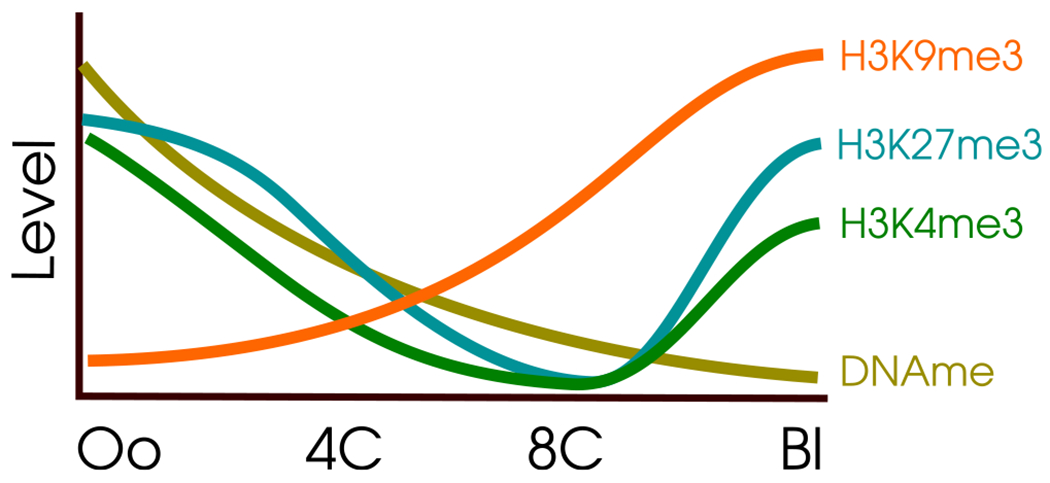
The X axis shows the developmental stage. The Y axis displays semiquantitative levels. H3K9me3 continually increases after fertilization, unlike other epigenetic marks. Adapted from Xu et al.’s Figure 1D. Oo, oocyte; 4C, 4-cell stage; 8C, 8-cell stage; Bl, blastocyst.
What function does this H3K9me3 deposition serve? Both groups found that H3K9me3 is deposited on retrotransposons and regulates their expression. While some retrotransposons were repressed by H3K9me3 throughout development, fascinatingly, others were regulated in a stage-specific manner. Yu et al. screened for possible 8C enhancers and identified a set of SINE-VNTR-Alu (SVA) elements on which H3K9me3 deposition reduces from the 4C to the 8C stage. Xu et al. demonstrated a stage-specific H3K9me3 deposition on distinct classes of LTR retrotransposons, often replacing repression mediated by the rapidly resetting DNA methylation. The authors found that this regulation leads to the transient expression of retrotransposons at the stage where DNA methylation is relaxed as part of the epigenetic reset, but H3K9me3 has not yet been deposited.
The next question to address was whether the H3K9me3-regulated retrotransposons have any developmental consequence. Yu et al. found that the reduced deposition of H3K9me3 on SVA elements during the transition from the 4C to the 8C stage facilitates the interaction of SVA elements with zygotic genome activation (ZGA) genes. They established that the H3K9me3 deposition is negatively correlated with expression levels of ZGA genes. These SVA elements also contain motifs for Dux, a known inducer of ZGA. These observations led the authors to hypothesize that SVA elements may be a mediator of ZGA induction. Indeed, they showed that CRISPR-KRAB-mediated silencing of SVA_D repeats (D chosen for its prevalence among SVA elements to be specifically derepressed at the 8C stage) results in defective ZGA and lowers rates of blastocyst formation. This finding strongly suggested a functional role for SVA elements in ZGA induction. The extent of the contribution of retrotransposons to human ZGA is remarkable. In the screen for 8C-specific enhancers, Yu et al. found that 79% of such enhancers derepressed at the 8C stage are repeat-element-derived, of which 40% are SVAs. Xu et al. also interrogated ZGA in a mouse model and found that Dux KO mice fail to properly establish H3K9me3 domains.
The function of H3K9me3 regulation in early development does not seem to be limited to regulating retrotransposons in ZGA. Both teams also found evidence that this regulation was involved in lineage segregation. Yu et al. demonstrated that H3K9me3 was differentially deposited on genes implicated in stem cell development and maintenance in the ICM and trophectoderm of blastocyst embryos. This result seemed to indicate that H3K9me3 was contributing to the segregation of the two lineages. Xu et al. found that loci marked with H3K9me3 specifically at the 8C stage were paradoxically more likely to become enhancers at later stages.
Finally, both studies touched on the evolution of epigenetic regulation in preimplantation development. Many retrotransposon families and insertions are highly species specific (Platt et al., 2018). SVA elements (as considered by Yu et al.) are hominoid specific, raising questions as to what human-specific changes to development might have evolved as a consequence of their insertion. Likewise, Xu et al. suggested, using bioinformatic analyses of binding sites and expression levels, that KRAB-ZNF proteins are responsible for the stage-specific retrotransposon repression they observed. As they note, the extraordinary diversity of ZNFs exists due to a host-versus-transposon “arms race” (Platt et al., 2018). This diversity suggests a broader role for the involvement of retrotransposons in the evolution of mammalian preimplantation development. One could speculate that species-specific retrotransposon insertions could lead to species-specific epigenetic regulation. For example, critical trophectoderm genes are wired by species-specific retrotransposons, which may explain the diversity of placental development in eutherians (Emera et al., 2012). The replicative nature of retrotransposons could also lend itself to rapid evolution of complex regulatory networks by dispersing regulatory sequences about the genome (Emera et al., 2012).
While both groups queried whole-genome H3K9me3 deposition in human embryos, they diverged in the focus of their analyses. It will be interesting to see if the results obtained by the two groups are reproducible using the data gathered by the opposite teams. This is of special interest because the data were obtained using different techniques: Cut&Run and low-input ChIP-seq.
Much about the epigenetic “reset” and regulation of human preimplantation development is still unknown, and the scarcity of embryos will likely continue to be a challenge. However, these two publications bring the field one step closer to answering some of the central questions of developmental biology.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Peaston AE, Evsikov AV, Graber JH, deVries WN, Holbrook AE, Solter D, and Knowles BB (2004). Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev. Cell 7, 597–606. 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Leung DC, and Lorincz MC (2012). Silencing of endogenous retroviruses: when and why do histone marks predominate? Trends Biochem. Sci 37, 127–133. 10.1016/j.tibs.2011.11.006. [DOI] [PubMed] [Google Scholar]
- Nicetto D, and Zaret KS (2019). Role of H3K9me3 heterochromatin in cell identity establishment and maintenance. Curr. Opin. Genet. Dev 55, 1–10. 10.1016/j.gde.2019.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton A, Brochard V, Galan C, Ruiz-Morales ER, Rovira Q, Rodriguez-Terrones D, Kruse K, Le Gras S, Udayakumar VS, Chin HG, et al. (2020). Heterochromatin establishment during early mammalian development is regulated by pericentromeric RNA and characterized by non-repressive H3K9me3. Nat. Cell Biol 22, 767–778. 10.1038/s41556-020-0536-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Kim JM, and Aoki F (2004). Regulation of histone H3 lysine 9 methylation in oocytes and early pre-implantation embryos. Development 131, 2269–2280. 10.1242/dev.01116. [DOI] [PubMed] [Google Scholar]
- Wang C, Liu X, Gao Y, Yang L, Li C, Liu W, Chen C, Kou X, Zhao Y, Chen J, et al. (2018). Reprogramming of H3K9me3-dependent heterochromatin during mammalian embryo development. Nat. Cell Biol 20, 620–631. 10.1038/s41556-018-0093-4. [DOI] [PubMed] [Google Scholar]
- Platt RN, Vandewege MW, and Ray DA (2018). Mammalian transposable elements and their impacts on genome evolution. Chromosome Res. 26, 25–43. [online] 26. 10.1007/s10577-017-9570-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Chen M, Hu Y, Ou S, Yu X, Liang S, Li N, Yang M, Kong X, Sun C, et al. (2022). The dynamic reprogramming of H3K9me3 at hominoid-specific retrotransposons during human preimplantation development. Cell Stem Cell 29, 1031–1050. [DOI] [PubMed] [Google Scholar]
- Xu R, Li S, Wu Q, Li C, Jiang M, Guo L, Chen M, Yang L, Dong X, Wang H, et al. (2022). Stage-specific H3K9me3 allocations ensure retrotransposon silencing in human preimplantation embryos. Cell Stem Cell 29, 1051–1066. [DOI] [PubMed] [Google Scholar]
- Emera D, and Wagner GP (2012). Transposable element recruitments in the mammalian placenta: impacts and mechanisms. Briefings in Functional Genomics 11, 267–276. 10.1093/bfgp/els013. [DOI] [PubMed] [Google Scholar]


