Abstract
Although BTB-zinc finger (BTB-ZF) transcription factors control the differentiation of multiple hematopoietic and immune lineages, how they function is poorly understood. The BTB-ZF factor Thpok controls intrathymic CD4+ T cell development and expression of most CD4+- and CD8+-lineage genes. Here, we identify the nucleosome remodeling and deacetylase (NuRD) complex as a novel Thpok cofactor. Using mass spectrometry and co-immunoprecipitation in primary T cells, we show that Thpok binds NuRD components independently of DNA association. We locate three amino-acid residues within the Thpok BTB domain that are required for both NuRD binding and Thpok functions. Conversely, a chimeric protein merging the NuRD component Mta2 to a BTB-less version of Thpok supports CD4+ T cell development, indicating that NuRD recruitment recapitulates the functions of the Thpok BTB domain. We found that NuRD mediates Thpok repression of CD8+-lineage genes, including the transcription factor Runx3, but is dispensable for Cd4 expression. We show that these functions cannot be performed by the BTB domain of the Thpok-related factor Bcl6, which fails to bind NuRD. Thus, cofactor binding critically contributes to the functional specificity of BTB-zinc finger factors, which control the differentiation of most hematopoietic subsets.
One-sentence summary:
Thpok association with the NuRD chromatin remodeling complex is necessary for CD4+ T cell differentiation in the thymus
Introduction
BTB-zinc finger (BTB-ZF) transcription factors contain, in addition to DNA binding zinc finger motifs that recognize specific DNA sequences, an amino-terminal BTB-POZ domain that notably serves for homodimerization (1, 2). BTB-ZF factors control multiple hematopoietic and immune differentiation processes. The BTB-ZF factor PLZF directs the differentiation of natural killer (NK) T cells, and LRF (encoded by Zbtb7a) represses fetal hemoglobin gene expression (3–5). Another BTB-ZF family member, Bcl6, controls the germinal center reaction, which drives the formation of high affinity antibodies essential for effective immune memory and vaccine responses (6, 7). In-depth analyses of Bcl6 functions have shown that it serves as an obligatory gene repressor (8). Transcriptional repression by sequence-specific DNA-binding proteins typically involves recruitment of co-repressors or co-repressor complexes, including molecules of the Groucho-TLE family, NCoR and related molecules, and the NuRD complex (9–11). Such co-repressors are thought to inhibit transcription through several mechanisms, including targeting of the general transcription machinery, post-translational histone modifications (e.g. histone deacetylation) or repositioning of nucleosomes.
Indeed, Bcl6 functions require recruitment of the corepressor NCoR, or the related protein Smrt, to its BTB domain (12–15). How general this paradigm is remains to be determined. Other BTB-ZF factors, including PLZF, also recruit NCoR, although possibly through distinct mechanisms (16, 17). Although other co-repressors or chromatin modifying complexes bind BTB domains of BTB-ZF proteins, the role of such binding in transcription factor function has not been established (5, 18–20). Thus, despite their physiological importance, it remains unclear how BTB-ZF factors control gene expression and how broadly the Bcl6-NCoR paradigm applies.
We addressed this question by studying Thpok (encoded by Zbtb7b, called Thpok here) (21, 22), a BTB-ZF factor controlling the differentiation and functions of CD4+ T cells and multiple processes outside of the immune system. These include collagen gene expression, mammary gland function, thermogenesis, and chromatin-lamina interactions in the nucleus (23–26). Furthermore, as with other members of the BTB-ZF family, including Bcl6 and LRF, dysregulated Thpok expression is involved in leukemia or lymphoma formation (7, 27, 28).
Together with CD8+ T cells, CD4+ T cells are essential for defenses against infections and differentiate in the thymus from precursors that express both CD4 and CD8 (double-positive, DP, thymocytes) (29–31). This process is controlled by the mutual antagonism between Thpok, expressed in CD4+-lineage cells, and Runx family transcription factors, of which Runx3 is expressed in CD8+-lineage cells (32, 33). Thpok inhibits expression of Runx3 (34–36), presumably through binding to cis-regulatory elements important for Runx3 expression (37). Furthermore, Thpok represses genes encoding CD8 molecules (Cd8a and Cd8b) (36, 38–40), binds to multiple genes expressed in cytotoxic T cells, and exerts a broad repressive function on the CD8+-lineage transcriptome (41–43).
Here, we combined biochemical and genetic approaches to investigate the mechanisms underpinning Thpok functions. We found that the Thpok BTB domain binds components of the NuRD (Nucleosome Remodeling and Deacetylase) chromatin remodeling complex (11, 44–47) rather than NCoR. Using both loss- and gain-of-function approaches, we show that such binding is instrumental for the function of Thpok in vivo. Accordingly, we found that the BTB domains of Thpok and Bcl6 are not functionally interchangeable in developing CD4+ T cells. These findings unveil new mechanisms used by BTB-zinc finger transcription factors and demonstrate that BTB domains are critical contributors to the functional specificity of BTB-zinc finger transcription factors.
Results
Thpok binds to the NuRD complex
To explore how Thpok regulates CD4+ T cell commitment, we searched for Thpok-interacting proteins using Liquid Chromatography-Mass Spectrometry (LC-MS). We used a version of Thpok (Thpok-Bio) subject to biotinylation in vivo by the BirA biotin ligase (48) (Fig. S1A), yielding a final product (Thpok-Biotin), which we previously showed supports CD4+ T cell development (42). To assess Thpok interactions in primary cells, we retrovirally expressed Thpok-Bio in in vitro activated CD4+ T cells from Thpokfl/fl Ox40-Cre+ mice carrying a Rosa26BirA allele. Expression of Ox40-Cre, initiated upon T cell activation (49, 50), causes the deletion of endogenous Thpokfl alleles (42, 51), so that transduced cells only express Thpok-Bio. Accordingly, flow cytometric analysis readily detected the BirA-biotinylated protein, Thpok-Biotin, in transduced cells (Fig. S1B). Streptavidin pull-down and LC-MS from such transduced CD4+ T cells identified peptides from 52 proteins (Fig. 1A and Table S1). Of these, 35 remained Thpok-bound despite lysate treatment with Ethidium Bromide (Thpok-EtBr), which impairs indirect associations mediated by DNA binding (52) (Fig. S1C and Table S1). This set of Thpok associated proteins (“Thpok 35-set” hereafter) comprised neither NCoR-family repressors nor Cullin 3, which both bind Bcl6 and PLZF (12, 13, 15, 20, 53). However, it included components of the Nucleosome Remodeling and Deacetylase (NuRD) complex (11), including the ATP dependent nucleosome remodeler Mi2β (encoded by Chd4), the histone deacetylases (HDAC) HDAC1 and HDAC2, and the metastasis-associated proteins (Mta) 1–3 (Fig. 1B). Interrogating previous RNAseq analyses (42, 43), we found expression of at least one member of each NuRD subfamily in developing αβ lineage thymocytes and in peripheral T cells (Fig. S1D).
Figure 1. Thpok binds NuRD.
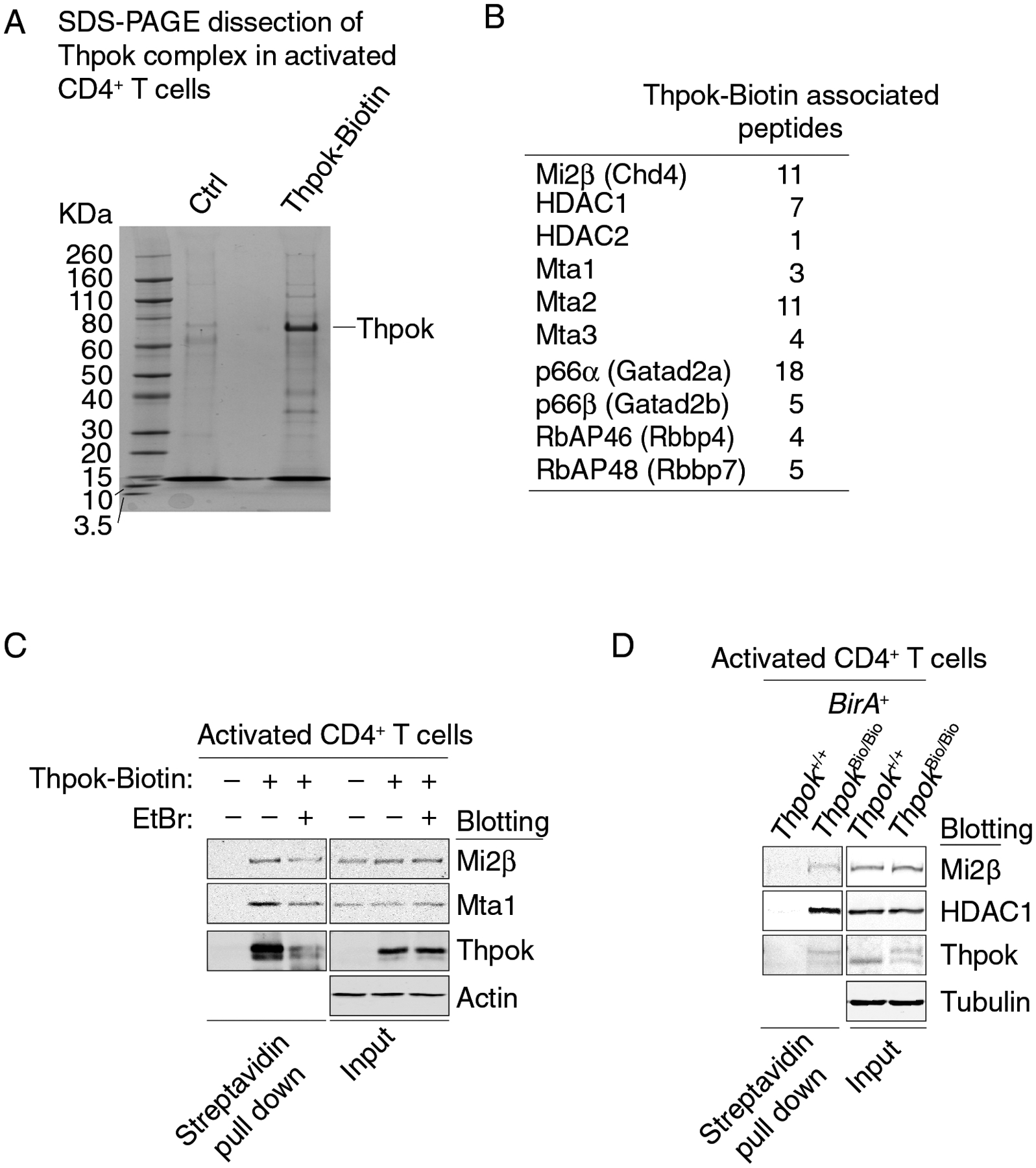
(A) SDS-PAGE and colloidal Coomassie blue staining of streptavidin pull down from activated Thpokfl/fl Ox40-Cre+ Rosa26BirA+ CD4+ T cells retrovirally transduced to express Thpok-Bio or with the empty retroviral vector (pMRx, Ctrl). Molecular markers are shown on the left.
(B) Thpok-associated NuRD-related peptides identified by mass spectrometry.
(C) Immunoblot analysis of streptavidin pull down (left panel) or whole cell lysates (right panel) from activated CD4+ T cells obtained from Thpokfl/fl Ox40-Cre+ Rosa26BirA+ that had been retrovirally transduced to express Thpok-Bio or with empty pMRx as a control vector (−). EtBr indicates treatment of cell lysates with Ethidium Bromide. Protein blots were probed with antibodies as indicated.
(D) Immunoblot analysis of streptavidin pull down (left panel) or whole cell lysates (right panel) from activated CD4+ T cells obtained from Thpok+/+ or ThpokBio/Bio Rosa26BirA+. Protein blots were probed with antibodies as indicated. Data are representative of three independent experiments.
We verified the association between Thpok and NuRD components by reciprocal co-immunoprecipitation from transfected HEK293T cells which express endogenous NuRD components (Fig. S1EF). Additionally, streptavidin pull-down from Thpok-Bio-transduced Thpokfl/fl Ox40-Cre+ Rosa26BirA+ activated CD4+ T cells detected EtBr-resistant Thpok-Biotin interaction with NuRD components Mi2β and Mta1, supporting the idea that the association of Thpok with NuRD was mediated by protein interactions independent of DNA binding (Fig. 1C). To verify that endogenously expressed Thpok interacts with NuRD, we analyzed activated CD4+ T cells from Rosa26BirA+ mice homozygous for a recombinant allele of Thpok expressing Thpok-Bio (ThpokBio), in which CD4+ T cells develop normally (42); we used cells from Thpok+/+ Rosa26BirA+ as controls. We found that endogenously expressed Thpok-Biotin recruits NuRD components Mi2β and HDAC1 (Fig. 1D). We conclude from these experiments that endogenous Thpok is associated with the NuRD complex.
Thpok controls chromatin organization at CD4+- and CD8+-lineage loci
Since NuRD affects both nucleosome positioning and covalent histone modifications (notably through deacetylases HDAC1 and HDCA2 and histone demethylases) (54–56), Thpok-NuRD complexes are expected to affect chromatin accessibility. However, we previously found that Thpok was dispensable for chromatin opening at, and initial upregulation of CD4+-lineage genes in MHC II-signaled thymocytes (43), questioning the relevance of NuRD binding for Thpok functions. To further address this question, we used single cell ATACseq (scATACseq) to compare chromatin accessibility in Thpok-sufficient (CD4+-differentiating) and -deficient (redirected to the CD8+-lineage) MHC II-restricted thymocytes.
We captured nuclei from Thpokfl/fl Cd4-Cre+ and Cd4-Cre− (Ctrl) mice expressing a ThpokGFP bacterial artificial chromosome (BAC) reporter that identifies MHC II-signaled cells in both strains (43, 57). From both strains, we sorted MHC II-signaled thymocytes (GFP+), and unsignaled CD69− DP thymocytes; we also sorted MHC I-restricted (GFP−) CD8+ SP thymocytes from Ctrl mice (Fig. S2AB). We integrated and analyzed two captures from each genotype using the Signac extension of the Seurat suite (58, 59). UMAP dimensional reduction showed cells segregating according to genotype rather than experimental replicate (Fig. S2C). Chromatin accessibility at Rag1 and S1pr1 identified unsignaled DP and mature SP thymocytes, respectively (43) (Fig. S2D). Unsupervised clustering and UMAP analysis distinguished unsignaled DP thymocytes, in clusters common to both genotypes, from all other cells which were grouped into clusters that were in part genotype-specific and provided limited resolution within each developmental trajectory (Table S2 and Fig. S2EF). In contrast, we previously showed that scRNAseq orders the developmental trajectories of post-DP Thpok-sufficient and -deficient thymocytes and identifies the “tipping point” at which Thpok-deficient cells switch from CD4+ to CD8+ features (Fig. 2A and Ref. 43). To leverage this higher resolution, we used the Signac integration feature to link each cell from the scATACseq data set to the transcriptomic cluster (43) that best matches its chromatin accessibility status, thereby defining cells groups linked to the same transcriptomic cluster (Fig. 2B).
Figure 2. Thpok enforces chromatin closure at CD8+-lineage genes.
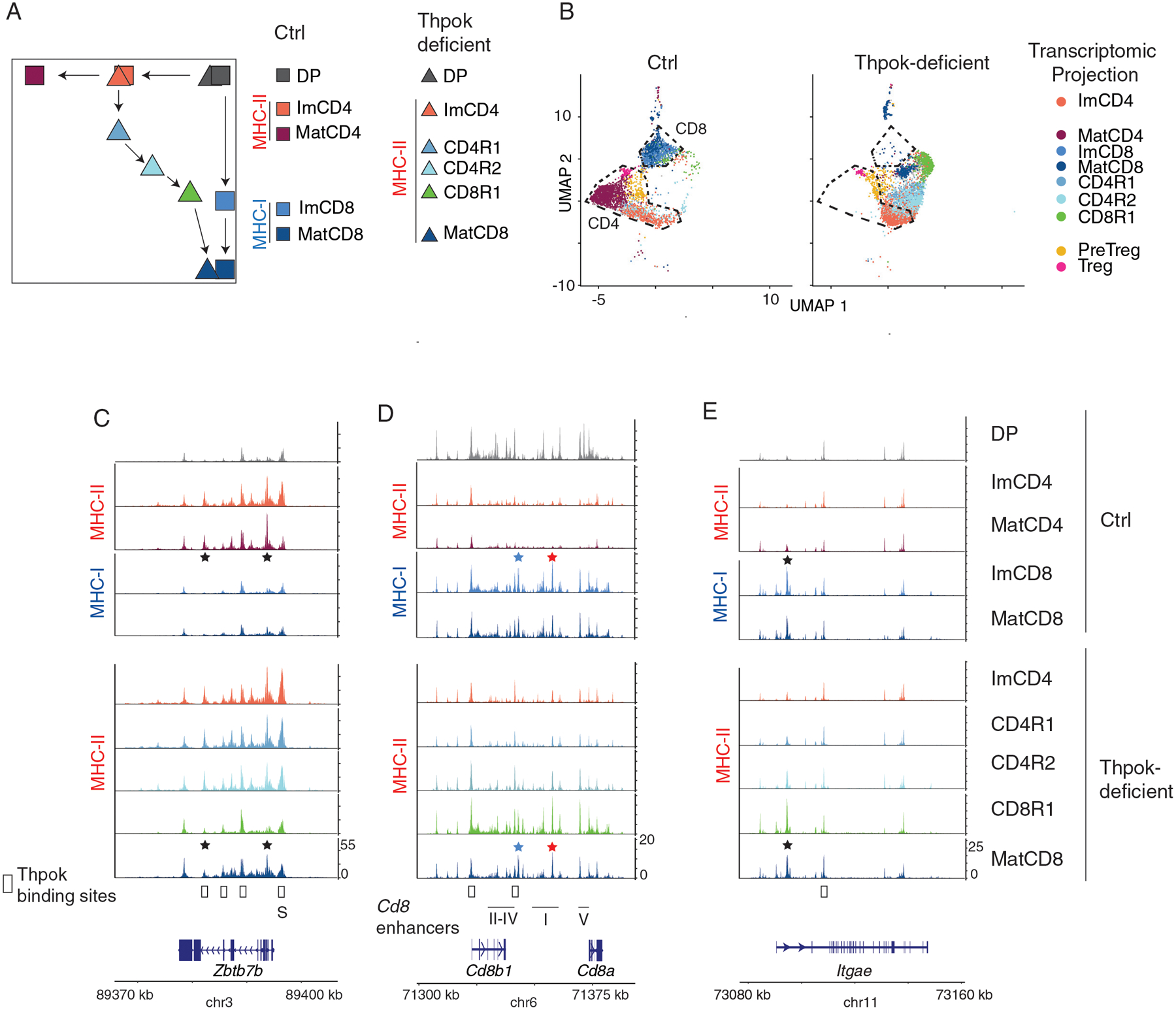
scATACseq comparison of (i) Ctrl (Thpokfl/fl Cd4-Cre−) ThpokGFP+, CD8+ SP and DP thymocytes and (ii) Thpok-deficient (Thpokfl/fl Cd4-Cre+) ThpokGFP+ and DP thymocytes, all from mice carrying a ThpokGFP BAC reporter. Data integrates two biological replicates from each genotype.
(A) Schematic CD4-CD8 expression plot showing developmental trajectories of Thpok-sufficient and -deficient thymocytes, and indicating previously identified scRNAseq transcriptomic clusters (43) used for scATACseq analyses. Note that the cluster order shown here, derived from pseudo-time analysis, is consistent with the experimentally determined CD8+-lineage potential of immature Thpok-deficient CD4+ SP thymocytes (109). Imm: immature; Mat: mature; R indicates MHC II-restricted thymocytes undergoing lineage redirection (43).
(B) UMAP dimensional reduction plot displaying cells separated by genotype and color-coded as in (A) according to the closest transcriptomic cluster match (43). DP thymocytes were omitted for clarity. Outlines indicate positions of Ctrl CD4+- and CD8+-lineage cells.
(C-E) Genome browser tracks show scATACseq signals at indicated genes (bottom), displayed as scaled sequence read density averaged for all cells sharing the indicated transcriptome cluster projection, separated by genotype (noted at the far right) and color-coded as in (A). DP thymocyte tracks are shown for Ctrl cells only. MHC restriction is indicated on the left side of each panel. The positions of Cd8 enhancers E8I-V and of the Thpok silencer (S) are indicated below the gene track (C, D). Stars indicate lineage specific peaks, including CD8+-lineage-specific peaks at Cd8 enhancers E8I (red) E8VI (blue). Open boxes above gene tracks indicate Thpok binding sites (GEO reference GSE148976) (43).
Along the CD4+-lineage trajectory, which was shared by both genotypes up to the Immature CD4+ SP group (ImCD4, at which Thpok is expressed), chromatin opening at CD4+- lineage genes Thpok and Cd40lg was independent of Thpok expression, consistent with population analyses (43) (Figs. 2C and S2G). Consistent with their Thpok-independent initial opening, CD4+-lineage specific accessible areas remained open in “redirected” mature CD8+ SP (MatCD8) Thpok-deficient cells, relative to their MHC I-restricted (Ctrl) counterparts (stars in Figs. 2C and S2G).
We next inquired how Thpok affected chromatin accessibility at the Cd8 locus. Consistent with CD8 expression pattern, chromatin at Cd8 was broadly accessible in DP thymocytes and MHC I-restricted CD8+ SP thymocytes (Fig. 2D). In Thpok-sufficient MHC II-restricted thymocytes, accessibility at Cd8 decreased as cells differentiated from DP to mature CD4+ SP (MatCD4). In Thpok-deficient MHC II-restricted thymocytes, accessibility at Cd8 followed a U-shaped pattern, initially decreasing before reverting to a pattern similar to that of Ctrl mature CD8+ SP cells. In these cells, we did not detect accessibility differences between genotypes at enhancers E8I and E8VI (Fig. 2D, red and blue stars), specific of mature CD8+ SP thymocytes and T cells (60, 61). At Itgae and Prf1, two CD8+-lineage specific genes not expressed in DP thymocytes, CD8+-specific chromatin accessibility progressed with similar developmental kinetics in Ctrl (MHC I-restricted) and redirected MHC II-restricted thymocytes, but not in Thpok-sufficient MHC II-restricted thymocytes (Figs. 2E and S2G). Since Thpok binds all three genes (43), these observations supported the conclusion that Thpok restrains chromatin opening at CD8+-lineage genes, prompting us to further study its association with NuRD.
The Thpok BTB domain binds NuRD and is needed for CD4+ T cell differentiation
We examined which domain of Thpok bound NuRD by generating deletions in the Thpok coding sequence (Fig. 3A). BTB domains promote homodimerization (62), which conceivably could affect Thpok function (21). To explore the role of the BTB domain without compromising homodimerization, we generated a Thpok mutant protein (ΔBTBLZ) lacking the BTB domain but containing the self-dimerizing leucine zipper from the chicken Basic Vitellogenin-promoter Binding Protein (B-VBP), which has no known dimerizing partner in mammalian cells (63) (Fig. S3A–C). Using immunofluorescence of transfected HEK293T cells, we verified these deletions did not affect nuclear localization (Fig. S3D). Transient transfections in HEK293T cells (Fig. 3B) showed that neither deletion of the carboxy-terminal domain (Thpok-ΔC) nor a point mutation in the second zing finger of Thpok, known to abolish Thpok activity (Thpok-HD) (22), disrupted NuRD binding. In contrast, despite efficient homodimerization, ΔBTBLZ failed to interact with NuRD components Mi2β, Mta2, and HDACs 1 and 2, demonstrating a requirement for the BTB domain (Fig. 3B). We conclude from these analyses that the Thpok BTB domains is needed for NuRD binding.
Figure 3. The Thpok BTB domain binds NuRD and is needed for CD4+ T cell development.
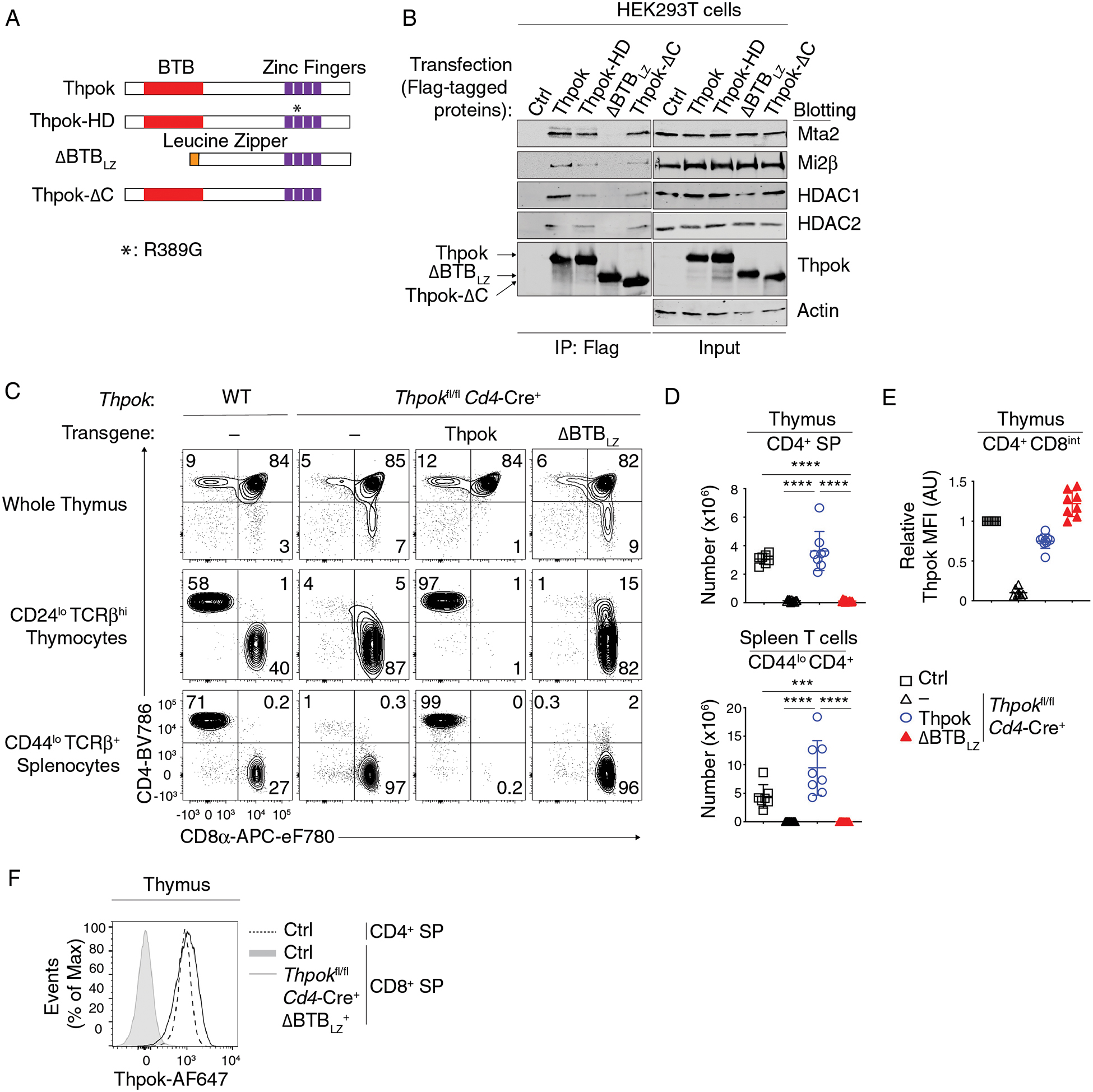
(A) Schematic diagram of Thpok and mutants thereof. The Thpok BTB domain and zinc finger motifs are depicted by red and purple boxes; the Leucine Zipper sequence (LZ) by the orange box. *: R389G mutation in the ThpokHD mutant (22).
(B) Immunoblot analysis of anti-Flag immunoprecipitates (left panel) or whole cell lysates (right panel) from HEK293T cells transfected with control vector (pcDNA3, Ctrl), Flag-tagged Thpok, Thpok-HD, ΔBTBLZ or Thpok-ΔC. Protein blots were probed with antibodies as indicated. Data are representative of more than three independent experiments.
(C) Expression of CD4 and CD8 in indicated thymocyte and splenocyte subsets from Thpokfl/fl Cd4-Cre+ mice expressing the indicated Thpok-derived transgene or no transgene (−).
(D) Numbers of CD44lo TCRβhi CD24lo CD4+ SP thymocytes (top) and of CD44lo TCRβhi CD4+ splenocytes (bottom) in mice shown in (C).
(E) Expression of transgenic Thpok or ΔBTBLZ in CD4+ CD8int TCRβhi CD69+ thymocytes was assessed by intra-cellular staining and flow cytometry and is presented relative to that of Thpok-expressing CD4+ CD8int TCRβhi CD69+ thymocytes in Thpokfl/fl Cd4-Cre− littermates (Ctrl), set as 1 in each experiment.
(F) Overlaid histograms show expression of transgenic ΔBTBLZ in TCRβhi CD24lo CD4−CD8+ thymocytes from Thpokfl/fl Cd4-Cre+ ΔBTBLZ+ mice (plain line) and of endogenous Thpok in CD4+ or CD8+ SP thymocytes from Thpokfl/fl Cd4-Cre− (Ctrl) littermates (dotted line and grey-shaded trace, respectively).
Data (C-F) are representative of 4 independent experiments totaling n= 7 (Thpokfl/fl Cd4-Cre−), 12 (Thpokfl/fl Cd4-Cre+), 8 (Thpokfl/fl Cd4-Cre+ Thpok transgenic) or 14 (Thpokfl/fl Cd4-Cre+ ΔBTBLZ+) mice. One-way ANOVA followed with Tukey multiple comparison tests. ***p<0.001, ****p<0.0001. Error bars indicate standard deviation.
To verify that the mutation did not prevent in vivo DNA recognition, we assessed binding of ΔBTBLZ to two previously identified Thpok binding regions in the Thpok and Cd4 silencers (35). Using chromatin immunoprecipitation (ChIP)-qPCR on CD4+ T cells expressing Thpok or ΔBTBLZ, we readily detected ΔBTBLZ binding to both sites (Fig. S3E).
To examine the impact of NuRD binding on Thpok functions, we generated a transgene expressing ΔBTBLZ using CD2-based cis-regulatory elements active in thymocytes and T cells. When expressed in otherwise wild-type mice, the transgene had no or little detectable effects on T cell development in three independent founder-derived lines (C5, A1, and E5), unlike a wild-type Thpok transgene which prevented CD8+ T cell development (21) (Fig. S3F–H). To examine if ΔBTBLZ could support CD4+ T cell development, we expressed ΔBTBLZ in Thpokfl/fl Cd4-Cre+ mice, which delete Thpok in DP thymocytes. We found that ΔBTBLZ failed to restore CD4+ T cell differentiation, as assessed on numbers of mature CD4+ SP thymocytes (with mature defined as CD44lo TCRβhi CD24lo throughout this study, Fig. S3F) and of CD4+ T cells in the spleen (Fig. 3CD). Given that similar results were observed with all three lines (Fig. S3I), we selected line E5 for subsequent analyses. To confirm proper expression of the transgenic ΔBTBLZ, we used an antibody which recognizes the linker region between the Thpok BTB domain and zinc finger motifs (Fig. S4A–C). We found equivalent expression of transgenic Thpok and ΔBTBLZ in intrathymically signaled (CD4+ CD8int TCRβhi CD69+) Thpokfl/fl Cd4-Cre+ thymocytes expressing either transgene (Fig. 3E, gating in Fig. S4D). In this subset, expression of the transgenic proteins was similar to that of endogenous Thpok molecules in signaled cells from wild type mice. Additionally, ΔBTBLZ was also detected in CD8+ thymocytes of transgenic mice (Fig. 3F), and its expression resulted in expression of CD4 in a subset of CD8+ mature thymocytes (Fig. S3J). We conclude from these analyses that, despite appropriate dimerization and expression, ΔBTBLZ failed to support CD4+ T cell differentiation.
The BTB domains of Thpok and Bcl6 are not functionally interchangeable
To further delineate the role of NuRD in Thpok functions, we swapped the BTB domain of Thpok for the distantly related BTB domain of Bcl6, generating a chimeric B-Thpok protein (Fig. 4A). B-Thpok efficiently dimerized but failed to bind NuRD (Figs. 4B and S5A). In contrast, a similar construct (L-Thpok) containing the BTB domain of Lrf, which is more closely related to that of Thpok, bound NuRD (Fig. 4B), consistent with previous results (5, 19). LC-MS analysis confirmed that the B-Thpok protein failed to recruit NuRD components, whereas, as expected, it was associated with NCoR1 (Fig. 4C). Of note, another domain of Bcl6, located between its BTB and zinc finger domains, binds NuRD (64); this region of Bcl6 is not included in the B-Thpok construct.
Figure 4. Functional specificity of BTB domains.
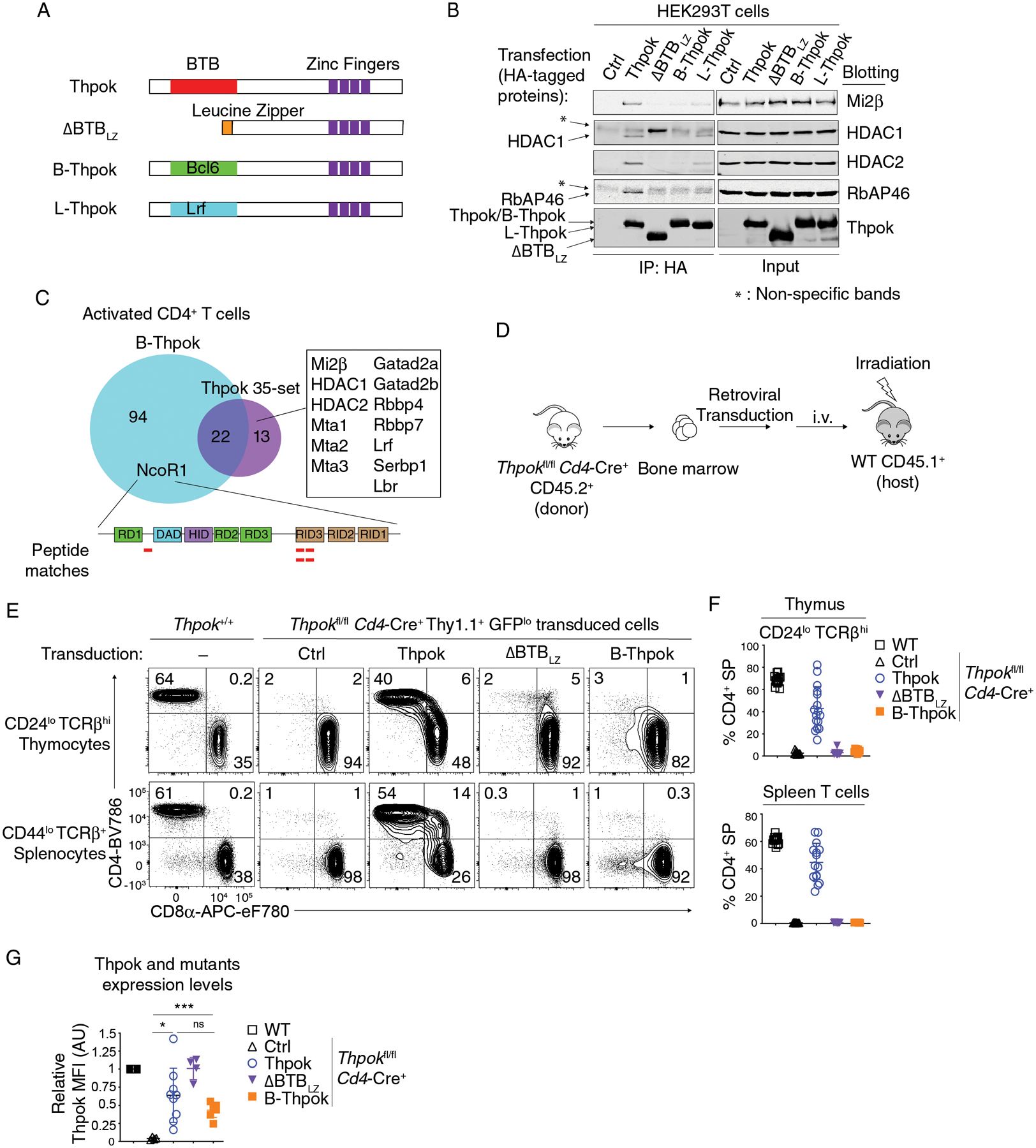
(A) Schematic diagram of Thpok and chimeric constructs. The Thpok BTB domain and zinc finger motifs are depicted by red and purple boxes; the Leucine Zipper sequence (LZ) by the orange box. The Bcl6 and Lrf BTB domain are depicted by green and cyan box, respectively.
(B) Immunoblot analysis of anti-HA immunoprecipitates (left panel) or whole cell lysates (right panel) from HEK293T cells transfected with control vector (pcDNA3, Ctrl), HA-tagged Thpok, ΔBTBLZ, B-Thpok or L-Thpok. Protein blots were probed with antibodies as indicated. * indicates non-specific bands. Data are representative of more than three independent experiments.
(C) Venn diagram showing B-Thpok binding partners and Thpok 35-set as detected by mass spectrometry after streptavidin pull down in activated CD4+ T cells. Proteins binding Thpok but not B-Thpok are listed in the box. Peptides from NCoR1 detected in the B-Thpok pull-down are depicted by red lines along the NCoR1 schematic (bottom). RD1–3: Repression Domain; DAD: Deacetylase Activity Domain; HID: Histone Interaction Domain; RID: Receptor Interacting Domain.
(D) Schematic diagram of retrogenic mice generation.
(E) Expression of CD4 and CD8 in indicated thymocyte and splenocyte subsets from retrogenic mice generated with Ctrl (empty pPMGfIT vector), or Thpok-, ΔBTBLZ- or B-Thpok-expressing pPMGfIT retroviruses. The contour plots are representative of three independent experiments totaling n= 13 (WT, C57BL/6 mice), 12 (Ctrl), 16 (Thpok), 9 (ΔBTBLZ) or 8 (B-Thpok) retrogenic mice.
(F) The percentages of CD4+ SP cells in CD44lo CD24lo TCRβhi thymocytes (top) and in CD44lo TCRβ+ splenocytes (bottom) in mice shown in (E).
(G) Flow cytometric expression of transduced Thpok, ΔBTBLZ and B-Thpok in CD4+ CD8int TCRβhi CD69+ thymocytes, expressed relative to that of Thpok-expressing CD4+ CD8int TCRβhi CD69+ in WT mice, set as 1 in each experiment. The data are summarized from two independent experiments totaling n= 8 (WT), 4 (Ctrl), 9 (Thpok), 4 (ΔBTBLZ) or 7 (B-Thpok) retrogenic mice. Two-way ANOVA *p<0.05, ***p<0.001, ns: p>0.05. Error bars indicate standard deviation.
We reasoned that, if NuRD recruitment is instrumental to Thpok functions, the B-Thpok protein, whose BTB domain does not bind NuRD, should fail to support the CD4+-lineage differentiation of Thpok-deficient thymocytes. To assess this prediction, we generated “retrogenic” mice, using a Cre-activated retroviral vector to target gene expression in DP thymocytes (65). In this vector, a floxed GFP open reading frame (ORF), including a stop codon, prevents translation of a downstream gene of interest (Thpok or derivative, Fig. S5BC). Cre expression excises the floxed GFP ORF, allowing translation of the gene of interest. A Thy1.1 reporter cDNA, downstream of an IRES, is expressed independently of Cre, identifying transduced cells. We transduced this vector into bone marrow progenitors from Thpokfl/fl Cd4-Cre+ CD45.2+ mice that were then transferred into irradiated congenic CD45.1+ host mice (Fig. 4D). In recipient mice, transduced cells expressed the donor marker CD45.2 and the retroviral Thy1.1 reporter. In line with the extended half-life of GFP in thymocytes (66), cells in which Cre expression had activated the retroviral expression cassette were identified by absent or residual GFP expression (Fig. S5D). In Thpokfl/fl Cd4-Cre+ cells, expression of wild-type Thpok from this vector resulted in the generation of mature CD4+ SP thymocytes and spleen T cells (Fig. 4EF). In contrast, retrogenic ΔBTBLZ and B-Thpok failed to restore CD4+ T cell development, despite expression levels comparable to retrogenic wild-type Thpok (Fig. 4G). These experiments indicated that the BTB domains of Thpok and Bcl6 are not functionally interchangeable, and that NuRD binding serves functions distinct from NCoR binding.
NuRD binding mediates Thpok BTB domain functions
To further delineate the role of NuRD in Thpok functions, we sought to identify amino acid residues critical for BTB-NuRD interactions. We compared the sequences of Thpok and Lrf BTB domains, which bind NuRD, with that of Bcl6, which does not. We considered regions with greater divergence between Thpok and Bcl6 than between Thpok and Lrf (Fig. S6A). We first focused on two segments mapping to putative β-sheet (β4) and α-helix (α6) motifs and facing outwardly, as inferred from the three-dimensional structure of the Lrf BTB domain (67, 68). To examine the role of these motifs in NuRD binding, we mutated GVCE (one-letter code) (β4) and MEI (α6) Thpok residues to the corresponding SVIN and RKF in Bcl6 (Figs. 5A and S6A). In transfected HEK293T cells, the Thpok-RKF mutant efficiently dimerized but failed to bind NuRD components Mi2β, RbAP46, and HDAC1 and HDAC2 (Figs. S6B and 5B), whereas the GVCE to SVIN mutation (Thpok-SVIN) had no detectable effect on NuRD components binding. Similar results were observed in Thpokfl/fl Ox40-Cre+ Rosa26BirA+ activated CD4+ T cells retrovirally transduced to express Thpok-Bio, ΔBTBLZ-Bio, or Thpok-RKF-Bio (Fig. S6C). LC-MS, performed from Thpok-RKF expressing CD4+ T cells using the same approach as for wild-type Thpok, showed that the mutation disrupted binding to all NuRD components and to Serbp1, a protein reported to associate with the NuRD component Mi2α (Chd3) (69) (Fig. 5C and Table S1). In contrast, the mutation had no effect on the other 24 non-NuRD interacting partners. Last, we examined if Thpok-RKF bound DNA in vivo, by subjecting Thpok-RKF-transduced CD4+ T cells to ChIP-qPCR. We found similar binding of Thpok and Thpok-RKF to Thpok binding regions in the Thpok and Cd4 silencers (35) (Fig. 5D), indicating that the RKF mutation did not affect DNA binding. We conclude from these results that Thpok MEI amino acids in the α6 segment are specifically required for NuRD binding. Although structural studies will be necessary to understand the geometry of the BTB-NuRD interactions, this motif is not spatially equivalent to the Smrt-binding groove of the Bcl6 BTB domain (12).
Figure 5. A NuRD-interacting segment of the Thpok BTB domain is needed for CD4 T cell development.
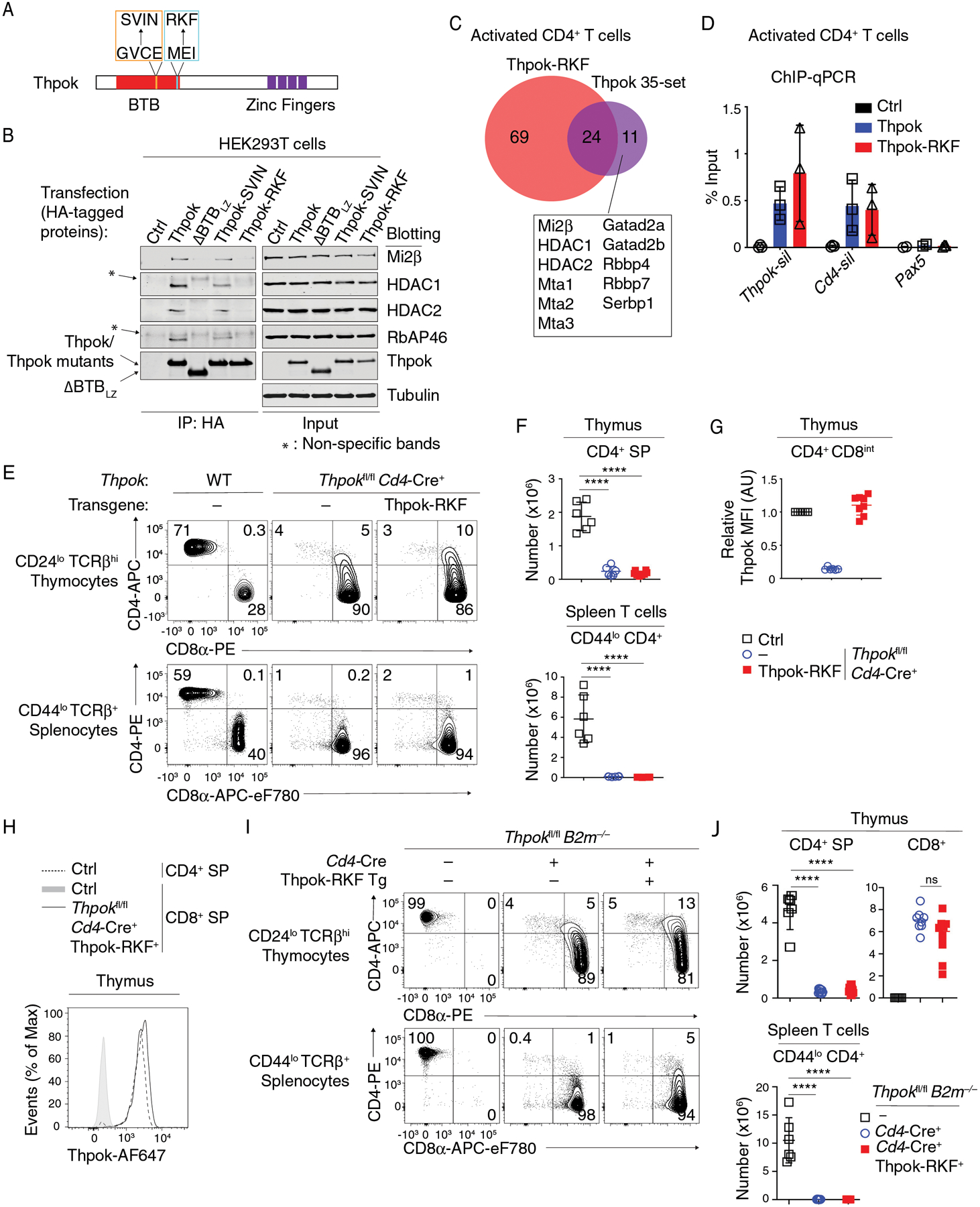
(A) Schematic diagram of Thpok-SVIN (orange line box) and Thpok-RKF (cyan line box) mutations.
(B) Immunoblot analysis of anti-HA immunoprecipitates (left) or whole cell lysates (right) from HEK293T cells transfected with control vector (pcDNA3, Ctrl), HA-tagged Thpok, ΔBTBLZ, Thpok-SVIN or Thpok-RKF. Protein blots were probed with antibodies as indicated. * indicates non-specific bands. Data are representative of three independent experiments.
(C) Venn diagram showing Thpok-RKF binding partners and Thpok 35-set as detected by mass spectrometry after streptavidin pull down in activated CD4+ T cells. Proteins binding Thpok but not Thpok-RKF complex are listed in the box.
(D) ChIP-qPCR was performed on Thpok and Cd4 silencer elements, and on a region of Pax5 (negative control), from in vitro activated CD4+ T cells from Thpokfl/fl Ox40-Cre+ Rosa26BirA+ that had been retrovirally transduced with empty pMRx (Ctrl), or pMRx expressing Thpok-Bio or Thpok-RKF-Bio. Data are expressed as percent of input DNA. Each symbol represents a separate determination, and the figure summarizes three independent experiments.
(E) Expression of CD4 and CD8 in indicated thymocyte and splenocyte subsets from indicated mice.
(F) Numbers of CD44lo CD24lo TCRβhi CD4+ SP thymocytes (top) and of CD44lo TCRβ+ CD4+ splenocytes (bottom) in mice shown in (E).
(G) Flow cytometric expression of transgenic Thpok-RKF in CD4+ CD8int TCRβhi CD69+ thymocytes, shown relative to that of Thpok-expressing CD4+ CD8int TCRβhi CD69+ in Thpokfl/fl Cd4-Cre− littermates (Ctrl), set as 1 in each experiment.
(H) Overlaid histograms show expression of transgenic Thpok-RKF in CD24lo TCRβhi CD4− CD8+ thymocytes from Thpokfl/fl Cd4-Cre+ Thpok-RKF+ mice (plain line) and of endogenous Thpok in CD4+ or CD8+ SP thymocytes from Thpokfl/fl Cd4-Cre− (Ctrl) littermates (dotted line and grey-shaded trace, respectively).
Data (E-H) are representative of three independent experiments totaling n= 6 (Thpokfl/fl Cd4-Cre− littermates), 6 (Thpokfl/fl Cd4-Cre+) or 8 (Thpokfl/fl Cd4-Cre+ Thpok-RKF+) mice. One-way ANOVA followed with Tukey multiple comparison tests ****p<0.0001. Error bars indicate standard deviation.
(I) Expression of CD4 and CD8 in indicated thymocyte and splenocyte subsets from Thpokfl/fl B2m−/− mice expressing or not the Thpok-RKF transgene.
(J) Numbers of CD44lo CD24lo TCRβhi thymocytes, CD4+ CD8− (top left) or CD8+ (including CD4+ and CD4−, top right) and of CD44lo TCRβ+ CD4+ CD8− splenocytes (bottom) in mice shown in (I).
Data (I-J) are representative three independent experiments totaling n= 6 (Cd4-Cre− littermates), 8 (Cd4-Cre+) or 10 (Cd4-Cre+ Thpok-RKF+) mice. One-way ANOVA followed with Tukey multiple comparison tests ****p<0.0001, ns: P>0.05. Error bars indicate standard deviation.
To investigate if the RKF mutation affected Thpok functions during CD4+ T cell development, we generated CD2-based Thpok-RKF transgenic lines. Similar to ΔBTBLZ, the Thpok-RKF transgene did not affect T cell development in wild-type mice and failed to restore CD4+ T cell development in Thpokfl/fl Cd4-Cre mice (Figs. 5EF and S6DE), despite being expressed at levels similar to wild-type Thpok in CD4+ CD8int TCRβhi CD69+ thymocytes and in CD8+ SP thymocytes (Fig. 5GH). To track the fate of MHC II-restricted precursors expressing Thpok-RKF, we expressed the transgene in Thpokfl/fl Cd4-Cre+ B2m−/− mice, in which neither CD4+ nor MHC I-restricted CD8+ T cells develop, because of Thpok and β2-microglobulin disruption, respectively (70). The development of CD8+ SP thymocytes and T cells in Thpok-RKF expressing Thpokfl/fl Cd4-Cre+ B2m−/− mice indicated that the RKF mutation prevented the CD4+ lineage differentiation, but not the development, of MHC II-restricted thymocytes (Fig. 5IJ). We conclude from these experiments that the Thpok BTB α6-MEI aminoacid motif is required for both Thpok-NuRD interactions and Thpok functions during CD4+ T cell development.
The Thpok BTB domain promotes CD4+ T cell development by binding NuRD
Detergent solubilization, necessary for LC-MS, could conceivably disrupt the interaction of NuRD-unrelated molecules to the α6 MEI motif. Indeed, other proteins had been reported to bind the Thpok BTB domain (39). To verify that the MEI motif actually serves by binding NuRD, we engineered a NuRD-binding but BTB-less version of Thpok. Mta subunits, of which Mta2 is the most abundantly expressed in CD4+-lineage thymocytes (Fig. S1D), serve as scaffolds for NuRD assembly (71, 72). Thus, we generated a ΔBTBLZ variant appended with the Mta2 coding sequence at its amino-terminal extremity (Mta2-ΔBTBLZ, Fig. 6A). Mta2-ΔBTBLZ homodimerized (Fig. S7A) and bound Mi2β and HDAC1 (Fig. 6B), supporting the conclusion that it can recruit NuRD.
Figure 6. NuRD recruitment recapitulates the Thpok BTB domain functions during CD4 T cell development.
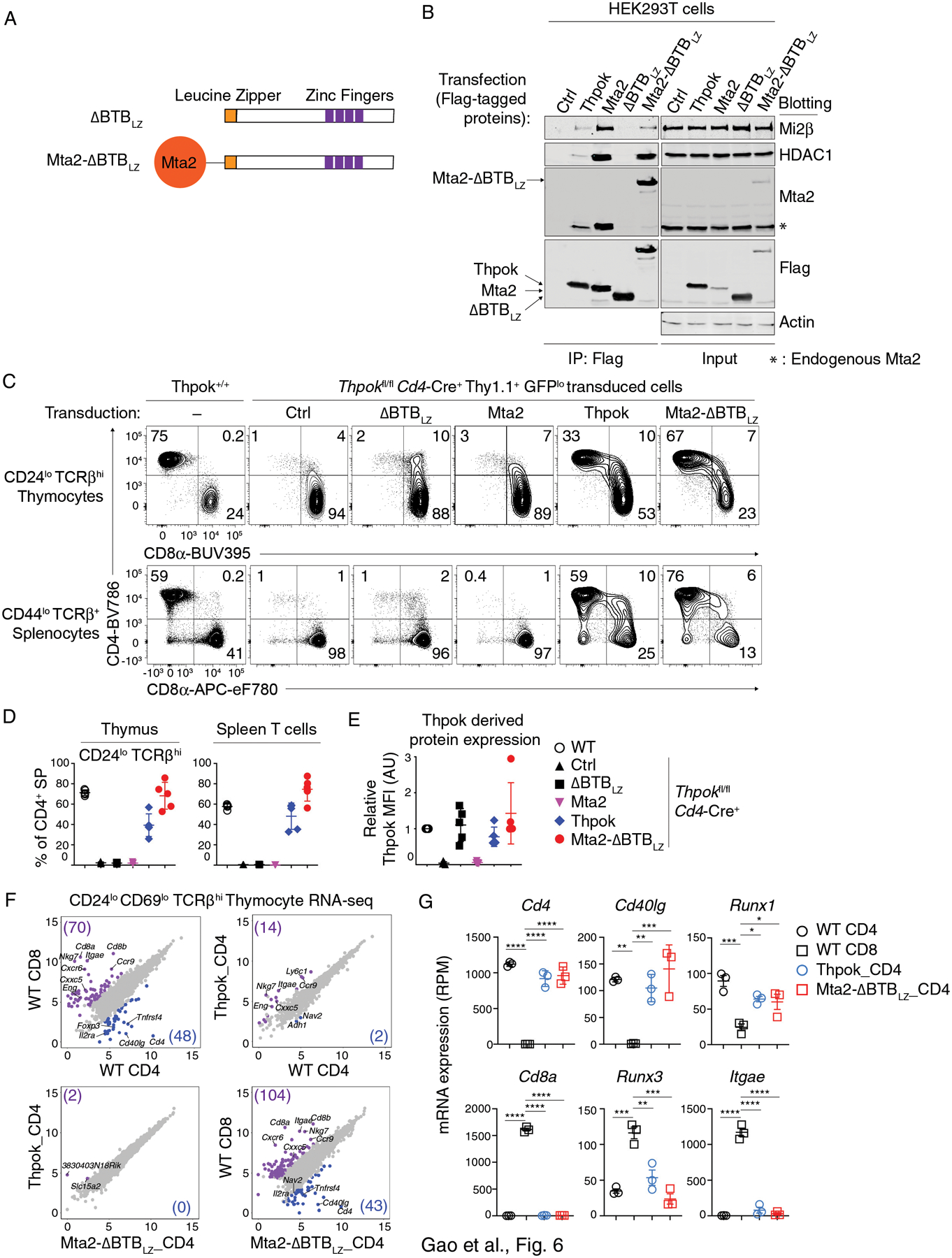
(A) Schematic diagram of ΔBTBLZ and Mta2-ΔBTBLZ.
(B) Immunoblot analysis of anti-Flag immunoprecipitates (left panel) or whole cell lysates (right panel) from HEK293T cells transfected with empty pcDNA3 (Ctrl), or vectors expressing Flag-tagged versions of Thpok, Mta2, ΔBTBLZ or Mta2-ΔBTBLZ. Protein blots were probed with antibodies as indicated. * indicates endogenous Mta2 protein. Data are representative of three independent experiments.
(C) Expression of CD4 and CD8 in indicated thymocyte and splenocyte subsets from retrogenic mice generated with empty pMGfIT (Ctrl), or pMGfIT expressing ΔBTBLZ, Mta2, Thpok, or Mta2-ΔBTBLZ. The contour plots are representative of three independent experiments with 5 mice per transduced vector in each experiment.
(D) Percentages of CD4+ CD8− (CD4+ SP) cells among CD44lo CD24lo TCRβhi thymocytes (left panel) and or CD44lo TCRβ+ splenocytes (right panel) from one of three independent experiments with 5 mice per group.
(E) Flow cytometric expression of retroviral ΔBTBLZ, Thpok or Mta2-ΔBTBLz in CD4+ CD8int TCRβhi CD69+ thymocytes, presented relative to that of Thpok-expressing CD4+ CD8int TCRβhi CD69+ in WT mice, set as 1 in each experiment.
(F) Scatter plots comparing gene expression (log2 RPM [Reads Per Million]) in CD4+ or CD8+ SP thymocytes from wild type mice (WT CD4 and WT CD8, respectively) and in CD4+ SP thymocytes from retrogenic mice (Thpok_CD4 and Mta2-ΔBTBLz_CD4), all purified as CD44lo CD24lo CD69lo TCRβhi as shown in Fig. S7B. Data is from three biological replicates processed separately up to sequencing. Reads mapping to Thpok (endogenous or retrovirally expressed) were omitted from the analyses. Genes with > 4-fold differential expression and FDR < 0.05 between indicated subsets are shown in purple or blue; numbers in corner indicate the count of differentially expressed genes (gene numbers in parentheses).
(G) Graphs display expression (RPM) of indicated genes in each cell subset analyzed in (F). Each symbol represents a distinct sample. One-way ANOVA followed with Tukey multiple comparison tests *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Error bars indicate standard deviation.
We tested the functions of Mta2-ΔBTBLZ in T cell development in vivo using the retrogenic approach. Mta2-ΔBTBLZ, but neither ΔBTBLZ nor Mta2, restored mature CD4+ SP thymocytes and spleen T cells populations (Fig. 6CD). We verified similar expression levels of ΔBTBLZ, Thpok and Mta2-ΔBTBLZ in CD4+ CD8int TCRβhi CD69+ thymocytes (Fig. 6E). To further characterize the effect of Mta2-ΔBTBLZ, we used RNA sequencing (RNAseq) to compare the transcriptomes of mature CD4+ SP thymocytes generated in retrogenic mice expressing Mta2-ΔBTBLZ or Thpok, to that of mature CD4+ and CD8+ SP thymocytes from wild-type mice as controls (Gating in Fig. S7B). We identified 118 genes differentially expressed between wild-type CD4+ and CD8+ SP cells (log2 Fold change > 2 and FDR < 0.05) (Fig. 6F, top left panel). We found only minimal differences between the transcriptomes of CD4+ SP cells expressing endogenous Thpok, retrogenic Thpok or Mta2-ΔBTBLZ, indicating that the fusion protein fully reconstituted CD4+ T cell development (Fig. 6F). This was notably illustrated by appropriate expression of Cd4, Cd8a, Cd40lg, and Runx1, and repression of Runx3 and Itgae (encoding CD103, a prototypical Runx3 target in the thymus) (73) (Fig. 6G). These experiments demonstrate that the BTB domain of Thpok supports CD4+ T cell development by binding the NuRD complex. Together with the strong ChIP DNA binding of ΔBTBLZ (Fig. S3E), the full restoration of CD4+ T cell differentiation by Mta2-ΔBTBLZ, which has the same leucine zipper dimerization interface as ΔBTBLZ, strongly supports the idea that both ΔBTBLZ and Mta2-ΔBTBLZ efficiently bind relevant Thpok target genes in vivo.
NuRD binding is needed for repression of Runx3
Thpok promotes CD4+ T cell differentiation at least in part by counteracting Runx activity, contributed in developing T cells by Runx1 and Runx3 (32, 33, 74). Such antagonism includes at least two components. First, Thpok inhibits Runx3 expression in MHC-II restricted thymocytes (34, 35). However, Runx3 disruption does not restore the CD4+ T cell differentiation of Thpok-deficient thymocytes (75), implying that at least some Thpok functions are independent from Runx3 repression. Indeed, Thpok also inhibits the repression of Cd4 by Runx proteins (35, 76), of which Runx1 is normally co-expressed with Thpok in CD4+ SP thymocytes and T cells. Thus, we assessed if NuRD recruitment mediated any of these Thpok functions.
We first examined if Thpok-RKF repressed Runx3. Consistent with previous results, intra-cellular staining of Thpok+/+ thymocytes detected Thpok but not Runx3 in CD4+ SP cells, and Runx3 but not Thpok in CD8+ SP cells (Fig. 7A, orange and blue traces, respectively). Runx3 but not Thpok was expressed in mature CD8+ SP thymocytes from B2m−/− Thpokfl/fl Cd4-Cre+ mice, indicating Runx3 de-repression. Mature CD8+ SP thymocytes from Thpok-RKF transgenic B2m−/− Thpokfl/fl Cd4-Cre mice co-expressed both Runx3 and the Thpok-RKF mutant protein, showing that Thpok-RKF failed to repress Runx3 (Fig. 7AB). Using ChIP-qPCR, we verified similar binding of Thpok and Thpok-RKF on a prominent Thpok binding site previously identified near the second Runx3 exon (Fig. 7C) (37, 42, 43). We conclude from these experiments that NuRD recruitment is needed for Thpok to repress Runx3.
Figure 7. Thpok-NuRD binding is needed for Runx3 repression.
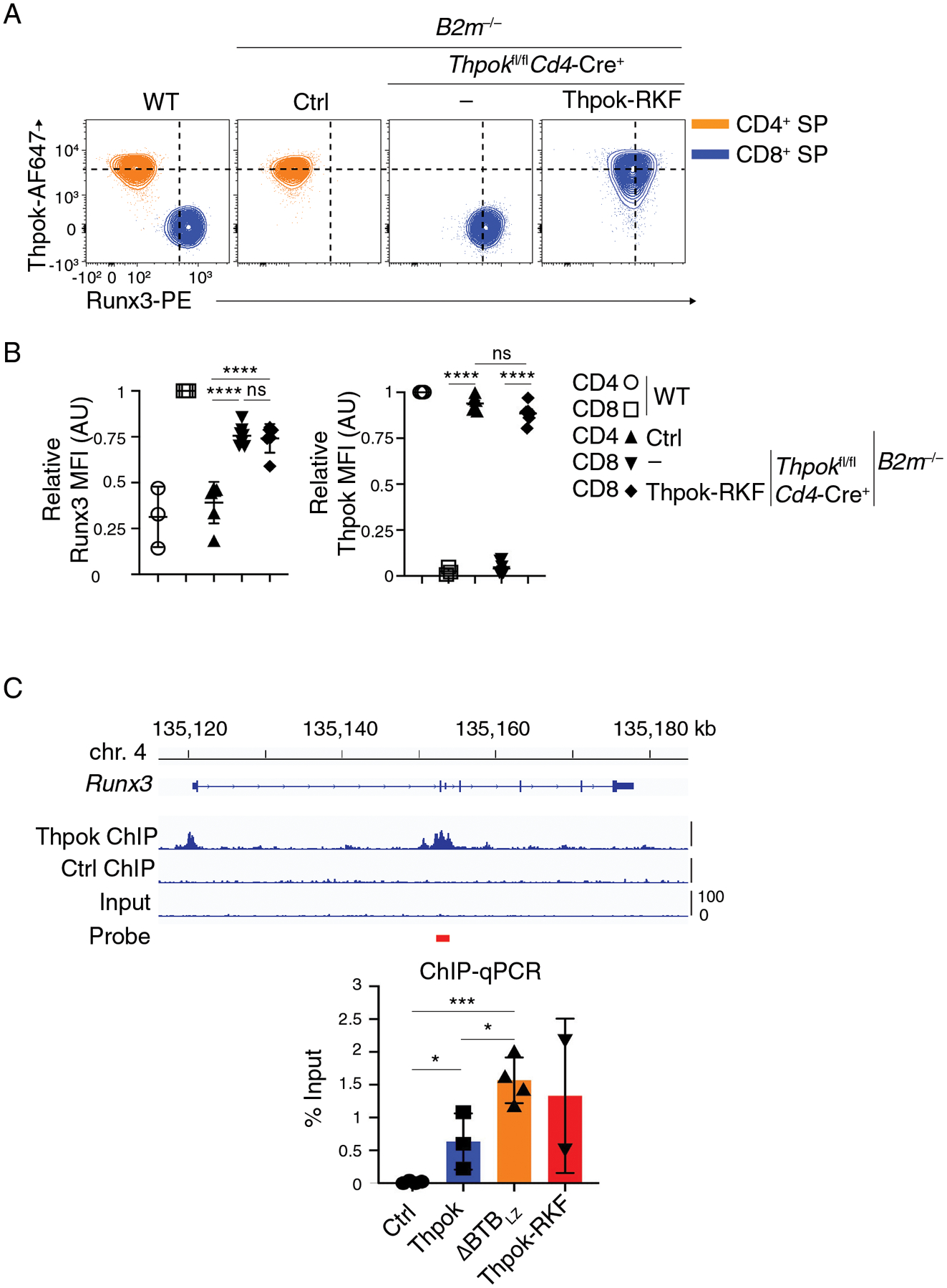
(A) Flow cytometric expression of intra-cellular Runx3 and Thpok in CD44lo CD24lo TCRβhi CD4+ SP (orange traces) or CD8+ SP (blue traces) thymocytes from indicated mice.
(B) (Left panel) Expression of Runx3 in CD44lo CD24lo TCRβhi CD4+ SP or CD8+ SP thymocytes is presented relative to that of CD44lo CD24lo TCRβhi CD8+ SP in WT mice, set as 1 in each experiment. (Right panel) Expression of transgenic Thpok-RKF in CD44lo CD24lo TCRβhi CD4+ SP or CD8+ SP thymocytes is presented relative to that of endogenous Thpok in CD44lo CD24lo TCRβhi CD4+ SP in WT mice, set as 1 in each experiment. One-way ANOVA followed with Tukey multiple comparison tests. ****p<0.0001, ns: p>0.05. Error bars indicate standard deviation.
Data (A, B) are representative of three independent experiments totaling n= 3 (WT), 6 (Cd4-Cre− Thpokfl/fl B2m−/−), 7 (Cd4-Cre+ Thpokfl/fl B2m−/−) or 6 (Cd4-Cre+ Thpokfl/fl B2m−/− Thpok-RKF+) mice.
(C) (Top) Thpok or Ctrl ChIP-seq traces on the Runx3 loci in activated CD4 T cells (42); the red bar schematizes the PCR probe in experiments below. (Bottom) Bar graph quantifies streptavidin-ChIP of activated CD4+ T cells from Thpokfl/fl Ox40-Cre+ Rosa26BirA+ mice that had been retrovirally transduced with empty pMRx (Ctrl) or with pMRx expressing Thpok-Bio, ΔBTBLZ-Bio or Thpok-RKF-Bio. Data shows the amount of PCR-amplified DNA, expressed as percent of input. Each symbol represents a separate determination, and the figure summarizes three distinct experiments. Unpaired t test. *p<0.05, ***p<0.001. Error bars indicate standard deviation.
NuRD binding is needed for of Runx3-independent functions of Thpok
We next examined if additional functions of Thpok, independent of Runx3 repression, required NuRD binding. We expressed Thpok-RKF in Thpokfl/fl Runx3fl/dYFP Cd4-Cre+ thymocytes, which express no endogenous Thpok and little or no Runx3 protein because the Runx3dYFP allele, which evaluates Runx3 expression from YFP fluorescence, is heavily hypomorphic (34) (experiment schematized in Fig. 8A). We reasoned that, if NuRD recruitment was dispensable for functions of Thpok other than Runx3 repression, Thpok-RKF, despite not binding NuRD, would restore the CD4+-lineage differentiation of Thpok-and Runx3-deficient thymocytes. Consistent with previous results (75), mature Thpokfl/fl Runx3fl/dYFP Cd4-Cre+ MHC II-restricted thymocytes were mostly CD8+ SP, with a smaller CD4+ (mostly CD4+ CD8+) component (Fig. 8B). Expression of Thpok-RKF in such cells failed to restore their differentiation into CD4+ SP thymocytes and T cells; instead, the resulting mature thymocytes were mostly CD4+ CD8+ (Fig. 8B–D). We performed RNAseq analysis of these cells and their transgene-negative counterparts (Fig. S8). Principal-component analysis (PCA) found that all Thpok-Runx3 double-deficient thymocyte subsets, regardless of Thpok-RKF expression, were closer to each other than to control CD4+ and CD8+ SP cells processed in parallel (Fig. 8E). Furthermore, Thpok-RKF had little or no effect on expression of CD4+-and CD8+-lineage signature genes identified in these experiments (Fig. 8FG). We conclude from these analyses that Thpok-RKF expression fails to support the CD4+-lineage differentiation of Runx3-deficient thymocytes and that NuRD binding is required for Thpok repression both of Runx3 and of CD8+-lineage genes expressed independently of Runx3. RNAseq also showed expression of Runx1 in Thpokfl/fl Runx3fl/dYFP Cd4-Cre+ thymocytes, regardless of Thpok-RKF expression (Fig. 8G). This was consistent with the idea that expression of CD8+-lineage genes in these Runx3-deficient cells was Runx1-dependent (32, 74).
Figure 8. Runx3-independent function of Thpok-NuRD complexes.
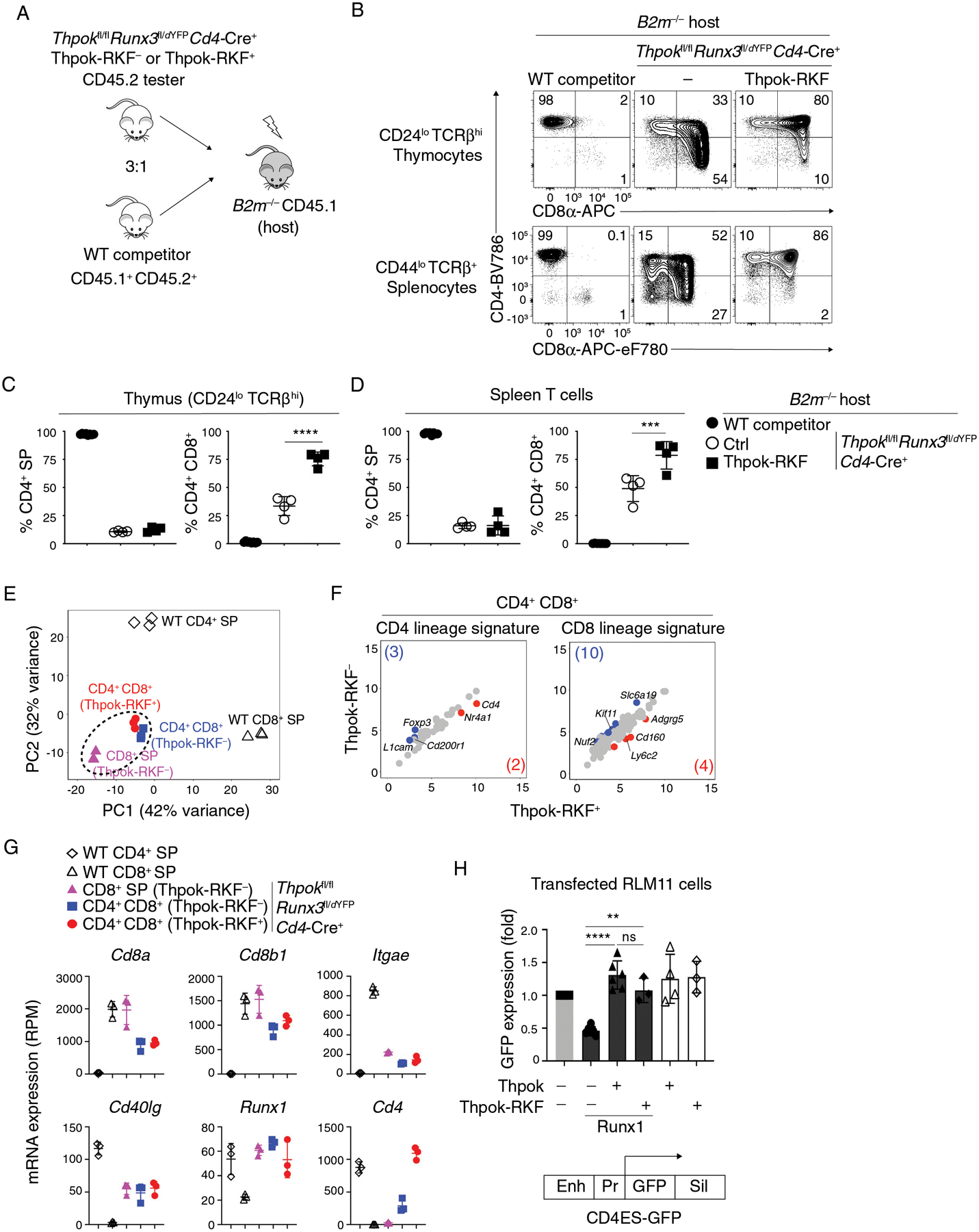
(A) Generation of mixed bone marrow chimeras from CD45.2+ tester (of indicated genotype) and CD45.1+ CD45.2+ (wild-type competitor) cells, analyzed in panels B-G.
(B) Expression of CD4 and CD8 in CD44lo TCRβhi CD24lo thymocytes from B2m−/− mixed chimeras generated as in (A).
(C, D) Percentage of CD4+ SP (left panel) or CD4+ CD8+ (right panel) cells among tester-derived CD44lo TCRβhi CD24lo thymocytes (C) or CD44lo TCRβ+ splenocytes (D).
Data (B-D) are from one set of 4 chimera and are representative of a total of 2 independently generated sets of chimeras with 4–5 mice per group. One-way ANOVA followed with Tukey multiple comparison tests ***p<0.001, ****p<0.0001. Error bars indicate standard deviation.
(E-G) RNA-seq analyses of CD44lo CD24lo CD69lo TCRβhi thymocytes of indicated genotypes and purified as in Fig. S8, that were CD4+ SP, CD8+ SP, or CD4+ CD8+. Data is representative of three separate RNA samples for each population (two biological replicates one of which split into two RNA samples subsequently processed separately).
(E) Principal-component analysis (PCA) displays cell subsets according to the first two components. Each symbol represents an individual RNAseq sample.
(F) Scatter plots comparing CD4 lineage (41 genes) and CD8 lineage signatures (121 genes) (defined as WT CD4+ SP vs WT CD8+ SP, log2 (Fold Change) >2 or <−2, FDR < 0.05, Table S2) in Thpok-RKF− and Thpok-RKF+ CD4+ CD8+ thymocytes. X-axis and y-axis present log2 RPM. Genes with 2-fold or greater differential expression between subsets (and FDR < 0.05) are shown in blue and red (gene numbers are shown in parentheses), respectively.
(G) Graphs display expression (RPM) of indicated genes in each analyzed cell subset (symbols on top). Each symbol represents a distinct sample. Error bars indicate standard deviation.
(H) GFP expression (indicative of Cd4 promoter activity) in RLM-11 cells transfected with (i) a GFP-based reporter plasmid for Cd4 gene expression (schematic at bottom) (ii) a vector expressing Cd8α as an internal control and (iii) expression vectors encoding Runx1, Thpok or Thpok-RKF as indicated. Enh, Pr and Sil indicate the Cd4 proximal enhancer, promoter and silencer, respectively (76, 110). Data are expressed relative to transfection with neither Thpok nor Runx1 vector (leftmost bar) and summarize more than three independent experiments. Each symbol represents a separate transfection. One-way ANOVA followed with Tukey multiple comparison tests. Unpaired t test. **p<0.01, ****p<0.0001, ns, p>0.05. Error bars indicate standard deviation.
In contrast, Thpok-RKF supported Cd4 expression in Thpok- and Runx3-deficient thymocytes (Fig. 8B, G), in line with its effect on CD4 expression in Thpok-deficient but Runx3-sufficient thymocytes (Fig. S6EF). Because the level of Runx1 in Thpok- and Runx3-deficient thymocytes was similar to that in wild-type CD4+ SP thymocytes (Fig. 8G), this suggested that the physiologically relevant Thpok antagonism of Runx1 in CD4+-lineage T cells did not require NuRD binding. To independently verify that Thpok-RKF, like Thpok, antagonizes the repression of Cd4 mediated by Runx1 (35, 76), we performed transient transfections in the RLM-11 thymoma cell line, in which Thpok prevents Runx1-mediated repression of a Cd4-GFP reporter cassette (76) (Fig. 8H). We found that transfected Thpok-RKF was as efficient as Thpok to antagonize Runx1 repression of the Cd4 reporter. Thus, the effect of Thpok on Cd4 expression is at least in part NuRD-independent.
In summary, we demonstrate that NuRD binding is both necessary and sufficient for the function of the Thpok BTB domain, which include repression of Runx3 and of CD8+-lineage genes. These experiments establish that the BTB domain is a critical component of the specificity of action of BTB-POZ zinc finger transcription factors.
Discussion
The present study identified the NuRD complex as an obligatory co-factor for Thpok-mediated repression of CD8+-lineage genes, including Runx3 and Cd8, which both are defining events during CD4+-lineage commitment. Identifying three aminoacid residues within the Thpok BTB domain required for NuRD binding, we show that NuRD is necessary for these functions. Conversely, enforcing NuRD binding makes the BTB domain dispensable for Thpok functions. These findings shed light both on our understanding of transcription factor function and on the mechanisms of CD4+-lineage commitment.
Analyses of transcription factor DNA binding with ever greater resolution, notably with ChIPseq and related technologies, has brought detailed insight into the in vivo distribution of transcription factor molecules onto their target genes. In contrast, there is much less understanding of how DNA-bound transcription factors control expression of their target genes. Such control is generally inferred as involving the binding of co-activators or co-repressors, that are thought to “bridge” transcription factor domains to the general transcription initiation machinery or to chromatin modifying enzymes. However, which co-factors actually mediate the in vivo functions of a given DNA-binding transcription factor is generally unknown.
These issues have special importance for BTB-ZF transcription factors, a family of approximately 50 proteins, many of which are involved in the differentiation or function of immune cells (1, 2, 77). Our current understanding of these factors function has emerged from studies of Bcl6, a factor involved in multiple differentiation processes in immune cells, most prominently in germinal center B and T cells. Bcl6 serves principally, if not exclusively, as transcriptional repressor (8), and such repressive functions are mediated by Bcl6 recruitment of the related co-repressors NCoR or Smrt (encoded by Ncor1 and Ncor2, respectively) to its BTB domain (12–15). Structural studies have found high conformation homology between BTB domains of otherwise distantly related transcription factors, including Bcl6 and Lrf, raising the question of whether such domains all carry similar activities because they recruit similar cofactors.
Similar to Bcl6, Thpok serves at least in part as a transcriptional repressor during CD4+-lineage commitment. However, we show here that this function of Thpok is mediated through recruitment of NuRD rather than NCoR, and that it cannot be fulfilled by the BTB domain of Bcl6, which binds NCoR but not NuRD. Thus, BTB domains are not functionally interchangeable among BTB-ZF transcription factors; rather, they contribute to the functional repertoire of these factors, together with target DNA recognition by the zinc finger domain and expression profile, and with the recruitment of other cofactors by additional protein domains (64, 78–80).
Although LRF was shown to bind NuRD (5, 19), the functional relevance of such interactions has not been investigated. Furthermore, Lrf had also been reported to recruit Smrt (81), although presumably not through interactions similar to those mediating Bcl6-Smrt binding (67, 68). Indeed, neither Thpok nor LRF were part of a set of 14 BTB-ZF factors identified by a recent yeast two-hybrid screen as binding the NuRD components Gatad2a or Gatad2b (18). The relevance of the NuRD-Thpok interaction, as demonstrated by our study, supports the idea that NuRD interactions with LRF and other BTB-ZF factors are important for their function.
NuRD binding implements Thpok-mediated repression on a broad array of CD8+ lineage genes, including Runx3 and CD8+-lineage genes that can be expressed independently of Runx3. The idea that Thpok acts on genes other than Runx3 is in line with our recent demonstration that Thpok binds most genes specifically expressed in CD8+-lineage thymocytes (43). It also fits with observations that Runx3 disruption is not sufficient to restore the CD4+ lineage differentiation of Thpok-deficient thymocytes (75). Because expression of Cd8 genes in Runx3-deficient CD8+ T cells is Runx1-dependent (74, 82), Cd8 repression by NuRD-bound Thpok indicates that NuRD components antagonize, directly or not, Runx1-induced gene transcription. This fits our previous observation that most genomic Thpok binding sites are in close proximity with Runx binding sites (43), as independently reported since (83).
In post-thymic CD4+ T cells, Thpok is essential to maintain the integrity of resting CD4+ T cells (which express Runx1), for the differentiation of Th2 CD4+ T cells, and for the functional fitness and memory differentiation of CD4+ T cells mounting Th1 responses to intra-cellular pathogens (41, 42, 51). We previously showed that these functions of Thpok were mediated in part or in totality by its inhibition of Runx3 expression or by antagonizing the function of Runx proteins (41, 42, 51), which are normally expressed in Th1 (Runx3) and Th2 (Runx1) effector cells. Thus, the identification of NuRD as required for Thpok- mediated Runx antagonism uncovers a mechanism essential for both CD4+ T cell development and responses. Reciprocally, Thpok is needed for T cell-induced inflammation in experimental models of colitis (84). Thus, because interactions between Bcl6 and NCoR are accessible to pharmacological intervention (85), our study suggests that similar approaches on the Thpok-NuRD interaction could be leveraged for treatment of inflammatory disease.
Previous studies had found that Thpok prevents Runx1- or Runx3-mediated repression of Cd4 (35, 76). We found here that, unlike for repression of CD8+-lineage genes, the effect of Thpok on Cd4 expression does not require NuRD. This observation contrasts with the need for Mi2β, a key component of NuRD, for Cd4 expression in DP thymocytes (86). It is possible that this function of Mi2β is specific to DP thymocytes; supporting this idea, Cd4 expression is controlled by distinct enhancers in DP and CD4 SP thymocytes (87–89). Alternatively, mature CD4+ SP thymocytes express the Mi2β-related Mi2α protein, unlike DP thymocytes which have minimal Mi2α expression (86); thus it is possible that expression of Cd4 is Mi2-dependent in all CD4+ thymocytes and T cells. Such Mi2 support of Cd4 expression would be independent of Thpok-Mi2 interactions, consistent with the fact that DP thymocytes do not express Thpok.
Which NuRD component(s) mediates Thpok functions remains to be determined. Because all canonical members of the NuRD complex (Mi2 helicases, HDAC, and proteins of the Mta, RbAp, and Gatad2 families) are the product of multigene families, the potential for functional redundancy has hampered analyses of their functions in T cell development and responses. However, two lines of evidence support the idea that histone deacetylase activity helps enforce Thpok-mediated repression of CD8+-lineage genes. First, histone H3 and H4 acetylation at the Cd8 locus was reported to be dependent on Thpok (39). Second, analyses of T cells carrying mutations in genes encoding HDAC1 and HDAC2, both NuRD components, suggest that these molecules serve to repress Runx3, Cd8, and cytotoxic genes in post-thymic CD4+ T cells, a function strikingly similar to that of Thpok (41, 90). Further suggesting that these enzymes mediate Thpok gene repression, HDAC1 and 2-deficient CD4+ T cells expressed CD8+-lineage genes despite conserved Thpok protein expression (90). While deletion of HDAC1 and 2 only had modest effects on Cd8 gene repression in thymocytes, this has been related to the extended half-life of these enzymes in non-dividing DP and SP thymocytes (90). Of note, our LC-MS analysis found no evidence for in vivo Thpok binding to class II HDACs, contrary to previous studies in transfected cell lines (39). This fits with the facts that NuRD is not associated with class II HDACs, and that enforcing Thpok-NuRD association bypasses the need for the Thpok BTB domain during CD4+ T cell development.
The observations that Thpok limits chromatin accessibility at CD8+-lineage genes and that NuRD binding is required for Thpok functions raise the possibility that Thpok serves by physically recruiting NuRD components (notably histone deacetylases, which promote chromatin compaction) to target genes. However, experimental evidence challenges such a simple idea. Previous ChIP analyses (90) have found no difference between CD4+ and CD8+ T cells for binding of HDAC1 and HDAC2 at Cd8, Runx3, and Eomes, even though Thpok is expressed and represses these genes in CD4+ T cell only. In line with these observations, ChIPseq analyses (55, 91) show Mta2 and Mi2β binding to the Cd8 locus in pre-DP and DP thymocytes, which do not express Thpok. Conversely, analyses from ChIPseq datasets (37, 91–95) show that CD8+-lineage-specific genes, e.g. Runx3 and Cd8, bind other NuRD-binding transcription factors, including Ikaros and Bcl11b (96, 97). Altogether, these findings indicate that NuRD components can interact with Thpok target genes independently of Thpok, presumably through other transcription factors.
We do not conclude that all functions of Thpok are mediated by NuRD binding, because our mass spectrometry analyses detected proteins that bind versions of Thpok unable to bind NuRD, and because our study did not assess Thpok functions in post-thymic cells (30, 41, 42, 70, 98). Throughout this study, we have used mitigation strategies to overcome generic technical limitations and potential confounders. For biochemical analyses, we verified key findings in primary cells to avoid relying on non-physiological expression conditions in transfected cells; we also considered the potential impact of detergent solubilization on the detection of relevant protein-protein interactions, notably in mass-spectrometry experiments. For genetic analyses, we have attempted to eliminate founder-specific effects in transgenic mouse lines by verifying several founder-derived lines for each transgenic strain we generated, and effects of premature retroviral gene expression using a stage-specific expression strategy. For scATACseq, current technical approaches cannot detect all open chromatin regions in every cell; furthermore, we established correspondences between scATACseq and scRNAseq clusters using a statistical computational treatment of separate scATACseq and scRNAseq data, rather than a combined scRNAseq-scATACseq measurement at the single-cell level.
In summary, we found that Thpok functions in CD4+ T cell differentiation require binding of NuRD to its BTB domain, and that, apart from dimerization, NuRD recruitment recapitulates the functional need for the BTB domain. NuRD recruitment is essential for Thpok to repress Runx3 and other CD8+-lineage genes, but not to support expression of Cd4. Our study also demonstrates the critical contribution that cofactor recruitment makes to the functional specificity of BTB-zinc finger transcription factors.
Methods
Study design
This study aimed at understanding the mechanisms of Thpok function in intrathymic CD4+ T cell differentiation. Our general approach was to (i) search for Thpok interacting proteins using Mass Spectrometry, (ii) generate Thpok mutants to identify the domains or amino-acid residues of Thpok responsible for such binding and (iii) use genetic approaches (generation of transgenic and retrogenic mice) to express such mutants to assess the physiological role of these domains in CD4+ T cell development. We generated both loss- (e.g. Thpok-RKF) and gain-of-function (e.g. Mta2-ΔBTBLZ) to verify the relevance of such interactions. We assessed the physiological relevance of all conclusions by evaluating CD4+ T cell development in in vivo genetic models. All experiments were performed at least twice (biological replicates), as indicated in figure legends. We did not used power analysis; sample sizes for experiments were based on previous experience from this and other laboratories. Because transgenic or retrogenic mice had to be identified before processing for experiments, we used neither blinding nor randomization.
Animals
Mice carrying Thpokfl (36), Ox40-Cre (49, 50), ThpokBio (42), Runx3dYFP (34), Rosa26BirA (99) alleles, or the Thpok transgene (21) have been described. B2m−/− (100) and Cd4-Cre mice (101) were from Taconic, H2-Ab1−/− (102) mice from JAX and CD45.1 and CD45.2 C57BL/6 mice from Charles River. All transgenic mice were heterozygous for the transgene they express. The Ox40-Cre allele was maintained heterozygous and only female Ox40-Cre+ mice were used for breeding. Except where specified otherwise, control mice included in experimental designs were Cre-negative animals from the same lines as experimental mice. Mice were housed and bred in specific pathogen-free facilities and analyzed between 6 and 20 weeks of age. Given the limited number of animals carrying relevant allelic combinations, mice of both sexes were used in experiments according to availability. Donor and recipient mice for bone marrow chimeras were sex matched. Animal procedures were approved by the NCI Animal Care and Use Committee. ΔBTBLZ or Thpok-RKF transgene was genotyped by following primers:
ΔBTBLZ-forward: 5’- CAGAGCTGCGCAAGGAAGTG-3’;
ΔBTBLZ-reverse: 5’- CCTTTGGTTTGAAGAAAAGCTTTCC-3’;
Thpok-RKF-forward: 5’-CGCTCTTGCTCTCTGTGTATG-3’;
Thpok-RKF-reverse: 5’- CCCACTGCCTTGTAGGAACTTCCT -3’.
Plasmid construction
DNA cloning was performed using restriction enzyme-ligase technology in E.coli DH5α strain. Single colonies were grown in appropriate volumes of LB Broth (1X) (Gibco) containing 100 μg/ml Ampicillin at 37 °C with shaking at 200~250 rpm overnight. Plasmid DNA was extracted with Qiagen QIApre Spin Miniprep or Midi kits. Thpok-HA, Thpok-Flag and Thpok deletion constructs were amplified from a Thpok cDNA (21) and were inserted between the EcoRI and XbaI sites in pcDNA3 vector (Invitrogen); we used empty pcDNA3 as control in all transient transfection experiments. ΔBTBLZ was amplified using the synthesized leucine zipper sequence (5’-CTC GAG ATT CGG GCA GCC TTC CTT GAG AAA GAG AAT ACG GCC CTG AGG ACG GAG GTT GCA GAG CTG CGC AAG GAA GTG GGG-3’) in the primer. Thpok-RKF and Thpok-SVIN were obtained by doing mutagenesis in the wild-type Thpok cDNA according to the manufacture’s protocol (QuickChangeII site-directed Mutagenesis kit from Agilent Technologies). pMGfIT vector was constructed from pMGfI4 (65), by replacing hCD4 with Thy1.1 between SnaBI and ClaI sites. cDNAs encoding Thpok and mutant thereof were inserted between EcoRI and MluI sites in pMGfIT vector. Thpok, Thpok-Bio, ΔBTBLZ-Bio or Thpok-RKF-Bio were amplified (42, 51) and were inserted between the EcoRI and NotI sites in pMRx vector (103) expressing mouse Thy1.1 as a reporter gene. For transgenic mice, bio-tag appended ΔBTBLZ or Thpok-RKF cDNAs were inserted between EcoRI and SalI sites of p29Δ2S− vector (104), respectively. Transgenic mice were obtained as described (105) and were identified by Southern hybridization of DNA obtained from tail tissue. The CD4ES GFP reporters, CMV promoter-driven expression vectors for Runx1 and CD8α were previously described (73, 76). All gene segments were amplified using conventional PCR techniques with high-fidelity PrimeSTAR HS DNA Polymerase (Takara) and verified by sequencing.
Cell culture and transfection
HEK293T cells were cultured in DMEM containing 10% heat-inactivated FBS, 1% penicillin/streptomycin/glutamine (Gibco, 10378) and 10 mM HEPES buffer (Corning). PlatE cells (106) used for retrovirus packaging were cultured in DMEM containing 10% heat-inactivated FBS, 1% penicillin/ streptomycin /glutamine supplemented (Gibco, 10378) and 10 mM HEPES buffer (Corning) with 1 μg/ml puromycin (Sigma) and 10 μg/ml blasticidine (Thermo Fisher). HEK293T and PlatE cells were transfected using Lipofectamine 2000 (Thermo Fisher) according to the manufacturer’s instruction. RLM-11 thymic lymphoma cells (76) were cultured in RPMI 1640 medium (Gibco) containing 10% heat-inactivated FBS, 1% penicillin /streptomycin /glutamine (Gibco, 10378) and 10 mM HEPES (Corning), and were transfected with the Nucleofector Kit L, program C-009 (Nucleofector II, Amaxa Biosystem). Cells were analyzed 24 hours post nucleofection by flow cytometry.
Immunofluorescence Microscopy
HEK293T cells were plated onto poly-L-Lysine-coated coverslips and then transfected with HA-tagged Thpok or Thpok mutants. 48 hours post transfection, cells were fixed with 2% paraformaldehyde for 30 min at room temperature. After three washes with PBS, the coverslips were incubated in blocking buffer (PBS, 3% BSA, 0.5% Triton X-100, 10% newborn calf serum) at room temperature for 1 hour followed by three washes with binding buffer (PBS, 3% BSA, 0.5% Triton X-100). Samples then were incubated with 2 μg/ml HA-probe (F-7, Santa Cruz Biotechnology) at room temperature for 1 hour. After three washes with binding buffer, samples were incubated with 10 μg/ml goat anti-mouse Alexa Fluor 555 (Thermo Fisher) at room temperature for 1 hour. Nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI) (Life Technologies). Samples were mounted on microscope slides using Mowiol (Sigma) and examined on Axioplan 2 imaging.
In vitro T cell activation and transduction
CD4+ T cells were purified from Thpokfl/fl Ox40-Cre+ or Thpokfl/fl Ox40-Cre+ Rosa26BirA+ spleen, with the mouse CD4-negative isolation kit (Dynal, Invitrogen) according to the manufacture’s protocol. Isolated CD4+ T cells were activated with anti-CD3 (1 μg/ml) and anti-CD28 (3 μg/ml) antibodies (BioXcell) and irradiated T-depleted (Pan T Dynal kit, Invitrogen) antigen-presenting cells (APCs, irradiated at 2500 rad) (36), supplied with IL12 (10 ng/ml) and anti-IL4 (10 μg/ml) (Peprotech). Activated CD4 cells were spinfected with retroviral supernatants 600 g, at 25 °C for 90 min 24 h after activation, resuspended and placed back at 37 °C in their original activation media for three days, after which the culture media was supplemented with 100 U/ml IL2 for 24 hours before harvesting for analysis 4 days after activation.
Streptavidin pull down, immunoprecipitation and immunoblot analysis
Retrovirally transduced CD4+ T cells or transfected 293T cells were collected, washed twice in ice-cold PBS, lysed by incubating 30 min in ice-cold RIPA buffer (50 mM Tris-HCl (pH 7.5), 1% NP-40, 150 mM NaCl, 1 mM EDTA and 10% Glycerol) supplemented with protease inhibitor cocktail (Roche). For the samples treated with Ethidium Bromide, 10 μg/ml Ethidium Bromide (Invitrogen) was added to the RIPA buffer at the beginning of cell lysis (52). Cell lysates were cleared by centrifuging at 12,000 rpm, 4 °C for 10 min. For immunoprecipitation, 1 μg antibody was incubated with 20 μl Protein A or Protein G Dynabeads (Thermo Fisher) in 500 μl ice-cold PBS, and rotated at 4 °C for 1 hour. Antibody-Protein A Dynabeads, antibody-Protein G Dynabeads (immunoprecipitation) or M-280 Streptavidin Dynabeads (Thermo Fisher, Streptavidin pull down) were washed once with PBS, then equilibrated with RIPA buffer. Equilibrated Dynabeads were incubated with cleared cell lysis and rotated at 4°C for 1–2 hours. After 4 washes with RIPA buffer, bead-bound immunocomplexes were suspended in loading buffer and boiled for 10 min at 100 °C. Proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) with Chameleon molecular weight markers (LI-COR Biosciences), and examined by Western blotting by standard procedures. Anti-actin or anti-tubulin used as the internal loading controls. Images were visualized using a ChemiDoc Touch Imaging system (Bio-Rad). The following antibodies were used for Immunoprecipitation and Western Blotting: anti-Flag antibody (clone M2, Sigma-Aldrich), anti-HA tag (Clone F-7, Santa Cruz), anti-CHD4/mi2 beta antibody (Bethyl Laboratories), NuRD Complex Antibody sampler Kit (Cell Signaling Technology), anti-MTA2 antibody (Abcam), anti-MTA1 Rabbit mAb (Clone D40D1, Cell Signaling Technology), anti-HDAC1 Mouse mAb (Clone 10E2, Cell Signaling Technology), anti-HDAC1 antibody (Rabbit polyclonal, Abcam), anti-HDAC2 Mouse mAb (Clone 3F3, Cell Signaling Technology), anti-RBAP46 antibody (Clone V415, Cell Signaling Technology), anti-α-Tubulin antibody mAb (Sigma-Aldrich), anti-β-Actin antibody mAb (Sigma-Aldrich), Alexa Fluor 680 AffiniPure Goat anti-mouse IgG (light chain specific, Jackson Immuno Research Laboratories), IRDye 800CW Goat anti-Rabbit IgG (H+L) (LI-COR Biosciences), Normal Rabbit IgG (EMD Millipore).
Mass-spectrometric analysis
Interacting proteins were fractionated by SDS-PAGE and each lane cut into 10 slices. The protein bands were then in-gel digested with trypsin (Thermo Fisher) overnight at 37 °C, as described (107). The peptides were extracted following cleavage and lyophilized, then solubilized for mass spectrometry analysis. They were trapped on a trapping column and separated on a 75 μm ×15 cm, 2 μm Acclaim PepMap reverse phase column (Thermo Fisher) using an UltiMate 3000 RSLCnano HPLC (Thermo Fisher) followed by online analysis by tandem mass spectrometry using a Thermo Orbitrap Fusion mass spectrometer. Parent full-scan mass spectra were collected in the Orbitrap mass analyzer set to acquire data at 120,000 FWHM resolution; ions were then isolated in the quadrupole mass filter, fragmented within the Higher-energy C-trap dissociation cell (HCD normalized energy 32%, stepped ± 3%), and the production analyzed in the ion trap. Proteome Discoverer 2.0 (Thermo Fisher) was used to search the data against murine proteins from the UniProt database using SequestHT (Thermo Fisher). The search was limited to tryptic peptides, with maximally two missed cleavages allowed. Cysteine carbamidomethylation was set as a fixed modification, and methionine oxidation set as a variable modification. The precursor mass tolerance was 10 ppm, and the fragment mass tolerance was 0.6 Da. The Percolator node was used to score and rank peptide matches using a 1% false discovery rate. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the MassIVE partner repository with the dataset identifier PXD028661.
Cell preparation, staining and sorting
Thymocytes and splenocytes were prepared and stained as described (42, 43, 51). Cell were stained with following antibodies and reagents: anti-B220-PerCpCy5.5 (Clone RA3–6B2, eBioscience), anti-CD4-APC (Clone GK1.5, eBioscience), anti-CD4 PerCpCy5.5 (CloneRM4–5, eBioscience), anti-CD4-eFluor 450 (Clone GK1.5, eBioscience), anti-CD4-BV786 (Clone RM4–4, BD Pharmingen), anti-CD4-PE (Clone GK1.5, BD Pharmingen), anti-CD8α-PE (Clone 53–6.7, eBioscience), anti-CD8α-APC (Clone 53–6.7, eBioscience), anti-CD8α-APC-eF780 (Clone 53–6.7, eBioscience), anti-CD8α-eFluor 450 (Clone 53–6.7, eBioscience), anti-CD8α-BUV395 (Clone 53–6.7, BD Pharmingen), anti-TCR beta-PE (Clone 57–597, eBioscience), anti-TCR beta-FITC (Clone 57–597, BD Pharmingen), anti-TCR beta-BV711 (Clone 57–597, BD Pharmingen), anti-TCR beta-BV650 (Clone 57–597, BD Pharmingen), anti-CD24-APC-eFlour 780 (Clone M1/69, eBioscience), anti-CD69-PE (Clone H1.2F3, BD Pharmingen), anti-CD69-BV711(Clone H1.2F3, BD Pharmingen), anti-CD44-Alexa Fluor 700 (Clone IM7, eBioscience), anti-CD45.1-PE-Cy7 (Clone A20, eBioscience), anti-CD45.1-BV650 (Clone A20, BD Pharmingen), anti-CD45.2-PerCpCy5.5 (Clone 104, BD Pharmingen), anti-CD45.2-BV786 (Clone 104, BD Pharmingen), anti-Thy1.1-APC (Clone HIS51, eBioscience), anti-Thy1.1-BUV737 (Clone ox-7, BD Pharmingen), anti-Thpok-Alexa Fluor 647 (Clone T43–94, BD Pharmingen), anti-Runx3-PE (Clone R3–5G4, BD Pharmingen), Streptavidin-Allophycocyanin (BD Pharmingen). Flow cytometry data were acquired on LSRFortessa, LSRFortessa X-20 or Symphony A5 (BD Biosciences) and analyzed with FlowJo (TreeStar) software. Dead cells were excluded by staining with 4’,6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI) (Life Technologies) (for live cell staining) or fixable blue fluorescence reactive dye (Invitrogen) (for intracellular staining). Purification of lymphocytes by cell sorting was done on a FACSAria II, a FACSViolet, or a FACSFusion (BD Biosciences). For sorting, cells were not stained with viability dye, but were gated on live cells determined by size.
ChIP-qPCR analysis
Splenic CD4+ T cells from Thpokfl/fl Ox40-Cre+ Rosa26BirA+ were activated and transduced as above with retrovirus expressing Bio-tagged wild-type Thpok or Thpok derivatives. Transduced cells were sorted on the basis of retrovirally encoded Thy1.1 expression, and ChIP was performed as described (42, 51). qPCR was conducted using PowerUp SYBR Green Master Mix (Thermo Fisher). The binding enrichments were shown as the percentages of ChIP input.
The primers used for qPCR quantification are as follows:
Cd4 silencer-forward: 5’-TGTAGGCACCCGAGGCAAAG-3’;
Cd4 silencer-reverse: 5’-GTTCCAGCACAGCAGCCCCA-3’.
Thpok silencer-forward: 5′-TGGTTTCGAGACTGGCTGGT-3′(40);
Thpok silencer-reverse: 5’-GACCGAGGAGCTGCTTTCAG-3’(40).
Pax5-forward: 5’-AGAACCTGTCCACCTTTCCTTC-3’(51);
Pax5-reverse: 5’-ATGTTCTCTGACCTCTGCAATG-3’(51).
Runx3-forward: 5’-CTTGGGGAGCACCTAGAAGTAG-3’;
Runx3-reverse: 5’- GTCTGGGCTCTTATTGCGGATCG-3’.
Mixed Bone Marrow Chimeras
Mixed bone marrow chimera was performed as described (42, 51). T cell-depleted (Pan T Dynal kit, Invitrogen) bone marrow cells were prepared from CD45 disparate donor mice (6–10 week old), mixed together at 3:1 ratio, and injected into lethally irradiated (950 rads) 8-week old CD45.1 recipients. Chimeric animals were analyzed 7–8 weeks post transplantation.
Generation of retrogenic mice
Bone marrow cells were harvested from Thpokfl/fl Cd4-Cre+ CD45.2+ donor mice 4 days after i.v. injection with 200 μl 5-FU (5-Fluorouracil, 25 mg/ml, Fresenius Kabi), washed with flush medium (DMEM with 2% FCS, 1% penicillin /streptomycin /glutamine and 10 mM HEPES buffer). Cells were cultured for 24 hours with Stem pro-34 SFM (Gibco, Thermo Fisher Scientific) supplemented with 1% penicillin /streptomycin /glutamine (Gibco, 10378) and cytokines cocktail (6 ng/ ml IL3, 10 ng/ml IL6, 100 ng/ ml SCF and 10 ng/ ml Flt3L) (Peprotech) before transduction with pMGfIT derivatives. Transduction was performed by spinning at 2500 rpm, 32 °C for 90 min with retroviral supernatants in the presence of cytokines cocktail (6 ng/ ml IL3, 10 ng/ml IL6, 100 ng/ ml SCF, 10 ng/ ml Flt3L) (Peprotech) and 4 μg/ml polybrene (Millipore). Transduction was repeated following the same procedure after a 24-hour culture. Cells were rested at 37 °C for 2–3 hours after the second transduction, collected, washed twice with ice-cold PBS, resuspended in ice-cold PBS, and i.v. injected into lethally irradiated (950 rads) CD45.1 recipients.
RNAseq
RNA from thymocytes sorted from indicated mice was isolated using the RNeasy Plus Micro kit (Qiagen). Quality control was performed by bioanalyzer (Agilent), and library was prepared by SMARTer Ultra Low Input reagent (Takara) and Nextera XT DNA (Illumina) library preparation kits. Libraries were sequenced with paired-end reads of 74 bp on a Nextseq sequencer (Fig. 6) or 100 bp on a Novaseq SP (Fig. 8) sequencer to reach 50 million read pairs per sample. Raw RNAseq fastq reads were aligned to mouse genome (mm10, Fig. 6; mm39, Fig. 8) using STAR. Gene-assignment and count of RNA reads were performed with HTseq. Further analyses were performed with R software and differentially expressed genes were identified using DEseq2. Expression of NuRD components in thymocytes (43) and LCMV-responding T cells (42) was determined from publicly available data reported in these studies (Gene Expression Omnibus references GSE148973 and GSE130474).
scATACseq
Thymocytes were sorted as indicated in Fig. S2 and processed for scATACseq as described, using the 10x Genomics chromium single Cell ATAC Solution (v1.0) according to the manufacturer’s instructions (43). Sequencing files were processed, and count matrixes generated using Cell Ranger ATACseq (v1.2.0). Replicate data integration, integration with scRNAseq data (transcriptome projection, Fig. 2) and subsequent analyses were performed in R using the Seurat (v4.0.4) and Signac (v1.4.0) packages (59, 108) as described (43). Peak traces were generated with the ConveragePlot function.
Statistical analyses
All statistical analyses were done with Prism software. Error bars in graphs indicate average ± standard deviation. Comparisons were done by one-way ANOVA followed with Tukey multiple comparison tests or two-way ANOVA.
Supplementary Material
Supplementary data file 1. Raw data file (Excel spreadsheet).
Supplementary data file 2. Unprocessed blots
Table S1. Peptides identified by LC-MS.
Table S2. Frequency of scATACseq clusters shown in Fig. S2E.
Table S3. CD4+ and CD8+ lineage signatures defined as in Fig. 8F.
Figure S1. NuRD-Thpok binding.
Figure S2. scATACseq analysis of Thpok impact on chromatin accessibility.
Figure S3. ΔBTBLZ properties and transgenic mouse lines.
Figure S4. Flow cytometric quantification of Thpok mutant expression.
Figure S5. B-Thpok dimerization and retrogenic expression.
Figure S6. Thpok-RKF protein dimerization and function.
Figure S7. Analyses of Mta2-ΔBTBLZ retrogenic mice.
Figure S8. Cell purification strategy for RNAseq of mixed bone marrow chimera.
Acknowledgments
We thank C. Li, S. Banerjee, S. Siddiqui, F. Livak and the CCR Flow Cytometry Core for expert cell sorting, J. Shetty, Y. Zhao and B. Tran for high-throughput DNA sequencing, P. Awasthi and the CCR Mouse Modeling & Cryopreservation Core for transgenic mouse generation, L. Conner, M. Wong and the CCR Genomics Core for DNA sequencing, M. Kelly and the CCR Single Cell Analysis Facility for technical guidance with scATACseq, T. Chen for assistance with RNA-seq analysis, and A. Bhandoola and J. Zhu for critical reading of the manuscript. High-throughput sequencing was performed by the CCR Genomics Core and the Frederick National Laboratory for Cancer Research sequencing facility. Sequence data processing analyses was performed on the NIH High performance Biowulf computing cluster.
Funding
This work was supported by the Intramural Research Program of the National Cancer Institute, Center for Cancer Research (CCR), National Institutes of Health and by a National Science Foundation (NSF) fellowship to L.B.C.
Footnotes
Competing Interests Statement
The authors have no competing interests.
Data and materials availability
All sequence data reported in this paper are publicly available on the NCBI Gene Expression Omnibus (GEO), from accession numbers GSE199739, GSE171610 and GSE185217. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References
- 1.Cheng ZY, He TT, Gao XM, Zhao Y, Wang J, ZBTB Transcription Factors: Key Regulators of the Development, Differentiation and Effector Function of T Cells. Front Immunol 12, 713294 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ellmeier W, Taniuchi I, The role of BTB-zinc finger transcription factors during T cell development and in the regulation of T cell-mediated immunity. Current topics in microbiology and immunology 381, 21–49 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, Bendelac A, The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity 29, 391–403 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, Chua K, Eidson M, Kim HJ, Im JS, Pandolfi PP, Sant’Angelo DB, The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nature immunology 9, 1055–1064 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masuda T, Wang X, Maeda M, Canver MC, Sher F, Funnell AP, Fisher C, Suciu M, Martyn GE, Norton LJ, Zhu C, Kurita R, Nakamura Y, Xu J, Higgs DR, Crossley M, Bauer DE, Orkin SH, Kharchenko PV, Maeda T, Transcription factors LRF and BCL11A independently repress expression of fetal hemoglobin. Science 351, 285–289 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crotty S, T Follicular Helper Cell Biology: A Decade of Discovery and Diseases. Immunity 50, 1132–1148 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basso K, Dalla-Favera R, BCL6: master regulator of the germinal center reaction and key oncogene in B cell lymphomagenesis. Adv Immunol 105, 193–210 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Choi J, Diao H, Faliti CE, Truong J, Rossi M, Bélanger S, Yu B, Goldrath AW, Pipkin ME, Crotty S, Bcl-6 is the nexus transcription factor of T follicular helper cells via repressor-of-repressor circuits. Nature immunology 21, 777–789 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Müller L, Hainberger D, Stolz V, Ellmeier W, NCOR1-a new player on the field of T cell development. J Leukoc Biol 104, 1061–1068 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Turki-Judeh W, Courey AJ, Groucho: a corepressor with instructive roles in development. Curr Top Dev Biol 98, 65–96 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Denslow SA, Wade PA, The human Mi-2/NuRD complex and gene regulation. Oncogene 26, 5433–5438 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Ahmad KF, Melnick A, Lax S, Bouchard D, Liu J, Kiang CL, Mayer S, Takahashi S, Licht JD, Prive GG, Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain. Molecular cell 12, 1551–1564 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Melnick A, Carlile G, Ahmad KF, Kiang CL, Corcoran C, Bardwell V, Prive GG, Licht JD, Critical residues within the BTB domain of PLZF and Bcl-6 modulate interaction with corepressors. Mol Cell Biol 22, 1804–1818 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhordain P, Albagli O, Lin RJ, Ansieau S, Quief S, Leutz A, Kerckaert JP, Evans RM, Leprince D, Corepressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL6 oncoprotein. Proc Natl Acad Sci U S A 94, 10762–10767 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huynh KD, Bardwell VJ, The BCL-6 POZ domain and other POZ domains interact with the co-repressors N-CoR and SMRT. Oncogene 17, 2473–2484 (1998). [DOI] [PubMed] [Google Scholar]
- 16.Melnick A, Ahmad KF, Arai S, Polinger A, Ball H, Borden KL, Carlile GW, Prive GG, Licht JD, In-depth mutational analysis of the promyelocytic leukemia zinc finger BTB/POZ domain reveals motifs and residues required for biological and transcriptional functions. Mol Cell Biol 20, 6550–6567 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Prive GG, Sequence and structural analysis of BTB domain proteins. Genome Biol 6, R82 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olivieri D, Paramanathan S, Bardet AF, Hess D, Smallwood SA, Elling U, Betschinger J, The BTB-domain transcription factor ZBTB2 recruits chromatin remodelers and a histone chaperone during the exit from pluripotency. J Biol Chem 297, 100947 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi WI, Jeon BN, Yoon JH, Koh DI, Kim MH, Yu MY, Lee KM, Kim Y, Kim K, Hur SS, Lee CE, Kim KS, Hur MW, The proto-oncoprotein FBI-1 interacts with MBD3 to recruit the Mi-2/NuRD-HDAC complex and BCoR and to silence p21WAF/CDKN1A by DNA methylation. Nucleic Acids Res 41, 6403–6420 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathew R, Seiler MP, Scanlon ST, Mao AP, Constantinides MG, Bertozzi-Villa C, Singer JD, Bendelac A, BTB-ZF factors recruit the E3 ligase cullin 3 to regulate lymphoid effector programs. Nature 491, 618–621 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun G, Liu X, Mercado P, Jenkinson SR, Kypriotou M, Feigenbaum L, Galera P, Bosselut R, The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nature immunology 6, 373–381 (2005). [DOI] [PubMed] [Google Scholar]
- 22.He X, He X, Dave VP, Zhang Y, Hua X, Nicolas E, Xu W, Roe BA, Kappes DJ, The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature 433, 826–833 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Galéra P, Musso M, Ducy P, Karsenty G, c-Krox, a transcriptional regulator of type I collagen gene expression, is preferentially expressed in skin. Proc Natl Acad Sci U S A 91, 9372–9376 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S, Mi L, Yu L, Yu Q, Liu T, Wang GX, Zhao XY, Wu J, Lin JD, Zbtb7b engages the long noncoding RNA Blnc1 to drive brown and beige fat development and thermogenesis. Proc Natl Acad Sci U S A 114, E7111–e7120 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang R, Ma H, Gao Y, Wu Y, Qiao Y, Geng A, Cai C, Han Y, Zeng YA, Liu X, Ge G, Th-POK regulates mammary gland lactation through mTOR-SREBP pathway. PLoS Genet 14, e1007211 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zullo JM, Demarco IA, Piqué-Regi R, Gaffney DJ, Epstein CB, Spooner CJ, Luperchio TR, Bernstein BE, Pritchard JK, Reddy KL, Singh H, DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell 149, 1474–1487 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Lee HO, He X, Mookerjee-Basu J, Zhongping D, Hua X, Nicolas E, Sulis ML, Ferrando AA, Testa JR, Kappes DJ, Disregulated expression of the transcription factor ThPOK during T-cell development leads to high incidence of T-cell lymphomas. Proc Natl Acad Sci U S A 112, 7773–7778 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lunardi A, Guarnerio J, Wang G, Maeda T, Pandolfi PP, Role of LRF/Pokemon in lineage fate decisions. Blood 121, 2845–2853 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taniuchi I, CD4 Helper and CD8 Cytotoxic T Cell Differentiation. Annual review of immunology 36, 579–601 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Vacchio MS, Bosselut R, What Happens in the Thymus Does Not Stay in the Thymus: How T Cells Recycle the CD4+-CD8+ Lineage Commitment Transcriptional Circuitry To Control Their Function. Journal of immunology 196, 4848–4856 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carpenter AC, Bosselut R, Decision checkpoints in the thymus. Nature immunology 11, 666–673 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woolf E, Xiao C, Fainaru O, Lotem J, Rosen D, Negreanu V, Bernstein Y, Goldenberg D, Brenner O, Berke G, Levanon D, Groner Y, Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc Natl Acad Sci U S A 100, 7731–7736 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, Ito Y, Littman DR, Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 111, 621–633 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Egawa T, Littman DR, ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nature immunology 9, 1131–1139 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muroi S, Naoe Y, Miyamoto C, Akiyama K, Ikawa T, Masuda K, Kawamoto H, Taniuchi I, Cascading suppression of transcriptional silencers by ThPOK seals helper T cell fate. Nature immunology 9, 1113–1121 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Wang L, Wildt KF, Castro E, Xiong Y, Feigenbaum L, Tessarollo L, Bosselut R, The zinc finger transcription factor Zbtb7b represses CD8-lineage gene expression in peripheral CD4+ T cells. Immunity 29, 876–887 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kojo S, Tanaka H, Endo TA, Muroi S, Liu Y, Seo W, Tenno M, Kakugawa K, Naoe Y, Nair K, Moro K, Katsuragi Y, Kanai A, Inaba T, Egawa T, Venkatesh B, Minoda A, Kominami R, Taniuchi I, Priming of lineage-specifying genes by Bcl11b is required for lineage choice in post-selection thymocytes. Nat Commun 8, 702 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jenkinson SR, Intlekofer AM, Sun G, Feigenbaum L, Reiner SL, Bosselut R, Expression of the transcription factor cKrox in peripheral CD8 T cells reveals substantial postthymic plasticity in CD4-CD8 lineage differentiation. J Exp Med 204, 267–272 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rui J, Liu H, Zhu X, Cui Y, Liu X, Epigenetic silencing of CD8 genes by ThPOK-mediated deacetylation during CD4 T cell differentiation. Journal of immunology 189, 1380–1390 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Mucida D, Husain MM, Muroi S, van Wijk F, Shinnakasu R, Naoe Y, Reis BS, Huang Y, Lambolez F, Docherty M, Attinger A, Shui JW, Kim G, Lena CJ, Sakaguchi S, Miyamoto C, Wang P, Atarashi K, Park Y, Nakayama T, Honda K, Ellmeier W, Kronenberg M, Taniuchi I, Cheroutre H, Transcriptional reprogramming of mature CD4(+) helper T cells generates distinct MHC class II-restricted cytotoxic T lymphocytes. Nature immunology 14, 281–289 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vacchio MS, Wang L, Bouladoux N, Carpenter AC, Xiong Y, Williams LC, Wohlfert E, Song KD, Belkaid Y, Love PE, Bosselut R, A ThPOK-LRF transcriptional node maintains the integrity and effector potential of post-thymic CD4+ T cells. Nature immunology 15, 947–956 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ciucci T, Vacchio MS, Gao Y, Tomassoni Ardori F, Candia J, Mehta M, Zhao Y, Tran B, Pepper M, Tessarollo L, McGavern DB, Bosselut R, The Emergence and Functional Fitness of Memory CD4(+) T Cells Require the Transcription Factor Thpok. Immunity 50, 91–105 e104 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chopp LB, Gopalan V, Ciucci T, Ruchinskas A, Rae Z, Lagarde M, Gao Y, Li C, Bosticardo M, Pala F, Livak F, Kelly MC, Hannenhalli S, Bosselut R, An Integrated Epigenomic and Transcriptomic Map of Mouse and Human alphabeta T Cell Development. Immunity 53, 1182–1201 e1188 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xue Y, Wong J, Moreno GT, Young MK, Côté J, Wang W, NURD, a Novel Complex with Both ATP-Dependent Chromatin-Remodeling and Histone Deacetylase Activities. Molecular cell 2, 851–861 (1998). [DOI] [PubMed] [Google Scholar]
- 45.Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL, Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature 395, 917–921 (1998). [DOI] [PubMed] [Google Scholar]
- 46.Wade PA, Jones PL, Vermaak D, Wolffe AP, A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol 8, 843–846 (1998). [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D, The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95, 279–289 (1998). [DOI] [PubMed] [Google Scholar]
- 48.Nakahashi H, Kwon KR, Resch W, Vian L, Dose M, Stavreva D, Hakim O, Pruett N, Nelson S, Yamane A, Qian J, Dubois W, Welsh S, Phair RD, Pugh BF, Lobanenkov V, Hager GL, Casellas R, A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep 3, 1678–1689 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klinger M, Kim JK, Chmura SA, Barczak A, Erle DJ, Killeen N, Thymic OX40 expression discriminates cells undergoing strong responses to selection ligands. Journal of immunology 182, 4581–4589 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu J, Min B, Hu-Li J, Watson CJ, Grinberg A, Wang Q, Killeen N, Urban JF Jr., Guo L, Paul WE, Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nature immunology 5, 1157–1165 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Vacchio MS, Ciucci T, Gao Y, Watanabe M, Balmaceno-Criss M, McGinty MT, Huang A, Xiao Q, McConkey C, Zhao Y, Shetty J, Tran B, Pepper M, Vahedi G, Jenkins MK, McGavern DB, Bosselut R, A Thpok-Directed Transcriptional Circuitry Promotes Bcl6 and Maf Expression to Orchestrate T Follicular Helper Differentiation. Immunity 51, 465–478 e466 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lai JS, Herr W, Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci U S A 89, 6958–6962 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dhordain P, Lin RJ, Quief S, Lantoine D, Kerckaert JP, Evans RM, Albagli O, The LAZ3(BCL-6) oncoprotein recruits a SMRT/mSIN3A/histone deacetylase containing complex to mediate transcriptional repression. Nucleic Acids Res 26, 4645–4651 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Q, Shi L, Gui B, Yu W, Wang J, Zhang D, Han X, Yao Z, Shang Y, Binding of the JmjC demethylase JARID1B to LSD1/NuRD suppresses angiogenesis and metastasis in breast cancer cells by repressing chemokine CCL14. Cancer Res 71, 6899–6908 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Klein BJ, Piao L, Xi Y, Rincon-Arano H, Rothbart SB, Peng D, Wen H, Larson C, Zhang X, Zheng X, Cortazar MA, Peña PV, Mangan A, Bentley DL, Strahl BD, Groudine M, Li W, Shi X, Kutateladze TG, The histone-H3K4-specific demethylase KDM5B binds to its substrate and product through distinct PHD fingers. Cell Rep 6, 325–335 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nishibuchi G, Shibata Y, Hayakawa T, Hayakawa N, Ohtani Y, Sinmyozu K, Tagami H, Nakayama J, Physical and functional interactions between the histone H3K4 demethylase KDM5A and the nucleosome remodeling and deacetylase (NuRD) complex. J Biol Chem 289, 28956–28970 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang L, Wildt KF, Zhu J, Zhang X, Feigenbaum L, Tessarollo L, Paul WE, Fowlkes BJ, Bosselut R, Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4(+) T cells. Nature immunology 9, 1122–1130 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R, Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R, Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taniuchi I, Ellmeier W, Transcriptional and epigenetic regulation of CD4/CD8 lineage choice. Adv Immunol 110, 71–110 (2011). [DOI] [PubMed] [Google Scholar]
- 61.Gulich AF, Preglej T, Hamminger P, Alteneder M, Tizian C, Orola MJ, Muroi S, Taniuchi I, Ellmeier W, Sakaguchi S, Differential Requirement of Cd8 Enhancers E8I and E8VI in Cytotoxic Lineage T Cells and in Intestinal Intraepithelial Lymphocytes. Front Immunol 10, 409 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen W, Zollman S, Couderc JL, Laski FA, The BTB domain of bric a brac mediates dimerization in vitro. Mol Cell Biol 15, 3424–3429 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.Moll JR, Olive M, Vinson C, Attractive interhelical electrostatic interactions in the proline- and acidic-rich region (PAR) leucine zipper subfamily preclude heterodimerization with other basic leucine zipper subfamilies. J Biol Chem 275, 34826–34832 (2000). [DOI] [PubMed] [Google Scholar]
- 64.Fujita N, Jaye DL, Geigerman C, Akyildiz A, Mooney MR, Boss JM, Wade PA, MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell 119, 75–86 (2004). [DOI] [PubMed] [Google Scholar]
- 65.Turner VM, Gardam S, Brink R, Lineage-specific transgene expression in hematopoietic cells using a Cre-regulated retroviral vector. J Immunol Methods 360, 162–166 (2010). [DOI] [PubMed] [Google Scholar]
- 66.McCaughtry TM, Wilken MS, Hogquist KA, Thymic emigration revisited. J Exp Med 204, 2513–2520 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stogios PJ, Chen L, Prive GG, Crystal structure of the BTB domain from the LRF/ZBTB7 transcriptional regulator. Protein Sci 16, 336–342 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schubot FD, Tropea JE, Waugh DS, Structure of the POZ domain of human LRF, a master regulator of oncogenesis. Biochem Biophys Res Commun 351, 1–6 (2006). [DOI] [PubMed] [Google Scholar]
- 69.Lemos TA, Passos DO, Nery FC, Kobarg J, Characterization of a new family of proteins that interact with the C-terminal region of the chromatin-remodeling factor CHD-3. FEBS letters 533, 14–20 (2003). [DOI] [PubMed] [Google Scholar]
- 70.Carpenter AC, Grainger JR, Xiong Y, Kanno Y, Chu HH, Wang L, Naik S, dos Santos L, Wei L, Jenkins MK, O’Shea JJ, Belkaid Y, Bosselut R, The transcription factors Thpok and LRF are necessary and partly redundant for T helper cell differentiation. Immunity 37, 622–633 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kumar R, Wang RA, Structure, expression and functions of MTA genes. Gene 582, 112–121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sher F, Hossain M, Seruggia D, Schoonenberg VAC, Yao Q, Cifani P, Dassama LMK, Cole MA, Ren C, Vinjamur DS, Macias-Trevino C, Luk K, McGuckin C, Schupp PG, Canver MC, Kurita R, Nakamura Y, Fujiwara Y, Wolfe SA, Pinello L, Maeda T, Kentsis A, Orkin SH, Bauer DE, Rational targeting of a NuRD subcomplex guided by comprehensive in situ mutagenesis. Nat Genet 51, 1149–1159 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grueter B, Petter M, Egawa T, Laule-Kilian K, Aldrian CJ, Wuerch A, Ludwig Y, Fukuyama H, Wardemann H, Waldschuetz R, Moroy T, Taniuchi I, Steimle V, Littman DR, Ehlers M, Runx3 regulates integrin alpha E/CD103 and CD4 expression during development of CD4-/CD8+ T cells. Journal of immunology 175, 1694–1705 (2005). [DOI] [PubMed] [Google Scholar]
- 74.Egawa T, Tillman RE, Naoe Y, Taniuchi I, Littman DR, The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J Exp Med 204, 1945–1957 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu X, Yin S, Cao W, Fan W, Yu L, Yin L, Wang L, Wang J, Runt-related transcription factor 3 is involved in the altered phenotype and function in ThPok-deficient invariant natural killer T cells. Cell Mol Immunol 11, 232–244 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wildt KF, Sun G, Grueter B, Fischer M, Zamisch M, Ehlers M, Bosselut R, The transcription factor Zbtb7b promotes CD4 expression by antagonizing Runx-mediated activation of the CD4 silencer. Journal of immunology 179, 4405–4414 (2007). [DOI] [PubMed] [Google Scholar]
- 77.Lee SU, Maeda T, POK/ZBTB proteins: an emerging family of proteins that regulate lymphoid development and function. Immunol Rev 247, 107–119 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mathew R, Mao AP, Chiang AH, Bertozzi-Villa C, Bunker JJ, Scanlon ST, McDonald BD, Constantinides MG, Hollister K, Singer JD, Dent AL, Dinner AR, Bendelac A, A negative feedback loop mediated by the Bcl6-cullin 3 complex limits Tfh cell differentiation. J Exp Med 211, 1137–1151 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McLoughlin P, Ehler E, Carlile G, Licht JD, Schafer BW, The LIM-only protein DRAL/FHL2 interacts with and is a corepressor for the promyelocytic leukemia zinc finger protein. J Biol Chem 277, 37045–37053 (2002). [DOI] [PubMed] [Google Scholar]
- 80.Mendez LM, Polo JM, Yu JJ, Krupski M, Ding BB, Melnick A, Ye BH, CtBP is an essential corepressor for BCL6 autoregulation. Mol Cell Biol 28, 2175–2186 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Choi WI, Kim Y, Kim Y, Yu MY, Park J, Lee CE, Jeon BN, Koh DI, Hur MW, Eukaryotic translation initiator protein 1A isoform, CCS-3, enhances the transcriptional repression of p21CIP1 by proto-oncogene FBI-1 (Pokemon/ZBTB7A). Cell Physiol Biochem 23, 359–370 (2009). [DOI] [PubMed] [Google Scholar]
- 82.Setoguchi R, Tachibana M, Naoe Y, Muroi S, Akiyama K, Tezuka C, Okuda T, Taniuchi I, Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science 319, 822–825 (2008). [DOI] [PubMed] [Google Scholar]
- 83.Basu J, Reis BS, Peri S, Zha J, Hua X, Ge L, Ferchen K, Nicolas E, Czyzewicz P, Cai KQ, Tan Y, Fuxman Bass JI, Walhout AJM, Grimes HL, Grivennikov SI, Mucida D, Kappes DJ, Essential role of a ThPOK autoregulatory loop in the maintenance of mature CD4(+) T cell identity and function. Nature immunology 22, 969–982 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reis BS, Rogoz A, Costa-Pinto FA, Taniuchi I, Mucida D, Mutual expression of the transcription factors Runx3 and ThPOK regulates intestinal CD4(+) T cell immunity. Nature immunology 14, 271–280 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cardenas MG, Yu W, Beguelin W, Teater MR, Geng H, Goldstein RL, Oswald E, Hatzi K, Yang SN, Cohen J, Shaknovich R, Vanommeslaeghe K, Cheng H, Liang D, Cho HJ, Abbott J, Tam W, Du W, Leonard JP, Elemento O, Cerchietti L, Cierpicki T, Xue F, MacKerell AD Jr., Melnick AM, Rationally designed BCL6 inhibitors target activated B cell diffuse large B cell lymphoma. J Clin Invest 126, 3351–3362 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Williams CJ, Naito T, Arco PG, Seavitt JR, Cashman SM, De Souza B, Qi X, Keables P, Von Andrian UH, Georgopoulos K, The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity 20, 719–733 (2004). [DOI] [PubMed] [Google Scholar]
- 87.Chong MM, Simpson N, Ciofani M, Chen G, Collins A, Littman DR, Epigenetic propagation of CD4 expression is established by the Cd4 proximal enhancer in helper T cells. Genes Dev 24, 659–669 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Henson DM, Chou C, Sakurai N, Egawa T, A silencer-proximal intronic region is required for sustained CD4 expression in postselection thymocytes. Journal of immunology 192, 4620–4627 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kojo S, Yasmin N, Muroi S, Tenno M, Taniuchi I, Runx-dependent and silencer-independent repression of a maturation enhancer in the Cd4 gene. Nat Commun 9, 3593 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Boucheron N, Tschismarov R, Goeschl L, Moser MA, Lagger S, Sakaguchi S, Winter M, Lenz F, Vitko D, Breitwieser FP, Muller L, Hassan H, Bennett KL, Colinge J, Schreiner W, Egawa T, Taniuchi I, Matthias P, Seiser C, Ellmeier W, CD4(+) T cell lineage integrity is controlled by the histone deacetylases HDAC1 and HDAC2. Nature immunology 15, 439–448 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Oravecz A, Apostolov A, Polak K, Jost B, Le Gras S, Chan S, Kastner P, Ikaros mediates gene silencing in T cells through Polycomb repressive complex 2. Nat Commun 6, 8823 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hu G, Cui K, Fang D, Hirose S, Wang X, Wangsa D, Jin W, Ried T, Liu P, Zhu J, Rothenberg EV, Zhao K, Transformation of Accessible Chromatin and 3D Nucleome Underlies Lineage Commitment of Early T Cells. Immunity 48, 227–242.e228 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lorentsen KJ, Cho JJ, Luo X, Zuniga AN, Urban JF Jr., Zhou L, Gharaibeh R, Jobin C, Kladde MP, Avram D, Bcl11b is essential for licensing Th2 differentiation during helminth infection and allergic asthma. Nat Commun 9, 1679 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Geimer Le Lay AS, Oravecz A, Mastio J, Jung C, Marchal P, Ebel C, Dembélé D, Jost B, Le Gras S, Thibault C, Borggrefe T, Kastner P, Chan S, The tumor suppressor Ikaros shapes the repertoire of notch target genes in T cells. Sci Signal 7, ra28 (2014). [DOI] [PubMed] [Google Scholar]
- 95.Schwickert TA, Tagoh H, Gültekin S, Dakic A, Axelsson E, Minnich M, Ebert A, Werner B, Roth M, Cimmino L, Dickins RA, Zuber J, Jaritz M, Busslinger M, Stage-specific control of early B cell development by the transcription factor Ikaros. Nature immunology 15, 283–293 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, Kingston R, Georgopoulos K, Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity 10, 345–355 (1999). [DOI] [PubMed] [Google Scholar]
- 97.Cismasiu VB, Adamo K, Gecewicz J, Duque J, Lin Q, Avram D, BCL11B functionally associates with the NuRD complex in T lymphocytes to repress targeted promoter. Oncogene 24, 6753–6764 (2005). [DOI] [PubMed] [Google Scholar]
- 98.Sujino T, London M, Hoytema van Konijnenburg DP, Rendon T, Buch T, Silva HM, Lafaille JJ, Reis BS, Mucida D, Tissue adaptation of regulatory and intraepithelial CD4(+) T cells controls gut inflammation. Science 352, 1581–1586 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Driegen S, Ferreira R, van Zon A, Strouboulis J, Jaegle M, Grosveld F, Philipsen S, Meijer D, A generic tool for biotinylation of tagged proteins in transgenic mice. Transgenic Res 14, 477–482 (2005). [DOI] [PubMed] [Google Scholar]
- 100.Zijlstra M, Bix M, Simister NE, Loring JM, Raulet DH, Jaenisch R, Beta 2-microglobulin deficient mice lack CD4–8+ cytolytic T cells. Nature 344, 742–746 (1990). [DOI] [PubMed] [Google Scholar]
- 101.Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, Cherry SR, Tsai JH, Tucker SM, Weaver WM, Kelso A, Jaenisch R, Wilson CB, A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 15, 763–774 (2001). [DOI] [PubMed] [Google Scholar]
- 102.Grusby MJ, Johnson RS, Papaioannou VE, Glimcher LH, Depletion of CD4+ T cells in major histocompatibility complex class II-deficient mice. Science 253, 1417–1420 (1991). [DOI] [PubMed] [Google Scholar]
- 103.Saitoh T, Nakano H, Yamamoto N, Yamaoka S, Lymphotoxin-beta receptor mediates NEMO-independent NF-kappaB activation. FEBS letters 532, 45–51 (2002). [DOI] [PubMed] [Google Scholar]
- 104.Love PE, Shores EW, Lee EJ, Grinberg A, Munitz TI, Westphal H, Singer A, Differential effects of zeta and eta transgenes on early alpha/beta T cell development. J Exp Med 179, 1485–1494 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bosselut R, Kubo S, Guinter T, Kopacz JL, Altman JD, Feigenbaum L, Singer A, Role of CD8beta domains in CD8 coreceptor function: importance for MHC I binding, signaling, and positive selection of CD8+ T cells in the thymus. Immunity 12, 409–418 (2000). [DOI] [PubMed] [Google Scholar]
- 106.Morita S, Kojima T, Kitamura T, Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther 7, 1063–1066 (2000). [DOI] [PubMed] [Google Scholar]
- 107.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M, In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1, 2856–2860 (2006). [DOI] [PubMed] [Google Scholar]
- 108.Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, Hoffman P, Stoeckius M, Papalexi E, Mimitou EP, Jain J, Srivastava A, Stuart T, Fleming LM, Yeung B, Rogers AJ, McElrath JM, Blish CA, Gottardo R, Smibert P, Satija R, Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e3529 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.He X, Park K, Wang H, He X, Zhang Y, Hua X, Li Y, Kappes DJ, CD4-CD8 lineage commitment is regulated by a silencer element at the ThPOK transcription-factor locus. Immunity 28, 346–358 (2008). [DOI] [PubMed] [Google Scholar]
- 110.Wang L, Bosselut R, CD4-CD8 lineage differentiation: Thpok-ing into the nucleus. Journal of immunology 183, 2903–2910 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data file 1. Raw data file (Excel spreadsheet).
Supplementary data file 2. Unprocessed blots
Table S1. Peptides identified by LC-MS.
Table S2. Frequency of scATACseq clusters shown in Fig. S2E.
Table S3. CD4+ and CD8+ lineage signatures defined as in Fig. 8F.
Figure S1. NuRD-Thpok binding.
Figure S2. scATACseq analysis of Thpok impact on chromatin accessibility.
Figure S3. ΔBTBLZ properties and transgenic mouse lines.
Figure S4. Flow cytometric quantification of Thpok mutant expression.
Figure S5. B-Thpok dimerization and retrogenic expression.
Figure S6. Thpok-RKF protein dimerization and function.
Figure S7. Analyses of Mta2-ΔBTBLZ retrogenic mice.
Figure S8. Cell purification strategy for RNAseq of mixed bone marrow chimera.
Data Availability Statement
All sequence data reported in this paper are publicly available on the NCBI Gene Expression Omnibus (GEO), from accession numbers GSE199739, GSE171610 and GSE185217. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.


