Abstract
Approximately 200 O-serogroups of Vibrio cholerae have already been identified; however, only 2 serogroups, O1 and O139, are strongly related to pandemic cholera. The study of non-O1 and non-O139 strains has hitherto been limited. Nevertheless, there are other clinically and epidemiologically important serogroups causing outbreaks with cholera-like disease. Here, we report a comprehensive genome analysis of the whole set of V. cholerae O-serogroup reference strains to provide an overview of this important bacterial pathogen. It revealed structural diversity of the O-antigen biosynthesis gene clusters located at specific loci on chromosome 1 and 16 pairs of strains with almost identical O-antigen biosynthetic gene clusters but differing in serological patterns. This might be due to the presence of O-antigen biosynthesis-related genes at secondary loci on chromosome 2.
Keywords: multi-chromosomal bacteria, O-antigen biosynthetic gene cluster, O-serogroup reference strain, Vibrio cholerae
Data Summary
Short-read sequence data were submitted to the DDBJ Sequenced Read Archive, and each accession number is listed in Table S1 (available in the online version of this article). The annotated sequences of O-antigen biosynthesis gene clusters have been deposited in GenBank/EMBL/DDBJ under accession numbers LC594800–LC595005. The high-quality finished genome assemblies with annotation of 10 Vibrio cholerae strains are also available in GenBank/EMBL/DDBJ under accession numbers AP023331–AP023332 and AP023369–AP023386. The authors confirm all supporting data, code and protocols have been provided within the article or through supplementary data files.
Impact Statement.
The O-antigen has been used epidemiologically to differentiate epidemic from non-epidemic Vibrio cholerae strains for decades. It has been used to infer the diversity of the species. Currently there are more than 200 types of reference strains, but there is no systematic analysis of V. cholerae strains and serotypes based on whole-genome analysis. Here we sequence and analyse all of the O-serogroup reference strains and elucidate the relations between these serogroups and the high genomic diversity of V. cholerae strains. Additionally, by combining serological analysis and genomic information of O-antigen biosynthetic genes, we reassess of the number of known O-serogroups. Our genomic insights give important clues for understanding of V. cholerae evolutionary processes as a representative of bacteria with multiple chromosomes.
Introduction
Vibrio cholerae is a member of the family Vibrionaceae , comprising curved, Gram-negative rods that are found in coastal waters and estuaries. O-specific polysaccharides (O-antigens) covering the outermost layer of Gram-negative bacteria are responsible for serological diversity. To date, 210 O-serogroups have been identified in V. cholerae , and O-serogroups have been used epidemiologically to classify strains within this species since the 1930s [1]. Only two serogroups, O1 and O139, are usually associated with epidemics of cholera, which is characterized by acute watery diarrhoea [2]. However, nonagglutinable vibrios, which are non-O1, non-O139 serogroup strains, have also been reported to cause cholera-like intestinal infections and are associated with a limited number of outbreaks [3, 4].
As a bacterium, V. cholerae has shown extraordinary genomic plasticity and ability to adapt to changing environments, a factor likely to have contributed to the emergence of the pathogenic serogroups. V. cholerae can acquire new genetic material by natural transformation during growth on chitin, a biopolymer that is abundant in aquatic habitats [5]. Examples of genetic traits linked to high virulence that can be transferred through this route include the CTX prophage, the type 3 secretion system genes and the lipopolysaccharide biosynthetic operon [6–10]. This raises the possibility that all strains, including non-O1 and non-O139 strains, could acquire functions that confer pandemic potential by the acquisition and exchange of genes through natural competence or other horizontal gene transfer mechanisms. Therefore, while multilocus sequence typing (MLST) offers a high level of discrimination between isolates of this species, whole-genome-level analysis is required to elucidate the genetic diversity and plasticity of the V. cholerae genome.
Currently, whole-genome sequencing-based analysis of V. cholerae has mainly been performed with serotypes O1 and O139, and the genetic diversity of the V. cholerae population is unclear. Since O-serogroup reference strains have shaped our view of this important bacterial pathogen, we performed comprehensive genome analyses on O-serogroup reference strains, including details of the O-antigen biosynthetic gene clusters (O-AGCs). Thus, we refined the number of O-serogroups according to serological analysis and the genomic information for O-AGCs and linked these data to the whole-genome phylogeny. We included 210 V . cholerae complex O-serogroup reference strains from the Sakazaki collection, comprising 194 V . cholerae strains, 14 Vibrio mimicus strains and 2 Vibrio metoecus strains [11]. The latter two species are included because they had previously been reported as biochemically atypical isolates of V. cholerae [12, 13]. We also determined 10 complete genome sequences of V. cholerae strains from different phylogenetic clusters to further investigate the genomic plasticity and chromosomal dynamics of this important reference collection. Our genomic insights provide important clues for understanding the evolutionary processes of V. cholerae , suggesting that new pandemic strains may emerge in the future.
Methods
V. cholerae O-serogroup reference strains
A total of 210 V . cholerae complex O-serogroup reference strains were used for whole-genome sequencing, which included 14 V . mimicus strains and 2 V . metoecus strains (Table S1). Among the O-serogroup reference strains, three strains (O167, O189 and O203) and one strain (O143) were identified as Aeromonas sp. and Vibrio fluvialis , respectively, based on conventional biochemical tests and 16S ribosomal DNA sequencing analysis. We excluded these four strains from further analysis.
Genome sequencing and read processing
Genomic DNA was extracted using the DNeasy Blood and Tissue kit (Qiagen); DNA concentrations were determined using a Qubit dsDNA HS assay kit (Thermo Fisher Scientific). A genomic library was prepared using the Nextera XT DNA Library Preparation kit (Illumina), and sequenced paired-end short reads were prepared on HiSeq 2500 or MiSeq sequencers (Illumina). The resultant reads were processed using the A5-miseq (v20160825) pipeline for trimming, correction and de novo assembly to generate contigs and scaffolds [14]. Genome annotation was performed using the Prokka (v1.13) pipeline with Prodigal for gene prediction, Aragorn for tRNA search and RNAmmer for rRNA searching [15]. We used 10 complete genome sequences for annotation instead of contigs in the draft genome. The assembled statistics and the general features of genomes used in this study are shown in Table S1. The resulting data were used for downstream analyses.
High-quality finished sequence of 10 V. cholerae strains
We selected 10 genetically distant V. cholerae strains on phylogenetic analysis to determine the high-quality finished sequence; 9 strains were from diarrhoea patients and 1 was from seawater (Table 1). A genomic library for P6-C4 chemistry was prepared using the RS II SMRTbell template preparation kit version 1.0 (Pacific Biosciences) and sequenced with the P6 version 2 single-molecule real-time sequencing platform (Pacific Biosciences). Sequencing reads were assembled de novo using Hierarchical Genome Assembly Process 3 [16]. This assembly was corrected with the Quiver consensus algorithm to obtain a high-accuracy genome assembly. The contig was further corrected using Pilon (v1.22) and the paired-end short reads [17].
Table 1.
|
General genome statistics |
N16961 |
VCSRO5 |
VCSRO17 |
VCSRO63 |
VCSRO77 |
VCSRO102 |
VCSRO207 |
VCSRO45 |
VCSRO51 |
VCSRO96 |
VCSRO162 |
|---|---|---|---|---|---|---|---|---|---|---|---|
|
Cluster 3 |
Cluster 3 |
Cluster 3 |
Cluster 3 |
Cluster 3 |
Cluster 3 |
Cluster 3 |
Cluster 2 |
Cluster 2 |
Cluster 2 |
Cluster 1 |
|
|
Chromosome 1 |
|||||||||||
|
Genome size (bp) |
2 961 149 |
2 952 352 |
2 939 341 |
2 869 733 |
3 064 657 |
2 874 693 |
2 868 058 |
3 021 501 |
2 967 527 |
2 887 793 |
2 966 062 |
|
No. of CDSs |
2775 |
2720 |
2703 |
2623 |
2801 |
2601 |
2592 |
2767 |
2737 |
2632 |
2691 |
|
No. of rRNA operon |
8 |
8 |
8 |
8 |
8 |
8 |
8 |
8 |
8 |
8 |
8 |
|
No. of tRNA and tmRNA |
95 |
99 |
100 |
101 |
101 |
96 |
101 |
100 |
102 |
97 |
103 |
|
GC content (%) |
47.70 |
47.90 |
47.69 |
48.08 |
47.76 |
47.99 |
48.01 |
47.87 |
47.93 |
48.09 |
47.68 |
|
No. of genomic island* |
6 |
5 |
5 |
2 |
5 |
4 |
3 |
6 |
5 |
3 |
6 |
|
No. of strain-specific genes |
147 |
139 |
96 |
114 |
198 |
66 |
93 |
170 |
163 |
82 |
273 |
|
Proportion of unique genes (%) |
5.30 |
5.11 |
3.55 |
4.35 |
7.07 |
2.54 |
3.59 |
6.14 |
5.96 |
3.12 |
10.14 |
|
No. of core genes |
1254 |
1254 |
1254 |
1254 |
1254 |
1254 |
1254 |
1254 |
1254 |
1254 |
1254 |
|
Proportion of core genes (%) |
45.19 |
46.10 |
46.39 |
47.81 |
44.77 |
48.21 |
48.38 |
45.32 |
45.82 |
47.64 |
46.60 |
|
Chromosome 2 |
|||||||||||
|
Genome size (bp) |
1 072 315 |
1 070 220 |
1 102 179 |
1 155 566 |
1 007 849 |
1 123 019 |
1 163 376 |
1 096 179 |
1 004 624 |
1 165 751 |
1 094 700 |
|
No. of CDSs |
1115 |
956 |
976 |
1035 |
916 |
1013 |
1017 |
990 |
895 |
1081 |
971 |
|
No. of rRNA operons |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
|
Number of tRNA and tmRNA |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
3 |
|
GC contents (%) |
46.92 |
47.20 |
47.28 |
46.95 |
47.17 |
46.87 |
46.85 |
46.66 |
47.06 |
46.62 |
46.53 |
|
No. of genomic island* |
1 |
3 |
3 |
3 |
3 |
2 |
5 |
5 |
2 |
4 |
3 |
|
No. of strain-specific genes |
144 |
89 |
87 |
123 |
153 |
153 |
139 |
83 |
67 |
184 |
222 |
|
Proportion of unique genes (%) |
12.91 |
9.31 |
8.91 |
11.88 |
16.70 |
15.10 |
13.67 |
8.38 |
7.49 |
17.02 |
22.86 |
|
No. of core genes |
196 |
196 |
196 |
196 |
196 |
196 |
196 |
196 |
196 |
196 |
196 |
|
Proportion of core genes (%) |
17.58 |
20.50 |
20.08 |
18.94 |
21.40 |
19.35 |
19.27 |
19.80 |
21.90 |
18.13 |
20.19 |
* The relevant characteristics of the genomic island identified in each strain are shown in Table S4.
Identification of O-antigen biosynthetic loci
We set the region from gmhD (VC0240 in V. cholerae O1 N16961 annotation) to rjg (VC0264 in V. cholerae O1 N16961 annotation) as an O-AGC, which was extracted from contigs of the draft genome [18]. The reference strains of O30, O32, O93, O116, O120 and O194 were found to lack rjg, and ybdG (VC0265 in V. cholerae O1 N16961 annotation) located downstream of rjg, was used for the right junction gene instead of rjg.
Detection of secondary loci of O-antigen biosynthesis gene was also performed by OrthoFinder (v2.3.7) to cluster the functional coding sequences (CDSs) of O-AGC and those of 10 complete genomes of O-serogroup reference strains, with the cut-off value set at 1e-25 to identify potential O-antigen biosynthesis genes [19].
Comprehensive genomic analysis
We carried out a pan-genome analysis using the 190 V . cholerae genomes from O-serogroup reference strains and the National Center for Biotechnology Information (NCBI) reference genome of seventh pandemic V. cholerae O1 strain N16961 [18]. We used the Roary pipeline, which generated core gene alignment [20]. To investigate the phylogeny of the V. cholerae genomes, we constructed a tree based on the alignment and pan-genome profiles were incorporated into phylogeny. We performed clusters of orthologous groups (COGs) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses for the functional classification of orthologous genes identified on the core or non-core genome. Further details are available in the Document S1.
Identification of genomic islands
In 10 high-quality finished V. cholerae genomes and the NCBI reference genome of strain N16961, genomic islands (GIs), which were defined as regions more than 15 kb length between the two loci of core genes or tRNAs, were determined and characterized (Document S1). The presence or absence of each GI in 190 V . cholerae O-serogroup reference strains and NCBI reference strain N16961 was confirmed by mapping reads to sequences of GIs using SRST2 (v0.2.0) with the minimum coverage cut-off set to 80 % [21].
Results
Genetic structures of O-antigens
Although only two serogroups, O1 and O139, have been known to cause repeated outbreaks and epidemics over the world, there are 210 O-serogroups of V. cholerae featured on the accredited O-antigen reference list. Considering the whole collection based on 16S ribosomal DNA taxonomy and conventional biochemical tests, three strains representing serogroups O167, O189 and O203, and one strain representing serogroup O143, were reclassified as Aeromonas sp. and V. fluvialis , respectively. Therefore, we deleted these four serogroups from the official V. cholerae complex accredited O-antigen reference list and completed the entire sequences of the O-AGCs for the rest of the 206 type strains (Table S1 and Fig. S1).
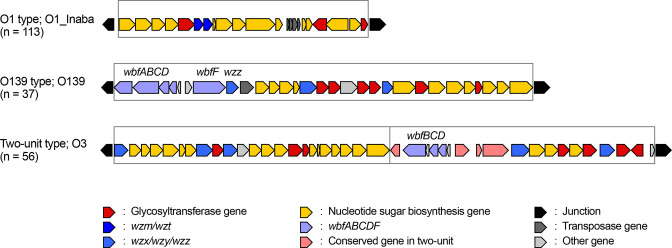
The sizes of O-AGCs ranged from 17.1 to 67.7 kb. We further investigated the size of O-AGCs and the number of their constitutive genes in V. cholerae complex and compared them with those of well-studied Escherichia coli . The median size of O-AGCs in V. cholerae complex and E. coli was 32.5 and 16.4 kb, respectively, an almost twofold difference (Fig. S2). In addition, the number of constitutive genes in V. cholerae O-AGCs was also approximately twofold higher than in E. coli , indicating the high genetic diversity of V. cholerae O-AGCs. An O-antigen synthesis unit requires three functional classes of proteins: nucleotide sugar biosynthesis, glycosyltransferases and O-antigen processing. We detected 262 O-antigen synthesis units in 206 strains, including 3 units that lacked the genes for O-antigen processing. Therefore, 150 strains possess 1 synthesis unit, and 56 strains possess 2 synthesis units. Among the 150 strains with 1 synthesis unit, 37 strains with different O-antigens shared 6 genes in the 5ʹ portion of the operon, which were previously reported as wbfABCDF, and wzz in the V. cholerae serogroup O139 genome [22]. We defined this O-AGC as the O139 type and the other O-AGCs with one synthesis unit as the O1 type. Of the 206, 56 strains possessed 2 O-antigen synthesis units named as belonging to the two-unit type, and the second unit conserved 7 genes at the 5ʹ end of the operon, of which 3 were represented by wbfBCD, but 4 genes differed (Fig. 1).
Fig. 1.
Classification of the O-antigen biosynthetic gene cluster. A representative of each type is enlarged from Fig. S1. An O-antigen synthesis unit, which contains genes related to nucleotide sugar biosynthesis, glycosyltransferases and O-antigen processing, is enclosed in a box. The O139 type of the O-antigen biosynthetic gene cluster possesses wbfABCDF and wzz in the 5′ region of the operon. The two-unit type of the O-antigen biosynthetic gene cluster possesses two synthesis units and conserved seven genes in the 5′ region of the second operon.
Overall, 1065 glycosyltransferase genes were identified and annotated in the O-antigen operons with units containing 1–7 glycosyltransferase genes (median=4). For the O-antigen processing gene, 46 units and 202 units carried gene pairs of wzm/wzt or wzx/wzy, respectively, and pglK, encoding putative ATP-binding cassette-type transporter of oligosaccharides, represented the candidate O-antigen processing gene in 11 units [23]. Importantly, this analysis also showed that of the 206 O-AGCs, 25 cluster pairs were almost identical in gene composition, and of these 9 strain pairs possessing them were also serologically identical according to the results of agglutinin absorption tests with each pair. Since we could not distinguish them either genetically or serologically, the strain from the pair that had been entered into the collection most recently (with the highest O-serogroup reference number; underlined below) was removed from the accredited O-antigen reference list: O5 and O185; O17 and O198; O18 and O136; O20 and O101; O31 and O84; O68 and O129; O74 and O200; O85 and O163; O87 and O119 (Table S1). Therefore, the number of strains in the current accredited list of V. cholerae complex O-antigen serotypes and genotypes is 197. However, these strains were still included for further analysis in this study.
Phylogenetic relations of V. cholerae O-serogroup reference strains
We investigated the phylogenetic relationships of these V. cholerae complex O-serogroup reference strains (n=206) in the context of pandemic strains (n=30) from a public database (Tables S1 and S3). Genome-wide phylogenetic analysis or MLST were performed to better understand more distant evolutionary relationships [12]. This showed that of the 206 V . cholerae complex O-serogroup reference strains, 190 strains clustered with known V. cholerae isolates, but the remaining 16 strains clustered more closely with genomes from V. mimicus (O20, O30, O32, O71, O101, O114-117, O135, O138, O194, O201 and O202) or V. metoecus (O154 and O195) isolate (Fig. S3). Considering the role of this reference collection, the following analysis was focused solely on the V. cholerae genomes. However, strains of V. mimicus and V. metoecus still remain in the V. cholerae complex O-serogroup reference collection for historical reasons (Table S1).
Core and pan-genome in V. cholerae and its intraspecies diversity
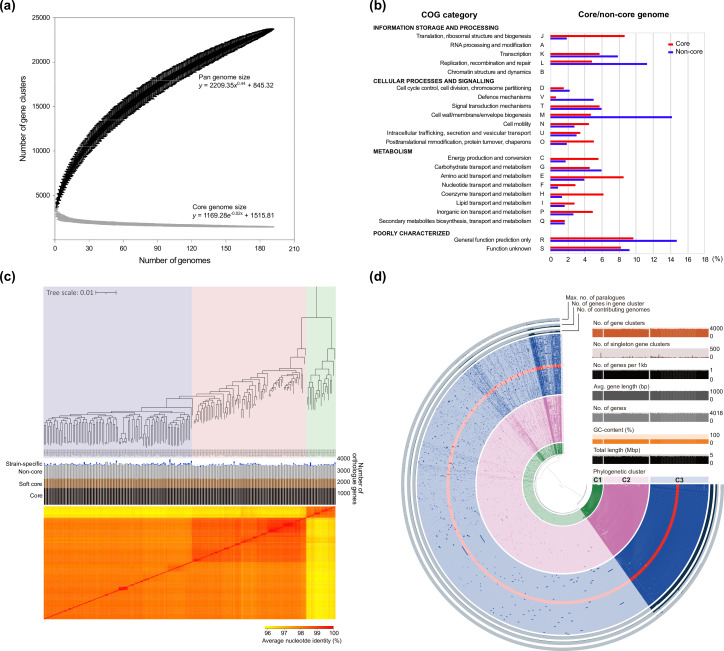
Pan-genome analysis revealed that there are 23 713 V . cholerae orthologous gene clusters, including 1450 core genes (present in ≥99 % of strains) and 822 soft-core genes (present in ≥95 % of strains), as calculated by maximum-likelihood methods (Table S4). The V. cholerae pan-genome can be considered ‘open’, with its size increasing logarithmically. This was supported by the parameter from Heaps’ law (γ=0.44) (Fig. 2a), indicating that the V. cholerae population displays a high level of genomic plasticity, consistent with the fact that it inhabits a broad set of complex environments.
Fig. 2.
Pan-genome profile and phylogenetic relation of the V. cholerae genomes. (a) A pan-genome curve for 191 V . cholerae was generated by plotting the total number of distinct gene families against the number of genomes considered using PanGP. Similarly, the number of shared gene families is plotted against the number of genomes to generate the core genome plot that depicts the trend in the contraction of the core genome size with sequential addition of more genomes. (b) Assignments of core and non-core genes to COG and KEGG, as predicted by their respective databases. The values in each category indicate the relative abundance of core or non-core gene sets identified in the pan-genome profile of 191 V . cholerae genomes. (c) The core gene-based phylogenetic tree classified into three groups (cluster 1, light green; cluster 2, pale pink; cluster 3, lavender) according to the statistical significance, as calculated by the hierBAPS clustering method. Heatmap shows the pairwise comparison of ANI values calculated on the whole-genome level by FastANI (v1.3). (d) Pan-genome profile and the relevant statistics are shown in the circular phylogram or bar plots. Orthologous gene clusters in the circular phylogram were organized by Euclidean distance and the Ward linkage algorithm in the anvi'o (v5) platform.
The distribution of COG functional categories in the core or dispensable (non-core) genes showed that several COG groups were overrepresented in the core genome when compared to the non-core genome (Fig. 2b). Conversely, the non-core genome carried a higher proportion of genes classified as ‘V’ (defence mechanisms), ‘M’ (cell wall/membrane/envelope biogenesis) and ‘L’ (replication, recombination and repair) than the core genome. It is important to note that the higher proportion of category M in non-core genes might be due to the various O-serogroup reference strains used in this study. In addition, a higher proportion of categories L and V is concordant with acquisition of foreign DNA that could contribute to survival under varied environmental niches.
Three phylogenetically distinct V. cholerae clusters
V. cholerae can mainly be separated into three statistically significant clusters using hierBAPS: cluster 1 (n=19), cluster 2 (n=75), and cluster 3 (n=96). Cluster 3 was assigned next to cluster 2, but its similarity to cluster 2 was weaker than that between cluster 1 and cluster 2. In addition to the ANI-based profile, the fixation index between cluster 2 and cluster 3 was the lowest (0.05812) among all the combinations, indicating that the genetic differentiation between clusters 2 and 3 was small (Fig. S4). This result implies that cluster 3 represents a more diverse genome cluster than the others in the V. cholerae population. Importantly, the pan-genome size showed that core and pan-genomes were similar between the three clusters (Fig. S5). We further performed COG and KEGG analyses for the functional classification of orthologous genes identified on the core or non-core genome in the three clusters (Fig. S5). In the COG analysis of the core or non-core genome, similar ratios of each functional category were observed among the three clusters. In the KEGG analysis of the core or non-core genome, the proportions of four categories (genetic information processing, environmental information processing, cellular processes and unclassified) were relatively high in all three clusters, which accounted for 12–21 % of total assignment, but there were no remarkable differences between clusters in the functional classification profiles.
Detailed genomic analysis of 10 V. cholerae genomes from the three species-wide phylogenetic clusters
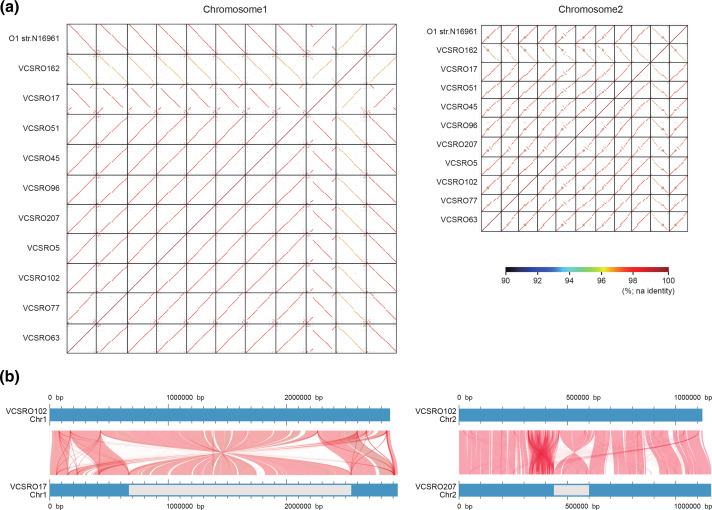
We generated a high-quality finished sequence of chromosome 1 (Chr1) and chromosome 2 (Chr2) from 10 strains randomly selected from the 3 V . cholerae clusters (1 strain from cluster 1, 3 strains from cluster 2, 6 strains from cluster 3). The genome sizes of Chr1 and Chr2 ranged from 2.87 to 3.06 Mb and 1.00 to 1.16 Mb, respectively (Table 1). A dot plot showing pairwise sequence alignment revealed that Chr1 exhibited high sequence conservation and genome synteny across the three clusters, except for O162 belonging to cluster 1 (Fig. 3a). Furthermore, there was a large inversion in the O17 genome in addition to several strain-specific deletions or insertions on Chr1 (Fig. 3b). However, sequence similarity on Chr2 was low between strains representing the different clusters with many insertions or deletions, compared to that on Chr1, even though the genome synteny was generally maintained, except for that in the superintegron (SI) region. Moreover, genomic regions with SIs adopted a mosaic structure as expected, and we found a large inversion neighbouring the SI region in O45 and O207 strains. To investigate its general traits within or across the cluster in V. cholerae , we added an additional 10 draft genome sequences randomly selected from all three clusters to the whole-genome comparative analysis. This analysis based on 21 V . cholerae genomes also demonstrated lower conservation of synteny in Chr2 due to the lower alignment of Chr2 sequences compared to Chr1 (Fig. S6). This result reflects the differing proportions of unique and core genes across chromosomes (Table 1). These results suggest that Chr2 may contribute to the genetic variation in the V. cholerae genome; meanwhile, Chr1 genetically or structurally maintained architectural stability. The numbers of CDSs or tRNA genes present in Chr1 or Chr2 were also similar to those reported previously [18].
Fig. 3.
Whole-genome alignment profile of 11 V . cholerae strains. (a) Dot plot representation of DNA sequence homology of Chr1 or Chr2 between strains. GenomeMatcher (v2.30) was used for blastn analysis and visualization of the results. (b) Linear maps of Chr1 (left panel) or Chr2 (right panel) with a large inversion were built using AliTV (v1.0.6) visualization software, based on the whole-genome alignments with Lastz aligner. The red plots represent the shared sequences showing >95 % similarity between two different genomes. The grey segments indicate the inverted region on Chr1 or Chr2.
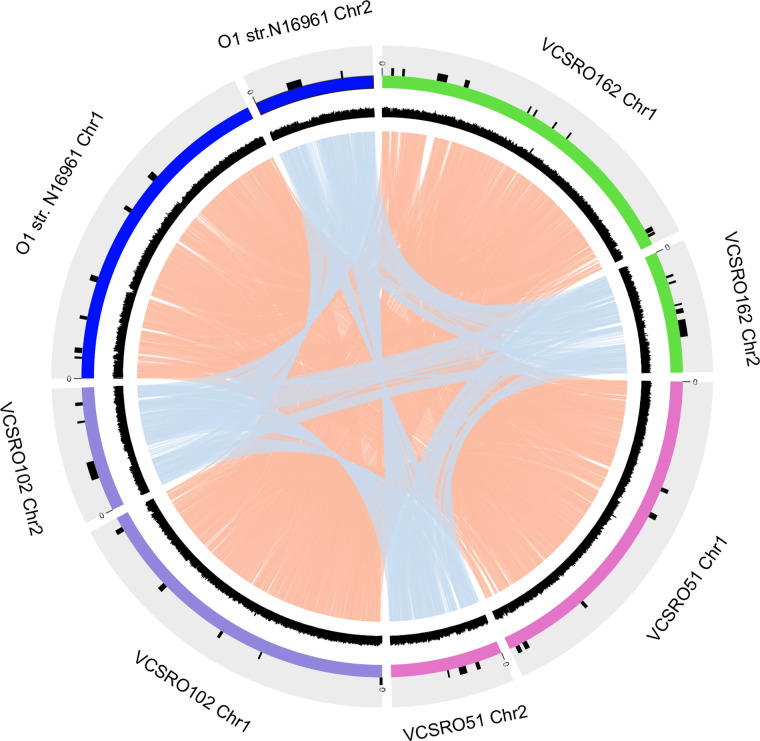
We showed that the core genome of V. cholerae comprises 1450 genes: 1254 and 196 genes were distributed on Chr1 and Chr2, respectively. Applying the same methods here, the proportion of unique genes in each strain was higher in Chr2 (7.5–22.9 %) than in Chr1 (2.5–10.1 %). While the proportion of core genes was higher in Chr1 (45.2–48.4 %) than in Chr2 (17.6–21.9 %), surprisingly, the exact number of core genes identified on Chr1 and Chr2 varied by only ~5 % between each strain. This implies that there was no or little exchange of core genes between two chromosomes. Therefore, we confirmed whether there is an inter-exchange of genes or any recombination event between Chr1 and Chr2 by analysing the chromosomal linkage with several complete genomes. Both Chr1 and Chr2 linkage maps showed that each chromosome was well conserved, even between different V. cholerae clusters, with the exception of GI regions. Furthermore, this profile revealed that inter-chromosome exchange of genes or recombination events was rare (Fig. 4), suggesting that the genome diversification and evolution of Chr1 and Chr2 were independent.
Fig. 4.
Linkage of representative genomes from each phylogenetic cluster in V. cholerae . The linkages of gene synteny in Chr1 or Chr2 were visualized using Circos (v0.69–7) and are shown by the lines coloured with orange and light blue, respectively. The outermost circles represent the GIs, chromosomes and GC contents of each reference genome. There was no synteny between Chr1 and Chr2 in any strain.
These results suggest that Chr1 and Chr2 may contribute to the stability and diversification of the V. cholerae genome, respectively. Chromosome-independent diversification could accelerate the populational genomic evolution of V. cholerae , reflecting their current phylogenetic relationship.
Characterization of genomic islands and their distributions in V. cholerae populations
GIs are crucial factors linked to genome diversity, plasticity and phylogenetic evolution in bacteria. Using 10 high-quality finished V. cholerae genomes and the NCBI reference genome for strain N16961, we identified 84 GIs in their genomes, where the 50 and 34 GIs were located on Chr1 and Chr2, respectively (Table S5). Some GIs, including CRISPR and CRISPR-associated genes (CRISPR/Cas), were detected on both chromosomes in different isolate genomes. The GIs on Chr1 included VSP-II, integrative conjugative elements, the type 3 secretion gene cluster and the auxiliary locus of the type 6 secretion system. Consistent with previous reports, the O-AGC was also identified as a GI on Chr1 [24].
We investigated the distribution of these GIs and the distribution across all V. cholerae genomes considered here or in each distinct cluster in the phylogeny (Fig. 5). Among the 84 GIs, (1) 61 GIs detected on <5 % of strain genomes were categorized as ‘specific’ GIs; (2) 5 GIs were considered to be ‘common’ GIs present in >50 % of strains analysed here; (3) the remaining 18 GIs, distributed among 5–50 % of strain genomes, were considered to be ‘moderately’ distributed. Of specific and moderate GIs, 63.3 % (50 out of 79) were detected on Chr1; meanwhile, all common GIs were detected on Chr2.
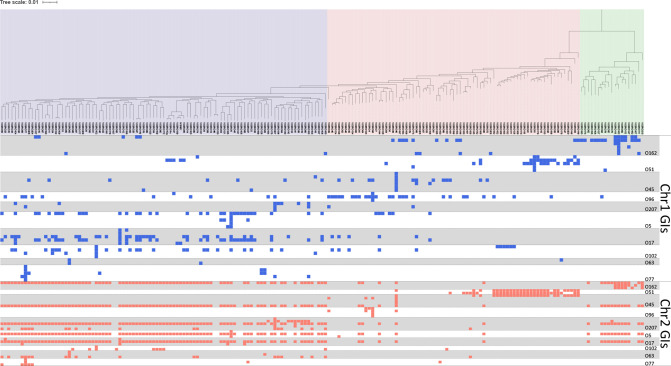
Fig. 5.
Distribution of GIs identified on Chr1 and Chr2 in 191 V . cholerae strains. The profile was plotted according to the phylogenetic tree shown in Fig. 2c. Blue and red dots indicate the presence of GIs identified on Chr1 and Chr2, respectively.
The most variable GI seen in all genomes was the SI located on Chr2. The dot plot analysis showed this SI region was highly variable (Fig. S7a, b). There were 1538 CDSs in SI regions, of which 191 formed orthologous groups (>2 CDSs) and 331 CDSs were unique to a single isolate genome. Among them, only two groups were shared in all SIs sequenced, and the highest number of shared orthologous groups seen between any two SIs was 66 (Fig. S7c–e).
Secondary loci of O-antigen biosynthetic gene on chromosome 2
The O-AGCs of all reference strains were found at a specific locus of Chr1. The pairwise genetic alignment analysis of O-AGCs revealed that 25 pairs were almost identical in gene composition and synteny, but 16 of the 25 pairs phenotypically showed different O-antigenic reactions. This suggests the involvement of O-antigen biosynthetic-related genes outside of the specific loci of Chr1. Orthologue analysis of the functional annotations of O-AGCs and 10 complete genome sequences of O-serogroup reference strains revealed the presence of additional genes homologous with those found in the main O-AGC but located outside of it on either Chr2 or Chr1, for which the average numbers were 5.0 (median: 5, range: 2–7) and 21.6 (median: 21, range: 18–25), respectively.
To investigate this further, we selected the reference strain for serogroup O63. Its O-AGC is almost identical to that of O131; nevertheless, they show different O-antigenic reactions. Comparing their genomes, we identified 20 orthologous groups of O-antigen biosynthesis-related genes outside of the main O-AGC. Of 20 orthologous groups, 17 were common in both O63 and O131. However, two of the other three were specific to O63, and one was specific to O131. Among them, one orthologous group was detected on the SI region on Chr2 of O63. The SI region is located on Chr2, suggesting that genes on not only Chr1 but also Chr2 are involved in O-antigen synthesis.
Discussion
Most genomic studies of V. cholerae have focused on serogroups O1 and O139 because of their role in human disease and global pandemics. This has meant that other V. cholerae serogroups have often been overlooked. An obvious starting point to link what we know about the V. cholerae population to genomic and phylogenetic information is the Sakazaki collection, which holds 206 serogroup reference strains. Our results indicated that the V. cholerae population has an ‘open’ pan-genome with a diverse composition of accessory genes. Genetic traits that might be correlated with the bacterial lifestyle include those for various ecological niches, environments, and external stressors, as shown in previous studies [25–27]. One of the bacterial components affected by the external environment is the O-antigen of the outermost cell envelope. Escherichia coli has association with host phylogenetic lineage and O-serogroup [28]. However, complete sequencing of the O-AGC from all the reference strains revealed 16 pairs of strains with almost identical O-AGCs but differing serological reactions. Based on complete sequencing of the 10 O-serogroup reference strains, O-AGCs were located on Chr1. However, we also identified the presence of putative O-antigen biosynthetic-related genes at secondary loci on Chr2. These findings could provide important evidence to understand the functional interaction of Chr1 and Chr2 in the ecological adaptation of V. cholerae .
Furthermore, GIs play an important role in the genome diversification of the V. cholerae population through acquisition of variable genes via mobile genetic elements for inhabitation or adaptation under various environments, which is consistent with our observations showing V. cholerae to have an open pan-genome [29]. In this study, we identified 84 known and novel GIs from 11 V . cholerae genomes. Differential distribution patterns of GIs were between Chr1 and Chr2, wherein Chr2 showed stepwise acquisition of foreign genetic elements from a common ancestor. SI, an important GI in V. cholerae , represents a potential gene capture system [30]. The pairwise comparisons of the SIs sequenced here showed how variable and complex their structure is, regardless of phylogenetic relations (Fig. S7). These results suggest that genetic variation of SIs might not be related to the stepwise evolutionary process but rather that this variation is a key factor contributing to the genome diversification for V. cholerae . We also detected GIs harbouring the CRISPR/Cas system in V. cholerae genomes. A recent study demonstrated that GIs with CRISPR/Cas provide recipient cells not only with a defence mechanism against maladaptive lateral gene transfer but also with a potential competitive advantage over bacteria lacking this GI and perhaps a novel virulence factor [31].
Multi-chromosome bacteria are thought to have originated from single-chromosome ancestors by transferring some essential genes from the chromosome to plasmids [32, 33]. Most genes required for growth and viability are located on Chr1, although some genes found only on Chr2 are also thought to be essential for normal cell function [18]. When considering the origin of multi-chromosomal bacteria, we infer that Chr1 is a ‘stable’ chromosome for the V. cholerae genome and Chr2 could be a ‘placeholder’ enabling the acquisition of massive external genes due to the lower number of core genes on Chr2.
In conclusion, our study showing the atlas of the V. cholerae pan-genome provides important clues allowing us to understand not only the genetic traits in V. cholerae but also the genomic plasticity in the evolution process in multi-chromosomal bacteria.
Supplementary Data
Funding information
This research was supported by Research Program on Emerging and Re-emerging Infectious Diseases (JP20fk0108139), Japan Agency for Medical Research and Development (E.A., H.I., M.O., M.M.) and Japan Initiative for Global Research Network on Infectious Diseases (JP20wm0125006), Japan Agency for Medical Research and Development (T.T.).
Acknowledgements
We thank Kanako Oba for her technical assistance.
Authors and contributors
E.A., M.O. and M.M. conceived the project. K.M., M.M., H.I. and M.O. designed the study. E.A. and M.M. participated in the sequencing experiments. K.M. and M.M. performed the bioinformatics analysis supervised by T.K. and M.O. E.A., A.I. and M.M. performed assembly and annotation of the O-antigen biosynthetic gene cluster. T.T. and M.M. performed characterization of genomic islands. K.M., M.M., H.I. and M.O. contributed to the interpretation of the results. I.N. and N.R.T. commented on the study. K.M. and M.M. prepared the manuscript and figures. N.R.T. revised the manuscript. All authors read and approved the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: CDS, coding sequence; Chr, chromosome; COG, Cluster of Orthologous Groups; GI, genomic island; KEGG, Kyoto Encyclopedia of Genes and Genomes; O-AGC, O-antigen biosynthetic gene cluster; SI, superintegron.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Supplementary material is available with the online version of this article.
References
- 1.Gardner AD, Venkatraman KV. The antigens of the cholera group of vibrios. J Hyg. 1935;35:262–282. doi: 10.1017/S0022172400032265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ryan ET. The cholera pandemic, still with us after half a century: time to rethink. PLoS Negl Trop Dis. 2011;5:e1003. doi: 10.1371/journal.pntd.0001003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Orginization Outbreak of gastro-enteritis by non agglutinable (NAG) vibrios = épidémie de gastro-entérite due a des vibrions non agglutinables. Weekly Epidemiological Record. 1969;44:10. [Google Scholar]
- 4.Tobin-D’Angelo M, Smith AR, Bulens SN, Thomas S, Hodel M, et al. Severe diarrhea caused by cholera toxin-producing Vibrio cholerae serogroup O75 infections acquired in the southeastern United States. Clin Infect Dis. 2008;47:1035–1040. doi: 10.1086/591973. [DOI] [PubMed] [Google Scholar]
- 5.Meibom KL, Blokesch M, Dolganov NA, Wu CY, Schoolnik GK. Chitin induces natural competence in Vibrio cholerae . Science. 2005;310:1824–1827. doi: 10.1126/science.1120096. [DOI] [PubMed] [Google Scholar]
- 6.Blokesch M, Schoolnik GK. Serogroup conversion of Vibrio cholerae in aquatic reservoirs. PLoS Pathog. 2007;3:e81. doi: 10.1371/journal.ppat.0030081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keymer DP, Miller MC, Schoolnik GK, Boehm AB. Genomic and phenotypic diversity of coastal Vibrio cholerae strains is linked to environmental factors. Appl Environ Microbiol. 2007;73:3705–3714. doi: 10.1128/AEM.02736-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Metzger LC, Blokesch M. Regulation of competence-mediated horizontal gene transfer in the natural habitat of Vibrio cholerae . Curr Opin Microbiol. 2016;30:1–7. doi: 10.1016/j.mib.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 9.Morita M, Yamamoto S, Hiyoshi H, Kodama T, Okura M, et al. Horizontal gene transfer of a genetic island encoding a type III secretion system distributed in Vibrio cholerae . Microbiol Immunol. 2013;57:334–339. doi: 10.1111/1348-0421.12039. [DOI] [PubMed] [Google Scholar]
- 10.Udden SMN, Zahid MSH, Biswas K, Ahmad QS, Cravioto A, et al. Acquisition of classical CTX prophage from Vibrio cholerae O141 by El Tor strains aided by lytic phages and chitin-induced competence. Proc Natl Acad Sci U S A. 2008;105:11951–11956. doi: 10.1073/pnas.0805560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brenner DJ, Davis BR, Kudoh Y, Ohashi M, Sakazaki R, et al. Serological comparison of two collections of Vibrio cholerae non O1. J Clin Microbiol. 1982;16:319–323. doi: 10.1128/jcm.16.2.319-323.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis BR, Fanning GR, Madden JM, Steigerwalt AG, Bradford HB, Jr, et al. Characterization of biochemically atypical Vibrio cholerae strains and designation of a new pathogenic species, Vibrio mimicus . J Clin Microbiol. 1981;14:631–639. doi: 10.1128/jcm.14.6.631-639.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirchberger PC, Turnsek M, Hunt DE, Haley BJ, Colwell RR, et al. Vibrio metoecus sp. nov., a close relative of Vibrio cholerae isolated from coastal brackish ponds and clinical specimens. Int J Syst Evol Microbiol. 2014;64:3208–3214. doi: 10.1099/ijs.0.060145-0. [DOI] [PubMed] [Google Scholar]
- 14.Coil D, Jospin G, Darling AE. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 2015;31:587–589. doi: 10.1093/bioinformatics/btu661. [DOI] [PubMed] [Google Scholar]
- 15.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 16.Chin C-S, Alexander DH, Marks P, Klammer AA, Drake J, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 17.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae . Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inouye M, Dashnow H, Raven L-A, Schultz MB, Pope BJ, et al. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014;6:11. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamasaki S, Shimizu T, Hoshino K, Ho ST, Shimada T, et al. The genes responsible for O-antigen synthesis of Vibrio cholerae O139 are closely related to those of Vibrio cholerae O22. Gene. 1999;237:321–332. doi: 10.1016/s0378-1119(99)00344-3. [DOI] [PubMed] [Google Scholar]
- 23.Nothaft H, Szymanski CM. Protein glycosylation in bacteria: sweeter than ever. Nat Rev Microbiol. 2010;8:765–778. doi: 10.1038/nrmicro2383. [DOI] [PubMed] [Google Scholar]
- 24.Chun J, Grim CJ, Hasan NA, Lee JH, Choi SY, et al. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae . Proc Natl Acad Sci U S A. 2009;106:15442–15447. doi: 10.1073/pnas.0907787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Maayer P, Chan WY, Rubagotti E, Venter SN, Toth IK, et al. Analysis of the Pantoea ananatis pan-genome reveals factors underlying its ability to colonize and interact with plant, insect and vertebrate hosts. BMC Genomics. 2014;15:404. doi: 10.1186/1471-2164-15-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donati C, Hiller NL, Tettelin H, Muzzi A, Croucher NJ, et al. Structure and dynamics of the pan-genome of Streptococcus pneumoniae and closely related species. Genome Biol. 2010;11:10. doi: 10.1186/gb-2010-11-10-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009;5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iguchi A, Iyoda S, Kikuchi T, Ogura Y, Katsura K, et al. A complete view of the genetic diversity of the Escherichia coli O-antigen biosynthesis gene cluster. DNA Res. 2015;22:101–107. doi: 10.1093/dnares/dsu043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Juhas M, van der Meer JR, Gaillard M, Harding RM, Hood DW, et al. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol Rev. 2009;33:376–393. doi: 10.1111/j.1574-6976.2008.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marin MA, Vicente ACP. Architecture of the superintegron in Vibrio cholerae: identification of core and unique genes. F1000Res. 2013;2:63. doi: 10.12688/f1000research.2-63.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Labbate M, Orata FD, Petty NK, Jayatilleke ND, King WL, et al. A genomic island in Vibrio cholerae with VPI-1 site-specific recombination characteristics contains CRISPR-Cas and type VI secretion modules. Sci Rep. 2016;6:36891. doi: 10.1038/srep36891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Egan ES, Waldor MK. Distinct replication requirements for the two Vibrio cholerae chromosomes. Cell. 2003;114:521–530. doi: 10.1016/s0092-8674(03)00611-1. [DOI] [PubMed] [Google Scholar]
- 33.Venkova-Canova T, Chattoraj DK. Transition from a plasmid to a chromosomal mode of replication entails additional regulators. Proc Natl Acad Sci U S A. 2011;108:6199–6204. doi: 10.1073/pnas.1013244108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.