Abstract
Analyzing radiology reports is a time-consuming and error-prone task, which raises the need for an efficient automated radiology report analysis system to alleviate the workloads of radiologists and encourage precise diagnosis. In this work, we present RadText, a high-performance open-source Python radiology text analysis system. RadText offers an easy-to-use text analysis pipeline, including de-identification, section segmentation, sentence split and word tokenization, named entity recognition, parsing, and negation detection. Superior to existing widely used toolkits, RadText features a hybrid text processing schema, supports raw text processing and local processing, which enables higher accuracy, better usability and improved data privacy. RadText adopts BioC as the unified interface, and also standardizes the output into a structured representation that is compatible with Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM), which allows for a more systematic approach to observational research across multiple, disparate data sources. We evaluated RadText on the MIMIC-CXR dataset, with five new disease labels that we annotated for this work. RadText demonstrates highly accurate classification performances, with a 0.91 average precision, 0.94 average recall and 0.92 average F-1 score. We also annotated a test set for the five new disease labels to facilitate future research or applications. We have made our code, documentations, examples and the test set available at https://github.com/bionlplab/radtext.
Index Terms—: Natural Language Processing, Text Analysis Systems, Radiology
I. Introduction
Radiology report analysis has long been a labor-some and error-prone process [1], which raises the need for accurate analysis tools to alleviate the workloads of radiologists and enhance accurate diagnosis. Though existing natural language processing (NLP) toolkits such as cTAKES [2], scispaCy [3], MedTagger [4], and CLAMP [5] have been widely used in text mining of clinical narratives in electronic health record (EHR), none of these tools on the use of NLP in EHRs is specific to radiology domain.
One recognized challenge is the requirement of proper radiology domain knowledge, without which the process of analyzing the structure of radiology text and interpreting the underlying meaning would be highly error-prone. For example, standardized terminology for each concept is important for NLP applications. Existing clinical NLP systems frequently use UMLS Methathesaurus as the medical lexicon [6]. However, few support RadLex, which offers radiology-specific terms such as devices and imaging techniques [7]. As a result, ambiguous terms (e.g., acronyms) can be interpreted differently. Another example is negation detection, which is also essential in radiology because diagnostic imagining is often used to rule out a condition. Systems in the clinical domain frequently implement this functionality by combining manually crafted rules with key terms based on the syntactic analysis [8], [9]. While they usually achieve good results in the general clinical domain, most cannot be directly applied to radiology reports mostly because sentences in radiology reports are usually telegraphic, with missing subjects and verbs. In addition, sentences in the radiology reports also contain long, complicated noun phrases. These obstacles pose a challenge to existing parsers that are modeled over well-formed sentences [10]. Therefore, the performance of negation detection algorithms significantly drops [11] in the case of radiology reports. In such cases, filling in the gaps requires additional rules to handle ill-formed sentences.
Another challenge is that every software intends to perform tasks on data in various formats. It thus remains challenging to seamlessly interchange data in and between different NLP tools. Such a bottleneck prevents combining these tools into a larger, more powerful, and more capable system in the clinical domain. To bridge this gap, the OMOP Common Data Model (CDM) is proposed to harmonize disparate observational databases of EHR [12]. The goal is to transform data contained within those databases into a common format (data model) and representation (terminologies, vocabularies, coding schemes) so that systematic analyses can be conducted in the common format. While OMOP CDM is an excellent schema to store structured data and provides a NOTE_NLP table to store NLP final results, it does not support representing complex, messy data between different NLP modules, such as hierarchical note structure (section, passage, sentence, token). Furthermore, it is almost impossible to store the parsing trees of each sentence in NOTE_NLP table. However, such text-preprocessing information is frequently reused in NLP algorithms and should be interchangeable and reusable. In addition, OMOP CDM must be realized in a relational database, which most of the common NLP tools do not support. These limitations result in the main barrier to the reuse of tools and modules and the development of text mining pipelines customized for different workflows. One alternative solution is the BioC format [13], an XML-based simple format to share text data and annotations. Unlike OMOP CDM, BioC emphasizes simplicity, interoperability, broad use and reuse of data interchange. It is thus suitable to represent, store and exchange the NLP results, especially complex intermediate results, in a simple manner. However, as initially designed for sharing different annotations relevant for biomedical research, BioC cannot be directly used for clinical notes. To overcome this issue, we propose to extend the BioC format with the OMOP CDM schema, called BioC-CDM, to store the results generated in the annotation process of clinical NLP that can be easily converted and imported into OMOP CDM.
In this work, we present RadText, an open-source Python radiology text analysis system. Unlike previous methods, RadText features a hybrid text analysis pipeline that utilizes high-performance third-party implementations, including machine learning-based methods and rule-based methods. As shown in Table I, compared to existing widely-used NLP toolkits, RadText has the following advantages:
Unified Interface. RadText uses BioC-CDM format as the unified interface throughout the system pipeline. BioC format simplifies data representation and data exchange and satisfies all the NLP tasks requirements in RadText.
Compatible with OMOP CDM. RadText standardizes its outputs into a structured representation compatible with OMOP CDM. This allows for transforming data into a common representation and further enables a systematic analysis of disparate observational data sources.
Easy to Use. RadText provides a user-friendly interface. RadText sequentially runs de-identification, section segmentation, sentence split and word tokenization, named entity recognition, parsing, and negation detection. Modular choice of design greatly improves flexibility, which enables users to adjust any module according to their specific use case, and to re-run each module if needed.
Raw Text Processing. RadText takes raw text as input, which means no text preprocessing (e.g., tokenization, annotation) is needed. This greatly enhances the usability and generalizability of RadText.
Local Machine. The entire system pipeline of RadText is running locally. Therefore no data will be uploaded to remote servers, greatly preserving user data privacy.
Open Source. To facilitate and drive future clinical NLP research and applications, RadText is fully open source and we make the source code, documentation, examples, and human-annotated test sets publicly available.
TABLE I.
Feature comparisons of RadText against other widely used NLP toolkits. Fully Neural: full neural network pipeline.
| System | Language | Raw-Text Processing | Locally Process | Fully Neural | Open Source |
|---|---|---|---|---|---|
| MetaMap | Prolog/Java | ✓ | Hybrid | ✗ | ✓ |
| cTakes | Java | ✓ | ✓ | ✗ | ✓ |
| MedSpaCy | Python | ✓ | ✓ | ✗ | ✓ |
| MedTagger | Java/C | ✓ | ✓ | ✗ | ✓ |
| CLAMP | Java | ✓ | ✓ | Hybrid | ✗ |
| RadText | Python | ✓ | ✓ | Hybrid | ✓ |
II. Related Work
Various NLP toolkits have been introduced to the clinical NLP community [14] and have been successfully applied to the information extraction task from clinical text. MetaMap [15] uses a knowledge-intensive approach based on symbolic, NLP, and computational-linguistic techniques to map the biomedical text into the Unified Medical Language System (UMLS) Metathesaurus [16]. Apache Clinical Text Analysis and Knowledge Extraction System (cTAKES) focuses on extracting clinical information from electronic health record free text, including processing clinical notes, and identifying clinical named entities [2]. Different from MetaMap and Apache cTAKES, which utilize machine learning methods to map words to medical concepts, MedTagger for indexing is built upon a fast string matching algorithm leveraging lexical normalization [4]. It thus requires rules designing and expert knowledge engineering. Instead of conducting sole information extraction, medspaCy [17] and Clinical Language Annotation, Modeling and Procssing (CLAMP) [5] are designed to be modularized so that users can choose from various choices of modular components for their individual applications. Medspacy features performing clinical NLP and text processing tasks with the popular spaCy [18] framework, which provides a robust architecture for building and sharing custom, high-performance NLP pipelines [17]. CLAMP also highlights enabling users to quickly build customized NLP pipelines for their clinical NLP tasks. Distinguished from these previous work, RadText aims to provide a high-performance clinical NLP toolkit in Python that focuses on radiology text analysis. RadText hence adopts a hybrid radiology text processing pipeline, bringing together a number of third-party analysis tools in the radiology domain, with each tool implementing one or more components of RadText’s working pipeline.
III. System Design and Architecture
A. BioC-CDM: BioC format compatible with OMOP CDM
We propose BioC-CDM to store the results generated in the annotation process of clinical NLP in the BioC format that can be easily converted and imported into OMOP CDM. A BioC-format file is an XML document as the basis of data class representation and data exchange, which can satisfy the needs of RadText’s NLP tasks throughout the entire pipeline [13]. OMOP CDM harmonizes disparate coding systems to a standardized vocabulary with minimal information loss. As a result, adopting BioC-CDM as RadText’s unified interface and using it as a common format representing all modular components’ output, eliminates the barrier of integration and greatly enhances RadText’s interoperability. Table II shows the current and our proposed mappings between OMOP CDM and BioC. Section IV-B1 shows how RadText can be used to implement mutual conversion between BioC format and OMOP CDM.
TABLE II.
Mapping radiology notes to the OMOP CDM and BioC using RadText.
| OMOP CDM field | BioC field | BioC class | Description |
|---|---|---|---|
| note_nlp_id | id | annotation | A unique identifier for each term extracted from a note. |
| note_id | doc | document | A foreign key to the Note table, uniquely identifying the note. |
| section_concept_id | section_concept_id | passage | A foreign key to the predefined Concept in the Standardized Vocabularies representing the section of the extracted term. |
| snippet | - | - | A small window of text surrounding the term. |
| offset | offset | Passage sentence annotation | Character offset of the extracted term in the input note. |
| lexical_variant | text | annotation | Raw text extracted by the NLP tool. |
| note_nlp_concept_id | lemma | annotation | A foreign key to a Concept table, representing the normalized concept of the extracted term. |
| note_nlp_source_concept_id | source_concept_id | annotation | A foreign key to a Concept table that refers to the code in the source vocabulary used by the NLP system. |
| nlp_system | nlp_system | collection | Name and version of the NLP system that extracted the term. |
| nlp_date,nlp_date_time | date | collection | The date of the note processing. |
| term_exists | exists1 | annotation | If the patient actually has or had the condition. |
| term_temporal | temporal | annotation | If a condition is “present” or just in the “past”. |
| term_modifiers | modifiers | annotation | Describes compactly all the modifiers extracted by the NLP system. |
currently called “negation”
B. Pipeline
The implementation of RadText is highly modular, where the modules range from de-identification to negation detection (see Figure 1). We highlight the details of each module in this section.
Fig. 1.
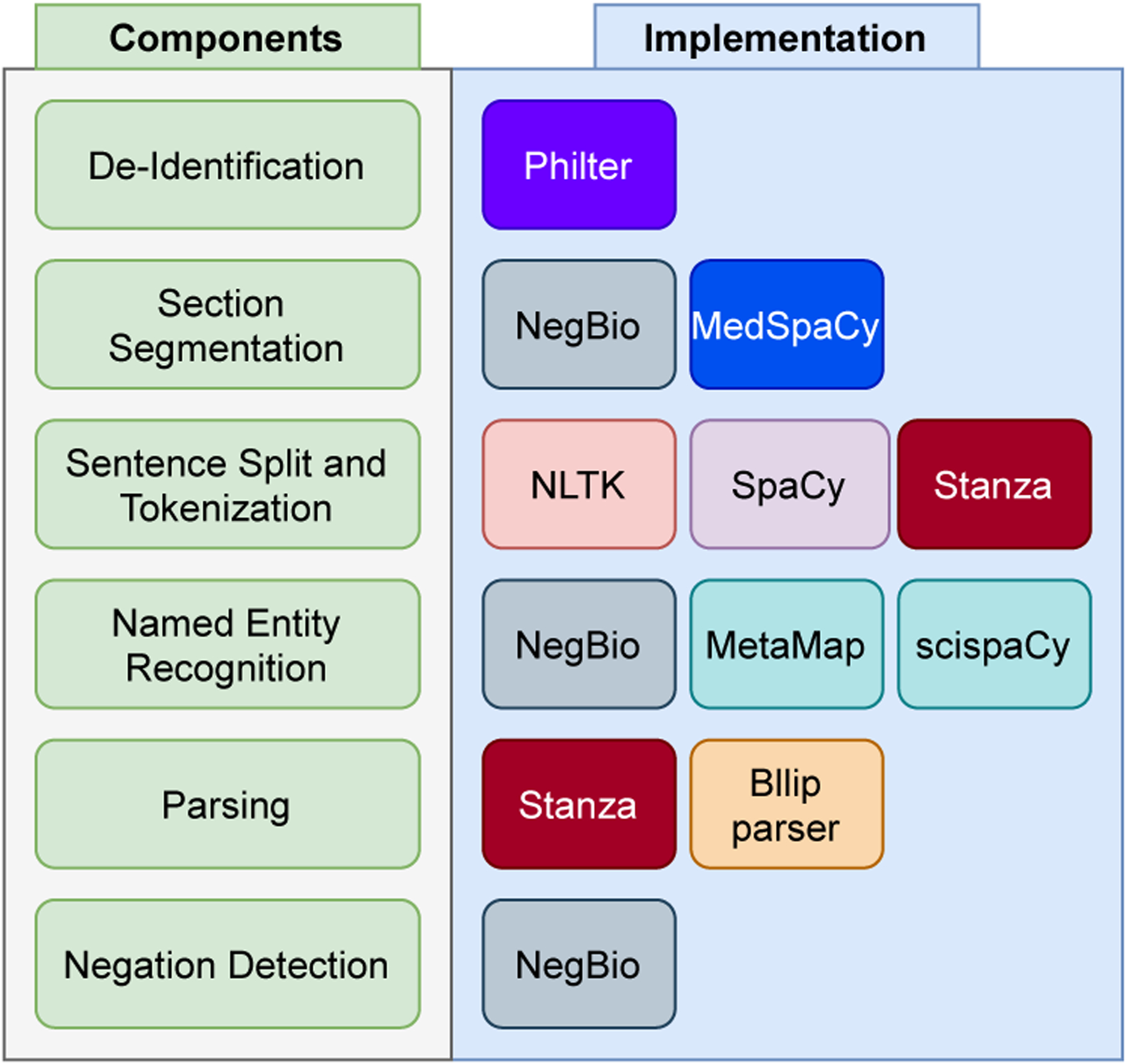
Overview of RadText’s NLP pipeline, main components and implementations.
1). De-Identification:
Radiology reports often contain protected health information (PHI), such as patient and provider names, addresses, and numbers [19]. Removal of PHI is important; otherwise, radiology reports remain largely unused for research. To address this issue, RadText uses Philter [19] for de-identification. It uses both rule-based and statistical approaches to remove identifiers defined in the HIPAA Safe Harbor guidelines [20].
The following code snippet shows an example of RadText’s de-identification output. The mentions of patient’s name, provider’s name, and dates belong to PHI. They are replaced with a sequence of “X”s respectively for de-identification purpose.


2). Section Segmentation:
Although radiology reports are in the form of free text, they are often structured in terms of sections, such as INDICATION, FINDINGS, and IMPRESSION. Identifying section types and section boundaries can help various successive processing steps to use a subset of sections or assign specific weights to the content of different sections [21]. For example, effusion and edema were mentioned in the INDICATION section of the sample report below. But we should not identify them as positive because the radiologist ruled them out in the FINDINGS section. Therefore, a named entity recognition tool that does not differentiate between sections will likely make errors.

In a preprocessing step, RadText splits each report into sections and provides two options: NegBio or MedSpaCy. Both approaches rely on hand-coded heuristics for section segmentation (boundary detection) and achieve good performances.
NegBio. The heuristics in NegBio are based on conventions like the capitalization of headers and the presence of colon and blank lines between headers and text. The set of heuristics was collected from the NIH Chest X-ray dataset [22] and the MIMIC-CXR dataset [23].
MedspaCy. MedspaCy includes an implementation of clinical section detection based on rule-based matching of the section titles with the default rules adapted from SecTag [24] and expanded through practice. The default rules were collected from different resources such as the Logical Observation Identifiers Names and Codes (LOINC) headers [25] and Quick Medical Reference (QMR) Findings Hierarchy [26] and were further revised based on the actual clinical notes from Vanderbilt EHR.
The following code snippet shows an example of the section segmentation output for the sample report above.

3). Sentence Split and Word Tokenization:
RadText tokenizes the input raw text and groups tokens into sentences as one part of preprocessing. RadText offers three options to tokenize and split reports into sentences, including NLTK [27], spaCy [18], and Stanza [28].
NLTK. The sentence tokenizer in NLTK uses an unsupervised algorithm to build a model for abbreviation words, collocations, and words that start sentences. It then uses that model to find the sentence boundaries [27].
spaCy. Sentence segmentation is part of spaCy’s English pipeline. It uses a variant of the non-monotonic arc-eager transition-system [29] with the addition of a “break” transition for sentence segmentation [18].
Stanza. Stanza combines tokenization and sentence segmentation from raw text as one single module in its pipeline. Stanza models it as a tagging task over character sequences, where the model predicts whether a given character is the end of a token, end of a sentence, or end of a multi-word token.
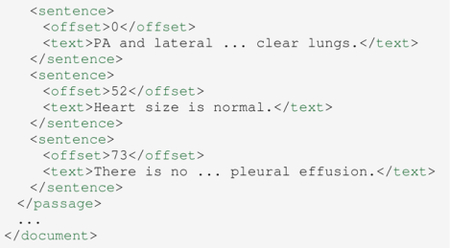
The following code snippet gives an example of RadText’s sentence split output. The input paragraph is split into three Sentence instances.

4). Named Entity Recognition:
Named entity recognition (NER) aims to determine and identify the words or phrases in text into predefined labels that describe the concepts of interest in a given domain [30]. To recognize the radiology-domain named entities (e.g., thoracic disorders) in each input sentence, RadText offers two options, spaCy-enabled rule-based method and MetaMap.
Rule-based Regular Expression. Rule-based NER methods use regular expressions that combine information from terminological resources and characteristics of the entities of interest manually constructed from report corpus. RadText adopts spaCy’s PhraseMatcher as part of this component. Rules defining concepts specify the text regular patterns to be matched and additional information about a concept, such as its unique id in the terminology.
MetaMap. UMLS is the most comprehensive standard terminology that is typically used as the basis for clinical concept extraction. Enabled by MetaMap, RadText is able to detect all the concepts in UMLS and map them to Concept Unique Identifier (CUI). In general, MetaMap is much more comprehensive than vocabulary-based patterns. But at the same time, MetaMap could be noisy and less accurate.
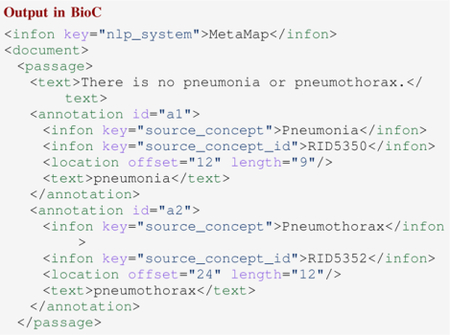
The following code snippet shows an example of RadText’s NER output, where “Pneumonia” and “Pneumothorax” are correctly recognized and their corresponding UMLS concept IDs are also identified.
![]()
5). Parsing:
RadText utilizes the universal dependency graph (UDG) to describe the grammatical relationships within a sentence in a way that can be understood by non-linguists and effectively used by downstream processing tasks [11]. UDG is a directed graph, which represents all universal dependency information within a sentence. The vertices in a UDG represent the information such as the word, part-of-speech and the word lemma. The edges in a UDG represent the typed dependencies from the governor to its dependent and are labeled with the corresponding dependency type. UDG effectively represents the syntactic head of each word in a sentence and the dependency relations between words. Figure 2 shows a UDG example of the sentence “There is no pleural effusion or pneumothorax” generated by Stanza [28]. In this example, “pleural” is the adjectival modifier of “effusion” and “effusion” and “pneumothorax” are coordinated findings.
Fig. 2.
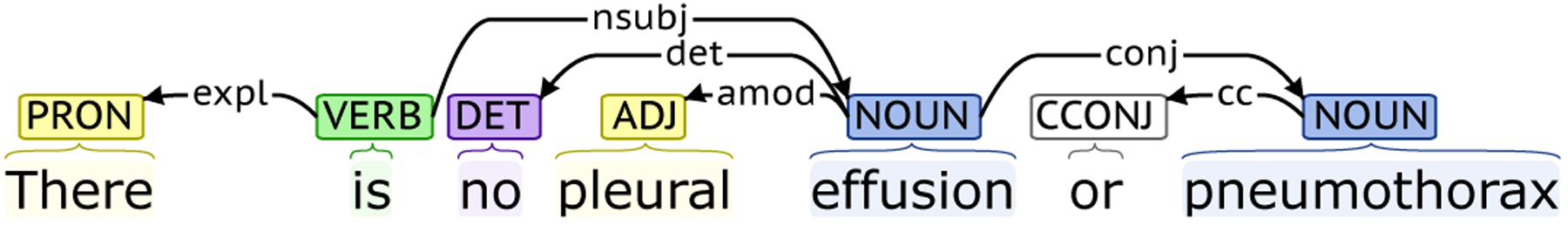
The obtained dependency graph of “There is no pleural effusion or pneumothorax” using Stanza [28].
To obtain the UDG of a sentence, RadText provides two options, Stanza or Bllip Parser with the Stanford dependencies converter [31].
Stanza. Stanza’s dependency parsing module builds a tree structure of words from the input sentence, representing the syntactic dependency relations between words. After tokenization, multi-word token (MWT) expansion, part-of-speech (POS) and morphological features tagging, and lemmatization, each sentence would have been directly parsed into the universal dependencies structure [28].
Bllip Parser with Stanford dependencies converter. RadText first parses each sentence to obtain the parse tree using the Bllip parser, which was trained with the biomedical model [31], [32]. It then applies the Stanford dependencies converter on the resulting parse tree with the CCProcessed and Universal option [33], [34] to derive the universal dependencies.
The following code snippet shows an example of RadText’s parsing result. In the sample sentence, “effusion” and “pneumothorax” are respectively assigned with node id of “T31” and “T33”. Derived from the universal dependency result, there is a conjunction relation between “T31” and “T33”.


6). Negation Detection:
Negative and uncertain medical findings are frequent in radiology reports [35]. Since they may indicate the absence of findings mentioned within the radiology report, identifying them is as important as identifying positive findings. For negation and uncertainty detection, RadText employs NegBio [11], [22], which utilizes universal dependencies for pattern definition and subgraph matching for graph traversal search so that the scope for negation/uncertainty is not limited to the fixed word distance [33].
The following code snippet shows an example of RadText’s negation detection output. In this sample sentence, “pneumothorax” is identified as negative according to NegBio’s internal negation rule of ID “nn180”.

IV. System Usage
RadText is designed to have a user-friendly interface and allow quick out-of-the-box usage for radiology text analysis. To achieve this, RadText provides automated pipeline usage and step-by-step modular choice of design. Therefore, Users can run RadText directly through the command line interface or import RadText as a Python library to use any functionality through RadText’s API.
A. Command Line Usage
The following command runs RadText’s entire pipeline in the sequential order of de-identification, section segmentation, sentence split and word tokenization, NER, parsing, and negation detection. The default section title vocabulary for the section segmentation module and concept vocabulary for the NER module are designed to be configurable. All intermediate result files will be generated and saved for use and reuse purpose. The automatic pipeline execution enables users to use RadText as an out-of-the-box toolkit without the need and efforts to figure out how each module of RadText works.
![]()
In addition to running RadText’s pipeline as a whole, users can also choose to run every single module of RadText by configuring the argument of the command above (see Table III). This enables users to re-run each single modular component to reproduce the result in case of any error, without the need of re-running RadText’s entire pipeline. All intermediate results are saved so that users can easily check the output of each module, which we believe will greatly facilitate error analysis and enhance RadText’s flexibility. The following code snippet shows a part of RadText’s pipeline.
TABLE III.
Command line arguments.
| Argument | Description |
|---|---|
| csv2bio | Transforms .csv text file into a BioC-format XML file. |
| deid | De-identifies all the reports. |
| split_section | Segments sections. |
| preprocess | Splits sentences and tokenizes words. |
| ner | Recognizes named entities. |
| parse | Parses to obtain the universal dependency graph. |
| neg | Detects negations. |
| collect_label | Collects and merges labels. |


B. Python API Usage
RadText can be directly imported as a Python library. Users can access all the functionalities of RadText through Python API.
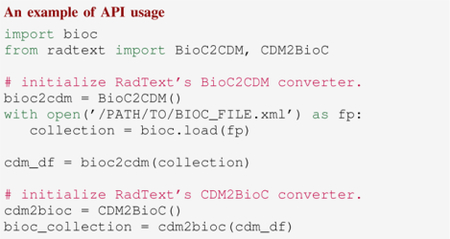
1). BioC-CDM Conversion:
RadText’s Python API supports the mutual conversion between BioC format and OMOP CDM. The following code snippet shows an example of converting BioC format to CDM and then converting CDM back to BioC format.
2). Pipeline Usage:
The following code snippet shows a minimal usage of RadText’s entire pipeline through Python API, which annotates a sample report and printing out all annotation results.

After running all modules, RadText returns a Collection instance that stores the final annotation results. Within a Collection instance, the annotations are stored in either Passage or Sentence classes. The following code snippet shows how we can access the detected disease findings and the corresponding negation status after obtaining the Collection instance.
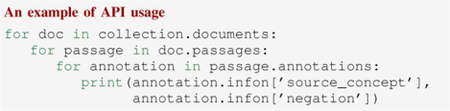
RadText’s Python API also allows partial pipeline execution. Therefore, users can pause after any module of RadText to access the intermediate NLP results. Since each module requires the output of the previous module, RadText’s API will automatically run every module until it finishes running the specified module.
The following code snippet shows an example of the partial execution of RadText. By specifying the annotator to be split_section, RadText will run de-identification and section segmentation sequentially. The output Collection instance will have the annotation results of section segmentation.

V. Evaluation
A. Dataset
We evaluated RadText on the MIMIC-CXR dataset [23]. MIMIC-CXR is a large publicly available dataset of radiographic studies performed at the Beth Israel Deaconess Medical Center. This dataset contains 227,827 radiology reports in total.
B. Experiments and Results
We evaluated RadText’s performance on five new disease findings that were not covered by previous works, including Calcification of the Aorta, Pneumomediastinum, Pneumoperitoneum, Subcutaneous Emphysema, Tortuous Aorta. We randomly selected 200 test reports from the MIMIC-CXR dataset and manually annotated the five new disease findings. We evaluated RadText by comparing the results of RadText with the manually-annotated gold standard. Precision, recall, and F1-score were computed accordingly based on the number of true positives, false positives and false negatives (see Table IV). The average precision score is 0.91, with the highest precision being 1.0 for Calcification of the Aorta and Tortuous Aorta; the average recall score is 0.94, with the highest recall being 1.0 for Pneumomediastinum and Pneumoperitoneum; and the average F-1 score is 0.92, with the highest F-1 score being 0.97 for Tortuous Aorta. RadText achieves an average precision of 0.91, an average recall of 0.94, and an average F-1 score of 0.92.
TABLE IV.
RadText performances on five new disease findings.
| Disease Finding | Precision | Recall | F-1 |
|---|---|---|---|
| Calcification of the Aorta | 1.00 | 0.87 | 0.93 |
| Pneumomediastinum | 0.70 | 1.00 | 0.82 |
| Pneumoperitoneum | 0.88 | 1.00 | 0.94 |
| Subcutaneous Emphysema | 0.95 | 0.91 | 0.93 |
| Tortuous Aorta | 1.00 | 0.94 | 0.97 |
| Macro Average | 0.91 | 0.94 | 0.92 |
VI. Conclusion and Future Work
In this work, we presented RadText, a high-performance Python radiology text analysis system. We highlighted that RadText features hybrid neural analysis, raw text processing and local processing, bringing higher accuracy, better usability and improved data privacy. RadText’s user-friendly user interface, easy-to-use command line usage and Python API allow users to have great flexibility on the radiology text analysis task. We evaluated RadText on the MIMIC-CXR dataset, especially on five new disease findings that were not covered by previous work, and the results demonstrated RadText’s superior performances on radiology report analysis. RadText employs BioC-CDM, which stores the results in the extended BioC format that is compatible with OMOP CDM. RadText’ compatibility with OMOP CDM supports collaborative research across disparate data sources.
In the future, RadText is going to be continuously maintained and expanded as new resources become available. For example, the NER module can be improved by incorporating scispaCy, developed for processing biomedical, scientific or clinical text [3]. By making RadText publicly available, we envision it can facilitate future research and applications in the healthcare informatics community.
Acknowledgment
This work is supported by the National Library of Medicine under Award No. 4R00LM013001 and the NIH Intramural Research Program, National Library of Medicine.
References
- [1].Brady, “Error and discrepancy in radiology: inevitable or avoidable?” Insights into Imaging, vol. 8, 12 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Savova G, Masanz J, Ogren P, Zheng J, Sohn S, Kipper-Schuler K, and Chute C, “Mayo clinical text analysis and knowledge extraction system (ctakes): Architecture, component evaluation and applications,” Journal of the American Medical Informatics Association : JAMIA, vol. 17, pp. 507–13, 09 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Neumann M, King D, Beltagy I, and Ammar W, “ScispaCy: Fast and Robust Models for Biomedical Natural Language Processing,” in Proceedings of the 18th BioNLP Workshop and Shared Task. Florence, Italy: Association for Computational Linguistics, Aug. 2019, pp. 319–327. [Online]. Available: https://www.aclweb.org/anthology/W19-5034 [Google Scholar]
- [4].Liu H, Bielinski SJ, Sohn S, Murphy S, Wagholikar KB, Jonnalagadda SR, Ravikumar K, Wu ST, Kullo IJ, and Chute CG, “An information extraction framework for cohort identification using electronic health records,” AMIA Joint Summits on Translational Science proceedings. AMIA Joint Summits on Translational Science, vol. 2013, p. 149–153, 2013. [Online]. Available: https://europepmc.org/articles/PMC3845757 [PMC free article] [PubMed] [Google Scholar]
- [5].Soysal E, Wang J, Jiang M, Wu Y, Pakhomov S, Liu H, and Xu H, “CLAMP – a toolkit for efficiently building customized clinical natural language processing pipelines,” Journal of the American Medical Informatics Association, vol. 25, no. 3, pp. 331–336, 11 2017. [Online]. Available: 10.1093/jamia/ocx132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bodenreider O, “The Unified Medical Language System (UMLS): Integrating biomedical terminology,” Nucleic Acids Research, vol. 32, no. Database issue, pp. D267–270, Jan. 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Langlotz CP, “RadLex: A new method for indexing online educational materials,” Radiographics: A Review Publication of the Radiological Society of North America, Inc, vol. 26, no. 6, pp. 1595–1597, 2006. Nov-Dec. [DOI] [PubMed] [Google Scholar]
- [8].Chapman WW, Hillert D, Velupillai S, Kvist M, Skeppstedt M, Chapman E, Conway M, Tharp M, Mowery DL, and Deleger L, “Extending the NegEx lexicon for multiple languages.” Studies in health technology and informatics, vol. 192, pp. 677–681, 2013. [PMC free article] [PubMed] [Google Scholar]
- [9].Chapman BE, Lee S, Kang HP, and Chapman WW, “Document-level classification of CT pulmonary angiography reports based on an extension of the ConText algorithm,” Journal of Biomedical Informatics, vol. 44, no. 5, pp. 728–737, Oct. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Fan J.-w., Yang EW, Jiang M, Prasad R, Loomis RM, Zisook DS, Denny JC, Xu H, and Huang Y, “Syntactic parsing of clinical text: Guideline and corpus development with handling ill-formed sentences,” Journal of the American Medical Informatics Association: JAMIA, vol. 20, no. 6, pp. 1168–1177, 2013. Nov-Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Peng Y, Wang X, Lu L, Bagheri M, Summers R, and Lu Z, “Negbio: a high-performance tool for negation and uncertainty detection in radiology reports,” in AMIA Joint Summits on Translational Science proceedings. AMIA Joint Summits on Translational Science, 2017, pp. 188–196. [PMC free article] [PubMed] [Google Scholar]
- [12].Voss E, Makadia R, Matcho A, Ma Q, Knoll C, Schuemie M, Defalco F, Londhe A, Zhu V, and Ryan P, “Feasibility and utility of applications of the common data model to multiple, disparate observational health databases,” Journal of the American Medical Informatics Association : JAMIA, vol. 22, 02 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Comeau D, Dogan R, Ciccarese P, Cohen K, Krallinger M, Leitner F, lu Z, Peng Y, Rinaldi F, Torii M, Valencia A, Verspoor K, Wiegers T, Wu C, and Wilbur W, “Bioc: a minimalist approach to interoperability for biomedical text processing,” Database : the journal of biological databases and curation, vol. 2013, p. bat064, 01 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Pons E, Braun LMM, Hunink MGM, and Kors JA, “Natural Language Processing in Radiology: A Systematic Review,” Radiology, vol. 279, no. 2, pp. 329–343, May 2016. [DOI] [PubMed] [Google Scholar]
- [15].Aronson A and Lang F-M, “An overview of metamap: Historical perspective and recent advances,” Journal of the American Medical Informatics Association : JAMIA, vol. 17, pp. 229–36, 05 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bodenreider O, “The unified medical language system (umls): Integrating biomedical terminology,” Nucleic acids research, vol. 32, pp. D267–70, 02 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Eyre H, Chapman AB, Peterson KS, Shi J, Alba PR, Jones MM, Box TL, DuVall SL, and Patterson OV, “Launching into clinical space with medspacy: a new clinical text processing toolkit in python,” in AMIA Annual Symposium Proceedings 2021, 2021. [Online]. Available: http://arxiv.org/abs/2106.07799 [PMC free article] [PubMed] [Google Scholar]
- [18].Honnibal M, Montani I, Van Landeghem S, and Boyd A, “spacy: Industrial-strength natural language processing in python,” 2020.
- [19].Norgeot B, Muenzen K, Peterson TA, Fan X, Glicksberg BS, Schenk G, Rutenberg E, Oskotsky B, Sirota M, Yazdany J, Schmajuk G, Ludwig D, Goldstein T, and Butte AJ, “Protected health information filter (philter): accurately and securely de-identifying free-text clinical notes,” NPJ Digital Medicine, vol. 3, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].O. f. C. Rights (OCR), “Guidance Regarding Methods for De-identification of Protected Health Information in Accordance with the Health Insurance Portability and Accountability Act (HIPAA) Privacy Rule,” https://www.hhs.gov/hipaa/for-professionals/privacy/special-topics/de-identification/index.html, Sep. 2012.
- [21].Tepper M, Capurro D, Xia F, Vanderwende L, and Yetisgen-Yildiz M, “Statistical Section Segmentation in Free-Text Clinical Records,” in Proceedings of the Eighth International Conference on Language Resources and Evaluation (LREC’12). Istanbul, Turkey: European Language Resources Association (ELRA), May 2012, pp. 2001–2008. [Google Scholar]
- [22].Wang X, Peng Y, Lu L, Lu Z, Bagheri M, and Summers RM, “Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases,” 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 3462–3471, 2017. [Google Scholar]
- [23].Johnson A, Pollard T, Berkowitz S, Greenbaum N, Lungren M, Deng C.-y., Mark R, and Horng S, “Mimic-cxr, a de-identified publicly available database of chest radiographs with free-text reports,” Scientific Data, vol. 6, p. 317, 12 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Denny JC, Miller RA, Johnson KB, and Spickard A, “Development and evaluation of a clinical note section header terminology,” in AMIA Annual Symposium proceedings. AMIA Symposium, 2008, pp. 156–160. [PMC free article] [PubMed] [Google Scholar]
- [25].McDonald CJ, Huff SM, Suico JG, Hill G, Leavelle D, Aller R, Forrey A, Mercer K, DeMoor G, Hook J, Williams W, Case J, and Maloney P, “LOINC, a universal standard for identifying laboratory observations: A 5-year update,” Clinical Chemistry, vol. 49, no. 4, pp. 624–633, Apr. 2003. [DOI] [PubMed] [Google Scholar]
- [26].Miller RA and Masarie FE, “Use of the Quick Medical Reference (QMR) program as a tool for medical education,” Methods of Information in Medicine, vol. 28, no. 4, pp. 340–345, Nov. 1989. [PubMed] [Google Scholar]
- [27].Bird S, Klein E, and Loper E, Natural Language Processing with Python, 1st ed. O’Reilly Media, Inc., 2009. [Google Scholar]
- [28].Qi P, Zhang Y, Zhang Y, Bolton J, and Manning CD, “Stanza: A Python natural language processing toolkit for many human languages,” in Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics: System Demonstrations, 2020. [Online]. Available: https://nlp.stanford.edu/pubs/qi2020stanza.pdf [Google Scholar]
- [29].Honnibal M and Johnson M, “An improved non-monotonic transition system for dependency parsing,” in Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing. Lisbon, Portugal: Association for Computational Linguistics, Sep. 2015, pp. 1373–1378. [Online]. Available: https://aclanthology.org/D15-1162 [Google Scholar]
- [30].Nadeau D and Sekine S, “A survey of named entity recognition and classification,” Lingvisticae Investigationes, vol. 30, 08 2007. [Google Scholar]
- [31].Charniak E and Johnson M, “Coarse-to-fine n-best parsing and MaxEnt discriminative reranking,” in Proceedings of the 43rd Annual Meeting of the Association for Computational Linguistics (ACL’05). Ann Arbor, Michigan: Association for Computational Linguistics, Jun. 2005, pp. 173–180. [Online]. Available: https://aclanthology.org/P05-1022 [Google Scholar]
- [32].Charniak E and McClosky D, “Any domain parsing: automatic domain adaptation for natural language parsing,” 2010.
- [33].de Marneffe M-C, Dozat T, Silveira N, Haverinen K, Ginter F, Nivre J, and Manning CD, “Universal Stanford dependencies: A cross-linguistic typology,” in Proceedings of the Ninth International Conference on Language Resources and Evaluation (LREC’14). Reykjavik, Iceland: European Language Resources Association (ELRA), May 2014, pp. 4585–4592. [Google Scholar]
- [34].Marneffe M-C and Manning C, “The stanford typed dependencies representation,” COLING Workshop on Cross-framework and Cross-domain Parser Evaluation, 01 2008. [Google Scholar]
- [35].Chapman WW, Bridewell W, Hanbury P, Cooper GF, and Buchanan BG, “Evaluation of negation phrases in narrative clinical reports,” Proceedings. AMIA Symposium, pp. 105–9, 2001. [PMC free article] [PubMed] [Google Scholar]


