Abstract
Sec20p is a component of the yeast Saccharomyces cerevisiae secretory pathway that does not have a close homolog in higher eukaryotic cells. To verify the function of Sec20p in other fungal species, we characterized the gene encoding a Sec20p homolog in the human fungal pathogen Candida albicans. The deduced protein has 27% identity with, but is missing about 100 N-terminal residues compared to S. cerevisiae Sec20p, which is part of the cytoplasmic tail interacting with the cytoplasmic protein Tip20p. Because a strain lacking both C. albicans SEC20 alleles could not be constructed, we placed SEC20 under transcriptional control of two regulatable promoters, MET3p and PCK1p. Repression of SEC20 expression in these strains prevented (MET3p-SEC20 allele) or retarded (PCK1p-SEC20 allele) growth and led to the appearance of extensive intracellular membranes, which frequently formed stacks. Reduced SEC20 expression in the PCK1p-SEC20 strain did not affect morphogenesis but led to a series of hypersensitivity phenotypes including supersensitivity to aminoglycoside antibiotics, to nystatin, to sodium dodecyl sulfate, and to cell wall inhibitors. These results demonstrate the occurrence and function of Sec20p in a fungal species other than S. cerevisiae, but the lack of the N-terminal domain and the apparent absence of a close TIP20 homolog in the C. albicans genome also indicate a considerable diversity in mechanisms of retrograde vesicle traffic in eukaryotes.
The fungal pathogen Candida albicans is able to effectively infect, survive within, and colonize the human host. Properties of its fungal cellular surface, which allow C. albicans to interact with host cells by adhering to and by invading cells and tissues, are essential for its virulence (6). Compositional changes of the cellular surface are associated with several cellular morphologies related to virulence, including the interconversion between a filamentous and a yeast growth form (dimorphism) and spontaneous phenotypic switching (reviewed in reference 12). Soluble secreted fungal proteins including proteases and lipases also regulate the course of infection (16, 18). Thus, an effective secretory machinery determining secretion of cellular surface and media proteins is pivotal for the role of C. albicans as the most successful human fungal pathogen.
Many of the details of eukaryotic secretion have been derived from studies of the apathogenic yeast Saccharomyces cerevisiae, and it has been shown that mechanisms of vesicular traffic are very similar in yeast and higher eukaryotic cells (17, 30). It has been shown that vesicles arising on a donor membrane contain v-SNARE membrane proteins, which recognize corresponding receptors, t-SNAREs, on target membranes (36, 41). In yeasts, endoplasmic reticulum (ER)-derived secretory vesicles carry a group of v-SNAREs, including Sec22p, which interact with the Golgi t-SNARE Sed5p (2, 40). Unexpectedly, Sec22p also acts as a v-SNARE in retrograde transport of Golgi-derived vesicles, whose function is the retrieval of proteins from Golgi compartments to the ER (21). Retrograde Golgi-ER transport requires Ufe1p as the t-SNARE in ER membranes and interacting proteins other than Sec22p, including the membrane protein Sec20p and a cytoplasmic protein, Tip20p (1, 9, 20, 21, 43, 44). Sec20p is a type II membrane protein containing a relatively small C-terminal lumenal domain of 91 amino acids ending in an HDEL motif, which is characteristic of soluble ER proteins (32, 43). Presence of the HDEL sequence on Sec20p is not essential, but it helps to maintain intracellular levels of Sec20p. Depletion of Sec20p leads to the accumulation of membranous structures and vesicles between the ER and the cytoplasmic membrane in the cell. A similar phenotype is found in tip20 mutants, which lack the Tip20 protein binding to the N-terminal (cytoplasmic) domain of Sec20p (14, 44). Sec20p is in the same membrane as Ufe1p and can be thought of as a partner t-SNARE, analogous to SNAP25 and syntaxin in the synapse (15, 41). However, Sec20p is unique among yeast secretory components because a structural homolog is not known to exist in mammalian cells; thus, Sec20p may be specific for fungi.
Some C. albicans homologs of essential S. cerevisiae secretion pathway genes have been characterized as able to complement the corresponding S. cerevisiae mutations and were found to be essential for C. albicans viability. Such genes include SEC18 (29), SEC14 (27, 35), and SEC4 (8, 26). Little is known about the functions of their gene products, although the accumulation of secretory vesicles in cells producing a dominant-negative version of Sec4p demonstrated a function of this protein in C. albicans analogous to its function in S. cerevisiae. Here we have characterized the C. albicans homolog to the S. cerevisiae Sec20 protein (ScSec20p). We show that the C. albicans SEC20 gene is essential and that depletion of Sec20p from cells leads to the accumulation of membranous structures as in S. cerevisiae. However, C. albicans Sec20p (CaSec20p) is not a close homolog of ScSec20p, because it lacks an extensive amino-terminal region and C. albicans SEC20 is not able to complement an S. cerevisiae sec20 mutant. We speculate that this lack of function may be related to the absence of a Tip20p homolog in C. albicans. Furthermore, because of the absence of Sec20p homologs in mammalian cells, we propose that Sec20p could constitute a target for future antifungal agents.
MATERIALS AND METHODS
Strains and growth conditions.
Yeast strains (Table 1) were grown in YPD or YP (YPD without glucose) medium or on supplemented SD minimal medium at 30°C (38). S. cerevisiae RSY275 (sec20-1) was cultured at 25°C and shifted to the nonpermissive temperature 37°C (17). Transformation of C. albicans was carried out by the spheroplast method (38). The PCK1 promoter (PCK1p) was induced in SCAA medium (SD medium with 2% Casamino Acids) and repressed in S4D medium (SD medium with 4% glucose) (19). To repress the MET3 promoter (MET3p), SDMC medium (SD medium containing 2.5 mM methionine and cysteine) (5) was used.
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Genotype or description | Reference |
|---|---|---|
| C. albicans strains | ||
| 3153 | Prototrophic | 45 |
| SC5314 | Prototrophic | 13 |
| CAI4 | ura3Δ::imm434/ura3Δ::imm434 | 13 |
| CA2 | As CAI4, but SEC20/sec20Δ::hisG-URA3-hisG | This work |
| CA10 | As CA2 | This work |
| CA2d | As CAI4, but SEC20/sec20Δ::hisG | This work |
| CA10a | As CA2d | This work |
| CAr2d110 | As CAI4, but sec20Δ::hisG/PCK1p-SEC20 | This work |
| CAr10a3 | As CAr2d110 | This work |
| CA2d1m | As CAI4, but sec20Δ::hisG/MET3p-SEC20 | This work |
| CA2d2m | As CA2d1m | This work |
| S. cerevisiae strain RSY275 | MATα sec20-1 ura3-52 his4-619 | 17 |
| S. cerevisiae transformation vectors | ||
| STM20 | URA3-marked 2 μm vector with TPI1p-SEC20 | 43 |
| pYW9 | URA3-marked CEN vector with PFK2p-SEC20 | This work |
| pYW10 | URA3-marked 2 μm vector with PFK2-SEC20 | This work |
| pYW11 | URA3-marked CEN vector with SEC20 | This work |
| C. albicans vectors | ||
| pBI-1 | PCK1 promoter in pRC2312 | 42 |
| pCaDis | URA3-marked vector with MET3 promoter | 5 |
| pYW7 | PCK1p-SEC20 fusion in pBI-1 | This work |
| pYW20 | MET3p-SEC20Δ fusion in pCaDis | This work |
To induce hyphal growth, 10 ml of an overnight culture was used to inoculate 100 ml of prewarmed YPD containing 10% bovine calf serum at 37°C. Control cultures were grown in YPD at 25°C with or without serum or at 37°C without serum. Alternatively, morphogenesis of C. albicans was regulated by different pH values as described elsewhere (3). The yeast form was grown at pH 4.5 and 25°C to the late exponential growth phase, and the hyphal form was induced at 37°C and pH 6.5. Control cultures were grown at 25°C and pH 4.5 or pH 6.5 or at 37°C and pH 4.5. Strain CAr2d110 (PCK1p-SEC20) was induced to form hyphae in S4D medium containing 5% horse serum at 37°C.
Isolation of SEC20.
To isolate the C. albicans SEC20 gene, a fosmid library (25) was screened with a 1.018-kb cDNA fragment of SEC20 (45). A 4.2-kb HindIII fragment of fosmid 14H1 was identified to contain SEC20 and was subcloned. A first subclone, pYW1S, was constructed containing a 2.321-kb SalI fragment inserted in pUC19. A second subclone (pYW2), in which SEC20 was completed, was constructed by inserting a 889-bp HindIII-XhoI fragment into pYW1S (HindIII/XhoI fragment). pYW2 contains the SEC20 coding region flanked by 1.42 and 275 bp of 5′ and 3′ untranslated regions, respectively. The 1.811-kb SEC20 region indicated in Fig. 1 was sequenced in both directions.
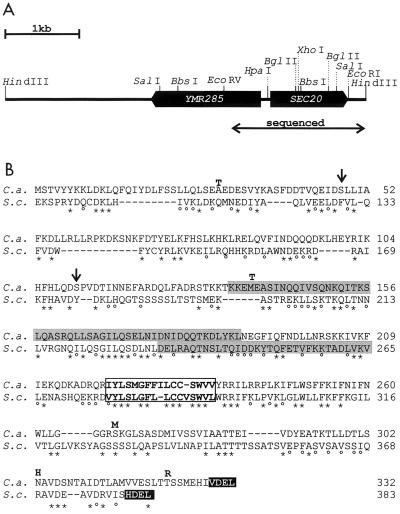
FIG. 1.
Sequences of SEC20 and the deduced protein. (A) Genomic configuration of the divergent SEC20 and YMR285 genes. (B) Comparison of C. albicans and S. cerevisiae Sec20 proteins (C.a. and S.c.) indicating identical (∗) and similar (°) residues. The framed region represents a transmembrane region, while regions predicted to adopt coiled-coil conformations are indicated by grey boxes. Arrows indicate the positions of serine residues encoded by the nonstandard CUG codon. Bold letters above the C. albicans sequence indicate residues present in the protein encoded by the cDNA of strain 3153 which do not exist in genomic DNA of strain 1161.
Overexpression of SEC20.
To overexpress SEC20 in C. albicans, we used pBI-1, a derivative of pRC2312 (42) containing a 1.5-kb BamHI-BglII promoter fragment of the C. albicans PCK1 gene (19). Into the BglII site downstream of PCK1p, the entire SEC20 coding region was cloned as a 1.072-kb BamHI fragment, which was generated by PCR using primers N-ATG-Bam (5′-CAGCGGATCCAATGTCAACAG-3′) (ATG start sequence underlined) and C-Stop-Bam (5′AACAACGGATCCAATGTCAAAG3′) (situated 70 bp downstream of the translational stop sequence), thereby generating plasmid pYW7. The PCR-generated SEC20 fragment was inserted into pUC18 (resulting in plasmid pYW1) and was verified by sequencing. The BamHI-EcoRI PCK1p-SEC20 fragment of pYW7 was also inserted into pUC18 to generate pYW6.
Gene disruptions.
SEC20 was deleted according to the Ura-blaster protocol (13). In preparation for disruption of SEC20, pYW2 was cut with BglII, and the 4-kb BamHI/BglII Ura-blaster fragment of plasmid p5921 was substituted for part of the SEC20 coding region. A 6.079-kb BbsI-HindIII disruption fragment was isolated from the resulting plasmid, pYW01, and used to transform strain CAI4 to uridine prototrophy.
A more extensive SEC20 deletion was constructed by first ligating the 4-kb SacI (filled in)/BamHI Ura-blaster fragment of plasmid p5921 to the large HpaI-BglII fragment of pYW2. A 5.727-kb BbsI-HindII fragment was isolated from the resulting plasmid, pYW02, and used to transform strain CAI4 to prototrophy. Correct insertion of the Ura-blaster fragment into SEC20 alleles was verified by PCR and Southern blot analyses of DNA of transformants, which was cut with SalI and probed with the 855-bp SalI/EcoRV SEC20 upstream fragment.
Transformants containing the sec20-Δ2 deletion derived from plasmid pYW01 were named CA2, and strains containing the sec20-Δ10 deletion derived from pYW02 were designated CA10. Heterozygous sec20Δ::hisG-URA3-hisG/SEC20 strains were plated on medium containing 0.02% 5-fluoroorotic acid (FOA), and FOA-resistant strains that had lost the URA3 sequence were analyzed by Southern blotting. Derivatives of CA2 and CA10, designated CA2d and CA10a (genotype sec20Δ::hisG/SEC20), were used for a second round of gene disruption using the original Ura-blaster disruption fragments. Although several transformants were tested by PCR or Southern blottings, none was found to have the sec20Δ::hisG/sec20Δ::hisG-URA3-hisG phenotype.
To replace the SEC20 promoter in the remaining wild-type SEC20 allele by the regulatable PCK1p (42), a plasmid carrying an appropriate disruption fragment was constructed. A 1.798-kb BamHI/BglII fragment of pYW2 containing SEC20 promoter sequences was inserted into the BamHI site of plasmid p1367/1 (containing URA3 23) to create plasmid pYW3. A 3.161-kb SmaI/PstI fragment of this plasmid was fused 5′ to the PCK1p-SEC20 fusion fragment (HindIII [filled in]-to-PstI fragment of pYW6) to construct pYW4. The 4.775-kb BbsI fragment of pYW4 was isolated and used to transform strains CA2d and CA10a to uridine prototrophy. The resulting strains were designated CAr10a3, CAr10a7, CAr2d110, and CAr2d107.
To replace the SEC20 promoter in one of the SEC20 alleles by the regulatable MET3p (5) a plasmid carrying an appropriate disruption fragment was constructed. First, the N-terminal region of the SEC20 coding region was amplified by PCR, using the primers N-ATG-Bam and C.Sec20/Met3-Bam530 (5′-GTTGTCAGGATCCAATTCACTCTGC-3′), by which BamHI sites (underlined) were placed 1 bp upstream of the ATG start codon and at position 516 bp of the SEC20 coding region. The BamHI PCR fragment was placed downstream of MET3p in plasmid pCaDis (5). The resulting plasmid, pYW20, was cut with BglII within the SEC20 coding region and used to transform strain CA2d to uridine prototrophy. Transformed strains were designated CA2d1m, CA2d2m, and CA2d6m.
Correct insertion of all disruption fragments was verified by Southern blotting of DNA transformants, which was cut with SalI and probed with an 855-bp SalI/EcoRV fragment derived from SEC20 upstream sequences.
RT-PCR.
Total RNA of C. albicans strains was isolated as described elsewhere (42) and treated with DNase I as instructed by the manufacturer (Gibco BRL). Reverse transcription-PCR (RT-PCR) was performed in a one-step reaction using an enhanced avian RT-PCR kit (Sigma). The reaction mixture was made up to a final volume of 50 μl with SEC20-specific primers C.Sec20/RT-100 (5′-CTGTCTACAAGGCCAGCTTTG-3′) and C.Sec20/RT-420 (5′-CTGCAATATACCTGCTGATAAC-3′) or EFB1-specific primers C.EFB/RT-70 (5′-ATTGAACGAATTCTTGGCTGAC-3′) and C.EFB/RT-970 (5′-CATCTTCAACAGCAGCTTG-3′) (37). cDNA synthesis was carried out at 45°C for 45 min. After the reaction was stopped by heating for 3 min at 94°C, 30 amplification cycles (94°C, 45 s; 56°C, 40 s; 72°C, 45 s) were conducted.
S. cerevisiae complementation tests.
Complementation tests were performed with different expression plasmids containing SEC20. The BamHI fragment of pYW1 was inserted into the BamHI site of pJJH70 (cen4, 2 μm ori) or pJJH461r (2 μm ori) to place SEC20 under transcriptional control of the S. cerevisiae PFK2p (33). The resulting plasmids were named pYW9 and pYW10. By insertion of the 2.482-kb BamHI-EcoRI fragment of pYW2 into YCplac33 (BamHI/EcoRI), a plasmid (pYW11) which contains the authentic SEC20 genomic promoter and coding regions was created. Plasmid STM20 contains the S. cerevisiae SEC20 gene under control of the TPI promoter (43). Complementation tests were performed with S. cerevisiae strain RSY275 (43), kindly provided by M. J. Lewis.
Quantitation of SEC20 transcript.
A radioactive SEC20 RNA probe was synthesized according to a commercial protocol (Promega), using 1 μg of PvuII-cut plasmid 1-8-1 (SEC20 cDNA [45]) as the template. RNA was synthesized in the presence of [32P]UTP, using T7 RNA polymerase. After phenol-chloroform purification, the probe was hybridized to the filters obtained in Northern blottings at 50°C [50% formamide, 50 mM sodium phosphate (pH 6.5), 0.8 M NaCl, 1 mM EDTA, 0.1% sodium dodecyl sulfate (SDS), 2.5× Denhardt's solution, salmon sperm DNA (250 μg/ml), poly(A) (10 μg/ml), yeast RNA (500 μg/ml)]. The filters were washed with 2× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7])–0.1% SDS at 60°C and if necessary with 50 mM NaCl–20 mM sodium phosphate (pH 6.5)–1 mM EDTA–0.1% SDS at 60°C. The signals were quantified directly by two-dimensional radioimaging with an AMBIS radioanalytic system (LabLogic, Sheffield, England) as described elsewhere (46).
Electron microscopy.
C. albicans strain CAr2d110 (sec20-Δ1::hisG/PCK1p-SEC20) was incubated in PCK1p-repressing minimal medium containing 4% glucose to logarithmic growth; the cells were harvested, fixed in 2.5% glutaraldehyde for 2 h, washed in buffer, and postfixed in 1% OsO4 in 0.1 M cacodylate buffer for 1 h. The material was dehydrated through a graded ethanol series and embedded in Epon 812 with the aid of epoxypropane as a linking agent. The resin mixture was equivalent to the 3A:1B formula of Luft (24), with catalyst present at all stages of infiltration. Sections were double stained in 2% aqueous uranyl acetate followed by Reynold's lead citrate. The preparations were examined in a Philips model 301 electron microscope.
C. albicans strain CA2d1m (sec20-Δ1::hisG/MET3p-SEC20) was incubated in MET3p-repressing minimal medium containing 2.5 mM methionine and cysteine to logarithmic growth; the cells were harvested, fixed in fresh 4% aqueous KMnO4, and incubated for 2 h at 4°C. Fixed cells were dehydrated in a graded series of ethanol and embedded in Epon 812. Sections were stained with Reynold's lead citrate for 15 min to increase the contrast of the membranes.
Nucleotide sequence accession number.
The sequence reported has been assigned DDBJ/EMBL/GenBank accession number AJ272499.
RESULTS
Isolation and sequence analysis of the C. albicans SEC20 gene.
A set of cDNAs of C. albicans strain 3153 that encode C. albicans proteins reacting with human sera was isolated previously (45). Among these clones, a 1,018-bp cDNA, 1-8-1, with homology to S. cerevisiae SEC20 was used for screening a fosmid gene library of strain 1161 (7). Fosmid 14H1, which is derived from chromosome 6, was found to contain SEC20, and the sequence of both strands of a 1.811-kb fosmid fragment was determined (DDBJ/EMBL/GenBank nucleotide sequence accession no. AJ272499). The sequence revealed a 996-bp intronless open reading frame (ORF) between nucleotide positions 556 and 1551 with the potential to encode a protein of 332 amino acids (36 kDa) with 27% identity to ScSec20p (43) (Fig. 1). Compared to ScSec20p, the hypothetical CaSec20p is N-terminally shortened by about 100 amino acids. CaSec20p is likely a type II integral membrane protein, as is ScSec20, with a possible transmembrane region situated between residues 220 and 234. The cytoplasmic domain is predicted to assume a coiled-coil structure; because of its presumed cytoplasmic localization, a potential N-linked glycosylation site at asparagine residue 199 is unlikely to be modified. The C-terminal VDEL sequence of CaSec20p has striking similarity to the HDEL ER retention signal of S. cerevisiae (32).
Genomic sequences encompassing the SEC20 region have been determined in the C. albicans genomic sequencing proj- ect (http://www-sequence.stanford.edu/group/candida/search.html). These sequences, which are derived from strain SC5314, show only two different nucleotides within the SEC20 coding region compared to the fosmid sequences of strain 1161. The SEC20 cDNA clone 1-8-1, on the other hand, contains 17 nucleotide differences compared to the fosmid sequence, of which 5 alter the protein sequence (Fig. 1). Thus, there is considerable divergence of SEC20 sequences between strains 3153 and strain 1161 or SC5314. The 5′ untranslated region of SEC20 contains three putative TATA sequences in positions −531 to −527, −325 to −321, and −265 to −261 relative to the ATG translational start sequence. Remarkably, an ORF divergently oriented to SEC20 commences only 138 bp upstream of the SEC20 translated region; the putative translation product of this ORF has 519 residues, which are 39 and 34% identical compared to the S. cerevisiae proteins encoded by ORFs YMR285 and YML118W, respectively, whose functions are unknown. Thus, the 138-bp SEC20 5′ untranslated region appears to contain two divergently oriented promoters. The 3′ untranslated region contains sequences identical to S. cerevisiae consensus sequences for efficient transcription termination and polyadenylation. Putative TTTTTATA termination sequences are located at positions +2, +41, and +144 following the SEC20 stop codon.
SEC20 is essential for growth.
To explore the function of SEC20, we generated strains in which individual SEC20 alleles were partially or entirely deleted. In the first approach, we generated deletion strains lacking the SEC20 coding region (deletions Δ2 and Δ10 [Fig. 2A]) by transformation with disruption fragments containing the Ura-blaster cassette (13). Two types of deletions were made, because the function of the divergently oriented YMR285 ORF had to be considered. In strains carrying deletion Δ10, the entire SEC20 coding region including 40 bp of its 5′ untranslated region was deleted; thus, the putative promoter region for ORF YMR285 was reduced to 98 bp. Deletion Δ2 maintains the entire intergenic region and still contains 330 bp of the SEC20 ORF, which we assumed would suffice for YMR285 promoter activity.
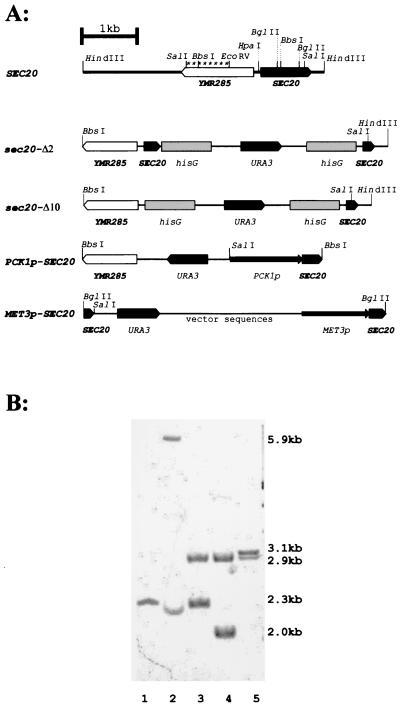
FIG. 2.
Disruption of SEC20 alleles. (A) Genomic structures of wild-type SEC20 and its disrupted derivative alleles. The Ura-blaster module (hisG-URA3-hisG) was used to disrupt SEC20 alleles partially (sec20-Δ2 and sec20-Δ10). In addition, the SEC20 coding region was placed under transcriptional control of PCK1p (PCK1p-SEC20) or MET3p (MET3p-SEC20). The SalI sites within the YMR285 gene and the disruption fragments which generate specific SalI fragments in Southern blottings of genomic DNA (B) are shown in boldface. Strains analyzed were CAI4 (lane 1), CA2 (lane 2), CA2d (lane 3), CA2d2m (lane 4), and CAr2d110 (lane 5). The probe used in the Southern blotting experiment (B) is indicated by asterisks (A). Relevant fragment sizes are indicated (B; see text).
Heterozygous strains carrying one wild-type plus one deleted allele of SEC20 (deletion Δ2 or Δ10) could be constructed without difficulty and were verified by Southern blotting (Fig. 2) and by PCR using gene-specific primers (data not shown). In Southern blotting of SalI-digested genomic DNA, the wild-type SEC20 allele yielded a 2.3-kb band (strain CAI4), while the sec20-Δ2::hisG-URA3-hisG allele generated a 5.9-kb band (strain CA2) (Fig. 2B). After FOA selection, we identified derivative strains containing the sec20-Δ2::hisG allele detectable as a 2.9-kb band (strain CA2d). Identical Southern results were obtained in transformants containing heterozygous strains containing the deleted sec20-Δ10 allele (data not shown). Surprisingly, attempts to delete the remaining wild-type SEC20 allele failed in strains carrying both types of deletions. Sixty-five transformants of strain CA2d transformed with the sec20-Δ2::Ura-blaster disruption fragment still retained at least one wild-type allele of SEC20. Likewise, 33 transformants obtained with the sec20-Δ10::Ura-blaster disruption fragment still contained an intact SEC20 allele. Thus, a complete sec20 null mutant was not obtained, suggesting that SEC20 is essential for viability.
To prove that cell growth depended on SEC20, we replaced the promoter region of the wild-type SEC20 allele remaining in the heterozygous strain CA2d by either the glucose-repressible PCK1p (19) or the methionine-repressed MET3p (5). Strain CAr2d110 (sec20Δ2::hisG/PCK1p-SEC20) was generated using a disruption cassette containing URA3 joined to PCK1p; strain CA2d2m (sec20Δ2::hisG/MET3p-SEC20) was constructed by integration of plasmid pYW20 (see Materials and Methods). The second disruption step led to disappearance of the remaining wild-type copy and to a new 3.1-kb fragment in CA2d110, or a 2.0-kb fragment in strain CA2d2m, as expected (Fig. 2B).
Growth of strain CAr2d110 containing SEC20 under control of PCK1p was identical to growth of nondisrupted control strains in media containing low concentrations of glucose (generation time of 90 min in SCAA medium). On the other hand, growth in S4D high-glucose medium significantly retarded growth of strain CAr2d110 (generation time, 120 min) compared to control strains (generation time, 90 min). This result suggested that SEC20 was essential for growth, but it was unclear if the SEC20 transcript was still present in this strain. Therefore, we examined the SEC20 transcript level in a control strain (SEC20/SEC20) and in the conditional PCK1p-SEC20 strain CAr2d110 during growth in low glucose (PCK1p induction) and high glucose (PCK1p repression). Because weak signals were obtained in conventional Northern experiments, we used RT-PCR with SEC20-specific primers to demonstrate the SEC20 transcript. To verify that the RNA preparation used was free of DNA, we also detected the transcript of the EFB1 gene, which contains an intron (37). None of the DNase-treated RNA preparation generated an EFB1 RT-PCR product of 0.7 kb characteristic of the unspliced gene, but all contained a 0.5-kb product indicating the spliced mRNA; thus, the RNA preparations were free of DNA. The 0.4-kb SEC20 RT-PCR product could be clearly detected at similar levels in all RNA preparations, including in the sample obtained from the PCK1p-SEC20 strain grown in repressing conditions (data not shown). The presence of the SEC20 transcript suggested that the slower growth of the PCK1p-SEC20 strain CAr2d110 was due to leaky expression of PCK1p in the presence of glucose.
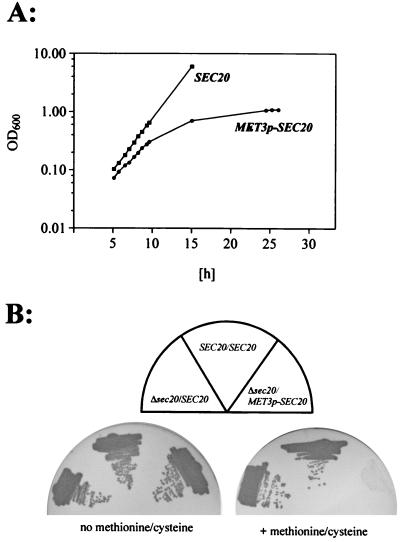
A very clear effect on growth was obtained using strain CA2d2m, in which SEC20 is under MET3p control. Addition of 2.5 mM methionine-cysteine completely blocked growth of this strain in liquid and on solid media (Fig. 3). On the other hand, the presence of 2.5 mM methionine or cysteine alone did not prevent growth, suggesting that repression of MET3p in such media was insufficient, as has been reported previously (5). These results strongly suggested that SEC20 is essential for growth.
FIG. 3.
SEC20 expression is required for growth. Growth of mutant strain CA2d2m, in which expression of SEC20 is controlled by MET3p (Δsec20/MET3p-SEC20), is compared to the SEC20/SEC20 strain CAI4[pBI-1] and the heterozygous strain CA2 (Δsec20/SEC20). (A) Stationary-phase cells were diluted to an optical density at 600 nm (OD600) of 0.05 in liquid repressing SD medium containing 2.5 mM methionine and 2.5 mM cysteine (time zero) and were grown at 30°C; 1:2 dilutions were carried out at 12 h to maintain logarithmic growth. (B) Cells were streaked on solid SD medium with or without 2.5 mM methionine-cysteine supplementation, and plates were incubated at 30°C.
C. albicans SEC20 is not functional in S. cerevisiae.
To test if the C. albicans SEC20 homolog was functional in S. cerevisiae, we constructed expression vectors in which the SEC20 coding region was under transcriptional control of S. cerevisiae PFK2p (33). Both a low-copy-number (pYW9) and a high-copy-number (pYW10) expression vector were constructed and used to transform a temperature-sensitive sec20-1 mutant (strain RSY275 17) at 25°C.
RSY275 transformants carrying either pYW9 or pYW10 could not grow at 37°C, nor could a transformant carrying plasmid pYW11, in which SEC20 was transcribed by its authentic promoter. However, complementation of the sec20-1 mutation was successful with plasmid STM20, which carries ScSEC20 (43). These results indicate that the C. albicans SEC20 allele is not functional in the heterologous yeast S. cerevisiae. Possible reasons for this failure include the N-terminal deletion of CaSec20p as well as residues incompatible with function, e.g., caused by two CUG codons encoding serine in C. albicans but leucine in S. cerevisiae.
Lowered SEC20 expression leads to antifungal supersensitivities.
Fungi defective in protein glycosylation are known to be supersensitive to several antifungal compounds, especially aminoglycoside antibiotics (10, 34, 48, 49). Although the reason for this supersensitivity is not known, we hypothesized that lack of glycosylation might cause defects in secretion and vice versa. Consequently, we used strain CAr2d110 to explore directly whether the decrease of secretion efficiency caused by low SEC20 expression on S4D high-glucose medium would cause antifungal supersensitivities.
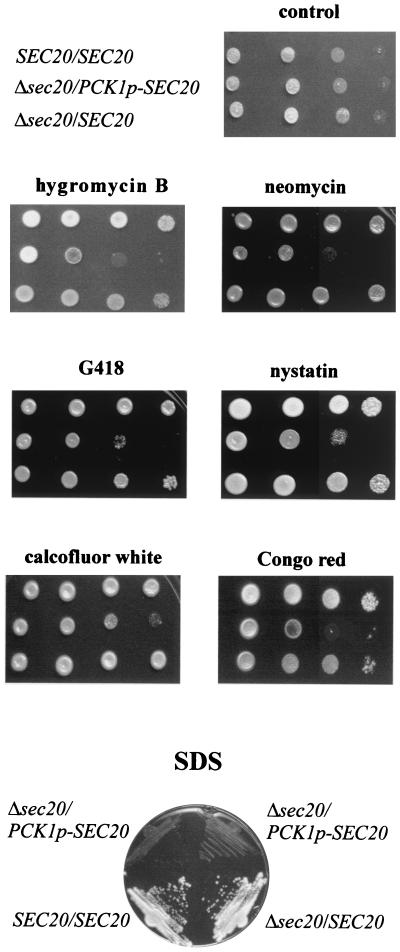
The aminoglycosides hygromycin B (0.25 mg/ml), G418 (0.6 mg/ml), and neomycin (30 mM) significantly blocked growth of CAr2d110 on repressing solid medium (Fig. 4). In addition, supersensitivity to the polyene antibiotic nystatin was observed. Because a reduced plating efficiency was observed on media containing calcofluor white (0.1 mg/ml) and cells did not grow on solid media containing low levels (0.09%) of the detergent SDS (Fig. 4), it appears that lowering of SEC20 expression causes cell wall defects (34, 48). The susceptibilities of strain CAr2d110 to vanadate (2.5 to 7.5 mM), clotrimazole (0.5 μg/ml), or caffeine (up to 20 mM) (data not shown) were unaltered, suggesting that a special subset of proteins involved in basal antifungal resistance is affected in strains with reduced SEC20 expression.
FIG. 4.
Lowering of SEC20 expression leads to supersensitivities. Five microliters of diluted cultures of strain CAI4[pBI-1] (SEC20/SEC20), the heterozygous strain CA2 (Δsec20/SEC20), and strain CAr2d110, in which SEC20 expression is under control of the glucose-repressible PCK1p (Δsec20/PCK1p-SEC20), were spotted on PCK1p-repressing YP4D medium containing various agents and were grown at 30°C. A series of 10-fold dilutions was applied, starting with a concentration of 5 × 105 cells/5 μl (from left to right). Plates contained no addition (control), hygromycin B (0.25 mg/ml), neomycin (30 mM), G418 (0.6 mg/ml), nystatin (1.5 μg/ml), calcofluor white (100 μg/ml), and Congo red (1 mg/ml). For the examination of SDS sensitivity, cells were streaked on YP4D plates containing 0.09% SDS and grown at 30°C.
Lowering of SEC20 expression does not affect morphogenesis.
It was reported that in the dimorphic yeast Yarrowia lipolytica, deletion of a SEC14 homolog did not prevent growth but affected the ability to form filaments (22). We therefore tested whether the lowering of SEC20 expression in strain CAr2d110 (PCK1p-SEC20), which was evidenced by the supersensitivity to some antifungals, also had an effect on hyphal formation. However, in PCK1p-repressing S4D medium, the addition of 5% serum stimulated hyphal formation of CAr2d110 at rates and to extents similar to those for control strains (data not shown). This result indicated that a slight reduction in SEC20 transcripts (and a considerable accumulation of intracellular membranes [see below]) had no effect on hyphal differentiation.
To examine if SEC20 expression was regulated by hyphal formation, we induced filamentation of strain 3153 by serum at elevated temperature (37°C) or neutral pH (3). In each induction condition, we calculated the percentage of cells forming hyphae and determined the SEC20 transcript level by direct quantitation of radioactive areas on a Northern blot by radioimaging. The blots used have been described previously (46) and were normalized to rRNA used as a standard. Since standard Northern blots showed only weak signals using SEC20 DNA probes, we improved detection of SEC20 mRNA by the use of a SEC20 RNA probe. In these blots, the SEC20 transcript was apparent as a single band of about 1.3 kb, which corresponds to the size of the SEC20 ORF (996 bp) (data not shown).
The addition of serum clearly stimulated hyphal formation, but the level of the SEC20 transcript did not increase proportionally, especially not in comparison with cells with lower and higher rates and levels of hyphal formation (data not shown). Similarly, use of a neutral pH in combination with an elevated temperature accelerated filamentation, but there was no clear correlation to the SEC20 transcript level. Thus, we conclude that the level of SEC20 expression, like the expression of many other genes (11, 47) does not directly reflect filament formation but may depend on other factors. We also determined that the SEC20 transcript was not induced by heat shock but increased during the early growth phase (data not shown).
Effects of reduced SEC20 expression on ultrastructural morphology.
In S. cerevisiae, the depletion of Sec20p in cells leads to the accumulation of sheets of membranes, which may arise by the fusion of retrograde vesicles with Golgi- instead of ER-target membranes (21, 43) or are derived from the ER (17). Using strains CA2d110 and CA2d2m, we performed a depletion experiment to investigate the consequences of a lowered Sec20p levels in C. albicans.
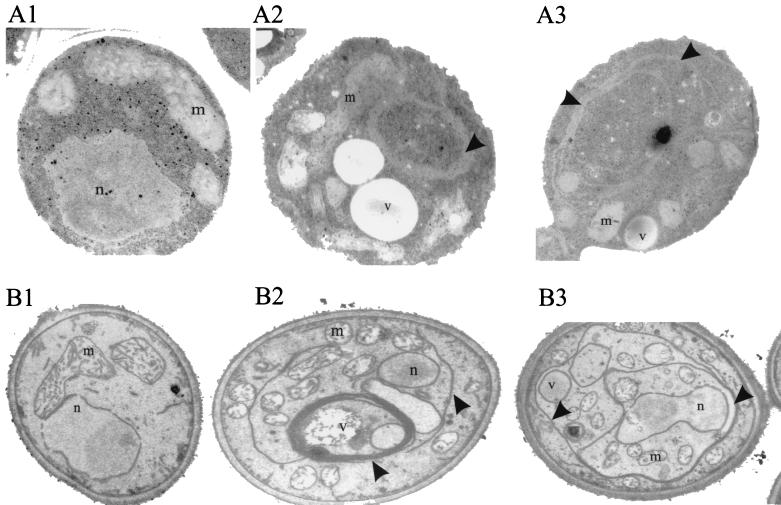
CA2d110 cells were grown to log phase in PCK1p-repressing S4D medium, fixed with glutaraldehyde and osmium tetroxide, and prepared for transmission electron microscopy (4). As shown in Fig. 5 (A2 and A3), a marked accumulation of membranes was detected in strain CA2d110 but not in control strains (A1). Accumulated membranes were apparent as long sheets, mostly in stacks of several (often triple) membranes. A similar phenotype occurred in strain CA2d2m containing the MET3p-SEC20 allele when cells were shifted to repressing medium containing methionine and cysteine. Again, extensive accumulation of membranes and stacking of accumulated membranes was observed (Fig. 5, B2 and B3). Because of the fixation with KMnO4, membranes were more preserved and more clearly visible in the latter experiment. Thus, because of a similar and even more drastic depletion phenotype compared to S. cerevisiae (43), the identified C. albicans SEC20 gene appears to fulfill a function in the secretory pathway similar to that of SEC20 in S. cerevisiae.
FIG. 5.
Electron microscopic images of strains with wild-type and reduced SEC20 expression levels. A wild-type strain (CAI4[pBI-1] (A1 and B1) and two conditional mutants, CAr2d110 (Δsec20/PCK1p-SEC20; A2 and A3) and CA2d2m (Δsec20/MET3p-SEC20; B2 and B3) were compared. Strains were grown in PCK1p-repressing S4D medium (A1 to A3) or in MET3p-repressing SDMC medium (B1 to B3). Cells were prepared for electron microscopy by fixation with glutaraldehyde (A1 to A3) or, to enhance the visualization of membranes, by KMnO4 (B1 to B3) fixation. Arrows indicate accumulated membranes. n, nucleus; m, mitochondria; v, vacuole.
DISCUSSION
The yeast Sec20 protein represents an exception to the universality of the secretory machinery in eukaryotes, because a homolog has not been identified in mammalian cells. This difference led to the question whether Sec20p was species specific for S. cerevisiae or whether other fungi contained a Sec20p homolog with an identical function. Here we describe for the first time a homolog of the S. cerevisiae Sec20 protein in another species, the most common fungal pathogen of humans, C. albicans (27% identity to ScSec20p). In the Schizosaccharomyces pombe genomic sequencing project (http://www.sanger.ac.uk/Projects/S_pombe), a sequence (EMBL SPAC23A1.15c) which possibly encodes another fungal Sec20p homolog (21% identity to ScSec20p) has been found, but not yet characterized, suggesting that Sec20p may occur in other fungal species. Our finding that in C. albicans Sec20p is essential and involved in the secretory machinery (as suggested by the accumulation of membranes in depleted strains) suggests Sec20p as a fungus-specific potential target for future antifungals.
The structure of the C. albicans Sec20 protein homolog is surprising, because sequences corresponding to about 100 amino-terminal residues of ScSec20p are missing. The amino end of ScSec20p is known to interact with an essential cytoplasmic protein, Tip20p, while the C-terminal end of ScSec20p, which is present in the ER lumen, together with the transmembrane region interacts with the t-SNARE Ufe1p; Ufe1p, in a complex with Sec20p and Tip20p, is able to bind the v-SNARE Sec22p present on retrograde vesicles originating from the Golgi (1, 9, 20, 21, 43, 44). Ufe1p is also instrumental in a Sec20p-independent function for fusion of ER compartments (31). Our computer searches did not reveal any gene or gene fragment with the potential to encode a Tip20p homolog in genomic sequences of C. albicans, which are almost completed (http://www-sequence.stanford.edu/group/candida/search.html). Thus, not only the potential interacting sequence in the Sec20p structure but also the expected interactor component is absent. It appears possible that part of the essential function of ScTip20p is to bind to ScSec20p. In C. albicans, the role of the amino end of Sec20p and of Tip20p may be assumed by other proteins, which may be C. albicans specific. Other structural features conserved in Sec20p are a single transmembrane region and a potential ER retention VDEL sequence at the carboxy end, which is similar to the HDEL sequence observed in ScSec20p (43). Although ER retention signals in C. albicans have not yet been defined, our computer searches identified in C. albicans genomic sequences a homolog of ScSec12p (Ca20C1.15c) which has the potential ER-targeting C-terminal sequence KDEL (32). ScSec12p is a type II membrane protein located in the ER (28), but it does not have a typical HDEL retention sequence. The retention signal on ScSec20p is not essential for its function but helps to maintain a high Sec20p concentration in the ER (43).
Sec20p is essential for the viability of C. albicans, which we demonstrated by placing SEC20 under transcriptional control of two different regulated promoters. The clearest result was obtained using MET3p, presumably because in the repressed state its expression is lower than that of PCK1p; repression of the MET3p-SEC20 allele prevented growth, while repression of the PCK1p-SEC20 allele in another strain significantly increased its generation time. This characteristic may be understood considering the low level of expression of SEC20, which became apparent in our Northern experiments. A shift of the MET3-SEC20 strain from inducing to repressing conditions led to a dramatic accumulation of intracellular membranes. A similar phenotype occurred in the PCK1p-SEC20 strain. This phenotype corresponds to but is even more dramatic than that of an S. cerevisiae strain in which SEC20 expression is lowered (43). The accumulated membranes in C. albicans appeared as extensive sheets, which frequently were stacked as triple-membrane layers. The nature of the accumulated membranes in S. cerevisiae and in C. albicans is unclear. It has been speculated for S. cerevisiae sec20 mutants that Golgi vesicles, which are prevented from returning to the ER, fuse with Golgi membranes, particularly because they carry a v-SNARE (Sec22p), which is used for forward and retrograde transport (21). Originally, because of the effect of sec20 mutations on forward ER-Golgi transport, it had been assumed that these membranes were ER derived (43). The primary function of Sec20p in C. albicans, including its function in secretion or in retrograde transport, remains to be proven.
Lowered expression of SEC20 in a strain carrying the PCK1p-SEC20 allele in its repressed state revealed new phenotypes, which were associated with suboptimal SEC20 expression. Such cells showed a supersensitivity to aminoglycoside antibiotics, including hygromycin B, neomycin, and G418, but also an increased susceptibility to the antifungal nystatin and cell wall-attacking agents such as calcofluor white, Congo red, and SDS. Conceivably, a suboptimal secretory machinery may lower levels of proteins required for cell surface stability. Such proteins may also include components of the protein glycosylation machinery, such as glycosyltransferases involved in O- and N-glycosylation, whose reduced activity leads to aminoglycoside sensitivity and cell wall fragility (10, 34, 48, 49). Interestingly, a sec20 allele was identified in a screen for aminoglycoside-supersensitive mutants of S. cerevisiae (39). Thus, even a mild reduction in intracellular Sec20p levels leads to significant sensitivity phenotypes. These findings provide new possibilities for the development of antifungal strategies.
ACKNOWLEDGMENTS
We thank M. Lewis and H. Pelham for generously supplying strains and plasmids. We thank N. Gow and A. Brown for helpful discussions and advice. We are grateful to S. Scherer and P. Magee for supplying the C. albicans fosmid library.
This work was supported by the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Ballensiefen W, Ossipov D, Schmitt H D. Recycling of the yeast v-SNARE Sec22p involves COPI-proteins and the ER transmembrane proteins Ufe1p and Sec20p. J Cell Sci. 1998;111:1507–1520. doi: 10.1242/jcs.111.11.1507. [DOI] [PubMed] [Google Scholar]
- 2.Banfield D K, Lewis M J, Pelham H R B. A SNARE-like protein required for traffic through the Golgi complex. Nature. 1995;375:806–809. doi: 10.1038/375806a0. [DOI] [PubMed] [Google Scholar]
- 3.Buffo J, Herman M A, Soll D R. A characterization of pH-regulated dimorphism in Candida albicans. Mycopathologia. 1984;85:21–30. doi: 10.1007/BF00436698. [DOI] [PubMed] [Google Scholar]
- 4.Byers B, Goetsch L. Electron microscopic observations on the meiotic karyotype of diploid and tetraploid Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1975;72:5056–5060. doi: 10.1073/pnas.72.12.5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Care R S, Trevethick J, Binley K M, Sudbery P E. The MET3 promoter: a new tool for Candida albicans molecular genetics. Mol Microbiol. 1999;34:792–798. doi: 10.1046/j.1365-2958.1999.01641.x. [DOI] [PubMed] [Google Scholar]
- 6.Chaffin W L, López-Ribot J L, Casanova M, Gozalbo D, Martinez J P. Cell wall and secreted proteins of Candida albicans: identification, function, and expression. Microbiol Mol Biol Rev. 1998;62:130–180. doi: 10.1128/mmbr.62.1.130-180.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chibana H, Magee B B, Grindle S, Ran Y, Scherer S, Magee P T. A physical map of chromosome 7 of Candida albicans. Genetics. 1998;149:1739–1752. doi: 10.1093/genetics/149.4.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clément M, Fournier H, de Repentigny L, Belhumeur P. Isolation and characterization of the Candida albicans SEC4 gene. Yeast. 1998;14:675–680. doi: 10.1002/(SICI)1097-0061(199805)14:7<675::AID-YEA252>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 9.Cosson P, Schröder-Köhne S, Sweet D S, Démollière C, Henneke S, Frigerio G, Letourneur F. The Sec20/Tip20p complex is involved in ER retrieval of dilysine-tagged proteins. Eur J Cell Biol. 1997;73:93–97. [PubMed] [Google Scholar]
- 10.Dean N. Yeast glycosylation mutants are sensitive to aminoglycosides. Proc Natl Acad Sci USA. 1995;92:1287–1291. doi: 10.1073/pnas.92.5.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delbrück S, Ernst J F. Morphogenesis-independent regulation of actin transcript levels in the pathogenic yeast Candida albicans. Mol Microbiol. 1993;10:859–866. doi: 10.1111/j.1365-2958.1993.tb00956.x. [DOI] [PubMed] [Google Scholar]
- 12.Ernst J F. Transcription factors in Candida albicans—environmental control of morphogenesis. Microbiology. 2000;146:1763–1774. doi: 10.1099/00221287-146-8-1763. [DOI] [PubMed] [Google Scholar]
- 13.Fonzi W A, Irwin M Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frigerio G. The Saccharomyces cerevisiae early secretion mutant tip20 is synthetic lethal with mutants in yeast coatomer and the SNARE proteins Sec22p and Ufe1p. Yeast. 1998;14:633–646. doi: 10.1002/(SICI)1097-0061(199805)14:7<633::AID-YEA267>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 15.Hayashi T, McMahon H, Yamasaki S, Binz T, Hata Y, Südhof T C, Niemann H. Synaptic vesicle membrane-fusion complex—action of clostridial neurotoxins on assembly. EMBO J. 1994;13:5051–5061. doi: 10.1002/j.1460-2075.1994.tb06834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hube B, Sanglard D, Odds F C, Hess D, Monod M, Schäfer W, Brown A J P, Gow N A R. Disruption of each of the secreted aspartyl proteinase genes SAP1, SAP2, and SAP3 of Candida albicans attenuates virulence. Infect Immun. 1997;65:3529–3538. doi: 10.1128/iai.65.9.3529-3538.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaiser C A, Schekman R. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell. 1990;61:723–733. doi: 10.1016/0092-8674(90)90483-u. [DOI] [PubMed] [Google Scholar]
- 18.Leidich S D, Ibrahim A S, Fu Y, Koul A, Jessup C, Vitullo J, Fonzi W, Mirbod F, Nakashima S, Nozawa Y, Ghannoum M A. Cloning and disruption of caPLB1, a phospholipase B gene involved in the pathogenicity of Candida albicans. J Biol Chem. 1998;273:26078–26086. doi: 10.1074/jbc.273.40.26078. [DOI] [PubMed] [Google Scholar]
- 19.Leuker C E, Sonneborn A, Delbrück S, Ernst J F. Sequence and regulation of the PCK1 gene encoding phosphoenolpyruvate carboxykinase of the fungal pathogen Candida albicans. Gene. 1997;192:235–240. doi: 10.1016/s0378-1119(97)00069-3. [DOI] [PubMed] [Google Scholar]
- 20.Lewis M J, Pelham H R B. SNARE-mediated retrograde traffic from the Golgi complex to the endoplasmic reticulum. Cell. 1996;85:205–215. doi: 10.1016/s0092-8674(00)81097-1. [DOI] [PubMed] [Google Scholar]
- 21.Lewis M J, Rayner J C, Pelham H R B. A novel SNARE complex implicated in vesicle fusion with the endoplasmic reticulum. EMBO J. 1997;16:3017–3024. doi: 10.1093/emboj/16.11.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez M C, Nicaud J M, Skinner H B, Vergnolle C, Kader J C, Bankaitis V A, Gaillardin C. A phosphatidylinositol/phosphatidylcholine transfer protein is required for differentiation of the dimorphic yeast Yarrowia lipolytica from the yeast to the mycelial form. J Cell Biol. 1994;125:113–127. doi: 10.1083/jcb.125.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Losberger C, Ernst J F. Sequence and transcript analysis of the C. albicans gene (URA3) encoding orotidine-5′-phosphate decarboxylase. Curr Genet. 1989;16:153–157. doi: 10.1007/BF00391471. [DOI] [PubMed] [Google Scholar]
- 24.Luft J H. Improvements in epoxy resin embedding methods. J Biophys Biochem Cytol. 1961;9:409–414. doi: 10.1083/jcb.9.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magee P T, Scherer S. Genome mapping and gene discovery in Candida albicans. ASM News. 1998;64:505–511. [Google Scholar]
- 26.Mao Y, Kalb V F, Wong B. Overexpression of a dominant-negative allele of SEC4 inhibits growth and protein secretion in Candida albicans. J Bacteriol. 1999;181:7235–7242. doi: 10.1128/jb.181.23.7235-7242.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monteoliva L, Sánchez M, Pla J, Gil C, Nombela C. Cloning of Candida albicans SEC14 homologue coding for a putative essential function. Yeast. 1996;11:1097–1105. doi: 10.1002/(SICI)1097-0061(19960915)12:11%3C1097::AID-YEA990%3E3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 28.Nakano A, Brada D, Schekman R. A membrane glycoprotein, Sec12p, required for protein transport from the endoplasmic reticulum to the Golgi apparatus in yeast. J Cell Biol. 1988;107:851–863. doi: 10.1083/jcb.107.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nieto A, Sanz P, Sentandreu R, Del Castillo L. Cloning and characterization of the SEC18 gene from Candida albicans. Yeast. 1993;8:875–887. doi: 10.1002/yea.320090808. [DOI] [PubMed] [Google Scholar]
- 30.Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- 31.Patel S K, Indig F E, Olivieri N, Levine N D, Latterich M. Organelle membrane fusion: a novel function for the syntaxin homolog Ufe1p in ER membrane fusion. Cell. 1998;92:611–620. doi: 10.1016/s0092-8674(00)81129-0. [DOI] [PubMed] [Google Scholar]
- 32.Pelham H R B. Control of exit from the endoplasmic reticulum. Annu Rev Cell Biol. 1989;5:1–23. doi: 10.1146/annurev.cb.05.110189.000245. [DOI] [PubMed] [Google Scholar]
- 33.Raben N, Exelbert R, Spiegel R, Sherman J B, Nakajima H, Plotz P, Heinisch J. Functional expression of human mutant phosphofructokinase in yeast: genetic defects in French Canadian and Swiss patients with phosphofructokinase deficiency. Am J Hum Genet. 1995;56:131–141. [PMC free article] [PubMed] [Google Scholar]
- 34.Ram A F J, Wolters A, ten Hoopen R, Klis F M. A new approach for isolating cell wall mutants in Saccharomyces cerevisiae by screening for hypersensitivity to calcofluor white. Yeast. 1994;10:1019–1030. doi: 10.1002/yea.320100804. [DOI] [PubMed] [Google Scholar]
- 35.Riggle P J, Slobodkin I V, Brown D H, Jr, Hanson M P, Volkert T L, Kumamoto C A. Two transcripts, differing at their 3′ ends, are produced from the Candida albicans SEC14 gene. Microbiology. 1997;143:3527–3535. doi: 10.1099/00221287-143-11-3527. [DOI] [PubMed] [Google Scholar]
- 36.Rothman J E, Wieland F T. Protein sorting by transport vesicles. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- 37.Schaller M, Schäfer W, Korting H C, Hube B. Differential expression secreted aspartyl proteinases in a model of human oral candidosis and in patient samples from the oral cavity. Mol Microbiol. 1998;29:605–615. doi: 10.1046/j.1365-2958.1998.00957.x. [DOI] [PubMed] [Google Scholar]
- 38.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 39.Shimma Y, Nishikawa A, bin Kassima B, Eto A, Jigami Y. A defect in GTP synthesis affects mannose outer chain elongation in Saccharomyces cerevisiae. Mol Gen Genet. 1997;256:469–480. doi: 10.1007/s004380050591. [DOI] [PubMed] [Google Scholar]
- 40.Sogaard M, Tani K, Ye R R, Geromanos S, Tempst P, Kirchhausen T, Rothman J E, Söllner T. A Rab protein is required for the assembly of SNARE complexes in the docking of transport vesicles. Cell. 1994;78:937–948. doi: 10.1016/0092-8674(94)90270-4. [DOI] [PubMed] [Google Scholar]
- 41.Söllner T, Bennett M K, Whiteheart S W, Scheller R H, Rothman J E. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell. 1993;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- 42.Stoldt V R, Sonneborn A, Leuker C E, Ernst J F. Efg1, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 1997;16:1982–1991. doi: 10.1093/emboj/16.8.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sweet D J, Pelham H R B. The Saccharomyces cerevisiae SEC20 gene encodes a membrane glycoprotein which is sorted by the HDEL retrieval system. EMBO J. 1992;11:423–432. doi: 10.1002/j.1460-2075.1992.tb05071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sweet D J, Pelham H R B. The TIP1 gene of Saccharomyces cerevisiae encodes an 80 kDa cytoplasmic protein that interacts with the cytoplasmic domain of Sec20p. EMBO J. 1993;12:2831–2840. doi: 10.1002/j.1460-2075.1993.tb05944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swoboda R K, Bertram G, Hollander H, Greenspan D, Greenspan J S, Gow N A R, Gooday G W, Brown A J P. Glycolytic enzymes of Candida albicans are nonubiquitous immunogens during candidiasis. Infect Immun. 1993;61:4263–4271. doi: 10.1128/iai.61.10.4263-4271.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swoboda R K, Bertram G, Budge S, Gooday G W, Gow N A R, Brown A J P. Structure and function of the HSP90 gene from the pathogenic fungus Candida albicans. Infect Immun. 1993;63:4506–4514. doi: 10.1128/iai.63.11.4506-4514.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swoboda R K, Bertram G, Delbrück S, Ernst J F, Gow N A R, Gooday G W, Brown A J P. Fluctuations in glycolytic mRNA levels during morphogenesis in Candida albicans reflect underlying changes in growth and are not a response to cellular dimorphism. Mol Microbiol. 1994;13:663–672. doi: 10.1111/j.1365-2958.1994.tb00460.x. [DOI] [PubMed] [Google Scholar]
- 48.Timpel C, Strahl-Bolsinger S, Ziegelbauer K, Ernst J F. Multiple functions of Pmt1p-mediated protein O-mannosylation in the fungal pathogen Candida albicans. J Biol Chem. 1998;273:20837–20846. doi: 10.1074/jbc.273.33.20837. [DOI] [PubMed] [Google Scholar]
- 49.Timpel C, Zink S, Strahl-Bolsinger S, Schröppel K, Ernst J. Morphogenesis, adhesive properties, and antifungal resistance depend on the Pmt6 protein mannosyltransferase in the fungal pathogen Candida albicans. J Bacteriol. 2000;182:3063–3071. doi: 10.1128/jb.182.11.3063-3071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]