Abstract
The human pathogen Eikenella corrodens expresses type IV pili and exhibits a phase variation involving the irreversible transition from piliated to nonpiliated variants. On solid medium, piliated variants form small (S-phase), corroding colonies whereas nonpiliated variants form large (L-phase), noncorroding colonies. We are studying pilus structure and function in the clinical isolate E. corrodens VA1. Earlier work defined the pilA locus which includes pilA1, pilA2, pilB, and hagA. Both pilA1 and pilA2 predict a type IV pilin, whereas pilB predicts a putative pilus assembly protein. The role of hagA has not been clearly established. That work also confirmed that pilA1 encodes the major pilus protein in this strain and showed that the phase variation involves a posttranslational event in pilus formation. In this study, the function of the individual genes comprising the pilA locus was examined using a recently developed protocol for targeted interposon mutagenesis of S-phase variant VA1-S1. Different pilA mutants were compared to S-phase and L-phase variants for several distinct aspects of phase variation and type IV pilus biosynthesis and function. S-phase cells were characterized by surface pili, competence for natural transformation, and twitching motility, whereas L-phase cells lacked these features. Inactivation of pilA1 yielded a mutant that was phenotypically indistinguishable from L-phase variants, showing that native biosynthesis of the type IV pilus in strain VA1 is dependent on expression of pilA1 and proper export and assembly of PilA1. Inactivation of pilA2 yielded a mutant that was phenotypically indistinguishable from S-phase variants, indicating that pilA2 is not essential for biosynthesis of functionally normal pili. A mutant inactivated for pilB was deficient for twitching motility, suggesting a role for PilB in this pilus-related phenomenon. Inactivation of hagA, which may encode a tellurite resistance protein, had no effect on pilus structure or function.
Eikenella corrodens is a gram-negative bacterium native to the oral cavity and gastrointestinal tract in humans. This bacterium can also be pathogenic, causing a variety of soft tissue and wound infections (6, 9, 10, 16), endocarditis (4, 9), and other opportunistic infections. E. corrodens has also been associated with periodontal diseases (2, 19, 21), although a causal role has not been clearly established. Like several gram-negative pathogens including Neisseria gonorrhoeae, N. meningitidis, and Moraxella bovis, E. corrodens exhibits a phase variation that results from altered synthesis of type IV pili and is reflected in colony morphology changes. On solid medium, small (S-phase) corroding and large (L-phase) noncorroding colonies are observed (7, 12, 15, 28). The L-phase variants arise irreversibly from S-phase variants at a frequency much greater than mutation rates. Colony morphology and phase variation correlates with the presence of pili on S-phase variants and the absence of pili on L-phase variants (11, 12). Because type IV pili can be determinants of pathogenesis (1, 5, 18, 29), the molecular basis of phase variation and the related phenomenon of antigenic variation are of considerable interest.
We recently isolated and characterized the pilA locus from an S-phase variant of E. corrodens strain VA1 (31). The pilA locus contains four tandemly arranged genes designated pilA1, pilA2, pilB, and hagA. Both pilA1 and pilA2 encode a type IV pilin, whereas pilB encodes a protein resembling the Dichelobacter nodosus FimB fimbrial assembly protein, and hagA encodes a putative hemagglutinin. Extensive DNA hybridization analyses indicated that pilA1 and pilA2 represent the only type IV pilin genes in this strain. In S-phase and L-phase cells, pilA1 is expressed as an abundant transcript initiating at an upstream promoter and terminating at a predicted hairpin structure between pilA1 and pilA2, whereas pilA2 and pilB are expressed as a low-abundance readthrough transcript. On the basis of protein and DNA sequence analyses, we determined that the pilA1-encoded pilin, designated PilA1, represents the major pilus protein for this strain. In contrast to the Neisseria and Moraxella species described above, the phase variation exhibited by E. corrodens strain VA1 does not involve a genomic recombination or mutagenic event that directly affects expression of the pilA locus. Both S-phase and L-phase cells similarly transcribe pilA1 and synthesize PilA1; however, S-phase cells export and assemble the PilA1 into pili whereas L-phase cells do not (31). The molecular basis of the presumed posttranslational alteration involving PilA1 export and assembly in L-phase variants remains to be determined.
We are examining the role of the strain VA1 pilA locus in type IV pilus biosynthesis and the related phenomena of competence for natural transformation and twitching motility. The earlier work established a dominant role for pilA1 in pilus biosynthesis; however, potential roles in this process for pilA2, pilB, or hagA remained to be defined. In this report, the function of the individual genes comprising the pilA locus was examined using a recently developed protocol for targeted interposon mutagenesis of E. corrodens. Analyses of different pilA mutants revealed that expression of pilA1, but not pilA2, is critical for synthesis of the pili responsible for the colony morphology of S-phase variants, competence, and twitching motility. This effort also suggests a role for pilB in twitching motility.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The strains used in this study are listed in Table 1. E. corrodens VA1 is a clinical isolate obtained from the Veterans Administration Medical Center, Kansas City, Mo. (8). Strain VA1-S1 is an S-phase isolate of strain VA1 that forms S-phase colonies and exhibits a typical frequency of phase variation to L-phase colonies on solid medium (31). Strain VA1-L2 is an L-phase isolate of VA1 that forms only L-phase colonies. E. corrodens was cultured aerobically at 35°C on chocolate agar plates (Remel, Lenexa, Kans.) supplemented with 6 ml of a 1.5% agar overlay containing 1% (wt/vol) l-ornithine monohydrochloride and 1% (wt/vol) decarboxylase base Moeller medium (Difco, Detroit, Mich.) to facilitate growth. For selection and maintenance of transformants, streptomycin or kanamycin was added to the overlay to achieve a final antibiotic concentration of 25 μg ml−1.
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| E. corrodens strains | ||
| VA1 | Clinical isolate | Laboratory collection |
| VA1-S1 | S-phase variant of VA1 | Laboratory collection |
| VA1-L2 | L-phase variant of VA1 | Laboratory collection |
| T18 | Derivative of VA1-S1; pilA1::Ω | This study |
| T6 | Derivative of VA1-S1; pilA2::Ω | This study |
| T11 | Derivative of VA1-S1; pilB::Ω | This study |
| T40 | Derivative of VA1-S1; hagA::Ω | This study |
| T99 | Derivative of VA1-S1; ΔpilA1 pilB::Ω | This study |
| E. coli DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) supE44 λ− thi-1 gyrA96 relA1 | Bethesda Research Labs |
| Plasmids | ||
| pHP45Ω | Source of aad gene (Ω cassette); Spr | 22 |
| pSKS101 | Source of aph gene; Kmr | 27 |
| pUC1819EcoRI | Modified pUC18 cloning vector; Apr | J. Golden |
| pUMC315 | pUC1819 with 2.0-kbp EcoRI fragment from pHP45Ω containing the Ω cassette | This study |
| pEC114 | pGEM3zf(+) containing the 3.9-kbp EcoRI fragment from VA1-S1 carrying the pilA locus (pilA1A2B hagA) | 31 |
| pEC207 | pEC114 deleted for 299-bp HindIII fragment (nucleotides 3654–3953) | This study |
| pEC211 | pEC114 with BamHI-digested Ω cassette inserted into BclI site at nucleotide 1327 in pilA2; pilA2::Ω | This study |
| pEC213 | pEC114 with BamHI site replacing BglII site at nucleotide 869 in pilA1 | This study |
| pEC216 | pEC213 with BamHI-digested Ω cassette inserted into BglII site at nucleotide 2008 in pilB; pilB::Ω | This study |
| pEC218 | pEC114 with BamHI site replacing BglII site at nucleotide 2008 in pilB | This study |
| pEC219 | pEC218 with BamHI-digested Ω cassette inserted into BglII site at nucleotide 869 in pilA1; pilA1::Ω | This study |
| pEC230 | pEC207 with 1575-bp SmaI-BglII fragment (nucleotides 432-2008) replaced with 1,035-bp PCR product from pEC114; ΔpilA1; pilA2 controlled by pilA1p | This study |
| pEC232 | pEC230 with BamHI-digested Ω cassette inserted into BglII site at nucleotide 2008 in pilB; ΔpilA1; pilA2 controlled by pilA1p; pilB::Ω | This study |
| pEC233 | pEC230 with BamHI-digested aph gene inserted into BglII site at nucleotide 2008 in pilB; ΔpilA1; pilA2 controlled by pilA1p; pilB::aph | This study |
| pEC235 | pEC230 with SalI-digested Ω cassette inserted into SalI site at nucleotide 3274 in hagA; ΔpilA1; pilA2B controlled by pilA1p; hagA::Ω | This study |
| pEC237 | pEC230 with SalI-digested aph gene inserted into SalI site at nucleotide 3274 in hagA; ΔpilA1; pilA2B controlled by pilA1p; hagA::aph | This study |
| pEC306 | pEC114 with SalI-digested Ω cassette inserted into SalI site at nucleotide 3274 in hagA; hagA::Ω | This study |
Nucleotide numbers correspond to pilA locus sequence deposited in GenBank (accession no. AF079304).
Escherichia coli strain DH5α was used as the host for cloning vectors. E. coli strains were propagated in liquid or on solid Luria-Bertani medium with antibiotics at standard concentrations (26).
DNA methods.
Restriction endonucleases and modifying enzymes were purchased from Promega (Madison, Wis.). DNA manipulations including restriction digestion, agarose gel electrophoresis, ligations, PCR amplifications, transformation of E. coli, and plasmid minipreparations were performed using established protocols (3, 26). E. corrodens genomic DNA was prepared as described for E. coli in reference 26 or using kits from Qiagen (Chatsworth, Calif.) or GenoTech (St. Louis, Mo.). For DNA hybridization analysis, digested DNA was transferred to Hybond-N+ (Amersham, Arlington Heights, Ill.) membrane by the method of Reed and Mann (25). DNA probes for the pilA locus (3.9-kbp EcoRI fragment from pEC114) or aad (2.0-kbp BamHI fragment from pHP45Ω) were generated from gel-purified fragments by digoxigenin labeling with a kit from Boehringer (Indianapolis, Ind.). DNA hybridizations were performed at 60°C as described by Sambrook et al. (26).
Construction of mutant pilA loci.
The plasmids and oligonucleotide primers used in this study are listed in Tables 1 and 2, respectively. All nucleotide numbers referenced below correspond to the pilA locus sequence deposited in the GenBank database (accession no. AF079304). A physical map for pilA is presented in Fig. 1. Physical maps for the described mutant pilA constructs are presented in Fig. 5. Plasmid pEC114 harbors the 3.9-kbp EcoRI fragment of strain VA1-S1 genomic DNA encompassing the pilA locus (31) and served as the DNA source for all mutant pilA constructs. Plasmid pEC207 is a derivative of pEC114 deleted for the 0.3-kbp HindIII fragment originating downstream of hagA (nucleotide 3654) and terminating at the HindIII site in the multiple cloning region; this deletion eliminates the restriction sites in the multiple cloning region. To facilitate subcloning of the aad gene (often referred to as the Ω cassette; confers resistance to the antibiotics streptomycin and spectinomycin), the 2.0-kbp EcoRI fragment containing aad from pHP45Ω (22) was ligated into vector pUC1819EcoRI digested with EcoRI. The product, designated pUMC315, provides for excision of aad with BamHI, SmaI, or SalI.
TABLE 2.
Primers used
| Primera | 5′→3′ sequenceb | Position,c restriction site |
|---|---|---|
| RH-1 (←) | TTCGggAtCcTTAGCAGCACCAGGAGCG | 888–915, BamHI site at 892 |
| RH-2 (←) | GGCAACTTGATGGCAAATATCCTAC | 1431–1454 |
| RH-3 (→) | GGCACCCAAACCCTTTACAAG | 1590–1610 |
| RH-10 (→) | CGGTCAGATCTCTACTTGGACTTGCGC | 864–890, BglII site at 869 |
| RH-7B (←) | GATAGgGATCcGCCTGCATGGAAGGGG | 1192–2018, BamHI site at 2008 |
| RH-8 (←) | CCGGTCgGATCcCTACTTGGACTTGC | 863–888, BamHI site at 868 |
| RH-9 (←) | GATAGAGATCTGCCTGCATGGAAGG | 1994–2018, BglII site at 2008 |
| RH-18 (→) | AGATCCCgggTGTTTGGCAAGGGGGAT | 967–993, SmaI site at 971 |
| RH-12(←) | AGCTTctcgaGGCAGAAACTATCTGCCTGC | 2128–2158, XhoI site at 2133 |
| RH-14 (←) | ATTCActCGaGCTTTTTCGCCAACATCGTC | 3299–3328, XhoI site at 3318 |
| 105-R1 (→) | TGTTATCGCCATTATCGG | 531–548 |
| 107-F3 (→) | AGAGCAACTCGCTTTACCC | 1079–1096 |
| 204-F2 (→) | CGGATACGATGTGCATG | 2885–2901 |
Arrows designate forward (→) or reverse (←) primer sequence.
Underscored letters correspond to specified restriction sites; lowercase letters designate introduced base substitutions.
Nucleotide numbers corresponding to pilA locus sequence deposited in GenBank (accession no. AF079304).
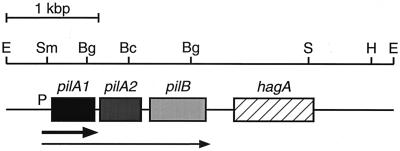
FIG. 1.
Physical map of the pilA locus for E. corrodens VA1-S1. Shaded and hatched boxes indicate sizes and positions of open reading frames as determined by sequence analysis. Arrows below the map designate mapped transcripts originating from the promoter upstream of pilA1. Flanking and internal restriction sites are shown for enzymes used in cloning and targeted mutagenesis. Bg, BglII; Bc, BclI; E, EcoRI; H, HindIII; S, SalI; Sm, SmaI.
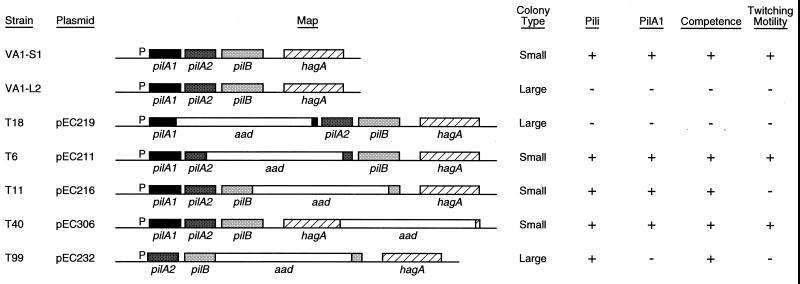
FIG. 5.
Physical maps and corresponding phenotypes of examined wild-type and pilA mutant strains of E. corrodens. Strains VA1-S1 and VA1-L2 are S-phase and L-phase variants, respectively, of the clinical isolate VA1. Mutant strains T18, T6, T11, T40, and T99 were isolated following transformation of strain VA1-S1 with the corresponding plasmid and selecting for double homologous recombinants. Each strain was assayed for colony morphology, surface pili, PilA1, competence for natural transformation, and twitching motility as described in Materials and Methods. Symbols indicate a positive (+) or negative (−) assay result.
The pilA1 gene was interrupted by insertion of aad following a minor modification of the pilA locus. The 1.14-kbp region encompassing the two internal BglII sites (nucleotides 869 to 2008) was amplified from pEC114 by PCR with primers RH-10 and RH-7b (substitutes BamHI site for BglII site at nucleotide 2008). The PCR product was digested with BglII and BamHI and ligated into pEC114 previously digested with BglII, effectively replacing the internal 1.14-kpb BglII fragment (nucleotides 869 to 2008) containing pilA1A2B sequences. The resulting plasmid, designated pEC218, harbors an otherwise intact pilA locus which lacks the BglII site in pilB. Subsequently, the 2.0-kbp BamHI fragment containing aad from pUMC315 was ligated into pEC218 previously digested with BglII, creating pEC219. The same strategy was used to interrupt pilB. In this case, the 1.14-kbp region encompassing the two internal BglII sites (nucleotides 869 to 2008) was amplified from pEC114 by PCR with primers RH-8 (substitutes BamHI site for BglII site at nucleotide 869) and RH-9. The PCR product was digested with BamHI and BglII and ligated into pEC114 previously digested with BglII as described above. The resulting plasmid, designated pEC213, harbors an otherwise intact pilA locus which lacks the BglII site in pilA1. The 2.0-kbp BamHI fragment containing aad from pUMC315 was ligated into pEC213 previously digested with BglII, creating pEC216.
To interrupt pilA2, the 2.0-kbp BamHI fragment containing aad from pUMC315 was ligated into the unique BclI site (nucleotide 1327) in pEC114, creating pEC211. Interruption of hagA was accomplished similarly by ligating the 2.0-kbp SalI fragment containing aad from pUMC315 into the unique SalI site (nucleotide 3274) in pEC114, creating pEC306.
To create a pilA locus lacking pilA1, pEC207 was digested with SmaI and BglII, and the larger digest product consisting of the original cloning vector and pilA flanking regions was gel purified using a kit from Qiagen (Valencia, Calif.). The 1.03-kbp fragment encompassing pilA2 and most of pilB (nucleotides 973 to 2008) was amplified from pEC114 by PCR with primers RH-18 (introduces SmaI site upstream of pilA2 coding region at nucleotide 973) and RH-9. The PCR product was digested with SmaI and BglII and ligated with the purified SmaI-BglII fragment from pEC207. The resulting plasmid, designated pEC230, places pilA2 and pilB under the control of pilA1p. Several selectable derivatives of this plasmid were used in this study. One, designated pEC232, was constructed by ligating the 2.0-kbp BamHI fragment containing aad from pUMC315 into the unique BglII site (nucleotide 2008) in pilB. A second, designated pEC233, was constructed by ligating the 2.0-kbp BamHI fragment containing aph (confers resistance to the antibiotic kanamycin) from pSKS101 (27) into the same BglII site. A third, designated pEC235, was constructed by ligating the 2.0-kbp SalI fragment containing aad from pUMC315 into the unique SalI site (nucleotide 3274) in hagA. A fourth, designated pEC237, was constructed by ligating the 2.0-kbp SalI fragment containing aph from pSKS101 into the same site.
Transformation and interposon mutagenesis of E. corrodens.
The protocol for transformation of E. corrodens was based on the procedure developed by Tonjum et al. (30). Cells of strain VA1-S1 were cultured on supplemented chocolate agar as described above. After 48 h, 10 S-phase colonies were harvested and resuspended in 1 ml of medium A (3.7% [wt/vol] brain heart infusion broth [BBL, Cockeysville, Md.], 0.05% [wt/vol] agar, 50 μM CaCl2, 0.2% [wt/vol] bovine serum albumin). For each transformation, a 10-μl aliquot of the resuspended cells was brought to 100 μl with medium A to achieve a cell density of approximately 2.5 × 106 CFU ml−1, and the suspension was provided linearized plasmid DNA (1 μg). Following incubation at 30°C for 45 min, the cells were plated onto supplemented chocolate agar and incubated at 35°C for 8 h. Selection was then applied by transferring the agar to a plate containing 6 ml of brain heart infusion broth supplemented with streptomycin (final concentration, 25 μg ml−1). Transformant colonies were isolated after 72 h and maintained on solid medium. Interposon mutagenesis of a targeted gene by insertion of aad via double homologous recombination between the genome of the recipient and the introduced DNA was confirmed for all mutants by PCR with the following primers: pilA1, 105-R1 and RH-1; pilA2, 107-F3 and RH-2; pilB, RH3 and RH12; hagA, 204-F2 and RH-14. For some mutants, interposon mutagenesis was also confirmed by DNA hybridization analysis using probes for pilA and aad. To assay competence for natural transformation, the wild-type and pilA mutant strains were subjected to the same protocol using linearized pEC233 (pEC237 for mutant T99) as the introduced DNA and kanamycin (final concentration, 25 μg ml−1) for selection and maintenance of transformants.
Electron microscopy.
Negative staining and immunogold electron microscopic examination of whole cells were performed as described elsewhere (14), using a polyclonal antiserum (1:1,000) prepared against pilin purified from strain VA1-S3.
Cell fractionation.
Cell fractionation was performed as described by Villar et al. (31). For the wild-type and pilA mutant strains, total cellular and surface protein fractions were isolated for analysis. All protein fractions were mixed with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and stored at −20°C.
SDS-PAGE and immunoblot analysis.
Protein samples were separated by SDS-PAGE on 20% polyacrylamide gels. Following electrophoresis, proteins were transferred to a nitrocellulose membrane (Nitrobind; Micron Separations Inc., Westborough, Mass.) as described by Ausubel et al. (3). The blots were blocked and incubated with a polyclonal antiserum (1:5,000) raised against a truncated PilA1 protein (31). Bound antibodies were visualized following incubation of the blots with goat anti-rabbit immunoglobulin G (1:5,000) conjugated to alkaline phosphatase (KLC Laboratories, Gaithersburg, Md.) according to the manufacturer's instructions.
Twitching motility.
Twitching motility by the wild-type and pilA mutant strains was assayed by the agar interface method described by McMichael (20) and by analysis of colony morphology. To facilitate microscopic examination of E. corrodens colonies in the latter procedure, cells were plated onto a sterilized dialysis membrane affixed to the agar surface of a supplemented chocolate agar plate and cultured as described above. After a 24-h incubation period, the membrane was peeled away from the agar and the colonies were examined under low magnification (×30 and ×50) for the convoluted edge and spreading that are characteristic of twitching motility (11).
RESULTS
Transformation of E. corrodens.
Earlier attempts by this and other laboratories to transform E. corrodens with circular plasmid DNA were not successful. In this work, an effective procedure for transformation of this species using linearized DNA was developed. Using the optimized conditions described above, transformation frequencies ranging from 1 × 10−5 to 3 × 10−5 were typically achieved, yielding 30 to 60 transformants per μg of transforming DNA. For interposon mutagenesis via double homologous recombination between the introduced DNA and the genome of the recipient, a minimum of 0.4 kbp of genomic sequences flanking the selectable marker (aad or aph) was required. All of the pilA mutants described below were generated by this protocol and confirmed for interposon mutagenesis of the targeted gene by DNA hybridization analysis and/or PCR (data not shown).
Phenotypes of wild-type and pilA mutant strains.
To examine the role of each gene constituting the pilA locus, different pilA mutants were compared to S-phase variant VA1-S1 and L-phase variant VA1-L2 for colony morphology, the presence of pili, the presence of PilA1 in surface and total cellular protein fractions, competence for natural transformation, and twitching motility. These phenotypes were chosen to represent several distinct aspects of phase variation and type IV pilus biosynthesis and function.
Strain VA1 exhibits an irreversible transition from S-phase to L-phase variants that is reflected in a colony morphology change. This phase transition is demonstrated by strains VA1-S1 and VA1-L2; on solid medium, strain VA1-S1 forms small colonies whereas strain VA1-L2 forms large colonies (compare Fig. 2A and B). As we reported earlier (31), the altered colony morphology of these phase variants correlates with the presence of pili on VA1-S1 cells and the absence of such pili on VA1-L2 cells (compare Fig. 3A and B). The detection of mature PilA1 in the total protein fraction but not the surface protein fraction of VA1-L2 cells (Fig. 4A and B, compare lanes 2 and 3) supports the hypothesis that a posttranslational event involving PilA1 export and/or assembly is responsible for the phase variation exhibited by strain VA1. In the assay for competence for natural transformation by linearized pEC233 DNA, strain VA1-S1 yielded the standard frequency of kanamycin-resistant colonies, whereas no resistant colonies were obtained for strain VA1-L2. In addition, two independent assays showed that only strain VA1-S1 is characterized by the phenomenon of twitching motility.
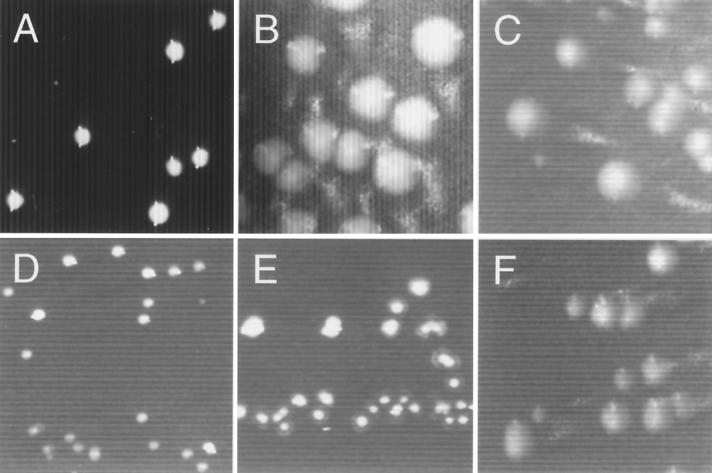
FIG. 2.
Colony morphologies of wild-type and pilA mutant strains of E. corrodens. Cells of strains VA1-S1 (A), VA1-L2 (B), T18 (C), T6 (D), T11 (E), and T99 (F) were cultured on chocolate agar. Magnification = ×12.5.
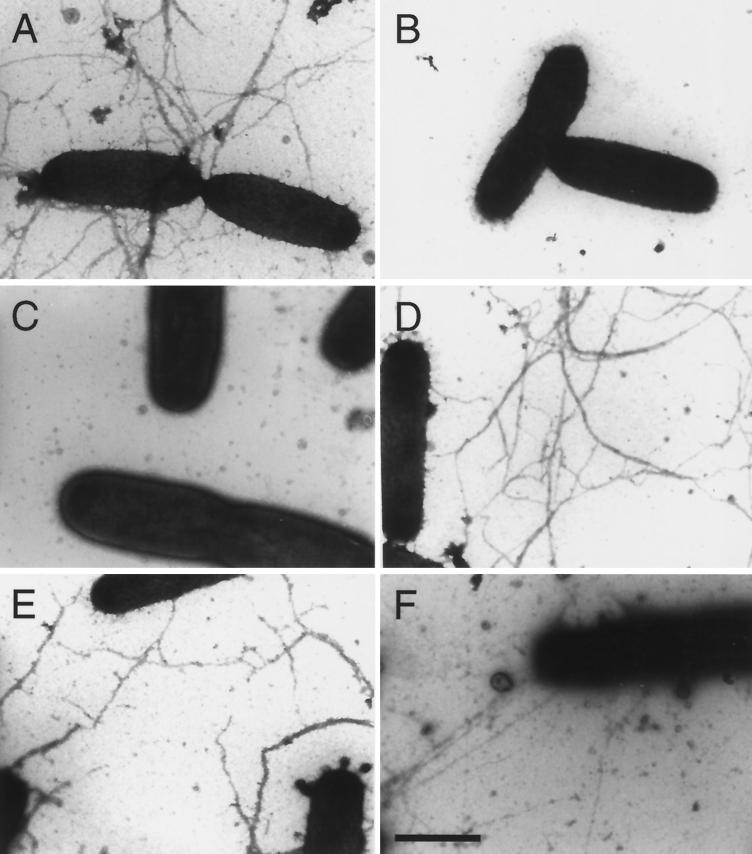
FIG. 3.
Differential piliation of wild-type and pilA mutant strains of E. corrodens. Cells of strains VA1-S1 (A), VA1-L2 (B), T18 (C), T6 (D), T11 (E), and T99 (F) were examined by immunogold electron microscopy. Bar = 100 nm.
FIG. 4.
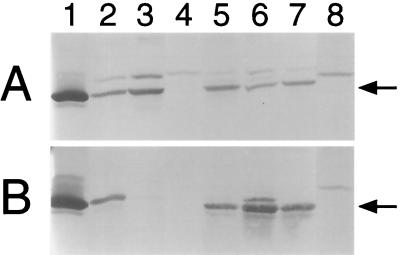
Localization of PilA1 in wild-type and pilA mutant strains of E. corrodens. (A) Total protein fraction; (B) surface protein fraction. Purified PilA1 (lane 1) and protein fractions from strains VA1-S1 (lane 2), VA1-L2 (lane 3), T18 (lane 4), T6 (lane 5), T11 (lane 6), T40 (lane 7), and T99 (lane 8) were subjected to immunoblot analysis with a polyclonal antiserum specific for PilA1. Each arrow marks the position of mature pilin.
A link between pilA1 activity and the type IV pilus-associated phenotypes was established with mutant strain T18, in which pilA1 was inactivated by insertion of aad (see Fig. 5). On solid medium, strain T18 formed large colonies that were indistinguishable from those of strain VA1-L2 (compare Fig. 2B and C). Electron microscopic examination showed that like cells of VA1-L2, cells of strain T18 lacked observable pili (compare Fig. 3B and C). In contrast to strain VA1-L2, PilA1 was not detected in the total protein fraction for strain T18 (Fig. 4A, compare lanes 3 and 4). Not surprisingly, strain T18 was not competent for transformation, nor did it exhibit twitching motility (Fig. 5). Thus, strain T18 closely resembles strain VA1-L2, and the collective phenotypes of this mutant suggest that expression of pilA1 is essential for pilus biosynthesis and related functions.
Inactivation of pilA2 or pilB resulted in strains that essentially exhibited the phenotypes of strain VA1-S1. By insertion of aad, strain T6 was inactivated for pilA2 whereas strain T11 was inactivated for pilB (Fig. 5). Both strains T6 and T11 formed small colonies on solid medium (Fig. 2D and E, respectively) and possessed PilA1-containing pili indistinguishable from those of strain VA1-S1 (compare Fig. 3D and E, respectively, with Fig. 3A). As for strain VA1-S1, mature PilA1 was detected in the total and surface protein fractions for both strains T6 and T11 (Fig. 4, lanes 5 and 6, respectively), and both mutants were competent for transformation by pEC233. However, strain T11 differed from strains VA1-S1 and T6 in that it was deficient for twitching motility, suggesting a possible role for pilB in this colony phenomenon. Although hagA was not predicted to play a role in pilus biosynthesis, strain T40, which was inactivated for hagA by aad (Fig. 5), was similarly examined for the pilus-associated phenotypes. Like strain VA1-S1, strain T40 was characterized phenotypically by small colony size (data not shown), PilA1-containing pili (data not shown), detectable PilA1 (Fig. 4, lane 7), competence for natural transformation, and twitching motility.
Strain T99 was generated to examine whether the pilin encoded by pilA2 could support pilus biosynthesis and the pilus-related phenotypes. The pilA locus in strain T99 lacks pilA1, contains pilA2 under the control of pilA1p, and is inactivated for pilB by aad (Fig. 5). On solid medium, strain T99 formed large colonies (Fig. 2F). Surprisingly, cells of strain T99 were found to possess pili that resembled those of strain VA1-S1 (Fig. 3F). Because the pili were not recognized by the PilA1 antisera (Fig. 3F) and no PilA1 was detected in any protein fraction for the strain (Fig. 4, lane 8), it was assumed that the pili of strain T99 were composed of PilA2. Strain T99 was competent for transformation by pEC237, suggesting that PilA1 is not essential for this process. However, in both assays for twitching motility, colonies of strain T99 did not exhibit any features characteristic of this pilus-associated phenomenon.
DISCUSSION
Genetic manipulation of the gram-negative pathogen E. corrodens has been compromised by the lack of an efficient transformation protocol for this species. In an earlier systematics analysis, Tonjum et al. demonstrated that several E. corrodens strains were naturally competent for genetic transformation by sheared genomic DNA (30). However, subsequent attempts by this and other laboratories to similarly transform E. corrodens with uncut plasmid vectors carrying host genomic sequences were not successful. Rao et al. developed a gene transfer system for E. corrodens strain ATCC 23834 that was based on conjugal transfer of a shuttle vector from E. coli (23). Although successful, this approach was limited by low frequencies of plasmid transfer and the requirement of a phage-based counterselection to inhibit growth of the donor. In this work, we have demonstrated that E. corrodens strain VA1-S1 is naturally competent for transformation by linearized plasmid vectors carrying host genomic sequences. As part of a mutational analysis of the four genes constituting the pilA locus, different plasmids on which the individual genes were interrupted by insertion of the selectable aad (or aph) marker were used to transform cells of strain VA1-S1. The interrupted pilA sequences were stably integrated into the genomic pilA locus, presumably via double homologous recombination, with transformation frequencies ranging from 1 × 10−5 to 3 × 10−5. To our knowledge, this effort represents the first application of both competence for natural transformation and interposon mutagenesis to study gene function in E. corrodens, and the results suggest that this organism should be amenable to standard genetic manipulation using these and related procedures.
Earlier work in our laboratory showed that for E. corrodens strain VA1, colony morphology and phase variation correlates with the presence of type IV pili on S-phase variants and the absence of such pili on L-phase variants. In this study, S-phase variant strain VA1-S1 and L-phase variant strain VA1-L2 were analyzed for competence for natural transformation and twitching motility, both of which have been associated with the expression of type IV pili in certain gram-negative bacteria. Not surprisingly, the piliated S-phase variant exhibited both competence and twitching motility whereas the nonpiliated L-phase variant exhibited neither, indicating that intact type IV pili are required for both processes in E. corrodens. This observation parallels the tight association between type IV pili and competence for natural transformation and twitching motility exhibited by N. gonorrhoeae (17). In contrast to N. gonorrhoeae and other type IV piliated pathogens, very little is known about the structure and function of the E. corrodens type IV pilus. However, recent work in our laboratory with S- and L-phase variants of strain VA1 indicates that the type IV pilus is essential for adherence to and cytotoxicity of human epithelial cells, suggesting that E. corrodens shares similar determinants of pilus structure and function with the better-characterized pathogens.
The type IV pilin gene pilA1 of strain VA1 plays a major role in pilus biosynthesis. Inactivation of pilA1 in S-phase variant strain VA1-S1 yielded mutant strain T18, which is phenotypically indistinguishable from L-phase variant strain VA1-L2; both strains T18 and VA1-L2 grow as large colonies and both lack pili, competence, and twitching motility. By design, interruption of pilA1 with aad would also have the polar effect of abolishing expression of pilA2 and pilB due to the intrinsic terminator encoded by the cassette (22). However, independent inactivation of pilA2 (strain T6) or pilB (strain T11) did not affect pilus formation, demonstrating that the T18 phenotype is dependent on the loss of pilA1 activity. The colony and piliation phenotypes of strain T18 corroborate earlier work showing that PilA1 is the major pilus protein for strain VA1. Recently we showed that both S-phase and L-phase cells transcribe pilA1 and synthesize PilA1; however, S-phase cells export and assemble the PilA1 into pili whereas L-phase cells do not, resulting in nonpiliated cells that grow as large colonies. Thus, despite the presence of the second type IV pilin gene pilA2, native biosynthesis of the type IV pilus in strain VA1 is dependent on expression of pilA1 and proper export and assembly of PilA1.
The type IV pilin gene pilA2 of strain VA1 does not play a major role in pilus biosynthesis. This was demonstrated by strain T6, which is inactivated for pilA2 and is phenotypically indistinguishable from S-phase variant strain VA1-S1, indicating that expression of pilA2 is not essential for biosynthesis of functionally normal pili. Earlier work showed that in both S- and L-phase variants of strain VA1, pilA1 is represented by an abundant transcript that terminates between pilA1 and pilA2, whereas pilA2 is represented by a much less abundant readthrough transcript encompassing pilA1, pilA2, and pilB. Whether pilin PilA2 is a minor pilus component in strain VA1-S1 is not known. In this study we showed that enhanced expression of pilA2 in a pilA1 null mutant background provided for synthesis of pili, presumably composed of PilA2, that share some features of the native pilus forms. This was demonstrated by mutant strain T99, in which pilA1 was deleted from the pilA locus in a manner that placed pilA2 adjacent to the native pilA1 promoter. Cells of strain T99 possessed pili indistinguishable from those of strain VA1-S1 and were naturally competent, suggesting that PilA2 is sufficient for synthesis of a functional pilus. Interestingly, strain T99 grew as large colonies, suggesting that the large-colony morphology of L-phase variants is due not to their lack of pili but rather to their lack of pili composed of PilA1. The structural features of PilA1 specific to the small-colony morphology of S-phase variants remain to be determined.
The pilB gene of the pilA locus is not essential for pili biosynthesis but may play a role in twitching motility by strain VA1. Inactivation of pilB in strain VA1-S1 yielded strain T11, which in these analyses was phenotypically indistinguishable from the parental strain except that it did not exhibit detectable twitching motility. A deficiency in twitching motility was also exhibited by strain T99, which possesses pili composed of PilA2 and is inactivated for pilB. Phenotypically, strain T11 resembles characterized pilT mutants of N. gonorrhoeae (34), Pseudomonas aeruginosa (33), and Myxococcus xanthus (35), which are piliated but lack twitching motility. Several lines of evidence suggest that pilT encodes a motor protein involved in pilus retraction (32). However, the predicted PilB protein does not show significant sequence identity to any reported PilT homologs. Instead, PilB shows greatest, albeit limited, sequence identity to the D. nodosus class I FimB protein, which is hypothesized to function in pilus assembly (13). Given that pilus biosynthesis was not noticeably impaired in strain T11, we favor the hypothesis that PilB represents a pilus structural component that provides for retraction of the filament, possibly by a mechanism that includes a PilT homolog.
The hagA gene of the pilA locus is not involved in pilus structure or function. In this study, inactivation of hagA yielded a strain that was phenotypically indistinguishable from the parental strain VA1-S1, which was not surprising given that hagA was thought to encode a hemagglutinin. The hemagglutinin gene designation for hagA was originally based on a BLAST analysis showing that the predicted HagA protein showed greater than 90% sequence identity to the protein predicted by the hae-1 gene of E. corrodens strain ATCC 23834 (31); correction of a presumed error in the deposited hae-1 sequence would render the two proteins nearly identical. The hae-1 gene product has been characterized as a hemagglutinin capable of inducing agglutination of neuraminidase-treated erythrocytes (24). However, a recent BLAST analysis suggests that the homologous hagA and hae-1 genes actually encode a tellurite resistance protein, bringing into question the earlier hemagglutinin gene designation for hae-1. Whether hagA encodes a hemagglutinin or a tellurite resistance protein would not seem to affect the results of this study but clearly needs to be resolved.
ACKNOWLEDGMENTS
We acknowledge P. Shubert, who constructed pEC306 and assisted in development of the transformation protocol for E. corrodens. We thank D. Viles for technical assistance and D. Law and staff for assistance with electron microscopy.
This research was partially supported by Public Health Service grant DE10439 (R.L.H.) from the National Institutes of Health.
REFERENCES
- 1.Alm R A, Mattick J S. Genes involved in the biogenesis and function of type-4 fimbriae in Pseudomonas aeruginosa. Gene. 1997;192:89–98. doi: 10.1016/s0378-1119(96)00805-0. [DOI] [PubMed] [Google Scholar]
- 2.Ashimoto A, Chen C, Bakler I, Slots J. Polymerase chain reaction detection of 8 putative periodontal pathogens in subgingival plaque of gingivitis and advanced periodontal lesions. Oral Microbiol Immunol. 1998;11:226–273. doi: 10.1111/j.1399-302x.1996.tb00180.x. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Klingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1989. [Google Scholar]
- 4.Berbari E F, Cockerill F R I, Steckelberg J M. Infective endocarditis due to unusual or fastidious microorganisms. Mayo Clin Proc. 1998;72:532–542. doi: 10.4065/72.6.532. [DOI] [PubMed] [Google Scholar]
- 5.Bieber D, Ramer S W, Wu C Y, Murray W J, Tobe T, Fernandez R, Schoolnik G K. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science. 1998;280:2114–2118. doi: 10.1126/science.280.5372.2114. [DOI] [PubMed] [Google Scholar]
- 6.Chen C K, Wilson M E. Eikenella corrodens in human oral and non-oral infections: a review. J Periodontol. 1992;63:941–953. doi: 10.1902/jop.1992.63.12.941. [DOI] [PubMed] [Google Scholar]
- 7.Chen C K, Wilson M E. Outer membrane protein and lipopolysaccharide heterogeneity among Eikenella corrodens isolates. J Infect Dis. 1990;162:664–671. doi: 10.1093/infdis/162.3.664. [DOI] [PubMed] [Google Scholar]
- 8.Cobb C M, Helber J T, Hirschberg R. Scanning electron microscopy of Eikenella corrodens colony morphology variants. J Periodontal Res. 1994;29:410–417. doi: 10.1111/j.1600-0765.1994.tb01242.x. [DOI] [PubMed] [Google Scholar]
- 9.Decker M D, Graham B S, Hunter E B, Liebowitz S M. Endocarditis and infections of intravascular devices due to Eikenella corrodens. Am J Med Sci. 1986;292:209–212. doi: 10.1097/00000441-198610000-00005. [DOI] [PubMed] [Google Scholar]
- 10.Griego R D, Rosen T, Orengo I F, Wolf J E. Dog, cat, and human bites: a review. J Am Acad Dermatol. 1995;33:1019–1029. doi: 10.1016/0190-9622(95)90296-1. [DOI] [PubMed] [Google Scholar]
- 11.Henrichsen J. The occurrence of twitching motility among gram-negative bacteria. Acta Pathol Microbiol Scand Sect B. 1975;83:171–178. doi: 10.1111/j.1699-0463.1975.tb00089.x. [DOI] [PubMed] [Google Scholar]
- 12.Henrichsen J, Blom J. Examination of fimbriation of some gram-negative rods with and without twitching and gliding motility. Acta Pathol Microbiol Scand Sect B. 1975;83:161–170. doi: 10.1111/j.1699-0463.1975.tb00088.x. [DOI] [PubMed] [Google Scholar]
- 13.Hobbs M, Dalrymple B P, Cox P T, Livingstone S P, Delaney S F, Mattick J S. Organization of the fimbrial gene region of Bacteroides nodosus: class I and class II strains. Mol Microbiol. 1991;5:543–560. doi: 10.1111/j.1365-2958.1991.tb00726.x. [DOI] [PubMed] [Google Scholar]
- 14.Hood B L, Hirschberg R. Purification and characterization of Eikenella corrodens type IV pilin. Infect Immun. 1995;63:3693–3696. doi: 10.1128/iai.63.9.3693-3696.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson F L, Goodman Y. Eikenella. In: Krieg N R, editor. Bergey's manual of systematic bacteriology. 9th ed. Vol. 1. Baltimore, Md: Williams & Wilkins; 1984. pp. 591–597. [Google Scholar]
- 16.Kentos A, De Vuyst P, Stuelens M J, Jacobs F, de Francquen P, Delaere B, Demaeyer P, Thys J P. Lung abscess due to Eikenella corrodens: three cases and review. Eur J Clin Microbiol Infect Dis. 1995;14:146–148. doi: 10.1007/BF02111877. [DOI] [PubMed] [Google Scholar]
- 17.Koomey M. Competence for natural transformation in Neisseria gonorrhoeae: a model system for studies of horizontal gene transfer. APMIS Suppl. 1998;84:56–61. doi: 10.1111/j.1600-0463.1998.tb05649.x. [DOI] [PubMed] [Google Scholar]
- 18.Krogfelt K A. Bacterial adhesion: genetics, biogenesis, and role in pathogenesis of fimbrial adhesins of Escherichia coli. Rev Infect Dis. 1991;13:721–735. doi: 10.1093/clinids/13.4.721. [DOI] [PubMed] [Google Scholar]
- 19.Lowenguth R A, Chin I, Caton J G, Cobb C M, Drisko C L, Killoy W J, Michalowicz B S, Pihlstrom B L, Goodson J M. Evaluation of periodontal treatments using controlled-release tetracycline fibers: microbiological response. J Periodontol. 1995;66:700–707. doi: 10.1902/jop.1995.66.8.700. [DOI] [PubMed] [Google Scholar]
- 20.McMichael J C. Bacterial differentiation within Moraxella bovis colonies growing at the interface of the agar medium with the Petri dish. J Gen Microbiol. 1992;138:2687–2695. doi: 10.1099/00221287-138-12-2687. [DOI] [PubMed] [Google Scholar]
- 21.Page R C. Current understanding of the aetiology and progression of periodontal disease. Int Dent J. 1986;36:153–161. [PubMed] [Google Scholar]
- 22.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 23.Rao V K, Whitlock J A, Progulske-Fox A. Development of a genetic system for Eikenella corrodens: transfer of plasmids pFM739 and pLES2. Plasmid. 1993;30:289–295. doi: 10.1006/plas.1993.1062. [DOI] [PubMed] [Google Scholar]
- 24.Rao V K, Whitlock J A, Proguske-Fox A. Cloning, characterization and sequencing of two haemagglutinin genes from Eikenella corrodens. J Gen Microbiol. 1993;139:639–650. doi: 10.1099/00221287-139-3-639. [DOI] [PubMed] [Google Scholar]
- 25.Reed K C, Mann D A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985;13:7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 27.Shapira S K, Chou J, Richaud F V, Casadaban M J. New versatile plasmid vectors for expression of hybrid proteins coded by a cloned gene fused to lacZ gene sequences encoding an enzymatically active carboxy-terminal portion of beta-galactosidase. Gene. 1983;25:71–82. doi: 10.1016/0378-1119(83)90169-5. [DOI] [PubMed] [Google Scholar]
- 28.Shiozu I, Shiozu J, Takazoe I, Okuda K. Corroding characteristics of Eikenella corrodens. Bull Tokyo Dent Coll. 1992;33:1–6. [PubMed] [Google Scholar]
- 29.Strom M S, Lory S. Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol. 1993;47:565–596. doi: 10.1146/annurev.mi.47.100193.003025. [DOI] [PubMed] [Google Scholar]
- 30.Tonjum T, Hagen N, Bovre K. Identification of Eikenella corrodens and Cardiobacterium hominis by genetic transformation. Acta Pathol Microbiol Immunol Scand Sect B. 1985;93:389–394. doi: 10.1111/j.1699-0463.1985.tb02907.x. [DOI] [PubMed] [Google Scholar]
- 31.Villar M T, Helber J T, Hood B, Schaefer M R, Hirschberg R L. Eikenella corrodens phase variation involves a posttranslational event in pilus formation. J Bacteriol. 1999;181:4154–4160. doi: 10.1128/jb.181.14.4154-4160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wall D, Kaiser D. Type IV pili and cell motility. Mol Microbiol. 1999;32:1–10. doi: 10.1046/j.1365-2958.1999.01339.x. [DOI] [PubMed] [Google Scholar]
- 33.Whitchurch C B, Hobbs M, Livingston S P, Krishnapillai V, Mattick J S. Characterisation of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialised protein export system widespread in eubacteria. Gene. 1991;101:33–44. doi: 10.1016/0378-1119(91)90221-v. [DOI] [PubMed] [Google Scholar]
- 34.Wolfgang M, Lauer P, Park H S, Brossay L, Hebert J, Koomey M. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol Microbiol. 1998;29:321–330. doi: 10.1046/j.1365-2958.1998.00935.x. [DOI] [PubMed] [Google Scholar]
- 35.Wu S S, Wu J, Kaiser D. The Myxococcus xanthus pilT locus is required for social gliding motility although pili are still produced. Mol Microbiol. 1997;23:109–121. doi: 10.1046/j.1365-2958.1997.1791550.x. [DOI] [PubMed] [Google Scholar]