Abstract
Previous studies have demonstrated that a proportion of Staphylococcus aureus isolates from bovine mastitis coproduce toxic shock syndrome toxin (TSST) and staphylococcal enterotoxin C (SEC). In this study, molecular genetic analysis of one such strain, RF122, revealed the presence of a 15,891-bp putative pathogenicity island (SaPIbov) encoding the genes for TSST (tst), the SEC bovine variant (sec-bovine), and a gene (sel) which encodes an enterotoxin-like protein. The island contains 21 open reading frames specifying hypothetical proteins longer than 60 amino acids including an integrase-like gene. The element is bordered by 74-bp direct repeats at the left and right junctions, and the integration site lies adjacent to the 3′ end of the GMP synthase gene (gmps) in the S. aureus chromosome. SaPIbov contains a central region of sequence identity with the previously characterized tst pathogenicity island SaPI1 (J. A. Lindsay et al., Mol. Microbiol. 29:527–543, 1998). A closely related strain, RF120, of the same multilocus enzyme electrophoretic type, random amplified polymorphic DNA type, and ribotype, does not contain the island, implying that the element is mobile and that a recent insertion/deletion event has taken place. TSST and TSST/SEC-deficient mutants of S. aureus strain RF122 were constructed by allele replacement. In vitro bovine Vβ-specific lymphocyte expansion analysis by culture supernatants of wild-type strains and of tst and sec-bovine allele replacement mutants revealed that TSST stimulates BTB13-specific T cells whereas SEC-bovine stimulates BTB93-specific T cells. This suggests that the presence of SaPIbov may contribute to modulation of the bovine immune response.
Staphylococcus aureus can cause many diseases in humans and animals. It is the most frequent cause of bovine mastitis and is a huge economic problem for the dairy industry worldwide (26). Typically, the disease is of a chronic nature, with subclinical mastitis being the most common form. The organisms may survive for long periods of time in the host without causing overt symptoms of disease. Often, antibiotic therapy merely converts a clinical infection to a subclinical form of the disease. The bacterial factors allowing persistence in the host are poorly understood.
S. aureus can produce several superantigens (SAgs) including toxic shock syndrome toxin 1 (TSST-1) and nine immunological variants (A to E and G to J) of staphylococcal enterotoxins (SEs) (6). These exotoxins are involved in modulating the host immune response and may contribute to evasion of host defenses and bacterial persistence (10). Genes encoding SAgs are often associated with mobile genetic elements such as pathogenicity islands, phages, and plasmids (5, 23, 34). Pathogenicity islands are accessory genetic elements that range in size from 10 to 200 kb, contain one or more genes associated with virulence, are bordered by directly repeated sequences, can be deleted en bloc, and may have integrase-like genes (15, 18). Recently Lindsay et al. (23) described a pathogenicity island (SaPI1) in a human clinical S. aureus isolate that contained the gene for TSST-1 (tst) and an open reading frame (ORF) with marked sequence similarity to those encoding SEs. The mobility of SaPI1 was demonstrated by phage-assisted excision, transduction, and site-specific integration into a recA mutant strain.
Previous studies (12, 19) showed that about 20% of bovine S. aureus strains coproduced TSST-1 and SEC. Since these toxins are rarely produced singly by bovine strains, their genes may be linked. This notion was supported by the observation that tst- and sec-specific probes hybridized to HindIII restriction fragments of the same size in Southern blot analysis.
In this study, we characterized the associated genetic element, named bovine staphylococcal pathogenicity island (SaPIbov), and analyzed the activity of these toxins on bovine lymphocytes.
MATERIALS AND METHODS
Bacterial strains.
Strains and plasmids are listed in Table 1. S. aureus strains were grown on tryptic soy agar or in tryptic soy broth and stored as glycerol stocks at −70°C. Where appropriate, the antibiotics erythromycin (10 μg/ml), tetracycline (2 μg/ml), and chloramphenicol (5 μg/ml) were incorporated.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmida | Properties | Reference(s) or source |
|---|---|---|
| S. aureus strains | ||
| RF122 | Wild-type strain from bovine mastitis (TSST+ SEC+) | 12 |
| RF122-1 | tst::Tcr mutant | This study |
| RF122-2 | tst::Tcrsec::Emr mutant | This study |
| RF120 | SaPIbov-negative strain of same clonal type as RF122 | 12 |
| KB103 | recA mutant | 3 |
| RN4220 | Restriction/modification− | 21 |
| Plasmids | ||
| pJRF | sec gene (150-bp internal deletion) with 1.4-kb TaqI Emr insert from pE194 cloned into pUC18 | This study |
| pJRF101 | 6.5-kb HindIII fragment in pBluescript KS(+) | This study |
| pJRF102 | 4-kb HindIII fragment from RF122 containing left-hand junction cloned into pUC19 | This study |
| pJRF103 | 1-kb PCR product from RF120 containing chromosomal insertion site of SaPIbov cloned into pCR2.1-TOPO | This study |
| pJRFsec::Emr | pJRF with pTS2 (Cmr) cloned into HindIII site flanking sec gene | This study |
| pRN6684 | tst::tmn (Tcr) Emr | 31 |
| pCR2.1-TOPO | Vector for cloning PCR products | Invitrogen |
| pE194 | 1.4-kb EmrTaqI fragment | 16 |
| pTS2 | Cmr, ts rep; derived from pTV1ts, MCSb from pBluescript | C. O'Connell (personal communication); 14, 33 |
| pBluescript KS(+) | Phagemid derived from pUC19; MCS, vector for blue/white colony selection | Stratagene |
Strains RF120 and RF122 were previously shown to be of the same clonal type by random amplified polymorphic DNA typing, multilocus enzyme electrophoretic typing, and ribotyping analysis (11).
MCS, multiple cloning site.
TSST-1 and SEC production.
Culture supernatant fluids of S. aureus were tested using reverse passive latex agglutination (RPLA) toxin detection kits for TSST-1 (TST-RPLA; Oxoid Ltd., Basingstoke, England) and SEC (SET-RPLA; Oxoid).
DNA manipulations.
Manipulations of DNA were performed by standard techniques (29).
Construction of plasmid pJRFsec::Emr.
The 5′ and 3′ parts of the sec gene including 400 and 250 bp, respectively, of flanking sequence were PCR amplified from plasmid pJRF101 using specific primers (Table 1). Primers were designed so that the resulting PCR products would include a single ClaI site at one end of both fragments. The PCR products were digested with restriction endonuclease ClaI and ligated together. The resulting single fragment contained a 150-bp internal deletion compared to the wild-type sec gene. This fragment was cut at natural XbaIII and HindIII restriction sites present internal to the 5′ and 3′ ends, respectively, and ligated into the multiple cloning site of pUC18. This resulted in a final insert size of 950 bp. A 1.4-kb TaqI fragment containing the erythromycin resistance determinant from pE194 (16) was cloned into the ClaI site in the middle of the sec gene to form pJRF. The temperature-sensitive plasmid vector pTS2 (14, 33), which confers chloramphenicol resistance, was cloned into the HindIII site flanking sec::Emr to create pJRFsec::Emr.
Plasmid pRN6684 for construction of the tst knockout carries an in vitro-constructed tst::Tcr mutation present in a similar temperature-sensitive plasmid (31).
Construction of allele replacement mutants.
Plasmids pRN6684 and pJRFsec::Emr were introduced by electroporation into S. aureus strain RN4220 (2). Once in strain RN4220, the plasmid was transduced into strain RF122 using phage 85 (13). Allele replacement was carried out as described previously (13). The temperature-sensitive phenotype of the plasmids facilitated integration by homologous recombination, and a double-crossover event resulting in a stable mutant was detected by plating on appropriate antibiotics. Loss of TSST-1 or SEC production was tested by RPLA analysis.
Stimulation of bovine lymphocytes.
To assess bovine Vβ (boVβ) expansion by proteins in staphylococcal cultures, peripheral blood mononuclear cells were obtained by gradient centrifugation of heparinized bovine venous blood according to standard procedures (7). For use in bVβ analysis, nonadherent lymphocyte-enriched cell suspensions were prepared as described by Deringer et al. (9) and adjusted to a concentration of 2.5 × 106 cells/ml.
Lymphocyte cultures (3 ml) were stimulated with protein preparations obtained from concentrated staphylococcal culture supernatants. Aerated S. aureus cultures (RF122 and mutant derivatives) were grown overnight in Todd-Hewitt broth to stationary phase. The culture supernatant fractions were precipitated with 4 volumes of ice-cold ethanol and incubated (−20°C) for several hours. The resulting precipitate was recovered by centrifugation, dried, and resolubilized in 500 μl of water. Following clarification by centrifugation, an aliquot (5 μl) of the protein concentrate was added to the lymphocyte cultures. Cell cultures were incubated for 4 days (37°C and 7% CO2). Control cultures without stimuli were used to quantify background levels of boVβ in each donor.
Isolation of lymphocyte RNA and cDNA production.
Following stimulation, RNA was isolated from cultures using Trizol reagent (Life Technologies, Gaithersburg, Md.). cDNA was generated from approximately 5 μg of RNA using Superscript II reverse transcriptase (Life Technologies) and random DNA hexamers.
Quantitative PCR to determine boVβ levels in stimulated lymphocyte cultures.
The method described by Kotb et al. (20) with the modifications of Deringer et al. (9) was used for assessment of boVβ expression. Primers used in PCR assays to analyze boVβ expression by SAgs were previously described (9). They were designed based on bovine gene sequences reported by Tanaka et al. (32). The bovine T-cell receptor (TCR) primers designed for this study were derived from sequence information for T-cell clones designated BTB10, BTB13, BTB18, BTB27, BTB35, and BTB93. The panel of primers used included six boVβ primers, each specific for a subfamily of boVβ genes. A primer specific for the constant region of the Cβ chain gene (boCβ primer) was also used for each PCR, in addition to two primers designed to amplify a region of the constant Cα chain (boCα) allowing normalization of the boVβ values. The 3′Cα and 3′Cβ primers were 32P labeled (5′ end labeled with T4 polynucleotide kinase [Life Technologies]) prior to use in PCRs. Thermocycling conditions consisted of an initial denaturation at 95°C for 3 min, followed by 30 cycles at 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min. PCR products were resolved on 2% agarose gels and then quantified by scanning dried gels using a GS525 molecular imager system (Bio-Rad, Hercules, Calif.). Each PCR product was quantified in pixel density units (PDU) using Molecular Analysis software (Bio-Rad). Each boVβ PDU value was normalized by dividing with its corresponding Cα control value. Results were expressed as an increase in expansion index compared to unstimulated control cultures. Values below or near 1.0 represent no significant increase in expression.
PCR analysis.
Genomic DNA was isolated as previously described (11). The PCR amplification conditions were an initial denaturation step at 94°C for 2 min, followed by 30 cycles of 94°C for 2 min of denaturation, different annealing temperatures depending on primer sequences for 1 min, and 72°C for 1 min. Long-template PCR was carried out using the Expand long-template PCR system (Roche Molecular Biochemicals) according to the manufacturer's instructions.
Southern blot analysis.
Genomic DNA isolated from S. aureus was digested with appropriate restriction endonucleases, resolved by electrophoresis on a 0.8% agarose gel, and transferred onto a nylon membrane (Hybond-N+; Amersham). Specific probes were labeled using the DIG (digoxigenin) system (Roche Molecular Biochemicals) during PCR or by random-primed labeling.
Identification and isolation of tst-encoding genetic element.
Initial Southern blot analysis identified a 6.5-kb HindIII restriction fragment which hybridized to both tst-specific and a sec-specific probes. A partial library of strain RF122 was made by size fractionation of a HindIII genomic digest in a sucrose gradient. The fraction containing the 6.5-kb restriction fragment size range was ligated with pBluescript KS(+), transformed into E. coli XL1-blue, and plated on L agar with ampicillin, X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), and IPTG (isopropyl-β-d-thiogalactopyranoside) (blue/white selection plates). Colony blotting using a tst-specific probe was used to identify positive colonies, which were confirmed by PCR using tst-specific primers (Table 2).
TABLE 2.
PCR primers used in this study
| Name | Sequence | Target or function (reference) |
|---|---|---|
| tn1 | ATCCCTTACTGCAACACAGG | tst probe amplification |
| tst2 | TTTCCAATAACCACCCGTTT | tst probe amplification |
| juncf3 | GGCTGGAAACCGCGTAATTA | Integration site, outward PCR |
| VL | GGAATTCTGTGAAGATAGAAGTCCACC | LHS 7-kb probe, outward PCR |
| VR | GCATCTGTTGCTTGATATAGG | RHS 4-kb probe (outward PCR) and 9-kb RHS region |
| JR1 | CAGAGTAGGTATCCGTGCGG | 9-kb RHS region |
| pBSr | AACAGCTATGACCATG | pJRFsec::Emr construction (5′ sec amplification) |
| sec2 | CCATCGATTAAGAAAAGTGTAACAGCTC | pJRFsec::Emr construction (5′ sec amplification) |
| sec3 | CCATCGATGTTATTCCTCCATACATACA | pJRFsec::Emr construction (3′ sec amplification), sec probe amplification |
| sec4 | AGCTAGTTCCTTATAACAGC | pJRFsec::Emr construction (3′ sec amplification), sec probe amplification |
| seg1 | CGTCTCCACCTGTTGAAGG | seg detection (27) |
| seg2 | CCAAGTGATTGTCTATTGGTCG | seg detection (27) |
| sei1 | CAACTCGAATTTTCAACAGGTAC | sei detection (27) |
| sei2 | CAGGCAGTCCATCTCCTG | sei detection (27) |
LHS, left-hand side; RHS, right-hand side.
Outward-directed PCR.
This was performed using a Vectorette II kit (Sigma-Genosys) according to the manufacturer's instructions. This system is used to amplify regions of unknown DNA sequence flanking a region of known DNA sequence. Briefly, the target DNA was digested with an appropriate restriction enzyme. Vectorette units were ligated onto the ends of the cleaved target DNA. PCR amplification was carried out with one primer directed to the known sequence (custom primer) and the other primer specific for the Vectorette unit (Vectorette primer). The amplified products were then cloned and sequenced or used as probes in Southern hybridization experiments.
DNA sequencing and analysis.
DNA sequencing analysis was carried out on both DNA strands by MWG-Biotech, Milton Keynes, United Kingdom. The BLAST algorithm (BLASTN and BLASTX) was used to search for sequence similarities (1).
Nucleotide sequence accession number.
The SaPIbov sequence shown in Fig. 3 has been assigned GenBank accession number AF217235.
FIG. 3.
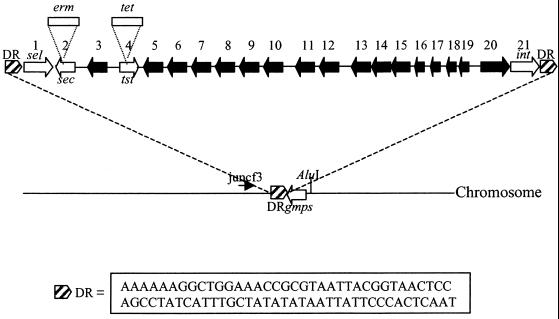
The 15,891-bp SaPIbov. Black arrows indicate ORFs of unknown function. White arrows indicate genes referred to in the text: sec, sel, int, tst, and gmps. The hatched boxes indicate positions of the direct repeats (DR), the sequence of which is given. Open boxes represent the erythromycin (erm) and tetracycline (tet) cassettes used to construct the TSST− and TSST− SEC− mutants. Primer juncf3 was used in Vectorette PCR to amplify DNA containing the SaPIbov integration site in strain RF120 after digestion with AluI.
RESULTS
Detection of the tst and sec genes.
Southern blot analysis was carried out on S. aureus RF122 genomic DNA digested with HindIII. Probes were generated by PCR amplification with primers specific for the tst and sec genes (Table 2). The probes hybridized to the same 6.5-kb restriction fragment (data not shown), suggesting that the genes lie adjacent to each other.
Cloning and sequencing of the tst/sec HindIII fragment.
The 6.5-kb HindIII fragment carrying tst and sec was cloned into pBluescript, forming pJRF101. DNA sequencing revealed that the sec and tst genes were approximately 2 kb apart and in opposite orientations (Fig. 1). Southern blot analysis using a PCR-generated probe specific for the region between the tst and sec genes indicated that this element was not present in a related strain (RF120) which did not produce TSST-1 or SEC (not shown). Use of plasmid pJRF101 as a probe showed the presence of the 6.5-kb hybridizing fragment in RF122 (Fig. 2) and, surprisingly, a second hybridizing fragment of 3 kb which was also present in RF120. Use of the plasmid vector only as a probe in Southern blot analysis did not demonstrate hybridization with genomic DNA from either strain RF120 or strain RF122 (not shown). This indicates that the 6.5-kb HindIII fragment contains sequences with homology elsewhere in the genome.
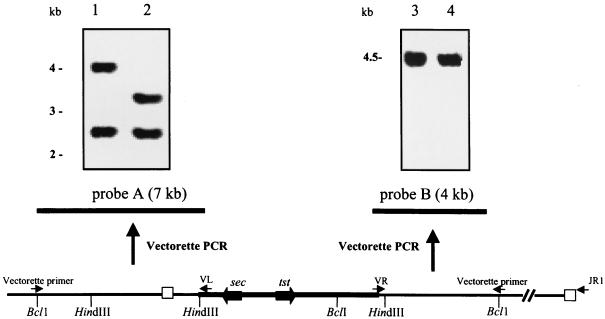
FIG. 1.
Mapping SaPIbov by PCR and Southern hybridization. Vectorette PCR was performed on DNA flanking the 6.5-kb HindIII fragment harboring tst and sec. DNA was cleaved with BclI, ligated with the Vectorette cassette, and subjected to PCR with outward-directed primer VL or VR and a primer specific for the Vectorette unit attached to the end of each fragment. These PCR products provided probes A and B, which were used to analyze genomic DNA of strain RF122 tst sec (lanes 1 and 3) and wild-type strain RF120 (lanes 2 and 4). Primers VR and JR1 were used to amplify the 9-kb right junction fragment. The left and right junctions of SaPIbov are denoted by open boxes.
FIG. 2.
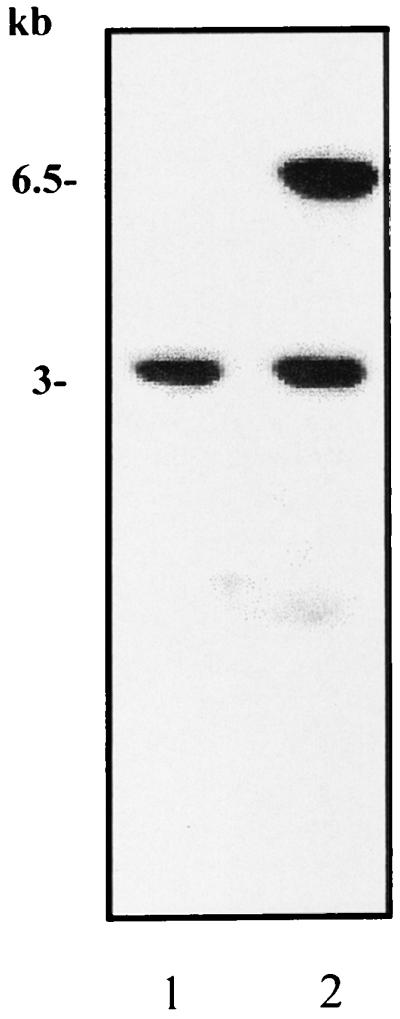
Southern blot analysis of HindIII-digested genomic DNA from strain RF120 (lane 1) and strain RF122 tst sec (lane 2) using pJRF101 containing the 6.5-kb HindIII fragment as a probe. The probe hybridized to a 6.5-kbp fragment of RF122 and also, unexpectedly, to a second 3-kb fragment of both strains, suggesting that the 6.5-kb tst/sec fragment contains homologous sequences elsewhere in the genome.
Characterization of the flanking regions.
To determine the extent of the putative insertion element containing the tst and sec genes, the sequences flanking the 6.5-kb HindIII fragment were analyzed by Southern hybridization. Probes specific for the flanking regions were constructed by outward-directed PCR using the Vectorette PCR system. Sequence analysis of pJRF101 identified a single BclI restriction site (Fig. 1) within the cloned 6.5-kb HindIII fragment. Southern blot analysis of strain RF122 genomic DNA cleaved with BclI and probed with pJRF101 indicated that BclI fragments of 11 and 5.5 kb overlapped the 6.5-kb tst/sec HindIII fragment. Vectorette PCR, outward directed from the 6.5-kb region of known sequence, was carried out on these BclI fragments with custom primers VL (left fragment) and VR (right fragment) and a primer specific for the Vectorette units that had been ligated to the ends of each fragment (Fig. 1). This resulted in PCR products of 7 kb (left) and 4 kb (right) (Fig. 1) which were DIG labeled and used as probes A and B in Southern blot analysis of strains RF122 and RF120, a related strain which does not contain the tst/sec element. This was done to determine if either of the Vectorette-amplified fragments contained a junction between the element and adjacent genomic DNA. If the probe contains sequence specific for the region containing the junction, it will hybridize to genomic DNA from both strains.
Southern hybridization analysis (Fig. 1) indicated that the TSST+ SEC+ strain RF122 and the related TSST− SEC− strain RF120 contained DNA sequences which hybridized with both probes A and B. Probe B hybridized to a single 4.5-kb HindIII fragment in both strains. This and Southern analysis using pJRF101 as a probe (Fig. 2) indicated that there were sequences present elsewhere in the genome with similarity to the tst/sec element. It appears that DNA within the 4.5-kb HindIII fragment is present in the tst/sec element in strain RF122 and also elsewhere in the genome in both strains RF122 and RF120. The absence of a second hybridizing band in RF122 is explained by the likelihood that the 4.5-kb fragment is duplicated elsewhere in the genome and so the two fragments appear as a single (more intense) band on the Southern blot. Accordingly, it was deemed that the right-hand junction may lie outside the 4-kb flanking region. Probe A hybridized to a 2.5-kb HindIII fragment in both strains RF122 and RF120 (Fig. 1). It also hybridized to a second fragment in strain RF122 (4 kb) and in strain RF120 (3.2 kb). This restriction fragment length polymorphism suggested that the left-hand junction lies within the 4-kb HindIII fragment of strain RF122 (Fig. 1). This fragment was cloned into pUC19 to form pJRF102 and sequenced. Southern analysis and sequence information identified where the left-hand junction was likely to be, and a primer (juncf3) was designed specific for a region to the left of the junction. Outward-directed Vectorette PCR was carried out on the TSST− SEC− RF120 DNA digested with AluI using this specific primer (Fig. 3), resulting in a 1-kb PCR product which was cloned into pCR2.1-TOPO (Invitrogen) to form pJRF103. Sequence analysis identified the point of divergence of RF120 DNA from RF122 DNA. This product contains the insertion site of the tst element and sequence to the right-hand side of this insertion site. Accordingly, a primer (JR1) specific for a region to the right of the diverged sequence in strain RF120 should be specific for the same region flanking the right-hand junction of the element in strain RF122. PCR primers VR and JR1 were then used to amplify the intervening sequencing of the tst element by long-range PCR (Fig. 1), resulting in a 9-kb PCR product which was sequenced directly.
Sequence analysis.
We propose that the inserted DNA element in S. aureus strain RF122 is a pathogenicity island referred to here as SaPIbov. Figure 3 shows a map of SaPIbov including 21 ORFs larger than 60 codons. The putative TSST protein showed up to 98% identity with proteins encoded by previously sequenced tst genes. It differs at three amino acid residues from tstO and seven amino acid residues from tst1 (22). The sec gene product is the SEC-bovine variant (24) which varies at three amino acid residues from SEC1 and is specific for bovine isolates of S. aureus. The hypothetical protein product of the staphylococcal enterotoxin (sel) gene, lying close to the left-hand junction, has 55% identity with SEI. In addition, there is an integrase-like gene just inside the right-hand junction, the protein product of which has 40% identity with a number of integrases including Streptococcus pyogenes bacteriophage T12 and phage T270, which carries the gene for streptococcal pyrogenic exotoxin A (SpeA). Products of other ORFs had low levels of similarity to proteins in the database. For example, ORF5 exhibited 34% identity with subunit 1 of the terminase enzyme of bacteriophage rho15.
Identification of the chromosomal integration site.
A 74-bp direct repeat occurs at the junction of the inserted element in strain RF122. This sequence occurs in the SaPIbov− strain RF120 and marks the chromosomal integration site which lies adjacent to the GMP synthase gene (gmps) in the S. aureus chromosome (Fig. 3).
Comparison with SaPI1.
Comparison of SaPIbov with the previously characterized SaPI1 (23) showed that although the elements appear to be related, they are quite distinct. There is a central region of sequence identity stretching from the tst gene to ORF11 containing six ORFs with up to 97% identity with ORFs from SaPI1. In addition, ORF15 has 71 and 100% identity with ORF12 and ORF13 from SaPI1, respectively. The direct repeats characteristic of pathogenicity islands are different in the two elements, with SaPI1 containing 17-bp repeats and SaPIbov containing 74-bp repeats. There was a single copy of the 74-bp repeat in strain RF120, which is closely related to strain RF122. BLAST analysis of the unfinished S. aureus genome databases of strains COL and 8325-4 found that only 24 of the 74 bases appeared to be present, while strain EMRSA-16 contained 27 of the 74 bases. Nonetheless, this still represents a recognition target which could potentially direct integration of the element into the genomes of these strains. The site of integration of SaPI1 lies near the tyrB gene, unlike that of SaPIbov, which lies at one end of the gmps gene. The deduced amino acid sequences of the integrase genes show about 40% identity. The five amino acids thought to be essential for integrase function are conserved (data not shown).
Mobility experiments.
To examine if phage could mobilize the pathogenicity island, we used transducing phages 80α, 85, and 11 to attempt to transduce SaPIbov marked with the tst::Tcr mutation to the recA strain KB103 (Table 1). No transductants were identified, which suggests that none of these phages could mobilize the element as has been reported for phages 13 and 80α with SaPI1.
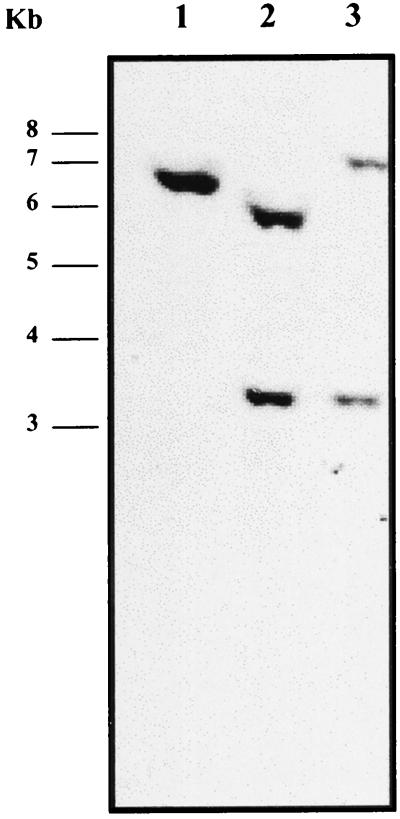
Confirmation of allele replacement by Southern hybridization.
The occurrence of a double-crossover event leading to the presence of a single copy of the mutated tst allele in strain RF122-1 and a single copy of the sec mutated allele in strain RF122-2 was confirmed by Southern hybridization (Fig. 4). A DIG-labeled PCR product specific for a region encompassing the SauI insertion site of the tet marker interrupting the tst gene was constructed and used to probe HindIII-cut genomic DNA from wild-type (RF122) and mutant (RF122-1 and RF122-2) strains. The presence of a HindIII site within the tet locus resulted in two hybridizing bands in the tst mutant (RF122-1) of 5.8 and 3.2 kb, compared to a single 6.5-kb hybridizing band in the wild type. In the tst sec double mutant (RF122-2), the 5.8-kb fragment was replaced by a fragment of 7.2 kb due to insertion of the 1.4-kb ermC fragment. This also confirmed that the genes are closely linked.
FIG. 4.
Southern blot hybridization analysis of tst and tst sec mutants. Genomic DNA from wild-type strain RF122 (lane 1), mutant RF122-1 tst (lane 2), and double mutant RF122-2 tst sec (lane 3) was cleaved with HindIII, separated by agarose electrophoresis, and transferred to a nylon membrane. It was hybridized with a probe specific for tst covering the site of insertion of the tet marker.
BoVβ analysis.
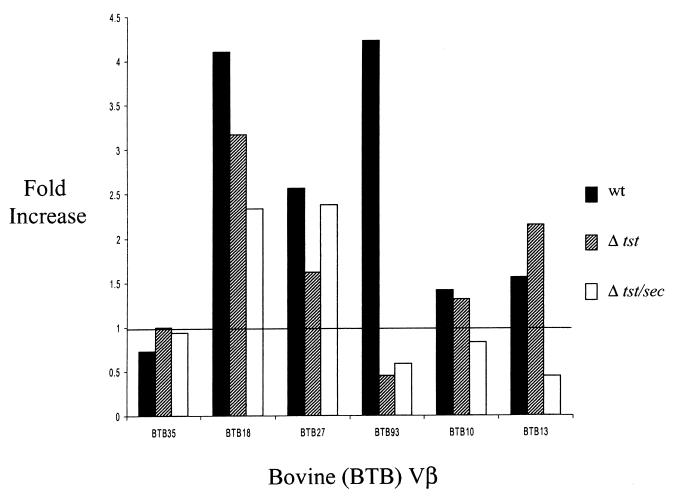
Initial screening of S. aureus RF122 indicated that the strain expressed SEC and TSST-1 (12). Therefore, it was of interest to determine whether this finding correlated with SAg properties of culture supernatant concentrates derived from this isolate. Culture supernatant concentrates from wild-type RF122 stimulated expression of boVβ BTB13 but not boVβ BTB35 (Fig. 5), typical of strains producing SEC-bovine as reported by Deringer et al. (9). Also, the sec mutant RF122-2 failed to activate expression of boVβ TB13 RNA. Interestingly, wild-type RF122 strongly activated expression of three additional boVβs (BTB18, BTB27, and BTB93) not shown previously to be associated with SEC-bovine. The activity of the RF122 tst::Tcr supernatant implicated TSST-1 as being responsible for expansion of boVβ BTB93 but not the other boVβs. Expression of boVβ BTB18 and boVβ BTB27 remained elevated even when cells were stimulated with the supernatant from the tst and the tst sec double mutants of strain RF122. This indicated that strain RF122 expresses SAgs in addition to SEC-bovine and TSST-1.
FIG. 5.
Representative analysis of boVβ expansion in bovine lymphocyte cultures. Bovine lymphocyte cultures were treated with protein preparations recovered from cultures of S. aureus RF122 (wt [wild-type]), S. aureus RF122-1 (Δtst), and S. aureus RF122-2 (Δtst/sec). Fold increase reflects the fold difference in measured PDU for each boVβ. This value was determined by quantifying reaction products (in PDU) generated from treated lymphocytes to those from an identical but untreated cell culture. All boVβ PDU values were normalized by quantifying bovine lymphocyte TCR Cα levels.
Screening for other SAgs.
Two additional SEs, SEG and SEI, have been recently identified (27). PCR analysis of strain RF122 and strain RF120 using gene-specific primers (Table 2) revealed that both isolates contained the seg and sei genes but they are not located within SaPIbov.
DISCUSSION
The screening procedure for staphylococcal SAgs used at the beginning of this project showed that strain RF122 produced both SEC and TSST-1. RF122 culture supernatants activated lymphocytes bearing Chothia subgroup 4 boVβ BTB13 (8), which is characteristic of SEC-bovine. The supernatants also demonstrated the unique inability of the SEC-bovine variant to activate boVβ BTB35 (9). Although the effect of TSST-1 on boVβs has not been reported previously, it was possible to attribute stimulation of boVβ BTB93 to this toxin. The wild-type strain activated boVβ BTB93, but the two mutants deficient in tst expression did not. This finding is entirely consistent with the effect of TSST-1 on human T cells. TSST-1 specifically activates human Vβ2, which belongs to Vβ subgroup 3. Interestingly, boVβ BTB93 is most related to human Vβs in the Chothia subgroup 3 (i.e., human Vβs 2 and 4) (9).
The residual SAg activity expressed by the RF122 tst and sec-bovine mutants suggested that the strains expressed SAgs other than SEC-bovine and TSST-1. The other expanded boVβs are only distantly related to those activated by SEC-bovine or TSST-1 and represent more divergent Vβ subgroups. For example, boVβ BTB18 and boVβ BTB27 TCRs are most related to human Vβs 1 and 7, which are classified in subgroups 1 and 2, respectively (9). Since expansion of neither of these could be attributed to either SEC-bovine or TSST-1, this activity was presumed to be caused by other SAgs. Molecular characterization of the pathogenicity island encoding the tst and sec-bovine genes revealed the presence of a novel enterotoxin-like gene (sel), the product of which is 55% identical to SEI. PCR analysis of RF122 genomic DNA revealed the presence of the recently identified seg and sei genes. We have shown that the recombinant SEL protein is expressed in S. aureus strain RN4220 and is mitogenic for bovine lymphocytes (unpublished data), suggesting that SEL is a SAg. Further work is in progress to confirm this. It is likely that the lymphocyte expansion unattributable to TSST-1 or SEC-bovine was caused by SEG, SEI, or SEL.
Lindsay et al. (23) demonstrated that SaPI1 could be mobilized by propagation of phages 13 and 80α. Propagation of the phages stimulated element excision and, in one case, replication as a circular plasmid-like element. The phages encapsidated the element and transduced it to other hosts, including a recA mutant. The element integrated at the att site, presumably directed by the element-encoded integrase. Although the phages used in this study were not effective at inducing mobilization, it is possible that the SaPIbov is mobilized in a similar fashion, and further screening may identify such a helper phage.
Strain RF122 is one of a group of clonally related strains, as identified by random amplified polymorphic DNA typing, multilocus enzyme electrophoretic typing, and ribotyping (11), which characteristically harbor SaPIbov and which produce TSST-1 and SEC (12). However, a few (12.5%) of the isolates in this group do not contain the element, suggesting that there has been a recent deletion event or, alternatively, that they represent the parent strain before the pathogenicity island was acquired. This supports the theory that the element is capable of deletion and/or horizontal transfer.
Comparison of SaPIbov with SaPI1 supports the statement of Lindsay et al. (23) that a family of related tst-containing elements exists in S. aureus. The elements show significant similarity clustered around the tst gene. However, outside this central core, the elements are quite distinct and appear to encode many different proteins.
SAgs act by binding specific Vβ elements on T cells, resulting in a proliferation of T cells and an overproduction of cytokines. This increased cytokine activity may have a detrimental effect on the host resulting in a suppressed immune response, shock, differential stimulation of CD4+ CD8+ cells, or T-cell unresponsiveness or deletion (25, 30). Through such immunomodulation, SAg action on the bovine host may facilitate persistence of S. aureus. The existence of variants of the SEC protein in staphylococci is presumably an adaptation to infection of different host species. Different SEC variants have been shown to interact with different T-cell Vβ elements (9). The deduced amino acid sequence of the product of the SaPIbov tst gene contains at least three divergent residues compared to other TSST sequences in the database. These differences could possibly be adaptations to the bovine immune system. It is clear from this study that TSST-1 activates T cells expressing specific Vβ elements. These Vβ elements are unique to the bovine host, and some amino acid residue differences may allow the toxin to bind specifically to them.
It appears to be a characteristic of superantigens that they are associated with mobile genetic elements. Both the sea and see genes are phage encoded (5), and the sed gene is plasmid encoded (4). It was previously reported that the seb and sec genes were associated with transmissible penicillin resistance plasmids. However, it is generally thought that these toxins are chromosomally encoded. This does not rule out the possibility that they are on mobile genetic elements. The seb gene is located on a DNA element that is at least 26.8 kb long (17). The present study reveals that the sec-bovine gene is associated with a pathogenicity island. This is the first study to characterize the genetic element encoding the sec gene. It is likely that the other sec variants may lie on related mobile genetic elements. Indeed, it will be interesting to ascertain the diversity of the elements encoding superantigens in S. aureus in general. In the near future, it is likely that a plethora of such elements will be characterized. The presence of superantigen genes on mobile elements facilitates their horizontal spread between strains of S. aureus. Moreover, the relatedness of the SaPIbov integrase to the integrase of S. pyogenes phage T270, which expresses SpeA, suggests that such elements may have crossed the genus barrier at some time.
Although not essential for virulence, SAgs may play an important role in host immune response evasion and survival. Overall, it is clear from this study that the presence of SaPIbov in strain RF122 enables it to produce SAgs which specifically activate bovine lymphocyte populations. Thus, through modulation of the immune response it may confer a survival advantage in the bovine host.
ACKNOWLEDGMENTS
This work was funded by a grant to J.R.F. from the Dairy Production Research Centre, Teagasc, and from the USDA (G.A.B.), Public Health Service (AI28401; G.A.B.), and Idaho Agricultural Experiment Station (G.A.B.). T.J.F. is supported by The Wellcome Trust.
REFERENCES
- 1.Altschul S F, Madden T L, Scaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Augustin J, Gotz F. Transformation of Staphylococcus epidermidis and other staphylococcal species with plasmid DNA by electroporation. FEMS Microbiol Lett. 1990;66:203–208. doi: 10.1016/0378-1097(90)90283-v. [DOI] [PubMed] [Google Scholar]
- 3.Bayles K W, Brunskill E W, Iandolo J J, Hruska L L, Huan S, Patee P A, Smiley B K, Yasbin R E. A genetic and molecular characterization of the recA gene from Staphylococcus aureus. Gene. 1994;147:13–20. doi: 10.1016/0378-1119(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 4.Bayles K, Iandolo J J. Genetic and molecular analyses of the gene encoding staphylococcal enterotoxin D. J Bacteriol. 1989;171:4799–4806. doi: 10.1128/jb.171.9.4799-4806.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Betley M J, Mekalanos J J. Staphylococcal enterotoxin A is encoded by phage. Science. 1985;229:185–187. doi: 10.1126/science.3160112. [DOI] [PubMed] [Google Scholar]
- 6.Bohach G A, Foster T J. Staphylococcus aureus exotoxins. In: Fischetti V, Novick R, Portnoy D, Rood J, editors. Gram-positive pathogens. Washington, D.C.: American Society for Microbiology; 1999. pp. 367–378. [Google Scholar]
- 7.Bøyum A. Isolation of mononuclear cells and granulocytes from human blood. Scand J Clin Lab Investig. 1968;21(Suppl. 97):77–89. [PubMed] [Google Scholar]
- 8.Chothia C, Boswell D R, Lesk A M. The outline structure of the T-cell αβ receptor. EMBO J. 1988;7:3745–3755. doi: 10.1002/j.1460-2075.1988.tb03258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deringer J R, Ely R J, Monday S R, Stauffacher C V, Bohach G A. Vβ-dependent stimulation of bovine and human T cells by host-specific staphylococcal enterotoxins. Infect Immun. 1997;65:4048–4054. doi: 10.1128/iai.65.10.4048-4054.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferens W F, Bohach G A. Persistence of Staphylococcus aureus on mucosal membranes: superantigens and internalization by host cells. J Lab Clin Med. 2000;135:225–230. doi: 10.1067/mlc.2000.105179. [DOI] [PubMed] [Google Scholar]
- 11.Fitzgerald J R, Meaney W J, Hartigan P J, Smyth C J, Kapur V. Fine-structure molecular epidemiological analysis of Staphylococcus aureus recovered from cows. Epidemiol Infect. 1997;119:261–269. doi: 10.1017/s0950268897007802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fitzgerald J R, Hartigan P J, Meaney W J, Smyth C J. Molecular population and virulence factor analysis of Staphylococcus aureus from bovine mastitis. J Appl Microbiol. 2000;88:1028–1038. doi: 10.1046/j.1365-2672.2000.01071.x. [DOI] [PubMed] [Google Scholar]
- 13.Foster T J. Molecular genetic analysis of staphylococcal virulence. Methods Microbiol. 1998;27:433–454. [Google Scholar]
- 14.Greene C, McDevitt D, Francois P, Vaudaux P E, Lew D P, Foster T J. Adhesion properties of mutants of Staphylococcus aureus defective in fibronectin-binding proteins and studies on the expression of fnb genes. Mol Microbiol. 1995;17:1143–1152. doi: 10.1111/j.1365-2958.1995.mmi_17061143.x. [DOI] [PubMed] [Google Scholar]
- 15.Hacker J, Blum-Oehler G, Mühldorfer I, Tschäpe H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 16.Horinouchi S, Weisblum B. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin B antibiotics. J Bacteriol. 1982;149:804–814. doi: 10.1128/jb.150.2.804-814.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johns M B, Khan S A. Staphylococcal enterotoxin B gene is associated with a discrete element. J Bacteriol. 1988;170:4033–4099. doi: 10.1128/jb.170.9.4033-4039.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaper J B, Hacker J. The concept of pathogenicity islands. In: Kaper J B, Hacker J, editors. Pathogenicity islands and other mobile virulence elements. Washington, D.C.: American Society for Microbiology; 1999. pp. 1–11. [Google Scholar]
- 19.Kenny K, Reiser R F, Bastida-Corcuera F D, Norcross N L. Production of enterotoxins and toxic shock syndrome toxin by bovine mammary isolates of Staphylococcus aureus. J Clin Microbiol. 1993;31:706–707. doi: 10.1128/jcm.31.3.706-707.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotb M, Watanabe-Ohnishi R, Wang B, Tomai M A, Le Gros L, Schlievert P M, El Demellawy M, Geller A M. Analysis of the TCR Vβ specificities of bacterial superantigens using PCR. Immunomethods. 1993;2:33–40. [Google Scholar]
- 21.Kreiswirth B N, Löhfdahl S, Betley M J, O'Reilly M, Schlievert P M, Bergdoll M S, Novick R P. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 22.Lee P K, Kreiswirth B, Deringer J, Projan S J, Eisner W, Smith B L, Carlson E, Novick R P, Schlievert P M. Nucleotide sequences and biologic properties of toxic shock syndrome toxin-1 from ovine and bovine-associated Staphylococcus aureus. J Infect Dis. 1993;165:1056–1063. doi: 10.1093/infdis/165.6.1056. [DOI] [PubMed] [Google Scholar]
- 23.Lindsay J A, Ruzin A, Ross H F, Kurepina N, Novick R P. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol Microbiol. 1998;29:527–543. doi: 10.1046/j.1365-2958.1998.00947.x. [DOI] [PubMed] [Google Scholar]
- 24.Marr J C, Lyon J D, Roberson J R, Lupher M, Davis W C, Bohach G A. Characterization of novel type C enterotoxins: biological and evolutionary implications. Infect Immun. 1993;61:4254–4262. doi: 10.1128/iai.61.10.4254-4262.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michie C A, Cohen J. The clinical significance of T-cell superantigens. Trends Microbiol. 1998;6:61–65. doi: 10.1016/S0966-842X(97)01193-1. [DOI] [PubMed] [Google Scholar]
- 26.Miles H, Lesser W, Sears P. The economic implications of bioengineered mastitis control. J Dairy Sci. 1992;75:596–605. doi: 10.3168/jds.S0022-0302(92)77797-2. [DOI] [PubMed] [Google Scholar]
- 27.Monday S R, Bohach G A. Use of multiplex PCR to detect classical and newly described pyrogenic toxin genes in staphylococcal isolates. J Clin Microbiol. 1999;37:3411–3414. doi: 10.1128/jcm.37.10.3411-3414.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munson S H, Tremaine M T, Betley M J, Welch R A. Identification and characterization of staphylococcal enterotoxin types G and I from Staphylococcus aureus. Infect Immun. 1998;66:3337–3348. doi: 10.1128/iai.66.7.3337-3348.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 30.Schlievert P M. Role of superantigens in human disease. J Infect Dis. 1993;167:997–1002. doi: 10.1093/infdis/167.5.997. [DOI] [PubMed] [Google Scholar]
- 31.Sloane R, de Azavedo J C, Arbuthnott J P, Hartigan P J, Kreiswirth B, Novick R, Foster T J. A toxic shock syndrome toxin mutant of Staphylococcus aureus isolated by allelic replacement lacks virulence in a rabbit uterine model. FEMS Microbiol Lett. 1991;62:239–244. doi: 10.1016/0378-1097(91)90164-6. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka A, Ishiguro N, Shinagawa M. Sequence and diversity of bovine T-cell receptor β-chain genes. Immunogenetics. 1990;32:263–271. doi: 10.1007/BF00187097. [DOI] [PubMed] [Google Scholar]
- 33.Youngman P. Plasmid vectors for recovering and exploiting Tn917 transpositions in Bacillus and other Gram-positive bacteria. In: Hardy K G, editor. Plasmids: a practical approach. Oxford, England: IRL Press; 1987. pp. 79–103. [Google Scholar]
- 34.Zhang S, Iandolo J J, Stewart G C. The enterotoxin D plasmid of Staphylococcus aureus encodes a second enterotoxin determinant (sej) FEMS Microbiol Lett. 1998;168:227–233. doi: 10.1111/j.1574-6968.1998.tb13278.x. [DOI] [PubMed] [Google Scholar]