Abstract
Diorganotin carboxylates have received much interest in past decades due to their rich structural chemistry and wide range of applications in various fields. This review study provides an in-depth analysis of carbon NMR data of dimethyltin complexes. The absorptions shown by the carbonyl carbon, methyl groups attached to tin and the other carbons present in the complexes were presented in this study. The effects of nature and the number of substituents attached are also described in this report.
Keywords: Dimethytin complexes, Dimethyltin carboxylates, Ligands, 13C NMR spectroscopy
Dimethytin complexes; Dimethyltin carboxylates; Ligands; 13C NMR spectroscopy.
1. Introduction
The significant rise in research activity in the chemistry of organotin complexes in past decades is probably because organotin compounds can be studied using various physical methods. The incredible structural and stereochemical diversity ability is an essential characteristic of organotin compounds. These compounds' synthesis, properties, industrial and biological applications were studied in all respect. The ambient nature of carboxylate ligands is due to the numerous structural motifs identified in this compound family [1, 2, 3]. Many factors, including the flexibility of coordination geometries, the number of coordinations, and the manner of coordination, all played a role in the structural diversity of organotin (IV) carboxylates [4]. In industry, diorganotin dicarboxylates are used as polymerization and transesterification catalysts [5, 6]. Dimethyltin derivatives have a wide range of properties, with anti-cancer, anti-tumour, and anti-bacterial prominent [7, 8]. In the α-glucosidase enzyme assay, some organotin complexes inhibited the enzyme's activity, while the standard drug acarbose inhibited poorly [9]. A surprising finding was that the 4-(diethylamino)benzic acid-derived dimethyltin (IV) complex outperformed the dibutyltin (IV) and triorganotin (IV) derivatives in terms of anti-bacterial activity [10]. Organotin carboxylates have a wide spectrum of applications and possible uses as indicated by the reports in the literature [11]. Organotin carboxylate compounds were used for their biocidal properties and effectiveness as medicinal compounds [12, 13]. Unique structural and diverse properties of organotin carboxylates make these compounds as significant class with applications as catalyst [14], anti-cancer [15], anti-fungal [16, 17], anti-bacterial [18], insecticidal [19], and wood preservatives agents [20].
The spectroscopic and technological developments enabled researchers from various disciplines to use these techniques not only for the complex structural purposes of molecules but also for wide-ranging applications. 13C Nuclear Magnetic Resonance (NMR) study has attained the status of one of the best spectroscopic methods for significant structural characterization, containing organic groups, among the currently useful spectroscopical techniques (Infrared, proton, carbon and tin NMR, Mössbauer spectroscopic studies and Mass spectrometry). In order to get a better insight of the structure chemistry of organotin carboxylates, the 13C NMR spectra of several dimethyltin carboxylates were investigated, and the results were presented in this paper.
2. 13C NMR data of dimethyltin carboxylates
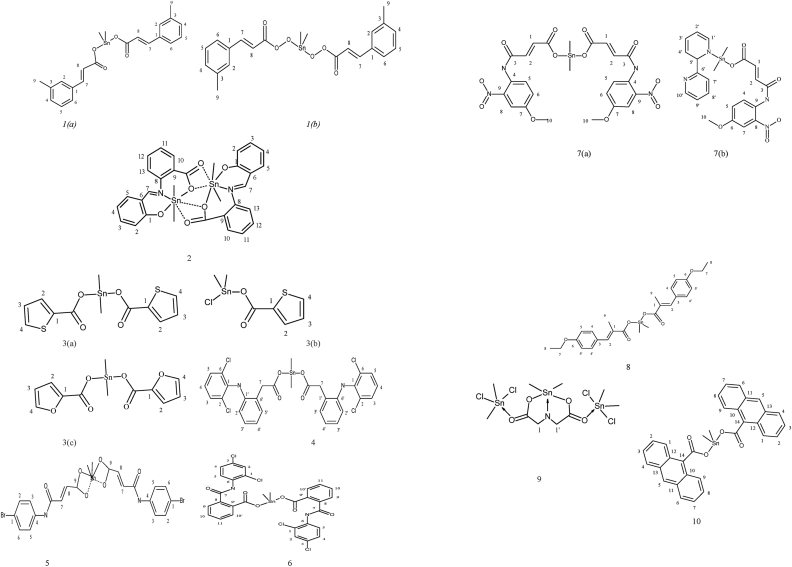
Two dimethyltin carboxylate complexes 1(a) and 1(b) of m-methyl trans-cinnamic acid ligand were synthesized, and their 13C spectroscopic characterization was carried out. The carbonyl carbon showed resonance at δ 176, and 173 ppm, the tin atom's methyl carbon, and the ligand's methyl group showed peaks at δ 5.2 and 9.8/6.6 ppm and δ 21.23 and 21.3 ppm respectively in different dimethyltin carboxylates [21]. A new dimethyl organotin complex (2) synthesized from ligand 2-(2-hydroxybenzylideneamino)benzoic acid showed carbon resonances at δ 173 ppm (COO); 147 (1, 6, 8, 9 carbons); 136, 130, 125 (2, 3, 4, 5, 10, 11, 12, 13 carbons), 115 (7 carbon), 10 (Sn–CH3) ppm respectively [22]. The carbonyl carbon in [Me2Sn(O2CC4H3E)2] complexes 3(a), 3(b) and 3(c) where E is oxygen or sulphur of 2-thiophene- or 2-furan-carboxylic acids showed resonance at δ 170 ppm while the ring carbons showed resonances between δ 127–134 ppm. In addition to finding the structure of methyltin (IV) compounds in solution, spectroscopic proof from the coupling constants of the methyl carbons and the tin hydrogen of [Me2Sn(O2CR)]2O2 (where R = Thiophene and furan) revealed that there was no interaction between tin and sulphur or oxygen atoms in the compound [23]. Based on spectroscopic evidence of {[Me2Sn(O2CR)]2O}2 (R = Thiophene and furan), the dimeric structure of dicarboxylato tetraorganodistannoxanes with R2Sn atoms upfield (for exocyclic) and downfield (for endocyclic) resonance. Deshielding of the respective free acids in the heterocyclic ring was shown [24]. A low-intensity peak at δ 178.5 ppm in the 13C NMR of dimethyltin (2-[2,6-dichlorophenylamino]phenylacetate) complex (4) demonstrated that the carboxylate ion was not in a free state. The shift in the exocyclic methylene carbon and carbon 6 field values towards lower field values revealed the bonding of the metal through the carboxylic group in the methylene carbon [25].
13C NMR results for Me2Sn(4-bromomaleanalate)2 (5), showed resonances caused by the carboxylate carbon after coordination has been shifted. During the process of complexation, the other carbons in the molecule did not move considerably. With the 1J value for the molecule, the projected C–Sn–C angle was 109.50°, indicating that the complex was in a tetragonal environment around the tin nucleus [26]. Table 1 gives the list and overview of the ligands. Shahzadi and co-workers reported carbon NMR of dimethyltin-2-[(2,4-dichloroanilino carbonyl)]benzoate complexes (6). The position of the carboxylate group in tin bonding was corroborated by the resonance attributed to C, which indicated a shift in the position of the carboxylate group following coordination. The carbon atoms of the methyl groups that are connected to the tin atom are discovered to be in the same location [27]. In dimethyltin complexes 7(a) and 7(b) derived from 4-((4-methoxy-2-nitrophenyl)amino)-4-oxobut-2-enoic acid ligand reported in a study. The carboxylate moiety resonances showed a noticeable shift to downfield concerning the Sn due to the electronegative Sn atom withdrawing electron density away from the ligand. In the Lockhart Equation, the C–Sn–C bond angle value for methyltin compounds calculated from the 2J (119Sn 1H) coupling was 117° [28]. Figure 1 explains the structures of complexes number 1–10. The 13C NMR of complexes (8) of formula [Me2SnL2] [where L = 3-(4-Ethoxyphenyl)-2-methylacrylate], showed 1J [119Sn, 13C], 703 Hz (146°) corresponding to 5-coordinated tin atoms in solutions comparable with literature results. The data was further supported five coordinated structure of the tin atom in solutions by C–Sn–C angles values using Lockhart Equation. The pattern of complexity ranging from six solid-state coordination to five solution coordination attributed to fluxion behaviour [29]. In a methanol solvent, dimethyltin dicarboxylates (9) were made by heating disodium iminodiacetate hydrate using dimethyltin dichloride. The carbon NMR of the complex showed resonances at δ 179 ppm due to carbonyl carbon and CH2 group attached to carbonyl carbon at δ 51.9 ppm [30]. The dimethyltin complexes (10) Me2Sn [OC(O)C14H9]2 CH3OH derived from the ligand anthracenecarboxylic acid, was reported to be characterized by elemental analysis, mass spectrometry, multinuclear nuclear magnetic resonance studies. The 13C NMR resonances due to different Carbons appeared at δ 5.4 ppm (CH3, 1J (119Sn–13C), 50.3 (CH3OH), 124.8, 124.9, 126.1, 126.7, 128.1, 128.4, 129.6, 130.5 (C14H9), and 178.4 (COO) ppm [31]. Table 2 depicts 13C NMR data of complexes number 1–10.
Table 1.
List and overview of ligands.
| Ligand no. | Ligand name | Structure | Double bond at position | Functional group/Substituents attached |
Reference | |
|---|---|---|---|---|---|---|
| Group | Position | |||||
| L1 | m-Methyl trans-cinnamic acid | 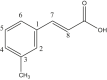 |
1, 3, 5, 7 | CH3 | 3 | [21] |
| L2 | 2-(2-Hydroxybenzylideneamino) benzoic acid | 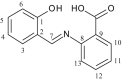 |
2, 4, 6, 7, 9, 11, 13 | OH | 1 | [22] |
| L3 | Thiophene-2-carboxylic acid | 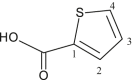 |
1, 3 | - | - | [23, 24] |
| L4 | Furan-2-carboxylic acid | 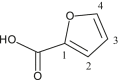 |
1, 3 | - | - | |
| L5 | (2-[2,6-Dichlorophenylamino] phenylacetic acid) | 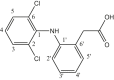 |
2, 4, 6, 1′, 3′, 5′ | Cl | 2, 6 | [25] |
| L6 | 4-Bromomaleanilic acid | 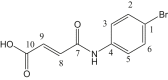 |
2, 4, 6, 8 | Br; C=O | 1; 7 | [26] |
| L7 | 2 [(2,4-DichloroanilinoCarbonyl)] benzoic acid) | 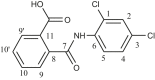 |
1, 3, 5, 8, 10, 9′ | Cl; C=O | 1, 3; 7 | [27] |
| L8 | 4-((4-methoxy-2-Nitrophenyl) amino)-4-oxobut-2-enoic Acid | 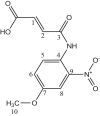 |
1, 4, 6, 8 | C=O; OCH3; NO2 | 3; 7; 9 | [28] |
| L9 | 3-(4-Ethoxyphenyl)-2-methylacrylic acid | 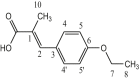 |
1, 3, 5′, 5 | CH3; OCH2CH3 | 1; 6 | [29] |
| L10 | Iminodiacetic acid | 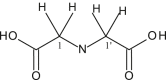 |
- | - | - | [30] |
| L11 | Di-(9- anthracene carboxylic acid) | 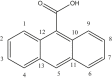 |
2, 4, 5, 6, 8, 10, 12 | - | - | [31] |
| L12 | 4-Phenylbutyric acid | 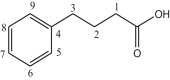 |
5, 7, 9 | - | - | [32] |
| L13 | 2-(2-Fluorobenzylidene) butanoic acid | 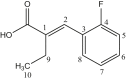 |
1, 4, 6, 8 | F; CH2CH3 | 4; 1 | [33] |
| L14 | (3,5-Di-tert-butyl-4-hydroxybenzoate) | 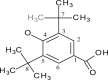 |
1, 3, 5 | C(CH3)3; OH | 3, 5; 4 | [34] |
| L15 | (3-(3,5-Di-tert-butyl-4-hydroxyphenyl) propionate) | 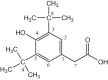 |
1, 3, 5 | C(CH3)3; OH | 3, 5; 4 | |
| L16 | 2-(N-Maleoyl)-3-phenylpropanoic acid | 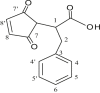 |
3, 5, 5′, 8 | C=O | 7, 7′ | [35] |
| L17 | 6-Nitropiperonylic acid | 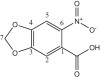 |
1, 3, 5 | NO2 | 6 | [36] |
| L18 | 3-[(3′,5′-Dimethylphenylamido)] propanoic acid | 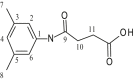 |
1, 3, 5 | C=O; CH3 | 9; 3, 5 | [37] |
| L19 | (S)-(+)-6-Methoxy-alpha-methyl-2-naphthaleneacetic acid | 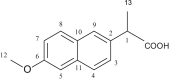 |
3, 11, 6, 8, 9 | CH3; OCH3 | 1; 6 | [38] |
| L20 | 2-[(2′,4′,6′-Tribromophenylamido)] benzoic Acid | 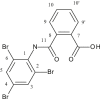 |
1, 3, 5, 9, 10′, 11 | C=O; Br | 7; 2, 4, 6 | [39] |
| L21 | 3-[(2′,4′,6′-Tribromophenylamido)] propanoic Acid | 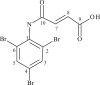 |
1, 3, 5, 8 | C=O; Br | 7; 2, 4, 6 | |
| L22 | 3-(4-Cyanophenyl)-2-methylacrylic acid | 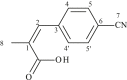 |
1, 3, 5′, 5 | CH3; CN | 1; 7 | [40] |
| L23 | 6-Chloro-3-pyridineacetic acid | 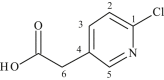 |
1, 3, 5 | Cl | 1 | [41] |
| L24 | Bis 2-(4-Methoxy-2-nitrophenylcarbamoyl) benzoic acid | 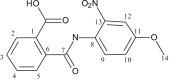 |
2, 4, 6, 8, 10, 12 | C=O; NO2; OCH3 | 7; 13; 11 | [42] |
| L25 | 3,4-Methylenedioxy 6-nitrophenylpropenoic Acid | 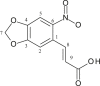 |
1, 3, 5, 8 | NO2 | 6 | [43] |
| L26 | 3-(4-Fluorophenyl) acrylic acid | 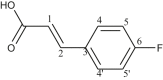 |
1, 3, 5′, 5 | F | 6 | [44] |
| L27 | 4-p (Chlorophenyl)-2-phenyl-5-thiazoleacetic acid | 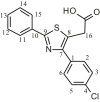 |
2, 4, 6, 7, 9, 10, 12, 14 | Cl | 4 | [45] |
| L28 | 2-{[5-(2-Nitrophenyl) furan-2-yl] methyleneamino} benzoic acid | 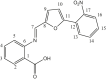 |
1, 3, 5, 7, 8, 10, 12, 14, 16 | NO2 | 17 | [46] |
| L29 | 1-Ethyl-7-methyl-4-oxo-1,4-dihydro-1,8-naphthyridine-3-carboxylic acid | 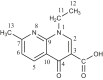 |
2, 10, 6, 8 | CH2CH3; C=O; CH3 | 1; 4; 7 | [47] |
| L30 | Phenyl acetylene carboxylic acid |  |
4, 6, 4' (C=C) 1 (C≡C) | - | - | [48] |
| L31 | 2-Phenyl-4-selenazole carboxylic acid |  |
3, 5, 7 | - | - | [49] |
| L32 | 2-Aminobenzoic acid |  |
1, 3, 5 | NH2 | 6 | [50] |
| L33 | 2,6-Pyridinedicarboxylic acid |  |
1, 2, 2′ | - | - | [51] |
Figure 1.
Structure of complex 1–10: (a) Dimethyl complexes 1 (a) and 1(b) with m-methyl trans-cinnamic acid ligand; 2 with 2-(2-hydroxybenzylideneamino)benzoic acid ligand; 3(a), 3(b) with thiophene-2-carboxylic acid and 3(c) with furan-2-carboxylic acid ligand; 4 complex (2-[2,6-dichlorophenylamino]phenylacetic acid) ligand; 5 with 4-bromomaleanilic acid ligand; 6 dimethyltin-2-[(2,4-dichloroanilinocarbonyl)]benzoate complex; 7(a) and 7(b) with 4-((4-methoxy-2-nitrophenyl)amino)-4-oxobut-2-enoic acid ligand; 8 with 3-(4-ethoxyphenyl)-2-methylacrylate ligand; 9 with iminodiacetate hydrate ligand; 10 with anthracene carboxylic acid ligand.
Table 2.
13C NMR data of complexes number 1–10.
| Complex | Complex formula | δ (13C) (ppm) |
J values (Hz) | C–Sn–C angle (º) | Reference | |||
|---|---|---|---|---|---|---|---|---|
| Carbons attached to Tin (Sn) |
Carbons of Ligand |
|||||||
| Position | Value | Position | Value | |||||
| 1 (a) | [Me2Sn(L1)2] | CH3 | 5.20 | COO | 176.0 | 1J [633.00] | 132.28 | [21] |
| 1 | 134.3 | |||||||
| 2 | 131.5 | |||||||
| 3 | 138.5 | |||||||
| 4 | 128.8 | |||||||
| 5 | 128.5 | |||||||
| 6 | 125.4 | |||||||
| 7 | 117.1 | |||||||
| 8 | 147.0 | |||||||
| 9 | 21.23 | |||||||
| 1 (b) | {[(Me2SnL1)2O]2} | CH3 | 9.80, 6.60 | COO | 173.0 | 1J [815.00], [756.00] | 148.25 | |
| 1 | 134.0 | |||||||
| 2 | 130.0 | |||||||
| 3 | 138.0 | |||||||
| 4 | 128.69 | |||||||
| 5 | 128.67 | |||||||
| 6 | 125.16 | |||||||
| 7 | 121.19 | |||||||
| 8 | 144.0 | |||||||
| 9 | 21.30 | |||||||
| 2 | [Me2Sn(L2)2] | CH3 | 10 | COO | 173 147 136, 130, 125 115 |
1J [960] | - | [22] |
| 1, 6, 8, 9 | ||||||||
| 2, 3, 4, 5, 10, 11, 12, 13 | ||||||||
| 7 | ||||||||
| 3 (a) | [Me2Sn(L3)2] | CH3 | 3.5 | COO | 170.7 | 1J [630] | 132 | [23, 24] |
| 1 | 133.7 | |||||||
| 2 | 133.4 | |||||||
| 3 | 128.0 | |||||||
| 4 | 134.6 | |||||||
| 3 (b) | [Me2SnCl(L3)] | CH3 | 3.5 | COO | 169.5 | - | - | |
| 1 | 132.6 | |||||||
| 2 | 133.6 | |||||||
| 3 | 127.8 | |||||||
| 4 | 134.8 | |||||||
| 3 (c) | [Me2Sn(L4)2] | CH3 | 5.1 | COO | 165.8 | - | - | |
| 1 | 144.7 | |||||||
| 2 | 118.9 | |||||||
| 3 | 111.8 | |||||||
| 4 | 146.4 | |||||||
| 4 | [Me2Sn(L5)2] | CH3 | 9.5, 6.6 | COO | 178.5 | - | 145, 141 | [25] |
| 1 | 137.9 | |||||||
| 2,6 | 129.7 | |||||||
| 3,5 | 128.8 | |||||||
| 4 | 123.9 | |||||||
| 5′ | 130.6 | |||||||
| 6′ | 125.4 | |||||||
| 7′ | 41.2 | |||||||
| 5 | [Me2Sn(L6)2] | CH3 | 4.5 | COO | 169.0 | - | - | [26] |
| 1 | 134.2 | |||||||
| 2,6 | 132.2 | |||||||
| 3,5 | 132.4 | |||||||
| 4 | 127.3 | |||||||
| 7 | 172.8 | |||||||
| 8 | 121.8 | |||||||
| 9 | 115.4 | |||||||
| 6 | [Me2Sn(L7)2] | CH3 | 14.12 | COO | 175.7 | - | - | [27] |
| 1 | 136.7 | |||||||
| 2,4 | 128.5 | |||||||
| 3,5 | 134.0 | |||||||
| 6 | 130.3 | |||||||
| 7 | 166.3 | |||||||
| 8,11 | 131.3 | |||||||
| 9,9′ | 124.0 | |||||||
| 10,10′ | 114.1 | |||||||
| 7 | [Me2Sn(L8)2] | CH3 | -0.7 | COO | 164.2 | 1J [833] | - | [28] |
| 1 | 131.4 | |||||||
| 2 | 133.8 | |||||||
| 3 | 170.2 | |||||||
| 4 | 125.2 | |||||||
| 5 | 126.8 | |||||||
| 6 | 121.1 | |||||||
| 7 | 156.3 | |||||||
| 8 | 109.5 | |||||||
| 9 | 142.7 | |||||||
| 10 | 56.6 | |||||||
| 8 | [Me2Sn(L9)2] | CH3 | 4.5 | COO | 179.1 | 2J [703] | 146.0 | [29] |
| 1 | 127.6 | |||||||
| 2 | 138.3 | |||||||
| 3 | 131.5 | |||||||
| 4,4′ | 130.9 | |||||||
| 5,5′ | 114.3 | |||||||
| 6 | 159.2 | |||||||
| 7 | 63.4 | |||||||
| 8 | 14.8 | |||||||
| 9 | 14.4 | |||||||
| 9 | {Me2SnL10 [Sn(Cl)2Me2]2} | CH3 | 23.5 | COO | 171.1 | - | 152.8, 134.3 | [30] |
| 1,1′ | 51.9 | |||||||
| 10 | [Me2Sn(L11)2] | CH3 | 5.4 | COO | 178.4 | 1J [642, 625] | 133 | [31] |
| 1–13 | 124.8–130.5 | |||||||
| 14 | 50.3 | |||||||
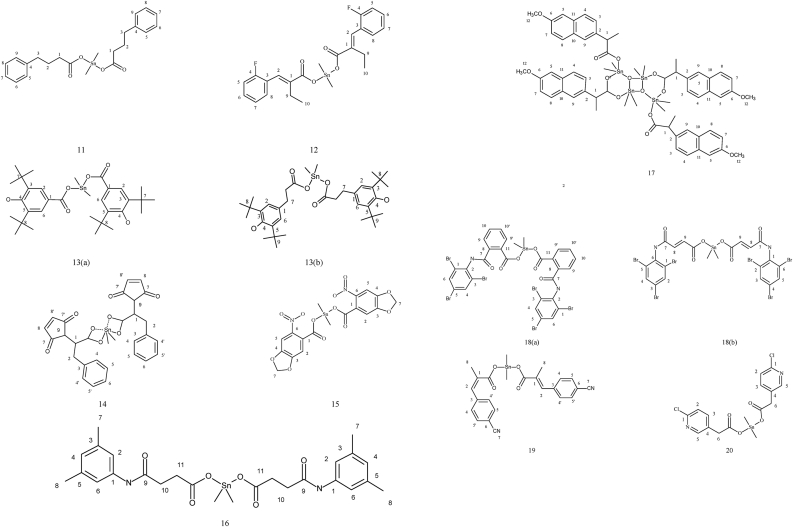
The reactions of Me2SnCl2 with NaO2C(CH2)3Ph, yielded carboxylates (11) of formula [{(Me2SnOC(CH2CH2CH2)2O}2]. The different carbon NMR peaks observed at δ 179.9, 35.7, 27.4, 34.4, 141.8–126.3, and 9.1 ppm [32]. The bis [-2-(2-fluorobenzylidene)butanoato] tetramethyldistannoxane complex (12) showed eleven peaks at δ 177.5, 130.2, 136.5, 123.7, 160.4, 115.7, 130.3, 130.02, 128.6, 21.5, 13.8 ppm due to carbons numbering from C1 to C-11 and at δ 4.8 ppm due to C–Sn [33]. The organotin (IV) carboxylates 13(a) and 13(b) based on 3,5-di-tert-butyl-4-hydroxyphenyl of formulae (RCOO)2SnMe2 showed peaks at δ 4.31 [Sn(CH3)2], 30.3 [С(СН3)3], 31.42 [ArCH2СH2COO], 34.31 [С(СН3)], 36.21 [ArCH2-СH2CO2], 124.88; 131.02; 135.87; 152.17 ppm due to Ar group and at δ 183.52 ppm due to carbonyl carbon [34]. The 13C NMR spectroscopy data of dimethyltin complexes (14) derived from 2-(N-maleoyl)-3-phenylpropanoic acid exhibited a number of signals representing magnetically different carbon atoms. The involvement of two C=O entities in the ligand produced two distinct, well-separated, regions ranging from δ 170–175 ppm. At δ 7.4 ppm, the moieties connected to the tin atom produced a signal [35]. The 13C NMR data of dimethytin compounds (15) derived from 6-nitropiperonylate, in CDCl3 solvent, showed resonances at δ 129.0, 108.5, 151.3, 149.8, 105.0, 143.2, 103.6 and 174.3 ppm attributed to carbons numbering from C1–C8 carbons. The methyl group attached to tin showed signal at δ 4.6 ppm [36]. Figure 2 depicts the structures of complex number 11–20. Dimethyltin complexes (16) developed from a new ligand synthesized by treating succinic anhydride and 3,5-dimethylaniline described in a report. The decrease in the 13C NMR shift upon coordination, in agreement with the results from the carboxylic carbon resonance, demonstrates that the carboxylate group plays a more significant role in the bonding to Sn. In the experimental findings, concerning reports in the literature, the carbon of alkyl groups bonded to tin is found in almost the same position [37]. A complex of formula {[Me2Sn(L)]2O}2 (17) reported in a study where ligand L was (S)-(+)-6-methoxy-alpha-methyl-2-naphthaleneaceto anion. A significant downfield shift in the 13C NMR spectrum, including all carbon resonances, was observed in comparison to the ligands, which was attributed to the outflow of electron density from the ligand to the metal atoms. At δ 174.8 ppm, the resonances are attributed to the carbonyl carbon [38]. In dimethyltin complexes 18(a) and 18(b) showed values of δ 163.0–166.8 and 170–176.8 ppm due to the –COO and –C=O group, respectively. Aromatic carbon resonances showed signals in the region reported in the literature [39]. The 13C NMR data of the complex (19) showed peaks of δ 177.6, 125.3, 140.3, 138.4, 128.2, 132.2, 111.8, 118.5, 14.4 ppm due to C1–C9 carbons and δ 4.6 ppm due to the methyl carbon attached to the tin [40]. The carbon NMR spectra of organotin complex (20) synthesized using 6-chloro-3-pyridineacetic acid ligand and dimethyltin dichloride showed signals at δ 173.3 ppm due to COO group, 124.1–150.2 ppm due to C6H5–C group, 37.7 ppm due to C6H5–CH2 group and δ 14.1 ppm due to CH3 carbon [41]. Table 3 presents 13C NMR data of complexes number 11–20.
Figure 2.
Structure of complex 11–20: dimethyl complexes; 11 with 4-phenylbutyric acid; 12 bis(-2-(2-fluorobenzylidene)butanoato) tetramethyldistannoxane complex; 13(a) with (3,5-di-tert-butyl-4-hydroxybenzoate) and 13 (b) with (3-(3,5-di-tert-butyl-4-hydroxyphenyl)propionate) ligand; 14 with 2-(N-maleoyl)-3-phenylpropanoic acid ligand; 15 with 6-nitropiperonylic acid ligand; 16 with 3-[(3′,5′-dimethylphenylamido)]propanoic acid ligand; 17 with (S)-(+)-6-methoxy-alpha-methyl-2-naphthaleneacetic acid ligand; 18(a) with 2-[(2′,4′,6′-tribromophenylamido)]benzoic acid and 18(b) with 3-[(2′,4′,6′-tribromophenylamido)]propenoic acid ligand; 19 with 3-(4-cyanophenyl)-2-methylacrylic acid ligand; 20 with 6-chloro-3-pyridineacetic acid ligand.
Table 3.
13C NMR data of complexes number 11–20.
| Complex | Compound | δ (13C) (ppm) |
J values (Hz) | C–Sn–C angle (º) | Reference | |||
|---|---|---|---|---|---|---|---|---|
| Carbons attached to Tin (Sn) |
Carbons of Ligand |
|||||||
| Position | Value | Position | Value | |||||
| 11 | [(Me2Sn(L12)2] | CH3 | 9.1 6.6 |
COO | 179.9 |
1J [802/767] 1J [758/722] |
147.1, 143.2 | [32] |
| 1 | 35.7 | |||||||
| 2 | 27.4 | |||||||
| 3 | 34.4 | |||||||
| 4–9 | 141.8–126.3 | |||||||
| 12 | {[Me2SnL13]2O}2 | CH3 | 4.8 | COO | 177.5 |
1J [14.3] 1J [248.2] 1J [21.8] 1J [8.2] 1J [3.7] 1J [3.0] |
- | [33] |
| 1 | 130.2 | |||||||
| 2 | 136.5 | |||||||
| 3 | 123.7 | |||||||
| 4 | 160.4 | |||||||
| 5 | 115.7 | |||||||
| 6 | 130.3 | |||||||
| 7 | 130.02 | |||||||
| 8 | 128.6 | |||||||
| 9 | 21.5 | |||||||
| 10 | 13.8 | |||||||
| 13 (a) | [Me2Sn(L14)2] | CH3 | 4.95 | COO | 176.42 | - | - | [34] |
| 1–6 | 120.53, 128.10, 135.75, 158.70, | |||||||
| 8 | 30.20 | |||||||
| 9 | 34.37 | |||||||
| 13 (b) | [Me2Sn(L15)2] | CH3 | 4.31 | COO | 183.52 | - | - | |
| 1–7 | 124.88, 131.02, 135.87, 152.17 | |||||||
| 8 | 31.42 | |||||||
| 9 | 36.21 | |||||||
| 14 | [Me2Sn(L16)2] | CH3 | 7.4 | COO | 170.1 | - | 131.3 | [35] |
| 2 | 54.3 | |||||||
| 3 | 34.9 | |||||||
| 4 | 137.6 | |||||||
| 5,5′ | 128.7 | |||||||
| 6,6′ | 133.7 | |||||||
| 7 | 126.6 | |||||||
| 8 | 175.2 | |||||||
| 9,9′ | 128.5 | |||||||
| 15 | [Me2Sn(L17)2] | CH3 | 4.6 | COO | 174.3 | - | - | [36] |
| 1 | 129.0 | |||||||
| 2 | 108.5 | |||||||
| 3 | 151.3 | |||||||
| 4 | 149.8 | |||||||
| 5 | 105.0 | |||||||
| 6 | 143.2 | |||||||
| 7 | 103.6 | |||||||
| 16 | [Me2Sn(L18)2] | CH3 | 29.6 | COO | 177.4 | 349–396 | 359.3 | [37] |
| 1 | 138.03 | |||||||
| 2/6 | 117.08 | |||||||
| 3/5 | 139.67 | |||||||
| 4 | 124.84 | |||||||
| 7/8 | 21.85 | |||||||
| 9 | 170.69 | |||||||
| 10 | 29.26 | |||||||
| 11 | 31.38 | |||||||
| 17 | [Me2Sn(L19)2] | CH3 | 18.90 | COO | 180.85 | - | - | [38] |
| 2–11 | 157.54–105.56 | |||||||
| 6 | 55.25 | |||||||
| 12 | 47.43 | |||||||
| 18 (a) | [Me2Sn(L20)2] | CH3 | 29.6 | COO | 174.8 | - | - | [39] |
| 1 | 136.7 | |||||||
| 2/4/6 | 128.6 | |||||||
| 3/5 | 134.5 | |||||||
| 7 | 166.7 | |||||||
| 8 | 131.0 | |||||||
| 9, 9′ | 124.1 | |||||||
| 10,10′ | 115.7 | |||||||
| 11 | 130.2 | |||||||
| 18 (b) | [Me2Sn(L21)2] | CH3 | 29.6 | COO | 175.7 | - | - | |
| 1 | 135.0 | |||||||
| 2/4/6 | 129.5 | |||||||
| 3/5 | 133.7 | |||||||
| 7 | 164.2 | |||||||
| 8 | 124.6 | |||||||
| 9 | 114.4 | |||||||
| 19 | [Me2Sn(L22)2] | CH3 | 4.6 | COO | 177.6 | 1J [704/670] | 138.5 | [40] |
| 1 | 125.3 | |||||||
| 2 | 140.3 | |||||||
| 3 | 138.4 | |||||||
| 4 | 28.2 | |||||||
| 5 | 132.2 | |||||||
| 6 | 111.8 | |||||||
| 7 | 118.5 | |||||||
| 8 | 14.4 | |||||||
| 20 | [Me2Sn(L23)2] | CH3 | 14.1 | COO | 173.3 | - | - | [41] |
| 1–5 | 124.1–150.2 | |||||||
| 6 | 37.7 | |||||||
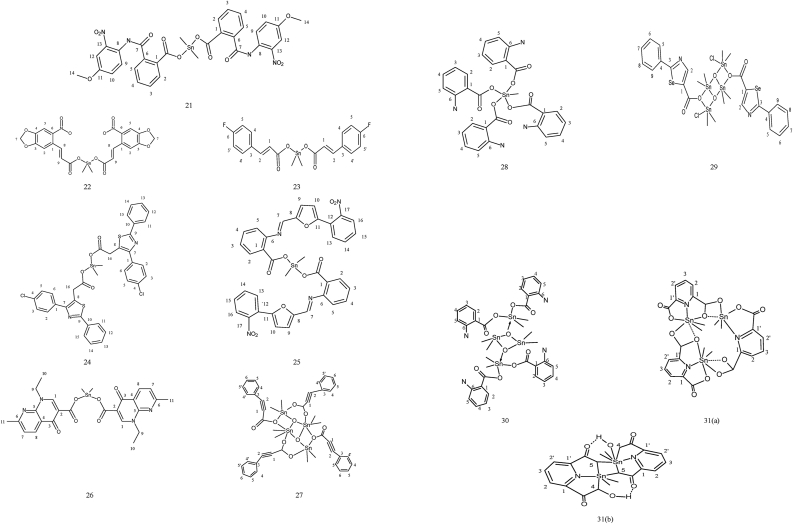
The dimethyltin complexes (21) derived from 2-(4-methoxy-2-nitrophenylcarbamoyl)benzoic acid ligand exhibited fifteen signals at δ 160.3, 129.6, 127.7, 132.9, 135.7, 124.4, 131.9, 167.0, 121.3, 128.7, 120.9, 156.6, 109.5, 142.5, and 56.0 ppm [C1–C15 carbons]. The Sn–CH3 carbon showed a signal at δ 0.6 ppm 1J [119Sn–13C], 800 Hz [42]. The complex of formula [Me2SnL2] (22), using 3,4-methylenedioxy-6-nitrophenylpropenoic acid ligand showed resonances due to carboxyl carbon moved to a lower region, indicating the coordination with the carboxyl group [43]. The dimethyltin complexes (23) derived from ligand 3-(4-fluorophenyl)acrylic acid ligand showed that C1 of the complexes were shifted downfield due to the drainage of electron density [44]. The NMR spectra of dimethyltin complexes (24) of 4-p-(chlorophenyl)-2-phenyl-5-thiazoleacetic acid showed signals due to carbonyl Carbon at δ 180 ppm [45]. A new dimethyltin complex (25) was synthesized general formula [Me2Sn(OBz)2] where OBz represents ligand named 2-{[5-(2-nitrophenyl)furan-2-yl]methyleneamino}benzoic acid and its spectroscopic data were analyzed. Coupling constants nJ (119Sn–13C) helps in the structural elucidation of the new complexes. Figure 3 depicts the structures of complex number 21–31.
Figure 3.
Structure of complex 21–31: dimethyl complexes; 21 with bis 2-(4-methoxy-2-nitrophenylcarbamoyl)benzoic acid; 22 3,4-methylenedioxy 6-nitrophenylpropenoic acid ligand; 23 with 3-(4-fluorophenyl)acrylic acid and 24 with 4-(p-chlorophenyl)-2-phenyl-5-thiazoleacetic acid ligand; 25 with 2-{[5-(2-nitrophenyl)furan-2-yl]methyleneamino}benzoic acid ligand; 26 with (1-ethyl-7-methyl-4-oxo-1,4-dihydro-1,8-naphthyridine-3-carboxylic acid) ligand; 27 with phenyl acetylene carboxylic acid acid; 28 with o-(p-dimethylaminobenzali- dine)benzoic acid ligand; 29 with 2-phenyl-4-selenazole carboxylic acid ligand; 30 with 2-aminobenzoic acid ligand; 31(a) and 31(b) with 2,6-pyridinedicarboxylic acid ligand.
13C NMR spectra of ligand (NaOBz) showed its shift as (δ ppm): 173.24 (COO); 112.61–178.71 (others) and of the complex as 175.30 (COO); 111.28–178.14 (others) respectively [46]. A new complex (26) derived from the ligand (1-ethyl-7-methyl-4-oxo-1,4-dihydro-1,8-naphthyridine-3-carboxylic acid). The spectra showed no appreciable change in the carbon resonances except the carbonyl carbon [47]. The [Me2Sn(pac)]4 complex (27) derived from the ligand phenyl acetylene carboxylic acid showed resonance values at δ 158.15 (COO); 83.20, 84.23 (C≡C); 120.22–132.95 (aromatic ring); 5.79, 10.14 ppm (methyl carbons attached directly to tin) [48]. The spectroscopic data of dimethyltin complex (28) derived from the ligand o-(p-dimethylaminobenzali- dine)benzoic acid exhibited signals at δ 175.0 (COO); 31.2 (NCH3); 110.7–150.3 (other carbons); 5.5 ppm (Sn–CH3) [49]. Another new dimethyltin complex (29) of 2-phenyl-4-selenazole carboxylic acid ligand showed two signals due to endocyclic/exocyclic tin atoms at δ 143 and 190 ppm, respectively, indicating dimer in solution [50]. Bis(μ3-oxo)bis (μ-O-aminobenzoato-O,O′)bis (O-aminobenzoato)tetrakis [di-methyltin (IV)] complex (30) showed the resonance signals at δ 175.1 (COO); 150.4–113.1 (aromatic carbons); 7.3 ppm (Sn–CH3) [51]. The spectroscopic data of two dimethyltin complexes 31(a) and 31(b) using ligand 2,6-pyridinedicarboxylic acid showed that the resonances due to carboxylate groups remained at almost the same resonance as that of the ligand. The methyl carbons attached directly to the tin also showed a point of difference between the two complexes [52]. Table 4 represents data related to 13C NMR data of complex number 21–31.
Table 4.
13C NMR data of complex number 21–31.
| Complex | Compound | δ (13C) (ppm) |
J values (Hz) | C–Sn–C angle (º) | Reference | |||
|---|---|---|---|---|---|---|---|---|
| Carbons attached to Tin (Sn) |
Carbons of Ligand |
|||||||
| Position | Value | Position | Value | |||||
| 21 | [Me2Sn(L24)2] | CH3 | 0.6 | COO | 160.3 | 1J [800] | 147 | [42] |
| 1 | 129.6 | |||||||
| 2 | 127.7 | |||||||
| 3 | 132.9 | |||||||
| 4 | 135.7 | |||||||
| 5 | 124.4 | |||||||
| 6 | 131.9 | |||||||
| 7 | 167.0 | |||||||
| 8 | 121.3 | |||||||
| 9 | 128.7 | |||||||
| 10 | 120.9 | |||||||
| 11 | 156.6 | |||||||
| 12 | 109.5 | |||||||
| 13 | 142.5 | |||||||
| 14 | 56.0 | |||||||
| 22 | [Me2Sn(L25)2] | CH3 | 4.34 | COO | 187.87 | - | - | [43] |
| 1 | 126.36 | |||||||
| 2 | 105.93 | |||||||
| 3 | 153.10 | |||||||
| 4 | 150.18 | |||||||
| 5 | 104.86 | |||||||
| 6 | 141.35 | |||||||
| 7 | 104.30 | |||||||
| 8 | 141.25 | |||||||
| 9 | 123.10 | |||||||
| 23 | [Me2Sn(L26)2] | CH3 | 4.6 | COO 1 2 3 4,4′ 5,5′ 6 |
171.3 | 1J [703] | 138.4 | [44] |
| 118.3 | ||||||||
| 145.4 | ||||||||
| 131.2 | ||||||||
| 129.5 | ||||||||
| 116.1, 115.8 | ||||||||
| 165.8, 162.4 | ||||||||
| 129.5 | ||||||||
| 24 | [Me2Sn(L27)2] | CH3 | 5.6 | COO | 170.0 | 1J [630] | - | [45] |
| 1 | 136.8 | |||||||
| 2,6 | 112.2 | |||||||
| 3,5 | 135.8 | |||||||
| 4 | 131.6 | |||||||
| 7 | 128.8 | |||||||
| 8 | 133.5 | |||||||
| 9 | 148.0 | |||||||
| 10 | 137.9 | |||||||
| 11,15 | 134.9 | |||||||
| 12,14 | 134.6 | |||||||
| 13 | 128.1 | |||||||
| 16 | 21.1 | |||||||
| 25 | [Me2Sn(L28)2] | CH3 | 14.95 | COO | 175.30 | - | - | [46] |
| 1 | 121.79 | |||||||
| 2 | 131.77 | |||||||
| 3 | 112.22 | |||||||
| 4 | 132.68 | |||||||
| 5 | 116.05 | |||||||
| 6 | 150.97 | |||||||
| 7 | 178.14 | |||||||
| 8 | 152.61 | |||||||
| 9 | 111.28 | |||||||
| 10 | 132.92 | |||||||
| 11 | 153.18 | |||||||
| 12 | 114.42 | |||||||
| 13 | 130.21 | |||||||
| 14 | 131.05 | |||||||
| 15 | 133.24 | |||||||
| 16 | 124.34 | |||||||
| 17 | 147.34 | |||||||
| 26 | [Me2Sn(L29)2] | CH3 | 1.5 | COO1234567891011 | 178.0 | - | - | [47] |
| 1 | 148.2 | |||||||
| 2 | 122.3 | |||||||
| 3 | 166.6 | |||||||
| 4 | 109.7 | |||||||
| 5 | 120.3 | |||||||
| 6 | 148.6 | |||||||
| 7 | 164.8 | |||||||
| 8 | 136.5 | |||||||
| 9 | 47.4 | |||||||
| 10 | 15.2 | |||||||
| 11 | 25.3 | |||||||
| 27 | [Me2Sn(L30)]4 | CH3 | 5.79, 10.14 |
COO | 158.15 | - | - | [48] |
| 1 | 83.20 | |||||||
| 2 | 84.23 | |||||||
| 3 | 132.95 | |||||||
| 4,4′ | 130.20 | |||||||
| 5,5′ | 120.22 | |||||||
| 6 | 128.54 | |||||||
| 29 | [(Me2Sn)4 (L31)2(Cl)2] | CH3 | - | COO | 176.21 | - | - | [49] |
| 1 | 169.13 | |||||||
| 2–7 | 127.93–151.37 | |||||||
| 30 | [Me2Sn(L32)2] | CH3 | 7.3 | COO | 175.1 | - | - | [50] |
| 1–6 | 150.4–113.1 | |||||||
| 31 (a) | [Me2Sn(L33)2] | CH3 | 10.1 | COO | 168.8 | - | 159.6–161.6 | [51] |
| 1,1′ | 141.3 | |||||||
| 2,2′ | 126.2 | |||||||
| 3 | 147.2 | |||||||
| 31 (b) | CH3 | 10.2 | COO | 166.8 | - | |||
| 1,1′ | 141.3 | |||||||
| 2,2′ | 126.2 | |||||||
| 3 | 147.2 | |||||||
3. Conclusion
The chemical shift of 13C nucleus is influenced by the electronic environment of the complex. The carbonyl carbon in these complexes showed resonances from δ 158–187 ppm. Some of the complexes showing absorption in higher ppm value were having electron donating groups and benzene rings in the structure. The complexes showing absorption in low ppm region were having electron withdrawing groups. As far as methyl groups attached with tin are concerned also showed absorption in different ppm values ranging from δ 0.6–29.6 ppm. The nitro groups containing complexes showed resonances in δ 0.6–1.5 ppm region. While the complexes contacting chloro, bromo and oxygen atoms showed absorption in δ 14–29.6 ppm region. The C–Sn–C bond angle ranges from 132-159⁰ in different dimethyltin complexes. The higher angle was seen in case of nitrogen containing complexes. This spectroscopic technique has contributed a lot in explaining the structural chemistry and in future also will play an important role in this area.
Declarations
Author contribution statement
All authors listed have significantly contributed to the development and the writing of this article.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
No data was used for the research described in the article.
Declaration of interest’s statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- 1.Kang W., Wu X., Huang J. Synthesis, crystal structure and biological activities of four novel tetranuclear di-organotin (IV) carboxylates. J. Organomet. Chem. 2009;694(15):2402–2408. [Google Scholar]
- 2.Dokorou V.N., Kovala-Demertzi D., Louloudi M., Silvestru A., Demertzis M.A. Synthesis, characterization and catalytic properties of diorganotin derivatives. Crystal and molecular structure of the first complex of 2-(2-methyl-3-nitroanilino) benzoic acid of 1, 2: 3, 4-di-μ2-2-(2-methyl-3-nitroanilino) benzoato-O, O-1, 3-bis-2-(2-methyl-3-nitroanilino) benzoato-O-1, 2, 4: 2, 3, 4-di-μ3-oxo-tetrakis [di-methyltin (IV)] J. Organomet. Chem. 2008;693(24):3587–3592. [Google Scholar]
- 3.Gómez E., Flores R., Huerta G., Alvarez-Toledano C., Toscano R.A., Santes V.c., Nava N., Sharma P. Dimethyltin (IV) 2, 6-disubstituted pyridine complexes. J. Organomet. Chem. 2003;672(1-2):115–122. [Google Scholar]
- 4.Abbas S.M., Ali S., Hussain S.T., Shahzadi S. Structural diversity in organotin (IV) dithiocarboxylates and carboxylates. J. Coord. Chem. 2013;66(13):2217–2234. [Google Scholar]
- 5.Evans C.J. Organotin compounds in modern technology. J. Organomet. Chem. Libr. 1985;16:1137–1138. [Google Scholar]
- 6.Lockhart T.P., Calabrese J.C., Davidson F. Structural studies of diorganotin (IV) carboxylates. X-ray and NMR structures of Me2Sn (OAc) 2 and a 7-coordinate tin anion, Me2Sn (OAc) 3NMe4. cntdot. 2CHCl3. Organometallics. 1987;6(12):2479–2483. [Google Scholar]
- 7.Zhang Q., Zhang M., Wang H., Tian X., Ma W., Luo L., Wu J., Zhou H., Li S., Tian Y. A series of two-photon absorption organotin (IV) cyano carboxylate derivatives for targeting nuclear and visualization of anti-cancer activities. J. Inorg. Biochem. 2019;192:1–6. doi: 10.1016/j.jinorgbio.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 8.Kumar M., Abbas Z., Tuli H.S., Rani A. Organotin complexes with promising therapeutic potential. Curr. Pharmacol. Rep. 2020:1–15. [Google Scholar]
- 9.Roy M., Roy S., Singh K.S., Kalita J., Singh S.S. Synthesis, characterization and anti-diabetic assay of diorganotin (IV) azo-carboxylates: crystal structure and topological studies of azo-dicarboxylic acid ligand and its cyclic tetranuclear dimethyltin (IV) complex. New J. Chem. 2016;40(2):1471–1484. [Google Scholar]
- 10.Win Y.-F., Teoh S.-G., Vikneswaran M., Ha S.-T., Ibrahim P. Synthesis and characterization of organotin (IV) complexes derived of 4-(diethylamino) benzoic acid: in vitro anti-bacterial screening activity. Int. J. Phys. Sci. 2010;5(8):1263–1269. [Google Scholar]
- 11.Tiekink E.R. Structural chemistry of organotin carboxylates: a review of the crystallographic literature. Appl. Organomet. Chem. 1991;5(1):1–23. [Google Scholar]
- 12.Cardarelli N.F. CRC Press; 2019. Tin as a Vital Nutrient: Implications in Cancer Prophylaxis and Other Physiological Processes. [Google Scholar]
- 13.Keppler B.K. Wiley VCH; 1993. Metal Complexes in Cancer Chemotherapy. [Google Scholar]
- 14.Sirajuddin M., Tariq M., Ali S. Organotin (IV) carboxylates as an effective catalyst for the conversion of corn oil into biodiesel. J. Organomet. Chem. 2015;779:30–38. [Google Scholar]
- 15.Wang H., Hu L., Du W., Tian X., Zhang Q., Hu Z., Luo L., Zhou H., Wu J., Tian Y. Two-photon active organotin (IV) carboxylate complexes for visualization of anti-cancer action. ACS Biomater. Sci. Eng. 2017;3(5):836–842. doi: 10.1021/acsbiomaterials.6b00786. [DOI] [PubMed] [Google Scholar]
- 16.Shah F.A., Sirajuddin M., Ali S., Abbas S.M., Tahir M.N., Rizzoli C. Synthesis, spectroscopic characterization, X-ray structure and biological screenings of organotin (IV) 3-[(3, 5-dichlorophenylamido)] propanoates. Inorg. Chim. Acta. 2013;400:159–168. [Google Scholar]
- 17.Bonire J.J., Ayoko G.A., Olurinola P.F., Ehinmidu J.O., Jalil N.S., Omachi A.A. Synthesis and antifungal activity of some organotin (IV) carboxylates. Met. Base. Drugs. 1998;5(4):233–236. doi: 10.1155/MBD.1998.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butt A.F., Ahmed M.N., Bhatti M.H., Choudhary M.A., Ayub K., Tahir M.N., Mahmood T. Synthesis, structural properties, DFT studies, antimicrobial activities and DNA binding interactions of two newly synthesized organotin (IV) carboxylates. J. Mol. Struct. 2019;1191:291–300. [Google Scholar]
- 19.Shahzadi S., Shahid K., Ali S., Mazhar M., Badshah A., Ahmed E., Malik A. Non-steroidal anti-inflammatory drugs (NSAIDs) as donor ligands in organotin (IV) derivatives: synthesis, spectroscopic characterization and biological applications. Turk. J. Chem. 2005;29(3):273–288. [Google Scholar]
- 20.Blunden S., Hill R. Bis (tributyltin) oxide as a wood preservative: its conversion to tributyltin carboxylates in Pinus sylvestris. Appl. Organomet. Chem. 1990;4(1):63–68. [Google Scholar]
- 21.Danish M., Ali S., Mazhar M., Badshah A., Choudhary M.I., Alt H.G., Kehr G. Mössbauer, multinuclear magnetic resonance and mass spectrometric studies of organotin carboxylates of m-methyltrans-cinnamic acid. Polyhedron. 1995;14(20-21):3115–3123. [Google Scholar]
- 22.Dey D.K., Saha M.K., Gielen M., Kemmer M., Biesemans M., Willem R., Gramlich V., Mitra S. Synthesis, spectroscopy and structure of [N-(2-carboxyphenyl) salicylideneiminato] dimethyltin (IV) J. Organomet. Chem. 1999;590(1):88–92. [Google Scholar]
- 23.Vatsa C., Jain V.K., Kesavadas T., Tiekink E.R. Structural chemistry of organotin carboxylates: XII. Synthesis and characterization of diorganotin (IV) carboxylates containing 2-thiophene-or 2-furan-carboxylic acid. Crystal structure of [Et2Sn (O2CC4H3S) 2] J. Organomet. Chem. 1991;410(2):135–142. [Google Scholar]
- 24.Vatsa C., Jain V., Das T., Tiekink E. Structural chemistry of organotin carboxylates VI. Characterization of {[R2Sn (O2CR′)] 2O} 2 (R= Me, Et, nPr and nBu; R′= thiophene and furan). X-ray crystal structure of {[nBu2Sn (O2CC4H3S)] 2O} 2. J. Organomet. Chem. 1990;396(1):9–18. [Google Scholar]
- 25.Kourkoumelis N., Demertzis M.A., Kovala-Demertzi D., Koutsodimou A., Moukarika A. Preparations and spectroscopic studies of organotin complexes of diclofenac. Spectrochim. Acta Mol. Biomol. Spectrosc. 2004;60(10):2253–2259. doi: 10.1016/j.saa.2003.11.027. [DOI] [PubMed] [Google Scholar]
- 26.Shahid K., Ali S., Shahzadi S., Badshah A., Khan K.M., Maharvi G.M. Organotin (IV) complexes of aniline derivatives. I. Synthesis, spectral and anti-bacterial studies of di-and triorganotin (IV) derivatives of 4-bromomaleanilic acid. Synth. React. Inorg. Met. Org. Chem. 2003;33(7):1221–1235. [Google Scholar]
- 27.Shahzadi S., Shahid K., Ali S., Mazhar M., Khan K. Organotin (IV) derivatives as biocides: an investigation of structure by IR, solution NMR, electron impact MS and assessment of structure correlation with biocidal activity. J. Iran. Chem. Soc. 2005;2(4):277–288. [Google Scholar]
- 28.Sirajuddin M., Ali S., McKee V., Akhtar N., Andleeb S., Wadood A. Spectroscopic characterizations, structural peculiarities, molecular docking study and evaluation of biological potential of newly designed organotin (IV) carboxylates. J. Photochem. Photobiol. B Biol. 2019;197 doi: 10.1016/j.jphotobiol.2019.111516. [DOI] [PubMed] [Google Scholar]
- 29.Tariq M., Sirajuddin M., Ali S., Khalid N., Shah N. Biological evaluations and spectroscopic characterizations of 3-(4-ethoxyphenyl)-2-methylacrylate based organotin (IV) carboxylates derivatives. Russ. J. Gen. Chem. 2017;87(11):2690–2698. [Google Scholar]
- 30.Hussain S., Ali S., Shahzadi S., Tahir M.N., Shahid M., Munawar K.S., Abbas S.M. Synthesis, spectroscopy, single crystal XRD and biological studies of multinuclear organotin dicarboxylates. Polyhedron. 2016;117:64–72. [Google Scholar]
- 31.Amini M.M., Azadmeher A., Khavasi H.R., Ng S. Synthesis, characterization, and molecular structures of di-and triorganotin (IV) complexes with 9-anthracenecarboxylic acid: the structural diversity in organotin 9-anthracenecarboxylates. J. Organomet. Chem. 2007;692(18):3922–3930. [Google Scholar]
- 32.Rocha C., de Morais B., Rodrigues B.L., Donnici C., de Lima G., Ardisson J.D., Takahashi J.A., Bitzer R.S. Spectroscopic and X-ray structural characterization of new organotin carboxylates and their in vitro antifungal activities. Polyhedron. 2016;117:35–47. [Google Scholar]
- 33.Iqbal M., Ali S., Muhammad N., Parvez M., Langer P., Villinger A. Synthesis, characterization, crystal structures and electrochemical studies of organotin (IV) carboxylates. J. Organomet. Chem. 2013;723:214–223. [Google Scholar]
- 34.Antonenko T., Shpakovsky D., Berseneva D., Gracheva Y.A., Dubova L., Shevtsov P., Redkozubova O., Shevtsova E., Tafeenko V., Aslanov L. Cytotoxic activity of organotin carboxylates based on synthetic phenolic antioxidants and polycyclic bile acids. J. Organomet. Chem. 2020;909 [Google Scholar]
- 35.Ahmed S., Ali S., Ahmed F., Bhatti M.H., Badshah A., Mazhar M., Khan K.M. Synthesis, spectroscopic characterization, and biological applications of organotin (IV) derivatives of 2-(N-maleoyl)-3-phenylpropanoic acid. Synth. React. Inorg. Met. Org. Chem. 2002;32(8):1521–1536. [Google Scholar]
- 36.Hanif M., Hussain M., Ali S., Bhatti M.H., Ahmed M.S., Mirza B., Stoeckli-Evans H. In vitro biological studies and structural elucidation of organotin (IV) derivatives of 6-nitropiperonylic acid: crystal structure of {[(CH2O2C6H2 (o-NO2) COO) SnBu2] 2O} 2. Polyhedron. 2010;29(1):613–619. [Google Scholar]
- 37.Shah F., Ali S., Shahzadi S., Rizzoli C., Ahmad S. Synthesis, spectral characterization and X-ray crystal structure of biologically active organotin (IV) 3-[(3′, 5′-dimethylphenylamido)] propanoates. J. Iran. Chem. Soc. 2012;9(6):923–932. [Google Scholar]
- 38.Ma C., Zhang B., Zhang S., Zhang R. Chiral organotin (IV) carboxylates complexes: Syntheses, characterization, and crystal structures with chiral (S)-(+)-6-methoxy-α-methyl-2-naphthaleneaceto acid ligand. J. Organomet. Chem. 2011;696(10):2165–2171. [Google Scholar]
- 39.Shahzadi S., Shahid K., Ali S. Coordination behavior of the carboxylate group in organotin (IV) derivatives of 2-[(2′, 4′, 6′-tribromophenylamido)] benzoic acid and 3-[(2′, 4′, 6′-tribromophenylamido)] propenoic acid: spectroscopic studies. Russ. J. Coord. Chem. 2007;33(6):403–411. [Google Scholar]
- 40.Tariq M., Ali S., Shah N.A., Muhammad N., Tahir M.N., Khalid N. Catalytic, biological and DNA interaction studies of 3-(4-cyanophenyl)-2-methylacrylate organotin (IV) carboxylates derivatives: synthesis, spectroscopic characterization and X-ray structures. Inorg. Chim. Acta. 2013;405:444–454. [Google Scholar]
- 41.Zhang J.-H., Zhang R.-F., Ma C.-L., Wang D.-Q., Wang H.-Z. New organotin carboxylates derived from 6-chloro-3-pyridineacetic acid exhibiting discrete molecular, drum-like, linear polymeric and ladder structures constructed from dimeric tetraorganodistannoxane units. Polyhedron. 2011;30(4):624–631. [Google Scholar]
- 42.Sirajuddin M., McKee V., Tariq M., Ali S. Newly designed organotin (IV) carboxylates with peptide linkage: synthesis, structural elucidation, physicochemical characterizations and pharmacological investigations. Eur. J. Med. Chem. 2018;143:1903–1918. doi: 10.1016/j.ejmech.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 43.Ahmad M.S., Hussain M., Hanif M., Ali S., Mirza B. Synthesis, chemical characterization and biological screening for cytotoxicity and antitumor activity of organotin (IV) derivatives of 3, 4-methylenedioxy 6-nitrophenylpropenoic acid. Molecules. 2007;12(10):2348–2363. doi: 10.3390/12102348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tariq M., Sirajuddin M., Ali S., Khalid N., Tahir M.N., Khan H., Ansari T.M. Pharmacological investigations and Petra/Osiris/Molinspiration (POM) analyses of newly synthesized potentially bioactive organotin (IV) carboxylates. J. Photochem. Photobiol. B Biol. 2016;158:174–183. doi: 10.1016/j.jphotobiol.2016.02.028. [DOI] [PubMed] [Google Scholar]
- 45.Ali S., Khokhar M.N., Bhatti M., Mazhar M., Masood M.T., Shahid K., Badshah A. 1H, 13C, and 119Sn NMR, IR, mass, thermal, and biological studies of organotin (IV) derivatives of 4-p-(Chlorophenyl)-2-phenyl-5-thiazoleacetic acid. Synth. React. Inorg. Met. Org. Chem. 2002;32(8):1373–1392. [Google Scholar]
- 46.Dias L.C., de Lima G.M., Takahashi J.A., Ardisson J.D. New di-and triorganotin (IV) carboxylates derived from a Schiff base: synthesis, characterization and in vitro antimicrobial activities. Appl. Organomet. Chem. 2015;29(5):305–313. [Google Scholar]
- 47.Ahmed S., Bhatti M.H., Ali S., Ahmed F. Organotin (IV) derivatives of 1-ethyl-1, 4-dihydro-7-methyl-4-oxo-1, 8-naphthyridine-3-carboxylic acid (nalidixic acid): synthesis, structural elucidation and biological activities. Turk. J. Chem. 2006;30(2):193–202. [Google Scholar]
- 48.Yousefi M., Tavakolinia F., Hassanzadeh S.M. International Conference on Biology. Environment and Chemistry (Singapoore); 2011. Comparison of anti-bacterial activities of di-and tri-tin (IV) carboxylate complexes. [Google Scholar]
- 49.Baul T.S.B., Tiekink E.R. Crystal and molecular structure of o-aminobenzoato (o-(p-dimethylaminobenzalidine) benzoato)(o-ammoniobenzoate) dimethyltin (IV), an example of a mixed carboxylate complex of tin. J. Chem. Crystallogr. 1996;26(6):393–397. [Google Scholar]
- 50.Nath M. Conventional and microwave-assisted synthesis, characterization, DFT calculations, in vitro DNA binding and cleavage studies of potential chemotherapeutic diorganotin (IV) mandelates. J. Photochem. Photobiol. B Biol. 2016;162:348–360. doi: 10.1016/j.jphotobiol.2016.06.040. [DOI] [PubMed] [Google Scholar]
- 51.Vieira F., Menezes D., De Lima G., Wardell J., Cortés M., Silva G., Vilas-Boas A., Maia J. da S. Effect of diorganotin (IV) carboxylate complexes,[N-(2-carboxyphenyl) salicylideneiminato] dimethyltin (IV), bis (μ3-oxo) bis (μ-O-aminobenzoato-O, O′) bis (O-aminobenzoato) tetrakis [dimethyltin (IV)] and bis (O-aminobenzoato-O, O′) di-n-butyltin (IV), on the membrane of Candida albicans cells—a mechanistic investigation of the antifungal activity of organotin complexes. Appl. Organomet. Chem. 2008;22(8):433–439. [Google Scholar]
- 52.Ma C., Li J., Zhang R., Wang D. Syntheses and crystal structures of dimethyltin (IV) derivatives with 2, 6-pyridinedicarboxylic acid. Inorg. Chim. Acta. 2005;358(15):4575–4580. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.