Summary
Poration of the outer mitochondrial membrane by the effector BCL-2 proteins BAK and BAX initiates apoptosis. BH3-only initiators BID and BIM trigger conformational changes in BAK and BAX transforming them from globular dormant proteins to oligomers of the apoptotic pores. Small molecules that can directly activate effectors are being sought for applications in cancer treatment. Here, we describe the small molecule SJ572946, discovered in a fragment-based screen that binds to the activation groove of BAK and selectively triggers BAK activation over that of BAX in liposome and mitochondrial permeabilization assays. SJ572946 independently kills BAK-expressing BCL2allKO HCT116 cells revealing on target cellular activity. In combination with apoptotic inducers and BH3 mimetics, SJ572946 kills experimental cancer cell lines. SJ572946 also cooperates with the endogenous BAK activator BID in activating a misfolded BAK mutant substantially impaired in activation. SJ572946 is a proof-of-concept tool for probing BAK-mediated apoptosis in preclinical cancer research.
Subject areas: Biochemistry, Small molecule, Cell biology, Structural biology
Graphical abstract

Highlights
-
•
BAK activator SJ572946 discovered in fragment-based screening
-
•
SJ572946 binds to the activation groove to activate BAK
-
•
SJ572946 cooperates with the endogenous BAK activator BID in apoptosis initiation
-
•
SJ572946 cooperates with pro-apoptotic therapeutics in tumor cell line killing
Biochemistry; Small molecule; Cell biology; Structural biology
Introduction
Mitochondrial apoptosis is fundamental to mammalian physiology (Czabotar et al., 2014; Singh et al., 2019). A key regulatory point of no return in this pathway is B cell lymphoma-2 (BCL-2)-regulated mitochondrial poration, which releases cytochrome c (cyt c) to activate apoptotic caspases that dismantle cells through intracellular proteolysis (Chipuk et al., 2010). Mitochondrial poration is executed by the pore-forming BCL-2 effectors BAK, BAX, and BOK (Moldoveanu and Czabotar, 2020). BAK and BAX are tightly regulated by BCL-2 protein binding partners (Kale et al., 2018; Singh et al., 2019). Pro-death BH3-only initiators (e.g. BID and BIM) bind to and trigger dormant effectors, which change shape, unfold, and auto-activate in trans to amplify the response (Iyer et al., 2020; Singh et al., 2022). The overall effector activation response is dictated through the combination of direct activation and auto-activation which can be separated through mutations that ablate the latter as shown recently for BAK (Singh et al., 2022). Active effectors dimerize (Birkinshaw et al., 2021; Brouwer et al., 2014; Czabotar et al., 2013), oligomerize (Aluvila et al., 2014; Dewson et al., 2009, 2012), bind lipids (Cowan et al., 2020), and permeabilize membranes forming toroidal pores (Flores-Romero et al., 2020). Pro-survival BCL-2 guardians (e.g. BCL-2, BCL-xL and MCL-1) prevent poration by binding to and sequestering the initiators (MODE 1) or the effectors (MODE 2) (Llambi et al., 2011). Mitochondrial poration ensues when active initiators and effectors overcome antagonism by active guardians. Physiologically, this occurs as cells respond to cellular stress including DNA damage, cytokine withdrawal, and ER stress (Singh et al., 2019).
High-resolution structures of dormant, directly activated, and auto-activated BAK have revealed the early changes in conformation associated with BAK activation (Brouwer et al., 2017; Moldoveanu et al., 2013; Singh et al., 2022). Activating BH3 ligands transiently engage the activation groove of dormant BAK (helices α2–α5) to destabilize the electrostatic network involving helix α1 at the bottom of the groove (Singh et al., 2022). Helix α1 dissociates in the presence of membranes, thereby facilitating major BAK unfolding, as demonstrated by epitope accessibility assessments using antibodies (Iyer et al., 2016, 2020), limited proteolysis (Llambi et al., 2011; Moldoveanu et al., 2013), crosslinking (Dewson et al., 2008, 2009; Leshchiner et al., 2013; Moldoveanu et al., 2013), hydrogen–deuterium exchange mass spectrometry (HDX-MS), and NMR spectroscopy (Sandow et al., 2021; Sperl et al., 2021). On helix α1 dissociation, BAK is thought to unfold into the α2–α5 core and the α6–α8 latch (Aluvila et al., 2014; Brouwer et al., 2014). The α2–α5 core subsequently associates into putative α2–α5 dimers as the building blocks of effector oligomers that mediate poration (Birkinshaw et al., 2021; Cowan et al., 2020). The α2–α5 dimers bind to, and can be bridged by, phospholipids (Cowan et al., 2020), but the mechanism of membrane permeabilize by effectors is poorly understood (Flores-Romero et al., 2020).
Deregulation of BCL-2 proteins prevents mitochondrial poration leading to apoptosis resistance in cancer cells (Hanahan and Weinberg, 2011). BCL-2 guardians are upregulated in many different tumors (Merino et al., 2018), whereas initiators and effectors are downregulated, leading to overall higher thresholds for apoptosis initiation (Czabotar et al., 2014; Moldoveanu et al., 2014; Singh et al., 2019). Consequently, BCL-2 proteins have been attractive drug targets (Ashkenazi et al., 2017). Rationally designed potent and selective therapeutic inhibitors of BCL-2 guardians have been successfully developed (Ashkenazi et al., 2017; Delbridge et al., 2016). In contrast, although small molecule modulators of the BCL-2 effectors have been discovered in preclinical studies, they have not been developed as therapeutic agents (Moldoveanu and Czabotar, 2020; Pogmore et al., 2021). Most of the published modulators of the BCL-2 effectors engage BAX (Moldoveanu and Czabotar, 2020). BKA-073 was recently reported to be a selective BAK activator (Park et al., 2021), whereas trifluoroperazine was reported to be a BAK inhibitor that blocks BAK-mediated liposome permeabilization (Song et al., 2014). Additionally, MSN-125 and OICR766A inhibit and induce apoptosis, respectively, in a BAK-dependent or BAX-dependent manner (Brahmbhatt et al., 2016; Niu et al., 2017). Moreover, the small molecule WEHI-9625 inhibits mouse BAK by forming a complex with VDAC2 (van Delft et al., 2019). The mechanisms of action of OICR766A and BKA-073 are more complex than direct BAK activation (see discussion) prompting us to seek additional chemical probes for this important system.
Here we present amino benzothiazole derivatives as proof-of-concept direct activators of BAK. These compounds engage multiple sites in the activation groove with weak affinity to destabilize and activate BAK, and they cooperate with the mechanism of BAK activation. Our lead compound, SJ572946, cooperates with BID to activate a BAK mutant that is severely impaired in activation. Furthermore, SJ572946 initiates mitochondrial apoptosis in tumor cells as a single agent and in combination with known inducers of apoptosis. Importantly, we present a powerful and rigorous platform for the discovery of BAK-selective small molecule modulators that we actively deploy to generate additional chemical matter for effector-mediated mitochondrial poration.
Results
Discovery of BAK-activating small molecules
Because of the importance of mitochondrial apoptosis initiation in pathophysiology we sought to validate and probe with small molecules the recently revised mechanism of BAK activation (Singh et al., 2022). Toward this goal we discovered BAK activators by screening of the St. Jude fragment library (∼5000 compounds) (Iconaru et al., 2015; Nishiguchi et al., 2021). Screening and initial validation at a single dose involved the following binding and functional analyses: 1) primary screening by ligand-based one-dimensional nuclear magnetic resonance spectroscopy (1D-NMR) performed on mixtures of 5 or 6 fragments; 2) ligand-based 1D-NMR validation of singles from mixtures containing BAK binders or “hits”; and 3) functional validation of hits in mitochondrial poration assays (Figure S1A). We identified 90 hits in 43 mixtures (Figures S1A and S1B), 22 of which released cyt c from purified mouse liver mitochondria of WT mice (Figure S1C). Some of the hits also release cyt c from mitochondria purified from livers of Bak1−/− mice, albeit less than was released from WT liver mitochondria (Figure S1C). Six of the BAK binders that activated BAK in mitochondrial poration assays are amino benzothiazole analogs (compounds B7, C3, F1, F7, G3, and SJ572946) (Figure S1B and Table S1). The high hit-rate of the fragment screen suggests that BAK is druggable according to hit rate predictions from NMR-based screening campaigns (Shuker et al., 1996).
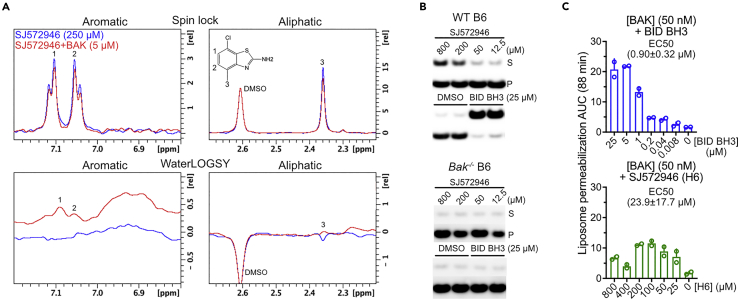
We repurchased compound SJ572946, also referred to as H6, and confirmed that it bound to BAK by spin lock (top) (Hajduk et al., 1997) and waterLOGSY (bottom) (Dalvit et al., 2001) ligand-based 1D-NMR (Figure 1A). These experiments measure the proton (1H) signals of SJ572946 ligand in the absence and presence of BAK. When the ligand binds, it takes the NMR properties of the target. In spin lock relaxation-based experiments, the ligand relaxed faster when bound to BAK resulting in signal attenuation (red peak heights are diminished compared to those of blue peaks in Figure 1A top). In waterLOGSY experiments, measuring magnetization transfer from water to ligand via nuclear Overhauser effects (NOEs), NOEs between water and fast tumbling SJ572946 are negative, whereas those between water and SJ572946 bound to BAK are positive (blue negative peaks are flipped, Figure 1A bottom). The peak for deuterated DMSO used as vehicle serves as internal control and it did not change height or phase ± BAK indicating that it does not bind BAK (Figure 1A). The waterLOGSY experiment is sensitive in detecting ligand aggregation when the negative peaks appear flat and it confirmed that SJ572946 has low solubility exhibiting low signal in the absence of BAK (the flat blue curve for the aromatic protons in Figure 1A, bottom left). Indeed, we noted visible precipitation in the NMR tubes containing SJ572946, consistent with poor solubility. SJ572946 induced dose-dependent release of cyt c from purified liver mitochondria of WT mice, but not from liver mitochondria of Bak1−/− mice, although the release was incomplete when compared to that induced by the prototypical direct BAK-activating BID BH3 peptide (Figure 1B). In addition, SJ572946 exhibited dose-dependent permeabilization of liposomes, measured as the BAK-mediated release of encapsulated fluorescent dye and quantified as the area under the curve (AUC) of the kinetic traces (Figures 1C and S2A). `
Figure 1.
SJ572946 activates BAK in membrane permeabilization
(A) Spin lock and waterLOGSY NMR spectra indicate SJ572946 binding to BAK judged by 1H signal attenuation and 1H signal flipping, respectively. The flatness of the waterLOGSY spectrum indicates poor solubility of SJ572946.
(B) Mitochondrial poration assays indicate that BAK-dependent cytochrome c release is induced by SJ572946 fragment from purified mouse mitochondria, as measured by immunoblotting. S and P, supernatant and pellet. B6, C57CL/6 mice.
(C) Liposome permeabilization measured as the area under the curve (AUC) for the normalized data in FigureS2A. Data are presented as the mean +SD from n = 2 experiments, each with n = 3 technical replicates. Because of the poor solubility of SJ572946, its activity decreases at concentrations exceeding 200 μM.
We determined that five commercially available analogs of SJ572946 (A1-A5) activated BAK in dose–response mitochondrial poration assays with purified WT and bak−/− mouse liver mitochondria (Figures S2C and S2D). At a concentration of 200 μM, the five analogs of SJ572946 induced liposome permeabilization exclusively in the presence of BAK (Figure S2E). A3, the closest analog to SJ572946, bound to BAK in ligand-based 1D-NMR experiments, exhibiting better solubility than SJ572946, as assessed by the superior signals in waterLOGSY spectra and no visible precipitation in the NMR tube (Figure S2F bottom). A3 induced dose-dependent liposome permeabilization with an EC50 of approximately 214 μM (Figures S2A, S2B, and S2G). Based on the promising properties of SJ572946 and A3, we sought to characterize their interactions with BAK.
Multipronged engagement of the activation groove of BAK by SJ572946 and A3
To determine the mechanism of action of the amino benzothiazoles as direct BAK activators we sought to investigate their binding to BAK further. A3 displaced the BID BH3 stabilized α-helix of BCL-2 protein (SAHB)-fluorescein from GST-BAK in a competitive time-resolved fluorescence resonance energy transfer (TR-FRET) assay, with IC50s of 570.1 and 1965.0 μM, respectively, indicating that they have lower affinity for the activation groove of BAK when compared with BID BH3 SAHB peptide, for which the IC50 was approximately 4.5 μM (Figure 2A). Remarkably robust liposome permeabilization ensued when a small fraction of the total BAK was activated by SJ572946 and A3 (with EC50s of approximately 24 μM and approximately 214 μM, respectively), and it is noteworthy that the EC50 was an order of magnitude lower than the IC50. Therefore, weak-affinity hit-and-run amino benzothiazole binding has major functional consequences, as we previously demonstrated with BAK-activating BH3 peptide ligands (Singh et al., 2022). We performed protein-based 2D-NMR titrations of 15N-BAK with SJ572946 and A3 and mapped their binding to the activation groove of BAK by chemical shift perturbation (CSP) analysis (Figures 2B, S3A, and S3B). At concentrations exceeding 250 μM, SJ572946 precipitated in the NMR tube; consequently, its binding at higher concentrations could not be detected by 2D NMR (Figures S3A and S3B). Weak binding precluded the detection of nuclear Overhauser effects for the interaction of SJ572946 and A3 with BAK; therefore, we could not determine the NMR structures of their complexes with BAK.
Figure 2.
SJ572946 binds the canonical groove of BAK to displace BID SAHB
(A) Competitive time-resolved fluorescence resonance energy transfer (TR-FRET) measurement of the displacement of fluorescein-labeled BID SAHB from GST-BAK. The concentration of peptide or fragment required for half-maximal inhibition (IC50) is summarized for one representative of n = 3 experiments, each comprising n = 3 technical replicates.
(B) Per-residue chemical shift perturbations (CSPs) induced by SJ572946 in the amide backbone of BAK, as calculated from the spectra shown in Figure S3A. The poor solubility of SJ572946 hampered similar analysis at the higher doses.
(C) Mapping of the CSPs in panel (B) onto the crystal structure of apo BAK identifies SJ572946 as binding to pockets (2) and (4)–(5) in the activation groove and to a cavity at the bottom of the activation groove near the electrostatic network involved in stabilizing helix α1. Docking scores for the top nine poses were calculated in Autodock Vina for SJ572946 binding to the three sites.
(D) Dye-free nano differential scanning fluorimetry thermal shift assay traces and melting temperatures (Tm) of BAK in the presence of DMSO or SJ572946 and A3. Data are mean Tm + SD of n = 1 replicate from n = 3 experiments, ∗∗∗∗p <0.0001, ∗∗p <0.0021; ∗p <0.0332; Tukey-Kramer one-way ANOVA.
To gain insights into the possible modes of binding of SJ572946 and A3 to BAK we performed molecular docking of SJ572946 and A3 to the apo-BAK structure (PDB 2imt) with AutoDock Vina (Trott and Olson, 2010). Our analysis revealed weak binding to pockets (2) and (4)–(5) at the top of the activation groove and possible stronger binding to a site at a previously recognized cavity at the bottom of the activation groove, based on the Vina docking scores (Figures 2C, S3C, and S3D). These sites overlap those exhibiting the largest CSP in 2D NMR analyses, thus supporting the binding of SJ572946 and A3 at the activation groove of BAK. The top nine poses of SJ572946 and A3 docked at each of the three binding sites in apo BAK are quite diverse yet have similar Vina docking scores, suggesting multiple possibilities for the binding of these hydrophobic compounds to the hydrophobic pockets in the activation groove of BAK.
Binding of SJ572946 and A3 induced modest destabilization of BAK in assays monitoring thermal denaturation by dye-free nano differential scanning fluorimetry (nanoDSF), reducing the melting temperature (Tm) of BAK by 0.8–1.3°C and 0.6–2.8°C, respectively (Figure 2D). Together, the results of our binding and docking analyses suggest that hit-and-run weak binding of SJ572946 and A3 at three sites in the activation groove (at the top and bottom) is responsible for BAK activation in mitochondrial and liposome permeabilization assays.
SJ572946-mediated direct activation cooperates with BAK auto-activation in vitro
To further validate the direct binding induced activation of BAK by SJ572946 we investigated BAK mutants previously characterized to uncouple direct activation by BH3 ligands and BAK auto-activation (Singh et al., 2022). BAK activation is a signal amplification process involving cooperation between direct activation, which initiates signaling, and auto-activation in trans, which propagates signaling (Figure 3A). These steps are mechanistically similar, involving BH3 ligand binding–induced destabilization of helix α1 at the bottom of the activation groove, which promotes BAK unfolding, exposure of the BAK BH3 domain, and activation of dormant BAK in trans to amplify signaling (Iyer et al., 2020; Singh et al., 2022). We reasoned that SJ572946 might cooperate with BAK activation. Accordingly, we docked SJ572946 in multiple poses to hydrophobic pocket (3) of the opened activation groove of BAK in complexes with activating BH3 ligands (PDB 7m5b). We concluded that SJ572946 binding to the active conformation of the groove exhibits better Vina docking scores than does binding to the occluded pockets in apo BAK (Figures 3B and 2C; BH3 peptide ligand was excluded from the docking to the M3W5 BID BH3-BAK activated complex). Our docking analysis of apo and BH3 ligand–opened BAK suggests that the binding of SJ572946 in multiple poses to hydrophobic sites (2)–(5) of the activation groove is the basis of activation of BAK by this small molecule ligand (Figures 2C and 3B).
Figure 3.
Direct activation by SJ572946 cooperates with BAK auto-activation
(A) Model of BAK activation involving a direct activation–auto-activation hierarchy. Like BID BH3, SJ572946 lowers the threshold level of BAK required for permeabilization by cooperating with the BAK auto-activation mechanism.
(B) Docking of SJ572946 at pocket (3) of the activation groove of BAK, modeled based on the peptide-excluded, opened activated complex M3W5 BID BH3–BAK.
(C) Location and effect of a panel of mutations used to probe BAK activation. SJ572946 activated R127A(s) BAK, which is refractory to auto-activation or activation by BID BH3, whereas BID BH3 activated I85A(4) BAK, which is refractory to auto-activation or activation by SJ572946. The numbering (1)–(6) indicates the positions of residues engaging the corresponding hydrophobic pockets in the activation groove. Residues involved in a salt bridge are marked with (s).
(D) The results of liposome permeabilization assays, quantified as the AUCs of the kinetic traces from Figure S4A, reveals impairment in auto-activation (top) and direct activation by BID BH3 and fragment SJ572946 for most mutants. Data are presented as the mean +SE of n = 2 experiments, each of n = 3 technical replicates, ∗∗∗∗p <0.0001, ∗∗∗p <0.0002, ∗∗p <0.0021, ∗p <0.0332, and ns: not significant; Tukey–Kramer one-way ANOVA.
Using a panel of previously described mutants in the BH3 domain and activation groove of BAK we used to uncouple direct activation and auto-activation (Singh et al., 2022), we tested the ability of SJ572946 to mimic direct activation by BID BH3. We previously showed that wild-type (WT) and “WT-like” V74A(1) BAK exhibited potent auto-activation at a concentration of 200 nM, whereas the other mutants in our panel were largely resistant to auto-activation, even at a concentration of 800 nM. Auto-activation-resistant mutants were partially activated by BID BH3 (10 μM), as previously reported (Singh et al., 2022), and SJ572946 (200 μM) also activated most of these mutants in liposome permeabilization assays (Figures 3D, S4A, and S4B). Explicitly, WT and “WT-like” V74A(1) BAK, both of which are competent in auto-activation, were similarly activated by BID BH3 and SJ572946 (Figures 3C, 3D, and S4A). In contrast, mutants impaired in auto-activation induced less liposome permeabilization than did WT BAK when activated by SJ572946 or BID BH3 (Figures 3C, 3D, and S4A). Interestingly, the I81A(3), I81R(3), I85A(4), and I85R(4) BAK mutants exhibited diminished to complete impairment [I85R(4)] in activation by SJ572946 when compared with BID BH3 in these assays, which supports SJ572946 binding to these residues at hydrophobic pockets (3) and (4), as predicted from NMR and docking analyses (Figures 3C, 3D, S4A, and S4B). Remarkably, SJ572946 activated R127A(s) BAK, a mutant fully impaired in auto-activation or direct activation by BID BH3 (Figures 3C, 3D, and S4A). Our data support the hypothesis that SJ572946 binds to the activation groove to promote BAK activation and that SJ572946 cooperates with BAK auto-activation.
SJ572946 cooperates with BID BH3 peptide ligands to activate BAK in vitro
Considering that SJ572946 and BH3 ligand activators of BAK target the activation groove we sought to investigate their cooperation in BAK activation using BID BH3 as the prototypical peptide ligand (Figure 4A). SJ572946 and BID BH3 bind to the activation groove of BAK with low affinity, and therefore we anticipated that their combination might exert additive effects in BAK activation. Indeed, compared to the calculated additivity in liposome permeabilization, the combination of SJ572946 (200 μM) with different doses of WT BID BH3 induced similar permeabilization at the highest peptide dose and a diminished response at lower peptide doses (Figures 4B and S5A). We noted a similar trend for the combination of A3 with WT BID BH3 (Figures 4B and S5A).
Figure 4.
SJ572946 and A3 cooperate with BID BH3 in BAK-mediated liposome permeabilization
(A) Model of direct BAK activation by BID BH3, compounds SJ572946 and A3, and their combination.
(B) Results of liposome permeabilization assays, quantified as the AUCs of the kinetic traces from Figure S5A, revealing the cooperation of WT BID BH3 and SJ572946 or A3 in direct BAK activation. Data are presented as the mean +SE of n = 2 experiments, each comprising n = 3 technical replicates, ∗∗p <0.0021, ∗p <0.0332; Tukey–Kramer one-way ANOVA.
(C) Docking of SJ572946 at pocket (3) of the activation groove of BAK, modeled based on the peptide-excluded, opened inactivated complex W3W5 BID BH3–BAK.
(D) Results of liposome permeabilization assays, quantified as the AUCs of kinetic traces from Figure S5B, revealing the cooperation of W3W5 BID BH3 and SJ572946 in direct BAK activation. W3W5 BID BH3 is inactivating and high doses and activating at low doses, as previously described (Singh et al., 2022). Data are presented as the mean +SE of n = 2 experiments, each comprising n = 3 technical replicates. ∗∗p <0.0021; Tukey–Kramer one-way ANOVA.
To test SJ572946 and BH3 ligand competition at the activation groove, we used the high-affinity W(3)W(5) BID BH3 peptide, which binds the activation groove to inactivate BAK by stabilizing the helix α1 electrostatic network at the bottom of the groove (Figure 4C) (Singh et al., 2022). Consistent with our earlier report, W(3)W(5) BID BH3 inactivated and activated BAK at concentrations of 10 and 2 μM, respectively (Figures 4D and S5B). Docking of SJ572946 at the open groove of the W(3)W(5) BID BH3:BAK complex (PDB 7m5a) indicates binding at hydrophobic pockets (2)–(3) overlapping the W(3) binding pocket and, thus, supports their competition for the activation groove (Figure 4C). We observed partial displacement by SJ572946 of 10 μM W(3)W(5) BID BH3 peptide and robust activation of BAK by SJ572946 at a concentration of 2 μM peptide in liposome permeabilization assays (Figure 4D). Our data are consistent with cooperation between SJ572946 (and A3) and BID BH3 ligands in BAK activation through mutually exclusive binding to overlapping sites in the activation groove of different BAK monomers. However, we do not exclude the possibility of SJ572946 and BID BH3 ligands cooperating by binding to BAK simultaneously.
The combination of SJ572946 and BID BH3 triggers activation of BAK mutant R127A(s) in liposomes but not in purified mitochondria
Intrigued by the ability of SJ572946 but not BID BH3 to activate R127A BAK in liposome permeabilization (Figure 3), we sought to characterize the mechanism of activation of this mutant further. The R127 side chain, located on the α5 side of hydrophobic pockets (2) and (3), forms a salt bridge to a conserved aspartate of the BH3 ligand in directly activated and auto-activated BH3–BAK complexes (Figure 3C); therefore, R127A(s) substitution was expected to lower the affinity of BAK for BH3 ligands severely. SJ572946 at concentrations of 125 and 250 μM weakly bound 15N-labeled R127A(s) BAK, as determined by 2D-NMR, inducing larger CSPs than were induced by its binding to WT BAK (Figures 2B, 2C, 5A, 5B, S3A, S3B, and S6A). CSP mapping onto the apo BAK structure revealed several possible binding sites, including the top of the activation groove, the bottom of this groove near the C-terminus of helix α1, and helices α6 and α7 near the N-terminus of helix α1 (Figure 5C). In contrast, WT BID BH3 at a concentration of 200 μM did not bind to 15N-labeled R127A(s) BAK showing no CSPs, as determined by 2D-NMR (Figure S6A). In thermal shift assays, SJ572946 and A3 modestly reduced the Tm of R127A(s) BAK, suggesting that it was destabilized (Figure S6B). SJ572946 cooperated additively with WT and W(3)W(5) BID BH3 peptide ligands in triggering R127A(s) BAK in liposome permeabilization assays (Figures 5D, S6C, and S6D). Remarkably, W(3)W(5) BID BH3 did not inhibit this mutant at a concentration of 10 μM, in contrast to the inhibition observed with WT BAK, being more potent alone than was WT BID BH3 in activating this mutant (Figures 5D and 4D).
Figure 5.
Auto-activation-impaired R127A BAK is extensively unfolded and degraded at the mitochondria
(A) Titration of 15N-labeled R127A(s) BAK with SJ572946, monitored by 15N-1H HSQC 2D-NMR.
(B) Per-residue CSPs induced by SJ572946 in the amide backbone of R127A(s) BAK, calculated from the spectra shown in panel (A). Dotted red line represents CSPs average +SD.
(C) Mapping of the CSPs in panel (B) onto the model of R127A(s) built using the crystal structure of apo BAK identifies SJ572946 binding to the activation groove.
(D) Results of liposome permeabilization assays, quantified as the AUCs of the kinetic traces from Figure S6C, revealing the cooperation of BID BH3 peptides with SJ572946 in direct activation of R127A(s) BAK. Data are presented as the mean +SE of n = 2 experiments, each comprising n = 3 technical replicates. ∗∗∗∗p <0.0001, ∗∗∗p <0.0002, ∗∗p <0.0021, ∗p <0.0332; Tukey–Kramer one-way ANOVA.
(E) Results of mitochondrial poration assays measuring cyt c release after incubation with WT BID BH3 ± SJ572946 (bottom), followed by limited proteolysis with calpain (middle) and BMH crosslinking (top) of mitochondria purified from BCL2allKO HCT116 cells constitutively expressing R127A(s) BAK. The aberrant patterns of proteolysis and crosslinking indicate that R127A(s) BAK is inactivated by adopting a conformation different from that of active BAK. Remarkably, R127A(s) BAK was susceptible to calpain proteolysis even in the absence of BH3 peptide activators. R127A(s) BAK was extensively degraded in these cells, as shown in the BMH crosslinking blots. The BMH crosslinking pattern did not change in the presence of BH3 peptide activators, consistent with BAK being in an altered unfolded conformation at the mitochondria.
Despite binding-induced activation of R127A(s) BAK in liposome permeabilization assays, mitochondria purified from BCL2allKO HCT116 cells constitutively expressing pMX-R127A(s) BAK-IRES-GFP were impaired in cyt c release on activation by WT BID BH3 ± SJ572946 (Figure 5E). In these cells, mitochondrial R127A(s) BAK was partially degraded into two fragments resembling those produced by limited proteolysis with m-calpain, suggesting that an endogenous protease may degrade this mutant. This mutant failed to respond to the thiol-targeting crosslinker BMH (bismaleimidohexane), which can detect oligomerization with activated WT BAK (Figure 5E) (Singh and Moldoveanu, 2019). Remarkably, adding WT BID BH3 ± SJ572946 did not change the banding pattern obtained with m-calpain proteolysis, suggesting that this mutant is spontaneously misfolded at the mitochondria and inactive in poration (Figure 5E). Purified Escherichia coli–expressed R127A(s) and WT MEAS-BAK-ΔTM-His6 were resistant to m-calpain proteolysis, suggesting that they were folded properly (Figure S6E). Together, our data suggest that proteolysis of misfolded BAK acts as a quality control measure in BCL2allKO HCT116 cells.
SJ572946 induces BAK-mediated apoptosis and cooperates with BID to activate R127A(s) BAK
SJ572946 exhibits in vitro properties of a bona fide BAK activator, and we wanted to investigate its ability to induce apoptosis in BAK-dependent cell lines. We assessed cell death using Incucyte imaging of the uptake over time of the cell impermeable DNA binding dye SYTOX Green. In bak−/− bax−/− HCT116 cells stably reconstituted with WT BAK by using the pMX-WT BAK-IRES-GFP retroviral system, SJ572946 induced residual apoptosis by itself, but it cooperated with an intrinsic apoptotic inducer (actinomycin D) and extrinsic apoptosis stimuli (TRAIL + cycloheximide) to induce dose-dependent and qVD-inhibited apoptosis (Figures 6A, 6B, S7A, and S7B). Additionally, in BCL2allKO HCT116 cells lacking the entire BCL-2 family repertoire (O'Neill et al., 2016) and reconstituted with Dox-inducible WT-like mCherry (mC)-V74A(1) BAK, SJ572946 induced BAK-dependent and dose-dependent apoptosis (Figures 6C, 6D, S7C, and S7D).
Figure 6.
SJ572946 alone or in combination with BID activates BAK in apoptosis
(A, C, and E) Cellular models of direct activation of BAK by SJ572946 and its cooperation with apoptosis inducers and BID.
(B and D) AUC analysis for cell death in bak−/−bax−/− HCT116 cells and BCL2allKO HCT116 cells reconstituted with constitutively expressed WT BAK and the Dox-inducible mC-V74A(1) BAK, respectively, and monitored by Incucyte imaging for SYTOX Green uptake in response to the apoptotic inducers actinomycin D (ActD), and TRAIL + cycloheximide (CHX). qVD inhibits caspases to block apoptosis. The kinetic traces are shown in Figures S7B and S7D.
(F) Immunoblot of BID and mCherry from BCL2allKO HCT116 cells reconstituted with Dox-inducible mC-R127A(s) BAK.
(G) Cell death in BCL2allKO HCT116 cells reconstituted with constitutively expressed BID and Dox-inducible mC-R127A(s) BAK, monitored for SYTOX Green uptake by Incucyte imaging in the presence and absence of SJ572946. AUCs of the kinetic traces at 24 h are shown. Cell death was blocked by qVD (40 μM), which is consistent with apoptosis. Data are presented as the mean +SD of one representative of n = 2 experiments, each comprising n = 3 technical replicates. ∗∗∗∗p <0.0001, ns not significant; Tukey–Kramer one-way ANOVA.
We next tested the apoptotic response in BCL2allKO HCT116 cells reconstituted with Dox-inducible mC-R127A(s) BAK (Figures 6E and 6F). This mutant was activated by SJ572946 ± BID BH3 ligands in liposome permabilization assays (Figure 5D) but not in mitochondrial permeabilization assays (Figure 5E). In the absence of BID, SJ572946 did not activate mC-R127A(s) BAK in these cells (Figure 6G, top panels). In the presence of constitutively expressed BID, Dox-mediated expression of mC-R127A(s) BAK resulted in residual apoptosis induction (Figure 6G, bottom panels). Remarkably, SJ572946 accelerated qVD-inhibited cell death induced by BID + mC-R127A(s) BAK, suggesting that the cell death resulted from apoptosis (Figure 6G, bottom panels). Our data support the existence of considerable cooperation between SJ572946 and BID in activating a BAK mutant with the propensity to misfold, inactivate, and degrade at the mitochondria; it also suggests a possible quality control mechanism that surveils BAK folding (Figures S7E and S7F).
SJ572946 preferentially activates BAK over BAX in membrane permeabilization and cooperates with intrinsic inducers of tumor apoptosis
To assess SJ572946 pro-apoptotic activity more broadly, we tested it in BAK-dependent and BAX-dependent in vitro functional assays and in a panel of human cancer cell lines. SJ572946 was selective for BAK-ΔTM-His6-mediated liposome permeabilization (mimicking endogenous TM-membrane-targeted BAK), exhibiting residual activation of full-length BAX in these assays (mimicking endogenous cytosolic BAX) (Figures 7A and S8A). In addition, BH3 profiling assays measuring cyt c release in digitonin-permeabilized cells showed that SJ572946 cooperated with BID and BIM BH3 peptides in WT and BAX−/− HeLa human cervical carcinoma cells, but not BAK−/− and BAX−/− BAK−/− HeLa cells (Figures 7B and S8C). In 72 h-long apoptosis assays, HeLa cells are very resistant to BH3 mimetics including the selective (ABT-199, A1331852) and the pan-acting (ABT-263) inhibitors of BCL-2 and BCL-xL, and the selective inhibitor of MCL-1 (S63845) exhibiting low percentage of staining for the markers of early apoptosis annexin V (AxV, binds to externalized phosphatidylserine), and late apoptosis propidium iodide (PI, cell impermeable DNA binding dye) as assessed by flow cytometry analysis (Figure S8D). Chemotherapeutic agents etoposide (a topoisomerase II and DNA synthesis inhibitor), doxorubicin (a topoisomerase II and DNA synthesis inhibitor), and staurosporine (a pan-kinase inhibitor) induced dose-dependent AxV and PI staining in up to 40–50% of the cells (Figure S8D). SJ572946 induced annexin V staining as a single agent and in combination with apoptosis inducers in WT and BAX−/− HeLa cells, but not in BAX−/− BAK−/− HeLa cells (Figure S8D). However, SJ572946 also promoted annexin V staining in BAK−/− HeLa cells, suggesting that in this cell line it may exhibit polypharmacologic properties (Figure S8D).
Figure 7.
SJ572946 selectivity in BAK-mediated membrane permeabilization and its cooperation with intrinsic apoptosis inducers
(A) Liposome permeabilization measured as the area under the curve (AUC) for the normalized data in Figure S8A. Data are presented as the mean +SD from n = 2 experiments, each comprising n = 3 technical triplicates. Because of the poor solubility of SJ572946, its activity decreases at concentrations higher than 200 μM.
(B) Cooperation of SJ572946 with BAK but not with BAX in HeLa cell BH3 profiling assays. Data are presented as the mean +SD from one representative of n = 1 (SU-DHL-10, SKNO-1 and Kasumi1 cell lines) or n = 3 experiments (all other cell lines) each of n = 1 technical replicates.
(C) Results of flow cytometry investigation of SJ572946 cooperation with intrinsic inducers of apoptosis in lymphoma cells (left panels) and leukemia cells (right panels). Data are presented as the mean +SD from n = 3 experiments, each of n = 1 technical replicates.
Supporting its selectivity for BAK-dependent apoptosis, in human lymphoma cell lines that express high levels of BAK protein (Raji Burkitt lymphoma cells), SJ572946 exhibited single-agent dose-dependent activity and cooperated with all tested chemotherapeutic agents in a dose-dependent manner (Figures 7C and S8E). In contrast, in lymphoma cells with high levels of BAX protein (U937 myeloid leukemia cells), SJ572946 cooperated with these agents only at the highest dose (Figures 7C and S8E). These lymphomas underwent apoptosis in response to S63845, etoposide, doxorubicin, and staurosporine as single agents, but they did not respond to the BH3 mimetics targeting BCL-2 and BCL-xL. Additionally, SJ572946 induced up to 50% cell death at only the highest dose in the SU-DHL-10 human B cell lymphoma cell line expressing low BAK and BAX protein levels, which was highly resistant to all the tested chemotherapeutic inducers of apoptosis (Figures 7C and S8E).
Furthermore, our analysis of five human leukemia cell lines that robustly respond to more than one intrinsic apoptosis inducers as single agents, indicated that SJ572946 did not promote cell death at doses below 100 μM, yet all the cell lines were susceptible to 300 μM SJ572946 (Figure S8E). Notably, only the Kasumi1 cell line exhibited dose-dependent cooperation of SJ572946 with BH3 mimetics, suggesting possible BAK dependence. Together, these data support the selectivity of SJ572946 for BAK in vitro and in human tumor cells and the dose-dependent cooperation of SJ572946 and known apoptotic inducers in lymphoma and some leukemia cell lines while suggesting that SJ572946 has polypharmacologic properties at high doses, exhibiting potent single-agent activity that is dependent on the mitochondrial pathway.
Discussion
The compound SJ572946 on which we report here is a readily available tool compound that is useful for engaging and probing the mechanism of BAK activation in apoptosis initiation. BAK activation is a multi-step mechanism initiated by a hit-and-run BH3 ligand binding mechanism that transforms dormant BAK from a globular folded protein to an open active conformation that has two roles: to propagate the activation through asymmetric BH3-in-groove auto-activation and to permeabilize membranes, possibly through α2–α5 symmetric BH3-in-groove core dimerization. The initiating event in BAK activation is BH3-in-groove triggering by the BH3-only direct activators BID, BIM, and others. SJ572946 mimics the role of the BH3 peptide ligands in BAK activation. It binds to multiple hydrophobic pockets in the activation groove, and possibly to a cavity at the bottom of the groove adjacent to the electrostatic network stabilizing helix α1, to destabilize BAK overall, as suggested by a decrease in the melting temperature relative to that of apo BAK. SJ572946 induces BAK-dependent membrane permeabilization of liposomes and purified mouse mitochondria, as well as BAK-dependent apoptosis in human tumor cells devoid of the entire BCL-2 repertoire (BCL2allKO HCT116 expressing BAK). It also additively cooperates with BID BH3 peptide ligands to activate BAK-mediated membrane permeabilization and displaces the inactivating W3W5 BID BH3 peptide ligand from the activation groove.
Using SJ572946 as a tool compound, we have revealed a new BAK activation paradigm: a misfolded, inactivated BAK mutant, which is resistant to activation by BID, SJ572946, or their combination in isolated mitochondria, may be activated in cells by a combination of SJ572946 and BID, possibly through interference with BAK misfolding. Future studies aided by SJ572946 will reveal more about this intriguing mechanism of inactivation.
SJ572946 exhibits selectivity for BAK over BAX in liposome permeabilization assays and mitochondrial permeabilization assays, cooperating with the BH3 activators BID and BIM in BH3 profiling assays. Of importance, SJ572946 does not permeabilize liposomes or mitochondria in the absence of BAK and BAX, thereby supporting its dependence on the mitochondrial pathway. Despite its selectivity for BAK over BAX in vitro, SJ572946 induces apoptosis in BAK−/− HeLa cells, suggesting polypharmacologic properties that manifest through activation of the mitochondrial pathway, since the BCL2allKO HCT116 and BAX−/− BAK−/− HeLa cells were resistant to apoptosis induced by SJ572946. The therapeutic window for SJ572946 is reached around 200 μM. At higher doses, SJ572946 mediated BAK-dependent or BAX-dependent apoptosis as a single agent. SJ572946 serves best as a useful tool in probing the mechanism of BAK activation in minimalistic in vitro and cellular systems, rather than as a first-in-class BAK selective drug.
To compare SJ572946 with currently available compounds that trigger BAK-mediated apoptosis, we note two published BAK activators. The first reported activator of BAK, OICR766A, is a direct BAX activator but indirect BAK activator discovered in a screen for BAX activators using liposome permeabilization assays (Brahmbhatt et al., 2016). OICR766A exhibits a narrow therapeutic window: it is inert at 1.25 μM in baby mouse kidney (BMK) cells, it induces apoptosis of BMK cells in a BAX- and/or BAK-dependent manner at 2.5 μM, yet it kills bak−/−bax−/− BMK cells at 10 μM (Brahmbhatt et al., 2016). The second reported BAK-selective activator, BKA-073, was recently discovered in a virtual screen (Park et al., 2021). At a concentration of 1 μM, BKA-073 was shown to induce complete cyt c release from purified mitochondria in a BAK-dependent but not BAX-dependent manner. In fluorescence polarization assays at the same concentration, BKA-073 apparently bound to BAK potently but did not bind to BAX, BCL-2, BCL-xL, BCL-w, or MCL-1 (Park et al., 2021). Because BKA-073 is not commercially available, we synthesized it at WuXi AppTec and report that it does not bind to apo BAK by ligand- or protein-based NMR (Figure S9). BKA-073 therefore activates BAK indirectly and therefore establishing its mechanism of action requires further investigation. We note that BKA-073 is a derivative of DNA-intercalating acridine, which may trigger apoptosis by interfering with DNA as reported for other acridine derivatives (Lang et al., 2013).
Mitochondrial apoptosis is central to mammalian physiology and its impairment is associated with many diseases. In cancer, cells become resistant to mitochondrial apoptosis and, as a result, they exhibit resistance to chemotherapy, radiotherapy, and immunotherapy. The BCL-2 family of proteins are established drug targets that are often dysregulated in cancer, either by oncogenic upregulation of the BCL-2 guardians or by downregulation of the pro-death BCL-2 proteins (Hanahan and Weinberg, 2011). The overarching goal of selective initiation of tumor apoptosis in cancer can theoretically be achieved by the combination of selective activators of BAK or BAX with selective inhibitors of one or more BCL-2 guardians. Accordingly, BH3 mimetic drugs have been developed against the BCL-2 guardians and are typically used in combination with other inducers of mitochondrial apoptosis that activate the pro-death BCL-2 proteins (e.g., chemotherapy). However, the toolkit of drugs and chemical probes targeting the BCL-2 family is incomplete (Ashkenazi et al., 2017; Diepstraten et al., 2022). The only FDA-approved drug targeting BCL-2 proteins is the BCL-2 selective inhibitor venetoclax (ABT-199), which is approved for treating several hematologic malignancies (Roberts, 2020; Souers et al., 2013). Exquisitely selective inhibitors of BCL-xL and MCL-1 have been developed, although their clinical development has been slowed to circumvent their associated toxicities (Kotschy et al., 2016; Leverson et al., 2015; Roberts et al., 2021). Potent pan-acting inhibitors of BCL-2, BCL-xL, and BCL-w have also been developed, but they are still in the early stages of clinical trials (Pullarkat et al., 2021; Tse et al., 2008). PROteolysis-TArgeting Chimera (PROTAC) degrader derivatives of BH3 mimetics have further refined their selectivity profiles and biological activity and show great potential in preclinical studies (Khan et al., 2019; Lv et al., 2021). The development of drugs against pro-death BCL-2 effectors is lagging. Although tool compounds that activate and inhibit BAK and BAX have been reported, their preclinical development is limited likely due to their polypharmacologic properties. SJ572946 is a readily available compound that directly activates BAK selectively over BAX, expanding the toolkit of effector-targeting small molecules for preclinical apoptosis and cancer research.
Limitations of the study
In this study, we have discovered a small molecule direct activator of BAK that selectively induces membrane permeabilization in a BAK-dependent manner in vitro. Despite this small molecule showing on target cellular activity in BAK-expressing BCL2allKO HCT116 cells devoid of the entire BCL-2 protein repertoire, one of its limitations is possible off-target cellular activities triggering apoptosis in the absence of BAK, through indirect activation of BAX. The off-target mechanisms of action have not been investigated here. Another limitation is the low affinity for BAK requiring relatively high doses for cellular activity, which is further hampered by its poor solubility.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-actin monoclonal antibody | Millipore Sigma | Cat# MAB1501; RRID:AB 2223041 |
| Anti-BAK (Ab-1) mouse monoclonal antibody | EMD Millipore | Cat# AM03; RRID:AB 2060945 |
| Anti-BAK mouse monoclonal antibody | Cell Signaling | Cat# 3814; RRID:AB 2290287 |
| Anti-human BID monoclonal antibody | Santa Cruz | Cat# SC-56025; RRID:AB 781628 |
| Anti-BAK | EMD Millipore | Cat# 06-536; RRID:AB 310159 |
| Anti-BAX rabbit monoclonal antibody | Cell Signaling | Cat# 2772S; RRID:AB 10695870 |
| Anti-mCherry monoclonal Antibody (16D7) | ThermoFisher Scientific | Cat# M11217; RRID:AB_2536611 |
| Anti-rat IgG, Horseradish Peroxidase whole antibody (from goat) | GE Lifesciences | Cat# NA935 |
| Anti-rabbit ECL antibody | GE Lifesciences | Cat# NA934 |
| Anti-mouse IgG ECL secondary antibody (from sheep) | GE Lifesciences | Cat# NA931 |
| Bacterial and virus strains | ||
| Escherichia coli strain: XL10-Gold | Agilent Technologies | Cat# C404010 |
| Escherichia coli strain: BL21 Star (DE3) | Thermo Fisher Scientific | Cat# C601003 |
| Escherichia coli strain: T7 Express | New England Biolabs | Cat# C2566I |
| Biological samples | ||
| None | N/A | N/A |
| Chemicals, peptides and recombinant proteins | ||
| 4,5,6,7-tetrahydrobenzo[d]thiazol-2-amine | Life Chemicals | Cat# F1040-0006 |
| N-(4-methoxy-7-methylbenzo[d]thiazol-2-yl)acetamide | Life Chemicals | Cat# F1813-0009 |
| N-(6-methylbenzo[d]thiazol-2-yl)acetamide | Life Chemicals | Cat# F0298-0133 |
| N-(3-cyano-5,6-dihydro-4H-cyclopenta[b]thiophen-2-yl)isobutyramide | Life Chemicals | Cat# F0098-0149 |
| 2-acetamido-6-methyl-4,5,6,7-tetrahydrobenzo[b]thiophene-3-carboxamide | Life Chemicals | Cat# F0325-0214 |
| 7-chloro-4-methylbenzo[d]thiazol-2-amine | Life Chemicals | Cat# F1911-0024 |
| 4,7-dimethoxybenzo[d]thiazol-2-amine | Life Chemicals | Cat# F1911-0031 |
| 2-amino-3-(2-ethoxy-2-oxoethyl)benzo[d]thiazol-3-ium | Life Chemicals | Cat# F2145-0549 |
| 4,7-dimethylbenzo[d]thiazol-2-amine | Life Chemicals | Cat# F1911-0027 |
| 6-(methylsulfonyl)benzo[d]thiazol-2-amine | Life Chemicals | Cat# F0901-0841 |
| 4,6-dichlorobenzo[d]thiazol-2-amine | ChemDiv | Cat# 2265-3111 |
| A-1331852 | MedKoo Biosciences, Inc. | Cat# 406841 |
| ABT-199 | MedKoo Biosciences, Inc. | Cat# 205807 |
| ABT-263 | MedKoo Biosciences, Inc. | Cat# 201970 |
| S63845 | MedKoo Biosciences, Inc. | Cat# 406849 |
| Etoposide | MedChemExpress | Cat# HY-13629 |
| Doxorubicin | MedChemExpress | Cat# HY-15142A |
| Staurosporine | MedChemExpress | Cat# HY-15141 |
| AlexaFluor488-conjugated Annexin V | Abcam | Cat# ab219916 |
| Propidium iodide | Abcam | Cat# ab14083 |
| DAPI | Abcam | Cat# ab228549 |
| AlexaFluor488-conjugated anti-cytochrome c antibody clone 6H2.B4 | BioLegend | Cat# 612310 |
| BMH crosslinker | Thermo Fisher Scientific | Cat# 22330 |
| Premium Select Fetal Bovine Serum | Atlanta Biologicals | Cat# S11550 |
| DMEM, high glucose, no phosphates | Thermo Fisher Scientific | Cat# 11971025 |
| RPMI 1640 | Gibco | Cat# 11875085 |
| Gibco L-glutamine | Thermo Fisher Scientific | Cat# 25030081 CAS# 56-85-9 |
| Gibco Penicillin-Streptomycin | Thermo Fisher Scientific | Cat# 15140122 CAS# 69-57-8, 57-92-1 |
| Gibco Sodium Pyruvate | Thermo Fisher Scientific | Cat# 11360070 CAS# 113-24-6 |
| Gibco MEM Non-Essential Amino Acids Solution | Thermo Fisher Scientific | Cat# 11140050 |
| Gibco 2-Mercaptoethanol | Thermo Fisher Scientific | Cat# 21985023 CAS# 60-24-2 |
| 0.25% Trypsin, 0.1% EDTA in HBSS w/o Calcium, Magnesium and Sodium Bicarbonate | Corning | Cat# 25053CI |
| 3-((3-cholamidopropyl)dimethylammonium)-1-propanesulfonate CHAPS | Gold Biotechnology | Cat# C-080-100 |
| hBID_BH3:WT EDIIRNIARHLAQVGDSMDRSI | (Singh et al., 2022) | N/A |
| hBID_BH3:W(3)W(5) EDIIRNIARHLAQWGDSMDRSW | (Singh et al., 2022) | N/A |
| Lipofectamine 3000 | Thermo Fisher Scientific | Cat# L3000015 |
| Doxycycline | Clontech | Cat# 631311 CAS# 24390-14-5 |
| SYTOX Green | Thermo Fisher Scientific | Cat# S7020 CAS# 163795-75-3 |
| Recombinant human TNF-α | Invivogen | Cat# Rhtnf-a |
| Recombinant human sTRAIL/Apo2L | Peprotech | Cat# 10770-798 |
| Cycloheximide | APExBIO | Cat# A8244 |
| Q-VD-Oph hydrate | APExBIO | Cat# A1901 |
| Ponceau S | G-Biosciences | Cat# 786-576 |
| cOmplete, Mini Protease Inhibitor Cocktail | Roche | Cat# 11836153001 |
| 15N Ammonium Chloride | Cambridge Isotope Laboratories | Cat# NLM-467-10 CAS# 12125-02-9 |
| 13C Glucose | Cambridge Isotope Laboratories | Cat# CLM-1396-1 CAS# 50-99-7 |
| Deuterated DTT | Cambridge Isotope Laboratories | Cat# DLM-2622-1 |
| Deuterium Oxide | Sigma Aldrich | Cat# 617385-1 CAS# 7789-20-0 |
| SYPRO™ Orange Protein Gel Stain (5,000X Concentrate in DMSO) | Thermo Fisher Scientific | Cat#S6650 |
| Critical commercial assays | ||
| Supersignal West Dura | Thermo Fisher Scientific | Cat# 34075 |
| QuikChange II XL Site-Directed Mutagenesis Kit | Agilent | Cat# 200517 |
| Experimental models: Cell lines | ||
| BCL2allKO HCT116 | (O’Neill et al., 2016) | N/A |
| WT, BAK−/−, BAX−/−, BAK−/− BAX−/− HeLa | (Fraser et al., 2019) | N/A |
| U937 | ATCC | Cat# CRL-1593.2 |
| Raji | ATCC | Cat# CCL-86 |
| SU-DHL-10 | ATCC | Cat# CRL-2963 |
| SKNO-1 | DSMZ | Cat# ACC 690 |
| Kasumi | ATCC | Cat# CRL-2724 |
| OCI-AML-2 | DSMZ | Cat# ACC 99 |
| MOLM13 | DSMZ | Cat# 554 |
| SKM-1 | ATCC | Cat# HTB-67 |
| Experimental models: Organism/strains | ||
| WT C57BL/6J mouse | The Jackson Laboratory | Strain#: 000664 |
| B6.129-Bak1tm1Thsn/J mouse | The Jackson Laboratory | Strain#: 004183 |
| Deposited data | ||
| Raw Western blots | This paper, Mendeley data | https://doi.org/10.17632/j6zg95s7nn.1 |
| Oligonucleotides | ||
| BAK R127A mutagenesis primers 5′-ggcatcaattggggcgctgtggtggctcttct-3′ 3′-ccgtagttaaccccgcgacaccaccgagaaga-5′ |
(Singh et al., 2022) | N/A |
| Recombinant DNA | ||
| pRetroX-TRE3G | N/A | Clontech |
| pCoofy3 | (Scholz et al., 2013) | Addgene #43983 |
| pNIC28-Bsa4 | (Strain-Damerell et al., 2014) | Addgene #26103 |
| pLZRS IRES GFP | (Moldoveanu et al., 2013) | Addgene #21961 |
| Human BAX-MxeGyrA-CBP fusion in pCoofy3 | This paper | N/A |
| Human BAK in pNIC28-Bsa4 | (Singh et al., 2022) | N/A |
| Human BAK and mutants in pRetroX-TRE3G | (Singh et al., 2022) | N/A |
| Human BID in pMX-IRES-GFP | (Singh et al., 2022) | N/A |
| Software and algorithms | ||
| Ccpnmr | (Skinner et al., 2016) | https://ccpn.ac.uk/ |
| CARA v1.9.1.7 | (Keller, 2004) | http://cara.nmr.ch/ |
| IncuCyte Zoom v2016A | Essen BioScience | https://www.essenbioscience.com |
| IncuCyte ZOOM 2016A Rev2 v20161.1.5932.22771 | Essen BioScience, Inc. | http://www.essenbioscience.com |
| MacPyMOL v1.7.6.3 | Schrödinger, LLC | http://www.pymol.org |
| Prism v8.0a | GraphPad Software, Inc. | http://www.graphpad.com |
| TopSpin v3.5 | Bruker | https://www.bruker.com/en/products-and-solutions/mr/nmr-software/topspin.html |
| Other | ||
| L-a-phosphatidylcholine (Egg, Chicken) | Avanti Polar Lipids | Cat# 840051C CAS# 97281-44-2 |
| L-a-phosphatidylinositol (Liver, Bovine) (sodium salt) | Avanti Polar Lipids | Cat# 840042C CAS# 383907-33-3 |
| L-a-phosphatidylserine (Brain, Porcine) (sodium salt) | Avanti Polar Lipids | Cat# 840032C CAS# 383907-32-2 |
| Cardiolipin (Heart, Bovine) (sodium salt) | Avanti Polar Lipids | Cat# 840012C CAS# 383907-10-6 |
| 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] (nickel salt) | Avanti Polar Lipids | Cat# 790404C CAS# 231615-77-3 |
| 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine | Avanti Polar Lipids | Cat# 850725C CAS# 4004-05-1 |
| ANTS (8-aminonpaphthalene-1,3,6-trisulfonic acid, disodium salt) | Molecular Probes | Cat# 1278701 CAS# 5398-34-5 |
| DPX (p-xylene-bis-pyridiunium bromide) | Molecular Probes | Cat# X1525 CAS# 14208-10-7 |
| Nickel agarose beads (high density) | Gold Biotechnology | Cat# H-320-500 |
| Amicon Ultra 15-mL 3K MWCO centrifugal filter | MilliporeSigma | Cat# UFC900324 |
| Amicon Ultra 15-mL 10K MWCO centrifugal filter | MilliporeSigma | Cat# UFC800324 |
| HiPrep Sephacryl S-100 HR column | GE Healthcare | Cat# 17116501 |
| Superdex 200 Increase 10/300 GL | GE Healthcare | Cat# 28990944 |
| Superdex 75 Increase 10/300 GL | GE Healthcare | Cat# 29148721 |
| MonoS 5/50 GL column | GE Healthcare | Cat# 17-5168-01 |
| HiPrep SP FF 16/10 | GE Healthcare | Cat# 28-9365-44 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Tudor Moldoveanu (tmoldoveanu@uams.edu).
Materials availability
There are MTA restrictions to the availability of BCL2allKO HCT116 cell line, which was obtained from the Xu Luo laboratory at University of Nebraska.
Experimental model and subject details
The HCT116 cell lines analyzed herein were derived from the HCT116 human colorectal carcinoma cell line from an adult male, including the BCL2allKO HCT116 cell line published by the Luo lab (O’Neill et al., 2016) and BAK−/− BAX−/− HCT116 cell line obtained from the Green lab at St. Jude Children’s Research Hospital. The HeLa panel of WT, BAK−/−, BAX−/−, BAK−/−, and BAX−/− cell lines were derived from the HeLa cervical carcinoma cell line in the Sarosiek Laboratory (Fraser et al., 2019). These cell lines have been authenticated by PCR and immunoblotting in the respective labs. The lymphoma and leukemia cell line panels were obtained from ATCC and DKMZ and they were authenticated by STR profiling by the respective vendors.
Method details
Recombinant protein expression and purification
WT and mutant MEAS-BAK-ΔTM-His6 were produced from the pRL574 vector, as previously described (Moldoveanu et al., 2006), and tevBAK (residues 22–186) by using the pNIC28-Bsa4 expression vector (Strain-Damerell et al., 2014) in T7 Express Competent E. coli cells (New England Biolabs). Bacteria were grown to OD 0.8–1.0 in Luria–Bertani medium at 37°C, and protein expression was induced overnight with 100 mg/L IPTG at 21°C. Bacteria were lysed by passing them through an Avestin EmulsiFlex-C3 microfluidizer. After centrifugation of the homogenate to separate the insoluble membranes, the protein was batch-purified by Ni2+-NTA affinity chromatography and by S-200 HR size-exclusion chromatography, using an ÄKTA Pure system (GE Healthcare). TevBAK was generated by cleaving off the histidine tag with TEV protease and purified by SPFF cation exchange chromatography (GE Healthcare). cBAK (residues 15–186) was generated by calpain proteolysis of MEAS-BAK-ΔTM-His6 as described previously (Moldoveanu et al., 2013). Briefly, to produce cBAK, MEAS-BAK-ΔTM-His6 was batch affinity purified on Ni2+-NTA resin then subjected to S-200 HR size-exclusion chromatography, digestion with μI–II calpain protease for 24–48 h to remove 14 residues at the N-terminus and the C-terminal His6 tag, and MonoQ anion exchange chromatography. BAK proteins were >95% pure, as assessed by sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE).
GST-BAK, used for competitive time-resolved fluorescence resonance energy transfer (TR-FRET) assays, was expressed from the pRL296 vector, batch affinity purified on glutathione agarose resin (Gold Biotechnology), and further enriched by S-200 HR size-exclusion chromatography.
Full-length BAX (BAX) was expressed and purified based on a modified published method (Dingeldein et al., 2019) as follows. BAX was expressed as a His6-GST-HRV3C-BAX-MxeGyrA-CBP fusion construct in pCoofy3 (Scholz et al., 2013); “His6” is the polyhistidine affinity tag, “GST” is the glutathione-S-transferase solubility tag, “HRV3C” is the cut site for the human rhinovirus type 14 3C protease, “BAX” is the full-length sequence of BAX, “MxeGyrA” is the GyrA intein from Mycobacterium xenopi, and CBP is the chitin binding domain from Bacillus circulans. A GeneBloc DNA fragment encoding Bax-MxeGyrA-CBP DNA (Figure S10, Integrated DNA Technologies, Coralville, IA) was ligated into the pCoofy3 vector by using an NEBuilder® HiFi DNA Assembly Kit (New England Biolabs). Recombinant protein expression was performed as described above. Cells were resuspended in 25 mM Tris, 150 mM NaCl, 5 mM imidazole (pH 7.6) and lysed by passing them twice through an Avestin EmulsiFlex-C3 microfluidizer. Centrifuged cell lysate was loaded onto 5 mL of Ni-NTA beads (Gold Biotechnology) and washed with 100 mL of Buffer N (25 mM Tris, 150 mM NaCl, 5 mM Imidazole, pH 7.6) and 20 mL of Buffer W (25 mM Tris, 150 mM NaCl, 20 mM imidazole, pH 7.6). MxeGyrA-CBP was cleaved from the fusion construct by a thiolysis reaction performed whereas the protein was bound to the beads by incubating the beads in 20 mL of 25 mM Tris, 150 mM NaCl, 5 mM imidazole, 200 mM β-mercaptoethanol, pH 7.6. Cleaved MxeGyrA-CBP fragment was washed away with Buffer N. The His6-GST-HRV3C-BAX fragment was eluted using 100 mL of 25 mM Tris, 150 mM NaCl, 250 mM imidazole. The eluted fraction was incubated with 1 mg of HRV3C protease and dialyzed overnight against Buffer N. The dialyzed protein was loaded onto 5 mL of Ni-NTA beads and washed with 10 mL of Buffer N (the BAX protein does not bind to the beads and was collected in the flow through). The fraction containing BAX was concentrated to a volume of 0.5 mL and loaded onto a Superdex 75 10/300 GL size-exclusion chromatography column (GE Healthcare, Chicago, IL). Fractions containing protein were collected and analyzed using SDS-PAGE and were found to contain >95% protein.
The buffers of all proteins were replaced with 20 mM HEPES (pH 6.8) or 20 mM phosphate (pH 6.8) ± DTT as needed through several rounds of concentration and dilution by centrifugation in Amicon Centrifugal filters (Millipore). Proteins were then flash frozen in liquid nitrogen and stored at −80°C.
Site-directed mutagenesis
A QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies) was used to introduce mutations into BAK.
Peptide synthesis
The BH3 peptides were produced by the Peptide Synthesis facility in the Hartwell Center for Biotechnology at St. Jude.
Primary screening by ligand-based NMR
The St. Jude “InHouse” fragment library subset (1222 compounds) (Iconaru et al., 2015; Nishiguchi et al., 2021) was screened as mixtures of 5 or 6 fragments, using pulse programs for regular 1D proton NMR, waterLOGSY, and T2 relaxation-based 1D NMR run as composite experiments on a Bruker Avance 600 MHz spectrometer, using a SampleJet autosampler, at 298K. Pools of 5 or 6 fragments were obtained from the Compound Management Center in the Department of Chemical Biology and Therapeutics at St. Jude. Next, 500 μL samples were prepared in 5-mm tubes for SampleJet racks by mixing fragments (each at a final concentration of 200 μM), purified unlabeled cBAK (10 μM), 20 mM phosphate buffer (pH 6.8), and 2 mM deuterated DTT, using a Gilson dispensing robot. Fragments in mixtures that exhibited BAK binding were validated by testing them as singles with the same composite ligand-based NMR method used with the mixtures.
NMR resonance assignments
Samples for chemical shift assignment contained 200 μM [13C, 15N] R127A(s) C166S G184C tevBAK, 20 mM sodium phosphate, 10% D2O, and 0.5% NaN3 (pH 6.8). Backbone assignments were obtained from 15N HSQC, HNCACB, CBCACONH, HNCO, and HNCACO experiments (Cavanagh et al., 2006). All spectra were acquired on a Bruker Avance 600 MHz spectrometer equipped with a TCI triple-resonance CryoProbe. Triple-resonance spectra were acquired using a 30% Non-Uniform Sampling (NUS) routine and reconstructed by using the compressed sensing method and MDDNMR software (Kazimierczuk and Orekhov, 2011). Sequential chemical shift assignment was performed using the CcpNmr Analysis software (Skinner et al., 2016). Chemical shift perturbation values were calculated using the following equation:
where and are the chemical shift differences for the amide 15N and 1H resonances, respectively, and = 0.16 and = 1 are the chemical shift weighting factors.
NMR titrations
15N-labeled cBAK, produced by calpain proteolysis of MEAS-BAK-ΔTM-His6, as described previously (Moldoveanu et al., 2013), and 15N-labeled tevBAK were produced in MOPS-based medium (Neidhardt et al., 1974) supplemented with 15NH4Cl in T7 Express Competent E. coli cells and were purified as described for unlabeled proteins above. Briefly, to produce cBAK (residues 15–186), MEAS-BAK-ΔTM-His6 was batch affinity purified on Ni2+-NTA resin then subjected to S-200 HR size-exclusion chromatography, digestion with μI–II calpain protease for 24–48 h to remove 14 residues at the N-terminus and the C-terminal His6 tag, and MonoQ anion exchange chromatography. 15N-1H TROSY NMR titrations of 50 μM 15N-labeled WT cBAK or R127A(s) C166S G184C tevBAK were performed with fragments dissolved in deuterated DMSO (DMSO-d) at six concentrations (50, 100, 200, 400, 600, and 800 μM), as well as titrations with DMSO-d control at the respective volumes used with the fragments. The NMR buffer contained 20 mM phosphate buffer, pH 6.8, 10% D20, and 2.5 μM deuterated DTT. NMR spectra were obtained with a Bruker Avance 600 MHz spectrometer at 298K, processed in TopSpin (Bruker), and analyzed using CARA (Keller, 2004).
Molecular docking
Molecular docking experiments were performed with the SJ572946 and A3 fragments and apo BAK (2imt), M(3)W(5) BID BH3-activated BAK (7m5b), and W(3)W(5) BID BH3-inactivated BAK (7m5a) using AutoDock Vina (Trott and Olson, 2010). Peptides were removed from the BH3 complexes with BAK. The center and size of each box were set as follows: for box 1, (3.35, 1.233, 19.008) and (22, 24, 22); for box 2, (3.35, −3.907, 19.008) and (22, 18, 22); and for box 3, (3.35, −3.105, 19.008) and (28, 20, 24).
Competitive time-resolved fluorescence resonance energy transfer assay
Time resolved fluorescence resonance energy transfer (TR-FRET) assays, measuring the displacement of the fluorescent peptide BID BH3 stabilized α-helix of BCL-2 protein–fluorescein (SAHB-F) from GST-BAK, were performed in 384-well polystyrene plates with flat bottoms and black lids (Corning, 3821BC). Master mix consisted of 50 mM Tris-HCl pH 7.5, 50 mM KCl, 0.1 mg/mL BSA, 2 mM DTT, 1 μM hBID BH3 SAHB-F, 5 nM GST-BAK, and 5 nM terbium (Tb)-labeled anti-GST antibody (Life Technologies, Carlsbad, CA). Peptides were pin-transferred with a Biomek FX workstation (Beckman Coulter Life Sciences, Indianapolis, IN) to achieve a final concentration of 1000 μM–0.2 μM in a final volume of 20 μL. Plates were gently centrifuged for 1 min at 1000 rpm to remove air bubbles then incubated for 90 min at room temperature. The TR-FRET signal of each well was measured using a PHERAstar Microplate Reader (BMG LABTECH) with the excitation wavelength set to 340 nm and the emission wavelengths to 520 nm (for BID BH3 SAHB-F) and 490 nm (for Tb). The TR-FRET signal is calculated as the ratio of the signals at 520 nm and 490 nm. Free peptide controls in each plate consisted of only BID BH3 SAHB-F and bound peptide controls containing BID BH3 SAHB-F + GST-BAK. The Z′-values were greater than 0.8, which suggested excellent assay performance (Zhang et al., 1999). Data were processed and visualized using the Robust Investigation of Screening Experiments (RISE) software, developed in-house, on the Pipeline Pilot platform (Accelrys, v.8.5.0). At least three independent dose–response experiments were performed for each peptide. IC50 values were estimated with RISE and GraphPad Prism v.8.0a.
Liposome permeabilization assay
Liposome permeabilization assays were performed as described previously (Asciolla et al., 2012; Henderson et al., 2007; Oh et al., 2010; Zheng et al., 2018). Briefly, lipid films were prepared using lipid ratios similar to those in mitochondrial membranes: 40.9% phosphatidylcholine, 26.6% phosphatidylethanolamine, 9.1% phosphatidylinositol, 8.3% phosphatidylserine, 7% cardiolipin, and 8.0% 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] (nickel salt) [DGS-NTA(Ni)] (Avanti Polar Lipids) at a final concentration of 2.5 μM in each assay. Also prepared was a solution containing 12.5 mM fluorophore 8-aminonaphthalene-1, 3, 6-trisulfonic acid, disodium salt (ANTS) and 45 mM quencher p-xylene-bis-pyridiniumbromide (DPX) in LUV buffer (10 mM HEPES pH 6.8, 200 mM KCl, and 5 mM MgCl2). The lipid film was mixed into the ANTS/DPX solution by vortex mixing and sonication (for approximately 15 min) followed by extrusion through a polycarbonate membrane with 2-μm pore size (Avanti Polar Lipids) to generate homogeneous large unilamellar vesicles (LUVs). LUVs were separated from free ANTS/DPX by Superdex 500 10/300 GL size-exclusion chromatography and stored at 4°C in the dark. Peptide and protein dilutions were prepared in 96-well black flat-bottom plates (Costar) kept on ice. The release of ANTS from permeabilized liposomes causes an increase in fluorescence, and this was monitored at 37°C for 90 min by using a CLARIOstar Microplate Reader (BMG LABTECH) with the excitation and emission wavelengths set to 360 nm and 530 nm, respectively. The data were normalized relative to the maximum and minimum fluorescence induced by 5% CHAPS and buffer controls, respectively. Liposome permeabilization was quantified by integrating the area under the curve of the normalized traces based on Simpson’s rule, and the data were analyzed with Excel and GraphPad Prism v8.0a. Variability in the permeabilization of different liposomes is common, and our analyses compared the permeabilization of the same liposomes under different conditions. Although the absolute permeabilization values may have differed, the trends for given conditions were always similar for different liposomes.
Thermal shift assay
Thermal shift assays were performed in capillaries containing 20 mM HEPES pH 7.0, 150 mM NaCl, 2 mM DTT, 0.5 mg/mL purified WT or R127A(s) BAK, and SJ572946 (0.25, 0.5 mM) or A3 (0.5, 2.0 mM) dissolved in DMSO, using a Prometheus NT.48 NanoDSF instrument (NanoTemper Technologies). Three experiments in triplicate were performed for each sample. The final data analysis was conducted in Excel and GraphPad Prism v8.0a.
Cloning and production of cells expressing a minimalist BCL-2 family repertoire
For mammalian cell culture experiments, the full-length human BAK gene was cloned into the pCR 2.1 TOPO vector, mutated as needed, and amplified by PCR for cloning into the BamHI and NotI sites of the pRetroX mCherry vector for retroviral transduction, as described previously (Singh et al., 2022). To test direct activation and autoactivation of BAK in cells, cells stably expressing mCherry-human BAK (mC-BAK) ± human BID (BID) were produced on a genetic background devoid of the entire BCL-2 family repertoire (all 17 BCL-2 genes), i.e., BCL2allKO HCT116 cells, previously produced (O'Neill et al., 2016). Mutant mC-BAK was expressed from the pRetroX-TRE3G-PuroR vector in BCL2allKO HCT116 cells expressing the Tet3G from pRetroX-Tet3G-BlastR by using the doxycycline (Dox)-based inducible Tet-On 3G retroviral system. Cells were selected with puromycin (2.5 μg/mL) and blasticidin (2 μg/mL) starting at 2 days after retroviral transduction. To enrich for populations expressing mC-BAK, cells were treated with a low dose of Dox (25 ng/mL) and the mCherry-positive cells were sorted by fluorescence-activated cell sorting (FACS). The enrichment was performed twice before the cell death analysis.
Stable, constitutive expression of human BID in mC-BAK–expressing BCL2allKO HCT116 cells was achieved by retroviral transduction based on pMX-human-BID-IRES-GFP. Two rounds of FACS gating on GFP-positive cells were performed to enrich for populations expressing BID.
Constitutively expressed BAK mutant R127A(s) was stably reconstituted in BCL2allKO HCT116 cells by pMX-human-BAK-IRES-GFP retroviral transduction. The cells were enriched for GFP-positive populations by two rounds of sorting. The cells were then used for mitochondrial poration assays.
Cell death analysis
Cells were seeded in 24-well plates (100,000 cells/well), 96-well plates (15,000 cells/well), or 384-well plates (10,000 cells/well) coated with 10 μg/mL fibronectin. After the cells had been left for ≥24 h to enable attachment and recovery, cell death was induced with Dox (100 ng/mL and 1000 ng/mL) and uptake of SYTOX Green (25 nM) was monitored by Incucyte imaging (Sartorius) in the presence or absence of compound SJ572946 ± qVD. This dye marks cells with compromised plasma membrane integrity. mC-BAK was expressed before 24 hr, which coincided with detectable cell death. Monitoring by Incucyte imaging was continued for up to 48 hr, then endpoint FACS analysis was performed to measure the extent of cell death based on the proportion of SYTOX Green–positive cells. Alternatively, at 24 hr, endpoint CytoTox-Glo luminescence analysis was performed according to the recommended protocol (Promega).
For co-expression of mC-R127A(s) BAK + BID, BID-expressing cells were treated with 1000 ng/mL Dox ± SJ572946 (200 μM) ± qVD (40 μM) and SYTOX Green uptake was recorded by Incucyte imaging for 24 hr. Even without activation of BID by TRAIL + CHX, SJ572946 was able to promote substantial cell death in these cells.
Cell culture
HeLa WT, HeLa BAX−/−, HeLa BAK1−/−, and HeLa BAX−/− BAK1−/− (DKO) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco) with 10% FBS (Life Technologies) and 1% penicillin-streptomycin (Life Technologies). U937, Raji, SU-DHL-10, Kasumi1, OCI-AML-2, MOLM13, and SKM-1 cells were cultured in RPMI 1640 medium (Gibco) with 10% FBS (Life Technologies) and 1% penicillin-streptomycin (Life Technologies). SKNO-1 were culture in RPMI 1640 medium (Gibco) with 10% FBS (Life Technologies), 1% penicillin-streptomycin (Life Technologies), and 10 ng/mL GM-CSF.
Chemosensitivity assays
To analyze apoptotic cell death in cancer cell lines on treatment with H6 and chemotherapeutics or other cell death inducers, treated cells were stained with Annexin V (AxV) and Propidium Iodine (PI). Cell lines were plated at 104 cells per well in 100 μL culture media on flat-bottom 96-well plates (Denville) and treated with of the following drugs at the concentrations specified: H6 300, 100, 30 μM, A-1331852 1, 0.1, 0.01 μM, ABT-199 1, 0.1, 0.01 μM, ABT-263 1, 0.1, 0.01 μM, S63845 1, 0.1, 0.01 μM, Etoposide 10, 1, 0.1 μM, Doxorubicin 1, 0.1, 0.01 μM, and Staurosporine 0.1, 0.01, 0.001 μM. DMSO (1%) was used as vehicle control. Cells were trypsinized (0.0025% Trypsin, Gibco) and collected after 72 h of treatment, then stained with viability markers AxV and PI with the following protocol. A staining solution was prepared with 10X Annexin binding buffer (0.1 M Hepes (pH 7.4), 1.4 M NaCl, and 25 mM CaCl2 solution, sterile filtered) and AxV/PI. AlexaFluor488-conjugated AxV and PI (Abcam) was added to solution at a dilution of 1:500. The staining solution was then added to the cells in solution at 1:10 dilution and the cells were stained for 20 min on ice. AxV/PI positivity was then measured on an Attune NxT flow cytometer equipped with an autosampler (Thermo Fisher).
Mitochondrial priming assays
Protocols for analyzing cytochrome c release in permeabilized cells on BH3 peptide treatment have been previously published (PMID: 30535998). Briefly, cells were suspended in mannitol experimental buffer (MEB; 10 mM HEPES (pH 7.5), 150 mM mannitol, 50 mM KCl, 0.02 mM EGTA, 0.02 mM EDTA, 0.1% BSA, and 5 mM succinate) with 0.001% digitonin and treated with compound H6 alone or in combination with BIM or BID peptides (sequences Ac-MRPEIWIAQELRRIGDEFNA-NH2 and Ac-EDIIRNIARHLAQVGDSMDRY-NH2 respectively; New England Peptide). Peptide treatment was carried out for 60 min at 28°C. After treatment, cells were fixed in 2% PFA, then quenched with a neutralization buffer [1.7 M tris base and 1.25 M glycine (pH 9.1)]. Cells were stained overnight with DAPI and an Alexa Fluor 647-conjugated anti-cytochrome c antibody (clone 6H2.B4, BioLegend) in a staining buffer (0.1% saponin; 1% BSA) and analyzed on an Attune NxT flow cytometer (Thermo Fisher) for cytochrome c positivity.
Immunoblot analysis
Expression of mC-BAK, BAK, and BID was monitored by immunoblotting with monoclonal antibodies against mCherry (16D7, Invitrogen), BAK (Ab-1, Calbiochem; 3814, Cell Signaling; 06-536, Millipore), BID (5C9, Santa Cruz), and BAX (2772S, Cell Signaling). Actin was used as a loading control and was detected with the C4 monoclonal antibody (Millipore). The secondary antibodies were goat anti-rat IgG (GE Healthcare) and sheep anti-mouse IgG (GE Healthcare).
Mitochondrial poration and limited proteolysis assays
Mitochondria were purified from BCL2allKO HCT116 cells constitutively expressing R127A(s) BAK from pMX-BAK-IRES-GFP by differential centrifugation according to published protocols (Singh and Moldoveanu, 2019). In poration assays, mitochondria were incubated for 2 h with WT BID BH3 peptide (1–50 μM) ± SJ572946 (200 μM) then centrifuged for 10 min at 4°C at the maximum speed achieved in a Sorvall HIGHPlate™ 6000 swinging-bucket rotor of a Sorvall Legend XTR tabletop centrifuge (Thermo Fisher). Cytochrome c release from mC-R127A(s) BAK ± BID mitochondria was monitored by immunoblotting, performed on a third of the pellet to assess cytochrome c release (7H8, Santa Cruz). Another third of the pellet was digested with m-calpain (100 nM) activated by 1 mM CaCl2 for 0.5 h, and the remaining third of the pellet was crosslinked with the amine crosslinker BMH (Pierce). Limited proteolysis and crosslinking of BAK was monitored by immunoblotting with an antibody against BAK (3814s, Cell Signaling). Purified E. coli–expressed WT BAK and R127A(s) BAK (1 mg/mL) were subjected to limited proteolysis with 1 mg/mL recombinant rat m-calpain in 50 mM Tris pH 7.6 and 1 mM CaCl2 for up to 80 min at room temperature, after which the reactions were quenched with SDS sample buffer. The results of the proteolysis were examined by SDS-PAGE.
Quantification and statistical analysis
For individual experiments, data were plotted as averages of measurements performed in triplicate or quadruplicate, with error bars indicating the standard deviation (SD). Data from two or three independent experiments were combined as the mean and standard error of the mean (SEM). Statistical analyses were performed as summarized in each figure by one-way ANOVA, using the Tukey–Kramer method for multiple comparisons with a two-tailed or multiplicity-adjusted, family-wise 0.05 confidence level in Prism v8.0a, or by using a t-test for comparisons of two groups. p values are indicated in figures as follows: ∗∗∗∗p < 0.0001, ∗∗∗p < 0.0002, ∗∗p < 0.0021, and ∗ p < 0.0332.
Acknowledgments
During the revision of the manuscript, we acknowledge the tragic death of co-author Geetika Singh. We would like to dedicate this work in her memory. We thank Keith A. Laycock for scientific editing of the manuscript. Funding for this study was supported by ALSAC. This work was supported by funding from the National Institutes of Healthgrants R37CA248565 (KAS) and R01DK125263 (KAS), Alex's Lemonade Stand Foundation for Childhood Cancers (KAS), and the Blavatnik Institute at Harvard (KAS).
Author contributions
Conceptualization: T.M., G.Se., and G.Si.; Investigation: T.M., G.S., X.Q., C.D.G., B.S., Z.I., C.R.G., W.Z., P.J.S., and W.L.; Formal analysis: T.M., G.Se., and G.Si.; Supervision: T.M., T.C., R.E.L., Z.R., and K.S.; Writing – original draft: T.M. and G.Se.; Writing – review and editing: T.M. All authors have provided feedback on the manuscript.
Declaration of interests
Author Tudor Moldoveanu is a member of the iScience Editorial Board.
Inclusion and diversity
One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
Published: October 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105064.
Supplemental information
Data and code availability
-
•
Western blots have been deposited in Mendeley. https://doi.org/10.17632/j6zg95s7nn.1. NMR data are available from the lead contact on request.
-
•
This article does not report original code.
-
•
Any additional information required to reanalyze the data reported in this article is available from the lead contact on request.
References
- Aluvila S., Mandal T., Hustedt E., Fajer P., Choe J.Y., Oh K.J. Organization of the mitochondrial apoptotic BAK pore: oligomerization of the BAK homodimers. J. Biol. Chem. 2014;289:2537–2551. doi: 10.1074/jbc.M113.526806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asciolla J.J., Renault T.T., Chipuk J.E. Examining BCL-2 family function with large unilamellar vesicles. J. Vis. Exp. 2012;68:4291. doi: 10.3791/4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazi A., Fairbrother W.J., Leverson J.D., Souers A.J. From basic apoptosis discoveries to advanced selective BCL-2 family inhibitors. Nat. Rev. Drug Discov. 2017;16:273–284. doi: 10.1038/nrd.2016.253. [DOI] [PubMed] [Google Scholar]
- Birkinshaw R.W., Iyer S., Lio D., Luo C.S., Brouwer J.M., Miller M.S., Robin A.Y., Uren R.T., Dewson G., Kluck R.M., et al. Structure of detergent-activated BAK dimers derived from the inert monomer. Mol. Cell. 2021;81:2123–2134.e5. doi: 10.1016/j.molcel.2021.03.014. [DOI] [PubMed] [Google Scholar]
- Brahmbhatt H., Uehling D., Al-Awar R., Leber B., Andrews D. Small molecules reveal an alternative mechanism of Bax activation. Biochem. J. 2016;473:1073–1083. doi: 10.1042/BCJ20160118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer J.M., Lan P., Cowan A.D., Bernardini J.P., Birkinshaw R.W., van Delft M.F., Sleebs B.E., Robin A.Y., Wardak A., Tan I.K., et al. Conversion of Bim-BH3 from activator to inhibitor of Bak through structure-based design. Mol. Cell. 2017;68:659–672.e9. doi: 10.1016/j.molcel.2017.11.001. [DOI] [PubMed] [Google Scholar]
- Brouwer J.M., Westphal D., Dewson G., Robin A.Y., Uren R.T., Bartolo R., Thompson G.V., Colman P.M., Kluck R.M., Czabotar P.E. Bak core and latch domains separate during activation, and freed core domains form symmetric homodimers. Mol. Cell. 2014;55:938–946. doi: 10.1016/j.molcel.2014.07.016. [DOI] [PubMed] [Google Scholar]
- Cavanagh J., Fairbrother W.J., Palmer A.J., III, Rance M., Skelton N.J. Second edition. Elsevier; 2006. Protein NMR Spectroscopy: Principles and Practices. [Google Scholar]
- Chipuk J.E., Moldoveanu T., Llambi F., Parsons M.J., Green D.R. The BCL-2 family reunion. Mol. Cell. 2010;37:299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan A.D., Smith N.A., Sandow J.J., Kapp E.A., Rustam Y.H., Murphy J.M., Brouwer J.M., Bernardini J.P., Roy M.J., Wardak A.Z., et al. BAK core dimers bind lipids and can be bridged by them. Nat. Struct. Mol. Biol. 2020;27:1024–1031. doi: 10.1038/s41594-020-0494-5. [DOI] [PubMed] [Google Scholar]
- Czabotar P.E., Lessene G., Strasser A., Adams J.M. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat. Rev. Mol. Cell Biol. 2014;15:49–63. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- Czabotar P.E., Westphal D., Dewson G., Ma S., Hockings C., Fairlie W.D., Lee E.F., Yao S., Robin A.Y., Smith B.J., et al. Bax crystal structures reveal how BH3 domains activate Bax and nucleate its oligomerization to induce apoptosis. Cell. 2013;152:519–531. doi: 10.1016/j.cell.2012.12.031. [DOI] [PubMed] [Google Scholar]
- Dalvit C., Fogliatto G., Stewart A., Veronesi M., Stockman B. WaterLOGSY as a method for primary NMR screening: practical aspects and range of applicability. J. Biomol. NMR. 2001;21:349–359. doi: 10.1023/a:1013302231549. [DOI] [PubMed] [Google Scholar]
- Delbridge A.R.D., Grabow S., Strasser A., Vaux D.L. Thirty years of BCL-2: translating cell death discoveries into novel cancer therapies. Nat. Rev. Cancer. 2016;16:99–109. doi: 10.1038/nrc.2015.17. [DOI] [PubMed] [Google Scholar]
- Dewson G., Kratina T., Czabotar P., Day C.L., Adams J.M., Kluck R.M. Bak activation for apoptosis involves oligomerization of dimers via their α6 helices. Mol. Cell. 2009;36:696–703. doi: 10.1016/j.molcel.2009.11.008. [DOI] [PubMed] [Google Scholar]
- Dewson G., Kratina T., Sim H.W., Puthalakath H., Adams J.M., Colman P.M., Kluck R.M. To trigger apoptosis, Bak exposes its BH3 domain and homodimerizes via BH3:groove interactions. Mol. Cell. 2008;30:369–380. doi: 10.1016/j.molcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Dewson G., Ma S., Frederick P., Hockings C., Tan I., Kratina T., Kluck R.M. Bax dimerizes via a symmetric BH3:groove interface during apoptosis. Cell Death Differ. 2012;19:661–670. doi: 10.1038/cdd.2011.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diepstraten S.T., Anderson M.A., Czabotar P.E., Lessene G., Strasser A., Kelly G.L. The manipulation of apoptosis for cancer therapy using BH3-mimetic drugs. Nat. Rev. Cancer. 2022;22:45–64. doi: 10.1038/s41568-021-00407-4. [DOI] [PubMed] [Google Scholar]
- Dingeldein A.P.G., Lindberg M.J., Ådén J., Zhong X., Stoll R., Gröbner G. Bax to the future - a novel, high-yielding approach for purification and expression of full-length Bax protein for structural studies. Protein Expr. Purif. 2019;158:20–26. doi: 10.1016/j.pep.2019.02.004. [DOI] [PubMed] [Google Scholar]
- Flores-Romero H., Ros U., Garcia-Saez A.J. Pore formation in regulated cell death. EMBO J. 2020;39:e105753. doi: 10.15252/embj.2020105753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser C., Ryan J., Sarosiek K. BH3 profiling: a functional assay to measure apoptotic priming and dependencies. Methods Mol. Biol. 2019;1877:61–76. doi: 10.1007/978-1-4939-8861-7_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajduk P.J., Olejniczak E.T., Fesik S.W. One-dimensional relaxation- and diffusion-edited NMR methods for screening compounds that bind to macromolecules. J. Am. Chem. Soc. 1997;119:12257–12261. [Google Scholar]
- Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Henderson M.P.A., Billen L.P., Kim P.K., Andrews D.W. Cell-free analysis of tail-anchor protein targeting to membranes. Methods. 2007;41:427–438. doi: 10.1016/j.ymeth.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Iconaru L.I., Ban D., Bharatham K., Ramanathan A., Zhang W., Shelat A.A., Zuo J., Kriwacki R.W. Discovery of small molecules that inhibit the disordered protein, p27(Kip1) Sci. Rep. 2015;5:15686. doi: 10.1038/srep15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer S., Anwari K., Alsop A.E., Yuen W.S., Huang D.C.S., Carroll J., Smith N.A., Smith B.J., Dewson G., Kluck R.M. Identification of an activation site in Bak and mitochondrial Bax triggered by antibodies. Nat. Commun. 2016;7:11734. doi: 10.1038/ncomms11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer S., Uren R.T., Dengler M.A., Shi M.X., Uno E., Adams J.M., Dewson G., Kluck R.M. Robust autoactivation for apoptosis by BAK but not BAX highlights BAK as an important therapeutic target. Cell Death Dis. 2020;11:268. doi: 10.1038/s41419-020-2463-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale J., Osterlund E.J., Andrews D.W. BCL-2 family proteins: changing partners in the dance towards death. Cell Death Differ. 2018;25:65–80. doi: 10.1038/cdd.2017.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazimierczuk K., Orekhov V.Y. Accelerated NMR spectroscopy by using compressed sensing. Angew. Chem. Int. Ed. Engl. 2011;50:5556–5559. doi: 10.1002/anie.201100370. [DOI] [PubMed] [Google Scholar]
- Keller R. CANTINA Verlag; Goldau, Switerland): 2004. The Computer Aided Resonance Assignment Tutorial. [Google Scholar]
- Khan S., Zhang X., Lv D., Zhang Q., He Y., Zhang P., Liu X., Thummuri D., Yuan Y., Wiegand J.S., et al. A selective BCL-XL PROTAC degrader achieves safe and potent antitumor activity. Nat. Med. 2019;25:1938–1947. doi: 10.1038/s41591-019-0668-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotschy A., Szlavik Z., Murray J., Davidson J., Maragno A.L., Le Toumelin-Braizat G., Chanrion M., Kelly G.L., Gong J.N., Moujalled D.M., et al. The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature. 2016;538:477–482. doi: 10.1038/nature19830. [DOI] [PubMed] [Google Scholar]
- Lang X., Li L., Chen Y., Sun Q., Wu Q., Liu F., Tan C., Liu H., Gao C., Jiang Y. Novel synthetic acridine derivatives as potent DNA-binding and apoptosis-inducing antitumor agents. Bioorg. Med. Chem. 2013;21:4170–4177. doi: 10.1016/j.bmc.2013.05.008. [DOI] [PubMed] [Google Scholar]
- Leshchiner E.S., Braun C.R., Bird G.H., Walensky L.D. Direct activation of full-length proapoptotic BAK. Proc. Natl. Acad. Sci. USA. 2013;110:E986–E995. doi: 10.1073/pnas.1214313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leverson J.D., Phillips D.C., Mitten M.J., Boghaert E.R., Diaz D., Tahir S.K., Belmont L.D., Nimmer P., Xiao Y., Ma X.M., et al. Exploiting selective BCL-2 family inhibitors to dissect cell survival dependencies and define improved strategies for cancer therapy. Sci. Transl. Med. 2015;7:279ra40. doi: 10.1126/scitranslmed.aaa4642. [DOI] [PubMed] [Google Scholar]
- Llambi F., Moldoveanu T., Tait S.W.G., Bouchier-Hayes L., Temirov J., McCormick L.L., Dillon C.P., Green D.R. A unified model of mammalian BCL-2 protein family interactions at the mitochondria. Mol. Cell. 2011;44:517–531. doi: 10.1016/j.molcel.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv D., Pal P., Liu X., Jia Y., Thummuri D., Zhang P., Hu W., Pei J., Zhang Q., Zhou S., et al. Development of a BCL-xL and BCL-2 dual degrader with improved anti-leukemic activity. Nat. Commun. 2021;12:6896. doi: 10.1038/s41467-021-27210-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino D., Kelly G.L., Lessene G., Wei A.H., Roberts A.W., Strasser A. BH3-mimetic drugs: blazing the trail for new cancer medicines. Cancer Cell. 2018;34:879–891. doi: 10.1016/j.ccell.2018.11.004. [DOI] [PubMed] [Google Scholar]
- Moldoveanu T., Czabotar P.E. BAX, BAK, and BOK: a coming of age for the BCL-2 family effector proteins. Cold Spring Harb. Perspect. Biol. 2020;12:a036319. doi: 10.1101/cshperspect.a036319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldoveanu T., Follis A.V., Kriwacki R.W., Green D.R. Many players in BCL-2 family affairs. Trends Biochem. Sci. 2014;39:101–111. doi: 10.1016/j.tibs.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldoveanu T., Grace C.R., Llambi F., Nourse A., Fitzgerald P., Gehring K., Kriwacki R.W., Green D.R. BID-induced structural changes in BAK promote apoptosis. Nat. Struct. Mol. Biol. 2013;20:589–597. doi: 10.1038/nsmb.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldoveanu T., Liu Q., Tocilj A., Watson M., Shore G., Gehring K. The X-ray structure of a BAK homodimer reveals an inhibitory zinc binding site. Mol. Cell. 2006;24:677–688. doi: 10.1016/j.molcel.2006.10.014. [DOI] [PubMed] [Google Scholar]
- Neidhardt F.C., Bloch P.L., Smith D.F. Culture medium for enterobacteria. J. Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiguchi G., Das S., Ochoada J., Long H., Lee R.E., Rankovic Z., Shelat A.A. Evaluating and evolving a screening library in academia: the St Jude approach. Drug Discov. Today. 2021;26:1060–1069. doi: 10.1016/j.drudis.2021.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu X., Brahmbhatt H., Mergenthaler P., Zhang Z., Sang J., Daude M., Ehlert F.G.R., Diederich W.E., Wong E., Zhu W., et al. A small-molecule inhibitor of Bax and Bak oligomerization prevents genotoxic cell death and promotes neuroprotection. Cell Chem. Biol. 2017;24:493–506.e5. doi: 10.1016/j.chembiol.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill K.L., Huang K., Zhang J., Chen Y., Luo X. Inactivation of prosurvival Bcl-2 proteins activates Bax/Bak through the outer mitochondrial membrane. Genes Dev. 2016;30:973–988. doi: 10.1101/gad.276725.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh K.J., Singh P., Lee K., Foss K., Lee S., Park M., Lee S., Aluvila S., Park M., Singh P., et al. Conformational changes in BAK, a pore-forming proapoptotic Bcl-2 family member, upon membrane insertion and direct evidence for the existence of BH3-BH3 contact interface in BAK homo-oligomers. J. Biol. Chem. 2010;285:28924–28937. doi: 10.1074/jbc.M110.135293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D., Anisuzzaman A.S.M., Magis A.T., Chen G., Xie M., Zhang G., Behera M., Sica G.L., Ramalingam S.S., Owonikoko T.K., et al. Discovery of small molecule Bak activator for lung cancer therapy. Theranostics. 2021;11:8500–8516. doi: 10.7150/thno.60349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogmore J.P., Uehling D., Andrews D.W. Pharmacological targeting of executioner proteins: controlling life and death. J. Med. Chem. 2021;64:5276–5290. doi: 10.1021/acs.jmedchem.0c02200. [DOI] [PubMed] [Google Scholar]
- Pullarkat V.A., Lacayo N.J., Jabbour E., Rubnitz J.E., Bajel A., Laetsch T.W., Leonard J., Colace S.I., Khaw S.L., Fleming S.A., et al. Venetoclax and Navitoclax in combination with chemotherapy in patients with relapsed or refractory acute lymphoblastic leukemia and lymphoblastic lymphoma. Cancer Discov. 2021;11:1440–1453. doi: 10.1158/2159-8290.CD-20-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A.W. Therapeutic development and current uses of BCL-2 inhibition. Hematology. Am. Soc. Hematol. Educ. Program. 2020;2020:1–9. doi: 10.1182/hematology.2020000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A.W., Wei A.H., Huang D.C.S. BCL2 and MCL1 inhibitors for hematologic malignancies. Blood. 2021;138:1120–1136. doi: 10.1182/blood.2020006785. [DOI] [PubMed] [Google Scholar]
- Sandow J.J., Tan I.K., Huang A.S., Masaldan S., Bernardini J.P., Wardak A.Z., Birkinshaw R.W., Ninnis R.L., Liu Z., Dalseno D., et al. Dynamic reconfiguration of pro-apoptotic BAK on membranes. EMBO J. 2021;40:e107237. doi: 10.15252/embj.2020107237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz J., Besir H., Strasser C., Suppmann S. A new method to customize protein expression vectors for fast, efficient and background free parallel cloning. BMC Biotechnol. 2013;13:12. doi: 10.1186/1472-6750-13-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuker S.B., Hajduk P.J., Meadows R.P., Fesik S.W. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- Singh G., Guibao C.D., Seetharaman J., Aggarwal A., Grace C.R., McNamara D.E., Vaithiyalingam S., Waddell M.B., Moldoveanu T. Structural basis of BAK activation in mitochondrial apoptosis initiation. Nat. Commun. 2022;13:250. doi: 10.1038/s41467-021-27851-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G., Moldoveanu T. Methods to probe conformational activation and mitochondrial activity of proapoptotic BAK. Methods Mol. Biol. 2019;1877:185–200. doi: 10.1007/978-1-4939-8861-7_13. [DOI] [PubMed] [Google Scholar]
- Singh R., Letai A., Sarosiek K. Regulation of apoptosis in health and disease: the balancing act of BCL-2 family proteins. Nat. Rev. Mol. Cell Biol. 2019;20:175–193. doi: 10.1038/s41580-018-0089-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner S.P., Fogh R.H., Boucher W., Ragan T.J., Mureddu L.G., Vuister G.W. CcpNmr AnalysisAssign: a flexible platform for integrated NMR analysis. J. Biomol. NMR. 2016;66:111–124. doi: 10.1007/s10858-016-0060-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S.S., Lee W.K., Aluvila S., Oh K.J., Yu Y.G. Identification of inhibitors against BAK pore formation using an improved in vitro assay system. Bull. Korean Chem. Soc. 2014;35:419–424. [Google Scholar]
- Souers A.J., Leverson J.D., Boghaert E.R., Ackler S.L., Catron N.D., Chen J., Dayton B.D., Ding H., Enschede S.H., Fairbrother W.J., et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat. Med. 2013;19:202–208. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- Sperl L.E., Rührnößl F., Schiller A., Haslbeck M., Hagn F. High-resolution analysis of the conformational transition of pro-apoptotic Bak at the lipid membrane. EMBO J. 2021;40:e107159. doi: 10.15252/embj.2020107159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strain-Damerell C., Mahajan P., Gileadi O., Burgess-Brown N.A. Medium-throughput production of recombinant human proteins: ligation-independent cloning. Methods Mol. Biol. 2014;1091:55–72. doi: 10.1007/978-1-62703-691-7_4. [DOI] [PubMed] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse C., Shoemaker A.R., Adickes J., Anderson M.G., Chen J., Jin S., Johnson E.F., Marsh K.C., Mitten M.J., Nimmer P., et al. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 2008;68:3421–3428. doi: 10.1158/0008-5472.CAN-07-5836. [DOI] [PubMed] [Google Scholar]
- van Delft M.F., Chappaz S., Khakham Y., Bui C.T., Debrincat M.A., Lowes K.N., Brouwer J.M., Grohmann C., Sharp P.P., Dagley L.F., et al. A small molecule interacts with VDAC2 to block mouse BAK-driven apoptosis. Nat. Chem. Biol. 2019;15:1057–1066. doi: 10.1038/s41589-019-0365-8. [DOI] [PubMed] [Google Scholar]
- Zhang J.H., Chung T.D., Oldenburg K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- Zheng J.H., Grace C.R., Guibao C.D., McNamara D.E., Llambi F., Wang Y.M., Chen T., Moldoveanu T. Intrinsic instability of BOK enables membrane permeabilization in apoptosis. Cell Rep. 2018;23:2083–2094.e6. doi: 10.1016/j.celrep.2018.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Western blots have been deposited in Mendeley. https://doi.org/10.17632/j6zg95s7nn.1. NMR data are available from the lead contact on request.
-
•
This article does not report original code.
-
•
Any additional information required to reanalyze the data reported in this article is available from the lead contact on request.