Abstract
The three-dimensional structure of DNA-dependent RNA polymerase (RNAP) from thermophilic Thermus aquaticus has recently been determined at 3.3 Å resolution. Currently, very little is known about T. aquaticus transcription and no genetic system to study T. aquaticus RNAP genes is available. To overcome these limitations, we cloned and overexpressed T. aquaticus RNAP genes in Escherichia coli. Overproduced T. aquaticus RNAP subunits assembled into functional RNAP in vitro and in vivo when coexpressed in E. coli. We used the recombinant T. aquaticus enzyme to demonstrate that transcription initiation, transcription termination, and transcription cleavage assays developed for E. coli RNAP can be adapted to study T. aquaticus transcription. However, T. aquaticus RNAP differs from the prototypical E. coli enzyme in several important ways: it terminates transcription less efficiently, has exceptionally high rate of intrinsic transcript cleavage, and is highly resistant to rifampin. Our results, together with the high-resolution structural information, should now allow a rational analysis of transcription mechanism by mutation.
Most bacterial RNA polymerase (RNAP) core enzymes consist of four core subunits (β′, β, and a dimer of identical α subunits). Binding of one of the several ς factors converts the core into the holoenzyme, which is able to initiate transcription from promoters. In vitro activity of several bacterial RNAPs can be recovered after separation of individual subunits in the presence of denaturing agents and subsequent mixing of the subunits and dialysis under controlled conditions (19). In vitro reconstitution of Escherichia coli RNAP from cloned and individually overexpressed and purified subunits provides a means of obtaining RNAP harboring lethal mutations in quantities sufficient for biochemical analyses (16). Structure-function studies of reconstituted recombinant E. coli RNAP mutants have provided crucial insights into the transcription mechanism and regulation and are primarily responsible for our increased understanding of this enzyme (see, for example, references 6, 8, and 17).
Recent structural analysis of RNAP from Thermus aquaticus provided the first tantalizing view of the bacterial RNAP core enzyme structure at 3.3 Å resolution (18). The availability of a relatively high-resolution structure qualitatively raises the importance of a rational structure-function analysis of RNAP. Despite obvious homology between RNAPs from T. aquaticus and E. coli, it would be highly desirable to perform mutational and structural studies using T. aquaticus RNAP rather than the better-studied E. coli counterpart. The important advantages of studying T. aquaticus RNAP include the ability to reduce assembly effects of mutations to a minimum and the ability to perform structural analysis of mutants. The disadvantages of the T. aquaticus system are the lack of functional assays, the general absence of data on gene transcription in this organism, and, more importantly, the inability to manipulate T. aquaticus RNAP subunit (rpo) genes. To overcome these limitations and to make full use of the available structural information, we cloned each of the T. aquaticus rpo genes in E. coli expression vectors and overproduced recombinant subunits. We show that recombinant T. aquaticus RNAP can be assembled in vitro and can be studied using assays developed for E. coli system.
MATERIALS AND METHODS
Cloning and expression of T. aquaticus rpo genes.
Fragments of genes coding for the core subunits of T. aquaticus RNAP were cloned previously (18). We used conventional cloning and PCR to assemble entire rpo genes in plasmid pT7Blue (Novagene), and we used site-directed mutagenesis to introduce an EcoRI site after the termination codon of every rpo gene and an NdeI site (CATATG) overlapping with the initiation ATG codon of every rpo gene (note that T. aquaticus rpo genes contain no NdeI or EcoRI sites 18). T. aquaticus rpo genes were recovered by NdeI-EcoRI treatment of pT7Blue-based recombinant plasmids and were subcloned into appropriately treated pET28 and/or pET21 T7 RNAP expression vectors. The resultant plasmids, pET28TaA, pET28TaB, pET28TaC, and pET28TaZ, express T. aquaticus RNAP α, β, β′, and ω, respectively, with N-terminal hexahistidine tags. Similar plasmids of the pET21 series express untagged subunits. E. coli BL21(DE3) cells transformed with the resulting plasmids overproduced individual T. aquaticus core RNAP subunits at high level on induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and the overexpressed proteins segregated into inclusion bodies.
A ∼600-bp fragment of T. aquaticus rpoD was amplified from T. aquaticus genomic DNA using degenerate primers specific for highly conserved regions 2 and 4, and its sequence was determined. The entire rpoD gene sequence was then obtained by applying inverse PCR to SacI- and BglI-digested T. aquaticus DNA. The entire rpoD gene was next subcloned into pET system vectors. Expressed RpoD was found in inclusion bodies.
Plasmid pET28TaABCZ, coexpressing T. aquaticus rpoA, rpoB, rpoC, and rpoZ genes, was constructed from pET28TaC, which was treated with HindIII and Klenow enzyme followed by EcoRI and ligated to a T. aquaticus rpoB-containing fragment prepared from pET21TaB by treating with BglII and Klenow followed by EcoRI. The resultant plasmid, pET28TaBC, has two genes, rpoB and rpoC, in opposite orientations, each being preceded by the T7 RNAP promoter. pET28TaBC was treated with EcoRI and ligated with the EcoRI-treated PCR fragment of pET21TaA. Oligonucleotides used for PCR were designed to anneal upstream of the T7 RNAP promoter of pET21TaA and downstream of the termination codon of rpoA, and each contained an EcoRI site. The resultant plasmid, pET28TaABC, has T. aquaticus rpoA and an upstream T7 RNAP promoter inserted between the rpoB and rpoC genes, with rpoA having the same orientation as rpoC. Plasmid pET28TaABCZ was constructed by ligating the SgrAI-treated pET28TaABC with the SgrAI-treated PCR fragment of pET21TaZ. Oligonucleotides used for PCR were designed to anneal upstream of the T7 RNAP promoter of pET21TaZ and downstream of the termination codon of rpoZ, and each contained an SgrAI site. The resultant plasmid, pET28TaABCZ, has three untagged genes, rpoZ, rpoB, and rpoA, in the same orientation, and C-terminally hexahistidine-tagged rpoC in the opposite orientation.
RNAP assembly and purification.
RNAP was reconstituted by a published procedure (2, 16). The molar ratio of α, β, and mutant β′ in the reconstitution reaction mixtures was 1:4:8. After reconstitution, RNAP preparations were either used directly or further purified by fast protein liquid chromatography and gel filtration on Superose-6 and Resource Q columns (Pharmacia-LKB Inc., Piscataway, N.J.) as described previously (2), concentrated by filtration through a C-100 concentrator (Amicon, Lexington, Mass.) to ∼1 mg/ml, and stored in 50% glycerol storage buffer at −20°C.
To purify the T. aquaticus core from E. coli cells coexpressing T. aquaticus rpo genes, BL21(DE3) cells harboring pET28TaABCZ were induced with 1 mM IPTG for 3 h. Cells were collected, and the cell pellet was resuspended in 20 volumes of buffer containing 50 mM Tris-HCl, 100 mM KCl, 0.1% Tween 20, 1 mM phenylmethylsulfonyl fluoride, and 1 mM EDTA (pH 7.9). The cells were lysed by sonication, and the cleared lysate was incubated at 65°C for 60 min. After centrifugation, the proteins in the supernatant were precipitated with ammonium sulfate (75% saturation), redissolved in IMAC starting buffer (10 mM Tris-HCl, 5% glycerol, 500 mM NaCl [pH 7.9]) and loaded on a 1-ml chelating HiTrap column (Pharmacia) loaded with Ni2+ and equilibrated in the same buffer. The column was washed with buffer containing 20 mM imidazole, and RNAP was eluted with 100 mM imidazole in the buffer. The fraction was diluted fivefold with TGE buffer (50 mM Tris-HCl, 5% glycerol, 50 mM NaCl, 1 mM β-mercaptoethanol) and loaded onto a Resource Q column equilibrated with the same buffer. The column was developed with a linear gradient of NaCl concentrations in TGE buffer. Chromatographically pure RNAP eluted at ca. 400 mM NaCl and was concentrated and stored as above.
Affinity labeling.
Reaction volumes of 10 μl contained 40 mM Tris-HCl (pH 7.9), 40 mM KCl, 10 mM MgCl2, ∼1.0 μg of RNAP core, 0.5 to 1.0 mM initiating AMP derivatized with an aldehyde group (3), and 100 ng of poly(dA-dT). The reaction mixtures were supplemented with 10 mM BH4 and incubated at 37°C for 10 min. [α-32P]UTP (3,000 Ci/mmol) was added to 0.3 mM (final concentration), and incubation was continued for 30 min at 37 or 65°C. Control experiments demonstrated that the resulting 32P labeling of RNAP depended on the addition of the template DNA (data not shown).
KMnO4 footprinting.
The 106-bp EcoRI DNA fragment containing the T7 A2 promoter (positions −84 to +32) was prepared as described previously (14). The fragment was 32P-end labeled by filling in EcoRI sticky ends with Klenow enzyme in the presence of [α-32P]dATP. The fragment was then treated with HincII (which cuts at position +22) to obtain bottom-strand-labeled fragment. Promoter complexes were formed in 20-μl reaction mixtures containing 200 nM RNAP holoenzyme, 100 nM 32P-end-labeled DNA fragment, 40 mM Tris-HCl (pH 7.9), 40 mM KCl, and 10 mM MgCl2. The reaction mixtures were preincubated for 15 min at 37°C (E. coli RNAP) or 65°C (T. aquaticus RNAP). Promoter complexes were then treated with 1 mM KMnO4 for 15 s at 37°C. The reactions were terminated by the addition of β-mercaptoethanol to 300 mM followed by phenol extraction, ethanol precipitation, and 10% piperidine treatment. Reaction products were analyzed by denaturing polyacrylamide gel electrophoresis (PAGE) (6% polyacrylamide).
In vitro transcription and transcript cleavage reactions.
Abortive initiation reaction mixtures (10 μl) contained 40 mM Tris-HCl (pH 7.9), 40 mM KCl, 10 mM MgCl2, 50 nM RNAP core enzymes, and 100 nM recombinant E. coli ς70 or T. aquaticus RpoD. The reaction mixtures were preincubated for 10 min at 45°C, and transcription was initiated by the addition of 100 mM CpA and 0.5 mM [α-32P]UTP (3,000 Ci/mmol) and allowed to proceed for an additional 10 min at 45°C. The reactions were terminated by the addition of an equal volume of urea-containing loading buffer, and the products were analyzed by denaturing gel electrophoresis (7 M urea, 20% polyacrylamide) and autoradiography.
To determine transcription elongation rates, elongation complexes stalled at position +20 (EC20) were prepared in 50-μl reaction mixtures containing 40 mM Tris-HCl (pH 7.9), 40 mM KCl, 10 mM MgCl2, 20 nM DNA fragment containing the T7 A1 promoter fused to the λ tR2 terminator (15), 50 nM RNAP, 0.05 mM CpApUpC, 50 μM (each) ATP and GTP, and 2.5 μM [α-32P]CTP (3,000 Ci/mmol). The reaction mixtures were incubated for 15 min at 23°C (E. coli) or 45°C (T. aquaticus). EC20 were synchronously restarted by the addition of nucleoside triphosphates (NTPs). Reactions proceeded for 0 to 600 s at 45°C. The reactions were terminated by the addition of formamide-containing loading buffer. Products were analyzed by urea-PAGE (7 M urea, 6% polyacrylamide), followed by autoradiography and PhosphorImager analysis.
To monitor transcript cleavage, EC21 was prepared by incubating immobilized purified EC20 with 25 μM UTP for 5 min at room temperature (E. coli RNAP) or 42°C (for T. aquaticus RNAP). Immobilized complexes were washed and left in ∼20 μl, in the presence or absence of recombinant E. coli GreA, for 10 min at 37°C (E. coli RNAP complexes) or 65°C (T. aquaticus RNAP complexes). Reactions were terminated by the addition of an equal volume of formamide-containing loading buffer and analyzed by denaturing gel electrophoresis (7 M urea, 20% polyacrylamide) and autoradiography.
Nucleotide sequence accession numbers.
The nucleotide sequences of the rpoA gene (encoding the α subunit of RNAP), the rpoB and rpoC genes (encoding the β and β′ subunits of RNAP, respectively), and the rpoZ gene (encoding the ω subunit of RNAP) have been deposited in GenBank under accession nos. Y19222, Y19223, and AJ295839, respectively.
RESULTS
T. aquaticus RNAP assembles in vitro.
As a preliminary feasibility experiment, we wished to establish that T. aquaticus RNAP assembles in vitro after denaturation. A highly pure T. aquaticus RNAP core sample prepared as described by Zhang et al. (18) was incubated in a denaturing buffer containing 7 M guanidine and then dialyzed in a low-salt buffer under conditions favoring RNAP reconstitution. The resulting preparation was assayed for nonspecific transcription on synthetic template poly(dA-dT). Transcription activity was recovered with high yield (data not shown), suggesting that T. aquaticus RNAP withstands the in vitro denaturation-renaturation cycle.
Cloning and overexpression of T. aquaticus RNAP core subunit genes, and in vitro assembly of the recombinant T. aquaticus RNAP core.
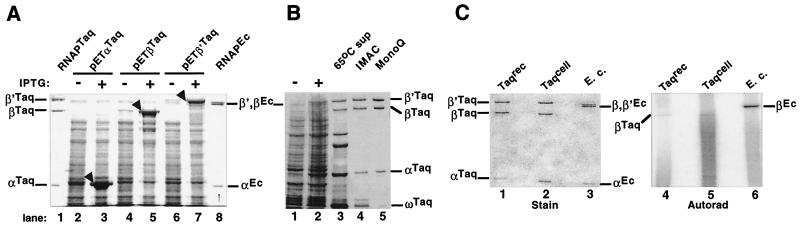
The sequences of T. aquaticus RNAP core subunit genes were determined previously (18). Each of the T. aquaticus rpo genes was cloned in E. coli pET expression vectors as described in Materials and Methods. Plasmids expressing rpo genes with or without affinity hexahistidine tags were constructed. E. coli cells harboring the expression plasmids overproduced high levels of recombinant T. aquaticus RNAP core subunits (Fig. 1A). Most of the overexpressed subunits were found in inclusion bodies and could be recovered by standard procedure (2).
FIG. 1.
Recombinant T. aquaticus RNAP. (A) Overexpression of T. aquaticus rpo genes in E. coli. T. aquaticus rpoA, rpoB, and rpoC genes coding for the α, β, and β′ RNAP subunits, respectively, were cloned in pET (Novagen) expression vectors. The plasmids were transformed in E. coli BL21(DE3) cells, the cells were induced with 1 mM IPTG, and proteins were separated on an SDS–8% polyacrylamide gel and revealed by Coomassie blue staining. Lanes 1 and 8 are controls and contain purified T. aquaticus and E. coli RNAP core enzymes, respectively. (B) Coexpression of T. aquaticus rpo genes in E. coli leads to assembled T. aquaticus RNAP. E. coli BL21(DE3) cells were transformed with pET28TaABCZ, expressing the T. aquaticus rpoA, rpoB, rpoCHis6, and rpoZ genes. Cells were induced with IPTG (lane 2) and lysed, and the extract was incubated at 65°C for 60 min. The supernatant (lane 3) was subjected to Ni2+-IMAC affinity chromatography, and material eluted from the Ni2+ sorbent in the presence of imidazole (lane 4) was further purified on a Resource Q column (lane 5). A stained SDS–12% polyacrylamide gel is presented. (C) Affinity labeling of recombinant T. aquaticus RNAP with a derivatized AMP initiator. In vitro-assembled T. aquaticus RNAP (Taqrec), RNAP purified from T. aquaticus biomass (18) (Taqcell), and control RNAP from E. coli (E. c.) were incubated with derivatized AMP (3) under conditions favoring cross-linking to E. coli β Lys1065, which is strictly conserved. Poly(dA-dT) template and [α-32P]UTP were added next, followed by additional incubations at 37°C (E. coli RNAP) or 65°C (T. aquaticus RNAPs). The reaction products were then resolved on an SDS-containing gel. The gel was stained (lanes 1 to 3) and autoradiographed (lanes 4 to 6). Radioactive labeling of RNAP β, which is template dependent (data not shown), testified that the enzymes are active.
A separate plasmid pET28TaABCZ, simultaneously expressing all four T. aquaticus rpo genes from T7 RNAP promoters, was also prepared. E. coli BL21(DE3) cells transformed with pET28TaABCZ expressed T. aquaticus RNAP subunits at low levels. The amount of overproduced T. aquaticus β and β′ was only slightly larger than the amount of the endogenous E. coli RNAP largest subunits, which form a characteristic double band on sodium dodecyl sulfate (SDS)-containing gels of whole-cell lysates (Fig. 1B, lanes 1 and 2). Coexpressed T. aquaticus subunits formed a complex that remained soluble after high-temperature treatment of E. coli extract (lane 3) and, when purified to homogeneity by IMAC and Resource Q chromatography, appeared indistinguishable from RNAP core enzyme purified from T. aquaticus cells (lane 5).
Recombinant T. aquaticus RNAP was also assembled from individually expressed subunits in vitro, as judged by the appearance of characteristic chromatographic peaks in the course of purification that separates assembled enzyme from assembly intermediates and unassembled subunits (reference 2 and data not shown). The catalytic proficiency of recombinant T. aquaticus RNAP was demonstrated by promoter-independent transcription of synthetic template poly(dA-dT) at the high temperature of 65°C (data not shown) and template-dependent affinity labeling with an initiating substrate derivative (Fig. 1C). In this reaction, recombinant, in vitro-reconstituted T. aquaticus RNAP was cross-linked to derivatized initiating AMP (3). The reaction mixtures were then supplemented with poly(dA-dT) template and [α-32P]UTP. As controls, labeling reactions were also performed with RNAP purified from T. aquaticus, as well as with E. coli RNAP. As explained elsewhere, in E. coli the affinity-labeling protocol results in covalent attachment of the radioactive pApU dinucleotide tag to the β subunit Lys1065 (3, 8). Previous studies demonstrated that the cross-linkable AMP derivative modifies β-like subunits in RNAP from bacterial, archaeal, and eucaryal systems and that residues homologous to the universally conserved E. coli β Lys1065 are cross-linked (3, 4, 12). Therefore, we expected that in T. aquaticus RNAP, the 1,119-amino-acid β subunit would be labeled. This expectation was fulfilled; as can be seen on the autoradiogram of the SDS-gel in Fig. 1C, the β subunit of in vitro-assembled T. aquaticus RNAP was labeled as efficiently as in the control enzyme purified from T. aquaticus cells and the larger, 1,342-amino-acid E. coli β subunit was absent from the recombinant T. aquaticus RNAP lane, establishing that little or no contaminating E. coli RNAP was present. On the basis of these results, we conclude that recombinant T. aquaticus RNAP assembled in vitro is active and free of contaminating E. coli RNAP. Similarly, T. aquaticus RNAP purified from E. coli cells coexpressing T. aquaticus rpo genes was also active and free of contaminating E. coli enzyme (data not shown). Below, we present an initial characterization of the recombinant T. aquaticus enzyme. Since the enzyme prepared by in vitro reconstitution was indistinguishable from that purified from E. coli cells (data not shown), all the data below are presented for in vitro-reconstituted T. aquaticus RNAP.
Promoter-specific transcription by recombinant T. aquaticus RNAP.
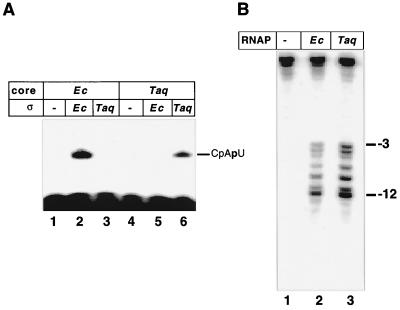
When recombinant T. aquaticus RNAP core was combined with E. coli ς70 at 45°C, a condition under which both T. aquaticus and E. coli enzymes are active, no promoter-specific initiation from strong, ς70-dependent T7 A1 promoter was observed (Fig. 2A, lane 5). Control experiments demonstrated that ς70 imparted promoter specificity to the E. coli core, as expected (lane 2). The result implies that E. coli ς70 is unable to form a productive complex with the T. aquaticus core. To overcome this problem, we cloned the homologue of the E. coli rpoD (ς70) gene from T. aquaticus (see Materials and Methods). Alignment of the deduced amino acid sequence revealed that the product of the T. aquaticus rpoD gene is very similar to the recently published sequence from T. thermophilus (11), as expected. T. aquaticus ς is also very similar to ς70 in conserved regions 2 and 4, responsible for promoter recognition. In striking contrast, T. aquaticus ς completely lacks the N-terminal conserved region 1. Instead, it contains a ∼100-amino-acid segment without homology to any of the published sequences.
FIG. 2.
Transcription initiation by recombinant T. aquaticus RNAP. (A) The indicated RNAP core enzymes were combined with recombinant E. coli (Ec) ς70 or T. aquaticus (Taq) RpoD in the presence of the T7 A1 promoter-containing DNA fragment, CpA primer, and [α-32P]UTP substrate. The reaction mixtures were incubated at 45°C for 15 min (lanes 1 to 3 and 4 to 6, respectively), and the products were resolved by denaturing PAGE and revealed by autoradiography. (B) Transcription complexes were formed at the indicated temperatures on the T7 A2 promoter-containing DNA fragment radioactively end labeled on the bottom strand, using recombinant E. coli (Ec) or T. aquaticus (Taq) RNAP holoenzymes, and probed with KMnO4. Reaction products were resolved on a 6% denaturing polyacrylamide gel and revealed by autoradiography. The permanganate-sensitive bands were assigned using G/A markers (not shown) (14).
T. aquaticus rpoD was cloned in the E. coli expression vector, and recombinant T. aquaticus ς was purified to homogeneity. The addition of T. aquaticus ς stimulated the synthesis of the abortive trinucleotide CpApU from the CpA initiator and UTP by the recombinant T. aquaticus core in the presence of a DNA fragment containing the T7 A1 promoter (Fig. 2A, lane 6). In contrast, the addition of T. aquaticus ς failed to stimulate transcription by the E. coli core enzyme (lane 3). The patterns of abortive products synthesized by E. coli and T. aquaticus holoenzymes on the T7 A1 promoter fragment in the presence of limiting sets of NTP substrates were identical, thus proving that the observed transcription initiation was promoter specific (data not shown). In addition, KMnO4 probing established that promoter complexes formed by the T. aquaticus holoenzyme on the well-studied T7 A2 promoter (14) are very similar or identical to the E. coli complexes (Fig. 2B).
In vitro transcription by the T. aquaticus RNAP holoenzyme.
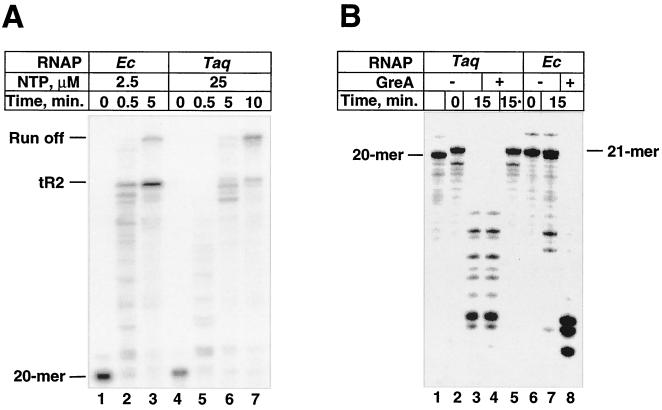
Having established conditions for promoter-specific transcription initiation by T. aquaticus RNAP, we wished to compare the transcription elongation, transcription termination, and transcript cleavage properties of T. aquaticus RNAP with those of the prototypic E. coli enzyme. Immobilized RNAP was used to obtain stalled elongation complexes containing 20-mer RNA on the T7 A1 promoter-containing DNA fragment (15). Purified elongation complexes were incubated in the presence of different concentrations of NTP, to monitor the transcription elongation rate and transcription termination on the rho-independent λ tR2 terminator located downstream. Alternatively, complexes containing 20-mer RNA were walked to position 21, purified, and incubated in the absence of NTP with or without E. coli GreA protein to monitor transcript cleavage. The most important conclusions from these experiments are summarized below. (i) T. aquaticus RNAP elongated RNA chains less efficiently than the E. coli enzyme did; in general a 10-fold-higher nucleotide concentration was required to achieve comparable elongation rates. Both enzymes exhibited transcription pausing, evident at shorter times (Fig. 3A, lanes 2 and 5); however, the pattern appeared to be distinct. (ii) T. aquaticus RNAP recognized the tR2 terminator less efficiently (transcription termination efficiency of ∼20%, compared to 86% with the E. coli enzyme). (iii) T. aquaticus RNAP actively cleaved 21-mer RNA in the stalled elongation complex in the absence of added cleavage factor. This activity was not further stimulated by E. coli GreA and was not due to RNase contamination, as shown by the control lane of Fig. 3B (lane 5) (in the control reaction, 21-mer RNA was prepared by phenol extraction of stalled complexes and pure RNA was then incubated with T. aquaticus RNAP before being loaded onto the gel).
FIG. 3.
Transcript elongation, termination, and cleavage by recombinant T. aquaticus RNAP. (A) Elongation complexes stalled at position 20 were prepared using DNA fragment containing the T7 A1 promoter and λ tR2 terminator as a template (lanes 1 and 4). Transcription was resumed at 45°C by the addition of the indicated concentrations of NTPs; at the times indicated, aliquots were withdrawn and reactions were terminated by the addition of formamide containing stop buffer. Reaction products were analyzed by 6% urea PAGE followed by autoradiography. (B) Immobilized elongation complexes stalled at position 20 (lane 1) were walked to position 21 (lane 2). The complexes were desorbed from Ni2+-nitrilotriacetic acid agarose and supplemented with recombinant E. coli GreA (0.04 μg), reaction mixtures were incubated for 15 min at 37 and 65°C (for E. coli and T. aquaticus RNAP, correspondingly), and the products were resolved by denaturing PAGE in a 20% urea gel. In lane 5, RNA was extracted from the stalled T. aquaticus elongation complex with phenol and then incubated in the presence of T. aquaticus RNAP and E. coli GreA.
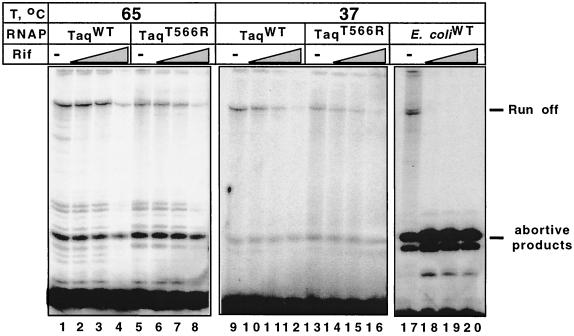
Structural analysis of the T. aquaticus core indicates that sites in subunit β that correspond to sites of known rifampin resistance mutations in E. coli form a cluster in the ceiling of DNA binding channel of the enzyme (18). Previous reports indicated that T. aquaticus RNAP is highly resistant to rifampin (5). With the exception of T. aquaticus β Thr566, which corresponds to E. coli β Arg687, which defines Rif cluster III (6), all T. aquaticus amino acids that can be involved in rifampin binding are identical to the corresponding amino acids in the wild-type, rifampin-sensitive E. coli β subunit. To test whether Thr566 is responsible for rifampin resistance, we substituted it for Arg, assembled the mutant enzyme in vitro, and determined its sensitivity to rifampin in a steady-state T7 A1 promoter transcription assay. The result, presented in Fig. 4, shows that in agreement with earlier data, T. aquaticus RNAP was highly resistant to rifampin and continued to synthesize full-sized transcripts even in the presence of 1,000 μg of rifampin per ml in the reaction mixture. In contrast, synthesis of the full-sized transcripts by the E. coli enzyme was completely inhibited in the presence of 10 μg of the drug per ml. T. aquaticus RNAP harboring the T566R substitution in subunit β was unaffected in its level of rifampin resistance. Therefore, the determinants of the resistance of T. aquaticus RNAP to rifampin must lie outside sites determined by genetic studies of resistance in E. coli and other sensitive bacteria.
FIG. 4.
The high level of rifampin resistance by T. aquaticus RNAP is not due to β Rif-cluster III Thr566. The indicated RNAPs were used to transcribe the T7 A1 promoter-containing DNA fragment (8) in a multiple-round transcription assay in the presence of three different rifampin (Rif) concentrations: 10, 100, and 1,000 μg/ml. Reaction products were resolved on a 20% denaturing polyacrylamide gel and revealed by autoradiography.
DISCUSSION
The recent advances in our understanding of the mechanism and regulation of bacterial transcription are largely due to recombinant E. coli RNAP technology that has allowed the preparation of sufficient quantities of RNAP mutants harboring lethal mutations for biochemical analysis. Until recently, this powerful approach had a significant limitation, since mutations that affected RNAP assembly could not be studied. The recent determination of the high-resolution structure of the T. aquaticus RNAP core allows one to perform a precise, structure-based analysis of bacterial RNAP function.
As our results demonstrate, recombinant T. aquaticus RNAP can be prepared in large amounts, and the discriminative transcription assays developed for the E. coli enzyme can be adapted to study this thermophilic enzyme. However, the T. aquaticus enzyme differs from the E. coli enzyme in several significant ways. Promoter complexes formed by the T. aquaticus holoenzyme are indistinguishable from E. coli RNAP complexes, suggesting that the promoter specificities of T. aquaticus RpoD and E. coli ς70 are the same. However, the two sigmas are not interchangeable. The functional specialization of sigmas is not due to the lack of formation of hybrid holoenzymes (data not shown). The unusual N-terminal extension present in T. aquaticus RpoD and/or the long (300-amino-acid) insertion in T. aquaticus β′, which is absent from E. coli counterpart and is located close to the conserved segment C involved in ς binding (1), may be responsible for the inability of hybrid holoenzymes to recognize promoters.
As with sigma, the E. coli transcript cleavage factor GreA did not alter the properties of T. aquaticus RNAP. In fact, T. aquaticus RNAP appears to be very active in factor-independent transcript cleavage even at low pH. High levels of intrinsic transcript cleavage were observed when T. aquaticus RNAP was purified from E. coli coexpressing T. aquaticus rpo genes or when the enzyme was prepared by in vitro reconstitution from isolated subunits (data not shown). Thus, the cleavage activity probably reflects the true properties of T. aquaticus RNAP rather than contamination with the E. coli Gre factor(s). High levels of cleavage activity may be essential for an enzyme which transcribes DNA at high temperature, a condition known to stimulate transcription arrest in E. coli (9). Despite the relatively slow elongation, T. aquaticus RNAP terminates transcription less efficiently, lending further support to the idea that the relationship between the transcription elongation rate and transcription termination is more complex than was previously thought (10).
The unusually high resistance of T. aquaticus RNAP to rifampin is not due to T. aquaticus β Thr655, the only T. aquaticus β amino acid that is different from E. coli residues known to be involved in rifampin resistance. This result suggests that additional amino acids weaken rifampin binding to T. aquaticus RNAP. It is not clear why mutational changes in these amino acids were not detected during intensive screens for rifampin resistance in E. coli and other rifampin-sensitive bacteria (7, 13). One possible explanation is that such changes lead to a lethal phenotype. Nevertheless, this result suggests that details of rifampin interaction with T. aquaticus RNAP may be significantly different from those of the interaction in rifampin-sensitive enzymes.
To summarize, the availability of recombinant T. aquaticus RNAP and discriminative transcription assays for this enzyme should make it possible to test, by means of genetic engineering and biochemical analysis, many of the predictions of the structure-functional model of transcription put forward by Zhang et al. (18). The ability to purify mutant T. aquaticus RNAP directly from E. coli cells should also allow a structural analysis of RNAP mutants.
ACKNOWLEDGMENTS
This work was supported by a Burroughs Wellcome Career Award to K.S. L.C. was supported by NIH grant 1 RO1 GM53759 to Seth A. Darst. L.M. is a recipient of a Charles and Johanna Busch Postdoctoral Fellowship.
We thank V. Nikifovov and A. Kulbachinskii for helpful discussions.
REFERENCES
- 1.Arthur T M, Burgess R R. Localization of a sigma70 binding site on the N terminus of the Escherichia coli RNA polymerase beta′ subunit. J Biol Chem. 1998;273:31381–31387. doi: 10.1074/jbc.273.47.31381. [DOI] [PubMed] [Google Scholar]
- 2.Borukhov S, Goldfarb A. Recombinant Escherichia coli RNA polymerase: purification of individually overexpressed subunits and in vitro assembly. Protein Exp Purif. 1993;4:503–511. doi: 10.1006/prep.1993.1066. [DOI] [PubMed] [Google Scholar]
- 3.Grachev M A, Kolocheva T I, Lukhtanov E A, Mustaev A. Studies on the functional topography of Escherichia coli RNA polymerase: highly selective affinity labeling by analogues of initiating substrates. Eur J Biochem. 1987;163:113–121. doi: 10.1111/j.1432-1033.1987.tb10743.x. [DOI] [PubMed] [Google Scholar]
- 4.Grachev M A, Mustaev A A, Zaychikov E F, Lindner A J, Hartmann G R. Localisation of the binding site for the initiating substrate on the RNA polymerase from Sulfolobus acidocaldarius. FEBS Lett. 1989;250:317–322. doi: 10.1016/0014-5793(89)80746-x. [DOI] [PubMed] [Google Scholar]
- 5.Fabry M, Sumegi J, Venetianer P. Purification and properties of the RNA polymerase of an extremely thermophilic bacterium: Thermus aquaticus T2. Biochim Biophys Acta. 1976;435:228–235. doi: 10.1016/0005-2787(76)90104-0. [DOI] [PubMed] [Google Scholar]
- 6.Igarashi K, Ishihama A. Bipartite functional map of the E. coli RNA polymerase alpha subunit: involvement of the C-terminal region in transcription activation by cAMP-CRP. Cell. 1991;65:1015–1022. doi: 10.1016/0092-8674(91)90553-b. [DOI] [PubMed] [Google Scholar]
- 7.Jin D J, Gross C A. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol. 1988;202:45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 8.Kashlev M, Lee J, Zalenskaya K, Nikiforov V, Goldfarb A. Blocking of the initiation-to-elongation transition by a transdominant RNA polymerase mutation. Science. 1990;248:1006–1009. doi: 10.1126/science.1693014. [DOI] [PubMed] [Google Scholar]
- 9.Komissarova N, Kashlev M. Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3′ end of the RNA intact and extruded. Proc Natl Acad Sci USA. 1997;94:1755–1760. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDowell J C, Roberts J W, Jin D J, Gross C. Determination of intrinsic transcription termination efficiency by RNA polymerase elongation rate. Science. 1994;266:822–825. doi: 10.1126/science.7526463. [DOI] [PubMed] [Google Scholar]
- 11.Nishiyama M, Kobashi N, Tanaka K, Takahashi H, Tanokura M. Cloning and characterization in Escherichia coli of the gene encoding the principal sigma factor of an extreme thermophile, Thermus thermophilus. FEMS Microbiol Lett. 1999;172:179–186. doi: 10.1111/j.1574-6968.1999.tb13467.x. [DOI] [PubMed] [Google Scholar]
- 12.Riva M, Schaffner A R, Sentenac A, Hartmann G R, Mustaev A A, Zaychikov E F, Grachev M A. Active site labeling of the RNA polymerases A, B, and C from yeast. J Biol Chem. 1987;262:14377–14380. [PubMed] [Google Scholar]
- 13.Severinov K, Soushko M, Goldfarb A, Nikiforov V. Rifampicin region revisited: new rifampicin-resistant and streptolydigin-resistant mutants in the β subunit of Escherichia coli RNA polymerase. J Biol Chem. 1993;268:14280–14825. [PubMed] [Google Scholar]
- 14.Severinov K, Darst S A. A mutant RNA polymerase that forms unusual promoter complexes. Proc Natl Acad Sci USA. 1997;94:13481–13486. doi: 10.1073/pnas.94.25.13481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zakharova N, Bass I A, Arsenieva E, Nikiforov V, Severinov K. Mutations in and monoclonal antibody binding to evolutionary hypervariable region of E. coli RNA polymerase β′ subunit inhibit transcript cleavage and transcript elongation. J Biol Chem. 1998;273:24912–24920. doi: 10.1074/jbc.273.38.24912. [DOI] [PubMed] [Google Scholar]
- 16.Zalenskaya K, Lee J, Gujuluva C N, Shin Y K, Slutsky M, Goldfarb A. Recombinant RNA polymerase: inducible overexpression, purification and assembly of Escherichia coli rpo gene products. Gene. 1990;89:7–12. doi: 10.1016/0378-1119(90)90199-2. [DOI] [PubMed] [Google Scholar]
- 17.Zaychikov E, Martin E, Denissova L, Kozlov M, Markovtsov V, Kashlev M, Heumann H, Nikiforov V, Goldfarb A, Mustaev A. Mapping of catalytic residues in the RNA polymerase active center. Science. 1996;273:107–109. doi: 10.1126/science.273.5271.107. [DOI] [PubMed] [Google Scholar]
- 18.Zhang G, Campbell L, Minakhin L, Richter C, Severinov K, Darst S A. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 19.Zillig W, Palm P, Heil A. Function and reassembly of subunits of DNA-dependent RNA polymerase. In: Losick R, Chamberlin M, editors. RNA polymerase. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1976. pp. 101–125. [Google Scholar]