Abstract
Synthesis of the O:54 O antigen of Salmonella enterica is initiated by the nonprocessive glycosyl transferase WbbE, assigned to family 2 of the glycosyl transferase enzymes (GT2). GT2 enzymes possess a characteristic N-terminal domain, domain A. Based on structural data from the GT2 representative SpsA (S. J. Charnock and G. J. Davies, Biochemistry 38:6380–6385, 1999), this domain is responsible for nucleotide binding. It possesses two invariant Asp residues, the first forming a hydrogen bond to uracil and the second coordinating a Mn2+ ion. Site-directed replacement of Asp41 (D41A) of WbbE, the analogue of the first Asp residue of SpsA, revealed that this is not required for activity. WbbE possesses three Asp residues near the position analogous to the second conserved residue. Whereas D95A reduced WbbE activity, activity in D93A and D96A mutants was abrogated, suggesting that either D93 or D96 may coordinate the Mn2+ ion. Our studies also identified a C-terminal region of sequence conservation in 22 GT2 members, including WbbE. SpsA was not among these. This region is characterized by an ED(Y) motif. The Glu and Asp residues of this motif were individually replaced in WbbE. E180D in WbbE had greatly reduced activity, and an E180Q replacement completely abrogated activity; however, D181E had no effect. E180 is predicted to reside on a turn. Combined with the alignment of the motif with potential catalytic residues in the GT2 enzymes ExoM and SpsA, we speculate that E180 is the catalytic residue of WbbE. Sequence and predicted structural divergence in the catalytic region of GT2 members suggests that this is not a homogeneous family.
Glycosyl transferases (GTs) are one of the most important classes of enzymes on earth, catalyzing the formation of a large proportion of the biomass on the planet. These are highly specific enzymes, but collectively represent a diversity of substrate and acceptor specificities. They catalyze the formation of a wide variety of molecules, including glycoproteins, glycolipids, oligosaccharides, and cell envelope and cell wall polysaccharides of the Bacteria, Archaea, fungi, plants, and crustaceans. In addition to their role in the formation of structural molecules, these enzymes play a role in numerous biological processes, such as immune recognition, protection of pathogens from host immune responses, intercellular signaling, and biofilm formation. GTs are of great interest in biotechnology because of their ubiquitous nature.
Despite their catalytic diversity, GTs are subdivided into only two main classes, either inverting or retaining enzymes, based on the stereochemistry of the substrate and product (reviewed in reference 31). While sequence analyses reveal that, in general, GTs exhibit relatively little identity in their primary structure, it has become evident that certain sequence motifs can be identified. Based on this, Campbell et al. (6) used basic local alignment search tool (BLAST) analysis (1) and hydrophobic cluster analysis (HCA) (13) to further classify 553 GTs into 26 families. These families have since been expanded and can be viewed at the following: http://afmb.cnrs-mrs.fr/∼pedro/CAZY/db.html. The sequence similarities observed within each family were concluded to be indicative of conserved tertiary structure, and therefore the three-dimensional fold within the conserved region is predicted to be the same among members of a given family. The largest of the 47 current families is family 2, which comprises GTs from all major groups of living organisms. GT family 2 (GT2) members are inverting GT enzymes using nucleotide-diphospho α-linked sugar donors to form β-linked products. Structural predictions and sequence alignments have revealed that these proteins are characterized by a region of alternating α-helices and β-sheets which form domain A, within which is a conserved DXD motif (29). Previous studies have revealed that the region C terminal to domain A diverges between at least some processive and nonprocessive GT2 members (24, 29).
The Salmonella O:54 O antigen has a number of unusual features. It is the only Salmonella O antigen containing N-acetylmannosamine (ManNAc), and it is the only known homopolymeric O antigen in this genus (25). The O:54 O antigen is also the only lipopolysaccharide (LPS) O antigen currently known to be synthesized by the synthase-dependent pathway (34). The O:54-specific GT WbbE (RfbA), the initiating enzyme for the O antigen, belongs to family 2. This enzyme is encoded by the plasmid-borne O:54 O-antigen biosynthesis genes (22, 23, 24). WbbE is a nonprocessive enzyme which has previously been shown to catalyze the addition of a single ManNAc (ManpNAc) residue to the undecaprenol (Und)-PP-GlcpNAc acceptor. O:54 polymerization and surface expression occur in a wbbEF-containing Escherichia coli host in which the entire O-antigen biosynthesis cluster of the host has been deleted. WbbF (RfbB) is therefore speculated to processively transfer all subsequent ManNAc residues in alternating β(1→3) and β(1→4) linkages, while simultaneously extruding the growing chain across the cytoplasmic membrane (24). WbbF has also been assigned to family 2 (6) and is one of a number of structurally homologous processive enzymes in this family (24).
To date, the crystal structures of only four GTs have been published: the DNA β-glucosyltransferase of bacteriophage T4 (33), the GT SpsA of Bacillus subtilis (7), the catalytic domain of a bovine β-galactosyltransferase (16), and most recently, the MurG GT of E. coli (19). The first exhibits little sequence similarity to any other GTs. The bovine β-galactosyl transferase has been classified into family 7 of the GTs, and MurG has been classified into family 28 (http://afmb.cnrs-mrs.fr/∼pedro/CAZY/db.html). SpsA has been shown to belong to the more predominant family 2 (6). Cocrystallization with UDP revealed that domain A in SpsA (and, by extension, in other family 2 members) represents the nucleotide binding fold. Unfortunately, the substrate of SpsA is not known, and while the crystallization data identified specific amino acid residues interacting with UDP and Mn2+, and a potential catalytic residue bound to a free glycerol, the identity of the catalytic residue remains speculative. Currently, it is believed that inverting GTs possess a catalytic residue which acts as a general base in a single nucleophile mechanism (10, 21). This residue is generally thought to be an aspartate or a glutamate.
The present communication provides further analysis of the WbbE ManpNAc transferase of Salmonella enterica serovar Borreze. Using site-directed mutagensis and in vivo activity assays, we confirm that the DXD motif of domain A does indeed play a critical role in catalysis in this enzyme, as suggested by the structural data of Charnock and Davies (7) and predicted by others (14, 17, 30). We identified a consensus sequence C terminal to domain A in a subset of monofunctional GT2 members, which is postulated to represent the catalytic domain. Sequence and predicted structural divergence within the region C terminal to domain A suggests a fundamental difference between some members of this family and a potential basis for further subdivision.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were grown at 37°C in Luria-Bertani broth supplemented, where appropriate, with antibiotics (ampicillin, 100 μg/ml; kanamycin, 50 μg/ml). For regulated overexpression of WbbE and WbbF, 100 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added to strains containing pET30a(+)-derived constructs (Novagen, Madison, Wis.) at a final concentration of 0.5 mM, and arabinose was added to those with pBAD24-derived constructs (18) at a final concentration of 0.02%.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant property | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | K-12 φ80d lacZΔM15 endA1 recA1 hsdR17(rK− mK−) supE44 thi-1 gyrA96 relA1 Δ(lacZYA0argF) U169 F− | 27 |
| E. coli BL21[λDE3] | F−ompT hsdSB(rB− mB−) gal dcm [λDE3] | Novagen |
| Plasmids | ||
| pBAD24 | Expression vector containing the catabolite-repressed PBAD promoter, the araC repressor gene, and bla; induced in the presence of arabinose | 17 |
| pET30a(+) | Expression vector containing an N-terminal His6 tag, lacI, and the T7 promoter | Novagen |
| pWQ835 | pBAD24 containing a 2.2-kb EcoRI-PstI insert containing wbbE and wbbF | This study |
| pWQ818 | Derivative of pWQ802 with deletion of 1.5-kb EcoRV-NcoI fragment containing the 3′ end of wbbF; wbbE+ | 21 |
| pWQ835-41 | pWQ835 mutant encoding D41A in WbbE | This study |
| pWQ835-93 | pWQ835 mutant encoding D93A in WbbE | This study |
| pWQ835-95 | pWQ835 mutant encoding D95A in WbbE | This study |
| pWQ835-96 | pWQ835 mutant encoding D96A in WbbE | This study |
| pWQ835-180 | pWQ835 mutant encoding E180D in WbbE | This study |
| pWQ835-180Q | pWQ835 mutant encoding E180Q in WbbE | This study |
| pWQ835-181 | pWQ835 mutant encoding D181E in WbbE | This study |
| pWQ835-181A | pWQ835 mutant encoding D181A in WbbE | This study |
| pWQ839 | pET30a(+) containing wbbE PCR amplified from pWQ835 on BglII-EcoRI fragment cloned in frame with N-terminal His6 tag | This study |
| pWQ839-93 | pET30a(+) containing wbbE PCR amplified from pWQ835-93 on BglII-EcoRI fragment cloned in frame with N-terminal His6 tag | This study |
| pWQ839-95 | pET30a(+) containing wbbE PCR amplified from pWQ835-95 on BglII-EcoRI fragment cloned in frame with N-terminal His6 tag | This study |
| pWQ839-96 | pET30a(+) containing wbbE PCR amplified from pWQ835-96 on BglII-EcoRI fragment cloned in frame with N-terminal His6 tag | This study |
| pWQ839-180 | pET30a(+) containing wbbE PCR amplified from pWQ835-180 on BglII-EcoRI fragment cloned in frame with N-terminal His6 tag | This study |
| pWQ839-180Q | pET30a(+) containing wbbE PCR amplified from pWQ835-180Q on BglII-EcoRI fragment cloned in frame with N-terminal His6 tag | This study |
| pWQ839-181A | pET30a(+) containing wbbE PCR amplified from pWQ835-181A on BglII-EcoRI fragment cloned in frame with N-terminal His6 tag | This study |
DNA manipulation and analyses.
Restriction enzyme digestions, ligations, and electroporations were performed as described by Sambrook et al. (28) and Binotto et al. (3). Inserts in pBAD24 (18) and pET30a(+) (Novagen) were obtained by PCR amplification using standard protocols. Primers were based on the pWQ799 sequences of wbbE and wbbF (23) (accession no. L39794) and were designed to incorporate novel restriction sites for cloning. PCR fragments were cleaned using the QIAquick DNA purification kit from QIAGEN Inc. (Chatsworth, Calif.). Plasmids were purified using QIAGEN spin columns. Nucleotide sequencing and oligonucleotide synthesis were performed by the Guelph Molecular Supercenter (University of Guelph, Guelph, Ontario, Canada). DNA sequence data were analyzed using AssemblyLIGN and MacVector software (International Biotechnologies Inc., New Haven, Conn.).
Analysis of protein structure.
Secondary structure predictions were done using HCA software (Doriane Informatique, Le Chesnay, France) as well as the method described by Garnier et al. (15). Protein homologies were detected using the Position Iterated (PSI)-BLAST search (2) for conserved motifs. Multiple sequence alignments were done using Align (http://vega.igh.cnrs.fr/bin/align-guess.cgi).
Site-directed mutagenesis.
Codons were altered using a method based on the QuikChange site-directed mutagenesis kit from Stratagene (La Jolla, Calif.). Mutagenic primers are listed in Table 2 and were used for mutagenesis of wbbE in plasmid pWQ835. Mutated DNA was sequenced (both strands) to confirm a single codon change and ensure no additional changes were introduced.
TABLE 2.
Oligonucleotide primers used for mutagenesis of wbbEa
| Primer | Sequence | Mutation |
|---|---|---|
| Mut-T (forward) | GAGGTATTGATTGTTGCTAATGGTGGA | D41A |
| Mut-L (reverse) | TCCACCATTAGCAACAATCAATACCTC | D41A |
| Mut-T1 (forward) | GCTGTTGCAGCTCCAGATGATATT | D93A |
| Mut-L1 (reverse) | AATATCATCTGGAGCTGCAACAGC | D93A |
| Mut-T2 (forward) | GCAGATCCAGCTGATATTAATGAG | D95A |
| Mut-L2 (reverse) | CTCATTAATATCAGCTGGATCTGC | D95A |
| Mut-T3 (forward) | GATCCAGATGCTATTAATGAGCCT | D96A |
| Mut-L3 (reverse) | AGGCTCATTAATAGCATCTGGATC | D96A |
| Mut-T4 (forward) | TTACATTTTGGAGACGACTATGATT | E180D |
| Mut-L4 (reverse) | AATCATAGTCCTCTCCAAAATGTAA | E180D |
| Mut-T5 (forward) | CATTTTGGAGAGGAGTATGATTTAG | D181E |
| Mut-L5 (reverse) | CTAAATCATACTCCTCTCCAAAATG | D181E |
| Mut-T6 (forward) | CATTTTGGAGAGGCCTATGATTTAG | D181A |
| Mut-L6 (reverse) | CTAAATCATAGGCCTCTCCAAAATC | D181A |
| Mut-T7 (forward) | CATTTTGGACAGGACTATGATTTAGT | E180Q |
| Mut-L7 (reverse) | ACTAAATCATAGTCCTGTCCAAAATG | E180Q |
The mutated nucleotide in each oligonucleotide is underlined.
Cell fractionation.
Bacterial cultures (50 ml) were grown to mid-exponential phase, and expression of the wbbE gene was then induced with IPTG. Expression was allowed to proceed for 2 h. Cells were then harvested by centrifugation, resuspended in 10 ml of residual medium, and lysed by ultrasonication. Unbroken cells were removed by centrifugation, and then the membranes were pelleted by ultracentrifugation (100,000 × g) for 1 h. An aliquot of the supernatant was kept as the cytosolic and periplasmic fraction, and the remainder was discarded. The pellet was then resuspended in 1 ml of 0.1 M Tris, 10 mM MgCl2, and 2% (wt/vol) Sarkosyl (N-lauroylsarcosine) in order to solubilize the inner membrane while precipitating the outer membrane (12). This solution was shaken for 30 min, after which the precipitated outer membrane was collected as a pellet by centrifugation at 10,000 × g for 30 min. The supernatant was concentrated approximately 20-fold, 10% sodium dodecyl sulfate (SDS) was added to a final concentration of approximately 7.5% (wt/vol), and the mixture was incubated at 100°C for 1 h. Any precipitate was removed by centrifugation at 10,000 × g for 10 min, and the cytoplasmic membrane-containing supernatant was combined with SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer.
Analysis of LPS.
LPS samples were prepared using the whole-cell lysate method of Hitchcock and Brown (20). Samples were separated using 18.5% polyacrylamide gels (9) and analyzed either by silver staining (32) or by Western blot analysis after electrotransfer to BioTrace nitrocellulose membrane (Gelman Sciences, Mississauga, Ontario, Canada). Blots were probed with absorbed polyclonal rabbit anti-O:54 antisera. The antisera were raised in New Zealand White rabbits immunized with formalin-killed whole cells of E. coli DH5α(pWQ802) (22). Cross-reacting antibodies were removed by absorbing the serum with whole cells of E. coli DH5α. The second antibody was goat anti-rabbit alkaline phosphatase-conjugated antibody (Jackson ImmunoResearch Laboratories Inc., West Grove, Pa.). Detection was achieved using 5-bromo-4-chloro-3-indolylphosphate (BCIP) (Roche Molecular Biochemicals, Laval, Quebec, Canada) and 4-nitroblue tetrazolium chloride (Sigma Chemical Co., St. Louis, Mo.).
Western blot analysis of proteins.
His-tagged protein samples were separated using 12% polyacrylamide gels, and then they were transferred to Biotrace nitrocellulose membranes. Blots were probed with QIAexpress anti-His6 antibody (QIAGEN Inc.) followed by goat anti-mouse alkaline phosphatase-conjugated secondary antibody (Jackson ImmunoResearch Laboratories Inc.) and detection according to instructions supplied by QIAGEN.
RESULTS AND DISCUSSION
Localization of WbbE in the cytoplasmic membrane.
The ManpNAc O:54 O antigen is produced through the action of two GTs, WbbE and WbbF (24). The WbbE GT has been identified as the first ManNAc transferase to act, transferring a single sugar to the GlcNAc-PP-Und carrier lipid. Based on polymerization and surface expression of O:54 LPS in an E. coli host strain with a deleted O-antigen biosynthesis cluster, as well as structural similarities with other synthases, WbbF is believed to processively transfer all subsequent ManNAc residues, while simultaneously transporting the growing polymer to the external face of the plasma membrane. The final step is ligation of the polymer to lipid A-core.
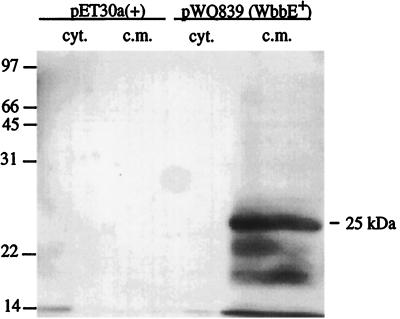
Previous studies indicated that WbbF is an integral membrane protein with four transmembrane domains as well as a periplasmic domain (24). In contrast, WbbE is predicted to possess only one potential transmembrane domain. To confirm the cellular localization of WbbE, cell fractionation was attempted using the pWQ835-containing expression clone. Although a variety of arabinose concentrations were used for induction, no unique protein was visible in any of the experimental samples, compared to the E. coli DH5α(pBAD24) negative control (data not shown). Cells expressing WbbE fused to an N-terminal His6 tag, encoded by plasmid pWQ839, were therefore used in cell fractionation studies to visualize the enzyme. Probing Western blots with an anti-His antibody revealed reactive bands in the inner membrane fraction of the O:54+ strain but not in the cytoplasmic fraction (Fig. 1). No protein was detected in the E. coli BL21/pET30a(+) negative control. The biggest protein in the WbbE-containing lane had an apparent molecular mass of approximately 25 kDa, smaller than the mass of 35.5 kDa predicted from sequence data for the His-tagged protein. The anomalous electrophoretic mobility of the enzyme is a common observation with membrane proteins, which tend to bind excess SDS and therefore display a higher electrophoretic mobility than predicted from sequence analysis (4). Other, faster migrating bands were also detected, suggesting proteolytic degradation. Once the cellular location of wild-type WbbE was determined, the various mutants employed in this study were assessed in the same manner. All of the mutant proteins displayed similar banding patterns and were produced at levels equivalent to that of the wild type (data not shown).
FIG. 1.
Western blot analysis of His-tagged WbbE in cytoplasmic and inner membrane fractions. Lanes: 1, cytoplasmic fraction from negative control BL21/pET30a(+); 2, cytoplasmic membrane fraction of negative control; 3, cytoplasmic fraction of BL21(pWQ839); 4, inner membrane fraction of BL21(pWQ839). Fusion proteins were detected with anti-His antibody.
To confirm that the WbbE fusion protein was functional, despite the presence of the His6 tag fusion and the anomalous electrophoretic mobility, E. coli BL21(pWQ839) was induced with IPTG and whole-cell lysates were prepared and analyzed by SDS-PAGE. The silver-stained gel revealed a modification to the core (data not shown), indicative of a core-plus-two-glycose size increase as shown previously for a K-12 strain harboring an active WbbE (24). This band has previously been shown to result from a single WbbE-specific ManNAc addition to the Und-PP-GlcNAc acceptor. The host Wzx (the so-called “flippase”) exports the lipid-linked disaccharide to the periplasmic face of the inner membrane (11), where it is ligated to lipid A-core by the WaaL ligase. The WbbE GT was therefore functional and localized in the cytoplasmic membrane, presumably forming a complex with WecA, the enzyme responsible for the initiating transfer of GlcNAc-1-P to Und-P (22, 27), and with WbbF.
Analysis of the catalytic role of the conserved aspartate residues in domain A of WbbE.
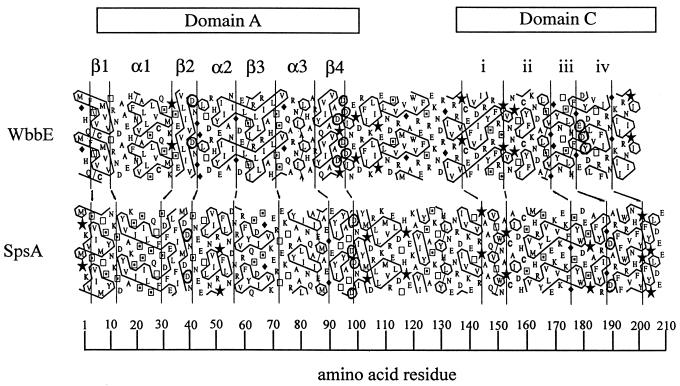
Using HCA, Saxena et al. first identified a region of homology, termed domain A, present in a number of inverting GTs of known or predicted function (29). Previous analyses identified the same domain in both WbbE (Fig. 2) and WbbF (24). Possession of this domain is the basis for inclusion in GT2 (6). SpsA is the only member of family 2 to be crystallized (7). The structural data revealed that SpsA is a metalloenzyme, and crystallization with UDP indicated that domain A is in fact a nucleotide-binding fold. Within this domain are a number of invariant aspartate residues. These have been shown to play a critical role in the catalysis of SpsA, based on cocrystallization with UDP and Mn2+ (7) and in other GT2 members, based on mutagenesis (cellulose synthase [30], KfiC [17], ExoM [14]). To confirm that these residues are also important in WbbE transferase activity, they were individually converted to alanines. To assess activity, LPS was prepared from arabinose-induced E. coli DH5α(pWQ835) cells expressing either the wild-type WbbE-WbbF or mutant pWQ835 derivatives. The ability of the mutant WbbE enzymes to participate with wild-type WbbF in O:54 O-antigen biosynthesis was assessed by SDS-PAGE and immunoblotting using polyclonal anti-O:54 antisera (Fig. 3). In this system, the host WecA function initiates polymerization, and then WbbE and WbbF complete the polymer and transport it to the periplasmic face of the cytoplasmic membrane, where it is ligated to lipid A-core and exported to the cell surface. All LPS preparations were made from the same number of cells; therefore, any change in the amount of reactive material must have arisen from either a change in the activity of the mutant WbbE enzyme or in the amount of the enzyme. Western blot analysis of the corresponding His6-tagged WbbE derivatives confirmed that there was no significant difference in the amounts of enzyme synthesized (data not shown). Asp41 in WbbE is analogous to Asp39 in SpsA, based on its position within domain A at the end of strand β2 (Fig. 2) (7, 24). Crystallization data revealed that this residue binds N-3 of the uracil base of UDP in SpsA. In WbbE, however, D41A had no obvious effect on the amount of O:54 O antigen synthesized. This is potentially due to a greater opportunity for interactions stabilizing the UDP-ManNAc substrate than would occur with UDP alone. Additionally, in SpsA, UDP has been shown to bind through aromatic stacking interactions as well as multiple hydrogen bonds. Therefore, eliminating one critical residue may not necessarily result in loss of substrate binding. In WbbE in particular, the equivalent conserved Asp does not appear to be critical for activity. These results are in contrast to those obtained for the analogous replacement in ExoM (14). In both in vitro and in vivo assays with the ExoM D44A mutant, activity was completely abrogated. The reason(s) for this discrepancy between the enzymes is unclear but may reflect the relative cumulative strengths of the substrate binding interactions.
FIG. 2.
HCA alignment of WbbE and SpsA. The HCA program writes protein sequences on a duplicated α-helical net and encloses clusters of hydrophobic amino acids in boxes. The plots are then visually compared for similarity in the hydrophobic cluster patterns, limiting analysis to the predicted globular portions of the proteins. Plots are aligned using traditional sequence alignments as a starting point. Hydrophobic clusters with obvious similarities were used as anchors for the structural alignment, as were regions containing Gly (♦) and Pro (★), which are often present in loops (13). Vertical lines were drawn to illustrate structurally conserved features. The prediction of β-strands and α-helices is based on the observed correlation between cluster shape and secondary structure (13). The single-letter code denotes amino acids except for proline, glycine, serine (⊡), and threonine (□). Conserved residues are circled.
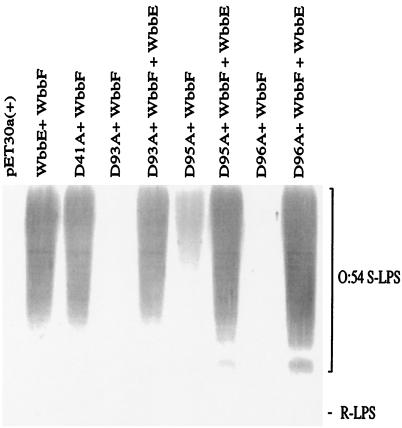
FIG. 3.
Analysis of the effect of replacements in domain A of WbbE on WbbE activity and O:54 LPS expression. Immunoblot of anti-O:54 reactive LPS from E. coli DH5α harboring wild-type or mutated WbbE and wild-type WbbF. Lanes: 1, pET30a(+) negative control; 2, pWQ835; 3, pWQ835-41; 4, pWQ835-93; 5, pWQ835-93, pWQ818; 6, pWQ835-95; 7, pWQ835-95, pWQ818; 8, pWQ835-96; 9, pWQ835-96, pWQ818. Polyclonal anti-O:54 antisera were adsorbed against E. coli DH5α, and therefore the R-LPS (lipid A-core) of the recombinants was not detected. The migration of S-LPS is shown.
The other invariant Asp in SpsA is Asp99, at the end of the β4 strand of domain A (Fig. 1) (7). This residue is associated with a conserved motif found in a number of nucleotide-binding GTs of various families (5, 10, 35). For example, in family 7, the motif occurs as DXD (16). In GT2 members, the motif may also occur as DXD; however, the exact number of Asp residues appears to vary between two and four (7). In WbbE, the sequence is DXDD (Fig. 2). Changing each Asp individually to Ala and analyzing the LPS profiles of the mutant strains revealed that the D93A (the first residue of the motif) and D96A replacements completely abrogated WbbE activity, while D95A led to significantly reduced activity (Fig. 3). All three mutant strains could be rescued to normal activity levels by the presence of the wild-type wbbE gene, confirming that the defect in synthesis was confined to the mutated wbbE gene product. The mutated wbbE genes were subcloned from the pWQ835 derivatives into the expression vector pET30a(+) to generate pWQ839-x. Cytoplasmic membrane extracts examined by Western blot analysis with anti-His antibody revealed no observable differences in mobilities or protein levels between the wild type and the mutant fusion (data not shown). The mutagenesis data therefore indicate that Asp95 of WbbE can be converted to a smaller, uncharged Ala with a reduction but not abrogation of activity. This suggests that Asp95 is unlikely to be the residue which coordinates the Mn2+ ion in WbbE. Alternative candidates are Asp93 and Asp96, both of which are critical for WbbE activity. It is clear from these results that sequence alignment in the absence of mutagenesis is not necessarily enough to identify critical residues even when comparing two GTs from the same family.
Identification of the ED(Y) motif in a subset of GT2 members.
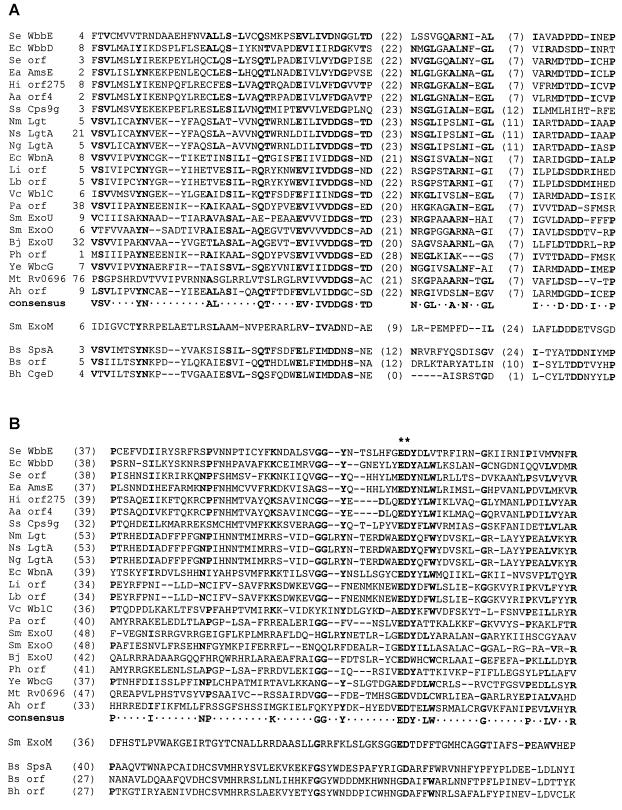
Our previous studies using conventional BLAST searches identified a region of sequence conservation C terminal to domain A in a number of nonprocessive GTs (Fig. 2). This domain, domain C, has a complex secondary structure with a conserved ED(Y) motif in region iv (24). To determine the relative distribution of this domain in GT2 enzymes, a PSI-BLAST search was done using the WbbE sequence and the resulting sequences were scanned for the ED(Y) motif. All of the sequences identified aligned minimally through domain A; most have been officially assigned to the GT2 family (http: //afmb.cnrs-mrs.fr/∼pedro/CAZY/db.html). While a number of sequences aligned only through domain A, 22 also possessed the ED(Y) motif (more than were identified by the conventional BLAST search). An alignment of all of the proteins identified by the database search using WbbE as the query sequence is shown in the top cluster of sequences in Fig. 4, along with a consensus. The sequences displayed belong to a broad spectrum of bacterial species from both the Bacteria and the Archaea (Pyrococcus abysii, Pyrococcus horikoshii). Interestingly, SpsA was not one of the proteins identified by the database search. An examination of the SpsA sequence, plus those of two of its homologues (identified by a BLAST search using the SpsA sequence), reveals that the proteins agree with the consensus for domain A (Fig. 4A) but show little to no sequence conservation through domain C (Fig. 4B). The ExoM sequence is included in the sequence alignment of Fig. 4 despite the fact that it too failed to be identified by either a BLAST or a PSI-BLAST database search. The reason for including this sequence is that it possesses an EDT motif in the position equivalent to that of EDY, and the results of mutagenesis studies suggest that this region contains the catalytic residue (14; see below). Interestingly, ExoM shows little homology to the consensus, even within domain A, with the exception of the conserved Asp residues.
FIG. 4.
Multiple-sequence alignment of proteins with homology to domains A and C of WbbE. Sequences in the upper cluster were identified by PSI-BLAST. All sequences were aligned using the Align algorithm (http://vega.igh.cnrs.fr/bin/align-guess.cgi). Partial sequences are shown, along with starting positions in parentheses after the protein names. The lengths of intervening sequences between aligned regions are indicated in parentheses, whereas gaps within the motif are indicated by dashes. Amino acids conserved in more than 50% of the proteins are indicated in bold and in the consensus sequence. Asterisks indicate strictly conserved residues. The origins and protein accession numbers for the various enzymes are indicated in order: Se, S. enterica serovar Borreze (accession no. AAC98401.1); Ec, E. coli O7 (AAC27537.1); Se, S. enterica (AF279620.1); Ea, Erwinia amylovora (Q46635); Hi, Haemophilus influenzae (C64175); Aa, Actinobacillus actinomycetemcomitans (BAA28129.1); Ss, Streptococcus suis (AAF18950.1); Nm, Neisseria meningitidis (CAB83816.1); Ns, Neissera subflava (AF240672.1); Ng, Neisseria gonorrhoeae (AAF14359); Ec, E. coli (AAD50485); Li, Leptospira interrogans (AAD52184.1); Lb, Leptospira borgpetersenii (AAD12967.1); Vc, Vibrio cholerae (BAA33634); Pa, Pyrococcus abyssi (G75005); S.m., Sinorhizobium (Rhizobium) meliloti (P33700); Sm, S. meliloti (P33697); Bj, Bradyrhizobium japonicum (AAC04822); Ph, P. horikoshii (A71157): Ye, Yersinia enterocolitica O:8 (AAC60770); Mt, Mycobacterium tuberculosis (CAB06459.1); Ah, Aeromonas hydrophila (AF146607.1); Sm, S. meliloti (P33695); Bs, B. subtilis (P39621); Bs, B. subtilis (CAB15817); Bh, Bacillus halodurans (BAB07092). Shown are domain A (A) and the region C terminal to domain A (B).
While the overall similarity is low among the 22 ED(Y)-containing sequences, there are a number of conserved residues. In addition, all but four of the sequences also possess a tyrosine after Glu-Asp. Given the identification of N-terminal domain A as the nucleotide-binding fold, one can speculate that the region C terminal to this would logically represent the catalytic domain. Due to the association of domain C with the ED(Y) motif, the 22 ED(Y)-containing proteins identified here might be expected to possess a conserved three-dimensional fold over this region. Possession of a common structure C terminal to domain A has already been suggested for a number of processive GT2 members. These proteins possess a region designated domain B, and this domain is speculated to possess the catalytic residue(s) (10, 24, 29). Perhaps not surprisingly, none of the known processive GT2 members aligned through the ED(Y) region with WbbE.
Recently, Garinot-Schneider et al. reported the identification of a putative catalytic residue within the EDT motif of ExoM (14) (Fig. 4B). In their study, Asp 187 of the EDT sequence was replaced with Glu, and subsequent in vivo and in vitro assays revealed a complete loss of activity. The analogous E186D mutation was not reported. The authors also reported that the D187 of ExoM aligned with D191 of SpsA, the residue speculated to represent the general base based on cocrystallization with glycerol (7). Despite an extremely low level of overall similarity, alignment of the SpsA sequence with those containing ED(Y) by using the Align algorithm (http://vega.igh.cnrs.fr/bin/align-guess.cgi) indicates that this residue also aligns with the Asp of the ED(Y) motif. The evidence therefore suggests that a catalytic residue may occur at the equivalent position in ExoM and SpsA and that the ED(Y) motif may also contain a catalytic base. The identification of a consensus in the ED(Y)-containing GT2 members suggests, however, that some GT2 enzymes diverge at least in sequence within the catalytic domain.
Analysis of the catalytic role of the conserved ED(Y) motif in domain C of WbbE.
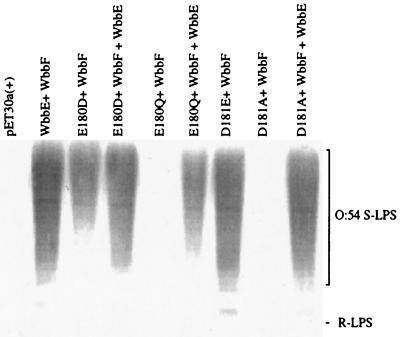
To pursue the possibility that ED(Y) contains a catalytic residue, site-directed replacement was performed on the strictly conserved Asp and Glu of the motif in WbbE. The ability of the mutant proteins to participate with WbbF in O:54 O-antigen synthesis was assessed (Fig. 5). All LPS preparations were made from the same number of cells. Analysis of the data revealed that the conservative change, E180D, resulted in a significant reduction in O-antigen synthesis, reflecting its effect on enzyme activity. This suggested that the size of the acidic residue is important. When the same residue was changed to the uncharged derivative (E180Q), activity was completely abrogated. Therefore, the charge on the residue is even more important. Interestingly, in contrast to the E180D replacement, in E. coli DH5α cells expressing wild-type WbbE in addition to the E180Q mutant enzyme, rescue of the mutant phenotype was not complete. Whether this is a true dominant-negative phenotype remains to be determined.
FIG. 5.
Analysis of the effect of replacements in the ED(Y) motif of WbbE on O:54 LPS expression. Immunoblot of anti-O:54 reactive LPS from E. coli DH5α harboring wild-type or mutated WbbE and wild-type WbbF. Lanes: 1, pET30a(+) (negative control); 2, pWQ835; 3, pWQ835-180; 4, pWQ835-180, pWQ818; 5, pWQ835-180Q; 6, pWQ835-180Q, pWQ818; 7, pWQ835-181; 8, pWQ835-181A; 9, pWQ835-181A, pWQ818. R-LPS was not detected due to the use of antisera adsorbed against E. coli DH5α. The migration of S-LPS is indicated.
When the contribution of D181 was assessed, the conservative replacement D181E had no effect, while the D181A replacement again abolished enzymatic activity. O-antigen levels were restored to that of the wild type in the presence of a functional WbbE, confirming that the reduction in O-antigen levels was due to the change in WbbE only. D181 is therefore required for activity; however, the size of the acidic residue at this position is not critical.
Manipulation of the ED(Y) motif indicated that the Glu and the Asp are both required for enzymatic activity. While only the acidic nature of D181 appeared to be important, both the size and charge of E180 were critical for activity. D181 may therefore contribute to the overall charge at that site. The observations for E180, in contrast, were those expected of a catalytic amino acid. Among the inverting glycoside hydrolases, the active site nucleophile may be either a Glu or an Asp, and mutagenesis indicates that both the size and the charge are critical for activity. In particular, withdrawal of even 1 Å may result in a virtually complete loss of activity in vitro (26). Although loss of activity of the E180D WbbE mutant was incomplete, one cannot directly compare glycoside hydrolases and GTs, as they are believed to use different enzymatic mechanisms (8, 10). While the geometrical position of the general base in a GT is obviously important, one can speculate that with a single catalytic residue instead of the two seen in glycoside hydrolases, the former may exhibit a greater degree of flexibility. The observation of incomplete loss of activity for E180D is in contrast to that of Garinot-Schneider et al. for the analogous replacement in ExoM (14). The reason for this is not apparent; however, the enzymes possess very little sequence homology over their entire lengths, and these discrepancies may simply be a reflection of this. In this context, it is interesting that similar discrepancies were observed between the two systems with the D-to-A replacement of the β2 Asp of domain A (14).
Catalytic residues are generally found on turns where they can move into position after the substrate and donor have entered the catalytic pocket or cleft. The hypothesis that E180 of WbbE may be the catalytic residue therefore has to be correlated with the predicted secondary structure of WbbE. Analyses of the WbbE sequence using HCA (Fig. 1) and the method of Garnier et al. (15) both predict that E180 is present on a turn.
Structural predictions for GT2 members.
While the ED(Y) motif is proposed to contain the catalytic residue in WbbE, and it appears to align with the proposed catalytic residues of ExoM and SpsA, it is E180 of WbbE and not D181 which is speculated to represent the catalytic residue. Based on sequence alignment alone, this would appear to be displaced by one residue from the equivalent ExoM and SpsA residues (Fig. 4B). However, results of the DXD mutagenesis, and the general observation that this motif can vary with respect to the number of Asp residues associated with it, suggest that sequence alignment is not enough to predict function. It is possible that E180 is in the same geometric position as the putative catalytic residues of ExoM and SpsA. Given that SpsA is the only GT2 member to be crystallized, the obvious question is whether, despite sequence divergence, the ED(Y)-containing members possess the same three-dimensional fold. HCA was therefore performed on the protein, and the plot was compared to that of WbbE (Fig. 1). While there may be some similarity, the data are equivocal.
Based on the combined evidence, the ED(Y) motif is proposed to contain the general base involved in catalysis in a subset of GT2 members. In the O:54 biosynthesis system, WbbE and WbbF are both ManNAc transferases and both belong to family 2. However, they differ significantly in their enzymatic activities, as the former is monofunctional whereas the latter is processive. Sequence and HCA comparisons of the two proteins revealed that the sequence conservation seen in domain A does not extend to the C-terminal region (24). The presence of domain C appears to correlate with a single glycosyl transfer activity (24), whereas at least some processive members appear to possess a distinctive domain B downstream of domain A (24, 29, 30). Although the mechanism of action of processive enzymes is the topic of much debate (10, 29), one can speculate that such an enzyme would possess a catalytic domain different from that of a monofunctional enzyme (i.e., domain B versus C). Combined with the observed sequence divergence between the ED(Y)-containing GTs and other family 2 members, it therefore appears that the GT2 enzymes, grouped together by virtue of their common N-terminal nucleotide-binding fold, are heterogeneous through the C-terminal region. This situation is reminiscent of the glycoside hydrolases. These enzymes have chimeric structures, possessing various combinations of substrate binding and catalytic domains (8). They are subdivided into families based on sequence conservation and into clans based on possession of a common catalytic domain structure. The strong homology within domain A of GT2 members, combined with homologies of subsets over the C-terminal region of these enzymes (domains C versus B), suggests that a similar subdivision may be necessary for this family of GTs.
ACKNOWLEDGMENTS
We thank Paul Amor for preparation of the anti-O:54 polyclonal antiserum and Sonia Bardy and Corin Forrester for technical assistance. We also thank the reviewers for their helpful comments.
This work was supported by grants to C.W. and A.J.C. from the Natural Sciences and Engineering Research Council (NSERC).
ADDENDUM IN PROOF
After acceptance of the manuscript, two reports of GT crystal structures were published. L. C. Pederson et al. (J. Biol. Chem. 275:34580–34585, 2000) reported crystallization of a GT2 member, and U. M. Ünligil et al. (EMBO J. 19:5269–5280, 2000) reported crystallization of a GT13 representative. Interestingly, both have domain A, and the catalytic residues identified were reported to be superimposable on D191 of SpsA and therefore also E180 of WbbE.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binotto J, MacLachlan P R, Sanderson P R. Electrotransformation of Salmonella typhimuriumLT2. Can J Microbiol. 1991;37:474–477. doi: 10.1139/m91-078. [DOI] [PubMed] [Google Scholar]
- 4.Bollag D M, Rozycki M D, Edelstein S J. Protein methods. 2nd ed. Toronto, Canada: John Wiley and Sons, Inc.; 1996. [Google Scholar]
- 5.Busch C, Hofmann F, Selzer J, Munro S, Jeckel D, Aktories K. A common motif of eukaryotic glysosyltransferases is essential for the enzyme activity of large clostridial cytotoxins. J Biol Chem. 1998;273:19566–19572. doi: 10.1074/jbc.273.31.19566. [DOI] [PubMed] [Google Scholar]
- 6.Campbell J A, Davies G J, Bulone V, Henrissat B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J. 1997;326:929–942. doi: 10.1042/bj3260929u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charnock S J, Davies G J. Structure of the nucleotide-diphospho-sugar transferase, SpsA, from Bacillus subtilis, in native and nucleotide-complexed forms. Biochemistry. 1999;38:6380–6385. doi: 10.1021/bi990270y. [DOI] [PubMed] [Google Scholar]
- 8.Clarke A J. Biodegradation of cellulose: enzymology and biotechnology. Lancaster, Pa: Technomic Press; 1996. [Google Scholar]
- 9.Darveau R P, Hancock R E W. Procedure for isolation of bacterial lipopolysaccharides from both smooth and rough Pseudomonas aeruginosa and Salmonella typhimurium. J Bacteriol. 1983;155:831–838. doi: 10.1128/jb.155.2.831-838.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies G J, Charnock S J. Structure of a nucleotide-diphospho-sugar transferase: implications for the synthesis of polysaccharides. In: Gilbert H J, Davies G J, Henrissat B, Svensson B, editors. Recent advances in carbohydrate bioengineering. Cornwall, Canada: MPG Books Ltd.; 1999. pp. 132–143. [Google Scholar]
- 11.Feldman M F, Marolda C L, Monteiro M A, Perry M B, Parodi A J, Valvano M A. The activity of a putative polyisoprenol-linked sugar translocase (Wzx) involved in Escherichia coliO antigen assembly is independent of the chemical structure of the O repeat. J Biol Chem. 1999;274:35129–35138. doi: 10.1074/jbc.274.49.35129. [DOI] [PubMed] [Google Scholar]
- 12.Filip C, Fletcher G, Wulff J L, Earhart C F. Solubilization of the cytoplasmic membrane of Escherichia coliby the anionic detergent sodium-lauryl sarcosinate. J Bacteriol. 1973;115:717–722. doi: 10.1128/jb.115.3.717-722.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaboriaud C, Bissery B, Benchetrit T, Mornon J P. Hydrophobic cluster analysis: an efficient new way to compare and analyse amino acid sequences. FEBS Lett. 1987;224:149–155. doi: 10.1016/0014-5793(87)80439-8. [DOI] [PubMed] [Google Scholar]
- 14.Garinot-Schneider C, Lellouch A C, Geremia R A. Identification of essential amino acids in the Sinorhizobium melilotiglucosyltransferase ExoM. J Biol Chem. 2000;275:31407–31413. doi: 10.1074/jbc.M004524200. [DOI] [PubMed] [Google Scholar]
- 15.Garnier J, Osguthorpe D J, Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978;120:97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- 16.Gastinel L N, Cambillau C, Bourne Y. Crystal structures of the bovine βgalactosyltransferase catalytic domain and its complex with uridine diphosphogalactose. EMBO J. 1999;18:3546–3557. doi: 10.1093/emboj/18.13.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffiths G, Cook N J, Gottfridson E, Lind T, Lidholt K, Roberts I S. Characterization of the glycosyltransferase enzyme from the Escherichia coliK5 capsule gene cluster and identification and characterization of the glucuronyl active site. J Biol Chem. 1998;273:11752–11757. doi: 10.1074/jbc.273.19.11752. [DOI] [PubMed] [Google Scholar]
- 18.Guzman L-M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBADpromoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ha S, Walker D, Shi Y, Walker S. The 1.9 A crystal structure of Escherichia coliMurG, a membrane-associated glycosyltransferase involved in peptidoglycan synthesis. Protein Sci. 2000;9:1045–1052. doi: 10.1110/ps.9.6.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hitchcock P J, Brown T M. Morphological heterogeneity among Salmonellalipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983;154:269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kapitonov D, Yu R K. Conserved domains of glycosyltransferases. Glycobiology. 1999;9:961–978. doi: 10.1093/glycob/9.10.961. [DOI] [PubMed] [Google Scholar]
- 22.Keenleyside W J, Perry M, MacLean L, Poppe C, Whitfield C. A plasmid-encoded rfbO:54 gene cluster is required for biosynthesis of the O:54 antigen in Salmonella entericaserovar Borreze. Mol Microbiol. 1994;11:437–448. doi: 10.1111/j.1365-2958.1994.tb00325.x. [DOI] [PubMed] [Google Scholar]
- 23.Keenleyside W J, Whitfield C. Lateral transfer of rfb genes: a mobilizable ColE1-type plasmid carries the rfb (O:54 antigen biosynthesis) gene cluster from Salmonella entericaserovar Borreze. J Bacteriol. 1995;177:5247–5253. doi: 10.1128/jb.177.18.5247-5253.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keenleyside W J, Whitfield C. A novel pathway for O-polysaccharide biosynthesis in Salmonella entericaserovar Borreze. J Biol Chem. 1996;271:28581–28592. doi: 10.1074/jbc.271.45.28581. [DOI] [PubMed] [Google Scholar]
- 25.Knirel Y A, Kochetkov N K. The structure of lipopolysaccharides of Gram-negative bacteria. III. The structure of O-antigens: a review. Biochemistry. 1994;59:1325–1383. [Google Scholar]
- 26.McCarter J D, Withers S G. Mechanisms of enzymatic glycoside hydrolysis. Curr Opin Struct Biol. 1994;4:885–892. doi: 10.1016/0959-440x(94)90271-2. [DOI] [PubMed] [Google Scholar]
- 27.Meier-Dieter U, Barr K, Starman R, Hatch L, Rick P D. Nucleotide sequence of the Escherichia coli rfegene involved in the synthesis of enterobacterial common antigen. J Biol Chem. 1992;267:746–753. [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 29.Saxena I M, Brown R M, Jr, Fevre M, Geremia R A, Henrissat B. Multidomain architecture of β-glycosyl transferases: implications for mechanism of action. J Bacteriol. 1995;177:1419–1424. doi: 10.1128/jb.177.6.1419-1424.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saxena I M, Brown R M. Identification of cellulose synthase(s) in higher plants: sequence analysis of processive, β-glycosyltransferases with the common motif “D,D,D35Q(R,Q)XRW.”. Cellulose. 1997;4:33–49. [Google Scholar]
- 31.Sinnott M L. Catalytic mechanisms of enzymic glycosyl transfer. Chem Rev. 1990;90:1171–1202. [Google Scholar]
- 32.Tsai G M, Frasch C E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- 33.Vrielink A, Ruger W, Driessen H P, Freemont P S. Crystal structure of the DNA modifying enzyme β-glucosyltransferase in the presence and absence of the substrate uridine diphosphoglucose. EMBO J. 1994;13:3413–3422. doi: 10.1002/j.1460-2075.1994.tb06646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitfield C, Amor P A, Koplin R. Modulation of the surface architecture of Gram-negative bacteria by the action of surface polymer: lipid A-core ligase and by determinants of polymer chain length. Mol Microbiol. 1997;23:629–638. doi: 10.1046/j.1365-2958.1997.2571614.x. [DOI] [PubMed] [Google Scholar]
- 35.Wiggins C A R, Munro S. Activity of the yeast MNN1 α 1,3-mannosyltransferase requires a motif conserved in many other families of glycosyltransferases. Proc Natl Acad Sci USA. 1998;95:7945–7950. doi: 10.1073/pnas.95.14.7945. [DOI] [PMC free article] [PubMed] [Google Scholar]