Abstract
Fat deposition is a vital factor affecting the economics of poultry production. Numerous studies on fat deposition have been done. However, the molecular regulatory mechanism is still unclear. In the present study, the whole-transcriptome RNA sequencing in abdominal fat, back skin, and liver both high- and low-abdominal fat groups was used to uncover the competitive endogenous RNA (ceRNA) regulation network related to chicken fat deposition. The results showed that differentially expressed (DE) genes in abdominal fat, back skin, liver were 1207(784 mRNAs, 330 lncRNAs, 41 circRNAs, 52 miRNAs), 860 (607 mRNAs, 166 lncRNAs, 26 circRNAs, 61 miRNAs), and 923 (501 mRNAs, 262 lncRNAs, 15 circRNAs, 145 miRNAs), respectively. The ceRNA regulatory network analysis indicated that the fatty acid metabolic process, monocarboxylic acid metabolic process, carboxylic acid metabolic process, glycerolipid metabolism, fatty acid metabolism, and peroxisome proliferator-activated receptor (PPAR) signaling pathway took part in chicken fat deposition. Meanwhile, we scan the important genes, FADS2, HSD17B12, ELOVL5, AKR1E2, DGKQ, GPAM, PLIN2, which were regulated by gga-miR-460b-5p, gga-miR-199-5p, gga-miR-7470-3p, gga-miR-6595-5p, gga-miR-101-2-5p. While these miRNAs were competitive combined by lncRNAs including MSTRG.18043, MSTRG.7738, MSTRG.21310, MSTRG.19577, and circRNAs including novel_circ_PTPN2, novel_circ_CTNNA1, novel_circ_PTPRD. This finding provides new insights into the regulatory mechanism of mRNA, miRNA, lncRNA, and circRNA in chicken fat deposition.
Key words: fat deposition, whole-transcriptome sequencing, ceRNA regulatory network, non-coding RNA, signaling pathway
INTRODUCTION
Chicken is one of the most important meat sources in the world, accounting for 32.63% of the total meat consumption (Tan et al., 2020). Modern commercial broilers have undergone extensive genetic selection to achieve rapid growth. Meanwhile, that also results in excessive deposition of subcutaneous and abdominal fat (Zuidhof et al., 2014). Excessive fat accumulation reduces meat production as well as feed efficiency, which causes a waste of resources and economic losses (Mir et al., 2017). In chicken, 70% fat is synthesized in the liver, then transported as the lipoproteins through the blood to the target tissues and stored as triglycerides. Five percent fat occurs in the adipose tissue mainly including in abdominal fat and subcutaneous, and 25% is acquired through diet (Wang et al., 2017). Fat accumulation is caused by excessive nutrient level and the genetic base of animals. Even though dietary inclusion of nutrients, such as proteins and fats can reduce the body fat storage, the genetic factors may be the most effective in reducing the fat accumulation in chickens (Crespo and Esteve-Garcia, 2002; Lotfi et al., 2011).
An increasing number of studies have shown that some non-coding RNAs such as long noncoding RNAs (lncRNAs), microRNAs (miRNAs), and circular RNAs (circRNAs), which play essential regulatory roles by forming a complex and precise post-transcriptional regulatory network(Ma et al., 2018; Wang et al., 2020b). Currently, most studies on non-coding RNAs related to fat deposition in chicken have been limited to the identification and expression profiling of miRNAs as well as exploring the functions of individual miRNAs (Wang et al., 2012; Ye et al., 2014; Fu et al., 2018). Several previous studies constructed the ceRNA regulation network for fat deposition traits in chicken. For example, one study found that circLCLAT1, circFNDC3AL, circCLEC19A, and circARMH1 can influence adipogenesis by regulating miRNAs via PPAR and fatty acid metabolism-related pathways (Zhang et al., 2020). Another study identified 3,881 lncRNAs and screened 235 differentially expressed (DE) lncRNAs, by constructing the ceRNA network identified 12 candidate lncRNA-miRNA-mRNA interactions in the chicken preadipocytes differentiation (Chen et al., 2019a). Zhai et al. (2021) identified the DElncRNAs and constructed the lncRNA-miRNA-mRNA interaction regulatory network related to the development of abdominal fat. However, ceRNA regulatory network among mRNA-miRNA-lncRNA-circRNA in chicken fat deposition is unclear.
Whole-transcriptome sequencing technologies which can capture a high-resolution picture of the transcriptomic landscape, have been widely utilized in the past decade to reveal the genetic origins of phenotypic traits (Shen et al., 2016; Xu et al., 2016). Guangxi Partridge chicken is a famous breed in Guangxi province, also included in Animal Genetic Resources in China: Poultry. It is well-known for the unique taste of its meat, but with excessive fat deposition (Wang et al., 2020a). In order to reveal the ceRNA regulatory network among mRNA-miRNA-lncRNA-circRNA in chicken fat deposition, the whole-transcriptome sequencing was employed and the interaction among mRNA-miRNA-lncRNA-circRNA was analyzed in abdominal fat, back skin, and liver of Guangxi Partridge chicken.
MATERIALS AND METHODS
Animals and Tissue Samples
One hundred fifty indigenous female Guangxi Partridge chickens were selected and bred according to the standard breeding program in the experimental farm of Guangxi University. At 120 days old, the chickens were slaughtered and ranked according to the abdominal fat rate. Three chickens were picked from the top 2-tail as the high- and low-abdominal fat group. The abdominal fat (H-AF1, H-AF2, H-AF3, L-AF1, L-AF2, L-AF3), back skin (H-BS1, H-BS2, H-BS3, L-BS1, L-BS2, L-BS3), and liver (H-Liver1, H-Liver2, H-Liver3, L-Liver1, L-Liver2, L-Liver3) were collected. These tissues were quickly frozen in liquid nitrogen and stored at −80° Celsius.
RNA Isolation, Library Construction, and Sequencing
Total RNA was extracted using the TRIzol Reagent (Invitrogen Life Technologies, Carlsbad, CA) according to the manufacturer's instructions and send to the Novogene Bioinformatics Institute (Novogene, Beijing, China) for library construction and sequencing. RNA purity and concentration were measured using the Nanophotometer R spectrophotometer (IMPLEN, Westlake Village, CA) and Qubit R RNA Assay Kit in Qubit R 2.0 Fluorometer (Life Technologies, Carlsbad, CA), RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA). For mRNA, lncRNA, and circRNA sequencing, 5 μg RNA from Liver, Back skin, and abdominal fat was used to prepare rRNA-depleted RNA by NEBNext R UltraTM Directional RNA Library Prep Kit for Illumina R (NEB, Ipswich, MA) following manufacturer's recommendations. There were three biological replicates per treatment, and a total of 18 libraries were prepared, the library preparations were sequenced on an Illumina Hiseq 4000 platform and 150 bp paired-end reads were generated. For small RNA sequencing, a total amount of 3 μg total RNA per sample was used as input material for the small RNA library. Following the manufacturer's recommendations, use the Illumina NEBNext Multiplex Small RNA Library Prep Set (NEB) to create a sequencing library and add the index code to the attribute sequence of each sample. Library preparation was sequenced on the Illumina Hiseq 2500/2000 platform to generate single-ended reads of 50 bp.
Read Mapping and Transcriptome Assembly
The paired-end raw reads were trimmed, and the quality was controlled by Trimmomatic (v0.39) (Bolger et al., 2014) and FastQC (v0.11.9) with default parameters. Then, clean reads were aligned to the chicken genome (downloaded from ensemble http://ftp.ensembl.org/pub/release-104/fasta/gallus_gallus/dna/Gallus_gallus.GRCg6a.dna.toplevel.fa.gz) using HISAT2 (v2.2.1) (Kim et al., 2015) software. Using samtools (v1.1.0) (Danecek et al., 2021) to convert the files from SAM file to BAM format. StringTie (v2.1.7) (Pertea et al., 2015) was used to assemble the transcripts and merge the transcripts among different samples in a reference-based approach (http://ftp.ensembl.org/pub/release-104/gtf/gallus_gallus/Gallus_gallus.GRCg6a.104.gtf.gz).
Differentially Expressed mRNA and Gene Functional Enrichment Analysis
Gene expression counts were obtained from BAM files using HTseq-count (Anders et al., 2015). To identify DEmRNAs between the liver, back skin, and abdominal fat, R package DEseq2 (version 3.14.0) (Love et al., 2014) was utilized for differential expression analysis with an absolute value of log2 fold change (log2FC) > 1 and P-value < 0.05. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathway enrichment analysis and gene set enrichment analysis (GSEA) of DEmRNAs were carried out using the clusterProfiler (v3.14.3) (Wu et al., 2021).
Identification of lncRNAs and Prediction of Target Genes
To identify the novel lncRNAs, the GFFcompare program (v0.11.2) was used to annotate the assembled transcripts by comparing them with the chicken reference annotation. To reduce the false‐positive rate, the following steps were performed to identify lncRNAs. 1) The transcript length shorter than 200 bp and only one exon reads were discarded. 2) Transcripts with read count coverage of less than five were deleted. 3) The remaining transcripts were identified as five kinds of lncRNAs according to class_code = ‘i’,‘j’,‘u’,‘x’,‘o’ (Lin and Mao, 2020). 4) We identified the lncRNAs overlapping among the four coding potential analysis tools including CPC2 (score > 0.5; Kang et al., 2017), CNCI (score > 0; Han et al., 2016), CPAT (score > 0.36; Wang et al., 2013), LGC (score > 0; Wang et al., 2019), and established them as the candidate novel lncRNA analysis data set. The transcripts were also used to blast the lncRNA sequences in the protein families (Pfam) database (http://pfam.xfam.org/). The Pfam database was used to search for protein domains in the Pfam HMM library to screen out transcripts with known protein domains. The lncRNAs genomic sequences were translated into protein sequences by the EMBOSS package (http://emboss.open-bio.org/). The genome sequence aligned to the Pfam database is removed, and other lncRNAs are used as the final novel lncRNA data set. The transcripts mapped to the Ensembl chicken reference annotation which gene_biotype = “lncRNA” were identified as known lncRNAs. HTseq-count was used to calculate lncRNA expression counts. DElncRNAs were extracted with |log2FC| > 1 and P-value < 0.05 by DEseq2. The potential cis- and trans-target mRNAs of DElncRNAs were predicted according to the gene expression and the position on the chromosome (Fu et al., 2019). The cis-targets gene was searched within a 10-kb window upstream or downstream of the lncRNA using bedtools (v2.30.0) (Quinlan and Hall, 2010). LncRNA transregulation analysis is based on the correlation coefficient between lncRNA and mRNA, and the value > 0.9 is considered to be trans-action.
Identification and Analysis of circRNAs
CIRI2 (version 2.0.6) and find_circ (version 1.2) were used to detect circular RNA in this study. First, the clean data were compared with the reference genome using BWA (Houtgast et al., 2018). Then, CIRI scans SAM files and detects junction reads with paired chiastic clipping (PCC) signals, paired end mapping (PEM) and GT-AG splicing signals as described by (Gao et al., 2018). Find_circ obtains junction reads via bowtie2 (Langmead and Salzberg, 2012) and samtools. The overlapped outputs from CIRI and find_circ were used for further analysis. We performed the spliced reads per billion mapping method to normalize circRNA expression. DEcircRNAs were extracted with |log2FC| > 1 and P-value < 0.05 by DEGseq2.
Identification and Analysis of miRNAs
The raw data in the fastq format is first processed using customized Perl and Python scripts. In this step, clean data were obtained by removing reads containing ploy N, with 5’ adapter contaminants, without 3’ adapter or the insert tag, containing ploy A or T or G or C and low-quality reads from raw data. miRNA lengths were determined by custom Perl scripts. Then, clean reads with a range of length from 15 to 30 bp were selected to do all the downstream analysis. The small RNA clean reads were mapped to the reference sequence by BWA to analyze their expression and distribution on the reference. Sequences that were successfully aligned in the previous step were extracted and realigned against the miRbase (https://www.mirbase.org/ftp.shtml) and Rfam (v14.6) database using Blast (Zhao and Chu, 2014). The results aligned to the mature miRNA coordinates from miRBase were transformed to read counts using custom Perl scripting. DEseq2 was used to perform differential expression analysis, and DEmiRNAs were extracted with |log2FC| > 1 and P-value < 0.05. TargetScan (Agarwal et al., 2015), and miRDB (Chen and Wang, 2020) were used for miRNA target‐gene prediction. To understand the miRNA target function, GO and KEGG enrichment analysis were performed on the predicted target-gene candidates.
Construction and Analysis of ceRNAs Regulatory Network
To discover the interactions of DEmRNAs, DElncRNAs, DEcircRNAs, and DEmiRNAs, we constructed a circRNA/lncRNA-miRNA-mRNA regulatory network based on the ceRNA hypothesis. Miranda and Targetscan were used to predict miRNA-lncRNA, miRNA-mRNA, and miRNA-circRNA pairs, and the correlation of these pairs was evaluated using the Spearman correlation coefficient (SCC) according to the expression. The interaction network was displayed using Cytoscape software (version 3.8.2) (Shannon et al., 2003).
QRT-PCR Validation of DEmRNAs, DElncRNAs, and DEmiRNAs
QRT-PCR was performed to validate the expression levels of DEmRNAs, DElncRNAs, and DEmiRNAs, 4 mRNAs, 2 lncRNAs, and 3 miRNAs were randomly selected of significantly different expression genes in abdominal fat, back skin, and liver. The QRT-PCR was conducted with RNA samples prepared for RNA-seq using 3 biological replicates. The primers were designed by Oligo7.0 software. RNA was reverse-transcribed into cDNA using RT Reagent Kit and Mir-X miRNA First-Strand Synthesis Kit (Takara, Dalian, China). The volume of the reaction mixture was 20 μL, with 2 μL of cDNA, 0.6 μL for each primer, 10 μL of SYBR (TaKaRa), and 6.8 μL of sterile water. The following QPCR reaction was performed as follows: 95°C for 30 s; 95°C for 5 s, 60°C for 30 s for 39 cycles; finally, melting curve collection at 65 to 95°C. β-actin was used as the endogenous control of lncRNA and mRNA, and U6 was used as the internal reference for miRNA. The relative gene expression was calculated by the 2−ΔΔCt method.
RESULTS
The Difference in Abdominal Fat Rate Between the High- and Low-Fat Groups
Microsoft Excel 2021 (Microsoft Corporation, Redmond, WA) was used for difference analysis (Student t test) of abdominal fat rate between the high- and low-fat groups. The results suggested that the abdominal fat rate in the high-fat group was significantly larger than those in the low-fat group (P < 0.01; Table 1).
Table 1.
Abdominal fat rate in high- and low-fat groups.
| Traits | High-fat group (n = 3) | Low-fat group (n = 3) |
|---|---|---|
| Abdominal fat rate (%) | 10.03 ± 1.53A | 4.21 ± 0.62B |
The different superscript uppercase letters represent a significant difference between the high-fat group and the low-fat group at the P = 0.01 level.
Global Responses of mRNA to Fat Deposition
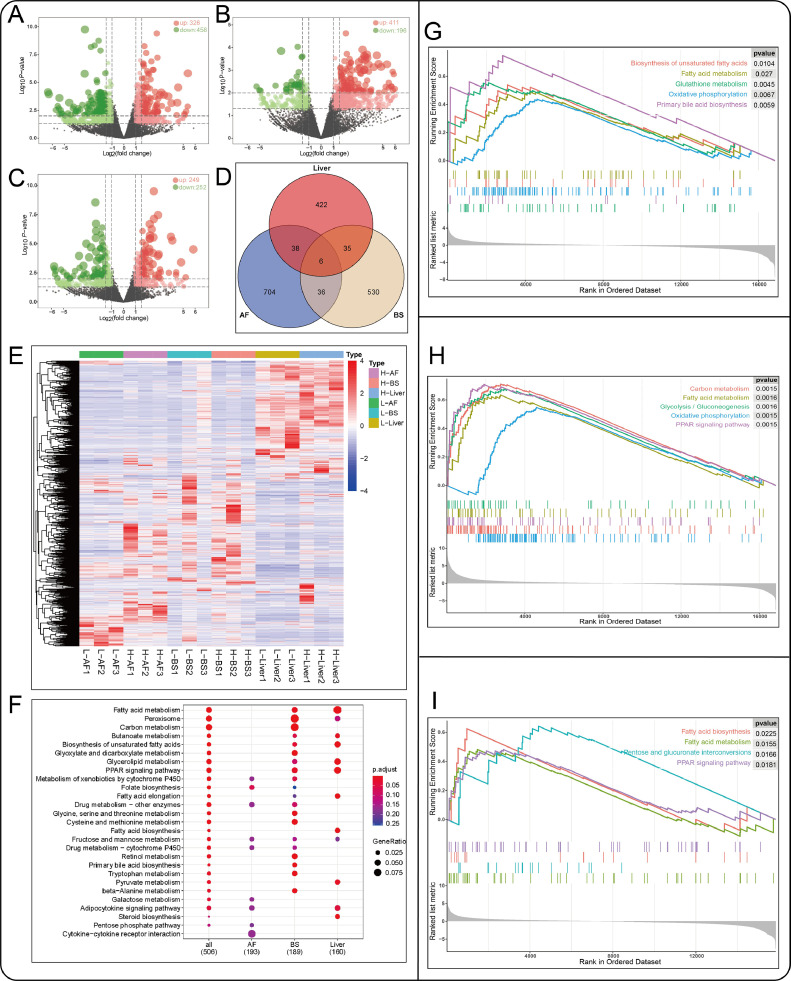
From RNAseq data, we identified 23,910 genes in the liver, back skin, and abdominal fat based on the chicken reference genome. After removing genes with low expression (FPKM < 1), 16,855, 16,863, and 15,852, genes in abdominal fat, back skin, and liver were considered for the downstream analysis. The log2FC and P-value of mRNA in are presented as volcano plot pictures in (Figures 1A–1C). The ranged of log2FC is from −7.65 to 9.75 in the liver, from −7.51 to 10.81 in back skin, and from −7.42 to 7.01 in abdominal fat. In all these genes, 784 (326 upregulated, 458 downregulated), 607 (411 upregulated, 196 downregulated), and 501 (249 upregulated, 252 downregulated) DEmRNAs were identified in abdominal fat, back skin, and liver, respectively (Table S1). Moreover, 6 DEmRNAs (SLC51B, IGFBP2, MSMO1, ENSGALG00000047326, ENSGALG00000042723, ENSGALG00000007080) were common identified in 3 tissues (Figure 1D). A heatmap showed the expression profiles of DEmRNAs in high- and low-fat groups (Figure 1E).
Figure 1.
DEmRNAs identify and analysis between high- and low-fat groups. A, B, C represented the volcano plot of mRNAs in abdominal fat, back skin, and liver. (D) Venn diagram shows the number of DEmRNAs identified in abdominal fat, back skin, and liver. (E) Heatmap of all DEmRNAs expression in each sample. (F) KEGG enrichment analysis compare of DEmRNAs in the abdominal fat, back skin, and liver. GSEA enrichment analysis in the abdominal fat (G), back skin (H), and liver (I). Abbreviation: KEGG, Kyoto Encyclopedia of Genes and Genomes.
To explore the functions of the DEmRNAs, the GO and KEGG analysis were performed. There are 11, 34, and 19 pathways that were significant enrichment in abdominal fat, back skin, and liver, respectively. DEmRNAs in abdominal were mainly significant enrichment to the folate biosynthesis (gga00790), fructose and mannose metabolism (gga00051), galactose metabolism (gga00052), glutathione metabolism (gga00480), and ECM-receptor interaction (gga04512). In back skin, DEmRNAs were most particularly enriched in peroxisome (gga04146), carbon metabolism (gga01200), glyoxylate and dicarboxylate metabolism (gga00630), fatty acid metabolism (gga01212), fatty acid degradation (gga00071), and PPAR signaling pathway (gga03320). As for DEmRNAs in liver, they were significantly enriched in fatty acid metabolism (gga01212), fatty acid biosynthesis (gga00061), PPAR signaling pathway (gga03320), glycerolipid metabolism (gga00561), and fatty acid elongation (gga00062) (Figure 1F). The detailed information of the enrichment analysis was shown in supplementary materials (Table S2). We further ranked the gene sets by log2FC and applied GSEA to identify functional gene sets. Similar to the KEGG enrichment analysis, GSEA found some genes participate in the PPAR signaling pathway, fatty acid biosynthesis, fatty acid metabolism, and oxidative phosphorylation, including SCD, MMP1, ELOVL2, ACSBG2, ACACA, and ACSL5 (Figure 1G-1I; Table S3).
Global Responses of lncRNA to Fat Deposition
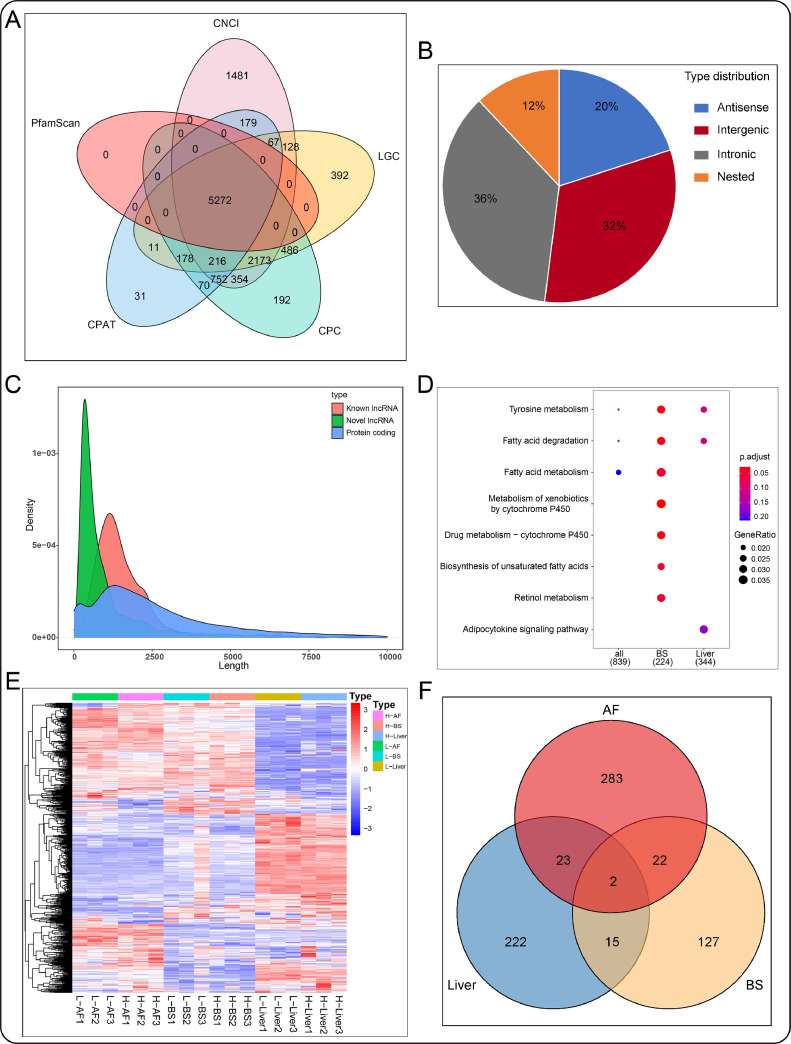
To identify the lncRNAs, we mapped RNA-seq data to the chicken reference genome where known lncRNA reads were removed. Other reads were analyzed by CNCI, CPC, CPAT, LGC, and PfamScan. Taking the intersection of the five tools, 5,272 lncRNAs were identified and included in the subsequent analysis (Figure 2A). Moreover, we found that novel and known lncRNAs were shorter than protein-coding transcripts (Figure 2C). lncRNA classification showed that 36% of intronic lncRNA, 32% of intergenic lncRNA, 20% of antisense lncRNA, and 12% of nested lncRNA (Figure 2B). The mean length of the novel and known lncRNAs was 825.4 bp and 745.6 bp, respectively, while protein-coding mRNA has an average length of 3,330 bp. At a cutoff with an absolute value of log2FC > 1 and P-value < 0.05, 330 (89 known, 241 novels), 166 (53 known, 113 novels), and 262 (86 known, 176 novels) DElncRNAs were identified in abdominal fat, back skin, liver, respectively (Table S4). Venn diagram of DElncRNAs among the three comparison groups was shown in Figure 2D. The heatmap of these DElncRNAs is presented in Figure 2E. Similar to DEmRNAs, the DElncRNAs of the high- and low-fat groups were clustered separately, while their 3 repeats were clustered together. To explore the potential functions of these DElncRNAs, their cis- and transtargeted mRNAs were predicted with bioinformatics methods. KEGG analysis was performed on the targets of DElncRNAs. The results showed that significantly enriched pathways were the fat acid degradation, tyrosine metabolism, fat acid metabolism (Figure 2F).
Figure 2.
lncRNAs identification and bioinformation analysis. (A) The lncRNA numbers detected by CPAT, CPC, LGC, PfamaScan, and CNCI software. (B) Identify the type of novel lncRNA. (C) Comparison of the transcription length of lncRNA and mRNA. (D) The DElncRNAs numbers in the abdominal fat, back skin, and liver. (E) DElncRNAs expression heatmap in each sample. (F) KEGG enrichment analysis of DElncRNAs in the abdominal fat, back skin, and liver. Abbreviation: KEGG, Kyoto Encyclopedia of Genes and Genomes.
Global Responses of circRNA to Fat Deposition
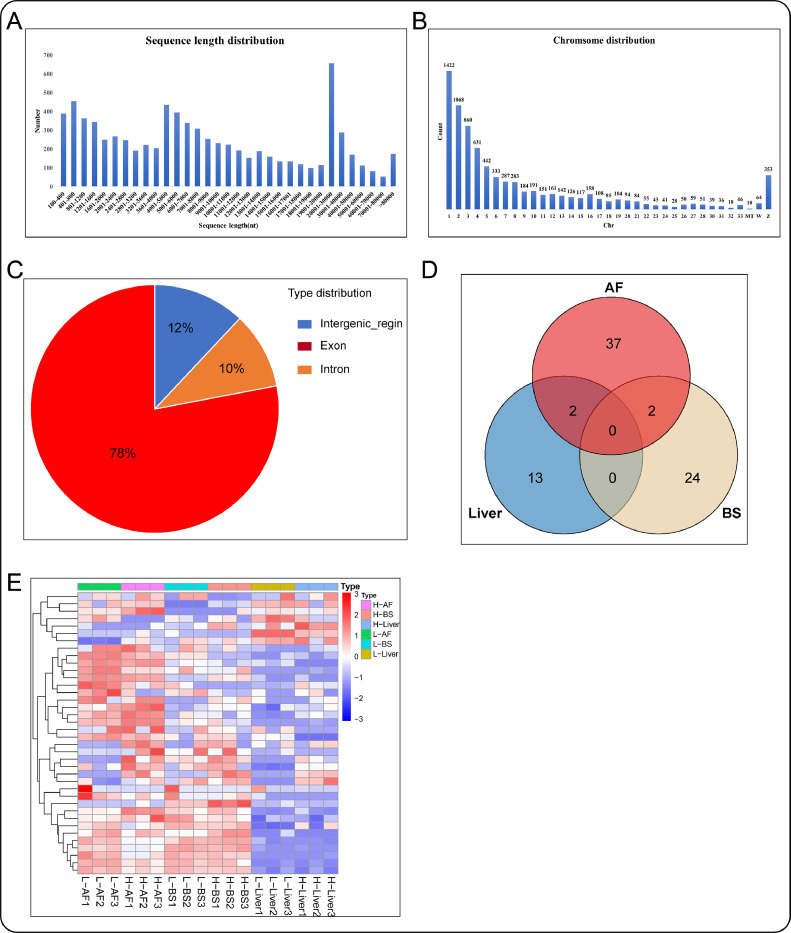
In order to improve the reliability of circRNA prediction, CIRI2 and find_circ were used to detect candidate circRNA. The intersection of these 2 tools total revealed 7,957 circRNAs in the abdominal fat, back skin, and liver, 78, 12, and 10% of them belong to the exon type, intergenic region type, and intron type, respectively (Figure 3C). The sequence length distribution of circRNAs is shown in Figure 3A, and most of them were shorter than 10,000 bp, and the longer the sequences, the fewer circRNAs’ numbers, and the longest circRNA sequence identified achieved 199kb. We identified maximum circRNAs on chromosome 1(chr1), followed by chr2, chr3, and chr4. The number of circRNA on the chromosome Z is significantly greater than that on the chromosome W (Figure 3B). After differential expression analysis, 15 (7 upregulated, 8 downregulated), 26 (15 upregulated, 11 downregulated), and 41 (25 upregulated, 16 downregulated) DEcircRNAs were identified in liver, back skin, and abdominal fat, respectively (Table S5). Two DEcircRNAs were common between liver and abdominal fat, 2 DEcircRNAs were common between back skin and abdominal fat. The heatmap showed the expression profiles of DEcircRNAs in each sample (Figure 3D).
Figure 3.
circRNAs identification and bioinformation analysis. (A–C) The characteristic of circRNAs in Sequence length, chromosome position, and type distribution. (D) DEcircRNAs numbers in the abdominal fat, back skin, and liver. (E) Heatmap of all DEcircRNAs.
Global Responses of miRNA to Fat Deposition
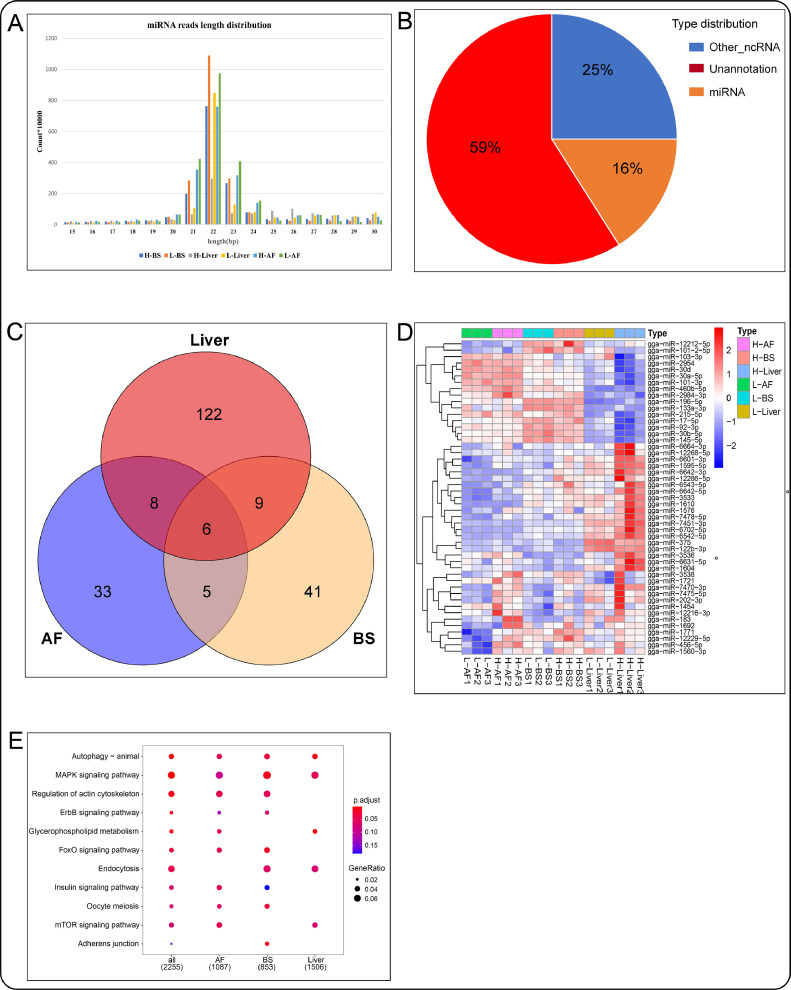
The lengths of most miRNA clean reads were 21 to 23 bp (Figure 4A) which conforms to the characteristics of miRNA and demonstrated the reliability of our data sets. Small RNA sequences classification showed that 59% of clean reads were unmatched, 25% was mapped to other non-coding RNA (rRNA, tRNA, snoRNA, etc.), and 16% was mapped to miRNA (Figure 4B). From 16% of clean reads, we finally identified 258 known miRNAs. After differential expression analysis, 52 (12 upregulated, 40 downregulated), 61 (15 upregulated, 46 downregulated), and 145 (44 upregulated, 101 downregulated) DEmiRNAs were identified in abdominal fat, back skin, and liver, respectively. By performing the intersection, we found that six miRNAs (gga-miR-6601-3p, gga-miR-7470-3p, gga-miR-1595-5p, gga-miR-12229-5p, and gga-miR-1721 gga-miR-375) are common in abdominal fat, back skin, and liver (Figure 4C). The expression profiles of DEmiRNAs in each sample are shown in (Figure 4D; Table S6). GO enrichment analysis of the target genes of DEmiRNAs showed that most of them were enriched in the alternative mRNA splicing, via spliceosome, regulation of alternative mRNA splicing, via spliceosome, transmembrane receptor protein tyrosine kinase signaling pathway, neuron differentiation, and neuron development. KEGG enrichment analysis showed that the significantly enriched KEGG signaling pathways of the targets of known DEmiRNAs were MAPK signaling pathway, FoxO signaling pathway, adherens junction, regulation of actin cytoskeleton, mTOR signaling pathway, insulin signaling pathway, and glycerophospholipid metabolism (Figure 4E).
Figure 4.
miRNAs identification and bioinformation analysis. (A) Length distribution of small RNA reads. (B) Percentage of different types of small RNAs. (C) Venn diagram showing the number of DEmiRNAs in the abdominal fat, back skin, and liver. (D) Heat map of DEmiRNAs. (E) KEGG enrichment of DEmiRNAs target genes in the abdominal fat, back skin, and liver. Abbreviation: KEGG, Kyoto Encyclopedia of Genes and Genomes.
Construction and Analysis of ceRNAs Regulatory Network
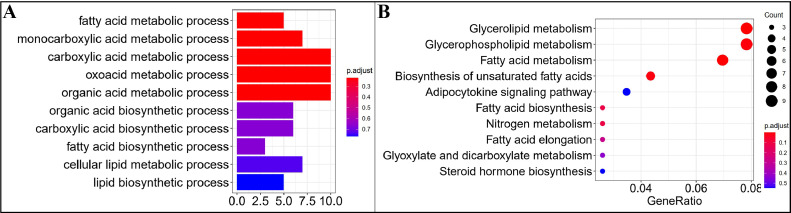
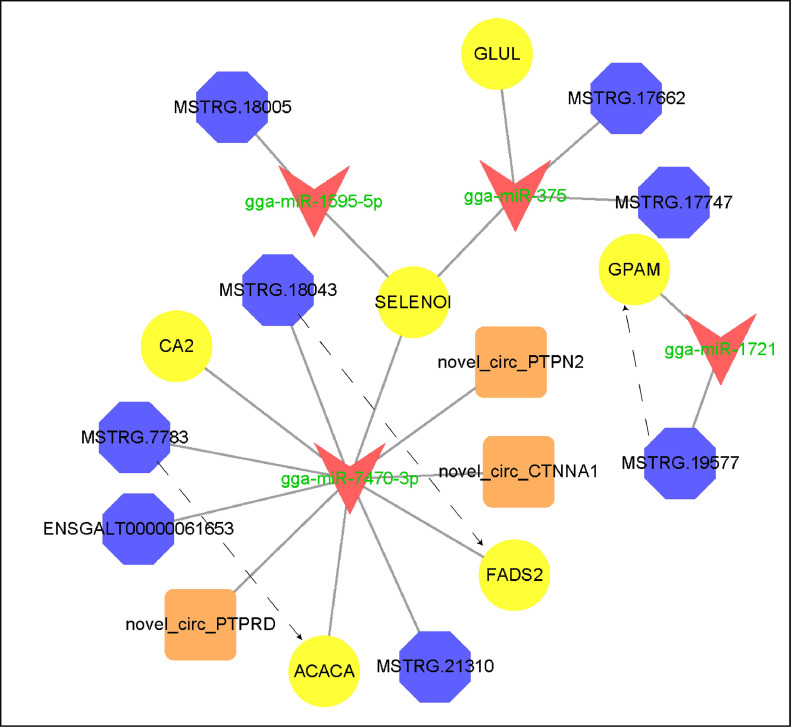
Based on the regulation relationship of DEmiRNA-DEmRNA, DEmiRNA-DElncRNA, and DEmiRNA-DEcircRNA, the lncRNA, mRNA, and circRNA which were significantly differentially expressed and regulated by the same miRNA were screened. In total, 2,663 lncRNA-miRNA-mRNA interactions, and 663 circRNA-miRNA-mRNA interactions were finally obtained in the liver, including 262 lncRNAs, 496 mRNAs, 15 circRNA, and 31 miRNAs. In the back skin, 418 lncRNA-miRNA-mRNA interactions, and 213 circRNA-miRNA-mRNA interactions were finally obtained, including 166 lncRNAs, 479 DEmRNAs, 26 circRNA, and 15 miRNAs. In the abdominal fat, 1,315 lncRNA-miRNA-mRNA interactions, and 461 circRNA-miRNA-mRNA interactions were finally obtained, including 19 miRNAs, 330 lncRNA, 634 mRNAs, and 41circRNA. The circRNA-miRNA-mRNA and lncRNA-miRNA-mRNA network were shown in (Figures 5–7). Based on the mRNAs involved in the DElncRNA/circRNA-DEmiRNA-DEmRNA regulatory network, GO and KEGG enrichment analysis were performed, and the results were shown in (Figure 8). The results showed that those genes related to miRNA, lncRNA, and circRNA were significantly enriched in fatty acid metabolic process, monocarboxylic acid metabolic process, carboxylic acid metabolic process, glycerolipid metabolism, fatty acid metabolism, and glycerophospholipid metabolism. To further identify the candidate ceRNA transcript-interaction relationship, we finally extracted 21 genes that were significantly enriched in GO and KEGG enrichment analysis. These genes were considered potential genes targets of fat deposition. These 21 genes include AKR1E2, CBR4, LPCAT2, FADS2, DGKQ, HSD17B12, GPAM, SELENOI, FADS1, LPIN1, ACSBG2, ELOVL5, PNPLA3, ACACA, ELOVL6, AGPAT2, AGPAT3, MOGAT1, ETNPPL, CA2, CA4. To further explore the candidate ceRNA subnetwork participate in fat deposition. Six miRNAs are shared by the liver, back skin, and abdominal fat, 21 genes included in the above significantly enriched pathway, and another candidate DEcircRNAs, DElncRNAs be extracted to the construction of the ceRNA regulatory network. At last, we established a ceRNA network model that included 4 miRNAs, 8 lncRNAs, 3 circRNAs, and 6 mRNAs. 22 interaction relationships were contained in the network (Figure 9).
Figure 6.
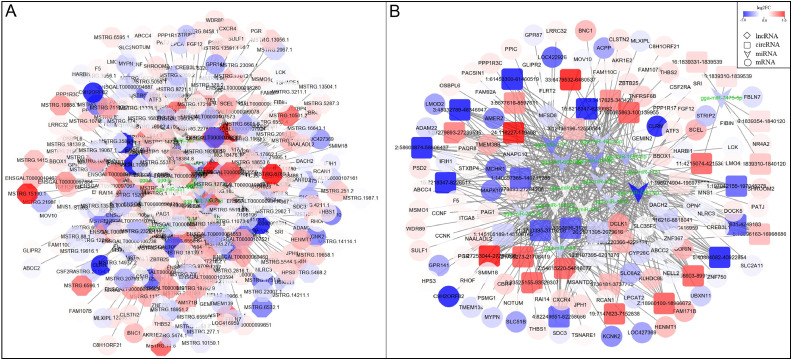
CeRNA network constructed by DEmRNAs, DElncRNAs, DEcircRNAs, and DEmiRNAs in the back skin. (A) (mRNA-miRNA-lncRNA ceRNA network) (B) (mRNA-miRNA-circRNA ceRNA network), the shades of the colors indicate the upregulated (red) and downregulated (blue) level.
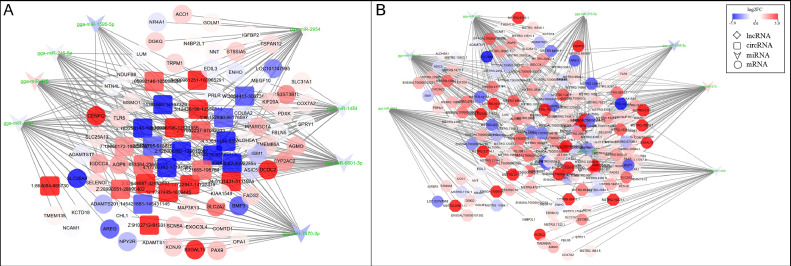
Figure 5.
CeRNA network constructed by DEmRNAs, DElncRNAs, DEcircRNAs, and DEmiRNAs in the abdominal fat. (A) (mRNA-miRNA-lncRNA ceRNA network) (B) (mRNA-miRNA-circRNA ceRNA network), the shades of the colors indicate the upregulated (red) and downregulated (blue) level.
Figure 7.
CeRNA network constructed by DEmRNAs, DElncRNAs, DEcircRNAs, and DEmiRNAs in the Liver. (A) (mRNA-miRNA-lncRNA ceRNA network) (B) (mRNA-miRNA-circRNA ceRNA network), the shades of the colors indicate the upregulated (red) and downregulated (blue) level.
Figure 8.
Functional enrichment analysis of mRNAs involved in the ceRNA network. (A) GO enrichment analysis, (B) KEGG enrichment analysis. Abbreviations: GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
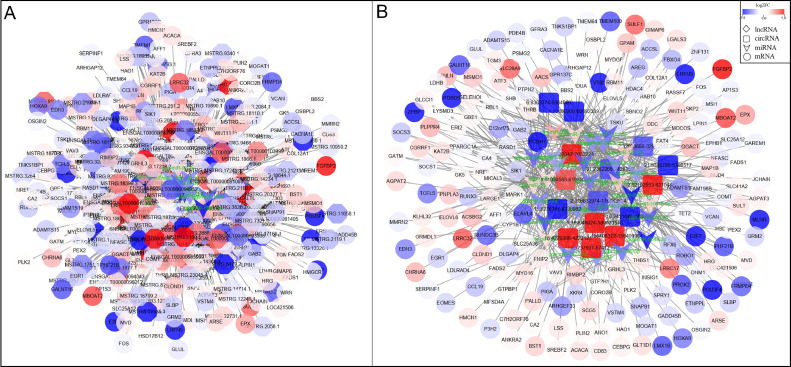
Figure 9.
The candidate ceRNA coregulation network relates to fat deposition. The yellow circle, red arrow, blue octagon, and orange square nodes represent co-differentially expressed mRNAs, miRNAs, lncRNAs, and circRNAs, respectively. The dotted line and the solid line indicate the coregulation between lncRNAs and mRNAs, and between miRNAs and other transcripts, respectively.
QRT-PCR Validation of DEmRNAs, DElncRNAs, and DEmiRNAs
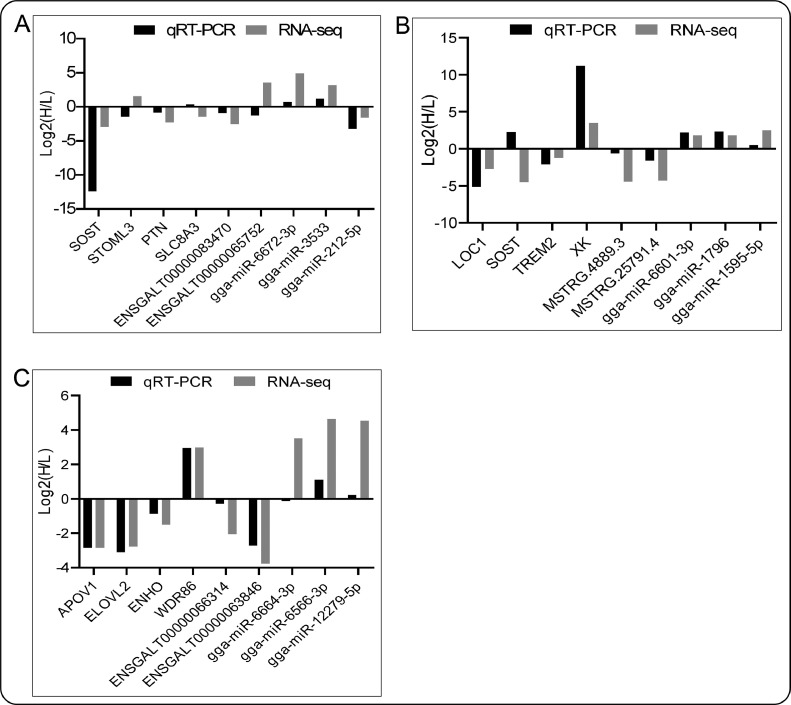
Ten transcripts were selected randomly from DEmRNAs, DElncRNAs, and DEmiRNAs for QRT-PCR validation (Figure 10). The results showed that some genes, including SOST, PTN, SLC8A3, STOML3, ENSGALT00000083470, gga-miR-212-5p, and gga-miR-10b-3p were highly expressed in high-fat group, while APOV1, ELOVL2, LOC1, MSTRG.25791.4, gga-miR-6672-3p, and gga-miR-3533 were highly expressed in low-fat group. Comparison of QRT-PCR and RNA-Seq data showed that 22 genes have consistent expression trends (over 80%), while five genes have different expression patterns.
Figure 10.
QRT-PCR analysis of mRNAs, lncRNA, and miRNA in abdominal fat (A), back skin (B), and liver (C).
DISCUSSION
The ceRNA hypothesis was first reported a few years ago, and ever since, it has been widely accepted and utilized for understanding various genetic mechanisms (Salmena et al., 2011). However, its application has been limited with respect to studies on indigenous chickens. In this study, we acquired high-throughput RNA-seq databases of the mRNAs, miRNAs, circRNAs, and lncRNAs of Guangxi Partridge chickens and compared their expressions between high- and low-fat groups. Subsequently, we screened the DE mRNAs, DEcircRNAs, DElncRNAs, and DEmiRNAs in abdominal fat, back skin, and liver tissues. Finally, based on the differential expression analysis, we predicted the circRNA-miRNA-mRNA and lncRNA-miRNA-mRNA networks that might be involved in the regulation of fat deposition.
Chicken liver is the main organ for adipogenesis, and it provides lipids to all the tissues (Nematbakhsh et al., 2021). In this study, the comparison between the high- and low-fat groups revealed 501 DEmRNAs, 262 DElncRNAs, 15 DEcircRNAs, and 145 DEmiRNAs in the liver. The GO and KEGG enrichment analysis of the DEmRNAs indicated that these genes were mainly involved in the PPAR signaling pathway, glycerolipid metabolism, fatty acid metabolism, biosynthesis of unsaturated fatty acids, insulin signaling pathway, and fatty acid elongation. The GSEA indicated that the difference gene sets significant enrichment in fatty acid biosynthesis, fatty acid metabolism, fentose and glucuronate interconversions, and PPAR signaling pathway. Some of these DEGs have been reported in the regulation of adipose deposition including ACACA, ELOVL6, SCD, FASN, FADS2, GPAM, and SREBPs. According to reports, ACACA, ELOVL6, SCD, and FASN, can encode the key enzymes that catalyze important reactions in the lipid metabolism of chicken. Specifically, acetyl-CoA carboxylase alpha (ACACA) and fatty acid synthase (FASN) are 2 important genes that encode major enzymes involved in the de novo fatty acid synthesis (Hicks et al., 2010; Laliotis et al., 2010). Similarly, Pirany et al. have reported that the ACACA expression is significant for fat deposition and leanness within the same strain of chicken (Pirany et al., 2020). Incidentally, FASN is also involved in the de novo synthesis of fatty acids during adipogenesis, specifically in the production of long-chain fatty acids in presence of malonyl-CoA (Wang et al., 2015b). Furthermore, the ELOVL fatty acid elongase (ELOVL) gene family can determine the overall fatty acid elongation, tissue distribution, and regulation, and it is a vital regulator of cellular lipid composition (Ma et al., 2017). ELOVL6 participates in the de novo lipogenesis in chicken liver, while ELOVL7 stimulates lipid accumulation in differentiated adipocytes (Loh et al., 2019) (Liu et al., 2019). Moreover, the studies reported increased ELOVL7 expression in chicken breast muscles with higher TG levels (Liu et al., 2019). The fatty acid desaturase 2 (FADS2) gene encode an enzyme that has a rate-limiting effect on long-chain polyunsaturated fatty acid synthesis (Yang et al., 2018). Additionally, glycerol-3-phosphate acyltransferase (GPAM) plays an important role in regulating cellular TG and phospholipid levels. Reportedly, transcription factors play important roles in controlling lipogenesis and lipolysis (D’Andre et al., 2013). Particularly, the sterol regulatory-element binding proteins (SREBPs) and PPARs are important in this regard since their expressions are critical for lipid biosynthesis. In our study, the sterol regulatory-element binding transcription factor 2 (SREBF2) was significantly differentially expressed (log2FC = 2.3, P-value = 0.005) in the liver. The results of DEmRNAs obtained in back skin and abdominal fat were similar in the liver. Enrichment analysis of miRNA-, lncRNA-, and circRNA-related target genes revealed that many DE transcripts participate in reactions associated with fat metabolism, such as tyrosine metabolism, fatty acid degradation, fatty acid metabolism, MAPK signaling pathway, and mTOR signaling pathway.
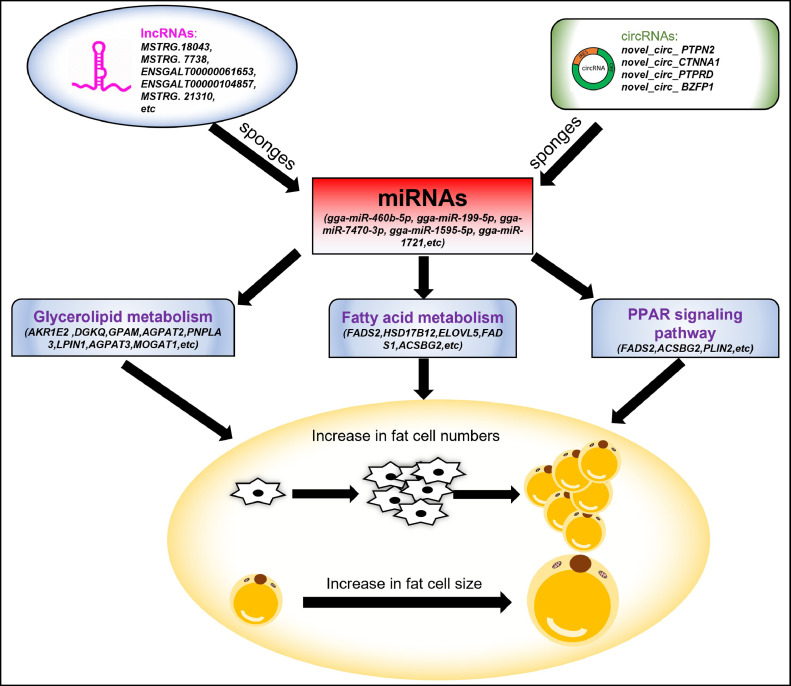
Based on the DEmRNAs, DElncRNAs, DEcircRNAs, and DEmiRNAs, we constructed the circRNA-miRNA-mRNA and lncRNA-miRNA-mRNA regulated networks for the liver, back skin, and abdominal fat tissues. To explore the potential functions of the ceRNA networks, we performed functional enrichment analysis for all DEmRNAs involved in these networks. Interestingly, the GO and KEGG analysis suggested that these genes participate extensively in signaling pathways associated with fat deposition, such as metabolism of monocarboxylic acids, fatty acids, carboxylic acids, and glycerolipids, fatty acid biosynthesis, fatty acid elongation, and PPAR signaling pathway. From the constructed ceRNA network, we identified several miRNAs, including gga-miR-92-3p, gga-miR-30d, gga-miR-30a-5p, gga-miR-17-5p, gga-miR-103-3p, gga-miR-101-3p, and gga-miR-101-2-5p, which have been previously reported to play a role in fat deposition. Among them, gga-miR-92-3p and gga-miR-17-5p are involved in regulating the accumulation of abdominal fat in Beijing You chickens (Huang et al., 2015), while gga-miR-30d and gga-miR-30a-5p play important roles in regulating adipogenesis in the abdominal adipose tissues of Chinese Gushi chickens (Chen et al., 2019b). Reportedly, gga-miR-30d also regulates the accumulation of abdominal fat (Li et al., 2016). There is study have also identified gga-miR-103-3p as a regulator of fat deposition in the abdominal and muscle (Fu et al., 2018). Moreover, gga-miR-101-3p is significantly differentially expressed in abdominal preadipocytes and differentiated adipocytes, which indicated its possible regulatory effect on adipogenesis (Wang et al., 2015a). Interestingly, previous research has shown gga-miR-196-5p might regulate different types of skeletal muscle fibers through PPAR, insulin, and adipocytokine signaling pathways (Liu et al., 2020). We identified gga-miR-196-5p is significant differential expression in high- and low-fat groups. This may suggest gga-miR-5p might be involved in intramuscular fat deposition. Alternatively, gga-miR-460b-5p can affect lipid and protein synthesis (Ge et al., 2021), thereby making it a potential candidate miRNA with respect to the regulation of fat deposition. Finally, the 6 notable DEmiRNAs shared by the liver, back skin, and abdominal fat were used as the core to establish a ceRNA network model that includes 4 miRNAs, 8 lncRNAs, 3 circRNAs, 6 mRNAs, and 22 interaction relationships (Figure 11). This ceRNA network can as a candidate regulation network in chicken fat deposition. Our results will be a useful reference to accelerate the understanding of the molecular mechanism for fat deposition in chicken.
Figure 11.
The proposed model in response to fat deposition in chicken. miRNAs act as the key regulators that directly target important signaling pathways related to fat deposition. Some lncRNAs and circRNAs can act as miRNA sponges and competitively bind the miRNAs, which may indirectly affect mRNA expression.
CONCLUSIONS
The current study reveals FADS2, HSD17B12, ELOVL5, AKR1E2, DGKQ, GPAM, and PLIN2 are important candidate genes of fat deposition in Guangxi Partridge chicken. While these genes are regulated by gga-miR-460b-5p, gga-miR-199-5p, gga-miR-7470-3p, gga-miR-6595-5p, gga-miR-101-2-5p, which were competitive combined by lncRNAs including MSTRG.18043, MSTRG.7738, MSTRG.21310, MSTRG.19577, and circRNAs including novel_circ_PTPN2, novel_circ_CTNNA1, novel_circ_PTPRD. This finding provides new insights into the ceRNA regulatory network of circRNA/lncRNA-miRNA-mRNA in chicken fat deposition.
ACKNOWLEDGMENTS
This work was supported by the Science and Technology Key Project of Guangxi (GK AB21075010) and the Science and Technology Major Project of Guangxi (GK AA17204027).
Authors' contributions: Cong Xiao: Data curation, Formal analysis, writing – original draft. Tiantian Sun: Validation, Investigation. Zhuliang Yang: Methodology, Data curation. Leqin Zou: Investigation, software. Jixian Deng: Supervision, Investigation, Resources. Xiurong Yang: funding acquisition, project administration, and writing – review, and editing.
Ethics approval and consent to participate: All animal experiments and methods in this study have been evaluated and approved by the Animal Ethics Committee of Guangxi University (GXU2018-058).
Consent for publication: All authors read and approved the submitted version.
Availability of data and material: The data sets analyzed during the current study are available from the corresponding author on reasonable request.
DISCLOSURES
The authors declare no competing financial interests.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2022.102121.
Appendix. Supplementary materials
Table S1 List of mRNA differential expression analysis in the abdominal fat, back skin, and liver.
Table S2 GO and KEGG functional enrichment analysis in the abdominal fat, back skin, and liver.
Table S3 GSEA enrichment analysis in the abdominal fat, back skin, and liver.
Table S4 List of lncRNA differential expression analysis in the abdominal fat, back skin, and liver.
Table S5 List of circRNA differential expression analysis in the abdominal fat, back skin, and liver.
Table S6 List of miRNA differential expression analysis in the abdominal fat, back skin, and liver.
REFERENCES
- Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Zhang T., Zhang S., Huang J., Zhang G., Xie K., et al. Identification of long non-coding RNA-associated competing endogenous RNA network in the differentiation of chicken preadipocytes. Genes (Basel) 2019;10 doi: 10.3390/genes10100795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic. Acids. Res. 2020;48:D127–D131. doi: 10.1093/nar/gkz757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zhao Y.L., Jin W.J., Li Y.F., Zhang Y.H., Ma X.J., et al. MicroRNAs and their regulatory networks in Chinese Gushi chicken abdominal adipose tissue during postnatal late development. BMC Genomics. 2019;20 doi: 10.1186/s12864-019-6094-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo N., Esteve-Garcia E. Nutrient and fatty acid deposition in broilers fed different dietary fatty acid profiles. Poult. Sci. 2002;81:1533–1542. doi: 10.1093/ps/81.10.1533. [DOI] [PubMed] [Google Scholar]
- D’Andre H.C., Paul W., Shen X., Jia X., Zhang R., Sun L., Zhang X. Identification and characterization of genes that control fat deposition in chickens. J. Anim. SciBiotechnol. 2013;4:43. doi: 10.1186/2049-1891-4-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O., Whitwham A., Keane T., McCarthy S.A., Davies R.M., Li H. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10 doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S., Zhao Y., Li Y., Li G., Chen Y., Li Z., Sun G., Li H., Kang X., Yan F. Characterization of miRNA transcriptome profiles related to breast muscle development and intramuscular fat deposition in chickens. J. Cell. Biochem. 2018;119:7063–7079. doi: 10.1002/jcb.27024. [DOI] [PubMed] [Google Scholar]
- Fu X.Z., Zhang X.Y., Qiu J.Y., Zhou X., Yuan M., He Y., Chun C., Cao L., Ling L., Peng L. Whole-transcriptome RNA sequencing reveals the global molecular responses and ceRNA regulatory network of mRNAs, lncRNAs, miRNAs and circRNAs in response to copper toxicity in Ziyang Xiangcheng (Citrus junos Sieb. Ex Tanaka) BMC Plant Biol. 2019;19:509. doi: 10.1186/s12870-019-2087-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Zhang J., Zhao F. Circular RNA identification based on multiple seed matching. Brief Bioinform. 2018;19:803–810. doi: 10.1093/bib/bbx014. [DOI] [PubMed] [Google Scholar]
- Ge P., Ma H., Li Y., Ni A., Isa A.M., Wang P., Bian S., Shi L., Zong Y., Wang Y., Jiang L., Hagos H., Yuan J., Sun Y., Chen J. Identification of microRNA-associated-ceRNA networks regulating crop milk production in pigeon (Columba livia) Genes. 2021;12 doi: 10.3390/genes12010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Liang Y., Li Y., Du W. Long roncoding RNA identification: comparing machine learning based tools for long noncoding transcripts discrimination [e-pub ahead of print] Biomed. Res. Int. 2016;2016 doi: 10.1155/2016/8496165. Accessed September 7, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks J.A., Trakooljul N., Liu H.C. Discovery of chicken microRNAs associated with lipogenesis and cell proliferation. Physiol. Genomics. 2010;41:185–193. doi: 10.1152/physiolgenomics.00156.2009. [DOI] [PubMed] [Google Scholar]
- Houtgast E.J., Sima V.M., Bertels K., Al-Ars Z. Hardware acceleration of BWA-MEM genomic short read mapping for longer read lengths. Comput. Biol. Chem. 2018;75:54–64. doi: 10.1016/j.compbiolchem.2018.03.024. [DOI] [PubMed] [Google Scholar]
- Huang H.Y., Liu R.R., Zhao G.P., Li Q.H., Zheng M.Q., Zhang J.J., Li S., Liang Z., Wen J. Integrated analysis of microRNA and mRNA expression profiles in abdominal adipose tissues in chickens. Sci. Rep. 2015;5 doi: 10.1038/srep16132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y.J., Yang D.C., Kong L., Hou M., Meng Y.Q., Wei L., Gao G. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic. Acids. Res. 2017;45:W12–W16. doi: 10.1093/nar/gkx428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laliotis G.P., Bizelis I., Rogdakis E. Comparative approach of the de novo fatty acid synthesis (Lipogenesis) between ruminant and non ruminant mammalian species: from bio-chemical level to the main regulatory lipogenic genes. Curr. Genomics. 2010;11:168–183. doi: 10.2174/138920210791110960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Ma Z., Jia L., Li Y., Xu C., Wang T., Han R., Jiang R., Li Z., Sun G., Kang X., Liu X. Systematic analysis of the regulatory functions of microRNAs in chicken hepatic lipid metabolism. Sci. Rep. 2016;6:31766. doi: 10.1038/srep31766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M., Mao Z.J. lncRNA-mRNA competing endogenous RNA network in IR-hepG2 cells ameliorated by APBBR decreasing ROS levels: a systematic analysis [e-pub ahead of print] Peerj. 2020;8 doi: 10.7717/peerj.8604. Accessed September 7, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Liu X., Cui H., Liu R., Zhao G., Wen J. Transcriptional insights into key genes and pathways controlling muscle lipid metabolism in broiler chickens. BMC Genomics. 2019;20 doi: 10.1186/s12864-019-6221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Zhang M., Shan Y., Ji G., Ju X., Tu Y., Sheng Z., Xie J., Zou J., Shu J. miRNA-mRNA network regulation in the skeletal muscle fiber phenotype of chickens revealed by integrated analysis of miRNAome and transcriptome. Sci. Rep. 2020;10:10619. doi: 10.1038/s41598-020-67482-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh H.Y., Norman B.P., Lai K.S., Rahman N., Alitheen N.B.M., Osman M.A. The regulatory role of microRNAs in breast cancer. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20194940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotfi E., Zerehdaran S., Ahani Azari M. Genetic evaluation of carcass composition and fat deposition in Japanese quail. Poult. Sci. 2011;90:2202–2208. doi: 10.3382/ps.2011-01570. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M.T., Cai B.L., Jiang L., Abdalla B.A., Li Z.H., Nie Q.H., Zhang X. lncRNA-Six1 is a target of miR-1611 that functions as a ceRNA to regulate Six1 protein expression and fiber type switching in chicken myogenesis. Cells. 2018;7 doi: 10.3390/cells7120243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Li H., Zheng H., Jiang K., Yan F., Tian Y., Kang X., Wang Y., Liu X. Hepatic ELOVL6 mRNA is regulated by the gga-miR-22-3p in egg-laying hen. Gene. 2017;623:72–79. doi: 10.1016/j.gene.2017.04.040. [DOI] [PubMed] [Google Scholar]
- Mir N.A., Rafiq A., Kumar F., Singh V., Shukla V. Determinants of broiler chicken meat quality and factors affecting them: a review. J. Food Sci. Technol.-Mysore. 2017;54:2997–3009. doi: 10.1007/s13197-017-2789-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nematbakhsh S., Pei Pei C., Selamat J., Nordin N., Idris L.H., Abdull Razis A.F. Molecular regulation of lipogenesis, adipogenesis and fat deposition in chicken. Genes (Basel) 2021;12 doi: 10.3390/genes12030414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirany N., Bakrani Balani A., Hassanpour H., Mehraban H. Differential expression of genes implicated in liver lipid metabolism in broiler chickens differing in weight. Br. Poult. Sci. 2020;61:10–16. doi: 10.1080/00071668.2019.1680802. [DOI] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Bai X., Xu J., Zhou M., Xu H., Nie Q., Lu M., Zhang X. Transcriptome sequencing reveals genetic mechanisms underlying the transition between the laying and brooding phases and gene expression changes associated with divergent reproductive phenotypes in chickens. Mol. Biol. Rep. 2016;43:977–989. doi: 10.1007/s11033-016-4033-8. [DOI] [PubMed] [Google Scholar]
- Tan X., Liu R., Xing S., Zhang Y., Li Q., Zheng M., Zhao G., Wen J. Genome-wide detection of key genes and epigenetic markers for chicken fatty liver. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21051800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Kim W.K., Cline M.A., Gilbert E.R. Factors affecting adipose tissue development in chickens: a review. Poult. Sci. 2017;96:3687–3699. doi: 10.3382/ps/pex184. [DOI] [PubMed] [Google Scholar]
- Wang G., Yin H., Li B., Yu C., Wang F., Xu X., Cao J., Bao Y., Wang L., Abbasi A.A., Bajic V.B., Ma L., Zhang Z. Characterization and identification of long non-coding RNAs based on feature relationship. Bioinformatics. 2019;35:2949–2956. doi: 10.1093/bioinformatics/btz008. [DOI] [PubMed] [Google Scholar]
- Wang H., Qin X., Li X., Wang X., Lei Y., Zhang C. Effect of chilling methods on the surface color and water retention of yellow-feathered chickens. Poult. Sci. 2020;99:2246–2255. doi: 10.1016/j.psj.2019.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Park H.J., Dasari S., Wang S., Kocher J.-P., Li W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model [e-pub ahead of print] Nucleic. Acids. Res. 2013;41 doi: 10.1093/nar/gkt006. Aaccessed September 7, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Du Z.Q., Cheng B., Wang Y., Yao J., Li Y., et al. Expression profiling of preadipocyte microRNAs by deep sequencing on chicken lines divergently selected for abdominal fatness [e-pub ahead of print] PLoS One. 2015;10 doi: 10.1371/journal.pone.0117843. Accessed September 7, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Shi Q.X., Wang S.C., Zhang H.F., Xu S.W. Ammonia regulates chicken tracheal cell necroptosis via the LncRNA-107053293/MiR-148a-3p/FAF1 axis. J. Hazardous Mater. 2020;386 doi: 10.1016/j.jhazmat.2019.121626. [DOI] [PubMed] [Google Scholar]
- Wang X.G., Yu J.F., Zhang Y., Gong D.Q., Gu Z.L. Identification and characterization of microRNA from chicken adipose tissue and skeletal muscle. Poult. Sci. 2012;91:139–149. doi: 10.3382/ps.2011-01656. [DOI] [PubMed] [Google Scholar]
- Wang Y., Viscarra J., Kim S.J., Sul H.S. Transcriptional regulation of hepatic lipogenesis. Nat. Rev. Mol. Cell Biol. 2015;16:678–689. doi: 10.1038/nrm4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., Fu X., Liu S., Bo X., Yu G. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation. 2021;2 doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Ji C., Zhang Y., Zhang Z., Nie Q., Xu J., Zhang D., Zhang X. Combination analysis of genome-wide association and transcriptome sequencing of residual feed intake in quality chickens. BMC Genomics. 2016;17:594. doi: 10.1186/s12864-016-2861-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Wang Y., Wang L., Shi Z., Ou X., Wu D., Zhang X., Hu H., Yuan J., Wang W., Cao F., Liu G. RNA-Seq reveals differentially expressed genes affecting polyunsaturated fatty acids percentage in the Huangshan Black chicken population. PLoS One. 2018;13 doi: 10.1371/journal.pone.0195132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Deng L., Liang M., Xu L., Zhang L., Ma Y., Li Y. MicroRNAs expression profiles in adipose tissues and liver from sex-linked dwarf and normal chickens. Acta Biochim. Biophys. Sin. (Shanghai) 2014;46:723–726. doi: 10.1093/abbs/gmu053. [DOI] [PubMed] [Google Scholar]
- Zhai B., Zhao Y., Fan S., Yuan P., Li H., Li S., Li Y., Zhang Y., Huang H., Li H., Kang X., Li G. Differentially expressed lncRNAs related to the development of abdominal fat in Gushi chickens and their interaction Regulatory Network. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.802857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Han Y., Zhai Y., Ma X., An X., Zhang S., Li Z. Integrative analysis of circRNAs, miRNAs, and mRNAs profiles to reveal ceRNAs networks in chicken intramuscular and abdominal adipogenesis. BMC Genomics. 2020;21:594. doi: 10.1186/s12864-020-07000-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Chu X. G-BLASTN: accelerating nucleotide alignment by graphics processors. Bioinformatics. 2014;30:1384–1391. doi: 10.1093/bioinformatics/btu047. [DOI] [PubMed] [Google Scholar]
- Zuidhof M.J., Schneider B.L., Carney V.L., Korver D.R., Robinson F.E. Growth, efficiency, and yield of commercial broilers from 1957, 1978, and 2005. Poult. Sci. 2014;93:2970–2982. doi: 10.3382/ps.2014-04291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 List of mRNA differential expression analysis in the abdominal fat, back skin, and liver.
Table S2 GO and KEGG functional enrichment analysis in the abdominal fat, back skin, and liver.
Table S3 GSEA enrichment analysis in the abdominal fat, back skin, and liver.
Table S4 List of lncRNA differential expression analysis in the abdominal fat, back skin, and liver.
Table S5 List of circRNA differential expression analysis in the abdominal fat, back skin, and liver.
Table S6 List of miRNA differential expression analysis in the abdominal fat, back skin, and liver.