Abstract
Cutaneous melanoma is a common cancer and cases have steadily increased since the mid 70s. For some patients, early diagnosis and surgical removal of melanomas is lifesaving, while other patients typically turn to molecular targeted therapies and immunotherapies as treatment options. Easy sampling of melanomas allows the scientific community to identify the most prevalent mutations that initiate melanoma such as the BRAF, NRAS, and TERT genes, some of which can be therapeutically targeted. Though initially effective, many tumors acquire resistance to the targeted therapies demonstrating the need to investigate compensatory pathways. Immunotherapies represent an alternative to molecular targeted therapies. However, inter-tumoral immune cell populations dictate initial therapeutic response and even tumors that responded to treatment develop resistance in the long term. As the protocol for combination therapies develop, so will our scientific understanding of the many pathways at play in the progression of melanoma. The future direction of the field may be to find a molecule that connects all of the pathways. Meanwhile, noncoding RNAs have been shown to play important roles in melanoma development and progression. Studying noncoding RNAs may help us to understand how resistance – both primary and acquired – develops; ultimately allow us to harness the true potential of current therapies. This review will cover the basic structure of the skin, the mutations and pathways responsible for transforming melanocytes into melanomas, the process by which melanomas metastasize, targeted therapeutics, and the potential that noncoding RNAs have as a prognostic and treatment tool.
Keywords: BRAF inhibitors, Checkpoint inhibitors, Drug resistance, Immunotherapy, Melanoma, Melanoma metastasis, Skin cancer, Targeted therapy, Therapeutic resistance
Introduction
Cutaneous melanoma is a skin neoplasm resulting from the transformation and uncontrolled proliferation of melanocytes in the stratum basale of the skin.1 The global incidence of melanoma is rising faster than any other solid tumor.2 In the United States alone, nearly 100,000 new cases are expected in 2022. More than 7000 of those patients are expected to die. What makes cutaneous melanoma more dangerous than other skin cancers—such as squamous cell carcinoma and basal cell carcinoma—is its ability to metastasize. As a fairly common cancer, melanoma is relatively easy to sample given its location. Having such easy access to patient samples has helped the scientific community really learn a lot about the disease and its process.
This review will cover the structural elements of the skin with their cellular components in order to highlight how melanoma progresses from initial transformation to metastasis. There is a clinical correlation between melanin production and developing melanoma. Thus, this review will also cover the epidemiological causes and risk factors associated with melanoma in addition to the role that melanin production plays in protecting patients from particular subtypes of the disease. More recently, the clinical classification of melanoma has been updated. So, we will cover those updates as well as the molecular pathways responsible for melanocyte transformation and metastasis. Several tools have been developed to treat melanoma. However, many of those treatments lack long term clinical efficacy. This review will explore the molecular undermining of why classical treatments fail while also suggesting novel components to target.
Biological characteristics and organization of the skin
Structural elements of the skin
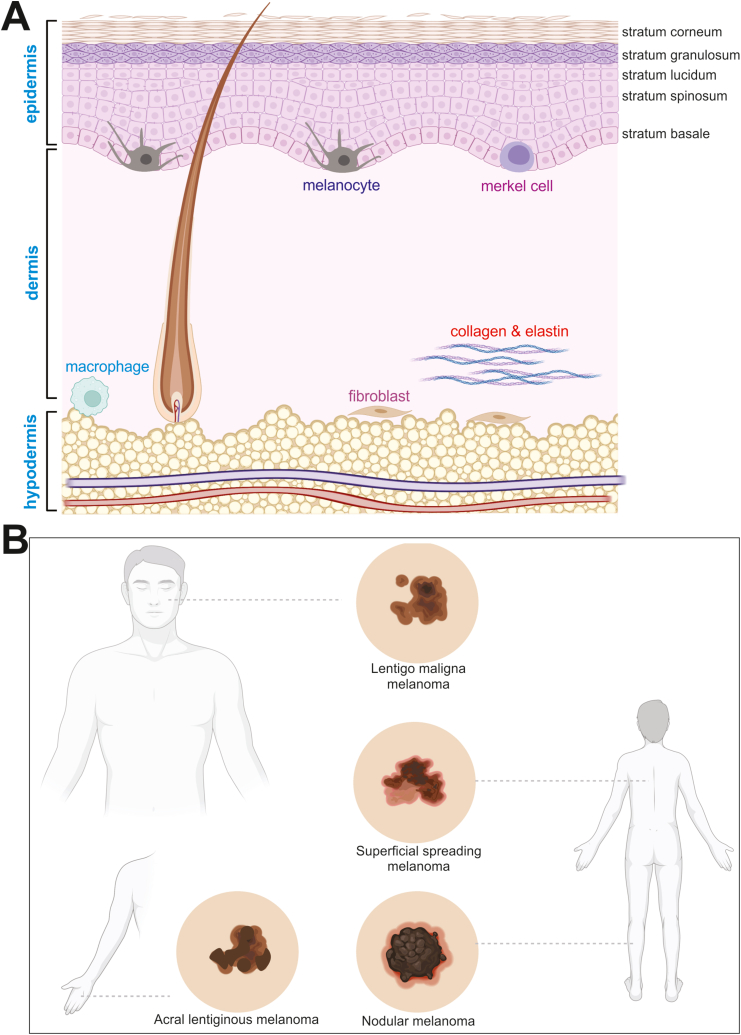
The skin is divided into three layers: the epidermis, the dermis, and the hypodermis (Fig. 1A).3 The hypodermis—the deepest of the three layers—is comprised of major branches of the skin's circulatory system, sweat glands, and the adipose tissue that is responsible for the production of vitamin D and triglycerides. Superficial to the hypodermis is the dermis. The deep layer of the dermis—also known as the reticular dermis—contains hair follicles and is comprised of a dense, irregular network of collagen and elastin, which is responsible for giving skin its strength and elasticity. Cellular components of the reticular dermis include fibroblasts, macrophages, and adipocytes. The papillary dermis rests superficial to the reticular dermis and is comprised of a loose connective tissue network and numerous capillaries. Though the papillary dermis represents about 20% of the dermis, it hosts all of the Meissners corpuscles (touch receptors) and free, non-myelinated nerve endings (temperature receptors). Sebaceous glands in the papillary dermis secrete sebum which helps keep skin flexible and prevents both excessive loss of and absorption of water on its surface. In thicker sections of skin, the apical layer of the dermis folds to form finger like projections known as dermal papillae, which improve nutrient delivery and adhesion to the more superficial epidermis. The epidermis is the most superficial layer of the skin and itself is comprised of four sublayers; the deepest of which—and the major focus of this review—is the stratum basale. The stratum basale is a single layer of cells that is comprised of keratinocytes, melanocytes, and Merkel cells.
Figure 1.
The structure and major components of the skin and gross melanoma lesions. (A) The skin consists of the cellular components of the three main layers, in addition to the five sublayers of the epidermis. Melanocytes are located in the stratum basale. (B) The locations and representative gross lesions of the three major subtypes of melanoma.
Cellular components of the skin
Keratinocytes make up the majority of cells in this single layer and as they proliferate and mature they give rise to the more superficial sublayers of the epidermis (Fig. 1A). Melanocytes produce melanin—the dark pigment responsible for skin color—and transport it to keratinocytes via special organelles known as melanosomes.4 Exogenously activated melanin production can be further stimulated by ultraviolet light (UV) exposure. Superficial to the stratum basale is an eight to ten cell thick layer known as the stratum spinosum. The stratum spinosum is characterized by an abundance of desmosomes anchoring neighboring cells to one another. The stratum granulosum is the three to five cell layer in which cells begin to produce lipid-rich granules and lose their nuclei. Cells in the most superficial layer of the skin—known as the stratum corneum—are flattened, dead scales filled with densely packed keratin. This layer is the protective sublayer responsible for water loss regulation. Altogether, the different layers of the skin work together to serve four major purposes: providing sensation, thermoregulation, protection, and metabolism.
The overwhelming majority of cells in the epidermis are keratinocytes. The newest cells undergo mitosis in the stratum basale. As cell division increases at the deepest level of the epidermis, those cells exert a force on older cells pushing them toward the surface of the skin. As keratinocytes move superficially, they begin to interlock at their protruding processes via desmosomes linked together with keratin. When this 8 to 10 cell layer is stained they exhibit a “spiny” phenotype which is why that sublayer of cells is known as the stratum spinosum. Superficial to this layer, keratinocytes adopt a slightly different phenotype. Though metabolically active, these cells are in the process of apoptosis and begin to flatten out. They allow skin to serve as a waterproof barrier. The next superficial layer of 3–4 keratinocytes are even flatter and more densely packed with keratin. Most of the cells in this layer have completed apoptosis. The dead cells no longer have nuclei or organelles, but the left behind keratin and cell membranes contribute to the protective quality of the epidermis.
Melanocyte, melanogenesis and skin pigmentation
Located in the stratum basale, melanocytes are neural crest derived cells responsible for the production of two distinct pigments, eumelanon and pheomelanin, via melanogenesis.5 Basal melanogenesis is responsible for skin pigmentation.6 Active melanogenesis is often initiated by exposure to UV radiation; most commonly through intense sun exposure. The purpose of such an increase in melanogenesis is a protective one. Melanin absorbs the UV light thereby preventing it from causing DNA damage to cells in the hypodermis. In both cases, melanin is produced in the melanocyte, packaged in the melanosome, and transported to keratinocytes where the melanin is exocytosed into the cytoplasm. Each melanocyte has an estimated 30 to 40 dendrites that allow for the transfer of melanosomes to keratinocytes.7 The phenotypic differences in skin pigmentation can be explained by the size and number of melanosomes, the amount and distribution of the types of melanin, and the distraction of keratinocytes.
Changes to skin pigmentation that take place immediately following exposure to intense UV radiation are due to oxidation of previously existing melanin.8 Whereas, longer term changes to skin pigmentation are due to increased eumelanin production. Melanosomes are strategically distributed in keratinocytes with increased UV exposure in order to efficiently protect those cells from UV exposure. Though UV based DNA damage has the ability to induce malignant transformation of melanocytes, only a small percentage of melanomas are related to extrinsic UV radiation.
Subtypes, classifications and epidemiology of melanoma
Subtypes of melanoma
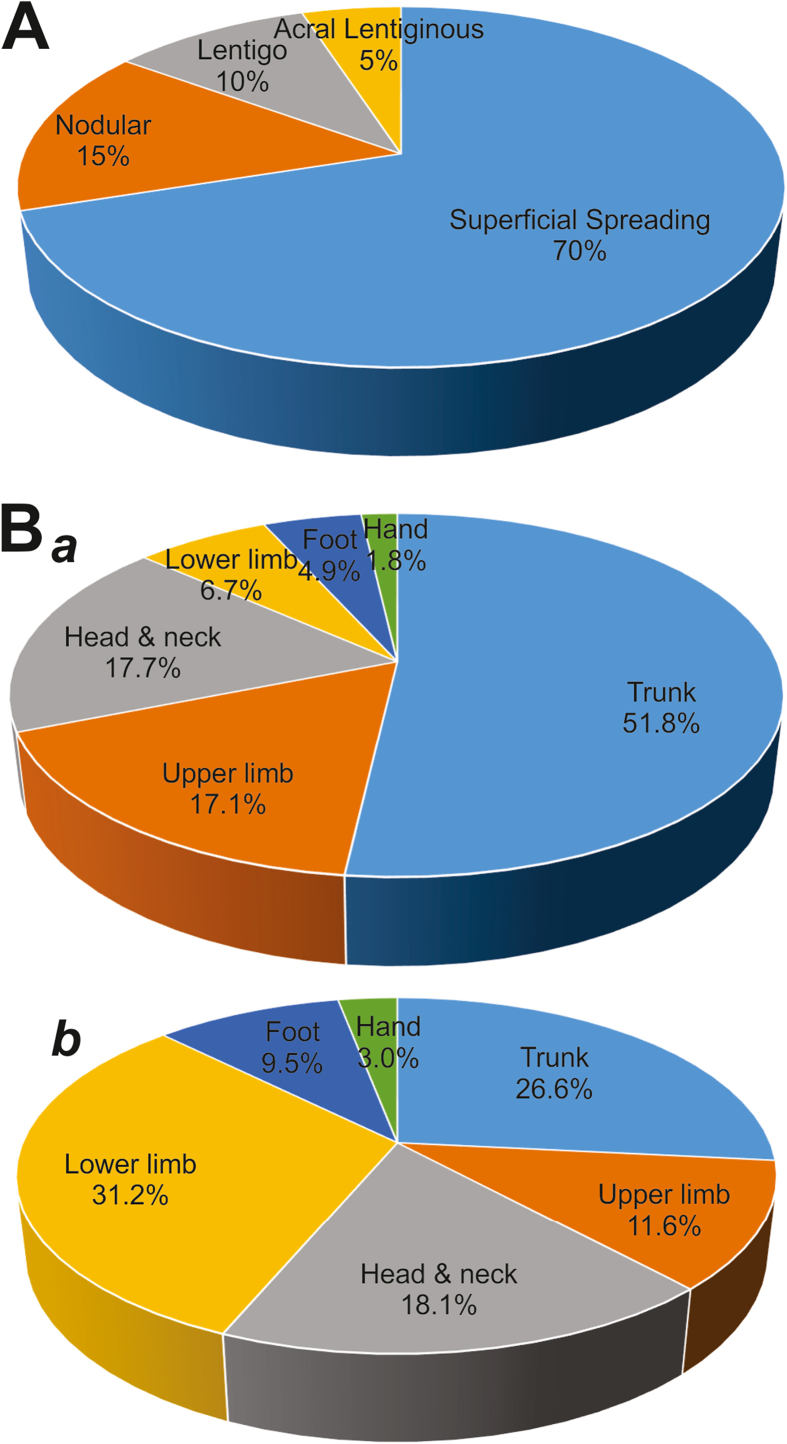
Melanoma is a skin neoplasm resulting from the transformation and uncontrolled proliferation of melanocytes in the stratum basale of the skin (Fig. 1A).1 There are four main subtypes of cutaneous melanoma: superficial spreading melanoma, lentigo melanoma, acral lentiginous melanoma, and nodular melanoma (Fig. 1B).9 Superficial spreading melanoma accounts for about 70% of melanoma cases and is characterized by a slowly growing patch of dark skin (Fig. 2A).10 This subtype usually occurs in fairer skinned patients and is typically found on the torso which is related to excessive UV exposure be it from the sun or tanning beds.10 When patients under 40 develop melanoma, it is usually the superficial spreading subtype.11 The next subtype, lentigo melanoma, typically develops in older patients with sun damaged skin on the face, ears, or neck. Acral lentiginous melanoma is typically found in patients of color and presents as a dark spot on the sole of the foot or palms. Acral lentiginous melanoma has its own subcategory called subungual melanoma that presents as dark vertical streaks under the nail beds of fingers and toes.12 It is the rarest form of melanoma and the only melanoma not associated at all with sun exposure.12 Nodular melanoma is the second most common form of melanoma making up about 15% of all cases.13 It is considered the most aggressive of all the subtypes because it grows the fastest.13 Furthermore, the anatomical locations of primary melanomas are quite different in men vs. women (Fig. 2B, panel a vs. b). Though it can occur in people of all ages and races, nodular melanoma is most common in fair-skinned patients over 65 who spend a lot of time in the sun or in tanning beds.14 It usually presents as a firm bump or node on the surface of the skin.
Figure 2.
The epidemiology of melanoma. (A) The schematic representation of the breakdown percentage of clinical cases for each subtype of melanoma. (B) The anatomical locations of primary melanomas in men (a) and women (b). The illustrations were inspired by BioRender.
Clinical classification of melanoma
Melanomas are clinically classified using scoring parameters set by the American Joint Committee on Cancer (AJCC) (Table 1).15 The thickness of the primary tumor (T) is scored from T0 which is the score given to an unidentified or completely regressed primary tumor to T4 which is the score give to a primary tumor with a thickness greater than 4 mm. Within the tumor thickness category is the subcategory of ulceration. Ulceration is the absence of an intact epithelium on the surface of the melanoma. Regional lymph node involvement (N) is determined using a combination of sentinel lymph node mapping and biopsy. Slightly different is the category of distant metastases (M). Distant metastasis scores range from M1a signifying metastatic lesions to superficial skin, subcutaneous lesions, or lymph node sites. On the other end of the spectrum, M1d tumors have metastasized to the central nervous system. The distant metastasis score is divided into two subcategories: low serum lactate dehydrogenase (LDH) levels (0) and elevated serum LDH levels (1).
Table 1.
American Joint Committee on Cancer (AJCC) TNM staging of melanoma.
| T Designation | Primary Tumor Thickness (mm) | N Designation | Regional Lymph Node | M Designation | Distant Metastasis |
|---|---|---|---|---|---|
| Tis | n/a | Nx | lymph node cannot be assessed | M0 | no metastasis |
| T1 | ≤1.0 | N1 | 1 lymph node | M1a | skin, cutaneous, distant lymph node |
| T2 | 1.0–2.0 | N2 | 2-3 lymph nodes | M1b | lung |
| T3 | 2.0–4.0 | N3 | 4+ lymph nodes | M1c | other visceral sites |
| T4 | ≥4.0 | M1d | central nervous system |
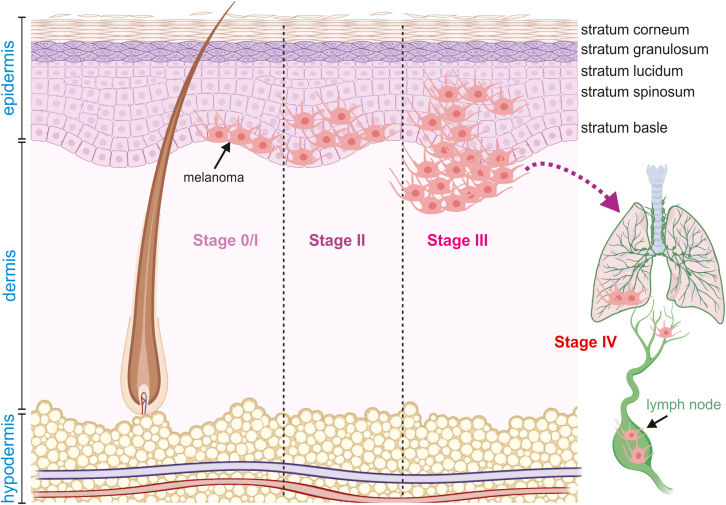
This complex staging system sheds light on the complexity of melanoma tumor biology. The aforementioned parameters are used to classify melanomas into four pathologic or surgical stages (Table 2). Stage 1 melanomas are marked by limited to low risk primary tumors with no evidence of regional or distant metastasis with distinguishing factors including tumor thickness and presence (or absence) of ulceration. Stage 4—the most advanced tumors—are marked by the most aggressive TMN parameters most notably the presence of distant metastasis to the central nervous system (Fig. 3). Mitotic rate, age, and gender are also valuable prognostic factors. Tumors with high mitotic rates are associated with poor outcomes; as is advanced age. However, younger patients tend to have tumors with more aggressive features. Women tend to have tumors with lower T scores and lower rates of ulceration.
Table 2.
Pathologic or surgical (0 - IV) staging of melanoma.
| Stage | Primary Tumor (T) | Regional Lymph Nodes (N) | Distant Metastasis (M) |
|---|---|---|---|
| 0 | Tis | N0 | M0 |
| I | T1a - T2a | N0 | M0 |
| II | T2b - T4b | N0 | M0 |
| III | T0 - T4b | N1a - N3c | M0 |
| IV | any T | any N | M1 |
Notes:
Tis: the melanoma cells are only in the very top layer of the skin surface. It is called melanoma in situ.
T0: no melanoma cells can be seen where the melanoma started (primary site).
T1: the melanoma is 1 mm thick or less. It is split into T1a and T1b.
T1a: the melanoma is less than 0.8 mm thick and the skin over the tumour does not look broken under the microscope (not ulcerated).
T1b: either: the melanoma is less than 0.8 mm thick but is ulcerated, or the melanoma is between 0.8 mm and 1.0 mm and may or may not be ulcerated.
T2: the melanoma is between 1 mm and 2 mm thick.
T3: the melanoma is between 2 mm and 4 mm thick.
T4: the melanoma is more than 4 mm thick.
T2 and T4 melanoma is further divided into a and b depending on whether it is ulcerated or not. A means without ulceration, b means with ulceration.
N0: there are no melanoma cells in the nearby lymph nodes.
N1: there are melanoma cells in one lymph node or there are in-transit, satellite or microsatellite metastases.
N2: there are melanoma cells in 2 or 3 lymph nodes or there are melanoma cells in one lymph node and there are also in-transit, satellite or microsatellite metastases.
N3: there are melanoma cells in 4 or more lymph nodes or there are melanoma cells in 2 or 3 lymph nodes and there are in-transit, satellite or microsatellite metastases or there are melanoma cells in any number of lymph nodes and they have stuck to each other (matted lymph nodes).
M0: the cancer hasn't spread to another part of the body.
M1: the cancer has spread to another part of the body. M1 can be further divided depending on which parts of the body the cancer has spread to and whether there are raised levels of a chemical in the blood called lactate dehydrogenase (LDH).
Figure 3.
The progression and pathologic staging of melanoma. Melanoma originates from its initial location in the stratum basale in Stage 0/1 disease and progresses to distant metastatic sites like the lungs in Stage 4 disease. The illustrations were inspired by BioRender.
Epidemiological causes
The two main causes of melanoma are extrinsic UV exposure and genetic predisposition. Early intermittent sun exposure in childhood is thought to cause DNA damage via UV irradiation that eventually manifest as melanomas on the thighs and trunk in adulthood. On the other hand, chronic sun exposure post childhood with accompanying damage to the skin (sunburn) is associated with melanomas developing on the arms, neck, and face. About 10% of melanoma cases are due to an inherited mutated gene.16
Risk factors
Having lighter skin is a risk factor for melanoma because skin cells produce less melanin, which is a protective pigment against UV damage. Additional phenotypical risk factors include having freckles and lighter colored hair. Some patients have several benign nevi (moles) which are typically harmless. However, the more nevi a patient has the more likely that patient is to develop melanoma in the future. It is important for those with multiple risk factors to undergo routine physical examinations to locate and track suspicious nevi.
Major mutations and the affected signaling pathways in melanoma development
BRAF mutations
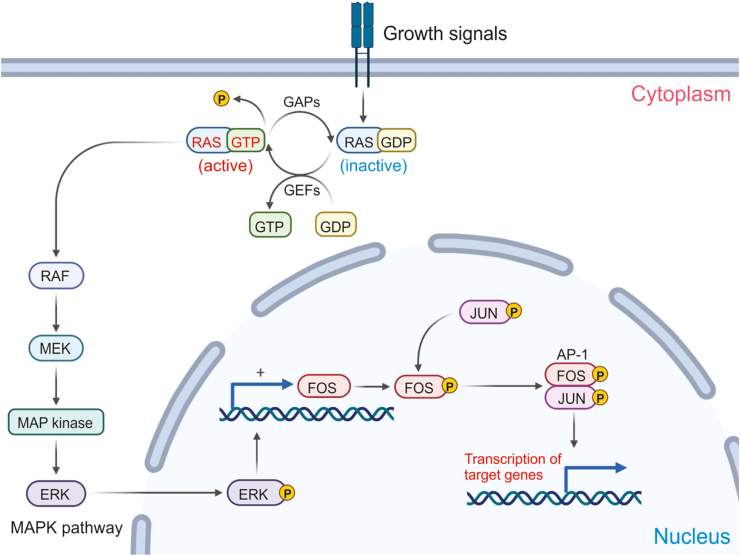
Aberrant activation of the BRAF kinase occurs in ∼50% of melanomas.17 Over 90% of those mutations are due to point mutations at codon 600 that result in the substitution of glutamic acid for valine.18 This constitutively active BRAF is not only implicated in tumor cell proliferation and survival through downstream MAPK effector proteins (Fig. 4), but it is also associated with increased tissue invasion, cell migration, metastasis, and evasion of the immune response.17 Furthermore, these mutations tend to be found in tumors that arise on skin that has not experienced chronic sun induced damage.
Figure 4.
The components of the mitogen activated protein kinase (MAPK) pathway. The signaling cascade begins with the binding of a mitogen or growth signal on a cell surface receptor. This binding process allows RAS to exchange GTP for GDP, thus continuing the phosphorylation cascade in the cytoplasm until ERK translocates into the nucleus. The illustrations were inspired by BioRender.
NRAS mutations
Mutations in NRAS—a small GTPase—are found in around 20% of all melanomas with wild type BRAF. 80% of those mutations are point mutations at position 61 that result in a glutamine–leucine substitution. Mutations at positions 12 and 13 are also prevalent though less so than the mutation at position 61. This leads to defective GTPase activity, accumulation of RAS-GTP, and insensitivity to physiological regulation by guanine nucleotide exchange factors (GEFs) and GTPase activating proteins (GAPs) (Fig. 4).19 Melanomas that harbor NRAS mutations tend to be associated with more aggressive tumors and poor health outcomes. This might be related to the fact that melanomas with NRAS mutations are more likely to have early initiation of the vertical growth phase.19 Patients with this variety of melanoma tend to be older (>55 years) and have a history of chronic UV exposure; though the UV exposure association is not as strong. Histologically, the lesions tend to be thicker and have higher mitotic activity with much lower rates of ulceration. However, there are higher rates of lymph node metastasis which may be related to increased cell motility and higher rates of epithelial mesenchymal transition (EMT) in NRAS mutated tumor cells.
Activation of MAPK pathway
MAPK signaling in NRAS mutated melanomas is slightly different than MAPK signaling through BRAF mutated melanomas (Fig. 4). NRAS mutated melanomas are reliant upon CRAF signaling instead of BRAF signaling.20 This occurs through two different mechanisms. The first mechanism involves BRAF inactivation via a negative feedback loop in which ERK limits BRAF signaling via phosphorylation. The second mechanism prevents CRAF inactivation by deregulating PKA via increased expression of phosphodiesterase IV. In spite of relying on a different RAF isoform, the downstream effectors MEK and ERK remain the same. The combination therapies mentioned in the BRAF section do show efficacy in NRAS mutated melanomas though, somewhat paradoxically, the impact is limited.
TERT promoter mutations
The aforementioned mutations in melanoma are in protein coding genes that are associated with the MAPK signaling pathway. Mutations in TERT are different because the mutations are found in a promoter gene. In fact, TERT mutations are found in around 70% of melanomas.21 The mutations—likely directly induced by UV damage—generate binding motifs for the ETS transcription factors that increase TERT transcriptional activity anywhere from two to fourfold. The increase in transcriptional activity essentially results in an increase in telomerase production. Though insufficient in bypassing oncogene-induced senescence, aberrant TERT activation may contribute to immortality in transformed melanocytes. Histologically, melanomas with TERT mutations tended to have thicker lesions with high mitotic activity and all of these characteristics were found in tumors of older patients. Interestingly enough, TERT mutations alone were not associated with tissue invasion rates, tumor ulceration, tumor necrosis, primary tumor location, or reduced survival.22 In one study, TERT mutations were associated with wild type BRAF, but the association was not statistically significant. One hypothesis is that TERT mutations allow neoplastic cells to survive long enough to develop other more important driver mutations as the disease progresses.
TERT expression is significantly higher in tumor cells than it is in normal cells, it is a promising therapeutic target.23 However, targeting telomerase proves to be a difficult task. Some of the challenge can be attributed to the fact that there is a very long lag time in order for such treatments to take effect. That lag time allows other mutations such as BRAF or NRAS to take over and drive carcinogenesis.
Metastatic program of melanoma
Biological characteristics of metastatic melanoma
Metastasis is a multistep process involving the progression from normal melanocytes to metastatic melanoma via the accumulation of genetic and epigenetic changes. In the Clark model, metastasis occurs late in the disease and metastatic cells emerge in a distant site.24 However, in the parallel progression model of metastasis,25,26 abnormal cells can spread earlier during primary tumor formation and can persist and subsequently transform to overt metastatic tumors in distant sites.27,28 The early mutations, such as BRAFV600E or NRASQ61 K/R/L, can play an important role in the dissemination of epithelial cells. In the parallel progression model, it is suggested that genetic and epigenetic signatures are distinct in metastases and primary tumors of individual patients. Prior studies have compared the DNA sequence of matched metastatic and primary melanomas and revealed both relative homogeneity29,30 and heterogeneity.31 These observations suggest that early dissemination with parallel progression and late dissemination with limited evolution at distant sites are both pathways with potential to give rise to clinically meaningful metastatic disease.
As previously highlighted, distant metastasis is associated with the worst health outcome for melanoma. In order for distant metastasis to occur the following steps are required: 1) tumor cells must dissociate from the primary tumor mass; 2) those cells must survive being transported to a new niche; and 3) the cells must respond to growth and proliferation signaling in the new niche in order to develop into a metastatic mass.32 Accomplishing those steps is no small feat and requires coordination of gene expression. Many of those genes, located on chromosome 6, were highlighted for their ability to suppress metastasis.33
Knockout of Kiss-1 resulted in neoplastic proliferation after the tumor cells seeded themselves in the metastatic niche.34 This is a significant gene because proliferation in the new niche is arguably the most important step of metastasis. As the size of the tumor increases, Kiss-1 expression generally decreases. Researchers are exploring the use of pharmaceuticals to prevent Kiss-1 secretion in order to suppress metastasis. However, a complex process like metastasis requires several genes. With the silencing of Pten, melanocytes undergo transformation and subsequent metastasis; thereby making Pten silencing a molecular marker for metastatic melanoma.35 The clinical impact of losing another tumor suppressor is huge as the 10-year survival rate for patients with metastatic melanoma can be less than 10%.36 The fact that tumor cells take residence in several different metastatic niches makes targeting metastasis with a pharmaceutical particularly challenging in spite of having direct molecular events associated with the process.
Metastatic progression of melanoma requires secondary genetic drivers
The metastatic progression of melanoma can also be guided by secondary genetic or phenotypic drivers (Fig. 5). For example, there is an increase in the TERT promoter mutations in metastases compared to primary melanomas, which represents an independent prognostic factor.21,37,38 A phenotypic driver of metastasis includes the stage-dependent regulation of anti-apoptotic proteins, such as the expression of BCL-2, which progressively decreases from the radial tumor growth phase to the vertical tumor growth phase while the expression levels of MCL1, BCL-XL, survivin (also known as BIRC5) and XIAP increase.39 The role of differentiation factors in metastasis is largely controlled by microphthalmia-associated transcription factor (MITF) and canonical WNT signaling. MITF is a transcription factor central to melanocyte survival, proliferation, and melanin-pigment production.40 MIFT amplification has been found in 21% of metastases compared to 10% of primary melanomas.41 The WNT signaling through β-catenin promotes melanocytic differentiation through direct transcriptional upregulation of MIFT.42 Oncogenic β-catenin mutations are thought to lead to constitutive activation of the canonical WNT pathway.43, 44, 45, 46 Recent studies suggest MITF and WNT signaling can display either protumorigenic or antitumorigenic properties depending on the microenvironment.47
Figure 5.
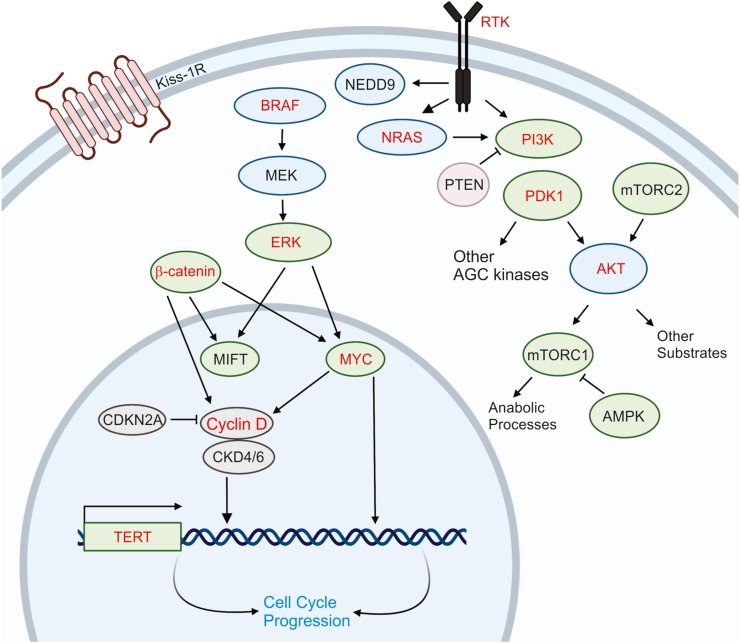
Crosstalk of major signaling pathways in melanoma metastasis. The interactions of several different signaling pathways, such as RAS/BRAF/MAPK pathway, PI3K/AKT/mTOR pathway, and β-catenin pathway are frequently involved in melanoma metastasis. Several of the components of the MAPK signaling pathway that is responsible for the transformation of melanocytes also play key roles in the melanoma metastasis. The illustrations were inspired by BioRender.
The mitogen-activated protein kinase (MAPK)/extracellular-signal-regulated kinase (ERK) pathway is the most mutated pathway in melanoma, with BRAF and NRAS being the most mutated components (Fig. 5).48 Epidemiologic observations from human melanomas suggest there is no difference in rates of metastasis between BRAF and NRAS mutant melanomas,49,50 though both may exhibit higher rates of metastasis compared to RAS/RAF wild type melanomas.51 The PI3K/AKT pathway is also frequently constitutively activated in melanoma through genetic or epigenetic inactivation of PTEN tumor suppressor52,53 and copy number gains or overexpression of AKT3 (Fig. 5).54,55 The loss of PTEN and subsequent activation of PI3K/AKT pathway drives the metastasis of melanomas initiated by activating RAS and RAF mutations.56,57 In these models, activation of PI3K/AKT is also critical in primary tumor formation, which makes it challenging to infer its specific role in metastasis (Fig. 5). Other genetic changes that have been more commonly identified in metastases compared to primary tumors include loss-of-function of CDKN2A mutations and chromosome 1 deletions causing loss of Kiss-1.50,58
A recent study by Deng et al showed the expression of Wnt-inducible signaling protein 1 (WISP1), a downstream effector of the Wnt/β-catenin pathway, is increased in melanoma and associated with reduced overall survival in patients diagnosed with primary melanoma.59 WISP1 is a soluble signal released by melanoma cells to reshape their microenvironment, enhanced tumor invasion, and metastasis by promoting an EMT-like process. WISP1 specifically upregulates EMT transcription factors and mesenchymal markers and represses E-cadherin and MITF. The role of WISP1 in melanoma has been previously studied in the context of fibroblasts and Notch signaling.60 It is suggested that WISP1 is a downstream target of Notch signaling, an intercellular signaling cascade that is activated in human melanoma cells and is essential for their growth and metastasis. Prior studies have concluded that Notch interacts with Wnt/β-catenin signaling in synergistic or antagonistic ways depending on the context.60
In the last decade, large scale approaches have been undertaken to identify metastasis regulators in melanoma. Functional assays have confirmed the pro-invasive properties of MET, ASPM, AKAP9, IMP3, PRKCA, RPA3, and SCAP2.61,62 Another study comparing metastatic and primary melanomas from an inducible mouse melanoma model identified gains in chromosome 6p and overexpression of NEDD9 in metastatic tumors. Functional assays have confirmed the overexpression of NEDD9 in tumors with activating BRAF or RAS mutations increased invasiveness in vitro and in vivo compared to BRAF or RAS melanomas lacking NEDD9 overexpression.63 Further studies are necessary to further explore the role of these genes and their interactions with other metastasis regulators.
Targeted therapies for melanoma
Targeting mutant BRAF
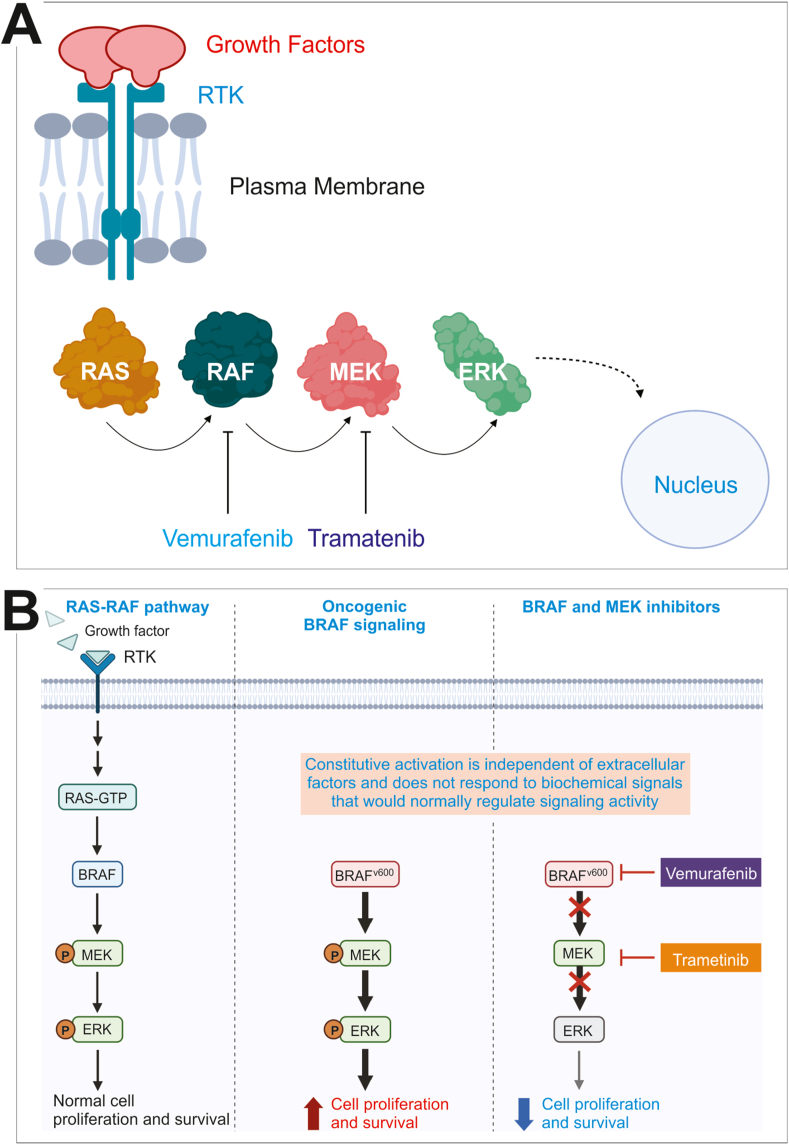
Inhibitors of BRAF (BRAFi) are the prototype targeted therapies that have met clinical successes, but also hampered by acquired resistance (Fig. 6A).64 The BRAFV600E mutation is associated with sensitivity to a class of agents that block hyperactive BRAFV600E activity. The most clinically effective BRAFi's are vemurafenib and dabrafenib. Although their significant clinical success is well documented, acquired resistance invariably occurs in most patients. Clinical data show acquired resistance develops as early as 2 months and as late as 18 months into treatment. It has been shown that resistance mechanism often involves reactivation of MAPK signaling. Though, BRAFV600E mutations account for more than 90% of BRAF mutations in melanoma, the drug that targets that mutant protein drives resistance by increasing activity of mutant BRAF monomers, increasing expression of PDGF receptors on the cell surface, increasing upstream NRAS mutations, and by increasing tumor cell sensitivity to HGF secreted by proximate stromal cells.65,66
Figure 6.
Targeted therapies and resistance for melanoma. (A) This panel depicts the major players in the MAPK signaling pathway along with two drugs used to inhibit the pathway. Vemurafenib is a RAF inhibitor, while Trametinib is an MEK inhibitor. (B) Combination therapy and targeted therapy resistance. BRAF and MEK inhibitors can be used together to overcome the resistance to BRAF inhibitors. However, tumors can use other mechanisms to escape BRAF/MEK inhibitor treatment, including up-regulating RAS and/or PI3K-AKT pathways. Some tumor cells have mutations in RAS, RAF, or MEK that allow them to escape canonical BRAF/MEK inhibitor treatment, while others are able to grow and proliferate due to stromal cells that secrete high levels of HGH (see the text). The illustrations were inspired by BioRender.
Targeting mutant NRAS
In spite of being the first oncogene identified in melanoma, treating melanomas with NRAS mutations has been extremely difficult. Farnesyl transferase inhibitors were designed to prevent post translational modification of RAS and its insertion into the plasma membrane thereby preventing RAS activation. Non-specificity might have contributed to such a low impact on NRAS mutated tumors because so many membrane bound proteins are farnesylated. Salirasib is a small molecule that disrupts RAS localization at the plasma membrane resulting in death of RAS transformed cells.19 This occurs via blocking RAS-GTP from binding galactin 1 on the plasma membrane. Salirasib has shown some promise at the preclinical level, though further investigation is required. Interfering RNAs are another tool used to target NRAS mutated melanomas. The approach has been validated in preclinical models, but delivery is a significant challenge due to the instability of nucleic acids in circulation.
Targeting the activated MAPK pathway
Targeting downstream components of the MAPK signaling pathway with MEK inhibitors decreased resistance, but only in the short term (Fig. 6B). Although MEK acts downstream of RAF, there is a relatively low incidence of MEK mutations in human cancers.67 MEK inhibitors—like trametinib—that allosterically bind both MEK1 and MEK2 increased progression free survival in most patients.68 However, acquired resistance to combination therapy eventually emerges. This suggests that tumor cells escape combination therapy downstream via ERK or upstream via the RAS-PI3K axis.
Immune checkpoint inhibitors and immunotherapies in melanoma treatment
In the early 1900s Paul Ehrlich suggested that cancer rates would be higher were it not for the immune system.69,70 More than a half century later, the immunosurveillance theory that suggested that the adaptive immune system is responsible for preventing cancer in immunocompetent hosts came to be. Since then, the hypotheses about how the role the immune system plays in cancer have become more developed and with that development came the advent of immunotherapy (Fig. 7); the most effective of which are checkpoint inhibitors.71 Generally speaking, checkpoint inhibitors are ligands that bind receptors that trigger signaling pathways that prevent or dampen an immune response to a pathogen.
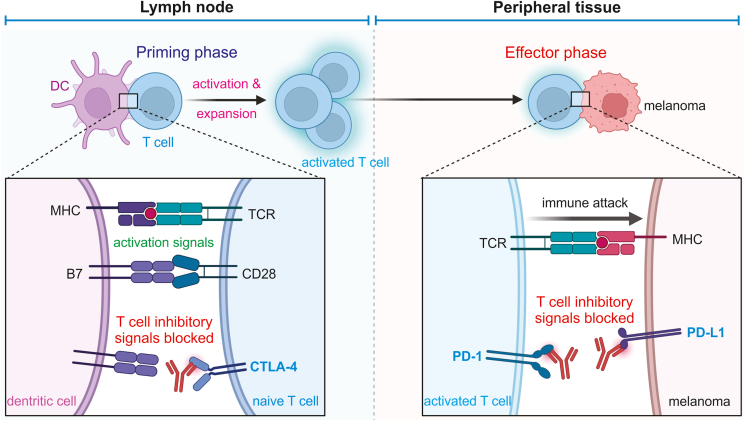
Figure 7.
Molecular immunology of immunotherapies for melanoma. Naïve T cells can be activated by interacting with dendritic cells (DC) through T-cell receptor (TCR), which is inhibited by B7/CTLA-4 interaction. Activated T cells launch immune attack on melanoma cells, which can be inhibited by PD-L1/PD-1 interaction. Therefore, targeted immunotherapeutic antibodies have been developed to inhibit CTLA-4, PD-L1 and/or PD-1 to treat melanoma and other types of cancer. The illustrations were inspired by BioRender.
First antibodies blocking CTLA-4 were developed (Fig. 7). The next generation of checkpoint inhibitors were PD-1 and PD-L1 monoclonal antibodies (Fig. 7). Generally speaking, checkpoint inhibitors were extremely effective at unlocking the ability of effector T cells to reduce tumor burden. In early clinical trials, treatment with ipilimumab, the CTLA-4 blockade molecule, resulted in 1- and 2-year survival rates of 46% and 24%, respectively. Treatment with PD-1 resulted in even better results. About 20% of melanoma patients treated with anti-PD-1 antibodies with and without the addition of anti-CTLA-4 antibodies experience complete remission. It is even understood that treatment can end after 6 months of therapy—thereby reducing side effects—because the chance of relapse is estimated as less than 5% over 5 years.
Though checkpoint inhibitors have shown great clinical success, the extent of that success is truly found in a minority of patients. Many patients experience either primary resistance or acquired resistance to those therapies. Some of the difference in patient outcomes can be attributed to variance in the density of immune cells in the tumor tissue. Patients with hot tumors or tumors with high immune cell infiltrate tend to respond better to checkpoint blockade therapy than patients with cold tumors. Elevated serum LDH levels are associated with primary resistance as well. Increased LDH expression in the tumor microenvironment results in increased activation of immunosuppressive cell types like myeloid derived suppressor cells and tumor associated macrophages. Much less is understood about acquired resistance to checkpoint inhibitors. One hypothesis is that checkpoint inhibitors act as a selective pressure.
Treatment eventually results in a tumor cell population that is resistant to the drugs. One marker of that resistant population is low beta-2-microglobulin (B2M). B2M is required for functional MHC-1 expression and MHC-1 is required to present tumor antigen to cytotoxic T cells. Additionally, acquired resistance can develop as the result of additional immune checkpoint marker upregulation such as TIM-3, LAG-3, and HAVCR-2. This type of acquired resistance shows that blocking one pathway toward immunosuppression might result in activation of other immunosuppressive pathways.
Potential roles of noncoding RNAs in melanoma development and treatment
As previously mentioned, both molecular targeted therapies and immunotherapies are initially quite efficacious when it comes to treating melanoma. However, acquired resistance is a major and, often, inevitable obstacle. Given that an estimated 2% of the genome is transcribed into mRNA, there is a high likelihood that noncoding RNA plays a crucial role in the epigenetic landscape of acquired drug resistance. This is especially significant given that dysregulation of noncoding RNA is linked to all cancers and affects all major cancer hallmarks.72
Noncoding RNAs can be separated into four large categories: ribosomal RNA, transfer RNA, short noncoding RNA, and long noncoding RNA.73 Ribosomal RNAs are molecular building blocks that bind ribosomal proteins resulting in small and large ribosomal subunits. Transfer RNAs are responsible for shuttling amino acids to the ribosome in the protein polymerization process. Long noncoding RNA molecules are typically longer than 200 nucleotides and make up the largest class of noncoding RNAs. Long noncoding RNAs can be divided into the following: long intergenic ncRNAs, antisense RNAs, pseudogenes, and circular RNAs. Short noncoding RNA are typically less than 200 nucleotides and can be divided into the following categories: microRNAs, siRNAs, snoRNAs, and piwi-interacting RNAs. Together, long and short noncoding RNAs interact with DNA, RNA, proteins, and micropeptides to induce epigenetic changes.
It is likely that short and long noncoding RNAs (lncRNAs) work in tandem to contribute to acquired targeted therapy resistance in melanoma. The miRNA, miR-211-5p, is the most differentially expressed miRNA between melanoma cell lines and normal melanocytes.74 Transformed melanocytes down regulate miR-211-5p, which results in decreased MITF, a transcription factor responsible for transcription of enzymes specific to melanogenesis. It has been demonstrated that melanocytes treated with vemurafenib express less pigmentation and that condition is reversed as the tumor cells develop resistance. Long noncoding RNAs such as BANCR further contribute to the development of melanoma by increasing cell proliferation via downstream ERK activation.75 One group demonstrated that inhibition of SAMMSON in addition to treatment with dabrafenib resulted in tumor regression in patient derived melanoma xenograft models.76 The interplay between short and long noncoding RNAs demonstrates that targeting both types of molecules will be necessary to combat acquired molecular targeted therapy resistance. Though promising, noncoding RNA therapeutics have familiar clinical obstacles such as trouble with off target interactions, inefficient delivery, and tolerability. Low clinical efficacy is the primary reason RNA cancer therapeutics have been discontinued in clinical studies.
Other treatment strategies for melanoma
Targeted therapies and immunotherapies have been widely utilized to treat melanomas of various types and stages. Other strategies, including surgical resection, chemotherapy, radiotherapy, and photodynamic therapy (PDT) have also been used as treatment.71 The use of these seemingly alternative treatment strategies has depended heavily on the features of the melanoma tumors and cells, including their relative locations, sizes, stages, and genomic sequences.71 Oftentimes, a combination of these strategies proves useful in addressing the symptoms of melanomas and the respective genetic and molecular causes.
Surgical resection of melanoma
Although the surgical removal of tumors has proved to be effective in treating melanoma, the success of the surgery is highly dependent on the stage of the cancer. In the earlier stages of melanoma, prior to metastasis, the surgical resection of the tumor can be a successful treatment.77 Once the cancer metastasizes, survival rates are significantly decreased when treated with surgical resection.77 Prior to the use of immunotherapy, the five-year survival rate for metastatic melanoma treated with surgical resection was 15–20%.78 Thus, the management of primary cutaneous melanomas can be handled by surgical resection whereas more advanced stages require additional therapeutic remedies.79 Surgery is the optimal treatment for patients with melanoma tumors in stages I–IIIB, although procedures for the surgery will differ depending on the clinical features of the tumors.71 Different excisions and safety margins exist for these surgeries; 0.5 cm for in situ melanomas, 1 cm for 2 mm thick tumors, and 2 cm for tumors thicker than 2 mm.71 Sentinel lymph node biopsy is also frequently done on patients who have melanoma tumors larger than 0.8 mm thick or thinner and ulcerated.77 In the cases that melanoma cells are present in the lymph nodes, the lymph nodes and surrounding area are often surgically removed.77 Surgical methodology similarly depends on clinical features of the tumor, although the use of Mohs micrographic surgery has been documented for the treatment of primary cutaneous melanomas.78
Chemotherapy and radiotherapy of melanoma
The use of chemotherapy for the treatment of metastatic melanoma began in the late 1960s with the use of melphalan, or 1-phenylalanine mustard.77 Melphalan proved to be both ineffective and highly toxic and did not make it past the clinical trials for its treatment of metastatic melanoma.77 The FDA then approved dacarbazine, or dimethyltriazeno-imidazole carboxamide, in 1975 and it has served as both the first and only chemotherapeutic drug approved by the FDA for melanoma.79 Dacarbazine works as an alkylating agent that generates methyl isothiocyanate (MITC) and exerts cytotoxic effects by preventing DNA replication.79 Other single agent chemotherapies, including the antimicrotubule agent vindesine, have shown a response rate of under 20% when used alone.79 The combination of dacarbazine and vindesine, however, have failed to show an increased response rate in metastatic melanoma patients.79 More recently, dacarbazine no longer serves as the standard of care for metastatic melanoma due to its median survival from 5 to 11 months and 1-year survival rate of 27%.77 To date, chemotherapy drugs have not been more effective in terms of survival rates, and none have been less toxic than dacarbazine.77 Therefore, chemotherapy use has decreased over time. Similarly, radiation therapy provides little clinical benefit as a treatment modality for melanoma due to the fact that melanoma is relatively radioresistant.36
Photodynamic therapy (PDT) of melanoma
Photodynamic therapy (PDT) is a relatively novel non-invasive treatment technique used to treat non-melanoma skin cancer, breast cancer, lung cancer, and other forms of cancer.80,81 Its use as a treatment for melanoma is not completely understood but studies examining the efficacy of PDT as a treatment for melanoma has proved promising.80 PDT works by using a non-toxic photoactivatable drug called photosensitizer to trigger photochemical reactions of tumor cells while minimizing the damage to normal tissues.80,82 In the case of localized and not malignant melanoma, surgery remains important in the treatment of melanoma. For more aggressive malignant tumors, the use of PDT can be effective due to its minimally invasive characteristics and relatively low side effect profile. Further research into PDT is required to determine efficacy.83
Conclusions and future directions
Melanoma is a fairly common and potentially fatal cancer that is relatively easy to sample given its location. Having such easy access to patient samples has helped the scientific community really learn a lot about the disease and its process. We have managed to identify the most prevalent mutations that initiate melanoma in BRAF, NRAS, and TERT genes. Targeted therapies were developed with this in mind. However, the prevalence of acquired resistance to those therapies demonstrates that those pathways are not as simple as we thought. Immunotherapies were supposed to be the answer to the issues of acquired resistance to targeted therapies. Arguably, they showed even greater initial success than targeted therapies. However, immunotherapies are successful in a minority of patients but many patients develop acquired resistance. It appears that treating melanoma requires combination therapies. As the protocol for combination therapies develop, so will our scientific understanding of the many pathways at play in the progression of melanoma. Perhaps the basic science will follow a similar trend. The future direction of the field may be to find a molecule that connects all of the pathways. It has been suggested that noncoding RNA is a strong candidate for understanding how all of the pathways of melanoma relate to one another. Studying the networks of noncoding RNA are particularly useful in understanding how resistance—both primary and acquired—develops. The more we understand the role of noncoding RNA in melanoma the more we will be able to harness the true potential of the therapies we already have.
Conflict of interests
The authors declare no competing conflicts of interests.
Funding
The reported work was supported in part by research grant from the National Institutes of Health (CA226303 to TCH, and DE030480 to RRR). WW was supported by the Medical Scientist Training Program of the National Institutes of Health (T32 GM007281). This project was also supported in part by The University of Chicago Cancer Center Support Grant (P30CA014599) and the National Center for Advancing Translational Sciences of the National Institutes of Health through Grant Number UL1 TR000430. TCH was also supported by the Mabel Green Myers Research Endowment Fund and The University of Chicago Orthopaedics Alumni Fund. Funding sources were not involved in the study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the paper for publication.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Contributor Information
Tong-Chuan He, Email: tche@uchicago.edu.
Rex C. Haydon, Email: rhaydon@bsd.uchicago.edu.
References
- 1.Miller A.J., Mihm M.C., Jr. Melanoma. N Engl J Med. 2006;355(1):51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- 2.Eggermont A.M., Spatz A., Robert C. Cutaneous melanoma. Lancet. 2014;383(9919):816–827. doi: 10.1016/S0140-6736(13)60802-8. [DOI] [PubMed] [Google Scholar]
- 3.Kanitakis J. Anatomy, histology and immunohistochemistry of normal human skin. Eur J Dermatol. 2002;12(4):390–399. [PubMed] [Google Scholar]
- 4.Kolarsick P., Kolarsick M., Goodwin C. Anatomy and physiology of the skin. J Dermatol Nurses Assoc. 2011;3(4):203–213. [Google Scholar]
- 5.Freedberg I., Eisen A., Wolff K., Austen K., Goldsmith L., Katz S. 6th ed. McGraw-Hill Education; 2003. Fitzpatrick's Dermatology in General Medicine. [Google Scholar]
- 6.D'Mello S.A., Finlay G.J., Baguley B.C., Askarian-Amiri M.E. Signaling pathways in melanogenesis. Int J Mol Sci. 2016;17(7):1144. doi: 10.3390/ijms17071144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cichorek M., Wachulska M., Stasiewicz A., Tymińska A. Skin melanocytes: biology and development. Postepy Dermatol Alergol. 2013;30(1):30–41. doi: 10.5114/pdia.2013.33376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maddodi N., Jayanthy A., Setaluri V. Shining light on skin pigmentation: the darker and the brighter side of effects of UV radiation. Photochem Photobiol. 2012;88(5):1075–1082. doi: 10.1111/j.1751-1097.2012.01138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halpern A.C., Marghoob A.A., Ofer R. The Skin Cancer Foundation; 2021. Melanoma. Melanoma Overview.https://www.skincancer.org/skin-cancer-information/melanoma/ [Google Scholar]
- 10.Bolognia J.L., Schaffer J.V., Cerroni L. 4th ed. Elsevier; 2017. Dermatology. [Google Scholar]
- 11.Superficial Spreading Melanoma. Memorial Sloan Kettering Cancer Center; 2021. https://www.mskcc.org/cancer-care/types/melanoma/types-melanoma/superficial-spreading-melanoma [Google Scholar]
- 12.Goydos J.S., Shoen S.L. Acral lentiginous melanoma. Cancer Treat Res. 2016;167:321–329. doi: 10.1007/978-3-319-22539-5_14. [DOI] [PubMed] [Google Scholar]
- 13.Menzies S.W., Moloney F.J., Byth K., et al. Dermoscopic evaluation of nodular melanoma. JAMA Dermatol. 2013;149(6):699–709. doi: 10.1001/jamadermatol.2013.2466. [DOI] [PubMed] [Google Scholar]
- 14.van der Meijden W.A., van Bruchem-Visser R.L., Thio H.B., van der Cammen T.J. Melanomas more serious in the elderly. Ned Tijdschr Geneeskd. 2010;154:A1535. [PubMed] [Google Scholar]
- 15.Keung E.Z., Gershenwald J.E. The eighth edition American Joint Committee on Cancer (AJCC) melanoma staging system: implications for melanoma treatment and care. Expert Rev Anticancer Ther. 2018;18(8):775–784. doi: 10.1080/14737140.2018.1489246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Potrony M., Badenas C., Aguilera P., et al. Update in genetic susceptibility in melanoma. Ann Transl Med. 2015;3(15):210. doi: 10.3978/j.issn.2305-5839.2015.08.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ribas A., Flaherty K.T. BRAF targeted therapy changes the treatment paradigm in melanoma. Nat Rev Clin Oncol. 2011;8(7):426–433. doi: 10.1038/nrclinonc.2011.69. [DOI] [PubMed] [Google Scholar]
- 18.Ascierto P.A., Kirkwood J.M., Grob J.J., et al. The role of BRAF V600 mutation in melanoma. J Transl Med. 2012;10:85. doi: 10.1186/1479-5876-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelleher F.C., McArthur G.A. Targeting NRAS in melanoma. Cancer J. 2012;18(2):132–136. doi: 10.1097/PPO.0b013e31824ba4df. [DOI] [PubMed] [Google Scholar]
- 20.Fedorenko I.V., Gibney G.T., Smalley K.S. NRAS mutant melanoma: biological behavior and future strategies for therapeutic management. Oncogene. 2013;32(25):3009–3018. doi: 10.1038/onc.2012.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang F.W., Hodis E., Xu M.J., Kryukov G.V., Chin L., Garraway L.A. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339(6122):957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hugdahl E., Kalvenes M.B., Mannelqvist M., Ladstein R.G., Akslen L.A. Prognostic impact and concordance of TERT promoter mutation and protein expression in matched primary and metastatic cutaneous melanoma. Br J Cancer. 2018;118(1):98–105. doi: 10.1038/bjc.2017.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reyes-Uribe P., Adrianzen-Ruesta M.P., Deng Z., et al. Exploiting TERT dependency as a therapeutic strategy for NRAS-mutant melanoma. Oncogene. 2018;37(30):4058–4072. doi: 10.1038/s41388-018-0247-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clark W.H., Jr., Elder D.E., Guerry D., 4th, Epstein M.N., Greene M.H., Van Horn M. A study of tumor progression: the precursor lesions of superficial spreading and nodular melanoma. Hum Pathol. 1984;15(12):1147–1165. doi: 10.1016/s0046-8177(84)80310-x. [DOI] [PubMed] [Google Scholar]
- 25.Klein C.A. Parallel progression of primary tumours and metastases. Nat Rev Cancer. 2009;9(4):302–312. doi: 10.1038/nrc2627. [DOI] [PubMed] [Google Scholar]
- 26.Stoecklein N.H., Klein C.A. Genetic disparity between primary tumours, disseminated tumour cells, and manifest metastasis. Int J Cancer. 2010;126(3):589–598. doi: 10.1002/ijc.24916. [DOI] [PubMed] [Google Scholar]
- 27.Podsypanina K., Du Y.C., Jechlinger M., Beverly L.J., Hambardzumyan D., Varmus H. Seeding and propagation of untransformed mouse mammary cells in the lung. Science. 2008;321(5897):1841–1844. doi: 10.1126/science.1161621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hüsemann Y., Geigl J.B., Schubert F., et al. Systemic spread is an early step in breast cancer. Cancer Cell. 2008;13(1):58–68. doi: 10.1016/j.ccr.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 29.Gartner J.J., Davis S., Wei X., et al. Comparative exome sequencing of metastatic lesions provides insights into the mutational progression of melanoma. BMC Genomics. 2012;13:505. doi: 10.1186/1471-2164-13-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turajlic S., Furney S.J., Lambros M.B., et al. Whole genome sequencing of matched primary and metastatic acral melanomas. Genome Res. 2012;22(2):196–207. doi: 10.1101/gr.125591.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pleasance E.D., Cheetham R.K., Stephens P.J., et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463(7278):191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee J.H., Miele M.E., Hicks D.J., et al. KiSS-1, a novel human malignant melanoma metastasis-suppressor gene. J Natl Cancer Inst. 1996;88(23):1731–1737. doi: 10.1093/jnci/88.23.1731. [DOI] [PubMed] [Google Scholar]
- 33.Goldberg S.F., Miele M.E., Hatta N., et al. Melanoma metastasis suppression by chromosome 6: evidence for a pathway regulated by CRSP3 and TXNIP. Cancer Res. 2003;63(2):432–440. [PubMed] [Google Scholar]
- 34.Nash K.T., Phadke P.A., Navenot J.M., et al. Requirement of KISS1 secretion for multiple organ metastasis suppression and maintenance of tumor dormancy. J Natl Cancer Inst. 2007;99(4):309–321. doi: 10.1093/jnci/djk053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Damsky W.E., Rosenbaum L.E., Bosenberg M. Decoding melanoma metastasis. Cancers (Basel) 2010;3(1):126–163. doi: 10.3390/cancers3010126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhatia S., Tykodi S.S., Thompson J.A. Treatment of metastatic melanoma: an overview. Oncology (Williston Park) 2009;23(6):488–496. [PMC free article] [PubMed] [Google Scholar]
- 37.Horn S., Figl A., Rachakonda P.S., et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339(6122):959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 38.Griewank K.G., Murali R., Puig-Butille J.A., et al. TERT promoter mutation status as an independent prognostic factor in cutaneous melanoma. J Natl Cancer Inst. 2014;106(9):dju246. doi: 10.1093/jnci/dju246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hartman M.L., Czyz M. Anti-apoptotic proteins on guard of melanoma cell survival. Cancer Lett. 2013;331(1):24–34. doi: 10.1016/j.canlet.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 40.Mitra D., Fisher D.E. Transcriptional regulation in melanoma. Hematol Oncol Clin North Am. 2009;23(3):447–465. doi: 10.1016/j.hoc.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 41.Garraway L.A., Widlund H.R., Rubin M.A., et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436(7047):117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 42.Widlund H.R., Horstmann M.A., Price E.R., et al. Beta-catenin-induced melanoma growth requires the downstream target Microphthalmia-associated transcription factor. J Cell Biol. 2002;158(6):1079–1087. doi: 10.1083/jcb.200202049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Demunter A., Libbrecht L., Degreef H., De Wolf-Peeters C., van den Oord J.J. Loss of membranous expression of beta-catenin is associated with tumor progression in cutaneous melanoma and rarely caused by exon 3 mutations. Mod Pathol. 2002;15(4):454–461. doi: 10.1038/modpathol.3880546. [DOI] [PubMed] [Google Scholar]
- 44.Forbes S.A., Tang G., Bindal N., et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38(Database issue):D652–D657. doi: 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Omholt K., Platz A., Ringborg U., Hansson J. Cytoplasmic and nuclear accumulation of beta-catenin is rarely caused by CTNNB1 exon 3 mutations in cutaneous malignant melanoma. Int J Cancer. 2001;92(6):839–842. doi: 10.1002/ijc.1270. [DOI] [PubMed] [Google Scholar]
- 46.Hodis E., Watson I.R., Kryukov G.V., et al. A landscape of driver mutations in melanoma. Cell. 2012;150(2):251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Damsky W.E., Theodosakis N., Bosenberg M. Melanoma metastasis: new concepts and evolving paradigms. Oncogene. 2014;33(19):2413–2422. doi: 10.1038/onc.2013.194. [DOI] [PubMed] [Google Scholar]
- 48.Lopez-Bergami P. The role of mitogen- and stress-activated protein kinase pathways in melanoma. Pigment Cell Melanoma Res. 2011;24(5):902–921. doi: 10.1111/j.1755-148X.2011.00908.x. [DOI] [PubMed] [Google Scholar]
- 49.Colombino M., Capone M., Lissia A., et al. BRAF/NRAS mutation frequencies among primary tumors and metastases in patients with melanoma. J Clin Oncol. 2012;30(20):2522–2529. doi: 10.1200/JCO.2011.41.2452. [DOI] [PubMed] [Google Scholar]
- 50.Rozenberg G.I., Monahan K.B., Torrice C., Bear J.E., Sharpless N.E. Metastasis in an orthotopic murine model of melanoma is independent of RAS/RAF mutation. Melanoma Res. 2010;20(5):361–371. doi: 10.1097/CMR.0b013e328336ee17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jakob J.A., Bassett R.L., Jr, Ng C.S., et al. NRAS mutation status is an independent prognostic factor in metastatic melanoma. Cancer. 2012;118(16):4014–4023. doi: 10.1002/cncr.26724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsao H., Zhang X., Benoit E., Haluska F.G. Identification of PTEN/MMAC1 alterations in uncultured melanomas and melanoma cell lines. Oncogene. 1998;16(26):3397–3402. doi: 10.1038/sj.onc.1201881. [DOI] [PubMed] [Google Scholar]
- 53.Zhou X.P., Gimm O., Hampel H., Niemann T., Walker M.J., Eng C. Epigenetic PTEN silencing in malignant melanomas without PTEN mutation. Am J Pathol. 2000;157(4):1123–1128. doi: 10.1016/S0002-9440(10)64627-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ko J.M., Velez N.F., Tsao H. Pathways to melanoma. Semin Cutan Med Surg. 2010;29(4):210–217. doi: 10.1016/j.sder.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 55.Stahl J.M., Sharma A., Cheung M., et al. Deregulated Akt3 activity promotes development of malignant melanoma. Cancer Res. 2004;64(19):7002–7010. doi: 10.1158/0008-5472.CAN-04-1399. [DOI] [PubMed] [Google Scholar]
- 56.Dankort D., Curley D.P., Cartlidge R.A., et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41(5):544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nogueira C., Kim K.H., Sung H., et al. Cooperative interactions of PTEN deficiency and RAS activation in melanoma metastasis. Oncogene. 2010;29(47):6222–6232. doi: 10.1038/onc.2010.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kotani M., Detheux M., Vandenbogaerde A., et al. The metastasis suppressor gene KiSS-1 encodes kisspeptins, the natural ligands of the orphan G protein-coupled receptor GPR54. J Biol Chem. 2001;276(37):34631–34636. doi: 10.1074/jbc.M104847200. [DOI] [PubMed] [Google Scholar]
- 59.Deng W., Fernandez A., McLaughlin S.L., Klinke D.J., 2nd WNT1-inducible signaling pathway protein 1 (WISP1/CCN4) stimulates melanoma invasion and metastasis by promoting the epithelial-mesenchymal transition. J Biol Chem. 2019;294(14):5261–5280. doi: 10.1074/jbc.RA118.006122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bedogni B. Notch signaling in melanoma: interacting pathways and stromal influences that enhance Notch targeting. Pigment Cell Melanoma Res. 2014;27(2):162–168. doi: 10.1111/pcmr.12194. [DOI] [PubMed] [Google Scholar]
- 61.Kabbarah O., Nogueira C., Feng B., et al. Integrative genome comparison of primary and metastatic melanomas. PLoS One. 2010;5(5):e10770. doi: 10.1371/journal.pone.0010770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yan J., Huang Q. Genomics screens for metastasis genes. Cancer Metastasis Rev. 2012;31(3–4):419–428. doi: 10.1007/s10555-012-9362-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim M., Gans J.D., Nogueira C., et al. Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell. 2006;125(7):1269–1281. doi: 10.1016/j.cell.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 64.Das Thakur M., Salangsang F., Landman A.S., et al. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature. 2013;494(7436):251–255. doi: 10.1038/nature11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Straussman R., Morikawa T., Shee K., et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487(7408):500–504. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Proietti I., Skroza N., Bernardini N., et al. Mechanisms of acquired BRAF inhibitor resistance in melanoma: a systematic review. Cancers (Basel). 2020;12(10):2801. doi: 10.3390/cancers12102801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao Y., Adjei A.A. The clinical development of MEK inhibitors. Nat Rev Clin Oncol. 2014;11(7):385–400. doi: 10.1038/nrclinonc.2014.83. [DOI] [PubMed] [Google Scholar]
- 68.Grimaldi A.M., Simeone E., Ascierto P.A. The role of MEK inhibitors in the treatment of metastatic melanoma. Curr Opin Oncol. 2014;26(2):196–203. doi: 10.1097/CCO.0000000000000050. [DOI] [PubMed] [Google Scholar]
- 69.Lugowska I., Teterycz P., Rutkowski P. Immunotherapy of melanoma. Contemp Oncol (Pozn) 2018;22(1A):61–67. doi: 10.5114/wo.2018.73889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robert C. A decade of immune-checkpoint inhibitors in cancer therapy. Nat Commun. 2020;11(1):3801. doi: 10.1038/s41467-020-17670-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Domingues B., Lopes J.M., Soares P., Pópulo H. Melanoma treatment in review. Immunotargets Ther. 2018;7:35–49. doi: 10.2147/ITT.S134842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Winkle M., El-Daly S.M., Fabbri M., Calin G.A. Noncoding RNA therapeutics - challenges and potential solutions. Nat Rev Drug Discov. 2021;20(8):629–651. doi: 10.1038/s41573-021-00219-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chan J.J., Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int J Mol Sci. 2018;19(5):1310. doi: 10.3390/ijms19051310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Díaz-Martínez M., Benito-Jardón L., Alonso L., Koetz-Ploch L., Hernando E., Teixidó J. miR-204-5p and miR-211-5p contribute to BRAF inhibitor resistance in melanoma. Cancer Res. 2018;78(4):1017–1030. doi: 10.1158/0008-5472.CAN-17-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu X., Zheng H., Tse G., Chan M.T., Wu W.K. Long non-coding RNAs in melanoma. Cell Prolif. 2018;51(4):e12457. doi: 10.1111/cpr.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Leucci E., Vendramin R., Spinazzi M., et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature. 2016;531(7595):518–522. doi: 10.1038/nature17161. [DOI] [PubMed] [Google Scholar]
- 77.Davis L.E., Shalin S.C., Tackett A.J. Current state of melanoma diagnosis and treatment. Cancer Biol Ther. 2019;20(11):1366–1379. doi: 10.1080/15384047.2019.1640032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Patel H., Yacoub N., Mishra R., et al. Current advances in the treatment of BRAF-mutant melanoma. Cancers (Basel) 2020;12(2):482. doi: 10.3390/cancers12020482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu Y., Sheikh M.S. Melanoma: molecular pathogenesis and therapeutic management. Mol Cell Pharmacol. 2014;6(3):228. [PMC free article] [PubMed] [Google Scholar]
- 80.Li X.Y., Tan L.C., Dong L.W., et al. Susceptibility and resistance mechanisms during photodynamic therapy of melanoma. Front Oncol. 2020;10:597. doi: 10.3389/fonc.2020.00597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hou Y.J., Yang X.X., Liu R.Q., et al. Pathological mechanism of photodynamic therapy and photothermal therapy based on nanoparticles. Int J Nanomedicine. 2020;15:6827–6838. doi: 10.2147/IJN.S269321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vera R.E., Lamberti M.J., Rivarola V.A., Rumie Vittar N.B. Developing strategies to predict photodynamic therapy outcome: the role of melanoma microenvironment. Tumour Biol. 2015;36(12):9127–9136. doi: 10.1007/s13277-015-4059-x. [DOI] [PubMed] [Google Scholar]
- 83.Baldea I., Giurgiu L., Teacoe I.D., et al. Photodynamic therapy in melanoma - where do we stand? Curr Med Chem. 2018;25(40):5540–5563. doi: 10.2174/0929867325666171226115626. [DOI] [PubMed] [Google Scholar]