Abstract
Sexual dimorphism has been reported in various human diseases including autoimmune diseases, neurological diseases, pulmonary arterial hypertension, and some types of cancers, although the underlying mechanisms remain poorly understood. The long noncoding RNA (lncRNA) X-inactive specific transcript (XIST) is involved in X chromosome inactivation (XCI) in female placental mammals, a process that ensures the balanced expression dosage of X-linked genes between sexes. XIST is abnormally expressed in many sex-biased diseases. In addition, escape from XIST-mediated XCI and skewed XCI also contribute to sex-biased diseases. Therefore, its expression or modification can be regarded as a biomarker for the diagnosis and prognosis of many sex-biased diseases. Genetic manipulation of XIST expression can inhibit the progression of some of these diseases in animal models, and therefore XIST has been proposed as a potential therapeutic target. In this manuscript, we summarize the current knowledge about the mechanisms for XIST-mediated XCI and the roles of XIST in sex-biased diseases, and discuss potential therapeutic strategies targeting XIST.
Keywords: Epigenetic regulation, Long noncoding RNA, Sex-biased diseases, X chromosome inactivation, XIST
Introduction
Noncoding RNAs (ncRNAs) are a class of transcripts that do not encode proteins. In mammalian transcriptomes, only a small fraction of transcripts have the capacity to encode proteins, and the vast majority of RNAs are noncoding, including ribosomal RNAs, transfer RNAs, small RNAs, long noncoding RNAs (lncRNAs), and circular RNAs. The large proportion of noncoding RNAs were once considered to be “transcriptional noise”, since their functions were unknown. It was later recognized that ncRNAs are indeed functional, and many of which are crucial for normal cell function. For example, some small RNAs, including small interfering RNAs (siRNAs) and microRNAs (miRNAs), are involved in post-transcriptional gene silencing. A special species of ncRNAs longer than 200 nucleotides known as lncRNAs,1 are abundant in mammalian transcriptomes and are involved in diverse biological processes ranging from transcriptional regulation, genome organization, genomic imprinting, dosage compensation, and cell differentiation, to tumorigenesis.2, 3, 4, 5, 6, 7, 8 The aberrant expression of many human lncRNAs has been documented in many diseases.9 For example, X-inactive specific transcript (XIST in humans and monkeys, Xist in mice) is involved in X chromosome inactivation (XCI) crucial for normal female development in placental mammals,10,11 and has been implicated in sex-biased diseases in recent years.12
LncRNA XIST and mechanisms for X chromosome inactivation
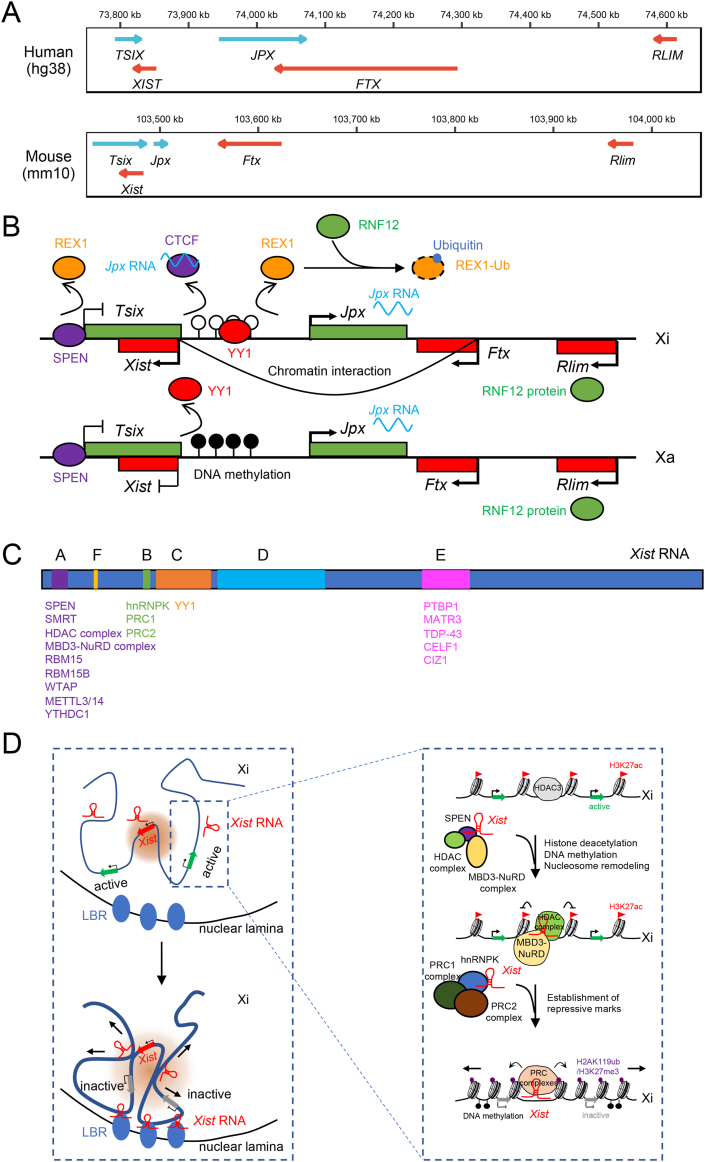
XIST is very important for sex determination and dosage compensation in mammals. In most mammals, sex is determined by the X and Y chromosomes, with females being XX and males XY.13 As a consequence, without dosage compensation, the extra X chromosome in females can lead to unbalanced X-linked gene expression. XCI produces dosage compensation by randomly inactivating one of the X chromosomes in females.14,15 Regulation of XCI in pre- and post-implantation development occurs differently. In pre-implantation, most mammalian embryos undergo XCI imprinting. Humans lack imprinted XCI and regulate gene expression by X chromosome dampening (XCD) instead.16,17 In post-implantation development, both human and other placental mammals undergo random XCI.16,17 The long noncoding transcript XIST (19 kb in humans, and 17 kb in mice) has been suggested to play vital roles in random XCI.17,18 Random XCI has three phases: initiation, establishment, and maintenance,19 and has been extensively investigated in mice. The XIST/Xist gene is located in the X inactivation center (XIC),20, 21, 22 and can be transcribed from the X chromosome to be inactivated (Xi), which functions in cis to coat Xi and nucleate dynamic protein complexes.23 Xist RNA-containing complexes gradually expand, allowing Xist to spread across Xi.23 Meanwhile, these complexes alter chromatin architecture and thus compact the chromosome, leading to progressive gene silencing along the Xi.23,24
Factors involved in transcriptional activation of XIST
The initiation stage of random XCI is a stochastic process involving X–X pairing, counting, and XIST/Xist activation.19,25, 26, 27 In early embryo development, XCI is regulated by XIST/Xist activators including Ftx,28 Jpx29,30 and RNF12 (encoded by Rlim),31 and inhibitors such as Tsix,32,33 which are located in XIC and are conserved between humans and mice (Fig. 1A). In mice, Jpx RNA activates Xist transcription in a dose-dependent manner by evicting CTCF30 (Fig. 1B), a DNA-binding insulator capable of repressing Xist expression.34,35 Ftx promotes the transcription of Xist through the proximity of their gene loci, which is independent of the Ftx RNA products28 (Fig. 1B). The X-encoded E3 ubiquitin ligase RNF1231 upregulates mouse Xist expression by targeting for degradation the pluripotency factor REX1,36 which normally activates Tsix and represses Xist expression through binding to regulatory regions36,37 (Fig. 1B). The autosomal transcription factor YY1 binds to the 5’ region of the Xist gene lacking DNA methylation and activates Xist expression, in competition with the Xist repressor REX1, whereas the methylated copy on the active X (Xa) cannot be bound38 (Fig. 1B). In addition, the chromatin remodeler SPEN (also known as SHARP) accumulates on the Xi early in mouse XCI to silence Tsix and activate Xist expression39 (Fig. 1B). In human pluripotent stem cells, XIST expression is silenced by the de novo DNA methyltransferases DNMT3A and DNMT3B.40
Figure 1.
The long noncoding Xist RNA and its roles in X chromosome inactivation. (A) Genomic arrangement of XIST/Xist and its regulators in X inactivation center (XIC) in human and mouse. (B) Factors involved in the transcriptional activation of mouse Xist. Jpx RNA transcribed from both X chromosomes interacts with CTCF insulator to release it from the Xist regulatory region. YY1 competes with REX1 repressor to bind the Xist regulatory region on Xi lacking DNA methylation, while the methylated copy is not bound, achieving selective activation of Xist. The repressor REX1 is further recognized by the E3 ubiquitin ligase RNF12, encoded by Rlim in XIC, leading to the ubiquitination and degradation of REX1. Ftx promotes Xist transcription through nuclear proximity of Xist and Ftx loci, independently of Ftx transcripts. However, active Ftx transcription is required for Xist accumulation. SPEN remodels the chromatin to silence Tsix and activate Xist. (C) Overview of repeat motifs in mouse Xist RNA. The proteins or complexes interacting with these repeats are indicated below. (D) The roles of Xist in the establishment of random XCI. As shown in the left panel, spatially, Xist RNA binds some locally confined loci on the Xi and nucleates local protein gradients to form SMACs (shown as light red circles). Xist RNA is tethered to the inactive X nucleation center by YY1. The Xi is recruited to the nuclear lamina through the Xist-LBR interaction, and the SMACs gradually expand to silence the whole X chromosome. Arrows indicate the expansion of the complex and the spreading of gene silencing on Xi. In the right panel, enlarged view of Xist-mediated chromatin dynamics near gene loci on Xi during XCI is shown. Xist RNA interacts with SPEN and further activates HDAC and MBD3-NuRD complexes, enabling removal of active histone marks, remodeling of nucleosomes, and DNA methylation. Xist also recruits PRC1 and PRC2 through hnRNPK to establish repressive histone marks, such as H2AK119ub and H3K27me3.
XIST-mediated polycomb recruitment, nuclear scaffolding and XCI
After Xist is activated, it establishes XCI through binding and recruiting proteins responsible for coating and spreading, histone modification, DNA methylation and chromatin compaction,12,41 that together form Barr bodies19 (Fig. 1D). During this process, active histone marks such as H3K4me1, H3K4me3, H3K9ac, H3K27ac, H4K5ac, H4K8ac, H4K12ac and H4K16ac are gradually lost, while repressive histone marks like H2AK119ub, H3K9me2, H3K9me3 and H3K27me3, and DNA methylation accumulate.24,42, 43, 44, 45, 46 Xist RNA contains multiple repeat motifs that can interact with various proteins (Fig. 1C). Upon Xist transcription, many Xist RNA-binding proteins immediately assemble on the multivalent E-repeat of Xist RNA,47 including PTBP1, MATR3, TDP-43 and CELF1. They form a condensate on the Xi via self-aggregation and protein interactions,47 restricting Xist to the Xi.23,48, 49, 50 Xist forms about 50 locally confined loci in open chromatin regions on Xi, each containing 2 Xist RNA molecules capable of nucleating supramolecular complexes (SMACs).23 The complexes gradually expand across the Xi and the dynamics create gradients of local proteins over broad genomic regions along the Xi23 (Fig. 1D). During this process, the Xist RNA's A-repeat binds to the corepressor SPEN/SHARP's C-terminal SPOC domain,41,51,52 which acts as a molecular integrator to bridge Xist RNA to the transcription machinery, nucleosome remodelers and histone deacetylases.52 The SPOC domain then interacts with the SMRT co-repressor and further activates pre-loaded histone deacetylase HDAC3 on Xi,24,53 resulting in the loss of active chromatin marks like H3K27ac.24 The B-repeat of Xist RNA recruits Polycomb repressive complexes PRC1 and PRC2 through directly binding with hnRNPK to establish the repressive chromatin marks H2AK119ub and H3K27me3 on Xi54, 55, 56, 57 and achieving selective X chromosome silencing.56,58 In humans, the E-repeat may also be required for PRC recruitment and H3K27me3 enrichment.59 In addition, XIST/Xist can recruit the m6A machinery to its transcript. In both humans and mice, the A-repeat interacts with the RNA-binding motif (RBM) proteins RBM15 and RBM15B.60,61 These RBM proteins further recruit the m6A methyltransferase METTL3/14 to specific sites in XIST/Xist through Wilms tumour-associated protein (WTAP), eventually resulting in m6A formation at adjacent sites.60,61 In humans, the m6A in XIST RNA is responsible for recruiting the m6A reader YTHDC1 to promote gene silencing,60 although the mechanisms remain elusive. In mice, YTHDC1 protein is recruited to Xist RNA through SPEN/SHARP's SPOC domain.52 The three-dimensional conformation further promotes Xist spreading to actively transcribed genes across Xi.62 During this process, many proteins are involved. The C-repeat of Xist RNA is bound by YY1, which tethers Xist to inactive X nucleation center on Xi.63 The A-repeat interacts with the Lamin B receptor (LBR) to recruit Xi to the nuclear lamina, enabling Xist to spread across Xi.64 In addition, the interaction between Xist and PRCs is also crucial for Xist spreading.65
XIST in XCI maintenance
The mechanism of XCI maintenance is less studied. During mouse embryonic stem cell differentiation, Xist expression is dispensable for XCI maintenance,66 whereas in human B cells, XIST is required for maintaining the X-inactivation of immune genes.12 A genome-wide RNAi screen in mouse embryonic fibroblasts identified 32 proteins involved in the maintenance of Xi silencing.67 One of those proteins, DNMT1, is responsible for CpG dinucleotide methylation maintenance, suggesting that DNA methylation is required for XCI maintenance in mice. A recent study also revealed that the condensate formed by many Xist RNA-binding proteins seeded by the Xist RNA's E-repeat, is crucial for gene silencing during the Xist-independent phase of XCI in differentiating mouse embryonic stem cells.47 In human B cells, comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-MS) uncovered the XIST-interacting proteome, which is different from the ones found in embryonic stem cells and myeloid cells.67 Some cell-specific XIST-interacting proteins may also contribute to XCI maintenance. For example, TRIM28, a cofactor of the H3K9me3-specific histone methyltransferase SETDB1,46,68 only binds XIST RNA in B cells.12 CRISPRi screening further indicates that TRIM28 is indispensable for XCI maintenance in B cells.12 Therefore, the XCI maintenance mechanisms may be tissue-specific.
Random XCI is crucial for normal female development in placental mammals. Under normal conditions, XCI can balance gene dosage between males and females in placental mammals. However, any mistakes occurring in this process may lead to cell dysfunction and disease. Since XIST function is related to sex chromosome gene expression, it has been hypothesized to be related to many sex-biased diseases.
XIST and sex-biased diseases
Sex disparities in disease are common. For example, most cancers show a large sex bias in incidence and mortality.69,70 The incidence of autoimmune and neurological diseases also differs between sexes.71 Even for COVID-19-associated illness, the severity and mortality is different for men and women.72, 73, 74 The difference might be attributed to sexual dimorphism75,76 and gender differences in attitudes and behavior.77,78 A growing body of evidence has revealed that the lncRNA XIST, an important regulator in X chromosome dosage compensation in placental mammals, can play pivotal roles in some sex-biased diseases.
XIST in autoimmune diseases
More than 80% of autoimmune diseases are female dominant, examples being systemic lupus erythematosus (SLE) and rheumatoid arthritis (RA). This female bias has been linked to X-linked immune gene dosage.71,79 The X chromosome is known to contain the largest number of immune response-related genes of the whole human genome.71,80, 81, 82 XCI has evolved to balance gene dosage between males and females. As a result, in female mammals, one of the X chromosomes is inactivated by XIST and most genes on the inactive X are silenced. However, a fraction of X-linked genes can still escape from X-inactivation and therefore have biallelic expression in both humans and mice,83, 84, 85 which leads to female-biased gene expression. In humans, about 15% of genes consistently escape from XCI and another 15% of genes vary between individuals or tissues.86
Most somatic cells maintain XCI with static enrichment of XIST and heterochromatin marks on the Xi, but immune cells exhibit a unique dynamic localization of XIST and epigenetic modifications to the Xi following stimulation.87, 88, 89, 90 Although female naïve and activated lymphocytes have similar high levels of XIST RNA, naïve lymphocytes lack canonical localization of XIST RNA transcripts on the Xi.90 In murine B cells, it has also been reported that Xist RNA disappears from the Xi at the pro-B cell stage with a gradual loss of heterochromatic modifications, while mature B cell activation restores Xist RNA localization and the heterochromatic modifications on Xi.91
Many studies have shown that altered XIST localization on Xi in lymphocytes may promote sex-biased autoimmune diseases. For example, in SLE patients, cellular imaging has shown that both B and T cells exhibit abnormal XIST RNA localization patterns without altered expression.87,88,90 Some recent studies uncovered more mechanistic details regarding autoimmune B cell dysregulation. Pyfrom et al showed that there is a complete lack of XIST localization on the Xi in human CD11c+ atypical memory B cells,87 a unique B cell population expanded in SLE and RA.92,93 Yu et al further showed that XIST is continually required in adult human B cells,12 finding that some X-linked immune genes in B cells lack promoter methylation, which requires XIST for silencing maintenance via enhancer H3K27ac deacetylation. For example, the X-linked Toll-like receptor 7 (TLR7), which recognizes single-strand RNA (ssRNA)-containing immune complexes involved in female-biased autoimmunity and ssRNA viral infection,94 is commonly overexpressed in SLE and RA patients and promotes the formation and activation of CD11c+ atypical memory B cells.92,93,95 Yu et al also found that in somatic cells, XIST complexes are tissue-specific.12 B cell-specific XIST cofactor TRIM28 may inhibit transcription elongation on immune genes such as TLR7, possibly through RING domain-mediated sumoylation of the transcription elongation kinase CDK9.12 SLE patients’ T cells also have dispersed XIST, altered XCI maintenance and aberrant overexpression of many X-linked genes, but the mechanisms are still unclear.88
In summary, in autoimmune disease, XIST localization on Xi of some immune cells is lost or changed, leading to altered XCI maintenance. Some XIST-dependent immune genes such as TLR7 can therefore be reactivated, which may be sufficient to promote isotype-switched immune cells and autoimmunity (Fig. 2A).
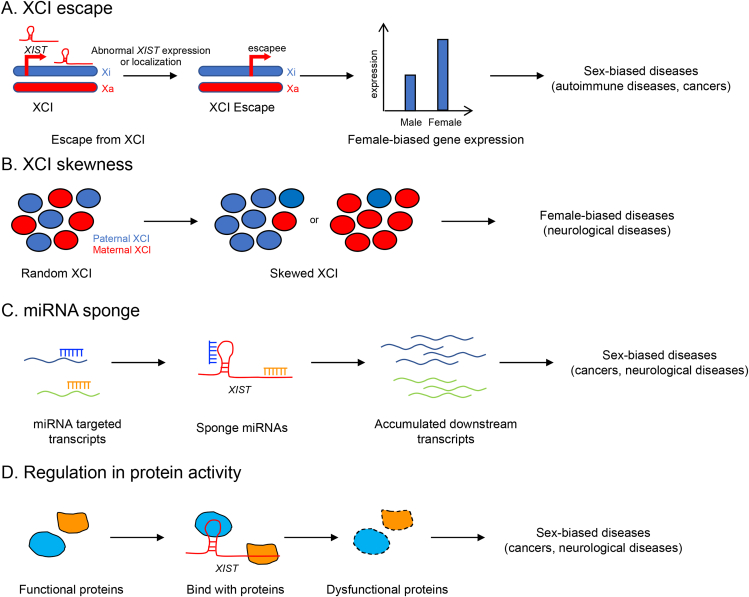
Figure 2.
Molecular mechanisms underlying roles of XIST in diseases. (A) X chromosome inactivation (XCI) escape. In female mammals, some X-linked genes can escape from XCI when XIST expression or localization is altered and therefore have biallelic expression, leading to female-biased gene expression, which might contribute to sex-biased diseases. (B) XCI skewness. In female mammals, the paternal and maternal X chromosomes generally have a similar opportunity to be silenced in somatic cells. Under specific condition, however, skewed XCI may occur, which may cause disease in females. (C) MiRNA sponge. The long noncoding transcript XIST can bind various miRNAs, resulting in derepression of miRNA targeted genes and pathology. (D) Regulation in protein activity. XIST can bind to developmentally critical proteins and affect their activity.
XIST in sex-biased cancers
For many cancers, the incidence, prevalence, prognosis, and mortality differ greatly between the sexes. For example, males have higher risks of bladder, colorectal, kidney, lung, liver and blood cancers, while females have higher risks of breast and thyroid cancers.69,70 In this section, we summarize current progress on the roles of XIST in some of these sex-biased cancers.
Some genes escaping from X-inactivation involve tumor suppressors
Tumors have significant numbers of genetic mutations. An investigation about the paired tumor-germline exome sequencing data across 21 tumor types identified six genes with higher loss-of-function mutation frequency in male-biased cancers.96 All six, ATRX, CNKSR2, DDX3X, KDM5C, KDM6A/UTX, and MAGEC3, are located in the non-pseudoautosomal region of the X chromosome.96 These genes can escape from X-inactivation, leading to female-biased expression, and are regarded as tumor suppressor genes as they are frequently mutated in cancer.97, 98, 99, 100, 101 They are therefore referred to as ‘escape from X-inactivation tumor-suppressor’ genes or EXITS genes.96 For example, the loss-of-function mutation in the H3K27me3 demethylase gene KDM6A/UTX mainly occurs in male-biased cancers.96,102,103 However, female cancers with KDM6A mutations usually require homozygous mutations, leading to lower female incidences.102 Other than disease incidence, gender-biased KDM6A expression also leads to different outcomes. The expression of KDM6A in female bladder cancer patients is significantly higher than in male patients and is correlated with longer survival and better prognosis of female patients.104 Conditional knockout of mouse Kdm6a increases bladder cancer risk and decreases overall female survival, which, however, does not significantly affect the survival or tumor burden of male mice.104 These results indicate that female-biased KDM6A expression exerts antitumor effects in bladder cancer leading to lower incidence and better prognosis in females.
Oncogenic role of XIST in male-biased cancers
XIST is normally not expressed in male somatic tissues. In the tissues involved in some male-biased cancers like bladder, colorectal, and lung tumors, however, XIST expression is abnormally elevated,105, 106, 107, 108, 109, 110, 111, 112, 113, 114 and the elevated XIST expression correlates with shorter survival and poor prognosis.105, 106, 107,115 In cell lines derived from these cancers, overexpression of XIST promotes cell proliferation, migration, invasion and epithelial–mesenchymal transition, and inhibits apoptosis,114,116,117 while knockdown of XIST has the opposite effect,105,106,109,112, 113, 114,116, 117, 118, 119, 120, 121, 122, 123, 124 irrespective of the sexual characteristics of cell lines (Table 1). Murine xenograft assays have also shown that for bladder,118 colorectal,109 or lung106,112, 113, 114,121, 122, 123 cancers, XIST silencing inhibits tumor growth in mice, and XIST overexpression promotes non-small cell lung cancer (NSCLC) tumor growth in mice.114 These suggest that XIST may play an oncogenic role in male-biased cancers.
Table 1.
Effects of manipulated XIST expression on cell proliferation and tumorigenesis in male-biased cancers.
| Cancer types | Cell lines | Gender of cell host | Genetic manipulation in XIST expression | Effects on cell proliferation and/or tumorigenesis | References |
|---|---|---|---|---|---|
| Bladder cancer | 5637 | Male | Overexpression | Promotion | 116 |
| Knockdown | Inhibition | 105,118 | |||
| 253J | Male | Knockdown | Inhibition | 119 | |
| T24 | Female | Knockdown | Inhibition | 105,116,118 | |
| RT112 | Female | Knockdown | Inhibition | 119 | |
| Colorectal cancer | HT29 | Female | Overexpression | Promotion | 117 |
| LoVo | Male | Knockdown | Inhibition | 117 | |
| SW480 | Male | Knockdown | Inhibition | 109 | |
| HCT116 | Male | Knockdown | Inhibition | 109 | |
| Non-small cell lung cancer | A549 | Male | Overexpression | Promotion | 114 |
| Knockdown | Inhibition | 106,112, 113, 114,120,122, 123, 124 | |||
| H1299 | Male | Overexpression | Promotion | 114 | |
| Knockdown | Inhibition | 112, 113, 114,122, 123, 124 | |||
| H522 | Male | Knockdown | Inhibition | 121 | |
| Calu3 | Male | Knockdown | Inhibition | 121 | |
| H226 | Male | Knockdown | Inhibition | 120 |
Mechanistically, XIST serves primarily as a miRNA molecular sponge to regulate the expression of miRNA targets in male-biased cancers (Fig. 2C). For example, upregulated XIST can bind miR-124 and promote the expression of Androgen Receptor (AR) to facilitate bladder cancer development.125 AR encodes a steroid hormone receptor functioning as a transcription factor to promote the progression of bladder cancer.126 Although androgen signaling promotes the progression of bladder cancer in both males and females, higher AR expression is observed in male patients127 and associated with higher bladder cancer risk.126 In mice, N-butyl-N-(4-hydroxybutyl)nitrosamine (BBN) can induce bladder cancer, with higher male incidence,128 possibly due to different AR expression levels. Knockout of the AR gene abolishes BBN-induced bladder carcinogenesis in both male and female mice,128 indicating that AR-mediated androgen signaling may play a crucial role in the progression of some male-biased cancers, at least in BBN-mediated bladder tumorigenesis. In the presence of androgen, AR can regulate the expression of downstream genes, many involved in bladder cancer outgrowth, including β-catenin,129 CD24,130 EGFR/ERBB2,131 and ELK1.132 AR protein accumulation is also seen in some male breast cancer patients, and high AR expression predicts inferior outcomes and poor tamoxifen treatment responses in male breast cancer.133 In addition, XIST can activate the Wnt/β-catenin signaling pathway to accelerate bladder and colon cancer progression by sponging miR-139-5p105 and miR-34a109 respectively. XIST targets miR-200b-3p to modulate the expression of ZEB1,134 sponges miR-132-3p to activate the MAPK1 signaling pathway,110 interacts with miR-137 to regulate the EZH2 signaling pathway,117 binds miR-486-5p to regulate the neuropilin-2 (NRP-2) pathway,108 inhibits miR-30a-5p to activate ROR1,135 suppresses miR-93-5p to modulate the HIF-1A/AXL signaling pathway,136 sponges miR-338-3p to regulate PAX5 expression,137 and targets miR-125b-2-3p to regulate the Wee1 signaling pathway,138 all of which promote colorectal cancer progression. XIST can also promote TGF-β-induced epithelial–mesenchymal transition through the miR-367/141-ZEB2120 and miR-137/Notch-1124 axes in non-small cell lung cancer. XIST promotes Bcl-2 expression through sponging miR-449a, thus exerting an anti-apoptotic effects in many cancers.113,139 XIST also targets miR-16 to activate CDK8,114 a member of the mediator complex acting as an oncogene,140 with XIST-mediated proliferation and migration of lung cancer cells reversed by miR-16 overexpression.114
Beside sponging miRNAs, the lncRNA XIST can also directly interact with proteins and affect their functions (Fig. 2D). For example, XIST binds to the DNA demethylase TET1 to reduce TET1-mediated demethylation on the tumor suppressor gene p53,141 thereby inhibiting p53 expression in bladder cancer,116 with XIST-mediated cell proliferation able to be reversed by expression of p53 in bladder cancer cells.116 XIST also directly binds with the H3K27me3-specific histone methyltransferase EZH2 to silence the expression of proposed tumor suppressor KLF2 in NSCLC cells.106 Methyltransferase-like14 (METTL14) can catalyze the N6-methyladenosine (m6A) on XIST transcript, which is further recognized by the m6A reader YTHDF2, leading to XIST degradation.142 METTL14 is downregulated and XIST expression is upregulated in colorectal cancer, and the loss of METTL14 has been shown to be associated with poor prognosis.142 Knockdown of METTL14 promotes colorectal cancer proliferation and invasion and substantially abolishes the m6A level of XIST, leading to augmented XIST expression,142 while METTL14 overexpression results in a remarkable decrease in XIST expression level, cell growth and invasion.142 It has therefore been proposed that METTL14 inhibits colorectal cancer progression by reducing XIST expression.142
XIST in female-biased and gynecologic cancers
Breast and thyroid cancers are more common in women than in men.70,143 The role of XIST in these female-biased cancers is complex. In normal female breast tissues, XIST is highly expressed, whereas in the cancer tissues or cells, XIST expression is downregulated relative to adjacent normal tissues or normal cell lines.144, 145, 146 XIST expression is also significantly reduced in brain-metastatic breast cancer, and decreased XIST expression promotes brain metastasis in breast cancer, while the dCas9-mediated overexpression does not.147 In breast cancer cells, XIST overexpression inhibits their proliferation, migration and invasion, and facilitates their apoptosis, while XIST silencing exerts the opposite effect.145,146 The xenograft tumor assay in BALB/c nude mice confirmed that XIST can retard breast tumor growth.146 XIST can activate CDX1 by sponging miR-155 to depress the growth, migration, and invasiveness of breast cancer.144 Interestingly, the expression of Jpx, an activator of XIST expression,29,30 has also been noted to be downregulated in breast cancer samples.145 Knockdown of XIST or SPEN/SHARP can promote the recruitment of HDAC3 to the promoter of PHLPP1.145 Since PHLPP1 encodes a phosphatase able to dephosphorylate AKT, knockdown of XIST leads to reduced PHLPP1 expression and increased AKT phosphorylation.145 As mentioned above, female-biased expression of KDM6A/UTX caused by escaping from XIST-mediated XCI can act as a tumor suppressor in some male-biased cancers. However, in other female-biased cancers like breast cancer, KDM6A/UTX may play an oncogenic role, since a study reported that KDM6A/UTX can cooperate with H3K4 methyltransferase MLL4 to promote the expression of many oncogenes and prometastatic genes, leading to cell proliferation and invasion in breast cancer cells, both in vitro and in a mouse xenograft model.148
In thyroid cancer, XIST may be oncogenic, contributing to female bias since females have higher expression of XIST compared to males. Compared to adjacent normal tissues or normal cell lines, XIST expression in thyroid cancer tissues and cell lines is upregulated,149, 150, 151 regardless of gender. In addition, XIST expression positively correlates with thyroid cancer progression.149,150 In both male-derived and female-derived thyroid cancer cells, XIST knockout inhibited cell proliferation, migration and invasion149, 150, 151 (Table 2), and its oncogenic role has been confirmed in the xenograft tumor assay in female nude mice.150 XIST regulates thyroid cancer progression by functioning as a competing endogenous RNA (ceRNA) to sponge miRNAs. For example, XIST can enhance the expression of the receptor tyrosine kinase MET by sponging miR-34a, resulting in increased phosphorylation of PI3K and AKT.150 XIST also upregulates CLDN1 expression through interaction with miR-101-3p, thereby promoting cell proliferation, migration, and invasion of thyroid cancer.151 In addition, in papillary thyroid carcinoma, XIST targets miR-141 to promote cell proliferation and invasion,149 although the downstream processes are yet to be elucidated.
Table 2.
Effects of manipulated XIST expression on cell proliferation and tumorigenesis for female-biased cancers.
| Cancer types | Cell lines | Gender of cell host | Genetic manipulation in XIST expression | Effects on cell proliferation and/or tumorigenesis | References |
|---|---|---|---|---|---|
| Breast cancer | MCF7 | Female | Overexpression | Inhibition | 145,146 |
| Knockdown | Promotion | 146,147 | |||
| MDA-MB-231 | Female | Overexpression | Inhibition | 146 | |
| Knockdown | Promotion | 146 | |||
| SKBR3 | Female | Knockdown | Promotion | 147 | |
| ZR75-1 | Female | Knockdown | Promotion | 147 | |
| MDA-MB231BrM2a | Female | Overexpression | Inhibition | 147 | |
| M10 | Female | Knockdown | Promotion | 145 | |
| Thyroid cancer | TPC-1 | Female | Knockdown | Inhibition | 149,151 |
| KAT18 | Unspecified | Knockdown | Inhibition | 150 | |
| FTC113 | Male | Knockdown | Inhibition | 150 |
As an important participant in XCI in female mammals, aberrant expression of XIST has also been implicated in ovarian and cervical cancers, two common gynecologic malignant tumors.70 Similarly to breast cancer, XIST expression in ovarian cancer tissues is downregulated compared to adjacent normal tissues.152 In addition, XIST expression correlates with ovarian cancer development, with downregulation in advanced stages, and higher expression is associated with better prognoses.152 In ovarian cancer cell lines, XIST overexpression suppresses cell proliferation,153, 154, 155 while XIST knockdown has the opposite effect.153 XIST suppresses cancer progression through sponging hsa-miR-214-3p154 and miR-106a.155 In recurrent ovarian tumors, XIST expression is also decreased compared to paired primary tumors and is associated with resistance to the anticancer agent Taxol.156 The loss of XIST also induces ovarian cancer stem cells to acquire Taxol resistance through modulation of the miR-93-5p/KMT2C axis.153 These findings indicate that XIST might not only be a biomarker for the diagnosis and prognosis of cancers, but also a potential therapeutic target for ovarian cancer. Notably, two recent studies have claimed that XIST promotes the proliferation, invasion, and migration of ovarian cancer cells by modulating the miR-335/BCL2L2 axis157 and regulating miR-149-3p.158 These findings require reconciliation with prior observations.
In cervical cancer, XIST expression is elevated,159, 160, 161 in contrast to breast or ovarian cancer. XIST knockdown in cervical cancer cell lines like SiHa, HeLa, C33A and Me180 cells inhibits cell proliferation, blocks the cell cycle, and promotes apoptosis.159, 160, 161 Reduced tumor growth is also observed in the murine xenograft assay after XIST silencing.161 In cervical cancer, XIST accelerates cancer progression by sponging various miRNAs, thereby derepressing the expression of oncogenic genes targeted by these miRNAs. For example, XIST interacts with miR-200a to upregulate Fus expression,159 binds miR-140-5p to promote ORC1 expression,160 and targets miR-889-3p to derepress SIX1 expression.161 It has been suggested that high expression of XIST is associated with unfavorable prognosis of cervical cancer patients.159 Taken together, these studies suggest that the role of XIST in tumorigenesis shows an organ-dependent pattern for female-biased and gynecologic cancers.
XIST in other sex-biased diseases
XIST in neurological disorders
Neurological disorders are nervous system-related and include those associated with neurodevelopment (autism spectrum disorders, schizophrenia, Rett syndrome, and Down syndrome) and neurodegeneration (Parkinson's, Alzheimer's and Huntington's diseases). Many neurological diseases show sex-bias. For example, autism spectrum disorder is a male-biased disease with three times more frequent observation in males than in females.162 Rett syndrome is a female-biased progressive neurodevelopmental disorder. Parkinson's disease (PD) is a male-biased neurodegenerative disorder, and Alzheimer's disease (AD) is a chronic neurodegenerative disease with higher prevalence in females.163 Some genes related to neuronal plasticity and cognitive process are located on the X chromosome. Nearly 20% of them, including MECP2, FMR1, and CDKL5, are correlated with neurodevelopmental diseases.164 It has therefore been suggested that XIST and XCI status may be responsible for the sex-bias of neurological diseases. In this section, we discuss XIST and its function in XCI in neurological diseases as a microRNA sponge, the phenomenon of XCI skewness (XIS) in some female patients and X-linked heterozygous mutation diseases related to XCI.
XIST or XCI with neurodevelopmental diseases
A common feature of neurodevelopmental disease patients is that most female patients show XCI skewness as compared to normal females. XCI skewness is a phenomenon in which one X chromosome is more active than the other.162 A study reported that a rare C-to-G mutation in the Xist promoter may lead to XCI skewness, and cells may prefer to inactivate X chromosome with this mutation.165 Interestingly, XCI skewness is increased in autistic females compared to normal females.166 However, this C-to-G mutation in Xist promoter was not detected and whether XCI skewness is responsible for the female-relative decreased susceptibility of autism spectrum disorder is still unclear.
Female-biased Rett syndrome is mainly caused by the heterozygous mutation of X-linked methyl-CpG-binding protein (MECP2).167 MeCP2 is a DNA methylation reader with both repressive and activating function with different cofactors.168 Rett syndrome is exclusively observed in females because MECP2 mutation is lethal in males during embryogenesis.169 Deletions and nonsense mutations of MECP2 are more severe than missense mutations and probably cause cells to preferentially inactivate X chromosomes.170 Although there is no known direct connection between XIST and Rett syndrome symptoms, downregulation of Xist caused by knocking down of bone morphogenetic protein (BMP)/TGF-β signaling pathway members can activate MECP2 gene expression on Xi allele in mouse embryonic fibroblasts.167 This study also showed that restoration of the wild-type allele of MeCP2 could be a promising therapeutic strategy of Rett syndrome in future.167
XIST in neurodegenerative diseases
XIST can serve as a miRNA sponge to regulate gene expression in neurodegenerative diseases. For the male-biased Parkinson's disease, XIST expression is generally upregulated and can sponge miR-199a-3p to enhance Sp1 gene expression. Sp1 promotes the transcription and translation of leucine-rich repeat kinase 2 (LRRK2), a key PD-related gene.171, 172, 173, 174 Overexpressing miR-199a-3p or knocking down XIST by shXIST can inhibit apoptosis and promote cell proliferation, which can rescue neurodegeneration.175 Furthermore, in vivo study in a PD mouse model showed that lentivirus vectors carrying shXIST or overexpression of miR-199a-3p mimics can alleviate Parkinson's disease-associated symptoms.175
AD involves the accumulation of β-amyloid (Aβ) peptide, which is a cleavage product of the amyloid precursor protein (APP). Xist expression was significantly upregulated in AD mice and cell models,176,177 where inflammation and injury of nerve cells occurred.177 Xist is a molecular sponge of miR-124 that targets BACE1, an enzyme crucial for the cleavage of APP and serving as a biomarker of AD. Xist silencing could reduce BACE1 expression through miR-124.176 Apart from sponging miRNA, Xist might also be involved in the progression of AD through its protein interaction. Xist could recruit the histone methyltransferase EZH2 to deposit H3K27me3 mark on the NEP1 promoter region to repress its expression. NEP1 is an enzyme responsible for Aβ degradation. Xist knockdown resulted in increased expression of NEP1 and alleviated Aβ-induced neuronal inflammation and damage.177 Therefore, XIST may be a potential therapeutic target for AD.
XIST in pulmonary arterial hypertension
Pulmonary arterial hypertension (PAH) is a female-biased disease characterized by the proliferation and overgrowth of dysfunctional pulmonary artery endothelial cells, leading to right heart failure.178 An epidemiology study showed that the approximate ratio of PAH females to male is 4:1.179 The higher incidence in female may be explained by the higher expression of XIST in females since upregulation of Xist can promote PAH related phenotypes in murine model cells of plexiform PAH.180 EHITSN (C-terminal protein fragments of intersectin-1) is generated during inflammation associated with PAH and can promote endothelial cell (EC) proliferation via activation of MAPK p38, ELK1 and FOS.181 Qin et al expressed EHITSN in pulmonary artery endothelial cells (PAECs) from both male and female donors, and observed that female EHITSN-transfected PAECs have a higher proliferation rate.180 Moreover, the Xist levels are also upregulated in both male and female EHITSN-transfected PAECs compared with controls, but female transfected PAECs showed more dramatic Xist increases. Treating female EHITSN-transfected PAECs with PenNPF, an EHITSN inhibitory peptide, can also reduce Xist levels and EC proliferation. Meanwhile, knockdown of Xist by siRNA can also impair the cell proliferation in female EHITSN-transfected PAECs. Increased Xist levels have also been detected in PAH patients with increased ELK1 and decreased KLF2, known targets of Xist with roles in EC proliferation and antiangiogenic effects only in female idiopathic PAH patients.180 In summary, higher XIST in females may explain the female-bias feature, and upregulation of XIST in PAH patients may operate through upregulation of ELK2 and downregulation of KLF2, both related to PAH.106,180,182
Although PAH is female-biased, the five-year survival rate from diagnosis in women is higher than in men.183 This higher survival rate may stem from either the protective effect of sex hormones or women's better response to current treatment.184 There is evidence that estrogen, which plays a vital role in the development of secondary sex characteristics, may attenuate the PAH phenotype in both males and females.185,186 As a result, since females have higher circulating estrogens than males, they may be better protected.183
Therapeutic strategies targeting XIST
XIST expression and/or its modification appear to be altered during the progression of many sex-biased diseases. It therefore could be used as a potential biomarker for the diagnosis and prognosis of several diseases. In addition, studies in cell and mouse models have shown that genetic manipulation of XIST expression can potentially inhibit the progression of many diseases including bladder, colorectal and lung cancers. XIST could therefore be regarded as an important therapeutic target for these diseases.
Some commercially available drugs can regulate XIST expression although detailed mechanisms remain ambiguous. 5-fluorouracil, cisplatin, mitomycin and adriamycin are effective chemotherapies for colorectal cancer. High level of XIST expression in colorectal cancer cells promotes resistance to these chemotherapies through the XIST/miR-30a-5p/ROR1 axis.135 Atractylenolide II, traditionally prescribed for melanoma treatment by Chinese medicine practitioners, is able to induce G1 cell-cycle arrest and apoptosis in B16 melanoma cells by modulating the expression of cell cycle-related genes or the phosphorylation level of related proteins.187 When applied to colorectal cells, atractylenolide II downregulates XIST expression and reverses the effect of XIST/miR-30a-5p/ROR1 axis in modulating the chemosensitivity of colorectal cancer cells.135 Platycodin D exerts anti-tumor effects in multiple cancers, including lung cancer,188 gastric cancer,189 hepatocellular carcinoma,190 and bladder cancer.191 In bladder cancer cells, platycodin D treatment can inhibit XIST expression and regulate the XIST/miR-335 axis to slow bladder cancer progression both in vitro and in vivo.191
Beside these drugs, some molecules specifically targeting the lncRNA XIST can also be designed. Multiple approaches targeting lncRNAs have been developed, including small interfering RNAs (siRNAs), antisense oligonucleotides (ASOs) and clustered regularly interspaced short palindromic repeats (CRISPR).6,192 siRNAs targeting specific lncRNA can trigger RNA-induced silencing complex to degrade the lncRNA, which has been adopted by many groups to knockdown XIST expression in various cancer cells such as the colorectal cancer cell line LoVo and NSCLC cell line A549. Clinically, some siRNA drugs have already been used to treat patients, such as Onpattro (patisiran) for the treatment of hereditary transthyretin amyloidosis with polyneuropathy.193 ASOs function through binding with specific RNA and recruiting RNase H to degrade the RNA of interest. Some ASO drugs have also been approved, including nusinersen to treat spinal muscular atrophy.194 CRISPR uses single guide RNAs to guide the Cas9 nuclease to cleave specific DNA sequences. However, its utilization for XIST needs further investigation and optimization, since XIST function is required for normal female mammals and off-target effects need to be considered.
In addition, the mechanisms underlying XIST-mediated XCI could be relevant to treatment of diseases like Down syndrome, caused by chromosome 21 trisomy, and associated with intellectual disability, hematopoietic disorders and early-onset Alzheimer's.195 Jiang et al proposed to use XIST to silence the whole extra chromosome 21. They transfected XIST on the gene-rich core of one chromosome 21 in stem cells from Down syndrome patients and successfully reduced chromosome 21 transcriptional outputs to near-normal levels.196 Chiang et al also found that XIST could rebalance chromosome 21 dosage in trisomic induced pluripotent stem cells (iPSCs).197
Concluding remarks
Sex disparities in disease are common, and traditional therapies without consideration of sex differences sometimes cause disparity of efficacy between sexes. XIST plays pivotal roles in modulating the progression of many sex-biased diseases, and it functions in these diseases through at least four different mechanisms: XCI escape, XCI skewness, miRNA sponge, and regulation of the activity of interacting proteins (Fig. 2). XIST-mediated XCI is crucial for normal female development in mammals, and any alteration in XIST expression or localization may cause escape from XCI, which might be a double-edged sword for females. On the one hand, the escaped gene expression might protect females from some diseases, such as male-biased cancers and even COVID-19. For example, female immune cells can have biallelic TLR7 expression due to XCI escape, which could in turn stimulate the cells to produce more type 1 interferon early in SARS-CoV-2 infection, therefore protecting females from COVID-19.198 On the other hand, the elevated expression of these escapees may also be detrimental to the immune response under normal conditions. Hence 80% of autoimmune disease patients are female.71 XCI skewness seems to be harmful to females, which is often observed in female-biased neurodevelopmental diseases. As a long transcript, XIST can bind large numbers of miRNAs and proteins and affect their function, which would promote or inhibit the progression of some diseases.
Our review also indicates that XIST expression or modification might be a biomarker for the diagnosis and prognosis of some diseases. Besides, XIST may be an excellent therapeutic target for some diseases, and several potential therapeutic strategies targeting XIST have been proposed. It should be noted that there remain many unknowns in the XIST field and sex-biased diseases. First, despite the fact that XIST expression is abnormally expressed in some sex-biased diseases and manipulated XIST expression can affect the progression of several diseases, whether and how XIST contributes to the sex disparity in the incidence and mortality of diseases need further elucidation. Most research to date has focused merely on the relationship between XIST expression and diseases without consideration of sex differences, as is done for most male-biased cancers. This has greatly limited interpretation. LncRNAs can regulate gene expression both in cis and in trans.199 The ectopic expression system used by some groups to study the effect of elevated XIST expression on disease progression may be useful to elucidate the trans-acting effects of XIST, which, however, may not always reflect the real effect of increased expression in endogenous XIST since its chromosomal localization may differ. For example, mislocalized (but unaltered) expression of XIST may contribute to female-biased autoimmunity.12,87,88,90 Second, conflicting results have been observed for XIST function by different research groups. For example, while most studies suggested that XIST has an anticancer effect on ovarian cancer,153, 154, 155, 156 some have claimed that XIST might exert an oncogenic role in its progression.157,158 This inconsistency may arise from many causes including different stages or subtypes of disease progression at sampling. Third, for some diseases, the relationship between XIST and disease progression was explored only by utilizing quantitative reverse transcription PCR to measure the expression of genes of interest, and therefore a global understanding of XIST function on transcriptome variation is lacking. Transcriptome-wide approaches, such as RNA-sequencing (RNA-Seq) or single cell RNA-Seq, could be applied to address the detailed mechanisms underlying the progression of XIST-mediated diseases in future.
Author contributions
JL, ZM, and LY studied the literature and drafted the manuscript under the supervision of QM. JL produced the figures. TW and GL assisted in manuscript collation and review. QM conceived the review, obtained funds and provided critical input and is the corresponding author. All authors contributed to the article and approved the submitted version.
Conflict of interests
The authors declare no conflicts of interest.
Funding
This work was supported by the National Natural Science Foundation of China (No. 32070870), Guangdong Basic and Applied Basic Research Foundation (No. 2021A1515010758), Guangdong Provincial Key Laboratory of Synthetic Genomics, Shenzhen Key Laboratory of Synthetic Genomics (No. ZDSYS201802061806209), Shenzhen Institute of Synthetic Biology Scientific Research Program (No. ZTXM20200008 and DWKF20210003) and the Strategic Priority Research Program of the Chinese Academy of Sciences (No. XDPB18).
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 2.Gil N., Ulitsky I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat Rev Genet. 2020;21(2):102–117. doi: 10.1038/s41576-019-0184-5. [DOI] [PubMed] [Google Scholar]
- 3.Quinodoz S.A., Jachowicz J.W., Bhat P., et al. RNA promotes the formation of spatial compartments in the nucleus. Cell. 2021;184(23):5775–5790. doi: 10.1016/j.cell.2021.10.014. e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Razin S.V., Gavrilov A.A. Non-coding RNAs in chromatin folding and nuclear organization. Cell Mol Life Sci. 2021;78(14):5489–5504. doi: 10.1007/s00018-021-03876-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tachiwana H., Yamamoto T., Saitoh N. Gene regulation by non-coding RNAs in the 3D genome architecture. Curr Opin Genet Dev. 2020;61:69–74. doi: 10.1016/j.gde.2020.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Wang T., Li J., Yang L., Wu M., Ma Q. The role of long non-coding RNAs in human imprinting disorders: prospective therapeutic targets. Front Cell Dev Biol. 2021;9:730014. doi: 10.3389/fcell.2021.730014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fatica A., Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 8.Bhan A., Soleimani M., Mandal S.S. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Batista P.J., Chang H.Y. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Penny G.D., Kay G.F., Sheardown S.A., Rastan S., Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379(6561):131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 11.Okamoto I., Nakamura T., Sasaki K., et al. The X chromosome dosage compensation program during the development of cynomolgus monkeys. Science. 2021;374(6570) doi: 10.1126/science.abd8887. [DOI] [PubMed] [Google Scholar]
- 12.Yu B., Qi Y., Li R., Shi Q., Satpathy A.T., Chang H.Y. B cell-specific XIST complex enforces X-inactivation and restrains atypical B cells. Cell. 2021;184(7):1790–1803. doi: 10.1016/j.cell.2021.02.015. e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng X., Berletch J.B., Nguyen D.K., Disteche C.M. X chromosome regulation: diverse patterns in development, tissues and disease. Nat Rev Genet. 2014;15(6):367–378. doi: 10.1038/nrg3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schulz E.G., Heard E. Role and control of X chromosome dosage in mammalian development. Curr Opin Genet Dev. 2013;23(2):109–115. doi: 10.1016/j.gde.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 15.Patrat C., Ouimette J.F., Rougeulle C. X chromosome inactivation in human development. Development. 2020;147(1):dev183095. doi: 10.1242/dev.183095. [DOI] [PubMed] [Google Scholar]
- 16.Okamoto I., Patrat C., Thépot D., et al. Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature. 2011;472(7343):370–374. doi: 10.1038/nature09872. [DOI] [PubMed] [Google Scholar]
- 17.Sahakyan A., Yang Y., Plath K. The role of Xist in X-chromosome dosage compensation. Trends Cell Biol. 2018;28(12):999–1013. doi: 10.1016/j.tcb.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sidorenko J., Kassam I., Kemper K.E., et al. The effect of X-linked dosage compensation on complex trait variation. Nat Commun. 2019;10(1):3009. doi: 10.1038/s41467-019-10598-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maduro C., de Hoon B., Gribnau J. Fitting the puzzle pieces: the bigger picture of XCI. Trends Biochem Sci. 2016;41(2):138–147. doi: 10.1016/j.tibs.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Brown C.J., Hendrich B.D., Rupert J.L., et al. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71(3):527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 21.Brown C.J., Ballabio A., Rupert J.L., et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349(6304):38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 22.Brockdorff N., Ashworth A., Kay G.F., et al. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature. 1991;351(6324):329–331. doi: 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- 23.Markaki Y., Chong J.G., Wang Y., et al. Xist nucleates local protein gradients to propagate silencing across the X chromosome. Cell. 2021;184(25):6174–6192. doi: 10.1016/j.cell.2021.10.022. e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Żylicz J.J., Bousard A., Žumer K., et al. The implication of early chromatin changes in X chromosome inactivation. Cell. 2019;176(1–2):182–197. doi: 10.1016/j.cell.2018.11.041. e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu N., Tsai C.L., Lee J.T. Transient homologous chromosome pairing marks the onset of X inactivation. Science. 2006;311(5764):1149–1152. doi: 10.1126/science.1122984. [DOI] [PubMed] [Google Scholar]
- 26.Monkhorst K., Jonkers I., Rentmeester E., Grosveld F., Gribnau J. X inactivation counting and choice is a stochastic process: evidence for involvement of an X-linked activator. Cell. 2008;132(3):410–421. doi: 10.1016/j.cell.2007.12.036. [DOI] [PubMed] [Google Scholar]
- 27.Jégu T., Aeby E., Lee J.T. The X chromosome in space. Nat Rev Genet. 2017;18(6):377–389. doi: 10.1038/nrg.2017.17. [DOI] [PubMed] [Google Scholar]
- 28.Furlan G., Gutierrez Hernandez N., Huret C., et al. The ftx noncoding locus controls X chromosome inactivation independently of its RNA products. Mol Cell. 2018;70(3):462–472. doi: 10.1016/j.molcel.2018.03.024. e8. [DOI] [PubMed] [Google Scholar]
- 29.Tian D., Sun S., Lee J.T. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143(3):390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun S., del Rosario B.C., Szanto A., Ogawa Y., Jeon Y., Lee J.T. Jpx RNA activates xist by evicting CTCF. Cell. 2013;153(7):1537–1551. doi: 10.1016/j.cell.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jonkers I., Barakat T.S., Achame E.M., et al. RNF12 is an X-Encoded dose-dependent activator of X chromosome inactivation. Cell. 2009;139(5):999–1011. doi: 10.1016/j.cell.2009.10.034. [DOI] [PubMed] [Google Scholar]
- 32.Lee J.T., Davidow L.S., Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet. 1999;21(4):400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 33.Lee J.T. Regulation of X-chromosome counting by Tsix and Xite sequences. Science. 2005;309(5735):768–771. doi: 10.1126/science.1113673. [DOI] [PubMed] [Google Scholar]
- 34.Bell A.C., West A.G., Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98(3):387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 35.Navarro P., Page D.R., Avner P., Rougeulle C. Tsix-mediated epigenetic switch of a CTCF-flanked region of the Xist promoter determines the Xist transcription program. Genes Dev. 2006;20(20):2787–2792. doi: 10.1101/gad.389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gontan C., Achame E.M., Demmers J., et al. RNF12 initiates X-chromosome inactivation by targeting REX1 for degradation. Nature. 2012;485(7398):386–390. doi: 10.1038/nature11070. [DOI] [PubMed] [Google Scholar]
- 37.Navarro P., Oldfield A., Legoupi J., et al. Molecular coupling of Tsix regulation and pluripotency. Nature. 2010;468(7322):457–460. doi: 10.1038/nature09496. [DOI] [PubMed] [Google Scholar]
- 38.Makhlouf M., Ouimette J.F., Oldfield A., Navarro P., Neuillet D., Rougeulle C. A prominent and conserved role for YY1 in Xist transcriptional activation. Nat Commun. 2014;5:4878. doi: 10.1038/ncomms5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robert-Finestra T., Tan B.F., Mira-Bontenbal H., et al. SPEN is required for Xist upregulation during initiation of X chromosome inactivation. Nat Commun. 2021;12(1):7000. doi: 10.1038/s41467-021-27294-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fukuda A., Hazelbaker D.Z., Motosugi N., et al. De novo DNA methyltransferases DNMT3A and DNMT3B are essential for XIST silencing for erosion of dosage compensation in pluripotent stem cells. Stem Cell Reports. 2021;16(9):2138–2148. doi: 10.1016/j.stemcr.2021.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chu C., Zhang Q.C., da Rocha S.T., et al. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161(2):404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grant M., Zuccotti M., Monk M. Methylation of CpG sites of two X-linked genes coincides with X-inactivation in the female mouse embryo but not in the germ line. Nat Genet. 1992;2(2):161–166. doi: 10.1038/ng1092-161. [DOI] [PubMed] [Google Scholar]
- 43.Jeppesen P., Turner B.M. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell. 1993;74(2):281–289. doi: 10.1016/0092-8674(93)90419-q. [DOI] [PubMed] [Google Scholar]
- 44.Chaumeil J., Okamoto I., Guggiari M., Heard E. Integrated kinetics of X chromosome inactivation in differentiating embryonic stem cells. Cytogenet Genome Res. 2002;99(1–4):75–84. doi: 10.1159/000071577. [DOI] [PubMed] [Google Scholar]
- 45.Heard E., Rougeulle C., Arnaud D., Avner P., Allis C.D., Spector D.L. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell. 2001;107(6):727–738. doi: 10.1016/s0092-8674(01)00598-0. [DOI] [PubMed] [Google Scholar]
- 46.Keniry A., Gearing L.J., Jansz N., et al. Setdb1-mediated H3K9 methylation is enriched on the inactive X and plays a role in its epigenetic silencing. Epigenetics Chromatin. 2016;9:16. doi: 10.1186/s13072-016-0064-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pandya-Jones A., Markaki Y., Serizay J., et al. A protein assembly mediates Xist localization and gene silencing. Nature. 2020;587(7832):145–151. doi: 10.1038/s41586-020-2703-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ridings-Figueroa R., Stewart E.R., Nesterova T.B., et al. The nuclear matrix protein CIZ1 facilitates localization of Xist RNA to the inactive X-chromosome territory. Genes Dev. 2017;31(9):876–888. doi: 10.1101/gad.295907.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sunwoo H., Colognori D., Froberg J.E., Jeon Y., Lee J.T. Repeat E anchors Xist RNA to the inactive X chromosomal compartment through CDKN1A-interacting protein (CIZ1) Proc Natl Acad Sci U S A. 2017;114(40):10654–10659. doi: 10.1073/pnas.1711206114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yue M., Ogawa A., Yamada N., Charles Richard J.L., Barski A., Ogawa Y. Xist RNA repeat E is essential for ASH2L recruitment to the inactive X and regulates histone modifications and escape gene expression. PLoS Genet. 2017;13(7) doi: 10.1371/journal.pgen.1006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Monfort A., di Minin G., Postlmayr A., et al. Identification of spen as a crucial factor for xist function through forward genetic screening in haploid embryonic stem cells. Cell Rep. 2015;12(4):554–561. doi: 10.1016/j.celrep.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dossin F., Pinheiro I., Żylicz J.J., et al. SPEN integrates transcriptional and epigenetic control of X-inactivation. Nature. 2020;578(7795):455–460. doi: 10.1038/s41586-020-1974-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McHugh C.A., Chen C.K., Chow A., et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521(7551):232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schoeftner S., Sengupta A.K., Kubicek S., et al. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 2006;25(13):3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Almeida M., Pintacuda G., Masui O., et al. PCGF3/5-PRC1 initiates Polycomb recruitment in X chromosome inactivation. Science. 2017;356(6342):1081–1084. doi: 10.1126/science.aal2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bousard A., Raposo A.C., Żylicz J.J., et al. The role of Xist-mediated Polycomb recruitment in the initiation of X-chromosome inactivation. EMBO Rep. 2019;20(10) doi: 10.15252/embr.201948019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pintacuda G., Wei G., Roustan C., et al. hnRNPK recruits PCGF3/5-PRC1 to the Xist RNA B-repeat to establish polycomb-mediated chromosomal silencing. Mol Cell. 2017;68(5):955–969. doi: 10.1016/j.molcel.2017.11.013. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee J.T., Bartolomei M.S. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152(6):1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 59.Dixon-McDougall T., Brown C.J. Independent domains for recruitment of PRC1 and PRC2 by human XIST. PLoS Genet. 2021;17(3) doi: 10.1371/journal.pgen.1009123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Patil D.P., Chen C.K., Pickering B.F., et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537(7620):369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Coker H., Wei G., Moindrot B., Mohammed S., Nesterova T., Brockdorff N. The role of the Xist 5' m6A region and RBM15 in X chromosome inactivation. Wellcome Open Res. 2020;5:31. doi: 10.12688/wellcomeopenres.15711.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Engreitz J.M., Pandya-Jones A., McDonel P., et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341(6147):1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jeon Y., Lee J.T. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011;146(1):119–133. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen C.K., Blanco M., Jackson C., et al. Xist recruits the X chromosome to the nuclear Lamina to enable chromosome-wide silencing. Science. 2016;354(6311):468–472. doi: 10.1126/science.aae0047. [DOI] [PubMed] [Google Scholar]
- 65.Colognori D., Sunwoo H., Kriz A.J., Wang C.Y., Lee J.T. Xist deletional analysis reveals an interdependency between xist RNA and polycomb complexes for spreading along the inactive X. Mol Cell. 2019;74(1):101–117. doi: 10.1016/j.molcel.2019.01.015. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wutz A., Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell. 2000;5(4):695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- 67.Chan K.M., Zhang H., Malureanu L., van Deursen J., Zhang Z. Diverse factors are involved in maintaining X chromosome inactivation. Proc Natl Acad Sci U S A. 2011;108(40):16699–16704. doi: 10.1073/pnas.1107616108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schultz D.C., Ayyanathan K., Negorev D., Maul G.G., Rauscher F.J., 3rd SETDB1:a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16(8):919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lopes-Ramos C.M., Quackenbush J., DeMeo D.L. Genome-wide sex and gender differences in cancer. Front Oncol. 2020;10:597788. doi: 10.3389/fonc.2020.597788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 71.Libert C., Dejager L., Pinheiro I. The X chromosome in immune functions: when a chromosome makes the difference. Nat Rev Immunol. 2010;10(8):594–604. doi: 10.1038/nri2815. [DOI] [PubMed] [Google Scholar]
- 72.Park R., Chidharla A., Mehta K., Sun W., Wulff-Burchfield E., Kasi A. Sex-bias in COVID-19-associated illness severity and mortality in cancer patients: a systematic review and meta-analysis. EClinicalMedicine. 2020;26:100519. doi: 10.1016/j.eclinm.2020.100519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brodin P. Immune determinants of COVID-19 disease presentation and severity. Nat Med. 2021;27(1):28–33. doi: 10.1038/s41591-020-01202-8. [DOI] [PubMed] [Google Scholar]
- 74.Wenham C., Smith J., Morgan R. Gender and COVID-19 Working Group. COVID-19:the gendered impacts of the outbreak. Lancet. 2020;395(10227):846–848. doi: 10.1016/S0140-6736(20)30526-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li C.H., Prokopec S.D., Sun R.X., et al. Sex differences in oncogenic mutational processes. Nat Commun. 2020;11(1):4330. doi: 10.1038/s41467-020-17359-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhu Y., Shao X., Wang X., Liu L., Liang H. Sex disparities in cancer. Cancer Lett. 2019;466:35–38. doi: 10.1016/j.canlet.2019.08.017. [DOI] [PubMed] [Google Scholar]
- 77.Galasso V., Pons V., Profeta P., Becher M., Brouard S., Foucault M. Gender differences in COVID-19 attitudes and behavior: panel evidence from eight countries. Proc Natl Acad Sci U S A. 2020;117(44):27285–27291. doi: 10.1073/pnas.2012520117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vlassoff C. Gender differences in determinants and consequences of health and illness. J Health Popul Nutr. 2007;25(1):47–61. [PMC free article] [PubMed] [Google Scholar]
- 79.Klein S.L., Flanagan K.L. Sex differences in immune responses. Nat Rev Immunol. 2016;16(10):626–638. doi: 10.1038/nri.2016.90. [DOI] [PubMed] [Google Scholar]
- 80.Youness A., Miquel C.H., Guéry J.C. Escape from X chromosome inactivation and the female predominance in autoimmune diseases. Int J Mol Sci. 2021;22(3):1114. doi: 10.3390/ijms22031114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fish E.N. The X-files in immunity: sex-based differences predispose immune responses. Nat Rev Immunol. 2008;8(9):737–744. doi: 10.1038/nri2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pinheiro I., Dejager L., Libert C. X-chromosome-located microRNAs in immunity: might they explain male/female differences? The X chromosome-genomic context may affect X-located miRNAs and downstream signaling, thereby contributing to the enhanced immune response of females. Bioessays. 2011;33(11):791–802. doi: 10.1002/bies.201100047. [DOI] [PubMed] [Google Scholar]
- 83.Berletch J.B., Ma W., Yang F., et al. Escape from X inactivation varies in mouse tissues. PLoS Genet. 2015;11(3):e1005079. doi: 10.1371/journal.pgen.1005079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Carrel L., Willard H.F. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434(7031):400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 85.Tukiainen T., Villani A.C., Yen A., et al. Landscape of X chromosome inactivation across human tissues. Nature. 2017;550(7675):244–248. doi: 10.1038/nature24265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Balaton B.P., Cotton A.M., Brown C.J. Derivation of consensus inactivation status for X-linked genes from genome-wide studies. Biol Sex Differ. 2015;6:35. doi: 10.1186/s13293-015-0053-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pyfrom S., Paneru B., Knox J.J., et al. The dynamic epigenetic regulation of the inactive X chromosome in healthy human B cells is dysregulated in lupus patients. Proc Natl Acad Sci U S A. 2021;118(24) doi: 10.1073/pnas.2024624118. e2024624118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Syrett C.M., Paneru B., Sandoval-Heglund D., et al. Altered X-chromosome inactivation in T cells may promote sex-biased autoimmune diseases. JCI Insight. 2019;4(7) doi: 10.1172/jci.insight.126751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Syrett C.M., Sindhava V., Sierra I., Dubin A.H., Atchison M., Anguera M.C. Diversity of epigenetic features of the inactive X-chromosome in NK cells, dendritic cells, and macrophages. Front Immunol. 2018;9:3087. doi: 10.3389/fimmu.2018.03087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang J., Syrett C.M., Kramer M.C., Basu A., Atchison M.L., Anguera M.C. Unusual maintenance of X chromosome inactivation predisposes female lymphocytes for increased expression from the inactive X. Proc Natl Acad Sci U S A. 2016;113(14):E2029–E2038. doi: 10.1073/pnas.1520113113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Syrett C.M., Sindhava V., Hodawadekar S., et al. Loss of Xist RNA from the inactive X during B cell development is restored in a dynamic YY1-dependent two-step process in activated B cells. PLoS Genet. 2017;13(10) doi: 10.1371/journal.pgen.1007050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cancro M.P. Age-associated B cells. Annu Rev Immunol. 2020;38:315–340. doi: 10.1146/annurev-immunol-092419-031130. [DOI] [PubMed] [Google Scholar]
- 93.Karnell J.L., Kumar V., Wang J., Wang S., Voynova E., Ettinger R. Role of CD11c + T-bet + B cells in human health and disease. Cell Immunol. 2017;321:40–45. doi: 10.1016/j.cellimm.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 94.Celhar T., Magalhães R., Fairhurst A.M. TLR7 and TLR9 in SLE: when sensing self Goes wrong. Immunol Res. 2012;53(1–3):58–77. doi: 10.1007/s12026-012-8270-1. [DOI] [PubMed] [Google Scholar]
- 95.Woodruff M.C., Ramonell R.P., Nguyen D.C., et al. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat Immunol. 2020;21(12):1506–1516. doi: 10.1038/s41590-020-00814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dunford A., Weinstock D.M., Savova V., et al. Tumor-suppressor genes that escape from X-inactivation contribute to cancer sex bias. Nat Genet. 2017;49(1):10–16. doi: 10.1038/ng.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pinto E.M., Chen X., Easton J., et al. Genomic landscape of paediatric adrenocortical tumours. Nat Commun. 2015;6:6302. doi: 10.1038/ncomms7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jiang L., Gu Z.H., Yan Z.X., et al. Exome sequencing identifies somatic mutations of DDX3X in natural killer/T-cell lymphoma. Nat Genet. 2015;47(9):1061–1066. doi: 10.1038/ng.3358. [DOI] [PubMed] [Google Scholar]
- 99.Dalgliesh G.L., Furge K., Greenman C., et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463(7279):360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ntziachristos P., Tsirigos A., Welstead G.G., et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature. 2014;514(7523):513–517. doi: 10.1038/nature13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shi B., Li W., Song Y., et al. UTX condensation underlies its tumour-suppressive activity. Nature. 2021;597(7878):726–731. doi: 10.1038/s41586-021-03903-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.van Haaften G., Dalgliesh G.L., Davies H., et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41(5):521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Van der Meulen J., Sanghvi V., Mavrakis K., et al. The H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia. Blood. 2015;125(1):13–21. doi: 10.1182/blood-2014-05-577270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kaneko S., Li X. X chromosome protects against bladder cancer in females via a KDM6A-dependent epigenetic mechanism. Sci Adv. 2018;4(6):eaar5598. doi: 10.1126/sciadv.aar5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hu Y., Deng C., Zhang H., Zhang J., Peng B., Hu C. Long non-coding RNA XIST promotes cell growth and metastasis through regulating miR-139-5p mediated Wnt/β-catenin signaling pathway in bladder cancer. Oncotarget. 2017;8(55):94554–94568. doi: 10.18632/oncotarget.21791. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 106.Fang J., Sun C.C., Gong C. Long noncoding RNA XIST acts as an oncogene in non-small cell lung cancer by epigenetically repressing KLF2 expression. Biochem Biophys Res Commun. 2016;478(2):811–817. doi: 10.1016/j.bbrc.2016.08.030. [DOI] [PubMed] [Google Scholar]
- 107.Liu J.L., Zhang W.Q., Zhao M., Huang M.Y. Upregulation of long noncoding RNA XIST is associated with poor prognosis in human cancers. J Cell Physiol. 2019;234(5):6594–6600. doi: 10.1002/jcp.27400. [DOI] [PubMed] [Google Scholar]
- 108.Liu A., Liu L., Lu H. LncRNA XIST facilitates proliferation and epithelial-mesenchymal transition of colorectal cancer cells through targeting miR-486-5p and promoting neuropilin-2. J Cell Physiol. 2019;234(8):13747–13761. doi: 10.1002/jcp.28054. [DOI] [PubMed] [Google Scholar]
- 109.Sun N., Zhang G., Liu Y. Long non-coding RNA XIST sponges miR-34a to promotes colon cancer progression via Wnt/β-catenin signaling pathway. Gene. 2018;665:141–148. doi: 10.1016/j.gene.2018.04.014. [DOI] [PubMed] [Google Scholar]
- 110.Song H., He P., Shao T., Li Y., Li J., Zhang Y. Long non-coding RNA XIST functions as an oncogene in human colorectal cancer by targeting miR-132-3p. J BUON. 2017;22(3):696–703. [PubMed] [Google Scholar]
- 111.Sun W., Zu Y., Fu X., Deng Y. Knockdown of lncRNA-XIST enhances the chemosensitivity of NSCLC cells via suppression of autophagy. Oncol Rep. 2017;38(6):3347–3354. doi: 10.3892/or.2017.6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang H., Shen Q., Zhang X., et al. The long non-coding RNA XIST controls non-small cell lung cancer proliferation and invasion by modulating miR-186-5p. Cell Physiol Biochem. 2017;41(6):2221–2229. doi: 10.1159/000475637. [DOI] [PubMed] [Google Scholar]
- 113.Zhang Y.L., Li X.B., Hou Y.X., Fang N.Z., You J.C., Zhou Q.H. The lncRNA XIST exhibits oncogenic properties via regulation of miR-449a and Bcl-2 in human non-small cell lung cancer. Acta Pharmacol Sin. 2017;38(3):371–381. doi: 10.1038/aps.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhou X., Xu X., Gao C., Cui Y. XIST promote the proliferation and migration of non-small cell lung cancer cells via sponging miR-16 and regulating CDK8 expression. Am J Transl Res. 2019;11(9):6196–6206. [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang X.T., Pan S.X., Wang A.H., Kong Q.Y., Jiang K.T., Yu Z.B. Long non-coding RNA (lncRNA) X-inactive specific transcript (XIST) plays a critical role in predicting clinical prognosis and progression of colorectal cancer. Med Sci Monit. 2019;25:6429–6435. doi: 10.12659/MSM.915329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hu B., Shi G., Li Q., Li W., Zhou H. Long noncoding RNA XIST participates in bladder cancer by downregulating p53 via binding to TET1. J Cell Biochem. 2019;120(4):6330–6338. doi: 10.1002/jcb.27920. [DOI] [PubMed] [Google Scholar]
- 117.Liu X., Cui L., Hua D. Long noncoding RNA XIST regulates miR-137-EZH2 axis to promote tumor metastasis in colorectal cancer. Oncol Res. 2018;27(1):99–106. doi: 10.3727/096504018X15195193936573. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 118.Xu R., Zhu X., Chen F., et al. LncRNA XIST/miR-200c regulates the stemness properties and tumourigenicity of human bladder cancer stem cell-like cells. Cancer Cell Int. 2018;18:41. doi: 10.1186/s12935-018-0540-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhou K., Yang J., Li X., Chen W. Long non-coding RNA XIST promotes cell proliferation and migration through targeting miR-133a in bladder cancer. Exp Ther Med. 2019;18(5):3475–3483. doi: 10.3892/etm.2019.7960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li C., Wan L., Liu Z., et al. Long non-coding RNA XIST promotes TGF-β-induced epithelial-mesenchymal transition by regulating miR-367/141-ZEB2 axis in non-small-cell lung cancer. Cancer Lett. 2018;418:185–195. doi: 10.1016/j.canlet.2018.01.036. [DOI] [PubMed] [Google Scholar]
- 121.Qiu H.B., Yang K., Yu H.Y., Liu M. Downregulation of long non-coding RNA XIST inhibits cell proliferation, migration, invasion and EMT by regulating miR-212-3p/CBLL1 axis in non-small cell lung cancer cells. Eur Rev Med Pharmacol Sci. 2019;23(19):8391–8402. doi: 10.26355/eurrev_201910_19150. [DOI] [PubMed] [Google Scholar]
- 122.Jiang Q., Xing W., Cheng J., Yu Y. Knockdown of lncRNA XIST suppresses cell tumorigenicity in human non-small cell lung cancer by regulating miR-142-5p/PAX6 axis. Onco Targets Ther. 2020;13:4919–4929. doi: 10.2147/OTT.S238808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rong H., Chen B., Wei X., et al. Long non-coding RNA XIST expedites lung adenocarcinoma progression through upregulating MDM2 expression via binding to miR-363-3p. Thorac Cancer. 2020;11(3):659–671. doi: 10.1111/1759-7714.13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang X., Zhang G., Cheng Z., et al. Knockdown of LncRNA-XIST suppresses proliferation and TGF-β1-induced EMT in NSCLC through the Notch-1 pathway by regulation of miR-137. Genet Test Mol Biomarkers. 2018;22(6):333–342. doi: 10.1089/gtmb.2018.0026. [DOI] [PubMed] [Google Scholar]
- 125.Xiong Y., Wang L., Li Y., Chen M., He W., Qi L. The long non-coding RNA XIST interacted with miR-124 to modulate bladder cancer growth, invasion and migration by targeting androgen receptor (AR) Cell Physiol Biochem. 2017;43(1):405–418. doi: 10.1159/000480419. [DOI] [PubMed] [Google Scholar]
- 126.Lombard A.P., Mudryj M. The emerging role of the androgen receptor in bladder cancer. Endocr Relat Cancer. 2015;22(5) doi: 10.1530/ERC-15-0209. R265-277. [DOI] [PubMed] [Google Scholar]
- 127.Laor E., Schiffman Z.J., Braunstein J.D., et al. Androgen receptors in bladder tumors. Urology. 1985;25(2):161–163. doi: 10.1016/0090-4295(85)90534-5. [DOI] [PubMed] [Google Scholar]
- 128.Miyamoto H., Yang Z., Chen Y.T., et al. Promotion of bladder cancer development and progression by androgen receptor signals. J Natl Cancer Inst. 2007;99(7):558–568. doi: 10.1093/jnci/djk113. [DOI] [PubMed] [Google Scholar]
- 129.Garg M., Maurya N. WNT/β-catenin signaling in urothelial carcinoma of bladder. World J Nephrol. 2019;8(5):83–94. doi: 10.5527/wjn.v8.i5.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Overdevest J.B., Knubel K.H., Duex J.E., et al. CD24 expression is important in male urothelial tumorigenesis and metastasis in mice and is androgen regulated. Proc Natl Acad Sci U S A. 2012;109(51) doi: 10.1073/pnas.1113960109. E3588–E3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Groenendijk F.H., de Jong J., Fransen, van de Putte E.E., et al. ERBB2 mutations characterize a subgroup of muscle-invasive bladder cancers with excellent response to neoadjuvant chemotherapy. Eur Urol. 2016;69(3):384–388. doi: 10.1016/j.eururo.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 132.Inoue S., Ide H., Mizushima T., Jiang G., Kawahara T., Miyamoto H. ELK1 promotes urothelial tumorigenesis in the presence of an activated androgen receptor. Am J Cancer Res. 2018;8(11):2325–2336. [PMC free article] [PubMed] [Google Scholar]
- 133.Zhao W., Li S., Tang D., et al. Androgen receptor expression in male breast cancer predicts inferior outcome and poor response to tamoxifen treatment. Eur J Endocrinol. 2014;171(4):527–533. doi: 10.1530/EJE-14-0278. [DOI] [PubMed] [Google Scholar]
- 134.Chen D.L., Chen L.Z., Lu Y.X., et al. Long noncoding RNA XIST expedites metastasis and modulates epithelial-mesenchymal transition in colorectal cancer. Cell Death Dis. 2017;8(8) doi: 10.1038/cddis.2017.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang R., Wang Z., Yu Q., et al. Atractylenolide II reverses the influence of lncRNA XIST/miR-30a-5p/ROR1 axis on chemo-resistance of colorectal cancer cells. J Cell Mol Med. 2019;23(5):3151–3165. doi: 10.1111/jcmm.14148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yang L.G., Cao M.Z., Zhang J., Li X.Y., Sun Q.L. LncRNA XIST modulates HIF-1A/AXL signaling pathway by inhibiting miR-93-5p in colorectal cancer. Mol Genet Genomic Med. 2020;8(4):e1112. doi: 10.1002/mgg3.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Li W., He Y., Cheng Z. Long noncoding RNA XIST knockdown suppresses the growth of colorectal cancer cells via regulating microRNA-338-3p/PAX5 axis. Eur J Cancer Prev. 2021;30(2):132–142. doi: 10.1097/CEJ.0000000000000596. [DOI] [PubMed] [Google Scholar]
- 138.Zeng Z.L., Lu J.H., Wang Y., et al. The lncRNA XIST/miR-125b-2-3p axis modulates cell proliferation and chemotherapeutic sensitivity via targeting Wee1 in colorectal cancer. Cancer Med. 2021;10(7):2423–2441. doi: 10.1002/cam4.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yip K.W., Reed J.C. Bcl-2 family proteins and cancer. Oncogene. 2008;27(50):6398–6406. doi: 10.1038/onc.2008.307. [DOI] [PubMed] [Google Scholar]
- 140.Firestein R., Bass A.J., Kim S.Y., et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455(7212):547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Marei H.E., Althani A., Afifi N., et al. p53 signaling in cancer progression and therapy. Cancer Cell Int. 2021;21(1):703. doi: 10.1186/s12935-021-02396-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yang X., Zhang S., He C., et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer. 2020;19(1):46. doi: 10.1186/s12943-020-1146-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sung H., Ferlay J., Siegel R.L., et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 144.Zheng R., Lin S., Guan L., et al. Long non-coding RNA XIST inhibited breast cancer cell growth, migration, and invasion via miR-155/CDX1 axis. Biochem Biophys Res Commun. 2018;498(4):1002–1008. doi: 10.1016/j.bbrc.2018.03.104. [DOI] [PubMed] [Google Scholar]
- 145.Huang Y.S., Chang C.C., Lee S.S., Jou Y.S., Shih H.M. Xist reduction in breast cancer upregulates AKT phosphorylation via HDAC3-mediated repression of PHLPP1 expression. Oncotarget. 2016;7(28):43256–43266. doi: 10.18632/oncotarget.9673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Liu B., Luo C., Lin H., Ji X., Zhang E., Li X. Long noncoding RNA XIST acts as a ceRNA of miR-362-5p to suppress breast cancer progression. Cancer Biother Radiopharm. 2021;36(6):456–466. doi: 10.1089/cbr.2019.3481. [DOI] [PubMed] [Google Scholar]
- 147.Xing F., Liu Y., Wu S.Y., et al. Loss of XIST in breast cancer activates MSN-c-Met and reprograms microglia via exosomal miRNA to promote brain metastasis. Cancer Res. 2018;78(15):4316–4330. doi: 10.1158/0008-5472.CAN-18-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kim J.H., Sharma A., Dhar S.S., et al. UTX and MLL4 coordinately regulate transcriptional programs for cell proliferation and invasiveness in breast cancer cells. Cancer Res. 2014;74(6):1705–1717. doi: 10.1158/0008-5472.CAN-13-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Xu Y., Wang J., Wang J. Long noncoding RNA XIST promotes proliferation and invasion by targeting miR-141 in papillary thyroid carcinoma. Onco Targets Ther. 2018;11:5035–5043. doi: 10.2147/OTT.S170439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Liu H., Deng H., Zhao Y., Li C., Liang Y. LncRNA XIST/miR-34a axis modulates the cell proliferation and tumor growth of thyroid cancer through MET-PI3K-AKT signaling. J Exp Clin Cancer Res. 2018;37(1):279. doi: 10.1186/s13046-018-0950-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Du Y.L., Liang Y., Cao Y., Liu L., Li J., Shi G.Q. LncRNA XIST promotes migration and invasion of papillary thyroid cancer cell by modulating miR-101-3p/CLDN1 axis. Biochem Genet. 2021;59(2):437–452. doi: 10.1007/s10528-020-09985-8. [DOI] [PubMed] [Google Scholar]
- 152.Hu Y., Mei X.Q., Tang D. Long non-coding RNA XIST is down-regulated and correlated to better prognosis in ovarian cancer. Math Biosci Eng. 2020;17(3):2070–2081. doi: 10.3934/mbe.2020110. [DOI] [PubMed] [Google Scholar]
- 153.Huang R., Zhu L., Zhang Y. XIST lost induces ovarian cancer stem cells to acquire taxol resistance via a KMT2C-dependent way. Cancer Cell Int. 2020;20:436. doi: 10.1186/s12935-020-01500-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wang C., Qi S., Xie C., Li C., Wang P., Liu D. Upregulation of long non-coding RNA XIST has anticancer effects on epithelial ovarian cancer cells through inverse downregulation of hsa-miR-214-3p. J Gynecol Oncol. 2018;29(6):e99. doi: 10.3802/jgo.2018.29.e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Guo T., Yuan D., Zhang W., et al. Upregulation of long noncoding RNA XIST has anticancer effects on ovarian cancer through sponging miR-106a. Hum Cell. 2021;34(2):579–587. doi: 10.1007/s13577-020-00469-w. [DOI] [PubMed] [Google Scholar]
- 156.Huang K.C., Rao P.H., Lau C.C., et al. Relationship of XIST expression and responses of ovarian cancer to chemotherapy. Mol Cancer Ther. 2002;1(10):769–776. [PubMed] [Google Scholar]
- 157.Meng Q., Wang N., Duan G. Long non-coding RNA XIST regulates ovarian cancer progression via modulating miR-335/BCL2L2 axis. World J Surg Oncol. 2021;19(1):165. doi: 10.1186/s12957-021-02274-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Jiang R., Zhang H., Zhou J., et al. Inhibition of long non-coding RNA XIST upregulates microRNA-149-3p to repress ovarian cancer cell progression. Cell Death Dis. 2021;12(2):145. doi: 10.1038/s41419-020-03358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhu H., Zheng T., Yu J., Zhou L., Wang L. LncRNA XIST accelerates cervical cancer progression via upregulating Fus through competitively binding with miR-200a. Biomed Pharmacother. 2018;105:789–797. doi: 10.1016/j.biopha.2018.05.053. [DOI] [PubMed] [Google Scholar]
- 160.Chen X., Xiong D., Ye L., et al. Up-regulated lncRNA XIST contributes to progression of cervical cancer via regulating miR-140-5p and ORC1. Cancer Cell Int. 2019;19:45. doi: 10.1186/s12935-019-0744-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Liu X., Xie S., Zhang J., Kang Y. Long noncoding RNA XIST contributes to cervical cancer development through targeting miR-889-3p/SIX1 axis. Cancer Biother Radiopharm. 2020;35(9):640–649. doi: 10.1089/cbr.2019.3318. [DOI] [PubMed] [Google Scholar]
- 162.Gong X., Bacchelli E., Blasi F., et al. Analysis of X chromosome inactivation in autism spectrum disorders. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(6):830–835. doi: 10.1002/ajmg.b.30688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Guo L., Zhong M.B., Zhang L., Zhang B., Cai D. Sex differences in Alzheimer's disease: insights from the multiomics landscape. Biol Psychiatr. 2022;91(1):61–71. doi: 10.1016/j.biopsych.2021.02.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Brand B.A., Blesson A.E., Smith-Hicks C.L. The impact of X-chromosome inactivation on phenotypic expression of X-linked neurodevelopmental disorders. Brain Sci. 2021;11(7):904. doi: 10.3390/brainsci11070904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Plenge R.M., Hendrich B.D., Schwartz C., et al. A promoter mutation in the XIST gene in two unrelated families with skewed X-chromosome inactivation. Nat Genet. 1997;17(3):353–356. doi: 10.1038/ng1197-353. [DOI] [PubMed] [Google Scholar]
- 166.Talebizadeh Z., Bittel D.C., Veatch O.J., Kibiryeva N., Butler M.G. Brief report: non-random X chromosome inactivation in females with autism. J Autism Dev Disord. 2005;35(5):675–681. doi: 10.1007/s10803-005-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sripathy S., Leko V., Adrianse R.L., et al. Screen for reactivation of MeCP2 on the inactive X chromosome identifies the BMP/TGF-β superfamily as a regulator of XIST expression. Proc Natl Acad Sci U S A. 2017;114(7):1619–1624. doi: 10.1073/pnas.1621356114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Good K.V., Vincent J.B., Ausió J. MeCP2:the genetic driver of rett syndrome epigenetics. Front Genet. 2021;12:620859. doi: 10.3389/fgene.2021.620859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Amir R.E., Van den Veyver I.B., Wan M., Tran C.Q., Francke U., Zoghbi H.Y. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23(2):185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 170.Xiol C., Vidal S., Pascual-Alonso A., et al. X chromosome inactivation does not necessarily determine the severity of the phenotype in Rett syndrome patients. Sci Rep. 2019;9(1):11983. doi: 10.1038/s41598-019-48385-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wang J., Song W. Regulation of LRRK2 promoter activity and gene expression by Sp1. Mol Brain. 2016;9:33. doi: 10.1186/s13041-016-0215-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lee B.D., Dawson V.L., Dawson T.M. Leucine-rich repeat kinase 2 (LRRK2) as a potential therapeutic target in Parkinson's disease. Trends Pharmacol Sci. 2012;33(7):365–373. doi: 10.1016/j.tips.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Cao J., Zhuang Y., Zhang J., et al. Leucine-rich repeat kinase 2 aggravates secondary brain injury induced by intracerebral hemorrhage in rats by regulating the P38 MAPK/Drosha pathway. Neurobiol Dis. 2018;119:53–64. doi: 10.1016/j.nbd.2018.07.024. [DOI] [PubMed] [Google Scholar]
- 174.Di Maio R., Hoffman E.K., Rocha E.M., et al. LRRK2 activation in idiopathic Parkinson's disease. Sci Transl Med. 2018;10(451):eaar5429. doi: 10.1126/scitranslmed.aar5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhou Q., Zhang M.M., Liu M., Tan Z.G., Qin Q.L., Jiang Y.G. LncRNA XIST sponges miR-199a-3p to modulate the Sp1/LRRK2 signal pathway to accelerate Parkinson's disease progression. Aging (Albany NY) 2021;13(3):4115–4137. doi: 10.18632/aging.202378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Du Y., Gao G., Li J., et al. Silencing of long noncoding RNA XIST attenuated Alzheimer's disease-related BACE1 alteration through miR-124. Cell Biol Int. 2020;44(2):630–636. doi: 10.1002/cbin.11263. [DOI] [PubMed] [Google Scholar]
- 177.Yan X.W., Liu H.J., Hong Y.X., Meng T., Du J., Chang C. lncRNA XIST induces Aβ accumulation and neuroinflammation by the epigenetic repression of NEP in Alzheimer's disease. J Neurogenet. 2022;36(1):11–20. doi: 10.1080/01677063.2022.2028784. [DOI] [PubMed] [Google Scholar]
- 178.Qin S., Predescu D.N., Patel M., Drazkowski P., Ganesh B., Predescu S.A. Sex differences in the proliferation of pulmonary artery endothelial cells: implications for plexiform arteriopathy. J Cell Sci. 2020;133(9):jcs237776. doi: 10.1242/jcs.237776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Batton K.A., Austin C.O., Bruno K.A., Burger C.D., Shapiro B.P., Fairweather D. Sex differences in pulmonary arterial hypertension: role of infection and autoimmunity in the pathogenesis of disease. Biol Sex Differ. 2018;9(1):15. doi: 10.1186/s13293-018-0176-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Qin S., Predescu D., Carman B., et al. Up-regulation of the long noncoding RNA X-inactive-specific transcript and the sex bias in pulmonary arterial hypertension. Am J Pathol. 2021;191(6):1135–1150. doi: 10.1016/j.ajpath.2021.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Patel M., Predescu D., Tandon R., et al. A novel p38 mitogen-activated protein kinase/Elk-1 transcription factor-dependent molecular mechanism underlying abnormal endothelial cell proliferation in plexogenic pulmonary arterial hypertension. J Biol Chem. 2013;288(36):25701–25716. doi: 10.1074/jbc.M113.502674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Atkins G.B., Jain M.K. Role of Krüppel-like transcription factors in endothelial biology. Circ Res. 2007;100(12):1686–1695. doi: 10.1161/01.RES.0000267856.00713.0a. [DOI] [PubMed] [Google Scholar]
- 183.Shapiro S., Traiger G.L., Turner M., McGoon M.D., Wason P., Barst R.J. Sex differences in the diagnosis, treatment, and outcome of patients with pulmonary arterial hypertension enrolled in the registry to evaluate early and long-term pulmonary arterial hypertension disease management. Chest. 2012;141(2):363–373. doi: 10.1378/chest.10-3114. [DOI] [PubMed] [Google Scholar]
- 184.Mair K.M., Johansen A.K., Wright A.F., Wallace E., MacLean M.R. Pulmonary arterial hypertension: basis of sex differences in incidence and treatment response. Br J Pharmacol. 2014;171(3):567–579. doi: 10.1111/bph.12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Xu D.Q., Luo Y., Liu Y., et al. Beta-estradiol attenuates hypoxic pulmonary hypertension by stabilizing the expression of p27kip1 in rats. Respir Res. 2010;11(1):182. doi: 10.1186/1465-9921-11-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Yuan P., Wu W.H., Gao L., et al. Oestradiol ameliorates monocrotaline pulmonary hypertension via NO, prostacyclin and endothelin-1 pathways. Eur Respir J. 2013;41(5):1116–1125. doi: 10.1183/09031936.00044112. [DOI] [PubMed] [Google Scholar]
- 187.Ye Y., Wang H., Chu J.H., et al. Atractylenolide II induces G1 cell-cycle arrest and apoptosis in B16 melanoma cells. J Ethnopharmacol. 2011;136(1):279–282. doi: 10.1016/j.jep.2011.04.020. [DOI] [PubMed] [Google Scholar]
- 188.Huang M.Y., Jiang X.M., Xu Y.L., et al. Platycodin D triggers the extracellular release of programed death Ligand-1 in lung cancer cells. Food Chem Toxicol. 2019;131:110537. doi: 10.1016/j.fct.2019.05.045. [DOI] [PubMed] [Google Scholar]
- 189.Peng Y., Fan J.Y., Xiong J., Lou Y., Zhu Y. miR-34a enhances the susceptibility of gastric cancer to platycodin D by targeting survivin. Pathobiology. 2019;86(5–6):296–305. doi: 10.1159/000502913. [DOI] [PubMed] [Google Scholar]
- 190.Lu J.J., Lu D.Z., Chen Y.F., et al. Proteomic analysis of hepatocellular carcinoma HepG2 cells treated with platycodin D. Chin J Nat Med. 2015;13(9):673–679. doi: 10.1016/S1875-5364(15)30065-0. [DOI] [PubMed] [Google Scholar]
- 191.Chen D., Chen T., Guo Y., Wang C., Dong L., Lu C. Platycodin D (PD) regulates LncRNA-XIST/miR-335 axis to slow down bladder cancer progression in vitro and in vivo. Exp Cell Res. 2020;396(1):112281. doi: 10.1016/j.yexcr.2020.112281. [DOI] [PubMed] [Google Scholar]
- 192.Chen Y., Li Z., Chen X., Zhang S. Long non-coding RNAs: from disease code to drug role. Acta Pharm Sin B. 2021;11(2):340–354. doi: 10.1016/j.apsb.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Setten R.L., Rossi J.J., Han S.P. The Current state and future directions of RNAi-based therapeutics. Nat Rev Drug Discov. 2019;18(6):421–446. doi: 10.1038/s41573-019-0017-4. [DOI] [PubMed] [Google Scholar]
- 194.Rinaldi C., Wood M.J.A. Antisense oligonucleotides: the next frontier for treatment of neurological disorders. Nat Rev Neurol. 2018;14(1):9–21. doi: 10.1038/nrneurol.2017.148. [DOI] [PubMed] [Google Scholar]
- 195.Gardiner K.J. Molecular basis of pharmacotherapies for cognition in Down syndrome. Trends Pharmacol Sci. 2010;31(2):66–73. doi: 10.1016/j.tips.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Jiang J., Jing Y., Cost G.J., et al. Translating dosage compensation to trisomy 21. Nature. 2013;500(7462):296–300. doi: 10.1038/nature12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Chiang J.C., Jiang J., Newburger P.E., Lawrence J.B. Trisomy silencing by XIST normalizes Down syndrome cell pathogenesis demonstrated for hematopoietic defects in vitro. Nat Commun. 2018;9(1):5180. doi: 10.1038/s41467-018-07630-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Spiering A.E., de Vries T.J. Why females do better: the X chromosomal TLR7 gene-dose effect in COVID-19. Front Immunol. 2021;12:756262. doi: 10.3389/fimmu.2021.756262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kopp F., Mendell J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]