Abstract
Transcription of the K+ transport operon kdp in Escherichia coli is induced during K+-limited growth by the action of a dual-component phosphorelay regulatory system comprised of a sensor kinase (integral membrane protein), KdpD, and a DNA-binding response regulator (cytoplasmic protein), KdpE. In this study, we screened for new dke (named dke for decreased kdp expression) mutations (in loci other than kdpDE) that led to substantially decreased kdp expression. One dke mutation was shown to be in hns, encoding the nucleoid protein H-NS. Another dke mutation was mapped to trxB (encoding thioredoxin reductase), and an equivalent reduction in kdp expression was demonstrated also for trxA mutants that are deficient in thioredoxin 1. Exogenously provided dithiothreitol rescued the kdp expression defect in trxB but not trxA mutants. Neither trxB nor trxA affected gene regulation mediated by another dual-component system tested, EnvZ-OmpR. Mutations in genes dsbC and dsbD did not affect kdp expression, suggesting that the trx effects on kdp are not mediated by alterations in protein disulfide bond status in the periplasm. Reduced kdp expression was observed even in a trxB strain that harbored a variant KdpD polypeptide bearing no Cys residues. A trxB hns double mutant was even more severely affected for kdp expression than either single mutant. The dke mutations themselves had no effect on strength of the signal controlling kdp expression, and constitutive mutations in kdpDE were epistatic to hns and trxB. These results indicate that perturbations in cytoplasmic thiol oxidation status and in levels of the H-NS protein exert additive effects, direct or indirect, at a step(s) upstream of KdpD in the signal transduction pathway, which significantly influence the magnitude of KdpD kinase activity obtained for a given strength of the inducing signal for kdp transcription.
Active uptake of K+ in Escherichia coli and other enterobacteria is mediated by an inducible high-affinity transport system, Kdp, and at least three lower-affinity transport systems (TrkD [also called Kup], TrkG, and TrkH) that are constitutively expressed (reviewed in reference 46). The Kdp transporter is a P-type ATPase comprised of four polypeptides encoded by genes of the kdpFABC operon. It appears that the physiological role of Kdp is to permit growth of E. coli in medium containing a sufficiently low concentration of extracellular K+ ([K+]e) that is not adequate for uptake through the constitutively expressed systems. The kdp operon is repressed under conditions of K+-replete growth and the Kdp transporter activity is also inhibited under these conditions.
Transcriptional control of the kdp operon has mainly been studied in strains carrying kdp-lac operon fusions, and it is mediated by KdpD and KdpE (37, 52), a protein pair that is a member of the family of dual-component regulatory systems found in various prokaryotes (for a review, see reference 36). KdpD (the sensor kinase) is an integral protein of the inner membrane which, during K+-limited growth, undergoes autophosphorylation on a cytoplasmic Asp residue; the phosphoryl group is then transferred to a His residue of the cytoplasmic response regulator protein KdpE, and phospho-KdpE binds to an operator site immediately upstream of the kdp operon promoter to activate transcription of the operon (21, 33, 34, 50). KdpD and KdpE are the products of an independent kdpDE operon situated immediately downstream of kdpFABC (37).
Even though the components of the signal transduction pathway downstream of KdpD autophosphorylation have been well characterized, the exact nature of the signal involved in kdp regulation is not clear. Among the alternatives that have been proposed as the signals determining KdpD kinase activity are intracellular K+ concentration ([K+]i), cell turgor, rate of transmembrane K+ flux, or the combination of [K+]e (or [K+]i) and osmotic strength of the medium (2, 12–14, 25, 27, 42, 49). Also not known is whether the signal acts directly on KdpD to modulate its kinase activity or indirectly via additional steps in the signal transduction pathway.
In this study, we employed approaches of insertional and localized mutagenesis to identify new loci that affect kdp-lac expression in trans. We found that mutations in trxA and trxB, encoding thioredoxin 1 and thioredoxin reductase, respectively, lead to a specific reduction in kdp-lac expression and that the reduction persists even in strains that express a cysteineless variant of the KdpD protein. We also found that a deficiency of nucleoid protein H-NS leads to down regulation of kdp. Data from epistasis experiments support the interpretation that the trx and hns mutations exert their effects on kdp regulation at a step(s) in the signal transduction pathway upstream of KdpD.
MATERIALS AND METHODS
Media and growth conditions.
Unless otherwise specified, cultures for determinations of growth rates and β-galactosidase activities were grown at 30°C in phosphate-buffered media with reciprocally varying concentrations of Na+ and K+ that were prepared, as described previously (9), by mixing together 115 mM K+-phosphate medium with 115 mM Na+-phosphate medium in the appropriate proportion so as to achieve the desired [K+]e. These media were supplemented with glucose and Casamino Acids (Difco) at 0.2 and 0.5%, respectively. Growth was monitored by measurement of absorbance at 600 nm. Medium KML (9) was used as the rich medium. Spectinomycin (Sp) was used at a final concentration of 50 μg/ml; other antibiotics and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) were added at concentrations as specified previously (2).
Bacterial strains and plasmids.
The E. coli K-12 strains employed in the study are listed in Table 1. The plasmids that were used included (i) pLG H-NS, a pSC101 replicon derivative which encodes Kanr and carries the cloned hns+ gene (54); (ii) pPV5-1 Cys+ and pPV5-1 Cys-less, which are both pMB9 (ColE1) replicon derivatives encoding Ampr and carrying variant versions of the kdpD gene under control of the tac promoter (the first has silent nucleotide substitutions that do not alter the amino acid sequence of the gene product and in the second, the codons for the six Cys residues in the native protein have all been altered to specify other amino acids [20]); and (iii) pBD-R511Q, which is also a pMB9 derivative encoding Ampr but which carries a kdpD variant (under control of a regulated ara promoter) with an altered codon 511 that specifies Gln instead of Arg (19). Additional plasmids pHYD704 and pHYD705 were constructed in this study from vector pCL1920 (pSC101 replicon, encoding Spr [26]) as described below; plasmid pHYD708 was constructed by the subcloning of a HindIII-SacI fragment carrying the lacIq gene from pMJR1560 (48) into the corresponding sites of vector pCL1920.
TABLE 1.
List of E. coli K-12 strainsa
| Strain | Genotype | Reference/source |
|---|---|---|
| MH225 | Δ(argF-lac)U169 rpsL150 relA1 araD139 flbB5301 deoC1 ptsF25 Φ(ompC-lacZ) 10-25 | 15 |
| TL1105A | thi rha nagA Δlac trkA405 trkD1 kdpA::[Mu lacZ(λ)] | 25 |
| GJ1426 | GJ1427 ΔtrxA | This study |
| GJ1427 | thi rha nagA lacZ trkA405 kdp-200::[λdlac(Ap)] | From GJ642 (reference 2) |
| GJ1428 | GJ1427 trxB30::Tn10dTet | This study |
| GJ1429 | GJ1427 ΔtrxA trxB30::Tn10dTet | This study |
| GJ1430 | GJ1427 trxB::kan | This study |
| GJ1431 | GJ1427 grxA::kan zbi::Tn10 | This study |
| GJ1438 | MH225 trxB30::Tn10dTet | This study |
| GJ1439 | MH225 trxB::kan | This study |
| GJ1441 | MH225 ΔtrxA | This study |
| GJ1442 | thi rha nagA lacZ trkA405 trkD1ΔkdpD kdp-204::λplacMu55(Kan) | From TK2240 (reference 11), in two steps |
| GJ1442H | GJ1442 hns-205::Tn10 | This study |
| GJ1442T | GJ1442 trxB30::Tn10dTet | This study |
| GJ1449 | thi rha nagA Δ(argF-lac)U169 trkA405 trkD1 kdp-200::[λdlac (Ap)] | From TK2205 (W. Epstein), in several steps |
| GJ1449H | GJ1449 hns-205::Tn10 | This study |
| GJ1449T | GJ1449 trxB30::Tn10dTet | This study |
| GJ1450 | GJ1449 kdp-205 | This study |
| GJ1450H | GJ1450 hns-205::Tn10 | This study |
| GJ1450T | GJ1450 trxB30::Tn10dTet | This study |
| GJ1451 | GJ1449 kdp-207 | This study |
| GJ1451H | GJ1451 hns-205::Tn10 | This study |
| GJ1451T | GJ1451 trxB30::Tn10dTet | This study |
| GJ1455 | TL1105A zci-3117::Tn10Kan hns-202 | This study |
| GJ1456 | TL1105A zci-3117::Tn10Kan | This study |
| GJ1458 | TL1105A zci-506::Tn10 hns-202 | This study |
| GJ1459 | TL1105A zci-506::Tn10 | This study |
| GJ1461 | TL1105A hns-205::Tn10 | This study |
| GJ1469 | GJ1427 hns-205::Tn10 | This study |
| GJ1470 | GJ1427 hns-205::Tn10 trxB::kan | This study |
| GJ1485 | TL1105A recB268::Tn10 | This study |
Genotype designations are as those in the work of Berlyn (4). All strains are F−. Allele numbers are given, where they are known. The trxB30 and hns-202 mutations are also referred to in the text as dke-1 and dke-2, respectively. In the strains listed, the following mutations were transduced from strains previously described: zci-3117::Tn10Kan and zci-506::Tn10 from CAG 18551 and CAG12169, respectively (47); hns-205::Tn10 from PD145 (8); ΔtrxA and trxB::kan from AD494 and WP570, respectively (7); grxA::kan and zbi::Tn10 from A407 (44); ΔkdpD from TKV2208 (20); recB268::Tn10 from JJC777 (5); and kdp-200, kdp-205, kdp-207, and kdp-204::λplacMu55(Kan) from GJ18, GJ618, GJ619, and GJ610, respectively (2).
Experimental techniques.
The procedures for P1 transduction (13), generation of Tn10dTet transpositions employing phage λ 1323 (22), and in vitro DNA manipulations and transformation (45) were as described previously. The procedure for making a strain ΔtrxA involved, first, the introduction of an ilv::Tn10 or ilv::Tn10Kan marker, followed by a second P1 transduction to Ilv+ with a lysate prepared on an ilv+ ΔtrxA strain; inheritance of ΔtrxA was assessed by scoring for resistance to phage T7 (28). The method of Murgola and Yanofsky (32) was followed for localized mutagenesis of the 28-min chromosomal region, in which P1 phage propagated on the zci-3117::Tn10Kan strain GJ1456 was treated with hydroxylamine and then used to transduce TL1105A to Kanr. The specific activity of β-galactosidase in cultures grown to mid-log phase was measured by the method of Miller (30), and the values are reported in Miller units.
RESULTS
Isolation of dke-1 and dke-2 mutants.
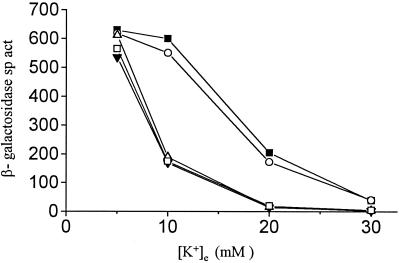
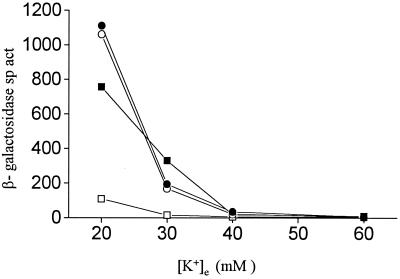
Strain TL1105A carries mutations in the kdp, trkA, and trkD genes (rendering it deficient in all the active transport systems for K+) and also a chromosomal kdp-lac fusion (25). Following whole-genome mutagenesis of a derivative of strain TL1105A with transposon Tn10dTet (22), we screened for clones that exhibited an altered lac expression phenotype on phosphate-buffered medium containing 20 mM [K+]e and X-Gal. One mutant exhibiting reduced lac expression under these conditions was identified, and preliminary P1 transduction experiments (data not shown) permitted the conclusions that the lac expression phenotype was (i) 100% linked to Tetr and (ii) unlinked to the kdpFABCDE locus. The mutation was designated dke-1 (named dke for decreased kdp expression). Comparison of the profiles of kdp-lac expression in an isogenic pair of strains, GJ1427 (dke+) and GJ1428 (dke-1::Tn10dTet), revealed that the reduction in kdp-lac expression in the latter was most pronounced (4- to 10-fold) at intermediate levels of [K+]e (Fig. 1). The further characterization of dke-1 is described below.
FIG. 1.
β-Galactosidase specific activities (sp. act., expressed in Miller units [30]) in kdp-lac strains with trx or grx mutations, as a function of [K+]e of the growth medium. ○, GJ1427 (parental); ▵, GJ1428 (dke-1; that is, trxB30::Tn10dTet); ▾, GJ1426 (ΔtrxA); ■, GJ1431 (grxA); □, GJ1429 (ΔtrxA trxB30::Tn10dTet).
The dke-2 mutant GJ1455 was also identified by screening on X-Gal-supplemented media derivatives of strain TL1105A, this time after localized mutagenesis of the 28-min region of the chromosome as described above. Our original rationale for undertaking this localized mutagenesis experiment was to examine whether missense mutations in kch, the gene encoding a putative K+ channel which maps to this chromosomal region (4, 43), could be identified that affect kdp-lac expression. However, the subsequent studies described below indicated that dke-2 is not in kch but is an hns mutation.
Characterization of dke-1 as a trxB::Tn10dTet insertion.
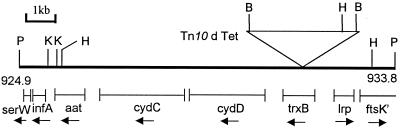
A PstI-digested chromosomal DNA library from a dke-1::Tn10dTet mutant derivative was established in the plasmid vector pCL1920. The Tn10dTet element is not digested with PstI, and hence plasmid clones bearing the dke-1::Tn10dTet insertion (with flanking chromosomal DNA) were obtained following Tetr selection. Two plasmids, with identical 12-kb inserts (comprising 3 kb of Tn10dTet and 9 kb of chromosomal DNA) but in opposite orientations relative to the vector backbone, were identified and designated pHYD704 and pHYD705. When radiolabeled pHYD704 DNA was used to probe the ordered E. coli genome library in λ phage constructed by Kohara et al. (24), intense hybridization signals were obtained for phage clones 213 and 214 along with weaker signals for the flanking clones 212 and 215 (data not shown). These results indicated that the Tn10dTet insertion is situated in the 19.9- to 20.1-centisome region (43). Restriction mapping of the insert DNAs in plasmids pHYD704 and pHYD705 permitted the inference that the Tn10dTet insertion had occurred at kb-coordinate 930.8 of the E. coli physical map, that is, approximately at the junction of the proximal and middle thirds of the trxB open reading frame, encoding thioredoxin reductase (Fig. 2). We were subsequently able to demonstrate that another well characterized trxB::kan insertion (7) is also associated with the phenotype of reduced kdp expression (see Fig. 3 and 6, curves for GJ1430). The new insertion mutation dke-1 obtained in this study has been designated trxB30::Tn10dTet.
FIG. 2.
Physical map of insert DNA in plasmids pHYD704 and pHYD705. Shown (with kilobase scale marked) is the restriction map of a PstI (P) fragment, inserted in the two orientations in plasmids pHYD704 and pHYD705, respectively, for the enzymes BamHI (B), HindIII (H), and KpnI (K). The line in bold represents the alignment to the physical map of the E. coli chromosomal PstI fragment that lies between kb coordinates 924.9 and 933.8 (43), and the inverted triangle represents the position of the dke-1::Tn10dTet insertion. The positions and transcriptional orientations of the different chromosomal genes that are carried on the insert are marked below the map.
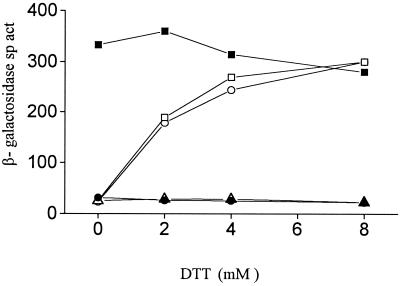
FIG. 3.
β-Galactosidase specific activities (sp. act., expressed in Miller units [30]) in kdp-lac strains with trx mutations, as a function of dithiothreitol (DTT) supplementation of growth medium containing 15 mM [K+]e. ■, GJ1427 (parental); ○, GJ1428 (trxB30::Tn10dTet); □, GJ1430 (trxB::kan); ▵, GJ1426 (ΔtrxA); ●, GJ1429 (ΔtrxA trxB30::Tn10dTet).
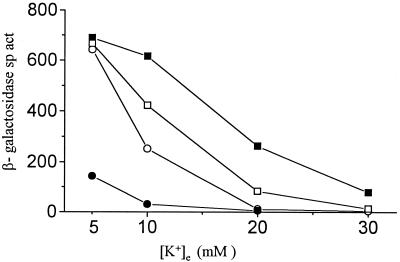
FIG. 6.
β-Galactosidase specific activities (sp. act., expressed in Miller units [30]) in kdp-lac strains with trxB or hns mutations as a function of [K+]e of the growth medium. ■, GJ1427 (parental); ○, GJ1430 (trxB::kan); □, GJ1469 (hns-205::Tn10); ●, GJ1470 (trxB::kan hns-205::Tn10).
Effects of other perturbations in cellular thiol oxidation status on kdp expression.
A characteristic feature of the E. coli cytoplasm is the absence of disulfide bonds in proteins. The reducing environment of the cytoplasm is maintained by the action of several reductant proteins, the three most effective of which are thioredoxin 1, thioredoxin 2, and glutaredoxin 1, which are encoded by trxA, trxC, and grxA, respectively (for a review, see reference 3). The first two proteins are substrates for thioredoxin reductase, while the last one derives its reducing potential from glutathione. Furthermore, of these three proteins, thioredoxin 1 and glutaredoxin 1 appear to be physiologically important during routine growth, whereas thioredoxin 2 is induced primarily under conditions of oxidative stress (41). Based on our identification of trxB as a dke locus, we tested the effects of other perturbations in cellular thiol oxidation status on kdp expression.
We found that a mutation in trxA, but not grxA, affected kdp-lac expression in a manner analogous to that described above for trxB (Fig. 1, curves for strains GJ1426 and GJ1431, respectively). A trxB trxA double mutant, GJ1429, showed a phenotype no more pronounced than either single mutant (Fig. 1). With increasing concentrations of dithiothreitol added to the culture medium, we noted a progressive restoration of kdp-lac expression in the trxB trxA+ strains GJ1428 and GJ1430 but not in the trxB+ trxA (GJ1426) or trxB trxA (GJ1429) derivatives (Fig. 3). The dithiothreitol supplementation experiment was done using concentrations of the reductant that were sublethal for the trxA and trxB strains (31).
We also examined whether the reported induction by oxidative stress of thioredoxin 2 (following the addition of H2O2 [41]) could rescue the kdp expression phenotype in a thioredoxin 1-deficient mutant, but the results were negative. The measured activities of β-galactosidase after growth in 20 mM [K+]e medium, without and with H2O2 supplementation (added as a 5 mM pulse to cultures in early log phase followed by continued incubation for 90 to 180 min), for a pair of isogenic kdp-lac strains were as follows: GJ1426 (ΔtrxA), 6.8 and 7.6 units, respectively; and GJ1427 (trxA+), 161 and 207 units, respectively.
Involvement of cytoplasmic, and not periplasmic, thiol oxidation status in kdp regulation.
Taken together, the above data indicated that the reducing potential of thioredoxin 1, which is generated either by the action of endogenous thioredoxin reductase or following exogenous dithiothreitol supplementation, is necessary for optimal regulation of the kdp operon in E. coli. Thioredoxin reductase and reduced thioredoxin 1 are involved in thiol-disulfide isomerization reactions not only in the cytoplasm, where they act directly, but also in the periplasm where they act indirectly via another disulfide bond isomerase, DsbC (for reviews, see references 3 and 38). Some of the features identified for the perturbation in kdp regulation, notably, dithiothreitol rescue and absence of grxA effect, have been shown for phenotypes that are periplasmically determined (39, 40), and we therefore tested such a possibility further. We found, however, that kdp-lac expression was unaffected in dsbC or dsbD mutants (data not shown), which are otherwise known to be perturbed in periplasmic thiol-disulfide redox reactions (3, 38). Our results therefore suggest that it is the cytoplasmic thiol oxidation status dictated by thioredoxin reductase and reduced thioredoxin 1 which may be important in kdp regulation.
trxB-determined phenotype is also seen in a strain with Cys-less KdpD.
The response regulator KdpE is a small protein located in the cytoplasm with a lone Cys residue. On the other hand, the membrane-localized sensor kinase KdpD has six Cys residues, and we considered the possibility that inappropriate disulfide bond formation within or between the monomer subunits of KdpD in trxB and trxA mutants results in the abnormal signal transduction for kdp expression.
Jung et al. have created a gene encoding a variant KdpD protein with no Cys residues, which is nevertheless normal for kdp signal transduction in vivo (20). We constructed strain GJ1442 (and also its trxB30 derivative, GJ1442T) that was chromosomally ΔkdpD kdpE+ and in which either the variant Cys-less KdpD protein or its normal counterpart could then be expressed (from the heterologous tac promoter) by introduction of the plasmids pPV5-1 Cys-less or pPV5-1 Cys+, respectively. All derivatives also carried the lacIq gene on plasmid pHYD708, in order to avoid the toxicity problems associated with otherwise massive overproduction of the KdpD proteins (reference 18 and data not shown).
The results presented in Fig. 4 indicate that the trxB::Tn10dTet mutation was associated with a reduction in kdp-lac transcription both in the strain that was expressing native KdpD and in the strain expressing the Cys-less variant. This provided conclusive evidence that the trxB effect on kdp is not mediated through the Cys residues of KdpD.
FIG. 4.
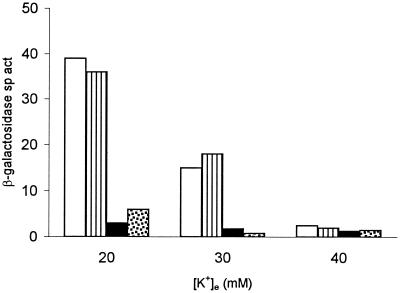
β-Galactosidase specific activities (sp. act., expressed in Miller units [30]) in kdp-lac ΔkdpD strain GJ1442 or its trxB30::Tn10dTet derivative, GJ1442T, each carrying plasmids pPV5-1 Cys+ or pPV5-1 Cys-less, encoding native KdpD or Cys-less KdpD, respectively, along with the lacIq-bearing plasmid pHYD708. Cultures were grown in media with the indicated [K+]e. Histogram symbols: open, GJ1442/pPV5-1 Cys+; striped, GJ1442/pPV5-1 Cys-less; solid, GJ1442T/pPV5-1 Cys+; stippled, GJ1442T/pPV5-1 Cys-less.
It may be noted that in the experiment shown in Fig. 4, the induced level of kdp-lac expression in the trxB+ control strains (with plasmid-borne kdpD) was itself lower than that normally obtained with haploid kdpD+E+ strains. Similar low values for kdp expression have been reported by Jung et al. (20), working with the same multicopy kdpD plasmids pPV5-1 Cys+ and PV5-1 Cys-less, and Jung and Altendorf (18) have suggested that an optimal level of KdpD protein is a critical factor in signal transduction.
Absence of effect of trxB or trxA on another dual-component regulatory system.
Osmolarity-dependent expression of the outer membrane protein gene ompC is under the control of a dual-component system consisting of the membrane-bound sensor kinase EnvZ and the cytoplasmic response regulator OmpR (reviewed in reference 36). In order to test whether the effects of perturbations in thiol-disulfide bond isomerization on kdp expression are specific to the particular dual-component regulatory system represented by KdpD and KdpE, we compared the levels of ompC-lac expression among trxB, trxA, and wild-type strains (Table 2). The basal level of ompC expression was unaffected in either of the mutant strains, and there was not any significant alteration in the magnitude of transcriptional induction of the promoter at elevated osmolarity. These data also indicated that the observed decrease in β-galactosidase activity in the kdp-lac fusion strains with the trxB or trxA mutation is not the consequence of inappropriate disulfide bond formation in the reporter enzyme. We have also obtained evidence that expression of a proU-lac fusion or of the wild-type lac operon is not affected in the mutants (data not shown). We therefore conclude that there is indeed a specificity associated with the reduction of kdp expression in trxB and trxA mutants.
TABLE 2.
ompC-lac regulation in trx mutantsa
| Strain (trx genotype) | β-Galactosidase sp act at NaCl concn (M) of:
|
||
|---|---|---|---|
| 0 | 0.1 | 0.3 | |
| MH225 (trx+) | 260 | 375 | 715 |
| GJ1438 (trxB30::Tn10dTet) | 281 | 398 | 648 |
| GJ1439 (trxB::kan) | 360 | 407 | 670 |
| GJ1441 (ΔtrxA) | 399 | 533 | 709 |
Specific activity (in Miller units [30]) of β-galactosidase was measured in cultures of the ompC-lac fusion strain MH225 and its trx mutant derivatives grown to mid-log phase in low-osmolarity K medium (13) supplemented with NaCl at the indicated concentrations.
The dke-2 mutation is an hns allele.
As described above, the dke-2 mutant was isolated following localized mutagenesis of the 28-min region of the chromosome. P1 transductional mapping experiments demonstrated that dke-2 is 95% cotransducible with each of the inserts zci-3117::Tn10Kan and zci-506::Tn10, which constitute a cognate pair in the collection of Singer et al. (47); this pair has subsequently been mapped to lie in the oppC gene (35). The induced level of kdp-lac expression in the mutant was 10- to 15-fold lower than that in the dke+ control (Fig. 5). The dke-2 mutation also conferred phenotypes of nonmotility as well as derepression of proU-lac expression (data not shown), which suggested (23, 54) that it is an allele of the hns gene, which maps close to oppC at 28 min and which encodes the nucleoid protein H-NS (4, 43). Introduction of the medium-copy-number plasmid pLG H-NS (carrying the hns+ gene) restored the kdp-lac expression profile in the mutant to that seen in the dke+ strain carrying the same plasmid (Fig. 5). Furthermore, a previously characterized hns-205::Tn10 mutation (8) conferred a phenotype similar to dke-2 on kdp-lac expression (Fig. 6; Table 3). These results support the conclusions that (i) dke-2 is in hns, and (ii) null mutations in hns serve to reduce kdp transcription. The dke-2 mutation has accordingly been designated hns-202.
FIG. 5.
β-Galactosidase specific activities (sp. act., expressed in Miller units [30]) in the isogenic kdp-lac strains GJ1459 (parental) and GJ1458 (dke-2; that is, hns-202) or their derivatives carrying the hns+-encoding plasmid pLG H-NS, as a function of [K+]e of the growth medium. ■, GJ1459; □, GJ1458; ○, GJ1459/pLG H-NS; ●, GJ1458/pLG H-NS.
TABLE 3.
Expression of kdp-lac in trx kdpDE double mutantsa
| dke mutation | β-Galactosidase sp act in presence of kdpDE mutation:
|
|||
|---|---|---|---|---|
| None | kdp-205 | kdp-207 | kdpD(R511Q) | |
| None | 250 | 713 | 890 | 423 |
| trxB30::Tn10dTet | 6 | 535 | 920 | 248 |
| hns-205::Tn10 | 9 | 540 | 919 | 236 |
The following sets of strains were used for assessment of kdp-lac expression (isogenic within each set and indicated, within parentheses, in the order of no dke mutation, trxB and hns): (i) no kdpDE mutation (GJ1449, GJ1449T, GJ1449H); (ii) with kdp-205 (GJ1450, GJ1450T, GJ1450H); (iii) with kdp-207 (GJ1451, GJ1451T, GJ1451H); and (iv) with kdpD-R511Q (derivatives of GJ1442, GJ1442T, and GJ1442H each transformed with plasmid pBD-R511Q). Cultures were grown to mid-log phase in medium containing 30 mM [K+]e (supplemented with ampicillin in the case of derivatives carrying plasmid pBD-R511Q). β-Galactosidase specific activities are reported in Miller units (30).
Expression of kdp-lac in the trxB::kan hns::Tn10 double mutant strain GJ1470 was reduced even more drastically than in either single mutant derivative (Fig. 6), suggesting that the two dke loci act additively in perturbing kdp regulation. A null mutation in stpA, the gene encoding the H-NS-like protein StpA that is believed to represent a molecular back-up of H-NS (55), by itself had no effect on kdp expression. An hns stpA double mutant was even more compromised for kdp regulation than was the hns single mutant (data not shown), but as explained below, interpretation of this finding is rendered difficult because of the poor growth rate observed with the double mutant strain (16, 55).
trxB and hns effects on kdp are unrelated to alterations in growth rates.
It is known (2) that for a given [K+]e, kdp expression in a strain decreases with decreasing growth rates, ostensibly because lower rates of K+ uptake suffice under these conditions (10). The following experiments demonstrated, however, that the dke nature of trxB may not be explained on this basis. Consistent with the findings of an earlier report (7), a trxB mutant GJ1428 grew just as well as its trxB+ parent GJ1427 (with doubling times of 40 min each) in medium with 20 mM [K+]e, that is, under the conditions where the mutation's effect on kdp-lac transcription is very pronounced (Fig. 1). Furthermore, supplementation of the cultures with 8 mM dithiothreitol led, as expected (31), to a reduction in growth rates of the two strains (with measured doubling times of 70 and 60 min, respectively), even as the level of kdp expression in the mutant was almost completely restored to that in the parent (data not shown; see also Fig. 3).
Mutations in hns are known to affect growth rate (55), and in order to examine whether the hns effect on kdp could be accounted for by such alterations we measured the doubling times and levels of kdp-lac expression in cultures of the parental strain TL1105A and of its derivatives carrying mutations in hns (GJ1461) or recB (GJ1485). The recB mutation was chosen as a control, as it effects a moderate growth rate reduction similar to that of the hns allele. The β-galactosidase activities (with culture doubling times in parentheses) for the parent, hns, and recB strains grown in 30 mM [K+]e were 306 units (45 min), 24 units (55 min), and 150 units (55 min), respectively. These results indicate that the two mutations each had equivalent effects in reducing the growth rate of the parental strain, but the reduction in kdp expression was very much more pronounced in the hns mutant than it was in the recB derivative. Therefore, the hns effect on kdp transcription is not solely because of a concomitant decrease in the growth rate.
kdpDE constitutive mutations are epistatic to trxB and hns.
In order to establish epistasis relationships, we examined the effects of the trxB::Tn10dTet or hns::Tn10 mutation on kdp-lac expression in strains carrying three different trans-acting mutations in the kdpDE locus. The latter included the kdp-205 and kdp-207 alleles described earlier (2), as well as a site-specific alteration in kdpD that results in an Arg511→Gln (R511Q) substitution in KdpD (19). Of these mutations, the kdp-205 mutant exhibits a reduced sensitivity for repression of the kdp operon by [K+]e, while the other two are fully constitutive. We found that the elevated levels of kdp-lac expression conferred by the kdpDE mutations were largely unaffected by the trxB or hns mutations (Table 3). As further discussed below, these results suggest that trxB and hns exert their effects on kdp expression at a step(s) upstream of KdpD in the signal transduction pathway.
DISCUSSION
The mechanism of transcriptional activation of the kdp operon in E. coli by the protein pair comprised of the sensor kinase KdpD and response regulator KdpE is well established, although the nature of the signal during K+-limited growth which leads to increased KdpD autophosphorylation is unclear. Different mutations could be expected to alter kdp-lac expression (for a given [K+]e) either by altering the strength of the environmental signal that is sensed by the cell in controlling kdp transcription (e.g., mutations in trkA or trkD) or by interfering with the signal transduction pathway (e.g., mutations in kdpD or kdpE). The hallmark of the former is that the change in kdp-lac expression in the mutant is inversely correlated with its growth ability in low-[K+]e media. By this criterion, the mutations that have been identified in this study as reducing kdp expression (trxB, trxA, and hns) appear to do so by interfering with signal transduction rather than signal strength, because there is no concomitant increase of K+-limited growth rates in the mutant cultures.
Cytoplasmic thiol oxidation status in kdp regulation.
The observations made in this study, concerning the trxB and trxA mutants as well as the effects of exogenous dithiothreitol supplementation, support the proposal that reduced thioredoxin 1 is required for appropriate signal transduction in kdp regulation in vivo. This requirement apparently cannot be substituted by thioredoxin 2 or the glutaredoxins, nor does it involve the thiol-disulfide isomerase DsbC in the periplasmic compartment (whose functioning is dependent on availability of reduced thioredoxin 1). We therefore suggest that this requirement is cytoplasmic. To our knowledge, this is the first example of a thiol oxidation status-determined function in the cytoplasmic compartment that is absolutely dependent only on reduced thioredoxin 1 and also one that is affected to an equivalent extent by trxB and trxA mutations.
That the trxB- or trxA-mediated reduction in reporter enzyme activity in the kdp-lac fusion strains is not a consequence, for example, of inappropriate disulfide bond formation in the cytoplasmically localized β-galactosidase was established in control experiments involving lacZ expression from other promoters, including its native promoter. Also, regulation was not affected in another system (EnvZ-OmpR) involving similar phosphotransfer (as in kdp) between an autophosphorylated sensor kinase and a cytoplasmic activator protein, hence arguing for a specificity in the reduced thioredoxin 1 requirement for kdp regulation.
As mentioned above, the signal controlling kdp expression is not known, but several models suggest that this signal acts directly on membrane-bound KdpD to determine the latter's autophosphorylation activity (25, 27, 42, 49). Our data, on the other hand, from the experiments employing strains with the Cys-less KdpD variant protein as well as those testing epistasis with kdpDE mutations, suggest that the absence of reduced thioredoxin 1 interferes with a step in the kdp signal transduction pathway upstream of KdpD function. (Implied also in such an interpretation is the notion that cellular thiol status does not exert its effect on kdp regulation via KdpE [by formation or breakage, for example, of a disulfide bridge between two monomer subunits], because KdpE is downstream of KdpD in the signal transduction pathway.) To that extent, therefore, we believe that alternative models may need to be considered in which the effect of the signal (which is generated during K+-limited growth) on KdpD activity is mediated or modulated by additional protein(s). Nevertheless, the exact mechanism by which reduced thioredoxin 1 participates in the signal transduction pathway remains to be determined.
Finally, it may be noted that the cytoplasmic thiol reductant glutathione has been shown in earlier studies both to accumulate during osmotic stress (when there is a concomitant cytoplasmic accumulation of K+) (29) and to mediate gating of the K+-efflux channels KefB and KefC (6). Glutathione-deficient strains, particularly those that are Kdp+, exhibit abnormalities in the maintenance of [K+]i (11). Cytoplasmic thioredoxin 1 has also been shown to leak out (through MscL channels) from cells subjected to an osmotic downshock (1). The relevance of any of these observations, however, to the findings described in this paper is unclear.
Nucleoid protein H-NS in kdp regulation.
Mutants in hns are known to be pleiotropic (for a review, see reference 53); this study has identified an additional phenotype, that of a significant reduction in the induced levels of kdp transcription, for these mutants. H-NS is known more for its role as a global repressor protein (53), and there are only a limited number of identified promoters whose transcription is reduced in an hns null mutant (16, 17, 23). Interestingly, there exists a bent-DNA motif (which is also a high-affinity binding site for H-NS) that overlaps the binding site for phospho-KdpE immediately upstream of the kdp operon promoter (51), and one could therefore envisage a direct role for H-NS in providing an optimal chromatin configuration for transcription activation by phospho-KdpE. However, our results from the epistasis experiments with kdpDE argue (as for trxB) that the effect of hns on kdp is upstream of KdpD and is, therefore, almost certainly indirect. The precise mechanism of this indirect effect of H-NS remains to be elucidated.
In conclusion, we have shown in this study that in addition to the KdpD and KpdE regulator proteins, factors such as cytoplasmic thiol oxidation status and the nucleoid protein H-NS can significantly affect in vivo expression of the kdp operon. It may therefore be necessary to accommodate the roles of these factors as well in models that seek to explain the mechanism of signal transduction in kdp operon regulation.
ACKNOWLEDGMENTS
We acknowledge Jon Beckwith, Erhard Bremer, Wolf Epstein, Carol Gross, Kirsten Jung, Nancy Kleckner, Bénédicte Michel, Sylvie Rimsky, Marjorie Russel, and Tom Silhavy for the various strains, phage, and plasmids used in the study.
A.A.S. was a recipient of Junior and Senior Research Fellowships of the Council of Scientific and Industrial Research. J.G. is an Honorary Faculty Member of the Jawaharlal Nehru Centre for Advanced Scientific Research.
REFERENCES
- 1.Ajouz B, Berrier C, Garrigues A, Besnard M, Ghazi A. Release of thioredoxin via the mechanosensitive channel MscL during osmotic downshock of Escherichia coli cells. J Biol Chem. 1998;273:26670–26674. doi: 10.1074/jbc.273.41.26670. [DOI] [PubMed] [Google Scholar]
- 2.Asha H, Gowrishankar J. Regulation of kdp operon expression in Escherichia coli: evidence against turgor as signal for transcriptional control. J Bacteriol. 1993;175:4528–4537. doi: 10.1128/jb.175.14.4528-4537.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Åslund F, Beckwith J. The thioredoxin superfamily: redundancy, specificity, and gray-area genomics. J Bacteriol. 1999;181:1375–1379. doi: 10.1128/jb.181.5.1375-1379.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berlyn M K B. Linkage map of Escherichia coli K-12, edition 10: the traditional map. Microbiol Mol Biol Rev. 1998;62:814–984. doi: 10.1128/mmbr.62.3.814-984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bidnenko V, Seigneur M, Penel-Colin M, Bouton M-F, Ehrlich S D, Michel B. sbcB sbcC null mutations allow RecF-mediated repair of arrested replication forks in rep recBC mutants. Mol Microbiol. 1999;33:846–857. doi: 10.1046/j.1365-2958.1999.01532.x. [DOI] [PubMed] [Google Scholar]
- 6.Booth I R, Jones M A, McLaggan D, Nikolaev Y, Ness L S, Wood C M, Miller S, Tötemeyer S, Ferguson G P. Bacterial ion channels. In: Konings W N, Kaback H R, Lolkema J S, editors. Handbook of biological physics. Vol. 2. Amsterdam, The Netherlands: Elsevier Science B.V.; 1996. pp. 693–729. [Google Scholar]
- 7.Derman A I, Prinz W A, Belin D, Beckwith J. Mutations that allow disulfide bond formation in the cytoplasm of Escherichia coli. Science. 1993;262:1744–1747. doi: 10.1126/science.8259521. [DOI] [PubMed] [Google Scholar]
- 8.Dersch P, Kneip S, Bremer E. The nucleoid-associated DNA-binding protein H-NS is required for the efficient adaptation of Escherichia coli to a cold environment. Mol Gen Genet. 1994;245:255–259. doi: 10.1007/BF00283274. [DOI] [PubMed] [Google Scholar]
- 9.Epstein W, Kim B S. Potassium transport loci in Escherichia coli K-12. J Bacteriol. 1971;108:639–644. doi: 10.1128/jb.108.2.639-644.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Epstein W, Schultz S G. Cation transport in Escherichia coli. V. Regulation of cation content. J Gen Physiol. 1965;49:221–234. doi: 10.1085/jgp.49.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferguson G P, Booth I R. Importance of glutathione for growth and survival of Escherichia coli cells: detoxification of methylglyoxal and maintenance of intracellular K+ J Bacteriol. 1998;180:4314–4318. doi: 10.1128/jb.180.16.4314-4318.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frymier J S, Reed T D, Fletcher S A, Csonka L N. Characterization of transcriptional regulation of the kdp operon of Salmonella typhimurium. J Bacteriol. 1997;179:3061–3063. doi: 10.1128/jb.179.9.3061-3063.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gowrishankar J. Identification of osmoresponsive genes in Escherichia coli: evidence for participation of potassium and proline transport systems in osmoregulation. J Bacteriol. 1985;164:434–445. doi: 10.1128/jb.164.1.434-445.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gowrishankar J. A model for the regulation of expression of the potassium-transport operon, kdp, in Escherichia coli. J Genet. 1987;66:87–92. [Google Scholar]
- 15.Hall M N, Silhavy T J. The ompB locus and the regulation of the major outer membrane porin proteins of Escherichia coli K-12. J Mol Biol. 1981;146:22–43. doi: 10.1016/0022-2836(81)90364-8. [DOI] [PubMed] [Google Scholar]
- 16.Johansson J, Balsalobre C, Wang S-Y, Urbonaviciene J, Jin D J, Sondén B, Uhlin B E. Nucleoid proteins stimulate stringently controlled bacterial promoters: a link between the cAMP-CRP and the (p)ppGpp regulons in Escherichia coli. Cell. 2000;102:475–485. doi: 10.1016/s0092-8674(00)00052-0. [DOI] [PubMed] [Google Scholar]
- 17.Johansson J, Dagberg B, Richet E, Uhlin B E. H-NS and StpA proteins stimulate expression of the maltose regulon in Escherichia coli. J Bacteriol. 1998;180:6117–6125. doi: 10.1128/jb.180.23.6117-6125.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung K, Altendorf K. Truncation of amino acids 12-128 causes deregulation of the phosphatase activity of the sensor kinase KdpD of Escherichia coli. J Biol Chem. 1998;273:17406–17410. doi: 10.1074/jbc.273.28.17406. [DOI] [PubMed] [Google Scholar]
- 19.Jung K, Altendorf K. Individual substitutions of clustered arginine residues of the sensor kinase KdpD of Escherichia coli modulate the ratio of kinase to phosphatase activity. J Biol Chem. 1998;273:26415–26420. doi: 10.1074/jbc.273.41.26415. [DOI] [PubMed] [Google Scholar]
- 20.Jung K, Heermann R, Meyer M, Altendorf K. Effect of cysteine replacements on the properties of the turgor sensor KdpD of Escherichia coli. Biochim Biophys Acta. 1998;1372:311–322. doi: 10.1016/s0005-2736(98)00070-4. [DOI] [PubMed] [Google Scholar]
- 21.Jung K, Tjaden B, Altendorf K. Purification, reconstitution, and characterization of KdpD, the turgor sensor of Escherichia coli. J Biol Chem. 1997;272:10847–10852. doi: 10.1074/jbc.272.16.10847. [DOI] [PubMed] [Google Scholar]
- 22.Kleckner N, Bender J, Gottesman S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- 23.Ko M, Park C. H-NS-dependent regulation of flagellar synthesis is mediated by a LysR family protein. J Bacteriol. 2000;182:4670–4672. doi: 10.1128/jb.182.16.4670-4672.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohara Y, Akiyama K, Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 25.Laimins L A, Rhoads D B, Epstein W. Osmotic control of kdp operon expression in Escherichia coli. Proc Natl Acad Sci USA. 1981;78:464–468. doi: 10.1073/pnas.78.1.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lerner C G, Inouye M. Low copy number plasmids for regulated low-level expression of cloned genes in Escherichia coli with blue/white insert screening capability. Nucleic Acids Res. 1990;18:4631. doi: 10.1093/nar/18.15.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malli R, Epstein W. Expression of the Kdp ATPase is consistent with regulation by turgor pressure. J Bacteriol. 1998;180:5102–5108. doi: 10.1128/jb.180.19.5102-5108.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mark D F, Richardson C C. Escherichia coli thioredoxin: a subunit of bacteriophage T7 DNA polymerase. Proc Natl Acad Sci USA. 1976;73:780–784. doi: 10.1073/pnas.73.3.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLaggan D, Logan T M, Lynn D G, Epstein W. Involvement of γ-glutamyl peptides in osmoadaptation of Escherichia coli. J Bacteriol. 1990;172:3631–3636. doi: 10.1128/jb.172.7.3631-3636.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. [Google Scholar]
- 31.Missiakas D, Georgopoulos C, Raina S. Identification and characterization of the Escherichia coli gene dsbB, whose product is involved in the formation of disulfide bonds in vivo. Proc Natl Acad Sci USA. 1993;90:7084–7088. doi: 10.1073/pnas.90.15.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murgola E J, Yanofsky C. Structural interactions between amino acid residues at positions 22 and 211 in the tryptophan synthetase alpha chain of Escherichia coli. J Bacteriol. 1974;117:444–448. doi: 10.1128/jb.117.2.444-448.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakashima K, Sugiura A, Kanamaru K, Mizuno T. Signal transduction between the two regulatory components involved in the regulation of the kdpABC operon in Escherichia coli: phosphorylation-dependent functioning of the positive regulator, KdpE. Mol Microbiol. 1993;7:106–116. doi: 10.1111/j.1365-2958.1993.tb01102.x. [DOI] [PubMed] [Google Scholar]
- 34.Nakashima K, Sugiura A, Momoi H, Mizuno T. Phosphotransfer signal transduction between two regulatory factors involved in the osmoregulated kdp operon in Escherichia coli. Mol Microbiol. 1992;6:1777–1784. doi: 10.1111/j.1365-2958.1992.tb01350.x. [DOI] [PubMed] [Google Scholar]
- 35.Nichols B P, Shafiq O, Meiners V. Sequence analysis of Tn10 insertion sites in a collection of Escherichia coli strains used for genetic mapping and strain construction. J Bacteriol. 1998;180:6408–6411. doi: 10.1128/jb.180.23.6408-6411.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ninfa A J. Regulation of gene transcription by extracellular stimuli. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 1246–1262. [Google Scholar]
- 37.Polarek J W, Williams G, Epstein W. The products of the kdpDE operon are required for expression of the Kdp ATPase of Escherichia coli. J Bacteriol. 1992;174:2145–2151. doi: 10.1128/jb.174.7.2145-2151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raina S, Missiakas D. Making and breaking disulfide bonds. Annu Rev Microbiol. 1997;51:179–202. doi: 10.1146/annurev.micro.51.1.179. [DOI] [PubMed] [Google Scholar]
- 39.Rietsch A, Belin D, Martin N, Beckwith J. An in vivo pathway for disulfide bond isomerization in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:13048–13053. doi: 10.1073/pnas.93.23.13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rietsch A, Bessette P, Georgiou G, Beckwith J. Reduction of the periplasmic disulfide bond isomerase, DsbC, occurs by passage of electrons from cytoplasmic thioredoxin. J Bacteriol. 1997;179:6602–6608. doi: 10.1128/jb.179.21.6602-6608.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ritz D, Patel H, Doan B, Zheng M, Åslund F, Storz G, Beckwith J. Thioredoxin 2 is involved in the oxidative stress response in Escherichia coli. J Biol Chem. 2000;276:2505–2512. doi: 10.1074/jbc.275.4.2505. [DOI] [PubMed] [Google Scholar]
- 42.Roe A J, McLaggan D, O'Byrne C P, Booth I R. Rapid inactivation of the Escherichia coli Kdp K+ uptake system by high potassium concentrations. Mol Microbiol. 2000;35:1235–1243. doi: 10.1046/j.1365-2958.2000.01793.x. [DOI] [PubMed] [Google Scholar]
- 43.Rudd K E. Linkage map of Escherichia coli K-12, edition 10: the physical map. Microbiol Mol Biol Rev. 1998;62:985–1019. doi: 10.1128/mmbr.62.3.985-1019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Russel M, Model P, Holmgren A. Thioredoxin or glutaredoxin in Escherichia coli is essential for sulfate reduction but not for deoxyribonucleotide synthesis. J Bacteriol. 1990;172:1923–1929. doi: 10.1128/jb.172.4.1923-1929.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 46.Silver S. Transport of inorganic cations. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 1091–1102. [Google Scholar]
- 47.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stark M J R. Multicopy expression vectors carrying the lac repressor gene for regulated high-level expression of genes in Escherichia coli. Gene. 1987;51:255–267. doi: 10.1016/0378-1119(87)90314-3. [DOI] [PubMed] [Google Scholar]
- 49.Sugiura A, Hirokawa K, Nakashima K, Mizuno T. Signal-sensing mechanisms of the putative osmosensor KdpD in Escherichia coli. Mol Microbiol. 1994;14:929–938. doi: 10.1111/j.1365-2958.1994.tb01328.x. [DOI] [PubMed] [Google Scholar]
- 50.Sugiura A, Nakashima K, Tanaka K, Mizuno T. Clarification of the structural and functional features of the osmoregulated kdp operon of Escherichia coli. Mol Microbiol. 1992;6:1769–1776. doi: 10.1111/j.1365-2958.1992.tb01349.x. [DOI] [PubMed] [Google Scholar]
- 51.Tanaka K, Muramatsu S, Yamada H, Mizuno T. Systematic characterization of curved DNA segments randomly cloned from Escherichia coli and their functional significance. Mol Gen Genet. 1991;226:367–376. doi: 10.1007/BF00260648. [DOI] [PubMed] [Google Scholar]
- 52.Walderhaug M O, Polarek J W, Voelkner P, Daniel J M, Hesse J E, Altendorf K, Epstein W. KdpD and KdpE, proteins that control expression of the kdpABC operon, are members of two-component sensor-effector class of regulators. J Bacteriol. 1992;174:2152–2159. doi: 10.1128/jb.174.7.2152-2159.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams R M, Rimsky S. Molecular aspects of the E. coli nucleoid protein, H-NS: a central controller of gene regulatory networks. FEMS Microbiol Lett. 1997;156:175–185. doi: 10.1111/j.1574-6968.1997.tb12724.x. [DOI] [PubMed] [Google Scholar]
- 54.Williams R M, Rimsky S, Buc H. Probing the structure, function, and interactions of the Escherichia coli H-NS and StpA proteins by using dominant negative derivatives. J Bacteriol. 1996;178:4335–4343. doi: 10.1128/jb.178.15.4335-4343.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang A, Rimsky S, Reaban M E, Buc H, Belfort M. Escherichia coli protein analogs StpA and H-NS: regulatory loops, similar and disparate effects on nucleic acid dynamics. EMBO J. 1996;15:1340–1349. [PMC free article] [PubMed] [Google Scholar]