Abstract
Delivery of exogenous heat shock protein 90α (Hsp90α) and/or its induced expression in neural tissues has been suggested as a potential strategy to combat neurodegenerative disease. However, within a neurodegenerative context, a pro-inflammatory response to extracellular Hsp90α (eHsp90α) could undermine strategies to use it for therapeutic intervention. The aim of this study was to investigate the biological effects of eHsp90α on microglial cells, the primary mediators of inflammatory responses in the brain. Transcriptomic profiling by RNA-seq of primary microglia and the cultured EOC2 microglial cell line treated with eHsp90α showed the chaperone to stimulate activation of innate immune responses in microglia that were characterized by an increase in NF-kB-regulated genes. Further characterization showed this response to be substantially lower in amplitude than the effects of other inflammatory stimuli such as fibrillar amyloid-β (fAβ) or lipopolysaccharide (LPS). Additionally, the toxicity of conditioned media obtained from microglia treated with fAβ was attenuated by addition of eHsp90α. Using a co-culture system of microglia and hippocampal neuronal cell line HT22 cells separated by a chamber insert, the neurotoxicity of medium conditioned by microglia treated with fAβ was reduced when eHsp90α was also added. Mechanistically, eHsp90α was shown to activate Nrf2, a response which attenuated fAβ-induced nitric oxide production. The data thus suggested that eHsp90α protects against fAβ-induced oxidative stress. We also report eHsp90α to induce expression of macrophage receptor with collagenous structure (Marco), which would permit receptor-mediated endocytosis of fAβ.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12192-022-01279-9.
Keywords: Hsp90, Extracellular HSPs, Nrf2, NF-kB, Inflammation, Microglia, Amyloid-beta, Marco
Introduction
Heat shock proteins (HSPs) promote protein homeostasis (proteostasis) via fulfilling several protein quality-control functions (Lang 2021). Collectively, the actions of HSPs and other molecular chaperones prevent the formation of protein aggregates and solubilize existing aggregates. Failure of these functions has particular significance in the emergence of neurodegenerative diseases caused by the build-up of amyloid deposits. Overexpression of various HSPs has been shown to protect against protein aggregation in vivo (Rampelt et al. 2012; Fujimoto et al. 2005). The observed decline of HSP expression in neural tissue with aging has further indicated their importance in maintaining proteostasis (Garigan et al. 2002; Hsu et al. 2003). This finding has indicated a potential opportunity for therapeutic intervention through increased local concentrations of HSPs as an approach to combatting protein aggregation in neurodegenerative disease. One such strategy utilizes heat shock response (HSR)–inducing compounds to increase tissue HSP levels, an approach that may be constrained by the relatively weak capacity for neurons to activate heat shock factor 1 (HSF1), the key transcription factor for stress-induced HSP expression (Calderwood and Murshid 2017). However, low levels of HSR activation within neurons may be augmented by uptake of extracellular HSPs (eHSPs) released by surrounding microglia and astrocytes (Guzhova et al. 2001; Tidwell et al. 2004). These findings have led to the concept that the HSR may not be totally cell-intrinsic and that an effective transcellular response may be involved (Taylor et al. 2007). Importantly for neurodegenerative disease, studies have shown HSPs to readily cross the blood–brain barrier and suggest that HSPs may be delivered exogenously and enter the brain space (Kirkegaard et al. 2016). Additionally, induction of HSP expression in other parts of the body may enable increased HSP concentrations within the brain (Oosten-Hawle and Morimoto 2014; Theriault et al. 2006). Canonically, delivery of HSPs increases cellular chaperone levels and also may influence cell phenotype on encountering receptors in target cells (Theriault et al. 2006). In mononuclear phagocytes, the cells under study here, HSPs have been shown to bind to the scavenger receptors. Indeed, a potential risk of HSP therapeutics for neurodegenerative disease is the possibility of inflammation (Zhang et al. 2018; Thawkar and Kaur 2019). Neuroinflammation is a key pathological factor in the etiology of Alzheimer’s disease. This response is largely mediated by microglial cells, the resident mononuclear phagocytes of the brain, as they attempt to detoxify beta-amyloid fibrils and necrotic cell bodies and adopt a toxic inflammatory phenotype (Clayton et al. 2017; Krasemann et al. 2017; Yamamoto et al. 2008).
The stress-inducible Hsp90α (encoded by HSP90AA1) can be detected in the extracellular space and has also been shown to be protective against protein aggregate formation and disease (Calderwood et al. 2016; Li et al. 2012). Hsp90α binds to the scavenger receptors, but it is not known whether it induces an inflammatory profile in microglial cells. This question has important implications for therapies seeking to increase concentrations of neuronal Hsp90. Our previous studies that examined the role of Hsp90 in immune and inflammatory responses as part of a program to employ the chaperones in cancer immune–based therapies found that although Hsp90 is internalized by target cells such as macrophages and dendritic cells and can facilitate immunity by transporting extracellular antigens into antigen processing cells, its effects were generally not inflammatory (DNA 2006; Hancock et al. 2015; Murshid et al. 2015). Indeed, when we compared the effects on macrophages of LPS with the inflammatory profile of eHsp90α to that of LPS, we found that although eHsp90α triggered many of the same changes in the macrophages as LPS, inflammatory gene expression was minimal. The inflammatory effects of eHsp90 were judged to be stalled by a homeostatic mechanism.
In the present study, we have examined the effects of eHsp90α on gene expression in microglia as part of a program to assess the role of Hsp90 in mediating a potential intercellular HSR in cells of the central nervous system (CNS). Our findings were that eHsp90, although modifying the expression of immune-related genes, also induced several immune-suppressive genes such as the scavenger receptor Marco and the immunometabolism regulator Acod1. Our data also pointed to the induction by Hsp90 of the transcription factor nuclear factor, erythroid-derived 2, like 2 (Nrf2), which is a central effector of the oxidative stress response and in addition to promoting cell survival, also exerts anti-inflammatory effects.
Materials and methods
Cell culture
The murine microglia cell line BV2, a line of immortalized mouse microglia (Stansley et al. 2012). BV2 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Carlsbad, CA, USA) supplemented by 10% fetal bovine serum (FBS, Gibco, Carlsbad, CA, USA) and penicillin–streptomycin (1000 units/ml, Invitrogen, Carlsbad, CA, USA), non-essential amino acids (Invitrogen, Carlsbad, CA, USA), HEPES (Corning, NY, USA), and monocyte colony–stimulating factor (M-CSF, 20 ng/ml, R&D Systems, Minnesota, MN, USA). The murine microglia cell line EOC2 (CRL-2467) was obtained from American Type Culture Collection (ATCC, Gaithersburg, MD, USA). EOC2 cultures were maintained in DMEM media supplemented with 10% HI FBS, 20% LADMAC conditioned media (ATCC, CRL-2420). Hippocampal neuronal cell line HT22 cells were sourced from ATCC and maintained in DMEM supplemented with 10% FBS, penicillin–streptomycin (1000 units/ml), 2 ml L-glutamine, and HEPES. Primary murine microglia were cultured and maintained according to Timmerman et al. (2018). Neonatal murine microglia were isolated from P0 CD-1 pups using CD11b microbeads (Ca #130–093-634, Miltenyi Biotec), and their purity was assessed by immunocytochemistry of myeloid cell markers (Iba-1 and CD11b), according to the published method (Ikezu 2020). All the cell cultures were maintained in a 5% CO2 humidified incubator at 37 °C.
Chemicals and reagents
Recombinant full-length human Hsp90α was expressed by baculovirus in Sf9 insect cells using a C-terminal His tag vector and purified by metal affinity chromatography, as previously described (Murshid et al. 2010). Hsp90 was therefore produced from a source free of endotoxin contamination. His-tag removal was performed using AcTEV protease and passing the proteolytically cleaved Hsp90 through a Ni–NTA purification system (Qiagen, Valencia, CA). The native Hsp90 is collected in the column eluate and used for generation of experimental data shown in Figs. 1, 2, 3A,D, 4, 5A-C,F and 6A; Suppl. Fig. 1; and Suppl. Fig. 2A. For all the other data, the experiments were performed using purchased human C-terminal His-tagged Hsp90α produced in baculovirus Sf9 insect cells (H36-50H, Signalchem, Richmond, BC, USA) and diluted in cell media to 10 μg/ml, and equimolar concentrations of His-tagged protein buffer (NP20-153–01, Signalchem, Richmond, BC) were added to control samples. Mouse Aβ1-42 and Aβ1-40, FITC tagged mouse Aβ1-42, and control peptides were purchased from American Peptides and AnaSpec. Lipopolysaccharide (LPS) was purchased from Sigma-Aldrich, St. Louis, MO, USA. The Nrf2 inhibitor, ML385 (Medchem Express, Monmouth Junction, NJ, USA) was used to inhibit the Nrf2 expression in BV2 cells. For experiments using ML385, BV2 cells were seeded (1 × 105 cells) and cultured in 35-mm dishes. The next day, 5 μM ML385 (ED50 = 1.9 μM) was added in the medium. On the 2nd day of post-ML385 addition, the cells were used for Hsp90α treatment assays.
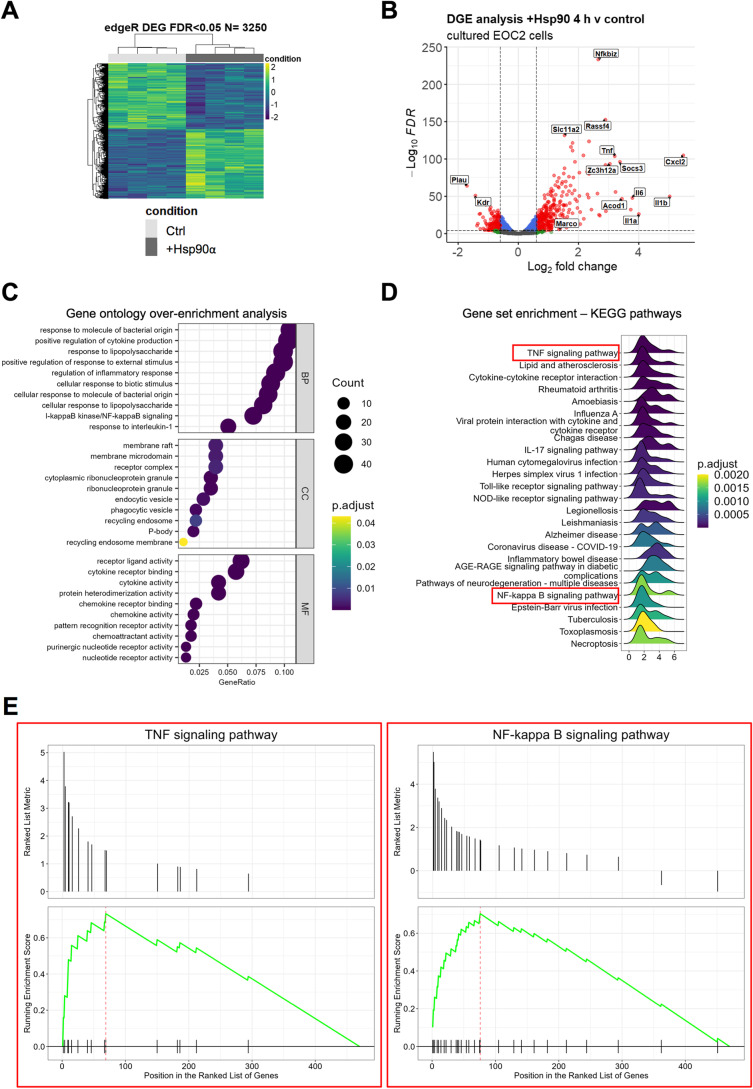
Fig. 1.
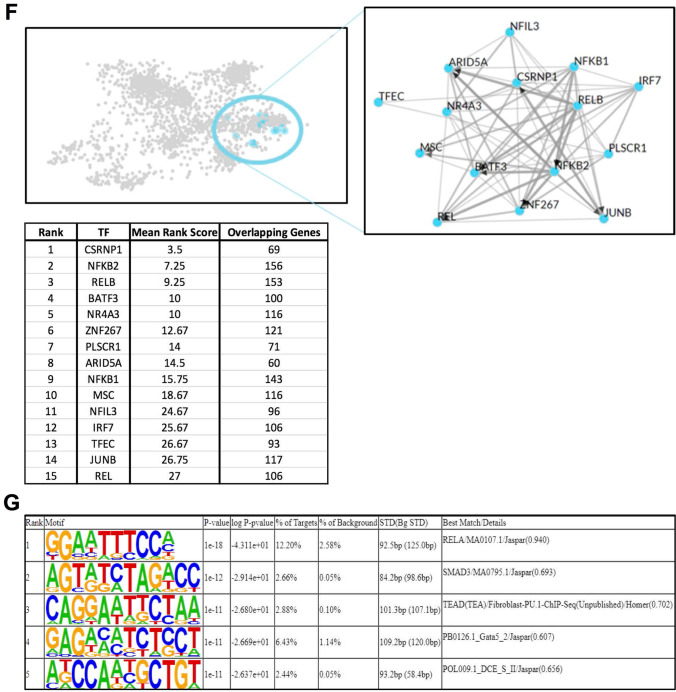
The transcriptional response to eHsp90α at 4 h is characterized by activation of NF-kB-regulated processes. A Heatmap representation of the 3250 DEG (FDR < 0.05) identified by RNA-seq analysis between EOC2 cells treated with 10 µg/ml Hsp90α for 4 h versus untreated control samples. B Volcanoplot representing the FDR and logFC values of changes in gene expression between EOC2 cells treated with 10 µg/ml Hsp90α for 4 h versus untreated control samples. Values for features that returned at least 3 counts per million (cpm) across 4 of the 8 samples with MGI symbols are shown. C Gene ontology over enrichment analysis was applied to the filtered EOC2 DEG list (FDR < 0.05, ± logFC 0.6) using the enrichGO function of the clusterProfiler package to return enriched descriptive terms belonging to the GO categories; biological process (BP), cellular component (CC), and molecular function (MF). D Ridgeplot of gene set enrichment of KEGG pathways performed on the same filtered EOC2 DEG list (FDR < 0.05, ± logFC 0.6) using the clusterProfiler function gseKEGG. Values for the directional expression distribution (x-axis) and Benjamini–Hochberg adjusted p-value (p.adjust) are represented. E GSEA plots of selected KEGG pathways representing the enrichment scores for the respective gene sets and the positions of genes within the gene set in the ranked DEG list ordered by descending logFC. F Results of ChEA3 TFEA analysis showing the top 15 transcription factors in the Global GTEx TF Network (left), the local network (right), and the top 15 TFs ranked by mean rank score (lower). The filtered EOC2 DEG list (FDR < 0.05, ± logFC 0.6) was used as input. G The top 5 TFs returned by HOMER TFEA analysis are shown. The filtered EOC2 DEG list (FDR < 0.05, ± logFC 0.6) was used as input with all features that returned at least 3 counts per million (cpm) across 4 of the 8 samples with MGI symbols as the background gene list
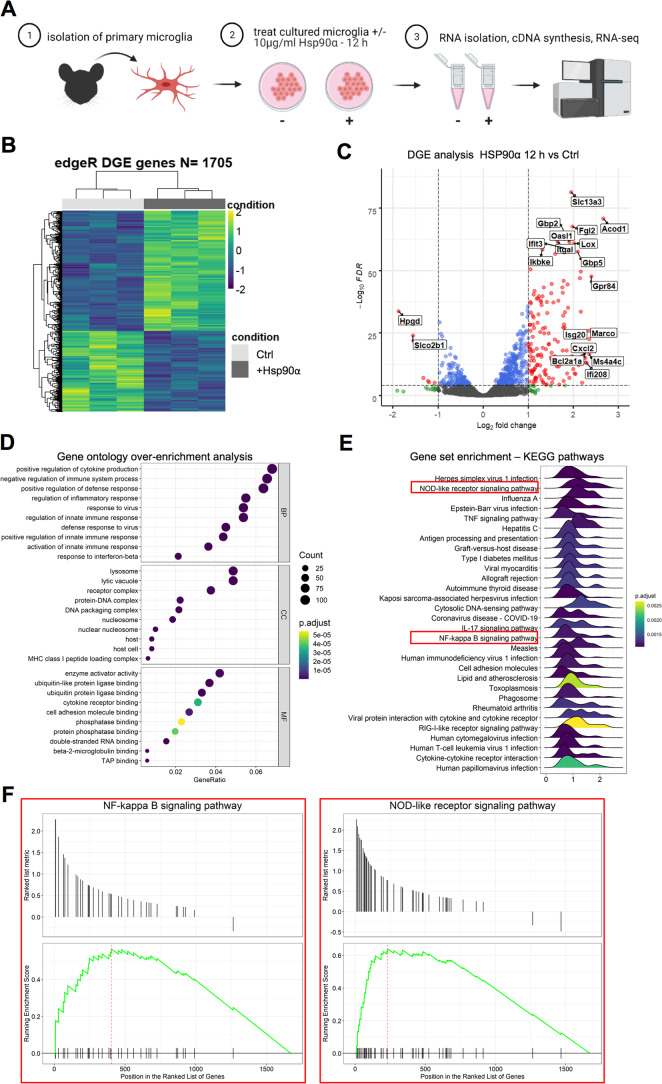
Fig. 2.
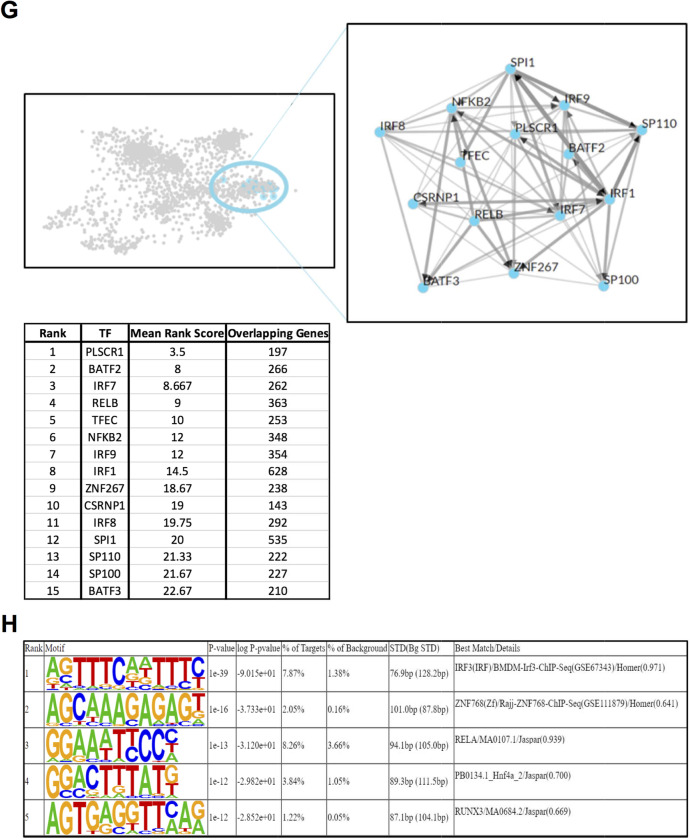
eHsp90α stimulates a transcriptional response upon immunological process-related genes in cultured murine primary microglial cells. A Schematic describing a simplified workflow employed for isolation and RNA-seq analysis of murine primary microglia treated with or without 10 µg/ml Hsp90α for 12 h. B Heatmap representation of the 1705 DEG (FDR < 0.05) identified by RNA-seq analysis between primary microglial cells treated with 10 µg/ml Hsp90α for 12 h versus untreated control samples. C Volcanoplot representing the FDR and logFC values of changes in gene expression between primary microglial cells treated with 10 µg/ml Hsp90α for 12 h versus untreated control samples. Values for features that returned at least 3 counts per million (cpm) across 3 of the 6 samples with MGI symbols are shown. D Gene ontology over enrichment analysis was applied to the primary microglia DEG list (FDR < 0.05) using the enrichGO function of the clusterProfiler package to return enriched descriptive terms belonging to the GO categories; biological process (BP), cellular component (CC), and molecular function (MF). E Ridgeplot of gene set enrichment of KEGG pathways performed on the primary microglia DEG list (FDR < 0.05) using the clusterProfiler function gseKEGG. Values for the directional expression distribution (x-axis) and Benjamini–Hochberg adjusted p-value (p.adjust) are represented. F GSEA plots of selected KEGG pathways representing the enrichment scores for the respective gene sets and the positions of genes within the gene set in the ranked DEG list ordered by descending logFC. G Results of ChEA3 TFEA analysis showing the top 15 transcription factors in the Global GTEx TF Network (left), the local network (right), and the top 15 TFs ranked by mean rank score (lower). The primary microglia DEG list (FDR < 0.05) was used as input. H The top 5 TFs returned by HOMER TFEA analysis are shown. The primary microglia DEG list (FDR < 0.05) was used as input with all features that returned at least 3 counts per million (cpm) across 3 of the 6 samples with MGI symbols as the background gene list
Fig. 3.
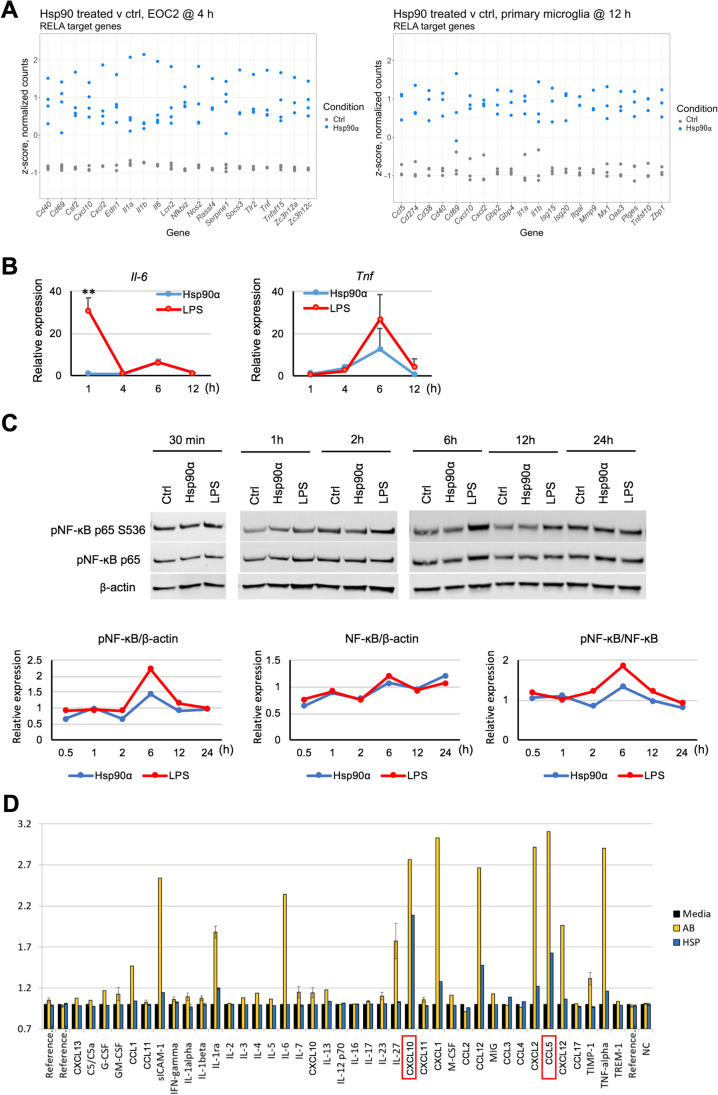
NF-kB target genes are induced in response to eHsp90α, with limited NF-KB activation compared to LPS. A Relative levels of mRNAs in EOC2 cells and primary microglia treated with or without 10 µg/ml Hsp90α for 4 h or 12 h, respectively, that were found to be differentially expressed and that have been identified as NF-κB p65 (RELA) regulatory targets within the curated datasets of the Chea3 TFEA software. The top 10 genes ranked by absolute logFC are shown. B BV2 were cultured in 2% FBS media and treated with Hsp90α (10 μg/ml) or LPS (1000 ng/ml) for 1, 4, 6, and 12 h at which time RNA was isolated and quantified by qPCR for the indicated mRNAs. Gene expression is shown relative to Actb and normalized to control untreated RNA samples collected at the same time points. Control samples were treated with an equimolar concentration of His-tagged protein buffer sourced from the same vendor as the His-tagged Hsp90α. C Under the same culture and treatment conditions as (B), BV2 cells were lysed with RIPA buffer at 30 min, 1, 2, 6, 12, and 24-h post-treatment and cell lysates were analyzed by western blot. Quantitative analyses of immunoblots for p-NF-κB p65 S536, NF-κB p65 are shown with p-NF-κB p65 S536 normalized to total p-NF-κB p65 levels and p-NF-κB p65 levels normalized to β-actin. Each expression ratio was normalized to control untreated samples collected at the same time points. D Cytokines released from primary microglia treated with Hsp90α (10 µg/ml) or fAβ (2 µM) were quantified after 12-h treatment using proteome profiler mouse cytokine array kit, according to manufacturer’s protocol. Cytokine spots’ image intensities are normalized to cytokine levels in culture media of untreated samples (normalized to 1)
Fig. 4.
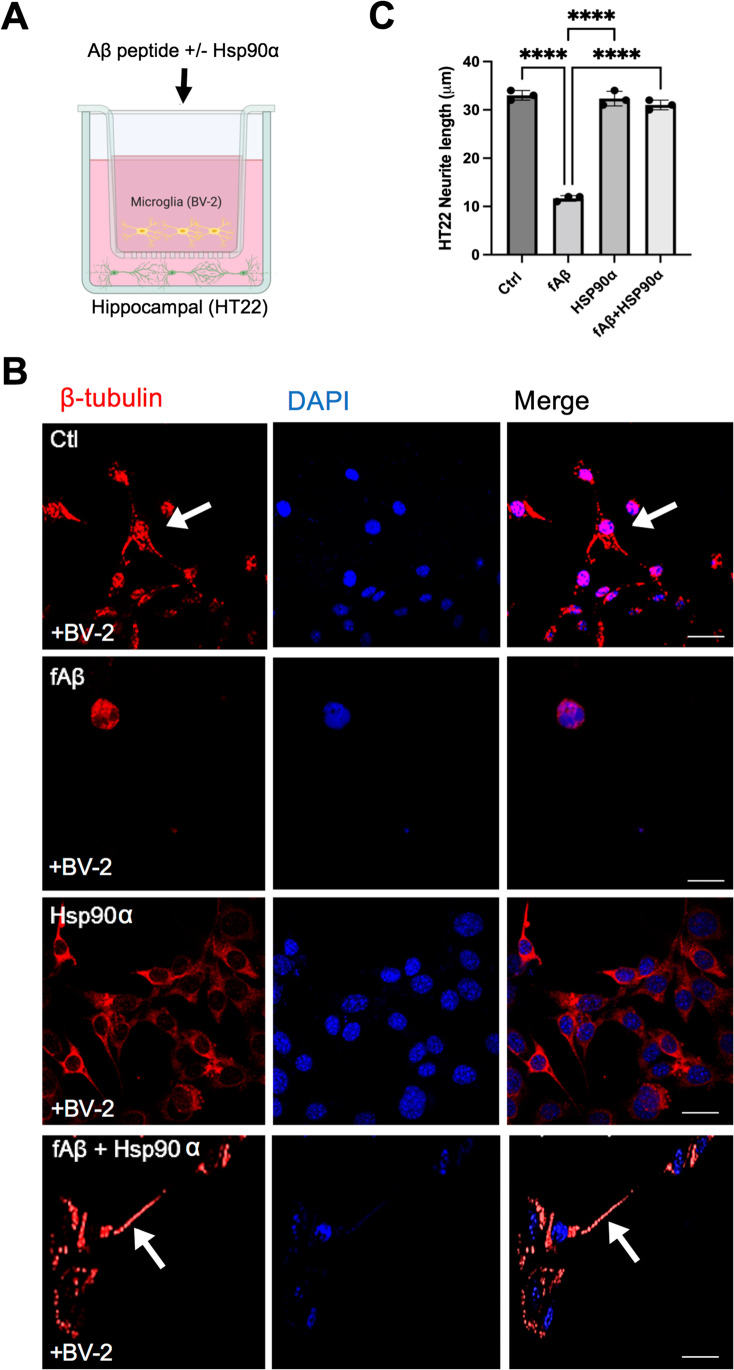
Extracellular Hsp90α mitigates fibrillar Aβ-induced neurotoxicity in vitro. A Schematic for BV2 microglia and HT22 hippocampal neuron co-culture. B HT22 hippocampal neuronal cells were grown on coverslips in the bottom layer of a transwell culture dish. BV2 cells were then added to the top layer of the transwell and incubated with no ligand (Ctrl) fibrillar f-Aβ1-42 (2 μM) (fAβ), Hsp90α (10 μg/ml), or f-Aβ1-42 + Hsp90α as indicated for 72 h. After the 72-h incubation, HT22 cells from the bottom wells were then fixed with 4% para formaldehyde and then permeabilized with 0.1% Triton X-100 before staining with anti-β-tubulin antibodies. Stained cells on coverslips were then examined by confocal microscopy. Scale bar = 5 μm. C β-tubulin-stained neurite outgrowth was measured using ImageJ. A total of 100 cells were counted in each sample. ****p < 0.0001 and n = 3. Cartoon created with BioRender.com. Experiments were repeated three times with similar results
Fig. 5.
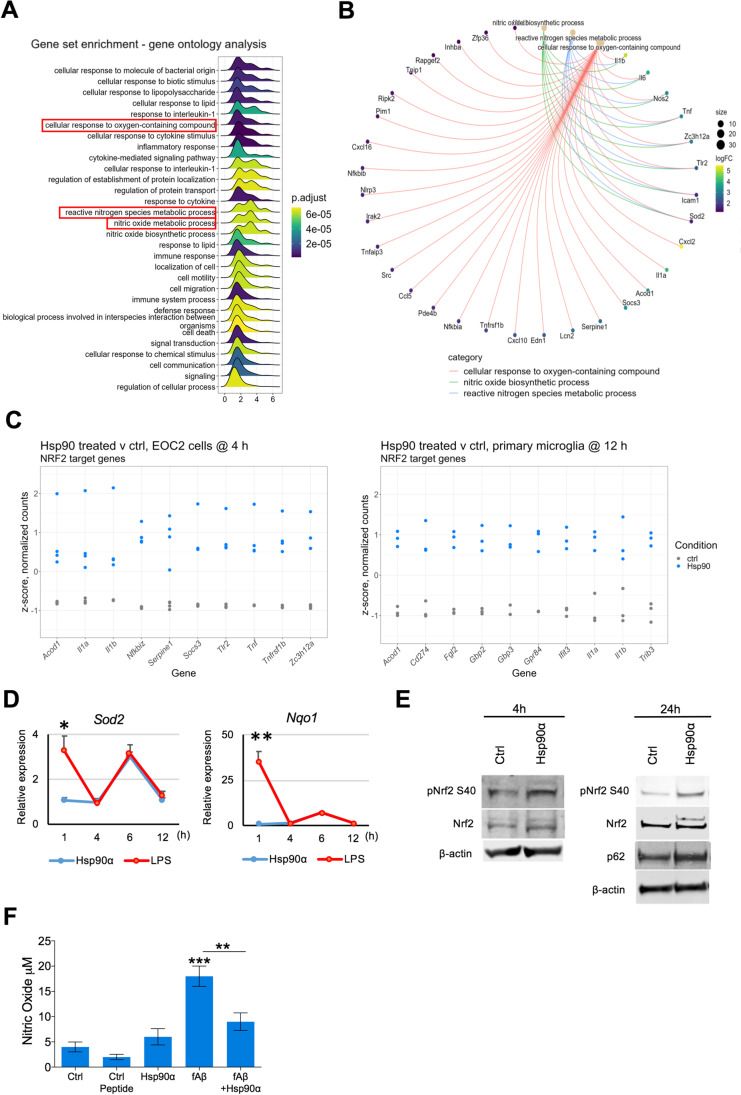
eHsp90α activates Nrf2 and promotes tolerance to oxidative stress. A Gene set enrichment GO analysis of DEG list of EOC2 cells treated with or without 10 µg/ml Hsp90α for 4 h. B ClusterProfiler cnetplot of DEG contributing to selected GO terms related to oxidative stress terms shown in (A). C Relative levels of mRNAs in EOC2 cells and primary microglia treated with or without 10 µg/ml Hsp90α for 4 h or 12 h, respectively, that were found to be differentially expressed and that have been identified as NRF2 regulatory targets within the curated datasets of the Chea3 TFEA software. The top 10 genes ranked by absolute logFC are shown. D BV2 cultured in 2% FBS media were treated with Hsp90α (10 μg/ml) or LPS (1000 ng/ml) for 1, 4, 6, and 12 h at which time RNA was isolated and quantified by qPCR for the indicated mRNAs. Gene expression is shown relative to Actb and normalized to untreated control RNA samples collected at the same time points. Control samples were treated with an equimolar concentration of His-tagged protein buffer sourced same vendor as the His-tagged Hsp90α. E Left: western blot analysis of pNrf2 S40, Nrf2 and β-actin in EOC2 cells 4 h after Hsp90α treatment. Right: western blot analysis of pNrf2 S40, Nrf2, p62, and β-actin levels in lysates of EOC2 cells at 24 h after Hsp90α treatment. F BV2 cells were incubated with the indicated ligands or with vehicle (ctrl) for 4–6 h. Control peptide was Aβ1-40. NO secretion to the medium was then quantitated using an Enzo nitric oxide quantitation assay kit, according to manufacturer’s protocol
Fig. 6.
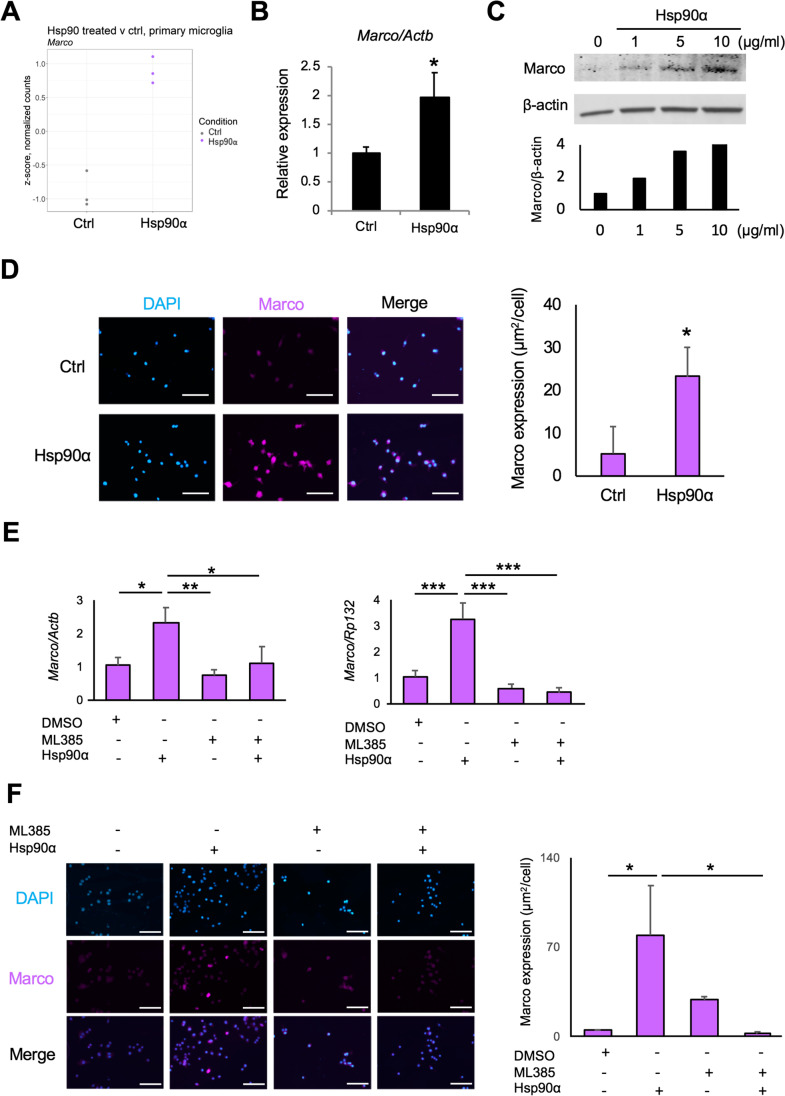
eHsp90α stimulates expression of the scavenger receptor Marco in murine microglial cells. A Relative levels of Marco mRNA in primary microglia treated with or without 10 µg/ml Hsp90α for 12 h, quantified by RNA-seq. Relative levels shown are z-scores of TMM-normalized counts. Fold change was determined using the edgeR statistical package. B RT-qPCR quantitation of Marco normalized to Actb in BV2 cells after 12 h. *p < 0.05, n = 4. C Upper: western blot analysis of Marco and β-actin protein levels from BV2 cells treated Hsp90α for 24 h. Lower: quantitative analyses of Western blot for Marco. The relative values of Marco to β-actin are shown. D Left: immunocytochemistry staining of Marco (Magenta) in BV2 cells treated with 10 µg/ml Hsp90α for 24 h. Blue, DAPI. Scale bars, 100 μm. Right: analysis of Marco expression in BV2 cells treated with Hsp90α. *p < 0.05, n = 3. E RT-qPCR analysis of Marco mRNA in BV2 cells treated with 10 µg/ml Hsp90α and/or 5 μM ML385. The relative mRNA levels were normalized to Actb (left) or Rpl32 (right), an internal control. *p < 0.05, **p < 0.01, ***p < 0.001, n = 3. F Left: immunocytochemistry of Marco (Magenta) in BV2 cells treated Hsp90α and/or ML385. Blue, DAPI. Scale bars, 100 μm. Right: analysis of Marco expression in BV2 cells treated Hsp90α and/or 5 μM ML385. *p < 0.05, n = 3
RNA-seq analysis
RNA-seq analysis was performed following a previously described workflow (Lang et al. 2018). EOC2 cells were treated with or without 10 µg/ml eHsp90α for 4 h at which time RNA was isolated using RNeasy kit (Qiagen). Primary microglia were treated with or without 10 µg/ml eHsp90α for 12 h, and RNA was isolated using the same method. RNA integrity was assessed by using a bioanalyzer (Agilent) with samples with RIN > 7 used for cDNA library synthesis. cDNA library synthesis was performed on ribosome RNA–depleted total RNA using a KAPA stranded RNA-seq with RiboErase (cat. no. KK8483) and Illumina adapters. cDNA libraries were pooled and sequenced with an Illumina HiSeq 2500 (50 cycles, paired-end) to give 10–20 million reads per sample. RNA integrity analysis, cDNA pooling, and NGS services were provided by Harvard Biopolymers Facility. Raw RNA-seq data was processed in a high-performance compute environment using Trimmomatic 0.36 to remove adapters and filter for high-quality reads (Bolger et al. 2014). Read quality was confirmed using FastQC 0.11.5 (Andrews n.d.), and aligned to the Mus musculus annotated genome GRCm38 (Frankish et al. 2019), using STAR 2.5.4a aligner with the –quantMode GeneCounts option (Dobin et al. 2013). Using R version > 3.6.2, feature counts were then filtered for features with at least 3 counts per million (cpm) across 4 (EOC2) or 3 (primary microglia) samples, which gave 10,790 and 12,866 remaining features, respectively. Feature counts were then TMM-normalized and differentially expressed genes (DEG) identified and fold changes quantified using the edgeR statistical package (Robinson et al. 2010). Gene ontology analysis was applied to the DEG list using the clusterProfiler package (Yu et al. 2012), which included representations of the enrichGO function for over-representation analysis and gseKEGG and gseGO functions for gene set enrichment analyses. TFEA analysis was performed using the ChEA3 API available at https://maayanlab.cloud/chea3/, where the DEG list (FDR < 0.05) was used as an input for ChEA3 TFEA analysis of the primary microglia dataset and a filtered DEG list (FDR < 0.05, ± logFC > 0.6) was used as input for the EOC2 ChEA3 TFEA analysis (Keenan et al. 2019). The same respective inputs were used for TFEA analysis by HOMER 4.11.1 (Heinz et al. 2010). The HOMER default settings were used with –cpg and –bg options, with the background gene lists specified to genes that had > 3 cpm across 4 (EOC2) or 3 (primary microglia) samples.
Western blot analysis
Western blotting was performed, as previously described (Gong et al. 2018). Briefly, cells were lysed in a RIPA buffer (Boston BioProducts, MA, USA) using 25-gauge syringes. The same protein amounts were subjected to 4–20% gradient gel (Genscript, NJ, USA), followed by transfer to a polyvinylidene fluoride (PVDF) membrane using wet methods where appropriate. The membranes were blocked in a blocking solution (LI-COR, Inc., Lincoln, NE, USA) for 60 min unless otherwise specified, and incubated overnight with a rabbit monoclonal anti-pNrf2 antibody (1/5000, ab76026, Abcam, Cambridge, MA), a mouse monoclonal anti-Nrf2 antibody (1/1000, MAB3925, Biotechne, Minnesota, MN, USA), a rabbit polyclonal anti-Nrf2 antibody (1/1000, NBP1-32,822, Biotechne, Minnesota, MN, USA), a rabbit polyclonal anti-Marco antibody (1/1000, orb6345, Biobyt, Cambridge, MA), a rabbit monoclonal anti-p-NF-κB p65 S536 antibody (1/1000, 3033S, Cell Signaling, Danvers, MA, USA), a rabbit monoclonal anti- NF-κB p65 antibody (1/1000, 8242S, Cell signaling, Danvers, MA, USA), a mouse monoclonal anti-p62 antibody (1/1000, ab56416, abcam), and a mouse monoclonal anti-actin antibody (1/20000, A5441, Sigma-Aldrich, MO, USA). The membranes were incubated for 1 h at room temperature with goat anti-rabbit IRDye 800 CW fluorescent secondary antibodies (1/15000, LI-COR, Inc., Lincoln, NE, USA) and 680 RD fluorescent secondary antibodies (1/15000, LI-COR, Inc., Lincoln, NE, USA). Blots were washed with Tris-buffered saline, 0.1% (w/v) Tween 20 (TBS-T), and visualized with the Odyssey Imaging System (LI-COR, Inc., Lincoln, NE, USA). The quantitative densitometric analysis was performed using Image Studio Lite Ver. 5.2 (LI-COR, Inc., Lincoln, NE, USA).
RT-qPCR analysis
Total RNA preparation and RT-qPCR were carried out, as described previously (Gong et al. 2018). The miRNeasy mini kit (Qiagen, Hilden, Germany) was used with DNase (Qiagen, Hilden, Germany). The total RNA concentration was measured by using a micro spectrometer Nanodrop one (Thermo Fisher, Waltham, MA, USA). cDNA synthesis was carried out by using iScript cDNA Synthesis Kit (Bio-Rad, Richmond, CA. USA). Specific primer pairs (Suppl. Table 3) were designed using Primer-BLAST or selected from PrimerBank, and the sequences were verified for the predicted target using Primer-BLAST to the Mus musculus RefSeq database (Wang and Seed 2003). Specificity for a single PCR product was confirmed by melt-curve analysis. Real-time qPCR was performed with Applied Biosystems PowerUp SYBR Green Master mix (Thermo Fisher, Waltham, MA, USA) using a StepOne Plus Instrument or 7500 Real-time PCR System (Applied Biosystems) with the cycling conditions 50 °C 2 min and 95 °C 2 min, (95 °C 3 s, 60 °C 30 s, × 40 cycles). Relative mRNA levels to Actb or Rpl32 mRNA levels were quantified by the ∆∆Ct method using the formula–fold change = 2 − ∆∆Ct. PCR reactions were carried out in triplicate, and the mean values were calculated with the mean ± S.D. of the biological triplicates presented.
Immunofluorescence
Cells were washed in ice-cold PBS, pH 7.4, fixed with 4% paraformaldehyde at room temperature for 10 min, and then permeabilized with 0.1% Triton X-100 for 5 min. The fixed cells were blocked in 3% normal goat serum solution for 30 min and then incubated overnight at 4 °C with rabbit anti-Marco antibody (1/100, orb6345, Biobyt, Cambridge, UK) in 3% normal goat serum solution. Cells were then incubated with anti- rabbit IgG AlexaFluor488 (Cell Signaling Technology, Danvers, MA, USA) for 1 h at room temperature. Cellular nuclei were stained with 4′, 6-diamidino-2-phenylindole (DAPI; Invitrogen, Carlsbad, CA, USA). HT22 neurites were stained using β-tubulin (ab15568, Abcam, Cambridge, MA), and fluorophore-tagged secondary antibodies were used to fluorescently stain the fixed cells, and nuclei were stained with DAPI. Coverslips were mounted with Prolong Gold medium. Slides were scanned using a Zeiss LSM confocal microscope with Zen software with the respective, appropriate filter sets, as previously described (Murshid et al. 2010; Okusha et al. 2020). Neurite growth in Fig. 5F is measured using ImageJ software. To quantify square micrometer–Marco positive signal/cell, positive signal was defined by the fluorescence intensity of the cells above that of cells stained without the primary antibodies added, which was subtracted as background signal.
Preparation of Aβ fibrils
Aβ1-42 was dissolved in DMSO (stock 500 μM) at room temperature and stored at − 20 °C. To this Aβ aliquot, we added 10 mM of HCl at RT, diluting to a final concentration of 100 μM of fAβ1-42. We mixed by vortex for 15 s, transferred the solution to 37 °C, and incubated for 24 h. The fAβ1-42 solution was then incubated for 24 h at 37 °C.
Quantification of nitric oxide production
Nitric oxide (NO) release from cells was measured (using the manufacturer’s protocol) in cells incubated with or without f-AB1-42 and in other control samples using a nitric oxide (total) detection kit, (Enzo Cat. no. ADI-917–020).
Cytokine array
Primary microglia culture was treated with or without eHsp90α (10 µg/ml) or fAβ (2 µM) for 12 h. About 100 µg of supernatant was used for each array. Cytokines released from primary microglia treated with Hsp90α/fAβ were quantified after 12-h treatment using proteome profiler mouse cytokine array kit, panel A (R&B Systems, Cat# ARY006), according to manufacturer’s protocol. Data shown are from a 5-min exposure of a film. Image analysis was performed using Western Vision Software, https://wvision.com/.
Cytokine array image analysis
Array images were collected using an X-ray film and analyzed using image processing software. A template was created to analyze pixel density in each spot of the array. Signal values were exported to a Microsoft Excel spread for plotting. Average signal (pixel density) intensity was measured for two spots where duplicate spots/dots represent one cytokine/chemokine. Averaged background signal was subtracted from each spot. We used a signal from a clear area of the array or negative control spots as a background value. Values are normalized to cytokine levels in the culture media of untreated samples (control).
Statistical analysis
Statistical significance was calculated using JMP Pro 15 and Microsoft Excel. Three or more mean values were compared using one-way analysis of variance (ANOVA), followed by Tukey’s post hoc test to determine p-values between experimental groups. Statistical significance between two experimental groups was performed using Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 was considered to indicate statistical significance. Data were expressed as mean ± SD, unless otherwise specified.
Results
The transcriptional response to eHsp90α is characterized by activation of NF-kB-regulated processes
To examine the immediate transcriptional response to eHsp90α, we performed RNA-seq analysis on murine EOC2 microglial cells cultured with or without eHsp90α for 4 h. Differential expression analysis identified 3250 genes to be significantly altered (FDR < 0.05) in the eHsp90α-treated group versus the non-treated control (Fig. 1A). While examining the most differentially expressed genes by logFC, it was observed that genes most sensitive to eHsp90α treatment tended to be induced (327 genes FDR < 0.05, + logFC 0.6) rather than repressed (164 genes FDR < 0.05, − logFC 0.6), with greatest effect-sizes generally observed in the induced genes (Fig. 1B; Suppl. Table 1). To gain insight into cellular processes most sensitive to eHsp90α, over-enrichment analysis was applied to the differential gene expression (DGE) list filtered for a bidirectional fold-change greater than 1.5 (FDR < 0.05, ± logFC 0.6), which returned a list of 470 differentially expressed genes (DEG) with MGI symbols. This analysis indicated that eHsp90α stimulates altered gene expression of mRNAs involved in various immunological and inflammatory processes (Fig. 1C). Similarly, KEGG pathway enrichment analysis also indicated the altered activity of pathways involved in immunity and inflammation (Fig. 1D), including the TNF and NF-κB signaling pathways (Fig. 1E). To further examine potential effector molecules responsive to eHsp90α, we next applied transcription factor enrichment analysis (TFEA) to the filtered DGE list (FDR < 0.05, ± logFC 0.6) using both ChEA3 (Fig. 1F) and HOMER (Fig. 1G) (Keenan et al. 2019; Heinz et al. 2010). Consistent with the KEGG pathway enrichment analysis, factors of the NF-κB family were identified by both TFEA tools to be among the most significantly altered activity in response to eHsp90α (NFKB2, RELB, NFKB1, REL) (Fig. 1F) and RELA (Fig. 1G). Taken together, the predominant transcriptional response to eHsp90α at 4 h was found to center upon NF-κB regulated processes including altered expression of genes with functional roles in inflammation and immunity.
eHsp90α stimulates a transcriptional response upon immunological process-related genes in primary murine microglial cells
To further characterize the transcriptional response of microglial cells to eHsp90α, we next compared the transcriptomic profiles of primary microglia treated with or without eHsp90α. We reasoned that the use of primary microglia may provide additional confidence in identification of eHsp90α-responsive genes in microglia when considered with data derived from EOC2 cells, while also allowing for the examination of the response to eHsp90α at a later time-point. The primary microglia were isolated, and the cultures were treated with or without 10 µg/ml eHsp90α for 12 h in triplicate, at which time RNA was isolated and analyzed by RNA-seq analysis (Fig. 2A). Differential gene expression analysis identified 1705 differentially expressed genes (FDR < 0.05) between the eHsp90α-treated and non-treated control conditions (Fig. 2B). Like changes observed in treated EOC2 cells, a cohort of genes exhibited a robust relative increase in mRNA levels in response to eHsp90α (Fig. 2C). Overrepresentation analysis revealed gene ontology terms again describing processes related to inflammation, infection, and immunity (Fig. 2D), together suggesting some level of innate activation of the eHsp90α-treated microglia. This was also indicated by KEGG pathway analysis, which identified pathways associated with contexts of immune activation (Fig. 2E). The NF-κB signaling pathway was again identified among processes to be altered in response to eHsp90α, in addition to the NOD-like receptor signaling pathway which is mechanistically connected to NF-κB (Fig. 2E, F). Components of the NF-κB transcriptional machinery were also among the top identified factors when TFEA analysis was applied to the DEG list (FDR < 0.05) (Fig. 2G, H). This data indicates that the predominant microglial response to eHsp90α is that of an increase in genes with functions in inflammation and immunity and that NF-κB is a likely driver of this transcriptional response.
NF-kB target genes are induced in response to eHsp90α, with moderate NF-κB activation compared to LPS
As significant research efforts have been directed towards understanding the inflammatory nature of extracellular HSPs (Calderwood et al. 2016), we next sought to further validate and characterize the apparent responsiveness of NF-κB to eHsp90α. We first identified the NF-κB-regulated genes most altered by eHsp90α in the RNA-seq analyses (Fig. 3A) and examined their responsiveness to eHsp90α in cultured murine BV2 microglial cells. RT-qPCR quantitation of NF-κB targets IL-6 (Il6) and TNF-α (Tnf) was performed on BV2 cultures treated with or without 10μg/ml eHsp90α treatment over a 12-h period (Fig. 3B). These qPCR analyses indicated both Il6 and Tnf mRNA responses to eHsp90α peaked at 6 h, while markedly more robust responses were seen in response to 1000 ng/ml LPS for Il-6 at 1 h and for Tnf at 6 h (Fig. 3B). The NF-κB p65 protein (encoded by RelA) forms heterodimers with the proteolytically processed forms of p105/p50 (Nfkb1) or p100/p52 (Nfkb2). The NF-κB p65/p50 dimer is considered the prototypical form; however, the other REL members RelB (RelB) or c-Rel (Rel) also form dimers with p50 or p52. To further characterize the relative activation of NF-κB by eHsp90α, we compared the levels of NF-κB p65 activating phosphorylation at serine 536 in BV2 cells treated with eHsp90α to levels of cells treated with LPS (Fig. 3C). NF-κB p65 becomes phosphorylated at Ser536 by the IKK kinases in response to various inflammatory stimuli and is considered indicative of NF-κB activation, reviewed in Huang et al. (2010). A modest increase in pNF-κB levels was detected in response to eHsp90α, the magnitude to which was lower than that stimulated by LPS at 6 h under the experimental conditions used (Fig. 3C). To further examine the possibility that eHsp90α leads to a moderated innate/inflammation response, we also measured the cytokine profile of cultured BV2 cells treated with eHsp90α or the inflammatory stimuli amyloid-beta (Aβ) (Fig. 3D). Increased levels of several cytokines were released in response to eHsp90α, although the magnitude to which eHsp90α stimulated cytokine release was notably lower than that stimulated by Aβ fibrils (Fig. 3D). In summary, we conclude that eHsp90α can stimulate microglia into an immune profile characterized by CXCL10 and CCL5 and an increase in the levels associated with NF-κB activation.
Extracellular Hsp90α mitigates fibrillary amyloid beta-induced neurotoxicity in vitro
We have previously reported a stalled inflammatory response in macrophages in the presence of eHsp90α (Murshid et al. 2015). As microglial inflammatory responses to Aβ fibrils are an important component of Aβ neurotoxicity, we next tested the hypothesis that eHsp90α protects against Aβ neurotoxicity associated with microglia-derived inflammation. To address this possibility, we co-treated murine microglial BV2 with purified Hsp90α and/or freshly prepared fibrillary Aβ1-42 (fAβ) and assessed the viability of adjacent neuronal HT22 cells potentially exposed to secreted microglial products via transwell culture preparations (Fig. 4A). BV2 cells pre-incubated with FITC fAβ-mediated toxicity towards the distant neuronal HT22 cells as indicated by extensive loss of microtubule-containing processes, a morphological measure of neuron cell viability (Hancock et al. 2015) (Fig. 4B). Within 72 h of fAβ incubation, many of the neuronal cells had lost their elongated processes (Fig. 4B). In contrast, when HT22 cells were co-cultured with BV2 cells that had been treated with both fAβ and eHsp90α in the top well of the transwell culture dish, the majority of the HT22 cells survived with neurite lengths comparable to those in the non-treated control, suggesting that addition of eHsp90α provided some protection against fAβ toxicity. eHsp90α treatment alone did not significantly impact neurite outgrowth (Fig. 4C). The extent of neurite outgrowth is quantified and is shown in Fig. 4C. Similar effects of exposure to fAβ with or without eHsp90α were also reported upon co-culture of HT22 neuronal cells with EOC2 microglial cells (A. Murshid, pre-print (Murshid et al. 2021)).
eHsp90α activates Nrf2 and promotes resistance to oxidative stress
In addition to gene ontology (GO) terms describing enrichment of processes related to inflammation and immunity, gene set enrichment GO analysis of the DEGs from EOC2 (Fig. 5A) and primary microglia (Suppl. Fig. 1A) treated with eHsp90α for 4 h or 12 h, respectively, also returned terms suggesting co-activation of an anti-oxidative stress response. When the genes contributing to these terms were examined (Fig. 5B; Suppl. Fig. 1B), many were also connected to the inflammatory signature described earlier; however, others such as upregulation of superoxide dismutase 2 (Sod2), indicated a possible activation of the anti-oxidative stress response. When we considered this data with our recent finding that eHsp90α can stimulate indicators of Nrf2 activation (Murshid et al. 2021), we hypothesized that some level of Nrf2 co-activation may be occurring in response to eHsp90α and that this may provide a potential mechanism by which eHsp90α was able to mitigate fAβ toxicity. To investigate this prospect, we identified the top-10 differentially expressed Nrf2-target genes in terms of bidirectional fold change in each of the RNA-seq datasets (Fig. 5C). All the genes identified were found to be increased in eHsp90α-treated samples compared to untreated control samples, suggesting eHsp90α may stimulate Nrf2 transcriptional activity. To determine whether this response is common across microglial cells from different sources, we measured the levels of canonical Nrf2 target genes Nqo1 and Sod2 in BV2 cells treated with eHsp90α relative to untreated controls by RT-qPCR (Fig. 5D). To gain additional insight into the events associated with eHsp90α-stimulated Nrf2 activation, we assessed Nrf2 activating phosphorylation at Serine 40 by western blot (Fig. 5E). Nrf2 is phosphorylated at Ser40 under oxidative conditions by PKC kinases, a modification that mediates Nrf2 release from its negative regulator Keap1 (Numazawa et al. 2003; Huang et al. 2002), followed by its nuclear translocation and increased transcriptional activity. Treatment of EOC2 cells with eHsp90α led to increased levels of Nrf2 phosphorylation at S40 after both 4 and 24 h, accompanied by increased protein levels of p62, encoded by Nrf2 target Sqstm1, also observed at 24 h (Fig. 5E). To determine whether these indicators of Nrf2 activation were paired with attenuated production of oxidative molecules in response to inflammatory stimuli, we measured nitric oxide (NO) release from BV2 cells cultured with fAβ, eHsp90α, or co-treated with both fAβ and eHsp90α (Fig. 5F). Consistent with eHsp90α possessing Nrf2-activating properties, cells co-treated with both fAβ and eHsp90α exhibited reduced levels of NO production compared to the increased NO release by cells treated with fAβ alone (Fig. 5F). These findings indicate that in addition to moderate activation of microglial inflammatory responses, eHsp90α also induces Nrf2 a process that may limit microglial production of NO in response to fAβ.
eHsp90α stimulates expression of the scavenger receptor Marco in murine microglial cells
Microglia play important roles in Aβ metabolism and the inflammatory response to Aβ in Alzheimer’s disease (Wilkinson and Khoury 2012). To gain some insight as to how eHsp90α may protect against the toxic effects of Aβ, we sought to identify eHsp90α — responsive mRNAs that may provide some basis for altered Aβ metabolism by BV2 microglial cells. We identified macrophage receptor with collagenous structure (Marco) as one such gene that was among the top-10 most differentially expressed genes in response to eHsp90α in the primary microglia RNA-seq dataset (Figs. 2C and 6A), and was also differentially expressed in the EOC2 dataset (Suppl. Fig. 2A). The encoded protein, Marco, is a scavenger receptor that has been identified as a microglial receptor for Aβ (Alarcon et al. 2005). Quantitation of Marco mRNA levels in eHsp90α-treated BV2 cells at 12 h found it to also be significantly upregulated compared to control untreated cells (Fig. 6B; Suppl. Fig. 2B), indicating Marco mRNA levels are induced by eHsp90α across multiple models of murine microglial cells. In eHsp90α treated BV2 cells, Marco protein levels were also observed to increase over a 24-h period in a concentration-dependent manner as measured by Western blot (Fig. 6C). This was consistent with significantly higher levels of Marco immunostaining observed by confocal microscopy in BV2 cells after 24-h treatment (Fig. 6D) and a trend towards higher levels in eHsp90α treated EOC2 cells (Suppl. Fig. 2C). As previous studies have found Marco expression to be dependent upon Nrf2 (Reddy et al. 2009), we tested the hypothesis that the observed increase in Marco expression may be a product of eHsp90α-mediated Nrf2 activation. To address this possibility, we employed the chemical inhibitor of Nrf2, ML385, which decreases Nrf2 expression at the mRNA and protein levels over an extended treatment (Suppl. Fig. 2D). Consistent with a possible role for Nrf2 in eHsp90α-stimulated Marco expression, BV2 cells treated with both eHsp90α and ML385 exhibited significantly reduced Marco mRNA levels compared to samples treated with eHsp90α alone (Fig. 6E). Similarly, ML385 was also found to reduce eHsp90α-induced Marco protein levels as measured by quantification of the immunofluorescent staining by confocal microscopy (Fig. 6F). We therefore conclude that increased levels of eHsp90α stimulate expression of the Aβ-binding receptor Marco at both the mRNA and protein levels and that this effect may be a product of the observed eHsp90α-stimulated Nrf2 activation. Hsp90 also led to induction of Acod1 (cis-aconitate decarboxylase), known to induce TNFAIP3, also upregulated in both EOC2 and primary microglia RNA-seq datasets (Suppl. Tables 1, 2), which contribute to inflammatory modulation and the slight increase of NO upon Hsp90 stimulation alone (Wu et al. 2020). In addition, Clec4e (C-type lectin 4e) was one of the major upregulated genes (Fig. 1). Clec4e is a receptor for damage-associated molecular patterns (DAMPs) and necrotic cell bodies and may contribute to the complex phenotype initiated by eHsp90α (Clement et al. 2016).
Discussion
Our data suggest that exposure of microglia to eHsp90α leads to an altered phenotype that includes a mild activation of inflammatory genes in microglial cells. The effects observed included induction of cytokine genes and other genes associated with innate immunity and inflammation (Figs. 1, 2, 3) (Hanisch 2002; Lee et al. 2002). Microglia are thought to exist in a number of activation states, and these may involve both pro-and anti-inflammatory cytokine synthesis, depending on the stimuli (Janda et al. 2018). Our previous data indicated that microglial activation was associated with increased capacity to phagocytose amyloid-beta indicating that eHsp90 was able to concomitantly increase phagocytosis in combination with cytokine induction (Figs. 1, 2, 3; (Murshid et al. 2021)). Activation of microglia to an M1 type phenotype is associated with reduced phagocytosis in contrast to the effects of Hsp90 (Cherry et al. 2014). eHsp90α appeared to induce a complex gene expression phenotype, including stimulation of expression of both NF-kB and Nrf2 target genes. In RNA-seq analysis, Acod1 was upregulated by Hsp90 stimulation in both EOC2 and primary microglia (Fig. 5C). Acod1 induces TNFAP3 and suppresses NF-kB signaling (Wu et al. 2020). Indeed, the increases of NO and NF-kB expressions were slight with Hsp90 stimulation alone. Thus, there was a concomitant induction of pro-inflammatory genes dependent on NF-kB and anti-oxidant and anti-inflammatory genes in the antioxidant response (AOR) including Sod2 and Marco. The AOR protects cells against reactive oxygen species (ROS) through induction of a multi-gene expression cascade (Nguyen et al. 2003). Marco is an emerging anti-inflammatory receptor in mononuclear phagocytes that is induced by NRF2 (Fig. 6) and operating by directly represses Toll-like receptor 4 and stimulates the secretion of anti-inflammatory cytokine IL-37 (Fleur et al. 2021; Kissick et al. 2014). Nrf2 activation may provide protection from oxidative stress associated with innate activation of microglial cells (Branca et al. 2017).
Inflammation in the confined spaces of the CNS is a potential hazard and is generally kept under strict control (Hanisch 2002). eHsp90α appears to induce a desirable activation state in microglia, in which cytokines and phagocytosis are activated along with protective Nrf2 activation and anti-inflammatory gene expression (Figs. 1, 2, 3, 4). Indeed, exposure to the chaperone was able to protect neighboring neurons from the oxidative burst accompanying internalization of fAβ (Fig. 4). Beneficial effects appeared to include suppression of NO secretion by microglia treated with fAβ (Fig. 4). eHsp90 may thus play key homeostatic effects by accumulating in microglia and protecting from proteotoxic stresses and/or by inducing the AOR and reducing effects of ROS during the oxidative burst accompanying phagocytosis. In addition, our previous studies showed that exposure to eHsp90 could direct internalized beta amyloid into the autophagy pathway which has been shown to reduce inflammatory effects (Murshid et al. 2021; Netea-Maier et al. 2016; Takahama et al. 2018).
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary Fig. 1 (PNG 705 KB) (A) Gene set enrichment GO analysis of DEG list of cultured primary microglia treated with or without 10 µg/ml Hsp90α for 12 h. B Relative levels of mRNAs in EOC2 cells and primary microglia treated with or without 10 µg/ml Hsp90α for 4 h or 12 h, respectively, that were found to be differentially expressed and that have been categorized under GO:1901701 ‘cellular response to oxygen-containing compound’. Gene expression levels were quantified by RNA-seq and the relative levels shown are z-scores of TMM-normalized counts.
Supplementary Fig. 2 (PNG 543 KB) (A) Relative levels of Marco mRNA in EOC2 cells treated with or without 10 µg/ml Hsp90α for 4 h, quantified by RNA-seq. Relative levels shown are z-scores of TMM-normalized counts. Fold change was determined using the edgeR statistical package. B RT-qPCR quantitation of Marco normalized to Rpl32 in BV2 cells after 12 h. * p < 0.05. n=4. C Quantitation of Immunofluorescence staining for Marco in paraformaldehyde fixed EOC2 cells treated with 10 µg/ml Hsp90α for 24 h, scale bar =100 µm. n=3. D BV2 cells were treated with Nrf2 inhibitor ML385 (ML) or vehicle control for the indicated periods at which time levels of Nrf2-encoding mRNA (Nfe2l2) were assessed.
Acknowledgements
We wish to thank the Department of Radiation Oncology for their continued support. Suraya Yasmine, Reeham Choudhury and Lay-Hong Ang are thanked for fine technical contributions. We thank Harvard Biopolymers Facility and Harvard Research Computing for their technical support. Figures 2A and 4A were made with Biorender.com.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
The original version of this article was revised: due to a technical error Figures 1 and 2 were incomplete. Figs 1F, 1G and Figs 2G, 2H have been added.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yuka Okusha, Benjamin J. Lang and Ayesha Murshid contributed equally to this work
Change history
7/13/2022
A Correction to this paper has been published: 10.1007/s12192-022-01285-x
Contributor Information
Yuka Okusha, Email: yokusha@bidmc.harvard.edu.
Stuart K. Calderwood, Email: scalderw@bidmc.harvard.edu
References
- Alarcon R, et al. Expression of scavenger receptors in glial cells. Comparing the adhesion of astrocytes and microglia from neonatal rats to surface-bound beta-amyloid. J Biol Chem. 2005;280(34):30406–15. doi: 10.1074/jbc.M414686200. [DOI] [PubMed] [Google Scholar]
- Andrews S (n.d.) FastQC: a quality control tool for high throughput sequence data. Available online at: 2010; Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branca C, et al. Genetic reduction of Nrf2 exacerbates cognitive deficits in a mouse model of Alzheimer’s disease. Hum Mol Genet. 2017;26(24):4823–4835. doi: 10.1093/hmg/ddx361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood SK, Gong J, Murshid A. Extracellular HSPs: the complicated roles of extracellular HSPs in immunity. Front Immunol. 2016;7:159. doi: 10.3389/fimmu.2016.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood SK, Murshid A. Molecular chaperone accumulation in cancer and decrease in Alzheimer’s disease: the potential roles of HSF1. Front Neurosci. 2017;11:192. doi: 10.3389/fnins.2017.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JD, Olschowka JA, O'Banion MK. Neuroinflammation and M2 microglia: the good, the bad, and the inflamed. J Neuroinflammation. 2014;11:98. doi: 10.1186/1742-2094-11-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton KA, Van Enoo AA, Ikezu T. Alzheimer’s disease: the role of microglia in brain homeostasis and proteopathy. Front Neurosci. 2017;11:680. doi: 10.3389/fnins.2017.00680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement M, et al. Necrotic cell sensor Clec4e promotes a proatherogenic macrophage phenotype through activation of the unfolded protein response. Circulation. 2016;134(14):1039–1051. doi: 10.1161/CIRCULATIONAHA.116.022668. [DOI] [PubMed] [Google Scholar]
- Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankish A, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47(D1):D766–D773. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto M, et al. Active HSF1 significantly suppresses polyglutamine aggregate formation in cellular and mouse models. J Biol Chem. 2005;280(41):34908–34916. doi: 10.1074/jbc.M506288200. [DOI] [PubMed] [Google Scholar]
- Garigan D, et al. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics. 2002;161(3):1101–1112. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Generation of anti-tumor immunity using mammalian heat shock protein 70 DNA (2006) 24(25) [DOI] [PubMed]
- Gong J, et al. Genotoxic stress induces Sca-1-expressing metastatic mammary cancer cells. Mol Oncol. 2018;12(8):1249–1263. doi: 10.1002/1878-0261.12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzhova I, et al. In vitro studies show that Hsp70 can be released by glia and that exogenous Hsp70 can enhance neuronal stress tolerance. Brain Res. 2001;914(1–2):66–73. doi: 10.1016/S0006-8993(01)02774-3. [DOI] [PubMed] [Google Scholar]
- Hancock MK et al (2015) A facile method for simultaneously measuring neuronal cell viability and neurite outgrowth. Curr Chem Genom Transl Med 9:6–16 [DOI] [PMC free article] [PubMed]
- Hanisch UK. Microglia as a source and target of cytokines. Glia. 2002;40(2):140–155. doi: 10.1002/glia.10161. [DOI] [PubMed] [Google Scholar]
- Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300(5622):1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- Huang B, et al. Posttranslational modifications of NF-kappaB: another layer of regulation for NF-kappaB signaling pathway. Cell Signal. 2010;22(9):1282–1290. doi: 10.1016/j.cellsig.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HC, Nguyen T, Pickett CB. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J Biol Chem. 2002;277(45):42769–42774. doi: 10.1074/jbc.M206911200. [DOI] [PubMed] [Google Scholar]
- Ikezu S et al (2020) Inhibition of colony stimulating factor 1 receptor corrects maternal inflammation-induced microglial and synaptic dysfunction and behavioral abnormalities. Mol Psychiatry [DOI] [PMC free article] [PubMed]
- Janda E, Boi L, Carta AR. Microglial phagocytosis and its regulation: a therapeutic target in Parkinson’s disease? Front Mol Neurosci. 2018;11:144. doi: 10.3389/fnmol.2018.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan AB, et al. ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res. 2019;47(W1):W212–W224. doi: 10.1093/nar/gkz446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard T, et al. Heat shock protein-based therapy as a potential candidate for treating the sphingolipidoses. Sci Transl Med. 2016;8(355):355ra118. doi: 10.1126/scitranslmed.aad9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissick HT, et al. The scavenger receptor MARCO modulates TLR-induced responses in dendritic cells. PLoS One. 2014;9(8):e104148. doi: 10.1371/journal.pone.0104148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasemann S, et al. The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity. 2017;47(3):566–581. e9. doi: 10.1016/j.immuni.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Fleur L, et al. Targeting MARCO and IL37R on immunosuppressive macrophages in lung cancer blocks regulatory T cells and supports cytotoxic lymphocyte function. Cancer Res. 2021;81(4):956–967. doi: 10.1158/0008-5472.CAN-20-1885. [DOI] [PubMed] [Google Scholar]
- Lang BJ, et al. A workflow guide to RNA-seq analysis of chaperone function and beyond. Methods Mol Biol. 2018;1709:233–252. doi: 10.1007/978-1-4939-7477-1_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang BJ et al (2021) The functions and regulation of heat shock proteins; key orchestrators of proteostasis and the heat shock response. Arch Toxicol 95(6):1943–1970 [DOI] [PubMed]
- Lee YB, Nagai A, Kim SU. Cytokines, chemokines, and cytokine receptors in human microglia. J Neurosci Res. 2002;69(1):94–103. doi: 10.1002/jnr.10253. [DOI] [PubMed] [Google Scholar]
- Li W, Sahu D, Tsen F. Secreted heat shock protein-90 (Hsp90) in wound healing and cancer. Biochim Biophys Acta. 2012;1823(3):730–741. doi: 10.1016/j.bbamcr.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshid A, et al. Scavenger receptor SREC-I mediated entry of TLR4 into lipid microdomains and triggered inflammatory cytokine release in RAW 264.7 cells upon LPS activation. PLoS One. 2015;10(4):e0122529. doi: 10.1371/journal.pone.0122529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshid A et al (2021) Extracellular Hsp90α Detoxifies β-amyloid fibrils through an NRF2 and autophagy dependent pathway. bioRxiv
- Murshid A, Gong J, Calderwood SK. Heat shock protein 90 mediates efficient antigen cross presentation through the scavenger receptor expressed by endothelial cells-I. J Immunol. 2010;185(5):2903–2917. doi: 10.4049/jimmunol.0903635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea-Maier RT, et al. Modulation of inflammation by autophagy: consequences for human disease. Autophagy. 2016;12(2):245–260. doi: 10.1080/15548627.2015.1071759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T, Sherratt PJ, Pickett CB. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu Rev Pharmacol Toxicol. 2003;43:233–260. doi: 10.1146/annurev.pharmtox.43.100901.140229. [DOI] [PubMed] [Google Scholar]
- Numazawa S, et al. Atypical protein kinase C mediates activation of NF-E2-related factor 2 in response to oxidative stress. Am J Physiol Cell Physiol. 2003;285(2):C334–C342. doi: 10.1152/ajpcell.00043.2003. [DOI] [PubMed] [Google Scholar]
- Okusha Y, et al. Rab11A functions as a negative regulator of osteoclastogenesis through dictating lysosome-induced proteolysis of c-fms and RANK surface receptors. Cells. 2020;9:2384. doi: 10.3390/cells9112384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampelt H, et al. Metazoan Hsp70 machines use Hsp110 to power protein disaggregation. EMBO J. 2012;31(21):4221–4235. doi: 10.1038/emboj.2012.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy NM, et al. Innate immunity against bacterial infection following hyperoxia exposure is impaired in NRF2-deficient mice. J Immunol. 2009;183(7):4601–4608. doi: 10.4049/jimmunol.0901754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansley B, Post J, Hensley K. A comparative review of cell culture systems for the study of microglial biology in Alzheimer’s disease. J Neuroinflammation. 2012;9:115. doi: 10.1186/1742-2094-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahama M, Akira S, Saitoh T. Autophagy limits activation of the inflammasomes. Immunol Rev. 2018;281(1):62–73. doi: 10.1111/imr.12613. [DOI] [PubMed] [Google Scholar]
- Taylor AR, et al. Regulation of heat shock protein 70 release in astrocytes: role of signaling kinases. Dev Neurobiol. 2007;67(13):1815–1829. doi: 10.1002/dneu.20559. [DOI] [PubMed] [Google Scholar]
- Thawkar BS, Kaur G. Inhibitors of NF-kappaB and P2X7/NLRP3/caspase 1 pathway in microglia: novel therapeutic opportunities in neuroinflammation induced early-stage Alzheimer’s disease. J Neuroimmunol. 2019;326:62–74. doi: 10.1016/j.jneuroim.2018.11.010. [DOI] [PubMed] [Google Scholar]
- Theriault JR, Adachi H, Calderwood SK. Role of scavenger receptors in the binding and internalization of heat shock protein 70. J Immunol. 2006;177(12):8604–8611. doi: 10.4049/jimmunol.177.12.8604. [DOI] [PubMed] [Google Scholar]
- Tidwell JL, Houenou LJ, Tytell M. Administration of Hsp70 in vivo inhibits motor and sensory neuron degeneration. Cell Stress Chaperones. 2004;9(1):88–98. doi: 10.1379/1466-1268(2004)009<0088:AOHIVI>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman R, Burm SM, Bajramovic JJ. An overview of in vitro methods to study microglia. Front Cell Neurosci. 2018;12:242. doi: 10.3389/fncel.2018.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oosten-Hawle P, Morimoto RI. Transcellular chaperone signaling: an organismal strategy for integrated cell stress responses. J Exp Biol. 2014;217(Pt 1):129–136. doi: 10.1242/jeb.091249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Seed B. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 2003;31(24):e154. doi: 10.1093/nar/gng154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson K, El Khoury J. Microglial scavenger receptors and their roles in the pathogenesis of Alzheimer’s disease. Int J Alzheimers Dis. 2012;2012:489456. doi: 10.1155/2012/489456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, et al. ACOD1 in immunometabolism and disease. Cell Mol Immunol. 2020;17(8):822–833. doi: 10.1038/s41423-020-0489-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, et al. Cytokine-mediated inhibition of fibrillar amyloid-beta peptide degradation by human mononuclear phagocytes. J Immunol. 2008;181(6):3877–3886. doi: 10.4049/jimmunol.181.6.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, et al. TREM2 modulates microglia phenotypes in the neuroinflammation of Parkinson’s disease. Biochem Biophys Res Commun. 2018;499(4):797–802. doi: 10.1016/j.bbrc.2018.03.226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1 (PNG 705 KB) (A) Gene set enrichment GO analysis of DEG list of cultured primary microglia treated with or without 10 µg/ml Hsp90α for 12 h. B Relative levels of mRNAs in EOC2 cells and primary microglia treated with or without 10 µg/ml Hsp90α for 4 h or 12 h, respectively, that were found to be differentially expressed and that have been categorized under GO:1901701 ‘cellular response to oxygen-containing compound’. Gene expression levels were quantified by RNA-seq and the relative levels shown are z-scores of TMM-normalized counts.
Supplementary Fig. 2 (PNG 543 KB) (A) Relative levels of Marco mRNA in EOC2 cells treated with or without 10 µg/ml Hsp90α for 4 h, quantified by RNA-seq. Relative levels shown are z-scores of TMM-normalized counts. Fold change was determined using the edgeR statistical package. B RT-qPCR quantitation of Marco normalized to Rpl32 in BV2 cells after 12 h. * p < 0.05. n=4. C Quantitation of Immunofluorescence staining for Marco in paraformaldehyde fixed EOC2 cells treated with 10 µg/ml Hsp90α for 24 h, scale bar =100 µm. n=3. D BV2 cells were treated with Nrf2 inhibitor ML385 (ML) or vehicle control for the indicated periods at which time levels of Nrf2-encoding mRNA (Nfe2l2) were assessed.