Abstract
Evidence is presented for an alternative to the superoxide dismutase (SOD)-catalase oxidative stress defense system in Desulfovibrio vulgaris (strain Hildenborough). This alternative system consists of the nonheme iron proteins, rubrerythrin (Rbr) and rubredoxin oxidoreductase (Rbo), the product of the rbo gene (also called desulfoferrodoxin). A Δrbo strain of D. vulgaris was found to be more sensitive to internal superoxide exposure than was the wild type. Unlike Rbo, expression of plasmid-borne Rbr failed to restore the aerobic growth of a SOD-deficient strain of Escherichia coli. Conversely, plasmid-borne expression of two different Rbrs from D. vulgaris increased the viability of a catalase-deficient strain of E. coli that had been exposed to hydrogen peroxide whereas Rbo actually decreased the viability. A previously undescribed D. vulgaris gene was found to encode a protein having 50% sequence identity to that of E. coli Fe-SOD. This gene also encoded an extended N-terminal sequence with high homologies to export signal peptides of periplasmic redox proteins. The SOD activity of D. vulgaris is not affected by the absence of Rbo and is concentrated in the periplasmic fraction of cell extracts. These results are consistent with a superoxide reductase rather than SOD activity of Rbo and with a peroxidase activity of Rbr. A joint role for Rbo and Rbr as a novel cytoplasmic oxidative stress protection system in D. vulgaris and other anaerobic microorganisms is proposed.
Although sulfate-reducing bacteria are classified as strict anaerobes, many species are aerotolerant and some species have been shown to accumulate at oxic-anoxic interfaces (28). Therefore, sulfate-reducing bacteria must deal with oxidative stress. Some sulfate-reducing bacteria have been shown to contain superoxide dismutases (SODs) and catalases, but other species do not demonstrate the activities expected for these enzymes (15, 18, 43). The apparent absence of SOD and catalase can be rationalized on the basis that their catalytic dismutation reactions (reactions 1 and 2) generate O2, which may be disadvantageous for strict anaerobes.
Evidence for an alternative oxidative stress protection mechanism in sulfate-reducing bacteria has begun to emerge. Rubredoxin oxidoreductase (Rbo), which has also been named desulfoferrodoxin (29), is the product of the rbo gene in several sulfate-reducing bacteria (7). A Desulfovibrio vulgaris strain with the rbo gene deleted was shown to be more air sensitive than the wild type (45). The Rbo from Desulfoarculus baarsii was shown to complement a sodA sodB defect in Escherichia coli (31). Rbo has no detectable sequence homologies to known SODs, and little or no SOD activity has been found for Rbos in vitro (26, 35). Nevertheless, the D. baarsii Rbo was reported to reduce the steady-state level of intracellular superoxide in the E. coli sodA sodB strain (25). This Rbo in vitro showed evidence for superoxide reductase (SOR) activity (26), i.e., reduction of O2−, presumably to H2O2, without dismutation (reaction 3).
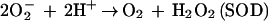 |
1 |
 |
2 |
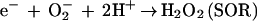 |
3 |
Rbo is a homodimeric protein, each subunit of which contains two mononuclear nonheme iron centers (11). Center I contains a distorted rubredoxin-type [Fe(SCys)4] coordination sphere and is presumably an electron transfer center. Center II contains a unique [Fe(NHis)4(SCys)] site that is rapidly oxidized by O2− and is, therefore, the likely site of superoxide reduction (26). The only iron site in a blue nonheme iron protein, neelaredoxin (Nlr) from Desulfovibrio gigas (10), closely resembles that of Rbo center II (11).
A second type of nonheme iron protein, rubrerythrin (Rbr), has also been implicated in oxidative stress defense in anaerobic microbes (1, 2, 24). Rbr is a homodimeric protein that contains both a rubredoxin-like [Fe(SCys)4] center and a nonsulfur, oxo-bridged diiron site (14). The best-characterized Rbr is that from the anaerobic sulfate-reducing bacterium D. vulgaris (strain Hildenborough). In vitro, D. vulgaris Rbr is able to function as the terminal component of an NADH peroxidase, catalyzing the reduction of hydrogen peroxide to water (reaction 4) (12, 13):
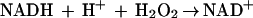 |
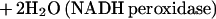 |
4 |
D. vulgaris contains a second Rbr-like protein, called nigerythrin (Ngr) (27, 32), which also exhibits NADH peroxidase activity (13).
In the present work we present evidence for the roles of Rbo and Rbr in the protection of D. vulgaris against exposure to air, superoxide, and hydrogen peroxide. We also describe a D. vulgaris gene with high sequence homology to bacterial Fe-SODs. A joint role for Rbo and Rbr as a novel oxidative stress protection system in anaerobic microorganisms is proposed.
MATERIALS AND METHODS
Molecular biology reagents and procedures.
Molecular biology procedures not explicitly described below followed those in references 3 or 36. Restriction enzymes were from either Boehringer Mannheim (Indianapolis, Ind.) or Promega, Inc. (Madison, Wis.).
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this work are listed in Table 1.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Genotype or relevant characteristic | Source or reference |
|---|---|---|
| Strains | ||
| D. vulgaris | ||
| Hildenborough | Wild type, Kmr | 27 |
| L2 | Δrbo Cmr Kmr | 45 |
| E. coli | ||
| DH5α | endA1 hsdR17(rK− mK+) supE44 thi-1 recA1 gyrA (Nalr) relA1 Δ(lacIZYA argF)U169 deoR [φ80dlac(lacZ)M15] | Laboratory stock |
| QC774 | GC4468 Φ(sodA-lacZ)49 Φ(sodB-kan)1-Δ2 Cmr Kmr | 9 |
| NC202 | ΔlacU169 rpsL Φ(katE::lacZ+)131 katG17::Tn10 | 39 |
| Plasmids | ||
| pCYB1 | Cloning/overexpression vector, Apr | New England BioLabs, Inc. (Beverly, Mass.) |
| pUC18 | Cloning vector, Apr | Laboratory stock |
| pT7-7 | Overexpression vector, Apr | 40 |
| pDK3-5 | pT7-7 derivative with D. vulgaris rbr gene in the NdeI/BamHI sites | 17 |
| pNS1 | pT7-7 derivative with D. vulgaris ngr gene in the NdeI/BamHI sites | 27 |
| pNLR119 | pUC18 derivative with a 2.2-kbp EcoRI/HindIII fragment of D. gigas chromosomal DNA containing the nlr gene | This work |
| pRbr | pCYB1 derivative with D. vulgaris rbr gene in the NdeI/BamHI sites | This work |
| pRbo | pCYB1 derivative with D. vulgaris rbo gene in the NdeI/SapI sites | This work |
| pNgr | pCYB1 derivative with D. vulgaris ngr gene in the NdeI/SapI sites | This work |
| pNlr | pCYB1 derivative with D. gigas nlr gene in the NdeI/HindIII restriction sites | This work |
Media and growth conditions.
D. vulgaris (Hildenborough) strains were maintained in liquid culture in Postgate's anaerobic medium C (33), supplemented with kanamycin (50 μg/ml) and chloramphenicol (10 μg/ml) when appropriate. Agar plates were poured aerobically using Postgate's medium E (33). After 24 h, the plates were transferred to a Coy anaerobic chamber (Coy Laboratory Products, Inc., Grass Lake, Mich.) containing an atmosphere of 5% hydrogen, 10% carbon dioxide, and 85% nitrogen and were allowed to equilibrate for 4 days prior to use. E. coli strains were grown aerobically at 37°C in Luria-Bertani (LB), M9, or M63 minimal medium containing ampicillin (100 μg per ml) when needed for plasmid maintenance. Where noted, M63 minimal medium was supplemented with 0.1% Casamino Acids and 0.5 mg of vitamin B1 per liter.
Cell extracts.
To prepare periplasmic and cytoplasmic fractions of D. vulgaris cells, 1-liter anaerobic cultures were grown overnight at 37°C to early stationary phase. All further workup was done under an aerobic atmosphere. Periplasmic and cytoplasmic fractions were prepared essentially as described previously (32, 42). The cells were harvested by centrifugation for 20 min at 4,000 × g and washed once with 50 mM Tris-HCl (pH 8.0). The cells were resuspended in 10 ml of 50 mM Tris-HCl–50 mM EDTA (pH 9.0) and were incubated on ice, with occasional stirring, for 45 min. After centrifugation, the red-colored supernatant, designated the periplasmic fraction, was removed. Cytochrome c3, a periplasmic protein, was measured by absorbance at 410 nm for oxidized samples and at 419 nm for dithionite reduced samples. The pellet was resuspended in 10 ml of 50 mM Tris-HCl–50 mM EDTA (pH 9.0) and was lysed to release the cytoplasmic fraction by three passes through a French pressure minicell at 18,000 psi. Desulfoviridin, a cytoplasmic protein, was detected in cytochrome fractions spectrophotometrically, as determined by A628 − (A608 + A648)/2 (21). The periplasmic and cytoplasmic fractions were dialyzed against 50 mM phosphate buffer (pH 7.8). Protein quantification was performed by the Bio-Rad DC protein assay using bovine serum albumin standards.
Superoxide dismutase activity.
A standard assay that relies on the inhibition of cytochrome c reduction by superoxide generated from xanthine/xanthine oxidase was used (6). Aliquots (∼100 μl) of cytoplasmic and periplasmic fractions were added to achieve ca. 50% inhibition of cytochrome c under standard assay conditions.
D. vulgaris oxidative stress assays.
For exposure to O2 and superoxide, 100-ml cultures of D. vulgaris wild type and strain L2 were grown in anaerobic medium C at 37°C until mid-log phase (optical density at 600 nm [OD600], ∼0.6). Forty-five-milliliter aliquots of the anaerobic cultures were transferred to sterile 250-ml flasks and continuously stirred with a magnetic stirrer in air at room temperature. For exposure to superoxide, paraquat (methyl viologen) (PQ) was added to a concentration of 10 μM. Aliquots (0.1 to 1 ml) of these air-exposed cultures were removed hourly, transferred to the anaerobic chamber, diluted (100 to 106) into anaerobic medium C, and immediately plated onto medium E plates in triplicate. After incubating the cultures for 5 days at 37°C, colonies were counted and the number of surviving CFU/ml was determined. As noted previously (45), the oxidatively stressed D. vulgaris cells produced variable sized colonies, making them difficult to enumerate. For the experiments reported in the present work, all colonies (large and small) were enumerated after 5 days of anaerobic incubation, and each reported CFU/ml number is the average from three plates.
To test for hydrogen peroxide sensitivity, anaerobic mid-log-phase cultures were spread onto medium E plates. Hydrogen peroxide (2.5 or 5.0 μmol in 10 μl of final volume) was spotted onto circular glass-fiber membranes placed on the cell lawns. The plates were incubated anaerobically for 3 days, and the diameters of growth inhibition surrounding the filters were measured to compare sensitivities.
Cloning of rbo, rbr, ngr, and nlr.
The D. vulgaris genes encoding Rbr, Ngr, and Rbo and the D. gigas gene encoding Nlr were amplified by PCR and cloned into the overexpression vector pCYB1. rbr and ngr were amplified from template plasmids pDK3-5 and pNS1, respectively. rbo was amplified from D. vulgaris chromosomal DNA. D. gigas nlr was cloned from D. gigas chromosomal DNA using a procedure similar to that described by Silva et al. (38), and pNLR119 (Table 1) was used as the template. Both D. vulgaris and D. gigas chromosomal DNAs had been isolated as previously described (27). Sequences of the oligonucleotide primers duplicated the N- and complementary C-terminal 14 bases of each gene using sequences from GenBank accession numbers M28848 (rbo), U82323 (rbr), U71215 (ngr), and AF034965 (nlr). An NdeI restriction site was incorporated into the start codon of each N-terminal primer. A stop codon (TGA) and an appropriate restriction site, either a SapI (rbo, ngr), BamHI (rbr), or HindIII (nlr) site, were added to the 3′ end of each C-terminal primer. All PCRs were carried out in 50-μl volumes in 1× PCR buffer (Boehringer Mannheim) containing 1.5 mM MgCl2, 200 mM deoxynucleoside triphosphate, 0.05 mM primers, and 2.5 U of Taq DNA polymerase (Promega). The amplification procedure consisted of a hot start of the template DNA and primers at 94°C for 10 min, followed by 30 cycles of 94°C for 1 min, 60°C for 1 min, and after addition of the remaining reaction components, 72°C for 2 min. The purified PCR products were digested with the appropriate restriction enzymes and ligated into corresponding restriction sites of pCYB1. Nucleotide sequences of the genes in the recombinant plasmids were verified by DNA sequencing, which was carried out in the Molecular Genetics Instrumentation Facility at the University of Georgia.
E. coli H2O2 survival assay.
All steps were carried out aerobically. Cultures of E. coli strain NC202 (katE katG) harboring the pCYB1-based vectors listed in Table 1 were grown at 37°C to an OD600 of 1.0 in LB-ampicillin medium. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to 2 mM and the culture was allowed to grow to an OD600 of 2.2. Aliquots of the cultures were diluted into 50 ml of LB medium in a 250-ml flask without antibiotics to an initial OD600 of 0.2. Hydrogen peroxide from a 30% aqueous stock was added to achieve a concentration of 2.5 mM H2O2. Aliquots of the cultures were removed immediately before and 30 min after the H2O2 challenge. These aliquots were diluted in LB medium containing catalase (100 μg/ml) and plated in triplicate on LB agar plates containing catalase (100 μg/ml). Colonies were enumerated after 3 or 4 days at 37°C.
SOD complementation assay.
E. coli QC774 (sodA sodB) cultures harboring one of the pCYB1-derived expression vectors were grown aerobically to saturation in M63-ampicillin medium supplemented with amino acids and vitamin B1 and were diluted to an initial OD600 of ca. 0.06 in aerobic M63-ampicillin medium without supplements. The cultures were incubated with shaking at 37°C, and the growth rate was monitored by measuring the OD600.
Cloning of a D. vulgaris SOD gene.
A conserved amino-acid-sequence region found in several bacterial Fe-SODs, DVWEHAYYLDYQN, was used to design a degenerate oligonucleotide probe with the sequence 5′-GAYGTNTGGGARCAYGCNTAYTAYCTNGAYTAYCARAAY-3′. The 3′ end of the probe was labeled with digoxigenin-ddUTP and then used in Southern blotting of D. vulgaris (Hildenborough) chromosomal DNA, both according to the protocol for the Genius system (Boehringer Mannheim). The probe hybridized to a 4.5-kbp EcoRI/HindIII restriction fragment of D. vulgaris chromosomal DNA. Therefore, EcoRI/HindIII restricted DNA fragments in the 4- to 5-kb range were isolated from an 0.8% low melting agarose gel by electroelution and were ligated into pUC18. The ligation mixture was transformed into E. coli DH5α and plated onto M9-ampicillin agar plates containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) at 2 mg/ml. White colonies were selected to inoculate 1.5-ml cultures grown in LB-ampicillin. Plasmids were purified by alkaline lysis from six combined cultures and were screened for the presence of the sod gene by Southern hybridization. Screening of 100 plasmid pools resulted in the isolation of a plasmid containing a 4.5-kbp EcoRI/HindIII fragment containing the D. vulgaris sod. Sequencing of the cloned gene was performed at the Molecular Genetics Instrumentation Facility at the University of Georgia.
Nucleotide sequence accession number.
The newly determined sequence was deposited in the GenBank database under accession no. AF034841.
RESULTS
Rbo protects D. vulgaris from exposure to superoxide but not to hydrogen peroxide.
Survival of the D. vulgaris Δrbo strain, L2, was previously shown to be lower than that of the wild type when anaerobically growing cells were diluted into aerobic medium and then exposed to air for several hours (45). PQ can be used to generate a steady-state level of superoxide within bacterial cells under aerobic conditions. We therefore compared the survival of D. vulgaris wild type with that of strain L2 after exposure to air + 10 μM PQ ([air + PQ]). The viability of log-phase D. vulgaris cells, as surviving CFU/ml exposed to [air + PQ] over a time course of 4 h, is shown in Fig. 1A, normalized to the air-only surviving CFU/ml. The unnormalized data are plotted in Fig. 1B. For cells of both the wild type and strain L2 that were exposed to air only, we observed a steeper drop-off in cell survival versus time than observed previously (45). This phenomenon can be attributed to the larger surface area for aeration used in our experiments. As can be seen in Fig. 1B, at 4 h exposure to air only, the surviving CFU/ml for strain L2 (1.03 ± 0.04 × 108) was greater than that of wild type (9 ± 4 × 106). This artifact could be due to the greater initial viable cell density we used for strain L2 than for wild type, thereby affording greater protection against oxidative stress at the earlier exposure times. By 8 h of air-only exposure, the surviving CFU/ml of strain L2 (9 ± 1 × 104) had decreased below that of wild type (2.1 ± 0.1 × 105), consistent with the greater air sensitivity of strain L2 previously observed (45). Strain L2 continued to exhibit lower cell survival at 12 h of air-only exposure (data not shown). By 24 h of air-only exposure neither the wild type nor strain L2 showed any survival under our conditions.
FIG. 1.
Viabilities of D. vulgaris wild type and L2 strains following exposure to either air or air plus 10 μM PQ. (A) Surviving CFU versus the time of the [air+PQ]-exposed cells normalized to the surviving CFU of the air-only exposed cells. (B) The actual CFU/ml versus the time for both air only- and [air + PQ]-exposed cells.
The novel observation in the present work occurred upon the addition of 10 μM PQ to the air-exposed D. vulgaris cells. Both the wild type and strain L2 lost viability faster when exposed to [air + PQ] than to air only but the effect of PQ was much greater on strain L2 (Fig. 1A). After 4 h the survival of wild-type cells exposed to [air + PQ] had decreased by a factor of ∼2 relative to those exposed to air only (9 ± 4 × 105 versus 4 ± 2 × 106), whereas the survival of the L2 [air + PQ]-exposed cells decreased by a factor of ∼500 (1.03 ± 0.04 × 108 versus 1.9 ± 0.2 × 105) relative to the cells exposed to air only. The much greater [air + PQ] versus air-only sensitivity of strain L2 compared to wild type was verified in two other experiments (data not shown). The addition of up to 100 μM PQ did not affect the anaerobic growth (measured as OD600) of either D. vulgaris wild type or L2 cultures (data not shown).
Anaerobic peroxide sensitivity was tested by growth inhibition on agar plates containing lawns of either wild type or L2 and filter disks saturated with hydrogen peroxide solutions. We found no significant difference in the zones of growth inhibition between wild type and L2 using two different amounts of hydrogen peroxide (Table 2).
TABLE 2.
H2O2 sensitivities and SOD activities of D. vulgaris (Hildenborough) strains
| D. vulgaris strain | H2O2 growth inhibition diam (mm) with:
|
SOD activities (U/mg of protein)a
|
||
|---|---|---|---|---|
| 2.5 μmol H2O2 | 5.0 μmol H2O2 | Periplasm | Cytoplasm | |
| Wild type | 3.00 ± 0.10 | 3.50 ± 0.10 | 5.1 ± 0.1 | 1.1 ± 0.1 |
| L2 (Δrbo) | 2.90 ± 0.10 | 3.70 ± 0.10 | 5.4 ± 0.1 | 1.3 ± 0.1 |
Subcellular fractions prepared as described in Materials and Methods.
SOD activity of D. vulgaris is not affected by the absence of Rbo and is concentrated in the periplasmic fraction.
A low level of SOD activity had previously been reported in D. vulgaris cell extracts (18). In order to localize and quantitate this activity we tested periplasmic and cytoplasmic fractions of D. vulgaris wild type and strain L2 for SOD activity using a standard assay (Table 2). The fidelity of periplasmic and cytoplasmic preparations were checked by comparing cytochrome c3 (a periplasmic protein) with desulfoviridin (a cytoplasmic protein) levels using a previously described spectrophotometric method (32, 42). No desulfoviridin could be detected in the periplasmic fractions, while more than 70% of the total cytochrome c3 detected was seen in the periplasmic fractions (data not shown). The specific SOD activities (per milligram of total protein) of these subcellular fractions were unaffected by the absence of the rbo gene. For both strains, the specific SOD activity was approximately five times greater in the periplasmic fractions than in the cytoplasmic fractions (Table 2), consistent with the presence of a periplasmic SOD in D. vulgaris.
D. vulgaris has an Fe or Mn SOD-like gene.
Using a degenerate oligonucleotide probe that was designed to encode a conserved region of Fe-SOD sequences, we cloned a D. vulgaris gene encoding a protein with 50% sequence identity (59% similarity) to that of E. coli Fe-SOD, when aligned as shown in Fig. 2. The N terminus for the D. vulgaris SOD was taken as the first methionine that codes for a continuous open reading frame and has a recognizable ribosome binding site (AGGAG) 6 bp upstream. Analysis of the amino acid sequence using SignalP (30) suggested the existence of a 34-residue export leader peptide with the most likely cleavage site between residues A34 and A35. As shown by the sequence alignments in Fig. 3, the putative leader peptide of the D. vulgaris SOD resembles those that are commonly used for export of bacterial redox proteins (5) with the notable exception that one of the two residues of a conserved arginine pair is replaced by a serine. We have independently confirmed this replacement by examination of the preliminary genome sequence for D. vulgaris (Hildenborough) (The Institute for Genomic Research, personal communication). The implication is that the D. vulgaris Fe- or Mn-SOD is periplasmic, in contrast to the Fe-SOD from E. coli, which is known to be cytoplasmic. Our database searches have not located a complete amino acid or nucleotide sequence of a recognizable SOD from any other sulfate-reducing bacterium. None of our attempts to express this D. vulgaris SOD gene in E. coli, including QC774, have resulted in an active SOD.
FIG. 2.
Amino acid sequence alignment of the putative sod gene product from D. vulgaris with the Fe-containing SOD from E. coli (GenBank accession no. J03511.1). The putative leader sequence cleavage site in the D. vulgaris SOD sequence is shown in italics between residues A34 and A35. Asterisks (∗) indicate iron ligand residues in E. coli SOD (22). +, amino acids are similar to each other.
FIG. 3.
Sequence alignment of export signal peptides from selected bacterial and archaeal redox proteins with the putative signal peptide of the D. vulgaris (Hildenborough) SOD (DvH_SOD). Alignment of the sequences follows that in reference 5. Sequence sources (GenBank accession numbers) are as follows: Db_HysB, Desulfomicrobium baculatum NiFeSe hydrogenase (P13063); Ws_HydA, Wolinella succinogenes NiFe hydrogenase (S33852); Sa_SoxF, Sulfolobus acidocaldarius Rieske iron-sulfur protein (S56156); Ec_HybA, E. coli hydrogenase (P37179); Ec_NrfC, E. coli putative iron-sulfur protein (P32708); Hi_NrfC, Haemophilus influenzae putative iron-sulfur protein (P45015); Sm_NosZ Sinorhizobium meliloti nitrous oxide reductase (Q59746); and Ps_NosZ, Pseudomonas stutzeri nitrous oxide reductase (P19573).
Rbo but not Rbr or Nlr can complement E. coli sodA sodB.
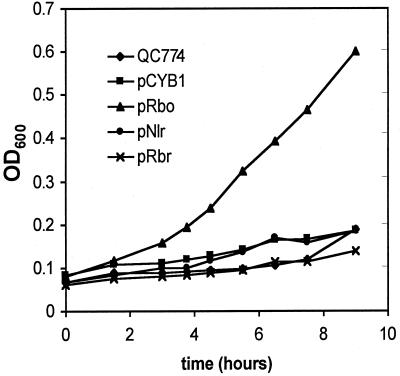
E. coli strain QC774 is deficient in both sodA and sodB encoding the Mn and Fe SODs, respectively (9). Under aerobic conditions this strain is unable to grow in minimal medium lacking amino acid supplements. This phenotype is attributed to superoxide damage to iron-sulfur enzymes in the biosynthetic pathway of branched chain amino acids and to Fenton chemistry resulting from increased free iron levels (41). Figure 4 shows that plasmid-borne expression of D. vulgaris Rbo is able to restore aerobic growth to this strain whereas expression of D. vulgaris Rbr and Ngr and D. gigas Nlr are not. Strain QC774 will grow aerobically in rich medium (LB) but unlike the wild type, this growth is inhibited by 10 μM PQ. Rbo expression completely restored growth to 10 μM PQ-stressed QC774 in LB, whereas once again expression of Rbr, Ngr, or Nlr did not (data not shown). Preinduction of protein overexpression by the addition of IPTG to 0.4 mM did not change these outcomes. The plasmid-expressed Rbr, Rbo, and Nlr have all been isolated from QC774 (E. D. Coulter, N. V. Shenvi, and D. M. Kurtz, Jr., unpublished data), demonstrating that these proteins can be expressed in this strain.
FIG. 4.
Aerobic growth versus the time at 37°C in M63 medium of E. coli QC774 (sodA sodB) containing pCYB1-based plasmids expressing D. vulgaris Rbo and Rbr and D. gigas Nlr.
D. vulgaris Rbr and Ngr but not Rbo are able to rescue E. coli kat from H2O2 stress.
E. coli strain NC202 lacks both katG and katE genes encoding catalase/peroxidase (HPI) and catalase (HPII), respectively (39). Plasmid-borne expression of D. vulgaris Rbr or Ngr increased the survival of this strain when exposed to H2O2 under aerobic conditions, as shown in Fig. 5. H2O2-exposed NC202 cells expressing Rbr showed at least 25% higher viability than those containing the parent plasmid, and cells expressing Ngr were fully protected against H2O2 exposure. Expression of Rbo failed to provide any protection to the H2O2-exposed NC202 strain and in fact reduced its viability to a few percent upon peroxide stress. Overexpression of these heterologous proteins in NC202 was verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown).
FIG. 5.
Viability of aerobically grown E. coli NC202 (katG katE) expressing plasmid-borne D. vulgaris Rbr, Ngr, or Rbo genes following a 30-min exposure to 2.5 mM H2O2. IPTG (0.4 mM) was added to induce overexpression of the genes prior to H2O2 exposure.
DISCUSSION
The results reported here indicate that D. vulgaris Rbo functions as an intracellular superoxide scavenger but not as an SOD, that D. vulgaris Rbr and Ngr protect against hydrogen peroxide stress in vivo, and that D. vulgaris contains a periplasmic Fe- (or possibly Mn-)SOD.
Sequence analysis of the D. vulgaris Fe-SOD gene indicated the presence of an export leader peptide commonly found in periplasmic redox proteins. The replacement of one of the two residues of a highly conserved arginine pair by a serine is unusual if not unique for redox protein leader peptides, including those previously reported in sulfate-reducing bacteria (44). Consistent with the implied periplasmic localization, we found that the majority of the SOD activity in D. vulgaris is associated with the periplasmic fraction of cell extracts, even in the rbo-deleted strain. We have found no data addressing the localizations of SODs in other sulfate-reducing bacteria. A few other periplasmic Fe/Mn-SODs have been reported, but only from aerobes (4, 16, 37).
A D. vulgaris Δrbo strain was previously found to be more air sensitive than wild type (45) but our results show that this air sensitivity is greatly magnified by PQ, a known intracellular superoxide generator. Presumably, the reduction in viability of the Δrbo strain exposed to air only is also due to intracellular generation of superoxide, but at a much slower rate than in the [air + PQ]-exposed cells. The D. vulgaris Δrbo strain showed no significant difference in the SOD activity of cellular extracts relative to wild type, indicating that Rbo is not an SOD. We therefore conclude that Rbo protects D. vulgaris against internal superoxide stress by functioning as an SOR (reaction 3). The reduction in viability of the aerobically grown E. coli catalase-deficient strain upon overexpression of Rbo can be explained by Rbo-catalyzed production of excess hydrogen peroxide from endogenously generated superoxide. A BLAST search of the preliminary D. vulgaris genome sequence (The Institute for Genomic Research, personal communication) revealed no other Rbo or Fe-SOD homologs. All available evidence indicates that Rbos are cytoplasmic proteins whereas our results strongly suggest that the D. vulgaris SOD is periplasmic. A proposed model for compartmentalized superoxide generation and scavenging in D. vulgaris is presented in Fig. 6.
FIG. 6.
Proposed model for an Rbo/Rbr oxidative stress protection system in D. vulgaris.
We found that neither D. vulgaris Rbr nor Ngr is capable of restoring growth to the E. coli sodA sodB strain in aerobic minimal medium. Rbr and Ngr are, however, capable of increasing the survival of an E. coli catalase-deficient strain that was exposed to hydrogen peroxide. This protection by Rbr and Ngr apparently occurs in competition with at least some of the 30 or more other proteins that are induced in response to peroxide stress in E. coli (34). As is the case for Rbo, the protection against hydrogen peroxide by Rbr and Ngr presumably requires an endogenous electron source, perhaps functioning analogously to the in vitro NADH peroxidase activity (reaction 4) (12, 13). Rbo and Rbr (and Ngr) may thus be complementary components of an alternative oxidative stress defense in D. vulgaris, as shown in Fig. 6.
Heme-containing catalases have been isolated from D. vulgaris (Hildenborough) and D. gigas (15, 18), and a catalase gene has previously been reported from D. vulgaris (Miyazaki F) (GenBank accession no. AB020341). Neither the Miyazaki nor the Hildenborough catalase genes (The Institute for Genomic Research, personal communication) contain a recognizable signal peptide sequence, which indicates that D. vulgaris catalase is cytoplasmic, as is Rbr (32). Since heme catalases typically have Kms for H2O2 in the millimolar to molar range (46), the D. vulgaris catalase might complement the intracellular Rbr-catalyzed hydrogen peroxide removal by diminishing high concentrations of extracellular hydrogen peroxide, which is freely diffusible across cell membranes (Fig. 6).
Endogenous electron donors to Rbo or Rbr have not been identified either when complementing the E. coli strains or in the native organism. The inability of plasmid-expressed D. gigas Nlr to restore aerobic growth to E. coli sodA sodB implies that center I {[Fe(SCys)4]}, which is presumably the electron transfer center of Rbo (and not present in Nlr), as well as center II, is required for complementation by Rbo. In D. vulgaris, one potential redox partner is rubredoxin, whose gene, rub, is cotranscribed with rbo (7). A rub gene is also adjacent to the rbo gene in D. baarsii (Fig. 7) (31). Based on genetic proximity, Rbr could also use rubredoxin-like proteins as redox partners. In D. vulgaris (Hildenborough), Rbr is cotranscribed with Rdl, yet another class of rubredoxin-type protein (23, 27).
FIG. 7.
Diagrams of open reading frames encoding adjacent rbo, rbr, and rub homologs from anaerobic bacteria and archaea. rub, rubredoxin gene; rbo, Rbo homolog gene; nlr, neelaredoxin homolog gene; rdl, rubredoxin-like protein gene; fur, Fur homolog gene. Horizontal distances between boxes are proportional to the relative spacings of the genes. Arrow-shaped orientations of boxes show putative transcription directions. Open reading frames were derived from sequences deposited in GenBank with the following accession numbers: X99543 (D. baarsii), AE001047 (Archaeoglobus fulgidus), AF156097 (P. furiosus), AJ248285 (Pyrococcus abysii), AE001739 (Thermotoga maritima), AF202316 (Moorella thermoacetica), U67520 (Methanococcus jannaschii), AE000854 (Methanobacterium thermoautotrophicum), M28848 [Desulfovibrio vulgaris (Hildenborough) rbo-rub] and U82323 [D. vulgaris (Hildenborough) fur-rbr-rdl].
The rbr gene in D. vulgaris (Hildenborough) is transcribed with a third gene designated fur (27), which encodes a protein showing highest sequence homology to PerR, a peroxide-responsive regulatory protein in Bacillus subtilis (8, 47). The product of the fur-like gene may, therefore, regulate the Rbo-Rbr oxidative stress response in D. vulgaris. The genes encoding Rbo and Rbr (and Ngr) occur in separate operons in D. vulgaris (7, 27). However, as diagrammed in Fig. 7, in other anaerobic microorganisms Rbo- and Rbr-like genes are located either in succession or within what appears to be the same operon.
To date, Rbo and Rbr (or their genes) have been reported only in anaerobic or microaerophilic bacteria and archaea, whereas we have found no Rbo or Nlr or Rbr homologs in sequenced genomes of aerobes (as of June 2000). We therefore propose that Rbo-Rbr constitutes an alternative oxidative stress protection system in anaerobes and microaerophiles operating similarly to that diagrammed in Fig. 6. Some aspects of our proposal resemble those recently suggested for oxidative stress protection in the anaerobic hyperthermophilic archaeon Pyrococcus furiosus (19). P. furiosus contains an Nlr-like protein with superoxide reductase activity as well as an Rbr, the genes for which are tandemly located (Fig. 7). The microorganismic segregation of SOD-catalase between aerobes and anaerobes appears to be less distinct than for Rbo-Rbr (20). The latter segregation suggests that the Rbo-Rbr oxidative stress protection system is well suited to anaerobic life in an aerobic world.
ACKNOWLEDGMENTS
This research was supported by NIH grant GM40388 to D.M.K. and a grant from the Natural Sciences and Engineering Research Council (NSERC) to G.V.
Sequence data for the D. vulgaris (Hildenborough) genome was obtained from The Institute for Genomic Research website at http://www.tigr.org. Sequencing of this genome was accomplished with support from the U.S. Department of Energy.
REFERENCES
- 1.Alban P S, Krieg N R. A hydrogen peroxide resistant mutant of Spirillum volutans has NADH peroxidase activity but no increased oxygen tolerance. Can J Microbiol. 1998;44:87–91. [Google Scholar]
- 2.Alban P S, Popham D L, Rippere K E, Krieg N R. Identification of a gene for a rubrerythrin/nigerythrin-like protein from Sprillum volutans by using amino acid sequence data from mass spectrometry and NH2-terminal sequencing. J Appl Microbiol. 1998;85:875–882. doi: 10.1046/j.1365-2672.1998.00602.x. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F A, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Green Publishing and Wiley-Interscience; 1990. [Google Scholar]
- 4.Barnes A C, Balebona M C, Horne M T, Ellis A E. Superoxide dismutase and catalase in Photobacterium damselae subsp. piscicida and their roles in resistance to reactive oxygen species. Microbiology. 1999;145:483–494. doi: 10.1099/13500872-145-2-483. [DOI] [PubMed] [Google Scholar]
- 5.Berks B C. A common export pathway for proteins binding complex redox cofactors? Mol Microbiol. 1996;22:393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- 6.Beyer W F, Fridovich I. Assaying for superoxide dismutase activity: some large consequences of minor changes in conditions. Anal Biochem. 1987;161:559–561. doi: 10.1016/0003-2697(87)90489-1. [DOI] [PubMed] [Google Scholar]
- 7.Brumlik M J, Voordouw G. Analysis of the transcriptional unit encoding the genes for rubredoxin (rub) and a putative rubredoxin oxidoreductase (rbo) in Desulfovibrio vulgaris (Hildenborough) J Bacteriol. 1989;171:4996–5004. doi: 10.1128/jb.171.9.4996-5004.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bsat N, Herbig A, Casillas-Martinez L, Setlow P, Helmann J D. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol Microbiol. 1998;29:189–198. doi: 10.1046/j.1365-2958.1998.00921.x. [DOI] [PubMed] [Google Scholar]
- 9.Carlioz A, Touati D. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J. 1986;5:623–630. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L, Sharma P, Le Gall J, Mariano A M, Teixeira M, Xavier A. A blue non-heme iron protein from Desulfovibrio gigas. Eur J Biochem. 1994;226:613–618. doi: 10.1111/j.1432-1033.1994.tb20087.x. [DOI] [PubMed] [Google Scholar]
- 11.Coehlo A V, Matias P, Fülop V, Thomson A, Gonzalez A, Carrondo M A. Desulfoferrodoxin structure determined by MAD phasing and refinement to 1.9-Å reveals a unique combination of a tetrahedral FeS4 centre with a square pyramidal FeSN4 centre. J Inorg Biochem. 1997;2:680–689. [Google Scholar]
- 12.Coulter E D, Shenvi N V, Beharry Z, Smith J J, Prickril B C, Kurtz D M., Jr Rubrerythrin-catalyzed substrate oxidation by dioxygen and hydrogen peroxide. Inorg Chim Acta. 2000;297:231–234. [Google Scholar]
- 13.Coulter E D, Shenvi N V, Kurtz D M., Jr NADH peroxidase activity of rubrerythrin. Biochem Biophys Res Commun. 1999;255:317–323. doi: 10.1006/bbrc.1999.0197. [DOI] [PubMed] [Google Scholar]
- 14.deMaré F, Kurtz D M, Jr, Nordlund P. The structure of Desulfovibrio vulgaris rubrerythrin reveals a unique combination of rubredoxin-like FeS4 and ferritin-like diiron domains. Nat Struct Biol. 1996;3:539–546. doi: 10.1038/nsb0696-539. [DOI] [PubMed] [Google Scholar]
- 15.Dos Santos W G, Pacheco I, Liu M Y, Teixeira M, Xavier A V, LeGall J. Purification and characterization of an iron superoxide dismutase and a catalase from the sulfate-reducing bacterium Desulfovibrio gigas. J Bacteriol. 2000;182:796–804. doi: 10.1128/jb.182.3.796-804.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geissdorfer W, Ratajczak A, Hillen W. Nucleotide sequence of a putative periplasmic Mn superoxide dismutase from Acinetobacter calcoaceticus ADP1. Gene. 1997;186:305–308. doi: 10.1016/s0378-1119(96)00728-7. [DOI] [PubMed] [Google Scholar]
- 17.Gupta N, Bonomi F, Kurtz D M, Jr, Ravi N, Wang D L, Huynh B H. Recombinant Desulfovibrio vulgaris rubrerythrin. Isolation and characterization of the diiron domain. Biochemistry. 1995;34:3310–3318. doi: 10.1021/bi00010a021. [DOI] [PubMed] [Google Scholar]
- 18.Hatchikian C E, LeGall J, Bell G R. Significance of superoxide dismutase and catalase activities in the strict anaerobes, sulfate-reducing bacteria. In: Michael A M, McCord J M, Fridovich I, editors. Superoxide and superoxide dismutase. New York, N.Y: Academic Press; 1977. pp. 159–172. [Google Scholar]
- 19.Jenney F E, Jr, Verhagen M J M, Cui X, Adams M W W. Anaerobic microbes: oxygen detoxification without superoxide dismutase. Science. 1999;286:306–309. doi: 10.1126/science.286.5438.306. [DOI] [PubMed] [Google Scholar]
- 20.Kirschvink J L, Gaidos E J, Bertani L E, Beukes N J, Gutzmer J, Maepa L N, Steinberger R E. Paleoproterozoic snowball earth: extreme climatic and geochemical global change and its biological consequences. Proc Natl Acad Sci USA. 2000;97:1400–1405. doi: 10.1073/pnas.97.4.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobayashi K, Takahashi E, Ishimoto M. Biochemical studies on sulfate-reducing bacteria: purification and some properties of sulfite reductase, desulfoviridin. J Biochem (Tokyo) 1972;72:879–887. doi: 10.1093/oxfordjournals.jbchem.a129982. [DOI] [PubMed] [Google Scholar]
- 22.Lah M S, Dixon M M, Pattridge K A, Stallings W C, Fee J A, Ludwig M L. Structure-function in Escherichia coli iron superoxide dismutase: comparisons with the manganese enzyme from Thermus thermophilus. Biochemistry. 1995;34:1646–1660. doi: 10.1021/bi00005a021. [DOI] [PubMed] [Google Scholar]
- 23.LeGall J, Liu M Y, Gomes C M, Braga V, Pacheco I, Regalla M, Xavier A V, Teixeira M. Characterisation of a new rubredoxin isolated from Desulfovibrio desulfuricans 27774: definition of a new family of rubredoxins. FEBS Lett. 1998;429:295–298. doi: 10.1016/s0014-5793(98)00610-3. [DOI] [PubMed] [Google Scholar]
- 24.Lehmann Y, Meile L, Teuber M. Rubrerythrin from Clostridium perfringens: cloning of the gene, purification of the protein, and characterization of its superoxide dismutase function. J Bacteriol. 1996;178:7152–7158. doi: 10.1128/jb.178.24.7152-7158.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liochev S I, Fridovich I. A mechanism for complementation of the sodA sodB defect in Escherichia coli by overproduction of the rbo gene product (desulfoferrodoxin) from Desulfoarculus baarsii. J Biol Chem. 1997;272:25573–25575. doi: 10.1074/jbc.272.41.25573. [DOI] [PubMed] [Google Scholar]
- 26.Lombard M, Fontecave M, Touati D, Niviere V. Reaction of the desulfoferrodoxin from Desulfoarculus baarsii with superoxide anion. Evidence for a superoxide reductase activity. J Biol Chem. 2000;275:115–121. doi: 10.1074/jbc.275.1.115. [DOI] [PubMed] [Google Scholar]
- 27.Lumppio H L, Shenvi N V, Garg R P, Summers A O, Kurtz D M., Jr A rubrerythrin operon and nigerythrin gene in Desulfovibrio vulgaris (Hildenborough) J Bacteriol. 1997;179:4607–4615. doi: 10.1128/jb.179.14.4607-4615.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marschall C, Frenzel P, Cypionka H. Influence of oxygen on sulfate reduction and growth of sulfate-reducing bacteria. Arch Microbiol. 1993;159:168–173. [Google Scholar]
- 29.Moura I, Tavares P, Moura J J, Ravi N, Huynh B H, Liu M Y, LeGall J. Purification and characterization of desulfoferrodoxin. A novel protein from Desulfovibrio desulfuricans (ATCC 27774) and from Desulfovibrio vulgaris (strain Hildenborough) that contains a distorted rubredoxin center and a mononuclear ferrous center. J Biol Chem. 1990;265:21596–21602. [PubMed] [Google Scholar]
- 30.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 31.Pianzzola M J, Soubes M, Touati D. Overproduction of the rbo gene product from Desulfovibrio species suppresses all deleterious effects of lack of superoxide dismutase in Escherichia coli. J Bacteriol. 1996;178:6736–6742. doi: 10.1128/jb.178.23.6736-6742.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pierik A J, Wolbert R B G, Portier G L, Verhagen M F J M, Hagen W R. Nigerythrin and rubrerythrin from Desulfovibrio vulgaris each contain two mononuclear iron centers and two dinuclear iron clusters. Eur J Biochem. 1993;212:237–245. doi: 10.1111/j.1432-1033.1993.tb17655.x. [DOI] [PubMed] [Google Scholar]
- 33.Postgate J C. Recent advances in the study of the sulfate-reducing bacteria. Bacteriol Rev. 1965;29:425–441. doi: 10.1128/br.29.4.425-441.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rocha E R, Smith C J. Characterization of a peroxide-resistant mutant of the anaerobic bacterium Bacteroides fragilis. J Bacteriol. 1998;180:5906–5912. doi: 10.1128/jb.180.22.5906-5912.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Romao C V, Liu M Y, Le Gall J, Gomes C M, Braga V, Pacheco I, Xavier A V, Teixeira M. The superoxide dismutase activity of desulfoferrodoxin from Desulfovibrio desulfuricans ATCC 27774. Eur J Biochem. 1999;261:438–443. doi: 10.1046/j.1432-1327.1999.00278.x. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Short K A, Blakemore R P. Periplasmic superoxide dismutases in Aquaspirillum magnetotacticum. Arch Microbiol. 1989;152:342–346. doi: 10.1007/BF00425171. [DOI] [PubMed] [Google Scholar]
- 38.Silva G, Oliveira S, Gomes C M, Pacheco I, Liu M Y, Xavier A V, Teixeira M, Legall J, Rodrigues-Pousada C. Desulfovibrio gigas neelaredoxin. A novel superoxide dismutase integrated in a putative oxygen sensory operon of an anaerobe. Eur J Biochem. 1999;259:235–243. doi: 10.1046/j.1432-1327.1999.00025.x. [DOI] [PubMed] [Google Scholar]
- 39.Smith T, Pitts K, McGarvey J A, Summers A O. Bacterial oxidation of mercury metal vapor, Hg(0) Appl Environ Microbiol. 1998;64:1328–1332. doi: 10.1128/aem.64.4.1328-1332.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tabor S. Expression using the T7 RNA polymerase/promoter system. In: Ausubel F A, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Green Publishing and Wiley-Interscience; 1990. pp. 16.2.1–16.2.11. [Google Scholar]
- 41.Touati D. Iron and oxidative stress in bacteria. Arch Biochem Biophys. 2000;373:1–6. doi: 10.1006/abbi.1999.1518. [DOI] [PubMed] [Google Scholar]
- 42.Van den Westen H S, Mayhew G, Veeger C. Separation of hydrogenase from intact cells of Desulfovibrio vulgaris. FEBS Lett. 1978;86:122–126. doi: 10.1016/0014-5793(78)80112-4. [DOI] [PubMed] [Google Scholar]
- 43.van Niel E W J, Gottschal J C. Oxygen consumption by Desulfovibrio strains with and without polyglucose. Appl Environ Microbiol. 1998;64:1034–1039. doi: 10.1128/aem.64.3.1034-1039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voordouw G. Molecular biology of sulfate-reducing bacteria. In: Odom J M, Singleton R J, editors. The sulfate-reducing bacteria: contemporary perspectives. New York, N.Y: Springer-Verlag; 1994. pp. 88–130. [Google Scholar]
- 45.Voordouw J K, Voordouw G. Deletion of the rbo gene increases the oxygen sensitivity of the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Appl Environ Microbiol. 1998;64:2882–2887. doi: 10.1128/aem.64.8.2882-2887.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zamocky M, Koller F. Understanding the structure and function of catalases: clues from molecular evolution and in vitro mutagenesis. Prog Biophys Mol Biol. 1999;72:19–66. doi: 10.1016/s0079-6107(98)00058-3. [DOI] [PubMed] [Google Scholar]
- 47.Zou P-J, Borovok I, Ortiz de Orué Lucana D, Müller D, Schrempf H. The mycelium-associated Streptomyces reticuli catalase-peroxidase, its gene and regulation by FurS. Microbiology. 1999;145:549–559. doi: 10.1099/13500872-145-3-549. [DOI] [PubMed] [Google Scholar]