Abstract
The two-component anthranilate 1,2-dioxygenase of the bacterium Acinetobacter sp. strain ADP1 was expressed in Escherichia coli and purified to homogeneity. This enzyme converts anthranilate (2-aminobenzoate) to catechol with insertion of both atoms of O2 and consumption of one NADH. The terminal oxygenase component formed an α3β3 hexamer of 54- and 19-kDa subunits. Biochemical analyses demonstrated one Rieske-type [2Fe-2S] center and one mononuclear nonheme iron center in each large oxygenase subunit. The reductase component, which transfers electrons from NADH to the oxygenase component, was found to contain approximately one flavin adenine dinucleotide and one ferredoxin-type [2Fe-2S] center per 39-kDa monomer. Activities of the combined components were measured as rates and quantities of NADH oxidation, substrate disappearance, product appearance, and O2 consumption. Anthranilate conversion to catechol was stoichiometrically coupled to NADH oxidation and O2 consumption. The substrate analog benzoate was converted to a nonaromatic benzoate 1,2-diol with similarly tight coupling. This latter activity is identical to that of the related benzoate 1,2-dioxygenase. A variant anthranilate 1,2-dioxygenase, previously found to convey temperature sensitivity in vivo because of a methionine-to-lysine change in the large oxygenase subunit, was purified and characterized. The purified M43K variant, however, did not hydroxylate anthranilate or benzoate at either the permissive (23°C) or nonpermissive (39°C) growth temperatures. The wild-type anthranilate 1,2-dioxygenase did not efficiently hydroxylate methylated or halogenated benzoates, despite its sequence similarity to broad-substrate specific dioxygenases that do. Phylogenetic trees of the α and β subunits of these terminal dioxygenases that act on natural and xenobiotic substrates indicated that the subunits of each terminal oxygenase evolved from a common ancestral two-subunit component.
Bioremediation efforts have focused attention on the bacterial degradation of xenobiotic methyl- and halogen-substituted benzoates by plasmid-encoded pathways of broad substrate specificity (2, 41). These catabolic routes may have evolved from narrow, substrate-specific, chromosomally encoded pathways for degrading natural compounds. The first step in the bacterial catabolism of benzoate or substituted benzoates is dihydroxylation of the aromatic ring (15). Subsequent metabolites are funneled into the tricarboxylic acid cycle. The aromatic ring-hydroxylating dioxygenases (ARHDOs) that initiate this sequence belong to a large enzyme family that operates on diverse substrates (4, 31). Although several dozen ARHDOs have been characterized, most of these enzymes were identified by their ability to dihydroxylate xenobiotic substrates.
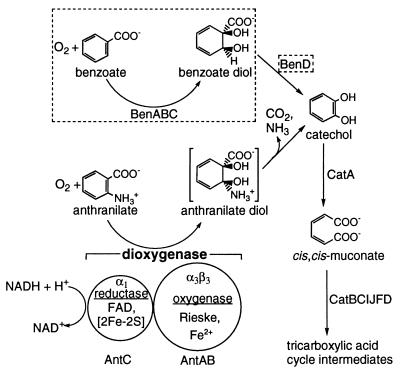
Anthranilate (2-aminobenzoate) is a naturally occurring compound that is formed during bacterial tryptophan degradation (20). In the early 1960s, it was shown that crude cell extracts of some Pseudomonas species convert anthranilate to catechol via insertion of both atoms of O2 into the aromatic ring (22, 25, 26, 40). Despite this long history and its metabolic significance, the enzyme responsible for this activity, anthranilate 1,2-DO (AntDO) (Fig. 1) has not been characterized beyond the studies cited above. The genes encoding AntDO, antABC, are known from only one microbe, namely, the soil bacterium Acinetobacter sp. strain ADP1 (6). A mutant able to grow on anthranilate at 23°C but not at 39°C was used to isolate antABC from the ADP1 chromosome. The temperature sensitivity of the mutant resulted from a point mutation in the 43rd codon of antA, resulting in a variant gene product with lysine rather than methionine (M43K AntA).
FIG. 1.
Degradation of anthranilate and benzoate in Acinetobacter sp. strain ADP1 via the β-ketoadipate pathway (19). Relevant compounds and roles of the component AntDO (AntAB and AntC) proteins are indicated. The corresponding BenDO (Ben) components and reactions are enclosed within dashed boxes.
The high degree of sequence similarity between antABC and benABC, the latter encoding benzoate 1,2-DO (BenDO) from the same organism (32), suggests a common evolutionary origin for both sets of genes. In fact, the pathways using these ARHDOs converge at catechol (Fig. 1). AntDO and BenDO are also closely related to several ARHDOs, encoded by mobile catabolic genes, which act on xenobiotics. These include the xylXYZ-encoded toluate (methylbenzoate) DO of Pseudomonas putida, the cbdABC-encoded halobenzoate DO of Burkholderia (formerly Pseudomonas) cepacia, the abs-encoded aminobenzene sulfonate DO from Alcaligenes sp. strain 0-1, and the tft-encoded 2,4,5-trichlorophenoxyacetic acid oxygenase of B. cepacia (Table 1) (9, 10, 13, 16, 17, 29, 44).
TABLE 1.
Classification of selected ARHDOs
| Enzymea (references) | Genes | Characteristics of components
|
||
|---|---|---|---|---|
| Oxygenase | Reductase | Ferredoxin | ||
| Class IB (two components) | ||||
| AntDO (6, 25) | antABC | Rieske type [2Fe-2S], mononuclear Fe(II) | Ferredoxin type [2Fe-2S], FAD | |
| BenDO (32, 45) | benABC | |||
| 2-Aminobenzenesulfonate DO (29) | absAb | |||
| 2-Halobenzoate 1,2-DO (13, 16) | cbdABC | |||
| Toluate 1,2-DO (17, 18) | xylXYZ | |||
| 2,4,5-Trichlorophenoxyacetic acid oxygenase (9, 10) | tftABb | |||
| Class IA (two components) | ||||
| Phthalate DO (3, 14, 43) | pht3,pht2 | Rieske type [2Fe-2S], mononuclear Fe(II) | Ferredoxin type [2Fe-2S], Flavin mononucleotide | |
| Class III (three components) | ||||
| NDO (24, 27) | ndoBACb | Rieske type [2Fe-2S], mononuclear Fe(II) | Ferredoxin type [2Fe-2S], FAD | Rieske type [2Fe-2S] |
Sequence comparisons with better-characterized ARHDOs (4, 31) led to the inferred functions and prosthetic groups for the reductase (AntC) and oxygenase (AntAB) components of AntDO (Fig. 1; Table 1). Each α subunit of the oxygenase component was predicted to contain one Rieske-type [2Fe-2S] center and one mononuclear center (Fe2+ in Fig. 1). The mononuclear center is presumed to be the site of O2 activation for insertion into the aromatic substrate. The reductase component was predicted to contain flavin adenine dinucleotide (FAD) and one [2Fe-2S] center (6).
As described in this report, we induced the Acinetobacter sp. strain ADP1 AntDO in Escherichia coli and purified the enzyme to homogeneity. The variant M43K AntDO was similarly purified and characterized in order to determine the basis for the temperature-sensitive phenotype of the mutant that produces it. The oligomeric state of AntDO was determined, and metal and prosthetic groups were identified. Previous studies suggest that in ADP1, AntDO and BenDO act efficiently in vivo only on anthranilate and benzoate, respectively, despite the similar structures of these substrates and the sequence similarity of the enzymes (6, 32). We compared anthranilate and benzoate as substrates for AntDO in vitro. The reaction products were identified to confirm that dihydroxylation of anthranilate results in spontaneous conversion to catechol by loss of ammonia and carbon dioxide, whereas dihydroxylation of benzoate yields a stable nonaromatic diol (Fig. 1) (6, 33). To help understand the relationship of AntDO to plasmid-encoded ARHDOs of broad substrate specificity, several halogenated and methylated benzoates were tested as substrates of AntDO. Recent sequencing of the Pseudomonas aeruginosa PAO1 genome also allowed us to identify the putative sequences of AntDO and BenDO from that bacterium and to compare sequence conservation among related enzymes.
MATERIALS AND METHODS
Reagents and general procedures.
Substrates were purchased from Sigma. Enzymatically produced (1R,2S)-1,2-dihydroxycyclohexa-3,5-diene carboxylate (benzoate 1,2-diol) was a gift from Albey M. Reiner (University of Massachusetts, Amherst). Oligonucleotides were synthesized by Integrated DNA Technologies. Nucleotide sequencing was done at the University of Georgia Molecular Genetics Instrumentation Facility (ABI373 sequencer; Applied Biosystems). Molecular biology procedures not detailed below followed standard protocols (36). Plasmid-bearing E. coli strains were cultured in Luria-Bertani medium with 100 μg of ampicillin/ml. Protein purity was judged by sodium dodecyl sulfate–12.5% polyacrylamide gel electrophoresis (SDS–12.5% PAGE) with Coomassie blue staining (37).
Plasmids for high-level ant gene expression.
Genes of Acinetobacter sp. strain ADP1 (also known as BD413 [23]) were PCR amplified with Pfu polymerase (Stratagene) from the template pBAC103 (6) and were placed under control of the inducible Ptac promoter of the pCYB1 vector (New England Biolabs). A 1,912-bp antAB amplicon was generated with the primers ANTAnde (5′-ggacttcatATGACTGCACGTAACCTCGC-3′) and ANTBtaa (5′-ataatagctcttccgcaTTAGACGTGATAGAAATCGAGTAC-3′) (sequences at the 5′ end of antA and the 3′ end of antB, respectively, are capitalized; added NdeI and SapI restriction sites are underlined). A 1,031-bp antC amplicon was similarly made with primers ANTCnde (5′-ggggaccatATGAATCATTCTGTTGCACTCAA-3′) and ANTCtaa (5-attatagctcttccgcaTTAAGTTTTTGCGGTATTACTTTG-3′). After digestion with NdeI and SapI, the amplicons were each ligated into pCYB1 to yield pBAC209 (antAB) and pBAC208 (antC). The nucleotide sequences of the plasmid-borne ant regions were confirmed to be the same as in GenBank AF071556.
Site-directed mutagenesis to produce M43K AntA.
An antA gene encoding lysine at residue 43 was constructed with the QuikChange Site Directed Mutagenesis kit (Stratagene). Plasmid pBAC209, which contains the wild-type antAB, was the template for the complementary mutagenic oligonucleotide primers 5′-TTTGAACTTGAAAAAGAACTCATTTTTG-3′ and 5′-CAAAAATGAGTTCTTTTTCAAGTTCAAA-3′ (the mutated codon is underlined). The mutated antAB DNA of the resulting plasmid, pDMK3, was verified by sequencing.
Induction of AntDO and preparation of cell extract.
One-liter cultures of E. coli strains DH5αF′ (Gibco BRL) or TOP10F′ (Invitrogen) carrying plasmids with antAB (pBAC209) or antC (pBAC208), respectively, were grown aerobically at 37°C until attaining an optical density at 600 nm of ∼0.6. The temperature was then reduced to 30°C, and ferrous ammonium sulfate (0.5 mg/ml) was added, followed by isopropyl-α-d-thiogalactopyranoside (100 μg/ml) to induce expression of the plasmid-borne ant genes. After overnight incubation at 30°C (∼18 h), cells were harvested by centrifugation and washed once with 20 mM potassium phosphate buffer (pH 7.5) (buffer A). Approximately 25 g of cells from six 1-liter cultures was suspended in 25 ml of buffer A containing 0.1 mM phenylmethylsulfonyl fluoride, 1 mg of DNase (Boehringer Mannheim), and 5% (vol/vol) glycerol. The suspended cells were sonicated on ice with a Branson sonifier cell disrupter 350 with a 0.5-inch probe tip for 2 min at 30-s intervals at 20 kHz. Cell debris was removed by centrifugation (12,000 × g for 30 min) at 4°C, resulting in red-brown or yellow-orange supernatants containing AntAB or AntC, respectively. Because M43K AntAB was predicted to be temperature sensitive, cells for its purification were grown at 23°C.
Purification of AntAB.
The procedure below was carried out at 4°C. The cell extract (35 ml) from 6 liters of cells was applied to a Whatman DE52 column; the anion exchange column (4 by 10 cm) was equilibrated in buffer A containing 100 mM NaCl (buffer B). The column was washed with 200 ml of buffer B, and bound AntAB was eluted with a 250-ml linear NaCl gradient (100 to 400 mM) in buffer A at a flow rate of 2.0 ml/min. Active fractions (assayed as described below) were pooled and concentrated by ultrafiltration in a 50-ml Amicon cell (YM-10 membrane) to 10 ml. Filtered 4 M (NH4)2SO4 was added with stirring to a final concentration of 1 M. The resulting precipitate was removed by centrifugation (12,000 × g for 15 min), and the supernatant was applied to a Butyl Sepharose column (2 by 11.5 cm; Pharmacia) equilibrated in buffer A containing 1 M (NH4)2SO4. AntAB was eluted at 2.0 ml/min with a 250-ml linear gradient of decreasing (NH4)2SO4 concentration (1 to 0 M) in buffer A. Active fractions were pooled and concentrated by ultrafiltration to 2 ml and equilibrated in 50 mM HEPES containing 200 mM KCl (pH 7.3) (buffer C). The concentrated sample was applied to a Sephacryl S300 column (1.6 by 60 cm; Pharmacia) equilibrated in buffer C and eluted in the same buffer at a flow rate of 0.5 ml/min. Active fractions were pooled and concentrated by ultrafiltration. The purified AntAB was frozen in 25-μl aliquots and stored at −80°C.
Purification of AntC.
Purification was carried out at 4°C under low-light conditions to minimize loss of flavin. Cell extract from 6 liters of AntC-induced cells (30 ml) was applied to the same DE52 anion exchange column as for AntAB purification. The column was washed with 200 ml of buffer A. Bound AntC was eluted with a 250-ml linear gradient of Na2SO4 (0 to 250 mM) in buffer A at 2.0 ml/min. Active fractions (assayed as described below) were pooled and concentrated by ultrafiltration to 10 ml. Filtered 4 M (NH4)2SO4 was added to a final concentration of 1 M. The precipitate was removed by centrifugation (12,000 × g for 15 min), and the supernatant was applied to a Butyl Sepharose column equilibrated in 100 mM potassium phosphate buffer (pH 7.5) (buffer D) containing 1 M (NH4)2SO4. AntC was eluted with a 250-ml linear gradient of decreasing (NH4)2SO4 concentration (1 to 0 M) at 2.0 ml/min in buffer D. Active fractions were pooled and concentrated by ultrafiltration to 10 ml and equilibrated in 50 mM morpholinepropanesulfonic acid (MOPS), pH 7.0. Samples were dialyzed against this same buffer containing 5% (vol/vol) glycerol and 1 mM FAD (4 liters for 12 h). The sample was concentrated by ultrafiltration, applied to a Sephacryl S200 column (1.6 by 60 cm; Pharmacia) equilibrated in buffer C, and eluted at a flow rate of 0.5 ml/min. Purified AntC was frozen in 25-μl aliquots and stored at −80°C.
Analyses.
Protein was quantitated by using the method described by Bradford (5), with bovine serum albumin as the standard (Bio-Rad). The native molecular weights of proteins were determined by gel filtration using a calibrated Sephacryl S300 column (flow rate, 0.5 ml/min) equilibrated in buffer C. The calibration proteins were ferritin (Mr, 450,000), catalase (Mr, 240,000), adolase (Mr, 158,000), bovine serum albumin (Mr, 68,000), hen egg albumin (Mr, 45,000), chymotrypsinogen A (Mr, 25,000), and cytochrome c (Mr, 12,500). Iron content was assessed with a colorimetric ferrozine method based on that described by Batie et al. (3). A 200-μl protein sample (approximately 10 μM) was added to a polystyrene tube containing 250 μl of 0.02% ascorbic acid, 30 μl of 6 M HCl, and 25 μl of a 5-mg/ml solution of ferrozine. The sample was mixed by vortexing, and 1 ml of 8 M guanidine–HCl was added. Saturated ammonium acetate (200 μl) was added, and A562 was measured. A standard curve with a ferrous ammonium sulfate solution was used to determine ɛ562 (28,000 M−1 cm−1) for the ferrous iron-ferrozine complex.
Flavin cofactor in AntC.
Flavin was extracted by boiling a concentrated 1-ml sample of AntC for 10 min. Denatured protein was removed by centrifugation. Thin-layer chromatography was used to identify the flavin, with silica gel-coated glass plates and a mobile phase of butanol–acetic acid–water (4:1:4, vol/vol/vol). Samples were visualized with a handheld UV light. After identification as FAD, the amount was quantitated spectrophotometrically (ɛ450, 11,300 M−1 cm−1; ɛ375, 9,300 M−1 cm−1) (13).
Oxygen consumption and NADH oxidation assays.
Substrate-dependent O2 consumption catalyzed by AntDO was monitored with a Yellow Springs Instruments Model 5300 biological oxygen monitor equipped with a Clark electrode. NADH oxidation was determined spectrophotometrically at 23°C by measuring the decrease in A340 (ɛ340, 6,300 M−1 cm−1). Unless stated otherwise, assays were carried out in 50 mM 2-(N-morpholino)ethanesulfonic acid (MES) (pH 6.3), 100 mM KCl, 0.5 mM substrate, 100 μM NADH, 0.5 μM purified AntAB (or a suitable amount of a crude enzyme preparation), and 0.18 μM reductase in a final volume of 1 ml for NADH oxidation assays or 2.5 ml for O2 consumption assays. Rates were corrected for a small background NADH oxidation or O2 consumption in the absence of substrate. In spectrophotometric assays, corrections were made for the absorbance of anthranilate (ɛ340, 1,050 M−1 cm−1). Reactions were monitored for 5 min, at which point 1,000 U of bovine liver catalase (Sigma) was added to the reaction mixture in the oxygraph. Hydrogen peroxide was quantified by the amount of O2 generated due to the disproportionation of H2O2 by catalase.
NADH-cytochrome c reductase activity.
The AntC-catalyzed reduction of cytochrome c by NADH was detected by an increase in A550 (ɛ550, 19,500 M−1 cm−1 for reduced cytochrome c minus oxidized cytochrome c). The assay mixture contained 20 μM cytochrome c (horse heart type 6; Sigma), 100 μM NADH, and 0.5 μM pure AntC (or a suitable amount of a crude enzyme preparation) in 100 mM potassium phosphate buffer, pH 7.
Optimization of AntDO activity.
A mixed-buffer system was used to determine the optimal conditions for assaying AntDO activity. This buffer system contained 50 mM total concentration of equimolar amounts of the following six buffers: MES (pKa, 6.1), MOPS (pKa, 7.2), HEPES (pKa, 7.5), N-tris(hydroxymethyl)methyl-3-aminopropanesulfonic acid (TAPS) (pKa, 8.4); 2-(N-cyclohexylamino)ethanesulfonic acid (CHES) (pKa, 9.3), and 3-(cyclohexylamino)-2-hydroxyl-1-propanesulfonic acid (CAPS) (pKa, 10.4). Individual NADH oxidation assays were carried out at pHs 5.5, 6.0, 6.5, 7.0, 8.0, and 9.0, using the conditions described above. After determining the optimal pH for NADH oxidation, assays were repeated in MES buffer (pH 6.3) at four different ionic strengths (0, 50, 100, and 150 mM KCl).
Monitoring reaction substrates and products by HPLC.
Samples from AntDO-catalyzed reactions (1 ml) were separated by high-performance liquid chromatography (HPLC) and were monitored by determining absorbance at 210 nm (automatic sampling system model AS-100, solvent delivery system 2800, detector UV-1806, and peak integration software; Bio-Rad). Compounds were eluted at a rate of 0.8 ml/min from a reverse-phase C18 column (250 by 4.6 mm; particle size, 5 μm; Columbus) with a mobile phase of 30% (vol/vol) acetonitrile-water containing 0.1% phosphoric acid. Substrates and products were identified and quantified by comparison to known standards.
Spectroscopy.
Electron paramagnetic resonance (EPR) spectra were recorded on a Bruker ESP-300E spectrometer equipped with an ER-4116 dual-mode cavity and an Oxford Instrument ESR-9 flow cryostat. UV-visible absorption spectra were obtained in 1-cm path-length quartz cuvettes on a Shimadzu UV2101-PC scanning spectrophotometer. To obtain absorption spectra of reduced [2Fe-2S] centers, oxidized proteins (250 μM in [2Fe-2S] centers) were reduced by anaerobic addition of an excess of sodium dithionite or catalytically by addition of AntC and excess NADH. Samples were diluted to concentrations appropriate for spectroscopy with 25 mM MOPS buffer, pH 7.3.
Putative AntDO and BenDO sequences of P. aeruginosa PAO1 and construction of phylogenetic trees.
The ADP1 BenDO and AntDO sequences were compared to six-phase translations of the complete genome sequence of P. aeruginosa PAO1 (http://www.pseudomonas.com) using the BLAST program of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). Putative antABC and benABC sequences at approximate positions 2829398 and 2835988 were identified on the basis of similarity to ADP1 sequences (52 to 70% identity between homologous deduced protein sequences), restriction site analyses of EcoRI and KpnI recognition sequences from previous studies, and the relative position of cat, ant, and ben genes determined by previous investigations of mutants and mapping studies of mutant loci (46, 47).
Deduced amino acid sequences of ARHDOs were aligned with the Pileup program of the Genetics Computer Group package (11). Trees were generated by applying the neighbor-joining method from a distance matrix created with PROTDIST using the Kimura algorithm of the Phylip program package (version 3.52c for UNIX; University of Washington, Seattle). The SeqBoot program was used for bootstrap analyses. Alternate algorithms for generating trees (Fitch and Kitch) and distance matrices (Dayhoff/PAM and parsimony) produced similar results.
RESULTS
Expression and purification of AntDO.
Genes encoding the ADP1 oxygenase (antAB) or reductase (antC) components were expressed from plasmids in E. coli, a bacterium with no known AntDO genes or activity. AntAB and AntC were each purified to homogeneity by anion exchange chromatography, hydrophobic interaction chromatography, and finally size exclusion chromatography. Active AntAB in E. coli could be detected by a color change upon addition of anthranilate and ferrous iron to the cultures. The medium gradually turned a dark purple, a color identical to that resulting from addition of catechol and iron to medium with no bacteria. As observed for other oxygenase components (30), AntAB appeared to use an endogenous source of electrons in E. coli to catalyze hydroxylation without its native reductase. This color development, presumably due to catechol formation, was a convenient visual indicator of AntAB activity. No dark color developed in the absence of anthranilate or when anthranilate and ferrous iron were added to medium without bacteria.
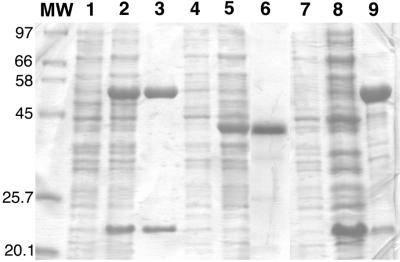
The antA and antB genes, adjacent in ADP1 (6), were coexpressed in E. coli. This yielded high levels of two proteins corresponding approximately in size to the deduced sequences of the α (54 kDa) and β (19 kDa) subunits, respectively, of AntDO (Fig. 2, lanes 2 and 3). Size exclusion chromatography of the pure AntAB revealed a single oligomer of approximately 220 kDa, which indicates that AntAB is an α3β3 hexamer (Table 2). Using ɛ454 (see below), approximately 12.5 mg of pure AntAB was obtained per liter of E. coli culture containing the antAB plasmid.
FIG. 2.
SDS-PAGE of AntAB, AntC, and the M43K AntAB variant. Lane 1, total protein of noninduced TOP10F′(pBAC209); lane 2, total protein of TOP10F′(pBAC209) with AntAB induced; lane 3, purified AntAB; lane 4, total protein of noninduced DH5αF′(pBAC208); lane 5, total protein of DH5αF′(pBAC208) with AntC induced; lane 6, purified AntC; lane 7, total protein of noninduced TOP10F′(pDMK3). Lanes 8 and 9 are the cell extract and pellet, respectively, following sonication and centrifugation of TOP10F′(pDMK3) with M43K AntAB induced. Numbers at left correspond to molecular masses (in kilodaltons) corresponding to the adjacent markers (lane MW).
TABLE 2.
Molecular and spectroscopic properties of recombinant Acinetobacter sp. strain ADP1 AntDO
| Component | Mol wt
(Mr)
|
Iron content (mol/mol of protein) | Flavin content (mol/mol of protein) | Absorption maxima, wavelengthf (ɛ [mM−1 cm−1]) | ||
|---|---|---|---|---|---|---|
| Sequence calculation | SDS-PAGE | Gel filtration | ||||
| AntAB | 53,942 (α), 19,333 (β) | 52,000 (α), 21,500 (β) | 220,000 | 8.8 ± 1a | 0 | 320 (31)ae 454 (14.4)a 555 (7)ae |
| M43K AntAB | 53,939 (α), 19,333 (β) | 52,000 (α), 21,500 (β) | 220,000 | 8.1 ± 0.2 | 0 | 320e 454 555e |
| AntC | 38,553 | 39,000 | 45,000 | 2.3 ± 0.5 | 0.7b, 1.5c | 330 (25)c 370 (23)c 450 (22)c 410, 462d |
Per α3β3 hexamer, as isolated (oxidized Rieske center, reduced mononuclear center).
As FAD from low-light isolation.
After reconstitution with FAD as described in Materials and Methods.
Low-flavin form; ɛ not determined (cf. Fig. 3).
Shoulder.
Wavelength is given in nanometers.
Independent expression of antC in E. coli resulted in abundant production of a protein of a size close to that predicted for AntC (Fig. 2, lanes 5 and 6). Initial attempts to isolate and purify AntC resulted in loss of flavin, a process that was visually evident during anion exchange chromatography as a yellow fraction eluting prior to the AntC protein. By minimization of its exposure to light during purification, AntC could be purified with retention of the majority of its flavin (see below). SDS-PAGE and gel filtration indicated that the purified AntC was a monomer of 39 kDa. Approximately 12.5 mg of pure AntC was obtained per liter of E. coli culture containing the antC plasmid.
Purification of the M43K variant DO.
A plasmid, pDMK3, was expressed in E. coli with the Acinetobacter wild-type antB downstream of the antA5024 allele (6), which encodes M43K AntA. The mutant antAB did not yield the color change indicative of catechol formation after anthranilate and iron were added to cultures grown at 23 or 37°C. Nevertheless, the variant DO could be purified by the same methods as for wild-type AntAB. In E. coli, the mutant antA yielded more insoluble AntA that localized to the pellet following sonication and centrifugation (Fig. 2, lane 9) than did the wild-type antA. The supernatant from which a low yield of the M43K oxygenase was purified contained more AntB relative to AntA than found with the wild-type oxygenase (Fig. 2, lane 8). The subunit sizes of the variant DO, however, were as expected (Fig. 2). The purified component migrated identically to the wild-type AntAB on the same size exclusion column and was, therefore, also inferred to be an α3β3 hexamer. SDS-PAGE of purified M43K AntAB showed α and β bands of approximately the same relative intensities as for the wild-type component (not shown).
Quantitation of the flavin in AntC.
The flavin in the recombinant AntC was identified as FAD by comparison to FAD and flavin mononucleotide standards using thin-layer chromatography. When isolated under low-light conditions, AntC contained up to 0.7 mol of FAD per mol of AntC monomer. The flavin content and reductase activity of AntC could be increased by reconstitution with excess FAD followed by passage over a Sephadex G-25 column to remove unbound FAD. Therefore, reconstitution was carried out prior to size exclusion chromatography during the purification of AntC, which yielded approximately 1.5 mol of FAD/mol of AntC (Table 2). Further FAD addition did not increase AntC activity. The AntC isolated in normal room light had less than 4% of the flavin in the low-light-isolated AntC on a per protein basis. The FAD-depleted AntC, when substituted for a comparable level of FAD-enriched AntC, had no detectable catalytic activity.
Iron content in AntDO.
AntC contained 2.3 ± 0.5 mol of iron/mol of AntC monomer, consistent with the prediction of one [2Fe-2S] center (7, 31). AntAB contained 8.8 ± 1 mol of iron/mol of protein based on a molecular weight of 220,000 (Table 2). Considering the α3β3 subunit composition, the iron analysis is consistent with the prediction of one [2Fe-2S] center and one mononuclear center per α subunit (7, 31). The purified M43K AntAB had 8.1 ± 0.2 mol of iron/mol of α3β3 hexamer, perhaps indicating one empty mononuclear site.
Spectroscopic properties of AntDO.
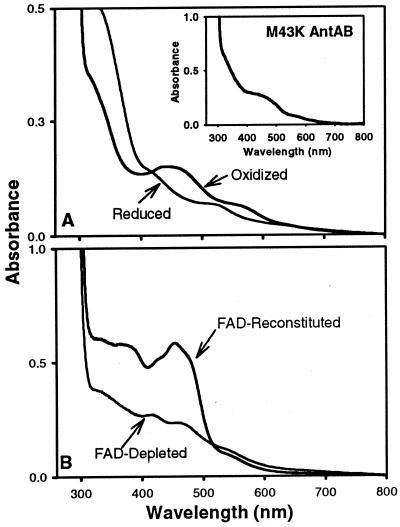
The absorption spectra of oxidized and enzymatically reduced wild-type AntAB are shown in Fig. 3A. The absorption maxima of the oxidized AntAB at 454 nm and the shoulder at 555 nm are typical of Rieske-type [2Fe-2S]2+ centers (3, 7, 12, 31). The ɛ454 was reproducibly determined to be 14,400 M−1 cm−1 per α3β3 hexamer. Upon anaerobic reduction with a catalytic amount of AntC and excess NADH, the absorption spectrum of AntAB resembled those of other reduced (i.e., [2Fe-2S]+) Rieske-type centers, with a general decrease in absorbance throughout the visible region and shoulders at ∼520 and 400 nm. Resting oxygenase components of other ARHDOs (7, 24, 43) invariably have their mononuclear centers in the Fe(II) oxidation state, and this reduced mononuclear center is not expected to contribute to either the oxidized or the reduced visible-absorption spectrum. Also shown in the inset of Fig. 3A is the absorption spectrum of the M43K AntAB. This spectrum is consistent with perturbation but not destruction of the oxidized Rieske center.
FIG. 3.
UV-visible absorption spectra of oxidized and enzymatically reduced AntAB (12 μM α3β3) and oxidized M43K AntAB (15 μM α3β3) (inset) (A) and oxidized AntC reconstituted with FAD or as isolated in normal room light (FAD depleted) (B).
The visible-absorption spectrum of low-light purified FAD-enriched AntC (Fig. 3B) and that of FAD-depleted AntC isolated under normal room light show that AntC has both FAD and a ferredoxin-type [2Fe-2S]2+ center (7, 21). The maxima at ∼425 and 460 nm and shoulder at ∼550 nm in the absorption spectrum of flavin-depleted AntC are characteristic of ferredoxin-type [2Fe-2S]2+ centers. The ratios of absorption intensities at 330 or 370 nm relative to that at 450 nm in the FAD-enriched reductase are higher than those for the phthalate 4,5-DO reductase component, which also contains one flavin and one [2Fe-2S]2+ chromophore (3, 14). Presumably, the excess FAD (1.5 mol/mol of AntC) is responsible for the altered absorption intensities of AntC.
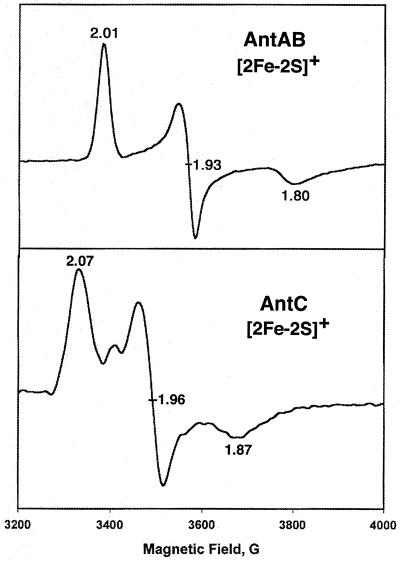
The EPR spectra (at 10 K) of the dithionite-reduced AntAB and AntC (Fig. 4) confirm the presence of [2Fe-2S] centers. The AntAB EPR spectrum (gaverage [gavg], 1.91) is characteristic of Rieske-type [2Fe-2S]+ centers (42), and the AntC spectrum (gavg, 1.97) is characteristic of ferredoxin-type [2Fe-2S]+ centers. EPR signals in (g = 4 to 6 regions), which are expected for a mononuclear high-spin Fe(III) site in a low-symmetry coordination sphere of oxygen and nitrogen ligands, were not observed for the recombinant wild-type AntAB in the absence of dithionite (spectrum not shown). Thus, the EPR-silent mononuclear site is most likely in the Fe(II) oxidation state.
FIG. 4.
EPR spectra of reduced AntAB and AntC. Purified samples of protein (250 μM) were reduced anaerobically with excess sodium dithionite (0.5 mM). Spectra were recorded under the following conditions: temperature, 10 K; microwave frequency, 9.59 GHz; modulation amplitude, 6.366 G; microwave power, 4 mW.
Catalytic properties of AntDO.
AntDO activity was measured by monitoring anthranilate-dependent consumption of NADH. The optimal pH and salt concentration for this activity were 6.3 and 100 mM KCl. This optimization was accomplished with saturating NADH, aromatic substrate, and O2 (typically 100 μM, 1 mM, and ∼0.25 mM), 0.5 μM AntAB (hexamer), and 0.18 μM AntC. These reagent concentrations were chosen for monitoring NADH consumption on a convenient time scale via absorbance at 340 nm. Unless otherwise noted, these conditions were used for all reported results. Under these nonsaturating AntC conditions, the turnover number of AntAB, measured as the rate of NADH consumption, was 63 min−1 on a per-Rieske-site basis.
The Km of AntAB for AntC was determined with 0.01 μM AntAB and variable concentrations of AntC from 0.1 to 2 μM. All other conditions were standard. A Lineweaver-Burk plot was used to estimate a Km for AntC of approximately 1 μM. With a saturating concentration of 2 μM AntC, 0.01 μM AntAB and 100 μM NADH yielded a turnover number of 1,300. Activity levels did not decrease when the anthranilate concentration was reduced to 1 μM. The small absorbance change for NADH consumption at anthranilate concentrations of <1 μM precluded accurate rate measurements below this concentration. Therefore, the Km of AntDO for anthranilate was inferred to be <1 μM. An analogous determination under the same conditions gave a Km for benzoate of ∼12 μM.
Activities of AntDO with anthranilate and benzoate.
Table 3 summarizes the activities of the purified recombinant AntDO and M43K AntDO with anthranilate and benzoate, as well as quantitations of substrate consumption and product formation. Several considerations circumscribe interpretation of the data in Table 3. These activities were measured under conditions that had been optimized for anthranilate as the substrate. The relative activities are for initial rates of NADH consumption only; for reasons discussed below, these activities do not necessarily correlate with or even signify consumption of the aromatic carboxylate (Table 3). A nonsaturating concentration of AntC was used to avoid significant background consumption of NADH and O2 and to allow measurements of substrate consumption on a reasonable time scale.
TABLE 3.
Activities and reaction stoichiometries of recombinant Acinetobacter sp. strain ADP1 AntDO and M43K AntDO with anthranilate and benzoatea
| Component | Aromatic substrate | Activity (%)b | Stoichiometries in
NADH-limited reactions (μM)c
|
|||
|---|---|---|---|---|---|---|
| Substrate consumed
|
Product formed, identity | |||||
| NADH | O2 | Aromatic substrate | ||||
| AntDO | Anthranilate | 100 | 95 (10) | 100 (7) | 91 (2) | 87 (2), catechol |
| Benzoate | 69 (4) | 89 (9) | 86 (8) | 83 (9) | NQ,d benzoate 1,2-diol | |
| M43K AntDO | Anthranilate | 24 (2) | 23 (4) | 24 (2) | None | 14 (3), H2O2 |
| Benzoate | 24 (3) | 23 (3) | 22 (5) | None | 16 (4), H2O2 | |
Assay conditions: 100 μM NADH, 250 μM O2, 500 μM aromatic carboxylate, 0.5 μM AntAB, 0.18 μM AntC, and in 50 mM MES, 100 mM KCl (pH 6.3) at room temperature (∼23°C). Values (with standard deviations in parentheses) listed are the averages of three determinations. Small background consumptions of NADH and O2 in the absence of aromatic carboxylates were subtracted in each case.
Initial rate of NADH oxidation (A340/min) expressed as a percentage of recombinant wild-type AntDO with anthranilate as the substrate (40 ± 1 μM NADH oxidized/min/μM AntAB hexamer) under the assay conditions listed in footnote a.
Measured by NADH (ɛ340, 6,300 M−1 cm−1), oxygen electrode (O2), or HPLC using known standards (aromatic substrate and product formed). Assay conditions are listed in footnote a.
NQ, not quantified because the compound was not well resolved by HPLC.
Under conditions optimized for anthranilate turnover, benzoate was a good substrate for AntDO, resulting in 70% of the activity rate as with anthranilate (Table 3). Benzoate allowed tight coupling of NADH and O2 and substrate consumption (i.e., 1:1:1 molar ratio) with no evidence for inactivation of the enzyme. No H2O2 was detected in these reactions. Together with the tight coupling, this indicates that all O2 consumed was incorporated into the substrate. The product of AntDO catalysis with benzoate was the same nonaromatic diol as that formed by the similar Acinetobacter sp. strain ADP1 BenDO (33). AntDO can efficiently convert benzoate to the nonaromatic diol under these conditions, but it may not do so in vivo (B. M. Bundy, L. S. Collier, and E. L. Neidle, unpublished data). Therefore, we investigated the effect of lowering the substrate concentrations. When substrate concentrations were decreased 100-fold to 5 μM, a value still saturating for anthranilate but not for benzoate, NADH oxidation with anthranilate remained identical to that previously observed. However, NADH oxidation with 5 μM benzoate decreased to a value 23% of that of anthranilate-dependent oxidation.
Activity of the variant M43K AntAB.
While the soluble yield was much lower, the characterization of the M43K AntAB described above indicated that its overall structure and metal centers are not grossly altered compared to those of the wild-type protein. The M43K AntAB had activity with either benzoate or anthranilate as the substrate that was approximately 25% of the NADH oxidation and O2 consumption activity of the recombinant wild type, using anthranilate as the substrate at 23°C. However, this activity, while substrate dependent, was completely uncoupled from substrate hydroxylation. No anthranilate or benzoate was consumed. All NADH and O2 consumed in the reaction could be accounted for by the generation of H2O2. Supplementing M43K AntAB with 5, 10, or 100 μM Fe(II) did not increase activity.
In our assays, neither wild-type nor M43K AntDO showed NADH consumption activity at 39°C. Samples of AntAB that were incubated at 39°C for 10 min and allowed to cool to room temperature exhibited 20% less activity than a non-heat-treated sample, and the stoichiometry of NADH consumption to O2 consumption was still 1:1 with no detectable H2O2. Heat-treated M43K AntDO had activities comparable to those of a non-heat-treated sample, but NADH consumption remained uncoupled from substrate hydroxylation, with H2O2 formation accounting for all the NADH and O2 consumed.
Substrate range of AntDO.
Since ADP1 cannot grow on halobenzoates or methylbenzoates, it seemed likely that AntDO would have a substrate range narrower than the broad-substrate-specific benzoate DOs. We tested o-fluorobenzoate, o-chlorobenzoate, and o-toluate as substrates for AntDO. All of these compounds, as well as anthranilate and benzoate, are good substrates for tightly coupled hydroxylation reactions catalyzed by the cbdABC-encoded 2-halobenzoate 1,2-DO from B. cepacia (13, 16). Under the assay conditions with 100 μM NADH shown in Table 3, HPLC analyses indicated that no o-chlorobenzoate and only 18 μM o-fluorobenzoate and 13 μM o-toluate were consumed. With each of these substrates, more than 40 μM NADH was consumed, indicating that electron transfer was either totally (o-chlorobenzoate) or partially (o-fluorobenzoate or o-toluate) uncoupled from substrate hydroxylation.
Putative AntDO and BenDO sequences from P. aeruginosa.
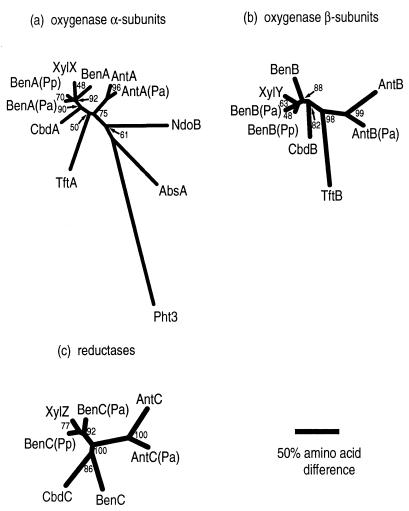
Sequence comparisons were used to address the evolutionary relationships among various ARHDOs. Previous studies of catabolic mutants and the relative positions of genetic loci (46, 47) allowed us to identify the putative AntDO and BenDO sequences of P. aeruginosa. In pairwise comparisons of the deduced sequences of AntDO from Acinetobacter sp. strain ADP1 and P. aeruginosa PAO1, identities and similarities were, respectively, 74 and 81% for the α subunits, 54 and 64% for the β subunits, and 60 and 68% for the reductases. These sequences were also compared to those of BenDO from P. putida PRS2000 (8). Phylogenetic trees (Fig. 5) were generated from multiple-sequence alignments of these ARHDOs and others that degrade xenobiotics, which have previously been characterized as class IB based on similarity among all components (Table 1). Sequences of the α subunits of two well-characterized ARHDOs of different classes were also aligned. These α subunits were NdoB of the class III naphthalene 1,2-DO (NDO) and Pht3 of the class IA phthalate DO (Table 1 and Fig. 5A). The sequences of other components of these latter two DOs were not included in the phylogenetic trees because of relatively poor alignment. The sequences are not known for the reductase of the 2,4,5-trichlorophenoxyacetic acid oxygenase or for that of the aminobenzene sulfonate DO. Only the sequence of the α subunit (AbsA) of the latter enzyme is available.
FIG. 5.
Phylogenetic trees of the α subunits (a), β subunits (b), and reductases (c) of known and putative class IB DOs. Also included were two α subunit components from different ARHDO classes (Table 1): NDO (NdoB, class III, M23914 [27]) and phthalate DO (Pht3, class IA, D13229 [34]). An outgroup (Pht3) was used only for panel a. Abbreviations: Abs, 2-aminobenzenesulfonate DO of Alcaligenes sp. strain AF109074) (29); Ant, AntDO of Acinetobacter sp. strain ADP1 (6); Ant(Pa), putative sequence of P. aeruginosa PAO1; Ben, BenDO of Acinetobacter sp. strain ADP1 (32); Ben(Pa), putative sequence of P. aeruginosa PAO1; Ben(Pp), sequence of P. putida PRS2000 (8); Cbd, 2-halobenzoate 1,2-DO of B. cepacia (X79076 [16]); Tft, 2,4,5-trichlorophenoxyacetic acid oxygenase of B. cepacia (U11420 [9]); Xyl, toluate 1,2-DO of P. putida (PIR A41659 [17]). The numbers at branch points indicate the confidence (in percent) as determined by bootstrap analysis with 100 replicates. The scale bar indicates the relative phylogenetic distances measured as number of amino acid substitutions per site.
DISCUSSION
At least three types of different enzymes can catalyze the first step in aerobic pathways for anthranilate catabolism. In eukaryotic microbes, anthranilate is first hydroxylated to form 2,3-dihydroxybenzoate by anthranilate hydroxylase. This enzyme is a flavoprotein monooxygenase in Trichosporon cutaneum (35) but contains only nonheme iron as a prosthetic group in Aspergillus niger (38, 39). Another enzyme, 2-aminobenzoate-coenzyme A ligase, initiates anthranilate degradation in a denitrifying Pseudomonas species (1). The AntDO described in this report is distinct from the previously mentioned enzymes and allows aerobic bacteria to use anthranilate as a source of carbon and energy. AntDO had not previously been purified to homogeneity from any organism.
Characterization of AntDO.
AntAB of Acinetobacter sp. strain ADP1 contains both mononuclear nonheme iron and Rieske-type [2Fe-2S] centers, as do all other known ARHDOs. The presumed ligands of the mononuclear and Rieske centers are well conserved among AntDO and other ARHDO oxygenase components (Fig. 6). The results reported here also confirm that AntDO can be classified as a two-component class IB ARHDO (4, 7, 31). The key features for this classification are the presence of both FAD and a [2Fe-2S] center in the reductase component (AntC) and the lack of a separate ferredoxin in the electron transfer chain (Table 1).
FIG. 6.
Sequence of ADP1 AntA and a consensus sequence derived from an alignment of the α subunits from nine class IB ARHDOs and the class III NDO (Table 1 and Fig. 5). The consensus sequence indicates residues that are identical in the nine class IB sequences. Bold residues are also identical in NdoB of NDO. Residues known to furnish ligands to the Rieske and mononuclear iron sites in NdoB are boxed. The italicized asparagine at position 214 of the consensus sequence is a possible ligand to the mononuclear iron site (24) and is conserved in nine of the sequences but not in AbsA.
Gel filtration chromatography indicates that AntDO is an α3β3 hexamer. This same oligomeric structure was found for the oxygenase component of NDO, a class III ARHDO (Table 1). NDO is the only such component for which an X-ray crystal structure has been solved (7a, 24), as discussed in reference 28. The oxygenase components of NDO and AntDO appear to share a common evolutionary origin (Fig. 5a). Presumably, structure-function relationships found in NDO (28) are similar to those in AntDO. Alignment of nine class IB α subunit sequences, including AntA, shows that all of the residues furnishing iron ligands in the class III NdoB are conserved (Fig. 6). Two class IB DOs, BenDO from P. putida (arvilla) C-1 and the 2-halobenzoate 1,2-DO from B. cepacia, were also shown to be α3β3 hexamers (13, 45). These latter two enzymes have pH optima in a range similar to that determined here for AntDO, pH 6.3 (13, 45). As found for AntDO, all of these analogous ARHDOs appear to have a monomeric reductase component.
Temperature sensitivity and the M43K AntDO variant.
The temperature-sensitive phenotype of the Acinetobacter mutant that encodes M43K AntDO (6) may result from instability of the α subunit prior to its assembly into the hexamer. This possibility is supported by the observation that the majority of the recombinant M43K AntA was insoluble. A low yield of a stable α3β3 hexamer of this variant enzyme could be purified, although the recombinant enzyme did not hydroxylate anthranilate at either the permissive (23°C) or nonpermissive (39°C) temperature for growth of the mutant on anthranilate. In the recombinant system, the M43K AntAB had a lower iron content than did the AntAB with a wild-type sequence, which raises the possibility that expressing the variant enzyme at high levels in E. coli prevents recovery of a hexameric enzyme with all of the mononuclear sites filled. Nevertheless, our studies of the soluble recombinant M43K AntDO suggest that temperature sensitivity in Acinetobacter does not result primarily from dissociation of the hexamer. The M43 residue is in a highly conserved region of the N-terminal domains of the class IB ARHDOs. In the alignment used to generate Fig. 5A, eight of the nine class IB sequences contained a methionine corresponding to the 43rd residue of the ADP1 AntA. The exception was AbsA, in which a lysine is normally present rather than methionine, consistent with active DOs tolerating this amino acid substitution under some conditions.
Substrate preferences of AntDO.
The purified AntDO catalyzed the conversion of anthranilate to catechol in a well-coupled reaction. The recombinant AntDO also catalyzed the conversion of benzoate to benzoate 1,2-diol, as does BenDO. Nevertheless, ADP1 strains lacking functional structural genes encoding BenDO do not grow on benzoate (Bundy, Collier, and Neidle, unpublished). This phenotype may reflect the inability of benzoate to induce AntDO, insufficient production of the benD-encoded benzoate diol dehydrogenase in vivo, and/or problems with the rate of benzoate diol formation by AntDO (6, 33). The lower apparent Km of AntDO for anthranilate than that for benzoate suggests that intracellular substrate levels also contribute to growth specificities. The apparent Km for anthranilate of <1 μM indicated a high affinity of AntDO for this substrate. To date, the best-characterized BenDO is from P. putida (arvilla) C-1, for which an apparent Km for benzoate of approximately 4 μM was reported (45). Since the latter enzyme also has a higher turnover number for its substrate, approximately fourfold higher than that of AntDO for anthranilate, the specificity constants (kcat/Km) for these two enzymes might be comparable. The ARHDO encoded by the cbdABC genes appears to have a lower affinity for its substrate with an apparent Km for 2-chlorobenzoate of 22 μM (13). This enzyme, unlike AntDO, has an extremely broad substrate range and can efficiently hydroxylate a variety of ortho-substituted benzoate analogs. The relatively low affinity of the halobenzoate 1,2-DO for 2-chlorobenzoate might indicate that the broad substrate specificity evolved at the expense of catalytic efficiency.
Evolution of the class IB ARHDOs.
In previous studies, evolutionary relationships among the α subunits of ARHDOs of different classes have been established (29, 30). In this report, the identification of the likely AntDO and BenDO sequences from P. aeruginosa PAO1 and the recent availability of the BenDO sequence from P. putida PRS2000 (8) made it possible to examine relationships among all components of the class IB ARHDOs. The branching patterns of the phylogenetic trees for the α and β subunits of the class IB terminal oxygenases are nearly identical. The matched branching patterns are consistent with the class IB ARHDOs having evolved from an ancestral two-subunit oxygenase component. The CbdAB component that hydroxylates halobenzoates may be more closely related to BenAB than to AntAB. The plasmid-encoded XylXY component, which hydroxylates methylbenzoates, appears to have diverged relatively recently from BenAB. The AbsA α subunit component was as distantly related to the other class IB ARHDOs as was the NdoB of the class III NDO, consistent with the Abs DO being an atypical Class IB enzyme (29).
Phylogenetic comparisons of the reductases (Fig. 5C) demonstrate that the Acinetobacter and Pseudomonas AntC homologs, like the corresponding AntA and AntB homologs, are closely related to each other. In contrast, the BenC reductase of ADP1 was not as closely related to its Pseudomonas homologs as were the BenA and BenB proteins. This greater divergence may reflect the relaxed requirement of oxygenase components for specific reductases. For example, the Acinetobacter AntAB oxygenase component does not require its cognate reductase for activity but appears to function both with BenC in vivo (6) and, as shown here, in a heterologous expression system with an unidentified E. coli reductase. During evolution, the reductases may tolerate more amino acid substitutions than do the oxygenases. Additionally, the recruitment and association of different oxygenase and reductase components may occur. The newly available BenDO sequences demonstrate the tight clustering of XylZ with the Pseudomonas BenC reductases. Thus the broad-substrate-specific toluate DO, encoded by the xylXYZ genes of a Pseudomonas plasmid (2, 18), may have evolved from a Pseudomonas BenDO rather than from the BenDO of ADP1 to which it has previously been compared (17).
ACKNOWLEDGMENTS
We thank Alison Buchan and Barny Whitman for helpful discussions on phylogenetic trees and Ish Dhawan for assistance in obtaining EPR spectra.
This work was supported by the National Institutes of Health Grant GM59818-01 (E.L.N. and D.M.K.), National Science Foundation Grant MCB-9808784 (E.L.N.), and National Science Foundation Research Training Grant BIR9413235 (D.M.E. and Z.M.B.).
REFERENCES
- 1.Altenschmidt U, Fuchs G. Novel aerobic 2-aminobenzoate metabolism. Eur J Biochem. 1992;205:721–727. doi: 10.1111/j.1432-1033.1992.tb16835.x. [DOI] [PubMed] [Google Scholar]
- 2.Assinder S J, Williams P A. The TOL plasmids: determinants of the catabolism of toluene and the xylenes. Adv Microb Physiol. 1990;31:1–69. doi: 10.1016/s0065-2911(08)60119-8. [DOI] [PubMed] [Google Scholar]
- 3.Batie C J, LaHaie E, Ballou D P. Purification and characterization of phthalate oxygenase and phthalate oxygenase reductase from Pseudomonas cepacia. J Biol Chem. 1987;262:1510–1518. [PubMed] [Google Scholar]
- 4.Bertini I, Cremonini M A, Ferretti S, Lozzi I, Luchionat C, Viezzoli M S. Arene hydroxylases: metalloenzymes catalysing dioxygenation of aromatic compounds. Coord Chem Rev. 1996;151:145–160. [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Bundy B M, Campbell A L, Neidle E L. Similarities between the antABC-encoded anthranilate dioxygenase and the benABC-encoded benzoate dioxygenase of Acinetobactersp. strain ADP1. J Bacteriol. 1998;180:4466–4474. doi: 10.1128/jb.180.17.4466-4474.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butler C S, Mason J R. Structure-function analysis of the bacterial aromatic ring-hydroxylating dioxygenases. Adv Microb Physiol. 1997;38:47–84. doi: 10.1016/s0065-2911(08)60155-1. [DOI] [PubMed] [Google Scholar]
- 7a.Carredano E, Karlsson A, Kauppi B, Choudhury D, Parales R E, Parales J V, Lee K, Gibson D T, Eklund H, Ramaswamy S. Substrate binding site of naphthalene 1,2-dioxygenase: functional implications of indole binding. J Mol Biol. 2000;296:701–712. doi: 10.1006/jmbi.1999.3462. [DOI] [PubMed] [Google Scholar]
- 8.Cowles C E, Nichols N N, Harwood C S. BenR, a XylS homologue, regulates three different pathways of aromatic acid degradation in Pseudomonas putida. J Bacteriol. 2000;182:6339–6346. doi: 10.1128/jb.182.22.6339-6346.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danganan C E, Ye R W, Daubaras D L, Xun L, Chakrabarty A M. Nucleotide sequence and functional analysis of the genes encoding 2,4,5-trichlorophenoxyacetic acid oxygenase in Pseudomonas cepaciaAC1100. Appl Environ Microbiol. 1994;60:4100–4106. doi: 10.1128/aem.60.11.4100-4106.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daubaras D L, Danganan C E, Hubner A, Ye R W, Hendrickson W, Chakrabarty A M. Biodegradation of 2,4,5-trichlorophenoxyacetic acid by Burkholderia cepaciastrain AC1100: evolutionary insight. Gene. 1996;179:1–8. doi: 10.1016/s0378-1119(96)00326-5. [DOI] [PubMed] [Google Scholar]
- 11.Devereaux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fee J A, Findling K L, Yoshida T, Hille R, Tarr G E, Hearshen D O, Dunham W R, Day E P, Kent T A, Munck E. Purification and characterization of the Rieske iron-sulfur protein from Thermus thermophilus. Evidence for a [2Fe-2S] cluster having non-cysteine ligands. J Biol Chem. 1984;259:124–133. [PubMed] [Google Scholar]
- 13.Fetzner S, Müller R, Lingens F. Purification and some properties of 2-halobenzoate 1,2-dioxygenase, a two-component enzyme system from Pseudomonas cepacia2CBS. J Bacteriol. 1992;174:279–290. doi: 10.1128/jb.174.1.279-290.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gassner G T, Ludwig M L, Gatti D L, Correll C C, Ballou D P. Structure and mechanism of the iron-sulfur flavoprotein phthalate dioxygenase reductase. FASEB J. 1995;9:1411–1418. doi: 10.1096/fasebj.9.14.7589982. [DOI] [PubMed] [Google Scholar]
- 15.Gibson D T, Subramanian V. Microbial degradation of aromatic hydrocarbons. In: Gibson D T, editor. Microbial degradation of organic compounds. New York, N.Y: Marcel Dekker, Inc.; 1984. pp. 181–252. [Google Scholar]
- 16.Haak B, Fetzner S, Lingens F. Cloning, nucleotide sequence, and expression of the plasmid-encoded genes for the two-component 2-halobenzoate 1,2-dioxygenase from Pseudomonas cepacia2CBS. J Bacteriol. 1995;177:667–675. doi: 10.1128/jb.177.3.667-675.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harayama S, Rekik M, Bairoch A, Neidle E L, Ornston L N. Potential DNA slippage structures acquired during evolutionary divergence of Acinetobacter calcoaceticus chromosomal benABC and Pseudomonas putida TOL pWW0 plasmid xylXYZ, genes encoding benzoate dioxygenases. J Bacteriol. 1991;173:7540–7548. doi: 10.1128/jb.173.23.7540-7548.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harayama S, Rekik M, Timmis K N. Genetic analysis of a relaxed substrate specificity aromatic ring dioxygenase, toluate 1,2-dioxygenase, encoded by TOL plasmid pWW0 of Pseudomonas putida. Mol Gen Genet. 1986;202:226–234. doi: 10.1007/BF00331641. [DOI] [PubMed] [Google Scholar]
- 19.Harwood C S, Parales R E. The β-ketoadipate pathway and the biology of self-identity. Annu Rev Microbiol. 1996;50:553–590. doi: 10.1146/annurev.micro.50.1.553. [DOI] [PubMed] [Google Scholar]
- 20.Hayaishi O, Stanier R Y. Bacterial oxidation of tryptophan. J Bacteriol. 1951;62:691–709. doi: 10.1128/jb.62.6.691-709.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hugo N, Armengaud J, Gaillard J, Timmis K N, Jouanneau Y. A novel [2Fe-2S] ferredoxin from Pseudomonas putidamt2 promotes the reductive reactivation of catechol 2,3-dioxygenase. J Biol Chem. 1998;273:9622–9629. doi: 10.1074/jbc.273.16.9622. [DOI] [PubMed] [Google Scholar]
- 22.Ichihara A, Adachi K, Hosokawa K, Takeda Y. Enzymatic hydroxylation of aromatic carboxylic acids; substrate specificities of anthranilate and benzoate oxidases. J Biol Chem. 1962;237:2296–2302. [PubMed] [Google Scholar]
- 23.Juni E, Janik A. Transformation of Acinetobacter calco-aceticus (Bacterium anitratum) J Bacteriol. 1969;98:281–288. doi: 10.1128/jb.98.1.281-288.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kauppi B, Lee K, Carredano E, Parales R E, Gibson D T, Eklund H, Ramaswamy S. Structure of an aromatic-ring-hydroxylating dioxygenase—naphthalene 1,2-dioxygenase. Structure. 1998;6:571–586. doi: 10.1016/s0969-2126(98)00059-8. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi S, Hayaishi O. Anthranilic acid conversion to catechol (Pseudomonas) Methods Enzymol. 1970;17A:505–510. [Google Scholar]
- 26.Kobayashi S, Kuno S, Itada N, Hayaishi O, Kozuka S, Oae S. O18studies on anthranilate hydroxylase—a novel mechanism of double hydroxylation. Biochem Biophys Res Commun. 1964;16:556–561. doi: 10.1016/0006-291x(64)90192-5. [DOI] [PubMed] [Google Scholar]
- 27.Kurkela S, Lehvaslaiho H, Palva E T, Teeri T H. Cloning, nucleotide sequence and characterization of genes encoding naphthalene dioxygenase of Pseudomonas putidastrain NCIB9816. Gene. 1988;73:355–362. doi: 10.1016/0378-1119(88)90500-8. [DOI] [PubMed] [Google Scholar]
- 28.Kurtz D M. Structure of an aromatic-ring-hydroxylating dioxygenase—naphthalene 1,2-dioxygenase. Chemtracts. 1998;6:571. doi: 10.1016/s0969-2126(98)00059-8. [DOI] [PubMed] [Google Scholar]
- 29.Mampel J, Ruff J, Junker F, Cook A M. The oxygenase component of the 2-aminobenzenesulfonate dioxygenase system from Alcaligenessp. strain O-1. Microbiology. 1999;145:3255–3264. doi: 10.1099/00221287-145-11-3255. [DOI] [PubMed] [Google Scholar]
- 30.Martin V J J, Mohn W W. A novel aromatic-ring-hydroxylating dioxygenase from the diterpeniod-degrading bacterium Pseudomonas abietaniphilaBKME-9. J Bacteriol. 1999;181:2675–2682. doi: 10.1128/jb.181.9.2675-2682.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mason J R, Cammack R. The electron-transport proteins of hydroxylating bacterial dioxygenases. Annu Rev Microbiol. 1992;46:277–305. doi: 10.1146/annurev.mi.46.100192.001425. [DOI] [PubMed] [Google Scholar]
- 32.Neidle E L, Hartnett C, Ornston L N, Bairoch A, Rekik M, Harayama S. Nucleotide sequences of the Acinetobacter calcoaceticus benABCgenes for benzoate 1,2-dioxygenase reveal evolutionary relationships among multicomponent oxygenases. J Bacteriol. 1991;173:5385–5395. doi: 10.1128/jb.173.17.5385-5395.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neidle E L, Shapiro M, Ornston L N. Cloning and expression in Escherichia coli of Acinetobacter calcoaceticusgenes for benzoate degradation. J Bacteriol. 1987;169:5496–5503. doi: 10.1128/jb.169.12.5496-5503.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nomura Y, Nakagawa M, Ogawa N, Harashima S, Oshima Y. Genes in PHT plasmid encoding the initial degradation pathway of phthalate in Pseudomonas putida. J Ferment Bioeng. 1992;74:333–344. [Google Scholar]
- 35.Powlowski J B, Dagley S, Massey V, Ballou D P. Properties of anthranilate hydroxylase (deaminating), a flavoprotein from Trichosporon cutaneum. J Biol Chem. 1987;262:69–74. [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 38.Subramanian V, Sugumaran M, Vaidyanathan C S. Anthranilate hydroxylase, an iron enzyme, from Aspergillus niger. Indian J Biochem Biophys. 1979;16:370–374. [PubMed] [Google Scholar]
- 39.Subramanian V, Vaidyanathan C S. Anthranilate hydroxylase from Aspergillus niger: new type of NADPH-linked nonheme iron monooxygenase. J Bacteriol. 1984;160:651–655. doi: 10.1128/jb.160.2.651-655.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taniuchi H, Hatanaka M, Kuno S, Hayaishi O, Nakajima M, Kurihara N. Enzymatic formation of catechol from anthranilic acid. J Biol Chem. 1964;239:2204–2211. [PubMed] [Google Scholar]
- 41.Timmis K N, Pieper D H. Bacteria designed for bioremediation. Trends Biotechnol. 1999;17:201–204. doi: 10.1016/s0167-7799(98)01295-5. [DOI] [PubMed] [Google Scholar]
- 42.Trumpower B L. Cytochrome bc1complexes of microorganisms. Microbiol Rev. 1990;54:101–129. doi: 10.1128/mr.54.2.101-129.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsang H-T, Batie C J, Ballou D P, Penner-Hahn J E. Structural characterization of the mononuclear iron site in Pseudomonas cepaciaDB01 phthalate dioxygenase using X-ray absorption spectroscopy. J Biol Inorg Chem. 1996;1:24–33. [Google Scholar]
- 44.Williams P A, Murray K. Metabolism of benzoate and the methylbenzoates by Pseudomonas putida (arvilla) mt-2: evidence for the existence of a TOL plasmid. J Bacteriol. 1974;120:416–423. doi: 10.1128/jb.120.1.416-423.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamaguchi M, Fujisawa H. Purification and characterization of an oxygenase component in benzoate 1,2-oxygenase system from Pseudomonas arvillaC-1. J Biol Chem. 1980;255:5058–5063. [PubMed] [Google Scholar]
- 46.Zhang C, Huang M, Holloway B W. Mapping of ben genes of Pseudomonas aeruginosa. FEMS Microbiol Lett. 1993;112:255–260. doi: 10.1016/0378-1097(93)90609-6. [DOI] [PubMed] [Google Scholar]
- 47.Zhang C, Huang M, Holloway B W. Mapping of the ben, ant, and cat genes of Pseudomonas aeruginosa and evolutionary relationship of the ben region of P. aeruginosa and P. putida. FEMS Microbiol Lett. 1993;108:303–310. doi: 10.1016/0378-1097(93)90560-o. [DOI] [PubMed] [Google Scholar]