Abstract
The association of IFN-γ receptor 1 (IFNGR1) gene polymorphisms with tuberculosis (TB) susceptibility has not been systematically studied. We therefore conducted a meta-analysis to assess their association. Literature search was conducted in PubMed, EMBASE, Web of Science, and the Cochrane Library. Odds ratio (OR) and 95% confidence interval (CI) was pooled by the random-effect model. Statistical analyses were performed using STATA 12.0 software. Fourteen studies involved 7,699 TB cases and 8,289 controls were included in this meta-analysis. A significant association was found between the IFNGR1 rs2234711 polymorphism and TB susceptibility among Africans in dominant model (OR = 1.24, 95%CI:1.01–1.52), and among Asians in allele model (OR = 0.89, 95%CI: 0.79–0.99), homozygote model (OR = 0.82, 95%CI: 0.70–0.98) and additive model (OR = 0.90, 95%CI: 0.83–0.97). In addition, a significant association was observed between the IFNGR1 rs7749390 polymorphism and TB susceptibility among Africans in allele model (OR = 0.89, 95%CI: 0.82–0.98). No significant association was found between the IFNGR1 rs1327474 polymorphism and TB susceptibility. In summary, IFNGR1 rs2234711 polymorphism was associated with increased TB susceptibility in Africans and decreased TB susceptibility in Asians, while IFNGR1 rs7749390 polymorphism was associated with decreased TB susceptibility in Africans.
Keywords: tuberculosis, interferon gamma receptor 1, polymorphism, susceptibility, metaanalysis
Introduction
Tuberculosis (TB), a chronic infectious disease caused by the bacterium mycobacterium tuberculosis (M.TB), affects several organs, but mostly attacks the lungs (1). According to the World Health Organization, TB is associated with an estimation of 10 million incident cases (2), among which 8.7 million cases are from 30 high-burden countries (3), and causes approximately 1.3 million deaths worldwide in 2018 (2). Over the past decade, the incidence of drug-resistant TB has continued to increase. Globally, 4.6% of TB cases are multidrug resistant, and in some areas, this proportion exceeds 25% (4). Therefore, TB remains a major public health concern worldwide, especially in developing countries.
Many studies have demonstrated that TB susceptibility is partly determined by the host genetic background (5–7). In support of this, differences in the rates of TB occurrence among ethnicities and families indicate a genetic predisposition to TB susceptibility (8). Interferon-gamma (IFN-γ) is well–known to play a critical role in the activation of macrophages and dendritic cells and influences the innate immune response to certain contagious agents, including M.TB (9). The interferon-gamma receptor 1 (IFNGR1) gene encodes the ligand-binding chain (alpha) of the IFN-γ receptor, which is located on chromosome 6q23.3 (10). After binding to IFN-γ, IFNGR1 could regulate the cytokine response in Natural killer and T cells to exert their microbicidal functions (11, 12). Animal and in vitro studies have demonstrated that IFNGR1 plays a pivotal role in the progression of TB (13, 14).
Previous studies have investigated the association between genetic variants of IFNGR1 and TB susceptibility based on the hypothesis that defects or variations in IFNGR1 may result in the increased susceptibility or accelerated progression of diverse diseases, such as inflammatory and virus-associated disorders (15, 16). Three potentially functional single nucleotide polymorphisms (SNPs) of the IFNGR1 gene (rs2234711, rs1327474, and rs7749390) have been investigated to explore their impact on TB susceptibility by several studies (17–30). Nonetheless, the results did not provide consistently significant associations between these SNPs possibly due to the use of sample sizes that did not allow the detection of small genetic effects. Therefore, this meta-analysis was conducted to detect the association of the IFNGR1 rs2234711, rs1327474, rs7749390 polymorphisms with TB susceptibility.
Methods
Literature search
A comprehensive literature search was conducted in PubMed, Embase, Web of Science, and the Cochrane Library of publications up to Apr 20th, 2022. The search terms used were as follows: (interferon-γ receptor OR interferon-γ receptor 1 OR IFNGR OR IFNGR1) AND tuberculosis. Further studies were identified from the reference lists, related articles and citation lists of each of papers identified from the initial searches.
The studies included in our meta-analysis met all the following criteria: (1) studies had to assess the association between IFNGR1 gene polymorphisms rs2234711, rs1327474, rs7749390, and TB susceptibility; (2) studies had a case-control or cohort design; (3) studies provided odds ratios (ORs) and the corresponding 95% confidence intervals (CIs) for at least for one model, or provided the allele frequencies to calculate the ORs and 95% CIs. Studies were excluded for the following criteria: (1) were reviews, meta-analysis, or case-reports; (2) provided no data about genotype distribution or allele frequency or ORs; (3) used duplicated data.
Data extraction and quality assessment
The following data were extracted from the included studies: first author, publication year, country, ethnicity of participants, genotyping method, diagnosis assays for detection of TB, sample sizes in case groups and control groups, genotype distributions and allele frequency or the most fully adjusted ORs and 95% CIs. Two co-authors independently extracted the data from each study and any discrepancies were resolved via discussion with the third author.
The methodological quality of the included studies was assessed using the Newcastle-Ottawa Scale (NOS) (31). Studies with a NOS star of 0–3, 4–6, and 7–9 were considered as low, moderate, and high quality, respectively. Departure from the Hardy Weinberg equilibrium (HWE) for the control group in each study was assessed with Pearson's goodness-of-fit chi-square test with one degree of freedom or was extracted from the studies which did not provide genotype distribution.
Statistical analysis
OR was used as a measure of the association between the IFNGR1 gene polymorphisms and TB susceptibility to combine the results. We used the most fully adjusted ORs and 95%CIs from each study for meta-analysis. When the adjusted ORs and 95%CIs were not provided by the included studies, we used the gene alle frequencies to compute the crude ORs and 95%CIs. The overall ORs and corresponding 95%CIs was pooled by the random-effect model. Heterogeneity across studies was detected by the Cochran Chi-square (significance level at P < 0.10) and Higgins I2 statistic (32, 33). The Begg's test (34) and the Egger's test (35) were used to assess publication bias. We pooled the ORs of associations between each SNP (for instance, the rs2234711) with TB susceptibility for six comparison models, including the dominant model (CC+CT vs. TT), allele model (C vs. T), homozygote model (CC vs. TT), heterozygote model (CT vs. TT), recessive model (CC vs. TT+CT), and additive model (CC vs. CT vs. TT). Sensitivity analysis was performed by individually removing the included studies to assess the stability of results. Subgroup analysis was conducted to determine whether there was an association between the IFNGR1 gene polymorphism and TB susceptibility in different ethnicities. All analyses were performed using STATA statistics software (Version 12.0, Stata Corporation, College Station, Texas 77, 845 United States). The statistical tests were all two-sided with a significance level of 0.05, unless otherwise specified.
Results
Literature search
Figure 1 presents the study selection flow. A total of 387 publications were retrieved from preliminary database search. Of these, 141 articles were removed as they were duplicates, leaving 246 articles for further review. By reading titles and abstracts, 223 were excluded as they were not relevant to our study. Through reading the full text of the remaining 23 articles, we further excluded 9 articles which did not meet the inclusion criteria. Finally, we included 14 articles with 15 case-control studies for the meta-analysis (17–30).
Figure 1.
Flow diagram for identification of eligible studies for this meta-analysis.
Characteristics of included studies
The general characteristics of each included study are summarized in Table 1. In total, 15,988 participants were included across all studies with 7,699 TB cases and 8,289 controls. Eight articles (21–23, 26–30) were performed in Asians, four (17, 18, 20, 24) were performed in Africans, and two (19, 25) were conducted in Europeans. Eleven (17–25, 27, 28), eight (17, 19, 21, 22, 26, 27, 29, 30), and seven (17, 21, 23–25, 29, 30) studies reported the rs2234711, rs1327474, and rs7749390 polymorphisms, respectively. For rs2234711, the genotype distributions in the controls deviated from the HWE in two studies (18, 20); for rs1237474, the genotype distributions in the controls deviated from the HWE in one study (30); the genotype distributions in the controls were consistent with the HWE for rs7749390. The average NOS score for included studies was 7.4 (standard deviation: 1.1) (Supplemental Table 1).
Table 1.
Basic information of studies included in this analysis.
| Study | Country, ethnicity | Sample size (case/control) | Diagnosed method | Genotyping method | Type | Specimen | SNPs | HWE |
|---|---|---|---|---|---|---|---|---|
| Awomoyi et al. (17) | Gambia, African | 320/320 | Sputum smear | PCR-RFLP | PTB | Blood | rs1327474, rs2234711, rs7749390 | >0.05 |
| Bulat-Kardum (18) | Croatia, European | 244/521 | Sputum smear and Chest x-ray | PCR | TB | Blood | rs1327474, rs2234711 | >0.05 |
| Cooke et al. (19) | Gambia, Guinea Bissau, and the Republic of Conakry, African | 682/619 | Sputum smear or culture | PCR-ARMS | PTB | Blood | rs2234711 | <0.05 |
| Hwang et al. (20) | Korea, Asian | 80/80 | Sputum smear | PCR-ARMS | PTB | Blood | rs1327474, rs2234711 | >0.05 |
| He et al. (21) | China, Asian | 222/188 | Sputum smear or culture; and/or Chest x-ray | PCR | PTB or EPTB | Blood | rs1327474, rs2234711, rs7749390 | >0.05 |
| Lü et al. (22) | China, Asian | 1,434/1,412 | Sputum smear or culture; and/or Chest x-ray | PCR | PTB | Blood | rs2234711, rs7749390 | >0.05 |
| Rudko et al. (24) | Russians, European | 238/265 | NA | PCR-RFLP | Primary lung TB and severe secondary TB | Blood | rs2234711, rs7749390 | >0.05 |
| Russians, European | 331/279 | NA | PCR-RFLP | Primary lung TB and severe secondary TB | Blood | rs2234711, rs7749390 | >0.05 | |
| Naderi et al. (23) | Iran, Asian | 173/163 | Clinical symptoms, sputum smear and Chest x-ray | PCR | PTB | Blood | rs1327474, rs7749390 | >0.05 (rs7749390), <0.05 (rs1327474) |
| Shin et al. (25) | Korean, Asian | 673/592 | Clinical symptom and culture | PCR | PTB | Blood | rs1327474, rs2234711 | >0.05 |
| Mayer (26) | Ghana, African | 1,999/2,589 | Sputum smear and/or culture | PCR-HRM | PTB | Blood | rs2234711, rs7749390 | >0.05 |
| Shamsi et al. (27) | Iran, Asian | 100/100 | Culture | PCR-RFLP | PTB | Blood | rs1327474 | >0.05 |
| He et al. (28) | China, Asian | 467/503 | Clinical symptoms, sputum smear and Chest x-ray | PCR | PTB | Blood | rs1327474, rs7749390 | >0.05 |
| Ali et al. (29) | Sudan, African | 100/50 | Sputum smear | PCR-RFLP | PTB | Sputum and blood samples | rs2234711 | <0.05 |
| Wu et al. (30) | China, Asian | 636/608 | Clinical symptoms, sputum smear and Chest x-ray | iMLDR | TB | Blood | rs2234711 | >0.05 |
TB, tuberculosis; PTB, pulmonary tuberculosis; EPTB, extrapulmonary tuberculosis; PCR, polymerase chain reaction; PCR-RFLP, PCR-restriction fragment length polymorphism; PCR-ARMS, PCR-Amplification refractory mutation system; PCR-HRM, PCR-high-resolution melt; iMLDR, improved multiplex ligation detection reaction.
Association of IFNGR1 rs2234711 polymorphism with TB susceptibility
Eleven studies with 12 case-control studies (17–25, 27, 28) (6,989 cases and 7,523 controls) were included in the meta-analysis on the association between the IFNGR1 rs2234711 polymorphism and TB susceptibility. The pooled ORs indicated no significant association in dominant model (CC + CT vs. TT) (OR = 1.06, 95% CI 0.90–1.24) (Figure 2), allele model (C vs. T) (OR = 0.99, 95% CI 0.90–1.08), recessive model (CC vs. TT + CT) (OR = 1.02, 95% CI 0.94–1.11), heterozygote model (CT vs. TT) (OR = 1.05, 95% CI 0.87–1.26), homozygote model (CC vs. TT) (OR = 0.98, 95% CI 0.84–1.16), and additive model (CC vs. CT vs. TT) (OR = 1.01, 95% CI 0.91–1.11). The association of the IFNGR1 rs2234711 polymorphism with TB susceptibility was significant in Africans (dominant model: OR = 1.24, 95% CI: 1.01–1.52) and Asians (homozygote model: OR = 0.82, 95% CI: 0.70–0.98), respectively (Table 2).
Figure 2.
Forest plot of association between IFNGR1 rs2234711 polymorphism and tuberculosis susceptibility (dominant model: CC + CT vs. TT).
Table 2.
Results of association between IFNGR1 gene polymorphisms and TB susceptibility.
| Polymorphisms | Genetic models | Subgroup | n | OR (95%CI) | I2 (%) | P for heterogeneity |
|---|---|---|---|---|---|---|
| rs1327474 | Dominant model | |||||
| Total | 8 | 0.83 (0.65, 1.05) | 47.8 | 0.063 | ||
| Asian | 6 | 0.78 (0.56, 1.09) | 58.4 | 0.034 | ||
| African | 1 | 0.83 (0.44, 1.55) | NA | NA | ||
| European | 1 | 1.01 (0.73, 1.40) | NA | NA | ||
| Allele model | ||||||
| Total | 7 | 0.81 (0.61, 1.08) | 66.2 | 0.007 | ||
| Asian | 5 | 0.74 (0.48, 1.15) | 73.4 | 0.005 | ||
| African | 1 | 0.84 (0.46, 1.53) | NA | NA | ||
| European | 1 | 1.03 (0.83, 1.28) | NA | NA | ||
| Recessive model | ||||||
| Total | 7 | 1.02 (0.73, 1.43) | 0 | 0.473 | ||
| Asian | 5 | 0.79 (0.34, 1.83) | 22.9 | 0.266 | ||
| African | 1 | 0.97 (0.06, 15.60) | NA | NA | ||
| European | 1 | 1.09 (0.74, 1.61) | NA | NA | ||
| Heterozygote model | ||||||
| Total | 7 | 0.81 (0.62, 1.07) | 42.9 | 0.105 | ||
| Asian | 5 | 0.76 (0.51, 1.14) | 57 | 0.054 | ||
| African | 1 | 0.82 (0.44, 1.54) | NA | NA | ||
| European | 1 | 0.98 (0.70, 1.38) | NA | NA | ||
| Homozygote model | ||||||
| Total | 6 | 0.92 (0.52, 1.63) | 24.8 | 0.248 | ||
| Asian | 4 | 0.72 (0.23, 2.31) | 51.9 | 0.1 | ||
| African | 1 | 0.95 (0.06, 15.18) | NA | NA | ||
| European | 1 | 1.08 (0.69, 1.69) | NA | NA | ||
| Additive model | ||||||
| Total | 6 | 0.83 (0.61, 1.12) | 68.5 | 0.007 | ||
| Asian | 4 | 0.76 (0.47, 1.22) | 77.1 | 0.004 | ||
| African | 1 | 0.85 (0.47, 1.53) | NA | NA | ||
| European | 1 | 1.03 (0.83, 1.28) | NA | NA | ||
| rs2234711 | Dominant model | |||||
| Total | 11 | 1.06 (0.90, 1.24) | 64.4 | 0.002 | ||
| Asian | 5 | 0.96 (0.75, 1.23) | 77 | 0.002 | ||
| African | 3 | 1.24 (1.01, 1.52) | 0 | 0.395 | ||
| European | 3 | 1.13 (0.83, 1.54) | 53.8 | 0.115 | ||
| Allele model | ||||||
| Total | 11 | 0.99 (0.90, 1.08) | 59.7 | 0.006 | ||
| Asian | 4 | 0.89 (0.79, 0.99) | 31 | 0.226 | ||
| African | 4 | 1.03 (0.87, 1.23) | 66.8 | 0.029 | ||
| European | 3 | 1.06 (0.89, 1.26) | 27.3 | 0.253 | ||
| Recessive model | ||||||
| Total | 11 | 1.02 (0.94, 1.11) | 0 | 0.53 | ||
| Asian | 5 | 0.99 (0.86, 1.13) | 35.4 | 0.185 | ||
| African | 3 | 1.10 (0.89, 1.36) | 0 | 0.868 | ||
| European | 3 | 1.00 (0.76, 1.31) | 0 | 0.37 | ||
| Heterozygote model | ||||||
| Total | 10 | 1.05 (0.87, 1.26) | 58.2 | 0.011 | ||
| Asian | 4 | 0.88 (0.71, 1.10) | 35.6 | 0.198 | ||
| African | 3 | 1.24 (1.00, 1.54) | 0 | 0.447 | ||
| European | 3 | 1.15 (0.79, 1.65) | 62.9 | 0.068 | ||
| Homozygote model | ||||||
| Total | 10 | 0.98 (0.84, 1.16) | 25.6 | 0.208 | ||
| Asian | 4 | 0.82 (0.70, 0.98) | 0 | 0.556 | ||
| African | 3 | 1.26 (0.98, 1.62) | 0 | 0.549 | ||
| European | 3 | 1.04 (0.7, 1.40) | 0 | 0.607 | ||
| Additive model | ||||||
| Total | 10 | 1.01 (0.92, 1.11) | 45 | 0.06 | ||
| Asian | 4 | 0.90 (0.83, 0.97) | 0 | 0.511 | ||
| African | 3 | 1.13 (0.99, 1.28) | 0 | 0.489 | ||
| European | 3 | 1.06 (0.89, 1.25) | 23.2 | 0.272 | ||
| rs7749390 | Dominant model | |||||
| Total | 7 | 1.02 (0.81, 1.27) | 61.9 | 0.015 | ||
| Asian | 4 | 1.00 (0.71, 1.41) | 78.1 | 0.003 | ||
| African | 1 | 0.96 (0.65, 1.42) | NA | NA | ||
| European | 2 | 1.15 (0.79, 1.67) | 0 | 0.361 | ||
| Allele model | ||||||
| Total | 8 | 0.98 (0.88, 1.09) | 58.5 | 0.018 | ||
| Asian | 4 | 1.07 (0.88, 1.29) | 72.4 | 0.012 | ||
| African | 2 | 0.89 (0.82, 0.98) | 0 | 0.481 | ||
| European | 2 | 0.89 (0.66, 1.20) | 55.8 | 0.132 | ||
| Recessive model | ||||||
| Total | 7 | 1.02 (0.85, 1.23) | 47.2 | 0.078 | ||
| Asian | 4 | 1.16 (0.92, 1.47) | 47.9 | 0.124 | ||
| African | 1 | 0.99 (0.68, 1.45) | NA | NA | ||
| European | 2 | 0.77 (0.57, 1.05) | 19.5 | 0.265 | ||
| Heterozygote model | ||||||
| Total | 7 | 1.00 (0.78, 1.27) | 62.5 | 0.014 | ||
| Asian | 4 | 0.93 (0.66, 1.31) | 76.1 | 0.006 | ||
| African | 1 | 0.96 (0.63, 1.47) | NA | NA | ||
| European | 2 | 1.30 (0.88, 1.27) | 0 | 0.64 | ||
| Homozygote model | ||||||
| Total | 7 | 1.05 (0.83, 1.34) | 50.8 | 0.058 | ||
| Asian | 4 | 1.14 (0.77, 1.69) | 73.2 | 0.011 | ||
| African | 1 | 0.97 (0.60, 1.56) | NA | NA | ||
| European | 2 | 0.97 (0.64, 1.46) | 0 | 0.324 | ||
| Additive model | ||||||
| Total | 7 | 1.03 (0.90, 1.19) | 64.5 | 0.01 | ||
| Asian | 4 | 1.07 (0.87, 1.30) | 75.1 | 0.007 | ||
| African | 1 | 1.21 (0.92, 1.60) | NA | NA | ||
| European | 2 | 0.89 (0.67, 1.19) | 52.9 | 0.145 |
Association of IFNGR1 rs1327474 polymorphism with TB susceptibility
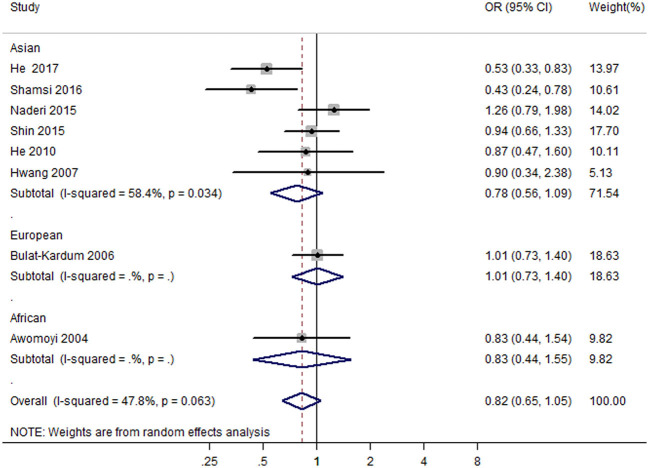
Eight case-control studies (17, 19, 21, 22, 26, 27, 29, 30) (2,279 cases and 2,467 controls) were included in the meta-analysis on the association between IFNGR1 rs1327474 polymorphism and TB susceptibility. The association of IFNGR1 rs1327474 polymorphism with TB susceptibility was not detected in the dominant model (GG + AG vs. AA) (OR = 0.83, 95% CI: 0.65–1.05) (Figure 3), allele model (G vs. A) (OR = 0.81, 95% CI 0.61–1.08), recessive model (GG vs. AG + AA) (OR = 1.02, 95% CI 0.73–1.43), heterozygote model (AG vs. AA) (OR = 0.81, 95% CI 0.62–1.07), homozygote model (GG vs. AA) (OR = 0.92, 95% CI 0.52–1.63), and additive model (GG vs. GA vs. AA) (OR = 0.83, 95% CI 0.61–1.12) (Table 2).
Figure 3.
Forest plot of association between IFNGR1 rs1327474 polymorphism and tuberculosis susceptibility (dominant model: GG + AG vs. AA).
Association of IFNGR1 rs7749390 polymorphism with TB susceptibility
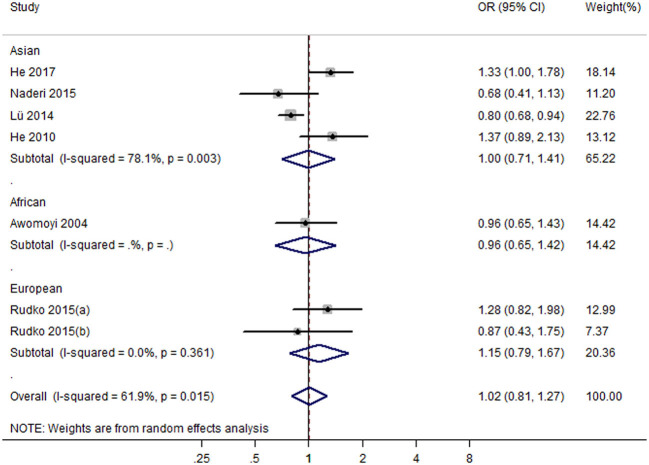
Seven studies with eight case-control studies (17, 21, 23–25, 29, 30) (5,184 cases and 5,719 controls) were included in the meta-analysis on the association between the IFNGR1 rs7749390 polymorphism and the TB susceptibility. The pooled ORs from the studies indicated no significant association in the dominant model (TT + TC vs. CC) (OR = 1.02, 95% CI 0.81–1.27) (Figure 4), allele model (T vs. C) (OR = 0.98, 95% CI 0.88–1.09), recessive model (TT vs. CC + TC) (OR = 1.02, 95% CI 0.85–1.23), heterozygote model (TC vs. CC) (OR = 1.00, 95% CI 0.78–1.27), homozygote model (TT vs. CC) (OR = 1.05, 95% CI 0.83–1.34), and additive model (TT vs. TC vs CC) (OR = 1.03, 95% CI 0.90–1.19). However, the association between the IFNGR1 rs7749390 polymorphism and TB susceptibility in the allele model was significant in Africans (OR = 0.89, 95% CI: 0.82–0.98) (Table 2).
Figure 4.
Forest plot of association between IFNGR1 rs7749390 polymorphism and tuberculosis susceptibility (dominant model: TT + TC vs. CC).
Sensitivity analysis
In the sensitivity analysis of the results from the dominant models, the influence of each individual data set on the pooled OR was assessed by removing each study individually. When excluding the results from Lu et al.'s (23) study, the pooled OR (1.13, 95% CI: 1.02–1.26) indicated an association between rs2234711 and TB susceptibility. When excluding the result from Naderi et al.'s study (30), rs1327474 was suggested to have a protective effect on TB susceptibility (OR = 0.77, 95% CI: 0.60, 0.99). For rs7749390, the sensitivity analysis demonstrated relatively robust results with an OR range from 0.95(95% CI: 0.76, 1.18) when excluding He et al.'s (21) study to 1.10 (95% CI: 0.89, 1.37) when omitting Lu et al's. (23) study.
To assess the possible effects of studies in which SNPs were not in HWE, we conducted a sensitivity analysis by omitting these studies and re-pooled the results. However, the overall ORs did not alter markedly in each model, except that rs1327474 seemed to be a protective polymorphism for TB risk in dominant model (OR = 0.77, 95% CI: 0.60–0.99).
Publication bias
The Begg's test and Egger's test were used to evaluate the publication bias of the included studies in this meta-analysis. The shape of Begg's funnel plot revealed no evidence of obvious asymmetry in all genetic models, which showed no potential publication bias (Supplementary Figures 1–3). Meanwhile, the statistical results from Egger's test and Begg's test also indicating that there was no publication bias across studies (all p > 0.05).
Discussion
To our knowledge, this is the first meta-analysis that systematically assessed the association of the IFNGR1 rs2234711, rs1327474, and rs7749390 polymorphisms with TB susceptibility. Based on 14 case-control studies with 15,988 participants, we found that IFNGR1 gene polymorphism was associated with TB susceptibility in some ethnic groups.
Various gene polymorphisms in the IFNGR1 are suggested to be associated with susceptibility of diverse diseases, such as inflammatory and virus-associated disorders (36–38). For the association of IFNGR1 gene polymorphisms with TB susceptibility, Wang et al. (39) conducted a meta-analysis focused on IFNGR1 rs2234711 polymorphism. Based on a total of six studies comprising 1,497 confirmed TB cases and 1,802 controls, Wang et al. (39) observed no significant association between IFNGR1 rs2234711 polymorphism and TB susceptibility. However, like the present meta-analysis, the association between IFNGR1 rs2234711 polymorphism and TB susceptibility was also found in Africans (dominant model: CC + TC vs. TT, OR = 1.24, 95% CI 1.02–1.51) in Wang et al.'s meta-analysis. Nevertheless, the results from the present meta-analysis were more reliable because of more included studies and larger sample size.
The rs2234711 polymorphism is located within the AP4 binding site in the 5' upstream region of the gene. This region has been associated with susceptibility to some infectious diseases, including TB (36, 37, 40). In this meta-analysis, significant association of rs2234711 polymorphism with TB susceptibility was detected in Africans (dominant model) and in Asians (homozygote model, allele model, and additive model). The result indicated that the c allele might be a potential risk factor for TB infection in Africans, whereas providing a protective effect against TB infection in Asians. Therefore, rs2234711 polymorphism may exert inversed effect on Africans and Asians. The rs7749390 polymorphism is located in an exon/intron splice site and likely to influence the intron–exon splicing process. Significant association were detected in the allele model among Africans in our study, which indicated that the T allele was associated with decreased risk for TB in the African population. The rs1327474 polymorphism, which is located in the promoter region of the IFNGR1 gene, has been shown to have higher transcriptional activity (21). He et al. reported that the rs1327474 variant was associated with a decreased risk of pulmonary TB (21). In this meta-analysis, however, similar association was only found in Africans (dominant model).
Gene polymorphisms are complicated and fluctuating, especially among different ethnicities and populations from different regions (41–43). Studies have revealed that discrepancy in distribution of IFNGR1 genotype may exist in different populations (44), which may contribute to the inconsistent associations of genetic polymorphisms with TB susceptibility. The minor allele frequency of the SNP s2234711 varies greatly in different areas, ranging from 0% in European populations to 60% in African–American populations (23). Furthermore, studies have shown that the genetic variation of the bacteria and the phylogeographic distribution of the TB-causing bacteria differed from ethnicity or populations, and patients of different ethnicity presents different TB phenotypes (45). Moreover, the burden of TB is much higher in Asia and Africa geographically than that in other regions (46–48). There is a necessity to further explore the possible effects of IFNGR1 gene polymorphisms on TB risk among more specifically defined ethnicities, such as the Caucasians, Africans, and Mongolians.
This meta-analysis indicated that Africans and Asians seem to be more sensitive to IFNGR1 gene polymorphisms compared with other ethnicities. It might be partially explained by the several factors. First, most of the original studies on TB were conducted in Asian and African populations, which provides more statistical power for detecting significant results. Second, the prevalence of TB was relatively high among Asians or Africans within the studies assessed here (49, 50), therefore more gene mutations could be detected. Third, environmental factors, such as diet, rainfall capacity, and social-economic factors in Asians or Africans may facilitate TB infection.
The present study systematically assessed the association between IFNGR1 gene polymorphisms and TB susceptibility. Strength of this meta-analysis was that the overall results were generally robust, as suggested by the subgroup analysis and sensitivity analysis. Nevertheless, some limitations should be noted. First, all included studies were case-control studies. Some included studies used matched controls (e.g., age and sex matched), while others did not. Confounding factors, such as, age, sex, HIV status, and TB severity, were not adjusted, which may lead to underestimation or overestimation of the risk estimates. Second, the number of included studies were relatively limited, and there was significant heterogeneity across studies, the geographic region, characteristics of participants, diagnosed method, genotyping method may all contribute to the heterogeneity across studies, the strength of our findings may be weakened. Third, this meta-analysis included studies (18, 20, 30) in which the SNPs were not in HWE, stability of the results could be affected, although sensitivity analysis omitting these studies (18, 20, 30) showed that the re-pooled ORs were consistent with the overall ORs.
In conclusion, this meta-analysis indicated that IFNGR1 rs2234711 polymorphism was associated with increased TB susceptibility in Africans and decreased TB susceptibility in Asians, while IFNGR1 rs7749390 polymorphism was associated with decreased TB susceptibility in Africans. This study provided evidence that IFNGR1 rs2234711 might be involved in the pathogenesis of TB in certain ethnic groups. However, further studies with larger sample size and more stringent design are warranted to confirm the present findings among different ethnic groups.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
XY and JC conceived and designed the experiments. LC, FZ, and YW performed the experiments. LC, JC, and FZ analyzed the data. LC, XY, and YW contributed reagents, materials, and analysis tools. LC and FZ wrote the paper. All authors reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2022.976221/full#supplementary-material
References
- 1.Glaziou P, Floyd K, Raviglione MC. Global epidemiology of tuberculosis. Semin Respir Crit Care Med. (2018) 39:271–85. 10.1055/s-0038-1651492 [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization . Global Tuberculosis Report 2019. Available online at: https://www.who.int/tb/publications/global_report/en/ (2019). (accessed May 23, 2022).
- 3.Furin J, Cox H, Pai M. Tuberculosis. Lancet. (2019) 393:1642–56. 10.1016/S0140-6736(19)30308-3 [DOI] [PubMed] [Google Scholar]
- 4.Lange C, Dheda K, Chesov D, Mandalakas AM, Udwadia Z, Horsburgh CR. Management of drug-resistant tuberculosis. Lancet. (2019) 394:953–66. 10.1016/S0140-6736(19)31882-3 [DOI] [PubMed] [Google Scholar]
- 5.Bellamy R. Susceptibility to mycobacterial infections: the importance of host genetics. Genes Immun. (2003) 4:4–11. 10.1038/sj.gene.6363915 [DOI] [PubMed] [Google Scholar]
- 6.Li C, Zhan Y, Li GZ, Liu SG. Association between severe preeclampsia and single nucleotide polymorphism of macrophage migration inhibitory factors−173G/C. Zhonghua Fu Chan Ke Za Zhi. (2012) 47:342–6. 10.3760/cma.j.issn.0529-567x.2012.05.006 [DOI] [PubMed] [Google Scholar]
- 7.Guo YL, Liu Y, Ban WJ, Sun Q, Shi GL. Association of mannose-binding lectin gene polymorphisms with the development of pulmonary tuberculosis in China. BMC Infect Dis. (2017) 17:210. 10.1186/s12879-017-2310-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dou HY, Chen YY, Kou SC, Su IJ. Prevalence of mycobacterium tuberculosis strain genotypes in Taiwan reveals a close link to ethnic and population migration. J Formos Med Assoc J. (2015) 114:484–8. 10.1016/j.jfma.2014.07.006 [DOI] [PubMed] [Google Scholar]
- 9.Cavalcanti YV, Brelaz MC, Neves JK, Ferraz JC, Pereira VR. Role of TNF-Alpha, IFN-Gamma, and IL-10 in the development of pulmonary tuberculosis. Pulm Med. (2012) 2012:745483. 10.1155/2012/745483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merlin G, van der Leede BJ, McKune K, Knezevic N, Bannwarth W, Romquin N, et al. The gene for the ligand binding chain of the human interferon gamma receptor. Immunogenetics. (1997) 45:413–21. 10.1007/s002510050223 [DOI] [PubMed] [Google Scholar]
- 11.Dupuis S, Döffinger R, Picard C, Fieschi C, Altare F, Jouanguy E, et al. Human interferon-gamma-mediated immunity is a genetically controlled continuous trait that determines the outcome of mycobacterial invasion. Immunol Rev. (2000) 178:129–37. 10.1034/j.1600-065X.2000.17810.x [DOI] [PubMed] [Google Scholar]
- 12.Johnson HM, Noon-Song E, Ahmed CM. Controlling nuclear jaks and stats for specific gene activation by ifn γ and other cytokines: a possible steroid-like connection. J Clin Cell Immunol. (2011) 2:1000112. 10.4172/2155-9899.1000113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jouanguy E, Lamhamedi-Cherradi S, Altare F, Fondanèche MC, Tuerlinckx D, Blanche S, et al. Partial interferon-gamma receptor 1 deficiency in a child with tuberculoid bacillus Calmette-Guérin infection and a sibling with clinical tuberculosis. J Clin Investig. (1997) 100:2658–64. 10.1172/JCI119810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altare F, Jouanguy E, Lamhamedi-Cherradi S, Fondanéche MC, Fizame C, Ribiérre F, et al. A causative relationship between mutant IFNgR1 alleles and impaired cellular response to IFNgamma in a compound heterozygous child. Am J Hum Genet. (1998) 62:723–6. 10.1086/301750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryberg A, Petersson F, Redeen S, Eriksson O, Borch K. Host gene polymorphisms in relation to helicobacter pylori infection and associated diseases in a population based cohort. Gastroenterol Res. (2013) 6:207–18. 10.4021/gr578w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhuo Y, Yang Y, Zhang M, Xu Y, Chen Z, Mu L, et al. Single nucleotide polymorphisms in IFN-γ signaling pathway associated with risk of hepatitis B virus infection in Chinese children. Can J Infect Dis Med Microbiol. (2020) 2020:8121659. 10.1155/2020/8121659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Awomoyi AA, Nejentsev S, Richardson A, Hull J, Koch O, Podinovskaia M, et al. No association between interferon-gamma receptor-1 gene polymorphism and pulmonary tuberculosis in a Gambian population sample. Thorax. (2004) 59:291–4. 10.1136/thx.2003.013029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bulat-Kardum L, Etokebe GE, Knezevic J, Balen S, Matakovic-Mileusnic N, Zaputovic L, et al. Interferon-gamma receptor-1 gene promoter polymorphisms (G-611A; T-56C) and susceptibility to tuberculosis. Scand J Immunol. (2006) 63:142–50. 10.1111/j.1365-3083.2005.01694.x [DOI] [PubMed] [Google Scholar]
- 19.Cooke GS, Campbell SJ, Sillah J, Gustafson P, Bah B, Sirugo G, et al. Polymorphism within the interferon-gamma/receptor complex is associated with pulmonary tuberculosis. Am J Respir Crit. (2006) 174:339–43. 10.1164/rccm.200601-088OC [DOI] [PubMed] [Google Scholar]
- 20.Hwang JH, Kim EJ, Kim SY, Lee SH, Suh GY, Kwon OJ, et al. Polymorphisms of interferon-gamma and interferon-gamma receptor 1 genes and pulmonary tuberculosis in Koreans. Respirology. (2007) 12:906–10. 10.1111/j.1440-1843.2007.01171.x [DOI] [PubMed] [Google Scholar]
- 21.He J, Wang J, Lei D, Ding S. Analysis of functional SNP in ifng/ifngr1 in Chinese Han population with tuberculosis. Scand J Immunol. (2010) 71:452–8. 10.1111/j.1365-3083.2010.02393.x [DOI] [PubMed] [Google Scholar]
- 22.Lü J, Pan H, Chen Y, Tang S, Feng Y, Qiu S, et al. Genetic polymorphisms of IFNG and IFNGR1 in association with the risk of pulmonary tuberculosis. Gene. (2014) 543:140–4. 10.1016/j.gene.2014.03.042 [DOI] [PubMed] [Google Scholar]
- 23.Naderi M, Hashemi M, Rezaei M, Safdari A. Association of genetic polymorphisms of IFNGR1 with the risk of pulmonary tuberculosis in Zahedan, Southeast Iran. Tuber Res Treat. (2015) 2015:292505. 10.1155/2015/292505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rudko AA, Garaeva AF, Bragina EY, Babushkina NP, Kolokolova OV, Lipaenkova ON, et al. Mutations in genes underlying atypical familial mycobacteriosis are not found in tuberculosis patients from Siberian populations. Tuberculosis (Edinburgh, Scotland). (2015) 95:204–7. 10.1016/j.tube.2015.01.002 [DOI] [PubMed] [Google Scholar]
- 25.Shin JG, Park BL, Kim LH, Namgoong S, Kim JO, Chang HS, et al. Association study of polymorphisms in interferon-γ receptor genes with the risk of pulmonary tuberculosis. Mol Med Rep. (2015) 12:1568–78. 10.3892/mmr.2015.3544 [DOI] [PubMed] [Google Scholar]
- 26.Meyer CG, Intemann CD, Förster B, Owusu-Dabo E, Franke A, Horstmann RD, et al. No significant impact of IFN-γ pathway gene variants on tuberculosis susceptibility in a West African population. Eur J Hum Genet. (2016) 24:748–55. 10.1038/ejhg.2015.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shamsi M, Zolfaghari MR, Farnia P. Association of IFN-γ and P2X7 receptor gene polymorphisms in susceptibility to tuberculosis among iranian patients. Acta Microbiol Immunol. (2016) 63:93–101. 10.1556/030.63.2016.1.7 [DOI] [PubMed] [Google Scholar]
- 28.He S, Wang B, Zhu X, Chen Z, Chen J, Hua D, et al. Association of IFNGR1 and IFNG genetic polymorphisms with the risk for pulmonary tuberculosis in the Chinese Tibetan population. Oncotarget. (2017) 8:98417–25. 10.18632/oncotarget.21413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ali AH, Omer AA, Saeed NS, Mansour EE, Elhassan MM. Influence of interferon-gamma Receptor 1 gene polymorphisms on the susceptibility to pulmonary tuberculosis among sudanese population. Int J Mycobacteriol. (2018) 7:26–31. 10.4103/ijmy.ijmy_206_17 [DOI] [PubMed] [Google Scholar]
- 30.Wu S, Wang Y, Zhang M, Wang M, He JQ. Genetic variants in IFNG and IFNGR1 and tuberculosis susceptibility. Cytokine. (2019) 123:154775. 10.1016/j.cyto.2019.154775 [DOI] [PubMed] [Google Scholar]
- 31.Wells GA, SB, O'Connell D, Peterson J, Welch V, Losos M, Tugwell P. The Newcastle-Ottawa Scale (NOS) for Assessing the Quality if Nonrandomized Studies in Meta-Analyses, 2012. Available online at: https://www.ohri.ca//programs/clinical_epidemiology/oxford.asp (accessed May 28, 2022).
- 32.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. (2002) 21:1539–58. 10.1002/sim.1186 [DOI] [PubMed] [Google Scholar]
- 33.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. (2003) 327:557–60. 10.1136/bmj.327.7414.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. (1994) 50:1088–101. 10.2307/2533446 [DOI] [PubMed] [Google Scholar]
- 35.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. (1997) 315:629–34. 10.1136/bmj.315.7109.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koch O, Awomoyi A, Usen S, Jallow M, Richardson A, Hull J, et al. IFNGR1 gene promoter polymorphisms and susceptibility to cerebral malaria. J Infect Dis. (2002) 185:1684–7. 10.1086/340516 [DOI] [PubMed] [Google Scholar]
- 37.Thye T, Burchard GD, Nilius M, Müller-Myhsok B, Horstmann RD. Genomewide linkage analysis identifies polymorphism in the human interferon-gamma receptor affecting Helicobacter pylori infection. Am J Hum Gemet. (2003) 72:448–53. 10.1086/367714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Canedo P, Corso G, Pereira F, Lunet N, Suriano G, Figueiredo C, et al. The interferon gamma receptor 1 (IFNGR1)−56C/T gene polymorphism is associated with increased risk of early gastric carcinoma. Gut. (2008) 57:1504–8. 10.1136/gut.2007.143578 [DOI] [PubMed] [Google Scholar]
- 39.Wang W, Ren W, Zhang X, Liu Y, Li C. Association between interferon gamma receptor 1-56C/T gene polymorphism and tuberculosis susceptibility: a meta-analysis. Chin Med J. (2014) 127:3782–8. [PubMed] [Google Scholar]
- 40.Martínez-Morales MC, Deswarte C, Castañeda-Casimiro J, Bustamante J, Blancas-Galicia L, Scheffler-Mendoza S. Disseminated infection by M. tuberculosis complex in patient with IFN-γ receptor 1 complete deficiency. Rev Alerg Mex. (2017) 64:499–504. 10.29262/ram.v64i4.329 [DOI] [PubMed] [Google Scholar]
- 41.Gelernter J, Kranzler H, Coccaro EF, Siever LJ, New AS. Serotonin transporter protein gene polymorphism and personality measures in African American and European American subjects. Am J Psychiatr. (1998) 155:1332–8. 10.1176/ajp.155.10.1332 [DOI] [PubMed] [Google Scholar]
- 42.Govan VA, Constant D, Hoffman M, Williamson AL. The allelic distribution of−308 Tumor Necrosis Factor-alpha gene polymorphism in South African women with cervical cancer and control women. BMC Cancer. (2006) 6:24. 10.1186/1471-2407-6-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rizzo SR, Gazito D, Pott-Junior H, Latini FR, Castelo A. Prevalence of IFNL3 gene polymorphism among blood donors and its relation to genomic profile of ancestry in Brazil. Braz J Infect Dis. (2016) 20:619–22. 10.1016/j.bjid.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosenzweig SD, Schäffer AA, Ding L, Sullivan R, Enyedi B, Yim JJ, et al. Interferon-gamma receptor 1 promoter polymorphisms: population distribution and functional implications. Clin Immunol. (2004) 112:113–9. 10.1016/j.clim.2004.03.018 [DOI] [PubMed] [Google Scholar]
- 45.Jagielski T, Minias A, van Ingen J, Rastogi N, Brzostek A, Zaczek A, et al. Methodological and clinical aspects of the molecular epidemiology of mycobacterium tuberculosis and other mycobacteria. Clin Microbiol Rev. (2016) 29:239–90. 10.1128/CMR.00055-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zielonka TM. Epidemiological situation of tuberculosis in the world based on a report of the World Health Organization 2013. Przegl Epidemiol. (2015) 69:687–91, 837–40. [PubMed] [Google Scholar]
- 47.Horton KC, MacPherson P, Houben RM, White RG, Corbett EL. Sex differences in tuberculosis burden and notifications in low- and middle-income countries: a systematic review and meta-analysis. PLoS Med. (2016) 13:e1002119. 10.1371/journal.pmed.1002119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Houben RM, Dodd PJ. The global burden of latent tuberculosis infection: a re-estimation using mathematical modelling. PLoS Med. (2016) 13:e1002152. 10.1371/journal.pmed.1002152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bishwajit G, Ide S, Ghosh S. Social determinants of infectious diseases in South Asia. Int Sch Res Notices. (2014) 2014:135243. 10.1155/2014/135243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Warren R, Ismail N, Chegou NN, Theron G, Walzl G, Malherbe ST, et al. Tuberculosis research in South Africa over the past 30 years: From bench to bedside. S Afr Med J. (2019) 109:45–52. 10.7196/SAMJ.2019.v109i11b.14249 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.