Abstract
The metabolic characteristics of Clostridium cellulolyticum, a mesophilic cellulolytic nonruminal bacterium, were investigated and characterized kinetically for the fermentation of cellulose by using chemostat culture analysis. Since with C. cellulolyticum (i) the ATP/ADP ratio is lower than 1, (ii) the production of lactate at low specific growth rate (μ) is low, and (iii) there is a decrease of the NADH/NAD+ ratio and qNADH produced/ qNADH used ratio as the dilution rate (D) increases in carbon-limited conditions, the chemostats used were cellulose-limited continuously fed cultures. Under all conditions, ethanol and acetate were the main end products of catabolism. There was no shift from an acetate-ethanol fermentation to a lactate-ethanol fermentation as previously observed on cellobiose as μ increased (E. Guedon, S. Payot, M. Desvaux, and H. Petitdemange, J. Bacteriol. 181:3262–3269, 1999). The acetate/ethanol ratio was always higher than 1 but decreased with D. On cellulose, glucose 6-phosphate and glucose 1-phosphate are important branch points since the longer the soluble β-glucan uptake is, the more glucose 1-phosphate will be generated. The proportion of carbon flowing toward phosphoglucomutase remained constant (around 59.0%), while the carbon surplus was dissipated through exopolysaccharide and glycogen synthesis. The percentage of carbon metabolized via pyruvate-ferredoxin oxidoreductase decreased with D. Acetyl coenzyme A was mainly directed toward the acetate formation pathway, which represented a minimum of 27.1% of the carbon substrate. Yet the proportion of carbon directed through biosynthesis (i.e., biomass, extracellular proteins, and free amino acids) and ethanol increased with D, reaching 27.3 and 16.8%, respectively, at 0.083 h−1. Lactate and extracellular pyruvate remained low, representing up to 1.5 and 0.2%, respectively, of the original carbon uptake. The true growth yield obtained on cellulose was higher, [50.5 g of cells (mol of hexose eq)−1] than on cellobiose, a soluble cellodextrin [36.2 g of cells (mol of hexose eq)−1]. The rate of cellulose utilization depended on the solid retention time and was first order, with a rate constant of 0.05 h−1. Compared to cellobiose, substrate hydrolysis by cellulosome when bacteria are grown on cellulose fibers introduces an extra means for regulation of the entering carbon flow. This led to a lower μ, and so metabolism was not as distorted as previously observed with a soluble substrate. From these results, C. cellulolyticum appeared well adapted and even restricted to a cellulolytic lifestyle.
Cellulose is of cardinal importance in the global carbon cycle: it accumulates in the environment due to its durable nature (5), and the main final products released during its fermentation are CH4 and CO2 (76). Bacteria are the major cellulose hydrolyzers in anaerobic cellulosic microbiota (35, 67), where cellulolytic clostridia play a key role (34).
The cellulose degradation process which occurs through cellulases has been studied extensively on cellulolytic clostridia, leading to the cellulosome concept (4, 6). The multienzymatic complexes found at the surface of the cells are responsible for adhesion of bacteria to cellulose fibers and allow a very efficient synergism of action of the different enzyme components (8). Genes encoding cellulases as well as the mechanism of action of the cellulosome are the subject of considerable research, while few studies have focused on the metabolic aspects of cellulose digestion by clostridia (27, 40).
Recent characterization of the carbohydrate catabolism of Clostridium cellulolyticum, a nonruminal mesophilic bacterium able to degrade crystalline cellulose, showed that (i) better control of catabolism occurred on a mineral salt-based medium (24, 48), (ii) carbon-limited and carbon-sufficient chemostats displayed major differences in regulatory responses of the carbon flow (25), and (iii) in nitrogen-limited conditions, glucose 6-phosphate (G6P) and glucose 1-phosphate (G1P) branch points play an important role in carbon flux divergence (22). These investigations, however, were performed with cellobiose, which is one of the soluble cellodextrins released during cellulolysis (56). In such investigations, the use of soluble sugars obviated the bacterial metabolic analysis on cellulose that was assumed difficult to undertake. Metabolic regulation processes found using cellobiose could differ or even be distorted from those with insoluble substrates.
While the first studies of cellulose focused mainly on C. cellulolyticum behavior, such as colonization or degradation with an insoluble substrate (19–21), recent investigations of cellulose fermentation in batch culture (12) have indicated that (i) metabolite yields depend strongly on the initial cellulose concentration and (ii) early growth arrest is linked to pyruvate overflow as in cellobiose batch culture (23).
In the last decade, efficient continuous-culture devices for growth on insoluble compounds have been developed (30, 31, 33, 37, 46, 63, 74) and used mainly to estimate the kinetics of cellulose degradation or colonization by various bacteria (1, 43, 58, 59, 71). Continuous culture is also a particularly useful and powerful tool for analyzing the physiology of microorganisms (42, 64).
The aim of this study was to investigate the carbon flow distribution and degradative characteristics of C. cellulolyticum when grown in mineral salt-based medium with cellulose, its natural substrate, in chemostat culture.
MATERIALS AND METHODS
Chemicals.
All chemicals were of highest-purity analytical grade. Unless mentioned otherwise, commercial reagents, enzymes, and coenzymes were obtained from Sigma Chemical Co., St. Louis, Mo. All gases used were purchased from Air Liquide, Paris, France.
Organism and medium.
C. cellulolyticum ATCC 35319 was originally isolated from decayed grass (52). Stocks of spores, stored at 4°C, were transferred to cellulose medium and heat shocked at 80°C for 10 min (12). Anaerobic cell cultures were subcultured once on cellulose before inoculation and growth in a bioreactor (12, 24). The defined medium used in all experiments was a modified CM3 medium (24) containing 0.37% cellulose MN301 (Macherey-Nagel, Düren, Germany).
Growth conditions.
C. cellulolyticum was grown on cellulose as the sole carbon and energy source in a mineral salt-based medium. All experiments were performed in a 1.5-liter-working-volume fermentor (LSL Biolafitte, St. Germain en Laye, France). The temperature was maintained at 34°C, and the pH was controlled at 7.2 by automatic addition of 3 N NaOH. Agitation was kept constant at 50 rpm. The inoculum was 10% by volume from an exponentially growing culture. Cells were grown in chemostat at various dilution rates, and each run was independent.
With cellulose, the chemostat system was a segmented gas-liquid continuous culture device as described by Weimer et al. (74). Modifications consisted of (i) sparging the culture medium with sterile oxygen-free N2; (ii) limiting oxygen entry and maintaining anaerobic culture conditions with connection of low-gas-permeability PharMed, Viton, or glass; (iii) setting up the T-fitting device directly into the feed reservoir to allow partitioning of the slurry into discrete liquid bubbles of N2 as soon as the medium was pumped, thus avoiding any cellulose sedimentation in the tube connecting the inside and outside of the reservoir of the cellulose-containing medium; (iv) permitting accurate and uniform dispensing of slurry by using cellulose MN301, which does not require any dry sieving prior to use due to its original small particle size (<45 μm). Microbial contamination was monitored regularly by microscopic observation. Achievement of steady-state values for both residual cellulose concentration and biomass required five to six dilutions. The cultures were maintained for an overall period of eight to nine residence times. Culture samples were removed at 6- to 30-h intervals; for each condition, the data were the average from at least three samples collected over 2- to 8-day periods in the steady state of the system.
Analytical procedures.
Biomass was estimated by bacterial protein measurement (46) using the Bradford dye method (10) as previously described (12).
Cellulose concentration was determined as described by Huang and Forsberg (29), using a washing procedure (69) and quantification by the phenol-sulfuric acid method (13) as already reported (12).
The relative crystallinity index of the cellulose was determined by the procedure of Shi and Weimer (59).
Hydrogen and carbon dioxide were analyzed on a gas chromatography unit as previously described (24).
Culture supernatants (10,000 × g, 15 min, 4°C) were stored at −80°C until analysis.
Extracellular proteins and amino acids were assayed as previously reported (22–25), using the Bradford dye method (10) and the procedure of Mokrasch (41), respectively.
Glucose was assayed enzymatically, using glucose oxidase and peroxidase with o-dianisidine as a chromophore.
Soluble cellodextrins were quantitatively assayed by high-performance liquid chromatography (HPLC) using refractive index detector and qualitatively using thin-layer chromatography (TLC) as already described (22).
Glycogen determination were performed using amyloglucosidase (EC 3.2.1.3) according to the procedure of Matheron et al. (38) as previously indicated (22).
Acetate, ethanol, lactate, and succinate were estimated by using the appropriate enzyme kits (Boehringer Mannheim, Meylan, France).
Extracellular pyruvate was assayed enzymatically by fluorometric detection of NADH as previously described (24).
Enzyme assays.
Cells were centrifuged (12,000 × g, 15 min, 0°C), and pellets were rapidly frozen with liquid nitrogen and stored at −80°C. Cells were resuspended in Tris-HCl buffer (50 mM, 2 mM dithiothreitol [DTT] [pH 7.4]) and then sonicated four times for 20 s each with a break of 60 s at a frequency of 20 kilocycles s−1. The supernatant was collected from the cell lysate following centrifugation (12,000 × g, 20 min, 4°C). The protein content of extracts was determined by the method of Bradford (10), using crystalline bovine serum albumin as the standard. Anaerobic conditions were maintained throughout the entire procedure, and all manipulations were performed under oxygen-free nitrogen atmosphere. All enzyme assays were performed at 34°C. Specific activity was determined in a range where linearity with protein concentration was established. For calculation of enzymatic activity, the molar extinction coefficients used for 5,5′-dithiobis-(2-nitrobenzoic acid), methyl viologen, and NAD(P)H were 13.6 (15), 7.71 (50), and 6.22 mM−1 cm−1 (55), respectively.
The phosphoglucomutase (PGM; EC 5.4.2.2) assay was based on the method of Lowry and Passonneau (36). The reaction mixture contained 50 mM Tris-HCl (pH 7.5), 20 mM DTT, 10 mM MgCl2, 1 mM AMP, 1 mM NAD+, 2 mM G1P, 3 μM glucose 1,6-diphosphate, and 4 U glucose 6-phosphate dehydrogenase (EC 1.1.1.49).
Glyceraldehyde 3-phosphate dehydrogenase (GAPDH; EC 1.2.1.12) activity was determined by the method of Ferdinand (16).
Pyruvate-ferridoxin oxidoreductase (PFO; EC 1.2.7.1) was assayed as described by Meinecke et al. (39). The reaction mixture contained 25 mM potassium phosphate buffer (pH 7.2), 0.14 mM sulfhydryl coenzyme A (CoA), 5 mM pyruvate, and 1 mM methyl viologen as the artificial electron acceptor.
Lactate dehydrogenase (LDH; EC 1.1.1.27) activity was determined as described elsewhere (32) in an assay mixture containing 20 mM potassium phosphate buffer (pH 7.4), 0.4 mM NADH, 1 mM fructose 1,6 diphosphate, and 20 mM pyruvate.
Phosphotransacetylase (PTA; EC 2.3.1.8) activity was measured as described by Andersch et al. (2).
Acetate kinase (AK; EC 2.7.2.1) activity was determined by coupling hexokinase (EC 2.7.1.1) and glucose 6-phosphate dehydrogenase (EC 1.1.1.49) and by following the NADPH-dependent oxidation of G6P to 6-phosphogluconate at 340 nm (32).
Acetaldehyde dehydrogenase (AADH; EC 1.2.1.10) was assayed as described by Dürre et al. (14). The reaction mixture contained 0.1 M Tris-HCl (pH 7.2), 2 mM DTT, 72 mM semicarbazide, 0.2 mM NADH, and 0.6 mM acetyl-CoA.
The alcohol dehydrogenase (ADH; EC 1.1.1.1) assay was based on the protocol described by Lamed and Zeikus (32). The assay mixture contained 100 mM potassium phosphate buffer (pH 7.4), 0.2 mM NADH, 0.2 mM DTT, and 40 mM acetaldehyde.
To determine the Km of PGM for G1P, a crude extract was dialyzed anaerobically against Tris-HCl buffer (50 mM, pH 7.5) containing 4 mM 2-mercaptoethanol. PGM activity was measured as described above; to determine Km, the G1P concentrations were varied from 0.02 to 20 mM.
Assay of intracellular compounds.
NAD(P)+ and NAD(P)H were first rapidly extracted with HCl and KOH, respectively, as described by Wimpenny and Firth (75). Levels of coenzymes were determined by fluorimetric measurements as previously described (24). Other intracellular compounds were extracted from a broth sample by HClO4, using a rapid extraction system (24).
ATP, ADP, and AMP were measured using a luciferin-luciferase luminescence system (microbial biomass test kit; Celsis Lumac, Landgraaf, The Netherlands) (22).
CoA and acetyl-CoA were measured by coupling appropriate enzyme assays (68) for fluorimetric determination of NADH. The assay mixture contained 50 mM phosphate buffer (pH 7.5), 1 mM MgCl2, 1 mM NAD+, 0.5 mM EDTA, 1 mM 2-ketoglutarate, and 0.1 U of 2-ketoglutarate dehydrogenase complex from pig heart to initiate CoA consumption. After complete depletion of the CoA in the extract, 2 U of citrate synthase (EC 4.1.3.7) and 4 mM oxaloacetate were added to measure the acetyl-CoA concentration.
Fluorimetric determination of G1P and G6P was based on the PGM assay described above, using glucose 6-phosphate dehydrogenase (EC 1.1.1.49) and PGM (EC 5.4.2.2) as previously described (22).
Calculations.
The metabolic pathways and equations for cellulose fermentation by C. cellulolyticum, expressed as n hexose equivalents corresponding to n glucose residues of the cellulose chain, were previously reported (12).
qcellulose is the specific rate of hexose residue fermented in millimoles per gram of cells per hour. qacetate, qethanol, and qlactate are the specific rates of product formation in millimoles per gram of cells per hour. qextracellular pyruvate is the specific rate of extracellular pyruvate formation in micromoles per gram of cells per hour. qpyruvate is the specific rate of pyruvate used in millimoles per gram of cells per hour, determined as follows: qpyruvate = qacetate + qethanol + qlactate + qextracellular pyruvate. qNADH produced and qNADH consumed, the specific rates of NADH production and NADH consumption, respectively, in millimoles per gram of cells per hour, were calculated as follows: qNADH produced = qpyruvate and qNADH consumed = 2 qethanol + qlactate.
The energetic yield of biomass (YATP) was calculated as follows: YATP = concnbiomass/(1.94 concnacetate + 0.94 concnethanol + 0.94 concnlactate + 0.94 concnextracellular pyruvate) (12). YATP is expressed in grams of cells per mole of ATP produced. qATP is the specific rate of ATP generation in millimoles per gram of cells per hour calculated by the following equation: qATP = 1.94 qacetate + 0.94 qethanol + 0.94 qlactate + 0.94 qextracellular pyruvate (12). The energetic efficiency (ATP-Eff) corresponding to ATP generation in cellulose catabolism is given by the ratio of qATP to qcellulose.
The energetic charge and oxidation/reduction (O/R) index were calculated according to Gottschalk (26).
The catabolic reduction charge (CRC) and anabolic reduction charge (ARC) were calculated as follows (3): CRC = NADH/(NADH + NAD+) and ARC = NADPH/(NADPH + NADP+).
The molar growth yield (YX/S) is expressed in grams of cells per mole of hexose equivalents fermented. The Pirt plot was used for determination of maximum yield and maintenance coefficient with the following equation (53): 1/Y = 1/Ymax + m tR, where Ymax is the true yield (in the absence of maintenance requirements), Y the observed yield, m is the maintenance coefficient, and tR is the retention time (i.e., tR = 1/D, where D is the dilution rate).
The first-order rate constant of cellulose removal was determined using the model equation (47) SR/S0 = k tR + x, where SR is the concentration of cellulose in the feed reservoir, S0 is the concentration of cellulose in the culture vessel, k corresponds to the rate constant of cellulose degradation, and x is equal to 1 since it corresponds to the y intercept (i.e., tR = 0 and thus SR/S0 = 1).
Mapping of the carbon flow.
Distribution of the carbon flow was determined by adapting the model developed by Holms (28) to C. cellulolyticum metabolism. In the steady state, the flux through each enzyme of the known metabolic pathway was determined as specified in Table 1. Carbon fluxes were expressed in milliequivalents of carbon per gram of cells per hour.
TABLE 1.
Calculations for analysis of the carbon flow during cellulose fermentation by C. cellulolyticum
| Stepa | Specific metabolic rate [meq of C (g of cells)−1 h−1] | Equationb |
|---|---|---|
| 1 | qcellulose | (S/X) × D |
| 2 | qlactate | (Clactate/X) × D |
| 3 | qacetate | (Cacetate/X) × D |
| 4 | qethanol | (Cethanol/X) × D |
| 5 | qextracellular pyruvate | (Cextracellular pyruvate/X) × D |
| 6 | qbiosynthesis from glycolysis | [(Cbiomass − Cglycogen + Camino acid + Cprotein)/X] × D |
| 7 | qCO2 | [(3) + (4)]/2 |
| 8 | qpyruvate | (2) + (3) + (4) + (5) + (7) |
| 9 | qacetyl-CoA | (3) + (4) |
| 10 | qG1P | (1) × 0.63 |
| 11 | qcellulose toward G6P | (1) × 0.37 |
| 12 | qG6P | (6) + (8) |
| 13 | qPGM | (12) − (11) |
| 14 | qglycogen | (Cglycogen/X) × D |
| 15 | qexopolysaccharide | (10) − (13) − (14) |
| 16 | qG1P toward glycogen and exopolysaccharides | (14) + (15) |
Step numbers are diagrammed in Fig. 3.
S is the concentration of cellulose consumed, expressed as millequivalents of C per liter; X is the biomass concentration at steady state, expressed as grams per liter; D is the dilution rate; C is milliequivalents of C per liter from by-products or biomass.
It was assumed that the intracellular β-glucan composed of n hexose residues was catabolized according to the model proposed by Strobel (62) as previously described (12). If n ≥ 2, then β-glucan (n) + Pi → G1P + β-glucan (n − 1), through cellodextrin and cellobiose phosphorylase. If n = 1, then glucose + ATP → G6P + ADP, via glucokinase. As a result, the entering carbon flow directed toward G6P was  qcellulose and that toward G1P was
qcellulose and that toward G1P was 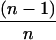 qcellulose. For example, if n = 1 (glucose) for the entering carbohydrate, then the qcellulose toward G6P is equal to qcellulose and that toward G1P is nil since no G1P can be formed. If n = 2 (cellobiose) for the entering β-glucan, then the qcellulose toward G6P is equal to
qcellulose. For example, if n = 1 (glucose) for the entering carbohydrate, then the qcellulose toward G6P is equal to qcellulose and that toward G1P is nil since no G1P can be formed. If n = 2 (cellobiose) for the entering β-glucan, then the qcellulose toward G6P is equal to 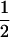 qcellulose and that toward G1P is
qcellulose and that toward G1P is 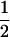 qcellulose since 1 glucose and 1 G1P are formed. If n = 3 (cellotriose) for the entering soluble biopolymer, then the qcellulose toward G6P is equal to
qcellulose since 1 glucose and 1 G1P are formed. If n = 3 (cellotriose) for the entering soluble biopolymer, then the qcellulose toward G6P is equal to 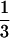 qcellulose and that toward G1P is
qcellulose and that toward G1P is 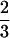 qcellulose since 1 glucose and 2 G1P are formed. For soluble β-glucans (n), where 1 ≤ n ≤ 7 (12, 51), average carbon flows directed toward G6P of 0.37 qcellulose and toward G1P of 0.63 qcellulose were calculated.
qcellulose since 1 glucose and 2 G1P are formed. For soluble β-glucans (n), where 1 ≤ n ≤ 7 (12, 51), average carbon flows directed toward G6P of 0.37 qcellulose and toward G1P of 0.63 qcellulose were calculated.
The turnover of a pool per hour was calculated from the specific rate and pool size expressed in moles or in carbon equivalents (22). It corresponded to the rate of input or output divided by the pool size, which is then the number of times the pool turns over every hour.
R corresponded to the ratio of specific enzyme activity to metabolic flux (28). Like specific enzyme activity, metabolic flux was expressed in micromoles per milligram of protein per minute from carbon flow calculated as described above.
Statistics.
Statistical analysis of the data was performed following analysis of variance and Student t test (9) with Excel.
RESULTS
Cellulose digestion and biomass formation.
Preliminary results in chemostats indicated that with between 2 and 6 g of cellulose liter−1 and at a dilution rate of 0.025 h−1, biomass formation increased proportionately to cellulose concentration (data not shown). C. cellulolyticum was then grown with 3.7 g of cellulose liter−1 at different steady-state combinations of D—equal to the microbial specific growth rate (μ)—which ranged from 0.014 to 0.083 h−1 (Table 2); steady-state growth on cellulose was attained neither at D < 0.01 h−1 nor at D > 0.09 h−1 since washout then occurred. The latter value corroborated the maximum growth rate (μmax) of 0.087 h−1 (i.e., a generation time of 8 h) estimated by following growth with the rate of tritiated thymidine incorporation into DNA during the exponential phase of batch growth on cellulose (20). Cell density was maximum at low D, i.e., 0.207 g liter−1 at 0.014 h−1, but slowly decreased with increasing D to reach 0.154 g liter−1 at 0.083 h−1 (Table 2). In all of the runs, microscopic examinations of the cultures revealed that almost all of the cellulose fibers were colonized by bacteria; the few particles that were not were most likely those that had been recently introduced into the bioreactor (74). Unattached cells were mostly observable under conditions of low D.
TABLE 2.
Fermentation parameters from continuous steady-state culturesa of C. cellulolyticum
| Parameter | Results obtained at D (h−1) ofb:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 0.014 | 0.027 | 0.035 | 0.041 | 0.055 | 0.064 | 0.076 | 0.083 | |
| Biomass (g liter−1) | 0.207 ± 0.019 | 0.212 ± 0.018 | 0.204 ± 0.014 | 0.199 ± 0.012 | 0.196 ± 0.015 | 0.185 ± 0.013 | 0.172 ± 0.011 | 0.154 ± 0.009 |
| Consumed cellulose (mmol of hexose eq) | 17.59 ± 0.85 | 12.29 ± 0.59 | 10.06 ± 0.52 | 8.81 ± 0.45 | 6.98 ± 0.33 | 6.18 ± 0.31 | 5.21 ± 0.25 | 4.70 ± 0.23 |
| qcellulose [mmol (g of cells)−1 h−1] | 1.19 | 1.57 | 1.73 | 1.82 | 1.96 | 2.14 | 2.30 | 2.53 |
| qpyruvate [mmol (g of cells)−1 h−1] | 1.94 | 2.49 | 2.62 | 2.74 | 2.84 | 2.96 | 3.15 | 3.39 |
| Product yield (%) of qpyruvate | ||||||||
| Acetate | 72.9 | 71.0 | 69.5 | 67.8 | 66.4 | 65.4 | 64.1 | 60.8 |
| Ethanol | 24.9 | 26.9 | 28.8 | 30.4 | 32.0 | 33.1 | 34.5 | 37.6 |
| Lactate | 1.9 | 1.8 | 1.5 | 1.6 | 1.4 | 1.4 | 1.3 | 1.3 |
| qextracellular pyruvate [μmol (g of cells)−1 h−1] | 5.97 | 6.24 | 6.87 | 7.84 | 7.32 | 5.16 | 4.27 | 12.06 |
| Glycogen [mg (g of cells)−1] | 58.8 ± 1.9 | 95.2 ± 2.8 | 108.7 ± 4.1 | 105.2 ± 3.8 | 99.5 ± 3.7 | 91.8 ± 3.4 | 84.5 ± 2.5 | 79.4 ± 2.1 |
| Extracellular proteins (mg liter−1) | 54.3 ± 2.7 | 44.8 ± 1.8 | 32.3 ± 1.6 | 19.4 ± 0.9 | 18.5 ± 0.8 | 17.4 ± 1.1 | 10.3 ± 0.7 | 13.8 ± 0.5 |
| Free amino acids (mg liter−1) | 138.9 ± 7.1 | 95.5 ± 4.9 | 88.1 ± 4.7 | 76.8 ± 4.1 | 63.2 ± 3.5 | 59.5 ± 3.7 | 48.4 ± 3.1 | 38.4 ± 2.2 |
| YX/S [g of cells (mol of hexose eq)−1] | 11.8 | 17.3 | 20.3 | 22.6 | 28.1 | 29.9 | 33.0 | 32.8 |
| Carbon recovery (%) | 96.5 | 98.3 | 97.1 | 97.6 | 98.8 | 97.2 | 97.7 | 95.8 |
The cellulose input was 0.37% (wt/vol), and ammonium was added at 15.13 mM.
Where indicated, values are averages of samples at steady-state ± standard deviations. All other values were determined with an average accuracy of ±10%.
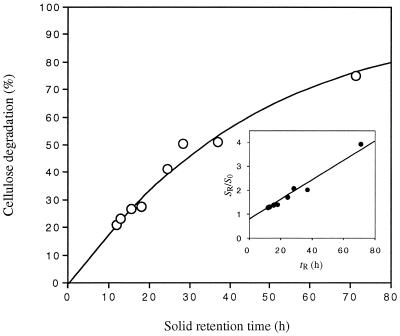
In continuous culture at 3.7 g of cellulose liter−1, C. cellulolyticum always left some undigested cellulose (Fig. 1). The longer was the solid retention time (tR = 1/D), the higher was the percentage of cellulose degradation: from 0.014 h−1 (i.e., tR = 71.4 h) to 0.083 h−1 (i.e., tR = 12.0 h), the percentages of digested cellulose at steady state were 75.0 and 20.9, respectively (Fig. 1). Regardless of D, glucose or cellodextrins could not be detected in the supernatant using enzymatic, HPLC, and TLC techniques, and so cellulose hydrolysis did not yield a significant pool of soluble sugars. Crystallinity measurements showed that the relative crystallinity index of cellulose in chemostat at 0.014 h−1 was 88.8, compared to 89.6 for the original cellulose MN301. Thus, even at long tR, the residual cellulose was not enriched in its crystalline content during the fermentation. Plots of SR/S0 versus tR were linear, and so cellulose digestion follows first-order kinetics where linear regression of the data (r2 = 0.976) gives a first-order rate constant for cellulose removal of 0.05 h−1 (Fig. 1).
FIG. 1.
Effect of solid tR on cellulose digestion by C. cellulolyticum. Inset, correlation between SR/S0 and tR.
The observed cell yields increased with increasing D and varied from 11.8 to 32.8 g of cells per mol of hexose eq consumed (Table 2). From the observed growth yield, which is affected by microbial endogenous metabolism and maintenance energy requirements, the true growth yield was calculated. A Pirt plot (r2 = 0.987) allowed determining the maintenance coefficient and YX/Smax, which were estimated at 0.9 mmol of hexose eq/g of cells/h and 50.5 g of biomass/mol of hexose eq consumed, respectively.
Metabolite production.
Acetate, ethanol, lactate, H2, and CO2 were the primary metabolic end products, and no succinate accumulation was observed (Table 2). The percentage of carbon flow toward fermentative metabolites, given by the ratio qpyruvate/qcellulose, indicated that 67.0 to 81.5% of the consumed cellulose was converted to extracellular pyruvate, lactate, CO2, acetate, and ethanol. The remaining carbon flow was oriented toward amino acid, protein, biomass, and exopolysaccharide formation. Exopolysaccharides could not be measured as previously described (48) because of significant interference with cellulose fibers leading to erroneous estimation of their concentrations, but they were readily observable by microscopic examination. The carbon balance, calculated by taking into account amino acids, proteins, fermentative end products, and biomass, was then in a range between 95.8 and 98.7%.
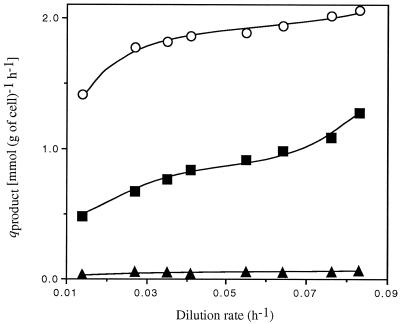
qacetate and qethanol increased 1.5- and 2.6-fold, respectively, with D (Fig. 2). Acetate production represented up to 72.9% of the carbon flowing toward catabolites (Table 2) and remained the higher specific production rate (Fig. 2). Levels of lactic acid and extracellular pyruvate formed were lower, reaching maxima of only 0.04 mmol (g of cells)−1 h−1 and 12.06 μmol (g of cells)−1 h−1, respectively (Fig. 2). Thus, the majority of the carbon flow was for acetate and ethanol production, with a progressive shift toward ethanol production with increasing D.
FIG. 2.
Influence of D on qacetate (○), qethanol (■), and qlactate (▴).
Fermentation balance.
Formulation of a reducing equivalent balance equation requires a fair knowledge of the biochemistry of carbon assimilation for a particular substrate (44, 45). The reactions leading to the formation of catabolites during the fermentation of cellulose by C. cellulolyticum were previously described (12). Cellulose hydrolysis liberates soluble sugars, which are then metabolized by bacteria and allow cell growth. The hexose residues of a β-1,4 polymer of glucose have a mean
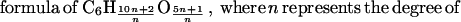 |
polymerization, i.e., the number ofglucose residues inside the biopolymer. From cellulose degradation, it was assumed that soluble cellodextrins with a degree of polymerization n between 1 and 7 could potentially be incorporated and fermented by the cells (12, 51, 56). This corresponds to an average formula of hexose equivalents from glucose to celloheptaose utilized by bacteria of C6.0H10.7O5.4.
The synthesis of biomass—with an elemental composition of C4H7NO2 (24)—from cellulose digestion can be represented by the overall scheme C6.0H10.7O5.4 + 1.5 NH3 → 1.5 C4H7NO2 + 2.4 H2O. Since no available hydrogen atoms were used to equilibrate the equation, this meant that when bacteria grow on cellulose, reducing equivalents NAD(P)H are well balanced by biomass synthesis. It was assumed that CO2 production through general decarboxylation enzymes and CO2 fixation in biomass gave a balance of almost nil (24, 44, 45).
Energetic and redox balance.
The stoichiometry of ATP generated over hexose equivalents fermented, i.e., ATP-Eff, was higher at low D since it was 2.72 and declined to 2.07 with the highest D value (Table 3) due to the decrease of the percentage of acetate production (Table 2). As D increased, qATP rose from 3.24 to 5.25 mmol (g of cells)−1 h−1 (Table 3). The apparent energetic yield increased with increasing D as well (Table 3). The low YATP obtained at low μ reflected an expenditure of energy due to the more pronounced maintenance requirement at low μ. The Pirt plot (r2 = 0.984) permitted determining a true energetic yield (YATPmax) of 30.3 g of cells (mol of ATP)−1 and a maintenance energy (mATP) estimated at 2.9 mmol of ATP (g of cells)−1 h−1.
TABLE 3.
Adenylate content and energetic balance of C. cellulolyticum cells at steady state
| Parameter | Results obtained at D (h−1) ofa:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 0.014 | 0.027 | 0.035 | 0.041 | 0.055 | 0.064 | 0.076 | 0.083 | |
| Pool ATP + ADP [μmol (g of cells)−1] | 4.41 | 4.55 | 5.18 | 5.83 | 5.42 | 6.05 | 5.98 | 6.91 |
| ATP/ADP ratio | 0.37 | 0.45 | 0.40 | 0.35 | 0.39 | 0.36 | 0.44 | 0.46 |
| AMP [μmol (g of cells)−1] | 0.10 ± 0.02 | 0.05 ± 0.01 | 0.09 ± 0.02 | 0.14 ± 0.03 | 0.12 ± 0.02 | 0.07 ± 0.01 | 0.11 ± 0.01 | 0.08 ± 0.02 |
| Energetic charge | 0.62 | 0.65 | 0.63 | 0.61 | 0.63 | 0.62 | 0.64 | 0.65 |
| qATP [mmol (g of cells)−1 h−1] | 3.24 | 4.11 | 4.29 | 4.44 | 4.56 | 4.72 | 4.98 | 5.25 |
| ATP-Eff | 2.72 | 2.62 | 2.48 | 2.44 | 2.33 | 2.21 | 2.16 | 2.07 |
| YATP [g of cells (mol of ATP)−1] | 4.3 | 6.6 | 8.2 | 9.2 | 12.1 | 13.6 | 15.3 | 15.8 |
See Table 2, footnote b.
Whatever the μ, the pool of ATP and ADP rose while AMP remained quite constant at ca. 0.10 μmol (g of cells)−1. The ratio ATP/ADP fluctuated between 0.37 and 0.46, while a mean value of 0.63 was obtained for the adenylate energy charge.
From the known catabolic pathways which produced and consumed reducing equivalents, the coenzyme balance could be calculated. qNADH produced increased with increasing D, from 1.94 to 3.39 mmol (g of cells)−1 h−1, as did qNADH used, which ranged from 0.99 to 2.59 mmol (g of cells)−1 h−1 (Table 4). qNADH produced/qNADH used, however, ranged from 1.96 to 1.31, indicating an excess of NADH since the ratio was always greater than 1. Despite this imbalance, the intracellular ratio NADH/NAD+ was always lower than 1 and the CRC was constant at around 0.27 (Table 4). This result correlated with H2/CO2 ratios which were always higher than 1. These data suggested that the regeneration of NADH to NAD+ was due to the NADH-ferredoxin (NADH-fd) reductase and hydrogenase activities, explaining the production of additional H2 and the intracellular NADH/NAD+ ratio lower than 1 (Table 4). Taking into account the gas production ratio, the O/R index was determined as very close to 1, indicating an efficient regulating system for the reoxidation of NADH via hydrogen production in addition to carbon fermentative pathways (23, 24).
TABLE 4.
Pyridine nucleotide content and redox balance for continuous steady-state cultures of C. cellulolyticum
| Parameter | Results obtained at D (h−1) ofa:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 0.014 | 0.027 | 0.035 | 0.041 | 0.055 | 0.064 | 0.076 | 0.083 | |
| NADH [μmol (g of cells)−1] | 3.39 ± 0.68 | 4.44 ± 0.89 | 4.65 ± 0.91 | 4.79 ± 0.95 | 4.83 ± 0.98 | 5.32 ± 1.07 | 5.68 ± 1.12 | 6.11 ± 1.19 |
| NAD+ [μmol (g of cells)−1] | 8.44 ± 1.73 | 10.05 ± 2.03 | 12.08 ± 2.41 | 13.11 ± 2.64 | 15.74 ± 2.78 | 16.16 ± 3.23 | 16.53 ± 3.32 | 19.45 ± 3.91 |
| NADPH [μmol (g of cells)−1] | 7.47 ± 1.68 | 8.04 ± 1.89 | 5.89 ± 1.19 | 5.65 ± 1.13 | 4.74 ± 0.94 | 4.43 ± 0.89 | 3.82 ± 0.87 | 4.26 ± 0.95 |
| NADP+ [μmol (g of cells)−1] | 0.11 ± 0.03 | 0.08 ± 0.02 | ND | ND | ND | ND | ND | ND |
| CRC | 0.29 | 0.31 | 0.28 | 0.27 | 0.23 | 0.25 | 0.26 | 0.24 |
| ARC | 0.99 | 0.99 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| qNADH produced [mmol (g of cells)−1 h−1] | 1.94 | 2.49 | 2.62 | 2.74 | 2.84 | 2.96 | 3.15 | 3.39 |
| qNADH used [mmol (g of cells)−1 h−1] | 0.99 | 1.38 | 1.55 | 1.71 | 1.86 | 1.99 | 2.21 | 2.59 |
| qNADH fd [mmol (g of cells)−1 h−1] | 0.94 | 1.11 | 1.07 | 1.03 | 0.98 | 0.96 | 0.94 | 0.80 |
| H2/CO2 ratio | 1.65 | 1.56 | 1.58 | 1.51 | 1.38 | 1.41 | 1.35 | 1.29 |
| O/R index | 0.93 | 0.95 | 0.92 | 0.94 | 0.99 | 0.96 | 0.98 | 0.97 |
See Table 2, footnote b. ND, not detectable.
Concerning the phosphopyridine nucleotides involved in biosynthesis pathways, the reduced form was in excess since the NADP+ pool was hardly detectable (Table 4). As a result, the ARC was constant and equal to 1.00, meaning that NADPH was largely available for biosynthesis reaction.
Kinetic analysis of cellulose fermentation.
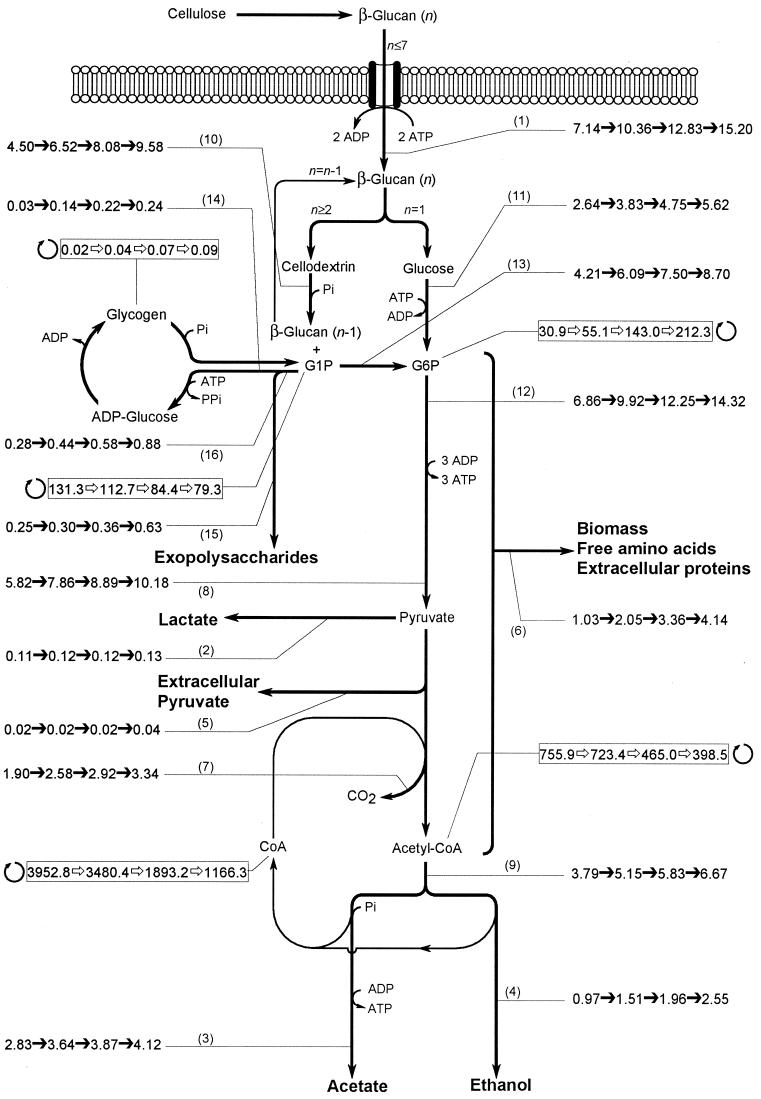
The effects of D on cellulose consumption and product formation are summarized in Fig. 3. The rate of cellulose consumption varied from 7.14 to 15.20 meq of C (g of cells)−1 h−1 with increasing D.
FIG. 3.
Distribution of carbon flux within the central metabolic pathways based on steady-state kinetics from continuous cultures of C. cellulolyticum grown on cellulose at D = 0.014, 0.035, 0.064, and 0.083 h−1. Carbon flow (➞) and turnover (➱) were calculated as specified in Materials and Methods and according to the values in Tables 1 and 2. Fluxes through metabolic pathways are expressed as millequivalents of C per gram of cells per hour and are indicated by thin lines where numbers in parentheses refer to step numbers in Table 1. Turnover per hour is indicated by a thin line connected to a box with a rolling circle symbolizing the turnover of the pool.
Carbon conversion to biomass, extracellular proteins, and free amino acids increased from 1.03 to 4.14 meq of C (g of cells)−1 h−1 (Fig. 3) and reached 27.3% of the original carbon uptake at D = 0.083 h−1. Most of the carbon used for these biosyntheses, however, was converted to biomass since regardless of D, both extracellular proteins and free amino acids represented a constant proportion of the original carbon, around 7.5%.
The cellulose catabolism leading to pyruvate from which fermentation end products were formed—i.e., acetate, CO2, extracellular pyruvate, ethanol, and lactate—increased from 5.82 to 10.18 meq of C (g of cells)−1 h−1 (Fig. 3), yet the percentage of the entering carbon used for the end product formation decreased from 81.5 to 67.0% with increasing growth rate. qacetate and qethanol increased from 2.83 to 4.12 and from 0.97 to 2.55 meq of C (g of cells)−1 h−1, respectively. However, when expressed as a percentage of qcellulose, the flux through the acetate pathway decreased from 39.6 to 27.1%, while through the ethanol production route this percentage rose from 13.5 to 16.8. qlactate increased only from 0.11 to 0.13 meq of C (g of cells)−1 h−1 and represented a small portion of the carbon uptake since it dropped from 1.5 to 0.9%. The specific rate of extracellular pyruvate formation ranged from 0.02 to 0.04 mmol (g of cells)−1 h−1, and this leak toward the outside of the cells bottomed out at 0.2% of the carbon uptake.
Another part of the entering carbon flow was directed toward exopolysaccharide synthesis (Fig. 3), which increased from 0.25 to 0.63 meq of C (g of cells)−1 h−1 and represented up to 4.2% of the specific consumption rate of cellulose.
Intracellular pool of hexose phosphate and CoA derivative.
The G6P pool was fueled by the carbon flow from glucokinase activity and by PGM activity from the G1P pool (Fig. 3). At this metabolic node, the G6P pool decreased with increasing growth rate (Table 5), indicating that the turnover of the pool increased and actually reached 212.3 h−1 at μ = 0.083 h−1 (Fig. 3). This variation was correlated with the increase of the carbon flow through glycolysis and biosynthesis metabolic pathways with higher μ. The qG6P increased from 6.86 to 14.32 meq of C (g of cells)−1 h−1 with higher D (Fig. 3); whatever the D, it represented a mean of 96.0% of the carbon uptake. The G1P pool rose from 5.71 to 20.12 μmol (g of cells)−1 with D (Table 5) and thus corresponded to a decrease in the turnover of the pool (Fig. 3). As a result, the G6P/G1P ratio ranged from 6.48 to 0.56 with increasing μ (Table 5).
TABLE 5.
Intracellular metabolite levels of continuous steady-state cultures of C. cellulolyticum
| Parameter | Results obtained at D (h−1) ofa:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 0.014 | 0.027 | 0.035 | 0.041 | 0.055 | 0.064 | 0.076 | 0.083 | |
| G6Pa [μmol (g of cells)−1] | 36.98 ± 1.14 | 35.76 ± 0.92 | 29.98 ± 0.79 | 26.31 ± 0.73 | 16.64 ± 0.58 | 14.28 ± 0.51 | 11.29 ± 0.62 | 11.24 ± 0.55 |
| G1Pb [μmol (g of cells)−1] | 5.71 ± 0.33 | 8.01 ± 0.40 | 9.65 ± 0.32 | 10.99 ± 0.54 | 14.16 ± 0.61 | 15.96 ± 0.57 | 18.23 ± 0.75 | 20.12 ± 0.68 |
| G6P/G1P ratio | 6.48 | 4.46 | 3.11 | 2.39 | 1.18 | 0.89 | 0.62 | 0.56 |
| Acetyl-CoA [μmol (g of cells)−1] | 2.51 ± 0.18 | 2.98 ± 0.21 | 3.56 ± 0.23 | 3.81 ± 0.25 | 5.75 ± 0.39 | 6.27 ± 0.42 | 7.06 ± 0.46 | 8.37 ± 0.56 |
| CoAc [μmol (g of cells)−1] | 0.48 ± 0.03 | 0.59 ± 0.04 | 0.74 ± 0.06 | 0.87 ± 0.05 | 1.29 ± 0.09 | 1.54 ± 0.11 | 2.12 ± 0.14 | 2.86 ± 0.19 |
| Acetyl-CoA/CoA ratio | 5.23 | 5.05 | 4.81 | 4.38 | 4.46 | 4.07 | 3.33 | 2.93 |
See Table 2, footnote b.
From the G1P junction, the carbon could be either stored as glycogen or converted to exopolysaccharide, or it could be directed more toward glycolysis via PGM (Fig. 3). Whatever the D, no cellotriose could be detected and G1P was metabolized as exopolysaccharides. With increasing D, the carbon flow via PGM increased from 4.21 to 8.70 meq of C (g of cells)−1 h−1, and the percentage of the original carbon flowing through this metabolic pathway remained quite constant, ranging from 59.0 to 57.2%. qglycogen increased from 0.03 to 0.24 meq of C (g of cells)−1 h−1 and represented up to 1.6% of the entering carbon flow. The turnover of the glycogen pool remained low since it increased from 0.02 to 0.09 times per hour with the highest D value and was correlated with increasing qpyruvate and qbiosynthesis.
The pool of CoA was formed from phosphotransacetylase and acetaldehyde dehydrogenase, and that of acetyl-CoA was formed from PFO activity (Fig. 3). At this branch point, the pools increased with μ and the acetyl-CoA/CoA ratio decreased slightly, from 5.23 to 2.93 (Table 5). The specific metabolic rate of acetyl-CoA ranged from 3.79 to 6.67 meq of C (g of cells)−1 h−1, while its turnover decreased from 755.9 to 398.5 times per hour. These facts are associated with (i) reorientation of the carbon fluxing through qbiosynthesis, increasing from 14.5 to 27.3%, at the expense of qpyruvate, decreasing from 81.5 to 67.0%; and (ii) decrease of the percentage of carbon flow through the acetate pathway, which was reoriented toward ethanol in different proportions since carbon conversion to acetate dropped from 39.6 to 27.1%, while it increased only from 13.6 to 16.8% through the ethanol pathway.
Enzymatic activities.
The effects of μ on specific activities of the enzymes are compiled in Table 6. In vitro enzyme activities were higher under conditions giving higher in vivo specific production rates. The level of GAPDH rose continuously with increasing carbon flow, indicating efficient hexose catabolism during glycolysis; from the lowest to the highest D, the GAPDH level increased 3.3-fold. From 0.014 to 0.083 h−1, PFO increased 1.7-fold, PTA increased 1.4-fold, AK increased 7.1-fold, AADH increased 3.2-fold, and ADH increased 2.9-fold.
TABLE 6.
Specific enzymatic activity and flux relative to available enzyme activity in C. cellulolyticum cell extract at steady-state growtha
| Enzymea | Results obtained at D (h−1) ofa:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 0.014
|
0.035
|
0.064
|
0.083
|
|||||
| Mean SEA [μmol min−1 (mg of protein)−1] ± SD | R | Mean SEA [μmol min−1 (mg of protein)−1] ± SD | R | Mean SEA [μmol min−1 (mg of protein)−1] ± SD | R | Mean SEA [μmol min−1 (mg of protein)−1] ± SD | R | |
| GAPDH (EC 1.2.1.12) | 1.13 ± 0.19 | 2.37 ± 0.28 | 2.82 ± 0.31 | 3.81 ± 0.45 | ||||
| PGM (EC 5.4.2.2) | 0.021 ± 0.006 | 1.3 | 0.028 ± 0.008 | 1.2 | 0.035 ± 0.013 | 1.1 | 0.044 ± 0.011 | 1.3 |
| LDH (EC 1.1.1.27) | 0.13 ± 0.01 | 150.6 | 0.26 ± 0.03 | 298.3 | 0.38 ± 0.04 | 402.9 | 0.43 ± 0.05 | 434.2 |
| PFO (EC 1.2.7.1) | 0.44 ± 0.04 | 10.2 | 0.57 ± 0.06 | 9.8 | 0.71 ± 0.07 | 10.9 | 0.74 ± 0.09 | 9.8 |
| PTA (EC 2.3.1.8) | 0.76 ± 0.08 | 23.9 | 0.95 ± 0.01 | 23.2 | 0.99 ± 0.13 | 22.8 | 1.03 ± 0.14 | 22.1 |
| AK (EC 2.7.2.1) | 0.22 ± 0.03 | 6.9 | 0.46 ± 0.04 | 11.3 | 1.11 ± 0.15 | 25.5 | 1.56 ± 0.21 | 33.6 |
| AADH (EC 1.2.1.10) | 0.09 ± 0.01 | 7.9 | 0.09 ± 0.02 | 5.4 | 0.24 ± 0.02 | 10.6 | 0.29 ± 0.03 | 10.2 |
| ADH (EC 1.1.1.1) | 0.17 ± 0.02 | 15.2 | 0.25 ± 0.03 | 14.9 | 0.41 ± 0.04 | 18.5 | 0.49 ± 0.06 | 17.2 |
SEA, specific activity of enzyme; R, ratio of specific enzymatic activity to metabolic flux through the metabolic pathway (see Results).
When the flux was expressed as micromoles per milligram per hour of protein from previously calculated values (Fig. 3), the ratio R (specific enzyme activity in biomass/metabolic flux) could be calculated (28). For metabolic pathways leading to acetate production via PTA and AK, R varied between 22.1 and 23.9 and from 6.9 to 33.6, respectively; with PFO, R was ca. 10.2. Similarly, R for enzymes of the ethanol pathway ranged from 5.4 to 10.6 and from 14.9 to 18.5 for AADH and ADH, respectively. However, with the metabolic route through PGM, this ratio was constant at only ca. 1.2. This indicated that flux via PGM utilized all of the available activity of the enzyme. The specific activity of PGM increased 2.1-fold from lowest to highest D (Table 6) and was thus correlated with higher specific metabolic rates. These results mean that the enzyme regulates activity in the cell to balance the flux from G1P to G6P whatever the μ. An apparent Km of 0.21 mM for G1P catalyzed by PGM was calculated from an Eadie-Hofstee plot (17) (data not shown). By assuming an internal volume of 1.67 ml (g of cells)−1 (24), the steady-state internal G1P concentration was calculated as 3.42 to 12.05 mM (Table 5); these data suggest that the PGM reaction was zero-order kinetics with respect to catalyzed substrate, which is in good agreement with the analysis of metabolic flux. In contrast, the ratio R for the lactate formation pathway was always very high, ranging from 150.6 to 434.2. This indicates that the enzyme concentration increases as catabolic carbon flow increases but was not correlated with a proportional rise of the specific production rate of lactate; LDH increased 3.5-fold, from 0.014 to 0.083 h−1 (Table 6), while qlactate varied only slightly (Fig. 3).
DISCUSSION
Based on microscopic observations, cellulose fibers were found to be covered by the microbial cell; such continuous cultures are generally regarded as cellulose limited (59, 71, 74). Yet cells which adhere to cellulose can also be considered to be in substrate-sufficient conditions. In fact, soluble sugars liberated from cellulose are the real substrate for growth and are incorporated by cells as rapidly as they are formed, since no glucose or cellodextrins could be detected in the supernatant. It may therefore be possible that bacteria display a metabolism under or near carbon-sufficient conditions and not carbon-limited conditions as previously suggested by microscopic examination. In cellobiose-sufficient conditions, the ATP/ADP ratios were as high as 7.21 and always higher than 1 (25), while in cellobiose limitation the ATP/ADP ratios ranged between 0.21 and 0.69 (E. Guedon, M. Desvaux, and H. Petitdemange, unpublished data). In the cellulose chemostat cultures carried out, the ATP/ADP ratios ranged from 0.37 to 0.46 and were always lower than 1 as in carbon-limited chemostats. In cellobiose limitation, few lactate molecules were produced at low D, contrary to what was observed in cellobiose-sufficient continuous culture (25). Moreover, on cellulose continuous culture, the ratios NADH/NAD+ and qNADH produced/qNADH used dropped as D increased, while in cellobiose-sufficient conditions these ratios rose (25). All of these results lead to the conclusion that the cellulose continuous cultures described here were performed under carbon limitation; it can thus be maintained that the chemostats were cellulose-limited continuous cultures.
In such conditions, the cellulose catabolism of C. cellulolyticum led to an acetate-ethanol fermentation maximizing ATP production. Concerning the distribution of the fluxes throughout the metabolic network, at least 94.2% of the carbon flow was used for generation of energy and biosynthetic precursors; the remaining carbon was converted to glycogen and exopolysaccharides. The proportion of carbon flowing through G6P remained quite constant regardless of D but differed between qbiosynthesis and qpyruvate as μ increased; more of the carbon uptake was directed toward biosynthesis pathways at high D values than at lower ones. Regardless of D, extracellular proteins and free amino acids represented around 7.5% of the entering carbon. The carbon flow was always directed mainly toward acetate production, which represented a minimum of 27.1% of the original carbon at the highest μ. Yet the carbon from glycolysis directed through biosynthesis and ethanol increased to 27.3 and 16.8%, respectively, at D = 0.083 h−1. Theorical calculation suggest on the one hand that with Trichoderma reesei used as a model, optimal growth of a cellulolytic anaerobe requires major allocation of ATP toward cellulase biosynthesis (70) and on the other hand that with low ATP production per mole of hexose equivalent, most of the carbon source is used for the generation of energy (57). From lowest to highest D, C. cellulolyticum diverted 7.8 to 21.6% of the cellulose to cell carbon, whereas 81.3 to 66.7% was used for ATP production.
On cellulose, G6P-G1P was an important branch point since the longer the soluble β-glucan uptake period is, the more G1P will be generated. The ratio of PGM specific activity to qPGM (close to 1) reflected the precision of the control exerted by this enzyme on the partition of the flux at this junction (28), where the conversion of G1P to G6P feeds further the Embden-Meyerhof pathway. A high R indicates that the fluxes are determined by the concentration of substrate available in the pool rather than the enzyme activity (28). However, intracellular concentrations of substrates, products, cofactors, or effector molecules as well as intracellular ionic strength, redox potential, or pH could influence the partition and regulation of flux at each step in the central metabolic pathways (28). In vitro enzyme assays could then differ from in vivo conditions, and the significance of a low R could be more difficult to establish. Control of the PGM pathway by the amount of PGM was reinforced by the finding that Km was more than 10-fold lower than the lowest intracellular G1P concentration determined. With higher D, the flow through glycolysis and biosynthesis increased, as did G6P turnover. The G1P pool, however, increased because the proportion of carbon flowing via PGM did not vary as much. G1P was thus directed toward exopolysaccharide formation, which reached a maximum of 4.2% of the original carbon uptake. The proportion of the flow through glycogen synthesis increased as well and represented up to 1.6% of the entering carbon flow. Here, glycogen turnover increased with D but remained low (maximum of 0.09 h−1), while the glycogen pool increased up to 108.7 mg (g of cells)−1 at 0.035 h−1 and then slowly decreased; glycogen biosynthesis could be adjusted as a function of the carbon flow. At the G1P-G6P branch point, the flux partitioning was then stabilized via PGM while the carbon surplus was dissipated by exopolysaccharide and intracellular glycogen synthesis. In fact, the percentage of G1P converted to G6P via PGM remained constant at around 93.7%, whereas the proportion of G1P converted to glycogen increased from 0.7 to 2.6% between D = 0.014 and 0.083 h−1. In the same time, exopolysaccharide represented around 5.3% of the G1P produced. These productions seem to buffer the increasing carbon flow from G1P, which could not be metabolized via PGM. On cellulose-limited chemostat cultures, carbon excess at this branch point was limited compared to ammonium-limited chemostat culture performed on cellobiose (which corresponded to an uncoupling between catabolism and anabolism), where exopolysaccharides and glycogen could represent up to 16.0 and 21.4%, respectively, of the specific rate of carbon consumed and where cellotriose was detected extracellularly (22). Glycogen was synthesized even in carbon-limited conditions and was present at all dilution rates. Such observations parallel those for Fibrobacter succinogenes, for which futile cycling of glycogen was reported (18, 38). These results could suggest that glycogen biosynthesis in cellulolytic bacteria in involved in carbon flow regulation (7) rather than in prolonging cell viability by providing energy storage (54) as for sporogenesis (49). Such a cycle could also be considered an energy-wasting reaction, as suggested by use of the term “futile cycle” for glycogen metabolism in Fibrobacter; however, here cellulose-fed continuous cultures were energy-limited cultures which do not normally waste ATP (57).
The flow of carbon was facilitated by the increase of specific enzymatic activity as μ increased. The fact that metabolic fluxes were much less than the available enzyme activities (except for PGM as specified above) indicated that specific enzyme activities were limited by the supply of substrate. The decline of acetyl-CoA turnover was corroborated by the analysis of metabolic flux distribution; the percentage of the carbon flowing through PFO diminished as D increased and the flux was directed away from acetate production. The acetyl-CoA/CoA ratio decreased and paralleled the decline of the ratios H2/CO2 and qNADH produced/qNADH used. These results are in agreement with the model of Decker et al. (11, 66) for the regulation of NADH-fd reductase activities which, through the consumption of NADH, direct the fluxes of metabolites in the direction of acetate for ATP production. On cellulose, the cells manage to maintain an equilibrated electron balance through the hydrogenase NADH-fd reductase activities and the ethanol pathway as indicated by the O/R index (23, 24). The pyruvate and acetyl-CoA branch points were parts of an independent two-node metabolic network (60). From acetyl-CoA, the main end product formed was acetate, but partitioning of the flux was modified in favor of ethanol production as D increased. Pyruvate was catabolized mainly via PFO. Whatever the D, the extracellular pyruvate formation rate was low and was correlated with the low lactate production which, moreover, represented less and less of the qpyruvate; the specific formation rates of both lactate and extracellular pyruvate represented a maximum of 1.8% of the original carbon uptake. Thus, whatever the D, there was no competition between PFO and LDH for the carbon flowing from glycolysis, contrary to what was observed on soluble sugar (24, 25). On cellobiose, lactate and extracellular pyruvate levels rose with increasing specific rates of consumed hexose higher than 2.82 mmol (g of cells)−1 h−1 (24), while on cellulose qhexose reached a maximum of 2.53 mmol (g of cells)−1 h−1.
The mATP obtained on cellulose, i.e., 2.9 mmol of ATP/g of cells/h, was very close to that obtained on cellobiose (r2 = 0.923), i.e., 2.2 mmol of ATP/g of cells/h; as well, the true energetic yield on cellobiose was 28.4 g of biomass per mol of ATP, near the YATPmax of 30.3 g of biomass per mol of ATP obtained on cellulose (values for the cellobiose-limited chemostat calculation were taken from a previous report 24). As for the true growth yield, the YX/Smax determined on cellobiose was lower than that on cellulose (Table 7). From statistical analysis of the Pirt plots, following regression analysis and Student t test (9) of the data, the maximum true growth yields were significantly different. This indicated a benefit for cells when grown on cellulose since a higher biomass could be reached for the same hexose equivalent quantity consumed. Such a result is consistent with the mechanism of carbohydrate uptake described by Strobel et al. (62), which is an energy-efficient transport system of soluble cellodextrins released from cellulolysis.
TABLE 7.
Kinetic and growth parameters of various species of cellulolytic bacteria determined in continuous culturesa
| Bacterial strain | Substrate | k (h−1) | m (mmol of hexose eq [g of cells]−1 h−1) | YX/Smax (g of cells [mol of hexose eq]−1 | Reference |
|---|---|---|---|---|---|
| Clostridium cellulolyticum ATCC 35319 | MN301 | 0.05 | 0.9 | 50.5 | This study |
| Cellobiose | 0.9b | 36.2b | 25 | ||
| C. thermocellum ATCC 27405 | Avicel | 0.17b | ND | 24.3b | 37 |
| Fibrobacter succinogenes S85 | Sigmacell 20 | 0.09c | 0.3bc | 56.7b | 73 |
| Ruminococcus albus 7 | Avicel | 0.05 | 0.6b | 20.1b | 47 |
| R. flavefaciens FD1 | Sigmacell 20 | 0.11 | 0.4b | 38.9b | 75 |
k is the rate constant of cellulose degradation; m is the growth maintenance coefficient; YX/Smax is the true growth yield. ND, not determined.
Calculated from previously reported data.
Average of previously reported data.
This investigation has also allowed characterization of the cellulose degradation properties of C. cellulolyticum from the consistent data summarized in Table 7. Our data were compiled along with those of other chemostat cultures performed with cellulose as described in the current literature. R. albus and C. cellulolyticum appear to be the bacteria with the lowest first-order cellulose hydrolysis rate constant; it is 3.4 times lower than that of C. thermocellum, which is the highest one so far determined, 0.17 h−1. Although a high cellulose degradation rate is often regarded as a general feature of thermophilic microorganisms relative to mesophilic species (37), such a difference in the rate constant of cellulose degradation could also result from the difference in cellulasic substrates used; in fact, the relative crystallinity index, porosity, allomorphs, capillary or gross surface area of the cellulose used could result in different kinetics of cellulose degradation (72, 73). Thus, the comparison of rate constant of cellulose digestion should not be overinterpreted, a caveat which points out the necessity of standardization of cellulose substrates used in such studies for further direct comparison. The true growth yield is among the highest reported, i.e., 50.5 g of cells/mol of hexose eq (Table 7). The growth maintenance coefficient reported in the literature could not be truly comparable since the rate of consumed hexose required for maintenance is related to the metabolic pathway which allowed the ATP production (57, 61, 65). Since the catabolic networks vary among the reported bacterial species (Table 7), differences in m values could be attributed to ATP generating a more or less efficient pathway that thus requires more or less hexose equivalents but can generate the same quantity of ATP per gram of cells per hour. Thus, comparison of mATP between cellulolytic bacteria would be more informative for evaluating the energy maintenance requirement.
The first step in investigating C. cellulolyticum metabolism was the use of a chemostat technique which leads to the accumulation of NADH (48). The use of a mineral salt-based medium permitted the induction of different metabolic regulatory responses and better control of the carbon and electron flows (24). Such findings led to the hypothesis of growth adapted to nutrient-poor conditions (23). These studies, however, were performed with soluble sugars, which facilitated study of the bacterial metabolism of cellulolytic bacteria. The next step was therefore investigation of the physiology of this microorganism on an insoluble substrate, more closely related to the natural ecological niche. In the present work with cellulose-limited continuous culture, no accumulation of NADH was observed and pyruvate overflow as high as on cellobiose chemostats did not occur (24). The insolubility and resistance of the cellulose to enzymatic hydrolysis physically prevents pyruvate overflow since μmax and carbon flow are inevitably lower than with a soluble substrate (24). Thus, on cellobiose-limited chemostats, some metabolic regulation such as the shift from acetate-ethanol fermentation to lactate-ethanol fermentation at high catabolic rates should be interpreted as a deregulation of the metabolism attributed to the growth of C. cellulolyticum on soluble sugars which represent conditions far from the physical nature of the cellulose. In cellulose continuous cultures, compared to cellobiose chemostats, a second regulation of the entering carbon was introduced by the depolymerization of the insoluble substrate into soluble sugars readily metabolized by bacteria for growth. This limitation led to a lower maximum specific growth rate reached on cellulose than on cellobiose; in turn, pyruvate leakage was limited. The observations with cellobiose chemostats should be interpreted as laboratory artifacts due to culture conditions far from those in which this bacterium has evolved in nature and emphasizes that the efficiency of catabolism is related to the degradative property of the bacterial cellulosome. In the course of evolution, the catabolic pathways are optimized as a function of the carbon flowing from cellulase activities and are not adapted to higher catabolic rates as in other clostridial bacteria (43). C. cellulolyticum appeared, therefore, well adapted and even restricted to a low carbon flow, which is characteristic of growth on its natural substrate, cellulose.
ACKNOWLEDGMENTS
This work was supported by the Commission of European Communities FAIR program (contract CT950191 [DG12SSMA]) and by the program Agrice (contract 9701041).
We thank J. Mejean for statistical analysis of the data, E. McRae for correcting the English and for critical reading of the manuscript, and G. Raval for technical assistance.
REFERENCES
- 1.Ahn H J, Lynd L R. Cellulose degradation and ethanol production by thermophilic bacteria using mineral growth medium. Appl Biochem Biotechnol. 1996;57–58:599–604. doi: 10.1007/BF02941740. [DOI] [PubMed] [Google Scholar]
- 2.Andersch W, Bahl H, Gottschalk G. Level of enzymes involved in acetate, butyrate, acetone and butanol formation by Clostridium acetobutylicum. Eur J Appl Microbiol Biotechnol. 1983;18:327–332. [Google Scholar]
- 3.Andersen K B, von Meyenburg K. Charges of nicotinamide adenine nucleotides and adenylate energy charge as regulatory parameters of the metabolism in Escherichia coli. J Biol Chem. 1977;252:4151–4156. [PubMed] [Google Scholar]
- 4.Bayer E A, Chanzy H, Lamed R, Shoham Y. Cellulose, cellulases and cellulosomes. Curr Opin Struct Biol. 1998;8:548–557. doi: 10.1016/s0959-440x(98)80143-7. [DOI] [PubMed] [Google Scholar]
- 5.Bayer E A, Lamed R. The cellulose paradox: pollutant par excellence and/or a reclaimable natural resource? Biodegradation. 1992;3:171–188. doi: 10.1007/BF00129082. [DOI] [PubMed] [Google Scholar]
- 6.Béguin P, Lemaire M. The cellulosome: an exocellular, multiprotein complex specialized in cellulose degradation. Crit Rev Biochem Mol Biol. 1996;31:201–236. doi: 10.3109/10409239609106584. [DOI] [PubMed] [Google Scholar]
- 7.Belanger A E, Hatfull G F. Exponential-phase glycogen recycling is essential for growth of Mycobacterium smegmatis. J Bacteriol. 1999;181:6670–6678. doi: 10.1128/jb.181.21.6670-6678.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boisset C, Chanzy H, Henrissat B, Lamed R, Shoham Y, Bayer E A. Digestion of crystalline cellulose substrates by Clostridium thermocellum cellulosome: structural and morphological aspects. Biochem J. 1999;340:829–835. [PMC free article] [PubMed] [Google Scholar]
- 9.Bouyer J. Méthodes statistiques: médecine-biologie. Paris, France: ESTEM Éditions INSERM; 1996. [Google Scholar]
- 10.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 11.Decker K, Rössle M, Kreusch J. The role of nucleotides in the regulation of the energy metabolism of Clostridium kluyveri. In: Schlegel H G, Gottschalk G, Pfennig N, editors. Microbial production and utilization of gases. Göttingen, Germany: Akademie der Wissenschaften zu Göttingen; 1976. pp. 75–83. [Google Scholar]
- 12.Desvaux M, Guedon E, Petitdemange H. Cellulose catabolism by Clostridium cellulolyticum growing in batch culture on defined medium. Appl Environ Microbiol. 2000;66:2461–2470. doi: 10.1128/aem.66.6.2461-2470.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubois M, Gilles K, Hamilton J K, Rebers P A, Smith F. A colorimetric method for the determination of sugars. Nature. 1951;168:167. doi: 10.1038/168167a0. [DOI] [PubMed] [Google Scholar]
- 14.Dürre P, Kuhn A, Gottwald M, Gottschalk G. Enzymatic investigations on butanol dehydrogenase and butyraldehyde dehydrogenase in extracts of Clostridium acetobutylicum. Appl Microbiol Biotechnol. 1987;26:268–272. [Google Scholar]
- 15.Ellman G L. Tissue sulfhydryl groups. Arch Biochem Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 16.Ferdinand W. The isolation and specific activity of rabbit muscle glyceraldehyde phosphate dehydrogenase. Biochem J. 1964;92:578–585. doi: 10.1042/bj0920578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fersht A. Enzyme structure and mechanism. W. H. New York, N.Y: Freeman and Co.; 1985. [Google Scholar]
- 18.Gaudet G, Forano E, Dauphin G, Delort A M. Futile cycling of glycogen in Fibrobacter succinogenes as shown by in situ 1H-NMR and 13C-NMR investigation. Eur J Biochem. 1992;207:155–162. doi: 10.1111/j.1432-1033.1992.tb17032.x. [DOI] [PubMed] [Google Scholar]
- 19.Gelhaye E, Gehin A, Petitdemange H. Colonization of crystalline cellulose by Clostridium cellulolyticum ATCC 35319. Appl Environ Microbiol. 1993;59:3154–3156. doi: 10.1128/aem.59.9.3154-3156.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gelhaye E, Petitdemange H, Gay R. Adhesion and growth rate of Clostridium cellulolyticum ATCC 35319 on crystalline cellulose. J Bacteriol. 1993;175:3452–3458. doi: 10.1128/jb.175.11.3452-3458.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giallo J, Gaudin C, Belaich J P. Metabolism and solubilization of cellulose by Clostridium cellulolyticum H10. Appl Environ Microbiol. 1985;49:1216–1221. doi: 10.1128/aem.49.5.1216-1221.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guedon E, Desvaux M, Petitdemange H. Kinetic analysis of Clostridium cellulolyticum carbohydrate metabolism: importance of glucose 1-phosphate and glucose 6-phosphate branch points for distribution of carbon fluxes inside and outside cells as revealed by steady-state continuous culture. J Bacteriol. 2000;182:2010–2017. doi: 10.1128/jb.182.7.2010-2017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guedon E, Desvaux M, Payot S, Petitdemange H. Growth inhibition of Clostridium cellulolyticum by an inefficiently regulated carbon flow. Microbiology. 1999;145:1831–1838. doi: 10.1099/13500872-145-8-1831. [DOI] [PubMed] [Google Scholar]
- 24.Guedon E, Payot S, Desvaux M, Petitdemange H. Carbon and electron flow in Clostridium cellulolyticum grown in chemostat culture on synthetic medium. J Bacteriol. 1999;181:3262–3269. doi: 10.1128/jb.181.10.3262-3269.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guedon E, Payot S, Desvaux M, Petitdemange H. Relationships between cellobiose catabolism, enzyme levels and metabolic intermediates in Clostridium cellulolyticum grown in a synthetic medium. Biotechnol Bioeng. 2000;67:327–335. doi: 10.1002/(sici)1097-0290(20000205)67:3<327::aid-bit9>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 26.Gottschalk G. Bacterial metabolism. New York, N.Y: Springer-Verlag; 1985. [Google Scholar]
- 27.Hazlewood G P, Gilbert H J. Xylan and cellulose utilization by the clostridia. In: Woods D R, editor. The clostridia and biotechnology. Stoneham, Mass: Butterworth-Heinemann; 1993. pp. 311–341. [PubMed] [Google Scholar]
- 28.Holms H. Flux analysis and control of the central metabolic pathways in Escherichia coli. FEMS Microbiol Rev. 1996;19:85–116. doi: 10.1111/j.1574-6976.1996.tb00255.x. [DOI] [PubMed] [Google Scholar]
- 29.Huang L, Forsberg C W. Cellulose digestion and cellulase regulation and distribution in Fibrobacter succinogenes subsp. succinogenes S85. Appl Environ Microbiol. 1990;56:1221–1228. doi: 10.1128/aem.56.5.1221-1228.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kistner A, Kornelius J H. A small-scale, three-vessel, continuous culture system for quantitative studies of plant fibre degradation by anaerobic bacteria. J Microbiol Methods. 1990;12:173–182. [Google Scholar]
- 31.Kleijntjens R H, De Boks P A, Luyben K C A M. A continuous thermophilic cellulose fermentation in an upflow reactor by a Clostridium thermocellum containing mixed culture. Biotechnol Lett. 1986;8:667–672. [Google Scholar]
- 32.Lamed R, Zeikus J G. Ethanol production by thermophilic bacteria: relationship between fermentation product yields of and catabolic enzyme activities in Clostridium thermocellum and Thermoanaerobium brockii. J Bacteriol. 1980;144:569–578. doi: 10.1128/jb.144.2.569-578.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee S F, Forsberg C W, Gibbins L N. Xylanolytic activity of Clostridium acetobutylicum. Appl Environ Microbiol. 1985;50:1068–1076. doi: 10.1128/aem.50.4.1068-1076.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leschine S B. Cellulose degradation in anaerobic environments. Annu Rev Microbiol. 1995;49:399–426. doi: 10.1146/annurev.mi.49.100195.002151. [DOI] [PubMed] [Google Scholar]
- 35.Ljungdahl L G, Eriksson K E. Ecology of microbial cellulose degradation. Adv Microb Ecol. 1985;8:237–299. [Google Scholar]
- 36.Lowry O H, Passonneau J V. Phosphoglucomutase kinetics with the phosphates of fructose, glucose, mannose, ribose and galactose. J Biol Chem. 1969;244:910–916. [PubMed] [Google Scholar]
- 37.Lynd L R, Grethlein H E, Wolkin R H. Fermentation of cellulosic substrates in batch and continuous culture of Clostridium thermocellum. Appl Environ Microbiol. 1989;55:3131–3139. doi: 10.1128/aem.55.12.3131-3139.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matheron C, Delort A M, Gaudet G, Forano E, Liptaj T. 13C and 1H nuclear magnetic resonance study of glycogen futile cycling in strains of the genus Fibrobacter. Appl Environ Microbiol. 1998;64:74–81. doi: 10.1128/aem.64.1.74-81.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meinecke B, Bertram J, Gottschalk G. Purification and characterization of the pyruvate-ferredoxin oxidoreductase from Clostridium acetobutylicum. Arch Microbiol. 1989;152:244–250. doi: 10.1007/BF00409658. [DOI] [PubMed] [Google Scholar]
- 40.Mitchell W J. Physiology of carbohydrate to solvent conversion by clostridia. Adv Microbiol Physiol. 1998;39:31–130. doi: 10.1016/s0065-2911(08)60015-6. [DOI] [PubMed] [Google Scholar]
- 41.Mokrasch L C. Use of 2,4,6-trinitrobenzenesulfonic acid for the coestimation of amines, amino acids, and proteins in mixtures. Anal Biochem. 1967;18:64–71. [Google Scholar]
- 42.Monod J. La technique de culture continue théorie et applications. Ann Inst Pasteur. 1950;79:390–410. [Google Scholar]
- 43.Morrison M, Mackie R I, Kistner A. 3-Phenylpropanoic acid improves the affinity of Ruminococcus albus for cellulose in continuous culture. Appl Environ Microbiol. 1990;56:3220–3222. doi: 10.1128/aem.56.10.3220-3222.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Papoutsakis E T. Equations and calculations for fermentations of butyric acid bacteria. Biotechnol Bioeng. 1984;26:174–187. doi: 10.1002/bit.260260210. [DOI] [PubMed] [Google Scholar]
- 45.Papoutsakis E T, Meyer C I. Equations and calculations of product yields and preferred pathways for butanediol and mixed-acid fermentations. Biotechnol Bioeng. 1985;27:50–66. doi: 10.1002/bit.260270108. [DOI] [PubMed] [Google Scholar]
- 46.Pavlostathis S G, Miller T L, Wolin M J. Fermentation of insoluble cellulose by continuous cultures of Ruminococcus albus. Appl Environ Microbiol. 1988;54:2655–2659. doi: 10.1128/aem.54.11.2655-2659.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pavlostathis S G, Miller T L, Wolin M J. Kinetics of insoluble cellulose fermentation by continuous cultures of Ruminococcus albus. Appl Environ Microbiol. 1988;54:2660–2663. doi: 10.1128/aem.54.11.2660-2663.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Payot S, Guedon E, Cailliez C, Gelhaye E, Petitdemange H. Metabolism of cellobiose by Clostridium cellulolyticum growing in continuous culture: evidence for decreased NADH reoxidation as a factor limiting growth. Microbiology. 1998;144:375–384. doi: 10.1099/00221287-144-2-375. [DOI] [PubMed] [Google Scholar]
- 49.Payot S, Guedon E, Desvaux M, Gelhaye E, Petitdemange E. Effect of dilution rate, cellobiose and ammonium availabilities on Clostridium cellulolyticum sporulation. Appl Microbiol Biotechnol. 1999;52:670–674. doi: 10.1007/s002530051577. [DOI] [PubMed] [Google Scholar]
- 50.Peck H D, Gest H A. A new procedure for assay of bacterial hydrogenases. J Bacteriol. 1956;71:70–80. doi: 10.1128/jb.71.1.70-80.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pereira A N, Mobedshashi M, Ladish M R. Preparation of cellodextrins. Methods Enzymol. 1988;160:26–45. [Google Scholar]
- 52.Petitdemange E, Caillet F, Giallo J, Gaudin C. Clostridium cellulolyticum sp. nov., a cellulolytic mesophilic species from decayed grass. Int J Syst Bacteriol. 1984;34:155–159. [Google Scholar]
- 53.Pirt S J. Principles of microbe and cell cultivation. Oxford, United Kingdom: Blackwell Scientific Publishers; 1975. [Google Scholar]
- 54.Preiss J. Regulation of glycogen synthesis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B Jr, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 1015–1024. [Google Scholar]
- 55.Rafter G W, Colowick S P. Enzymatic preparation of DPNH and TPNH. Methods Enzymol. 1957;3:887–899. [Google Scholar]
- 56.Russell J B. Fermentation of cellodextrins by cellulolytic and noncellulolytic rumen bacteria. Appl Environ Microbiol. 1985;49:572–576. doi: 10.1128/aem.49.3.572-576.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Russell J B, Cook G M. Energetics of bacterial growth: balance of anabolic and catabolic reactions. Microbiol Rev. 1995;59:48–62. doi: 10.1128/mr.59.1.48-62.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi Y, Odt C L, Weimer P J. Competition for cellulose among three predominant ruminal cellulolytic bacteria under substrate-excess and substrate-limited conditions. Appl Environ Microbiol. 1997;58:2583–2591. doi: 10.1128/aem.63.2.734-742.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shi Y, Weimer P J. Response surface analysis of the effects of pH and dilution rate on Ruminococcus flavefaciens FD-1 in cellulose-fed continuous culture. Appl Environ Microbiol. 1992;58:2583–2591. doi: 10.1128/aem.58.8.2583-2591.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stephanopoulos G, Vallino J J. Network rigidity and metabolic engineering in metabolite overproduction. Science. 1991;252:1675–1681. doi: 10.1126/science.1904627. [DOI] [PubMed] [Google Scholar]
- 61.Stouthamer A H. Determination and significance of molar growth yields. Methods Microbiol. 1969;1:629–663. [Google Scholar]
- 62.Strobel H J, Caldwell F C, Dawson K A. Carbohydrate transport by the anaerobic thermophile Clostridium thermocellum LQRI. Appl Environ Microbiol. 1995;61:4012–4015. doi: 10.1128/aem.61.11.4012-4015.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taguchi F, Yamada K, Hasegawa K, Taki-Saito T, Hara K. Continuous hydrogen production by Clostridium sp. strain no. 2 from cellulose hydrolysate in an aqueous two-phase system. J Ferment Bioeng. 1996;82:80–83. [Google Scholar]
- 64.Tempest D W. The continuous cultivation of microorganisms. I. Theory of the chemostat. Methods Microbiol. 1970;2:259–276. [Google Scholar]
- 65.Tempest D W, Neijssel O M. The status of YATP and maintenance energy as biologically interpretable phenomena. Annu Rev Microbiol. 1984;38:459–486. doi: 10.1146/annurev.mi.38.100184.002331. [DOI] [PubMed] [Google Scholar]
- 66.Thauer R, Jungermann C, Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev. 1977;41:100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tomme P, Warren R A J, Gilkes N R. Cellulose hydrolysis by bacteria and fungi. Adv Microb Physiol. 1995;37:1–81. doi: 10.1016/s0065-2911(08)60143-5. [DOI] [PubMed] [Google Scholar]
- 68.Tubbs P K, Garland P B. Assay of coenzyme A and some acyl derivates. Methods Enzymol. 1969;13:535–551. [Google Scholar]
- 69.Updegraff D M. Semimicro determination of cellulose in biological materials. Anal Biochem. 1969;32:420–424. doi: 10.1016/s0003-2697(69)80009-6. [DOI] [PubMed] [Google Scholar]
- 70.Walsum G P, Lynd L R. Allocation of ATP synthesis of cells and hydrolytic enzymes in cellulolytic fermentative microorganisms: bioenergetics, kinetics, and bioprocessing. Biotechnol Bioeng. 1998;58:316–320. [PubMed] [Google Scholar]
- 71.Weimer P J. Effects of dilution rate and pH on the ruminal cellulolytic bacterium Fibrobacter succinogenes S85 in cellulose-fed continuous culture. Arch Microbiol. 1993;160:288–294. doi: 10.1007/BF00292079. [DOI] [PubMed] [Google Scholar]
- 72.Weimer P J, Lopez-Guisa J M, French A D. Effect of cellulose fine structure on kinetics of its digestion by mixed ruminal microorganisms in vitro. Appl Environ Microbiol. 1990;56:2421–2429. doi: 10.1128/aem.56.8.2421-2429.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weimer P J, French A D, Calamari T A. Differential fermentation of cellulose allomorphs by ruminal cellulolytic bacteria. Appl Environ Microbiol. 1991;57:3101–3106. doi: 10.1128/aem.57.11.3101-3106.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weimer P J, Shi Y, Odt C L. A segmented gas/liquid delivery system for continuous culture of microorganisms on insoluble substrates and its use for growth of Ruminococcus flavefaciens on cellulose. Appl Microbiol Biotechnol. 1991;36:178–183. [Google Scholar]
- 75.Wimpenny J W T, Firth A. Levels of nicotinamide adenine dinucleotide and reduced nicotinamide adenine dinucleotide in facultative bacteria and the effect of oxygen. J Bacteriol. 1972;111:24–32. doi: 10.1128/jb.111.1.24-32.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wolin M J, Miller T L. Bioconversion of organic carbon to CH4 and CO2. Geomicrobiol J. 1987;5:239–259. [Google Scholar]