Highlights
-
•
Computed tomography imaging contains quantifiable information to characterize colorectal liver metastases.
-
•
Shape, texture, and intensity statistical features quantified the computed tomography liver volume.
-
•
An artificial intelligence model to predict local progression from radiomic features was developed with high accuracy.
-
•
Maximum dosage and textural coarseness of liver volume were features with highest predictive value.
Keywords: Radiomics, Artificial intelligence, Machine learning, Computer vision, Survival analysis
Abstract
Background and Purpose
Prognostic assessment of local therapies for colorectal liver metastases (CLM) is essential for guiding management in radiation oncology. Computed tomography (CT) contains liver texture information which may be predictive of metastatic environments. To investigate the feasibility of analyzing CT texture, we sought to build an automated model to predict progression-free survival using CT radiomics and artificial intelligence (AI).
Materials and Methods
Liver CT scans and outcomes for N = 97 CLM patients treated with radiotherapy were retrospectively obtained. A survival model was built by extracting 108 radiomic features from liver and tumor CT volumes for a random survival forest (RSF) to predict local progression. Accuracies were measured by concordance indices (C-index) and integrated Brier scores (IBS) with 4-fold cross-validation. This was repeated with different liver segmentations and radiotherapy clinical variables as inputs to the RSF. Predictive features were identified by perturbation importances.
Results
The AI radiomics model achieved a C-index of 0.68 (CI: 0.62–0.74) and IBS below 0.25 and the most predictive radiomic feature was gray tone difference matrix strength (importance: 1.90 CI: 0.93–2.86) and most predictive treatment feature was maximum dose (importance: 3.83, CI: 1.05–6.62). The clinical data only model achieved a similar C-index of 0.62 (CI: 0.56–0.69), suggesting that predictive signals exist in radiomics and clinical data.
Conclusions
The AI model achieved good prediction accuracy for progression-free survival of CLM, providing support that radiomics or clinical data combined with machine learning may aid prognostic assessment and management.
1. Introduction:
Patients with colorectal cancer develop colorectal liver metastases (CLM) in approximately 50 % of cases [1] with 40 % recurring within 12-months. Surgery is the standard of care for patients that present with liver-limited resectable CLM, with reported 5-year survival ranging from 28 to 58 % [2]. Non-surgical liver-directed local therapy for CLM, such as thermal ablation, can be effective, but is invasive [3]. External beam radiation therapy (EBRT) has emerged as an alternative, non-invasive approach for localized therapy of CLM in patients who are ineligible for other treatment options. Numerous clinical factors have been shown to influence the local control of CLM associated with EBRT, including dose delivered to the lesion and the size of the lesion [4]. Prognosis of local tumor control is essential to determine appropriate treatment for CLM, motivating the development of prediction models to aid clinical decision making.
Several models exist for predicting clinical outcomes in CLM patients. A common approach utilizes multivariate Cox proportional hazard regression using clinically relevant variables and selecting high hazard ratio variables for a scoring system [5], [6], [7], [8], [9], [10], [11], [12], [13]. Fong et al. [6], for instance, utilized five clinicopathological variables (node-positive primary, interval from primary to metastases, number of hepatic tumors, if largest hepatic tumor > 5 cm, and carcinoembryonic antigen level > 200 ng/ml) for their scoring system. Wang et al. [14] evaluated the accuracy of nine different survival prediction scoring systems with six of the scoring systems resulting in a concordance index (C-index) crossing below the 0.50 threshold in its 95 % confidence interval. A C-index crossing 0.50 would indicate prediction no better than random chance. There are limitations with these scoring systems, namely that scoring systems require manual thresholding and that other data, such as CT imaging, may provide features predictive of outcome.
CT has been standard of care for characterizing tumor response and radiomics is an emerging field that shows promise in analyzing complex details in CT scans. Radiomic features are computed textural attributes of which quantitatively characterize shape, intensity statistics, and gray-level relationships within the anatomy of interest. Specific to liver metastases, Fiz et al. [15] report 32 different studies up to June 2020 evaluating the association of radiomics to overall survival, tumor size, or response evaluation criteria, however the studies assess for association and did not measure predictive accuracy. Ganeshan et al. [16] observed that intensity and entropy of a liver CT volume of CLM patients significantly changed after contrast injection, indicating that radiomics can capture textural changes from enhancement. Miles et al. [17] investigated radiomics in relation to CLM survival by computing intensity and uniformity features from a CT liver volume and observing that textural uniformity was significantly associated with increased survival. Creasy et al. [18] and Simpson et al. [19] also observed that increased homogeneity in liver CT volumes were associated with increased risk of hepatic recurrence.
Artificial intelligence (AI) methods have shown potential in survival prediction in previous studies [20], [21]. AI, specifically machine learning, initializes models with parameters that can be optimized as more training data is available. This allows for the initialization of complex model architecture which may be more suitable for interdependent variables than a linear models in previous scoring systems.
To address the limitations of current methods, we set out to evaluate whether an automated prediction system can predict progression-free survival for CLM patients treated with RT. Specifically, we aimed to develop prediction model utilizing existing radiomic libraries to extract features from liver volumes as input data to machine learning models to predict patient outcomes.
2. Materials and methods
2.1. Data collection and equipment
This retrospective analysis was approved by the institutional review board with a waiver of informed consent at Memorial Sloan Kettering Cancer Center (MSK) (New York, NY). The MSK database was queried to obtain pre-treatment CT scans data for patients receiving radiation treatment for CLM between February 2006 and February 2019. Images were included if taken under a contrast enhancement protocol where 150 mL of intravenous iohexol contrast was administered and images were acquired at the portal venous phase, 75 s after the start of injection. Liver and gross tumor volumes (GTV) were segmented by radiation oncologists at MSK, as part of standard of care. This created volume subsets of liver volumes only, GTV only, and liver and GTV volumes. The MSK database was also queried to obtain dosimetric treatment parameters and right-censored time-to-event data for the outcome of progression in the treated tumor (local progression).
The query resulted in obtaining CT imaging and chart data for N = 97 patients, with 129 lesions identified. Of the 129 lesions, 55 resulted in local progression, 67 in no progression, and seven undetermined. The baseline distribution of clinical variables is summarized in Supplementary Table S1 (grouped by lesion) and Supplementary Table S2 (grouped by patient). The mean freedom from local progression was 10.5 months. Dosage and fractions are summarized in Supplementary Table S3. Liver metastases were treated with total dose range of 24–80 Gy (mean: 52.6 Gy) and number of fractions from 3 to 50 (mean: 8.6).
2.2. Image analysis
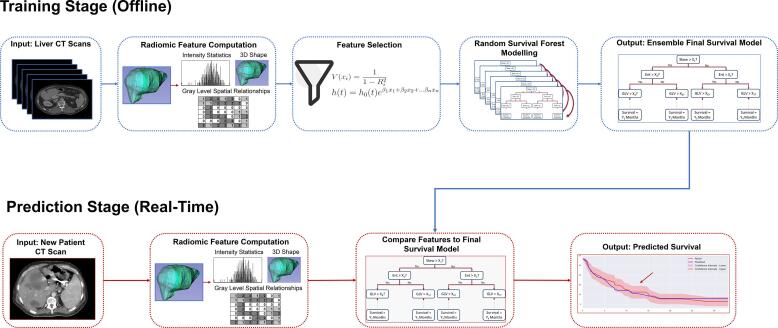
The task for the AI model was to predict the primary endpoint, defined as time from CLM radiation therapy until local tumor progression. To accomplish this, an AI survival prediction model, visualized in Fig. 1, was developed, consisting of an offline training component and a real-time prediction component. The input to the training component is a set of liver CT scans. Radiomic features are extracted from the liver and/or tumor volumes to train a survival model, which learns to predict a survival time interval for patients in the training dataset. After training, new patient CT scans can be used as an input to the finalized model to compute a real-time survival prediction. The training stage contains three main components: radiomic feature extraction, feature selection, and random survival forest modelling. The AI model was programmed in Python, utilizing the PyRadiomics [22] and PySurvival libraries [23]. Concordance indices were programmed in R with the Hmisc library [24].
Fig. 1.
A visualization of the survival prediction system. The system contains two stages. The first is a training stage, where radiomic features are extracted from a set of computed tomography liver scans. Variance inflation factor and hazard ratio ranking is then used to filter out low information yielding features. The remaining features are used to train a random survival forest prediction model. Once the survival model has been built, it can be exported to a real-time prediction environment, where liver scans of new patients can be fed as input to the survival model to obtain a predicted survival for the new patient. In this way, most of the computation required is done beforehand to build the model and prediction can occur in real-time for new patients.
2.3. Radiomic feature extraction
In the first stage, 108 radiomic features were computed for a liver volume extracted from a CT scan. This includes computations related to shape, intensity statistics, gray-level co-occurrence matrices, gray level run-length matrices, gray level dependence matrices, and gray tone difference matrices. A full list of radiomic features is available in Supplementary Table S4. The majority of radiomic features follow the Image Biomarker Standardisation Initiative (IBSI) guidelines. Deviations from IBSI are listed in Supplementary Table S5. Radiomic feature settings were selected used with the PyRadiomics application programming interface. Specifically, resampling was not performed, intensities were discretized with a fixed bin width of 25, and texture matrices were computed by aggregated from averaging the 3-dimensional directions from each individual 3-dimensional matrix. A set of radiomic features was computed for each lesion. Lesions were grouped together so that when they are shuffled into validation sets that no patient will have lesions both in the training and validation subset. An example of predictive radiomic features and associated outcomes is displayed in Supplementary Fig. S6.
2.4. Feature selection
Retaining all 108 radiomic features would likely result in overfitting due the dimensionality of the feature space being too large for the sample size [25]. Redundant features were removed using a variance inflation factor threshold of ten as an indicator of collinearity [26]. We then ranked remaining features using the hazard ratios predicted for each variable in a Cox proportional hazards model (CPH) [27], removing features until the ratio of features to samples was less than 1:10.
2.5. Random survival forest model
To predict survival from the filtered feature set, the random survival forest (RSF) algorithm was used [28]. The algorithm creates ensemble decision tree with nodes representing features with a threshold value. The features used and the threshold values are iteratively optimized to maximize the log-rank statistic between two child nodes. The full algorithm is listed in Supplementary Equation S7. A template RSF was instantiated using the PySurvival library [23] and then hyperparameters of number of trees, maximum number of patients for a terminal node, and maximum depth were optimized with a gridsearch algorithm. After optimization, feature importances were computed by error rates between the perturbed and unperturbed model for that feature.
2.6. Validation and statistical analysis
A 4 k-fold cross-validation scheme was used to provide multiple estimates of the performance of the model. The data was partitioned into four subsets of equal size and proportion of recurrences. The survival model was built by performing feature selection, training the RSF model, and hyperparameter optimization on three of the subsets and then evaluated with the remaining subset. This was repeated 4 times with a different testing subset. The concordance index (C-index), computed by Somers’ Dxy rank correlation [29], and integrated Brier score (IBS) were averaged over four k-folds with confidence intervals computed by using the standard error of the distribution of C-indices. One limitation of this method is that the sample size may not allow larger k-fold splits for a more accurate measurement of the confidence interval [30]. All analysis was programmed with Python.
Ablation analysis was performed to investigate the performance of the model when adjustments to individual components were made. First, we defined 11 different feature sets:
-
1.
Non-imaging and non-treatment clinical data: baseline patient variables not related to treatment information or tumor geometry from CT imaging.
-
2.
Treatment clinical data: variables related to treatment parameters, including dosimetric variables.
-
3.
Imaging clinical data: variables related to tumor geometry measured in CT imaging.
-
4.
All pre-treatment clinical data: All clinical data except treatment clinical data. This represents variables that are not based on physician judgment for treatment planning.
-
5.
All clinical data: The union of feature sets 1–3.
-
6.
Radiomics: tumor volume: radiomic features computed from the tumor volume only.
-
7.
Radiomics: liver parenchyma: radiomic features computed from the liver parenchyma only.
-
8.
Radiomics: liver parenchyma + tumor: radiomic features computed from the union of the tumor volume and liver parenchyma.
-
9.
Treatment clinical data and radiomics from liver parenchyma + tumor: the union of feature sets 2. and 8.
-
10.
Non treatment clinical data and radiomics from liver parenchyma + tumor: the union of feature sets 4. and 8.
-
11.
All clinical data and radiomics from liver parenchyma + tumor: the union of feature sets 5. and 8.
Table 1 displays a list of categorized clinical variables.
Table 1.
The categorization of clinical variables to imaging, treatment, and other (non-imaging and non-treatment) clinical variables. The goal of this categorization was to observe if different subsets of clinical data performed better at prediction progression in the absence of other subsets.
| Category | Variables |
|---|---|
| Imaging Clinical Data | Number of lesions at radiotherapy Other sites at radiotherapy Lesion dimension 1 Lesion dimension 2 PTV (cm3) |
| Treatment Clinical Data | Biologically effective dose (Gy) Minimum dose for planning target volume (Gy) Maximum dose (Gy) Dose for 95 % of target volume (% of intended prescribed dose) Systemic treatment before radiotherapy Lines of chemotherapy Hepatic arterial infusion pump before radiotherapy Reirradiation Surgery before radiotherapy Ablation before radiotherapy Yttrium-90 embolization before radiotherapy Arterial embolization before radiotherapy |
| Other Clinical Data | Primary tumor subsite Metastasis at diagnosis Number of liver lesions at diagnosis Other sites at diagnosis Liver location Carcinoembryonic antigen Kirsten rat sarcoma virus mutation |
Each feature set was used to build a RSF survival model with feature selection, without feature selection, and with a CPH model with gridsearch optimization of the regularization parameter. The goal was to evaluate the performance of radiomics compared to clinical data, whether the combination of both enhance performance, whether different radiomic volumes are more predictive, whether the lack of feature selection will result in overfitting, and whether using a CPH model is sufficient.
3. Results
The averaged cross-validation accuracies for the radiomic RSF models in Table 2 demonstrate that nearly all input dataset variations resulted in a C-index greater than 0.50 within 95 % confidence interval ranges. The highest average prediction accuracy occurred when combining both radiomics of the liver parenchyma and tumor volume with treatment data (C-index: 0.73 [0.64, 0.82]). This was not statistically significantly different from models utilizing only clinical data. Utilizing only radiomic data from the liver parenchyma and tumor volume resulted in a C-index of 0.68 [0.62, 0.74]. The IBS of all radiomic RSF models were below 0.25.
Table 2.
A summary of accuracy results for each input combination to the model. The artificial intelligence model achieved good, nonrandom C-indices and feature selection decreased the variance of the cross-validation accuracies.
| Input Features | Concordance Index (95 % CI) | Integrated Brier Score (95 % CI) |
|---|---|---|
| (No Feature Selection, Local Progression as Outcome) | ||
| Other Clinical Data | 0.64 [0.54, 0.75] | 0.18 [0.15, 0.22] |
| Imaging Clinical Data | 0.66 [0.61, 0.71] | 0.17 [0.14, 0.20] |
| Treatment Clinical Data | 0.69 [0.62, 0.77] | 0.17 [0.14, 0.20] |
| All Pre-treatment Clinical Data | 0.63 [0.55, 0.71] | 0.22 [0.19, 0.25] |
| All Clinical Data | 0.67 [0.58, 0.75] | 0.16 [0.15, 0.18] |
| Radiomics: Tumor Volume | 0.64 [0.52, 0.76] | 0.18 [0.17, 0.18] |
| Radiomics: Liver Parenchyma | 0.61 [0.53, 0.69] | 0.21 [0.19, 0.23] |
| Radiomics: Liver Parenchyma + Tumor | 0.66 [0.58, 0.74] | 0.20 [0.17, 0.22] |
| Treatment Clinical Data + Radiomics from Liver Parenchyma and Tumor | 0.66 [0.59, 0.73] | 0.19 [0.18, 0.21] |
| All Pre-treatment Clinical Data + Radiomics from Liver Parenchyma and Tumor | 0.66 [0.55, 0.77] | 0.21 [0.17, 0.25] |
| All Clinical Data and Radiomics from Liver Parenchyma + Tumor | 0.64 [0.60, 0.68] | 0.19 [0.16, 0.22] |
| (With Feature Selection, Local Progression as Outcome) | ||
| Other Clinical Data | 0.66 [0.56, 0.76] | 0.19 [0.16, 0.22] |
| Imaging Clinical Data | 0.61 [0.56, 0.66] | 0.17 [0.14, 0.19] |
| Treatment Clinical Data | 0.72 [0.64, 0.79] | 0.18 [0.15, 0.21] |
| All Pre-treatment Clinical Data | 0.65 [0.58, 0.72] | 0.21 [0.18, 0.24] |
| All Clinical Data | 0.62 [0.56, 0.69] | 0.19 [0.16, 0.22] |
| Radiomics: Tumor Volume | 0.58 [0.51, 0.84] | 0.19 [0.16, 0.24] |
| Radiomics: Liver Parenchyma | 0.66 [0.60, 0.72] | 0.20 [0.18, 0.22] |
| Radiomics: Liver Parenchyma + Tumor | 0.68 [0.62, 0.74] | 0.20 [0.16, 0.25] |
| Treatment Clinical Data + Radiomics from Liver Parenchyma and Tumor | 0.73 [0.64, 0.82] | 0.18 [0.15, 0.20] |
| All Pre-treatment Clinical Data + Radiomics from Liver Parenchyma and Tumor | 0.66 [0.57, 0.75] | 0.20 [0.17, 0.23] |
| All Clinical Data and Radiomics from Liver Parenchyma + Tumor | 0.69 [0.65, 0.74] | 0.23 [0.21, 0.26] |
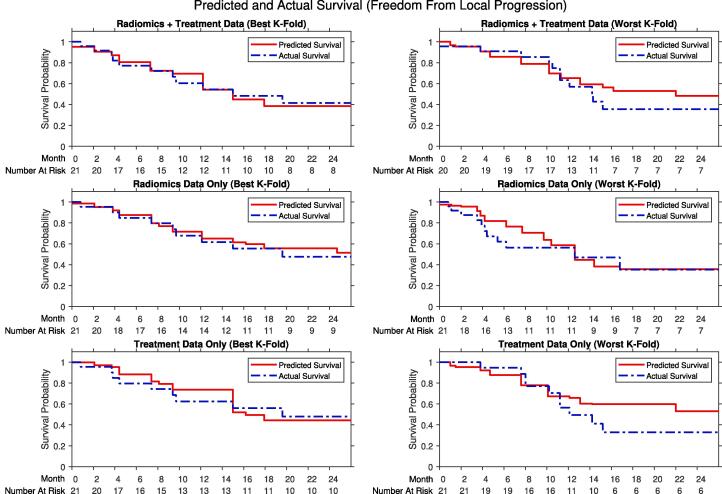
Accuracies for the radiomic CPH models, summarized in Table 3, demonstrate that all models crossed the 0.50 threshold. However, the variance of the confidence interval was such that the upper bound of the accuracies overlap with the radiomic RSF models. The predicted survival and IBS curves compared to ground truth in Fig. 2 demonstrate similarity between the prediction model and actual outcomes.
Table 3.
A summary of accuracy results for each input combination to the model which utilized radiomic features as input to a Cox proportional hazards model. All models cross the 0.50 concordance index threshold, indicating that random prediction cannot be ruled out. However, the upper bound for most models overlaps with the random survival forest models, indicating high variance in Cox modeling.
| Input Features | Concordance Index (95 % CI) | Integrated Brier Score (95 % CI) |
|---|---|---|
| With Cox Proportional Hazards Model | ||
| Other Clinical Data | 0.53 [0.50, 0.56] | 0.20 [0.18, 0.22] |
| Imaging Clinical Data | 0.56 [0.45, 0.67] | 0.25 [0.22, 0.28] |
| Treatment Clinical Data | 0.50 [0.48, 0.52] | 0.24 [0.20, 0.28] |
| All Pre-treatment Clinical Data | 0.54 [0.48, 0.60] | 0.19 [0.15, 0.23] |
| All Clinical Data | 0.57 [0.48, 0.66] | 0.21 [0.16, 0.26] |
| Radiomics: Tumor Volume | 0.47 [0.42, 0.52] | 0.22 [0.17, 0.27] |
| Radiomics: Liver Parenchyma | 0.49 [0.42, 0.56] | 0.24 [0.22, 0.26] |
| Radiomics: Liver Parenchyma + Tumor | 0.43 [0.40, 0.46] | 0.25 [0.21, 0.29] |
| Treatment Clinical Data + Radiomics from Liver Parenchyma and Tumor | 0.53 [0.45, 0.61] | 0.19 [0.15, 0.23] |
| All Pre-treatment Clinical Data + Radiomics from Liver Parenchyma and Tumor | 0.55 [0.49, 0.61] | 0.20 [0.17, 0.23] |
| All Clinical Data and Radiomics from Liver Parenchyma + Tumor | 0.58 [0.47, 0.67] | 0.22 [0.19, 0.25] |
Fig. 2.
Comparison of the predicted local progression-free survival (red), defined as freedom from local progression, from the random survival forest compared to the actual survival (red) from a Kaplan-Meier model of the outcome data. Comparisons include the best k-fold (left) and worst k-fold (right) during cross-validation from using radiomics using liver and tumor volumes and treatment data (top), radiomics data only (middle), or treatment data only (bottom). All models a higher C-index greater than 0.50 and the usage of radiomic features enhances the accuracy of the model compared to with treatment data alone. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The final models were uploaded to a public repository linked in Supplementary Material S8.
Feature importance computation results in Table 4 identifies that the most predictive radiomic feature was the neighboring gray tone difference matrix (NGTDM) strength. The most predictive clinical variable was maximum dose, significantly greater than any other clinical variable.
Table 4.
The feature importances for the random survival forest model utilizing treatment data only, radiomics data only, or the combination of both. Maximum dose was observed to be the most predictive feature, significantly with more information gain than any other treatment feature. Gray tone difference matrix computations were the most predictive when only using radiomics data. Both gray tone difference matrices and maximum dose features resulted in high predictive value in the combined model. However, the importance of maximum dose was decreased compared to when using only treatment data, indicating that the model is still able to predict survival with the remaining radiomic features.
| Feature (Treatment Data Only) | Importance Score (95 % CI) |
|---|---|
| Maximum Dose | 10.84 [6.35, 15.34] |
| Carcinoembryonic Antigen at Radiotherapy | 2.69 [−0.43, 5.81] |
| Lines of Chemotherapy | 2.53 [1.16, 3.9] |
| Pump Before Radiotherapy | −0.81 [−1.57, −0.05] |
| Feature (Radiomics on Liver Plus Tumor Volume Only) | |
| Neighborhood Gray Tone Difference Matrix Strength | 3.74 [2.25, 5.22] |
| Neighborhood Gray Tone Difference Matrix Busyness | 3.32 [2.5, 4.15] |
| Kurtosis | 1.97 [1.58, 2.37] |
| Maximum 2D Diameter Slice | 1.45 [0.20, 2.69] |
| Gray Level Size Zone Matrix Low Gray Level Emphasis | 0.33 [−0.75, 1.42] |
| Neighborhood Gray Tone Difference Matrix Contrast | 0.02 [−0.78, 0.82] |
| Skewness | −0.25 [−0.81, 0.31] |
| Gray Level Co-occurrence Matrix Cluster Shade | −0.88 [−2.78, 1.01] |
| Feature (Treatment Data and Radiomics on Liver Plus Tumor Volume) | |
| Maximum Dose | 3.83 [1.05, 6.62] |
| Neighborhood Gray Tone Difference Matrix Strength | 1.90 [0.93, 2.86] |
| Lines of Chemotherapy | 1.36 [0.38, 2.35] |
| Gray Level Size Zone Matrix Low Gray Level Emphasis | 1.01 [−0.37, 2.39] |
| KRAS Mutation | 0.65 [0.10, 1.19] |
| Carcinoembryonic Antigen at Radiotherapy | 0.48 [−1.11, 2.08] |
| Gray Level Size Zone Matrix Nonuniformity | 0.48 [−0.32, 1.27] |
| Gray Level Co-occurrence Matrix Cluster Shade | 0.17 [−0.98, 1.32] |
| Pump Before Radiotherapy | −0.08 [−1.21, 1.04] |
| Skewness | −0.29 [−0.73, 0.15] |
4. Discussion
The goal of the study was to develop a method utilizing radiomics and machine learning to predict time until local progression of CLM patients. A prediction pipeline was developed to extract radiomic features from a CT image to compute predictions with an RSF model. Prediction accuracies greater than most previous studies were achieved utilizing either clinical or imaging data.
The IBS of every dataset combination was below the threshold of 0.25, indicating that the predictions by the RSF model is non-random [31]. This suggests that there is predictive texture within the liver parenchyma and tumor volume. This is consistent with Simpson et al. [19], who observed that radiomic features were associated with recurrence and are potentially reflective of tissue abnormalities that create a metastatic environment.
There are several opportunities we aimed to address to improve on existing methods. First, CPH modeling in theory is parameterized with lower complexity than RSF and may be unable to capture nonlinear dependencies [32]. However, from our results, this is indeterminate as although the CPH did not perform better than random chance, there was a wide confidence interval overlapping with the RSF model. The IBS of the CPH model was not greater than the 0.25 threshold for only the combined radiomics and clinical subsets. Recent studies modelling survival with radiomics show no significant difference between CPH and RSF [33], [34]. Comparison of our model may require a larger sample size and to evaluate the feature selection and optimization methods other studies have used. Secondly, existing studies performing linear mapping of hazard ratios to prediction scores and may oversimplify nonlinear dependencies between variables, particularly when relying on rounding to integer scores. Thirdly, there may be predictive information missed if only analyzing clinicopathological variables. As tumor progression results in changes in tissue, there may be observable structural changes in the liver associated with survival.
Most prior studies reported a C-index under 0.60 when tested on external datasets, with one model by achieving a C-index of 0.64 [14]. However, we did not have access to all variables used, which is required for a fairer comparison between manual scoring systems and automated RSF methods.
The radiomic model from the union of the liver parenchyma and tumor with feature selection enabled achieved a C-index (95 % CI) of 0.68 (0.62–0.74). Utilizing tumor or liver parenchyma volumes only performed within the same confidence interval range. This suggests that both liver parenchyma and tumor contain textural features predictive of local control. As the features are computed as a single point-data characteristic value for the volume, it is difficult to localize the exact regions of abnormal texture. Future studies that isolate patches of the liver can be conducted to localize regions with abnormal radiomic values.
Without feature selection, there was a larger variance across the cross-validation folds. This is likely due to overfitting as the number of input variables defines the dimensionality in the optimization problem for the machine learning model. The optimized solution may be too specific to the training data, resulting in lower testing accuracy.
Utilizing only clinical data did not result in a statistically significant decrease in accuracy than with radiomics alone. In the combined mode, the two most predictive features were similarly maximum dose with a feature importance score (95 % CI) of 3.83 (1.05–6.62) and NGTDM strength with a feature importance score (95 % CI) of 1.90 (0.93–2.86). Moreover, the feature importance (95 % CI) of maximum dose decreased from 10.84 (6.35, 15.34) in the treatment data only model to 3.83 (1.05, 6.62) in the combined model, indicating that the radiomic features contribute to prediction even when treatment data is available. There are variables similar to maximum dose, such as dose covering 95 % of the planning target volume, that were removed by the feature selection due to collinearity. It should be noted that dosage is increased for tumors that may have shown radioresistance, hence some expert prior knowledge is required for this variable whereas the radiomic features are dependent only on the image.
Further validation with a diverse patient population from different centers for instance is required to evaluate generalizability and larger sample sizes may allow for less aggressive feature selection [35]. As the samples are limited to patients treated with primary or adjuvant RT, future studies may include patients before and after radiotherapy, as texture in CT scans may change after treatment. Another exclusion is of patients who are deceased. We were unable to evaluate the effect of death on the recurrence prediction model, which may require reparameterization with competing risks. Further validation requires adherence to reproducibility principles, which has been a reported challenge in expanding radiomic studies as this requires reporting of imaging acquisition settings and standardizing cutoff values for feature selection [36].
It has been a reported challenge of radiomics that these is no standardized cutoff or clinical interpretation of features [15]. For instance, positive skewness mathematically indicates asymmetric intensity distribution biased for higher intensities. However, the cause of increased skewness is indeterminate. Hypotheses include fresh blood having greater attenuation than denatured blood or high intensity occurring due to greater distribution of contrast, which is expected to be high density [37]. The observation that radiomic features are predictive motivates further studies to associate with structural changes. In future studies, histological analysis comparing regions of different skewness may reveal cellular changes that represent progression of disease.
In this work, we have developed a tumor progression prediction model for CRM treated with primary or adjuvant RT utilizing radiomic features from CT scans and AI RSF modeling. As a proof of concept, this study provides support that radiomic AI methods may be developed to aid prognostic decision making in radiation oncology. This can be extended to existing initiatives to integrate radiomics analysis to hospital picture archiving and communications systems [38] to provide new data for clinicians. Radiomic features determined to be predictive may be investigated in the future to understand structural changes reflected in radiomic observations in the CT scan for new data in analysis of liver texture.
Author Contributions
Ricky Hu: Conceptualization, Formal analysis, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. Ishita Chen: Conceptualization, Data curation, Formal analysis, Methodology, Writing – review & editing. Jacob Peoples: Formal analysis, Methodology, Software, Supervision, Writing – review & editing. Jean-Paul Salameh: Formal analysis, Writing – review & editing. Mithat Gönen: Methodology, Supervision, Writing – review & editing. Paul B. Romesser: Conceptualization, Data curation, Funding acquisition, Investigation, Project administration, Resources, Methodology, Formal analysis, Supervision, Writing – review & editing. Amber L. Simpson: Conceptualization, Data curation, Funding acquisition, Investigation, Project administration, Resources, Methodology, Formal analysis, Supervision, Writing – review & editing. Marsha Reyngold: Conceptualization, Data curation, Funding acquisition, Investigation, Project administration, Resources, Methodology, Formal analysis, Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Paul B. Romesser is a EMD Serono consultant and reports support for travel from Elekta and Philips healthcare and prior research funding from EMD Serono.
Acknowledgments
We acknowledge funding supported by the National Institutes of Health/National Cancer Institute Support Grant P30 CA008748, the National Institutes of Health/National Cancer Institute early career development award K08 CA255574 and National Cancer Institute R01 CA233888. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.phro.2022.09.004.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Abdalla E.K., Vauthey J.N., Ellis L.M., Ellis V., Pollock R., Broglio K., et al. Recurrence and outcomes following hepatic resection, radiofrequency ablation, and combined resection/ablation for colorectal liver metastases. Ann Surg. 2004;239:818–827. doi: 10.1097/01.sla.0000128305.90650.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leung U., Gönen M., Allen P.J., Kingham T.P., DeMatteo R.P., Jarnagin W.R., et al. Colorectal cancer liver metastases and concurrent extrahepatic disease treated with resection. Ann Surg. 2017;265:158–165. doi: 10.1097/SLA.0000000000001624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruers T, Van Coevorden F, Punt CJ, Pierie JE, Borel-Rinkes I, Ledermann JA, et al. Local treatment of unresectable colorectal liver metastases: results of a randomized phase II trial. J Natl Cancer Inst. 2017;109:djx015. https://doi.org/10.1093/jnci/djx015. [DOI] [PMC free article] [PubMed]
- 4.Mahadevan A., Blanck O., Lanciano R., Peddada A., Sundararaman S., D'Ambrosio D., et al. Stereotactic Body Radiotherapy (SBRT) for liver metastasis - clinical outcomes from the international multi-institutional RSSearch® Patient Registry. Radiat Oncol. 2018;13:26. doi: 10.1186/s13014-018-0969-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nordlinger B., Guiguet M., Vaillant J.C., Balladur P., Boudjema K., Bachellier P., et al. Surgical resection of colorectal carcinoma metastases to the liver. A prognostic scoring system to improve case selection, based on 1568 patients. Association Française de Chirurgie. Cancer. 1996;77:1254–1262. [PubMed] [Google Scholar]
- 6.Fong Y., Fortner J., Sun R.L., Brennan M.F., Blumgart L.H. Clinical score for predicting recurrence after hepatic resection for metastatic colorectal cancer: analysis of 1001 consecutive cases. Ann Surg. 1999;230:309–321. doi: 10.1097/00000658-199909000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwatsuki S., Dvorchik I., Madariaga J.R., Marsh J.W., Dodson F., Bonham A.C., et al. Hepatic resection for metastatic colorectal adenocarcinoma: a proposal of a prognostic scoring system. J Am Coll Surg. 1999;189:291–299. doi: 10.1016/s1072-7515(99)00089-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Konopke R., Kersting S., Distler M., Dietrich J., Gastmeier J., Heller A., et al. Prognostic factors and evaluation of a clinical score for predicting survival after resection of colorectal liver metastases. Liver Int. 2009;29:89–102. doi: 10.1111/j.1478-3231.2008.01845.x. [DOI] [PubMed] [Google Scholar]
- 9.Nagashima I., Takada T., Matsuda K., Adachi M., Nagawa H., Muto T., et al. A new scoring system to classify patients with colorectal liver metastases: proposal of criteria to select candidates for hepatic resection. J Hepatobiliary Pancreat Surg. 2004;11:79–83. doi: 10.1007/s00534-002-0778-7. [DOI] [PubMed] [Google Scholar]
- 10.Imai K., Allard M.A., Castro Benitez C., Vibert E., Sa Cunha A., Cherqui D., et al. Nomogram for prediction of prognosis in patients with initially unresectable colorectal liver metastases. Br J Surg. 2016;103:590–599. doi: 10.1002/bjs.10073. [DOI] [PubMed] [Google Scholar]
- 11.Sasaki K., Morioka D., Conci S., Margonis G.A., Sawada Y., Ruzzenente A., et al. The tumor burden score: A new, “metro-ticket” prognostic tool for colorectal liver metastases based on tumor size and number of tumors. Ann Surg. 2018;267:132–141. doi: 10.1097/SLA.0000000000002064. [DOI] [PubMed] [Google Scholar]
- 12.Rees M., Tekkis P.P., Welsh F.K., O'Rourke T., John T.G. Evaluation of long-term survival after hepatic resection for metastatic colorectal cancer: a multifactorial model of 929 patients. Ann Surg. 2008;247:125–135. doi: 10.1097/SLA.0b013e31815aa2c2. [DOI] [PubMed] [Google Scholar]
- 13.Brudvik K.W., Jones R.P., Giuliante F., Shindoh J., Passot G., Chung M.H., et al. RAS mutation clinical risk score to predict survival after resection of colorectal liver metastases. Ann Surg. 2019;269:120–126. doi: 10.1097/SLA.0000000000002319. [DOI] [PubMed] [Google Scholar]
- 14.Wang K., Liu W., Yan X.L., Li J., Xing B.C. Long-term postoperative survival prediction in patients with colorectal liver metastasis. Oncotarget. 2017;8:79927–79934. doi: 10.18632/oncotarget.20322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiz F., Viganò L., Gennaro N., Costa G., La Bella L., Boichuk A., et al. Radiomics of liver metastases: A systematic review. Cancers. 2020;12:2881. doi: 10.3390/cancers12102881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ganeshan B., Burnand K., Young R., Chatwin C., Miles K. Dynamic contrast-enhanced texture analysis of the liver: initial assessment in colorectal cancer. Invest Radiol. 2011;46:160–168. doi: 10.1097/RLI.0b013e3181f8e8a2. [DOI] [PubMed] [Google Scholar]
- 17.Miles K.A., Ganeshan B., Griffiths M.R., Young R.C., Chatwin C.R. Colorectal cancer: texture analysis of portal phase hepatic CT images as a potential marker of survival. Radiology. 2009;250:444–452. doi: 10.1148/radiol.2502071879. [DOI] [PubMed] [Google Scholar]
- 18.Creasy J.M., Cunanan K.M., Chakraborty J., McAuliffe J.C., Chou J., Gonen M., et al. Differences in liver parenchyma are measurable with CT radiomics at initial colon resection in patients that develop hepatic metastases from stage II/III colon cancer. Ann Surg Oncol. 2021;28:1982–1989. doi: 10.1245/s10434-020-09134-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simpson A.L., Doussot A., Creasy J.M., Adams L.B., Allen P.J., DeMatteo R.P., et al. Computed tomography image texture: a noninvasive prognostic marker of hepatic recurrence after hepatectomy for metastatic colorectal cancer. Ann Surg Oncol. 2017;24:2482–24490. doi: 10.1245/s10434-017-5896-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim D.W., Lee S., Kwon S., Nam W., Cha I.H., Kim H.J. Deep learning-based survival prediction of oral cancer patients. Sci Rep. 2019;9:6994. doi: 10.1038/s41598-019-43372-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Field M., Hardcastle N., Jameson M., Aherne N., Holloway L. Machine learning applications in radiation oncology. Phys Imaging Radiat Oncol. 2021;19:13–24. doi: 10.1016/j.phro.2021.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Griethuysen J.J.M., Fedorov A., Parmar C., Hosny A., Aucoin N., Narayan V., et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77:e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fotso, S. PySurvival: Open source package for survival analysis modeling, https://square.github.io/pysurvival/; 2019 [accessed 15 April 2022].
- 24.Harrell FE, Dupont C. Hmisc: Harrell miscellaneous R package version 4.6-0, https://cran.r-project.org/web/packages/Hmisc/index.html; 2021 [accessed 15 April 2022].
- 25.Liu R., Gillies D.F. Overfitting in linear feature extraction for classification of high-dimensional image data. Pattern Recogn. 2016;53:73–86. doi: 10.1016/j.patcog.2015.11.015. [DOI] [Google Scholar]
- 26.Salmeron R., Garcıa C.B., Garcıa J. Variance inflation factor and condition number in multiple linear regression. J Stat Comput Simul. 2018;88:2365–2384. doi: 10.1080/00949655.2018.1463376. [DOI] [Google Scholar]
- 27.Bourgon R., Gentleman R., Huber W. Independent filtering increases detection power for high-throughput experiments. Proc Natl Acad Sci U S A. 2010;107:9546–9551. doi: 10.1073/pnas.0914005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishwaran H., Kogalur U.B., Blackstone E.H., Lauer M.S. Random survival forests. Ann Appl Stat. 2008;2:841–860. doi: 10.1214/08-AOAS169. [DOI] [Google Scholar]
- 29.Newson R. Confidence intervals for rank statistics: Somers’ D and extensions. Stata J. 2006;6:309–334. doi: 10.1177/1536867X0600600302. [DOI] [Google Scholar]
- 30.Rodriguez J.D., Perez A., Lozano J.A. Sensitivity analysis of k-fold cross validation in prediction error estimation. IEEE PAMI. 2009;32:569–575. doi: 10.1109/TPAMI.2009.187. [DOI] [PubMed] [Google Scholar]
- 31.Steyerberg E.W., Vickers A.J., Cook N.R., Gerds T., Gonen M., Obuchowski N., et al. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology. 2010;21:128–138. doi: 10.1097/EDE.0b013e3181c30fb2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin D.Y., Wei L.J. The robust inference for the cox proportional hazards model. J Am Stat Assoc. 1989;84:1074–1078. doi: 10.2307/2290085. [DOI] [Google Scholar]
- 33.Leger S., Zwanenburg A., Pilz K., Lohaus F., Linge A., Zöphel K., et al. A comparative study of machine learning methods for time-to-event survival data for radiomics risk modelling. Sci Rep. 2017;7:13206. doi: 10.1038/s41598-017-13448-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang E., Joel M.Z., Chang H.Y., Du J., Khanna O., Omuro A., et al. Comparison of radiomic feature aggregation methods for patients with multiple tumors. Sci Rep. 2021;11:9758. doi: 10.1038/s41598-021-89114-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Menze B.H., Kelm B.M., Masuch R., Himmelreich U., Bachert P., Petrich W., et al. A comparison of random forest and its Gini importance with standard chemometric methods for the feature selection and classification of spectral data. BMC Bioinform. 2009;10:213. doi: 10.1186/1471-2105-10-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfaehler E., Zhovannik I., Wei L., Boellaard R., Dekker A., Monshouwer R, et al. A systematic review and quality of reporting checklist for repeatability and reproducibility of radiomic features. Phys Imaging Radiat Oncol. 2021;20:69–75. doi: 10.1016/j.phro.2021.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miles K.A., Ganeshan B., Hayball M.P. CT texture analysis using the filtration-histogram method: what do the measurements mean? Cancer Imaging. 2013;13:400–406. doi: 10.1102/1470-7330.2013.9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhovannik I., Pai S., da Silva Santos T.A., van Driel L.L.G, Dekker A., Fijten R., et al. Radiomics integration into a picture archiving and communication system. Phys Imaging Radiat Oncol. 2021;20:30–33. doi: 10.1016/j.phro.2021.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.