Abstract
The Tat system is a recently discovered protein export pathway that serves to translocate folded proteins, often containing redox cofactors, across the bacterial cytoplasmic membrane. Here we report that tat strains are associated with a mutant cell septation phenotype, where chains of up to 10 cells are evident. Mutant strains are also hypersensitive to hydrophobic drugs and to lysis by lysozyme in the absence of EDTA, and they leak periplasmic enzymes, characteristics that are consistent with an outer membrane defect. Both phenotypes are similar to those displayed by strains carrying point mutations in the lpxC (envA) gene. The phenotype was not replicated by mutations affecting synthesis and/or activity of all known or predicted Tat substrates.
Approximately 20% of all proteins synthesized by Escherichia coli are predicted to be located outside the cytoplasmic compartment. Most proteins destined for export are synthesized with N-terminal signal sequences that direct translocation by the general secretory (Sec) apparatus (17, 21). Sec-dependent signal sequences lack sequence similarity but have similar overall physical-chemical properties (32).
A subset of periplasmic and periplasmically oriented proteins are exported by a mechanism distinct from the Sec pathway (3, 25, 26, 34). Such proteins are synthesized with, or in some cases associate with partner proteins synthesized with, signal sequences harboring the (S/T)RRxFLK “twin-arginine” motif (2). These distinctive signal sequences target substrate proteins to the twin-arginine signal peptide-dependent protein translocase (Tat translocase) which is structurally and mechanistically related to the ΔpH-dependent protein translocase of plant thylakoid membranes (8, 29). Many Tat-dependent substrate proteins bind redox cofactors and are involved in bacterial energy metabolism. Cofactor acquisition is a cytoplasmic event and is a prerequisite for export, suggesting that proteins are translocated by the Tat apparatus in a folded conformation (22, 25).
Minimally the E. coli Tat translocase comprises four probable membrane proteins, encoded by the tatA, tatB, tatC, and tatE genes. The tatA, tatB, and tatC genes are cotranscribed with a fourth gene, tatD, encoding a soluble protein with no discernible role in Tat-dependent protein export (35). TatA, TatB, and TatE are sequence-related proteins, each of which is predicted to comprise a single N-terminal transmembrane α-helix, immediately followed by an amphipathic α-helix at the cytoplasmic side of the membrane (7, 26). TatA and TatE are more than 50% identical at the amino acid level and have overlapping functions in Tat-dependent protein export. Thus, comutation of both tatA and tatE is necessary to observe a complete block in export by the Tat pathway (26). TatB, which is more divergently related to TatA/E (approximately 25% amino acid identity), is a distinct and essential component of the Tat apparatus (27). The tatC gene encodes a highly hydrophobic protein, predicted to contain six membrane-spanning α-helices, which is critical for Tat function (5). In each case, a block in Tat-dependent protein export results in the mislocalization of Tat-dependent substrate proteins to the cytoplasmic compartment. Tat mutant strains are unable to grow anaerobically with either dimethyl sulfoxide or trimethylamine-N-oxide as sole terminal electron acceptor, reflecting the failure to correctly localize the terminal reductases. The mutant strains otherwise display no discernible growth phenotype.
In this paper, we report that strains lacking genes encoding essential Tat components form chains of up to 10 cells, which appear to be defective in cell separation. Electron microscopy reveals that the phenotype is similar to that of an lpxC (envA) mutant, which has a defect in the synthesis of outer membrane lipid A. Consistent with this, tat mutant strains are hypersensitive to hydrophobic drugs and to lysozyme-induced lysis in the absence of EDTA. We show that this phenotype is not due to the mislocalization of any single Tat substrate protein.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains used during this study are listed in Table 1. In-frame deletions of genes sufI, yacK, ydhX, ydcG, wcaM, ycdB, and yaeI were constructed based on a strategy described previously (26) and using the method of Hamilton et al. (11). The primer sequences used for construction of these mutations are available on request.
TABLE 1.
Strains used
| Strain | Genotype | Reference |
|---|---|---|
| MC4100 | F− ΔlacU169 araD139 rpsL150 relA1 ptsF rbs flbB5301 | 6 |
| RK4353 | araD139 Δ(argF-lac)U169 deoC1 flhD5301 gyrA219 non-9 ptsF25 relA1 rpsL150 | 31 |
| LCB320 | thi-1 thr-1 leu-6 lacY1 supE34 rpsL175 | 4 |
| LCB2048 | LCB320 nar25(narG-H) narU-Z::Kanr | 4 |
| JCB387 | E. coli RV ΔnirB | 9 |
| BØD | MC4100 ΔtatB | 27 |
| B1LK0 | MC4100 ΔtatC | 5 |
| JARV15 | MC4100 ΔtatAE | 26 |
| HDJ123 | Hfr Δ(gpt-lac)5 relA1 spoT1 thi-1 Δhya(Kanr) Δhyb(Kanr) ΔhycB-H(Cmr) | 28 |
| LP200 | LCB2048 napG-ccmA::Ery | 20 |
| JCB3878 | JCB378 ΔnrfC | 9 |
| RK5221 | RK4353 moa::Mucts | 31 |
| DSS501 | MC4100 torA::Mud1(Apr-lac) Δdms (Kmr) | 23 |
| JCB20480 | LCB2048 napA::Ery | 20 |
| NRS-3 | MC4100 ΔsufI | This work |
| NRS-4 | MC4100 ΔyacK | This work |
| NRS-5 | MC4100 ΔyacK ΔsufI | This work |
| TAN-1 | MC4100 ΔydhX | This work |
| TAN-2 | MC4100 ΔydcG | This work |
| TAN-3 | MC4100 ΔwcaM | This work |
| TAN-4 | MC4100 ΔycdB | This work |
| TAN-5 | MC4100 ΔyaeI | This work |
Strains were cultured either aerobically or anaerobically on Luria-Bertani (LB) medium (24), either at 37°C or (for strains carrying Mu insertions) at 30°C. Concentrations of antibiotics were as described previously (26).
Cell fixation and microscopic analysis.
Aerobically grown cells at early exponential phase (optical density at 600 nm [OD600] of 0.4) were fixed in a final concentration of 2.5% glutaraldehyde in 10 mM EDTA. The cells were subsequently harvested and suspended in 50 mM Tris-HCl (pH 8.0). The cells were treated for nucleoid analysis using DAPI (4′,6-diamidino-2-phenylindole) staining as described elsewhere (13). The fixed cells were examined using a Zeiss Axioplot microscope under both fluorescent and natural light. Photographs were taken using the microscope internal camera on technical pan film (Kodak).
Electron microscopy.
Prior to fixation, the bacteria were embedded in 1% agarose by mixing equal volumes of the E. coli liquid cultures with 2% (wt/vol) type VII low-gelling-temperature agarose (Sigma) in water at 37°C. These mixtures were plunged onto ice to set the agarose into a firm gel. Then 1-mm3 pieces of agarose containing the cells were cut out of the blocks and immediately placed in 2.5% (vol/vol) glutaraldehyde in 0.05 M sodium cacodylate (pH 7.3) and left overnight at 4°C to fix the cells.
The fixative was washed out by three successive 10-min washes in 0.05 M sodium cacodylate, and then this wash buffer was replaced with 1% (wt/vol) OsO4 in 0.05 M sodium cacodylate for 1 h at room temperature. The osmium fixation was followed by three 10-min washes in distilled water before initiation of the ethanol dehydration series. Once into 100% ethanol, the samples were infiltrated with LR White resin (London Resin Company, Reading, United Kingdom) by increasing the ratio of resin to ethanol every hour: 1:1, 2:1, and 3:1 and finally 100% resin. After remaining in resin for 24 h at room temperature, during which time there were two further changes of fresh resin, the samples were transferred into Beem capsules with more fresh LR White and placed at 60°C for 16 h to polymerize the resin.
The material was sectioned with a glass knife using a Reichert ultramicrotome (Leica, Milton Keynes, United Kingdom). Ultrathin sections of approximately 90 nm were picked up on 200-mesh copper grids which had been pyroxylin and carbon coated. The sections were stained with 2% (wt/vol) uranyl acetate for 1 h and 1% (wt/vol) lead citrate for 1 min, washed in water, and air dried. The grids were viewed in a JEOL 1200 EX transmission electron microscope at 80 kV, and photographs were taken on Kodak electron image film.
Sensitivity assays.
Antibiotic and detergent sensitivity assays were performed by seeding LB top agar with 50-μl aliquots of stationary-phase cultures of the strains of interest. After solidification, disks containing either 300 μg of erythromycin, 800 μg of rifampin, 25 μg of ampicillin, 10% sodium dodecyl sulfate (SDS) or 20% Triton X-100 were placed on the medium and incubated at 37°C for 16 h. Lysozyme sensitivity assays were performed as described elsewhere (18). Efficiency of plaquing (EOP) assays against bacteriophages P1 and λ were performed with LB medium. Cells grown overnight in LB medium were incubated with the bacteriophage for 5 min at room temperature prior to being seeded in LB top agar and incubated at 37°C.
Qualitative RNase assay.
RNase I (ribonucleate 3′-pyrimidino-oligonucleotidohydrolase; EC 3.1.4.22) leakage on agar plates was observed by the method of Weigand and Rothfield (33) except that LB medium was used.
RESULTS
Tat mutant strains show a defect in cell division.
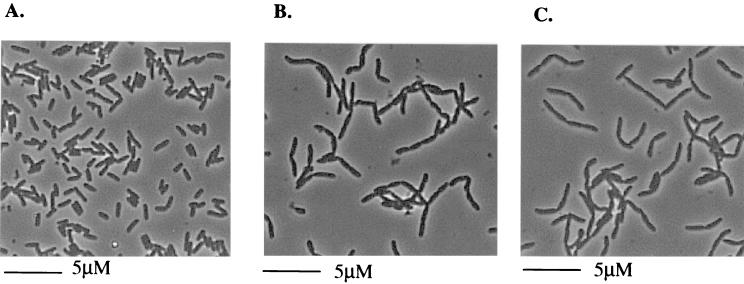
Initial light microscopic analysis of exponentially growing cultures of tat mutant strains (Fig. 1) indicated that the cells display an apparent defect in cell division. Mutant strains formed chains up to 10 cells long. Chain formation was observed irrespective of whether strains were grown aerobically or anaerobically and was observed for cells cultured at either 30 or 37°C. Strains with deletion mutations in genes tatAE (Fig. 1B), tatC (Fig. 1C), tatB or tatA to E (data not shown), all of which completely block export of Tat-dependent substrates, displayed the most severe chain-forming phenotype. Consistent with the incomplete block in Tat-dependent protein export observed in a tatA mutant (26), this strain showed only a mild chain-forming phenotype (maximum of four to five cells per chain). The tatE mutant did not form chains of cells (results not shown). The phenotype is specific to the deletion of the tat genes, since introduction of plasmid-encoded wild-type tat genes to the mutant strains reverted the morphology of the cells to that of the parental strain, MC4100 (not shown).
FIG. 1.
Light microscope analysis of MC4100 (parental strain) (A), JARV15 (ΔtatAE) (B), and B1LK0 (ΔtatC) (C). The cells were grown aerobically in LB medium to an OD600 of 0.4.
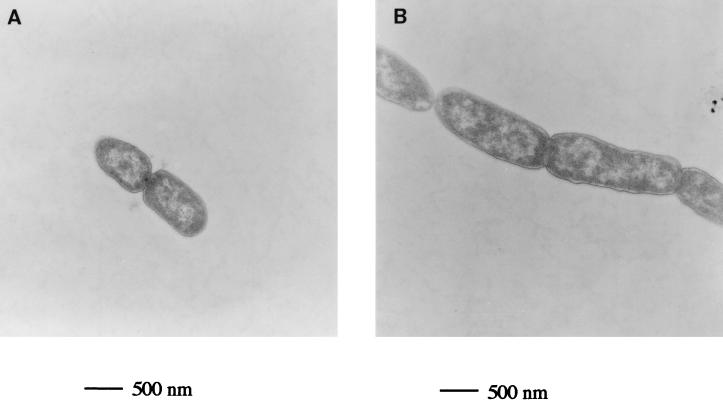
Transmission electron microscopy of the parental and ΔtatC mutant strains (Fig. 2) shows that the tat mutant strain is blocked in a late stage of cell division, since the division septum is clearly visible. This is consistent with light microscopy of DAPI-stained cells, which indicated that nucleoid replication and partitioning, both early events in cell division, were not compromised (results not shown). The tat cells are also slightly elongated compared to the parental strain. The overall morphology of the tatC strain is strikingly similar to that described for a point mutation in lpxC (envA), which also forms chains of cells separated by a division septum (18, 19).
FIG. 2.
Transmission electron micrograph of tat+ and tat deletion E. coli strains grown aerobically to an OD600 of 0.4 in LB medium. (A) MC4100 (parental strain). The averaged cell length was 0.9 μM, and the averaged breadth was 0.6 μM. (B) B1LK0 (ΔtatC). The averaged cell length was 1.8 μM, and the average breadth was 0.6 μM.
Antibiotic and detergent hypersensitivity of tat mutant strains.
The lpxC gene encodes UDP-3-O-acyl-N-acetylglucosamine deacetylase, which is the second enzyme, and catalyzes the first committed step, of lipid A biosynthesis (30, 38). Although the gene is essential for cell viability (1), a number of point mutations have been described, at least one of which decreases but does not abolish the deacetylase activity (19, 38). A feature of lpxC mutants is that they are associated with decreased levels of outer membrane lipid A and are supersensitive to antibiotics. To further explore the similarities between tat and lpxC strains, we tested the sensitivity of tat strains to the antibiotics erythromycin, rifampin, and ampicillin. As shown in Table 2, both the tatB and tatC mutants were much more sensitive to these antibiotics than the parental strain, suggesting that the permeability barrier of the outer membrane is compromised. This inference is supported by the observation that the tat strains are also sensitive to the anionic detergent SDS but not the nonionic surfactant Triton X-100 (Table 2).
TABLE 2.
Antibiotic sensitivities and EOP conferred by tat mutant strains
| Relevant genotype (strain) | Zone of inhibition (mm)a
|
EOPb
|
|||||
|---|---|---|---|---|---|---|---|
| E | R | A | S | T | P1 | λ | |
| tat+ (MC4100) | 3 | 9 | 4 | 0 | 0 | 1 | 1 |
| ΔtatB (BØD) | 7 | 14 | 8 | 7 | 0 | <10−8 | 0.79 |
| ΔtatC (B1LK0) | 7 | 13 | 9 | 7 | 0 | <10−8 | 0.73 |
Measured as diameter on a 13-mm-diameter disk. E, R, A, S, and T disks contained erythromycin (300 μg), rifampin (800 μg), ampicillin (25 μg), SDS (10%), and Triton X-100 (20%), respectively (n = 3 to 4).
The EOP of the parental strain MC4100 was taken as 1, and the mutant EOP values were normalized accordingly.
tat mutants are periplasmically leaky.
A further feature of lpxC strains is that they leak periplasmic enzymes into the growth medium (37). We tested whether tat mutants also showed periplasmic leakage by looking for the release of periplasmic RNase I into agar plates impregnated with RNA. The tatB and tatC mutant strains leaked RNase, while the parental strain did not (data not shown).
Lysozyme sensitivity of tat mutants.
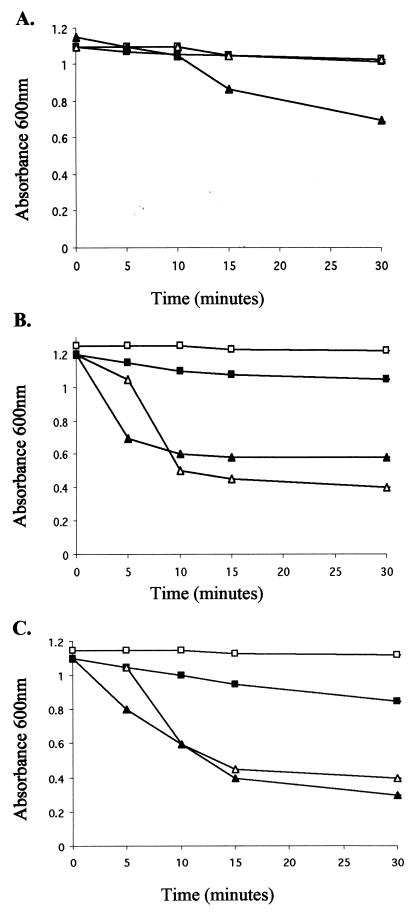
We next tested whether the tat strains were sensitive to cell lysis by lysozyme in the absence of EDTA. In wild-type strains, EDTA is required to chelate divalent metal cations that serve to stabilize the structure of the lipopolysaccharide (LPS), in order to allow access of lysozyme to the underlying murein sacculus. This is confirmed in Fig. 3A, where significant time-dependent lysis of the parental strain, MC4100, is observed only in the presence of both EDTA and lysozyme. In marked contrast, strains deleted for tatAE (JARV15 [Fig. 3B]) and tatC (B1LK0 [Fig. 3C]) show very rapid and dramatic lysis upon addition of lysozyme alone. This result is consistent with the observation that the lpxC strain D22 is also highly sensitive to lysozyme-induced cell lysis in the absence of EDTA (18) and further supports the contention that tat strains have a defect in the outer membrane.
FIG. 3.
Lysozyme and EDTA treatment of MC4100 (A), JARV15 (ΔtatAE) (B), and B1LK0 (ΔtatC) (C). The cells were grown to an OD600 of 1.0 to 1.3, harvested, washed in 50 mM Tris-HCl (pH 7.4), and then treated with either no additions (□), 0.25 mM EDTA (pH 8.0) (■), 100 μg of lysozyme/ml (▴), or 0.25 mM EDTA and 100 μg of lysozyme/ml (▵). Absorbance of the cells was monitored at 600 nm.
tat mutants are highly resistant to infection by P1 phage.
During the course of experiments with tat mutants, we noted that they were extremely difficult to transduce with P1 phage (27). A titration of P1 against the parental, tatB mutant, and tatC mutant strains (Table 2) indicates that the tat mutants are at least 108 times more resistant to lysis by P1 than the tat+ strain. Resistance to P1 infection has not been reported for lpxC strains, but high levels of resistance are associated with a mutation in the galU gene encoding UDP-glucose pyrophosphorylase which adds glucose residues to the LPS core (10). In contrast, the tat strains were not significantly resistant to lysis by phage λ, which infects cells by binding to LamB, the maltose receptor, situated in the outer membrane. These observations suggest that tat strains specifically have a defect in the LPS component of the outer membrane.
The cell envelope defect in tat strains is unlikely to result from mislocalization of any single Tat substrate.
One possible explanation for the observed cell envelope defect of tat strains is that there is a block in the translocation of a Tat-dependent substrate responsible for outer membrane assembly and/or cell separation. Substrates of the E. coli Tat pathway are invariably synthesized as preproteins with distinctive N-terminal twin-arginine signal peptides (2). Tat signal peptides can be readily identified by virtue of both conserved sequence motifs and overall physicochemical properties. In terms of physicochemical properties, Tat signal peptides encompass a positively charged N terminus followed by a hydrophobic region (rich in glycine residues) and are usually punctuated by a short, positively charged C-terminal domain (2, 3). The conserved (S/T)RRxFLK twin-arginine motif is present at the N-terminal/hydrophobic region boundary, and an AxA signal peptidase recognition sequence can usually be identified within the C-terminal region (2). Using these criteria, analysis of the E. coli genome sequence reveals 23 open reading frames encoding proteins with plausible twin-arginine signal sequences. Of these, at least 16 bind or are predicted to bind redox cofactors, and many play characterized roles in anaerobic respiration.
In an attempt to ascertain whether any one of these substrates was required for cell separation, we undertook a systematic screen of cellular morphology in either strains with null mutations in genes encoding Tat-dependent proteins or in strains deficient in molybdenum cofactor biosynthesis and therefore in export and function of Tat-dependent molybdoenzymes. Null mutant strains for the molybdoenzymes TorA, DmsA, and NapA as well as the molybdenum cofactor mutant RK5221, which in addition would be expected to have mislocalized BisZ, FdnG, FdoG, YnfE, YnfF, and YagT, appeared wild type when analyzed by light microscopy (results not shown). Further, strains deleted for both uptake hydrogenases, HYD1 and HYD2, the iron sulfur proteins NapG and NrfC, the putative multicopper oxidase YacK and its non-copper binding homologue SufI, and uncharacterized proteins YdhX, WcaM, YdcG, YcdB, and YaeI all displayed wild-type cellular morphology (results not shown). Thus, it is unlikely that the observed phenotype arises from the cellular mislocalization of any one of these proteins alone. We stress that the observed tat phenotype may, however, arise from the inability of the cell to properly localize an entire set of proteins. We did not construct a mutation in the gene encoding AmiA since although it has a reasonable twin-arginine signal sequence, pulse-chase analysis indicates that it is not a substrate of the Tat pathway (N. R. Stanley, B. C. Berks, and T. Palmer, unpublished data). It remains a possibility that the phenotype is due to a defect in the translocation of the putative iron-dependent hydrolase YahJ. This has yet to be tested since we have been unable to construct a deletion mutation in the encoding gene.
DISCUSSION
In this paper, we report the observation that E. coli strains defective in components of the Tat protein export pathway are impaired in the cell separation stage of cell division. It is likely that this phenotype is linked to a defect in the biosynthesis of the outer membrane, since tat mutant strains are supersensitive to killing by hydrophobic drugs and to lysis by lysozyme in the absence of EDTA and are resistant to infection by P1 phage. An obvious explanation to account for the observations is that a protein substrate normally exported by the Tat pathway is required for one of the stages of outer membrane assembly and/or cell separation. In this context, it should be noted that the gene encoding SufI, a previously characterized Tat substrate, was first identified as a multicopy suppressor of the cell division phenotype of an ftsI mutant (16). This suggests a role for SufI in the division process. However, strains deleted for genes encoding both SufI and its homologue YacK showed wild-type morphology. Moreover, strains affected in the synthesis and/or assembly of a further 19 putative Tat substrate proteins did not exhibit the chain-forming phenotype. It remains a possibility that the observed phenotype is due to the mislocalization of a hitherto unsuspected Tat substrate protein, a nonprotein substrate, or a combination of several previously characterized protein substrates. It should also be considered that the loss of the Tat translocase itself might have a direct effect on LPS biosynthesis, although a mechanism by which such a situation could arise is unclear.
The cell separation phenotype observed with tat mutant strains is strikingly similar to that previously described for a point mutation within the lpxC (envA) gene. However, it should be noted that although lpxC is essential for cell viability, the tat strains described here show no growth defect (other than with dimethyl sulfoxide or trimethylamine-N-oxide as sole terminal electron acceptor). LpxC is a cytoplasmic metal-containing deacetylase which catalyzes the first committed step of lipid A biosynthesis, one of the major components of the E. coli outer membrane (14, 15, 38). In addition to a defect in cell separation, lpxC mutations are associated with increased permeability of the outer membrane, reflected in an increased sensitivity to hydrophobic drugs and other compounds. Due to the similar phenotypes shown by these different mutant strains, we investigated whether the LpxC protein was destabilized in tat strains by Western blotting. However, cellular levels of LpxC were similar in both the wild-type and tat mutant backgrounds (results not shown), indicating that the observed phenotype is probably not due to a direct effect on LpxC.
The lpxC mutation is also associated with a sixfold decrease of N-acetylmuramyl-l-alanine amidase activity. The amidase enzyme is believed to cleave the division septum and probably accounts for the chain-forming phenotype of lpxC strains (35). The genome of E. coli codes for three putative periplasmic N-acetylmuramyl-l-alanine amidases (13). Of these, the AmiA protein bears a signal sequence that harbors a reasonable twin-arginine motif, differing only in the substitution of a valine at the consensus phenylalanine position. However, pulse-chase experiments indicate that AmiA is not a substrate for the Tat pathway, and therefore it is unlikely that tat mutants fail to export enzymes responsible for septal murein cleavage.
In conclusion, E. coli tat mutant strains display an unexpected cell separation morphology and cell envelope defect, the reason for which is unclear. It would be interesting to ascertain whether this phenotype is also associated with tat mutations in other bacteria.
ACKNOWLEDGMENTS
We thank Katherine Young and Merck Research Laboratories for providing anti-LpxC antiserum. We thank J.-V. Höltje for helpful discussions.
We acknowledge the Norwich Research Park (N.R.S.) and the Royal Society (T.P.) for support.
REFERENCES
- 1.Beall B, Lutkenhaus J. Sequence analysis, transcriptional organization, and insertional mutagenesis of the envA gene of Escherichia coli. J Bacteriol. 1987;169:5408–5415. doi: 10.1128/jb.169.12.5408-5415.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berks B C. A common export pathway for proteins binding complex redox cofactors? Mol Microbiol. 1996;22:393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- 3.Berks B C, Sargent F, Palmer T. The Tat protein export pathway. Mol Microbiol. 2000;35:260–274. doi: 10.1046/j.1365-2958.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- 4.Blasco F, Nunzi F, Pommier J, Brasseur R, Chippaux M, Giordano G. Involvement of the narJ or narW gene product in the formation of active nitrate reductase in Escherichia coli. Mol Microbiol. 1992;6:209–219. doi: 10.1111/j.1365-2958.1992.tb02003.x. [DOI] [PubMed] [Google Scholar]
- 5.Bogsch E G, Sargent F, Stanley N R, Berks B C, Robinson C, Palmer T. An essential component of a novel bacterial export system with homologues in plastids and mitochondria. J Biol Chem. 1998;273:18003–18006. doi: 10.1074/jbc.273.29.18003. [DOI] [PubMed] [Google Scholar]
- 6.Casadaban M J, Cohen S N. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci USA. 1979;76:4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chanal A, Santini C-L, Wu L-F. Potential receptor function of three homologous components, TatA, TatB and TatE, of the twin-arginine signal sequence-dependent metalloenzyme translocation pathway in Escherichia coli. Mol Microbiol. 1998;30:673–678. doi: 10.1046/j.1365-2958.1998.01095.x. [DOI] [PubMed] [Google Scholar]
- 8.Dalbey R E, Robinson C. Protein translocation into and across the bacterial plasma membrane and the plant thylakoid membrane. Trends Biochem Sci. 1999;24:17–22. doi: 10.1016/s0968-0004(98)01333-4. [DOI] [PubMed] [Google Scholar]
- 9.Darwin A, Tormay P, Page L, Griffiths L, Cole J. Identification of the formate dehydrogenases and genetic determinants of formate-dependent nitrite reduction by Escherichia coli K12. J Gen Microbiol. 1993;139:1829–1840. doi: 10.1099/00221287-139-8-1829. [DOI] [PubMed] [Google Scholar]
- 10.Franklin N C. Mutation in the gal U gene of E. coli blocks phage P1 infection. Virology. 1969;38:189–191. doi: 10.1016/0042-6822(69)90144-5. [DOI] [PubMed] [Google Scholar]
- 11.Hamilton C M, Aldea M, Washburn B K, Babitzke P, Kushner S R. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiraga S, Niki H, Ogura T, Ichinose C, Mori H, Ezaki B, Joffe A. Chromosome partitioning in Escherichia coli: novel mutants producing anucleate cells. J Bacteriol. 1989;171:1496–1505. doi: 10.1128/jb.171.3.1496-1505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Höltje J-H. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hyland S A, Eveland S S, Anderson M S. Cloning, expression and purification of UDP-3-O-acyl-GlcNAc deacetylase from Pseudomonas aeruginosa: a metalloamidase of the lipid A biosynthesis pathway. J Bacteriol. 1997;179:2029–2037. doi: 10.1128/jb.179.6.2029-2037.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackman J E, Fierke C A, Tumey L N, Pirrung M, Uchiyama T, Tahir S H, Hindsgaul O, Raetz C R H. Antibacterial agents that target lipid A biosynthesis in Gram-negative bacteria. J Biol Chem. 1999;275:11002–11009. doi: 10.1074/jbc.275.15.11002. [DOI] [PubMed] [Google Scholar]
- 16.Kato J-I, Nishimura Y, Yamada M, Suzuki H, Hirota Y. Gene organization in the region containing a new gene involved in chromosome partition in Escherichia coli. J Bacteriol. 1988;170:3967–3977. doi: 10.1128/jb.170.9.3967-3977.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manting E H, Driessen A J M. Escherichia coli translocase: the unravelling of a molecular machine. Mol Microbiol. 2000;37:226–238. doi: 10.1046/j.1365-2958.2000.01980.x. [DOI] [PubMed] [Google Scholar]
- 18.Normark S, Boman H G, Bloom G D. Cell division in a chain-forming envA mutant of Escherichia coli K12. Acta Pathol Microbiol Scand Sect B. 1971;79:651–664. doi: 10.1111/j.1699-0463.1971.tb00093.x. [DOI] [PubMed] [Google Scholar]
- 19.Normark S, Boman H, Matsson E. Mutant of Escherichia coli with anomalous cell division and ability to decrease episomally and chromosomally mediated resistance to ampicillin and several other antibiotics. J Bacteriol. 1969;97:1334–1342. doi: 10.1128/jb.97.3.1334-1342.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Potter L C, Cole J A. Essential roles for the products of the napABCD genes but not napFGH, in periplasmic nitrate reduction by Escherichia coli K12. Biochem J. 1999;344:69–76. doi: 10.1042/0264-6021:3440069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pugsley A G. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodrigue A, Chanal A, Beck K, Müller M, Wu L-F. Co-translocation of a periplasmic enzyme complex by a hitchhiker mechanism through the bacterial Tat pathway. J Biol Chem. 1999;274:13223–13228. doi: 10.1074/jbc.274.19.13223. [DOI] [PubMed] [Google Scholar]
- 23.Sambasivarao D, Weiner J H. Differentiation of the multiple S- and N-oxide reducing activities of Escherichia coli. Curr Microbiol. 1991;23:105–110. [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Santini C-L, Ize B, Chanal A, Müller M, Giordano G, Wu L-F. A novel Sec-independent periplasmic translocation pathway in Escherichia coli. EMBO J. 1998;17:101–112. doi: 10.1093/emboj/17.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sargent F, Bogsch E G, Stanley N R, Wexler M, Robinson C, Berks B C, Palmer T. Overlapping functions of components of a Sec-independent protein export pathway. EMBO J. 1998;17:3640–3650. doi: 10.1093/emboj/17.13.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sargent F, Stanley N R, Berks B C, Palmer T. Sec-independent protein translocation in Escherichia coli: a distinct and pivotal role for the TatB protein. J Biol Chem. 1999;274:36073–36083. doi: 10.1074/jbc.274.51.36073. [DOI] [PubMed] [Google Scholar]
- 28.Sauter M, Böhm R, Böck A. Mutational analysis of the operon (hyc) determining hydrogenase 3 formation in Escherichia coli. Mol Microbiol. 1992;6:1523–1532. doi: 10.1111/j.1365-2958.1992.tb00873.x. [DOI] [PubMed] [Google Scholar]
- 29.Settles A M, Yonetani A, Baron A, Bush D R, Cline K, Martienssen R. Sec-independent protein translocation by the maize Hcf106 protein. Science. 1997;278:1467–1470. doi: 10.1126/science.278.5342.1467. [DOI] [PubMed] [Google Scholar]
- 30.Sorensen P G, Lutkenhaus J, Young K, Eveland S S, Anderson M S, Raetz C R H. Regulation of UDP-3-O-[R-3-hydroxymyristoyl]-N-acetylglucosamine deacetylase in Escherichia coli. J Biol Chem. 1996;271:25898–25905. doi: 10.1074/jbc.271.42.25898. [DOI] [PubMed] [Google Scholar]
- 31.Stewart V, MacGregor C H. Nitrate reductase in Escherichia coli K-12: involvement of the chlC, chlE, and chlG loci. J Bacteriol. 1982;151:788–799. doi: 10.1128/jb.151.2.788-799.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- 33.Weigand R A, Rothfield L I. Genetic and physiological classification of periplasmic-leaky mutants of Salmonella typhimurium. J Bacteriol. 1976;125:340–345. doi: 10.1128/jb.125.1.340-345.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiner J H, Bilous P T, Shaw G M, Lubitz S P, Frost L, Thomas G H, Cole J A, Turner R J. A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins. Cell. 1998;93:93–101. doi: 10.1016/s0092-8674(00)81149-6. [DOI] [PubMed] [Google Scholar]
- 35.Wexler M, Sargent F, Jack R L, Stanley N R, Bogsch E G, Robinson C, Berks B C, Palmer T. TatD is a cytoplasmic protein with DNAse activity. No requirement for TatD-family proteins in Sec-independent protein export. J Biol Chem. 2000;275:16717–16722. doi: 10.1074/jbc.M000800200. [DOI] [PubMed] [Google Scholar]
- 36.Wolf-Watz H, Normark S. Evidence for a role of N-acetylmuramyl-l-alanine amidase in septum separation in Escherichia coli. J Bacteriol. 1976;128:580–586. doi: 10.1128/jb.128.2.580-586.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young K, Silver L L. Leakage of periplasmic enzymes from envA1 strains of Escherichia coli. J Bacteriol. 1991;173:3609–3614. doi: 10.1128/jb.173.12.3609-3614.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Young K, Silver L L, Bramhill D, Cameron P, Eveland S S, Raetz C R H, Hyland S A, Anderson M S. The envA permeability/cell division gene of Escherichia coli encodes the second enzyme of lipid A biosynthesis. J Biol Chem. 1995;270:30384–30391. doi: 10.1074/jbc.270.51.30384. [DOI] [PubMed] [Google Scholar]