Abstract
Previous research has indicated an intimate functional communication between mast cells (MCs) and neutrophils during inflammatory conditions, but the nature of such communication is not fully understood. Activated neutrophils are known to release DNA-containing extracellular traps (neutrophil extracellular traps [NETs]) and, based on the known ability of tryptase to interact with negatively charged polymers, we here hypothesized that tryptase might interact with NET-contained DNA and thereby regulate NET formation. In support of this, we showed that tryptase markedly enhances NET formation in phorbol myristate acetate-activated human neutrophils. Moreover, tryptase was found to bind vividly to the NETs, to cause proteolysis of core histones and to cause a reduction in the levels of citrullinated histone-3. Secretome analysis revealed that tryptase caused increased release of numerous neutrophil granule compounds, including gelatinase, lactoferrin, and myeloperoxidase. We also show that DNA can induce the tetrameric, active organization of tryptase, suggesting that NET-contained DNA can maintain tryptase activity in the extracellular milieu. In line with such a scenario, DNA-stabilized tryptase was shown to efficiently degrade numerous pro-inflammatory compounds. Finally, we showed that tryptase is associated with NET formation in vivo in a melanoma setting and that NET formation in vivo is attenuated in mice lacking tryptase expression. Altogether, these findings reveal that NET formation can be regulated by MC tryptase, thus introducing a novel mechanism of communication between MCs and neutrophils.
Keywords: Mast cells, Neutrophils, Tryptase, Neutrophil extracellular traps, Histones, Melanoma
Introduction
Mast cells (MCs) are hematopoietic immune cells with an impact on numerous physiological and pathological settings. For example, MCs are major players in allergic conditions and can also contribute to the pathology of numerous other pathological settings, including rheumatoid arthritis, cardiovascular complications, bone disease, fibrosis, and various inflammatory conditions of the skin [1, 2, 3, 4, 5, 6]. On the other hand, MCs are also known to have beneficial functions, including a major role in degrading various toxins and also in the host defense against parasitic and bacterial insults [7, 8].
There is considerable evidence that MCs can have an impact on neutrophils. This was originally proposed from models of sepsis, where that lack of MCs was associated with reduced recruitment of neutrophils into the peritoneal cavity of mice, and it was suggested that the ability of MCs to recruit neutrophils is dependent on their release of TNF [9, 10]. This notion has recently gained further support by showing that MCs can directly release TNF into the circulation, thereby promoting neutrophil recruitment [11]. Further, it has been suggested that the ability of MCs to recruit neutrophils is dependent on their release of neutrophil-attracting CXCL1/CXCL2 chemokines [12], whereas another study linked the neutrophil-attracting capacity of MCs to their expression of IL-33 [13]. It has also been shown that MCs can enhance neutrophil recruitment in the context of allograft rejection [14] and in several other settings [15, 16, 17, 18, 19, 20]. However, adding complexity to this issue, it has been shown that MCs can have a dampening impact on neutrophil recruitment in a setting of acute ischemic kidney injury [21], and MC products have been shown to be cytotoxic for neutrophils [22] and to limit neutrophil recruitment by degrading TNF [23].
When neutrophils have been attracted to the site of bacterial challenge, they will participate in the clearance of the invading bacteria. This can occur by phagocytosis and by the release of antibacterial compounds. Additionally, it is now established that neutrophils can neutralize bacteria by releasing extracellular traps (neutrophil extracellular traps [NETs]), a meshwork containing DNA, other nuclear constituents and compounds derived from the neutrophil secretory granules [24, 25, 26].
Although the collective evidence from previous research has firmly linked MCs to processes in which neutrophils are recruited, there is limited knowledge of whether MCs can have a more direct impact on neutrophil function. Under inflammatory conditions, MCs are prone to undergo degranulation, in response to various secretagogues that might be released during such conditions [27]. When MCs degranulate, they release large quantities of preformed mediators that are stored within the MC secretory granules. These include histamine and other biogenic amines, various cytokines (such as TNF), growth factors, proteoglycans as well as remarkably large amounts of neutral proteases, the latter including tryptase, chymase, and carboxypeptidase A3 (CPA3) [28, 29, 30]. Hence, neutrophils invading the inflammatory tissues will be exposed to these granule mediators. A plausible scenario could thus be that the function of neutrophils might be influenced through their interaction with the preformed mediators that are released by degranulating MCs in the inflammatory milieu.
MC tryptase is a tetrameric serine protease, in which all of the active sites are facing a narrow central pore. Due to this organization, tryptase is resistant to all known macromolecular protease inhibitors and has a relatively narrow substrate cleavage profile [31, 32]. It is established that the tryptase tetramer is stabilized by interaction with heparin proteoglycan, carrying a high negative charge density due to extensive sulfation and carboxylation [33]. However, in a recent study, we showed that tryptase can additionally be stabilized by DNA [34]. Considering that NET formation is a prominent event during inflammatory conditions and that such conditions also are associated with a massive release of tryptase from MCs, we here hypothesized that MC tryptase may interact with the DNA component of NETs and that tryptase potentially might affect neutrophils through such an interaction. Indeed, the findings presented here indicate that tryptase can have a major impact on NETs, introducing a novel mechanism by which MCs can influence the function of neutrophils under inflammatory conditions.
Materials and Methods
Reagents
Recombinant human (rh) cytokines and chemokines were purchased from ImmunoTools (Friesoythe, Germany). rh β-tryptase was prepared as described [35]. Porcine intestinal mucosa heparin, phorbol myristate acetate (PMA), calcium ionophore A23187, and lipopolysaccharide from Escherichia coli O111:B4 (lipopolysaccharide [LPS]) were from Sigma-Aldrich (Steinheim, Germany). N-formyl-Met-Leu-Phe (fMLP) was from Tocris (Abingdon, UK). NucBlueTM live ReadyProbesTM reagent (Hoechst-33342), NucGreenTM dead 488 ReadyProbesTM reagent (Sytox-green), ProLongTM diamond antifade mountant, goat anti-rat Alexa-647 antibody, goat anti-rabbit Alexa-555 antibody and BS3 (bis[sulfosuccinimidyl]suberate) were from Thermo Fisher Scientific (Waltham, MA, USA). Rat anti-mouse Ly-6G and Ly-6C antibody were from BD-Pharmingen (San Jose, CA, USA), rabbit anti-human histone histone 2A (H2A), H2B, H3, and H4 antibodies were from Abcam (Cambridge, UK) and rabbit anti-human histone H3 citrulline Arg2, −8 and −17 antibody was from Novus (Centennial, CO, USA).
Cleavage of Cytokines by Recombinant Human β-Tryptase
rh tryptase (1 ng/μL) was prepared in PBS (pH 7.2) alone, or either with DNA (∼10 ng/μL) or heparin (∼10 ng/μL) and incubated for 1 h on a shaker at room temperature. Cytokines and chemokines were dissolved in sterile water or PBS, following the instructions from the supplier (ImmunoTools); the final concentrations were 0.1–0.25 μg/μL. Subsequently, 5 μL (∼1.25 μg) of the cytokine in a 1.5 mL tube was mixed with either rh β-tryptase (∼50 ng) alone, DNA-stabilized β-tryptase or heparin-stabilized β-tryptase, followed by incubation at 37°C for 2.5 h. Controls were prepared by replacing β-tryptase with sterile water or PBS. After incubation for 2.5 h, the reactions were terminated by the addition of 3 μL of 4× SDS-PAGE sample buffer (including 0.5 μL of β-mercaptoethanol) to each sample, followed by heating for 7 min at 85°C. The reaction mixtures were then analyzed on 4–20% mini-protean tgx stain-free gels (Bio-Rad, Hercules, CA, USA). To visualize the proteins, the gels were stained overnight in colloidal Coomassie staining solution and washed with H2O for 2–4 h.
Tryptase Activity
rh β-tryptase (1 ng/μL), in PBS (pH 7.2; 100 μL total volume) was incubated either alone or in the presence of heparin or DNA (double-stranded). Incubations were performed in 96-well plates. The plates were incubated for 1 h at 37°C, followed by measurement of absorbance after adding 20 μL from a stock solution (10 mM in H2O) of the chromogenic substrate S-2288 (Chromogenix, Milano, Italy). Tryptase activity was monitored by reading the absorbance at 405 nm over 60 min using a microplate reader (M200-TECAN Infinite). Assays were performed in triplicates.
DNA Isolation
DNA was isolated from HMC-1 cells using the QIAamp DNA mini kit, following the manufacturer's instructions.
Isolation of Human Neutrophils
Fresh human blood was collected from healthy donors in tubes containing preservative-free ACD and mixed 1:1 with RPMI medium. The mixture was carefully layered on the top of 15 mL of 67% Percoll in a 50-mL tube. Tubes were centrifugated at 1,000 g for 30 min at 4°C without brake. Mononuclear cells were removed, and the tube was rinsed twice with PBS. The cell pellet was resuspended in 10 mL ice-cold PBS and kept on ice. Red blood cells were lysed by adding red blood cell lysis buffer (Roche, Mannheim, Germany). Cells were centrifugated at 400 g for 5 min at 4°C and lysis was repeated once or twice until no red blood cells were visible. The remaining pellet was resuspended to 1 × 106 cells/mL in RPMI and kept on ice. The experiments on human neutrophils were approved by the relevant Ethical Committee (Etikprövningsmyndigheten Dnr 2020-05080).
Neutrophil Activation
Freshly isolated human neutrophils (0.2 × 106 cells/mL in RPMI) were distributed in triplicates into 24-well plates. PMA was used at a final concentration of 50 nM, N-fMLP at 0.1 μM, A23187 at 4 μM, and LPS at 10 μg/mL. rh β-tryptase was used at a final concentration of 100 nM. The plates were incubated at 37°C (5% CO2) for different time periods. Supernatants were collected and cells were harvested by adding 100 μL of SDS Laemmli buffer.
Western Blot
Western blot was performed using 4–20% mini-protean tgx stain-free gels, followed by transfer with Bio-Rad trans-blot turbo system using trans-blot turbo mini 0.2 μm PVDF transfer packs. Membranes were blocked for 1 h with intercept (PBS) blocking buffer from Li-Cor (Lincoln, NE, USA), followed by 3× washing with TBS with 0.1% tween. Membranes were incubated with primary antibodies overnight at 4°C, followed by 3× washing with TBS with 0.1% tween and incubation for 1 h at room temperature with Li-Cor secondary antibody (diluted 1:15,000 in PBS). Membranes were scanned using the Odyssey CLx fluorescence imaging system (Li-Cor).
Tryptase Binding to NETs
Freshly isolated human neutrophils (0.2 × 106 cells/mL in RPMI) were distributed in duplicates into 8-chamber polystyrene vessel tissue culture-treated glass slides (Corning, Big Flats, NY, USA). PMA was used at a final concentration of 50 nM, and slides were incubated at 37°C (5% CO2) for different time periods. After incubation, supernatants were removed and samples were washed 3× with PBS. 100 nM of β-tryptase labeled with Alexa-488 (in PBS) was added, followed by incubation for 15 min (37°C; 5% CO2). Supernatants were removed and samples were washed 3× with PBS, fixed with 4% paraformaldehyde, and stained with NucBlueTM live ReadyProbesTM. Samples were washed 3× with PBS and mounted using ProLongTM diamond antifade mountant. Images were acquired using 63× NA 1.40 oil objective on Zeiss LSM700 confocal microscope. Images were analyzed using Imaris software (Bitplane, Zurich, Switzerland).
Immunohistochemistry
Freshly isolated human neutrophils (0.2 × 106 cells/mL in RPMI) were distributed in duplicates into 8-chamber polystyrene vessel tissue culture treated glass slides. PMA was used at a final concentration of 50 nM and β-tryptase at 100 nM. The slides were incubated at 37°C (5% CO2) for 2 h. Supernatants were removed, samples were fixed and permeabilized with methanol for 10 min, washed 3× with PBS and blocked for 15 min at room temperature with 10% goat serum in PBS. Primary antibody was incubated overnight at 4°C; slides were washed 3× with PBS and incubated with the secondary antibody for 1 h at room temperature. After 3× washing with PBS, slides were mounted using ProLongTM Diamond Antifade Mountant. Images were acquired using 63× NA 1.40 oil objective on a Zeiss LSM700 confocal microscope and a Zeiss LSM710 SIM-Elyra super-resolution microscope with automatic segmentation (Zeiss, Oberkochen, Germany), or a Nikon 90i fluorescent microscope with ×20 objective (Nikon, Melville, NY). Images were analyzed using ImageJ open source image processing software and Imaris software (Bitplane).
B16F10 Tumor Sections
B16F10 tumors from wild-type and Mcpt6 knockout mice were obtained as described [36]. Tissue sections from paraffin-embedded tissue (5 μm) were deparaffinized and first stained with NucGreenTM Dead 488 ReadyProbes, followed by NucBlueTM Live ReadyProbes. Samples were then fixed and permeabilized with methanol for 10 min, washed 3× with PBS and blocked for 15 min at room temperature with 10% goat serum in PBS. Tissue sections were then incubated overnight at 4°C with rat anti-mouse Ly-6G and Ly-6C antibody diluted 1:500 in PBS. Sections were washed 3 times in PBS and incubated for 1 h at room temperature with goat anti-rat Alexa-647 antibody (1:1,000 in PBS), washed 3× with PBS and incubated at room temperature for 2 h with rabbit anti-Mcpt6 immune serum (1:500 in PBS). Sections were washed 3× with PBS and incubated for 1 h at room temperature with goat anti-rabbit Alexa-555 antibody (1:1,000 in PBS). Samples were washed 3× with PBS and mounted using ProLongTM Diamond Antifade Mountant. Images were acquired using a Zeiss LSM700 confocal microscope and were analyzed using Imaris software. The animal experiments were approved by the relevant Ethical Committee (Uppsala djurförsöksetiska nämnd Dnr 5.8.18-04096/2019).
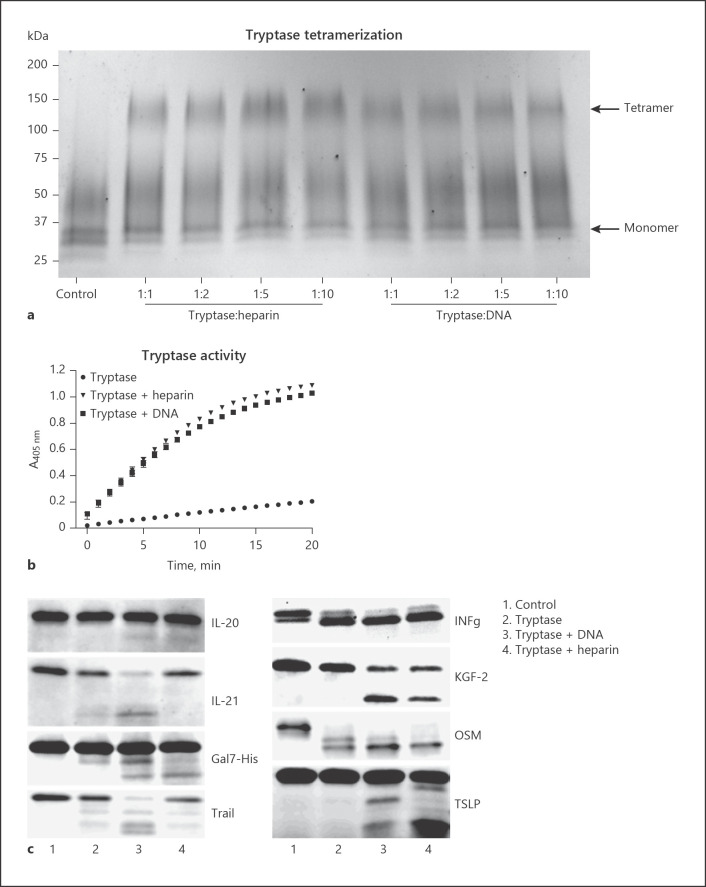
Tryptase Tetramerization
1.5-mL tubes containing rh β-tryptase (1 μg) alone or with increasing concentrations of heparin or DNA in PBS were incubated for 15 min at 37°C in an orbital shaker. Samples were adjusted to 30 μL final volume. After incubation, 10 μL of BS3 cross-linker (10 mg/mL) was added, followed by incubation for 5 min at room temperature. Ten microliters of 5× concentrated Laemmli buffer was added, and samples were analyzed by PAGE.
Mass Spectrometry
Supernatants from untreated neutrophils or neutrophils treated with 50 nM PMA and/or 100 nM β-tryptase were collected. The samples were reduced, alkylated, and digested with trypsin according to a standard operating procedure. Next, the collected peptides were vacuum centrifuged to dryness using a SpeedVac system. Thereafter, the samples were resolved in 30 μL 0.1% formic acid and further diluted 4×. The resulting peptides were separated in reversed-phase on a C18-column and electrosprayed on line to a QExactive Plus Orbitrap mass spectrometer (Thermo Finnigan) with 35 min gradient. Tandem mass spectrometry was performed applying HCD. Database searches were performed using the Sequest algorithm, embedded in Proteome Discoverer 1.4 (Thermo Fisher Scientific) against the database consisted of Homo sapiens proteome extracted from Uniprot, Release 2019. The search parameters were set to enzyme: Trypsin. Fixed modification was carbamidomethyl, and variable modifications were oxidation (M) and deamidated (NQ). The search criteria for protein identification were set to at least 2 matching peptides of 95% confidence level per protein.
Statistics
All analyses were performed in GraphPad Prism 7.0 software (GraphPad Software Inc., San Diego, CA, USA) using unpaired t test. Results shown are presented as mean values ± SEM. A p value ≤0.05 was considered statistically significant.
Results
PMA Induces NET Formation in Human Peripheral Blood-Derived Neutrophils
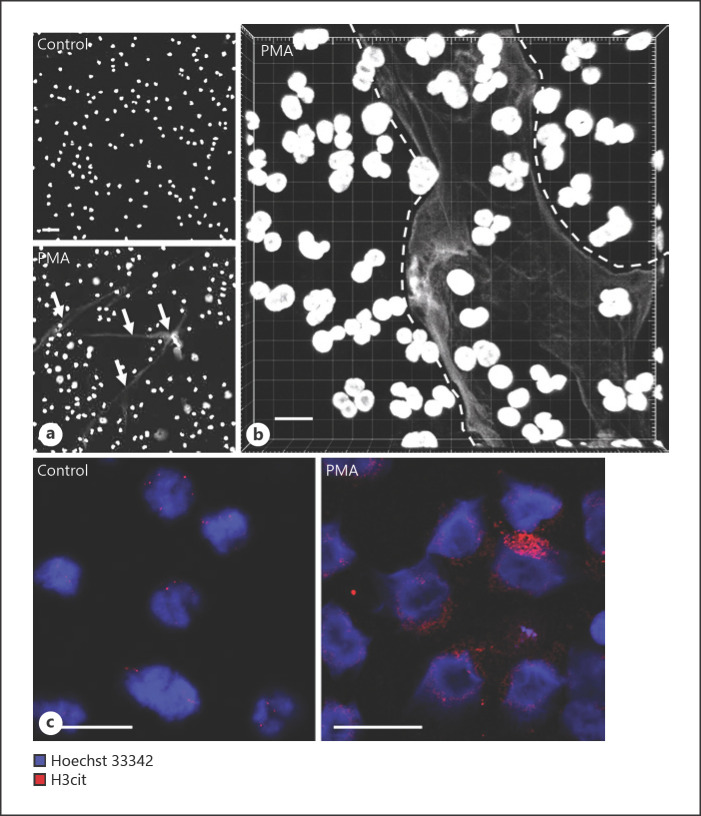
Neutrophils were purified from human peripheral blood by density gradient centrifugation. To induce NET formation, the neutrophils were treated with PMA, an established method to induce NET formation [24]. As seen in Figures 1a and 3, addition of PMA to the purified neutrophils caused the cells to attach to the substratum, indicating cellular activation. Moreover, PMA induced a time-dependent release of DNA-containing structures with a morphological appearance strongly reminiscent of NETs (Fig. 1a, b, 3). Previous studies have revealed that citrullination of histone-3 (H3) at Arg2, −8, and −17 is a hallmark event during NET formation [37]. To further substantiate that the morphological effects observed after PMA treatment of the neutrophils represents NET formation, we stained the activated cells with an antibody-recognizing citrullinated H3 (H3cit). Indeed, PMA-treatment of the neutrophils caused a marked increase in H3 citrullination (Fig. 1c).
Fig. 1.
PMA induces NET formation and H3 citrullination (H3cit) in human peripheral blood-derived neutrophils. Human neutrophils were left untreated (control) or treated with 50 nM PMA for 2 h. a Fluorescence microscopy images showing Hoechst-33342 DNA staining and NET formation in PMA-treated neutrophils; NETs are indicated by white arrows. b Z-stack confocal microscopy image showing Hoechst-33342 staining of PMA-treated neutrophils. Note the presence of NETs in between dashed lines. c Confocal microscopy images showing increased H3cit after PMA activation of neutrophils. Scale bars, 10 μm. PMA, phorbol myristate acetate.
Tryptase Causes Exaggerated NET Formation in Human Neutrophils
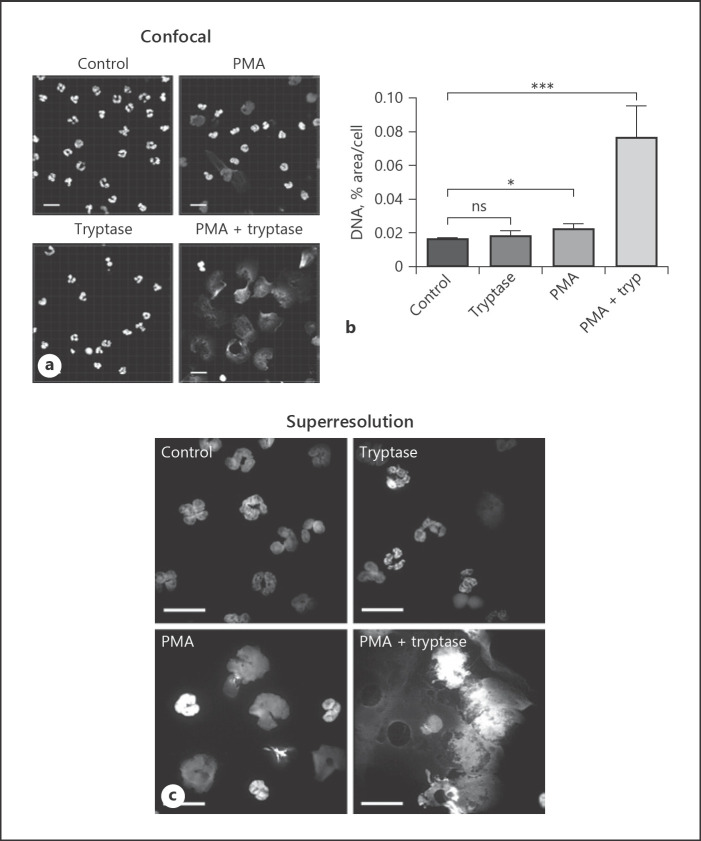
To assess whether tryptase can affect NET formation, we conducted experiments in which NETs were induced in the absence or presence of rh β-tryptase. Tryptase alone did not induce any major morphological effects on the neutrophils, and no apparent NET formation could be observed (Fig. 2a-c). However, when tryptase was added together with PMA, we noted that NET formation was markedly more pronounced than neutrophils that had been treated with PMA only (Fig. 2a-c; online suppl. Fig. 1; see www.karger.com/doi/10.1159/000520972 for all online suppl. material). Moreover, whereas neutrophils treated with PMA only retained considerable nuclear integrity, a striking observation was that neutrophils treated with PMA + tryptase displayed a completely disintegrated nuclear architecture (Fig. 2a-c). Taken together, these experiments suggest that tryptase has a strong potentiating effect on NET formation induced by PMA-treatment of human neutrophils.
Fig. 2.
Tryptase causes exaggerated NET formation in human neutrophils. Neutrophils isolated from human peripheral blood were left untreated (control) or were treated with 50 nM PMA and/or 100 nM β-tryptase for 2 h. a Representative z-stack confocal microscopy image showing Hoechst-33342 staining. Scale bars, 10 μm. b Analysis of the nuclear DNA area, based on fluorescence microscopy images, using ImageJ software (n = 5). Data are given as pixel area/cell; data represent mean values ± SEM; *p ≤ 0.05, ***p ≤ 0.001; n.s., not significant. c Super-resolution images from neutrophils treated with 50 nM PMA and/or 100 nM β-tryptase for 2 h and stained with Hoechst-33342. Note the massive NET formation in PMA + tryptase-treated cells. Scale bars, 10 μm. PMA, phorbol myristate acetate.
NET formation was also seen in response to a calcium ionophore (A23187) and in response to N-fMLP, the latter representing a bacteria-derived neutrophil-activating peptide, and tryptase was shown to augment NET formation in response to both of these agents (online suppl. Fig. 2). In contrast, NET formation was not seen in response to bacterial LPS, either in the absence or presence of tryptase (online suppl. Fig. 2).
Tryptase binds strongly to heparin within the granules of MCs and is important for activation and tetramer formation of tryptase [38]. We thus considered the possibility that heparin might affect the ability of tryptase to promote NET formation. However, the ability of tryptase to promote NET formation was not affect by the presence of heparin (online suppl. Fig. 2).
Tryptase Binds to NETs
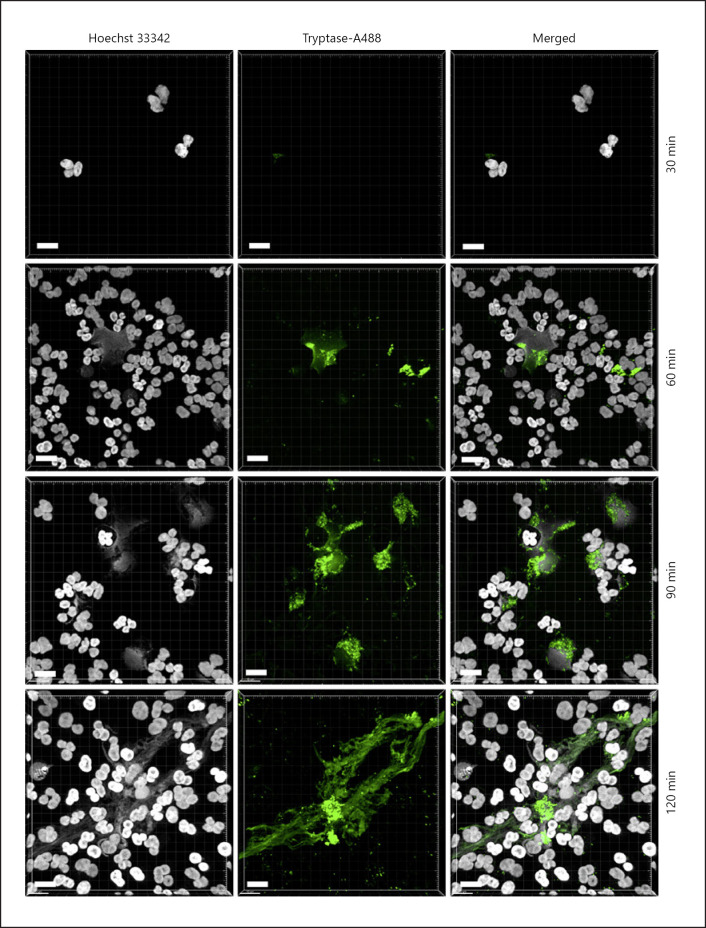
The findings above suggest that MC tryptase has a strong potentiating effect on NET formation. To approach the mechanism behind this, we first investigated whether tryptase has the ability to interact with the NETs. To this end, we labeled recombinant β-tryptase with Alexa-488, and then added the labeled tryptase to PMA-activated human neutrophils followed by confocal microscopy analysis. As seen in Figure 3, tryptase consistently associated with PMA-induced NETs. In fact, the labeled tryptase largely overlapped with NETs throughout the neutrophil population, suggesting that tryptase binds efficiently to the NETs. However, we noted that tryptase positivity was not uniformly distributed within the individual NETs, being concentrated in distinct areas of the NET structures whereas adjacent regions within the NETs were tryptase-negative (Fig. 3).
Fig. 3.
Tryptase binds to NETs. Human neutrophils were treated with 50 nM PMA for different time periods as indicated, followed by 15 min incubation with 100 nM β-tryptase labeled with Alexa-488, and Hoechst-33342 staining. Three dimensional-images were generated from z-stack confocal sections. Scale bars, 10 μm.
Tryptase Causes Core Histone Truncation, Reduction of Citrullinated H3, and Release of Neutrophil Granule Constituents in PMA-Activated Human Neutrophils
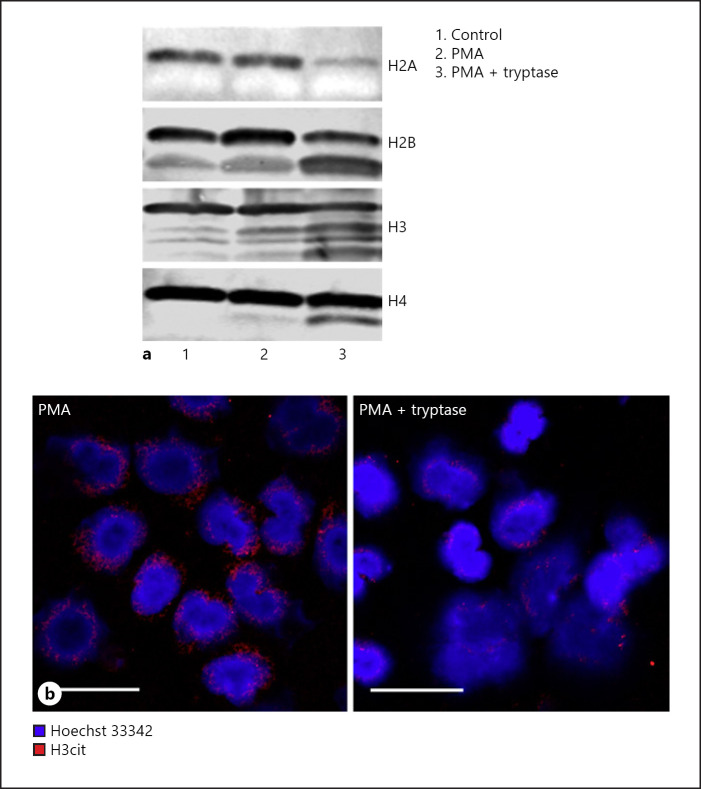
To further elucidate the mechanism behind the impact of MC tryptase on NETs, we considered the possibility that tryptase achieves this effect by executing proteolytic effects. Based on our previous observation that tryptase can execute core histone truncation in the nucleus of MCs [39], we assessed whether tryptase could cause proteolysis of core histones in the PMA-treated human neutrophils. As seen in Figure 4a, all of the core histones (H2A, H2B, H3, and H4) showed minimal signs of degradation in the absence of PMA. Similarly, the core histones were largely intact also after PMA activation of the neutrophils. In contrast, marked core histone truncation was seen after treatment of the PMA-treated neutrophils with tryptase. Distinct proteolytic fragments were observed for H2B, H3, and H4 after treatment with tryptase, along with a reduced intensity of the bands corresponding to the intact histones. For H2A, a marked reduction in the levels of intact protein was seen after treatment with tryptase, whereas distinct proteolytic fragments could not be seen (Fig. 4a). Most likely, the latter observation is explained by degradation into smaller fragments that were not detected by the antibody.
Fig. 4.
Tryptase causes core histone truncation in PMA-activated human neutrophils. a Equal amounts of human neutrophils were left untreated (control) or treated with 50 nM PMA and/or 100 nM β-tryptase for 2 h. Western blot analyses of core histones were performed using equal number of cells from different treatments. b Confocal microscopy images showing H3cit in PMA-activated neutrophils versus neutrophils that were PMA-activated in the presence of tryptase. Note the remarkable reduction of H3cit when neutrophils were co-treated with β-tryptase. PMA, phorbol myristate acetate.
Having shown that tryptase can regulate NET formation, we next asked whether this could be associated with effects on the levels of H3cit. As shown in Figure 4b, whereas extracellular traps from PMA-activated neutrophils were positive for H3cit, the levels of H3cit were markedly reduced after treatment with tryptase. This suggests that tryptase has the ability to cleave off H3 N-terminal tails carrying these modifications.
To provide further insight into the mechanism by which tryptase affects the NETs, we assessed whether tryptase affected the release of proteins from PMA-treated neutrophils. To this end, we analyzed the conditioned media from untreated, PMA-treated, tryptase treated and PMA + tryptase-treated neutrophils for alterations in the proteome. This analysis revealed little release of neutrophil granule compounds in untreated neutrophils, and also only minimal release after treatment with PMA only (Table 1). Under these conditions, defensin-1 appeared to be the major neutrophil product released. In contrast, we noted that the treatment with tryptase (only) resulted in a more substantial release of neutrophil granule proteins, including lactoferrin, gelatinase, and lysozyme. An even more pronounced release of the corresponding granule proteins was seen after treatment with tryptase combined with PMA (Table 1). When cells were treated with PMA + tryptase in the presence of a tryptase inhibitor (Nafamostat), the pattern of released neutrophil proteins was similar as after treatment with PMA only (Table 1), indicating that the ability of tryptase to cause the release of neutrophil granule compounds is dependent on its proteolytic activity. Taken together, these findings suggest that tryptase has the capacity to modify the secretome of PMA-activated neutrophils.
Table 1.
Secretome analysis
| Treatment | Protein description | Score | Coverage | |
|---|---|---|---|---|
| Control | Neutrophil defensin 1 | 4.39 | 19.15 | |
|
| ||||
| Tryptase | Lactoferrin | 13.54 | 17.46 | |
| Neutrophil defensin 1 | 6.84 | 20.21 | ||
| Neutrophil gelatinase | 2.65 | 13.13 | ||
| Lysozyme C | 2.16 | 12.84 | ||
|
| ||||
| PMA | Neutrophil defensin 1 | 9.51 | 19.15 | |
| Lactoferrin | 1.98 | 3.66 | ||
|
| ||||
| PMA + tryptase | Lactoferrin | 21.10 | 15.35 | |
| Neutrophil defensin 1 | 13.80 | 20.21 | ||
| Neutrophil gelatinase | 4.95 | 20.71 | ||
| Myeloperoxidase | 3.97 | 2.95 | ||
| Cystatin | 1.74 | 30.61 | ||
|
| ||||
| PMA + tryptase + nafamostat | Lactoferrin | 14.45 | 15.63 | |
| Neutrophil defensin 1 | 7.16 | 20.21 | ||
Mass spectrometry analysis of supernatants from PMA-activated human neutrophils. PMA, phorbol myristate acetate.
DNA Maintains the Active Tetrameric State of Tryptase and Promotes Tryptase-Mediated Proteolysis of Inflammatory Cytokines
Our findings suggest that tryptase has the capacity to bind to NETs, most likely as a result of electrostatic interactions with the DNA moiety of these structures. Next, we evaluated whether DNA can affect the function of tryptase. To this end, we first assessed whether DNA can maintain the tetrameric and active organization of tryptase. Indeed, as shown by native gel electrophoresis, the tryptase tetramer was preserved in the presence of DNA, and as expected also in the presence of heparin (Fig. 5a). In contrast, tetrameric tryptase was undetectable in the absence of DNA or heparin (Fig. 5a). Stabilization of tryptase by the respective compounds was also verified by measurements of enzymatic activity (Fig. 5b).
Fig. 5.
DNA maintains the active tetrameric state of tryptase and promotes tryptase-mediated proteolysis of inflammatory cytokines. a rh β-tryptase was incubated alone or in the presence of increasing concentrations of heparin or DNA (15 min; 37°C), followed by cross-linking with BS3 and SDS-PAGE analysis. Tryptase monomers and tetramers are indicated by arrows. b rh β-tryptase (1 ng/μL) was incubated alone or in the presence of either heparin (10 ng/μL) or DNA (10 ng/μL; room temperature; 1 h). Residual tryptase enzymatic activity was then measured using the chromogenic substrate S-2288. c rh cytokines/chemokines were incubated either alone (control) or in the presence of tryptase, DNA-stabilized tryptase or heparin-stabilized tryptase as indicated, followed by SDS-PAGE analysis.
To assess whether the association of tryptase with DNA affects its ability to act proteolytically on macromolecular targets, we assessed whether DNA-stabilized tryptase can degrade a number of cytokines and other pro-inflammatory compounds. Out of 73 tested candidates (online suppl. Table 1), we noted proteolytic effects of tryptase on only 8 different compounds (IFNγ, TSLP, TRAIL, IL-20, Gal7-His, KGF-2, IL-21, and OSM) (Fig. 5c). Notably, DNA and heparin differed somewhat in their ability to promote tryptase-catalyzed degradation of the respective compounds. Thus, whereas DNA-stabilized tryptase readily degraded TRAIL and IL-21, heparin-stabilized tryptase was less efficient in this respect. Conversely, heparin-stabilized tryptase appeared more effective in the degradation of TSLP versus tryptase stabilized by DNA (Fig. 5c).
Tryptase Promotes NET Formation in vivo
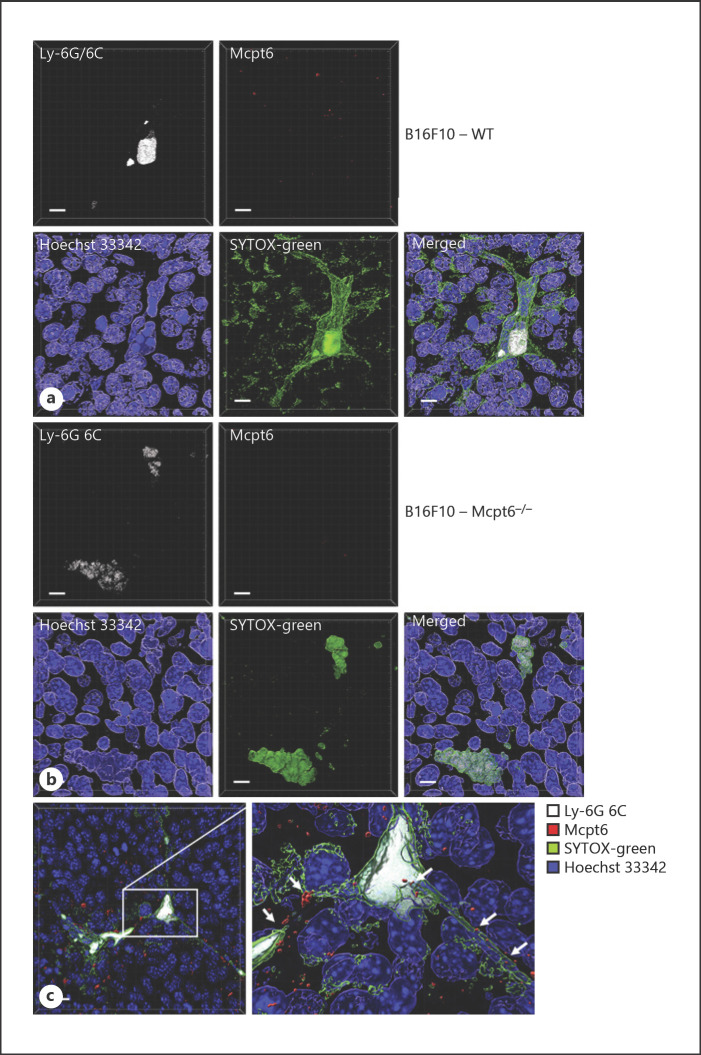
In the next set of experiments, we asked whether tryptase is associated with NETs formed in vivo, and whether our observed effect of tryptase on NET features can be translated into in vivo conditions. Melanomas are known to be associated with extensive inflammatory infiltrates, consisting of various immune cell types including, among others, MCs and neutrophils [36, 40]. Moreover, we showed recently that MCs populating melanoma tumors show extensive signs of activation and can release tryptase into the tumor milieu [36]. We thus reasoned that tryptase, released from tumor-associated MCs, may engage in contacts with NETs that are possibly formed in the inflammatory context of the tumor. As shown in Figure 6a and online supplementary Figure 3, we indeed identified an abundance of neutrophils in the tumor stroma, as judged by Gr1 (Ly-6G/6C) staining. Moreover, we observed that the tumor-associated neutrophils frequently showed signs of NET formation, as judged by the presence of extracellular DNA arranged as NET-like fibers (assessed with a cell nonpermeable DNA probe; SYTOX-green) (Fig. 6a, c). We also noted that tryptase was frequently associated with the NETs (Fig. 6a, c). To evaluate the possible functional impact of tryptase om NET formation in vivo, we analyzed corresponding tumors from tryptase-deficient (Mcpt6−/−) mice. This analysis revealed that NET formation was dramatically reduced in tumor tissue from tryptase-deficient versus wild-type animals (Fig. 6b; online suppl. Fig. 3). In fact, NET formation could not be detected in any of 4 investigated sections taken from tumors of Mcpt6−/− mice. Hence, these data suggest that tryptase interacts with NETs in vivo and that tryptase has a major impact on NET formation in a melanoma setting in vivo.
Fig. 6.
Tryptase is associated with NETs in vivo and NET formation in vivo is attenuated in mice lacking tryptase expression. Sections from subcutaneous B16F10 melanoma tumors were stained for neutrophils (Ly-6G/6C), Mcpt6, extracellular DNA (SYTOX-green; cell-impermeable) and with Hoechst 33342 (depicts nuclear DNA; cell-permeable). The images represent 3D images generated from z-stacks. Sections from wild-type (WT) (a) and tryptase-deficient (Mcpt6−/−) (b) mice were analyzed. a–c Sections were first stained with SYTOX-green, followed by Hoechst-33342. Cells were then permeabilized and stained for Ly-6G/6C (Gr-1) and Mcpt6. a Tumor section from WT mice showing the presence of activated neutrophils and extensive NET formation (green). Note the presence of Mcpt6 in close contact with NETs. b Tumor section from Mcpt6−/− mice showing neutrophils with attenuated NET formation. Note the absence of Mcpt6 staining. c Higher magnification of tumors from WT mice showing neutrophils and NET formation in association with Mcpt6. Scale bars, 10 μm. Mcpt6, MC tryptase; WT, wild-type.
Discussion
Previous research has suggested that there is an intimate relationship between MCs and neutrophils, as manifested by the known ability of MCs to secrete cytokines and chemokines having the capacity to recruit neutrophils [9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20]. With this work, we introduce an additional level of communication between these 2 cell types, by demonstrating that MC tryptase, a major component of the MC secretory granules, has the ability to promote the formation of NETs. Importantly, tryptase alone did not induce extracellular trap formation in neutrophils. However, when tryptase was added in the presence of neutrophil-activating stimulus, NET formation was markedly more pronounced than if neutrophil-activating stimulus was added alone.
The mechanism underlying the ability of tryptase to potentiate NET generation is intriguing. NET formation is a complex process, with a hallmark event being chromatin decondensation [25]. Such chromatin decondensation is thought to depend on the citrullination of H3 at Arg2, −8, and −17 [37]. This causes neutralization of the positive charge at these positions, leading to weakened histone-DNA interactions, which will promote NET formation. Here, we showed that tryptase has the capacity to truncate the NET-contained core histones, which is in line with our previous observations where tryptase was shown to execute cell-intrinsic core histone truncation in the nucleus of MCs [41]. In our previous studies, we showed that tryptase cleaves the corresponding core histones in the N-terminal regions, and it is notable that the proteolytic histone fragments observed in the previous study closely resemble those seen in the present work (see [41] and Fig. 4a). Based on these observations, we can therefore propose that the core histone truncation observed in the present study represents cleavages in the N-terminal portions of the core histones. Most likely, this will result in the proteolytical removal of Arg residues that are sites for citrullination, which may serve to reduce the positive charge of H3, in turn promoting NET formation. In support of this scenario, we show that tryptase causes a reduction in the levels of N-terminal histone citrullination (at Arg2, −8, and −17). Altogether, we may thus propose that tryptase has the capacity to cleave off N-terminal ends of core histones, which can lead to decreased interaction of histones with DNA, in turn contributing to DNA decondensation and thereby promoting NET formation. Notably, this scenario resembles the mechanism by which neutrophil elastase has been shown to promote NET formation [42].
Intriguingly, it has been shown that MCs can form extracellular traps with antimicrobial activity [43]. Based on the present study, we may thus propose that tryptase might have the capacity to regulate the generation of such MCs-derived extracellular traps. However, this remains to be investigated.
Notably, tryptase did not localize to the nuclei of the neutrophils. This suggests that tryptase acts on core histones after their exit from the nucleus, rather than by entering the neutrophil nuclei and carrying out histone proteolysis within this compartment. It is also notable that tryptase was associated with distinct subregions of the NETs, rather than being uniformly distributed over the entire NET structures. It is known from previous studies that tryptase binds strongly to polyanions of glycosaminoglycan type [38], and we recently showed that tryptase also can bind to and be stabilized by DNA [34]. Hence, a likely scenario could be that tryptase binds to the NETs through electrostatic interactions with the DNA moiety of the NETs. Through this interaction, tryptase may become stabilized and may gain access to proteolytic targets such as the core histones.
The impact of tryptase on NETs could have multiple pathophysiological consequences. As shown here, DNA-stabilized tryptase has a strong capacity to carry out proteolysis of various inflammatory cytokines; in fact, DNA-associated tryptase was even more efficient at cleaving TRAIL and IL-21 than were heparin-stabilized tryptase. Hence, a likely scenario could be that NET-associated tryptase can regulate the cytokine profile in pathological contexts where both MCs and neutrophils are present and are activated. We also noted that tryptase has the ability to cause increased release of neutrophil granule compounds into the surrounding milieu, with possible consequences related to the impact of such compounds on the inflammatory outcome. Another likely scenario is that promotion of NET formation by tryptase can result in a more efficient clearance of bacteria through NET-dependent mechanisms.
On a different angle, we here show that tryptase is indispensable for NET formation in the inflammatory milieu of melanoma tumors in vivo. Of note, we have recently shown that tryptase has a protective impact in a melanoma model [36], and there are also clinical studies pointing to a positive association between high tryptase levels and favorable clinical outcome in melanoma [44]. Although the exact mechanisms behind these findings are not clear, we may, based on the present investigation, speculate that the impact of tryptase on melanoma outcome could be related to its ability to regulate NET formation and functionality.
Statement of Ethics
The experiments on human neutrophils were approved by the relevant Ethical Committee (Etikprövningsmyndigheten Dnr 2020-05080). The animal experiments were approved by the relevant Ethical Committee (Uppsala djurförsöksetiska nämnd Dnr 5.8.18-04096/2019).
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
Funding Sources
This study was supported by grants from the Swedish Heart and Lung Foundation, Swedish Research Council, Swedish Cancer Foundation, and Knut & Alice Wallenberg Foundation.
Author Contributions
G.P. designed and planned the study, interpreted data and wrote the manuscript; S.A. performed experiments and wrote parts of the article; M.G. performed experiments; J.A. performed imaging analyses; A.-K.O. contributed to the design of the study, and contributed to the writing of the article; C.P.S. expressed, purified and labeled tryptase, and contributed to the writing of the article; F.R.M. designed the study, planned, and performed most of the experiments, and wrote parts of the manuscript.
Data Availability Statement
All data generated or analyzed during this study are included in this article. Further inquiries can be directed to the corresponding authors.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Supplementary data
References
- 1.Harvima IT, Nilsson G, Suttle MM, Naukkarinen A. Is there a role for mast cells in psoriasis? Arch Dermatol Res. 2008 Oct;300((9)):461–78. doi: 10.1007/s00403-008-0874-x. [DOI] [PubMed] [Google Scholar]
- 2.Dudeck A, Dudeck J, Scholten J, Petzold A, Surianarayanan S, Kohler A, et al. Mast cells are key promoters of contact allergy that mediate the adjuvant effects of haptens. Immunity. 2011 Jun 24;34((6)):973–84. doi: 10.1016/j.immuni.2011.03.028. [DOI] [PubMed] [Google Scholar]
- 3.Schubert N, Dudeck J, Liu P, Karutz A, Speier S, Maurer M, et al. Mast cells promote T cell driven antigen-induced arthritis despite being dispensable in T cell bypassing antibody-induced arthritis. Arthritis Rheumatol. 2015;67((4)):903–13. doi: 10.1002/art.38996. [DOI] [PubMed] [Google Scholar]
- 4.Kroner J, Kovtun A, Kemmler J, Messmann JJ, Strauss G, Seitz S, et al. Mast cells are critical regulators of bone fracture-induced inflammation and osteoclast formation and activity. J Bone Miner Res. 2017 Dec;32((12)):2431–44. doi: 10.1002/jbmr.3234. [DOI] [PubMed] [Google Scholar]
- 5.Bradding P, Pejler G. The controversial role of mast cells in fibrosis. Immunol Rev. 2018 Mar;282((1)):198–231. doi: 10.1111/imr.12626. [DOI] [PubMed] [Google Scholar]
- 6.Maurer M, Eyerich K, Eyerich S, Ferrer M, Gutermuth J, Hartmann K, et al. Urticaria: collegium internationale allergologicum (CIA) update 2020. Int Arch Allergy Immunol. 2020;181((5)):321–33. doi: 10.1159/000507218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnzon CF, Rönnberg E, Pejler G. The role of mast cells in bacterial infection. Am J Pathol. 2016 Oct 15;186:4–14. doi: 10.1016/j.ajpath.2015.06.024. [DOI] [PubMed] [Google Scholar]
- 8.Mukai K, Tsai M, Starkl P, Marichal T, Galli SJ. IgE and mast cells in host defense against parasites and venoms. Semin Immunopathol. 2016 Sep;38((5)):581–603. doi: 10.1007/s00281-016-0565-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Echtenacher B, Männel DN, Hültner L. Critical protective role of mast cells in a model of acute septic peritonitis. Nature. 1996;381((6577)):75–7. doi: 10.1038/381075a0. [DOI] [PubMed] [Google Scholar]
- 10.Malaviya R, Ikeda T, Ross E, Abraham SN. Mast cell modulation of neutrophil influx and bacterial clearance at sites of infection through TNF-alpha. Nature. 1996;381((6577)):77–80. doi: 10.1038/381077a0. [DOI] [PubMed] [Google Scholar]
- 11.Dudeck J, Kotrba J, Immler R, Hoffmann A, Voss M, Alexaki VI, et al. Directional mast cell degranulation of tumor necrosis factor into blood vessels primes neutrophil extravasation. Immunity. 2021 Mar 9;54((3)):468–83.e5. doi: 10.1016/j.immuni.2020.12.017. [DOI] [PubMed] [Google Scholar]
- 12.De Filippo K, Dudeck A, Hasenberg M, Nye E, van Rooijen N, Hartmann K, et al. Mast cell and macrophage chemokines CXCL1/CXCL2 control the early stage of neutrophil recruitment during tissue inflammation. Blood. 2013 Jun 13;121((24)):4930–7. doi: 10.1182/blood-2013-02-486217. [DOI] [PubMed] [Google Scholar]
- 13.Enoksson M, Moller-Westerberg C, Wicher G, Fallon PG, Forsberg-Nilsson K, Lunderius-Andersson C, et al. Intraperitoneal influx of neutrophils in response to IL-33 is mast cell-dependent. Blood. 2013 Jan 17;121((3)):530–6. doi: 10.1182/blood-2012-05-434209. [DOI] [PubMed] [Google Scholar]
- 14.Ngo Nyekel F, Pacreau E, Benadda S, Msallam R, Abrink M, Pejler G, et al. Mast cell degranulation exacerbates skin rejection by enhancing neutrophil recruitment. Front Immunol. 2018;9:2690. doi: 10.3389/fimmu.2018.02690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakae S, Suto H, Berry GJ, Galli SJ. Mast cell-derived TNF can promote Th17 cell-dependent neutrophil recruitment in ovalbumin-challenged OTII mice. Blood. 2007 May 1;109((9)):3640–8. doi: 10.1182/blood-2006-09-046128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamura Y, Kambe N, Saito M, Nishikomori R, Kim YG, Murakami M, et al. Mast cells mediate neutrophil recruitment and vascular leakage through the NLRP3 inflammasome in histamine-independent urticaria. J Exp Med. 2009 May 11;206((5)):1037–46. doi: 10.1084/jem.20082179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pimentel TA, Sampaio AL, D'Acquisto F, Perretti M, Oliani SM. An essential role for mast cells as modulators of neutrophils influx in collagen-induced arthritis in the mouse. Lab Invest. 2011 Jan;91((1)):33–42. doi: 10.1038/labinvest.2010.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie Y, Zhang H, Liu S, Chen G, He S, Li Z, et al. Mast cell activation protects cornea by promoting neutrophil infiltration via stimulating ICAM-1 and vascular dilation in fungal keratitis. Sci Rep. 2018 May 30;8((1)):8365. doi: 10.1038/s41598-018-26798-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Almeida AD, Silva IS, Fernandes-Braga W, LimaFilho ACM, Florentino ROM, Barra A, et al. A role for mast cells and mast cell tryptase in driving neutrophil recruitment in LPS-induced lung inflammation via protease-activated receptor 2 in mice. Inflamm Res. 2020 Oct;69((10)):1059–70. doi: 10.1007/s00011-020-01376-4. [DOI] [PubMed] [Google Scholar]
- 20.Elbasiony E, Mittal SK, Foulsham W, Cho W, Chauhan SK. Epithelium-derived IL-33 activates mast cells to initiate neutrophil recruitment following corneal injury. Ocul Surf. 2020 Oct;18((4)):633–40. doi: 10.1016/j.jtos.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madjene LC, Danelli L, Dahdah A, Vibhushan S, Bex-Coudrat J, Pacreau E, et al. Mast cell chymase protects against acute ischemic kidney injury by limiting neutrophil hyperactivation and recruitment. Kidney Int. 2020;97((3)):516–27. doi: 10.1016/j.kint.2019.08.037. [DOI] [PubMed] [Google Scholar]
- 22.Sutherland RE, Olsen JS, McKinstry A, Villalta SA, Wolters PJ. Mast cell IL-6 improves survival from Klebsiella pneumonia and sepsis by enhancing neutrophil killing. J Immunol. 2008 Oct 15;181((8)):5598–605. doi: 10.4049/jimmunol.181.8.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piliponsky AM, Chen CC, Rios EJ, Treuting PM, Lahiri A, Abrink M, et al. The chymase mouse mast cell protease 4 degrades TNF, limits inflammation, and promotes survival in a model of sepsis. Am J Pathol. 2012 Sep;181((3)):875–86. doi: 10.1016/j.ajpath.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, et al. Neutrophil extracellular traps kill bacteria. Science. 2004 Mar 5;303((5663)):1532–5. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 25.Brinkmann V, Zychlinsky A. Beneficial suicide: why neutrophils die to make NETs. Nat Rev Microbiol. 2007 Aug;5((8)):577–82. doi: 10.1038/nrmicro1710. [DOI] [PubMed] [Google Scholar]
- 26.Pham CT. Neutrophil serine proteases: specific regulators of inflammation. Nat Rev Immunol. 2006 Jul;6((7)):541–50. doi: 10.1038/nri1841. [DOI] [PubMed] [Google Scholar]
- 27.Galli SJ, Nakae S, Tsai M. Mast cells in the development of adaptive immune responses. Nat Immunol. 2005 Feb;6((2)):135–42. doi: 10.1038/ni1158. [DOI] [PubMed] [Google Scholar]
- 28.Pejler G, Abrink M, Ringvall M, Wernersson S. Mast cell proteases. Adv Immunol. 2007;95:167–255. doi: 10.1016/S0065-2776(07)95006-3. [DOI] [PubMed] [Google Scholar]
- 29.Pejler G, Rönnberg E, Waern I, Wernersson S. Mast cell proteases: multifaceted regulators of inflammatory disease. Blood. 2010 Jun 17;115((24)):4981–90. doi: 10.1182/blood-2010-01-257287. [DOI] [PubMed] [Google Scholar]
- 30.Wernersson S, Pejler G. Mast cell secretory granules: armed for battle. Nat Rev Immunol. 2014 Jul;14((7)):478–94. doi: 10.1038/nri3690. [DOI] [PubMed] [Google Scholar]
- 31.Pereira PJ, Bergner A, Macedo-Ribeiro S, Huber R, Matschiner G, Fritz H, et al. Human beta-tryptase is a ring-like tetramer with active sites facing a central pore. Nature. 1998;392((6673)):306–11. doi: 10.1038/32703. [DOI] [PubMed] [Google Scholar]
- 32.Fu Z, Akula S, Thorpe M, Hellman L. Highly selective cleavage of TH2-promoting cytokines by the human and the mouse mast cell tryptases, indicating a potent negative feedback loop on TH2 immunity. Int J Mol Sci. 2019 Oct 17;20((20)):5147. doi: 10.3390/ijms20205147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ronnberg E, Melo FR, Pejler G. Mast cell proteoglycans. J Histochem Cytochem. 2012 Dec;60((12)):950–62. doi: 10.1369/0022155412458927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alanazi S, Grujic M, Lampinen M, Rollman O, Sommerhoff CP, Pejler G, et al. Mast cell beta-tryptase is enzymatically stabilized by DNA. Int J Mol Sci. 2020 Jul 17;21((14)):5065. doi: 10.3390/ijms21145065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sommerhoff CP, Bode W, Pereira PJ, Stubbs MT, Sturzebecher J, Piechottka GP, et al. The structure of the human betaII-tryptase tetramer: fo(u)r better or worse. Proc Natl Acad Sci U S A. 1999;96((20)):10984–91. doi: 10.1073/pnas.96.20.10984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grujic M, Hellman L, Gustafson AM, Akula S, Melo FR, Pejler G. Protective role of mouse mast cell tryptase Mcpt6 in melanoma. Pigment Cell Melanoma Res. 2020 Jan 1;33((4)):579–90. doi: 10.1111/pcmr.12859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Li M, Stadler S, Correll S, Li P, Wang D, et al. Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J Cell Biol. 2009 Jan 26;184((2)):205–13. doi: 10.1083/jcb.200806072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hallgren J, Pejler G. Biology of mast cell tryptase. An inflammatory mediator. FEBS J. 2006 May;273((9)):1871–95. doi: 10.1111/j.1742-4658.2006.05211.x. [DOI] [PubMed] [Google Scholar]
- 39.Melo FR, Wallerman O, Paivandy A, Calounova G, Gustafson AM, Sabari BR, et al. Tryptase-catalyzed core histone truncation: a novel epigenetic regulatory mechanism in mast cells. J Allergy Clin Immunol. 2017 Aug;140((2)):474–85. doi: 10.1016/j.jaci.2016.11.044. [DOI] [PubMed] [Google Scholar]
- 40.Ladanyi A. Prognostic and predictive significance of immune cells infiltrating cutaneous melanoma. Pigment Cell Melanoma Res. 2015 Sep;28((5)):490–500. doi: 10.1111/pcmr.12371. [DOI] [PubMed] [Google Scholar]
- 41.Melo FR, Vita F, Berent-Maoz B, Levi-Schaffer F, Zabucchi G, Pejler G. Proteolytic histone modification by mast cell tryptase, a serglycin proteoglycan-dependent secretory granule protease. J Biol Chem. 2014 Mar 14;289((11)):7682–90. doi: 10.1074/jbc.M113.546895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Papayannopoulos V, Metzler KD, Hakkim A, Zychlinsky A. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J Cell Biol. 2010 Nov 1;191((3)):677–91. doi: 10.1083/jcb.201006052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.von Kockritz-Blickwede M, Goldmann O, Thulin P, Heinemann K, Norrby-Teglund A, Rohde M, et al. Phagocytosis-independent antimicrobial activity of mast cells by means of extracellular trap formation. Blood. 2008 Mar 15;111((6)):3070–80. doi: 10.1182/blood-2007-07-104018. [DOI] [PubMed] [Google Scholar]
- 44.Crincoli E, Moliterni E, Catania F, Didona D, Calvieri S, Paolino G. Correlation of serum tryptase levels with total number of nevi, Breslow thickness, ulceration, and mitotic index in melanoma patients: evaluation of a promising prognostic marker. Melanoma Res. 2019;29((6)):621–5. doi: 10.1097/CMR.0000000000000561. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Data Availability Statement
All data generated or analyzed during this study are included in this article. Further inquiries can be directed to the corresponding authors.