Summary
Background
Nasopharyngeal carcinoma (NPC) exhibits significant familial aggregation; however, its susceptibility genes are largely unknown. Thus, this study aimed to identify germline mutations that might contribute to the risk of familial NPC, and explore their biological functions.
Methods
Whole-exome sequencing was performed in 13 NPC pedigrees with multiple cases. Mutations co-segregated with disease status were further validated in a cohort composed of 563 probands from independent families, 2,953 sporadic cases, and 3,175 healthy controls. Experimental studies were used to explore the functions of susceptibility genes and their disease-related mutations.
Findings
The three rare missense mutations in POLN (DNA polymerase nu) gene, P577L, R303Q, and F545C, were associated with familial NPC risk (5/576 [0·87%] in cases vs. 2/3374 [0·059%] in healthy controls with an adjusted OR of 44·84 [95% CI:3·91-514·34, p = 2·25 × 10−3]). POLN was involved in Epstein-Barr virus (EBV) lytic replication in NPC cells in vitro. POLN promoted viral DNA replication, immediate-early and late lytic gene expression, and progeny viral particle production, ultimately affecting the proliferation of host cells. The three mutations were located in two pivotal functional domains and were predicted to alter the protein stability of POLN in silico. Further assays demonstrated that POLN carrying any of the three mutations displayed reduced protein stability and decreased expression levels, thereby impairing its ability to promote complete EBV lytic replication and facilitate cell survival.
Interpretation
We identified a susceptibility gene POLN for familial NPC and elucidated its function.
Funding
This study was funded by the National Key Research and Development Program of China (2021YFC2500400); the National Key Research and Development Program of China (2020YFC1316902); the Basic and Applied Basic Research Foundation of Guangdong Province, China (2021B1515420007); the National Natural Science Foundation of China (81973131); the National Natural Science Foundation of China (82003520); the National Natural Science Foundation of China (81903395).
Keywords: Familial nasopharyngeal carcinoma, Whole-exome sequencing, POLN gene, Rare germline mutations, Epstein-Barr virus
Research in context.
Evidence before this study
Previous linkage studies have demonstrated the contribution of certain chromosomal regions, such as 6p22, 4p15·1–q12, 3p21·31–21·2, and 5p13·1, to the risk of familial NPC. To date, only a few studies have reported the susceptibility genes for familial NPC in the Southern Chinese population, including rare mutations in MST1R and 12 other genes.
Added value of this study
This study identified a susceptibility gene POLN for familial NPC, supporting that germline mutations in DNA repair factors are associated with NPC risk. Functional studies have uncovered the substantial function of POLN in the course of EBV lytic replication and have provided experimental evidence for the involvement of germline mutations in EBV-related oncogenesis.
Implications of all the available evidence
Identification of susceptibility genes for NPC screening in high-risk populations could contribute to earlier diagnosis and improved prognosis, especially in those with a familial background. Clarifying the mechanism by which the susceptibility gene contributes to NPC development is helpful to deepen our understanding of the aetiology of NPC.
Alt-text: Unlabelled box
Introduction
Nasopharyngeal carcinoma (NPC) is a malignancy arising from the epithelium of the nasopharynx,1 characterised by an extraordinary ethnic and geographical distribution. It is rare in most parts of the world but is endemic in Southern China (particularly in Guangdong), Southeast Asia, Northern Africa, and Alaska.2 Familial clustering has been widely reported,3 and more than 10% of patients have a positive family history of NPC in endemic regions.4,5 Familial NPC is defined as a family with one or more NPC patients, in addition to the proband within three generations.4 The risk dramatically increased 4 to 10-fold among individuals with a first-degree relative with NPC compared with those without a family history.3 Familial NPC cases also have a significantly younger age of onset than sporadic cases.6 The geographical distribution and familial clustering of NPC suggest that genetic factors may play a critical role in its aetiology. Therefore, it is important to identify genetic susceptibility genes for NPC screening in high-risk populations, which would contribute to earlier diagnosis and better prognosis.
Linkage studies have demonstrated the contribution of some loci to the risk of familial NPC, including 6p22 (human leukocyte antigen locus),7 4p15·1–q12,8 3p21·31–21·2,9 and 5p13·1.10 Recent efforts to identify additional cancer predisposition genes have emphasised the importance of rare mutations.11 According to the hypothesis that high penetrance and highly pathogenic rare genetic mutations tend to be enriched in families and co-segregate with cases,12 additional causative mutations have been identified in a variety of tumors in high-risk multiplex cancer families, including BRCA1 and BRCA2-negative breast cancer, colorectal cancer, pancreatic cancer, non-medullary thyroid cancer and Hodgkin lymphoma families.13, 14, 15, 16, 17 Until now, only a few studies performed in NPC endemic areas have reported the susceptibility genes for familial NPC, including rare mutations in MST1R gene18 and 12 other genes.19 Additional susceptibility genes remain to be identified, and the underlying mechanisms require further investigation.
The default infection programme of Epstein-Barr virus (EBV) in pharyngeal epithelial cells is lytic infection.20, 21, 22 During lytic infections, almost all viral genes are expressed in an ordered cascade. First, the immediate-early (IE) genes BZLF1 and BRLF1 are expressed, which activates the expression of early genes. Following viral DNA replication, late genes are expressed.23 However, the burden of producing a large number of progeny virus particles during lytic reactivation may induce cell death.24 Therefore, the role of EBV life cycle in NPC tumorigenesis is complex. We recently found that the EBV loads at the nasopharyngeal site are correlated with each other among blood-relative pairs, and a significantly high heritability was also observed25 within families with multiplex NPC cases, indicating that host genetics interacts with EBV in the development of NPC.
In this study, to identify germline mutations that might contribute to the risk of familial NPC, we conducted whole-exome sequencing (WES) of 13 NPC pedigrees, followed by validation in a cohort of familial cases, sporadic cases, and healthy controls (n=6,890). We found that rare POLN mutations are associated with familial NPC risk. POLN encodes a DNA polymerase with translesion DNA synthesis activity,26 and participates in cross-linking repair and homologous recombination.27 We further demonstrated the functional relevance of POLN in EBV lytic replication and the possible mechanism by which rare functional mutations influence familial NPC risk.
Methods
Study populations and samples collection
The NPC multiplex families were recruited from “High-risk Nasopharyngeal Carcinoma Family Screening Program” in Sun Yat-sen University Cancer Center (SYSUCC, Guangzhou, Guangdong, China). Patients and their unaffected relatives from 13 high-risk families were enrolled in this study. Each family had whole blood or saliva samples available from at least two NPC patients and at least one unaffected first-degree relative (Figure S8). WES was conducted on available samples, including 35 familial cases and 34 unaffected relatives (Table S1).
A total of 351 additional familial cases, defined as having at least one family member within third-degree relatives suffering NPC, were recruited from SYSUCC from 2008 to 2015 for Sanger sequencing. In addition to the public database, an in-house WES database, including 199 healthy individuals recruited from Guangdong, China, was also used for mutation filtering (Table S2).
For independent replication, 212 unrelated familial cases and 2,953 sporadic cases were recruited from SYSUCC from 2004 to 2017, and 3,175 healthy controls were recruited from Guangdong China from 2009 to 2016 (Table S2). All patients were Han Chinese and were histopathologically diagnosed.
Additionally, mRNA samples from 84 NPC tissues and 12 non-cancerous nasopharyngeal tissues were obtained from the SYSUCC biobank. Protein samples were obtained from 8 additional NPC tissues and 5 non-cancerous nasopharyngeal tissues. All NPC tissues were pathologically classified as undifferentiated non-keratinising tumours.
Sequencing and genotyping
Genomic DNA was extracted from peripheral blood and saliva using an automated workstation (Chemagic Star, Bonaduz, Switzerland) and quantified using the PicoGreen reagent (Invitrogen, Cat#P11495, Carlsbad, CA, USA). 2·5 μg of DNA from each individual was fragmented using Covaris ultrasonicator (Woburn, MA, USA). Indexed paired-end libraries were prepared using the SureSelectXT Human All Exon Kit (V6) (Agilent, Cat#5190-8863, Santa Clara, CA, USA). paired-end 150-bp read-length sequencing was performed using Illumina HiSeq 2500 technology (San Diego, CA, USA).
The WES data were analysed according to the recommendations of GATK Best Practices.28 The sequence reads were aligned to the human reference genome (hg19/GRCh37) using the Burrows-Wheeler aligner29 and Picard tools (http://broadinstitute.github.io/picard/) were used to sort the bam files and mark duplicates. GATK was then used to conduct indel realignment, base quality score recalibration, variant calling, and variant quality score recalibration (VQSR).28 The sensitivity thresholds for single-nucleotide polymorphisms (SNPs) and indels were chosen at 99·5% and 99%, respectively. The quality of the mutations was checked by GATK and plink to remove abnormal samples or contamination. Genotypes with quality scores < 20 or read depths < 8 were treated as missing values. Heterozygotes with alternative allele frequencies > 0·75 or < 0·25 were also considered ambiguous calling. Mutations with missing rates of > 20% were excluded from further analyses. ANNOVAR30 was used to annotate the qualified mutations.
For Sanger sequencing of POLN gene, we used the Primer-BLAST online database (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) to design 26 pair primers (Table S7) for all exons and exon-intron boundaries of POLN. High fidelity DNA polymerase (Genestar, Beijing, CHN) was used for PCR. Amplified DNA fragments were sent for Sanger sequencing (Majorbio, Shanghai, CHN). All sequencing results were aligned to the POLN RefSeq sequence (NG_046934.1) using the DNAMAN software for mutation detection. We applied Sanger sequencing to validate the mutations identified by WES and TaqMan genotyping assays (Figure S1), and partial samples carried wild-type POLN identified by TaqMan genotyping assay (Figure S9).
TaqMan SNP Genotyping Assay (Applied Biosystems, ABI, Carlsbad, CA, USA) followed by the LightCycler Real-Time PCR 480 system (Roche, Basel, Switzerland) was used for genotyping, according to the manufacturer's instructions. Blank and positive controls were used for each experiment. Probe sequences are listed in Table S8. The 5 μl reaction system included 0·25 μl primer plus probe, 1 μl DNA (25 ng), 2·5 μl TaqMan universal MMIX II with UNG (ABI, Cat#4440046) and 1·25 μl H2O. The reaction conditions were as follows: incubation at 50 °C for 2 min, polymerase activation at 95 °C for 10 min, and 40 cycles of amplification at 95 °C for 15 s and 60 °C for 1 min.
Mutation selection
To identify candidate mutations that may play important roles on NPC development, we first conducted a co-segregation analysis on the 13 families to identify mutations segregating with NPC onset, that is, shared by all NPC cases but absent in the unaffected relatives in the family. We then filtered the non-synonymous rare mutations with minor allele frequency (MAF) < 0·1% in all public databases: 1000 Genomes (all population and East Asians), gnomAD (all population and East Asians with genome and exome data, respectively), ESP6500, ExAC (all population and East Asians with and without TCGA data, respectively), the Kaviar database, and our in-house WES database with 199 healthy individuals. Among the resulting 145 rare non-synonymous co-segregating mutations, we used the dbNSFP database in ANNOVAR to conduct functional prediction. The mutations predicted as “damaging” in SIFT, polyphen2 HVAR, and FATHMM and located in the protein domain were further filtered. We performed a literature search for target gene(s) with functional relevance to cancer. A detailed mutation selection flowchart is shown in Figure 1a.
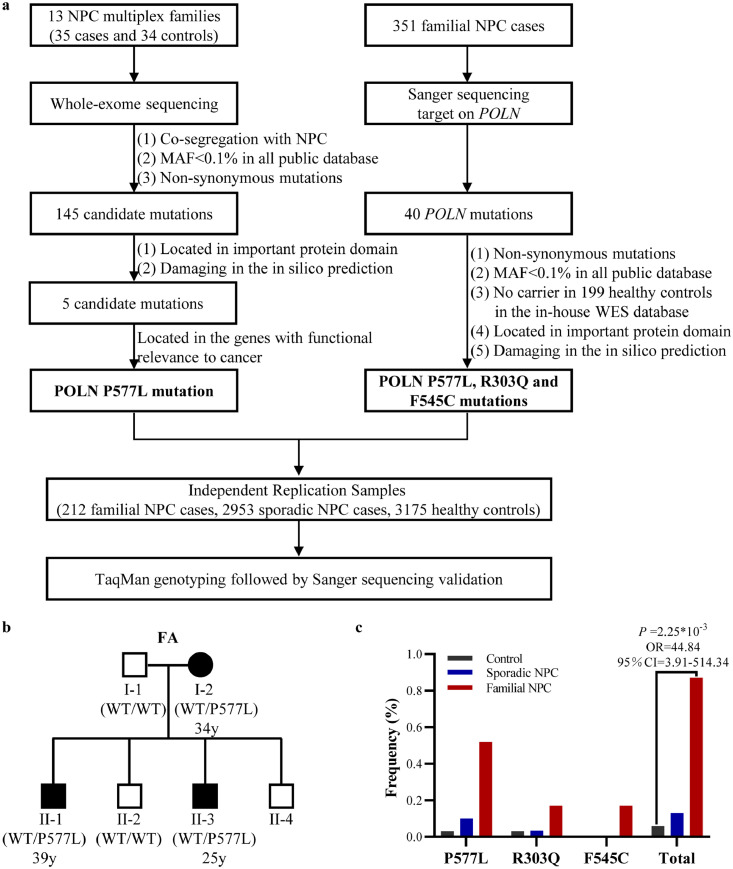
Figure 1.
Identification of three POLN mutations associated with familial NPC. (a) Workflow for the identification and validation of the association between POLN and familial NPC. (b) Pedigree of FA NPC family. DNA samples of individuals I-1, I-2, II-1, II-2, and II-3 were extracted for WES. (c) The frequencies of the POLN P577L, R303Q, and F545C rare mutations in familial cases, sporadic cases, and controls, respectively. Logistic regression analysis was conducted to compare the frequencies of all three mutations in familial NPC cases with healthy controls after adjusting for age and sex.
NPC, Nasopharyngeal carcinoma; WES, whole-exome sequencing.
The mutations identified in 351 unrelated familial NPC cases were filtered by population frequency using the selection strategies aforementioned. Those located in the protein domain and predicted as “damaging” in SIFT, polyphen2 HVAR, and Mutation Taster were further selected for the functional study.
Cell culture
HK1 EBV+ and Tet-BZLF1/CNE2 EBV+ NPC cells were obtained from Professor Mu-Sheng Zeng (Sun Yat-sen University, China).31,32 Tet-BZLF1/CNE2 EBV+ cells conditionally expressed the exogenous protein Zta under the control of a tetracycline-inducible promoter (Figure S10). Both NPC cell lines were cultured in RPMI 1640 medium (Gibco, Cat# 11875093, Carlsbad, CA, USA) supplemented with 10% foetal calf serum (Gibco, Cat#16140071). Geneticin (700 μg/ml) (G418) (Sigma, Cat#G5013) was added to the culture medium to maintain the EBV-positive rate. Human embryonic kidney 293T (HEK-293T) cells used for virus packaging were purchased from ATCC (ATCC, CRL-3216). HEK-293T cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) (Gibco, Cat# 11965092) with 10% foetal calf serum. All cells were cultured at 37 °C in an incubator with a humidified atmosphere of 5% CO2 and tested negative for mycoplasmas (Figure S11).
Plasmids, mutagenesis
POLN full-length cDNA (NCBI: NM_181808.3) was obtained from Vigene Biosciences (Rockville, MD, USA) and cloned into the pCDH-CMV-MCS-EF1-Puro lentiviral vector using the ClonExpress Entry One Step Cloning Kit (Vazyme, Cat#C112, Nanjing, Jiangsu, CHN) for wild-type (WT) POLN expression plasmid construction. We performed polymerase chain reaction (PCR) to generate POLN P577L, F545C, and R303Q mutants by site-directed mutagenesis of the WT POLN expression plasmid with specific primers (Table S7) and used Dpn1 restriction enzyme (Thermo Scientific, Cat#ER1701, Waltham, MA, USA) to digest the WT template. HA-tags were fused to the C-terminus of the WT and mutant POLN plasmids. All plasmids were verified using DNA sequencing.
Lentivirus assembly and transduction
For lentiviral production, HEK-293T cells were co-transfected with PMD2G, psPAX2, and pCDH POLN WT or mutant plasmids using polyethyleneimine (PEI) (YEASEN, Cat#40816ES02, Shanghai, CHN). The supernatant medium was refreshed 12 h after transfection. Lentivirus particles were collected at 48 h and 72 h after transfection. NPC cells were seeded in 6-well plates and cultured until they reached approximately 70% confluency. Then, cells were transduced with the negative control, POLN WT, or mutant lentivirus particles at the same multiplicity in the presence of 8 μg/ml of polybrene (Solarbio, Cat# H8761, Beijing, CHN). Real-time quantitative polymerase chain reaction (RT-qPCR) and western blotting were used to examine POLN expression (Figure S12, S5, S3 and S7).
RNA interference of POLN
The siRNA duplexes used to knockdown POLN were purchased from RiboBio (Guangzhou, CHN). The commercial transfection reagent Lipofectamine 3000 (Cat # L3000075; Invitrogen) was used for cell transfection. The siRNA sequences used are listed in Table S9. siRNA efficacy was confirmed by RT-qPCR and western blotting (Figure S13 and S4).
RNA extraction, reverse transcription, and real-time quantitative polymerase chain reaction
Total RNA was extracted from cells and tissues using TRIzol reagent (Invitrogen, Cat#15596026) following the manufacturer's protocol. Total RNA was reverse-transcribed using the PrimeScript RT reagent Kit with gDNA Eraser (TaKaRa, Cat#RR047A, Japan). RT-qPCR was performed using a LightCycler 480 SYBR Green I Master (Roche, Cat#04887352001) on a LightCycler 480 System (Roche). The primers used for POLN, BZLF1, BRLF1, BKRF4, BOLF1, BLLF1, and GAPDH are listed in Table S7. GAPDH was used as an internal control to normalise the relative expression of the genes. The relative mRNA expression was calculated using the comparative Ct method 2−ΔΔCt.
Protein Extraction and Western Blot analysis
Proteins were extracted using a whole-cell lysis assay (KeyGEN BioTECH, Cat#KGP2100, Nanjing, Jiangsu, CHN). BCA Assay Reagent (Cwbiotech, Cat#CW0014S, Beijing, CHN) was used to measure protein concentration. Protein lysates (20 μg) were electrophoresed on a 10% sodium dodecyl sulfate (SDS) (Aladdin, Cat#S108346, Shanghai, CHN) polyacrylamide gel and transferred onto PVDF membranes (Merck Millipore, Cat#IPVH00010, Billerica, MA, USA). Membranes were blocked with TBST (Solarbio, Cat#T1085) containing 5% non-fat dried milk (BIO-RAD, Cat#1706404, Hercules, CA, USA) and incubated overnight at 4 °C with POLN (1:1,000) (lifeSpan bioSciences, Lsbio, Rabbit, Cat#LS-C163843, Seattle, WA, USA), HA (1:1,000) (Cell Signalling Technology, CST, Mouse, Cat#2367, RRID:AB_10691311, Boston, MA, USA), BZLF1 (1:250) (Santa Cruz Biotechnology, Mouse, Cat# sc-53904, RRID:AB_783257, Dallas, TX, USA), BRLF1 (1:500) (Bioss, Rabbit, Cat#bs-4542R, Boston, MA, USA) and α-Tublin (1:1,000) (Proteintech, Mouse, Cat#66031-1-Ig, Wuhan, Hubei, CHN) antibodies. The membranes were subsequently incubated with rabbit (1:5,000) (ZSGB-BIO, Cat#ZB-2301, Beijing, CHN) or mouse (1:5,000) (ZSGB-BIO, Cat#ZB-2305) secondary antibodies conjugated to horseradish peroxidase and incubated with ECL reagents (Merck Millipore, Cat#WBKLS). Band intensities were quantified using Image Lab (Bio-Rad).
Cycloheximide (CHX) chase assay
HK1 EBV+ and Tet-BZLF1/CNE2 EBV+ NPC cells were seeded in 10 cm dishes and transduced with POLN WT or mutant lentivirus, respectively. After 72 h, the cells were seeded in 6-well plates and treated with 100 μg/ml cycloheximide (CHX) (MCE, Cat#HY-12320, Shanghai, CHN). The protein was collected at four time points: 0 h, 2 h, 4 h and 8 h, and the changes in POLN were analysed by western blotting.
EBV lytic replication induction
To induce EBV lytic replication, 12-O-Tetradecanoylphorbol-13-Acetate (TPA) (20 ng/mL) (CST, Cat#4174S) with sodium butyrate (NaB) (3 mM) (Macklin, Cat#156-54-7, Shanghai, CHN) (TPA/NaB) or doxycycline (DOX) (0·2μg/ml) (Sigma, Cat#D3072) was added to the culture medium of HK1 EBV+ or Tet-BZLF1/CNE2 EBV+ NPC cells, respectively. The cell assays were conducted 24 h after induction.
Cell Immunofluorescence
Cells were washed twice with phosphate buffered saline (PBS) (Gibco, Cat#10010023), fixed with 4% paraformaldehyde (G-CLONE, Cat#PN4204, Beijing, CHN) for 30 min at room temperature (RT), and neutralised with 2 mg/ml glycine (Sigma, Cat#G8898). Subsequently, the cells were permeabilised with 0·5% Triton X-100 (Solarbio, Cat# P1080) for 10 min and blocked with 5% BSA (MP, Cat#9048-46-8, Santa Ana, CA, USA) for 1 h, followed by incubation with POLN (1:250) (Thermo Scientific, Rabbit, Cat# PA5-65162, RRID: AB_2663861) and BMRF1 (1:250) (Abcam, mouse, Cat# ab30541, RRID: AB_732194, Cambs, UK) primary antibodies at 4 °C overnight. After three washes in TBST, the coverslips were incubated with Alexa Fluor 555 goat anti-mouse IgG (1:1,000) (CST, Cat# 4409, RRID: AB_1904022) and Alexa Fluor Plus 647 donkey anti-rabbit IgG (1:1,000) (Thermo Scientific, Donkey, Cat# A32795, RRID: AB_2762835) for 1 h in a dark room. The coverslips were washed three times and stained with DAPI (Solarbio, Cat#C0065) for 5 min at RT. After drying and mounting with ProLong Gold Antifade Mountant (Invitrogen, Cat#P36930), the subcellular localisation of POLN and BMRF1 proteins was analysed using a confocal microscope (ZEISS, LSM880 with fast airyscan, Oberkochen, Germany). ZEN Blue software was used for the quantification of colocalisation between BMRF1 and POLN.
Flow Cytometry
The cells were collected and washed twice with PBS. The percentage of GFP -positive cells was evaluated by flow cytometry (CytoFLEX 2; Becman, Brea, CA, USA) using signal wavelength analysis.
EBV DNA extraction and copy number detection
Cell culture media used for determining extracellular genome copies were treated with DNase I (Vazyme, Cat#EN401/402) before DNA extraction to digest cell-free DNA which was not detected in the DNase-resistant viral particles. Total DNA was extracted from cells and culture media simultaneously using the Blood/Cell/Tissue Genomic DNA Extraction Kit (TIANGEN, Cat#DP304, Beijing, CHN), following the manufacturer's protocol. EBV DNA copy numbers were determined by RT-qPCR using primers targeting the EBV BALF5 and β-Globin genes of the human genome (Table S7). Standard sample ladders (102, 103, 104, 105 and 106 copies/μl) were used to generate a standard curve. The relative EBV DNA copy number in cells was expressed as the ratio of the copy number of the EBV genome to that of the β-Globin DNA.
Cell Viability Assay
One thousand NPC cells were seeded in each well of a 96-well plate and cultured for one to five days. Ten microlitres of Cell Counting Kit-8 (CCK8) (APExBIO, Cat#K1018, Houston, TX, USA) solution was added to each well. After incubation for 2 h, the absorbance was measured at 450 nm using a spectrophotometre (BIO-Tek epoch, Winooski, VT, USA).
Statistical analysis
Statistical analyses were performed using R (version 3.5.0) and GraphPad Prism (version 9.0.1). Logistic regression adjusted for age and sex was used to compare the mutation frequencies of mutations in familial NPC cases or sporadic NPC cases with those in healthy controls. Student's t-test was conducted to compare the age of onset between P577L carriers and non-carriers in familial NPC cases. Experimental data are presented as the mean ± standard deviation (SD) from three separate experiments. A two-sided Student's t-test was used to compare data points between the two groups. Two-way analysis of variance (ANOVA) was used to test the statistical significance of the growth difference between the two groups. The results were considered statistically significant at p < 0·05.
Ethics statement
This study was approved by the ethical review committees of Sun Yat-sen University Cancer Center, Guangzhou, China (YB2020-001-01). All subjects signed informed consent. Our study conforms to the Regulation of the People's Republic of China on the Administration of Human Genetic Resources (CJ1259).
Role of funding source
The funders had no role in collection, analysis, and interpretation of the data or in the writing of this publication.
Results
POLN P577L mutation co-segregates with NPC onset in FA family
WES was conducted on the available samples from 13 families recruited through “High‐risk Nasopharyngeal Carcinoma Family Screening Program” (see methods), including 35 familial NPC patients and 34 healthy relatives. The data quality and characteristics of the samples are summarised in Tables S1 and S2, respectively. The mean age at disease onset of 35 familial NPC patients was 40·03 years (SD 8·47, range 24-65), which is younger than that reported for sporadic cases (46·6 years)6. The average sequencing coverage is 107X.
A total of 219,053 SNPs and indels were identified. By focusing on mutations that were present in all cases but not in any unaffected relatives within a family, co-segregation analysis was performed using a dominant model. As shown in Figure 1a, after filtering out common and synonymous mutations, we identified 145 rare disease-segregating mutations (MAF < 0·1% in public databases) in 141 genes (Table S3), including 129 nonsynonymous mutations, 11 INDELs mutations, and 5 stop-gain mutations. We then narrowed down the candidate mutations to five mutations (Table 1), which are located within important protein domains and were predicted to be damaging mutations. rs146182235 in POLN (NM_181808.4: exon16: c.C1730T: p.P577L) had the highest rank score in all three in silico tools (SIFT converted rankscore =0·91, Polyphen2 HVAR rankscore =0·97, FATHMM converted rankscore = 0·97) (Table 1). This mutation was carried by all affected individuals in the FA family (Figure 1b; the ages of onset were 34, 39, and 25 years, respectively). The genotypes of the POLN P577L mutation were validated using Sanger sequencing (Figure S1). In addition, FA family members did not carry any previously reported germline mutations in genes associated with familial NPC18,19 (Table S4). POLN encodes type A DNA polymerase, which is responsible for DNA replication and repair,27 and mutations in this gene were associated with an increased risk of familial melanoma,33 lung cancer,34 breast cancer,35 and prostate cancer.36 Therefore, we selected POLN as the candidate gene associated with the development of familial NPC.
Table 1.
Five candidate rare mutations identified in the co-segregation analysis.
| MAFd in public database |
In silico prediction |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID | Chra | Position | Refb | Mutc | Gene | Variant | 1000 genomes |
gnomAD_exome |
ESP6500 | ExAC_nonTCGA |
SIFT | Polyphen2 HVAR | FATHMM | ||||
| Alle | EASf | Alle | EASf | Alle | Alle | EASf | Prediction | Rank Score | Rank Score | Rank Score | |||||||
| FA | 4 | 2158516 | G | A | POLN | p.P577L | . | . | 0.0002 | 0.0003 | 0.0002 | 0.0001 | 0.0001 | Damaging | 0.91 | 0.97 | 0.97 |
| FA | 13 | 20987480 | C | A | CRYL1 | p.G227V | . | . | 4.07E-06 | 0 | . | 9.49E-06 | 0 | Damaging | 0.91 | 0.97 | 0.91 |
| F058 | 17 | 74765828 | G | A | MFSD11 | p.G250D | . | . | 1.77E-05 | 0.0002 | . | 1.89E-05 | 0.0003 | Damaging | 0.91 | 0.97 | 0.82 |
| F080 | 12 | 57642540 | G | C | STAC3 | p.I127M | . | . | 4.06E-06 | 0 | . | . | . | Damaging | 0.72 | 0.82 | 0.93 |
| F025 | 2 | 170129490 | C | T | LRP2 | p.R688H | . | . | 4.07E-05 | 0 | 7.70E-05 | 4.71E-05 | 0 | Damaging | 0.68 | 0.92 | 0.87 |
Chromosomal region.
Reference allele.
Mutant allele.
Minor allele frequency.
All population.
East Asians.
POLN mutations identified in additional familial NPC cases
Sanger sequencing of all POLN exons and exon-intron boundaries was performed in 351 additional familial NPC cases. The clinical characteristics of these samples are described in Table S2. To investigate other mutations in POLN, we integrated the 351 familial NPC cases with the WES data of the 13 families, excluding the FA family, and found a total of 40 mutations in POLN among these 363 familial NPC cases (Table S5). Three rare mutations were predicted to be damaging. We detected one more POLN P577L mutation carrier (female, age at onset: 42 years). Additionally, two additional POLN rare mutations, R303Q (exon6: c.G908A) carried by a male patient diagnosed at 41 years of age and F545C (exon15: c.T1634G) carried by a male patient diagnosed at 42 years of age were found. Sanger sequencing was performed to validate these mutations (Figure S1).
POLN mutations are associated with an increased risk of familial NPC
Genotyping of three POLN mutations (P577L, F545C, and R303Q) was performed in an independent cohort of 212 familial NPC cases, 2,953 sporadic NPC cases, and 3,175 healthy controls (Table S2). The P577L mutation was detected in five individuals, including one patient with familial NPC (diagnosed at 43 years of age), three sporadic patients (diagnosed at 68, 42, and 40 years of age respectively), and one healthy control (60-year-old). The R303Q mutation was detected in one sporadic patient (diagnosed at 65 years of age) and one healthy control (59-year-old). No additional F545C mutation was detected in these samples. The genotypes of all mutation carriers were validated using Sanger sequencing (Figure S1).
Combining all the independent samples included in this study (576 independent familial cases [13 for WES, 563 for validation], 2,953 sporadic cases, and 3,374 healthy controls), the POLN rare damaging mutations showed a significantly higher frequency in familial NPC cases (5/576, 0·87%) than in healthy controls (2/3374, 0·059%) with an adjusted OR of 44·84 (95% CI:3·91-514·34, logistic regression, p = 2·25 × 10−3) (Figure 1c). Additionally, these rare damaging mutations showed a relatively higher frequency in sporadic NPC cases (4/2953, 0·135%) than in healthy controls (2/3374, 0·059%) with an adjusted OR of 2·41 (95% CI:0·44-13·21, logistic regression, p = 0·31). We also observed a younger age of onset in the familial patients carrying P577L (mean age:39·67 years) than in those not carrying this mutation (mean age: 47·95 years, Student's t-test, p = 0·096).
We further analysed WES data from publicly available NPC studies18,19,37, 38, 39, 40 to investigate the mutations of POLN in NPC cases (Table S6). POLN P577L, R303Q, and F545C mutations were not detected in these studies. However, we found 14 additional POLN rare mutations in 19 NPC cases, including one synonymous mutation, six missense mutations, one frameshift mutation, and six intronic mutations.
POLN is involved in the completion of EBV lytic replication
POLN was reported to interact with DNA repair factors Rad51 and PCNA,27 which have been shown to be involved in EBV DNA replication.41,42 To investigate the function of POLN in EBV DNA replication, we examined the subcellular localisation of POLN and BMRF1 (DNA processivity factor, completely coincided with the newly synthesized EBV DNA43) in two EBV-positive NPC cell lines, HK1 EBV+ and Tet-BZLF1/CNE2 EBV+. After lytic replication induction, we found that POLN was colocalised with the BMRF1 protein (Manders' Colocalization Coefficients: HK1 EBV+: 0·91, Tet-BZLF1/CNE2 EBV+: 0·87) (Figure 2a and Figure S2a), suggesting that POLN may tend to be located in EBV replication compartments. Thus, we examined whether POLN expression affected EBV DNA replication. The two cell lines contained an exogenous recombinant EBV strain that expressed enhanced green fluorescent protein (GFP). We overexpressed POLN in both cell lines and observed that cells overexpressing POLN showed a higher percentage of GFP-positive cells than the control, with and without lytic replication induction (Figure 2b and Figure S2b). We further confirmed this result by quantifying the intracellular EBV DNA copy number using RT-qPCR. Overexpression of POLN increased the intracellular EBV genome copy number (Figure 2c and Figure S2c). Next, we used two siRNAs to knock down the expression of POLN in the two cell lines, and found that the knockdown of POLN decreased the percentage of GFP-positive cells (Figure 2d and Figure S2d) and the intracellular EBV genome copy number (Figure 2e and Figure S2e). These results suggested that POLN plays a substantial role in EBV DNA replication.
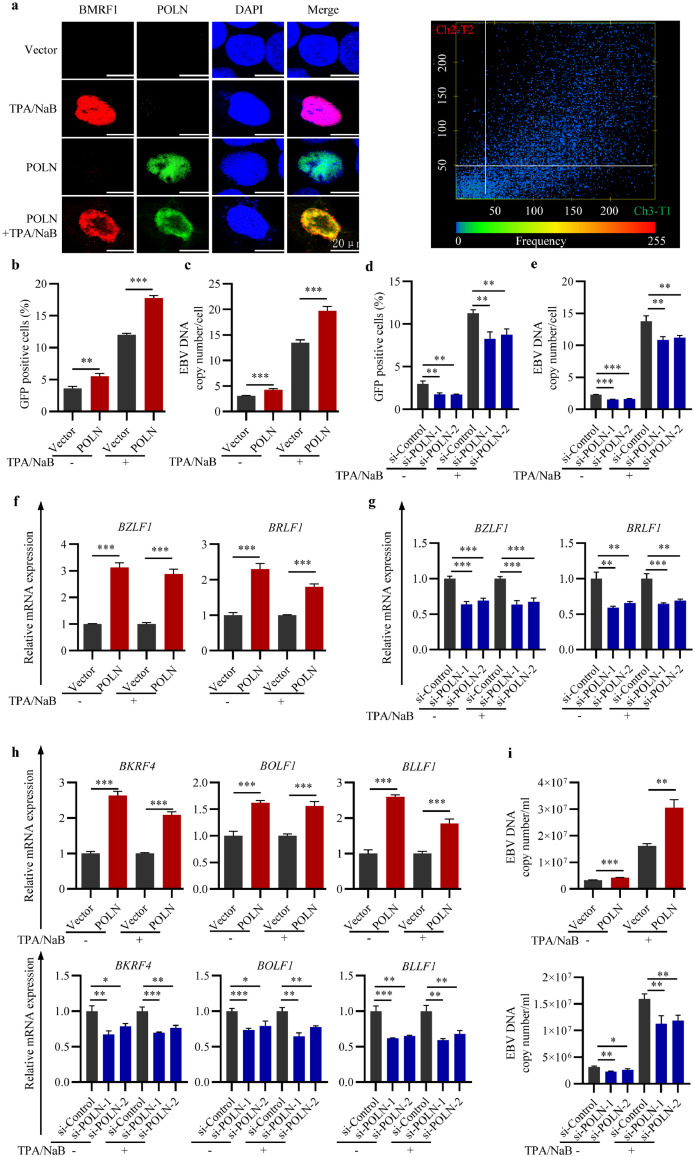
Figure 2.
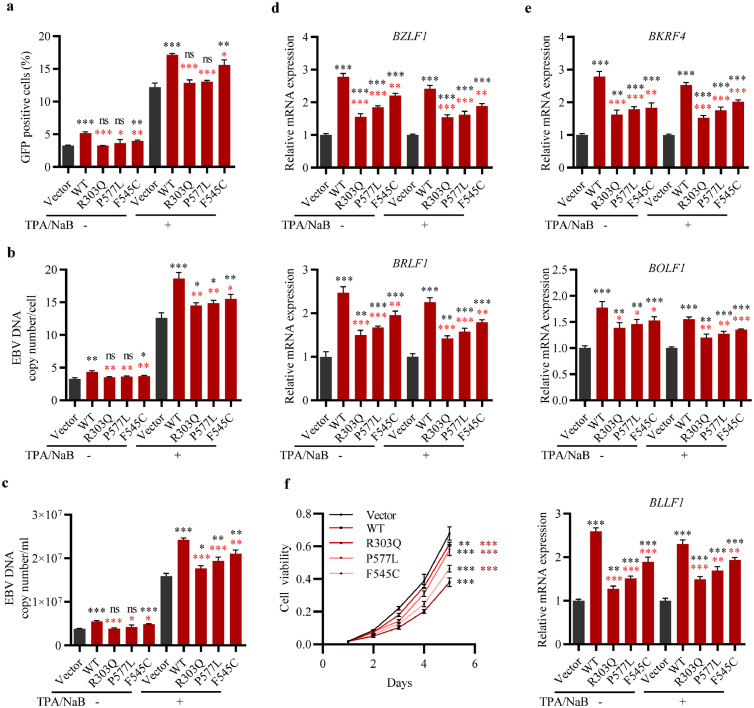
POLN is involved in the completion of EBV lytic replication. POLN was overexpressed or knocked down in HK1 EBV+ cells, twenty-four hours after induction of EBV lytic replication by TPA/NaB, cells were harvested. (a) Immunofluorescence (left panel) was performed to show the location of BMRF1 (red) and POLN (green) in HK1 EBV+ cells, DAPI (blue). The picture on the right panel showed the scatterplot of colocalisation between BMRF1 and POLN. (b) The effect of POLN overexpression on the proportion of GFP-positive cells. (c) The effect of POLN overexpression on the intracellular EBV genome copy numbers (d) The effect of POLN knockdown on the proportion of GFP-positive cells. (e) The effect of POLN knockdown on the intracellular EBV genome copy numbers. (f) The effect of POLN overexpression on the mRNA expression level of EBV IE genes BZLF1 and BRLF1. (g) The effect of POLN knockdown on the mRNA expression level of EBV IE genes BZLF1 and BRLF1. (h) The effect of POLN overexpression or knockdown on the mRNA expression level of EBV true late genes BKRF4, BOLF1, and BLLF1. (i) The effect of POLN overexpression or knockdown on the extracellular EBV genome copy numbers. (b-i) Data were presented as mean ± SD (N = 3), ***p < 0·001, **p < 0·01, *p < 0·05, ns: not significant (Student's t-test).
EBV, Epstein-Barr virus; SD, Standard deviation; WT, wild type.
Viral IE genes BZLF1 and BRLF1 play critical roles in the initiation of lytic replication of EBV,44 both of which were upregulated in POLN-overexpressing cells (Figure 2f and Figure S2f, S3), and downregulated in POLN knockdown cells (Figure 2g and Figure S2g, S4). The expression of EBV true late genes is strictly dependent on viral DNA replication.23 Our results showed that the expression of true late genes BKRF4, BOLF1, and BLLF1 was up-regulated at least 1.5-fold in both cells which overexpressed POLN with and without EBV reactivation, and the knockdown of POLN decreased their expression (Figure 2h and Figure S2h). POLN promotes EBV DNA replication and increases viral true late gene expression, which encouraged us to investigate whether POLN affects progeny viral particle production. The amount of DNase-resistant extracellular viral particles in cell culture supernatants was evaluated and the results showed that the extracellular EBV DNA copy number was increased when POLN was overexpressed and decreased when POLN was knocked down (Figure 2i and Figure S2i). These findings revealed that POLN was involved in the completion of EBV lytic replication.
POLN inhibits the proliferation of NPC cells and shows lower expression in NPC tissues
In epithelial cells, the latent infection and abortive/incomplete lytic infection of EBV in host cells have been shown to contribute to NPC carcinogenesis.24,45 In complete lytic cycle, productively infected cells eventually lyse, resulting in the inhibition of cell proliferation.24 Based on the finding that POLN plays a role in complete lytic reactivation and viral production, we subsequently evaluated the effect of POLN on NPC cell proliferation. CCK8 assay showed that overexpression of POLN significantly inhibited the proliferation of NPC cells (Figure 3a), whereas knockdown of POLN promoted the proliferation of NPC cells (Figure 3b).
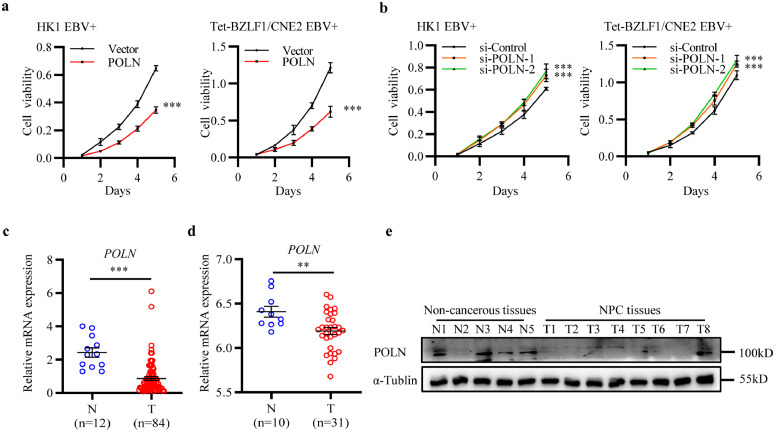
Figure 3.
POLN inhibited the proliferation of NPC cells and showed lower expression in NPC tissues. (a) The effect of POLN overexpression on cell proliferation in HK1 EBV+ and Tet-BZLF1/CNE2 EBV+ cells. Data were presented as mean ± SD (N = 4), ***p < 0·001 (two-way ANOVA). (b) The effect of POLN knockdown on cell proliferation in HK1 EBV+ and Tet-BZLF1/CNE2 EBV+ cells. Data were presented as mean ± SD (N = 4), ***p < 0·001 (two-way ANOVA). (c) The mRNA expression level of POLN was measured by RT-qPCR in non-cancerous nasopharyngeal tissues (N, n=12) and NPC tissues (T, n=84), data were presented as mean ± SEM. (d) Expression analysis of POLN mRNA in normal nasopharyngeal tissues (N, n=10) and NPC tissues (T, n=31) from GSE12452, data were presented as mean ± SEM. (c, d) Student's t-test was used for the statistical analysis. ***p < 0·001, **p < 0·01. (e) The protein expression level of POLN in non-cancerous nasopharyngeal tissues (N, n=5) and NPC tissues (T, n=8) was measured by western blot.
SEM, Standard error of mean; ANOVA, Analysis of variance.
Next, we examined POLN mRNA expression in 12 non-cancerous nasopharyngeal tissues and 84 NPC tissues using RT-qPCR and found lower POLN expression in NPC tissues (Figure 3c). We also obtained gene expression data from GEO datasets (https://www.ncbi.nlm.nih.gov/geo/) (GSE12452).46 Consistently, POLN was lower levels in NPC tissues (n=10) than in normal nasopharyngeal tissues (n=31) (Figure 3d). To further confirm this finding, we examined POLN protein expression in five non-cancerous nasopharyngeal samples and eight clinical NPC samples by western blotting, and found lower POLN protein expression in NPC tissues (Figure 3e).
The three rare damaging mutations impaired the POLN protein stability and decreased the protein expression
POLN is a 100.3-kDa protein with 900 amino acids and two functional domains, DnaQ_like_exo and DNA_pol_A_theta (Figure 4a). The three identified amino acids were located within the functional domains and showed high evolutionary conservation in five orthologous proteins (Figure 4b). We used INPS (http://inps.biocomp.unibo.it) to predict the thermodynamic free energy change for the three mutations and found decreased stability in the mutant protein. We further performed molecular modelling of the 194-859 amino acid region of POLN (PDB 4XVK) (https://www.rcsb.org/structure/4XVK) using the PyMOL software to evaluate the potential structural changes induced by the mutations (Figure 4c). Arg 303 (basic) was thought to be involved in a salt bridge interaction with Asp 335 (acidic) (distance: 2·5 Å), and the Arg to Gln (R303Q) mutation was predicted to impair this interaction (Figure 4d). Substitution of Pro with Leu (P577L) was predicted to induce the loss of conformational constraint ability and, therefore, may destabilise the protein structure. P577L and F545C are located near the DNA-binding site (residues 573-576 and 543-544, respectively) and were predicted to weaken the interaction with DNA (8·5 Å for Pro577 versus 9·3 Å for Leu577; 4·6 Å for Phe545 versus 6·9 Å for Cys545) (Figure 4e-4f). Phe545 is involved in the formation of π-π stacking interactions with DNA, and the substitution of Phe with Cys was predicted to impair this interaction. In summary, the three identified amino acid substitutions were predicted to impair POLN protein stability and slightly decrease its DNA-binding ability.
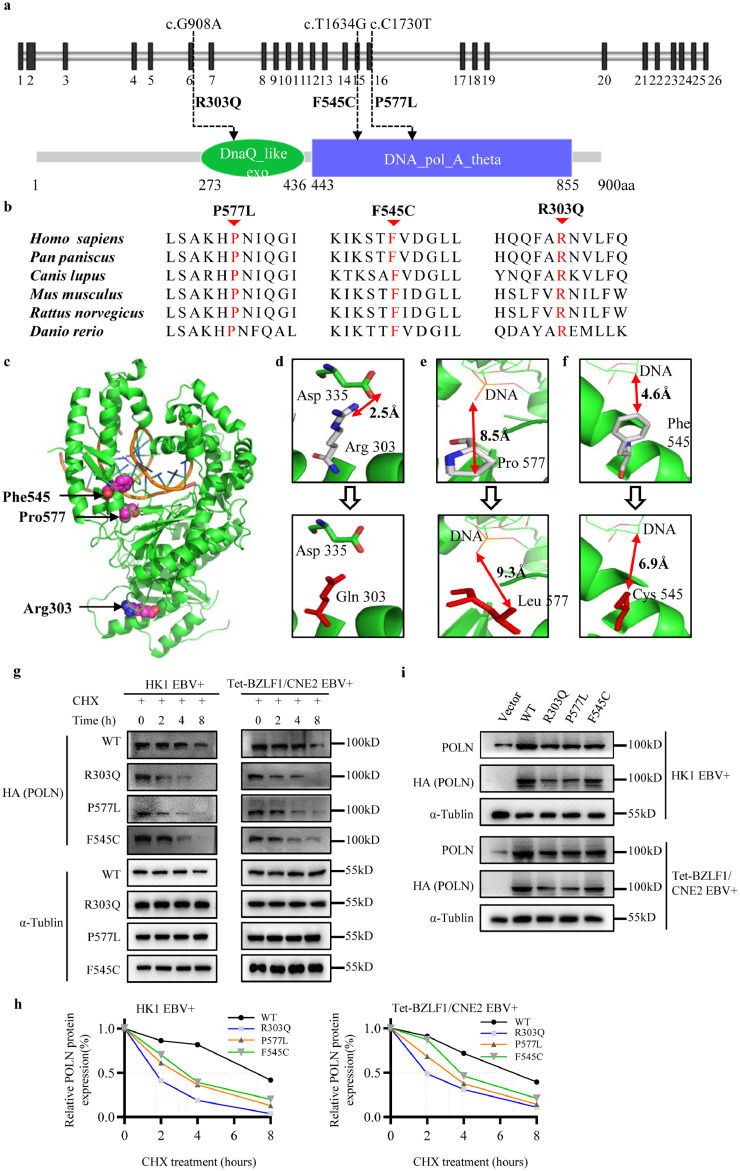
Figure 4.
The efficacy of mutations on POLN protein. (a) Top, schematic representation of POLN gene (reference sequences: NM_181808.4), which was composed of 26 exons, the solid black rectangles indicated exons and the gray horizontal bars introns. Bottom, schematic of the POLN protein (reference sequences: NP_861524.2) showing its different domains, DnaQ_like_exo (green) and DNA_pol_A_theta (blue). Dashed lines indicated the location of mutations. (b) Amino acid conservation analysis. Multiple sequence alignment of POLN ortholog protein across six species (ClustalW, BioEdit software) showed evolutionary conservation at the site of the identified amino acid substitution (indicated by the red arrow). (c) 3D representation of the crystal structure of the 194-859 amino acid region of POLN (PDB 4XVK). The position of mutated residues was highlighted in pink ball, DNA in orange, PyMOL software (Version 1.7.0.0) was used for visualization. (d-f) The interaction and distance modification resulting from the P577L, R303Q, and F545C mutations. The WT and mutated residues were colored by gray and red, respectively. The red double arrow line showed the interaction distance. (g-i) HK1 EBV+ and Tet-BZLF1/CNE2 EBV+ cells were transduced with vector, WT POLN lentivirus or mutant POLN lentivirus harbouring R303Q, P577L, and F545C mutations. (g-h) Cells were treated with CHX for 0 h, 2 h, 4 h and 8 h, and the protein was collected. (g) The changes of HA (POLN) protein were analyzed by western blot. (h) Quantification of CHX chase assays. Image J was used to analyze the grayscale of each band. Dotted line indicated 50% of normalized HA (POLN) protein. (i) In western blot assay, endogenous and exogenous POLN expressions were detected using POLN or HA antibodies, respectively.
We performed a cycloheximide (CHX) chase experiment to analyse the protein stability of the WT and mutant POLN. Consistent with the results of the above in silico analysis, the three mutants showed shorter degradation half-lives than WT POLN (Figure 4g-4h). Moreover, we performed a western blot assay to assess the effect of mutations on protein expression and found a reduction in POLN protein expression levels in all three mutant types, compared to WT POLN (Figure 4i). Moreover, the R303Q and P577L mutated POLN protein showed lower expression levels than those of the F545C mutation. Both bioinformatics and experimental results suggested that the three mutations impaired POLN protein stability by shortening its degradation half-life.
The three rare damaging mutations impair the function of POLN in EBV lytic replication and NPC cells proliferation
We have observed that POLN is involved in EBV lytic cycle, including the process of EBV DNA replication, IE genes and true late genes expression, as well as progeny EBV particles production. As the three mutations have been shown to decrease the protein stability and expression of POLN, we further investigated whether these mutations impair its function. We overexpressed WT POLN and mutant POLN harbouring R303Q, P577L, and F545C in HK1 EBV+ and Tet-BZLF1/CNE2 EBV+ cell lines, using lentiviruses at the same multiplicity. The mRNA expression levels of the WT and mutant POLN were similar (Figure S5). Twenty-four hours after lytic replication induction, we examined the percentage of GFP-positive cells and measured intracellular EBV DNA titrations together with DNase-resistant extracellular viral particles. Expression of each of the three mutant POLN resulted in a lower percentage of GFP-positive cells (Figure 5a and Figure S6a) and lower intracellular EBV DNA copy numbers (Figure 5b and Figure S6b) than those detected in WT POLN. Consistently, a decreased extracellular viral particle EBV genome copy number was detected in NPC cells overexpressing mutant POLN (Figure 5c and Figure S6c). Compared to WT POLN, mutant POLN decreased the expression of IE genes BZLF1 and BRLF1 (Figure 5d and Figure S6d, S7), and true late genes BKRF4, BOLF1 and BLLF1 (Figure 5e and Figure S6e). The results of the cell viability assay showed that the mutations weakened the inhibitory effect of POLN on the proliferation of NPC cells (Figure 5f and Figure S6f). The P577L and R303Q mutations showed a greater effect than the F545C mutation. These results suggest that the three rare mutations could impair the function of POLN in promoting EBV lytic replication and inhibiting cell proliferation, which indicates the important role of these mutations in NPC development.
Figure 5.
Mutations impaired the function of POLN in EBV lytic replication and cell proliferation. (a-e) HK1 EBV+ cells were transduced with vector, WT POLN lentivirus and mutant POLN lentivirus harbouring R303Q, P577L, and F545C mutations, 24 h after induction of viral lytic replication by TPA/NaB, cells were harvested. (a) The effect of POLN mutations on the proportion of GFP-positive cells. (b) The effect of POLN mutations on the intracellular EBV genome copy numbers (c) The effect of POLN mutations on the extracellular EBV genome copy numbers. (d) The effect of POLN mutations on the mRNA expression level of EBV IE genes BZLF1 and BRLF1. (e) The effect of POLN mutations on the mRNA expression level of EBV true late genes BKRF4, BOLF1, and BLLF1. (a-e) Data were presented as mean ± SD (N = 3), ***p < 0·001, **p < 0·01, *p < 0·05, ns: not significant (Student's t-test). (f) CCK8 assay was used to detect the proliferation of HK1 EBV+ cells expressing vector, WT POLN, and mutant POLN. Data were presented as mean ± SD (N = 4), ***p < 0·001, **p < 0·01 (two-way ANOVA). Black asterisk: compared with vector, red asterisk: compared with WT POLN.
Discussion
Using co-segregation and association analyses in multiplex NPC families and an independent validation cohort of familial cases, sporadic cases, and healthy controls (N=6,890), we identified three germline mutations, P577L, R303Q, and F545C of POLN, which confer susceptibility to familial NPC. By integrating bioinformatics analysis and functional assays, we showed that compared with WT POLN, the three mutations decreased the protein expression and stability of POLN, impaired the function of POLN in EBV lytic replication, and promoted cell proliferation.
We and others have identified genetic mutations in some DNA repair genes that are associated with NPC susceptibility, such as those involved in base excision repair (XRCC1,47,48 OGG1,49, 50, 51 APEX152), homologous recombination repair (XRCC3,53 RAD51L1,54 XRCC255), nucleotide excision repair (ERCC1,56,57 XPC,57,58 XPD57,59,60). Previous studies have reported that DNA repair factors, such as DNA mismatch repair factors,42 homologous recombinational factors,41 and translesion DNA synthesis factors61, 62, 63 are involved in EBV lytic replication. Interestingly, EBV hijacks DNA damage response transducers to orchestrate its life cycle.64 POLN is also a DNA repair factor and is mainly responsible for translesion DNA synthesis26 and DNA cross-link repair.65 The cellular condition of the S-phase cell cycle appears to favour viral lytic replication.66 The expression level of POLN is the highest in the S-phase cell cycle.27 In addition, EBV lytic replication-related DNA repair factors Rad51 and PCNA can interact with POLN, and this interaction only occurs in the S phase.27 These findings indicate that the DNA repair factor POLN may participate in EBV lytic replication.
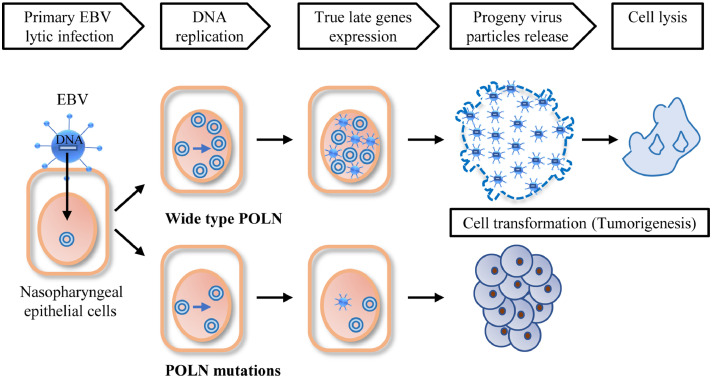
Lytic or latent infection or abortive lytic infection that contributes to EBV-associated oncogenesis remains obscure. In EBV-associated lymphoma, lytic infection may contribute to oncogenesis, and deletions in the viral genome, such as the BART region, were found to promote EBV lytic reactivation.67 However, for EBV-associated epithelial cancers, deletions in the EBV genome are rare, and BART-encoding miRNAs are highly expressed, which promotes latent EBV infection. Traditionally, EBV in NPC was classified as latency II, in which EBNA1, LMP1, and LMP2A are expressed at the protein level, but antibody levels for lytic antigens (Zta, Rta, and VCA) increased in NPC sera, suggesting that lytic infection may contribute to NPC carcinogenesis. In addition to IHC which detects protein expression, mRNA detection by RT-PCR and recent single-cell RNA sequencing has revealed EBV antigen expression in NPC. In one study of NPC biopsies, all eight samples were positive for the IE gene BZLF1, but only five of the eight samples tested positive for the early gene BMLF1 expression, and none tested positive for another early gene BBLF2/3 by RT-PCR assays.68 In NPC single-cell sequencing, both latent genes (LMPs and EBNA1) and a few lytic genes (A73, BARF0, BNFR1, and BALF4) were expressed, but no mRNAs of late lytic genes were detected.69 Thus, NPC tumours harbour cells undergoing an incomplete or abortive lytic phase, in which some early lytic genes are expressed but no viral particles are produced. Therefore, abortive/incomplete lytic infection of epithelial cells is highly likely to contribute to NPC carcinogenesis. The gene we identified, POLN, facilitated complete lytic reactivation and viral production, and its mutations failed to support EBV production, which is in line with the in vivo scenario. For individuals carrying these three mutations, EBV may also be prone to latent infection or abortive lysis in epithelial cells, facilitating the survival and malignant transformation of EBV-infected epithelial cells (Figure 6).
Figure 6.
A proposed schematic for the molecular mechanism of POLN and the rare damaging mutations. Primary EBV infection of nasopharyngeal cells harbouring WT POLN finally resulted in host cell death, however, when EBV primarily infect nasopharyngeal cells harbouring three mutations, the reduced EBV DNA replication, viral structural proteins expression and progeny viral particles release would contribute to the survival of EBV-infected host cells, which may promote the tumorigenesis of NPC.
As a multifactorial disease, a complex interplay between host genetics, EBV infection, and environmental factors has been shown to be involved in NPC. Many lines of evidence indicate that host heredity interacts with EBV infection and plays a critical role in disease development. We previously reported that human genetic variants of homologous recombination repair genes are associated with EBV antibody titres in healthy Cantonese.70 Recently, we found that nasopharyngeal EBV loads are positively correlated between relative pairs from NPC multiplex families and have significantly high heritability.25 EBV infection status could modulate the role of genetic polymorphism in NPC.71 However, laboratory evidence of the interaction between the etiological factors of NPC is lacking. Our functional experiments validated that POLN participates in the completion of EBV lytic replication. In spontaneous lytic infection (low-level lytic activation) or induced lytic infection (high-level lytic activation), POLN function was impaired by the deleterious mutations identified in familial NPC.
We performed an integrated study combining bioinformatics analysis and functional experiments to explore germline mutations in NPC and demonstrated how rare mutations in the POLN exon region contribute to NPC susceptibility. Our results provide experimental evidence of the interaction between host genetics and EBV infection in NPC carcinogenesis, highlighting the importance of DNA repair factors in EBV lytic replication and carcinogenesis. However, this study had several limitations. First, whether POLN mutations are linked to NPC in other populations remains unknown. Our analyses were restricted to participants of Han Chinese ancestry, which limits the generalisability of our findings to diverse populations. Second, we did not have direct evidence to support POLN as a bona fide polymerase for EBV, as in vitro viral DNA replication assays were lacking; alternatively, whether POLN participates in EBV lytic activation in an enzymatic activity-independent manner was unclear, as none of the mutation sites were located in DNA-binding or catalytic sites of POLN but greatly affected POLN protein stability. Third, whether POLN mutations also involved in other EBV-associated cancers, such as EBV-positive gastric cancer and EBV-related lymphomas like Burkitt's lymphoma and NK/T cell lymphoma, were currently unknown. Finally, to clarify whether somatic mutations in POLN affect NPC development through the “two-hit” model, paired blood and NPC tissue samples are needed to analyse the impact of POLN somatic mutations on gene expression and tumour characteristics.
In summary, our study identified a susceptibility gene POLN for familial NPC, and highlighted its function in regulating EBV lytic replication. Our study supports the assumption that DNA repair factors play an important role in NPC development and that the interaction between the etiological factors of NPC is important for tumourigenesis.
Contributors
Jia W.H. conceived and supervised the study. Feng L. co-supervised, designed functional experiments and interpreted the results. Xiao R.W. and Wang F. performed key laboratory assays, including functional experiments, Sanger sequencing, TaqMan genotyping assays and data analysis. Wang T.M. worked on sequencing data, bioinformatic and statistics analysis. Zhang J.B. coordinated part workflow. Zhang J.B., Wu Z.Y., Deng C.M., Liao Y. and Dong S.Q. constructed DNA libraries for WES, genotyping, Sanger sequencing. Wu Z.Y., Deng C.M., Liao Y., Zou T., Yang D.W., He Y.Q., Xue W.Q., Liu Y.Y. and Xia Y. F. recruited and collected blood samples of NPC family members, collected the clinical information of participants. Zheng X.H., Li X.Z., Zhang P.F., Zhang S.D. and Hu Y.Z. extracted DNA. Mu J.B. gave important consultation, critical revision and manuscript edition. Gao S. consulted for protein structure prediction in silico. Jia W.H. and Xiao R.W. wrote the manuscript. Jia W.H. and Feng L. revised the manuscript with the assist of Wang F., Xiao R.W. and Wang T.M.
Jia W.H., Feng L., Xiao R.W. and Wang F. directly accessed and verified the underlying data reported in the manuscript. All authors read and approved the final version of the manuscript.
Data sharing statement
The datasets used and/or analyzed during the current study are available from the corresponding author (Prof. Wei-Hua Jia:jiawh@sysucc.org.cn) on reasonable request. The key raw data of this research have been deposited in the Research Data Deposit public platform (www.researchdata.org.cn, accession number: RDDB2022136179).
Declaration of interests
No conflicts of interest exist for any of the authors.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2021YFC2500400); the National Key Research and Development Program of China (2020YFC1316902); the Basic and Applied Basic Research Foundation of Guangdong Province, China (2021B1515420007); the National Natural Science Foundation of China (81973131); the National Natural Science Foundation of China (82003520); the National Natural Science Foundation of China (81903395). We acknowledge the support of cancer prevention center. We appreciate the help of endoscopist and nurses. We thank all participants in this study.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104267.
Contributor Information
Lin Feng, Email: fengl@sysucc.org.cn.
Wei-Hua Jia, Email: jiawh@sysucc.org.cn.
Appendix. Supplementary materials
References
- 1.Chen YP, Chan ATC, Le QT, Blanchard P, Sun Y, Ma J. Nasopharyngeal carcinoma. Lancet. 2019;394(10192):64–80. doi: 10.1016/S0140-6736(19)30956-0. [DOI] [PubMed] [Google Scholar]
- 2.Tang LL, Chen WQ, Xue WQ, et al. Global trends in incidence and mortality of nasopharyngeal carcinoma. Cancer Lett. 2016;374(1):22–30. doi: 10.1016/j.canlet.2016.01.040. [DOI] [PubMed] [Google Scholar]
- 3.Chang ET, Adami HO. The enigmatic epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers Prev. 2006;15(10):1765–1777. doi: 10.1158/1055-9965.EPI-06-0353. [DOI] [PubMed] [Google Scholar]
- 4.Cao SM, Chen SH, Qian CN, Liu Q, Xia YF. Familial nasopharyngeal carcinomas possess distinguished clinical characteristics in southern China. Chin J Cancer Res. 2014;26(5):543–549. doi: 10.3978/j.issn.1000-9604.2014.10.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loh KS, Goh BC, Lu J, Hsieh WS, Tan L. Familial nasopharyngeal carcinoma in a cohort of 200 patients. Arch Otolaryngol Head Neck Surg. 2006;132(1):82–85. doi: 10.1001/archotol.132.1.82. [DOI] [PubMed] [Google Scholar]
- 6.Zeng YX, Jia WH. Familial nasopharyngeal carcinoma. Semin Cancer Biol. 2002;12(6):443–450. doi: 10.1016/s1044579x02000871. [DOI] [PubMed] [Google Scholar]
- 7.Lu SJ, Day NE, Degos L, et al. Linkage of a nasopharyngeal carcinoma susceptibility locus to the HLA region. Nature. 1990;346(6283):470–471. doi: 10.1038/346470a0. [DOI] [PubMed] [Google Scholar]
- 8.Feng BJ, Huang W, Shugart YY, et al. Genome-wide scan for familial nasopharyngeal carcinoma reveals evidence of linkage to chromosome 4. Nat Genet. 2002;31(4):395–399. doi: 10.1038/ng932. [DOI] [PubMed] [Google Scholar]
- 9.Xiong W, Zeng ZY, Xia JH, et al. A susceptibility locus at chromosome 3p21 linked to familial nasopharyngeal carcinoma. Cancer Res. 2004;64(6):1972–1974. doi: 10.1158/0008-5472.can-03-3253. [DOI] [PubMed] [Google Scholar]
- 10.Hu LF, Qiu QH, Fu SM, et al. A genome-wide scan suggests a susceptibility locus on 5p 13 for nasopharyngeal carcinoma. Eur J Hum Genet. 2008;16(3):343–349. doi: 10.1038/sj.ejhg.5201951. [DOI] [PubMed] [Google Scholar]
- 11.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freimer N, Sabatti C. The use of pedigree, sib-pair and association studies of common diseases for genetic mapping and epidemiology. Nat Genet. 2004;36(10):1045–1051. doi: 10.1038/ng1433. [DOI] [PubMed] [Google Scholar]
- 13.Shahi RB, De Brakeleer S, Caljon B, et al. Identification of candidate cancer predisposing variants by performing whole-exome sequencing on index patients from BRCA1 and BRCA2-negative breast cancer families. BMC Cancer. 2019;19(1):313. doi: 10.1186/s12885-019-5494-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toma C, Diaz-Gay M, Franch-Exposito S, et al. Using linkage studies combined with whole-exome sequencing to identify novel candidate genes for familial colorectal cancer. Int J Cancer. 2020;146(6):1568–1577. doi: 10.1002/ijc.32683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts NJ, Norris AL, Petersen GM, et al. Whole genome sequencing defines the genetic heterogeneity of familial pancreatic cancer. Cancer Discov. 2016;6(2):166–175. doi: 10.1158/2159-8290.CD-15-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Liyanarachchi S, Miller KE, et al. Identification of rare variants predisposing to thyroid cancer. Thyroid. 2019;29(7):946–955. doi: 10.1089/thy.2018.0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rotunno M, McMaster ML, Boland J, et al. Whole exome sequencing in families at high risk for Hodgkin lymphoma: identification of a predisposing mutation in the KDR gene. Haematologica. 2016;101(7):853–860. doi: 10.3324/haematol.2015.135475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai W, Zheng H, Cheung AK, et al. Whole-exome sequencing identifies MST1R as a genetic susceptibility gene in nasopharyngeal carcinoma. Proc Natl Acad Sci U S A. 2016;113(12):3317–3322. doi: 10.1073/pnas.1523436113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu G, Hsu WL, Coghill AE, et al. Whole-exome sequencing of nasopharyngeal carcinoma families reveals novel variants potentially involved in nasopharyngeal carcinoma. Sci Rep. 2019;9(1):9916. doi: 10.1038/s41598-019-46137-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lau R, Middeldorp J, Farrell PJ. Epstein-Barr virus gene expression in oral hairy leukoplakia. Virology. 1993;195(2):463–474. doi: 10.1006/viro.1993.1397. [DOI] [PubMed] [Google Scholar]
- 21.Frangou P, Buettner M, Niedobitek G. Epstein-Barr virus (EBV) infection in epithelial cells in vivo: rare detection of EBV replication in tongue mucosa but not in salivary glands. J Infect Dis. 2005;191(2):238–242. doi: 10.1086/426823. [DOI] [PubMed] [Google Scholar]
- 22.Laichalk LL, Thorley-Lawson DA. Terminal differentiation into plasma cells initiates the replicative cycle of Epstein-Barr virus in vivo. J Virol. 2005;79(2):1296–1307. doi: 10.1128/JVI.79.2.1296-1307.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Djavadian R, Chiu YF, Johannsen E. An Epstein-Barr virus-encoded protein complex requires an origin of lytic replication in cis to mediate late gene transcription. PLoS Pathog. 2016;12(6) doi: 10.1371/journal.ppat.1005718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosemarie Q, Sugden B. Epstein-Barr virus: how its lytic phase contributes to oncogenesis. Microorganisms. 2020;8(11) doi: 10.3390/microorganisms8111824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng MQ, Wang TM, Liao Y, et al. Nasopharyngeal Epstein-Barr virus DNA loads in high-risk nasopharyngeal carcinoma families: Familial aggregation and host heritability. J Med Virol. 2020 doi: 10.1002/jmv.26198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamanaka K, Minko IG, Takata K, et al. Novel enzymatic function of DNA polymerase nu in translesion DNA synthesis past major groove DNA-peptide and DNA-DNA cross-links. Chem Res Toxicol. 2010;23(3):689–695. doi: 10.1021/tx900449u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moldovan GL, Madhavan MV, Mirchandani KD, McCaffrey RM, Vinciguerra P, D'Andrea AD. DNA polymerase POLN participates in cross-link repair and homologous recombination. Mol Cell Biol. 2010;30(4):1088–1096. doi: 10.1128/MCB.01124-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H, Li Y, Wang HB, et al. Ephrin receptor A2 is an epithelial cell receptor for Epstein-Barr virus entry. Nat Microbiol. 2018;3(2):1–8. doi: 10.1038/s41564-017-0080-8. [DOI] [PubMed] [Google Scholar]
- 32.Xia TL, Li X, Wang X, et al. N(6)-methyladenosine-binding protein YTHDF1 suppresses EBV replication and promotes EBV RNA decay. EMBO Rep. 2021;22(4):e50128. doi: 10.15252/embr.202050128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang XS, Pfeiffer RM, Wheeler W, et al. Genetic variants in DNA repair genes and the risk of cutaneous malignant melanoma in melanoma-prone families with/without CDKN2A mutations. Int J Cancer. 2012;130(9):2062–2066. doi: 10.1002/ijc.26231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kazma R, Babron MC, Gaborieau V, et al. Lung cancer and DNA repair genes: multilevel association analysis from the International Lung Cancer Consortium. Carcinogenesis. 2012;33(5):1059–1064. doi: 10.1093/carcin/bgs116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silvestrov P, Maier SJ, Fang M, Cisneros GA. DNArCdb: A database of cancer biomarkers in DNA repair genes that includes variants related to multiple cancer phenotypes. DNA Repair. 2018;70:10–17. doi: 10.1016/j.dnarep.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu J, Wei Y, Pan J, et al. Prevalence of comprehensive DNA damage repair gene germline mutations in Chinese prostate cancer patients. Int J Cancer. 2021;148(3):673–681. doi: 10.1002/ijc.33324. [DOI] [PubMed] [Google Scholar]
- 37.Lin DC, Meng X, Hazawa M, et al. The genomic landscape of nasopharyngeal carcinoma. Nat Genet. 2014;46(8):866–871. doi: 10.1038/ng.3006. [DOI] [PubMed] [Google Scholar]
- 38.Zheng H, Dai W, Cheung AK, et al. Whole-exome sequencing identifies multiple loss-of-function mutations of NF-kappaB pathway regulators in nasopharyngeal carcinoma. Proc Natl Acad Sci U S A. 2016;113(40):11283–11288. doi: 10.1073/pnas.1607606113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li YY, Chung GT, Lui VW, et al. Exome and genome sequencing of nasopharynx cancer identifies NF-kappaB pathway activating mutations. Nat Commun. 2017;8:14121. doi: 10.1038/ncomms14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.You R, Liu YP, Lin DC, et al. Clonal mutations activate the NF-kappaB pathway to promote recurrence of nasopharyngeal carcinoma. Cancer Res. 2019;79(23):5930–5943. doi: 10.1158/0008-5472.CAN-18-3845. [DOI] [PubMed] [Google Scholar]
- 41.Kudoh A, Iwahori S, Sato Y, et al. Homologous recombinational repair factors are recruited and loaded onto the viral DNA genome in Epstein-Barr virus replication compartments. J Virol. 2009;83(13):6641–6651. doi: 10.1128/JVI.00049-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daikoku T, Kudoh A, Sugaya Y, et al. Postreplicative mismatch repair factors are recruited to Epstein-Barr virus replication compartments. J Biol Chem. 2006;281(16):11422–11430. doi: 10.1074/jbc.M510314200. [DOI] [PubMed] [Google Scholar]
- 43.Daikoku T, Kudoh A, Fujita M, et al. Architecture of replication compartments formed during Epstein-Barr virus lytic replication. J Virol. 2005;79(6):3409–3418. doi: 10.1128/JVI.79.6.3409-3418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Countryman J, Miller G. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc Natl Acad Sci U S A. 1985;82(12):4085–4089. doi: 10.1073/pnas.82.12.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Munz C. Latency and lytic replication in Epstein-Barr virus-associated oncogenesis. Nat Rev Microbiol. 2019;17(11):691–700. doi: 10.1038/s41579-019-0249-7. [DOI] [PubMed] [Google Scholar]
- 46.Sengupta S, den Boon JA, Chen IH, et al. Genome-wide expression profiling reveals EBV-associated inhibition of MHC class I expression in nasopharyngeal carcinoma. Cancer Res. 2006;66(16):7999–8006. doi: 10.1158/0008-5472.CAN-05-4399. [DOI] [PubMed] [Google Scholar]
- 47.Laantri N, Jalbout M, Khyatti M, et al. XRCC1 and hOGG1 genes and risk of nasopharyngeal carcinoma in North African countries. Mol Carcinog. 2011;50(9):732–737. doi: 10.1002/mc.20754. [DOI] [PubMed] [Google Scholar]
- 48.Lin J, Ye Q, Wang Y, Wang Y, Zeng Y. Association between XRCC1 single-nucleotide polymorphisms and susceptibility to nasopharyngeal carcinoma: An update meta-analysis. Medicine (Baltimore. 2018;97(32):e11852. doi: 10.1097/MD.0000000000011852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho EY, Hildesheim A, Chen CJ, et al. Nasopharyngeal carcinoma and genetic polymorphisms of DNA repair enzymes XRCC1 and hOGG1. Cancer Epidemiol Biomarkers Prev. 2003;12(10):1100–1104. [PubMed] [Google Scholar]
- 50.Ban EZ, Lye MS, Chong PP, Yap YY, Lim SYC, Abdul Rahman H. Haplotype CGC from XPD, hOGG1 and ITGA2 polymorphisms increases the risk of nasopharyngeal carcinoma in Malaysia. PLoS One. 2017;12(11) doi: 10.1371/journal.pone.0187200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zou H, Li Q, Xia W, Liu Y, Wei X, Wang D. Association between the OGG1 Ser326Cys polymorphism and cancer risk: evidence from 152 case-control studies. J Cancer. 2016;7(10):1273–1280. doi: 10.7150/jca.15035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu Z, Li S, Ning S, et al. Association of the rs1760944 polymorphism in the APEX1 base excision repair gene with risk of nasopharyngeal carcinoma in a population from an endemic area in South China. J Clin Lab Anal. 2018;32(2):e22238. doi: 10.1002/jcla.22238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu JC, Tsai CW, Hsu CM, et al. Contribution of double strand break repair gene XRCC3 genotypes to nasopharyngeal carcinoma risk in Taiwan. Chin J Physiol. 2015;58(1):64–71. doi: 10.4077/CJP.2015.BAD279. [DOI] [PubMed] [Google Scholar]
- 54.Qin HD, Shugart YY, Bei JX, et al. Comprehensive pathway-based association study of DNA repair gene variants and the risk of nasopharyngeal carcinoma. Cancer Res. 2011;71(8):3000–3008. doi: 10.1158/0008-5472.CAN-10-0469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh SA, Ghosh SK. Polymorphisms of XRCC1 and XRCC2 DNA repair genes and interaction with environmental factors influence the risk of nasopharyngeal carcinoma in northeast India. Asian Pac J Cancer Prev. 2016;17(6):2811–2819. [PubMed] [Google Scholar]
- 56.Yang ZH, Dai Q, Kong XL, Yang WL, Zhang L. Association of ERCC1 polymorphisms and susceptibility to nasopharyngeal carcinoma. Mol Carcinog. 2009;48(3):196–201. doi: 10.1002/mc.20468. [DOI] [PubMed] [Google Scholar]
- 57.Keppen C, Barooah P, Borthakur P, et al. Genetic polymorphisms along with dietary and environmental factors enhance the susceptibility to nasopharyngeal carcinoma in Nagaland of northeast India. Biochem Genet. 2020;58(4):533–550. doi: 10.1007/s10528-020-09954-1. [DOI] [PubMed] [Google Scholar]
- 58.Yang ZH, Liang WB, Jia J, Wei YS, Zhou B, Zhang L. The xeroderma pigmentosum group C gene polymorphisms and genetic susceptibility of nasopharyngeal carcinoma. Acta Oncol. 2008;47(3):379–384. doi: 10.1080/02841860701558815. [DOI] [PubMed] [Google Scholar]
- 59.Yang ZH, Du B, Wei YS, et al. Genetic polymorphisms of the DNA repair gene and risk of nasopharyngeal carcinoma. DNA Cell Biol. 2007;26(7):491–496. doi: 10.1089/dna.2006.0537. [DOI] [PubMed] [Google Scholar]
- 60.Ban EZ, Lye MS, Chong PP, Yap YY, Lim SYC, Abdul Rahman H. Association of hOGG1 Ser326Cys, ITGA2 C807T, TNF-A -308G>A and XPD Lys751Gln polymorphisms with the survival of Malaysian NPC patients. PLoS One. 2018;13(6) doi: 10.1371/journal.pone.0198332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whitehurst CB, Vaziri C, Shackelford J, Pagano JS. Epstein-Barr virus BPLF1 deubiquitinates PCNA and attenuates polymerase eta recruitment to DNA damage sites. J Virol. 2012;86(15):8097–8106. doi: 10.1128/JVI.00588-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kumar R, Whitehurst CB, Pagano JS. The Rad6/18 ubiquitin complex interacts with the Epstein-Barr virus deubiquitinating enzyme, BPLF1, and contributes to virus infectivity. J Virol. 2014;88(11):6411–6422. doi: 10.1128/JVI.00536-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dyson OF, Pagano JS, Whitehurst CB. The translesion polymerase pol eta is required for efficient Epstein-Barr virus infectivity and is regulated by the viral deubiquitinating enzyme BPLF1. J Virol. 2017;91(19):e00600-17. doi: 10.1128/JVI.00600-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hau PM, Tsao SW. Epstein-Barr virus hijacks DNA damage response transducers to orchestrate its life cycle. Viruses. 2017;9(11):341. doi: 10.3390/v9110341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zietlow L, Smith LA, Bessho M, Bessho T. Evidence for the involvement of human DNA polymerase N in the repair of DNA interstrand cross-links. Biochemistry. 2009;48(49):11817–11824. doi: 10.1021/bi9015346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou J, Deng Z, Norseen J, Lieberman PM. Regulation of Epstein-Barr virus origin of plasmid replication (OriP) by the S-phase checkpoint kinase Chk2. J Virol. 2010;84(10):4979–4987. doi: 10.1128/JVI.01300-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Okuno Y, Murata T, Sato Y, et al. Defective Epstein-Barr virus in chronic active infection and haematological malignancy. Nat Microbiol. 2019;4(3):404–413. doi: 10.1038/s41564-018-0334-0. [DOI] [PubMed] [Google Scholar]
- 68.Martel-Renoir D, Grunewald V, Touitou R, Schwaab G, Joab I. Qualitative analysis of the expression of Epstein-Barr virus lytic genes in nasopharyngeal carcinoma biopsies. J Gen Virol. 1995;76(Pt 6):1401–1408. doi: 10.1099/0022-1317-76-6-1401. [DOI] [PubMed] [Google Scholar]
- 69.Jin S, Li R, Chen MY, et al. Single-cell transcriptomic analysis defines the interplay between tumor cells, viral infection, and the microenvironment in nasopharyngeal carcinoma. Cell Res. 2020;30(11):950–965. doi: 10.1038/s41422-020-00402-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shen GP, Pan QH, Hong MH, et al. Human genetic variants of homologous recombination repair genes first found to be associated with Epstein-Barr virus antibody titers in healthy Cantonese. Int J Cancer. 2011;129(6):1459–1466. doi: 10.1002/ijc.25759. [DOI] [PubMed] [Google Scholar]
- 71.Zheng J, Liu B, Zhang L, et al. The protective role of polymorphism MKK4-1304 T>G in nasopharyngeal carcinoma is modulated by Epstein-Barr virus' infection status. Int J Cancer. 2012;130(9):1981–1990. doi: 10.1002/ijc.26253. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.