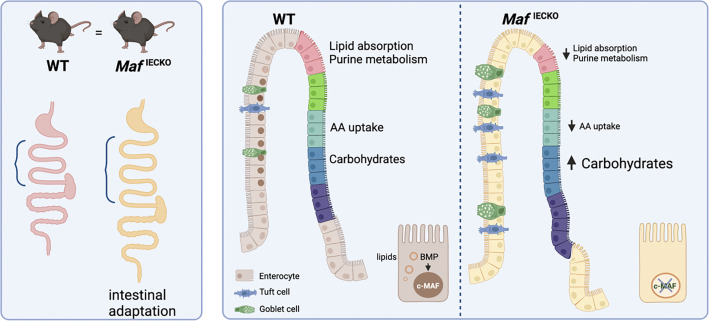
c-MAF, highly expressed in villus small intestinal enterocytes, coordinates transcriptional programs for nutrient absorption. Enterocyte-specific c-MAF inactivation impairs dietary lipid handling but also leads to tuft cell expansion and adaptive gut lengthening, revealing crosstalk between enterocytes and tuft cells during intestinal adaptation.
Abstract
Small intestinal villi are structural and functional units present in higher vertebrates and uniquely adapted to nutrient absorption. Villus enterocytes are organized in transcriptional “zones” dedicated to specialized tasks such as absorption of specific nutrients. We report that the transcription factor c-MAF is expressed in differentiated lower and mid-villus enterocytes and is a target of BMP signaling. Maf inactivation perturbed the villus zonation program by increasing carbohydrate-related transcripts while suppressing transcripts linked to amino-acid and lipid absorption. The formation of cytoplasmic lipid droplets, shuttling dietary fat to chylomicrons, was impaired upon Maf loss indicating its role in dietary lipid handling. Maf inactivation under homeostatic conditions expanded tuft cells and led to compensatory gut lengthening, preventing weight loss. However, delayed Maf−/− enterocyte maturation impaired weight recovery after acute intestinal injury, resulting in reduced survival. Our results identify c-MAF as a regulator of the intestinal villus zonation program, while highlighting the importance of coordination between stem/progenitor and differentiation programs for intestinal regeneration.
Graphical Abstract
Introduction
The intestinal epithelium continuously regenerates to replace damaged cells and ensure absorption of nutrients and water. Crypt-housed intestinal stem cells give rise to enterocytes, responsible for nutrient absorption; Paneth cells, producing antimicrobial peptides and proteins; mucus-producing goblet cells, tuft cells, and enteroendocrine cells, which secrete hormones (Barker, 2014; Sato et al., 2009). Disrupted intestinal epithelial regeneration is a frequent and severe side effect of anti-cancer therapies (Yu, 2013).
Much attention has been paid to the characterization of the intestinal crypt compartment (Barker et al., 2007; Clevers, 2013; Guiu et al., 2019). However, the discovery of functional compartmentalization of enterocytes along the intestinal villus axis has highlighted the heterogeneity and complexity of differentiated epithelial cells (Moor et al., 2018). This zonation extends to nutrient absorption pathways as mid-villus enterocytes absorb amino acids and carbohydrates, while villus tip enterocytes take up lipids and produce chylomicrons (Moor et al., 2018). The villus tip enterocytes are also enriched in transcripts encoding anti-inflammatory regulators of purine metabolism, such as 5′ nucleotidase (NT5E) and adenosine deaminase (ADA), which may be important for decreasing leukocyte infiltration and inflammatory cytokine release (Blume et al., 2012; Bynoe et al., 2012; Mabley et al., 2003). Bmpr1a deletion in enterocytes impairs their zonation program, indicating the key role of bone morphogenetic protein (BMP) pathway in villus enterocyte specialization (Beumer et al., 2022).
The MAF family of transcription factors harbors a conserved basic region-leucine zipper domain recognizing unusually long DNA stretches called Maf recognition elements (Kerppola and Curran, 1994). c-MAF regulates transcriptional programs for lens fiber, tubular renal cell, and chondrocyte differentiation (Hong et al., 2011; Imaki et al., 2004; Kawauchi et al., 1999). Since its identification as a regulator of Th2 lymphocytes (Ho et al., 1996), the role of c-MAF in immunity has been under intense investigation. c-MAF is expressed in innate immune cells (Parker et al., 2020; Pokrovskii et al., 2019), including type-2 macrophages (Daassi et al., 2016), B lymphocytes, and multiple T cell subsets (Andris et al., 2017; Giordano et al., 2015; Hiramatsu et al., 2010; Pot et al., 2009). In gut T cells, c-MAF regulates the balance between T helper type 17 cell and regulatory T cell numbers to maintain intestinal homeostasis (Imbratta et al., 2019). Here, we report a novel role of c-MAF in differentiated enterocytes of the small intestine.
Results and discussion
c-MAF is a BMP target expressed in small intestinal enterocytes
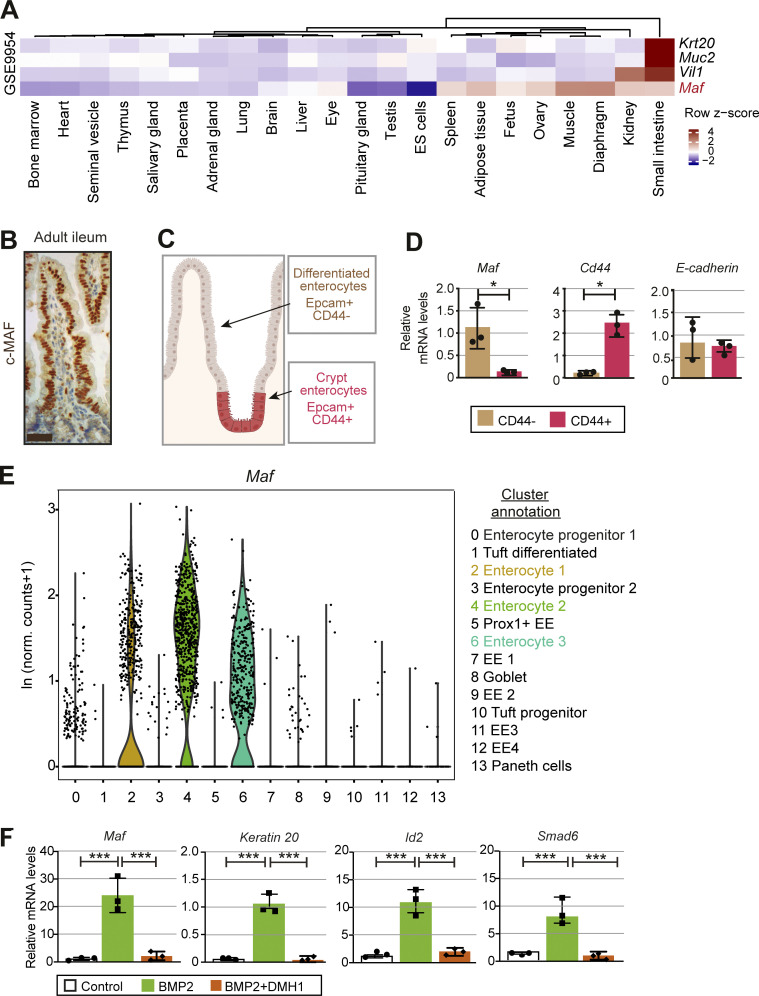
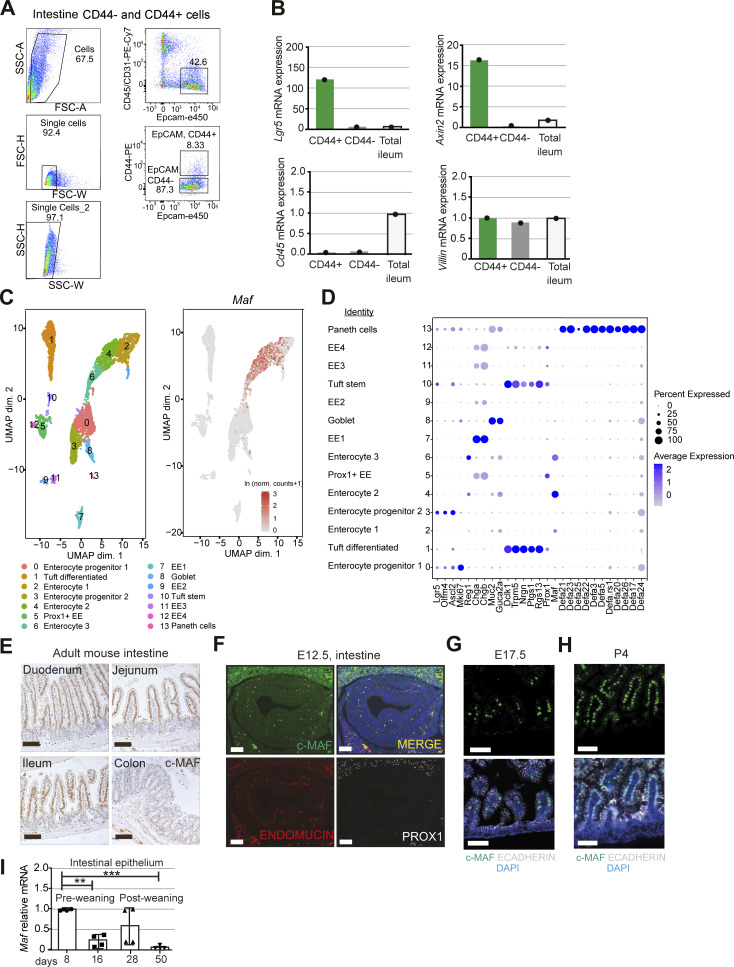
Maf is highly expressed in murine eye, spleen, fat, ovary, muscle, diaphragm, kidney, and small intestine (Fig. 1 A). To identify Maf expressing cells in the gut, we stained intestinal sections for c-MAF protein and analyzed Maf levels in sorted EPCAM+/CD44+ progenitors and mature EPCAM+/CD44− enterocytes (Snippert et al., 2009; Zeilstra et al., 2008). We observed high levels of Maf mRNA and protein in mature villus CD44− enterocytes but not in CD44+ progenitors or secretory cells (Fig. 1, B–E; and Fig. S1, A and B). Maf expression in differentiated enterocytes was confirmed in a single-cell RNA-sequencing (scRNAseq) dataset of mouse intestinal epithelium (Fig. 1 E and Fig. S1, C and D; Yan et al., 2017). Along the gut, c-MAF was highly expressed in small intestinal enterocytes, while c-MAF levels were low in the colonic epithelium (Fig. S1 E).
Figure 1.
c-MAF is expressed in small intestinal enterocytes and induced upon differentiation. (A) Expression profile of Maf and intestinal epithelial genes Krt20, Muc2, and Vil1 in mouse tissues (GSE9954). A high z-score (dark red) indicates higher expression of the genes in specific tissues. (B) Staining for c-MAF (brown) in adult mouse ileum. Scale bar, 25 μm. (C) Schematic representation of the location of Epcam+CD44+ and Epcam+CD44− cells along the villus and sorted from WT small intestine, created with Biorender.com. (D) Maf is expressed in Epcam+CD44− cells. RT-qPCR analysis of the indicated genes. Data shown as mean ± SD. Two-tailed unpaired Student’s t test; *, P < 0.05; n = 3 WT. (E) Maf is expressed in mature enterocytes. Expression level per cell (ln[normalized counts + 1]) of Maf overlaid on scRNAseq dataset of Yan et al. (2017). (F) BMP2 induces Maf. WT small intestinal organoids were treated for 24 h with vehicle, 100 ng/ml recombinant BMP2 or BMP2+ 3 µm DMH1. Relative expression levels of Maf, Krt20, Id2, Smad6 are shown as mean ± SD. Two-tailed unpaired Student’s t test; ***, P < 0.001; n = 3 different organoid WT samples used.
Figure S1.
c-MAF is expressed in adult mouse enterocytes. (A) Sorting strategy for isolation of differentiated Epcam+ CD45−CD31− CD44− and Epcam+ CD45− CD31− CD44+ enterocytes from 6-wk-old mice. (B) Sorted CD44+ and CD44− intestinal epithelial cell purity analysis. mRNA levels of Lgr5, Axin2, Cd45, and Villin normalized to 18S. (C) UMAP plot of Yan et al. (2017) dataset. 13 major clusters of gut epithelial cells were identified. Maf expression per cell (ln[normalized counts + 1]) overlaid on UMAP plot. (D) Maf is expressed in enterocyte clusters. Dot plot of markers in all clusters from Yan et al. (2017) dataset. The color code indicates scaled average expression level in each cluster, and the dot size denotes the percent of cells in each cluster expressing the given gene. Annotation of all clusters based on specific cell type markers (Yan et al., 2017). (E) Chromogenic staining for c-MAF along murine small intestine and colon. Scale bar, 50 μm. (F) c-MAF is absent in the murine gut epithelium at E12.5. Staining for c-MAF (green), endomucin (red), PROX1 (white), and DAPI (blue). Scale bar, 200 μm. (G) c-MAF is expressed in the murine gut epithelium at E17.5. Staining for c-MAF (green), E-cadherin (white), and DAPI (blue). Scale bar, 50 μm. (H) c-MAF is expressed in the murine gut at postnatal stage P4. Staining for c-MAF (green), E-cadherin (white), and DAPI (blue). Scale bar, 50 μm. (I) Maf mRNA relative levels in murine gut epithelium from postnatal day 8 to 50. One-way ANOVA with Tukey post hoc test. Day 8 vs. day 16 **, P < 0.01; day 8 vs. day 50 ***, P < 0.001; N = 4 WT mice. Data shown as mean ± SD.
c-MAF was absent in embryonic day 12.5 (E12.5) gut, and sporadic c-MAF–producing gut epithelial cells were observed at E17.5 (Fig. S1, F and G). All enterocytes expressed c-MAF at postnatal day 4 (P4) and thereafter (Fig. 1 B and Fig. S1, E, H, and I). Treatment with broad-spectrum antibiotics did not change epithelial c-MAF levels (data not shown), indicating that its expression is driven by an intrinsic epithelial maturation program.
BMP signaling is important for intestinal epithelial differentiation (Clevers, 2006). Given high expression of c-MAF in differentiated enterocytes in vivo, we treated cultured small intestinal organoids with BMP2. BMP2 induced expression of target genes Id2 and Smad6 (Hollnagel et al., 1999; Takase et al., 1998) and Krt20, a marker of differentiated intestinal epithelial cells (Moll et al., 1990; Fig. 1 F). BMP2 also strongly promoted the expression of Maf, and this induction was abolished upon treatment with the selective BMP inhibitor DMH-1 (Fig. 1 F). Collectively, our data establish that c-MAF is expressed in mature small intestinal enterocytes in vivo and that its expression is induced upon BMP-driven differentiation in vitro.
Maf loss perturbs the enterocyte zonation transcriptional program
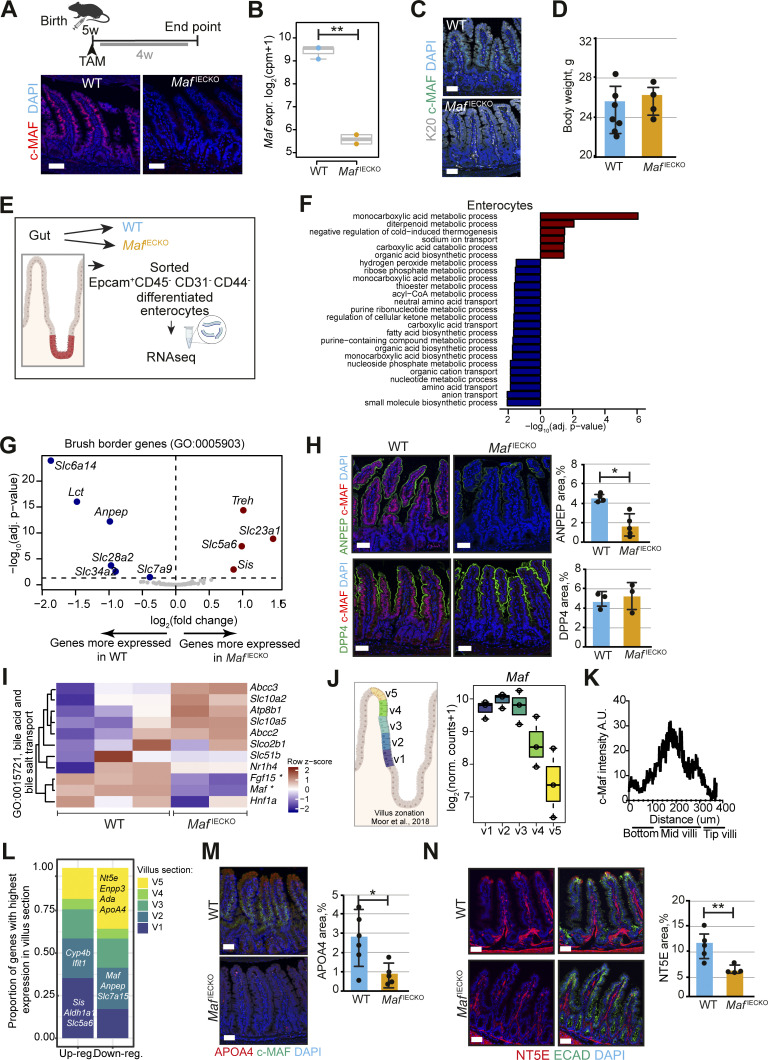
To study the role of intestinal epithelial c-MAF, we generated Maf flox/flox; Villin-CreERT2 mice (MafIECKO), in which tamoxifen administration leads to Maf inactivation in the entire intestinal epithelium (el Marjou et al., 2004). Despite the loss of c-MAF protein and mRNA, and no compensatory increase in other MAF family members (Fig. 2, A and B; and Fig. S2 A; Kataoka, 2007), neither enterocyte differentiation nor stem-cell proliferation were affected, as determined by the analysis of KRT20 staining and crypt EdU+ epithelial cells, respectively (Fig. 2 C and Fig. S2 B; Chan et al., 2009). The body weights of MafIECKO mice and WT littermates were comparable (Fig. 2 D), indicating globally unaltered nutrient absorption capacity.
Figure 2.
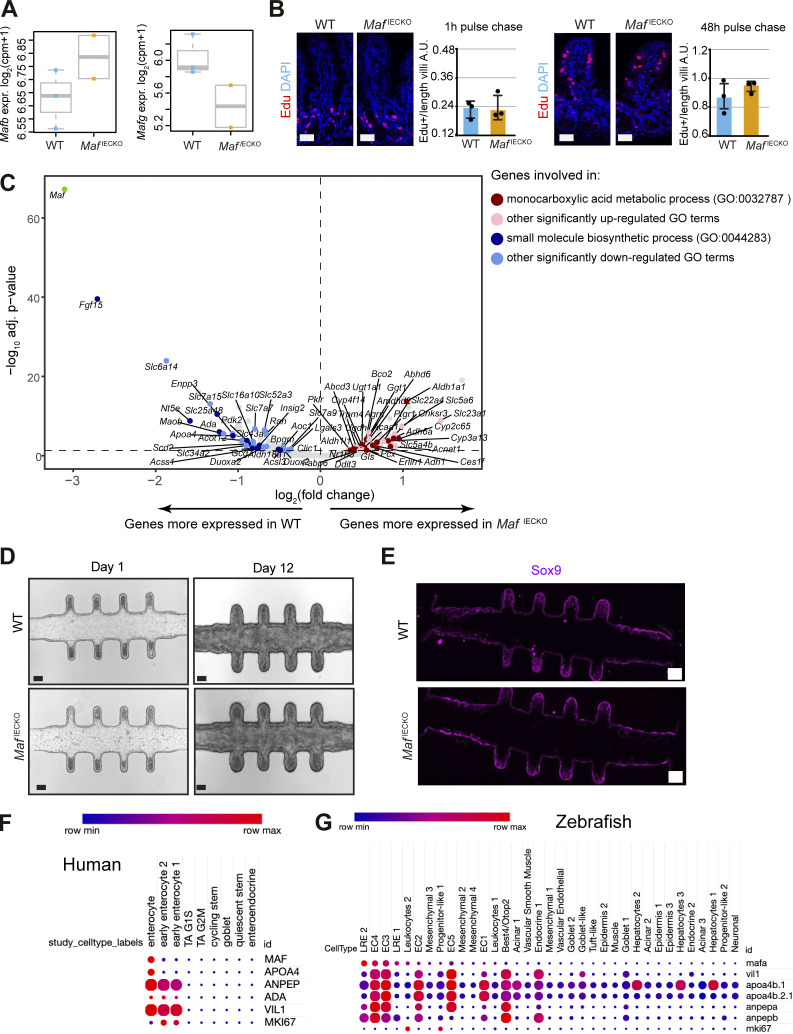
Loss of Maf in adult enterocytes alters their transcriptional zonation program. (A) Maf depletion in MafIECKO mice. Staining for c-MAF (red), and DAPI (blue). WT and MafIECKO mice 1 mo after the initiation of depletion. n = 10 WT, n = 11 MafIECKO analyzed from three different cohorts. Scale bar, 50 μm. (B) Maf levels are significantly reduced in MafIECKO mice. n = 3 WT; n = 2 MafIECKO. Wald test in DESeq2; **, P < 0.01. (C) Maf depletion does not change overall enterocyte differentiation. Staining for Krt20 (K20, white), c-MAF (green) and DAPI (blue). Data analyzed from two different cohorts. Scale bar, 50 μm. (D) Maf depletion does not affect mouse weight. Two-tailed unpaired Student’s t test; n = 7 WT; n = 4 MafIECKO. n.s., not significant. Data analyzed from two different cohorts and shown as mean ± SD. (E) Sorting strategy for isolation of differentiated enterocytes from WT and MafIECKO mice for RNAseq, created with Biorender.com. (F) Signaling pathways enriched in MafIECKO vs. WT mouse enterocytes. Bar plots of GO gene sets significantly (adjusted P value <0.05) over-represented (red bars) or under-represented (blue bars) in MafIECKO vs. WT mice after 1 wk of Maf depletion. n = 3 WT, n = 2 MafIECKO. One-sided Fisher’s exact test followed by Benjamini–Hochberg P value adjustment. (G) Volcano plot of differentially expressed brush border genes (GO: 0005903) in MafIECKO mice (Wald test in DESeq2). (H) ANPEP but not DPP4 is reduced upon Maf depletion. Staining for ANPEP or DPP4 (green), c-MAF (red), and DAPI (blue). Scale bar, 50 μm. Percentage of area per tissue in WT and MafIECKO mice. Data shown as mean ± SD. Two-tailed unpaired Student’s t test; *, P < 0.05. ANPEP n = 4 WT; n = 5 MafIECKO; DPP4 n = 3 WT; n = 3 MafIECKO. (I) Heatmap of bile acid regulators, Maf and Fgf15 were significantly different in MafIECKO versus WT mice (Wald test in DESeq2). (J) Maf expression is zonated along small intestinal villus, as observed along villus zones described in Moor et al. (2018), created with Biorender.com. (K) c-MAF protein intensity along villi; n = 4 WT mice. (L) Proportion of genes differentially expressed upon Maf loss with highest expression in the five different villus zones described in Moor et al. (2018). Most genes induced in the absence of Maf have their highest expression level in proximal zone v1, while most decreased genes in the absence of Maf are highly expressed in villus tip zone v5. Examples of transcripts characteristic of each zone are indicated. (M) Marker of v4 and v5 zones APOA4 is reduced in MafIECKO mice. Staining for c-MAF (green), APOA4 (red), and DAPI (blue). Scale bar, 50 μm. Percentage of area in WT and MafIECKO mice. Two-tailed unpaired Student’s t test; *, P < 0.05. n = 6 WT; n = 5 MafIECKO. Data analyzed from two different cohorts and shown as mean ± SD. (N) NT5E is downregulated after Maf depletion. Staining for NT5E (red), E-cadherin (green), and DAPI (blue). The percentage of NT5E in the epithelial area is represented as mean ± SD. Two-tailed unpaired Student’s t test; **, P < 0.01. N = 5 WT, n = 4 MafIECKO. Scale bar, 50 µm. Data analyzed from two different cohorts and shown as mean ± SD.
Figure S2.
Characterization of the role of Maf in adult intestine. (A) No compensation by other large or small Maf family members upon Maf depletion. Mafb and Mafg expression levels in Epcam+ CD44− RNA samples from WT and MafIECKO mice. Wald test in DESeq2. N = 3 WT; n = 2 MafIECKO mice. (B) Epithelial cell proliferation and migration are unchanged in MafIECKO mice. Staining for EdU (red) and DNA (blue). Scale bar, 50 μm. Quantification of EdU + epithelial cells per length of villi. Two-tailed unpaired Student’s t test. n = 3 WT; n = 3 MafIECKO 1 and 48 h after EdU administration. Data shown as mean ± SD. (C) Volcano plot of differentially expressed transcripts in MafIECKO versus WT enterocytes. Color scale indicates genes included in different GO pathways; Wald test in DESeq2. (D) Maf loss does not impact growth characteristics or morphology of mini-guts in vitro. Appearance of mini-guts derived from WT and MafIECKO mice organoids at day 1 and 12 after seeding in the microfluidic chip device (Nikolaev et al., 2020). n = 7 WT or MafIECKO mini-intestines analyzed. Scale bar, 70 µm. (E) Mini-intestines stem cell compartment is not affected in the absence of c-MAF. Sox9 stained mini-intestines derived from WT and MafIECKO mice organoids. n = 7 WT or MafIECKO mini-intestines analyzed. Scale bar, 100 µm. (F) MAF is highly expressed in differentiated human ileal enterocytes but not in progenitor/stem cells or secretory lineages. Dot plot of the indicated transcripts from human ileum dataset (https://singlecell.broadinstitute.org/single_cell/study/SCP817/comparison-of-ace2-and-tmprss2-expression-in-human-duodenal-and-ileal-tissue-and-organoid-derived-epithelial-cells#study-summary). The color code indicates the expression level in each cluster, and the dot size denotes the percent of cells in each cluster expressing the given gene. (G) Zebrafish lysosome-rich enterocytes (LRE2) involved in protein catabolism and intestinal epithelial cells involved in nutrient absorption express high levels of mafa (Park et al., 2019). Dot plot of indicated transcripts from GSE161855 (Willms et al., 2022). The color code indicates the expression level in each cluster, and the dot size denotes the percent of cells in each cluster expressing the given gene. EC, intestinal epithelial cell.
We next compared the transcriptomes of differentiated CD44− small intestinal enterocytes sorted from WT or MafIECKO mice. Loss of Maf resulted in 106 differentially upregulated genes and 77 downregulated genes (adjusted P value <0.05, mean expression level >500 normalized counts; Table S1). Analysis of signaling pathways using Gene Ontology terms (Ashburner et al., 2000; The Gene Ontology Consortium, 2019) indicated defective pathways for amino and fatty acid metabolism, while carbohydrate metabolism pathways were increased (Fig. 2, E and F and Table S1).
The brush border is a hallmark of enterocyte specialization essential for digestion and absorption of nutrients (Carboni et al., 1987). In the absence of Maf, production of brush-border-specific carbohydrate metabolism transcripts such as Treh or Sis (Hauri et al., 1979) was enhanced, whereas the amino acid transporters Slc7a9 and Slc6a14 (Karunakaran et al., 2011), the dipeptide degrading aminopeptidase Anpep (Luan and Xu, 2007), and the purine transporter Slc28a2 (Young et al., 2013) were diminished (Fig. 2 G). Other enterocyte differentiation transcripts such as Krt20, Fabp1, or Dpp4 were not affected (Table S1; Bass, 1988; Darmoul et al., 1992). Alanyl aminopeptidase (ANPEP) but not dipeptidyl peptidase 4 (DPP4) protein was reduced in MafIECKO enterocytes confirming RNAseq results (Fig. 2 H). In addition to changes in the nutrient transporters, the atypical fibroblast growth factor 15 (Fgf15), which represses bile acid synthesis (Inagaki et al., 2005), was reduced in Maf-depleted enterocytes (Fig. S2 C). Consistently, the expression of the main bile acid transporter Slc10a2 was upregulated (Fig. 2 I). Thus, loss of Maf does not impact overall enterocyte differentiation but fine tunes the transcriptional programs for nutrient and bile acid absorption.
Mature enterocytes are organized in transcriptionally specialized zones along the villus axis (Moor et al., 2018). We analyzed Maf expression along the villus length based on the intestinal zonation pattern described by Moor et al. (2018). As observed by immunohistochemistry (Fig. 2 A), the lower and mid-villus areas v1, v2, and v3 expressed the highest levels of Maf, while Maf expression decreased in zones v4 and v5 at the villus tip (Fig. 2, J and K). We deconvoluted the zonation pattern of genes differentially expressed in MafIECKO vs. WT enterocytes based on their preferential expression in the lower, mid, or tip of villi in the Moor et al. dataset. We observed significant transcriptional changes in the Maf-expressing v2 zone, which also expressed the main transporters including amino acid transporters such as Slc16a10, Slc43a2, and Slc7a7 (Bröer, 2008) and peptidases such as Anpep (Table S1).
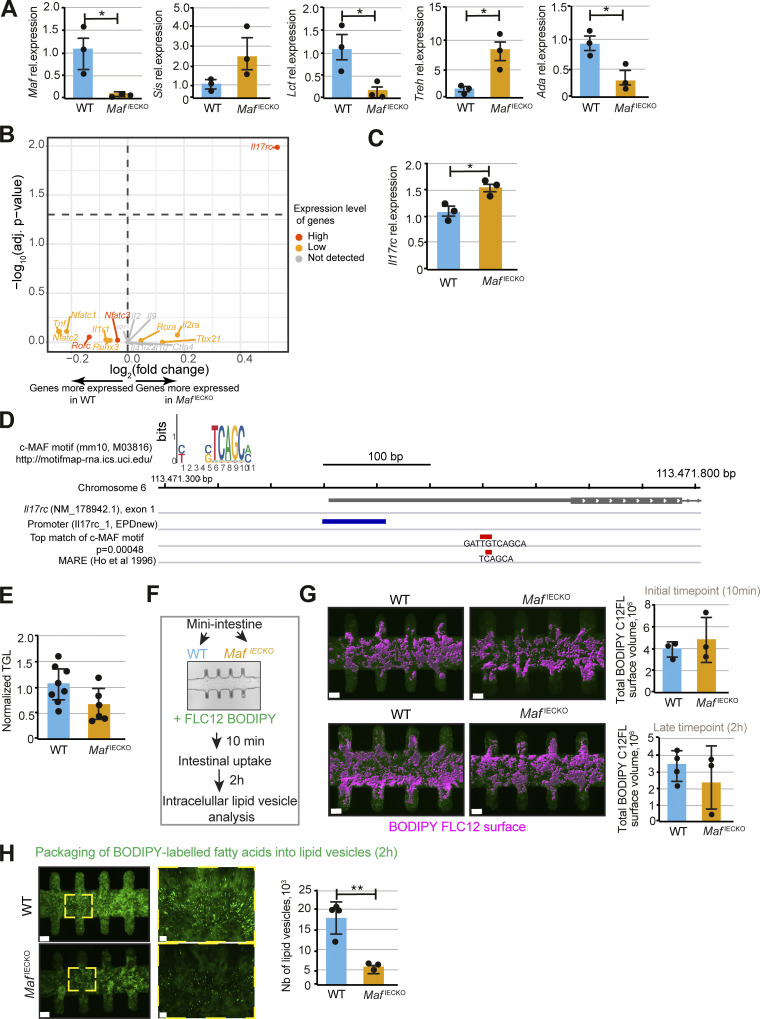
Surprisingly, loss of Maf decreased many transcripts located in zone v5 at the tip of the villus, where Maf levels are low. We observed downregulation of Apoa4, needed for assembly of chylomicrons (Kohan et al., 2015) and purine catabolism genes Nt5e, Ada, Npp3, and Slc28a2 (Huber-Ruano et al., 2010; Moor et al., 2018; Robson et al., 2006; Fig. 2 L and Table S1). In line with mRNA analysis, APOA4 and NT5E proteins were significantly lower at the tip of the villi of MafiECKO mice (Fig. 2, M and N). We also confirmed changes in levels of Sis, Lct, Treh, and Ada in MafiECKO enterocytes by quantitative PCR (qPCR; Fig. 3 A). These data indicate that Maf−/− enterocytes migrating toward the villus tip are unable to undergo further specialization.
Figure 3.
Altered dietary lipid handling upon loss of Maf in adult intestinal epithelium. (A) Relative expression of Maf, Sis, Lct, Treh, Ada, in Epcam+CD44− cells. RT-qPCR analysis of the indicated genes. Data shown as mean ± SD. Two-tailed unpaired Student’s t test; *, P < 0.05; Sis P = 0.16; n = 3 WT, n = 3 MafIECKO mice. (B) Volcano plot showing expression of genes regulated by c-MAF in immune cells (Gabryšová et al., 2018) in WT vs. MafIECKO enterocytes. Genes labeled in gray were not detected in the RNAseq data. Genes labeled in orange had average normalized counts below 500, and genes labeled in red had average normalized counts above 500. Genes located above the horizontal dashed line have adj. P value <0.05 (Wald test in DESeq2). (C) Relative expression of Il17rc in Epcam+CD44− cells. RT-qPCR analysis. Two-tailed unpaired Student’s t test; *, P < 0.05; n = 3 WT, n = 3 MafIECKO mice. Data shown as mean ± SD. (D) Genome browser view of the location of the c-MAF binding site motif (in red) in close vicinity of the IL17rc promoter (in dark blue). The binding motif is shown as the best match of the position weight matrix (using the FIMO tool, which computes a log-likelihood ratio score for each position in a sequence and converts the score to a P value using dynamic programming methods) and as the location of the c-MAF response element (MARE) described by Ho et al. (1996). The figure was made using the Integrative Genomics Viewer (v.2.8.0). (E) Normalized triglyceride (TGL) levels in systemic blood of n = 8 WT and n = 6 MafIECKO mice, two-tailed unpaired Student’s t test; P = 0.08. Data analyzed from two different cohorts and shown as mean ± SD. (F) Workflow of the analysis of lipid uptake and packaging in WT and MafIECKO mini-guts in vitro. (G) Initial BODIPY FLC12-labeled lipid uptake is not altered in the absence of c-MAF. Quantification of the total BODIPY FLC12 surface volume at initial timepoint (10 min) or latest timepoint (2 h) in WT and MafIECKO mini-guts. Pictures of BODIPY FLC12 surface volume (magenta) and its quantification. Scale bar, 70 µm. 10 min: n = 3 WT, n = 3 MafIECKO, 2 h: n = 4 WT, n = 3 MafIECKO mice organoid–derived mini-guts. Data shown as mean ± SD. (H) Reduced number of intracellular lipid droplets in MafIECKO organoid-derived mini-guts. Images and quantification of BODIPY FLC12-labeled lipid droplets 2 h after lipid addition to mini-intestines. Scale bar, 70 µm. **, P < 0.01; n = 4 WT, n = 3 MafIECKO mini-intestines. Data shown as mean ± SD.
We asked whether epithelial c-MAF controls expression of c-MAF targets identified in other cell types. Most c-MAF target genes, such as IL10, CTLA4, IL22, and IFNγ, were discovered in immune cells (Aschenbrenner et al., 2018; Cao et al., 2005; Xu et al., 2009). We analyzed the expression of canonical immune c-MAF targets from Gabryšová et al. (2018) in our RNAseq dataset. Most such transcripts were lowly expressed or absent in enterocytes (Fig. 3 B), except for Nfatc3, Rorc, and Il17rc (Fig. 3 B). Maf loss did not affect the expression of Nfatc3 or Rorc, while Il17rc was upregulated (Fig. 3, B and C). We have found a putative c-MAF–binding site in the vicinity of the Il17rc promoter, indicating that c-MAF may directly repress Il17rc transcription (Fig. 3 D). Collectively, these data indicate that, with the exception of Il17rc, enterocyte c-MAF target genes are distinct from other cell types.
Loss of Maf perturbs dietary lipid handling
RNAseq data suggested impaired nutrient processing in MafiECKO mice. Based on loss of Apoa4 expression, we studied lipid transport by analyzing circulating triglycerides levels, which were downregulated in MafiECKO mice (Fig. 3 E). To further test the role of c-MAF in lipid absorption, we took advantage of the “mini-gut” devices, which have better defined in vivo–like crypt and villus-like domains in comparison with organoids cultured in Matrigel (Nikolaev et al., 2020). Seeding efficiency, morphological appearance, and expression of the stem-cell marker Sox9 (Blache et al., 2004) were similar between Maf-proficient and -deficient epithelial cells (Fig. S2, D and E), confirming that c-MAF promotes enterocyte zonation rather than stem-cell differentiation.
Next, we analyzed the uptake of boron dipyrromethene (BODIPY)–labeled lipids in control and Maf-depleted mini-guts (Fig. 3, F and G). Despite comparable initial lipid uptake by control and Maf-depleted cells (Fig. 3 G), the formation of cytoplasmic lipid droplets, previously shown to shuttle dietary lipids to chylomicrons (Zembroski et al., 2021), was significantly reduced in the absence of Maf (Fig. 3 H). This result is consistent with our observation of APOA4 downregulation in Maf-deficient enterocytes, as APOA4 is a component of both cytoplasmic lipid droplets and chylomicrons (Wang et al., 2015; Fig. 2 M). Collectively, our data show that c-MAF is dispensable for the transition from progenitor to differentiated enterocytes. However, c-MAF maintains a transcriptional program both ensuring an enterocyte zone dedicated to purine catabolism and bile acid transport while also promoting dietary lipid handling.
c-MAF expression in enterocytes is evolutionary conserved
To study the evolutionary conservation of c-MAF function in intestinal epithelium, we analyzed human MAF and zebrafish ortholog mafa expression in publicly available scRNAseq datasets. MAF was highly expressed in differentiated human ileal enterocytes, which also expressed high levels of ANPEP and APOA4, but not in progenitor/stem cells or secretory lineages (Fig. S2 F). Among zebrafish intestinal populations, mafa was high in enterocytes 3 and 4, involved in lipid and carbohydrate absorption (Willms et al., 2022), while progenitor cells or secretory lineages were negative. Of interest, lysosome-rich enterocytes involved in protein catabolism had the highest levels of mafa (Fig. S2 G; Park et al., 2019). These results indicate conservation of c-MAF function in the regulation of nutrient absorption transcriptional programs in vertebrates.
Maf ablation expands tuft cells and increases gut length at steady state
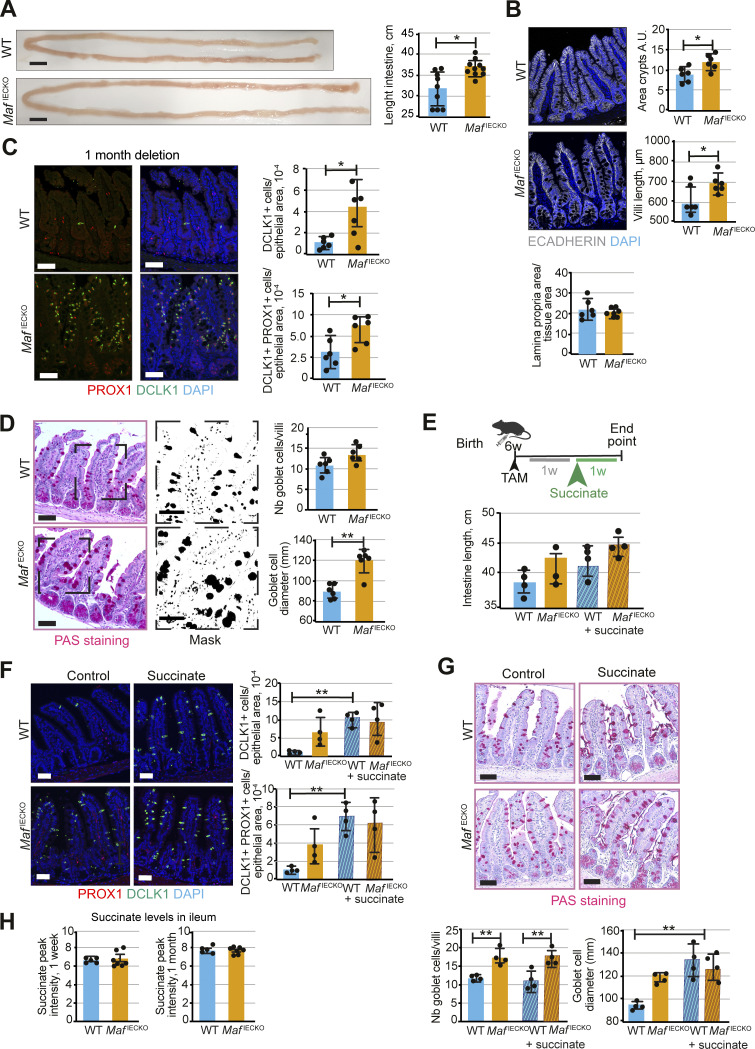
We were puzzled that despite prominent transcriptional changes in enterocytes, MafIECKO mouse weight was not affected. We, therefore, compared the gut length and sizes of crypts and villi in Maf-deficient and WT mice. The small intestine, but not colon, of MafIECKO mice was significantly longer compared to WT mice (Fig. 4 A and Fig. S3 A), indicating a compensatory enlargement of the absorptive surface. Accordingly, small intestinal crypt area and villus length were significantly greater in MafIECKO animals, while there were no changes in the underlying stroma (Fig. 4 B). Maf inactivation did not increase crypt fission (Fig. S3 B), which could alternatively increase the intestinal surface (Dehmer et al., 2011).
Figure 4.
Loss of Maf in adult intestinal epithelium leads to gut remodeling and tuft cell expansion. (A) Increased length of small intestine in MafIECKO mice. Scale bar, 1 cm. Two-tailed unpaired Student’s t test; *, P < 0.05. n = 9 WT, n = 10 MafIECKO. Data analyzed from three different cohorts and shown as mean ± SD. (B) Increased villi length and crypt area upon loss of Maf. Staining for E-cadherin (white) and DAPI (blue). Quantification of crypt area, villi length, and lamina propria area of small intestine. Two-tailed unpaired Student’s t test; *, P < 0.05. n = 6 WT, n = 6 MafIECKO. Data analyzed from two different cohorts and shown as mean ± SD. (C) Tuft cell number is increased upon Maf depletion. Staining for PROX1 (red), DLCK1 (green), and DAPI (blue). Number of PROX1+ DCLK1+ cells per epithelial area. Two-tailed unpaired Student’s t test; *, P < 0.05. n = 6 WT, n = 6 MafIECKO. Data analyzed from three different cohorts and shown as mean ± SD. Scale bar, 50 μm. (D) Goblet cell hyperplasia upon loss of Maf. PAS staining and mask. Quantification of number of goblet cells per villi and their diameter following periodic acid-Schiff staining. Scale bar, 50 μm. Two-tailed unpaired Student’s t test. Number of goblet cells/villi P = 0.06; goblet cell diameter **, P < 0.01. n = 6 WT; n = 6 MafIECKO. Data analyzed from two different cohorts and shown as mean ± SD. (E) Succinate treatment scheme and intestine length. n = 4 WT, n = 4 MafIECKO n = 4 WT + succinate, n = 4 MafIECKO + succinate. Data shown as mean ± SD. WT vs. WT succinate. One-way ANOVA with Tukey post hoc test. P = 0.06; MafIECKO vs. MafIECKO + succinate, P = 0.11. w, week. TAM, tamoxifen. (F) Tuft cell numbers upon succinate treatment. Staining for PROX1 (red), DCLK1 (green), and DAPI (blue). Scale bar, 50 μm. Number of tuft cells per epithelial area. One-way ANOVA with Tukey post hoc test. **, P < 0.01. n = 4 WT, n = 4 MafIECKO n = 4 WT + succinate, n = 4 MafIECKO + succinate. Data shown as mean ± SD. (G) Succinate treatment induces goblet cell hyperplasia. PAS staining quantification of number and size of goblet cells. Scale bar, 50 μm. One-way ANOVA with Tukey post hoc test, goblet diameter WT vs. MafIECKO P = 0.08 and WT vs. WT + succinate and MafIECKO vs. MafIECKO + succinate **, P < 0.01. n = 4 WT, n = 4 MafIECKO n = 4 WT + succinate, n = 4 MafIECKO + succinate. Data shown as mean ± SD. (H) Relative abundance of succinate in ileum tissues. Intensities of m/z 117.0192, annotated as succinate, were plotted for ileum tissues of WT and MafIECKO mice 1 wk and 1 mo after Maf depletion. Two-tailed unpaired Student’s t test. n = 5 WT, n = 7 MafIECKO. Data analyzed from two different cohorts and shown as mean ± SD.
Figure S3.
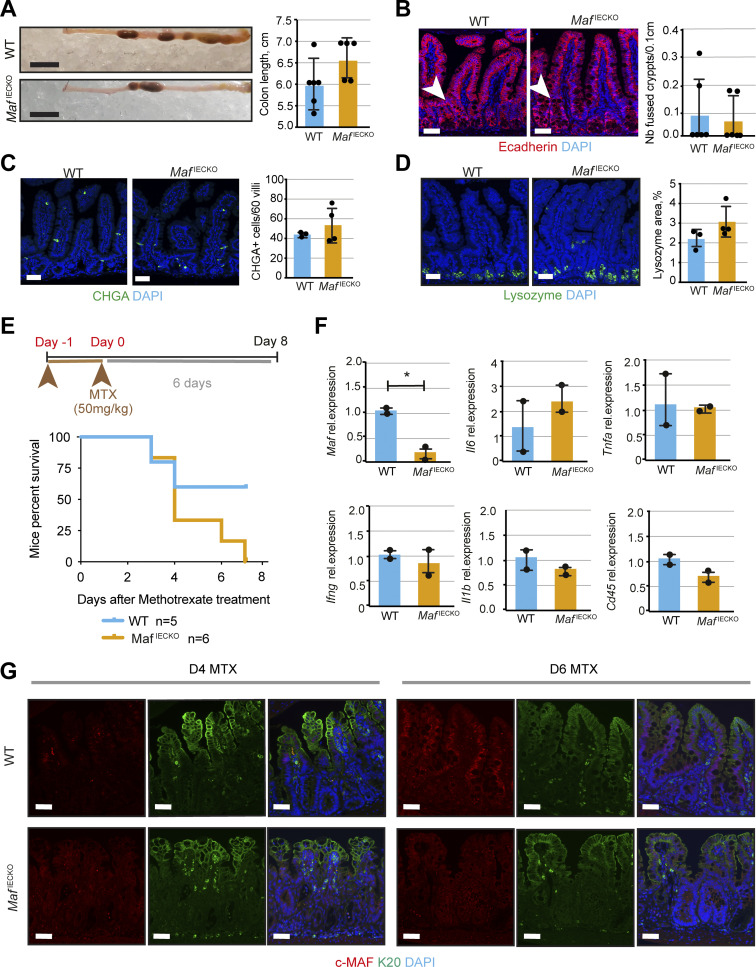
Intestinal Maf loss does not affect other secretory lineages but prevents recovery after anti-metabolite treatment. (A) Colon length in MafIECKO mice. Data shown as mean ± SD. Scale bar, 1 cm. Two-tailed unpaired Student’s t test. n = 6 WT; n = 5 MafIECKO. (B) No differences in the number of crypt fission events upon Maf loss. Staining for E-cadherin (red) and DAPI (blue). Scale bar, 50 μm. Quantification of number of fused crypts per 0.1 cm of tissue. Two-tailed unpaired Student’s t test. n = 6 WT; n = 6 MafIECKO. Data shown as mean ± SD. (C) Maf loss does not affect enteroendocrine cells. Staining for chromogranin A (CHGA, green) and DAPI (blue). Scale bar, 50 μm. Number of CHGA+ cells in 60 villi analyzed. Two-tailed unpaired Student’s t test. n = 3 WT; n = 4 MafIECKO. Data shown as mean ± SD. (D) Loss of Maf does not alter Paneth cells. Staining for lysozyme (green) and DAPI (blue). Scale bar, 50 μm. Lysozyme + area per tissue area of WT and MafIECKO mice. Two-tailed unpaired Student’s t test. n = 3 WT; n = 4 MafIECKO. Data shown as mean ± SD. (E) Loss of Maf reduces survival of mice after MTX treatment. Two consecutive doses of MTX were used and mice followed for a week. Mice percent survival; log-rank (Mantel Cox) test. P = 0.09, n = 5 WT; n = 6 MafIECKO mice. (F) Immune status is not altered upon Maf loss at day 6 after MTX. Relative mRNA levels for Maf, Il6, Tnfa, Ifng, Il1b, Cd45 normalized to 18S. Data expressed as mean ± SD. Two-tailed unpaired Student’s t test; *, P < 0.05. n = 2 WT; n = 2 MafIECKO mice. (G) Krt20 (K20) levels do not change upon Maf loss after MTX treatment. Staining for c-MAF (red), K20 (green), and DAPI (blue). D, day. Scale bar, 50 μm.
Tuft cells are chemosensory epithelial cells important for gut immune functions and intestinal regeneration (Howitt et al., 2016; Schneider et al., 2019; von Moltke et al., 2016; Westphalen et al., 2014). Upon helminth infection, tuft cell expansion induces adaptive gut lengthening to preserve its nutrient extraction capacity (Nadjsombati et al., 2018; Schneider et al., 2018). Mucus-producing goblet cells also participate in defense and repair of the intestinal mucosa (Renes et al., 2002). Staining for the tuft cell marker DCLK1, tuft/enteroendocrine cell marker PROX1, enteroendocrine marker CHGA, or Paneth cell marker lysozyme (Cetin et al., 1989; Gerbe et al., 2009; Peeters and Vantrappen, 1975; Yan et al., 2017) revealed an expansion of tuft cells, but not enteroendocrine or Paneth cells in MafIECKO small intestine 1 mo after Maf inactivation (Fig. 4 C and Fig. S3, C and D). We also observed hyperplasia and a tendency toward increased goblet cell number in MafIECKO mice as determined by Periodic Acid–Schiff (PAS) staining (Fig. 4 D).
The microbial metabolite succinate is a potent inducer of tuft cell expansion (Banerjee et al., 2020; Haber et al., 2017; Lei et al., 2018; Nadjsombati et al., 2018; Schneider et al., 2018); therefore, we used this model to explore the mechanism promoting tuft cell expansion in MafIECKO mice. Succinate administration increased gut length and the number of tuft cells and promoted goblet cell hyperplasia in WT mice. However, no further induction of tuft or goblet cell number was observed in succinate-treated Maf-deficient mice (Fig. 4, E–G). We speculated that Maf depletion elevates gut succinate levels; however, WT and Maf-deficient intestines harbored comparable levels of this metabolite (Fig. 4 H). Although additional studies are necessary, based on these results we propose that c-MAF and succinate share common downstream mechanisms for tuft cell expansion and gut remodeling. Loss of c-MAF increases epithelial Il17rc (Fig. 3, B and C), thus enhanced IL17R signaling, previously shown to be important for secretory lineage commitment (Lin et al., 2022) may also contribute to the tuft expansion in MafIECKO mice.
BMP signaling promotes zonation of enterocytes and restricts tuft cell numbers at steady state (SS) and during infection (Beumer et al., 2022; Lindholm et al., 2022). Our results designate c-MAF as a downstream transcriptional mediator that controls both functions. Tuft cell expansion is a response to intestinal injury (May et al., 2014; Middelhoff et al., 2017) and we propose that a “tuft cell phase” is one step of intestinal regeneration toward re-establishment of enterocyte zonation and absorptive function. Upon epithelial-specific Maf depletion, the gut adopts a chronic injury response highlighted by loss of enterocyte zonation and absorptive capacity but also increased tuft cell numbers and gut lengthening (Fig 2, G–N; and Fig. 4, A and C), which preserves a global metabolic balance of the organism.
Epithelial Maf loss impedes recovery after acute intestinal injury
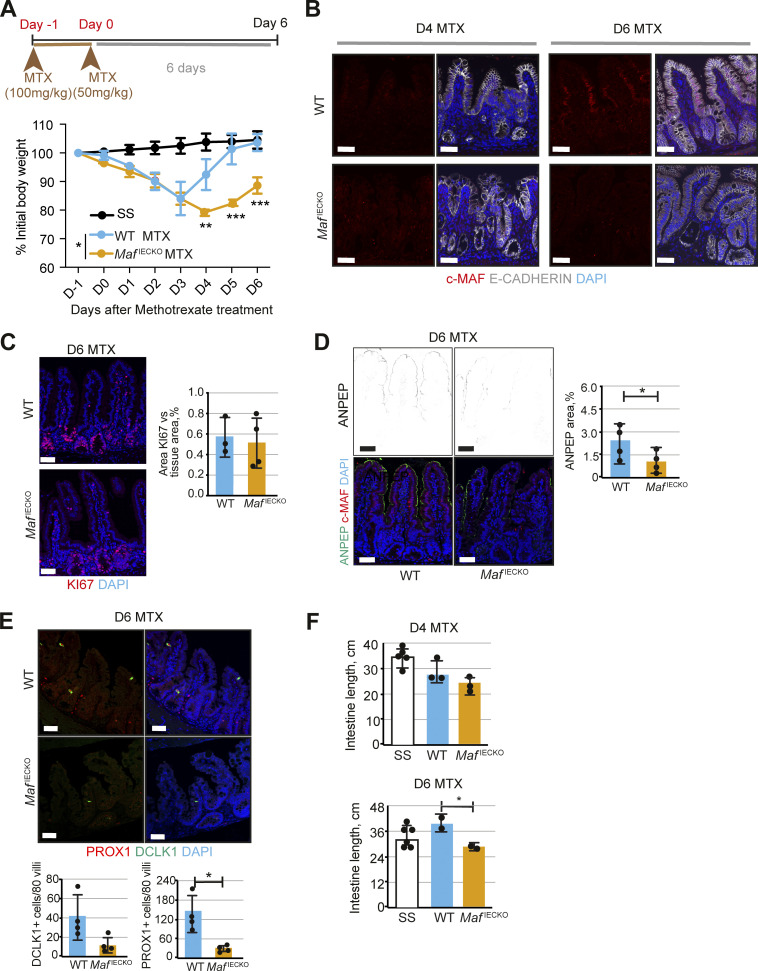
Since loss of Maf under homeostatic conditions led to intestinal adaptation, we tested whether epithelial Maf inactivation affects responses to acute intestinal injury. Administration of the anti-metabolite methotrexate (MTX) depletes proliferative progenitor cells, resulting in villus degeneration, transient malabsorption, and body weight loss (Sonis et al., 2001; Visentin et al., 2012). The intestinal epithelium is rapidly regenerated from columnar base stem cells leading to recovery of pre-treatment body weight (Tian et al., 2011). MTX-treated WT and MafIECKO mice displayed a similar decrease in body weight until day 3 (Fig. 5 A). As expected (Aparicio-Domingo et al., 2015), weight recovery was observed in WT mice after the peak of damage. However, MafIECKO mice failed to gain weight leading to a tendency to decreased animal survival (Fig. 5 A and Fig. S3 E).
Figure 5.
Defective expression of nutrient transporters and tuft cell expansion upon Maf loss impedes recovery after anti-metabolite treatment. (A) Loss of Maf prevents recovery after MTX treatment. Percentage of initial body weight, mean ± SD. Two-way ANOVA (Šídák test). Day (D) 4 WT MTX vs. MafIECKO MTX **, P < 0.01; Day 5 and 6 WT MTX vs. MafIECKO MTX ***, P < 0.001; n = 5 WT; n = 5 MafIECKO MTX mice treated and n = 4 WT and n = 4 MafIECKO control mice at the start of the treatment. SS, steady state. Data analyzed from two different cohorts and shown as mean ± SD. (B) c-MAF is downregulated at the peak of MTX treatment (day 4) and recovered at day 6 after treatment initiation. Staining for c-MAF (red), E-cadherin (white), and DAPI (blue). Scale bar, 50 μm. (C) No differences in epithelial cell proliferation in MafIECKO mice. Staining for Ki67 (red) and DAPI (blue). Scale bar, 50 μm. Two-tailed unpaired Student’s t test. n = 3 WT; n = 3 MafIECKO. Data shown as mean ± SD. (D) ANPEP is reduced at day 6 after MTX upon Maf depletion. Staining for ANPEP (green), c-MAF (red), and DAPI (blue). Scale bar, 50 μm. Percentage of area per tissue in WT and MafIECKO mice. *, P < 0.05. n = 4 WT; n = 4 MafIECKO. Data shown as mean ± SD. (E) Tuft cells are reduced upon Maf loss at day 6 after MTX treatment. Staining for PROX1 (red), DLCK1 (green), and DAPI (blue). Scale bar, 50 μm. Number of PROX1+ and DCLK1+ cells in 80 villi. DCLK1+ cells/80 villi P = 0.05; PROX1+ cells/80 villi. Two-tailed unpaired Student’s t test *, P < 0.05. n = 4 WT, n = 4 MafIECKO. Data shown as mean ± SD. (F) Decreased length of small intestine in Maf depleted mice at day 6 after MTX treatment. Length of small intestine (cm) in WT and MafIECKO at day 4 and 6 after MTX initiation. One-way ANOVA with Tukey post hoc test. *, P < 0.05. D4 MTX, n = 5 SS (PBS), n = 3 WT, n = 3 MafIECKO; D6 MTX, n = 6 SS (PBS), n = 2 WT, n = 2 MafIECKO. Data shown as mean ± SD.
In WT mice, MTX treatment decreased epithelial c-MAF expression, which was regained during the regeneration phase (Fig. 5 B). As MTX treatment targets proliferative cells, we analyzed epithelial cells during the recovery phase by staining for Ki67. In agreement with an absence of c-MAF in this compartment (Fig. 1 B), we did not observe differences in regenerating crypt proliferation between WT and MafIECKO mice (Fig. 5 C). The intestinal inflammatory response assessed by the expression of Il6, Tnfa, Ifng, Il1b, Cd45 (Fig. S3 F), villus regeneration or differentiation of the intestinal epithelium, as determined by KRT20 staining (Fig. S3 G), was similar in WT vs. MafIECKO mice. However, levels of ANPEP (Fig. 5 D) were diminished at day 6 after MTX in MafIECKO mice as compared to WT animals. Unlike at SS, the small intestines of MTX-treated MafIECKO harbored significantly fewer tuft cells and the gut length was shorter as compared to WT mice (Fig. 5, E and F; and data not shown). These results indicate that Maf loss impairs the recovery after acute intestinal epithelial damage due to a diminished nutrient transport capacity of the regenerating epithelium and the inability to mount tuft cell–mediated compensatory intestinal lengthening.
In summary, our study sheds light on the novel evolutionary conserved role of c-MAF for maintaining enterocyte zonation and dietary lipid homeostasis. It also highlights a mechanistic role for c-MAF in promoting differentiated epithelial cell recovery after acute intestinal injury and shows the importance of the crosstalk between differentiated enterocytes and tuft cells to promote long-term intestinal adaptation.
Materials and methods
Mice
All experiments were approved by the Animal Ethics Committee of Vaud, Switzerland. Maf fl/fl mice (Giordano et al., 2015) were obtained from the laboratory of Gregory Verdeil. Mice were crossed with Villin-CreERT2 mice (el Marjou et al., 2004) to obtain Maffl/fl; Villin-CreERT2 mice for Maf deletion in the intestinal epithelium. Maffl/fl mice were used as controls (WT). To activate the CreERT2 recombinase, 4–5-wk-old Maffl/fl and Maffl/fl; Villin-CreERT2 received three intraperitoneal injections of tamoxifen (50 μg/g body weight) in Kolliphor EL (Sigma-Aldrich) every other day. For RNAseq analysis, mice were sacrificed 1 wk after tamoxifen initiation. We validated Maf deletion by RT-qPCR and/or immunostaining in all samples used in this study (e.g., Fig. 2 A). For all other experiments, mice were dissected after 1-mo deletion. For intestinal injury experiments, Maffl/fl and Maffl/fl; Villin-CreERT2 were injected with 100 mg/kg of MTX at day −1 and with 50 mg/kg at day 0 as previously described (Aparicio-Domingo et al., 2015). Body weight was monitored daily, and the intestine was collected at days 4 and 6 after the last MTX injection. EdU (10 mg/kg mouse; Invitrogen) was injected intraperitoneally 1 h or 48 h prior to dissection. Maffl/fl; Villin-CreERT2 were administered 150 mM succinate in drinking water for 7 d as described previously (Schneider et al., 2018), 1 wk after Maf deletion. All experiments were performed in females, as they displayed more pronounced gut lengthening and tuft-cell expansion phenotype.
Staining procedures and image acquisition
Paraffin sections
5-μm sections were deparaffinized and subjected to heat-induced epitope retrieval using low or high pH retrieval solutions (DAKO), incubated with antibodies and mounted in Fluoromount-G mounting medium supplemented with DAPI (eBioscience). The details of antibodies are provided in Table S2. Alternatively, sections were stained using H&E (Harris modified H&E solution; Sigma-Aldrich) and slides were mounted in Aquatex (Merck). Chromogenic detection was performed as previously described (Ragusa et al., 2014).
All images were taken using confocal Zeiss Zen v11.0.0.0 (Carl Zeiss) and analyzed using Imaris 8 (v.8.0.2; Bitplane) and Photoshop CC v2015.1.2 (Adobe) software. Bright-field microscope images were taken using a Leica microscope MZ16 with a Leica Microsystems DFC 295 camera and LAS v4.2 software (Leica Microsystems).
Image quantifications
For histological analyses, at least two to three pictures per sample were taken with a Zeiss Zen v11.0.0.0 confocal microscope with 20× objective and quantified using ImageJ Fiji software. Keratin 20 (Krt20), ANPEP, DPP4, and APOA4, lysozyme-stained areas were normalized to total tissue area. For the quantification of epithelial cell proliferation, we measured the overlap of EdU/Ki67 signal over the E-cadherin selection. The images were transformed into 8-bit using ImageJ Fiji software and a threshold was set up. The same threshold was kept for all the pictures. The number of PROX1+, DCLK1+, CHGA+ cells was quantified manually per epithelial area or in a total of 60 villi in Maffl/fl and Maffl/fl; Villin-CreERT2 mice. Goblet cell number per villus was manually quantified by following PAS staining. Goblet cell diameter was analyzed staining using particle analysis mask in Fiji. For quantification of crypt area, the region of interest of crypts was quantified and measured. For lamina propria analysis, the non-epithelial area was quantified versus total tissue area. The pictures taken of every mouse were analyzed, and the average value of those was plotted. GraphPad Prism 8 program was used to make the graphs, and each dot represents a single mouse.
Intestinal epithelial cell sorting
The second half of jejunum and the complete ileum were dissected and flushed with cold PBS. Peyer’s patches and mesenteric fat were removed, and the remaining small intestine was cut longitudinally and then washed with cold PBS. 0.5–1 cm intestinal pieces were incubated for 20 min at 37°C in digestion buffer containing DMEM, 2% serum, Liberase TL (0.2 mg/ml; Roche), DNase I (1 U/ml; Invitrogen), and 1% gentamicin with gentle stirring. To favor tissue dissociation, the pieces were mixed by pipetting up/down several times after 10 and 20 min. Isolated cells in the supernatant were harvested and kept on ice with a 1:1 volume of complete DMEM until the end of the digestion process. In total, two identical digestions were performed for the complete dissociation of the intestine. The cell suspension was passed through a 40-μm strainer, centrifuged for 6 min at 150 g at 4°C, and resuspended in FACS buffer (PBS, 2% serum, and 2 mM EDTA). Cells were incubated with Fc block (antibody to CD16/32) for 20 min on ice and stained with conjugated antibodies in FACS buffer for 30 min. Following exclusion of dead cells using DAPI and gating on single cells, differentiated intestinal epithelial cells were selected as single Epcam+CD45−CD31−CD44− or Epcam+CD45−CD31−CD44+, gated and sorted directly in lysis buffer RLT containing β-mercaptoethanol (RNeasy micro kit, Qiagen). Cells were sorted on a BD FACSAria II (SORP) v8.0.1 cell sorter with BD FACSDiva software (BD Biosciences). Gating strategy is presented in Fig. S1. Antibodies used for flow cytometry are listed in Table S2.
Tissue nucleic acid isolation and RT-qPCR
RNA isolation
Sorted cells, cultured organoids, or total intestinal epithelium were transferred into RLT lysis buffer with β-mercaptoethanol (Qiagen) and snap-frozen on dry ice. RNA was isolated using Qiagen RNeasy Plus Micro Kit (Qiagen).
RT-qPCR
We used reverse-transcriptor First Strand cDNA Synthesis Kit (Roche Diagnostics) and StepOnePlus Real-Time PCR instrument (Applied Biosystems) and SensiFAST SYBR Lo-Rox kit (Bio94020; Meridian) for the qPCR reaction analyzed in QuantStudio 3 Real-Time PCR System (Applied Biosystems) using 40 cycles and 60°C annealing temperature. Data were analyzed using the comparative Ct method as described by the manufacturer. Gene expression was normalized to Actb or 18S (Fig. 1 D and F; Fig. 3 A; and Figs. S1, S2, and S3) levels. RT-qPCR results are shown as fold change over controls.
The nucleotide sequences of the real-time qPCR primers used in this study are described in Table S2.
Bulk RNAseq
RNA quality control, library preparations, and sequencing reactions were conducted at GENEWIZ, LLC. Total RNA samples were quantified using Qubit 2.0 Fluorometer (Life Technologies) and RNA integrity was checked with 4,200 TapeStation (Agilent Technologies). RNAseq library preparation used NEBNext Ultra RNA Library Prep Kit for Illumina by following the manufacturer’s recommendations (NEB). Briefly, enriched RNAs were fragmented for 15 min at 94°C. First-strand and second-strand cDNAs were subsequently synthesized. cDNA fragments were end-repaired and adenylated at 3′ ends, and universal adapter was ligated to cDNA fragments, followed by index addition and library enrichment with limited cycle PCR. Sequencing libraries were validated on the Agilent TapeStation (Agilent Technologies) and quantified using Qubit 2.0 Fluorometer (Invitrogen) as well as by qPCR (Applied Biosystems). The sequencing libraries were clustered on two lanes of a flowcell. After clustering, the flowcell was loaded on the Illumina HiSeq instrument according to manufacturer’s instructions. The samples were sequenced using a 2 × 150 paired-end configuration. Image analysis and base calling were conducted by the HiSeq Control Software. Raw sequence data (bcl files) generated from Illumina HiSeq was converted into fastq files and de-multiplexed using Illumina’s bcl2fastq v.2.17 software. One mismatch was allowed for index sequence identification.
Bulk RNAseq preprocessing and analysis
Sequencing reads were pseudoaligned using kallisto (v. 0.44.0; Bray et al., 2016) against the Mus musculus reference transcriptome obtained from Ensembl (GRCm38.p6). Kallisto was run with default settings plus sequence bias correction.
Transcript-level abundances quantified by kallisto were imported into R (v. 3.5.3). Conversion of Ensembl transcript identity to Ensembl gene identity, gene symbols, gene biotype, and description was performed using biomaRt (v. 2.38.0; Durinck et al., 2009). Counts for gene-level analysis were estimated using the “tximport” function of the tximport package (v. 1.10.0; Soneson et al., 2015), with abundance estimates scaled using the average transcript length and library size (arguments set to: countsFromAbundance = “lengthScaledTPM” and dropInfReps = T). Next, we filtered out very lowly expressed genes by only retaining genes that were expressed at more than 0.45 counts per million in at least two samples. Differential gene expression analysis between Maf knockout and WT samples was performed using the “DESeq” function (with betaPrior = T) of the DESeq2 package (v. 1.14.1; Love et al., 2014), and extracting the results using the function “results” with arguments addMLE = T and α = 0.05. This yielded differential expression results for 15,558 genes, 6,751 of which had average normalized counts above 500.
Over-representation analysis of Gene Ontology (GO) terms (Ashburner et al., 2000; The Gene Ontology Consortium, 2019) was performed on genes with adjusted P value <0.05 and average normalized counts above 500, separately for the upregulated or downregulated genes, using the “enrichGO” function of the clusterProfiler package (v.3.18.1 loaded in R v.4.0.2; Yu et al., 2012) with argument minGSSize = 30.
Expression of Maf in publicly available datasets
We obtained microarray data of mouse tissues from Gene Expression Omnibus (GEO; GSE9954; Thorrez et al., 2008). CEL files were imported into R (v. 3.5.3) using the “justRMA” function of the affy package (v. 1.60.0; Gautier et al., 2004) and were converted to log2 (normalized expression values). Annotation of microarray probes to gene symbols was obtained through the mouse4302.db package (v. 3.2.3), and we selected the probe for Maf with highest overall average expression (1456060_at) to create a heatmap using ComplexHeatmap (v. 1.20.0; Gu et al., 2016).
We obtained scRNAseq data of mouse intestinal epithelial cells from GEO (GSE99457; Yan et al., 2017). The three output files of the cell ranger pipeline of all 10 samples were imported into R (v. 4.0.2) and pre-processed with Seurat (v. 4.0.4; Hao et al., 2021). Counts were normalized with the logNormalize method and a scale factor of 10,000. A principal component analysis was generated on the scaled values of the top 2,000 variable genes selected with the variance-stabilizing method. A uniform manifold approximation and projection (UMAP) was generated using the first 20 principal components (PCs), followed by clustering of cells using the “FindNeighbors” and “FindClusters” functions with PCs 1–20 and a resolution parameter of 0.3. Clusters were annotated to cell types using known marker genes. To determine in which clusters the expression of Maf was highest, differential gene expression analysis among clusters was performed using the “FindAllMarkers” function with default parameters.
For the analysis of zonation, we obtained RNAseq data of mouse enterocytes from laser-micro-dissected villus sections from GEO (GSE109413; Moor et al., 2018). Kallisto pseudo-alignment results were imported into R (v. 3.5.3) and pre-processed the same way as for the Maf KO and WT samples described above with tximport and biomaRt and normalized with DESeq2.
Maf expression in human and zebrafish intestinal epithelial cells was analyzed using single-cell portal analysis tool (https://singlecell.broadinstitute.org/single_cell) and pre-publication dataset of human duodenum and ileum (https://singlecell.broadinstitute.org/single_cell/study/SCP817/comparison-of-ace2-and-tmprss2-expression-in-human-duodenal-and-ileal-tissue-and-organoid-derived-epithelial-cells#study-summary) and dataset of intestine from zebrafish cultured under conventional conditions (GSE161855; Willms et al., 2022).
Using our RNAseq fold change results, we determined whether genes regulated by Maf in immune cells were significantly differentially expressed in MafLECKO mice. These genes were compiled from the literature (Gabryšová et al., 2018) and contained genes that are known to be regulated or that are potentially regulated by Maf, as well as interleukin-related genes (including interleukins and interleukin receptors).
c-MAF promoter analysis
We searched for the c-MAF–binding site motif in a 700-bp-long region spanning the Il17rc promoter, expanding 500 bp upstream and 200 bp downstream of the transcription start site of Il17rc. The extended promoter sequence was obtained from the Eukaryotic Promoter Database (mm10, https://epd.epfl.ch/mouse/mouse_database.php?db=mouse).
We obtained the mouse c-MAF-binding site motif from the MotifMap database (Liu et al., 2017; http://motifmap-rna.ics.uci.edu/, motif M03816) and searched for its best match using the FIMO tool (Grant et al., 2011) of the online MEME Suite (v.5.4.1; https://meme-suite.org/meme/tools/fimo).
Intestinal organoid culture
The organoids for studies of Maf regulation were isolated from intestinal stem cells of WT mice and cultured as previously described (O’Rourke et al., 2016). For cell harvesting, Matrigel was disrupted manually. For subsequent passaging, the organoids were dissociated by manual pipetting, the cells were washed, counted, and 1,000 cells/50 μl of Matrigel were cultured again as above. The organoids were treated 2 d later after plating with vehicle, mouse recombinant Bmp2 (100 ng/ml; R&D) or BMP inhibitor DMH-1 (3 µM; Tocris) for 24 h and RNA isolated.
Organoids culture for mini-intestine generation
Mouse distal intestinal organoids and mini-intestines were in base medium (BM) prepared from Advanced DMEM/F12 medium (Gibco) and supplemented with 1× Glutamax (Gibco), 10 mM Hepes (Gibco), 100 μg/ml penicillin-streptomycin (Gibco), 1× B27 supplement (Gibco), 1× N2 supplement (Gibco), 500 μM N-acetylcysteine (Sigma-Aldrich), and 100 µg/ml Primocin (Invivogen). For differentiation, the medium was supplemented with 50 ng/ml EGF (R&D), 100 ng/ml Noggin (EPFL Protein Expression Core Facility), and 500 ng/ml R-Spondin 1 (EPFL Protein Expression Core Facility). For organoid expansion, the differentiation medium was further supplemented with 3 μM CHIR99021 (Stemgent) and 1 mM valproic acid (Sigma-Aldrich). For the mini-intestine expansion medium, the differentiation medium was supplemented with 1 mM valproic acid, 2.5 μM Thiazovivin (Stemgent), and 1 μg/ml WNT3A (Time Bioscience).
Organoids were grown in 20 μl drops of Matrigel (Corning; growth factor–reduced, phenol red–free formulation) and organoid expansion medium. The medium was refreshed every other day. Organoids were sub-cultured every fourth day. For passaging, Matrigel drops were disrupted and organoids were extracted by centrifugation (3 min, 2× 300 g, 4°C) followed by the mechanical dissociation of organoids into small fragments, which were subsequently re-embedded in Matrigel.
Mini-intestines were cultured according to Nikolaev et al. (2020). Organoids were isolated from Matrigel and dissociated into single cells on day 3 after passaging using TrypLE (Gibco) supplemented with 2,000 U/ml DNaseI (Roche), 1 mM N-acetylcysteine (Sigma-Aldrich), and 10 μM Y27632 (Stemgent). Isolated single cells were seeded at a density of ∼106 cells/ml into the microfluidic chip device described in Nikolaev et al. (2020). After 3 min of adherence, residual, non-adhered cells were washed away, and the cells were cultured in mini-intestine expansion medium. The medium was changed daily to supply fresh medium and remove dead cells and debris from the apical lumen of the microfluidic channel. On days 4 and 5 of the mini-intestine culture, the mini-intestine expansion medium was gradually replaced by differentiation medium on the apical side. Subsequently, the basolateral medium was gradually replaced by differentiation medium on days 6 to 8.
Lipid uptake in mini-intestines
For lipid uptake experiments in mini-intestines, a 200 µM BODIPY FL C12 (Invitrogen) solution in chemically defined lipid concentrate (Gibco) was prepared. Before the treatment, the apical channel of 14-d-old mini-intestines was perfused with BM to remove dead cells and debris. Subsequently, mini-intestines were treated apically with BM supplemented with 10% of the BODIPY FL C12 solution (20 µM) for 10 min. After the treatment, mini-intestines were washed three times with 1× PBS, immediately fixed or incubated with growth medium for two more hours, and subsequently fixed using 4% paraformaldehyde in PBS overnight.
For fatty acid uptake experiments, whole-mount fluorescent imaging of fixed mini-intestines for fatty acid uptake experiments was conducted on a Zeiss LSM980 confocal microscope system (Bioimaging and Optics Core Facility, École polytechnique fédérale de Lausanne) equipped with a 25×/0.80 NA water-immersion objective, which was operated using the ZEN Blue Software v. 3.4.91.
Fluorescent images of cryosectioned stained mini-intestine samples were acquired on a Leica SP8 upright confocal microscope system (Bioimaging and Optics Core Facility, École polytechnique fédérale de Lausanne) equipped with a 20×/0.75 NA air objective, operated by the LAS-X software (Leica).
Fatty acid uptake was assessed by quantifying the BODIPY FL C12 fluorescent signal in mini-intestines fixed immediately after the treatment using the surface creation tool of IMARIS v. 9.9.1. The detection settings were adjusted separately for each sample so that all cells showing a fluorescent signal were detected. Subsequently, the total volume of all detected surfaces in one sample was used as one data point.
Lipid droplet vesicle numbers were detected using the spots detection tool of IMARIS. The detection settings were separately adjusted for each data set so that all vesicles were detected in each sample.
Lipid droplets were quantified from samples incubated for 2 h after treatment before fixation.
Mini-intestine staining and imaging
Bright-field imaging of living mini-intestines was performed using a Nikon Eclipse Ti2 inverted micro-scope system equipped with a 10×/0.30 NA air objective controlled by NIS-Elements AR v.5.11.02 (Nikon) software. The extended depth of field images of bright-field images were calculated from a 60,100-µm z-stack image using the built-in NIS-Elements function.
Fixed mini-intestine samples were incubated in a mixture of cryomatrix and 30% sucrose (vol/vol 50-50%). Subsequently, mini-intestines were incubated in 100% cryomatrix overnight before embedding the tissues in cryomatrix in a tissue mold and freezing them on dry ice. Finally, the samples were cut into sections of 40-µm thickness at −20°C using a CM3050S cryostat (Leica). Cryosectioned mini-intestine samples were permeabilized using 0.2% Triton X-100 in PBS containing 10% FBS (PBS-FBS) for 1 h at room temperature (RT). Blocking was performed for 1 h at RT using a blocking buffer (0.01% Triton X-100 in PBS-FBS). Primary antibodies were diluted in blocking buffer, added to the sample, and incubated overnight at 4°C. The samples were washed with blocking buffer three times for 15 min at RT. After overnight incubation in secondary antibody solutions at 4°C, the cryosections were washed with blocking buffer, mounted in Fluoromount-G (SouthernBiotech), and analyzed by confocal microscopy.
The details of the antibody used are provided in Table S2.
Serum triglyceride analysis
Serum was isolated after centrifugation at +4°C and then stored at −80°C. Serum parameters were measured on two times diluted samples (1:1 ratio of serum to diluent) using DimensionXpand Plus (Siemens Healthcare Diagnostics AG). The biochemical tests were performed according to the manufacturer kit for triglycerides (DF69A; Siemens Healthcare).
Metabolomics
For polar metabolite extraction of tissue samples, 20 mg tissue was mixed with 200 μl acetonitrile:methanol:water (40:40:20) and homogenized with a TissueLyser for 2 min. Samples were centrifuged at 15,000 g for 30 s at 4°C; the supernatants were transferred to a new tube and used for mass spectometry analysis. Analysis of metabolite extracts was performed in flow injection analysis–time-of-flight mass spectrometry on an Agilent 6550 Q-TOF instrument as previously described (Fuhrer et al., 2011). Mass spectra were recorded from a mass/charge ratio of 50–1,000 in high-resolution negative ionization mode. Ions were annotated by matching their measured mass with reference compounds derived from the Human Metabolome Database (HMDB 4.0), allowing a tolerance of 1 mD. Intensities of m/z 117.0192, annotated as succinate, were plotted for ileum tissues of WT and MafIECKO mice.
Statistical analysis
Statistical analysis was performed using Graphpad Prism 8 for MacOS (GraphPad). The number of mice analyzed is indicated for each image in the figure legends. For in vivo experiments comparing WT and Maffl/fl; Villin-CreERT2 animals, two-tailed unpaired Student’s t test was performed to determine statistical significance between two means. For in vivo experiments comparing control and MTX-treated WT and Maffl/fl; Villin-CreERT2 animals and control and succinate treated mice, we used one-way ANOVA with Tukey post hoc test. For body weight analysis along MTX treatment, two-way ANOVA (Šídák test) was used. Scattered dot plot data are shown as mean ± SD, where each single dot represents an individual mouse. P value is stated in each figure. Differences were considered statistically significant at P < 0.05.
Resource availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Tatiana Petrova (tatiana.petrova@unil.ch).
Materials availability
This study did not generate new unique reagents.
Online supplemental material
Fig. S1 shows pattern of c-MAF expression in embryonic, postnatal, and adult enterocytes. Fig. S2 shows additional analyses of c-MAFiECKO mice and evolutionary conservation of c-MAF expression in human and zebrafish enterocytes. Fig. S3 shows effects of c-MAF inactivation on secretory lineages and intestinal regeneration after anti-metabolite treatment. Table S1 shows differentially expressed genes and pathways in Cmaf-deficient enterocytes and distribution of such differentially expressed genes according to the zonation pattern from Moor et al. (2018). Table S2 shows main reagents used.
Supplementary Material
shows differentially expressed genes and pathways in Cmaf-deficient enterocytes and distribution of such differentially expressed genes according to the zonation pattern from Moor et al. (2018).
shows main reagents used.
Acknowledgments
We thank A. Moor and D. Velin for useful discussions, S. Nassiri for the initial analysis of RNAseq data, C. Beauverd for mouse genotyping, colony maintenance and immunostainings, and J. Huelsken for providing mouse Rspo1 (Swiss Institute for Experimental Cancer Research, École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland). Mouse Pathology, Flow Cytometry, Animal, Cellular Imaging, and Genomic Technology Facilities of the University of Lausanne are gratefully acknowledged.
This work was supported by the Swiss National Science Foundation (CRSII5_177191, 31ER30_160674, 31003A-156266 and CRSK-3_190200 to T.V. Petrova) and an interdisciplinary grant of the Faculty of Biology and Medicine of University of Lausanne (to T.V. Petrova).
Author contributions: A. González-Loyola and T.V. Petrova planned the study and wrote the paper. T.V. Petrova supervised the research and acquired funding. A. González-Loyola performed the mouse studies and analyzed the data. J. Bernier-Latmani contributed to Fig. 4 H. I. Roci contributed to Fig. 3, A and C; and Fig. 4 H. T. Wyss contributed to Fig. 1, A and E; Fig. S1, F and G; Fig. 2, G and H; FIg. 3, F and H; and Fig. S2, B–D. J. Langer contributed to Fig. 3, F–H; and Fig. S2, D and E. S. Durot contributed to Fig. 4 H. O. Munoz provided samples for Figs. 2 and 3. B. Prat-Luri provided samples for Fig. S1. M. Delorenzi, M.P. Lutolf, N. Zamboni, and G. Verdeil provided reagents and/or intellectual contribution. All authors read, commented on, and approved the manuscript.
Data availability
Bulk RNAseq raw sequence data and a table of counts per gene per sample reported in this paper have been deposited in the GEO repository with the accession number GSE189852.
References
- Andris, F., Denanglaire S., Anciaux M., Hercor M., Hussein H., and Leo O.. 2017. The transcription factor c-Maf promotes the differentiation of follicular helper T cells. Front. Immunol. 8:480. 10.3389/fimmu.2017.00480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio-Domingo, P., Romera-Hernandez M., Karrich J.J., Cornelissen F., Papazian N., Lindenbergh-Kortleve D.J., Butler J.A., Boon L., Coles M.C., Samsom J.N., and Cupedo T.. 2015. Type 3 innate lymphoid cells maintain intestinal epithelial stem cells after tissue damage. J. Exp. Med. 212:1783–1791. 10.1084/jem.20150318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschenbrenner, D., Foglierini M., Jarrossay D., Hu D., Weiner H.L., Kuchroo V.K., Lanzavecchia A., Notarbartolo S., and Sallusto F.. 2018. An immunoregulatory and tissue-residency program modulated by c-MAF in human TH17 cells. Nat. Immunol. 19:1126–1136. 10.1038/s41590-018-0200-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner, M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. 2000. Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat. Genet. 25:25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, A., Herring C.A., Chen B., Kim H., Simmons A.J., Southard-Smith A.N., Allaman M.M., White J.R., Macedonia M.C., Mckinley E.T., et al. 2020. Succinate produced by intestinal microbes promotes specification of tuft cells to suppress ileal inflammation. Gastroenterology. 159:2101–2115.e5. 10.1053/j.gastro.2020.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker, N. 2014. Adult intestinal stem cells: Critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 15:19–33. 10.1038/nrm3721 [DOI] [PubMed] [Google Scholar]
- Barker, N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., and Clevers H.. 2007. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 449:1003–1007. 10.1038/nature06196 [DOI] [PubMed] [Google Scholar]
- Bass, N.M. 1988. The cellular fatty acid binding proteins: Aspects of structure, regulation, and function. Int. Rev Cytol. 111:143–184. 10.1016/s0074-7696(08)61733-7 [DOI] [PubMed] [Google Scholar]
- Beumer, J., Puschhof J., Yengej F.Y., Zhao L., Martinez-Silgado A., Blotenburg M., Begthel H., Boot C., van Oudenaarden A., Chen Y.G., and Clevers H.. 2022. BMP gradient along the intestinal villus axis controls zonated enterocyte and goblet cell states. Cell Rep. 38:110438. 10.1016/j.celrep.2022.110438 [DOI] [PubMed] [Google Scholar]
- Blache, P., van de Wetering M., Duluc I., Domon C., Berta P., Freund J.N., Clevers H., and Jay P.. 2004. SOX9 is an intestine crypt transcription factor, is regulated by the Wnt pathway, and represses the CDX2 and MUC2 genes. J. Cell Biol. 166:37–47. 10.1083/jcb.200311021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blume, C., Felix A., Shushakova N., Gueler F., Falk C.S., Haller H., and Schrader J.. 2012. Autoimmunity in CD73/Ecto-5′-nucleotidase deficient mice induces renal injury. PLoS One. 7:e37100. 10.1371/journal.pone.0037100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray, N.L., Pimentel H., Melsted P., and Pachter L.. 2016. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34:525–527. 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- Bröer, S. 2008. Apical transporters for neutral amino acids: Physiology and pathophysiology. Physiology. 23:95–103. 10.1152/physiol.00045.2007 [DOI] [PubMed] [Google Scholar]
- Bynoe, M.S., Waickman A.T., Mahamed D.A., Mueller C., Mills J.H., and Czopik A.. 2012. CD73 is critical for the resolution of murine colonic inflammation. J. Biomed. Biotechnol. 2012:260983. 10.1155/2012/260983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, S., Liu J., Song L., and Ma X.. 2005. The protooncogene c-Maf is an essential transcription factor for IL-10 gene expression in macrophages. J. Immunol. 174:3484–3492. 10.4049/jimmunol.174.6.3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carboni, J.M., Howe C.L., West A.B., Barwick K.W., Mooseker M.S., and Morrow J.S.. 1987. Characterization of intestinal brush border cytoskeletal proteins of normal and neoplastic human epithelial cells. A comparison with the avian brush border. Am. J. Pathol. 129:589–600 [PMC free article] [PubMed] [Google Scholar]
- Cetin, Y., Müller-Köppel L., Aunis D., Bader M.F., and Grube D.. 1989. Chromogranin A (CgA) in the gastro-entero-pancreatic (GEP) endocrine system. II. CgA in mammalian entero-endocrine cells. Histochemistry. 92:265–275. 10.1007/BF00500540 [DOI] [PubMed] [Google Scholar]
- Chan, C.W.M., Wong N.A., Liu Y., Bicknell D., Turley H., Hollins L., Miller C.J., Wilding J.L., and Bodmer W.F.. 2009. Gastrointestinal differentiation marker Cytokeratin 20 is regulated by homeobox gene CDX1. Proc. Natl. Acad. Sci. USA. 106:1936–1941. 10.1073/pnas.0812904106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers, H. 2006. Wnt/beta-catenin signaling in development and disease. Cell. 127:469–480. 10.1016/j.cell.2006.10.018 [DOI] [PubMed] [Google Scholar]
- Clevers, H. 2013. The intestinal crypt, a prototype stem cell compartment. Cell. 154:274–284. 10.1016/j.cell.2013.07.004 [DOI] [PubMed] [Google Scholar]
- Daassi, D., Hamada M., Jeon H., Imamura Y., Nhu Tran M.T., and Takahashi S.. 2016. Differential expression patterns of MafB and c-Maf in macrophages in vivo and in vitro. Biochem. Biophys. Res. Commun. 473:118–124. 10.1016/j.bbrc.2016.03.063 [DOI] [PubMed] [Google Scholar]
- Darmoul, D., Lacasa M., Baricault L., Marguet D., Sapin C., Trotot P., Barbat A., and Trugnan G.. 1992. Dipeptidyl peptidase IV (CD 26) gene expression in enterocyte-like colon cancer cell lines HT-29 and Caco-2. Cloning of the complete human coding sequence and changes of dipeptidyl peptidase IV mRNA levels during cell differentiation. J. Biol. Chem. 267:4824–4833. 10.1016/s0021-9258(18)42906-7 [DOI] [PubMed] [Google Scholar]
- Dehmer, J.J., Garrison A.P., Speck K.E., Dekaney C.M., Van Landeghem L., Sun X., Henning S.J., and Helmrath M.A.. 2011. Expansion of intestinal epithelial stem cells during murine development. PLoS One. 6:e27070. 10.1371/journal.pone.0027070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck, S., Spellman P.T., Birney E., and Huber W.. 2009. Mapping identifiers for the integration of genomic datasets with the R/bioconductor package biomaRt. Nat. Protoc. 4:1184–1191. 10.1038/nprot.2009.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- el Marjou, F., Janssen K.P., Chang B.H., Li M., Hindie V., Chan L., Louvard D., Chambon P., Metzger D., and Robine S.. 2004. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 39:186–193. 10.1002/gene.20042 [DOI] [PubMed] [Google Scholar]
- Fuhrer, T., Heer D., Begemann B., and Zamboni N.. 2011. High-throughput, accurate mass metabolome profiling of cellular extracts by flow injection-time-of-flight mass spectrometry. Anal. Chem. 83:7074–7080. 10.1021/ac201267k [DOI] [PubMed] [Google Scholar]
- Gabryšová, L., Alvarez-Martinez M., Luisier R., Cox L.S., Sodenkamp J., Hosking C., Pérez-Mazliah D., Whicher C., Kannan Y., Potempa K., et al. 2018. c-Maf controls immune responses by regulating disease-specific gene networks and repressing IL-2 in CD4+ T cells. Nat. Immunol. 19:497–507. 10.1038/s41590-018-0083-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier, L., Cope L., Bolstad B.M., and Irizarry R.A.. 2004. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 20:307–315. 10.1093/bioinformatics/btg405 [DOI] [PubMed] [Google Scholar]
- Gerbe, F., Brulin B., Makrini L., Legraverend C., and Jay P.. 2009. DCAMKL-1 expression identifies Tuft cells rather than stem cells in the adult mouse intestinal epithelium. Gastroenterology. 137:2179–2180. 10.1053/j.gastro.2009.06.072 [DOI] [PubMed] [Google Scholar]
- Giordano, M., Henin C., Maurizio J., Imbratta C., Bourdely P., Buferne M., Baitsch L., Vanhille L., Sieweke M.H., Speiser D.E., et al. 2015. Molecular profiling of CD8 T cells in autochthonous melanoma identifies Maf as driver of exhaustion. EMBO J. 34:2042–2058. 10.15252/embj.201490786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, C.E., Bailey T.L., and Noble W.S.. 2011. FIMO: Scanning for occurrences of a given motif. Bioinformatics. 27:1017–1018. 10.1093/bioinformatics/btr064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Z., Eils R., and Schlesner M.. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 32:2847–2849. 10.1093/bioinformatics/btw313 [DOI] [PubMed] [Google Scholar]
- Guiu, J., Hannezo E., Yui S., Demharter S., Ulyanchenko S., Maimets M., Jørgensen A., Perlman S., Lundvall L., Mamsen L.S., et al. 2019. Tracing the origin of adult intestinal stem cells. Nature. 570:107–111. 10.1038/s41586-019-1212-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber, A.L., Biton M., Rogel N., Herbst R.H., Shekhar K., Smillie C., Burgin G., Delorey T.M., Howitt M.R., Katz Y., et al. 2017. A single-cell survey of the small intestinal epithelium. Nature. 551:333–339. 10.1038/nature24489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, Y., Hao S., Andersen-Nissen E., Mauck W.M., Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., et al. 2021. Integrated analysis of multimodal single-cell data. Cell. 184:3573–3587.e29. 10.1016/j.cell.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauri, H.P., Quaroni A., and Isselbacher K.J.. 1979. Biogenesis of intestinal plasma membrane: Posttranslational route and cleavage of sucrase-isomaltase. Proc. Natl. Acad. Sci. USA. 76:5183–5186. 10.1073/pnas.76.10.5183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramatsu, Y., Suto A., Kashiwakuma D., Kanari H., Kagami S.i., Ikeda K., Hirose K., Watanabe N., Grusby M.J., Iwamoto I., and Nakajima H.. 2010. c-Maf activates the promoter and enhancer of the IL-21 gene, and TGF-beta inhibits c-Maf-induced IL-21 production in CD4+ T cells. J. Leukoc. Biol. 87:703–712. 10.1189/jlb.0909639 [DOI] [PubMed] [Google Scholar]
- Ho, I.C., Hodge M.R., Rooney J.W., and Glimcher L.H.. 1996. The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell. 85:973–983. 10.1016/s0092-8674(00)81299-4 [DOI] [PubMed] [Google Scholar]
- Hollnagel, A., Oehlmann V., Heymer J., Rüther U., and Nordheim A.. 1999. Id genes are direct targets of bone morphogenetic protein induction in embryonic stem cells. J. Biol. Chem. 274:19838–19845. 10.1074/jbc.274.28.19838 [DOI] [PubMed] [Google Scholar]
- Hong, E., Di Cesare P.E., and Haudenschild D.R.. 2011. Role of c-Maf in chondrocyte differentiation: A review. Cartilage. 2:27–35. 10.1177/1947603510377464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitt, M.R., Lavoie S., Michaud M., Blum A.M., Tran S.V., Weinstock J.V., Gallini C.A., Redding K., Margolskee R.F., Osborne L.C., et al. 2016. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science. 351:1329–1333. 10.1126/science.aaf1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber-Ruano, I., Pinilla-Macua I., Torres G., Casado F.J., and Pastor-Anglada M.. 2010. Link between high-affinity adenosine concentrative nucleoside transporter-2 (CNT2) and energy metabolism in intestinal and liver parenchymal cells. J. Cell. Physiol. 225:620–630. 10.1002/jcp.22254 [DOI] [PubMed] [Google Scholar]
- Imaki, J., Tsuchiya K., Mishima T., Onodera H., Kim J.I., Yoshida K., Ikeda H., and Sakai M.. 2004. Developmental contribution of c-maf in the kidney: Distribution and developmental study of c-maf mRNA in normal mice kidney and histological study of c-maf knockout mice kidney and liver. Biochem. Biophys. Res. Commun. 320:1323–1327. 10.1016/j.bbrc.2004.05.222 [DOI] [PubMed] [Google Scholar]
- Imbratta, C., Leblond M.M., Bouzourène H., Speiser D.E., Velin D., and Verdeil G.. 2019. Maf deficiency in T cells dysregulates Treg–TH17 balance leading to spontaneous colitis. Sci. Rep. 9:6135. 10.1038/s41598-019-42486-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki, T., Choi M., Moschetta A., Peng L., Cummins C.L., McDonald J.G., Luo G., Jones S.A., Goodwin B., Richardson J.A., et al. 2005. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metabol. 2:217–225. 10.1016/j.cmet.2005.09.001 [DOI] [PubMed] [Google Scholar]
- Karunakaran, S., Ramachandran S., Coothankandaswamy V., Elangovan S., Babu E., Periyasamy-Thandavan S., Gurav A., Gnanaprakasam J.P., Singh N., Schoenlein P.V., et al. 2011. SLC6A14 (ATB0, +) protein, a highly concentrative and broad specific amino acid transporter, is a novel and effective drug target for treatment of estrogen receptor-positive breast cancer. J. Biol. Chem. 286:31830–31838. 10.1074/jbc.M111.229518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka, K. 2007. Multiple mechanisms and functions of maf transcription factors in the regulation of tissue-specific genes. J. Biochem. 141:775–781. 10.1093/jb/mvm105 [DOI] [PubMed] [Google Scholar]
- Kawauchi, S., Takahashi S., Nakajima O., Ogino H., Morita M., Nishizawa M., Yasuda K., and Yamamoto M.. 1999. Regulation of lens fiber cell differentiation by transcription factor c-Maf. J. Biol. Chem. 274:19254–19260. 10.1074/jbc.274.27.19254 [DOI] [PubMed] [Google Scholar]
- Kerppola, T.K., and Curran T.. 1994. A conserved region adjacent to the basic domain is required for recognition of an extended DNA binding site by Maf/Nrl family proteins. Oncogene. 9:3149–3158 [PubMed] [Google Scholar]
- Kohan, A.B., Wang F., Lo C.M., Liu M., and Tso P.. 2015. ApoA-IV: Current and emerging roles in intestinal lipid metabolism, glucose homeostasis, and satiety. Am. J. Physiol. Gastrointest. Liver Physiol. 308:G472–G481. 10.1152/ajpgi.00098.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, W., Ren W., Ohmoto M., Urban J.F., Matsumoto I., Margolskee R.F., and Jiang P.. 2018. Activation of intestinal tuft cell-expressed Sucnr1 triggers type 2 immunity in the mouse small intestine. Proc. Natl. Acad. Sci. USA. 115:5552–5557. 10.1073/pnas.1720758115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, X., Gaudino S.J., Jang K.K., Bahadur T., Singh A., Banerjee A., Beaupre M., Chu T., Wong H.T., Kim C.K., et al. 2022. IL-17RA-signaling in Lgr5+ intestinal stem cells induces expression of transcription factor ATOH1 to promote secretory cell lineage commitment. Immunity. 55:237–253.e8. 10.1016/j.immuni.2021.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm, H.T., Parmar N., Drurey C., Campillo Poveda M., Vornewald P.M., Ostrop J., Díez-Sanchez A., Maizels R.M., and Oudhoff M.J.. 2022. BMP signaling in the intestinal epithelium drives a critical feedback loop to restrain IL-13-driven tuft cell hyperplasia. Sci. Immunol. 7:eabl6543. 10.1126/sciimmunol.abl6543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Sun S., Bredy T., Wood M., Spitale R.C., and Baldi P.. 2017. MotifMap-RNA: A genome-wide map of RBP binding sites. Bioinformatics. 33:2029–2031. 10.1093/bioinformatics/btx087 [DOI] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan, Y., and Xu W.. 2007. The structure and main functions of aminopeptidase N. Curr. Med. Chem. 14:639–647. 10.2174/092986707780059571 [DOI] [PubMed] [Google Scholar]
- Mabley, J.G., Pacher P., Liaudet L., Soriano F.G., Haskó G., Marton A., Szabo C., and Salzman A.L.. 2003. Inosine reduces inflammation and improves survival in a murine model of colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 284:G138–G144. 10.1152/ajpgi.00060.2002 [DOI] [PubMed] [Google Scholar]
- May, R., Qu D., Weygant N., Chandrakesan P., Ali N., Lightfoot S.A., Li L., Sureban S.M., and Houchen C.W.. 2014. Brief report: Dclk1 deletion in tuft cells results in impaired epithelial repair after radiation injury. Stem Cells. 32:822–827. 10.1002/stem.1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middelhoff, M., Westphalen C.B., Hayakawa Y., Yan K.S., Gershon M.D., Wang T.C., and Quante M.. 2017. Dclk1-expressing tuft cells: Critical modulators of the intestinal niche? Am. J. Physiol. Gastrointest. Liver Physiol. 313:G285–G299. 10.1152/ajpgi.00073.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll, R., Schiller D.L., and Franke W.W.. 1990. Identification of protein IT of the intestinal cytoskeleton as a novel type I cytokeratin with unusual properties and expression patterns. J. Cell Biol. 111:567–580. 10.1083/jcb.111.2.567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moor, A.E., Harnik Y., Ben-Moshe S., Massasa E.E., Rozenberg M., Eilam R., Bahar Halpern K., and Itzkovitz S.. 2018. Spatial reconstruction of single enterocytes uncovers broad zonation along the intestinal villus axis. Cell. 175:1156–1167.e15. 10.1016/j.cell.2018.08.063 [DOI] [PubMed] [Google Scholar]
- Nadjsombati, M.S., McGinty J.W., Lyons-Cohen M.R., Jaffe J.B., DiPeso L., Schneider C., Miller C.N., Pollack J.L., Nagana Gowda G.A., Fontana M.F., et al. 2018. Detection of succinate by intestinal tuft cells triggers a type 2 innate immune circuit. Immunity. 49:33–41.e7. 10.1016/j.immuni.2018.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev, M., Mitrofanova O., Broguiere N., Geraldo S., Dutta D., Tabata Y., Elci B., Brandenberg N., Kolotuev I., Gjorevski N., et al. 2020. Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nature. 585:574–578. 10.1038/s41586-020-2724-8 [DOI] [PubMed] [Google Scholar]
- O’Rourke, K.P., Ackerman S., Dow L.E., and Lowe S.W.. 2016. Isolation, culture, and maintenance of mouse intestinal stem cells. Bio-protocol. 6:e1733. 10.21769/bioprotoc.1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J., Levic D.S., Sumigray K.D., Bagwell J., Eroglu O., Block C.L., Eroglu C., Barry R., Lickwar C.R., Rawls J.F., et al. 2019. Lysosome-rich enterocytes mediate protein absorption in the vertebrate gut. Dev. Cell. 51:7–20.e6. 10.1016/j.devcel.2019.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, M.E., Barrera A., Wheaton J.D., Zuberbuehler M.K., Allan D.S.J., Carlyle J.R., Reddy T.E., and Ciofani M.. 2020. c-Maf regulates the plasticity of group 3 innate lymphoid cells by restraining the type 1 program. J. Exp. Med. 217:e20191030. 10.1084/jem.20191030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters, T., and Vantrappen G.. 1975. The paneth cell: A source of intestinal lysozyme. Gut. 16:553–558. 10.1136/gut.16.7.553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokrovskii, M., Hall J.A., Ochayon D.E., Yi R., Chaimowitz N.S., Seelamneni H., Carriero N., Watters A., Waggoner S.N., Littman D.R., et al. 2019. Characterization of transcriptional regulatory networks that promote and restrict identities and functions of intestinal innate lymphoid cells. Immunity. 51:185–197.e6. 10.1016/j.immuni.2019.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pot, C., Jin H., Awasthi A., Liu S.M., Lai C.Y., Madan R., Sharpe A.H., Karp C.L., Miaw S.C., Ho I.C., and Kuchroo V.K.. 2009. Cutting edge: IL-27 induces the transcription factor c-Maf, cytokine IL-21, and the costimulatory receptor ICOS that coordinately act together to promote differentiation of IL-10-producing Tr1 cells. J. Immunol. 183:797–801. 10.4049/jimmunol.0901233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragusa, S., Cheng J., Ivanov K.I., Zangger N., Ceteci F., Bernier-Latmani J., Milatos S., Joseph J.M., Tercier S., Bouzourene H., et al. 2014. PROX1 promotes metabolic adaptation and fuels outgrowth of Wnt(high) metastatic colon cancer cells. Cell Rep. 8:1957–1973. 10.1016/j.celrep.2014.08.041 [DOI] [PubMed] [Google Scholar]
- Renes, I.B., Verburg M., Van Nispen D.J.P.M, Buller H.A., Dekker J., and Einerhand A.W.C.. 2002. Distinct epithelial responses in experimental colitis: implications for ion uptake and mucosal protection. Am. J. Physiol. Gastrointest. Liver Physiol. 283:G169-179. 10.1152/ajpgi.00506.2001 [DOI] [PubMed] [Google Scholar]
- Robson, S.C., Sévigny J., and Zimmermann H.. 2006. The E-NTPDase family of ectonucleotidases: Structure function relationships and pathophysiological significance. Purinergic Signal. 2:409–430. 10.1007/s11302-006-9003-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., and Clevers H.. 2009. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 459:262–265. 10.1038/nature07935 [DOI] [PubMed] [Google Scholar]
- Schneider, C., O’Leary C.E., and Locksley R.M.. 2019. Regulation of immune responses by tuft cells. Nat. Rev. Immunol. 19:584–593. 10.1038/s41577-019-0176-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, C., O’Leary C.E., von Moltke J., Liang H.E., Ang Q.Y., Turnbaugh P.J., Radhakrishnan S., Pellizzon M., Ma A., and Locksley R.M.. 2018. A metabolite-triggered tuft cell-ILC2 circuit drives small intestinal remodeling. Cell. 174:271–284.e14. 10.1016/j.cell.2018.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert, H.J., van Es J.H., van den Born M., Begthel H., Stange D.E., Barker N., and Clevers H.. 2009. Prominin-1/CD133 marks stem cells and early progenitors in mouse small intestine. Gastroenterology. 136:2187–2194.e1. 10.1053/j.gastro.2009.03.002 [DOI] [PubMed] [Google Scholar]
- Soneson, C., Love M.I., and Robinson M.D.. 2015. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Res. 4:1521. 10.12688/f1000research.7563.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonis, S.T., Oster G., Fuchs H., Bellm L., Bradford W.Z., Edelsberg J., Hayden V., Eilers J., Epstein J.B., LeVeque F.G., et al. 2001. Oral mucositis and the clinical and economic outcomes of hematopoietic stem-cell transplantation. J. Clin. Oncol. 19:2201–2205. 10.1200/JCO.2001.19.8.2201 [DOI] [PubMed] [Google Scholar]
- Takase, M., Imamura T., Sampath T.K., Takeda K., Ichijo H., Miyazono K., and Kawabata M.. 1998. Induction of Smad6 mRNA by bone morphogenetic proteins. Biochem. Biophys. Res. Commun. 244:26–29. 10.1006/bbrc.1998.8200 [DOI] [PubMed] [Google Scholar]
- The Gene Ontology Consortium . 2019. The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 47:D330–D338. 10.1093/nar/gky1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorrez, L., Van Deun K., Tranchevent L.C., Van Lommel L., Engelen K., Marchal K., Moreau Y., Van Mechelen I., and Schuit F.. 2008. Using ribosomal protein genes as reference: A tale of caution. PLoS One. 3:e1854. 10.1371/journal.pone.0001854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, H., Biehs B., Warming S., Leong K.G., Rangell L., Klein O.D., and de Sauvage F.J.. 2011. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 478:255–259. 10.1038/nature10408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visentin, M., Zhao R., and Goldman I.D.. 2012. The antifolates. Hematol. Oncol. Clin. North Am. 26:629–648. 10.1016/j.hoc.2012.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moltke, J., Ji M., Liang H.E., and Locksley R.M.. 2016. Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature. 529:221–225. 10.1038/nature16161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F., Kohan A.B., Lo C.M., Liu M., Howles P., and Tso P.. 2015. Apolipoprotein A-IV: A protein intimately involved in metabolism. J. Lipid Res. 56:1403–1418. 10.1194/jlr.R052753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphalen, C.B., Asfaha S., Hayakawa Y., Takemoto Y., Lukin D.J., Nuber A.H., Brandtner A., Setlik W., Remotti H., Muley A., et al. 2014. Long-lived intestinal tuft cells serve as colon cancer-initiating cells. J. Clin. Invest. 124:1283–1295. 10.1172/JCI73434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willms, R.J., Jones L.O., Hocking J.C., and Foley E.. 2022. A cell atlas of microbe-responsive processes in the zebrafish intestine. Cell Rep. 38:110311. 10.1016/j.celrep.2022.110311 [DOI] [PubMed] [Google Scholar]
- Xu, J., Yang Y., Qiu G., Lal G., Wu Z., Levy D.E., Ochando J.C., Bromberg J.S., and Ding Y.. 2009. c-Maf regulates IL-10 expression during Th17 polarization. J. Immunol. 182:6226–6236. 10.4049/jimmunol.0900123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, K.S., Gevaert O., Zheng G.X.Y., Anchang B., Probert C.S., Larkin K.A., Davies P.S., Cheng Z.F., Kaddis J.S., Han A., et al. 2017. Intestinal enteroendocrine lineage cells possess homeostatic and injury-inducible stem cell activity. Cell Stem Cell. 21:78–90.e6. 10.1016/j.stem.2017.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, J.D., Yao S.Y.M., Baldwin J.M., Cass C.E., and Baldwin S.A.. 2013. The human concentrative and equilibrative nucleoside transporter families, SLC28 and SLC29. Mol. Aspects Med. 34:529–547. 10.1016/j.mam.2012.05.007 [DOI] [PubMed] [Google Scholar]
- Yu, J. 2013. Intestinal stem cell injury and protection during cancer therapy. Trans. Cancer Res. 2:384–396 [PMC free article] [PubMed] [Google Scholar]
- Yu, G., Wang L.G., Han Y., and He Q.Y.. 2012. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics. 16:284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeilstra, J., Joosten S.P.J., Dokter M., Verwiel E., Spaargaren M., and Pals S.T.. 2008. Deletion of the WNT target and cancer stem cell marker CD44 in Apc(Min/+) mice attenuates intestinal tumorigenesis. Cancer Res. 68:3655–3661. 10.1158/0008-5472.CAN-07-2940 [DOI] [PubMed] [Google Scholar]
- Zembroski, A.S., Xiao C., and Buhman K.K.. 2021. The roles of cytoplasmic lipid droplets in modulating intestinal uptake of dietary fat. Annu. Rev. Nutr. 41:79–104. 10.1146/annurev-nutr-110320-013657 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows differentially expressed genes and pathways in Cmaf-deficient enterocytes and distribution of such differentially expressed genes according to the zonation pattern from Moor et al. (2018).
shows main reagents used.
Data Availability Statement
Bulk RNAseq raw sequence data and a table of counts per gene per sample reported in this paper have been deposited in the GEO repository with the accession number GSE189852.