Abstract
Background
Tuberculosis (TB) is a disease with worldwide presence and a major cause of death in several developing countries. Current diagnostic methodologies often lack specificity and sensitivity, whereas a long time is needed to obtain a conclusive result.
Methods
In an effort to develop better diagnostic methods, this study aimed at the discovery of a biomarker signature for TB diagnosis using a Nuclear Magnetic Resonance based metabolomics approach. In this study, we acquired 1H NMR spectra of blood serum samples of groups of healthy subjects, individuals with latent TB and of patients with pulmonary and extra-pulmonary TB. The resulting data were treated with uni- and multivariate statistical analysis.
Results
Six metabolites (inosine, hypoxanthine, mannose, asparagine, aspartate and glutamate) were validated by an independent cohort, all of them related with metabolic processes described as associated with TB infection.
Conclusion
The findings of the study are according with the WHO Target Product Profile recommendations for a triage test to rule-out active TB.
Keywords: Tuberculosis, Diagnosis, NMR Metabolomics, Biomarkers, Serum
At a glance of commentary
Scientific background on this subject
Tuberculosis is a disease with worldwide presence and a major cause of death in developing countries. Current diagnostic methodologies often lack specificity and sensitivity, whereas a long time is needed to obtain a conclusive result. Untargeted metabolomics is a suitable strategy for a broad discovery of meaningful diagnosis biomarkers.
What this study adds to this field?
A metabolite signature for tuberculosis diagnosis was validated by an independent cohort. All metabolites are related with metabolic processes described as associated with TB infection. The findings of the study are according with the WHO Target Product Profile recommendations for a triage test to rule-out active TB.
Tuberculosis (TB), caused by Mycobacterium tuberculosis (Mtb), despite being completely curable, has reemerged as a global pandemic. Failure of effective vaccine protection, lack of early detection of the disease, emergence of drug resistance, deadly synergism with HIV infection and increasing global international migration flows have limited the success of TB disease management. WHO estimated the occurrence of around 10 million new TB cases in 2018 [1]. In order to overcome these frightful numbers, the End TB Strategy defined by this organization states as a priority the “early diagnosis of TB (…), and systematic screening of latent and high-risk groups” [1].
The diagnosis methods currently in use have major drawbacks. Microbiological detection of Mtb in sputum smear lacks sensitivity leading to late TB diagnosis [2]. Culture methods, the gold standard for TB diagnosis, require several weeks and developed laboratory capacity [3]. Most clinical features of TB and abnormalities on chest X-ray or histology results have low specificity, leading to false positive diagnoses. There are few rapid diagnostics recommended by WHO, being Xpert MTB/RIF assay the most widespread [4]. However this test requires instrumentation not broadly available in endemic settings [5].
As specified recently, the highest priorities in TB diagnosis are a rapid biomarker-based, non-sputum test for detecting active TB with the purpose of initiating treatment (1). Active TB patients, including HIV co-infected individuals and children, often do not present Mtb positive sputum, with blood and urine being the more convenient samples for diagnosis [4]. This may be based on the quantification of Mtb and its products or on biomarkers of the host's state [4]. Among pathogen compounds, the quantification of lipoarabinomannan (LAM) has been widely explored. However it suffers from poor sensitivity [6]. The same problem arises for Mtb DNA detection [4] and variability is associated with detection of proteins showing mycolyl transferase activity [7]. On the side of the host, several biomolecules detected in pulmonary TB (PTB) patients (specific transcriptional signature, micro-RNAs, antibodies and interferon inducible protein) were tested, but the simultaneous measurement of a large set of genes, variable accuracy, and the heterogeneity to antibody response proved to be sub-optimal [5].
Lately, the urgent need for new non-DNA biomarkers has been consistently advocated [2,4,7]. Besides, diagnostic performance will be improved using multi-biomarker signatures instead of a single biomarker [2,7,8]. Untargeted approaches, like metabolomics, are the more suitable strategy for a broad discovery and evaluation of meaningful biomarkers, contributing also to solve one of the major difficulties in establishing an accurate TB diagnosis test, and the partial understanding of the complex host–pathogen interaction [[8], [9], [10]]. Relevant findings for TB diagnosis using these strategies are summarized in recent reviews [8,9,11]. However, their importance is diminished by cohort and methodological variabilities. Moreover, though publications of new biomarkers are common, follow up studies on the refinement, validation and independent confirmation of them are not taken. A recent trans-African metabolomic study involving different populations determined a TB metabolic biosignature prior to TB diagnostic [10]. In another recent study, the authors determine 13 metabolites in serum that were altered in multidrug-resistant TB patients when compared with healthy controls and drug-susceptible TB patients [12]. Recently, we conducted a serum metabolomic study using an Indian TB cohort. The results suggested significant alterations in serum metabolites from TB patients when compared with healthy controls and asymptomatic house-hold contacts of active TB patients [13].
Here, we present a serum untargeted metabolomic study using Nuclear Magnetic Resonance (NMR) among Portuguese population comprising of healthy subjects, individuals with latent TB and patients with pulmonary and extra-pulmonary TB that show the potentiality of metabolomics to find new biomarkers for TB diagnosis.
Materials and methods
Ethical statement
The collection of human blood samples was approved by the National Commission for Data Protection (Lisbon, Portugal) (N. 5985/2014) and the Heath Regional Administration of Lisbon and Tagus Valley (ARSLTV) Ethical Commission (Lisbon, Portugal) (Proc.057/CES/INV/2014). All subjects provided written informed consent for participation in this study.
Cohort and collection of sera samples
Three experimental categories were established with a subdivision of the patients group: 1)Patients with pulmonary and extra-pulmonary primary diagnosed TB infection; 2)Latent TB population: Healthy individuals, asymptomatic for TB infection with positive IGRA test; 3) Control population: Healthy individuals with negative IGRA test. General inclusion criteria were age ≥18 and ≤65 years, HIV negative and absence of history of organ transplant, diabetes, chronic kidney failure or any other respiratory illness; patients were excluded if under treatment with anti-TB therapy >1.5 months or if other co-morbidity was detected during the study.
No fasting peripheral blood samples were collected, between 2014 and 2016, by venipuncture to clot activator tubes containing silica microparticles at the Centres for Pneumological Diagnosis of Venda Nova and of Almada-Seixal (Lisbon region, Portugal) and stored at 4 °C. Three hours after blood collection, samples were centrifuged and passed through 0.2 μm filters for serum decontamination. All serum aliquots were immediately frozen at −80 °C and never thawed until assayed. Detection of latent TB was performed for samples from asymptomatic subjects using the IGRA test (QuantiFERON®-TB Gold IT, QIAGEN). Samples showing hemolysis or with undetermined IGRA results were excluded.
Sample preparation for NMR metabolomics analysis
Each serum sample was thawed for 1 h and passed through a centrifugal filter with a 5 kDa cut-off to remove macromolecules shown to interfere with the NMR metabolite signals. First, the centrifugal filters were washed four times with 100 mM NaCl and twice with MiliQ water, prior to sample treatment to remove glycerol. Filtered serum was transferred to NMR tubes and proportionally added to phosphate buffer prepared in D2O containing an internal standard (4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS) 0.25 mM). The solution in the NMR tube was homogenized by repeated inversions avoiding the formation of air bubbles. This protocol was established after optimization of several steps, namely serum thawing time, sample stability and ultrafiltration conditions with the main goal of improving reproducibility and minimizing time dependent sample degradation. To evaluate sample stability NMR spectra were acquired immediately after sample preparation and after 4, 9, 13, 16 and 27 h. Comparison of NMR spectra profiles has shown deviations in chemical shifts, in particular for histidine, due to changes in sample pH, which was minimized using a phosphate buffer (50 mM pH 7.0) and keeping 3 h timing between sample preparation and NMR data acquisition.
NMR data acquisition and processing
Proton (1H) spectroscopy was performed on an 800 MHz Bruker AvanceII+ spectrometer equipped with a room temperature triple resonance HCN Z-gradient probe at 298 K, at the Magnetic Resonance Centre António Xavier at ITQB NOVA. 1D 1H -NOESY spectra (spectral width: 30.04 ppm; mixing time: 0.01 s; relaxation delay: 4 s; acquisition time: 1.36 s and 128 repetitions) were collected for each serum sample using the ‘‘noesygppr1d’’pulse sequence. From each experimental group one of the most concentrated samples was used for the collection of 2D NMR spectra to assist with assignment, namely J-resolved, 1H–1H COSY, 1H–1H TOCSY, and 1H–13C HSQC. The spectral acquisition conditions used were different from those suggested by Chenomx NMR Suite 8.1 identification and integration software (Chenomx). However, comparison of the collected data with tests acquired under reference conditions revealed no significant differences in the determination of metabolite concentrations and thus were used without corrections. Spectra were processed and analysed using TopSpin 3.2 software (Bruker).
Compound identification and quantification
Metabolite identification and quantification were performed using Chenomx NMR Suite 8.1 software, using its internal reference library (Version 10) and resorting to the information available in the Human Metabolome Data Base (HMDB 3.6), in the Biological Magnetic Resonance Bank (BMRB) and published work. 2D 1h–13c HSQC spectra, when applicable, were used to aid metabolite identification. Determination of each metabolite concentration was based on the known concentration of the internal standard, DSS, considering its dilution in each sample.
The NMR metabolomics data were deposited to the MetaboLights repository with the dataset identifier MTBLS2318.
Statistical analysis
From the set of metabolites assigned, potential sample contaminants (methanol, acetone and glycerol), urea and DSS were excluded. Additionally, metabolites corresponding to the anti-TB drugs were not considered for this analysis. Once the complete set of metabolomic features has been generated, univariate and multivariate data analysis methods were applied to investigate: (a) the general structure of the metabolomics data in the dataset, and (b) how the different metabolic features are related with the phenotypic data associated with the samples. Shortly, a Principal Component Analysis (PCA) analysis of metabolite concentrations is performed to identify outliers. A normality test is performed for each metabolite by Shapiro test. Metabolites concentrations that exceed 6 times the interquantile range (IQR) were excluded and boxplot plots were performed for the metabolites that showed statistical significance determined using the two-tailed unpaired Wilcox test considering p < 0.05 to reject the null-hypothesis using the adjusted p value for multiple testing (Benjamini-Hochberg). Univariate analysis was performed using a home-made R script. Multivariate analysis was performed in SIMCA software (PCA and Principal Least Square-Discriminant Analysis, PLS-DA) and in Metaboanalyst [14] (receiver operating characteristic curve (ROC) curves) using Vast and autoscaling methods for data scaling, respectively. Metabolite amounts were considered significantly different between experimental groups if Variable Influence in Projection (VIP) > 1.5.
Indian validation cohort
An Indian cohort that was already object of a scientific publication [13] was used here for validation of the obtained statistical model after profiling of their NMR spectra (cohort data in Supplementary Table 1). The selection criteria of the study participants, their categorization, the protocols for sample treatment and NMR data processing were standardized according with those used for the Portuguese cohort. For the validation twelve healthy controls with negative Mantoux test and an equal number of PTB patients were selected. The model was built with 6 metabolites that showed significant differences in the Portuguese cohort and using ROC curves analysis implemented in Metaboanalyst [14]. Metabolite concentrations were normalized by quantile and scaled by autoscaling.
Pathway analysis and biomarkers functional analysis
Pathway Analysis using all quantified metabolites was carried out using MetaboAnalyst [14], integrating pathway enrichment analysis and pathway topology analysis. Metabolite concentrations were normalized by quantile and scaled by autoscaling. Selected pathways have negative logarithm of p-value higher than 10 (y axis) and impact above 0, or pathway impact higher than 0.5 (x axis).
Results
Cohort
Thirty-seven TB patients which includes 25 patients with PTB and 12 with extra pulmonary TB (EPTB) were enrolled in the study (EPTB, [Table 1]). Two other asymptomatic experimental groups were considered: healthy controls (n = 57) and latent TB (n = 10) including individuals with a negative or a positive IGRA test, respectively. Gender, age, weight and smoking habits which are describe to be important in TB were also collected [[15], [16], [17], [18], [19]].
Table 1.
Cohort characterization.
| Tuberculosis Patients |
Latent | Controls | ||
|---|---|---|---|---|
| Pulmonary | Extra-pulmonary | |||
| Total individuals (n) | 25 | 12 | 10 | 57 |
| Age (A/B/C) | 10/12/2 | 3/2/7 | 2/5/3 | 23/28/7 |
| Gender (F/M) | 7/17 | 9/3 | 8/2 | 29/29 |
| Body Mass Index (BMI) (L/N/H) | 9/14/1 | 0/5/6 | 0/7/3 | 3/41/12 |
| Smoking habits (Y/N/Ex) | 6/8/5 | 0/8/0 | 0/4/0 | 11/32/5 |
Categorization for age is < 30 (A), 30–50 (B) and >50 years (C), for BMI is low (L), normal (N) and high (H), and for present smoking habits yes (Y), non (N), ex-smoker (Ex). A discrepancy between total number of individuals for the several criteria is due to missing information.
Evaluation of differential metabolic profiles between experimental groups
Different serum sample preparation protocols were tested, and the one that showed better results was with ultrafiltration (see Supplementary Fig. S1). Therefore, although ultrafiltration is an additional step in the sample preparation procedure it was deemed necessary, as corroborated in previous studies [20,21].
Sixty metabolites were assigned in the 1H NMR spectra using 2D NMR data. The signal intensity of 59 metabolites were quantified and used for the metabolomics analysis. The concentration of five identified metabolites could be influenced by sample handling (e.g. acetone, ethanol, which are volatile metabolites; glycerol, a preservative of ultrafiltration devices; propylene glycol, a probable exogenous contaminant; and urea whose ability to be quantified is incompatible with the data acquisition protocol used) or by subject habits (as in the case of caffeine). Beside these, π-methylhistidine was excluded from the analysis as its assignment is not clear (see Supplementary Table 2). The main metabolite classes assigned were amino acids, organic acids, carbohydrates and nucleoside intermediates. An estimate of the extent of spectral assignment was made from the ratio of the areas of the assigned and the residual spectral lines, and was found to be 93.4% of the observable spins.
The metabolite concentrations quantified for the patient and control experimental groups are depicted in Supplementary Table 2. According with The Human Metabolome Database (HMDB), all these metabolites have been already detected in blood. The number of detected and quantified metabolites was in accordance with previously published results [22]. The mean concentration for each metabolite from the control group was compared using a two-tailed t-test with those reported in the literature, and for 26 metabolites similar concentrations were found with 95% significance. Any discrepancies found was mainly attributed to differences in blood sample treatment, in particular if the analysis was performed in serum or plasma and to the protocol used for removing macromolecules.
From the quantified metabolites only three present a % of occurrence below 100%, formate (98.2%), ascorbate (96.5%) and fructose (31.6%). All the metabolites present in all control individuals have also 100% occurrence in the TB patients’ group, except for trigonelline, compound with the lowest determined concentration (See Supplementary Table 2).
PTB and EPTB groups were not discriminated by multi-variate analysis (see Supplementary Fig. 2). Therefore, further analysis was carried out with data involving TB patient group comprising the two types of the disease.
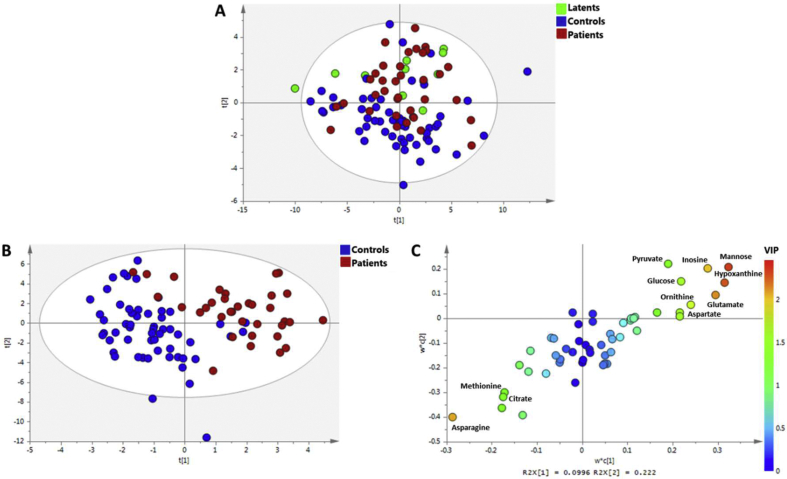
In the PCA analysis considering the three groups (Control, Latent and Patient), it was possible to observe a tendency for the separation between Controls and Patients, being the Latents samples distributed between them [Fig. 1 A]. The respective PLS-DA model was not discriminative, having a Q2 = 0.306 (see Supplementary Fig. 3). The pairwise PLSDA analysis of the different groups, lead to discriminant models for Controls vs Patients (see [Fig. 1B] and Supplementary Fig. 4), Latent vs Patients and a no discriminant model in the case of Controls vs Latent (see Supplementary Figs. 5 and 6). For Controls vs Patients, the metabolites more important for the separation are mannose, hypoxanthine, glutamate, inosine and aspartate that are increased in Patients, while asparagine is decreased in this group [Fig. 1C]. For Latents vs Patients, the levels of hypoxantine, inosine, valine and fucose, citrate, creatine and fructose were significantly elevated but less abundant in Patients (Supplementary Fig. 5C).
Fig. 1.
Discrimination of TB Patients using quantified serum metabolites. (A) Principal Component Analysis score plot using serum metabolites of the three groups: Control, Patient and Latent. In the lower panels is presented the PLS-DA model for the Controls and Patients comparison; (B) score plot of the two first components (acc = 94%, R2 = 0,75, Q2 = 0.51) and (C) loadings plot with the metabolites colored by its variance importance in projection (VIP) in the first component. Node color changes from blue to red with increasing VIP. Metabolites with VIP value > 1.5 are depicted in the plot.
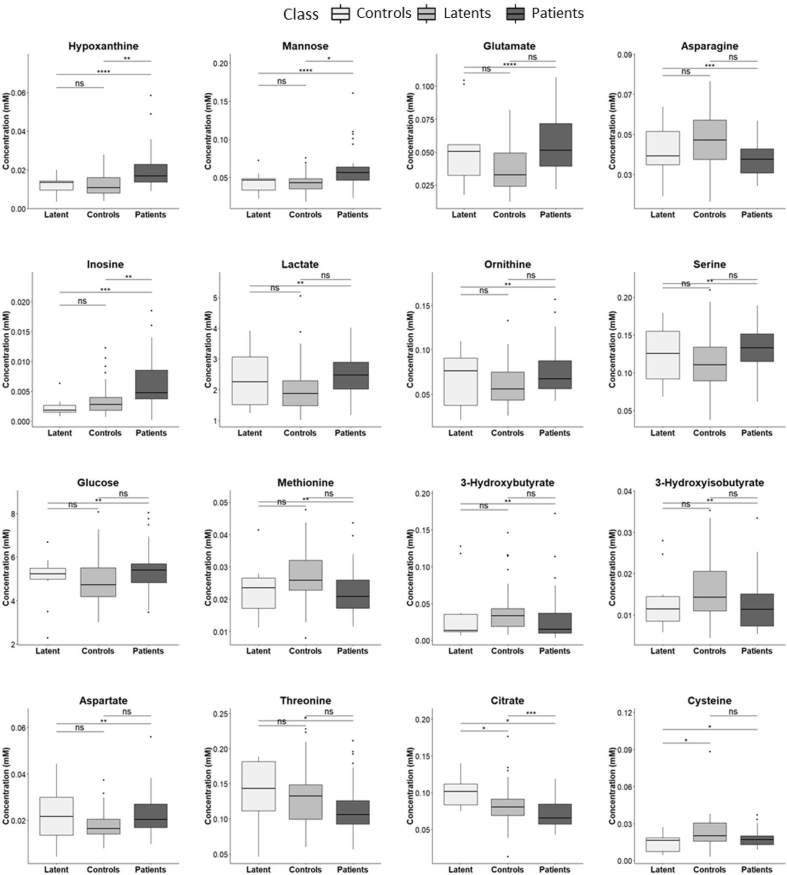
Most discriminant metabolites in the PLS-DA models for both pairwise comparisons were also found significantly different by univariate analysis ([Fig. 2] and Supplementary Table S2). For Controls versus Patients, besides mannose, hypoxanthine, glutamate, inosine and aspartate the levels of lactate, ornithine, glucose, serine and cysteine were increased in patients serum, while asparagine, 3-hydroxyisobutyrate, 3-hydroxybutyrate, citrate, methionine and threonine levels were decreased. Between Latent and Patients, beside the metabolites found significantly different by multivariate analysis, mannose and 2-aminobutyrate are increased in patients. Although no discrimination was obtained between Latents and Controls by supervised multivariate analysis, 10 discriminant metabolites were determined by the Wilcox test, three of them (acetate, malonate and cystine) only for this pairwise comparison. The levels of 2-aminobutyrate, cysteine, fucose and valine were lowest, while citrate, creatine and fructose (Supplementary Table S2) were highest in Latents.
Fig. 2.
Serum metabolite concentrations significantly different between Controls and Patients comparison. Boxplots from Wilcox test (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001).
Metabolic pathways altered in patients
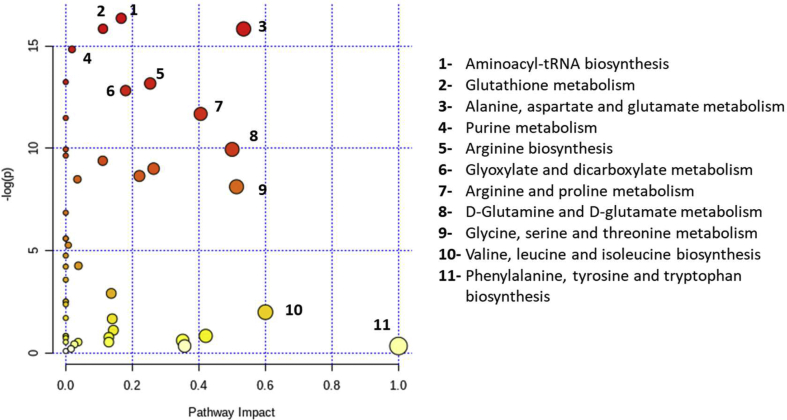
Concentrations determined for all metabolites were used to perform Pathway Analysis between Control and Patient groups, indicating what are the metabolic pathways affected by TB [Fig. 3]. Amino acids are among the metabolites which levels were more different between the two groups, then it is not unexpected that pathways related with amino acid metabolism are among the most altered, with special focus on those related with glutamate, a key metabolite in several pathways, as is the case of glutathione (GSH); alanine, aspartate and glutamate; arginine and proline or d-glutamine and d-glutamate metabolisms. Beside the alterations in amino-acid metabolism, other show also relevant changes, like purine metabolism, glyoxylate and dicarboxylate metabolism and aminoacyl-tRNA biosynthesis.
Fig. 3.
Amino acid metabolic pathways are among the most altered in TB patients. Differential pathway analysis between Controls and TB Patients using metabolite levels by MetaboAnalyst. Depicted pathways have negative logarithm of p-value higher than 10 (y axis) and pathway impact above 0, or pathway impact higher than 0.5 (x axis). Node color changes from white to red with decreasing p-value; and node radius correlates with pathway impact values. Pathways are ordered by their respective p-values.
Integrated analysis combining metabolomics profiles from Indian and Portuguese cohorts
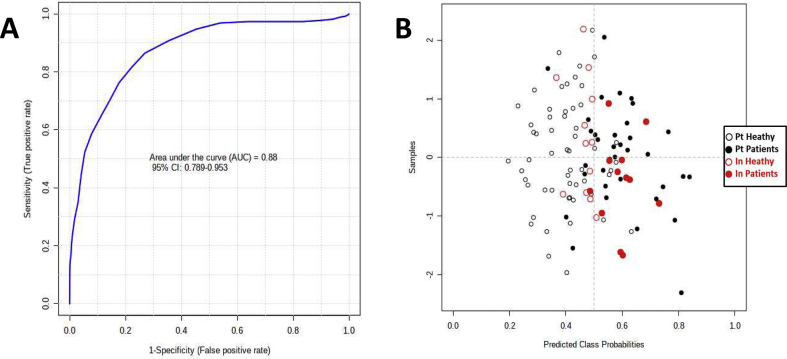
To validate our model, samples from a previous Indian cohort [13] were used. In this case, 12 samples from healthy individuals and 12 from TB patients (information on samples is depicted in Supplementary Table S1) were treated and analyzed following the same protocol used for the Portuguese cohort. Thirty three metabolites whose concentrations were found differential in this study or in Albors-Vaquer [13] were assigned to the NMR spectra. For the ROC curve were only selected the metabolites found differentially abundant both by uni- and multivariate analysis and with an area under the curve (AUC) > 0.6 (hypoxanthine, asparagine, mannose, aspartate, glutamate and inosine). ROC analysis using the Portuguese cohort and the combination of the above metabolites gives a good predictive value for a 95% confidence interval (CI = 0.79–0.95, AUC = 0.88, accuracy = 0.80, specificity = 0.88, sensitivity = 0.68) using a PLS algorithm with one latent variable [Fig. 4]. In the case of the Indian samples, the model predicts correctly 11 patients and 11 controls, for a total of 12 individuals per each group. Indian samples were predicted with an AUC and accuracy of 0.90 and 0.92 (95% CI) using the Portuguese cohort (sensitivity = 0.92, specificity = 0.92).
Fig. 4.
Receiver operating characteristic curve (ROC) curve and Indian samples prediction, considering hypoxanthine, asparagine, mannose, aspartate, glutamate and inosine levels. (A) ROC analysis using Controls and Patients from the Portuguese cohort. (B) Prediction of the samples from the Indian cohort. In black the Portuguese samples and in red the Indian samples, filled and outline circles for Patients and Controls, respectively.
Discussion
Characterization of the Portuguese cohort
A total of 37 patients were included in our cohort. From those 32% were diagnosed with EPTB, value unexpectedly high when compared with the percentage of this class of TB patients in Eastern Mediterranean countries in 2015 (23%) [23]. The distribution of patients by gender for both patient groups is in accordance with several studies [[15], [16], [17]]. Garcia-Rodriguez et al. [17] reported that the mean age for EPTB is higher than that for PTB, in our cohort 58% of EPTB patients are over 50 years, while 92% PTB patients have less than 50 years old. Leung et al. [24] concluded that obesity is associated with a lower risk of PTB in the older Hong Kong population. Consistently, in our study, only 4% of the PTB patients present a high Body Mass Index (BMI). By opposition, this level was found in 55% of the extra-pulmonary patients and in 21% of the control group. In this group, the majority of the individuals (73%) have a normal BMI. Several studies suggest the existence of an association between tobacco smoking and PTB [25], in particular related to the delay in diagnosis [26,27]. Fitly, in our PTB patients group, 74% were smokers or ex-smokers, contrasting with the absence of EPTB patients smoking habits. Although our cohort is not suitable for epidemiologic evaluations, it shares several similarities with published epidemiologic studies confirming its representativeness.
A metabolite biomarkers signature
Comparison of metabolites concentrations of Controls vs Patients by uni- and multivariate analysis found sixteen metabolites with significant differential amounts. Glucose, mannose, lactate, hypoxanthine, inosine, ornithine, glutamate, aspartate and serine are increased in Patients, while 3-hydroxyisobutyrate, 3-hydroxybutyrate, citrate, asparagine, methionine, threonine and cysteine are decreased. Interestingly, between Latent and Patients, for the metabolites significantly different hypoxanthine, mannose, inosine and citrate are common to the previous showing the same type of variation. Moreover, 2-aminobutyrate, fucose and valine are increased in patients and 2-fructose and creatine are decreased. These results suggest the existence of some similar affected biological/metabolic processes in individuals with active and latent TB infection. By univariate analysis, seven of the discriminant metabolites between patients and each healthy group were also found discriminant between both healthy groups. Creatine, fructose, lactate, citrate are increased in Latents and 2-aminobutyrate, fucose, valine and cysteine are decreased. Additionally, acetate is increased and cystine and malonate are decreased.
Similar variations (metabolite increased/decreased amount) of ten metabolites were reported for metabolome comparison between TB patients and healthy controls. In one study for citrate, hypoxanthine, inosine and ornithine, in two studies for asparagine, aspartate, cysteine, mannose and threonine, in three studies for glutamate and in five for lactate differential amounts were determined [13,[28], [29], [30], [31], [32], [33], [34]]. Moreover, glucose was found increased in infected mice and guinea-pigs relative to non-infected animals [35,36]. For the comparison between TB patients and Latents it was determined by Weiner et al. [28] changes similar to our data for creatine, hypoxanthine, inosine, mannose and citrate amounts. This last was also reported by Albors-Vaquer et al. [13]. Metabolites differential amounts between Controls and Latents were evaluated in few publications [13,28,29]. Only for acetate a match with our results was found [13].
Functional analysis of potential biomarkers
Considering the defined acceptance criteria for Pathway Analysis, thirty metabolites, including eighteen amino acids, were assigned to eleven pathways as detailed in [Fig. 3]. Metabolomic studies reporting changes in amino acid metabolism in TB infection are ubiquitously described in the literature [28,31,33,35,37] although some discrepancies were found for six proteogenic amino acids, reasonable consistency was determined for asparagine, aspartate, cysteine, glutamate and threonine. In our work, only the proteogenic amino acids aspartate, glutamate and serine have an increased concentration in sera of TB patients. These results reproduce those previously reported in Refs. [28,29,31,33] and agree with the characteristic protein turnover pattern previously described. TB infection is a chronic wasting disease that, by opposition to undernutrition, results in a diversion of ingested amino acids away from utilization for protein synthesis and thus towards oxidation with consequent loss from the body protein pool, suggesting the occurrence of an “anabolic block” [38]. Additionally, it was found in our study, as previously in Weiner et al. [28], that the ratio glutamine/glutamate is lower in TB patients compared with controls, which also indicates a risk of loss of body mass [39].
The excess amount of free amino acids, in contrast with glucose and fatty acids, cannot be stored or excreted. Rather the α-amino group of the amino acids is removed and the resulting carbon skeleton is converted into acetyl-CoA, acetoacetyl-CoA, pyruvate, or one of the intermediates of the citric acid cycle. It follows that amino acids can originate glucose, fatty acids and ketone bodies. In our results, four amino acids decreased in TB patients corrocorate an increase in amino acid oxidation during TB infection. The surplus of amino acids is also converted into urea. In fact, ornithine, an intermediate of the urea cycle, has its concentration significantly increased in TB patients. 3-hydroxybutyrate and 3-hydroxyisobutyrate are ketone bodies resulting from partial-degradation products of branched-chain amino acids. Besides, 3-hydroxybutyrate synthesis from acetoacetate also occurs in human liver during a fasting state [40], we observed its decrease, together with that of 3-hydroxyisobutyrate, in Patients compared to Controls. However, Weiner et al. [10] also detected a decrease of 3-hydroxybutyrate at the onset of the disease. Surprisingly, comparing Patients with Latents, valine, the only increased proteogenic amino acid, is the metabolite with higher increment.
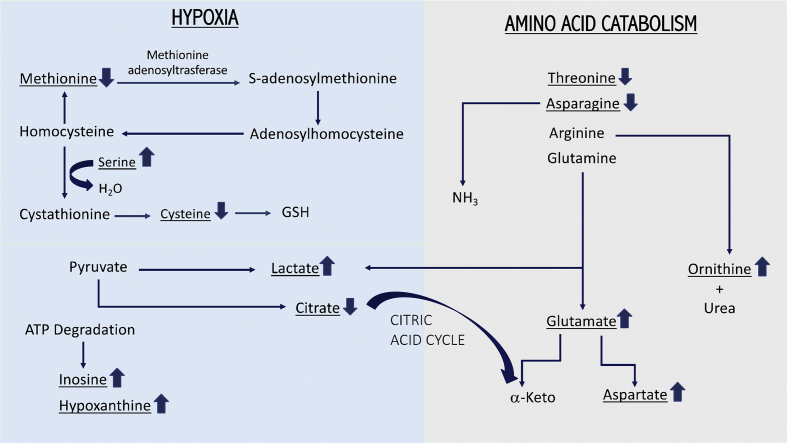
On the other hand, changes in amino acid concentrations observed between healthy and infected individuals can also be explained based on the virulence mechanisms of Mtb and host immunometabolism [9]. This intracellular pathogen thrives inside macrophages, where it has to face an acidic environment with limited availability of essential nutrients. It was hypothesized that asparagine retrieved by Mtb or indirectly from the host could be used as a nitrogen source. Gouzy et al. [41] reported that Mtb employs an asparagine transporter and a secreted asparaginase to assimilate nitrogen and resist acidic stress through asparagine hydrolysis and ammonia release. In this context, results from a recent metabolomics study in Mtb under microbicidal stresses suggested that high levels of ammonia were released as an adaptive response to acidic stress due to increased flux through l-asparaginase rather than urease activity [42]. Additionally, the hydrolysis of arginine to ornithine and urea by macrophages expressed arginase in hypoxic and necrotic regions of TB granulomas, plays a crucial role in controlling both Mtb growth and TB pathology [43] and contributes for the increase of ornithine levels in sera. In fact, our results show a decrease in asparagine and an increase in ornithine concentrations in TB patients sera. Moreover, glutamine can be converted to α-ketoglutarate through glutamate to fuel the tricarboxylic acid cycle with carbon. Koeken et al. [44] concluded that this pathway is implicated in effective host response against Mtb, since involved genes are differentially expressed in Mtb-infected macrophages and blood transcriptomic profiles of individuals with latent or active TB infection. Else than glutamate, glutaminolysis generates aspartate and lactate, metabolites we found increased in Patients. Zhou et al. [33], Zhou et al. [32], Weiner et al. [28], Jain et al. [34] and Albors-Vaquer et al. [13] have also reported an increase of lactate levels in sera of TB patients, that is consistent with increased anaerobic glycolysis. It is known that TB infection induces granulomatous inflammation in the lung with central necrosis and tissue hypoxia [45]. Therefore, the accumulation of lactate could be an index of tissue hypoxia and extent of necrosis as the infection progresses. Shi et al. [46] hypothesized that transition from acute to chronic TB infection in mouse lungs is accompanied by a metabolic shift from oxidative phosphorylation toward enhanced glucose uptake, glycolysis and formation and secretion of lactate. This pattern, known as the Warburg effect, is characteristic of cancer cell metabolism and is facilitated by monocarboxylate transporters. These transmembrane proteins allow maintaining the optimal concentration of pyruvate, the substrate of lactate dehydrogenase, while exporting lactate to keep cytosolic pH around physiological levels [46]. This assumption is consistent with the fact that although lactate is systematically found increased in TB patients blood, pyruvate is less frequently detected. Moreover, the association of the Warburg effect to TB infection explains the reduction of citrate levels observed in our study and by Weiner et al. [28], since pyruvate is mainly diverted to lactate production. Other metabolites, as inosine and hypoxanthine, also found increased in TB active and latent infection here, by Weiner et al. [28] and Huang et al. [30], have been indicated as biomarkers of hypoxia associated with cardiac ischemia [47]. At the onset of hypoxia, tissues are rapidly depleted of ATP leading to the accumulation of its catabolic by-products, namely inosine and hypoxanthine. Furthermore, it is known that methionine adenosyltransferase, the enzyme that converts methionine to S-adenosylmethionine and subsequently to cysteine, is down regulated by hypoxia [48]. Furthermore, cysteine could be consumed to form GSH to overcome the oxidative stress produced by Mtb. Allen et al., have shown that GSH has contributed in inhibiting the growth of intracellular Mtb through bacteriostatic mechanisms [49]. The pathway that connects these several intermediates is depicted in [Fig. 5]. In opposition the levels of serine, which condensates with homocysteine originating cystathionine, an intermediate for cysteine synthesis, are increased. These amino acid concentrations differences match with those observed in our results and are corroborated by Weiner et al. [10,28] and Albors-Vaquer et al. [13].
Fig. 5.
Biological relevance of biomarkers. Metabolites with differential amounts are underlined. The arrows on the right side of the name define their increased or decreased levels in Tuberculosis infection.
In accordance with Weiner et al. [10,28], we detected an increase of mannose in sera of TB patients and Latents. This aldohexose is a critical component of the Mtb cell wall glycan lipoarabinomannan. This finding together with an observed increase of phosphatidylinositol and of the glycolipid trehalose-6-mycolate, a constituent of the mycolic acid layer of the cell wall membranes [31] are consistent, suggesting they are derived from cell walls of the infecting Mtb.
Two monosaccharides, fructose and fucose, although not showing significant differences between Patients and Controls, are, respectively decreased and increased in Patients compared to Latents, being fructose the metabolite with higher reduction. Although, we could not find an explanation for these results, it is interesting to point that levels of fucose in plasma-derived IgG are lower in individuals with latent than in those with active TB [50].
Final remarks
This study results are according with the WHO Target Product Profile recommendations for a triage test to rule-out active TB [51], since it is the first to report a blood metabolite signature with a minimal target sensitivity and specificity for the validation of an independent cohort. The determination of this metabolite signature can be translated to a point-of-care inexpensive assay.
Metabolites identified with significant different levels in Tb infection using metabolomic approaches [10,13,[28], [29], [30], [31], [32], [33], [34], [35], [36], [37],52], besides showing several similarities also evidence some discrepancies. These are either related with the set of metabolites identified, either with the type of variation, concentration increase or decrease. These disparities can be easily explained based on the diversity of the protocols used for sample treatment, methodologies used for metabolite quantification and characteristics of the cohorts. Anyhow, functional metabolomics analysis has led to important common conclusions.
Variations in serum metabolome related to TB infection with higher relevance are associated to amino acid catabolism pathways, hypoxia and Mtb specific constituents, with some of the processes superimposing to others for some metabolites [Fig. 5]. Protein degradation associated to TB infection generates a disturbance in sera levels of free amino acids. Some were consistently found increased and others with lower levels in Patients. Increase of the concentration levels of the products related with amino acid catabolism (acetate, acetoacetate, pyruvate, urea or ornithine) was unanimous. Due to the known hypoxia conditions induced by granulomatous inflammation in the lungs, it was possible to explain the accumulation of several metabolites associated with this condition (lactate, inosine, hypoxanthine and serine) and the depletion of others (cysteine, methionine and citrate). Compounds found in sera and probably originated from the mycobacterium, due to its biological processes or specific biomolecular composition, if constantly detected should be considered relevant biomarkers for TB diagnosis. This is the case for mannose.
Based on others and our results, namely the validation of a set of six discovered metabolites by an independent cohort, all of them related with the metabolic processes associated with TB infection, our proposed signature for TB diagnosis includes inosine, hypoxanthine, mannose, asparagine, aspartate and glutamate. As previously stated [28], most likely a specific signature for TB diagnosis should include several metabolites.
At this moment, a set of potential candidates for TB diagnosis biomarkers is already available. The next step will be to extend the evaluation of their specificity and selectivity for TB, namely in relation to other pulmonary infectious diseases. Target experimental approaches will be the better choice at this stage for the validation of these biomarkers. They should be quantified for an extensive cohort, ideally including a diversified population selected based on clear standardized criteria.
Funding
This research was funded to the project “TBomics - an OMICS approach for diagnosing tuberculosis”, New INDIGO Partnership Programme on Biotechnology applied to Human Health (INDIGO-DBT2-062), ERA-NET Project supported by Fundação para a Ciência e Tecnologia (FCT), Portugal and Department of Biotechnology, Govt. of India. Short–term European Molecular Biology Organization fellowship (STF–7009) to AR covering for a stay at ITQB NOVA, Portugal is acknowledged.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgments
To the nurse teams at CDPs Almada-Seixal and Venda Nova.
Footnotes
Peer review under responsibility of Chang Gung University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bj.2021.07.006.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.World Health Organization. Global tuberculosis report 2019. https://www.who.int/publications/i/item/9789241565714/; 2019 [Accessed 30 July 2021].
- 2.Goletti D., Petruccioli E., Joosten S.A., Ottenhoff T.H.M. Tuberculosis biomarkers: from diagnosis to protection. Infect Dis Rep. 2016;8:6568. doi: 10.4081/idr.2016.6568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tucci P., González-Sapienza G., Marin M. Pathogen-derived biomarkers for active tuberculosis diagnosis. Front Microbiol. 2014;5:549. doi: 10.3389/fmicb.2014.00549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Acharya B., Acharya A., Gautam S., Ghimire S.P., Mishra G., Parajuli N., et al. Advances in diagnosis of Tuberculosis: an update into molecular diagnosis of Mycobacterium tuberculosis. Mol Biol Rep. 2020;47:4065–4075. doi: 10.1007/s11033-020-05413-7. [DOI] [PubMed] [Google Scholar]
- 5.Gardiner J.L., Karp C.L. Transformative tools for tackling tuberculosis. J Exp Med. 2015;212:1759–1769. doi: 10.1084/jem.20151468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bulterys M.A., Wagner B., Redard-Jacot M., Suresh A., Pollock N.R., Moreau E., et al. Point-of-care urine LAM tests for tuberculosis diagnosis: a status update. J Clin Med. 2019;9:111. doi: 10.3390/jcm9010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yerlikaya S., Broger T., MacLean E., Pai M., Denkinger C.M. A tuberculosis biomarker database: the key to novel TB diagnostics. Int J Infect Dis. 2017;56:253–257. doi: 10.1016/j.ijid.2017.01.025. [DOI] [PubMed] [Google Scholar]
- 8.Preez I.D., Luies L., Loots D.T. Metabolomics biomarkers for tuberculosis diagnostics: current status and future objectives. Biomark Med. 2017;11:179–194. doi: 10.2217/bmm-2016-0287. [DOI] [PubMed] [Google Scholar]
- 9.Bisht D., Sharma D., Sharma D., Singh R., Gupta V.K. Recent insights into Mycobacterium tuberculosis through proteomics and implications for the clinic. Expert Rev Proteomics. 2019;16:443–456. doi: 10.1080/14789450.2019.1608185. [DOI] [PubMed] [Google Scholar]
- 10.Weiner 3rd, J., Maertzdorf J., Sutherland J.S., Duffy F.J., Thompson E., Suliman S., et al. Metabolite changes in blood predict the onset of tuberculosis. Nat Commun. 2018;9:5208. doi: 10.1038/s41467-018-07635-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haas C.T., Roe J.K., Pollara G., Mehta M., Noursadeghi M. Diagnostic “omics” for active tuberculosis. BMC Med. 2016;14:37. doi: 10.1186/s12916-016-0583-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang H., Han Y.S., Chen J., Shi L.Y., Wei L.L., Jiang T.T., et al. Highlight article: the novel potential biomarkers for multidrug-resistance tuberculosis using UPLC-Q-TOF-MS. Exp Biol Med. 2020;245:501–511. doi: 10.1177/1535370220903464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albors-Vaquer A., Rizvi A., Matzapetakis M., Lamosa P., Coelho A., Patel A.B., et al. Active and prospective latent tuberculosis are associated with different metabolomic profiles: clinical potential forthe identification of rapid and non-invasive biomarkers. Emerg Microbes Infect. 2020;9:1131–1139. doi: 10.1080/22221751.2020.1760734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chong J., Wishart D.S., Xia J. Using metaboanalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr Protoc Bioinformatics. 2019;68:e86. doi: 10.1002/cpbi.86. [DOI] [PubMed] [Google Scholar]
- 15.Guler S.A., Bozkus F., Inci M.F., Kokoglu O.F., Ucmak H., Ozden S., et al. Evaluation of pulmonary and extrapulmonary tuberculosis in immunocompetent adults: a retrospective case series analysis. Med Princ Pract. 2015;24:75–79. doi: 10.1159/000365511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sunnetcioglu A., Sunnetcioglu M., Binici I., Baran A.I., Karahocagil M.K., Saydan M.R. Comparative analysis of pulmonary and extrapulmonary tuberculosis of 411 cases. Ann Clin Microbiol Antimicrob. 2015;14:34. doi: 10.1186/s12941-015-0092-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.García-Rodríguez J.F., Álvarez-Díaz H., Lorenzo-García M.V., Mariño-Callejo A., Fernández-Rial Á., Sesma-Sánchez P. Extrapulmonary tuberculosis: epidemiology and risk factors. Enferm Infecc Microbiol Clín. 2011;29:502–509. doi: 10.1016/j.eimc.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 18.Bates M.N., Khalakdina A., Pai M., Chang L., Lessa F., Smith K.R. Risk of tuberculosis from exposure to tobacco smoke: a systematic review and meta-analysis. Arch Intern Med. 2007;167:335–342. doi: 10.1001/archinte.167.4.335. [DOI] [PubMed] [Google Scholar]
- 19.Casha A.R., Scarci M. The link between tuberculosis and body mass index. J Thorac Dis. 2017;9:E301–E303. doi: 10.21037/jtd.2017.03.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daykin C.A., Foxall P.J., Connor S.C., Lindon J.C., Nicholson J.K. The comparison of plasma deproteinization methods for the detection of low-molecular-weight metabolites by (1)H nuclear magnetic resonance spectroscopy. Anal Biochem. 2002;304:220–230. doi: 10.1006/abio.2002.5637. [DOI] [PubMed] [Google Scholar]
- 21.Tiziani S., Emwas A.H., Lodi A., Ludwig C., Bunce C.M., Viant M.R., et al. Optimized metabolite extraction from blood serum for 1H nuclear magnetic resonance spectroscopy. Anal Biochem. 2008;377:16–23. doi: 10.1016/j.ab.2008.01.037. [DOI] [PubMed] [Google Scholar]
- 22.Nagana Gowda G.A., Raftery D. Quantitating metabolites in protein precipitated serum using NMR spectroscopy. Anal Chem. 2014;86:5433–5440. doi: 10.1021/ac5005103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.World Health Organization. Global tuberculosis report 2016. geneva, switzerland, https://www.afro.who.int/publications/global-tuberculosis-report-2016/; 2016 [accessed 30 July 2021].
- 24.Leung C.C., Lam T.H., Chan W.M., Yew W.W., Ho K.S., Leung G., et al. Lower risk of tuberculosis in obesity. Arch Intern Med. 2007;167:1297–1304. doi: 10.1001/archinte.167.12.1297. [DOI] [PubMed] [Google Scholar]
- 25.Kolappan C., Gopi P.G. Tobacco smoking and pulmonary tuberculosis. Thorax. 2002;57:964–966. doi: 10.1136/thorax.57.11.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aziza R., Sanae H., Hatim K., Bourkadi J.E. Pulmonary tuberculosis specificities in smokers. Egypt J Chest Dis Tuberc. 2015;64:929–932. [Google Scholar]
- 27.Afanas’ev I.V., Ivanoskiĭ V.B. [Smoking and tuberculosis] Probl Tuberk. 1979:45–48. [PubMed] [Google Scholar]
- 28.Weiner 3rd, J., Parida S.K., Maertzdorf J., Black G.F., Repsilber D., Telaar A., et al. Biomarkers of inflammation, immunosuppression and stress with active disease are revealed by metabolomic profiling of tuberculosis patients. PloS One. 2012;7 doi: 10.1371/journal.pone.0040221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho Y., Park Y., Sim B., Kim J., Lee H., Cho S.N., et al. Identification of serum biomarkers for active pulmonary tuberculosis using a targeted metabolomics approach. Sci Rep. 2020;10:3825. doi: 10.1038/s41598-020-60669-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang H., Shi L.Y., Wei L.L., Han Y.S., Yi W.J., Pan Z.W., et al. Plasma metabolites Xanthine, 4-Pyridoxate, and d-glutamic acid as novel potential biomarkers for pulmonary tuberculosis. Clin Chim Acta. 2019;498:135–142. doi: 10.1016/j.cca.2019.08.017. [DOI] [PubMed] [Google Scholar]
- 31.Frediani J.K., Jones D.P., Tukvadze N., Uppal K., Sanikidze E., Kipiani M., et al. Plasma metabolomics in human pulmonary tuberculosis disease: a pilot study. PloS One. 2014;9 doi: 10.1371/journal.pone.0108854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou A., Ni J., Xu Z., Wang Y., Zhang H., Wu W., et al. Metabolomics specificity of tuberculosis plasma revealed by (1)H NMR spectroscopy. Tuberculosis (Edinb) 2015;95:294–302. doi: 10.1016/j.tube.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou A., Ni J., Xu Z., Wang Y., Lu S., Sha W., et al. Application of (1)h NMR spectroscopy-based metabolomics to sera of tuberculosis patients. J Proteome Res. 2013;12:4642–4649. doi: 10.1021/pr4007359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jain A., Kumar A., Singh H., Rai M.K., Chaturvedi S., Guleria A., et al. 114 Nuclear magnetic resonance (NMR) based serum metabolomics in sarcoidosis and tuberculosis: search for a biomarker. Rheumatology. 2018;57 lssue suppl_3:key075.338. [Google Scholar]
- 35.Shin J.H., Yang J.Y., Jeon B.Y., Yoon Y.J., Cho S.N., Kang Y.H., et al. (1)H NMR-based metabolomic profiling in mice infected with Mycobacterium tuberculosis. J Proteome Res. 2011;10:2238–2247. doi: 10.1021/pr101054m. [DOI] [PubMed] [Google Scholar]
- 36.Somashekar B.S., Amin A.G., Tripathi P., MacKinnon N., Rithner C.D., Shanley C.A., et al. Metabolomic signatures in Guinea pigs infected with epidemic-associated W-Beijing strains of Mycobacterium tuberculosis. J Proteome Res. 2012;11:4873–4884. doi: 10.1021/pr300345x. [DOI] [PubMed] [Google Scholar]
- 37.Sun L., Li J.Q., Ren N., Qi H., Dong F., Xiao J., et al. Utility of novel plasma metabolic markers in the diagnosis of pediatric tuberculosis: a classification and regression tree analysis approach. J Proteome Res. 2016;15:3118–3125. doi: 10.1021/acs.jproteome.6b00228. [DOI] [PubMed] [Google Scholar]
- 38.Macallan D.C., McNurlan M.A., Kurpad A.V., de Souza G., Shetty P.S., Calder A.G., et al. Whole body protein metabolism in human pulmonary tuberculosis and undernutrition: evidence for anabolic block in tuberculosis. Clin Sci. 1998;94:321–331. doi: 10.1042/cs0940321. [DOI] [PubMed] [Google Scholar]
- 39.Kinscherf R., Hack V., Fischbach T., Friedmann B., Weiss C., Edler L., et al. Low plasma glutamine in combination with high glutamate levels indicate risk for loss of body cell mass in healthy individuals: the effect of N-acetyl-cysteine. J Mol Med. 1996;74:393–400. doi: 10.1007/BF00210633. [DOI] [PubMed] [Google Scholar]
- 40.Newman J.C., Verdin E. β-hydroxybutyrate: much more than a metabolite. Diabetes Res Clin Pract. 2014;106:173–181. doi: 10.1016/j.diabres.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gouzy A., Larrouy-Maumus G., Bottai D., Levillain F., Dumas A., Wallach J.B., et al. Mycobacterium tuberculosis exploits asparagine to assimilate nitrogen and resist acid stress during infection. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1003928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rizvi A., Shankar A., Chatterjee A., More T.H., Bose T., Dutta A., et al. Rewiring of metabolic network in Mycobacterium tuberculosis during adaptation to different stresses. Front Microbiol. 2019;10:2417. doi: 10.3389/fmicb.2019.02417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duque-Correa M.A., Kühl A.A., Rodriguez P.C., Zedler U., Schommer-Leitner S., Rao M., et al. Macrophage arginase-1 controls bacterial growth and pathology in hypoxic tuberculosis granulomas. Proc Natl Acad Sci USA. 2014;111:E4024–E4032. doi: 10.1073/pnas.1408839111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koeken V.A.C.M., Lachmandas E., Riza A., Matzaraki V., Li Y., Kumar V., et al. Role of glutamine metabolism in host defense against Mycobacterium tuberculosis infection. J Infect Dis. 2019;219:1662–1670. doi: 10.1093/infdis/jiy709. [DOI] [PubMed] [Google Scholar]
- 45.Via L.E., Lin P.L., Ray S.M., Carrillo J., Allen S.S., Eum S.Y., et al. Tuberculous granulomas are hypoxic in Guinea pigs, rabbits, and nonhuman primates. Infect Immun. 2008;76:2333–2340. doi: 10.1128/IAI.01515-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi L., Salamon H., Eugenin E.A., Pine R., Cooper A., Gennaro M.L. Infection with Mycobacterium tuberculosis induces the Warburg effect in mouse lungs. Sci Rep. 2015;5:18176. doi: 10.1038/srep18176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Farthing D.E., Farthing C.A., Xi L. Inosine and hypoxanthine as novel biomarkers for cardiac ischemia: from bench to point-of-care. Exp Biol Med (Maywood) 2015;240:821–831. doi: 10.1177/1535370215584931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Avila M.A., Carretero M.V., Rodriguez E.N., Mato J.M. Regulation by hypoxia of methionine adenosyltransferase activity and gene expression in rat hepatocytes. Gastroenterology. 1998;114:364–371. doi: 10.1016/s0016-5085(98)70489-5. [DOI] [PubMed] [Google Scholar]
- 49.Allen M., Bailey C., Cahatol I., Dodge L., Yim J., Kassissa C., et al. Mechanisms of control of Mycobacterium tuberculosis by NK cells: role of glutathione. Front Immunol. 2015;6:508. doi: 10.3389/fimmu.2015.00508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu L.L., Chung A.W., Rosebrock T.R., Ghebremichael M., Yu W.H., Grace P.S., et al. A functional role for antibodies in tuberculosis. Cell. 2016;167:433–443. doi: 10.1016/j.cell.2016.08.072. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.MacLean E., Broger T., Yerlikaya S., Fernandez-Carballo B.L., Pai M., Denkinger C.M. A systematic review of biomarkers to detect active tuberculosis. Nat Microbiol. 2019;4:748–758. doi: 10.1038/s41564-019-0380-2. Erratum in: Nat Microbiol 2019 Apr:30804546. [DOI] [PubMed] [Google Scholar]
- 52.Che N., Cheng J., Li H., Zhang Z., Zhang X., Ding Z., et al. Decreased serum 5-oxoproline in TB patients is associated with pathological damage of the lung. Clin Chim Acta. 2013;423:5–9. doi: 10.1016/j.cca.2013.04.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.