Abstract
Preproinsulin entry into the endoplasmic reticulum yields proinsulin, and its subsequent delivery to the distal secretory pathway leads to processing, storage, and secretion of mature insulin. Multiple groups have reported that treatment of pancreatic beta cell lines, rodent pancreatic islets, or human islets with proteasome inhibitors leads to diminished proinsulin and insulin protein levels, diminished glucose-stimulated insulin secretion, and changes in beta-cell gene expression that ultimately lead to beta-cell death. However, these studies have mostly examined treatment times far beyond that needed to achieve acute proteasomal inhibition. Here, we report that although proteasomal inhibition immediately downregulates new proinsulin biosynthesis, it nevertheless acutely increases beta-cell proinsulin levels in pancreatic beta cell lines, rodent pancreatic islets, and human islets, indicating rescue of a pool of recently synthesized WT INS gene product that would otherwise be routed to proteasomal disposal. Our pharmacological evidence suggests that this disposal most likely reflects ongoing endoplasmic reticulum–associated protein degradation. However, we found that within 60 min after proteasomal inhibition, intracellular proinsulin levels begin to fall in conjunction with increased phosphorylation of eukaryotic initiation factor 2 alpha, which can be inhibited by blocking the general control nonderepressible 2 kinase. Together, these data demonstrate that a meaningful subfraction of newly synthesized INS gene product undergoes rapid proteasomal disposal. We propose that free amino acids derived from proteasomal proteolysis may potentially participate in suppressing general control nonderepressible 2 kinase activity to maintain ongoing proinsulin biosynthesis.
Keywords: insulin production, endoplasmic reticulum, secretory pathway, cell stress, ERAD
Abbreviations: CHX, cycloheximide; DMEM, Dulbecco's modified Eagle's medium; eIF2α, eukaryotic initiation factor 2 alpha; ER, endoplasmic reticulum; ERAD, ER-associated degradation; FBS, fetal bovine serum; GCN2, general control nonderepressible 2; GSIS, glucose-stimulated insulin secretion; VCP, valosin-containing protein
Preproinsulin, the initial INS gene translation product in pancreatic β-cells, is the single most-expressed gene product in pancreatic β-cells (1). Preproinsulin is an evanescent protein (2), most of which has been reported to undergo turnover with an estimated half-life of ∼1 min (3). Preproinsulin turnover may represent two distinct activities: (a) predominant translocation across the membrane of the endoplasmic reticulum (ER) with successful conversion to proinsulin (4) or (b) failed translocation of a subset of molecules across the ER membrane (5, 6, 7, 8, 9) with subsequent degradation (10). The extent to which WT preproinsulin in WT β-cells is routed to proteasomal degradation (e.g., via pre-emptive ER quality control (11, 12, 13)) has, to our knowledge, never been studied.
Beyond preproinsulin, proinsulin, contained within the ER lumen (14), is the most abundantly detected newly made translation product in β-cells. Previous studies have suggested that the life span of the entire population of β-cell proinsulin molecules is on the order of 4 h (15). A subset of proinsulin molecules might undergo ER-associated degradation (ERAD), which is a process that ultimately involves substrate delivery for proteasomal disposal (16). Indeed, ERAD function is now recognized as essential to the maintenance of the differentiated state of β-cells (17). Nevertheless, ERAD of proinsulin has primarily been identified for misfolded mutant proinsulin molecules (18, 19, 20, 21, 22, 23) that are responsible for the syndrome of mutant INS gene–induced diabetes of youth (14, 24); much less is known about proteasomal disposal of WT proinsulin in WT β-cells (25).
Several groups working in pancreatic β-cell lines have reported that proteasome inhibition for 12 to 24 h leads to diminished β-cell insulin content and glucose-stimulated insulin secretion (GSIS) (26, 27); these effects have been observed even within 6 h (28). Indeed, extended proteasomal inhibition causing impaired GSIS has been replicated in human islets and is accompanied by changes in β-cell gene expression (29). Thereafter, continued proteasomal inhibition in both β-cell lines and human islets leads to β-cell death (30, 31).
Shorter treatment of mouse pancreatic islets with lactacystin (for 2 h) has been reported to cause new synthesis of proinsulin to decline (measured by metabolic pulse labeling with radioactive amino acid (32)), presumably accounting for the decreased insulin content and GSIS noted previously. Confusingly, however, immunoreactive intracellular proinsulin does not appear to be decreased after short-term treatment (25), a period that does not involve any change of insulin mRNA level (32)—indeed, GSIS might even increase upon 1 to 2 h of proteasome inhibition (33, 34). As proinsulin content is a key factor in insulin biogenesis (14), the extent to which ongoing proteasome activity in unstressed WT β-cells contributes to proinsulin content is an important consideration of which little is known. In this study, we have set out to clarify the acute effects of proteasome inhibition on proinsulin content in pancreatic β-cells.
Results
Effect of proteasome inhibition on proinsulin biosynthesis in pancreatic β-cells
Although there are many studies treating pancreatic β-cells with MG132 for hours (or even days), the primary action of MG132 to inhibit proteasomes can be seen within 5 min (35). This has led us to consider the possibility of time-dependent responses in which the earliest impact of proteasome inhibition may represent that for degradative substrates, whereas later responses may reflect secondary effects brought about by activation or inhibition of downstream signaling pathways. Given the short half-life of proinsulin (and even shorter for preproinsulin), it seemed appropriate to begin by examining the impact of acute proteasome inhibition on proinsulin biosynthesis as well as steady-state level. In our experiments, a fresh medium change was applied to all β-cell lines or islets just a few hours before each experiment, in order to ensure that in all cases β-cells had ample access to fresh nutrients, thereby eliminating this as a potential variable.
To examine proinsulin biosynthesis, the mouse pancreatic β-cell line, Min6, was pulse labeled with 35S-amino acids in the presence or the absence of MG132 (10 μM) for 10 min, and nascent proinsulin was immunoprecipitated and resolved by SDS-PAGE and phosphorimaging (Fig. 1A). Proteasomal inhibition significantly decreased nascent proinsulin biosynthesis in Min6 cells (normalized to trichloroacetic acid–precipitable counts per minute, Fig. 1, A and B), and essentially identical findings were obtained in the rat pancreatic β-cell line, INS1E (Fig. 1C). Moreover, this effect was apparent by immunofluorescence after washout of cycloheximide (CHX) to visually examine newly synthesized proinsulin molecules (Fig. S1E). The molecular basis for the immediate inhibition of proinsulin biosynthesis upon addition of proteasome inhibitor is currently unknown, but it is not readily explained by general control nonderepressible 2 (GCN2) activation (as it could not be reversed by coincubation with a GCN2 inhibitor [Fig. 1D; discussed later]).
Figure 1.
Effect of acute proteasome inhibition on proinsulin biosynthesis in pancreatic β-cells.A, Min6 cells were pulse labeled with 35S-Met/Cys for 10 min in the absence or the presence of 10 μM MG132. Cell lysates (normalized to TCA-precipitable counts) were subjected to immunoprecipitation with anti-insulin antibody and analyzed by reducing SDS-PAGE and phosphorimaging. B, quantitation (mean ± SD) of proinsulin synthesis normalized to TCA-precipitable counts is shown from replicate experiments like those shown in A (Student's t test, ∗∗p < 0.01). C, quantitation (mean ± SD) of newly synthesized proinsulin is shown from replicate experiments in INS1E cells, following the same protocol as that shown in A (Student's t test, ∗p < 0.05). D, INS1E cells in the presence of MG132 were pulse labeled with 35S-Met/Cys for 10 min ± GCN2 inhibitor (HY-112654, 5 μM). Cell lysates (normalized to TCA-precipitable counts) were subjected to immunoprecipitation with anti-insulin and analyzed by by reducing SDS-PAGE and phosphorimaging. TCA, trichloroacetic acid.
Effect of proteasome inhibition on proinsulin protein level in pancreatic β-cells
MG132 inhibits the proteasomal degradation of ubiquitylated protein substrates; indeed, Min6 cells treated with 10 μM MG132 did accumulate ubiquitylated proteins as detected in transfected cells that express Ub-FLAG (Fig. 2A) (and similar accumulation was observed upon treatment with the valosin-containing protein (VCP) inhibitor CB-5083 [Fig. 2A] (36, 37, 38)). Moreover, some of the ubiquitylated protein in MG132-treated Min6 cells is likely to include proinsulin, as detected by proinsulin immunoblotting after immunoprecipitation of FLAG-containing proteins (Fig. 2B). Interestingly, despite the immediate drop in proinsulin biosynthesis (Fig. 1), total proinsulin protein level in pancreatic β-cells promptly increased upon MG132 addition (Fig. 2C, quantified in Fig. 2D), and similar accumulation was observed upon VCP inhibition with CB-5083 (Fig. 2C, quantified in Fig. 2D). As ERAD involves retrotranslocation from the ER lumen and extraction from ER membrane-to-cytosol via the VCP/p97 AAA-ATPase for delivery to proteasomes (39), these data do support the notion that a fraction of WT (pre)proinsulin undergoes ERAD (16, 18, 22, 23).
Figure 2.
Acute proteasome inhibition increases proinsulin protein level in pancreatic β-cells.A, Min6 cells were transfected to express FLAG-tagged ubiquitin, and the transfected cells trypsinized and replated into different wells. At 48 h post-transfection, cells were incubated in fresh medium with either 10 μM MG132, 10 μM CB-5083, or vehicle (DMSO) alone for 4 h and lysed with SDS-free lysis buffer containing 25 mM NEM. Cell lysates (6% of total) were analyzed by reducing SDS-PAGE and immunoblotting with anti-FLAG. Actin (below) is a loading control. B, the remaining cell lysates (after sample removal for analysis as in A) were immunoprecipitated with anti-FLAG-Ub antibody overnight and analyzed by reducing SDS-PAGE and immunoblotting with anti-proinsulin. C, INS1E cells were incubated in fresh RPMI1640 medium (11.1 mM glucose) for 3.5 h plus either 10 μM MG132, 10 μM VCP inhibitor (CB-5083), or vehicle (DMSO) alone for an additional 30 min before cell lysis with analysis by SDS-PAGE and immunoblotting with anti-proinsulin. Actin (below) is a loading control. D, quantitation (mean ± SD) of proinsulin protein levels is shown from replicate experiments like those shown in C (one-way ANOVA, ∗p < 0.05). DMSO, dimethyl sulfoxide; NEM, N-ethylmaleimide; VCP, valosin-containing protein.
If proteasome inhibition immediately decreases proinsulin biosynthesis yet increases proinsulin protein abundance, then it stands to reason that this must involve proinsulin molecules that were already synthesized prior to the addition of MG132. With this in mind, we treated INS1E β-cells ± MG132 during a 30 min chase following a 10 min pulse labeling with 35S-amino acids. In this case, recovery of newly synthesized proinsulin was notably increased (Fig. 3A), and this was quantified both in INS1E (rat) and Min6 (mouse) β-cells (Fig. 3B). Similar accumulation was observed upon VCP inhibition with CB-5083 (Fig. 3A, quantified in Fig. S2). By immunofluorescence in INS1E cells, during the first 15 min after synthesis, proinsulin resides exclusively in the ER, and even after 30 min (although some proinsulin advances to the Golgi complex and colocalizes with GM130), much newly made proinsulin still resides within the ER (Fig. S1, B–D). In independent experiments when CHX was added to “tighten the chase” after pulse labeling of Min6 cells, the recovery of newly synthesized proinsulin was still clearly increased in cells chased in the presence of MG132 (Fig. 3C), and to a smaller degree, preproinsulin was also increased (Fig. 3C). The MG132 data are consistent with an inhibition of ERAD, which has been reported to have the untoward effect of promoting the accumulation of disulfide-linked secretory protein complexes within the ER (40, 41). Indeed, within 30 min of MG132 addition, accompanying the increase in total proinsulin (seen by immunoblotting of reducing SDS-PAGE) was an increase in disulfide-linked complexes of proinsulin detected upon nonreducing SDS-PAGE (Fig. S3).
Figure 3.
Recovery of recently synthesized proinsulin is increased by acute proteasome inhibition.A, INS1E cells were pulse labeled with 35S-Met/Cys for 10 min, followed by a 30 min chase in the presence of either MG132 (10 μM), CB-5083 (10 μM), or vehicle alone (DMSO) added to the chase medium. Cell lysates (normalized to TCA-precipitable counts) were immunoprecipitated with anti-insulin and analyzed by reducing SDS-PAGE and phosphorimaging. B, quantitation (mean ± SD) of the recovery of newly synthesized proinsulin is shown from replicate experiments like those shown in A, in INS1E cells (left graph) and Min6 cells (right graph) (Student's t test, ∗p < 0.05). C, a pulse-labeling protocol in Min6 cells similar to those shown in A and B, except that cycloheximide (CHX, 100 μg/ml) was added during the chase. DMSO, dimethyl sulfoxide; TCA, trichloroacetic acid.
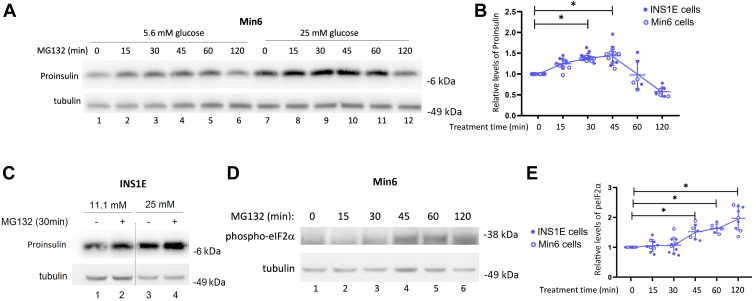
In a time course following MG132 addition, we noted that the total abundance of proinsulin protein (as measured by immunoblotting) rose upon proteasomal inhibition within 15 min (Figs. 4A and S3); however, the effect peaked by ∼45 min and began to decline thereafter (Fig. 4A, lanes 7–12, quantified in Fig. 4B). Min6 cells are ordinarily grown in medium containing 25 mM glucose; when the experiment testing acute proteasome inhibition was conducted instead at 5.6 mM glucose, overall cellular proinsulin levels were lower, but the impact of MG132 treatment was similar (Fig. 4A, lanes 1–6). The effect of MG132 to acutely increase proinsulin levels in INS1E cells was also observed at two different glucose concentrations (Fig. 4C). The eventual decline of cellular proinsulin levels followed a rise in the level of phospho–eukaryotic initiation factor 2 alpha (eIF2α) (Fig. 4, D and E), which can act as a negative regulator of proinsulin biosynthesis (42, 43).
Figure 4.
Time-dependent effects of acute proteasome inhibition in pancreatic β-cells.A, Min6 cells were incubated in fresh DMEM (5.6 mM glucose lanes 1–6 or 25 mM glucose lanes 7–12) for 4 h, during which MG132 (10 μM) was added for the times indicated immediately prior to cell lysis at 4 h. Cell lysates were analyzed by reducing SDS-PAGE and immunoblotting with antiproinsulin. Tubulin (below) is a loading control. B, quantitation (mean ± SD) of the recovery of proinsulin (normalized to tubulin) is shown from replicate experiments like those shown in A, in INS1E cells (11.1 mM glucose, closed symbols) and Min6 cells (25 mM glucose, open symbols) (one-way ANOVA, ∗p < 0.05). C, INS1E cells were incubated in fresh RPMI1640 medium (11.1 mM glucose, lanes 1 and 2, or 25 mM glucose, lanes 3 and 4) for 4 h during which MG132 (10 μM) was added for the last 30 min prior to cell lysis at 4 h. Cell lysates were analyzed by reducing SDS-PAGE and immunoblotting with anti-proinsulin. Tubulin (below) is a loading control. D, immunoblotting of phospho-eIF2α in INS1E cells (11.1 mM glucose) following a protocol similar to that described in A. Tubulin (below) is a loading control. E, quantitation (mean ± SD) of phospho-eIF2α (normalized to tubulin) in INS1E cells is shown from replicate experiments like those shown in D, in INS1E cells (11.1 mM glucose, closed symbols) and Min6 cells (25 mM glucose, open symbols) (one-way ANOVA, ∗p < 0.05). DMEM, Dulbecco's modified Eagle's medium; eIF2α, eukaryotic initiation factor 2 alpha.
If recently made molecules are subject to “protection” by proteasomal inhibition, then inhibiting the synthesis of these recently made proinsulin molecules should limit the rise in proinsulin levels caused by MG132 treatment. With this in mind, we inhibited protein synthesis (with high-dose CHX) for 30 min during concurrent proteasome inhibition. Without CHX, acute MG132 treatment increased proinsulin level and initiated an increase in phospho-eIF2α (Fig. 5A, lane 2 versus 1), whereas CHX treatment itself lowered total β-cell proinsulin levels and limited the extent to which MG132 could increase these levels (Fig. 5A, lanes 3 and 4). Moreover, if CHX was added 5 min before the addition of the proteasome inhibitor, this treatment essentially abolished the ability of MG132 to acutely increase β-cell proinsulin levels (shown and quantified in Fig. 5B). Similarly, MG132 treatment could not increase β-cell proinsulin levels when longer times (45 min) were used for cotreatment with CHX—or with two alternative inhibitors that use entirely different mechanisms of action to block protein synthesis (Fig. 5C). Altogether, these data establish that ongoing proinsulin biosynthesis contributes importantly to its steady-state protein level in β-cells, and a meaningful fraction of recently made WT proinsulin (and perhaps also some preproinsulin) is routinely routed to proteasomal disposal.
Figure 5.
Acute proteasome inhibition increases the recovery of only recently made proinsulin molecules.A, INS1E cells were incubated in fresh medium (11.1 mM glucose) for 4 h, and MG132 (10 μM) ± cycloheximide (CHX, 100 μg/ml) was added during the last 30 min before cell lysis. Actin (below) is a loading control. B, the gels at left utilized the same protocol as A except that when CHX was used, it was added 5 min before the addition of MG132. Cell lysates were analyzed by SDS-PAGE under reducing conditions, followed by immunoblotting. Actin (below) is a loading control. The graph at right shows quantitation (mean ± SD) from replicate experiments like those shown at left (one-way ANOVA, ∗p < 0.05). C, INS1E cells were incubated as in A except that MG132 treatment for 45 min before cell lysis was performed in the absence or presence of cotreatment with different protein synthesis inhibitors: harringtonine (“HT,” 2 μg/ml), CHX (100 μg/ml), puromycin (50 μM), or vehicle alone (DMSO). Cell lysates were analyzed by reducing SDS-PAGE gels and immunoblotting for proinsulin. Tubulin (below) is a loading control. DMSO, dimethyl sulfoxide.
Secondary effects of proteasome inhibition in pancreatic β-cells
As noted in the aforementioned introduction section, extended proteasomal deficiency in pancreatic β-cells leads to diminished insulin content and GSIS (26, 27, 28) and other adverse sequelae (29, 30, 31). Indeed as noted previously, after acute proteasomal inhibition, proinsulin protein levels peak rapidly but then begin to decline (Fig. 4B), accompanied by increased intracellular phospho-eIF2α (Fig. 4, D and E). Proteasomal protein degradation releases free amino acids available to generate charged tRNAs, which are sensed in the cytosol resulting in diminished activity of the GCN2 protein kinase that phosphorylates eIF2α (44). In contrast, proteasomal inhibition reduces cytosolic availability of amino acids, and this can contribute to the activation of GCN2 (45, 46).
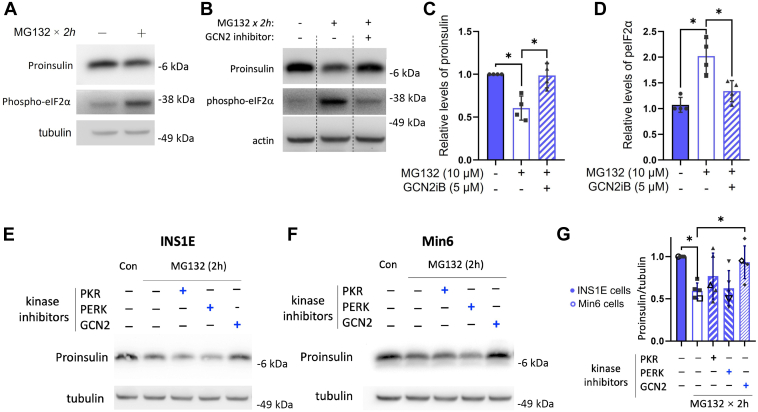
The inhibition of nascent proinsulin biosynthesis in the presence of MG132 was not reversed by concurrent addition of GCN2 inhibitor (Fig. 1D), but at longer MG132 treatment times, the increase of phospho-eIF2α (Fig. 6A) was inhibited by concurrent addition of GCN2 inhibitor (Fig. 6, B and D). Accompanying this, β-cell proinsulin protein levels were largely restored (Fig. 6, B and C). Indeed, when β-cells were cotreated with individual inhibitors of three of the four distinct eIF2α kinases: PKR, PERK, or GCN2 (HRI inhibitor was not available), it was the GCN2 inhibitor that selectively prevented the diminution of proinsulin protein level triggered by proteasome inhibition (with MG132) in INS1E cells (Fig. 6E), Min6 cells (Fig. 6F), or the combination thereof (Fig. 6G). Altogether, the data suggest a biphasic response of proinsulin protein in pancreatic β-cells subjected to proteasome inhibition: beginning with a rapid rise but accompanying activation of the GCN2 protein kinase, leading to a subsequent fall in proinsulin levels (Fig. 4B).
Figure 6.
Secondary effects of proteasome inhibition in pancreatic β-cells.A, INS1E cells were incubated in fresh medium (11.1 mM glucose) for 2 h followed by another 2 h incubation ± MG132 (10 μM) before cell lysis, reducing SDS-PAGE, and immunoblotting for proinsulin or phospho-eIF2α. Tubulin (below) is a loading control. B, the same protocol as that shown in A, except that one sample receiving MG132 was cotreated with GCN2 inhibitor (HY-112654, 5 μM). Actin (below) is a loading control. C and D, quantitation (mean ± SD) of proinsulin and phospho-eIF2α (p-eIF2α) protein levels from replicate experiments like those in B is shown in C and D, respectively (one-way ANOVA, ∗p < 0.05). E, INS1E cells were incubated in fresh medium (11.1 mM glucose) for 3.5 h followed by a 2 h treatment with MG132 (10 μM) in the absence or the presence of inhibitors of various eIF2α kinases, including PKR inhibitor (C16, 5 μM), PERK inhibitor-II (GSK2656157, 2.5 μM), and GCN2 inhibitor (HY-112654, 5 μM). Tubulin (below) is a loading control. F, the same protocol as that shown in E was applied to Min6 cells (grown in DMEM with 25 mM glucose). G, quantitation (mean ± SD) of proinsulin protein levels from replicate experiments like those in E and F (one-way ANOVA, ∗p < 0.05). DMEM, Dulbecco's modified Eagle's medium; eIF2α, eukaryotic initiation factor 2 alpha; GCN2, general control nonderepressible 2.
Acute effect of proteasome inhibition in pancreatic islets
Mature insulin levels are higher in the β-cells of animal islets than in β-cell lines, but the synthetic regulation of proinsulin appears quite similar between these systems. We therefore examined the acute effect of MG132 treatment in isolated pancreatic islets of normal mice and observed an immediate increase of proinsulin protein levels (Fig. 7A, lane 2 versus 1, quantified in Fig. 7B). Cotreatment of the mouse islets with CHX limited the ability of acute proteasome inhibition to increase proinsulin abundance (Fig. 7A, lane 4 versus 3, quantified in Fig. 7B), and pretreatment with CHX dropped proinsulin protein levels even further, eliminating any effect of MG132 (Fig. 7A, lanes 5 and 6, quantified in Fig. 7B). Similarly, human pancreatic islets acutely treated with MG132 also increased proinsulin levels (normalized to islet actin, Fig. 7C, lane 2 versus 1), and the effects of pretreatment or cotreatment with CHX eliminated the ability of proteasome inhibition to boost proinsulin levels above that of the control (Fig. 7, C and D). Altogether all these data, consistent from human islets, rodent islets, and pancreatic β-cell lines, support that a meaningful but unsuspected subfraction of recently synthesized INS gene product is routinely routed for proteasomal disposal, most likely by ERAD.
Figure 7.
Acute effect of proteasome inhibition in pancreatic islets.A, mouse (C57BL/6J) pancreatic islets were incubated in fresh medium containing 20 mM glucose for 4 h; when used, MG132 was added for the last 30 min before cell lysis. For the islets shown in lanes 3 and 4, cycloheximide (CHX, 100 μg/ml) was also added for the last 30 min (CHX cotreated ± MG132). For the islets shown in lanes 5 and 6, CHX was added 20 min before MG132 addition and continued during the MG132 treatment (CHX pretreated ± MG132). Actin (below) is a loading control. B, quantitation (mean ± SD) of proinsulin protein levels in mouse islets from replicate experiments like those shown in A (one-way ANOVA, ∗p < 0.05); the second bar is statistically higher than all other bars shown on the graph. C, pancreatic islets from nondiabetic humans were treated essentially identically to the mouse islet experiments shown in A, except that CHX pretreatments and cotreatments with MG132 (lanes 3 and 4) were directly compared with the effect of proteasome inhibition in the absence of CHX (lane 2). Actin (below) is a loading control. D, quantitation (mean ± SD) of proinsulin protein levels in human islets from replicate experiments like those shown in C (one-way ANOVA, ∗p < 0.05); the second bar is statistically higher than all other bars shown on the graph.
Discussion
As noted previously, several prior publications have argued that proteasome inhibition suppresses proinsulin protein levels in pancreatic β-cells. In the current study, we have focused our efforts primarily within the first 45 min after acute proteasome inhibition in β-cells, in order to be unencumbered by secondary cellular responses. We find that although proinsulin biosynthesis is immediately suppressed, proinsulin protein levels are promptly increased by acute proteasome inhibition in β-cell lines, rodent islets, and human islets. Such a result uncovers a contribution of ongoing proteasomal disposal of proinsulin in the background of normal β-cell function that has gone largely unsuspected.
Each preproinsulin takes ∼20 s to synthesize, and it has been estimated that a β-cell generates ∼6000 new proinsulin molecules per second (47). Here, we observe that inhibiting new protein synthesis for 45 min causes β-cell proinsulin levels to drop precipitously, which indicates that the steady-state level of proinsulin in β-cells is largely maintained by continuously ongoing proinsulin biosynthesis (15). Moreover, our current findings emphasize that for the maintenance of total proinsulin protein level (which may be estimated on the order of ∼20 million molecules per cell in a rodent β-cell line), ongoing proinsulin biosynthesis must offset the loss of roughly one-third of proinsulin that occurs via proteasomal disposal.
The simplest hypothesis for the observed proteasomal disposal of proinsulin in β-cells is ERAD; however, this process has been primarily identified for misfolded mutant proinsulin, with considerably less information available about the degradation of WT proinsulin molecules (19, 20, 22, 23). Nevertheless, there has been suggestive evidence that the fate of WT proinsulin may be affected by ERAD function in β-cells in vivo and in vitro (18, 25). Indeed, we find that the inhibition of VCP/p97 (which is required for ERAD-mediated proteasomal degradation (38, 39)) can similarly increase β-cell proinsulin levels.
Interestingly, we find that proteasome inhibition during a 10 min pulse of β-cells with 35S-amino acids consistently decreases rather than increases the recovery of newly synthesized proinsulin. However, shortly thereafter, a fraction of WT proinsulin does become available for degradation as seen by proinsulin protection in pulse-labeled β-cells chased in the presence of inhibitors of either proteasomes or VCP/p97. Reciprocally, blocking protein synthesis beginning 5 min before acute MG132 addition largely eliminates the ability of proteasome inhibitor to increase β-cell proinsulin levels. Both results suggest that a portion of WT proinsulin molecules can begin to be targeted for proteasomal disposal within a number of minutes after their initial synthesis. This is a time when young proinsulin molecules are susceptible to misfolding, and we find that acute proteasomal inhibition leads to increased accumulation of misfolded disulfide-linked proinsulin complexes.
Loss of β-cell ERAD function has fairly profound consequences on downstream signaling pathways, leading to a loss of β-cell differentiation/maturity (17), and long-term loss of proteasome activity leads to β-cell death (30). Here, we find that one secondary signal that relatively rapidly follows proteasome inhibition in β-cells is an increase in phospho-eIF2α. The phospho-eIF2α signal precedes a decline in proinsulin levels and continues to be elevated for a period even after proinsulin protein has fallen below initial control levels. We hypothesize that proteasomal function provides an ongoing contribution to free amino acid levels in the cytosol of β-cells, and these levels are sensed, indirectly, by the GCN2 protein kinase. As sustained proteasomal inhibition limits the generation of free cytosolic amino acids, a starvation response by GCN2 can be expected. We speculate that this can explain the ability of GCN2 inhibition to diminish phospho-eIF2α levels in β-cells under sustained proteasomal blockade, accompanied by a notable increase in proinsulin protein levels. The fact that this behavior cannot be replicated by inhibitors of two other eIF2α kinases suggests a specific link that allows ongoing proteasome activity to limit the activity of GCN2 kinase in pancreatic β-cells.
Importantly, the impact of proteasome inhibition to acutely increase β-cell proinsulin levels was consistently observed in both murine and human islets. The biosynthetic rate of proinsulin protein (at a fixed extracellular glucose concentration) has never been directly compared between human islets, rodent pancreatic islets, and rodent β-cell lines (this would not be straightforward because different antiproinsulin antibodies show different degrees of immunoreactivity to human proinsulin, rodent proinsulin-1, and rodent proinsulin-2). However, the effect of acute proteasome activity to increase proinsulin levels can occur in β-cells at both higher extracellular glucose and normal extracellular glucose conditions. In each of the β-cell systems studied, proinsulin biosynthesis must be sufficient to account not only for the secretion of proinsulin as well as the conversion to insulin for production and maintenance of the insulin storage pool (14) but also (as highlighted in the current work) for ongoing intracellular degradative pathways for WT proinsulin that are likely to include ERAD-mediated proteasomal disposal (as well as, perhaps, ER-phagy (48, 49, 50)).
Experimental procedures
Antibodies
Mouse anti-rodent proinsulin (CCI-17) antibody was from Novus Biologicals (catalog no.: NB100-73013). Mouse anti-beta actin antibody was from Proteintech (catalog no.: 66009-1-Ig). Mouse anti-tubulin antibody was from Sigma (catalog no.: T5168). Rabbit anti-phosphorylation eIF2α antibody was from Cell Signaling Technology (catalog no.: 9721S). Rabbit anti-GM130 antibody was from Abcam (catalog no.: ab52649). DTT, MG-132, CHX, and N-ethylmaleimide were from Sigma. GCN2 inhibitor was from MedChemExpress (catalog no.: HY-112654). CB-5083 was from APExBIO (catalog no.: B6032). Met/Cys-deficient Dulbecco's modified Eagle's medium (DMEM) and all other tissue culture reagents were from Invitrogen. 35S-amino acid mixture was from PerkinElmer. Protein A-Agarose was from Repligen.
Cell culture
INS1E rat insulinoma cells (American Type Culture Collection) were cultured in RPMI1640 medium (11.1 mM glucose) supplemented with 10% fetal bovine serum (FBS), penicillin–streptomycin, 1 mM sodium pyruvate, 10 mM Hepes, and 0.05 mM 2-mercaptoethanol (Sigma). Min6 cells were cultured in DMEM (25 mM glucose) supplemented with 10% FBS, 0.1 mM β-mercaptoethanol, and penicillin–streptomycin.
Metabolic pulse labeling
Rat INS1E cells or mouse Min6 cells were pulse labeled with [35S] Met/Cys. Briefly, cells were incubated in complete medium for 3.5 h, followed by an additional 30 min in Met/Cys-deficient DMEM containing 25 mM glucose. Cells were then pulse labeled in the same medium with [35S] Met/Cys for 10 min. In some experiments, MG132 (10 μM) was added during the pulse; in other experiments, it was added only during the chase. Cell lysates were normalized to trichloroacetic acid–precipitable counts and were precleared with zysorbin for 1 h before immunoprecipitation with anti-insulin antibody overnight at 4 °C. Samples were subjected to 4 to 12% NuPage gradient gels under reducing condition, dried, and examined by phosphorimaging.
CHX washout and immunofluorescence
INS1E cells were pretreated with CHX (100 μg/ml) for 4 h, washed with cold PBS three times, and incubated in fresh medium for the time indicated. Cells were fixed with 4% formaldehyde for 20 min at room temperature and permeabilized with 0.2% Triton X-100 for an additional 15 min. After blocking with 3% bovine serum albumin for 1 h, cells were incubated with anti-proinsulin and anti-GM130 antibodies at 4 °C overnight. Fluorophore-conjugated secondary antibody staining was performed at room temperature for 1 h. Sections were treated with Prolong Gold 4′,6-diamidino-2-phenylindole and imaged by Nikon A1 confocal microscope.
Western blotting
Cells were lysed in radioimmunoprecipitation assay buffer with protease inhibitor cocktail on ice for 15 min and centrifuged at 12,000 rpm at 4 °C for 15 min. Total protein lysates were boiled in sample buffer containing 100 mM DTT for 5 min, separated by 4 to 12% gradient Nu-page gels, and electrotransferred to nitrocellulose membrane and blotted with primary antibodies (1:1000 dilution, diluted in Tris-buffered saline with Tween-20 with 5% bovine serum albumin) at 4 °C overnight. Horseradish peroxidase–conjugated secondary antibody (1:5000 dilution) incubation was performed at room temperature for 1 h. Imaging was acquired using the Clarity Western ECL substrate.
Mouse islets
All experiments performed with mice were in compliance with and approved by the University of Michigan Institutional Animal Care and Use Committee (PRO00009936). Murine islets were isolated by collagenase digestion via the common bile duct, followed by pancreatic digestion ex vivo. The digest was washed and spun on a Histopaque (catalog no.: 1077; Sigma–Aldrich) gradient (900g for 20 min without brake). Islets were collected, washed, handpicked, and incubated overnight in RPMI1640 medium plus 10% FBS, at 37 °C.
Human islets
Isolated islets from nondiabetic human donors were purchased from Prodo Labs. Modest clinical data were made available by the supplier, including HbA1c values in the donors <5.7%, and all routine blood tests were said to be normal, and patients negative for coronavirus disease 2019. In our studies, 30 to 35 human islet equivalents were analyzed for each experimental condition (i.e., per tube).
Statistics
Statistical analysis was assessed by Student’s t test and one-way ANOVA test to determine the differences between groups using GraphPad Prism 8 software (GraphPad Software, Inc). Data are presented as means ± SD. p Value <0.05 was considered as statistically significant.
Data availability
All data are contained within the manuscript, with primary data available upon request (Dr Peter Arvan, University of Michigan, email: parvan@umich.edu).
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We acknowledge assistance from the Michigan Diabetes Research Center Morphology Core (National Institutes of Health grant P30 DK020572) and the University of Michigan Protein Folding Diseases Initiative.
Author contributions
L. Q., B. T., M. L., and P. A. conceptualization; X. X., A. A., L. H., M. A., and S. J. methodology; X. X., A. A., L. H., M. A., and S. J. validation; X. X., A. A., and L. H. formal analysis; X. X. and P. A. writing–original draft; X. X., A. A., L. H., M. A., S. J., L. Q., B. T., M. L., and P. A. writing–review & editing; X. X., A. A., and L. H. visualization; P. A. supervision; X. X., M. L., and P. A. funding acquisition.
Funding and additional information
This work was supported by the National Institutes of Health R01 DK111174 (to L. Q., B. T., and P. A.); DK48280 (to P. A.); and also from the National Natural Science Foundation of China (grant no.: 82000796), the PUMC Youth Fund supported by the Fundamental Research Funds for the Central Universities (grant no.: 3332020080), the Tianjin Municipal Science and Technology Commission (grant no.: 18JCQNJC82100), and the China Scholarship Council (grant no.: 201806940006) to X. X.; as well as the National Natural Science Foundation of China (grant nos.: 81620108004 and 81830025), and the National Key R&D Program (grant no.: 2019YFA0802502) to M. L. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Ronald Wek
Contributor Information
Ming Liu, Email: mingliu@tmu.edu.cn.
Peter Arvan, Email: parvan@umich.edu.
Supporting information
References
- 1.Vasiljevic J., Torkko J.M., Knoch K.-P., Solimena M. The making of insulin in health and disease. Diabetologia. 2020;63:1981–1989. doi: 10.1007/s00125-020-05192-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu M., Wright J., Guo H., Xiong Y., Arvan P. Proinsulin entry and transit through the endoplasmic reticulum in pancreatic beta cells. Vitam. Horm. 2014;95:35–62. doi: 10.1016/B978-0-12-800174-5.00002-8. [DOI] [PubMed] [Google Scholar]
- 3.Patzelt C., Labrecque A.D., Duguid J.R., Carroll R.J., Keim P.S., Heinrikson R.L., et al. Detection and kinetic behavior of preproinsulin in pancreatic islets. Proc. Natl. Acad. Sci. U. S. A. 1978;75:1260–1264. doi: 10.1073/pnas.75.3.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu M., Lara-Lemus R., Shan S.O., Wright J., Haataja L., Barbetti F., et al. Impaired cleavage of preproinsulin signal peptide linked to autosomal-dominant diabetes. Diabetes. 2012;61:828–837. doi: 10.2337/db11-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo H., Xiong Y., Witkowski P., Cui J., Wang L.J., Sun J., et al. Inefficient translocation of preproinsulin contributes to pancreatic beta cell failure and late-onset diabetes. J. Biol. Chem. 2014;289:16290–16302. doi: 10.1074/jbc.M114.562355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X., Itani O.A., Haataja L., Dumas K.J., Yang J., Cha J., et al. Requirement for translocon-associated protein (TRAP)alpha in insulin biogenesis. Sci. Adv. 2019;5:1–9. doi: 10.1126/sciadv.aax0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang Y., Xu X., Arvan P., Liu M. Deficient endoplasmic reticulum translocon-associated protein complex limits the biosynthesis of proinsulin and insulin. FASEB J. 2021;35 doi: 10.1096/fj.202002774R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu X., Huang Y., Li X., Arvan P., Liu M. The role of TRAPgamma/SSR3 in preproinsulin translocation into the endoplasmic reticulum. Diabetes. 2022;71:440–452. doi: 10.2337/db21-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kriegler T., Kiburg G., Hessa T. Translocon-associated protein complex (TRAP) is crucial for co-translational translocation of pre-proinsulin. J. Mol. Biol. 2020;432:166694. doi: 10.1016/j.jmb.2020.10.028. [DOI] [PubMed] [Google Scholar]
- 10.Guo H., Sun J., Li X., Xiong Y., Wang H., Shu H., et al. Positive charge in the n-region of the signal peptide contributes to efficient post-translational translocation of small secretory preproteins. J. Biol. Chem. 2018;293:1899–1907. doi: 10.1074/jbc.RA117.000922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang S.W., Rane N.S., Kim S.J., Garrison J.L., Taunton J., Hegde R.S. Substrate-specific translocational attenuation during ER stress defines a pre-emptive quality control pathway. Cell. 2006;127:999–1013. doi: 10.1016/j.cell.2006.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kadowaki H., Nagai A., Maruyama T., Takami Y., Satrimafitrah P., Kato H., et al. Pre-emptive quality control protects the ER from protein overload via the proximity of ERAD components and SRP. Cell Rep. 2015;13:944–956. doi: 10.1016/j.celrep.2015.09.047. [DOI] [PubMed] [Google Scholar]
- 13.Kadowaki H., Satrimafitrah P., Takami Y., Nishitoh H. Molecular mechanism of ER stress-induced pre-emptive quality control involving association of the translocon, Derlin-1, and HRD1. Sci. Rep. 2018;8:7317. doi: 10.1038/s41598-018-25724-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu M., Huang Y., Xu X., Li X., Maroof A., Arunagiri A., et al. Normal and defective pathways in biogenesis and maintenance of the insulin storage pool. J. Clin. Invest. 2021;131 doi: 10.1172/JCI142240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haataja L., Snapp E., Wright J., Liu M., Hardy A.B., Wheeler M.B., et al. Proinsulin intermolecular interactions during secretory trafficking in pancreatic beta cells. J. Biol. Chem. 2013;288:1896–1906. doi: 10.1074/jbc.M112.420018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoelen H., Zaldumbide A., van Leeuwen W.F., Torfs E.C., Engelse M.A., Hassan C., et al. Proteasomal degradation of proinsulin requires Derlin-2, HRD1 and p97. PLoS One. 2015;10 doi: 10.1371/journal.pone.0128206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shrestha N., Liu T., Ji Y., Reinert R., Torres M., Li X., et al. Sel1L-Hrd1 ER-associated degradation maintains beta-cell identity via TGFbeta signaling. J. Clin. Invest. 2020;130:3499–3510. doi: 10.1172/JCI134874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu Y., Gao Y., Zhang M., Deng K.Y., Singh R., Tian Q., et al. Endoplasmic reticulum-associated degradation (ERAD) has a critical role in supporting glucose-stimulated insulin secretion in pancreatic beta-cells. Diabetes. 2019;68:733–746. doi: 10.2337/db18-0624. [DOI] [PubMed] [Google Scholar]
- 19.Xu B., Allard C., Alvarez-Mercado A.I., Fuselier T., Kim J.H., Coons L.A., et al. Estrogens promote misfolded proinsulin degradation to protect insulin production and delay diabetes. Cell Rep. 2018;24:181–196. doi: 10.1016/j.celrep.2018.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tiwari A., Schuiki I., Zhang L., Allister E.M., Wheeler M.B., Volchuk A. SDF2L1 interacts with the ER-associated degradation machinery and retards the degradation of mutant proinsulin in pancreatic beta-cells. J. Cell Sci. 2013;126:1962–1968. doi: 10.1242/jcs.117374. [DOI] [PubMed] [Google Scholar]
- 21.Sugawara T., Kano F., Murata M. Rab2A is a pivotal switch protein that promotes either secretion or ER-associated degradation of (pro)insulin in insulin-secreting cells. Sci. Rep. 2014;4:6952. doi: 10.1038/srep06952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cunningham C.N., He K., Arunagiri A., Paton A.W., Paton J.C., Arvan P., et al. Chaperone-driven degradation of a misfolded proinsulin mutant in parallel with restoration of wild type insulin secretion. Diabetes. 2017;66:741–753. doi: 10.2337/db16-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He K., Cunningham C.N., Manickam N., Liu M., Arvan P., Tsai B. PDI reductase acts on Akita mutant proinsulin to initiate retrotranslocation along the Hrd1/Sel1L-p97 axis. Mol. Biol. Cell. 2015;26:3413–3423. doi: 10.1091/mbc.E15-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu M., Hodish I., Haataja L., Lara-Lemus A.R., Rajpal G., Wright J., et al. Proinsulin misfolding and diabetes: mutant INS gene-induced diabetes of youth. Trends Endocrinol. Metab. 2010;21:652–659. doi: 10.1016/j.tem.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X., Yuan Q., Tang W., Gu J., Osei K., Wang J. Substrate-favored lysosomal and proteasomal pathways participate in the normal balance control of insulin precursor maturation and disposal in beta-cells. PLoS One. 2011;6 doi: 10.1371/journal.pone.0027647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baldeon M.E., Neece D.J., Nandi D., Monaco J.J., Gaskins H.R. Interferon-gamma independently activates the MHC class I antigen processing pathway and diminishes glucose responsiveness in pancreatic beta-cell lines. Diabetes. 1997;46:770–778. doi: 10.2337/diab.46.5.770. [DOI] [PubMed] [Google Scholar]
- 27.Hofmeister-Brix A., Lenzen S., Baltrusch S. The ubiquitin-proteasome system regulates the stability and activity of the glucose sensor glucokinase in pancreatic beta-cells. Biochem. J. 2013;456:173–184. doi: 10.1042/BJ20130262. [DOI] [PubMed] [Google Scholar]
- 28.Kawaguchi M., Minami K., Nagashima K., Seino S. Essential role of ubiquitin-proteasome system in normal regulation of insulin secretion. J. Biol. Chem. 2006;281:13015–13020. doi: 10.1074/jbc.M601228200. [DOI] [PubMed] [Google Scholar]
- 29.Bugliani M., Liechti R., Cheon H., Suleiman M., Marselli L., Kirkpatrick C., et al. Microarray analysis of isolated human islet transcriptome in type 2 diabetes and the role of the ubiquitin-proteasome system in pancreatic beta cell dysfunction. Mol. Cell. Endocrinol. 2013;367:1–10. doi: 10.1016/j.mce.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 30.Broca C., Varin E., Armanet M., Tourrel-Cuzin C., Bosco D., Dalle S., et al. Proteasome dysfunction mediates high glucose-induced apoptosis in rodent beta cells and human islets. PLoS One. 2014;9 doi: 10.1371/journal.pone.0092066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Litwak S.A., Wali J.A., Pappas E.G., Saadi H., Stanley W.J., Varanasi L.C., et al. Lipotoxic stress induces pancreatic beta-cell apoptosis through modulation of Bcl-2 proteins by the ubiquitin-proteasome system. J. Diabetes Res. 2015;2015:280615. doi: 10.1155/2015/280615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitiphongspattana K., Mathews C.E., Leiter E.H., Gaskins H.R. Proteasome inhibition alters glucose-stimulated (pro)insulin secretion and turnover in pancreatic beta-cells. J. Biol. Chem. 2005;280:15727–15734. doi: 10.1074/jbc.M410876200. [DOI] [PubMed] [Google Scholar]
- 33.Lopez-Avalos M.D., Duvivier-Kali V.F., Xu G., Bonner-Weir S., Sharma A., Weir G.C. Evidence for a role of the ubiquitin-proteasome pathway in pancreatic islets. Diabetes. 2006;55:1223–1231. doi: 10.2337/db05-0450. [DOI] [PubMed] [Google Scholar]
- 34.Liu C.Y., Hao Y.N., Yin F., Zhang Y.L., Liu J.H. Geniposide accelerates proteasome degradation of Txnip to inhibit insulin secretion in pancreatic beta-cells. J. Endocrinol. Invest. 2017;40:505–512. doi: 10.1007/s40618-016-0591-9. [DOI] [PubMed] [Google Scholar]
- 35.Meiners S., Laule M., Rother W., Guenther C., Prauka I., Muschick P., et al. Ubiquitin-proteasome pathway as a new target for the prevention of restenosis. Circulation. 2002;105:483–489. doi: 10.1161/hc0402.102951. [DOI] [PubMed] [Google Scholar]
- 36.Anderson D.J., Le Moigne R., Djakovic S., Kumar B., Rice J., Wong S., et al. Targeting the AAA ATPase p97 as an approach to treat cancer through disruption of protein homeostasis. Cancer Cell. 2015;28:653–665. doi: 10.1016/j.ccell.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang E.Y., To M., Tran E., Dionisio L.T.A., Cho H.J., Baney K.L.M., et al. A VCP inhibitor substrate trapping approach (VISTA) enables proteomic profiling of endogenous ERAD substrates. Mol. Biol. Cell. 2018;29:1021–1030. doi: 10.1091/mbc.E17-08-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang F., Li S., Houerbi N., Chou T.F. Temporal proteomics reveal specific cell cycle oncoprotein downregulation by p97/VCP inhibition. Cell Chem. Biol. 2022;29:517–529.e5. doi: 10.1016/j.chembiol.2021.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rabinovich E., Kerem A., Frohlich K.U., Diamant N., Bar-Nun S. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol. Cell. Biol. 2002;22:626–634. doi: 10.1128/MCB.22.2.626-634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi G., Somlo D.R.M., Kim G.H., Prescianotto-Baschong C., Sun S., Beuret N., et al. ER-associated degradation is required for vasopressin prohormone processing and systemic water homeostasis. J. Clin. Invest. 2017;127:3897–3912. doi: 10.1172/JCI94771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim G.H., Shi G., Somlo D.R., Haataja L., Song S., Long Q., et al. Hypothalamic ER-associated degradation regulates POMC maturation, feeding, and age-associated obesity. J. Clin. Invest. 2018;128:1125–1140. doi: 10.1172/JCI96420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheuner D., Vander Mierde D., Song B., Flamez D., Creemers J.W.M., Tsukamoto K., et al. Control of mRNA translation preserves reticulum function in beta cells and maintains glucose homeostasis. Nat. Med. 2005;11:757–764. doi: 10.1038/nm1259. [DOI] [PubMed] [Google Scholar]
- 43.Back S.H., Scheuner D., Han J., Song B., Ribick M., Wang J., et al. Translation attenuation through eIF2alpha phosphorylation prevents oxidative stress and maintains the differentiated state in beta cells. Cell Metab. 2009;10:13–26. doi: 10.1016/j.cmet.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Sousa-Coelho A.L., Marrero P.F., Haro D. Activating transcription factor 4-dependent induction of FGF21 during amino acid deprivation. Biochem. J. 2012;443:165–171. doi: 10.1042/BJ20111748. [DOI] [PubMed] [Google Scholar]
- 45.Jiang H.Y., Wek R.C. Phosphorylation of the alpha-subunit of the eukaryotic initiation factor-2 (eIF2alpha) reduces protein synthesis and enhances apoptosis in response to proteasome inhibition. J. Biol. Chem. 2005;280:14189–14202. doi: 10.1074/jbc.M413660200. [DOI] [PubMed] [Google Scholar]
- 46.Harding H.P., Ordonez A., Allen F., Parts L., Inglis A.J., Williams R.L., et al. The ribosomal P-stalk couples amino acid starvation to GCN2 activation in mammalian cells. Elife. 2019;8 doi: 10.7554/eLife.50149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schuit F.C., Kiekens R., Pipeleers D.G. Measuring the balance between insulin synthesis and insulin release. Biochem. Biophys. Res. Commun. 1991;178:1182–1187. doi: 10.1016/0006-291x(91)91017-7. [DOI] [PubMed] [Google Scholar]
- 48.Chen Y.J., Knupp J., Arunagiri A., Haataja L., Arvan P., Tsai B. PGRMC1 acts as a size-selective cargo receptor to drive ER-phagic clearance of mutant prohormones. Nat. Commun. 2021;12:5991. doi: 10.1038/s41467-021-26225-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Y.-J., Williams J.M., Arvan P., Tsai B. Reticulon protects the integrity of the ER membrane during ER escape of large macromolecular protein complexes. J. Cell Biol. 2020;219 doi: 10.1083/jcb.201908182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parashar S., Ferro-Novick S. Architecture of the endoplasmic reticulum plays a role in proteostasis. Autophagy. 2022;18:937–938. doi: 10.1080/15548627.2022.2030175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the manuscript, with primary data available upon request (Dr Peter Arvan, University of Michigan, email: parvan@umich.edu).